Abstract
The microbial assemblages of Lake Cisó and Lake Vilar (Banyoles, northeast Spain) were analyzed in space and time by microscopy and by performing PCR-denaturing gradient gel electrophoresis (DGGE) and sequence analysis of 16S rRNA gene fragments. Samples obtained from different water depths and at two different times of the year (in the winter during holomixis and in the early spring during a phytoplankton bloom) were analyzed. Although the lakes have the same climatic conditions and the same water source, the limnological parameters were different, as were most of the morphologically distinguishable photosynthetic bacteria enumerated by microscopy. The phylogenetic affiliations of the predominant DGGE bands were inferred by performing a comparative 16S rRNA sequence analysis. Sequences obtained from Lake Cisó samples were related to gram-positive bacteria and to members of the division Proteobacteria. Sequences obtained from Lake Vilar samples were related to members of the Cytophaga-Flavobacterium-Bacteroides phylum and to cyanobacteria. Thus, we found that like the previously reported differences between morphologically distinct inhabitants of the two lakes, there were also differences among the community members whose morphologies did not differ conspicuously. The changes in the species composition from winter to spring were also marked. The two lakes both contained sequences belonging to phototrophic green sulfur bacteria, which is consistent with microscopic observations, but these sequences were different from the sequences of cultured strains previously isolated from the lakes. Euryarchaeal sequences (i.e., methanogen- and thermoplasma-related sequences) also were present in both lakes. These euryarchaeal group sequences dominated the archaeal sequences in Lake Cisó but not in Lake Vilar. In Lake Vilar, a new planktonic population related to the crenarchaeota produced the dominant archaeal band. The phylogenetic analysis indicated that new bacterial and archaeal lineages were present and that the microbial diversity of these assemblages was greater than previously known. We evaluated the correspondence between the abundances of several morphotypes and DGGE bands by comparing microscopy and sequencing results. Our data provide evidence that the sequences obtained from the DGGE fingerprints correspond to the microorganisms that are actually present at higher concentrations in the natural system.
Identification and quantification of organisms, which provide the key parameters in diversity studies, are routinely performed operations in macroecology but are still difficult tasks in microbial ecology (5, 38). Measurements of bacterial metabolic processes yield valuable ecological information but, most of the time, give no clue as to which species are involved (15). As a result, our knowledge of the taxonomic compositions of microbial communities and of the factors which control the abundance and distribution of microbial populations is extremely limited.
Over the last 10 years several molecular techniques have been developed in order to study natural samples (33). These techniques can help identify microorganisms without isolation (2, 22) and have revealed the enormous extent of microbial diversity (43). Moreover, new molecular approaches have been proposed recently in order to link microbial processes with the organisms involved (3, 9). However, it is very probable that molecular techniques provide a biased view of microbial diversity. For example, many of the procedures rely on PCR, a technique in which biases have been shown to exist, and on cloning, which can act in a selective way (58). Likewise, it is not clear whether bacterial cells in nature exhibit different degrees of resistance to cell breakage, which is necessary for nucleic acid extraction. Altogether, it is difficult to ascertain whether the collection of sequences obtained from an environment represents the natural assemblage accurately. Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA genes is a molecular technique that is used to study the dynamic behavior of complex microbial assemblages (33, 36) and to isolate microorganisms in pure culture (53, 57). In this study we tested the performance of a widely used technique, DGGE, in an environment for which double checking with microscopic techniques is at least partially possible.
To do this, we used microbial assemblages that inhabit sulfurous karstic lakes. These assemblages include a few photosynthetic populations that are very large (44) and are easily distinguished and quantified on the basis of morphology and pigment content at the genus level and even at the species level (47). Lake Cisó and Lake Vilar are two karstic lakes that have been studied over the last 20 years by using classical methods (46). These two lakes are about 1 km apart and have the same climatic conditions and groundwater sources (8). However, the phototrophic bacteria, algae, ciliates, rotifers, and crustaceans in Lake Vilar differ markedly from those in Lake Cisó (15, 16, 20, 29, 45, 46). The differences have tentatively been attributed to the different limnological parameters and light environments in the two lakes (18–20). The identities of other populations of the microbial assemblages were not known previously.
Here we describe the identities of such populations as determined by sequencing of PCR-amplified, DGGE separated, 16S rRNA gene fragments. A detailed microscopic quantification analysis of photosynthetic microorganisms was performed, and the results obtained were compared to the 16S rRNA gene sequences recovered. This analysis provided a double check for the possible biases and sensitivity of the molecular approach. Moreover, the information obtained directly from the DGGE band patterns was validated after we performed a sequence analysis of single DGGE bands; this study addressed the potential and limitations of fingerprint analysis for studying natural communities.
MATERIALS AND METHODS
Description of the lakes.
Lake Cisó and Lake Vilar belong to the Banyoles karstic system located in Girona in northeast Spain (42°8′N, 2°45′E). The high concentrations of dissolved sulfate in the lake water (up to 10 mM) result in high concentrations of sulfide after microbial activity (20). Lake Cisó is a small holomictic lake with a surface area of approximately 650 m2 and a maximum depth of 6.5 m. Physicochemical properties of this lake and the distribution and activity of microorganisms in it have been extensively studied by using classical limnological and microbiological methods (46). Lake Cisó becomes anoxic during winter holomixis (complete mixing), and high sulfide concentrations (up to 0.5 mM) are present in the entire water column from the bottom of the lake to the surface. Under these conditions all eukaryotic organisms disappear, and the microbial assemblage is almost exclusively prokaryotic; dense populations of photosynthetic sulfur bacteria are distributed throughout the lake. Light penetration is severely limited by the abundant populations, and light is extinguished at a depth of only a few centimeters. At the beginning of thermal stratification (which lasts from April until September), the epilimnion becomes oxic, and a morphologically more diverse microbial assemblage (including eukaryotic microorganisms) develops. In the hypolimnion, the sulfide concentrations increase (up to 1.2 mM), and a discrete zone where oxygen and sulfide coexist is established in the metalimnion (depth, about 1.5 m). Here, a stratified microbial community develops coinciding with the well-established gradient of physicochemical conditions (29, 46).
Lake Vilar is about 1 km away from Lake Cisó and consists of two basins with a maximum depth of 9 m and a surface area of approximately 11,000 m2. In contrast to holomictic Lake Cisó, Lake Vilar is meromictic. Sulfide is present during the entire year, although it is restricted to the deeper, high-conductivity waters. A stable chemocline exists at 4.5 m between the oxygenated surface and the sulfide-rich bottom water. Here, dense populations of photosynthetic sulfur bacteria can develop when enough light penetrates (18, 31).
Sampling procedure.
The two lakes were sampled on the same day within a 2- to 3-h interval. Samples were collected on 19 February 1996 during the winter mixing and on 9 April 1996 during a phytoplankton spring bloom. Depth profiles for water temperature, conductivity, and oxygen concentration were determined in situ by using a portable multisensor probe (model Hydrolab DS-3; Hydrolab Instruments). Light penetration was measured with a submersible spherical quantum meter (model QSP-170; Biospherical Instruments). Samples used for biological and chemical analyses were taken from different depths by using a battery-driven pump connected with tubing to a conical polyvinyl chloride structure to improve laminar sampling at the interface. For sulfide measurements, 10-ml subsamples were first alkalinized by adding 100 μl of 10 N NaOH and then chemically fixed by adding zinc acetate to a final concentration of 0.1 M (17). To determine total cells counts, 10-ml subsamples were fixed by adding formaldehyde to a final concentration of 4% (vol/vol). Samples used for molecular biological analyses were kept in the dark on ice until further processing in the laboratory, which was always started within 2 to 6 h after collection.
Chemical and biological analyses.
Sulfide contents were measured spectrophotometrically by the methylene blue colorimetric method (17). DAPI (4′,6′-diamidino-2-phenylindole)-stained cells (48) were counted by using an epifluorescence Axiophot II microscope (Zeiss, Jena, Germany) and previously described statistical recommendations (23). In each case the standard deviation was less than 10% of the cell count. Morphologically distinguishable phototrophic bacteria were identified as described by Pfennig and Trüper (47). Some taxonomically valuable characteristics, such as motility and the presence of gas vesicles, were observed by using phase-contrast microscopy and live samples. For nucleic acid analysis, 1 to 5 liters of lake water was concentrated by using a refrigerated centrifuge operated at 8,000 × g (Sorvall Instruments). The cell pellet was kept frozen at −80°C until it was used. Microscopic observation of the supernatants revealed that between 1 and 3% of the cells were not recovered by this method.
Nucleic acid extraction and PCR amplification of 16S rDNA.
DNA was extracted with hot sodium dodecyl-sulfate-phenol and was purified by using phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol), followed by ethanol precipitation; about 50 mg (fresh weight) of bacterial cells and the protocol of Oelmüller et al. (40) were used. We determined microscopically that phototrophic bacteria were effectively broken by this method. Fragments of the 16S ribosomal DNA (rDNA) suitable for DGGE analysis were obtained by using three different primer combinations for different phylogenetic lineages (Table 1); one combination was used for members of the domain Bacteria (34, 35), another combination was used for the oxygenic phototrophs and targeted the 16S rRNA genes of cyanobacteria and algal chloroplasts (39), and the third combination was used for members of the domain Archaea (49, 55). The lengths of the PCR products were 585, 446, and 624 bp, respectively. Recently, researchers have shown that the bacterial primer combination consisting of primers 341F and 907R used here provides the most reliable results (21, 24). The PCR conditions used for the bacterial and cyanobacterial primer sets have been described previously by Muyzer et al. (34) and Nübel et al. (39), respectively. To amplify the archaeal genes, we used a touchdown protocol for 20 cycles with temperatures ranging from 71 to 61°C; the annealing temperature was reduced 1°C every two cycles. This procedure was followed by 15 additional cycles at an annealing temperature of 61°C. Except for the initial denaturation step (94°C, 5 min), denaturation and annealing phase steps were 1 min long, while most of the polymerization phase steps were 3 min long (the only exception was the final cycle, which was 10 min long).
TABLE 1.
Sequences, target sites, and specificities of the primers used in this study
| Primera | Target siteb | Sequence (5′ to 3′) | Specificity | Reference |
|---|---|---|---|---|
| 341F-GCc | 341-357 | CCT ACG GGA GGC AGC AG | Most Bacteria | 35 |
| 907R | 907-926 | CCG TCA ATT CMT TTG AGT TT | Most known organisms | 37 |
| CYA359F-GC | 359-378 | GGG GAA TYT TCC GCA ATG GG | Cyanobacteria and chloroplasts | 39 |
| CYA781R(a)d | 781-805 | GAC TAC TGG GGT ATC TAA TCC CAT T | Cyanobacteria and chloroplasts | 39 |
| CYA781R(b)d | 781-805 | GAC TAC AGG GGT ATC TAA TCC CTT T | Cyanobacteria and chloroplasts | 39 |
| ARC344F-GC | 344-363 | ACG GGG YGC AGC AGG CGC GA | Most Archaea | 49 |
| ARC915R | 915-934 | GTG CTC CCC CGC CAA TTC CT | Most Archaea | 55 |
F (forward) and R (reverse) indicate the orientation of the primers in relation to the rRNA sequence.
Escherichia coli numbering of Brosius et al. (6).
GC is a 40-nucleotide GC-rich sequence attached to the 5′ end of the primer as described by Muyzer et al. (34). The GC sequence is 5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G-3′.
Reverse primer CYA781R is an equimolar mixture of CYA781R(a) and CYA781R(b).
DGGE analysis of PCR products and sequencing.
DGGE was performed as described previously (34) for 3.5 h at a constant voltage of 200 V. The archaeal primer set gave unsatisfactory melting behavior results with one of the four cultures of methanogens tested. The PCR product obtained from this culture did not stop at a fixed position in the gel but kept migrating until electrophoresis was stopped. If this also happened with the PCR products from a complex community, the faster DGGE band would include a mixture of sequences. However, sequencing of this band showed that the problem did not occur with our natural samples. About 600 ng of PCR product was deposited in each well. After electrophoresis, the gels were stained with ethidium bromide and photographed with UV transillumination (wavelength, 314 nm) with a Polaroid camera. The photographs were scanned, and the digitized images were processed with the NIH Image software (National Institutes of Health, Bethesda, Md.) in order to measure relative band intensities. A band was defined as an ethidium bromide signal whose intensity was more than 0.2% of the total intensity for each lane.
Prominent bands were excised from the gels, reamplified, and electrophoresed again in a DGGE gel as previously described (34). The new PCR products were purified by using a Qiaquick PCR purification kit (Qiagen Inc.). A Taq Dyedeoxy terminator cycle sequencing Ready Reaction kit (Applied Biosystems, Forster City, Calif.) was used to sequence the 16S rDNA fragments with the appropriate forward PCR primer without the GC clamp. The sequencing reactions were performed with an Applied Biosystems model 373S DNA sequencer.
Comparative sequence analysis and construction of phylogenetic trees.
A BLAST search (1) was used to get a first indication of what sequences were retrieved. New sequences were aligned with about 5,400 homologous complete prokaryotic 16S rRNA primary structures (28) by using the automated aligning tool of the ARB program package (http://www.mikro.biologie.tu-muenchen.de). A similarity matrix was constructed in order to obtain sequence similarity values. Then partial sequences were inserted into the optimized tree derived from the complete sequence data by using the maximum-parsimony criterion and a special ARB parsimony tool that did not affect the initial tree topology (27). The resulting tree was pruned to save space, and the closest relatives were retained.
Nucleotide sequence accession numbers.
A total of 26 partial sequences (lengths, 500 to 600 bp) have been deposited in the EMBL nucleotide sequence database under the following accession numbers: AJ239988 to AJ239990 (CIARC-1 to CIARC-3), AJ239991 to AJ240001 (CIBAC-1, -2, -3, -6, -7, -9, -10, -11, -12, -13, and -15), AJ240002 and AJ240003 (CICYA-1 and CICYA-2), AJ240004 to AJ240006 (VIARC-1 to VIARC-3), AJ240007 to AJ240011 (VIBAC-1, -3, -4, -6, and -7), and AJ240012 and AJ240013 (VICYA-1 and VICYA-2). These sequences are the sequences shown in Fig. 4 and 5. Each band designation includes, in addition to a number (see Fig. 3), a code for the lake (CI, Lake Cisó; VI, Lake Vilar) and a code for the primer set used (ARC, Archaea; BAC, Bacteria; CYA, cyanobacteria).
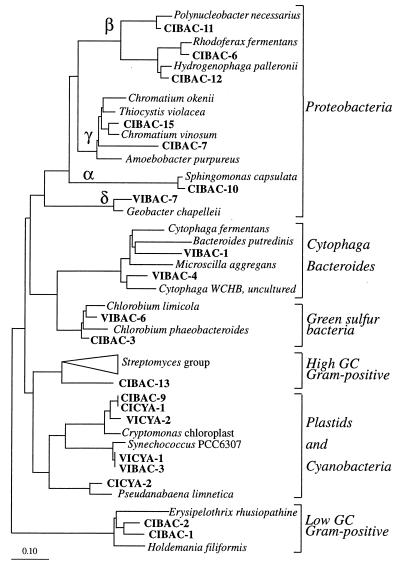
FIG. 4.
Phylogenetic affiliations within the domain Bacteria of the excised bands obtained from the gels in Fig. 3A and 3B. The tree was constructed by adding by parsimony analysis the partial sequence data to a previously validated and optimized tree. Scale bar = 0.10 mutation per nucleotide position.
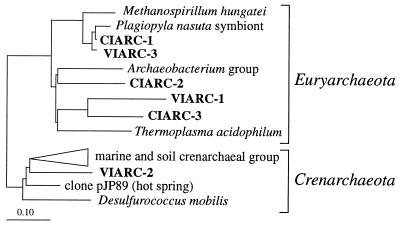
FIG. 5.
Phylogenetic affiliations within the domain Archaea of excised bands obtained from the gel in Fig. 3C. The tree was constructed by adding by parsimony analysis the partial sequence data to a previously validated and optimized tree. Scale bar = 0.10 mutation per nucleotide position.
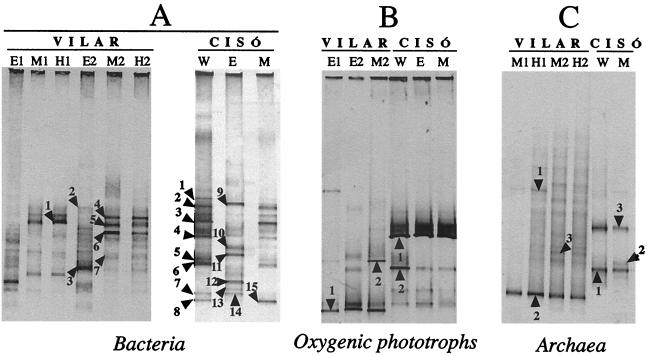
FIG. 3.
Negative images of ethidium bromide-stained DGGE gels containing PCR-amplified segments of 16S rRNA genes obtained by using three primer sets. The band patterns were obtained with bacterial primers (A), cyanobacterium-chloroplast primers (B), and archaeal primers (C) from the oxic epilimnion (lanes E) and from the anoxic metalimnion (lanes M) and hypolimnion (lanes H). Lanes E1, M1, and H1 and lanes E2, M2, and H2 correspond to winter and spring, respectively, in Lake Vilar. Lane W and lanes E and M correspond to winter and spring, respectively, in Lake Cisó. Bands were designated as described in the text.
RESULTS
Physicochemical parameters of the lakes.
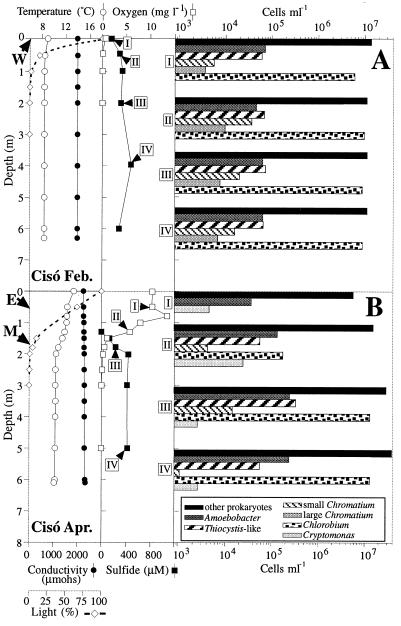
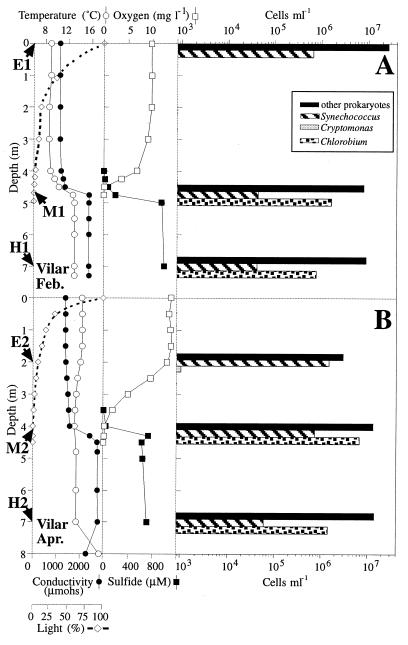
Figures 1 and 2 show the vertical profiles for temperature, conductivity, light, and concentrations of oxygen and sulfide for Lake Cisó and Lake Vilar, respectively. Figures 1A and 2A show the data obtained in February 1996 during the winter holomixis, and Fig. 1B and 2B show the data obtained in April 1996 during the early spring. In Lake Cisó the conductivity did not differ along the vertical profiles, whereas in Lake Vilar it did, reflecting the meromictic nature of this lake. The water temperature in both lakes ranged from 8 to 15°C, but due to its smaller volume and sheltered environment, Lake Cisó became thermally stratified during the first weeks of spring. The patterns of incident light extinction differed between lakes and between seasons. No light penetrated further than a depth of 2 m in Lake Cisó, whereas light penetrated to a depth of 5 m in Lake Vilar. Opposite oxygen and sulfide gradients were observed in both lakes. During the winter, the water column in Lake Cisó was anaerobic (Fig. 1A), whereas the maximal oxygen concentration was found in the spring (Fig. 1B), mainly due to the photosynthetic activity of the alga Cryptomonas sp. However, no oxygen was detected at depths below 1.5 m. In Lake Vilar, the maximal oxygen concentration was also found in the spring due to the activity of different algae and cyanobacteria. High sulfide concentrations were found in both lakes, and these concentrations ranged from 0.4 mM in Lake Cisó to 1.0 mM in Lake Vilar.
FIG. 1.
Vertical profiles for temperature, conductivity, oxygen concentration, sulfide concentration, light penetration, and cell counts on two different days in Lake Cisó. (A) Winter mixing (19 February 1996). (B) Spring stratification (9 April 1996). The sampling depths used for the molecular survey are indicated by arrows. Cell counts are on a logarithmic scale.
FIG. 2.
Vertical profiles for temperature, conductivity, oxygen concentration, sulfide concentration, light penetration, and cell counts on two different days in Lake Vilar. (A) Winter (19 February 1996). (B) Spring (9 April 1996). The sampling depths used for the molecular survey are indicated by arrows. Cell counts are on a logarithmic scale.
Vertical distribution of microbial populations.
The vertical distribution of microorganisms was assessed by microscopy. The most abundant group in all of the samples was the group designated “other prokaryotes” (Fig. 1 and 2). This group included all of the morphologically indistinguishable prokaryotes. The bacteria with conspicuous morphologies are shown in Table 2. In Lake Cisó five such bacterial populations were distinguished on the basis of shape, size, and autofluorescence (Fig. 1). These populations were populations of green sulfur bacteria of the Chlorobium type (autofluorescent, nonmotile, small rods), whose concentrations reached 107 cells ml−1, and purple sulfur bacteria similar to both Amoebobacter and Thiocystis-like cells, whose concentrations reached 105 cells ml−1. Other populations identified in Lake Cisó were populations of small and large Chromatium cells (Table 2) whose concentrations were between 103 and 104 cells ml−1. In Lake Vilar, purple sulfur bacteria were not detected during the time that samples were collected, and the sulfur bacteria present were similar to Chlorobium phaeobacteroides (maximum concentration, 7 × 106 cells ml−1).
TABLE 2.
Morphotypes identified by microscopy in Lake Cisó and Lake Vilar, including some phenotypic characteristics and mean cell sizes
| Morphotype | Morphology | Cell size (μm) | Presence in:
|
Reference(s) | Related sequence(s)a | |
|---|---|---|---|---|---|---|
| Lake Cisó | Lake Vilar | |||||
| Amoebobacter | Coccus | 1.5-2.5 | + | − | 46 | CIBAC-7 |
| Thiocystis class | Ovoid | 3.0-4.0 × 5.0-6.0b | + | − | 46 | CIBAC-15 |
| Large Chromatium | Rod | 4.0-6.0 × 8.0 | + | − | 47 | None |
| Small Chromatium | Rod | 2.0 × 4.0 | + | − | 47 | None |
| Chlorobium | Rod | 0.8 × 2.0-2.5 | + | + | 47 | CIBAC-3, VIBAC-6 |
| Synechococcus | Coccus | 1.5-2.0 | − | + | This study | VIBAC-3, VICYA-1 |
| Pseudanabaena | Long filament | + | − | This study | CICYA-2 | |
| Plagiopyla-Metopus | Autofluorescent, rod-shaped endosymbiont cells | + | + | 12, 30 | CIARC-1, VIARC-3 | |
| Cryptomonas | Chloroplast | + | + | 46 | CIBAC-9, CICYA-1, VICYA-2 | |
| Other prokaryotes | No distinctive morphological traits | + | + | 29; this study | More than 20c | |
Oxygenic phototrophic populations were also identified. In Lake Vilar, populations of algae (Cryptomonas and Crucigenia species; concentrations, up to 2 × 103 cells ml−1 each) and cyanobacteria (Synechococcus-shaped cells; concentration, 106 cells ml−1) were detected, whereas Cryptomonas cells (concentrations, up to 2.4 × 104 cells ml−1) were the dominant algae in Lake Cisó. The cyanobacterium Pseudanabaena sp. was observed sporadically in Lake Cisó but not in Lake Vilar. Several ciliates previously reported to occur in these ecosystems, such as Plagiopyla sp. at anaerobic depths and Coleps sp. at aerobic depths (30), were detected microscopically as well.
DGGE fingerprint analyses.
DGGE analyses were performed with samples obtained at selected depths (indicated by arrows in Fig. 1 and 2) by using primer sets for members of the Bacteria (Fig. 3A), for oxygenic phototrophs (Fig. 3B), and for members of the Archaea (Fig. 3C). Each microbial assemblage produced a reproducible DGGE fingerprint in the different analyses performed. For Lake Cisó, one sample was analyzed for the winter profile and was used as a representative of the entire water column (lanes W) since the lake is homogeneously mixed at this time of the year. In the spring, a sample from the oxygenated epilimnion (lanes E) and another sample from the top of the anaerobic layer (metalimnion, lanes M) were analyzed. Samples from the bottom of the lake produced band patterns similar to those of samples from the metalimnion (data not shown). For Lake Vilar, we analyzed the epilimnion, metalimnion, and hypolimnion in the winter (lanes E1, M1, and H1, respectively) and in the spring (lanes E2, M2, and H2, respectively). Thus, we were able to monitor qualitative differences in the communities between lakes, between two seasons, and among depths.
First, we found that there were marked differences in the mobilities of the DGGE bands when we compared lakes; the most intense bands for the two lakes did not coincide with any of the primer sets tested (Fig. 3). Therefore, we expected the PCR-recovered populations in Lake Cisó and Lake Vilar to be different. Despite the difference in composition, the overall numbers and relative intensities of the bands were similar for the two lakes; i.e., the total number of bands detected ranged from 8 to 17 for both lakes, and just a few bands (one to three bands) appeared to be the most intense bands in all cases.
Second, when we compared seasons, the winter bacterial community in Lake Cisó (Fig. 3A, lane W) shared more bands with the anaerobic spring community (Fig. 3A, lane M) than with the aerobic spring community (Fig. 3A, lane E). This shift in the community fingerprint was expected as a result of growth of cyanobacteria or algae (the bacterial primer recovered DNA from chloroplasts) in the epilimnion. However, no changes in the fingerprint resulting from oxygenic phototrophs were detected (Fig. 3B, lanes W, M, and E). Therefore, the shift was probably due to the appearance of new populations of members of the Bacteria. A very weak PCR product was obtained for members of the Archaea in the aerobic samples, and this product did not yield a DGGE band pattern (data not shown). Archaeal bands from the anaerobic zone of Lake Cisó migrated to the same position in the winter and in the spring (Fig. 3C, lanes W and M), indicating that the archaeal assemblages were very similar.
In Lake Vilar, the change of season resulted in a marked shift in the bacterial populations both in the aerobic zones and in the anaerobic zones (Fig. 3A). Bands that appeared to be dominant in the winter were less intense or disappeared completely in the spring (Fig. 3A, lanes E1 and E2). Moreover, new dominant DGGE bands were obtained with the spring samples (Fig. 3A, lane M2, bands 4 and 5). When the oxygenic phototrophs were analyzed, new bands were detected in the spring (Fig. 3B, lane E2), and these bands might have been partially responsible for the shift observed with the bacterial primers. As occurred in Lake Cisó, the archaeal pattern did not change markedly (Fig. 3C).
Finally, the DGGE patterns of samples obtained from different depths reflected substantial differences between the aerobic and anaerobic assemblages.
Phylogenetic affiliations of predominant community members.
Most bands (40 bands) in the DGGE fingerprints were excised, reamplified, purified, and sequenced. A total of 26 of the 40 bands yielded sequences without ambiguous positions and were included in phylogenetic trees (Fig. 4 for members of the Bacteria and Fig. 5 for members of the Archaea). These bands were designated as described above. Of the remaining 14 bands, 6 bands (mainly weak bands) did not yield a reamplification product or sequence, and 2 did yield such a product but there were many ambiguous positions (more than 20% of the partial sequence). For the last six bands (labeled in Fig. 3 but not included in Fig. 4 and 5) some positions (about 10% of the partial sequence) were ambiguous, and these bands were not included in the tree. However, the affiliations of some of these sequences provided results that were consistent with microscopic observations (e.g., VIBAC-2 with Cryptomonas sp. and CIBAC-4 with Chlorobium limicola) and with the affiliations of bands exhibiting the same melting behavior in the gels (CIBAC-14 with CIBAC-7 and CIBAC-8 with CIBAC-15).
The resulting parsimony trees are shown in Fig. 4 (Bacteria) and Fig. 5 (Archaea). Parallel analyses in which we used the neighbor-joining method with different correction coefficients (the Kimura and Jukes-Cantor coefficients) resulted in the same tree topology (data not shown). The 16S rRNA-defined groups corresponding to predominant DGGE bands were distributed throughout both phylogenetic trees, indicating that the level of genetic diversity in the lakes studied was high. Gram-positive organisms and members of the Proteobacteria (alpha, beta, and gamma subdivisions) dominated the PCR-amplified, 16S rRNA-defined, bacterial populations in Lake Cisó, whereas members of the Cytophaga-Flavobacterium-Bacteroides phylum and Cyanobacteria dominated the populations in Lake Vilar. Methanogen- and thermoplasma-related sequences were present in both lakes; however, whereas in Lake Cisó the sequences of members of these groups were the dominant recovered archaeal sequences, in Lake Vilar a planktonic crenarchaeota-like population appeared to be the predominant archaeal population.
Table 3 shows the physiology of the closest relatives for organisms that exhibit levels of rRNA sequence identity of ≥95% (genus level rRNA difference). All of the closest relatives had metabolism that was compatible with the environment. Most of the sequences from members of the Archaea were not included in Table 3 because they were too distantly related to any cultured organism. Sequences affiliated with the Cytophaga phylum or with gram-positive organisms (both high- and low-G + C-content organisms) also lacked close relatives and are not included in Table 3.
TABLE 3.
Ecophysiology of the closest relatives of the 16S rDNA sequences that exhibited levels of similarity of ≥95% (genus level identity)
| Phylogenetic group | Sequence | Related sequence | % Similarity | Ecophysiology |
|---|---|---|---|---|
| Algae | CIBAC-9 | Cryptomonas chloroplast | 96 | Oxygenic photosynthesis |
| CICYA-1 | Cryptomonas chloroplast | 96 | Oxygenic photosynthesis | |
| VICYA-2 | Cryptomonas chloroplast | 95 | Oxygenic photosynthesis | |
| Cyanobacteria | VIBAC-3 | Synechococcus sp. strain PCC 6307 | 97 | Oxygenic photosynthesis |
| VICYA-1 | Synechococcus sp. strain PCC 6307 | 97 | Oxygenic photosynthesis | |
| CICYA-2 | Pseudoanabaena limnetica | 97 | Oxygenic photosynthesis | |
| Green sulfur bacteria | CIBAC-3 | Chlorobium phaeobacteroides | 95 | Anoxygenic sulfur photosynthesis |
| VIBAC-6 | Chlorobium phaeobacteroides | 96 | Anoxygenic sulfur photosynthesis | |
| Alpha-Proteobacteria | CIBAC-10 | Sphingomonas | 98 | Highly versatile |
| Beta-Proteobacteria | CIBAC-6 | Rhodoferax fermentans | 96 | Highly versatile |
| CIBAC-12 | Hydrogenophaga | 96 | Fermenter | |
| Euryarchaea | CIARC-1 | Ciliate endosymbiont | 95 | Methanogenesis |
| VIARC-3 | Ciliate endosymbiont | 95 | Methanogenesis |
A sequence alignment performed with ARB showed that the same sequences were recovered with different primer sets and the amplification conditions employed. Thus, in Lake Cisó, bands CIBAC-9 (amplified with the bacterial primer set) and CICYA-1 (amplified with the cyanobacterium-chloroplast primer set) were identical and closely related to Cryptomonas bands (Fig. 4). The same thing occurred in Lake Vilar for bands VIBAC-3 and VICYA-1 in relation to Synechococcus bands.
Interestingly, the phylotypes related to Cryptomonas sp. recovered from Lake Cisó (CICYA-1) and Lake Vilar (VICYA-2) were not identical; they exhibited 97% similarity. Likewise, the brown-pigmented Chlorobium-like phylotypes from the two lakes (CIBAC-3 and VIBAC-6) exhibited 96.9% similarity. Thus, the phylogenetic affiliations revealed that different phylotypes were present in Lake Cisó and Lake Vilar for at least two different microorganisms.
Microscopic observations and recovered 16S rRNA-defined populations.
Several populations recovered by PCR and sequencing confirmed microscopic observations. Cryptomonas-like algae and Synechococcus-like cyanobacteria were dominant phytoplankton components as determined by both microscopic counting (Fig. 1 and 2) and analysis of PCR products recovered with specific primers (Fig. 3B and 4). Also, Pseudanabaena-like cyanobacteria were recovered from Lake Cisó but not from Lake Vilar, which was in agreement with microscopic observations. Several common ciliates found previously in these ecosystems, such as Plagiopyla sp. at anaerobic depths and Coleps sp. at aerobic depths (30), were detected microscopically. It has been shown that such organisms carry endosymbiontic microbial populations (12). The CIARC-1 and VIARC-1 bands most likely corresponded to archaeal endosymbionts of the ciliate Plagiopyla sp. (Fig. 5). Among the photosynthetic bacteria (i.e., members of the Chlorobiaceae and Chromatiaceae), organisms present at higher cell concentrations were recovered from the sequenced DGGE bands, but organisms present at lower concentrations (e.g., large Chromatium okenii-like cells) were not. Tables 4 and 5 show the relationship between cell counts and the signal intensities of the PCR-amplified bands.
TABLE 4.
Cell abundance and signal intensity for bacterial populations in Lake Cisó, as determined by microscopy and DGGE sequencing, respectively
| Sample pattern | % of Amoebobacter morphotype | CIBAC-7 signal intensity (%) | % of Thiocystis morphotype | CIBAC-15 signal intensity (%) |
|---|---|---|---|---|
| W | 0.4 | 1.1 | 0.3 | 2.0 |
| E | 0.8 | 6.0 | NDa | ND |
| M | 1.1 | 1.5 | 1.7 | 16.0 |
ND, not detected.
TABLE 5.
Cell abundance and signal intensity for bacterial populations in Lake Vilar, as determined by microscopy and DGGE sequencing, respectively
| Sample pattern | % of Synechococcus morphotype | VIBAC-3 signal intensity (%) | % of Chlorobium morphotype | VIBAC-6 signal intensity (%) |
|---|---|---|---|---|
| E1 | 2.4 | 8.1 | NDa | ND |
| M1 | 0.4 | ND | 13.9 | 6.7 |
| H1 | 0.4 | 2.6 | 7.6 | 3.1 |
| E2 | 30.0 | 28.1 | ND | ND |
| M2 | 3.5 | 0.5 | 30.1 | 15.6 |
| H2 | 0.5 | 4.2 | 9.8 | 12.0 |
ND, not detected.
DISCUSSION
Interpretation of data resulting from molecular approaches for studying microbial diversity in natural environments presents uncertainties, and several difficulties may arise in the long process from natural samples to sequences (11, 13, 51, 56). Some of the problems are intrinsic to PCR amplification kinetics, and, therefore, they are shared by all of the approaches that use such a step (PCR cloning and PCR fingerprinting techniques). Other difficulties are specific for the DGGE technique which we used in this study. In the present study we used independently obtained microscopy-based information for some of the organisms present in Lake Cisó and Lake Vilar to investigate the magnitude of the problems. When DGGE is used, the limitations can be probed to a certain extent by excising bands and sequencing. Heteroduplex formation (10, 13, 32) seemed not to be a significant problem in our analysis. We excised, reamplified, reelectrophoresed, and sequenced a substantial number of bands (40 bands), and only 4 of these bands (10%) yielded multiple bands after reamplification. The multiple bands (presumably representing homo- and heteroduplexes) were clearly separated in the gel when we performed a new DGGE analysis, and, therefore, they cannot be related to the quality of gel separation or the precision of excision. The possibility that in a single organism there were multiple 16S rRNA operons with different migration behaviors on DGGE gels was also eliminated after sequencing. The main problem seemed to be the presence of different sequences at the same or very similar positions in the fingerprint. A cloning step is needed in such cases in order to determine the composition of the DNA mixture, but in addition the quality of gel separation and band excision can be improved. Two of the bands resulted in a large number of sequence uncertainties (e.g., Fig. 3A, main band in lane E), and six bands resulted in a lower number of ambiguous positions (see above). The latter sequences were affiliated with some of the populations detected microscopically, reflecting the fact that these probably mixed sequences were dominated by one PCR product and provided valuable information. At any rate, most of the bands in our analysis yielded a clean sequence, indicating that each band represented a different microbial population.
The molecular methods which we used should recover the microscopically recognizable populations in Lake Cisó and Lake Vilar, and each band should have an intensity proportional to its abundance. We recovered most such populations, and, in addition, the PCR products were dominant when the populations were predominant community members. The differences between microscopic counts and signal intensity were always within a factor of 10 (Tables 4 and 5). Thus, variations in the signal intensity revealed variations in the abundance of a population in the environment at the order-of-magnitude level.
Another question to consider is what percentage of the populations present in situ can be retrieved by PCR-DGGE analysis (detectability). Working with mixtures of pure cultures, Muyzer et al. (35) found that DGGE could not detect a population whose abundance was less than approximately 1% of the total cell count. We observed a similar threshold with our natural samples. At least four morphotypes of purple sulfur bacteria were identified visually in Lake Cisó (Fig. 1 and Table 2), but only two types of sequences corresponding to members of the Chromatiaceae were recovered. Only sequences corresponding to the dominant members of the Chromatiaceae (Amoebobacter and Thiocystis-like species), whose concentrations were more than 0.3 to 1% of the total DAPI counts, were recovered. Sequences corresponding to other Chromatium-like cells (Chromatium okenii, Chromatium weissei, and small Chromatium spp.) always accounted for less than 0.1% of total DAPI counts and were not detected by DGGE. The detection threshold for Synechococcus spp. was 0.4% (Table 5). However, we cannot eliminate the possibility that some of the minor bands that were not excised from the gel corresponded to the other Chromatiaceae morphotypes, that they were masked if they exhibited the same melting behavior as other populations (41) or that their concentrations were less than the detection limit of the staining solution. More information might be obtained from the weak bands by performing Southern hybridization with specific probes (34).
The final step in the process is affiliation of the sequences obtained from the DGGE fingerprints with taxa. Although complete 16S rDNA sequences should be used for phylogenetic reconstruction (26), partial sequences may be sufficient to determine the closest relatives of unknown sequences and assign them to well-established phylogenetic groups (27, 37, 54). However, a combination of partial sequences and poor representation of a given microbial group in databases (as in the case of the Chromatiaceae) may prevent accurate allocation of sequences. For instance, the CIBAC-15 sequence was more closely related to Chromatium vinosum than to Thiocystis sp. Microscopy and pigment composition showed that Thiocystis-like organisms were present in Lake Cisó (Thiocystis class in Table 2). Specific pigments from C. vinosum, on the other hand, were not detected in the lake. Probably, the sequence recovered from Lake Cisó is the sequence of a new organism that would be easier to identify if the complete 16S rDNA sequence was available.
All of the problems described above need to be considered before conclusions concerning the natural assemblages themselves are reached. However, the following questions can be addressed with a high level of confidence.
Does the molecular approach recover the main organisms known to be present as determined by microscopy, and does this approach reveal additional differences between the assemblages in the two lakes studied for other prokaryotic populations that were not identified by microscopy?
We retrieved, as PCR-amplified 16S rRNA genes, most of the algae, cyanobacteria, and photosynthetic sulfur populations detected microscopically in Lake Cisó and Lake Vilar. The only morphologies that were not retrieved by DGGE were those corresponding to Chromatium populations found at low levels (less than 0.3% of the total cell counts). The method which we used revealed that that there are differences in the bacterial populations of the two lakes that could not be revealed by microscopy. In addition, different archaeal sequences were recovered from the two lakes. Interestingly, populations that were present in both lakes, such as the C. phaeobacteroides populations, belonged to different phylotypes. A similar result was derived from a study of Chromatium minus isolates obtained from three lakes in Spain (7). This may indicate that the anaerobic layers of stratified lakes may act as islands in an aerobic world for anaerobic microorganisms.
Are the identified sequences identical (or similar) to the sequences isolated in pure culture from the same lakes?
Over time, many microorganisms have been isolated in pure culture from the two lakes in order to carry out physiological studies. From an ecological perspective, it is very important to determine whether such isolates are the same organisms that are dominant in the environment or whether a minor component of the natural assemblage was selected by the isolation procedure. Several sequences related to Chlorobium spp. were recovered in the present study, but they did not match exactly any of the sequences deposited in the 16S rRNA database. This is especially remarkable because most of the Chlorobium culture sequences available in the database were obtained from the lakes in the area around Banyoles, including Lake Cisó, Lake Vilar, and Lake Banyoles (the latter is only 10 m away from Lake Vilar) (14). Thus, the ecologically relevant Chlorobium populations (as detected by DGGE and sequencing) and the populations isolated in pure culture from the same environments were not the same.
Are the potential metabolic characteristics of the sequences recovered consistent with the environmental conditions?
The activities and functional roles of the phylotypes recovered cannot be known until the corresponding microorganisms are isolated in pure culture and characterized. However, the known metabolic characteristics of the closest relatives were generally consistent with the environmental conditions present in the lakes (i.e., anaerobiosis, simultaneous presence of light and sulfide). Surprisingly, other potentially dominant organisms at oxic-anoxic interfaces, such as sulfur or ammonia oxidizers, were not recovered. Sulfate reducers were not recovered from the water column of Lake Cisó either. Again, some of the weak bands that were not sequenced could have corresponded to these organisms, the organisms might have been present at low levels at the time of sampling, or the organisms might be sharply stratified in the water column and thus might have escaped detection because of the sampling resolution.
The presence of several Cytophaga-related organisms that dominate the anaerobic, sulfide-rich water of Lake Vilar remains intriguing, because most representatives of this large group of bacteria are aerobic or microaerophilic (50). Recent molecular studies, however, have shown that Cytophaga-related organisms are dominant components of the microbial assemblages in anaerobic marine sediments (25, 52). These results and our results suggest that the Cytophaga-like bacteria available in culture collections do not account for the full range of metabolic capabilities present in the group. More extensive studies are needed to establish the ecological importance of these organisms.
Are there archaeal groups living in these environments?
Culturable members of the Archaea found in freshwater plankton are restricted to methanogens and sulfur-metabolizing thermophiles (4). We found methanogen-related sequences in the anaerobic sulfide-rich waters of Lake Cisó and Lake Vilar, and archaeal PCR products were obtained from the aerobic depths as well. However, one of the most interesting findings of this study was the existence of a population related to the Crenarchaeota in the sulfide-rich water column of Lake Vilar. Whereas all of the available pure cultures of members of the Crenarchaeota are sulfur-metabolizing thermophiles, sequences related to members of this group have recently been recovered from a wide variety of temperate and cold environments (4), although there have been no studies which have reported the presence of such populations in the plankton of freshwater lakes. Obviously, members of this group are able to thrive in a wide range of nonextreme environments. The phylogenetic affiliation indicates that the crenarchaeal sequence obtained from Lake Vilar (VIARC-2) does not exhibit a high level of similarity with the sequence of any cultured organism (level of similarity to Desulfurococcus mobilis, 81%) and that it is included in the so-called freshwater cluster (4). This finding and the ecological distribution suggest that VIARC-2 is a novel type of organism. Recently, another sequence related to crenarchaeotal sequences was recovered from the sulfide-rich waters of a Norwegian meromictic lake (42), suggesting that this potential phenotype is widely distributed. Unfortunately, the organisms have not been recovered in pure culture, and their physiology and metabolism remain unknown. Their distinct vertical distribution (they accumulate in sulfide-rich layers) suggests that they have an ecological role in the anaerobic biogeochemical activity of the lake, but their relative abundance remains to be determined. Some of the sequences recovered in our study were closely related to the sequences of endosymbiontic organisms (bacterial endosymbionts of aerobic ciliates, CIBAC-11, and methanogenic endosymbionts of the anaerobic ciliate Plagiopyla sp.). VIARC-2 could also correspond to the sequence of an endosymbiontic organism. In order to determine the actual environment of VIARC-2, in situ hybridization or selective prefiltration of fresh samples should be carried out.
Overall, the evidence obtained with microscopically identifiable populations indicates that the dominant DGGE bands corresponded to predominant populations in the ecosystems studied. Although the data resulting from a comparison of DGGE band patterns were limited, they allowed us to monitor community changes that were consistent with the microscopic results and with the shift in environmental conditions. The microscopic control results and the sequencing data led us to conclude that the number and intensity of DGGE bands provide valuable information concerning variations in species evenness. The number of DGGE bands is related to the number of populations that account for more than 0.3 to 0.4% of the total cell counts. Thus, the species richness or total microbial diversity in the system cannot be accurately estimated with this method. However, a general pattern was recognized when the different DGGE gels were analyzed, independent of the lake, season, or depth; there were a few very abundant populations (accounting for more than 10% of the total cell count) and a few populations that accounted for between 1 and 10% of the total cell count. There are probably many more populations that account for less than 1% of the total cell count, but they cannot be retrieved by DGGE. Thus, the ecosystems studied may be very species rich, but the evenness is certainly low. This may be a general pattern for aquatic microbial communities (44).
ACKNOWLEDGMENTS
This work was financed by the Max-Planck-Gesellschaft of Germany and by DGICyT grant PB95-0222 from the Spanish Ministerio de Educación y Cultura. Parts of this work were funded by project MIDAS (Microbial Diversity in Aquatic Systems; grant MAS3-CT97-0154) from the European Union.
R. Amann is acknowledged for his generous and continuous support. R. Rosselló-Mora was extremely helpful with the phylogenetic analysis, and U. Nübel helped with the cyanobacterial PCR-DGGE analysis. E.O.C. benefited from the CSIC-MPG exchange program. Mica and Jordi from Can Masó d'Olives and “Milqui” are thanked for their support.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreasen K, Nielsen P H. Application of microautoradiography to the study of substrate uptake by filamentous microorganisms in activated sludge. Appl Environ Microbiol. 1997;63:3662–3668. doi: 10.1128/aem.63.9.3662-3668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravalli R N, She Q X, Garrett R A. Archaea and the new age of microorganisms. Trends Ecol Evol. 1998;13:190–194. doi: 10.1016/S0169-5347(98)01343-3. [DOI] [PubMed] [Google Scholar]
- 5.Brock T D. The study of microorganisms in situ: progress and problems. Symp Soc Gen Microbiol. 1987;41:1–17. [Google Scholar]
- 6.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Bacteriol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 7.Casamayor E O, Calderón-Paz J I, Mas J, Pedrós-Alió C. Identification of phototrophic sulfur bacteria through the analysis of lmwRNA band patterns. Arch Microbiol. 1998;170:269–278. doi: 10.1007/s002030050642. [DOI] [PubMed] [Google Scholar]
- 8.Casamitjana C, Roget E. Resuspension of sediment by focused groundwater in Lake Banyoles. Limnol Oceanogr. 1993;38:643–656. [Google Scholar]
- 9.Chen F, González J M, Dustman W A, Moran M A, Hodson R E. In situ reverse transcription, an approach to characterize genetic diversity and activities of prokaryotes. Appl Environ Microbiol. 1997;63:4907–4913. doi: 10.1128/aem.63.12.4907-4913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart E L, Herring B, Rodrigo A G, Mullins J I. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. Genome Res. 1995;4:216. doi: 10.1101/gr.4.5.s202. [DOI] [PubMed] [Google Scholar]
- 11.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenchel T, Finlay B J. Synchronous division of an endosymbiotic methanogenic bacterium in the anaerobic ciliate Plagiopyla frontata Kahl. J Protozool. 1991;38:22–28. [Google Scholar]
- 13.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueras J B, García-Gil L J, Abellà C A. Phylogeny of the genus Chlorobium based on 16S rDNA sequence. FEMS Microbiol Lett. 1997;152:31–36. doi: 10.1111/j.1574-6968.1997.tb10405.x. [DOI] [PubMed] [Google Scholar]
- 15.García-Cantizano J, Calderón-Paz J I, Pedrós-Alió C. Thymidine incorporation in Lake Cisó: problems in estimating bacterial secondary production across oxic-anoxic interfaces. FEMS Microbiol Ecol. 1994;14:53–64. [Google Scholar]
- 16.Gasol J M, Guerrero R, Pedrós-Alió C. Seasonal variations in size structure and procaryotic dominance in sulfurous Lake Cisó. Limnol Oceanogr. 1991;36:860–872. [Google Scholar]
- 17.Golterman H L, Clymo R S, Ohnstad M A M. Methods for physical and chemical analysis of fresh waters. IBP Handbook no. 8. Oxford, United Kingdom: Scientific Publications; 1978. [Google Scholar]
- 18.Guerrero R, Montesinos E, Esteve I, Abellà C. Physiological adaptation and growth of purple and green sulfur bacteria in a meromictic lake (Vilà) as compared to a holomictic lake (Sisó) In: Dokulil M, Metz H, Jewson D, editors. Shallow lakes. Contribution to their limnology. Developments in hydrobiology. Vol. 3. The Hague, The Netherlands: Dr. W. Junk Publishers; 1980. pp. 161–171. [Google Scholar]
- 19.Guerrero R, Montesinos E, Pedrós-Alió C, Esteve I, Mas J, van Gemerden H, Hofman P A G, Bakker J F. Phototrophic sulfur bacteria in two Spanish lakes: vertical distribution and limiting factors. Limnol Oceanogr. 1985;30:919–931. [Google Scholar]
- 20.Guerrero R, Pedrós-Alió C, Esteve I, Mas J. Communities of phototrophic sulfur bacteria in lakes of the Spanish Mediterranean region. Acta Acad Abo. 1987;47:125–151. [Google Scholar]
- 21.Hansen M C, Tolkernielsen T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 22.Hugenholtz P, Pace N R. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 23.Kirchman D, Sigda J, Kapuscinski R, Mitchell R. Statistical analysis of the direct count method for enumerating bacteria. Appl Environ Microbiol. 1982;44:376–82. doi: 10.1128/aem.44.2.376-382.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llobet-Brossa E, Rosselló-Mora R, Amann R. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig W, Schleifer K H. Bacterial phylogeny based on 16S and 23S rDNA sequence analysis. FEMS Microbiol Rev. 1994;15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 28.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massana R, García-Cantizano J, Pedrós-Alió C. Components, structure and fluxes of the microbial food web in a small, stratified lake. Aquat Microb Ecol. 1996;11:279–288. [Google Scholar]
- 30.Massana R, Pedrós-Alió C. Role of anaerobic ciliates in planktonic food webs: abundance, feeding, and impact on bacteria in the field. Appl Environ Microbiol. 1994;60:1325–1334. doi: 10.1128/aem.60.4.1325-1334.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montesinos E, Guerrero R, Abellà C, Esteve I. Ecology and physiology of the competition between Chlorobium limicola and Chlorobium phaeobacteroides. Appl Environ Microbiol. 1983;46:1007–1016. doi: 10.1128/aem.46.5.1007-1016.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muyzer G. Structure, function and dynamics of microbial communities: the molecular biological approach. In: Carvalho G R, editor. Advances in molecular ecology. NATO Science Series. Amsterdam, The Netherlands: IOS Press; 1998. pp. 87–117. [Google Scholar]
- 34.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3.4.4. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 35.Muyzer G, De Waal E D, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 37.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 38.Nübel U, García-Pichel F, Kühl M, Muyzer G. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids from oxygenic phototrophs in microbial mats. Appl Environ Microbiol. 1999;65:422–430. doi: 10.1128/aem.65.2.422-430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nübel U, García-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oelmüller U, Krüger N, Steinbüchel A, Friedrich C G. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J Microbiol Methods. 1990;11:73–84. [Google Scholar]
- 41.Overmann J, Tuschak C. Phylogeny and molecular fingerprinting of green sulfur bacteria. Arch Microbiol. 1997;167:302–309. doi: 10.1007/s002030050448. [DOI] [PubMed] [Google Scholar]
- 42.Øvreas L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 44.Pedrós-Alió C. Diversity of bacterioplankton. Trends Ecol Evol. 1993;8:86–90. doi: 10.1016/0169-5347(93)90057-V. [DOI] [PubMed] [Google Scholar]
- 45.Pedrós-Alió C, García-Cantizano J, Calderón-Paz J I. Bacterial production in anaerobic water columns. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Current methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 519–530. [Google Scholar]
- 46.Pedrós-Alió C, Guerrero R. Microbial ecology in Lake Cisó. Adv Microb Ecol. 1993;13:155–209. [Google Scholar]
- 47.Pfennig N, Trüper H G. Anoxygenic phototrophic bacteria. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams and Wilkins; 1989. pp. 1635–1653. [Google Scholar]
- 48.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 49.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reisenbach H. The order Cytophagales. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 4. Berlin, Germany: Springer-Verlag; 1991. pp. 3631–3675. [Google Scholar]
- 51.Reysenbach A L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosselló-Mora R, Thamdrup B, Schäfer H, Weller R, Amann R. The response of the microbial community of marine sediment to organic carbon input under anaerobic conditions. Syst Appl Microbiol. 1999;22:237–248. doi: 10.1016/S0723-2020(99)80071-X. [DOI] [PubMed] [Google Scholar]
- 53.Santegoeds C M, Nold S C, Ward D M. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl Environ Microbiol. 1996;62:3922–3928. doi: 10.1128/aem.62.11.3922-3928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stackebrandt E, Rainey F A. Partial and complete 16S rDNA sequences, their use in generation of 16S rDNA phylogenetic trees and their implications in molecular ecological studies. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3.1.1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–17. [Google Scholar]
- 55.Stahl D A, Amann R I. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N. Y.: Wiley; 1991. pp. 205–248. [Google Scholar]
- 56.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teske A, Sigalevich P, Cohen Y, Muyzer G. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl Environ Microbiol. 1996;62:4210–4215. doi: 10.1128/aem.62.11.4210-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wintzingerode F V, Goebel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]