Abstract
The X chromosome of therian mammals shows strong conservation among distantly related species, limiting insights into the distinct selective processes that have shaped sex chromosome evolution. We constructed a chromosome-scale de novo genome assembly for the Siberian dwarf hamster (Phodopus sungorus), a species reported to show extensive recombination suppression across an entire arm of the X chromosome. Combining a physical genome assembly based on shotgun and long-range proximity ligation sequencing with a dense genetic map, we detected widespread suppression of female recombination across ∼65% of the Phodopus X chromosome. This region of suppressed recombination likely corresponds to the Xp arm, which has previously been shown to be highly heterochromatic. Using additional sequencing data from two closely related species (P. campbelli and P. roborovskii), we show that recombination suppression on Xp appears to be independent of major structural rearrangements. The suppressed Xp arm was enriched for several transposable element families and de-enriched for genes primarily expressed in placenta, but otherwise showed similar gene densities, expression patterns, and rates of molecular evolution when compared to the recombinant Xq arm. Phodopus Xp gene content and order was also broadly conserved relative to the more distantly related rat X chromosome. These data suggest that widespread suppression of recombination has likely evolved through the transient induction of facultative heterochromatin on the Phodopus Xp arm without major changes in chromosome structure or genetic content. Thus, substantial changes in the recombination landscape have so far had relatively subtle influences on patterns of X-linked molecular evolution in these species.
Keywords: sex chromosome evolution, sex-biased genes, transposable element accumulation, faster-X evolution
Significance.
Sex chromosome evolution represents a dynamic process of genomic specialization that is thought to be dependent on evolution of recombination. Here, we use genome sequencing and genetic mapping to show that one arm comprising over half of the X chromosome in a species of dwarf hamster has largely lost the ability to recombine in males and females. Although these dramatic shifts in recombination frequencies might eventually lead to sex chromosome degeneration, loss of recombination on this arm is associated with relatively minor changes in chromosome structure and gene contents in this species. These results underscore the conservation of the X chromosome across mammals, and allow us to test predictions about how genetic recombination influences sex chromosome evolution.
Introduction
The recurrent evolution of sex chromosomes represents one of the most extreme examples of genomic specialization. One hallmark of sex chromosome evolution is that the chromosome present only in the heterogametic sex often evolves a highly reduced, repetitive structure consisting primarily of a sex determination switch and genes involved in reproduction (reviewed in Bachtrog et al. 2014; Wright et al. 2016; Charlesworth 2021). A common outcome of this process is an orthologous but heteromorphic sex chromosome pair with a single, degenerated sex-limited chromosome and a largely intact, recombining chromosome found in both sexes. Heteromorphic sex chromosomes are found in several of the most commonly studied organisms, including XY male heterogametic systems (most mammals and Drosophila), and ZW female heterogametic systems (birds and Lepidoptera). However, examination of these relatively ancient sex chromosomes reveals only the late stages of a presumably dynamic and complex process of structural evolution, functional specialization, and gene loss (Bachtrog et al. 2014).
As sex chromosomes have been surveyed in more taxa, it has become clear that the amount of degeneration on the sex-limited chromosome (e.g., the Y or W) can be decoupled from chromosome age (Darolti et al. 2019) and can vary between very closely related species (Bracewell et al. 2017; Darolti et al. 2019; Peichel et al. 2020). Recombination is the primary determinant of this dynamic process. Once a locus with sex-specific effects arises on a chromosome with a sex determination gene, sexually antagonistic selection should favor maintaining linkage between the two loci through suppressed recombination (Charlesworth 2021). As this process continues, regions with suppressed recombination are expected to lose functional genes and accumulate repetitive elements (Charlesworth et al. 1986) and ultimately increase the proportion of the chromosome packaged as heterochromatin (Zhou et al. 2013). Chromosomal inversions are thought to be a primary mechanism for recombination suppression, as they impede pairing and synapsis between homologous chromosomal regions in heterozygotes (Kirkpatrick 2010) and thereby link sexually antagonistic alleles to sex determination loci (Roberts et al. 2009). Consistent with this model, complex structural rearrangements are a common feature differentiating heteromorphic sex chromosomes. However, inversions are only one possible mechanism for suppressed recombination (Furman et al. 2020), and it remains unclear if the rapid accumulation of inversions on sex chromosomes are a cause or consequence of broader recombination suppression (Wright et al. 2016).
In contrast, the X or Z chromosomes usually undergo free recombination in the homogametic sex (e.g., XX females or ZZ males). Consequently, shared sex chromosomes often maintain some similarity to their inferred ancestral (autosomal) form and tend to be more conserved but also have the potential to evolve rapidly. For example, X chromosomes tend to show broad conservation of gene content and order across placental mammals, reflecting a common origin and strong purifying selection related to dosage compensation (Ohno 1967; Pessia et al. 2012) and likely other functional dynamics (Furman et al. 2020; Brashear et al. 2021). At the same time, the X chromosome also shows several signatures of rapid evolution relative to the autosomes, including more rapid protein-coding evolution (i.e., faster-X evolution; reviewed in Meisel and Connallon 2013) and enrichment for genes with both male- and female-specific functions (Wang et al. 2008; Reinius et al. 2012). Faster-X evolution is often thought to reflect accelerated adaptive change due to immediate exposure of recessive beneficial mutations in males (Meisel and Connallon 2013; Kousathanas et al. 2014), with adaptive faster-X evolution predicted when beneficial mutations are recessive on average (Charlesworth et al. 1987). Effective population size (Ne) is predicted to differ between the X chromosome and the autosomes (i.e., ¾ autosomal Ne in mammals given equal sex ratios and reproductive success), which can influence fixation rates of deleterious and beneficial mutations (Vicoso and Charlesworth 2009; Mank et al. 2010 a). The X chromosome also tends to have a lower recombination rate in mammals, which could reduce effective population sizes and the efficacy of X-linked selection due to negative or positive selection at linked sites (i.e., Hill–Robertson interference; Hill and Robertson 1966). Both the strength of faster-X evolution and the pattern of sex-biased gene content varies considerably across taxa (Bachtrog et al. 2014), likely reflecting variation in effective population sizes, the degree of female-biased transmission of the X, mechanisms of dosage compensation (Mank et al. 2010b), and epigenetic inactivation during certain developmental processes (e.g., meiotic sex chromosome inactivation [MSCI] in mammalian testis; Khil et al. 2004). Understanding how all these processes shape mammalian X chromosome evolution has been limited by the relatively ancient common origin and general structural conservation shared among taxa with sequenced genomes.
A rich history of mammalian cytogenetic research points to many potential exceptions to a conserved X paradigm that might help advance understanding of sex chromosome evolution (Veyrunes et al. 2008; Couger et al. 2021). One such exception may occur within the Cricetid rodent genus Phodopus, comprised of three species: Phodopus sungorus (Siberian or Djungarian hamster), the closely related sister taxon P. campbelli (Campbell’s dwarf hamster), and P. roborovskii (Desert hamster). All three species are endemic to xeric steppe and desert habitats of central Asia (Ross 1994, 1995, 1998). Phodopus sungorus and P. campbelli have been developed as laboratory models to understand circadian and circannual plasticity in physiology, reproduction, and behavior (e.g., Steinlechner et al. 1983; Wynne-Edwards 1995; Surbhi et al. 2019). Phodopus species are also noteworthy for their unusually large sex chromosomes. The submetacentric P. sungorus X chromosome comprises ∼10% of the haploid female karyotype (Haaf et al. 1987), approximately 2-fold more than the mammalian standard karyotype (Pathak and Stock 1974), with the karyotypically shorter (Xp) arm appearing largely as condensed constitutive (C-banding) heterochromatin (Gamperl et al. 1977; Haaf et al. 1987). The heterochromatic Xp arm is presumed to represent a massive expansion of highly repetitive (presumably non-genic) sequences in Phodopus (Haaf et al. 1987), with a short distal pseudoautosomal region (PAR) where all male-specific recombination occurs. Subsequent immunostaining (Bikchurina et al. 2018) and genetic crossing (Brekke et al. 2021) experiments have confirmed extensive suppression of female recombination on the presumed Xp arm in P. sungorus, P. campbelli, and/or their F1 hybrids. However, the genic content of Xp is unknown and X-linked incompatibilities are also primary determinants of hybrid male sterility (Bikchurina et al. 2018) and disrupted hybrid placental development (Brekke et al. 2021) in intercrosses between P. sungorus and P. campbelli, suggesting that functionally relevant components of the Xp arm may be rapidly evolving.
Here we combine shotgun and long-range proximity ligation sequencing with genetic mapping data to construct the first chromosome-scale de novo genome assembly for the Siberian dwarf hamster (P. sungorus). We then use multi-tissue transcriptomes, shotgun genome sequencing from P. campbelli and P. roborovskii, and comparative genome data from other mammals to examine the evolution of chromosome structure, genetic content, and protein-coding sequences across the Phodopus X chromosome.
Results
Phodopus Chromosome-Scale Genome Assembly and Annotation
We generated an initial de novo assembly for a single P. sungorus female based on 742 million paired Illumina reads (150 bp PE, Illumina HiSeq X), resulting in near complete assembly of the estimated non-repetitive genome and 91% of the estimated total genome (estimated size 2,310 Megabases or Mb). The total assembly covered 2,113.3 Mb (98,297 contigs, contig N50 = 51.5 kilobases or kb) collected into 69,381 scaffolds (scaffold N50 = 79.7 kb). These data were then augmented with long-range proximity ligation data using complementary Dovetail Chicago and Hi-C methods. The end result was a highly contiguous scaffolded assembly (scaffold N50 = 165.75 Mb; scaffold N90 = 30.61 Mb) with ∼90% of the build contained within 14 scaffolds (supplementary table S1, Supplementary Material online); a dramatic improvement relative to a previously published draft Illumina shotgun assembly for this species (contig N50 = 2.2 kb, scaffold N50 = 4.8 kb; Bao et al. 2019). We then evaluated the quality of the genome assembly by re-mapping the short reads and comparing diploid genotype likelihoods for matched and mismatched bases relative to the haploid assembly of the longest scaffold for each chromosome as implemented with Referee (Thomas and Hahn 2019). We estimated a per-base error rate of 4.22 × 10−5, with 67.6% of evaluated bases receiving a quality score over 80 (supplementary fig S1, Supplementary Material online).
We previously generated a genetic map for Phodopus (Brekke et al. 2021) composed of 14 major linkage groups based on a backcross mapping experiment (F1 female [P. campbelli female×P. sungorus male]×P. campbelli male). Both P. sungorus and P. campbelli have 13 mostly metacentric autosomes and a sex chromosome pair (2N = 28; FN = 51; Ross 1995, 1998), suggesting recovery of one linkage group per chromosome. To combine our genetic and physical maps, we mapped the ordered markers used to construct the genetic map to the genome assembly and anchored 37 scaffolds (99% of the total genome assembly) into 14 chromosomal linkage groups. Nine of the large scaffolds corresponded to single linkage groups, likely representing eight of the 13 autosomes and the X chromosome. Genotypes on the X chromosome were consistent with hemizygous male inheritance. The X chromosome scaffold was 119 Mb, which corresponds to ∼5.6% of the total assembly; results that seemingly contradict reports of an unusually large X based on karyotype data (i.e., ∼10% of the haploid female karyotype; Haaf et al. 1987). Four autosomes were recovered in two large scaffolds, while chromosome 5 was more fragmented with six assembled scaffolds (supplementary table S2, Supplementary Material online; numbering based on de novo genetic map lengths from Brekke et al. 2021). Scaffolds on chromosome 5 shared homology with at least four different chromosomes in rat (chromosomes 2, 13, 17, 19; supplementary fig S2, Supplementary Material online). It is possible that the less contiguous assembly of 5 reflects assembly issues related to structural evolution (fissions and fusions).
Finally, we used available mouse protein data, Phodopus transcriptome data sampled from 10 tissues, and ab initio predictions to generate and refine gene models. After filtering, we annotated 27,906 protein-coding and non-coding genes, including 87% (264 of 303) of the single-copy orthologs from the set of eukaryotic benchmarking genes (BUSCO, odb9; Simao et al. 2015). Of the annotated genes, 23,736 corresponded to protein-coding transcripts in the house mouse (mm10), of which 14,411 were 1:1 orthologs.
Widespread Suppression of X-Linked Recombination on the Xp Arm
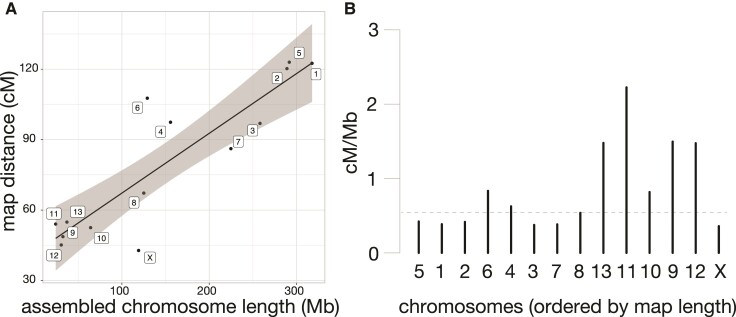
Our previous mapping experiment indicated map compression on one end of the X chromosome (Brekke et al. 2021). To quantify per-chromosome recombination rates and the magnitude of X-linked compression, we compared the number of base pairs anchored on each chromosome to its genetic length on the linkage map. Genome build size (Mb) predicted 76.6% of the variance in recombination frequencies (map distance in centimorgans or cM), with 11 chromosomes falling within the 95% confidence interval of the linear model (fig. 1A). The X chromosome was ∼1.7× the expected physical length predicted by genetic map distances (71.6 cM expected vs 41.3 cM observed; fig. 1). Recombination rates vary considerably across mammalian chromosomes and species, with X-linked rates typically lower in rodents in sex-averaged studies (Dumont and Payseur 2008). We estimated an X-linked female recombination rate in dwarf hamsters to be 0.36 cM/Mb (fig. 1B), compared to a genome-wide recombination rate of 0.55 cM/Mb which is similar to genome-wide rates in mice (0.52 cM/Mb) and rats (0.55 cM/Mb; rates from Dumont and Payseur 2008). The Mouse Genome Informatics genetic map reports a sex averaged X chromosome recombination rate of 0.43 cM/Mb.
Fig. 1.
Assembled length vs. genetic map distance for all chromosomes. (A) Assembled chromosome length, determined by the total length of scaffolds anchored to the chromosome level, compared with the length of each chromosome in the genetic map. Black line shows the linear relationship between genetic and physical distance, with the gray shading indicating the 95% confidence interval of the linear model. (B) Recombination rate (cM/Mb) for all chromosomes, ordered by genetic length in the anchoring map. Genome-wide recombination rate is indicated with a dashed line. Chromosome numbers are retained from Brekke et al (2021), which named linkage groups based on genetic length from a de novo map.
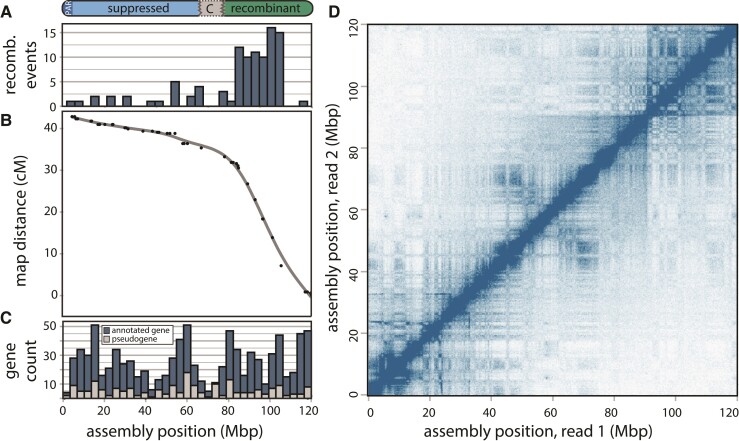
Centromeres are dynamic chromosomal features that include diverse families of repeats (Hartley and O'Neill 2019) and can have functional epi-alleles that vary even within populations (Purgato et al. 2015), making it difficult to localize centromeres in genome assemblies. Therefore, we examined crossover events across the assembled chromosomes and used the inherent reduction in recombination that results from condensed pericentric heterochromatin during meiosis to approximate the location of the centromeres in our assembly. Of the 13 autosomes, 10 displayed characteristic sinusoidal patterns with increased recombination near the distal regions of the chromosome arms (supplementary fig S3, Supplementary Material online), as seen in other mammals with metacentric centromeres (Jensen-Seaman et al. 2004). Two chromosomes showed patterns consistent with an assembly error (chromosomes 9 and 11), which were localized by the bounds of the markers in the genetic map but were unable to be further resolved without generating additional higher coverage long-read data for P. sungorus. In contrast, the X chromosome showed a partial sinusoidal pattern only for the first third (∼42 Mb) of the chromosome, with female recombination showing a reduction at the presumed centromere and then remaining suppressed along the rest of the X (fig. 2). Based on previous observations that recombination is mostly absent from the condensed Xp (Bikchurina et al. 2018), we used the inflection point of recombination suppression to partition the X chromosome assembly into two arms, ∼42 Mb (Xq) and ∼77 Mb (Xp) long respectively. Under this model, the Xp arm (i.e., the shorter arm under standard convention) actually comprises 65% of the assembled X Phodopus chromosome (77 of 119 Mb).
Fig. 2.
Recombination suppression, gene content, and chromatin configuration of the Phodopus X chromosome. (A) Recombination events are predominantly localized to a single arm of the X chromosome, which results in (B) an arm-specific map compression due to suppressed recombination. (C) Genes with complete gene models (blue) and putative pseudogenes with only expressed transcript support (gray) are distributed uniformly across the chromosome, despite the varying recombination levels. (D) Hi-C chromatin interaction plot, showing short- and long-range interactions between points on the X chromosome with a 250 Kbp resolution and a square root coverage normalization.
In the P. sungorus karyotype, the ‘shorter’ Xp arm usually appears as condensed chromatin, which obscures its physical length relative to the less condensed Xq arm. The distal end of the Xp also harbors the PAR, which is the location of X-Y homology, synapsis, and all X-linked recombination in males. To verify that the longer assembled arm was indeed the Xp, we generated whole genome shotgun (WGS) sequence data for one outcrossed male P. sungorus and two reciprocal hybrids between P. sungorus and P. campbelli. We then used normalized patterns of female versus male sequencing coverage (supplementary fig S4A, Supplementary Material online) and SNP density (supplementary fig S4B, Supplementary Material online) to localize the PAR to the distal 3 Mb of the longer arm. Hereafter, we refer to the suppressed and recombinant regions as the Xp and Xq chromosomal arms, respectively, with the caveat that additional work is needed to verify the exact location of the centromere (see Haenel et al. 2018). The overwhelming majority of observed female crossover events in our mapping panel occurred along the Xq arm, with an elevated estimated recombination rate of 0.78 cM/Mb relative to the Phodopus genome-wide rate of 0.55 cM/Mb. Conversely, female recombination was severely suppressed across the entire Xp arm (0.13 cM/Mb), with 74 Mb of 77 Mb presumably experiencing little or no recombination in either sex.
The observation of considerably reduced recombination across Xp is broadly consistent with heterochromatic suppression of recombination in females (Bikchurina et al. 2018). However, our recombination data derived from F1 females between P. sungorus and P. campbelli and, therefore, could also reflect species-specific structural differences. To address this alternative hypothesis, we evaluated short- and long-read mapping data and compared Hi-C chromatin configuration maps between the species. Comparisons of breakpoints identified by mapping of short read pairs support a ∼20 Mb inversion on the distal region of Xp (spanning 95–115 Mp on the P. sungorus assembly, supplementary fig S5, Supplementary Material online), which was further supported by 45 Nanopore long reads spanning the inferred breakpoints. Species differences in chromosome order were also detected from reductions in close chromosomal contacts in the P. campbelli Hi-C data at 115 Mb, compared to the P. sungorus assembly (supplementary fig S6C, Supplementary Material online). This apparent inversion only encompassed 26% of the Xp arm (20 of 77 Mb) and was insufficient to explain the broad recombination suppression extending well outside of this region.
Conservation of Ancestral Gene Content and Order Despite the Evolution of Extensive Repressive Heterochromatin
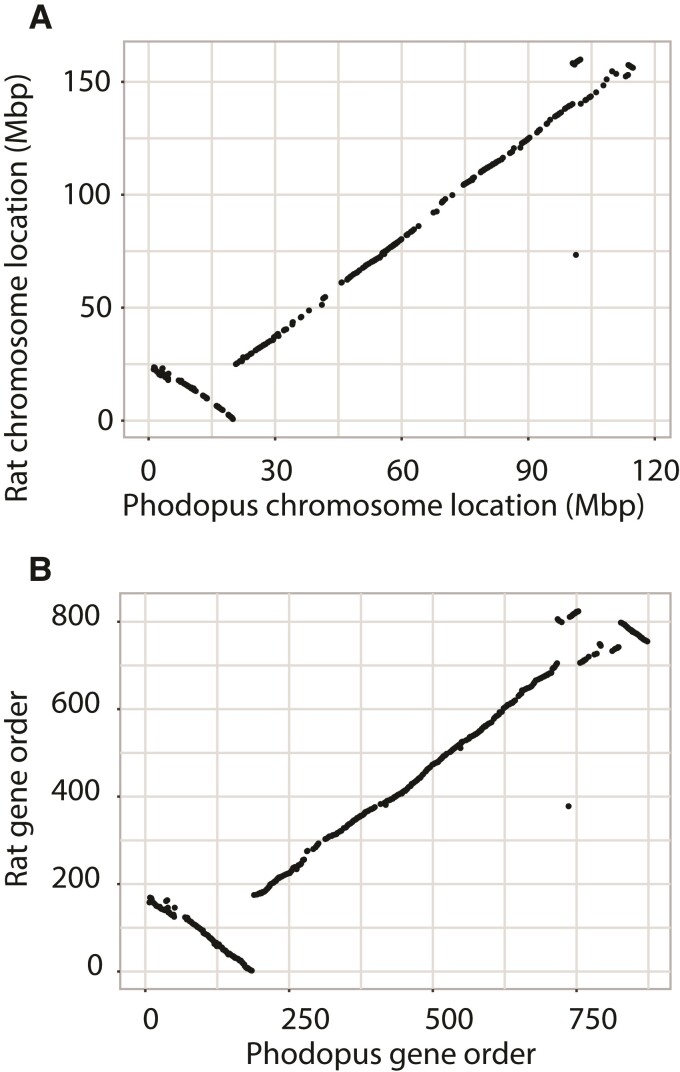
Extraordinary conservation of gene orders is one of the hallmarks of mammalian X chromosome evolution (Ohno 1973) and may reflect strong selection preserving three-dimensional chromosomal structure of the X (Brashear et al. 2021). To test whether dwarf hamsters have broadly maintained X chromosome synteny, we aligned the dwarf hamster X chromosome to the rat, mouse, dog, and human reference genomes. We also compared the relative order of our annotated genes to orthologous genes on the rat X chromosome. Genome-wide comparisons between the structure of rat and dwarf hamster support previous observations of elevated conservation of X synteny (supplementary fig S2, Supplementary Material online). Indeed, the X is the only chromosome that retains extensive synteny of gene content and order between these species. The distal end of the dwarf hamster Xp showed a similar pattern of rearrangement relative to both the dog and human references (supplementary fig S7, Supplementary Material online). We detected several mouse-specific rearrangements across the X relative to other species, consistent with previous reports (Gibbs et al. 2004; Brashear et al. 2021). However, we found high conservation of X-linked gene orders between the P. sungorus dwarf hamster reference and rat, with an apparent inversion on the distal end of Xp and slight re-ordering of genes on the distal end of Xq (fig. 3). The inversion breakpoints on Xp match up with the inversion detected between Phodopus species (supplementary fig S5, Supplementary Material online), suggesting that P. campbelli retains the same, presumably ancestral, gene order as rat.
Fig. 3.
Conservation of chromosome location and gene order between dwarf hamster and rat. Gene start position (Mb) (A) and relative order (B) are largely conserved between dwarf hamster and rat along the X chromosome.
Constitutive heterochromatic chromosomal regions are generally assumed to be depauperate of expressed genes, reflecting the repressive effects of condensed chromatin on transcription and enrichment of tandem repeats near pericentromeric and telomeric chromatin (Saksouk et al. 2015). Given this and the relatively large size of the Phodopus X chromosome, the condensed and darkly staining Xp arm has been assumed to be highly repetitive and largely non-genic (Haaf et al. 1987). Contrary to these predictions, we found that the Xp arm harbors approximately 54% of expressed genes on the Phodopus X chromosome (379 of 697 genes, supplementary table S3, Supplementary Material online), which is similar to the proportion of genes found on the orthologous and largely collinear arm in rat (57%, 472 of 824, supplementary table S3, Supplementary Material online). The Xp region did have fewer intact genes than expected given its proportional length in the dwarf hamster assembly (379 genes observed vs 432 genes expected, Fisher’s exact P < 0.0001). However, this pattern was also apparent in the orthologous region of the rat X (Fisher’s exact P < 0.0001), suggesting an ancestral reduction in gene density on Xp rather than gene loss subsequent to recombination suppression.
Even though we found many intact genes on the dwarf hamster Xp, it is possible that the presence of heterochromatin could still impact gene function by broadly suppressing levels of gene expression. When we considered all the tissues except for testis (supplementary fig S8A, Supplementary Material online), there was a slight reduction in expression on Xq relative to autosomes (pairwise Wilcoxon P = 0.018) but no difference in the expression levels of Xp relative to the autosomes (pairwise Wilcoxon P = 0.702) or Xq (pairwise Wilcoxon P = 0.126). Testis was examined separately (supplementary fig S8B, Supplementary Material online), as the X chromosome likely undergoes MSCI and postmeiotic repression during spermatogenesis, which is expected to reduce expression of many X-linked genes. Reductions in testis expression consistent with MSCI were apparent for both Xq (pairwise Wilcoxon P < 0.0001) and Xp (pairwise Wilcoxon P < 0.0001), with no difference in expression levels between the arms (pairwise Wilcoxon P = 0.44).
When we included transcribed pseudogenes in the counts of gene distribution across arms, rat no longer showed a deficit of genes on the region orthologous to hamster Xp (660 of 1084; Fisher’s exact P = 0.1194), indicating more ancient loss of functional genes in this region in rodents that pre-dates recombination suppression in Phodopus. When we included potential pseudogenes from dwarf hamster (i.e., assembled transcripts without a complete annotated gene model), Phodopus continued to have slightly fewer genes than expected based on length (511 of 896; Fisher’s exact P < 0.0001). This may indicate that dwarf hamsters have lost pseudogene expression relative to the rat, though it is more likely that this difference merely reflects more complete rat annotation.
Finally, if Xp were comprised of constitutive heterochromatin we should detect a clear signal of insulation from within-arm, long-range interactions from our Hi-C data as is seen in the inactive mouse X (Giorgetti et al. 2016). In contrast, our Hi-C results from liver tissue of both P. sungorus (fig. 2D) and P. campbelli (supplementary fig S6B, Supplementary Material online) revealed smaller interspersed blocks of topographically associated domains that indicate active transcription (Giorgetti et al. 2016). Thus, there was relatively little insulation on the arm with suppressed recombination, indicating that the heterochromatic state seen in metaphase karyotypes is tissue-dependent or perhaps transient in at least some transcriptionally active tissues.
Expansion of Transposable Element Content on Xp
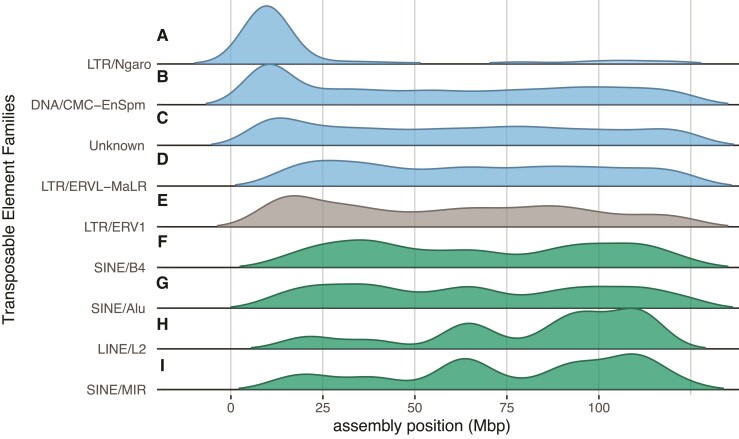
Another potential consequence of reduced recombination is the accumulation of transposable elements (TEs) due to a reduced efficacy of selection (Dolgin and Charlesworth 2008). To test for an accumulation of TEs and other repeats on Xp, we annotated repetitive sequences in our genome build and the rat genome (version rn6; Gibbs et al. 2004). If suppressed recombination leads to the accumulation of TEs and repeats, we might expect to see a relative enrichment of repeat families on the dwarf hamster Xp relative to the homologous arm in rat. We found support for this hypothesis, as repeat families enriched on Xq were shared between the two species, but repeat families enriched on Xp were found enriched predominantly in hamster only. Dwarf hamster and rat shared patterns of TEs enriched on the female-recombining Xq and several TE families (e.g., short and long interspersed nuclear elements [SINEs and LINEs]) were de-enriched on Xp and the homologous arm in rat (fig. 4F–I; supplementary table S4, Supplementary Material online), likely reflecting the shared evolutionary history of this region of the X chromosome. However, three of the four TE families enriched on Xp in dwarf hamster showed no evidence for repeat expansions in rat. Only one long terminal repeat (LTR) family of endogenous retroviruses (ERV1) was also enriched in the orthologous region of the rat X (fig. 4E). Two families (DNA/EnSpm and LTR/MaLR, fig. 4B, D) enriched on Xp in dwarf hamster were significantly de-enriched in rat (supplementary table S4, Supplementary Material online). We also found that the Ngaro family of LTRs (fig. 4A) was highly enriched on only the recombination-reduced Xp (170 of 179 copies, Fisher’s exact P < 0.0001). In addition to TEs, we identified enrichment of other repetitive element classes (simple repeats, low complexity sequence, and repeat families of unknown identity, fig. 4C) on the Phodopus Xp, but not the recombining orthologous arm in rat. Thus, suppression of recombination appears associated with the accumulation of some repetitive sequences on the Phodopus X chromosome.
Fig. 4.
Spatial distributions of the classes transposable elements significantly enriched on an arm of the Phodopus X chromosome. Distributions of TE families across the Phodopus X assembly, position in Mbp. The top four classes (A–D; blue) are enriched on Xp only in dwarf hamster, ERV1 (E, gray) is enriched on Xp in both dwarf hamster and rat, and the bottom four classes (F–I; green) are enriched on Xq in both dwarf hamster and rat. Significance determined by binomial test, see supplementary table S4, Supplementary Material online for values.
Molecular Evolution of the X Chromosome
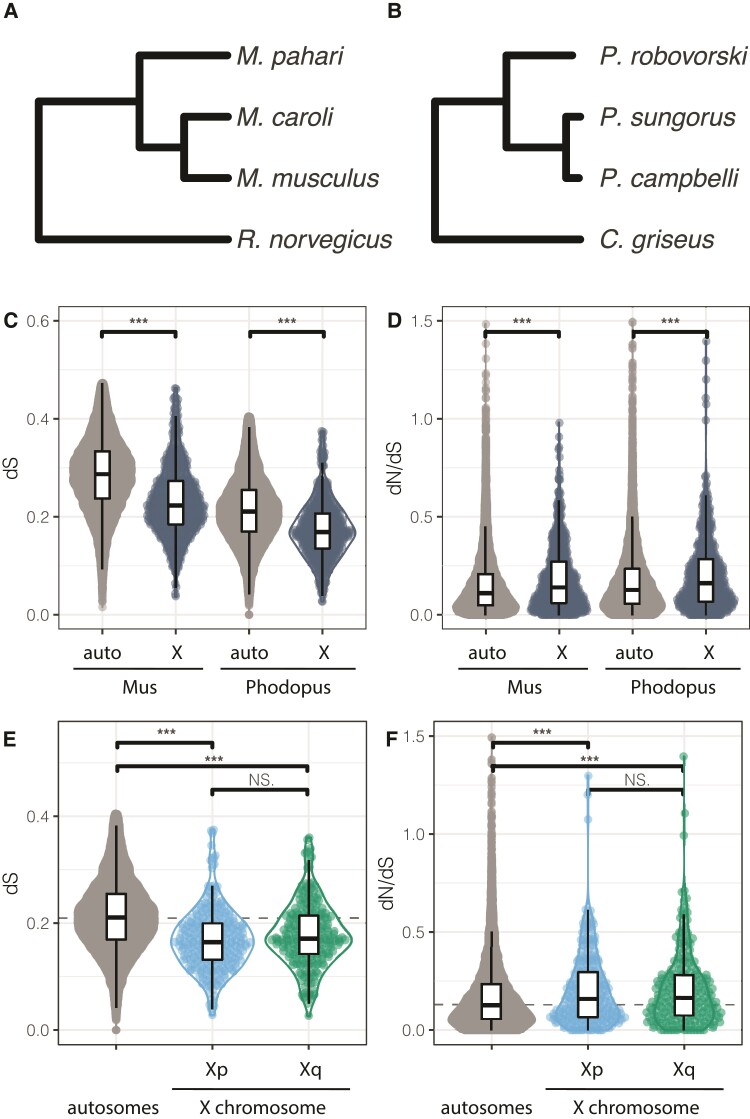
To quantify patterns of molecular evolution on the X chromosome and autosomes in dwarf hamsters, we evaluated the ratio of per site nonsynonymous (dN) and synonymous (dS) nucleotide changes in protein coding sequences (dN/dS). We estimated gene-wide rates in two parallel sets of four hamster or mouse species, each spanning similar evolutionary timescales (fig. 5A, B). Consistent with previous results (Kousathanas et al. 2014; Larson et al. 2016), we found faster-X protein-coding evolution for both mice (Mus dN/dSauto = 0.15; dN/dSX = 0.19, Wilcoxon P < 0.0001) and hamsters (Phodopus dN/dSauto = 0.17; dN/dSX = 0.21; fig. 5D, Wilcoxon P < 0.0001). We also found a decrease in the rate of synonymous changes on the X chromosome for both groups (Wilcoxon Mus P < 0.0001, Phodopus P < 0.0001) without a corresponding reduction in non-synonymous changes (fig. 5C). Interestingly, dS was similar between the Xp and Xq arms (fig. 5E) and we did not observe differences in dN/dS between the two arms (fig. 5F).
Fig. 5.
Evolutionary rates on autosomes and X chromosome arms in Phodopus. Relationships between the Murid species (including Mus, A) and Cricetid species (including Phodopus, B) are used to evaluate rates of molecular evolution between the autosomes and X chromosome, as determined by branch-specific rates of change in synonymous nucleotides (dS; C) and the ratio of rates of change in non-synonymous to synonymous nucleotides (dN/dS; D) for coding genes on the autosomes and X chromosome. For the Phodopus hamsters, the pattern of dS (E and dN/dS (F) was also evaluated by X chromosome arm. Asterisks indicate significant differences within panel at P < 0.05, Wilcoxon nonparametric multiple comparisons test. Dashed lines indicate genome-wide median.
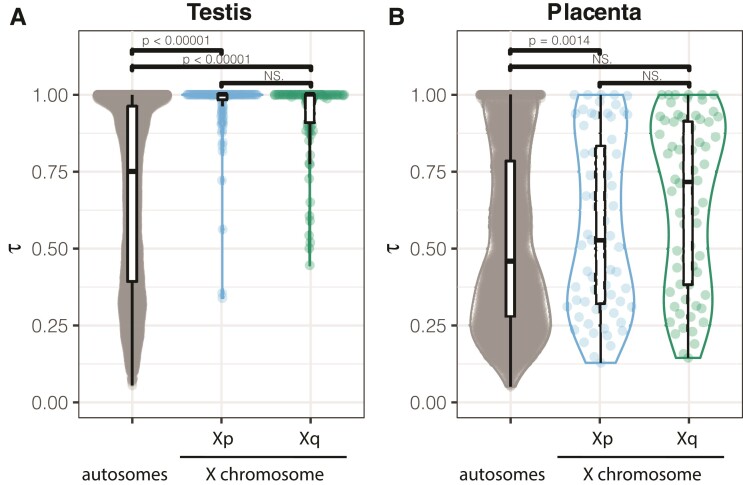
To test whether dwarf hamsters display sex-biased gene expression on the X chromosome, we examined gene expression across nine tissues (brain, heart, kidney, liver, muscle, spleen, uterus, placenta, and testis) using a specificity index (τ) to identify genes with tissue-enriched expression (supplementary table S5, Supplementary Material online; fig. 6). Approximately the same number of genes were expressed in each tissue (average = 17,904 ± 542 genes, range 15,117–19,693 genes), and a majority of these genes were annotated as protein-coding based on orthology with Mus (93%, 23,736 of 25,506 genes). The testis was enriched for tissue-specific expression; nearly one-third of all genes with a tissue specificity index (τ) greater than 0.8 were expressed in the testis (2,918 of 8,895 genes, supplementary table S5, Supplementary Material online), consistent with previous studies (Kryuchkova-Mostacci and Robinson-Rechavi 2017). We also detected enrichment on the X chromosome for testis-specific genes (164 genes, hypergeometric P < 0.0001, fig. 6A) and placenta-specific genes (50 genes, hypergeometric P < 0.0001, fig. 6B). When considered separately, each arm of the X chromosome was enriched for testis-specific expression (Xq, 67 genes, hypergeometric P = 0.0005; Xp, 97 genes, hypergeometric P < 0.0001, fig. 6A). However, only the female-recombining Xq arm was enriched for placenta-specific genes (29 genes, hypergeometric P < 0.0001, fig. 6B).
Fig. 6.
Tissue specificity in expressed genes. Tissue specificity score (τ) for genes with highest expression in testis (A) and placenta (B), by X chromosome region. Higher values of τ indicate higher tissue specificity in expression pattern.
When considering all genes expressed in the testis, rather than those with testis-specific expression, we no longer found an enrichment on the X chromosome (447 observed, 489 expected; Fisher’s exact P = 0.0572). The X showed a general de-enrichment of all placenta expressed genes (375 observed, 445 expected; Fisher’s exact P = 0.0037), which was likely driven by a greater reduction of expressed genes on Xp (199 observed, 241 expected; Fisher’s exact P = 0.0048) compared to Xq (186 observed, 202 expected; Fisher’s exact P = 0.1016).
We next evaluated protein-coding sequence evolution in the four tissues with at least ten tissue-specific genes on each arm of the hamster X chromosome (brain, testis, placenta, and uterus, supplementary fig S9, Supplementary Material online). In Mus, previous work has shown increased dN/dS of genes with male-biased expression on both the X and the autosomes, but no evidence for faster evolution in genes with female-biased expression (Kousathanas et al. 2014). In hamsters, we detected faster evolution of testis-biased genes across the autosomes and X compared to genome-wide values (Wilcoxon p <0.0001), with dN/dS for these genes on Xp slightly elevated relative to testis-biased autosomal genes (Wilcoxon P = 0.037, supplementary fig S9B, Supplementary Material online). We also found elevated dN/dS for X-linked placenta-specific genes, with placenta-biased genes evolving faster on Xq (Wilcoxon P = 0.012, supplementary fig S9C, Supplementary Material online). Tissue-specific genes are often associated with elevated dN/dS relative to the genome-wide baselines (Winter et al. 2004), presumably reflecting reduced evolutionary constraints. However, in our study only a small proportion of the variance in dN/dS was explained by overall expression levels (supplementary fig S10A, Supplementary Material online), and tissue specificity was a significant predictor of dN/dS only for some tissues (e.g., testis but not brain [supplementary fig S10B, Supplementary Material online]). In this limited survey, we detected significantly reduced rates for autosomal brain genes (Wilcoxon P < 0.0001) and an increase in dN/dS for a given τ on the X relative to the autosomes for all genes (supplementary fig S10C, Supplementary Material online), suggesting that tissue-specificity per se does not account for increased rates of X-linked evolution in dwarf hamsters.
Discussion
X chromosomes are often characterized by specialized gene contents that also tend to be highly conserved between species, reflecting the late stages of a presumably dynamic process shaped by diverse selective pressures. Here, we show that recombination suppression has evolved along most of an entire arm (∼65%) of the X chromosome in Phodopus dwarf hamsters. These findings provide an opportunity to evaluate the causes and evolutionary consequences of widespread recombination suppression on a mammalian sex chromosome.
The Evolution of Recombination Suppression on the Phodopus X Chromosome
Most studies that have examined recombination suppression in natural systems have focused on segregating structural inversions, which prevent the inverted region from forming successful Holliday junctions between alternate haplotypes. Pairing of sequence between sister chromatids is critical to successful crossing over during homologous recombination (Hunter 2015). On autosomes, segregating inversions have been found to prevent recombination between adaptive loci co-localized in the genome, linking together sets of genes for local adaptation (i.e., ‘supergenes’ in birds, butterflies, and Mimulus; reviewed in Thompson and Jiggins 2014). Inversions are also generally assumed to play a critical role in the evolution of heteromorphic sex chromosomes (Furman et al. 2020; Olito and Abbott 2020), including stratified divergence between the mammalian X and Y chromosomes through a series of rearrangements (Lahn and Page 1999). Indeed, inversions have been shown to maintain a heritable unit consisting of sexually antagonistic alleles and a sex determination gene in young, homomorphic sex chromosomes in cichlid fishes (Roberts et al. 2009). New inversions arise even on relatively old and conserved sex chromosomes—the mammalian X and avian Z chromosomes are enriched for inversions relative to autosomes (Connallon et al. 2018). Theory predicts that the higher efficacy of selection on the X should limit the accumulation of deleterious mutations in inversions in the short term, which ultimately should facilitate higher inversion fixation rates in the long term when compared with autosomes (Connallon et al. 2018).
Despite the intuitive link between structural changes and reduced recombination, we did not find large chromosomal rearrangements that easily explain the widespread suppression of recombination observed on the dwarf hamster Xp. Although our recombination map was generated with interspecies hybrids (Brekke et al. 2021), the lack of support for an inversion model is unsurprising in this instance. The Xp usually appears to be highly condensed within Phodopus species, with clear evidence for suppression of female recombination on the presumed Xp arm within both P. sungorus and P. campbelli based on immunolocalization of the mismatch repair protein MLH1 (Bikchurina et al. 2018). Thus, there are multiple lines of evidence for recombination suppression in crosses both within and between Phodopus species.
Karyotypes in P. sungorus and P. campbelli show strong C-banding of the Xp arm (Gamperl et al. 1977; Haaf et al. 1987), a hallmark of constitutive heterochromatin. However, these early studies also noted some variation between individuals in the condensation of Xp in different chromosome spreads, suggesting that the ‘constitutive’ heterochromatin in this region was unlike the condensed, gene-poor regions of chromosomes found predominantly near centromeres and telomeres. Our findings confirm that the heterochromatic state of Xp is likely transient rather than classically constitutive. Phodopus Xp shows a chromatin state consistent with that of at least one transcriptionally active X chromosome in female liver tissue (fig. 2; supplementary fig S6, Supplementary Material online), and the arm also has many genes expressed across tissues with no broad reduction in gene expression (supplementary fig S8, Supplementary Material online). One possible explanation is that Xp is condensed using canonical constitutive chromatin mechanisms, but in an inconsistent manner. Pericentromeric constitutive heterochromatin has been historically viewed as unchanging in the face of tissue type and environmental factors. However, research has increasingly uncovered dynamic complexity in the activity of the constitutive (H3K9me3) histone modification machinery (Saksouk et al. 2015), including expression of pericentromeric repeats during development and in actively proliferating tissues (Rudert et al. 1995). The Phodopus X chromosome appears to have maintained broad conservation of gene order over the last ∼23 million years since it shared a common ancestor with murine rodents (fig. 3; Schenk et al. 2013). Therefore, many interspersed repeats associated with constitutive chromatin would presumably have had to have evolved along the Xp without major changes in synteny.
Perhaps more likely, condensation of Xp may be occurring via facultative heterochromatin pathways, such as that used in X chromosome inactivation (XCI) in females (Zylicz and Heard 2020). These mechanisms are often context-dependent (e.g., sensitive to developmental timing and environmental cues) in addition to being sequence-dependent. XCI is initiated at the X inactivation center, where long noncoding RNA (lncRNA) Xist coats the X chromosome that will undergo inactivation (Penny et al. 1996), and then recruits the universal machinery for the formation of facultative heterochromatin in an X-specific manner as XCI progresses (Pintacuda et al. 2017). The X chromosome is prepared for heterochromatin formation even before Xist marks the inactive X, where enzymes critical to chromosome condensation are pre-loaded at specific enhancers along the X chromosome (Zylicz and Heard 2020). Thus, the X chromosome has sequences across its length that prime the formation of condensed heterochromatin in females. It is plausible such mechanisms could have been co-opted or expanded from the standard XCI pathway, inducing the transient heterochromatin state observed on Xp. Future studies examining histone marks indicative of facultative and constitutive heterochromatin are needed to test the epigenetic mechanisms underlying the formation of transient heterochromatin on Xp.
Considerable literature has focused on the role that inversions play in generating recombination suppression (e.g., Charlesworth 2017; Fuller et al. 2020; Olito and Abbott 2020). However, there remains relatively little direct evidence that rearrangements act as the primary proximate mechanism underlying the suppression of recombination on sex chromosomes (Furman et al. 2020). Regardless of the precise mechanisms, our results point to the relatively recent evolution of epigenetic suppression of recombination across a large portion of a rodent X chromosome. It is also possible that other sequence changes, such as smaller inversions, nucleotide divergence (Lukacsovich and Waldman 1999), or TE accumulation (Furman et al. 2020), generally contribute to reduced X-linked recombination frequencies between P. sungorus and P. cambelli. However, such mechanisms cannot explain suppression within these species (Bikchurina et al. 2018). In other eukaryotic taxa, reports suggesting that non-inversion mechanisms may be at play in suppressed recombination have been more limited. Sylvioidea songbirds have a ‘neo Z’ region (i.e., an autosomal fusion on the Z chromosome) with a history of complex, progressive recombination suppression and repeat accumulation (Sigeman et al. 2021). Perhaps more relevant, recombination suppression in a group of Ascomycete fungi pre-dates inversions between species (Sun et al. 2017), with a proposed mechanism involving increased DNA methylation generated via the fungus-specific repeat-induced point (RIP) mutation repair pathway (van Wyk et al. 2020).
Evolutionary Consequences of Recombination Suppression
In principle, a long-term reduction of recombination on Xp should lead to different evolutionary trajectories for the two arms of the Phodopus X and provide insights into general models of sex chromosome differentiation. First, the accumulation of repetitive elements and structural variants (i.e., inversions, insertion-deletions; Wright et al. 2016) are common features of sex chromosome evolution, and may be enriched in regions of historically low recombination (Rifkin et al. 2020). The accumulation of sex-linked repetitive elements is generally thought to result from recombination suppression (Charlesworth et al. 1986; Kent et al. 2017; but see Furman et al. 2020). Assuming our inference of a non-rearrangement mechanism of suppression is correct, the enrichment for TEs on Xp between P. campbelli and P. sungorus is likely a consequence of a change in the recombination landscape. Some genomic sequences are predisposed to TE insertions (i.e., TE hotspots; Levy et al. 2010). However, we observed expansions of several TE families on the Phodopus Xp relative to the largely syntenic rat X chromosome, suggesting the differential accumulation of lineage-specific repeats independent of the ancestral sequence landscape of the rodent X chromosome. Another prominent feature of X chromosome repetitive content is the evolution of ampliconic genes (Mueller et al. 2008) that are likely involved in genomic conflict during spermatogenesis (Cocquet et al. 2012). Unfortunately, these expansions are highly lineage-specific in rodents and our assembly was of insufficient quality to detect novel highly repetitive ampliconic regions in Phodopus.
The Phodopus system may also inform how recombination influences the dynamics of molecular evolution of the X chromosome. One outstanding question about the generation and maintenance of genetic diversity is the extent to which recombination itself is a mutagenic process. This question has been difficult to answer because recombination rate is correlated with other features such as centromere location (Jensen-Seaman et al. 2004), gene density, and hotspot activity (Paigen et al. 2008). In humans, sequencing evidence from sperm (Arbeithuber et al. 2015) and parent-offspring trios (Halldorsson et al. 2019) demonstrates that recombination leads to mutation. However, it is less clear the extent to which this phenomenon contributes to sequence divergence across taxa (Rousselle et al. 2020). Our finding of similar levels of synonymous sequence divergence on the Xp and Xq arms suggests that changes in recombination frequencies have so far been a negligible contributor to substitution rates since the origin of Xp suppression in the common ancestor of extant Phodopus species.
The mammalian X chromosome generally shows lower synonymous substitution rates, presumably due to higher mutation rates in males (i.e., male-driven molecular evolution; Cioppi et al. 2019) and female-biased transmission of the X chromosome (Miyata et al. 1987). Despite a reduction in baseline mutation, theory predicts more efficient selection in males on the hemizygous X chromosome (Rice 1984; Charlesworth et al. 1987), which may increase the fixation rate for recessive beneficial alleles (i.e., the faster-X effect), especially in the testis and other male-specific tissues. Consistent with this, the X chromosomes of several mammal species show relatively higher rates of protein-coding evolution (dN/dS). Our finding of elevated X-linked dN/dS in dwarf hamsters parallels results in mice and rats (fig. 5) and indicates that faster-X protein evolution is likely a general feature of rodent genomes.
Though far from settled, faster-X evolution in mammals has been interpreted as likely reflecting more effective positive selection for recessive beneficial mutations (reviewed in Meisel and Connallon 2013) including statistical evidence for more frequent positive directional selection on X-linked genes in mice (Kousathanas et al. 2014). It is generally difficult to ascribe adaptive versus non-adaptive processes to differences in rates of protein evolution on the X chromosome and autosomes (Mank et al. 2010b; Charlesworth et al. 2018); however, our analysis of protein-coding evolution on the Phodopus X, and Xp versus the Xq in particular, revealed some interesting patterns worthy of speculation. In general, faster-X protein evolution can occur through increased fixation of slightly deleterious variants when Ne on the X is much lower than the theoretical prediction of ¾ autosomal Ne (Vicoso and Charlesworth 2009). In mammals, however, Ne on the X is more likely to be elevated relative to theoretical predictions (i.e., >¾ autosomal Ne) due to higher variance in male reproductive success manifest from pre- and postcopulatory sexual selection (reviewed in Vicoso and Charlesworth 2009). Under these conditions, faster-X adaptive evolution is predicted over a much broader range of dominance levels, while X-linked fixations rates for deleterious variants should be much lower than on the autosomes (i.e., slower-X evolution; Vicoso and Charlesworth 2009). However, lower X-linked recombination can also enhance the linked effects of background purifying selection and sweeps due to positive selection or female meiotic drive, generally reducing Ne and the efficacy of natural selection on the X (i.e., Hill-Robertson interference; Hill and Robertson 1966). In Phodopus, if faster-X protein evolution was predominantly caused by elevated fixation of slightly deleterious mutations because of linked selection, then we would predict that this signal would be magnified on the largely non-recombinant Xp. We did not detect such differences, with both arms showing similarly elevated dN/dS relative to the autosomes (fig. 5).
We also observed elevated X-linked evolution for placental genes but only on Xq. Notably, Xq shows considerably higher recombination rates in females relative to the autosomes (0.78 cM/Mb versus the autosomal median rate of 0.63 cM/Mb) which should enhance the efficacy of selection on Xp, including the removal of deleterious mutations and the fixation of beneficial mutations. In general, female-limited genes are not predicted to show faster-X evolution as there is no X versus autosomal difference in dominance in females. However, hamster placentas show epigenetic silencing of the paternal X chromosome (Brekke et al. 2016), resulting in functional hemizygosity for genes expressed in these tissues (i.e., similar to XY males). Given these considerations, the elevated dN/dS signal in females is most consistent with more efficient positive selection.
The X chromosome is also predicted to accumulate genes with sexually antagonistic variation (Rice 1984), which can lead to an enrichment of sex-biased genes. Similar to predictions for faster-X evolution, the outcomes of selection for sex-biased gene contents are predicted to depend on the dominance of sexually antagonistic mutations. Genes with male-biased expression are predicted to accumulate on the X if male-beneficial mutations are on average recessive. Likewise, given female-biased transmission, female-biased genes are predicted to accumulate on the X if female-beneficial mutations are on average dominant. Interestingly, these theoretical predictions play out differently in different systems (Gurbich and Bachtrog 2008). In Drosophila, the X appears to be strongly de-enriched for genes with male-biased expression (Meiklejohn et al. 2003; Parisi et al. 2003), while mammals have been reported to be enriched for both male- and female-biased genes with some variation across studies (Lercher et al. 2003; Khil et al. 2004). In hamsters, we observed enrichment for both testis- and placenta-specific genes, but not for some other sex-specific tissues (i.e., uterus and ovary). These potentially conflicting patterns may be explained by inactivation of the paternal X chromosome in the placenta and other extraembryonic tissues of rodents (Okamoto and Heard 2006; Brekke et al. 2016) resulting in predominantly maternal expression of X-linked placental genes. Thus, placental genes are functionally hemizygous, similar to X-linked genes in males. Notably, we found that testis-specific genes were enriched on both arms of the X, while enrichment of placental-specific genes was restricted to the female-recombining Xq arm. These patterns are based on small numbers of genes but suggest that higher recombination may enhance the efficacy of sexually antagonistic selection in females. Functional enrichment of X-linked testis and placental expressed genes is not likely to occur due to alternative models of relaxed constraint and is more likely a reflection of long-term adaptive dynamics.
Finally, the unusual evolutionary dynamics on the Phodopus X chromosome may also be influencing the rapid evolution of reproductive barriers between dwarf hamster species. Male hybrid sterility between female P. campbelli and male P. sungorus has been attributed to asynapsis of the sex chromosomes during meiosis (Bikchurina et al. 2018), indicating that pairing may be inhibited by sequence or structural divergence that has accumulated on the PAR-containing Xp. Crosses between female P. sungorus and male P. campbelli result in extreme hybrid placental overgrowth caused by a large-effect X-linked quantitative trait locus (QTL) that disrupts of a network of autosomal genes expressed in the placenta (Brekke et al. 2021). With the availability of a genome assembly, we can now localize this QTL to near the P. sungorus Xq-Xp boundary (supplementary fig S11, Supplementary Material online). This signature could, for example, reflect centromeric incompatibilities perhaps related to female meiotic drive, although we have not observed evidence for drive in hybrids in this system (Brekke et al. 2021). Understanding if and how the evolution of widespread recombination suppression is related to speciation awaits a more detailed dissection of the genetic and mechanistic bases of these hybrid incompatibilities.
Materials and Methods
Animal Sampling
A single P. sungorus female was chosen for de novo genome sequencing and assembly from a breeding colony at the University of Montana (UM), originally established in 2011 descending from wild-caught animals collected 1981–1990 (Scribner and Wynne-Edwards 1994; see Brekke and Good 2014 for further details on P. sungorus and P. campbelli resources). The UM colony was maintained to maximize outbreeding, but levels of inbreeding are nonetheless elevated in these lines (Brekke et al. 2018). In addition to collecting liver from the reference individual for DNA sequencing, we collected seven snap-frozen tissues for use in transcriptome-based genome annotation (brain, heart, kidney, liver, muscle, spleen, and uterus). Testes from a single P. sungorus male from the same UM line were also collected to expand the annotation tissue panel. Augmenting the annotation panel, we collected and snap froze skin biopsies from molting P. sungorus females as part of an ongoing study on gene expression during seasonal molts. We sampled liver tissue from three additional males for shotgun genome sequencing: one outbred male P. sungorus was derived from a cross between a male from our original UM colonies and a female from supplemental stock kindly provided by Matthew Paul (University of Buffalo) in 2019, and two hybrid male embryos from reciprocal F1 crosses between P. sungorus and P. campbelli. Finally, we sequenced liver tissue from one female P. campbelli, derived from the UM research colony, and one male P. roborovskii sourced from the pet trade. All animal care and breeding was performed at UM with oversight of the University of Montana Institutional Animal Care and Use Committee (animal use protocols 039-13JGDBS-090413, 050-16JGDBS-082316 & 035-19JGDBS-062519).
Genome and Transcriptome Sequencing
Liver tissue was sent to Dovetail Genomics (Santa Cruz, CA) for generation of all sequencing data used in the de novo assembly, including Illumina whole genome sequencing (150 bp PE, Illumina HiSeq X) augmented with Dovetail's complementary Chicago (relying on artificial chromatin constructed in vitro) and Hi-C (relying on intact chromosomes in situ) long-range proximity ligation data. The libraries were prepared using Dovetail kits according to the manufacturer’s protocol. For Hi-C libraries, chromatin was fixed in place with formaldehyde in the nucleus and then extracted. Fixed chromatin was digested with DpnII, the 5′ overhangs filled with biotinylated nucleotides, and free blunt ends ligated. After ligation, cross-linking was reversed and DNA was purified. Following purification, biotin not contained in the center of the ligated fragment was removed. Chicago libraries were prepared similarly, except DNA was removed from the nucleus and formed into artificial chromatin rather than fixed in situ. For all libraries, DNA was sheared to ∼350 bp mean size and libraries were generated using Illumina-compatible adaptors.
We also generated an additional Hi-C library from P. campbelli liver tissue, processed following the same Dovetail protocols as the libraries generated for genome assembly. Whole genome sequencing libraries from P. campbelli, P. roborovskii, and three hybrid males were constructed using the NEBNext Ultra II DNA kit (New England Biolabs, E7645S). RNAseq libraries were constructed from all reference tissues at the University of Montana using an Illumina TruSeq Stranded mRNA library kit (Illumina, 20020595) following manufacturer’s instructions, with the exception of the testis library, which was constructed at Novogene (Davis, CA). All tissues were sequenced on 3 lanes of 150 bp PE Illumina Hiseq 4000 (reference transcriptomes) and 3 lanes of 150 bp PE Illumina Hiseq X (HiC and whole genome libraries) at Novogene.
High molecular weight DNA for long read sequencing was extracted from the sequenced P. sungorus and P. campbelli females using the MagAttract HMW DNA kit (Qiagen, 67563) and assessed using a genomic DNA ScreenTape (Agilent 5067–5365) on a 2200 TapeStation (Agilent, G2965AA). Libraries were constructed and sequenced in the UM Genomics Core (Missoula, MT) using the MinION flowcell platform (Oxford Nanopore, sequencing kit SQK-LSK109 and flow cell FLO-MIN106D).
Genome Assembly and Annotation
An initial shotgun sequence-based assembly was generated with 150 bp PE Illumina Hiseq data. Reads were trimmed with Trimmomatic (v0.39, ILLUMINACLIP to remove adaptors, LEADING:20, SLIDINGWINDOW:13:20, MINLEN:23). The hybrid read-based/k-mer assembler Meraculous was used to generate a de novo assembly, with a constrained heterozygous (II) model with 79-mers and homozygous peak depth 40.0 selected for depth of coverage and balance between repetitive and heterozygous fractions (Chapman et al. 2011). Reads from the additional Chicago and Hi-C libraries were used in conjunction with the de novo assembly as input for Dovetail’s HiRise genome assembly pipeline (Putnam et al. 2016). The first assembly iteration aligned Chicago library sequences to the draft assembly with a modified SNAP mapper (Zaharia et al. 2011). HiRise analyzed the distance between Chicago read pairs within scaffolds using a likelihood model, which was used to identify joins and misjoins for scaffolding. The second assembly iteration used the same strategy, scaffolding with the proximity-ligation HiC data.
To assess assembly quality on a per-site basis, we re-mapped the raw reads that went into the assembly and ran Referee (Thomas and Hahn 2019) on the resulting BAM file to calculate genotype likelihoods while accounting for mapping quality. Sites in which the called base in the assembly does not match one of the two alleles in the most likely genotype are inferred as errors and can be corrected by inserting one of the alleles from that genotype.
We generated a reference-guided genetic map using data from our previously published Phodopus genetic map based on high-throughput genotypes (ddRAD tags) from 189 backcross individuals (F1 female [P. campbelli female x. P. sungorus male]×P. campbelli male; Brekke et al. 2021). Processed RAD tags were aligned to the reference genome using the Burrows-Wheeler aligner (BWA) (Li and Durbin 2009), and variants were called using HaplotypeCaller (gatk, v4; McKenna et al. 2010). The genetic map was constructed using rQTL as described previously (Brekke et al. 2021), with the exception that duplicate markers were only dropped from the analysis if they were mapped to the same genomic scaffold. While these markers were redundant for identifying recombination events in the genetic map, they were informative for linking unanchored scaffolds and retained for scaffold placement. Integration of genetic mapping data onto the Phodopus build also allowed to re-estimate and localize the X-linked single QTL associated with variation in hybrid backcross placental weight (see Brekke et al. 2021 for analytical details).
We used the MAKER3 pipeline (v3.01.03; Holt and Yandell 2011) to annotate gene models using ab initio and transcript-based evidence. Initial reference-guided de novo transcriptome assemblies were built with the Tuxedo pipeline (bowtie v2.3.4.3; Langmead and Salzberg 2012, tophat v2.1.1 and cufflinks v2.2.1; Trapnell et al. 2012) including data from all reference tissues and published data previously used for de novo P. sungorus placental transcriptomes (Brekke et al. 2016). These transcriptomes were given to the MAKER pipeline, which combined the Phodopus expression data, SNAP and Augustus ab initio gene prediction models, and protein homology to Mus to predict the location of genes and genic features. These genes were then matched with Mus (mm10) and Rattus (rnor6) identity through reciprocal best hit basic local alignment search tool (BLAST) searches (Camacho et al. 2009), using a max e-value cutoff of 1e-3 and then taking the longest remaining match between the two species.
Identification of the PAR and X-Linked Structural Variation
For each all WGS samples, reads were trimmed (Trimmomatic v0.39; Bolger et al. 2014) and aligned to the P. sungorus assembly using BWAmem (v0.7.17; Li and Durbin 2009). Read depth was called using ‘samtools depth’ and averaged across 2500 bp sliding windows (Li et al. 2009). For the X chromosome, average coverage in each window was normalized by median coverage, and adjusted to reflect ploidy (ie. female ploidy is 2N). Next, we filtered out all windows that included bases overlapping with any annotated repeat (Repeatmasker; Smit et al. 2013–2015). We excluded regions annotated for STRs/low complexity sequences and all windows where the P. sungorus female had coverage over the 95% quantile of coverage (1.68X the median) in order to remove artifacts resulting from collapsed repeats or paralogous genes in the assembly. We used the same custom library to identify repetitive regions in both Phodopus and Rattus norvegicus (rnor6) as was used to repeat mask the genome for annotation.
We used four approaches to evaluate structural variation on the Phodopus X chromosome. First, we identified putative interspecific inversions using mapping discordance between P. campbelli and P. sungorus Illumina paired-end reads mapped to the X chromosome assembly (Breakdancer v1.3.6; Chen et al. 2009). For each region of the genome with an identified structural rearrangement, Breakdancer outputs Read 1 and Read 2 locations along the assembly. We used analysis based on P. sungorus reads to control for any signal due to genome assembly error or repetitive content, and evaluated the difference between the reference and P. campbelli reads to find species-specific inversions by normalizing all counts to the reference female, which was sequenced to higher sequence coverage. Second, we mapped Oxford Nanopore ultralong reads from P. sungorus and P. campbelli to the assembly using minimap2 (-ax map-ont; v2.17; Li 2018) and then used IGV (v2.7.2; Robinson et al. 2011) to visually verify the large distal inversion by identifying that mapped across the inversion breakpoint, comparing these to a list of 45 Nanopore reads with regions mapped to either side of the inversion. Third, raw HiC reads from both P. sungorus and P. campbelli were processed with the Juicer pipeline to align and call HiC contact points (Durand et al. 2016b), normalized using the Knight-Ruiz method. These matrices were then visualized and compared using Juicebox (v1.11.08; Durand et al. 2016a). Fourth, we evaluated gene order/synteny across mammals. To assess synteny, we downloaded X chromosome assemblies from Ensembl (release 100) for Rattus norvegicus (rat, rnor6), Mus musculus (mouse, mm10), Homo sapiens (human, GrCh38), and Canis familiaris (domestic dog, CanFam3.1). For each species, we used minimap2 (Li 2018) to align conserved segments from the P. sungorus X chromosome to the target reference (-asm20). Gene order was also evaluated by comparing the locations of Phodopus genes with those of the rat genes identified through reciprocal BLAST (described above). Recombination rate was evaluated using the RAD markers used to anchor the genome to the genetic map, using only the sequence length between markers to avoid including sequence without sampled recombination data.
Gene Expression
We performed gene expression analysis using Hisat2 (v 2.2.0; Sirén et al. 2014) to map reads to the genome assembly, and then used the MAKER-generated annotation to count reads with FeatureCounts (v2.0.1; Liao et al. 2014). We allowed for multiple mapping but proportionally distributed multiply mapped reads across target genes. Counts were normalized using transcripts per million (TPM), and log2 transformed for analysis. All placenta samples were quantified following reference alignment, and the average TPM across samples was used to evaluate tissue specificity. We calculated the tissue specificity metric τ as described, using the established cutoff of τ ≥ 0.8 (Kryuchkova-Mostacci and Robinson-Rechavi 2017). Expressed genes were also evaluated for overlap protein-coding molecular evolution metrics (see below).
Molecular Evolution
We downloaded orthologous protein coding sequences for five species from Ensembl (release 100): R. norvegicus, M. musculus, M. caroli, M. pahari, and C. griseus. We used these sequences in conjunction with the P. sungorus genome and reference-guided assemblies of P. campbelli and P. roborovskii, constructed using iterative mapping to the P. sungorus to reduce reference bias. These iterative assemblies were as implemented with a modified version of pseudo-it (v.beta 3.1; Sarver et al. 2017) that incorporates insertion-deletion variation. For each species, we ran pseudo-it for four iterations and used the UCSC liftOver tool (University of California, Santa Cruz Genomics Institute) to transfer P. sungorus gene annotations to each pseudo-assembly.
We split the eight species into two groups of four species that spanned approximate similar evolutionary timescales. For the hamster group (Cricitidae: P. sungorus, P. campbelli, P. roborovskii, and C. griseus), we used BLAST (Camacho et al. 2009) to obtain orthologous protein coding transcripts between the C. griseus genome and P. sungorus by setting a max e-value cutoff of 1e-3 and then taking the longest remaining match between the two species. This resulted in 16,569 single-copy orthologs between these five species. For the mouse/rat group (Muridae: M. musculus, M. caroli, M. pahari, and R. norvegicus), we used Ensembl annotation to identify 15,772 single-copy orthologs. We aligned orthologous coding-sequences in both groups using MACSE (Ranwez et al. 2011), which produces in-frame codon alignments while accounting for frameshifts. First, sequences that contain non-homologous sequences were trimmed with the trimNonHomologousFragments sub-program. In some cases, this removed an entire sequence within the orthogroup, and this entire group was removed from subsequent steps. In total, 2,498 genes were removed from the hamster group and 266 were removed from the mouse group. Next, we aligned sequences with the main MACSE program, alignSequences (Ranwez et al. 2011). We followed this with a post-alignment trimming step using the trimAlignment sub-program, after which we calculated summary statistics for each alignment and removed those that were short (<100 bp), contained a high proportion of sites with gaps or missing data (>20%), contained identical sequences, or contained premature stop codons. In total, using these filters we removed 527 genes from the hamsters and 79 genes from the mice. Following all alignment, trimming and filtering steps, we recovered 13,546 in-frame codon alignments for the hamsters and 15,429 from the mice.
Using these alignments, for both groups, we estimated maximum-likelihood gene trees using IQ-Tree (Minh et al. 2020) and estimated dN and dS for each gene using the corresponding gene trees in order to minimize the potential effects of discordance (Mendes and Hahn 2016). Species level phylogenies were also verified from maximum likelihood analysis of a concatenated alignment (fig. 5A, B), but were not used for the analysis of molecular evolution. We used HyPhy (Pond et al. 2004) to estimate dN/dS using the FitMG94.bf model (https://github.com/veg/hyphy-analyses), estimating a single dN/dS value per gene and summarizing dN and dS separately by summing their values over all branches (fig. 5). For each dataset, we flagged genes with dS values above the top 98th percentile of all genes as having potentially poor alignments and filtered them from subsequent analysis (supplementary fig S12, Supplementary Material online; 271 Cricetidae genes and 309 Muridae genes).
Statistical Analyses
All basic statistical tests were performed in R v 4.1.0 (RCoreTeam 2017). Non-parametric models were performed with the Kruskal Wallace test function (kruskal.test) and posthoc p-values were evaluated with pairwise.wilcox.test. Linear models we performed using lm function for standard models, and lmer for mixed models. Enrichment was tested either with hypergeometric test (for enrichment of categories, phyper) or Fisher’s exact binomial test (for enrichment based on expected distributions from length, binom.test). Multiple testing was corrected using the Bonferroni Holm method.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgements
We thank the Good lab for helpful comments on data analysis and interpretation. Two anonymous reviewers provided thoughtful comments that improved the manuscript. Ned Place, Matthew Paul, and Robert Johnston provided animal stocks. Kelly Carrick, Jess Wexler, and the University of Montana LAR staff assisted with animal care. Mark Daly, Thomas Swale, and Dovetail Genomics LLC performed genome sequencing and assembly. Denghui David Xing performed Nanopore sequencing at the UM Genomics Core. This research was supported by grants from the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health (R01-HD094787 to JMG), a Matching Funds Award from Dovetail Genomics, a grant from the National Science Foundation (DEB 1754096 to JMG), and the National Science Foundation Graduate Research Fellowship Program (EEKK: DGE-1313190, and KEH DGE-2034612). This study included research conducted in the University of Montana Genomics Core, supported by a grant from the M. J. Murdock Charitable Trust (to JMG). Computational resources and support from the University of Montana’s Griz Shared Computing Cluster (GSCC), supported by grants from the National Science Foundation (CC-2018112 and OAC-1925267, JMG co-PI), contributed to this research.
Contributor Information
Emily C. Moore, Division of Biological Sciences, The University of Montana, Missoula, Montana, 59812, USA
Gregg W. C. Thomas, Division of Biological Sciences, The University of Montana, Missoula, Montana, 59812, USA
Sebastian Mortimer, Division of Biological Sciences, The University of Montana, Missoula, Montana, 59812, USA.
Emily E. K. Kopania, Division of Biological Sciences, The University of Montana, Missoula, Montana, 59812, USA
Kelsie E. Hunnicutt, Department of Biological Sciences, The University of Denver, Denver, Colorado, 80208, USA
Zachary J. Clare-Salzler, Division of Biological Sciences, The University of Montana, Missoula, Montana, 59812, USA
Erica L. Larson, Department of Biological Sciences, The University of Denver, Denver, Colorado, 80208, USA
Jeffrey M. Good, Division of Biological Sciences, The University of Montana, Missoula, Montana, 59812, USA
Author Contributions
E.C.M., G.W.C.T., and J.M.G. conceived of the analyses; E.C.M., E.E.K.K., K.E.E.H., Z.J.C.S., and E.L.L. collected tissues and constructed sequencing libraries; S.M. and E.C.M. performed genome annotation; E.C.M. and G.W.C.T. conducted all other analyses; and E.C.M., G.W.C.T., and J.M.G. wrote the manuscript with feedback from all authors.
Data Availability
Data available through SRA BioProject number PRJNA786909 and PRJNA787539.
Literature Cited
- RCoreTeam . 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Arbeithuber B, Betancourt AJ, Ebner T, Tiemann-Boege I. 2015. Crossovers are associated with mutation and biased gene conversion at recombination hotspots. Proc Natl Acad Sci. 112:2109–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao R, et al. 2019. Genome sequencing and transcriptome analyses of the Siberian hamster hypothalamus identify mechanisms for seasonal energy balance. Proc Natl Acad Sci. 116:13116–13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikchurina TI, et al. 2018. Chromosome synapsis and recombination in male-sterile and female-fertile interspecies hybrids of the dwarf hamsters (Phodopus, Cricetidae). Genes (Basel). 9:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracewell RR, Bentz BJ, Sullivan BT, Good JM. 2017. Rapid neo-sex chromosome evolution and incipient speciation in a major forest pest. Nat Commun. 8:1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brashear WA, Bredemeyer KR, Murphy WJ. 2021. Genomic architecture constrained placental mammal X chromosome evolution. Genome Res. 31:1353–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke TD, et al. 2021. X chromosome-dependent disruption of placental regulatory networks in hybrid dwarf hamsters. Genetics. 218:iyab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke TD, Good JM. 2014. Parent-of-origin growth effects and the evolution of hybrid inviability in dwarf hamsters. Evolution. 68:3134–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke TD, Henry LA, Good JM. 2016. Genomic imprinting, disrupted placental expression, and speciation. Evolution. 70:2690–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke TD, Steele KA, Mulley JF. 2018. Inbred or outbred? Genetic diversity in laboratory rodent colonies. G3: Genes Genomes Genet. 3:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinform. 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, et al. 2011. Meraculous: de novo genome assembly with short paired-end reads. PLoS One. 6:e23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2017. Evolution of recombination rates between sex chromosomes. Philos Trans R Soc B Biol Sci. 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2021. When and how do sex-linked regions become sex chromosomes? Evolution. 75:569–581. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Campos JL, Jackson BC. 2018. Faster-X evolution: theory and evidence from Drosophila. Mol Ecol. 27:3753–3771. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. 1987. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 130:113–146. [Google Scholar]
- Charlesworth B, Langley C, Stephan W. 1986. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics. 112:947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, et al. 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 6:677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioppi F, Casamonti E, Krausz C. 2019. Age-dependent de novo mutations during spermatogenesis and their consequences. Adv Exp Med Biol. 1166:29–46. [DOI] [PubMed] [Google Scholar]
- Cocquet J, et al. 2012. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 8:e1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, et al. 2018. Local adaptation and the evolution of inversions on sex chromosomes and autosomes. Philos Trans R Soc B Biol Sci. 373:20170423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG. 2014. Evolutionary inevitability of sexual antagonism. Proc R Soc B Biol Sci. 281:20132123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couger MB, et al. 2021. Sex chromosome transformation and the origin of a male-specific X chromosome in the creeping vole. Science. 372:592–600. [DOI] [PubMed] [Google Scholar]
- Darolti I, et al. 2019. Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers. Proc Natl Acad Sci. 116:19031–19036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin ES, Charlesworth B. 2008. The effects of recombination rate on the distribution and abundance of transposable elements. Genetics. 178:2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. 2008. Evolution of the genomic rate of recombination in mammals. Evolution. 62:276–294. [DOI] [PubMed] [Google Scholar]
- Durand NC, et al. 2016a. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Systems. 3:99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, et al. 2016b. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Systems. 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, et al. 2020. Extensive recombination suppression and epistatic selection causes chromosome-wide differentiation of a selfish sex chromosome in Drosophila pseudoobscura. Genetics. 216:205–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman BLS, et al. 2020. Sex chromosome evolution: so many exceptions to the rules. Genome Biol Evol. 12:750–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamperl R, Vistorin G, Rosenkranz W. 1977. New observations on the karyotype of the Djungarian hamster, Phodopus sungorus. Experientia. 33:1020–1021. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 428:493–521. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature. 535:575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurbich TA, Bachtrog D. 2008. Gene content evolution on the X chromosome. Curr Opin Genet Dev. 18:493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Weis H, Schmid M. 1987. A comparative cytogenetic study on the mitotic and meiotic chromosomes in hamster species of the genus Phodopus (Rodentia, Cricetinae). Z Säugetierkund. 52:281–290. [Google Scholar]
- Haenel Q, Laurentino TG, Roesti M, Berner D. 2018. Meta-analysis of chromosome-scale crossover rate variation in eukaryotes and its significance to evolutionary genomics. Mol Ecol. 27:2477–2497. [DOI] [PubMed] [Google Scholar]
- Halldorsson BV, et al. 2019. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science. 363. [DOI] [PubMed] [Google Scholar]
- Hartley G, O'Neill RJ. 2019. Centromere repeats: hidden gems of the genome. Genes (Basel). 10:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. 1966. The effect of linkage on limits to artificial selection. Genetics Res. 8:269–294. [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 7:a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, et al. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14:528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent TV, Uzunovic J, Wright SI. 2017. Coevolution between transposable elements and recombination. Philos Trans R Soc B Biol Sci. 372:20160458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 36:642–646. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8:e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A, Halligan DL, Keightley PD. 2014. Faster-X adaptive protein evolution in house mice. Genetics. 196:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryuchkova-Mostacci N, Robinson-Rechavi M. 2017. A benchmark of gene expression tissue-specificity metrics. Brief Bioinform. 18:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 1999. Four evolutionary strata on the human X chromosome. Science. 286:964–967. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, Keeble S, Vanderpool D, Dean MD, Good JM. 2016. The composite regulatory basis of the large X-effect in mouse speciation. Mol Biol Evol. 34:282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. 2003. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol Biol Evol. 20:1113–1116. [DOI] [PubMed] [Google Scholar]
- Levy A, Schwartz S, Ast G. 2010. Large-scale discovery of insertion hotspots and preferential integration sites of human transposed elements. Nucleic Acids Res. 38:1515–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. [DOI] [PubMed] [Google Scholar]
- Lukacsovich T, Waldman AS. 1999. Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics. 151:1559–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Nam K, Ellegren H. 2010a. Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol. 27:661–670. [DOI] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. 2010b. Effective population size and the faster-X effect: emperical results and their interpretation. Evolution. 64:663–674. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Parsch J, Ranz JM, Hartl DL. 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc Natl Acad Sci. 100:9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T. 2013. The faster-X effect: integrating theory and data. Trends Genet. 29:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes FK, Hahn MW. 2016. Gene tree discordance causes apparent substitution rate variation. Syst Biol. 65:711–721. [DOI] [PubMed] [Google Scholar]
- Minh BQ, et al. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. 1987. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol. 52:863–867. [DOI] [PubMed] [Google Scholar]
- Mueller JL, et al. 2008. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nature Genetics. 40:794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1967. Sex chromosomes and sex-linked genes. Berlin: Springer Verlag. [Google Scholar]
- Ohno S. 1973. Ancient linkage groups and frozen accidents. Nature. 244:259–262. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Heard E. 2006. The dynamics of imprinted X inactivation during preimplantation development in mice. Cytogenet Genome Res. 113:318–324. [DOI] [PubMed] [Google Scholar]
- Olito C, Abbott JK. 2020. The evolution of suppressed recombination between sex chromosomes by chromosomal inversions. bioRxiv:2020.2003.2023.003558. [DOI] [PubMed]
- Paigen K, et al. 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet. 4:e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, et al. 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 299:697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak S, Stock AD. 1974. The X chromosomes of mammals: karyological homology as revealed by banding techniques. Genetics. 78:703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel CL, et al. 2020. Assembly of the threespine stickleback Y chromosome reveals convergent signatures of sex chromosome evolution. Genome Biol. 21:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature. 379:131–137. [DOI] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GAB. 2012. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci. 109:5346–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G, et al. 2017. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol Cell. 68:955–969.e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. 2004. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 21:676–679. [DOI] [PubMed] [Google Scholar]
- Ponnikas S, Sigeman H, Abbott JK, Hansson B. 2018. Why do sex chromosomes stop recombining? Trends Genet. 34:492–503. [DOI] [PubMed] [Google Scholar]
- Purgato S, et al. 2015. Centromere sliding on a mammalian chromosome. Chromosoma. 124:277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. 2011. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS One. 6:e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius B, et al. 2012. Abundance of female-biased and paucity of male-biased somatically expressed genes on the mouse X-chromosome. BMC Genomics. 13:607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 38:735–742. [DOI] [PubMed] [Google Scholar]
- Rifkin JL, et al. 2020. Widespread recombination suppression facilitates plant sex chromosome evolution. Mol Biol Evol. 38:1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RB, Ser JR, Kocher TD. 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 326:998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PD. 1994. Phodopus roborovskii. Mamm Species. 459:1–4. [Google Scholar]
- Ross PD. 1995. Phodopus campbelli. Mamm Species. 503:1–7. [Google Scholar]
- Ross PD. 1998. Phodopus sungorus. Mamm Species. 595:1–9. [Google Scholar]
- Rousselle M, et al. 2020. Is adaptation limited by mutation? A timescale-dependent effect of genetic diversity on the adaptive substitution rate in animals. PLoS Genet. 16:e1008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudert F, Bronner S, Garnier J-M, Dolle P. 1995. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome. 6:76–83. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Simboeck E, Dejardin J. 2015. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin. 8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver BAJ, et al. 2017. Phylogenomic insights into mouse evolution using a pseudoreference approach. Genome Biol Evol. 9:726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk JJ, Rowe KC, Steppan SJ. 2013. Ecological opportunity and incumbency in the diversification of repeated continental colonizations by muroid rodents. Syst Biol. 62:837–864. [DOI] [PubMed] [Google Scholar]
- Scribner SJ, Wynne-Edwards KE. 1994. Thermal constraints on maternal behavior during reproduction in dwarf hamsters (Phodopus). Physiol Behav. 55:897–903. [DOI] [PubMed] [Google Scholar]
- Sigeman H, et al. 2021. Avian neo-sex chromosomes reveal dynamics of recombination suppression and W degeneration. Mol Biol Evol: MSAB. 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Sirén J, Välimäki N, Mäkinen V. 2014. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans Comput Biol Bioinform. 11:375–388. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. < http://www.repeatmasker.org >. [Google Scholar]
- Steinlechner S, Heldmaier G, Becker H. 1983. The seasonal cycle of body weight in the Djungarian hamster: photoperiodic control and the influence of starvation and melatonin. Oecologia. 60:401–405. [DOI] [PubMed] [Google Scholar]
- Sun Y, Svedberg J, Hiltunen M, Corcoran P, Johannesson H. 2017. Large-scale suppression of recombination predates genomic rearrangements in Neurospora tetrasperma. Nat Commun. 8:1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surbhi SK, Kyne RF, Nelson RJ, Paul MJ. 2019. Photoperiod regulates hypothalamic miR-155 gene expression in female, but not male, Siberian hamsters (Phodopus sungorus). Behav Neurosci. 133:240–246. [DOI] [PubMed] [Google Scholar]
- Thomas GWC, Hahn MW. 2019. Referee: reference assembly quality scores. Genome Biol Evol. 11:1483–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, Jiggins CD. 2014. Supergenes and their role in evolution. Heredity. 113:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk S, Wingfield BD, De Vos L, van der Merwe NA, Steenkamp ET. 2020. Genome-wide analyses of repeat-induced point mutations in the Ascomycota. Front Microbiol. 11:622368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrunes F, et al. 2008. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18:965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. 2009. Effective population size and the faster-X effect: an extended model. Evolution. 63:2413–2426. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 3:e3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. 2004. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 14:54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Dean R, Zimmer F, Mank JE. 2016. How to make a sex chromosome. Nat Commun. 7:12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne-Edwards K. 1995. Biparental care in Djungarian but not Siberian dwarf hamsters (Phodopus). Anim Behav. 50:1571–1585. [Google Scholar]
- Zaharia M, et al. 2011. Faster and more accurate sequence alignment with SNAP. In. arXiv.
- Zhou Q, et al. 2013. The epigenome of evolving Drosophila neo-sex chromosomes: dosage compensation and heterochromatin formation. PLoS Biol. 11:e1001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz JJ, Heard E. 2020. Molecular mechanisms of facultative heterochromatin formation: an X-chromosome perspective. Ann Rev Biochem. 89:255–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available through SRA BioProject number PRJNA786909 and PRJNA787539.