Abstract
Methionine is the initiator amino acid for protein synthesis, the methyl source for most nucleotide, chromatin, and protein methylation, and the carbon backbone for various aspects of the cellular antioxidant response and nucleotide biosynthesis. Methionine is provided in the diet and serum methionine levels fluctuate based on dietary methionine content. Within the cell, methionine is recycled from homocysteine via the methionine cycle, which is linked to nutrient status via one-carbon metabolism. Unlike normal cells, many cancer cells, both in vitro and in vivo, show high methionine cycle activity and are dependent on exogenous methionine for continued growth. However, the molecular mechanisms underlying the methionine dependence of diverse malignancies are poorly understood. Methionine deprivation initiates widespread metabolic alterations in cancer cells that enable them to survive despite limited methionine availability, and these adaptive alterations can be specifically targeted to enhance the activity of methionine deprivation, a strategy we have termed “metabolic priming”. Chemotherapy-resistant cell populations such as cancer stem cells, which drive treatment-resistance, are also sensitive to methionine deprivation, suggesting dietary methionine restriction may inhibit metastasis and recurrence. Several clinical trials in cancer are investigating methionine restriction in combination with other agents. This review will explore new insights into the mechanisms of methionine dependence in cancer and therapeutic efforts to translate these insights into enhanced clinical activity of methionine restriction in cancer.
Keywords: Methionine, metabolism, one-carbon, epigenetics, oxidative stress, nutrition, cancer therapy
Methionine cycle and one-carbon metabolism: linking carbon flux to nutrient status
Methionine is an essential amino acid for protein synthesis as well as many biochemical reactions required for cell viability and growth (Figure 1). S-adenosylmethionine (SAM) is the universal methyl donor for RNA, DNA, and chromatin methylation, and is synthesized from methionine via methionine adenosyltransferase (MAT) in the first step of the methionine cycle (Figure 2A) [1,2]. MAT I (a tetramer) and MAT III (a dimer) are generally expressed in the liver, where high SAM synthesis occurs, and are encoded by MAT1A, while MAT II (a dimer) is expressed in most other cell types and is encoded by MAT2A [3-5]. SAM is then converted to S-adenosylhomocysteine (SAH) through various transmethylation reactions. SAH is hydrolyzed to homocysteine by SAH hydrolase (AHCY or SAHH), which can then be re-methylated to methionine by methionine synthase (5-methyltetrahydrofolate-homocysteine methyltransferase; MTR or MS) or betaine-homocysteine methyltransferase (BHMT) to complete the methionine cycle [1,2]. Alternatively, homocysteine can be diverted into the transsulfuration pathway by cystathionine-β-synthase (CBS) to become cystathionine, which is then converted to cysteine by cystathionase (CTH) for use in glutathione production and maintenance of redox homeostasis [1,3]. Additionally, SAM is utilized for polyamine synthesis, which produces S-methyl-5’-thioadenosine (MTA) as a byproduct that can then be recycled back to methionine in the methionine salvage pathway [1,3]. The immediate precursor to methionine in the salvage pathway, 2-keto-4-methylthiobutyrate (MTOB), can also be converted to methional, which is capable of activating the apoptotic cascade, linking methionine metabolism to cell fate decisions [1].
Figure 1.
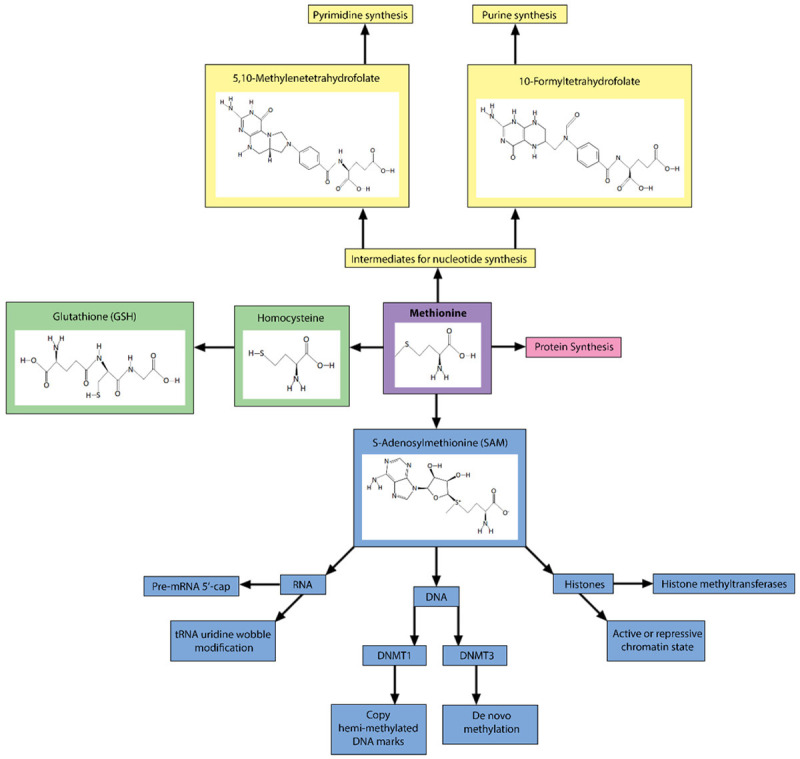
Diverse cellular functions of methionine in cell survival, growth and division. Methionine is an essential amino acid, meaning it must be provided through diet. The main cellular functions for methionine are protein synthesis, generation of intermediates and cofactors for nucleotide and glutathione synthesis, and generation of S-adenosylmethionine (SAM), the universal methyl donor for methylation of RNA, DNA, and histones.
Figure 2.
Methionine and Folate Cycles are Linked by One-Carbon Metabolism. A. The methionine and folate cycles functionally interact to generate intermediates and cofactors for various anabolic processes. Available methionine largely depends on dietary input. Methionine cycle intermediates can be diverted into the methionine salvage pathway to promote purine synthesis and into the transsulfuration pathway to aid in the production of reduce glutathione. B. The methionine and folate cycles interact to divert intermediates based on methionine availability. SAM normally inhibits MTHFR and BHMT and promotes CBS to divert intermediates towards nucleotide and glutathione synthesis when methionine and SAM are plentiful. During times of low SAM, this inhibition in released, increasing the synthesis of methionine from homocysteine as well as allowing 5-methyltetrahydrofolate to begin to accumulate. 5-methyltetrahydrofolate blocks GNMT to allow methyltransferases to utilize available SAM to maintain RNA, DNA, and histone methylation. MAT2A selectively increases during low methionine, further increasing SAM synthesis. Together, this regulation maintains intermediates around the folate and methionine cycle in times of low methionine input. Abbreviations: TS, tetrahydrofolate synthase; MTHFR, methylenetetrahydrofolate reductase; SHMT, serine hydroxymethyltransferase; MS, methionine synthase; MAT, methionine adenosyltransferase; MAT2A, methionine adenosyltransferase 2A; GNMT, glycine N, methyltransferase; SAHH/AHCY, adenosylhomocysteinase; BHMT, betaine-homocysteine S-methyltransferase; CBS, cystathionine-β-synthase; MTAP, S-methyl-5’-thioadenosine phosphorylase.
The methionine and folate cycles are directly linked via so-called one-carbon metabolism to stabilize metabolite levels, particularly during times of low methionine availability to maintain flux through the methionine cycle instead of diverting intermediates towards other pathways [6] (Figure 2B). When SAM is abundant, indicative of high methionine levels, SAM inhibits the enzymes methylenetetrahydrofolate reductase (MTHFR) and BHMT to limit conversion of homocysteine to methionine, allowing homocysteine to be diverted for transsulfuration [5,6]. SAM also activates CBS, further diverting homocysteine towards transsulfuration [5-7]. When methionine concentrations are low, SAM levels decline, releasing inhibition of MTHFR and BHMT, and suppressing activation of CBS to maintain flux through the methionine cycle. Low methionine synthesis leads to accumulation of 5-methyltetrahydrofolate (5-mTHF), which inhibits glycine N-methyltransferase (GNMT) to direct SAM usage towards DNA methyltransferases (DNMTs) [6]. Moreover, MAT2A expression is selectively increased when methionine levels are low [8-10], suggesting that SAM biosynthesis is regulated in an effort to maintain cellular SAM levels. These coordinated interactions allow a cell to buffer declining methionine and SAM levels in times of low methionine input to maintain protein and polyamine synthesis as well as methylation reactions, all of which are necessary for cell growth and survival.
Cancer metabolic vulnerabilities: an addiction to methionine
Oncogenic alterations in cancer cells modify their nutrient intake and utilization, leading to altered metabolic states that fuel their enhanced proliferation and growth [11]. One such tumor-specific metabolic alteration is methionine dependence, namely, the requirement of exogenous methionine for tumor growth and the inability of transformed cells to grow when methionine is replaced by homocysteine in cell media or the diet [12-15]. This effect appears to be specific to transformed cells, as normal cells maintain growth and viability, even on prolonged methionine deprivation when supplemented with homocysteine [12-14]. When human sarcoma HOS-1A cells were co-cultured with fibroblast FS-3 cells, methionine deprivation selectively suppressed growth of the cancer cells while maintaining normal growth of the fibroblasts [16]. To date, many cancer cell lines evaluated, including prostate, breast, bladder, colon, glioma, kidney, melanoma, and leukemia cells, along with fresh patient tumors cultured ex vivo, have been found to exhibit some degree of methionine dependence [1,13,14,17-19]. Methionine deprivation has also been found to decrease invasion and wound healing, suggesting it may be useful in targeting metastatic subclones [20,21]. Viewed from this context, cancer cells are methionine auxotrophs “addicted” to exogenous methionine, a metabolic dependence intrinsic to the transformed phenotype that is not manifested by most normal cells [22-24].
One exception to the lack of methionine-dependence in normal cells was observed in healthy human lymphocytes cultured in methionine-free media. One study cultured both normal and malignant B and T lymphocytes in methionine-free media and all cell lines with the exception of one failed to grow and colony formation was abolished [25]. However, other studies conducted since have raised concern that the level of homocysteine used was not sufficient to support cell growth. Another study used lymphocytes isolated from fasting blood samples of healthy volunteers in methionine-free media and found a range of methionine-dependence, with approximately a third of participant’s lymphocytes growing at half the rate observed in full methionine media [26]. This study found that methionine-dependence did not relate to micronucleus frequency or mutations in either MTHFR or methionine synthase [26]. A more recent study found that the MTHFR C677T allele (rs1801133) significantly reduced the methionine-dependence index of lymphocytes, a measure of viable cell number between methionine-complete and methionine-free media [27]. Further evaluation of the methionine-dependent phenotype in lymphocytes, the underlying mechanisms, and the potential impact of a methionine restricted diet on lymphocyte growth in vivo is clearly warranted.
In animal models, dietary methionine restriction has been demonstrated to have antitumor activity against a broad spectrum of tumors in vivo and extend survival [1,12,15]. More recently, these findings have been extended to patient-derived xenograft models of cancer, which more faithfully capture the genetic heterogeneity of human cancer than established cancer cell line, including two chemotherapy-resistant KRas-driven colorectal cancer PDX models [28]. Methionine deprivation or restriction also inhibits metastasis in diverse murine models including sarcoma, triple-negative breast cancer and melanoma [20,29,30]. These antitumor effects of methionine restriction have been attributed to cell-cycle blockade and/or apoptosis induction in tumor cells [16,17,31,32]. Dietary methionine restriction reduces methionine metabolite levels in serum, liver, and tumor cells [28,33], however, some tissue-specific alterations in methionine cycle metabolites were found. One study using dietary methionine restriction in mice did not find a reduction in liver methionine levels [34]. Other studies found an initial decrease in liver methionine and methionine metabolites, which in one was maintained for the duration of the study [28] while in another recovered within 4 days [35]. A study evaluating sex difference in response to methionine restriction found liver methionine declined in female mice but not male mice, while liver SAM levels only decreased in male mice [36]. These studies suggest the impact of dietary methionine restriction on liver metabolites may depend on several factors, including time since diet initiation and gender. Considering the liver is normally the largest consumer of exogenous methionine, it is possible there are liver-specific compensatory mechanisms to maintain methionine levels [5,37]. It is also important to note that in most mammals, including rodents, SAM availability is regulated by threonine metabolism via threonine dehydrogenase (TDH), however in humans TDH is a non-functional pseudogene, suggesting human cells may be more reliant on methionine metabolism than other mammalian cells [8].
Given their dependence on exogenous methionine, many tumors have acquired mechanisms to take up methionine more efficiently than surrounding normal tissue in the tumor microenvironment [38]. Methionine and other essential amino acids are transported into cells by L-Type Amino Acid Transporters, LAT1, LAT2, LAT3, and LAT4 [39]. LAT1 is commonly overexpressed in various types of cancer [39,40], and LAT1 expression is often associated with poor survival [40]. The expression levels of LAT1 also correlate with intracellular availability of SAM and the level of H3K9me3 within cells [39], suggesting LAT1 expression affects methionine availability or aspects of methionine metabolism. Moreover, LAT1 knockdown reduces the viability of cancer cell lines [40]. Intriguingly, overexpression of LAT4/SLC43A2 in tumor cells enables them to preferentially take up methionine in the tumor microenvironment and deplete methionine in T cells, leading to loss of key epigenetic drivers of cytokine signaling and impaired T cell immunity, while inhibition of LAT4 increased basal and checkpoint-induced tumor immunity [41]. Based on the increased uptake of methionine into rapidly growing tumor tissue, the radiotracer L-methyl-11C-methionine (11C-methionine) has also been used to measure treatment responsiveness [42] and to differentiate tumor margins from necrotic areas from previous radiation treatment to more accurately measure residual disease [43].
Mechanisms of methionine dependence in cancer: oxidative stress derails methionine recycling
Rapidly proliferating cancer cells have an increased demand for methionine for protein synthesis, for maintenance and de novo methylation of DNA and chromatin, which requires SAM, and for the methionine cycle to generate intermediates and cofactors used in glutathione and nucleotide biosynthesis [2]. Methionine deprivation in transformed cells has been found to lower available methionine and SAM [9,44-48]. Methionine still appears to incorporate into newly synthesized proteins at a rate comparable to healthy cells, suggesting endogenous methionine is preferentially utilized or mobilized for protein synthesis in methionine limitation [1,46,49,50]. Many enzymes within the methionine cycle, including MTR and AHCY, as well as enzymes involved in transmethylation, polyamine synthesis, and transsulfuration have been shown to be highly active in many types of tumor cells, suggesting many aspects of methionine metabolism may influence the growth capacity of cancer cells [49,51]. In culture, methionine deprivation has also been found to induce apoptosis and increase p53-positive cells; however, this induction does not appear to fully depend on p53 status, as p53 knockout only led to a partial rescue of cell death in response to methionine deprivation [8,17,52]. Interestingly, wild-type p53 interacts with DNA (cytosine-5)-methyltransferase (DNMT1) to increase the de novo DNA methylation and gene silencing capacity of DNMT1, suggesting functional p53 may use DNMT1 and epigenetic plasticity to evade apoptosis in low methionine stress conditions [53].
A common feature of methionine-dependent cancer cells is their increased demand for exogenous methionine for transmethylation reactions [1,19,48,51,54]. When 18 tumor cell lines and 4 fibroblast cell lines were compared, all tumor cell lines showed elevated rates of transmethylation relative to fibroblasts in both normal and methionine-depleted media [55]. Methionine-dependent mammary adenocarcinoma and lymphoma cell lines were found to accumulate SAH five times faster than methionine-independent cell lines in response to AHCY inhibition, suggesting higher rates of transmethylation [49]. The same study isolated a methionine-independent-revertant from the mammary adenocarcinoma cell line and found the major difference between dependent and independent status was decreased transmethylation rates in the independent revertant [49]. Notably, the methionine-independent revertant also lost several hallmarks of malignancy, including colony-formation and increased anchorage-independence [46,56,57]. Moreover, methionine deprivation in cancer stem cells inhibits diverse histone methylation marks and these effects are largely abrogated by SAM or methionine supplementation but not homocysteine [10,44].
Many studies have also found a lower SAM:SAH ratio in methionine-dependent versus independent cells in response to methionine deprivation, with a methionine-independent-revertant exhibiting a normal SAM:SAH ratio [46,49,50]. Furthermore, SAM supplementation has also been found to rescue the effects of methionine deprivation in a variety of cell types [45,58], including cancer stem cells [10,44]. Interestingly, transmethylation and SAM may play a role in the development of cancer, as global DNA hypomethylation is commonly found in many types of cancer and pre-cancerous cells, and has been hypothesized to result from unbalanced global methylation, leading to the gradual loss of DNA methylation, particularly in genomic areas copied late in replication [46,59,60]. SAM is also used in the synthesis of polyamines, and various cancer cells have been found to have hyperactive polyamine synthesis to support rapid rates of growth and division; however, spermidine supplementation during methionine deprivation did not rescue cell growth defects [56].
While the transsulfuration pathway for glutathione synthesis is largely limited to the liver, pancreas, and kidneys in healthy adults, cancer cells appear to have a heightened dependency on transsulfuration of homocysteine to buffer oxidative stress that accumulates as a result of rapid cell growth paired with metabolic dysfunction [2,61]. In humans, the conversion of homocysteine to cystathionine is reduced 20% under dietary methyl group restriction [5]. The methionine-dependent phenotype can be partially reversed by over-expression of the cystine transporter xCT, and loss of the xCT transporter leads to a significant decrease in intracellular glutathione [61,62]. Indeed, oncogenic PI3KCA confers methionine dependence by reducing xCT expression and cystine import, thereby diverting homocysteine to the transsulfuration pathway to buffer oxidative stress rather than recycling methionine [61]. Moreover, our group and others have demonstrated that methionine deprivation transiently depletes glutathione levels and induces oxidative stress [28,63-65], which is critical for the cytotoxic effects of methionine deprivation because antioxidants such as N-acetylcysteine (NAC) attenuate its cytotoxicity [28,65]. Collectively, these findings strongly suggest that at least part of the methionine dependence of cancer cells results from the need to divert homocysteine from the methionine cycle into the transsulfuration pathway to promote glutathione synthesis and buffer oxidative stress, thereby creating a dependence on exogenous methionine.
Methionine deprivation also activates nuclear factor erythroid 2-related factor 2 (NRF2) and enhances complex formation at antioxidant response elements (ARE) [66]. Interestingly, MAT II has been found to associate with MAFK-NRF2 and affect repression of target genes [67]. As noted above, the transient oxidative stress induced by methionine deprivation may partially result from glutathione depletion. One study found glutathione showed a time-dependent reduction during methionine deprivation, followed by an increase in nuclear NRF2 [68]. However, data on serum and intracellular glutathione levels in response to methionine deprivation have been variable [69-72], possibly due to ATF4-NRF2 activating multiple genes involved in glutathione biosynthesis [73] and likely reflecting temporal and/or tissue-specific differences. Indeed, a study of long-term methionine restriction in F344 rats found short-term methionine restriction depletes glutathione [74], while adaptive mechanisms, including diminished mitochondrial ROS production [75], lead to increased blood glutathione when methionine restriction is prolonged [74]. One consequence of diverting homocysteine away from the methionine cycle is reduced available intermediates for SAM synthesis needed to maintain methylation reactions and polyamine synthesis for cell growth and division [56]. Oxidative stress increases the activity of CBS while MAT I and MAT III activity is reduced, creating a further deficit in serum methionine and SAM availability [7,76]. Increased uptake in exogenous methionine commonly seen in cancer may contribute to cancer cell dependence on transsulfuration through high intracellular sulfur load and the generation of cytotoxic sulfur intermediates that can interfere with cellular detoxification proteins and mitochondrial electron transport chain function [7,38]. Interestingly when methionine-dependent and methionine-independent-revertant mammary carcinoma cell lines were compared, methionine-dependent cells showed an increase in total SAH as well as increased conversion of homocysteine back to SAH compared to the revertant, further implicating oxidative stress with methionine dependence, as homocysteine itself directly contributes to oxidative damage [56]. Homocysteine also inhibits AHCY to further increase SAH levels [77]. SAH binds and inhibits methyltransferase activity [78], and intracellular SAH concentrations have been found to correlate with DNA hypomethylation [79]. In healthy year-old mice, methionine restriction decreased liver SAH levels 50% compared to control-fed mice; methionine restricted mice also maintained global DNA methylation in the liver comparable to young animals, while control-fed mice gradually lost global DNA methylation marks, similar to what is commonly seen in human aging [73]. Healthy cells in aging animals and humans may benefit from the reduced SAH burden seen during methionine restriction, while cancer cells, which have a generally heightened need for methionine, SAM, and redox buffering, may suffer from methyl shortage leading to cell cycle arrest or cell death.
Another factor evaluated for its role in methionine dependence is deletion of the methionine salvage enzyme gene, methylthioadenosine phosphorylase (MTAP). The MTAP gene is commonly deleted in a variety of cancer types, often due to co-deletion with the adjacent tumor suppressor gene CDKN2A which encodes p16 (INK4A) [1,54]. MTAP uses MTA, a byproduct from the utilization of SAM in polyamine synthesis, to regenerate methionine. While MTAP is frequently mutated or deleted in cancer, cancer cell lines with intact methionine cycle enzymes can still be methionine dependent, suggesting MTAP deletion may facilitate, but is not crucial for, methionine-dependent status [80]. Furthermore, MTAP deletion or re-introduction in various human cancer cell lines did not affect methionine-dependent growth defects [81]. However, MTAP deletion does uncover a druggable dependence on the protein arginine methyltransferase PRMT5, which is directly inhibited by the resultant high levels of MTA [82,83]. Moreover, MTAP deletion sensitize tumor cells to depletion of MAT2A, which is needed to produce SAM for PRMT5-mediated methylation [82,83]. PRMT5 has a much higher affinity for MTA than SAM, making its catalytic activity sensitive to the SAM/MTA ratio [83], suggesting methionine restriction may further promote PRMT5 dependence in MTAP-deficient cells.
Another metabolic vulnerability of transformed cells exposed by methionine restriction is their intrinsic demand for nucleotide synthesis to fuel tumor growth. The methionine cycle is inextricably linked to the folate cycle via one-carbon metabolism (Figure 2) and perturbations of methionine availability impact folate cycle activity and outputs such as redox buffering and thymidine and purine synthesis for nucleotides [2]. Indeed, the folate cycle was the target of the first class of cytotoxic agents (e.g., 5-fluorouracil and methotrexate) that were developed [84]. Although the critical role of the pentose phosphate pathway (PPP) in producing NADPH for reductive synthesis and redox homeostasis is well established, the folate cycle was recently shown to be a key source of cellular NADPH synthesis from NADP via the conversion of methylene tetrahydrofolate to 10-formyl-tetrahydrofolate [85]. Intriguingly, the PPP also plays a key role in folate cycle activity: inhibition of PPP increases NADP levels which inhibit the folate cycle enzyme dihydrofolate reductase (DHFR) [86], thereby directly linking the PPP to one-carbon metabolism. In addition to redox buffering, the folate cycle plays a key role in nucleotide synthesis by generative thymidine and purine precursors. Notably, methionine restriction rapidly depletes multiple nucleotide precursors in the serum of humans and in murine tumors in vivo, while supplementation of nucleosides partly attenuates the cytotoxicity of methionine restriction in cultured cancers cells [28]. Metastatic melanoma cells are under greater oxidative stress than primary tumor cells and are readily suppressed by folate inhibition [87], suggesting that one-carbon metabolism may play a particularly prominent role in cancer progression/metastasis. Overall, these findings point to the interconnected nature of the folate and methionine cycles and the role of nucleotide depletion in the effects of methionine restriction.
Methionine-sensing and the cellular stress response: living with less (Methionine)
The integrated stress response (ISR) is a highly conserved adaptive response to disparate cellular stressors mediated by protein kinase R (PKR) in response to double stranded RNA, protein kinase R-like endoplasmic reticulum kinase (PERK) in response to endoplasmic reticulum (ER) stress, heme-regulated inhibitor (HRI) in response to heme depletion, and general control nonderepressible 2 (GCN2) in response to amino acid deprivation and ultraviolet (UV) damage [69,88]. These kinases phosphorylate eukaryotic initiation factor 2α (eIF2α), which blocks translation re-initiation by reducing GDP exchange for GTP on eIF2, inhibiting its function. Although global protein translation is suppressed, activating transcription factor (ATF) 4 mRNA is preferentially translated during times of cell stress when complete assembly of the translational machinery on ATF4 mRNA is slowed so that the 40S ribosomal subunit bypasses the upstream open reading frame, uORF2, that is translated in the absence of stress [89]. When the 60S ribosomal subunit attaches downstream of uORF2, the coding sequence of ATF4 is translated, allowing protein levels to rapidly increase in response to eIF2α inactivation, including amino acid limitation [88,90]. ATF4 is a member of the ATF/cyclic AMP response element binding protein (ATF/CREB) family that dimerizes and binds to DNA at CRE motifs. A component of the methionine deprivation-specific transcriptional response induced by ATF4 may depend on its dimerization partner [91-93].
Our lab observed that ATF4 and Sestrin2 (SESN2), an ATF4 target, are rapid induced in triple-negative breast cancer cells in response to methionine deprivation [94]. Methionine deprivation also induced phosphorylation of eIF2α; however, this phosphorylation was not dependent on GCN2 or PERK, as knockdown of either or both did not eliminate phosphorylation of eIF2α or induction of ATF4 or SESN2 in response to methionine deprivation [94]. The potential roles of PKR and HRI, other known kinases capable of phosphorylating eIF2α, have not been investigated during methionine deprivation. However, ATF4 and the ISR can be activated without eIF2α phosphorylation [95] and at least part of the methionine deprivation-specific response may be eIF2α phosphorylation-independent [96]. Methionine deprivation in eIF2α-knockout mouse embryonic fibroblasts (MEFs) resulted in an increase in ATF4 mRNA (albeit blunted response), and the authors postulated ATF4 is induced via lack of initiator tRNAmet instead of phosphorylation of eIF2α [96]. Another group reported that hepatic ATF4-target gene induction in response to methionine restriction was not dependent on GCN2 or phosphorylation of eIF2α [97]. They also found glutathione levels were decreased and NRF2 targets were strongly induced, suggesting ATF4-NRF2 may be induced regardless of phosphorylation of eIF2α in response to methionine restriction [97]. GCN2- and eIF2α phosphorylation-independent induction of ATF4 and ATF4-target genes in response to methionine restriction were also observed in mouse liver, along with glutathione depletion and NRF2 induction [98]. NRF2-ATF4 have been found to interact with each other at stress responsive elements [93], possibly in response to phosphoinositide 3-kinase (PI3K) [99] or protein kinase C (PKC) [100] pathway activation. Clearly, additional studies are needed to clarify the precise mechanisms by which methionine restriction activates the ISR and induces ATF4 transcription. Although GCN2 and phosphorylated eIF2α play key roles in the canonical amino acid deprivation response [101], the methionine deprivation-specific response appears to be at least partly independent of GCN2 and eIF2α phosphorylation to activate ATF4.
Another stress-activated pathway is autophagy, which functions to recycle macromolecules and organelles within a cell during stress or when nutrients are limited [102]. Methionine restriction has been found to extend the life- and health-span of various organisms [103,104], and this effect has been attributed to increased autophagic flux [105,106]. Interestingly, methionine restriction led to a decrease in Reactive Oxygen Species (ROS) [105], suggesting the increase in autophagy may promote mitochondrial function to prevent ROS leak. In line with these findings, a recent study found methionine restriction elevated mitophagy specifically, which is the autophagic recycling of mitochondria [107]. Furthermore, cells restricted of methionine no longer showed lifespan extension when key mitophagy genes were deleted or when mitochondrial function was blocked by the mitochondrial toxin, paraquat [107]. Mitochondrial peroxide production, indicative of mitochondrial dysfunction, was reduced approximately in half by methionine restriction compared to controls, and this reduction was not seen in cells with impaired mitophagy [107], suggesting increased mitophagy may sustain functional mitochondria. Although methionine deprivation has been demonstrated to activate autophagy in cancer cells [108,109], the role of autophagy in the antitumor effects of methionine restriction is poorly understood.
Methionine restriction and the cancer epigenome: methylation matters
The bulk of genomic methylation is determined early in development to aid in cell differentiation and repress genes not needed for tissue-specific function. These methylation patterns often remain static throughout life, with changes mainly occurring in response to stress and injury [110,111]. Epigenetic plasticity is seen in the normal cellular response to injury; however, when prolonged this plasticity can contribute to the hallmarks of cancer [112]. Gene mutations and truncations can also alter gene methylation patterns at specific loci, leading to heritable epigenetic alterations [110]. Global hypomethylation has been implicated in the acquisition of senescent cells, aging, and various age-related diseases [113]. Cancer cells in general exhibit genome-wide loss of epigenetic stability, often exhibiting global hypomethylation [114,115] and promoter-specific hyper-methylation [116], which has been implicating in oncogene activation and tumor suppressor silencing respectively [117,118]. These epigenetic alterations are also observed in pre-cancerous cells [112,119], normal tissue adjacent to tumor tissue [120,121], and hereditary human cancers [122]. Loss of heterochromatin, particularly loss of H3K9me2, has also been associated with the epithelial-to-mesenchymal (EMT) transition used by cancer cells for migration and metastasis, further implicating epigenetic dysfunction with malignant progression [123-125]. Multiple studies evaluating the cancer cell epigenome have found a correlation between epigenetic dysfunction and tumor progression, particularly for sustained growth in suboptimal conditions [123,126,127].
Methionine deprivation, through reduced available SAM, decreases the histone methylation marks H3K4me2, H3K4me3, H3K9me2, and H3K27me3 in vitro and in vivo [8,34,45]. Bulk screening for methylation at CpG island promoters or long interspersed nuclear element-1 (LINE-1) repetitive DNA sequences found no significant alterations, indicating cells respond to methionine deprivation by salvaging methyl groups preferentially from existing histone marks instead of DNA methylation [45]. Widespread DNA hypomethylation in cancer is associated with gene silencing due to the formation of repressive chromatin structures at these sites, suggesting cancer cells with epigenetic dysfunction may rely more heavily on histone methylation than DNA methylation to maintain viability [59,128]. DNA methylation marks are also generally more stable than histone methylation [129-131], implicating histone methylation as more dynamic and/or replaceable for cell survival than DNA methylation [110,112,132]. Interestingly, MAT II directly interacts with histone methyltransferases and this interaction can alter histone methylation dynamics [133]. Further work evaluating the allele-specific methylation response to methionine deprivation over time may better elucidate the role DNA methylation plays in methionine deprivation, as bulk methylome sequencing may overlook changes in individual allele methylation patterns.
“Epigenetic persistence” is defined as the ability of a cell to reestablish its epigenetic signature following environmental alterations or metabolic stress [134]. In response to methionine deprivation both di- and tri-methyl histone marks are preferentially removed while H3K9 mono-methylation marks are preserved to maintain heterochromatin stability during times of low SAM availability [134]. When H3K9 mono-methylation is blocked and cells are subjected to low SAM availability, they are no longer capable of epigenetic persistence when returned to full methionine media [134], suggesting H3K9 mono-methylation is necessary for a cell to maintain its heterochromatin state. The specific role of epigenetic persistence, and H3K9 mono-methylation in mediating the effects of methionine deprivation on transformed cells will require further investigation.
Additionally, SAM is utilized for RNA modifications to generate functional mRNA, control mRNA degradation, and aid in tRNA base-pairing under stress conditions [135-137]. The most abundant modification of eukaryotic mRNA is methylation of adenosine by guanine 7-methyltransferase (RNMT), which uses SAM to create the 5’-methyl cap on pre-mRNA, where eukaryotic initiation factor 4α (eIF4α) binds and recruits the 40S ribosomal subunit to begin translation and control m6A-dependent mRNA decay [136,138,139]. In response to genotoxic stress, the uridine of tRNAs that encode lysine, glutamine, and glutamic acid is modified to enhance the wobble of the third position for less exact base pairing in translation, allowing base-pair mutated transcripts to be translated [2,137]. Both uridine modifications, methoxycarbonylmethyluridine and thiolation of uridine, consume SAM, and the abundance of tRNA uridine thiolation correlates with sulfur amino acid availability [106,137]. Lysine, glutamine, and glutamic acid codons are prevalent in ribosome biogenesis genes, suggesting altered tRNA thiolation may impact general translation under reduced SAM conditions [106]. Ribosome thiolation also alters complex I ROS production, suggesting reducing thiolation may alter cellular redox status [140]. This may be another mechanism by which methionine deprivation preferentially affects cancer cells that require mRNA methylation to generate mature mRNA transcripts and may depend more heavily on tRNA uridine modifications to translate damaged mRNA transcripts, both of which may contribute to translation inhibition in cancer cells in response to methionine limitation.
Cancer stem cells: a dependency on SAM biosynthesis
Pluripotent embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) both appear more methionine-dependent than differentiated cells, and self-renewal capacity correlates with SAM concentration [8]. Methionine deprivation in ESCs and iPSCs primes cells for differentiation, and once differentiated, total cellular methionine demand decreases [8]. This suggests that part of the methionine-dependence of transformed cells may be due to the stem-like characteristics of some cancer cells. It also suggests that normal stem cell maintenance may be adversely affected by prolonged methionine deprivation. However, a study by Altundag et al. [141] comparing bone marrow mesenchymal stem cells, umbilical cord stem cells, and cancer stem cells found differing responses of each cell type to varying levels of methionine. Notably in cancer stem cells OCT3/4 and NANOG were nearly undetectable at 0 and 10 µM methionine, while levels of both were maintained in other stem cells, suggesting cancer stem cells may have a selective vulnerability to low methionine conditions.
Cancer stem cells or tumor-initiating cells commonly exhibit resistance to a variety of cytotoxic agents and undergo latent growth periods, making them treatment-resistant and critical to eliminate to reduce the risk of recurrence [142]. Redox state contributes to cancer stem cell therapeutic resistance, with quiescent cancer stem cell populations exhibiting low ROS levels due to enhanced antioxidant pathways and reliance on glycolysis [143-145]. Tumorspheres, cancer cells grown on ultra-low-attachment plates, are a common method to propagate tumor-initiating and cancer stem cells [142,146]. Work from our lab using triple-negative breast cancer (TNBC) tumorspheres showed methionine deprivation robustly decreased the number of tumorspheres as well as the population of cancer cells with CD44hi/CD24low cancer stem cell immunophenotype [10]. These effects were partly rescued by SAM supplementation, underscoring that SAM, rather than methionine per se, is the critical metabolite for cancer stem cell survival. Methionine deprivation also increased mRNA and protein levels of MAT2A as previously reported [8,9] and led to H3K4me3 demethylation and suppression of Sox9 expression [10], a regulator of plasticity in cancer stem cells [147]. Moreover, inhibition of MAT2A by siRNAs and/or the amino acid analogue cycloleucine [148] augmented the effects of methionine deprivation on tumorspheres and these molecular markers, respectively [10]. These results underscore that methionine metabolism, specifically SAM biosynthesis, plays a critical role in cancer stem cell survival, although the functional role of these epigenetic alterations in the observed effects of methionine deprivation on cancer stem cells has not been delineated.
Lung cancer tumorspheres are also enriched for SAM and SAH as well as methionine cycle activity and intermediates relative to isogenic, adherent cells, suggesting cancer stem cells may have a heightened requirement for methionine and SAM when compared to their more differentiated counterparts [44]. Following 48 hours of methionine deprivation, lung cancer tumorspheres showed reduced methionine cycle activity and a 30-fold reduction in SAM accompanied by decreased histone methylation, suggesting exogenous methionine deprivation greatly affects cancer stem cell methionine metabolism [44]. Furthermore, when tumorspheres were injected into NOD Scid Gamma (NSG) mice following methionine deprivation they greatly lost their tumor-forming abilities, decreasing the tumor burden 94% compared to mice injected with tumorspheres grown in control media, and this effect was specific to methionine deprivation [44]. Inhibition of MAT2A with the small molecule FIDAS-5 [149] phenocopied many of the cellular and in vivo effects of methionine deprivation and these effects were attenuated by SAM supplementation [44]. Collectively, these studies underscore that cancer stem cells rely on methionine and SAM biosynthesis for their self-renewal and tumor-forming capacity, pointing to this metabolic vulnerability as a druggable target to eliminate these chemotherapy-resistant cells.
Methionine restriction: priming cancer cells to die
Despite preclinical activity against diverse malignancies, methionine restriction has been largely disappointing in clinical trials to date (see section below). We postulated we could enhance the activity of methionine restriction by targeting molecular vulnerabilities uncovered by low methionine stress, a strategy we have termed “metabolic priming”. To this end, we performed a proteomics analysis of triple-negative breast cancer cells cultured in methionine-free media [150]. One of the proteins induced by methionine deprivation is the proapoptotic TRAIL receptor-2 (TRAIL-R2/DR5). Methionine deprivation, either through methionine-free media or the addition of methioninase, sensitized cells to the agonistic TRAIL-R2 monoclonal antibody lexatumumab by increasing cell surface expression of TRAIL-R2 [150]. At least part of this increase in TRAIL receptor expression in response to methionine deprivation is attributed to a loss of MAGE family member D2 (MAGED2) protein, which normally inhibits TRAIL receptor expression [150]. Interestingly, non-transformed breast epithelial cells were largely resistant to the combination of methionine deprivation and lexatumumab likely due to the lack of cell surface induction of TRAIL-R2. Mice fed a methionine-free diet and treated with lexatumumab for 3 weeks had smaller primary tumors and reduced lung metastases compared to mice treated with a methionine-deficient diet or lexatumumab alone in a triple-negative mCherry-MDA-MB-468 tumor model [150]. Similar results were recently reported for oral methioninase and the agonistic TRAIL-R2 mAb tigatuzumab in a mouse model of pancreatic cancer [151].
A second intriguing protein identified in our proteomic screen is MAT2A [150], the widely expressed enzyme that converts methionine into SAM. Methionine deprivation increases MAT2A mRNA stability [139] as well as protein and mRNA levels in response to declining SAM availability [10,152]. The MAT inhibitor cycloleucine [148] augmented the effects of methionine deprivation on tumorsphere formation and demethylation of H3K4me3 [10]. Moreover, the combination methionine deprivation with cycloleucine was more effective than either treatment alone at suppressing lung metastases in immunodeficient mice with orthotopic triple-negative breast tumors [10]. These effects are unlikely to be off-target effects of cycloleucine because siRNAs specifically targeting MAT2A phenocopy the effects of cycloleucine on tumorspheres [10]. Both lexatumumab and cycloleucine had previously been abandoned as anti-cancer treatments due to lack of efficacy [153-157] and unacceptable toxicity [158,159] respectively in clinical trials. When paired with methionine deprivation, both drugs showed enhanced cytotoxicity at much lower doses, suggesting methionine deprivation may be a useful strategy to enhance efficacy and lower toxicity of various anti-cancer drugs [10,150]. Additionally, newer MAT2A inhibitors such as FIDAS-5 [149] and the allosteric inhibitor PF-9366 [37] may have improved therapeutic indices.
Another potentially attractive biological target that emerged from the initial proteomics screen of triple-negative breast cancer cells treated with methionine deprivation is thioredoxin reductase 1 (TXNRD1) [150]. TXNRD1 is the cytosolic isoform of the enzyme that catalyzes the rate-limiting step in the thioredoxin antioxidant pathway [160]. Tumor cells rely on the glutathione and thioredoxin pathways to buffer and survive the high levels of ROS they encounter in the tumor microenvironment [161]. Although tumor cells can survive defects in either single pathway, dual inhibition of both antioxidant pathways is synthetic lethal due to overwhelming oxidative stress [162-164]. Consistent with methionine being metabolized to glutathione via the transsulfuration of homocysteine, methionine restriction transiently depleted reduced glutathione, triggering oxidative stress and inducing TXNRD1 mRNA/protein via a NRF2/ATF4-dependent mechanism [65]. Strikingly, methionine restriction primed triple-negative breast cancer cells, but not untransformed breast epithelial cells, to respond to TXNRD1 silencing or the pan-TXNRD inhibitor Auranofin [65], a gold-containing anti-rheumatic drug [165]. Notably, methionine restriction or TXNRD inhibition alone had modest or no effects on cell viability. Furthermore, the synergy between methionine restriction and TXNRD inhibition was dependent on oxidative stress and was abrogated by N-acetylcysteine treatment [65]. Dietary methionine restriction also increased TXNRD1 expression in mammary tumors and enhanced the antitumor activity of Auranofin in metastatic and Patient-derived Xenograft (PDX) models of breast cancer in mice [65]. Overall, these studies provide proof-of-concept preclinical evidence that methionine restriction can be used to metabolically prime tumor cells to respond to rationally selected agents that target vulnerabilities exposed by methionine stress.
Methionine restriction in combination with cytotoxic agents
Many studies have examined combining methionine deprivation or restriction with various cytotoxic agents to enhance their therapeutic activity in preclinical models (Table 1). Methionine deprivation induces a reversible growth arrest in late S/G2 phase of the cell cycle [46,166] and/or G1 arrest [19,167], sensitizing cancer cells to cell cycle phase-specific chemotherapies such as DNA-damaging and microtubule-targeted agents [1,16]. Furthermore, DNA and chromatin methylation have been implicated in resistance to DNA-damaging agents by limiting access to DNA [38]. Methionine deprivation or restriction reduces various histone methylation marks [10,34,44,134], potentially facilitating DNA access to enhance the activity of DNA-damaging drugs such as doxorubicin and cisplatin. Indeed, the methylation inhibitor decitabine enhanced the antitumor activity of enzymatic methionine depletion in a soft tissue sarcoma PDX model [168], while another methylation inhibitor (azacitidine) augmented the activity of enzymatic methionine depletion in a PDX model of osteosarcoma [169]. Moreover, multi-drug-resistant colon cancer xenografts were sensitive to the combination of cisplatin, methionine deprivation, and the methionine analog ethionine to potentiate methionine deprivation [63]. Similarly, the combination of doxorubicin, methionine deprivation and ethionine resulted in 51% growth inhibition and 1.7-fold longer survival in a xenograft model of small cell lung cancer resistant to doxorubicin alone [63]. Similarly, methionine deprivation in combination with doxorubicin and vincristine prolonged survival of Yoshida sarcoma-bearing rats despite only partially depleting circulating methionine (18.4 μM in methionine-depleted TPN-treated rats versus 87.7 μM in control rats) [170]. Notably, the combination of methionine deprivation with doxorubicin and vincristine has tumor-selective cytotoxic effects on a broad spectrum of cancer cell types, including methionine-independent revertant cancer cells, with minimal effects on cocultured fibroblasts [16]. These preclinical findings support the combination of methionine deprivation with DNA-damaging and microtubule-targeted agents.
Table 1.
Methionine restriction or deprivation in combination with various chemotherapies in preclinical models
| Methionine restriction to metabolically prime cancer cells | ||
|
| ||
| Chemotherapy | Drug Target | Reference(s) |
|
| ||
| Lexatumumab | Monoclonal agonist antibody for the death receptor TRAIL receptor 2 | Strekalova et al. 2015 [150] |
| Cycloleucine | Methionine adenosyltransferase (MAT) inhibitor | Strekalova et al. 2019 [10] |
| Auranofin | Thioredoxin reductase (TXNRD) inhibitor | Malin et al. 2021 [65] |
|
| ||
| Methionine restriction and standard of care chemotherapy | ||
|
| ||
| Chemotherapy | Drug Target | Reference(s) |
|
| ||
| 5-Fluorouracil | Thymidylate synthase (TS) inhibitor | Gao et al. 2019 [28] |
| Yoshioka et al. 1998 [52] | ||
| Cisplatin | Binds DNA and interferes with repair | Poirson-Bichat et al. 2000 [63] |
| Tan et al. 1999 [210] | ||
| Doxorubicin | Binds and inhibits DNA Topoisomerase II | Poirson-Bichat et al. 2000 [63] |
| Stern et al. 1986 [16] | ||
| Nagahama et al. 1998 [170] | ||
| Vincristine | Interacts with tubulin to prevent mitotic spindle formation | Stern et al. 1986 [16] |
| Nagahama et al. 1998 [170] | ||
| Decitabine | DNA demethylating agent | Higuchi [168] |
| Azacitidine | DNA demethylating agent | Higuchi [169] |
| Gemcitabine | Pyrimidine antagonist | Kawaguchi [171] |
Some of the earliest chemotherapy drugs that were developed target folate metabolism (e.g., 5-fluorouracil (FU) and methotrexate) [84], suggesting that they may act synergistically with methionine restriction to disrupt one-carbon metabolism. Indeed, a recent study demonstrated that doses of 5-FU that were inactive against a colorectal cancer patient-derived xenograft model in mice synergized with dietary methionine restriction to cause oxidative stress, disrupt nucleotide metabolism and inhibit tumor growth [28]. Notably, the antiproliferative effects of methionine restriction and 5-FU were partially rescued by N-acetylcysteine and/or nucleosides [28], pointing to the critical role of ROS and disrupted nucleotide metabolism, respectively, in the observed effects. Similarly, the combination of the nucleoside analog gemcitabine with methionine depletion inhibited tumor growth and induced regression in a mouse model of gemcitabine-resistant pancreatic cancer [171]. The synergistic effects of methionine restriction with antifolate agents may at least partly reflect the reduced protein level and activity of thymidylate synthase (TS) observed during methionine deprivation [172]. SAM supplementation has been found to increase resistance to 5-fluorouracil through upregulation of DNMTs [173], suggesting methionine deprivation may synergize with 5-fluorouracil administration by maintaining low intracellular SAM levels. Other chemotherapy drugs that target the folate cycle, such as methotrexate and pemetrexed, inhibit dihydrofolate reductase, leading to accumulation and diversion of 5,10-methylene tetrahydrofolate into thymidylate synthesis, reducing available intermediates for one-carbon biosynthesis and to re-methylate homocysteine into methionine [174]. Methotrexate has also been reported to inhibit MAT2A expression and activity [175]. Additionally, methionine restriction enhances the antitumor activity of radiation in a sarcoma mouse model and alters redox and nucleotide metabolism [28]. Overall, these and other studies point to the ability of methionine restriction to augment the activity of chemotherapy drugs that target folate metabolism.
Methionine contributes to the production of glutathione via its transsulfuration to cysteine [176] and elevated glutathione has also been implicated in the development of drug resistance [17]. Since methionine restriction lowers intracellular glutathione levels in tumor cells [28], methionine restriction may further prove to be a useful strategy to enhance the sensitivity of cancer cells to various chemotherapeutic agents that augment oxidative stress [17,63,65]. Many commonly used classes of chemotherapy, such as anthracyclines, alkylating agents and platinum compounds as well as radiation generate oxidative stress via production of ROS and superoxide radicals [177,178]. However, moderate levels of oxidative stress induced by these drugs can decrease drug effectiveness by activating cytoprotective antioxidant systems [177]. Hence, the combined use of these chemotherapy drugs with methionine restriction may enhance ROS by diminishing glutathione levels, resulting in lethal oxidative stress. Consistent with this idea, studies have demonstrated that the enhanced activity of chemotherapy drugs in combination with methionine deprivation or restriction is dependent on oxidative stress and that this synergy is abrogated by various antioxidants [28,63]. Additionally, we recently demonstrated that methionine restriction depletes glutathione, activates TXNRD1 by an NRF2/ATF4-depedent mechanism, and renders tumors dependent on TXNRD1 [65], strongly implicating this protein as a promising therapeutic target in combination with methionine restriction. Consistent with this idea, TXNRD1 expression in diverse malignancies correlates with tumor grade and/or poor prognosis [179-181].
Methionine restriction in humans and clinical trials
Human fasting plasma methionine concentration ranges from 3-30 micromolar and correlates with SAM levels [34]. This variation in plasma methionine is partially explained by diet (~30%), as well as clinical variables such as age, gender and body composition (~30%), and genetic variables [34]. A study with six healthy, middle-aged individuals on a low methionine diet (approximately 2.92 mg/kg/day) resulted in lower serum methionine and methionine cycle metabolites, as well as diminished reduced glutathione and N-acetyl cysteine (NAC) [28]. Dietary methionine restriction (2 mg per kg per day) in 12 patients with diverse metastatic solid tumors led to a 58% reduction in plasma methionine by 2 weeks, with only mild losses in body weight that were regained upon return to a full methionine diet [182]. However, another study of twenty-six obese adults with metabolic syndrome randomized to a diet restricted to 2 mg methionine/kg body weight per day and were provided capsules containing placebo or methionine (33 mg/kg body weight per day) resulted in only a 13.8% reduction in plasma methionine concentrations and both groups had similar weight loss and improved metabolic indices (decreased plasma insulin and triglycerides), while the methionine restricted group had significantly higher fat oxidation and reduced hepatic lipid content [183]. In each of these studies, dietary methionine restriction was accomplished by providing 75% of total protein through a methionine-free medicinal beverage (Hominex-2, Abbott Nutrition) with the remaining 25% of protein through low-methionine foods such as fruits, vegetables, and grains [28,182,183]. These studies support the tolerability and feasibility of methionine restricted diets even in patients with advance malignancies, but they also suggest that variability in dietary adherence due to poor diet palatability or other variables [184] may affect the impact on systemic methionine levels, an important consideration when assessing its potential antitumor effects in clinical trials.
To overcome the palatability issue of existing methionine restriction diets, Fang et al. [185] explored the option of subjecting protein to oxidizing conditions to reduce methionine and cysteine while still leaving the protein intact. To do this, casein was oxidized by hydrogen peroxide to remove essentially all methionine and cysteine without impacting other amino acids (tryptophan and lysine were impacted but were added back) [185]. Mice were fed either an elemental methionine restriction diet, an oxidized casein methionine restriction diet or one of the respective control diets for 8 weeks, and several behavioral, physiological, endocrine and transcriptional endpoints were evaluated [185]. Both methionine restriction diets performed comparably in almost every measure studied including body weight, fat mass, energy expenditure, lipogenic gene expression, and gene expression signatures in line with ISR activation [185]. While these results are promising, future work is necessary to evaluate the feasibility of subjecting other proteins to oxidizing conditions as well as the palatability of these oxidized proteins in humans.
Based on preclinical studies demonstrating that a methionine-free diet [63] or methionine-free total parenteral nutrition (TPN) [186] sensitized drug-refractory solid tumors to alkylating chemotherapies, clinical studies investigating methionine deprivation in combination with the alkylating agent, cystemustine were initiated. In phase I and phase II clinical trials of glioma and melanoma patients on 24-hour methionine deprivation in combination with cystemustine (every two weeks), adherence to the diet was acceptable, there was no significant loss of body weight, and blood and inflammation markers remained stable, while plasma methionine fell by 41% or 53% on the day of the dietary intervention in these studies [187,188]. In the phase I study, 8 of 10 patients showed disease stabilization after the second cycle of treatment, 2 of 7 showed stabilization after the fourth cycle of treatment, and the median overall survival was 6.5 months [187]. In the phase II trial, of the 22 patients treated, 3 showed disease stabilization and the median survival time was 4.6 months [188]. There were no complete or partial responses.
Methionine deprivation in preclinical models inhibits O(6)-methylguanine-DNA methyltransferase (MGMT), an enzyme linked to resistance to alkylating agents [189,190]. Consistent with these observations, methionine deprivation in combination with cystemustine in 6 glioma and melanoma patients resulted in a 13% decrease in MGMT activity in peripheral blood mononuclear cells [191]. The decreases in MGMT activity as well as plasma methionine were not significantly different when methionine deprivation was extended beyond 24-hours, suggesting brief methionine deprivation may be sufficient to re-sensitize cancer cells to chemotherapy [191]. The purpose of this study was only to evaluate MGMT activity and plasma methionine levels, so response was not evaluated [191].
Due to the success of methionine deprivation in combination with 5-fluorouracil and platinum agents [28,38,63,150,172,192,193] the FOLFOX combination of 5-fluorouracil, leucovarin, and oxaloplatin, was tested in combination with three consecutive days of dietary methionine restriction in 11 metastatic colorectal cancer patients, and found to lower plasma methionine by 58% while maintaining good compliance and tolerance to the diet [194]. Of the 11 patients, 4 were evaluated for a response, 3 of whom showed a partial response and the other one showed stabilization [194].
Based on preclinical data [195] and small clinical studies showing synergy between a methionine-depleting TPN and 5-FU [196,197], a methionine- and cystine-free TPN infusion, was given in combination with 5-FU to 7 advanced gastric cancer patients and the results compared to 7 control gastric cancer patients given standard TPN and 5-FU [196]. The activity of thymidylate synthase, the primary molecular target of 5-fluorouracil, was reduced in the methionine-free TPN group (1.12 pmol/g) compared to control (2.35 pmol/g) [196]. Methionine-free TPN also enhanced necrotic area within tumors, shown by higher histological grades, without any major complications [196]. Since then, other groups have evaluated the combination of a methionine-depleting TPN and 5-FU on cancer [198,199], including a clinical study that found this combination suppressed proliferation of tumor cells in gastric cancer patients [200].
Overall, these small clinical studies have shown dietary methionine restriction lowers circulating methionine and is well tolerated even in combination with various chemotherapy drugs in patients with advanced cancer. To date, none of these studies have demonstrated objective response. Considering many are pilot studies of small patient size, larger trials are necessary to more accurately evaluate anti-cancer outcomes in response to methionine restriction. However, given the lack of clear efficacy in any of these studies, we have proposed to study methionine restriction in combination with agents that target pathways activated by low methionine stress (“metabolic priming”), while others have combined methionine restriction with cytotoxic agents that act synergistically with methionine deprivation.
There are currently multiple clinical trials ongoing and/or recently ended that may help to better elucidate the therapeutic potential of methionine restriction in cancer (Table 2) [201-208]. One trial (NCT03733119) was designed to examine whether methionine restriction metabolically primes metastatic triple-negative breast cancer to respond to a novel TRAIL agonist (ONC201) based on preclinical findings from our lab and others that methionine deprivation induces expression of TRAIL-R2 and sensitizes tumors to TRAIL-R2 agonists [150,151]. Another trial (NCT03186937) was designed to evaluate the impact of methionine restriction on TRAIL receptor expression and its impact on cancer stem cells based on preclinical findings that methionine deprivation targets many properties of cancer stem cells [10,150]. Methionine restriction was also tested in combination with radiation therapy in lung, prostate and breast cancer patients (NCT03574194) to determine if dietary methionine restriction can be used to radio-sensitize tumors. Another study (NCT00508456) in patients with glioblastoma multiforme was conducted to test the safety and efficacy of combination methionine restriction with temozolomide, an alkylating agent commonly used to treat glioblastoma. To better understand the impacts of methionine metabolism on life- and health-span, two studies were conducted to evaluate energy and glucose metabolism in overweight or diabetic adults using dietary methionine restriction (NCT00640757) or dietary methionine and cysteine restriction (NCT03629392), and a new study using sulfur amino acid (SAA) restriction has recently started recruiting (NCT04701346). One study also evaluated metabolic changes during combination methionine-cysteine restriction in healthy adults (NCT02192437). It remains to be determined what has been learned from these small studies in patients with diverse malignancies and/or metabolic disorders. Moreover, many of these studies have been hindered by difficulties recruiting patients and challenges with the tolerability of the methionine restricted diet for more than a few cycles, a shortcoming addressed by using non-dietary approaches to deplete methionine (discussed in the next paragraph). Clearly, methionine restriction is likely to have direct antitumor effects by depleting tumor methionine levels as well as indirect systemic effects by improving metabolic health that may be challenging to tease apart in clinical studies. Additionally, the potential adverse effects of methionine restriction on immune cells [209] in the tumor microenvironment, which are outcompeted for methionine uptake by tumor cells when methionine levels are limiting [41] will have to be investigated.
Table 2.
Recent clinical trials utilizing methionine restriction
| Identifier | Status | Recruiting Location | Study Title | Condition(s) | Study Goals |
|---|---|---|---|---|---|
| NCT03186937 | Terminated | University of Wisconsin Madison Carbone Cancer Center | A window of opportunity study of methionine deprivation in triple negative breast cancer | Triple negative breast cancer (TNBC) | • Effect of methionine restriction on TRAIL R2 cell surface expression |
| • Effect on cancer stem cells and metabolic health | |||||
| NCT03574194 | Suspended | West Virginia University Cancer Institute | Methionine-restricted diet to potentiate the effects of radiation therapy | Lung cancer | • Safety and adherence of methionine-restricted diet to radio-sensitize tumors |
| Prostate cancer | |||||
| Breast cancer | |||||
| NCT03733119 | Terminated | University of Wisconsin Madison Carbone Cancer Center | ONC201 with and without methionine-restricted diet in patients with metastatic triple negative breast cancer | Metastatic triple negative breast cancer (TNBC) | • Phase II trial combination methionine restriction and ONC201 (Akt/ERK inhibitor) for response rate |
| NCT00508456 | Terminated | U.T.M.D. Anderson Cancer Center Houston Texas | Dietary Methionine Restriction Plus Temozolomide for Recurrent GBM | Glioblastoma Multiforme | • Safety and tolerability of methionine restriction in combination with temozolomide |
| • Correlate response and survival with serum methionine and peripheral blood lymphocyte methylation | |||||
| NCT00640757 | Completed | Pennington Biomedical Research Center | Methionine-Restriction Diet (MRD) in Obese Adults with Metabolic Syndrome | Metabolic syndrome | • Impact of methionine restricted diet on weight loss and glucose metabolism/tolerance |
| NCT02192437 | Completed | Penn State University State College Milton S. Hershey Medical Center | Dietary Methionine and Cysteine Restriction in Healthy Adults | Healthy Individuals | • Evaluate short- and long-term metabolic changes associated with health-span in response to dietary methionine or methionine-cysteine restriction |
| NCT03629392 | Completed | University of Oslo, Norway | Methionine and Cysteine Restriction in Humans | Overweight Individuals | • Effect of methionine and cysteine restriction on energy and macronutrient metabolism in overweight humans |
| NCT04701346 | Recruiting | University of Oslo, Norway | Sulfur Amino Acids, Energy Metabolism and Obesity (STAY) | Overweight and Obese Individuals | • Effect of dietary SAA restriction on body weight/composition and energy expenditure in humans |
| • Comparison of high SAA and low SAA diet |
Alternatives to dietary methionine restriction
L-Methionine-γ-lyase or methioninase, a bacterially derived enzyme that degrades sulfur-containing amino acids including methionine, depletes methionine in both media and serum and may be a more readily translatable approach to clinical methionine restriction [54,210,211]. Co-culture of human embryonic lung fibroblasts and acute lymphoblastic leukemia cells treated with methioninase plus homocysteine eliminated malignant cells but had little impact on fibroblasts. Serum methionine levels in mice were > 95% depleted following a single injection of 300 units methioninase and this decrease was maintained for 8 hours [52]. Methioninase also depleted serum homocysteine, showing it effectively reduces methionine cycle intermediates [212]. Additionally, methioninase acts synergistically with 5-FU and other cytotoxic drugs in preclinical models [52,210]. Methioninase did not significantly alter body weight following 21 days of treatment and showed minimal antigenicity in immune-competent mice following repeat exposures [52]. However, bacterial methioninase has a short half-life in macaque monkeys (approximately 2 hours) and resulted in anaphylactic reactions [213]. In humans, methioninase depleted serum methionine within 30 minutes and was maintained for up to 4 hours after infusion had stopped without outward signs of toxicity [214].
More recently, oral recombinant methioninase has been demonstrated to have antitumor activity in diverse murine models of cancer, including PDX models [215,216]. In an orthotopic model of TNBC, oral recombinant methioninase reduced recurrent tumor weight and the number of metastatic lung nodules (5.3 in control versus 1.3 in treated mice) [217]. Oral methioninase has also been showed to act synergistically with 5-FU, platinum agents and other cytotoxic drugs in preclinical models [218,219]. Additionally, three patients with advanced prostate cancer have been treated with oral methioninase resulting in reduction [220] or stabilization [221] of PSA levels.
To enhance immune tolerance and prolong catalytic activity, bacterial methioninase has also been encapsulated in human erythrocytes [222]. Human erythrocyte encapsulated methioninase maintained lower serum methionine levels with 34% depletion compared to control at 9 days after administration [223]. Human erythrocyte encapsulated methioninase inhibited the growth of human glioblastoma tumor xenografts by 85% compared to control at day 45 [223]. Additionally, human cystathionine-γ-lyase, which normally catalyzes the conversion of cystathionine to cysteine, has been engineered to degrade methionine instead of cystathionine by three amino acid substitutions and shown to robustly reduce methionine levels for 72 hours, exerting antitumor effects in murine models without overt toxicity [108,224]. This engineered human methioninase depleted methionine and SAM, enhanced ROS, induced autophagy and activated apoptosis in cultured cancer cells [108]. Collectively, these studies suggest methioninase, particularly humanized or encapsulated versions, may be a safe and effective way to reduce circulating and intracellular methionine levels without altering diet, thereby potentially accelerating clinical translation of methionine restriction.
Conclusions
Due to their intrinsic hyperproliferation and oxidative stress fueled by oncogenic alterations, cancer cells and cancer stem cells have a high demand for methionine for protein synthesis and SAM generation, which is used for RNA, DNA, and histone methylation, as well as glutathione and nucleotide synthesis. When this augmented demand for methionine is paired with limited availability of methionine, the ensuing oxidative stress and deficiency of nucleotide precursors exposes a metabolic vulnerability termed “methionine-dependence” that is a hallmark of transformed cells. Although the underlying molecular mechanisms of this metabolic vulnerability are incompletely understood, numerous groups have explored its therapeutic potential in diverse preclinical models of cancer and small clinical studies in patients with advanced malignancies. Rationally designed combination therapies with synergistic cytotoxic agents such as antifolates or exposed metabolic liabilities (e.g., TRAIL or thioredoxin pathways) are likely to be needed to enhance the clinical activity of methionine restriction. Moreover, the tolerability of dietary methionine restriction particularly for prolonged therapy as well as other barriers to patient recruitment have hindered clinical investigation to date. The use of methionine-degrading enzymes, especially humanized or encapsulated versions, or the use of methionine oxidation of protein-rich foods to deplete methionine levels may partly address this issue in future studies. Finally, the elucidation of biomarkers of methionine-dependent tumors such as key epigenetic signatures as well a more complete molecular understanding of the cellular response to methionine restriction in tumor cells may point to improved strategies to target this unique cancer metabolic vulnerability.
Acknowledgements
This work was supported in part by grants from the V Foundation for Cancer Research (VLC), Wisconsin Partnership Program (VLC) and Breast Cancer Research Foundation (VLC).
Disclosure of conflict of interest
None.
References
- 1.Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev. 2012;38:726–736. doi: 10.1016/j.ctrv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Sanderson SM, Gao X, Dai Z, Locasale JW. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat Rev Cancer. 2019;19:625–637. doi: 10.1038/s41568-019-0187-8. [DOI] [PubMed] [Google Scholar]
- 3.Zuhra K, Augsburger F, Majtan T, Szabo C. Cystathionine-beta-synthase: molecular regulation and pharmacological inhibition. Biomolecules. 2020;10:697. doi: 10.3390/biom10050697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosnan JT, da Silva RP, Brosnan ME. The metabolic burden of creatine synthesis. Amino Acids. 2011;40:1325–1331. doi: 10.1007/s00726-011-0853-y. [DOI] [PubMed] [Google Scholar]
- 5.Mato JM, Corrales FJ, Lu SC, Avila MA. S-adenosylmethionine a control switch that regulates liver function. FASEB J. 2002;16:15–26. doi: 10.1096/fj.01-0401rev. [DOI] [PubMed] [Google Scholar]
- 6.Nijhout HF, Reed MC, Anderson DF, Mattingly JC, James SJ, Ulrich CM. Long-range allosteric interactions between the folate and methionine cycles stabilize DNA methylation reaction rate. Epigenetics. 2006;1:81–87. doi: 10.4161/epi.1.2.2677. [DOI] [PubMed] [Google Scholar]
- 7.Stipanuk MH, Ueki I. Dealing with methionine/homocysteine sulfur: cysteine metabolism to taurine and inorganic sulfur. J Inherit Metab Dis. 2011;34:17–32. doi: 10.1007/s10545-009-9006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Chantar ML, Latasa MU, Varela-Rey M, Lu SC, Garcia-Trevijano ER, Mato JM, Avila MA. L-methionine availability regulates expression of the methionine adenosyltransferase 2A gene in human hepatocarcinoma cells: role of S-adenosylmethionine. J Biol Chem. 2003;278:19885–19890. doi: 10.1074/jbc.M211554200. [DOI] [PubMed] [Google Scholar]
- 10.Strekalova E, Malin D, Weisenhorn EMM, Russell JD, Hoelper D, Jain A, Coon JJ, Lewis PW, Cryns VL. S-adenosylmethionine biosynthesis is a targetable metabolic vulnerability of cancer stem cells. Breast Cancer Res Treat. 2019;175:39–50. doi: 10.1007/s10549-019-05146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 12.Mecham JO, Rowitch D, Wallace CD, Stern PH, Hoffman RM. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem Biophys Res Commun. 1983;117:429–434. doi: 10.1016/0006-291x(83)91218-4. [DOI] [PubMed] [Google Scholar]
- 13.Kreis W, Goodenow M. Methionine requirement and replacement by homocysteine in tissue cultures of selected rodent and human malignant and normal cells. Cancer Res. 1978;38:2259–2262. [PubMed] [Google Scholar]
- 14.Halpern BC, Clark BR, Hardy DN, Halpern RM, Smith RA. The effect of replacement of methionine by homocystine on survival of malignant and normal adult mammalian cells in culture. Proc Natl Acad Sci U S A. 1974;71:1133–1136. doi: 10.1073/pnas.71.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugimura T, Birnbaum SM, Winitz M, Greenstein JP. Quantitative nutritional studies with water-soluble, chemically defined diets. VII. Nitrogen balance in normal and tumor-bearing rats following forced feeding. Arch Biochem Biophys. 1959;81:439–447. doi: 10.1016/0003-9861(59)90224-3. [DOI] [PubMed] [Google Scholar]
- 16.Stern PH, Hoffman RM. Enhanced in vitro selective toxicity of chemotherapeutic agents for human cancer cells based on a metabolic defect. J Natl Cancer Inst. 1986;76:629–639. doi: 10.1093/jnci/76.4.629. [DOI] [PubMed] [Google Scholar]
- 17.Lu S, Epner DE. Molecular mechanisms of cell cycle block by methionine restriction in human prostate cancer cells. Nutr Cancer. 2000;38:123–130. doi: 10.1207/S15327914NC381_17. [DOI] [PubMed] [Google Scholar]
- 18.Guo HY, Herrera H, Groce A, Hoffman RM. Expression of the biochemical defect of methionine dependence in fresh patient tumors in primary histoculture. Cancer Res. 1993;53:2479–2483. [PubMed] [Google Scholar]
- 19.Guo H, Lishko VK, Herrera H, Groce A, Kubota T, Hoffman RM. Therapeutic tumor-specific cell cycle block induced by methionine starvation in vivo. Cancer Res. 1993;53:5676–5679. [PubMed] [Google Scholar]
- 20.Jeon H, Kim JH, Lee E, Jang YJ, Son JE, Kwon JY, Lim TG, Kim S, Park JH, Kim JE, Lee KW. Methionine deprivation suppresses triple-negative breast cancer metastasis in vitro and in vivo. Oncotarget. 2016;7:67223–67234. doi: 10.18632/oncotarget.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graziosi L, Mencarelli A, Renga B, D’Amore C, Bruno A, Santorelli C, Cavazzoni E, Cantarella F, Rosati E, Donini A, Fiorucci S. Epigenetic modulation by methionine deficiency attenuates the potential for gastric cancer cell dissemination. J Gastrointest Surg. 2013;17:39–49. doi: 10.1007/s11605-012-1996-1. discussion p. 49. [DOI] [PubMed] [Google Scholar]
- 22.Parkhitko AA, Jouandin P, Mohr SE, Perrimon N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell. 2019;18:e13034. doi: 10.1111/acel.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman RM, Erbe RW. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc Natl Acad Sci U S A. 1976;73:1523–1527. doi: 10.1073/pnas.73.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lien EC, Vander Heiden MG. A framework for examining how diet impacts tumour metabolism. Nat Rev Cancer. 2019;19:651–661. doi: 10.1038/s41568-019-0198-5. [DOI] [PubMed] [Google Scholar]
- 25.Kano Y, Sakamoto S, Kasahara T, Kusumoto K, Hida K, Suda K, Ozawa K, Miura Y, Takaku F. Methionine dependency of cell growth in normal and malignant hematopoietic cells. Cancer Res. 1982;42:3090–3092. [PubMed] [Google Scholar]
- 26.Crott J, Thomas P, Fenech M. Normal human lymphocytes exhibit a wide range of methionine-dependency which is related to altered cell division but not micronucleus frequency. Mutagenesis. 2001;16:317–322. doi: 10.1093/mutage/16.4.317. [DOI] [PubMed] [Google Scholar]
- 27.Beetstra S, Suthers G, Dhillon V, Salisbury C, Turner J, Altree M, McKinnon R, Fenech M. Methionine-dependence phenotype in the de novo pathway in BRCA1 and BRCA2 mutation carriers with and without breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2565–2571. doi: 10.1158/1055-9965.EPI-08-0140. [DOI] [PubMed] [Google Scholar]
- 28.Gao X, Sanderson SM, Dai Z, Reid MA, Cooper DE, Lu M, Richie JP Jr, Ciccarella A, Calcagnotto A, Mikhael PG, Mentch SJ, Liu J, Ables G, Kirsch DG, Hsu DS, Nichenametla SN, Locasale JW. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature. 2019;572:397–401. doi: 10.1038/s41586-019-1437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, Tan Y, Kubota T, Moossa AR, Hoffman RM. Methionine depletion modulates the antitumor and antimetastatic efficacy of ethionine. Anticancer Res. 1996;16:2719–2723. [PubMed] [Google Scholar]
- 30.Miousse IR, Tobacyk J, Quick CM, Jamshidi-Parsian A, Skinner CM, Kore R, Melnyk SB, Kutanzi KR, Xia F, Griffin RJ, Koturbash I. Modulation of dietary methionine intake elicits potent, yet distinct, anticancer effects on primary versus metastatic tumors. Carcinogenesis. 2018;39:1117–1126. doi: 10.1093/carcin/bgy085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman RM, Yano S. Tumor-specific S/G 2-phase cell cycle arrest of cancer cells by methionine restriction. Methionine Dependence of Cancer and Aging: Springer; 2019. pp. 49–60. [DOI] [PubMed] [Google Scholar]
- 32.Fu YM, Zhang H, Ding M, Li YQ, Fu X, Yu ZX, Meadows GG. Selective amino acid restriction targets mitochondria to induce apoptosis of androgen-independent prostate cancer cells. J Cell Physiol. 2006;209:522–534. doi: 10.1002/jcp.20766. [DOI] [PubMed] [Google Scholar]
- 33.Hens JR, Sinha I, Perodin F, Cooper T, Sinha R, Plummer J, Perrone CE, Orentreich D. Methionine-restricted diet inhibits growth of MCF10AT1-derived mammary tumors by increasing cell cycle inhibitors in athymic nude mice. BMC Cancer. 2016;16:349. doi: 10.1186/s12885-016-2367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, Nichenametla SN, Locasale JW. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 2015;22:861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stone KP, Ghosh S, Kovalik JP, Orgeron M, Wanders D, Sims LC, Gettys TW. The acute transcriptional responses to dietary methionine restriction are triggered by inhibition of ternary complex formation and linked to Erk1/2, mTOR, and ATF4. Sci Rep. 2021;11:3765. doi: 10.1038/s41598-021-83380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallis KF, Melnyk SB, Miousse IR. Sex-specific effects of dietary methionine restriction on the intestinal microbiome. Nutrients. 2020;12:781. doi: 10.3390/nu12030781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan CL, Kaiser SE, Bolanos B, Nowlin D, Grantner R, Karlicek-Bryant S, Feng JL, Jenkinson S, Freeman-Cook K, Dann SG, Wang X, Wells PA, Fantin VR, Stewart AE, Grant SK. Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A. Nat Chem Biol. 2017;13:785–792. doi: 10.1038/nchembio.2384. [DOI] [PubMed] [Google Scholar]
- 38.Cellarier E, Durando X, Vasson MP, Farges MC, Demiden A, Maurizis JC, Madelmont JC, Chollet P. Methionine dependency and cancer treatment. Cancer Treat Rev. 2003;29:489–499. doi: 10.1016/s0305-7372(03)00118-x. [DOI] [PubMed] [Google Scholar]
- 39.Salisbury TB, Arthur S. The regulation and function of the L-type amino acid transporter 1 (LAT1) in cancer. Int J Mol Sci. 2018;19:2373. doi: 10.3390/ijms19082373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hafliger P, Charles RP. The L-type amino acid transporter LAT1-an emerging target in cancer. Int J Mol Sci. 2019;20:2428. doi: 10.3390/ijms20102428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, Nwosu ZC, Zhang L, Czerwonka A, Pawlowska A, Xia H, Li J, Liao P, Yu J, Vatan L, Szeliga W, Wei S, Grove S, Liu JR, McLean K, Cieslik M, Chinnaiyan AM, Zgodzinski W, Wallner G, Wertel I, Okla K, Kryczek I, Lyssiotis CA, Zou W. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature. 2020;585:277–282. doi: 10.1038/s41586-020-2682-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huovinen R, Leskinen-Kallio S, Nagren K, Lehikoinen P, Ruotsalainen U, Teras M. Carbon-11-methionine and PET in evaluation of treatment response of breast cancer. Br J Cancer. 1993;67:787–791. doi: 10.1038/bjc.1993.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Momose T, Nariai T, Kawabe T, Inaji M, Tanaka Y, Watanabe S, Maehara T, O’da K, Ishii K, Ishiwata K, Yamamoto M. Clinical benefit of 11C methionine PET imaging as a planning modality for radiosurgery of previously irradiated recurrent brain metastases. Clin Nucl Med. 2014;39:939–43. doi: 10.1097/RLU.0000000000000561. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Yip LY, Lee JHJ, Wu Z, Chew HY, Chong PKW, Teo CC, Ang HY, Peh KLE, Yuan J, Ma S, Choo LSK, Basri N, Jiang X, Yu Q, Hillmer AM, Lim WT, Lim TKH, Takano A, Tan EH, Tan DSW, Ho YS, Lim B, Tam WL. Methionine is a metabolic dependency of tumor-initiating cells. Nat Med. 2019;25:825–837. doi: 10.1038/s41591-019-0423-5. [DOI] [PubMed] [Google Scholar]
- 45.Tang X, Keenan MM, Wu J, Lin CA, Dubois L, Thompson JW, Freedland SJ, Murphy SK, Chi JT. Comprehensive profiling of amino acid response uncovers unique methionine-deprived response dependent on intact creatine biosynthesis. PLoS Genet. 2015;11:e1005158. doi: 10.1371/journal.pgen.1005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffman RM. The wayward methyl group and the cascade to cancer. Cell Cycle. 2017;16:825–829. doi: 10.1080/15384101.2017.1304330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman RM. Is the Hoffman effect for methionine overuse analogous to the warburg effect for glucose overuse in cancer? Methods Mol Biol. 2019;1866:273–278. doi: 10.1007/978-1-4939-8796-2_21. [DOI] [PubMed] [Google Scholar]
- 48.Stern PH, Wallace SD, Hoffman RM. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J Cell Physiol. 1984;119:29–34. doi: 10.1002/jcp.1041190106. [DOI] [PubMed] [Google Scholar]
- 49.Judde JG, Ellis M, Frost P. Biochemical analysis of the role of transmethylation in the methionine dependence of tumor cells. Cancer Res. 1989;49:4859–4865. [PubMed] [Google Scholar]
- 50.Coalson DW, Mecham JO, Stern PH, Hoffman RM. Reduced availability of endogenously synthesized methionine for S-adenosylmethionine formation in methionine-dependent cancer cells. Proc Natl Acad Sci U S A. 1982;79:4248–4251. doi: 10.1073/pnas.79.14.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bera S, Wallimann T, Ray S, Ray M. Enzymes of creatine biosynthesis, arginine and methionine metabolism in normal and malignant cells. FEBS J. 2008;275:5899–5909. doi: 10.1111/j.1742-4658.2008.06718.x. [DOI] [PubMed] [Google Scholar]
- 52.Yoshioka T, Wada T, Uchida N, Maki H, Yoshida H, Ide N, Kasai H, Hojo K, Shono K, Maekawa R, Yagi S, Hoffman RM, Sugita K. Anticancer efficacy in vivo and in vitro, synergy with 5-fluorouracil, and safety of recombinant methioninase. Cancer Res. 1998;58:2583–2587. [PubMed] [Google Scholar]
- 53.Esteve PO, Chin HG, Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc Natl Acad Sci U S A. 2005;102:1000–1005. doi: 10.1073/pnas.0407729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaturvedi S, Hoffman RM, Bertino JR. Exploiting methionine restriction for cancer treatment. Biochem Pharmacol. 2018;154:170–173. doi: 10.1016/j.bcp.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 55.Stern PH, Hoffman RM. Elevated overall rates of transmethylation in cell lines from diverse human tumors. In Vitro. 1984;20:663–670. doi: 10.1007/BF02619617. [DOI] [PubMed] [Google Scholar]
- 56.Borrego SL, Fahrmann J, Datta R, Stringari C, Grapov D, Zeller M, Chen Y, Wang P, Baldi P, Gratton E, Fiehn O, Kaiser P. Metabolic changes associated with methionine stress sensitivity in MDA-MB-468 breast cancer cells. Cancer Metab. 2016;4:9. doi: 10.1186/s40170-016-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffman RM, Jacobsen SJ, Erbe RW. Reversion to methionine in dependence in simian virus 40-transformed human and malignant rat fibroblasts is associated with altered ploidy and altered properties of transformation. Proc Natl Acad Sci U S A. 1979;76:1313–1317. doi: 10.1073/pnas.76.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Booher K, Lin DW, Borrego SL, Kaiser P. Downregulation of Cdc6 and pre-replication complexes in response to methionine stress in breast cancer cells. Cell Cycle. 2012;11:4414–4423. doi: 10.4161/cc.22767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, Camargo AA, Stevenson BJ, Ecker JR, Bafna V, Strausberg RL, Simpson AJ, Ren B. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liteplo RG, Kerbel RS. Reduced levels of DNA 5-methylcytosine in metastatic variants of the human melanoma cell line MeWo. Cancer Res. 1987;47:2264–2267. [PubMed] [Google Scholar]
- 61.Lien EC, Ghisolfi L, Geck RC, Asara JM, Toker A. Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci Signal. 2017;10:eaao6604. doi: 10.1126/scisignal.aao6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu Y, Albuquerque CP, Braas D, Zhang W, Villa GR, Bi J, Ikegami S, Masui K, Gini B, Yang H, Gahman TC, Shiau AK, Cloughesy TF, Christofk HR, Zhou H, Guan KL, Mischel PS. mTORC2 regulates amino acid metabolism in cancer by phosphorylation of the cystine-glutamate antiporter xCT. Mol Cell. 2017;67:128–138. e127. doi: 10.1016/j.molcel.2017.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poirson-Bichat F, Bras Goncalves RA, Miccoli L, Dutrillaux B, Poupon MF. Methionine depletion enhances the antitumoral efficacy of cytotoxic agents in drug-resistant human tumor xenografts. Clin Cancer Res. 2000;6:643–653. [PubMed] [Google Scholar]
- 64.Tsai CW, Lin AH, Wang TS, Liu KL, Chen HW, Lii CK. Methionine restriction up-regulates the expression of the pi class of glutathione S-transferase partially via the extracellular signal-regulated kinase-activator protein-1 signaling pathway initiated by glutathione depletion. Mol Nutr Food Res. 2010;54:841–850. doi: 10.1002/mnfr.200900083. [DOI] [PubMed] [Google Scholar]
- 65.Malin D, Lee Y, Chepikova O, Strekalova E, Carlson A, Cryns VL. Methionine restriction exposes a targetable redox vulnerability of triple-negative breast cancer cells by inducing thioredoxin reductase. Breast Cancer Res Treat. 2021;190:373–387. doi: 10.1007/s10549-021-06398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hensen SM, Heldens L, van Enckevort CM, van Genesen ST, Pruijn GJ, Lubsen NH. Activation of the antioxidant response in methionine deprived human cells results in an HSF1-independent increase in HSPA1A mRNA levels. Biochimie. 2013;95:1245–1251. doi: 10.1016/j.biochi.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 67.Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ito T, Ohta M, Kera Y, Noda T, Igarashi K. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell. 2011;41:554–566. doi: 10.1016/j.molcel.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 68.Lin AH, Chen HW, Liu CT, Tsai CW, Lii CK. Activation of Nrf2 is required for up-regulation of the pi class of glutathione S-transferase in rat primary hepatocytes with L-methionine starvation. J Agric Food Chem. 2012;60:6537–6545. doi: 10.1021/jf301567m. [DOI] [PubMed] [Google Scholar]
- 69.Jonsson WO, Margolies NS, Anthony TG. Dietary sulfur amino acid restriction and the integrated stress response: mechanistic insights. Nutrients. 2019;11:1349. doi: 10.3390/nu11061349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCarty MF, Barroso-Aranda J, Contreras F. The low-methionine content of vegan diets may make methionine restriction feasible as a life extension strategy. Med Hypotheses. 2009;72:125–128. doi: 10.1016/j.mehy.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 71.Najim N, Podmore ID, McGown A, Estlin EJ. Methionine restriction reduces the chemosensitivity of central nervous system tumour cell lines. Anticancer Res. 2009;29:3103–3108. [PubMed] [Google Scholar]
- 72.Maddineni S, Nichenametla S, Sinha R, Wilson RP, Richie JP Jr. Methionine restriction affects oxidative stress and glutathione-related redox pathways in the rat. Exp Biol Med (Maywood) 2013;238:392–399. doi: 10.1177/1535370213477988. [DOI] [PubMed] [Google Scholar]
- 73.Mattocks DA, Mentch SJ, Shneyder J, Ables GP, Sun D, Richie JP Jr, Locasale JW, Nichenametla SN. Short term methionine restriction increases hepatic global DNA methylation in adult but not young male C57BL/6J mice. Exp Gerontol. 2017;88:1–8. doi: 10.1016/j.exger.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Richie JP Jr, Leutzinger Y, Parthasarathy S, Malloy V, Orentreich N, Zimmerman JA. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994;8:1302–1307. doi: 10.1096/fasebj.8.15.8001743. [DOI] [PubMed] [Google Scholar]
- 75.Caro P, Gomez J, Sanchez I, Naudi A, Ayala V, Lopez-Torres M, Pamplona R, Barja G. Forty percent methionine restriction decreases mitochondrial oxygen radical production and leak at complex I during forward electron flow and lowers oxidative damage to proteins and mitochondrial DNA in rat kidney and brain mitochondria. Rejuvenation Res. 2009;12:421–434. doi: 10.1089/rej.2009.0902. [DOI] [PubMed] [Google Scholar]
- 76.Mosharov E, Cranford MR, Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39:13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- 77.Poirson-Bichat F, Bras Goncalves RA, Dutrillaux B, Poupon MF. Growth of methionine dependent human prostate cancer (PC-3) is inhibited by ethionine combined with methionine starvation. Br J Cancer. 1997;75:1605–12. doi: 10.1038/bjc.1997.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deguchi T, Barchas J. Inhibition of transmethylations of biogenic amines by S-adenosylhomocysteine. Enhancement of transmethylation by adenosylhomocysteinase. J Biol Chem. 1971;246:3175–3181. [PubMed] [Google Scholar]
- 79.Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, Santos-Guzman J, Swendseid ME, Cogger EA, James SJ. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr. 2001;131:2811–2818. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- 80.Tisdale MJ. Changes in tRNA methyltransferase activity and cellular S-adenosylmethionine content following methionine deprivation. Biochim Biophys Acta. 1980;609:296–305. doi: 10.1016/0005-2787(80)90241-5. [DOI] [PubMed] [Google Scholar]
- 81.Tang B, Li YN, Kruger WD. Defects in methylthioadenosine phosphorylase are associated with but not responsible for methionine-dependent tumor cell growth. Cancer Res. 2000;60:5543–5547. [PubMed] [Google Scholar]
- 82.Mavrakis KJ, McDonald ER 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, Ruddy DA, Venkatesan K, Yu J, McAllister G, Stump M, deBeaumont R, Ho S, Yue Y, Liu Y, Yan-Neale Y, Yang G, Lin F, Yin H, Gao H, Kipp DR, Zhao S, McNamara JT, Sprague ER, Zheng B, Lin Y, Cho YS, Gu J, Crawford K, Ciccone D, Vitari AC, Lai A, Capka V, Hurov K, Porter JA, Tallarico J, Mickanin C, Lees E, Pagliarini R, Keen N, Schmelzle T, Hofmann F, Stegmeier F, Sellers WR. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 83.Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, Konteatis Z, Murtie J, Blake ML, Travins J, Dorsch M, Biller SA, Marks KM. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 2016;15:574–587. doi: 10.1016/j.celrep.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 84.Zarou MM, Vazquez A, Vignir Helgason G. Folate metabolism: a re-emerging therapeutic target in haematological cancers. Leukemia. 2021;35:1539–1551. doi: 10.1038/s41375-021-01189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, Rabinowitz JD. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. 2019;1:404–415. [PMC free article] [PubMed] [Google Scholar]
- 87.Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 92.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, Cantley LC. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AM, Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 94.Rajanala SH, Ringquist R, Cryns VL. Methionine restriction activates the integrated stress response in triple-negative breast cancer cells by a GCN2- and PERK-independent mechanism. Am J Cancer Res. 2019;9:1766–1775. [PMC free article] [PubMed] [Google Scholar]
- 95.Chen H, Song R, Wang G, Ding Z, Yang C, Zhang J, Zeng Z, Rubio V, Wang L, Zu N, Weiskoff AM, Minze LJ, Jeyabal PV, Mansour OC, Bai L, Merrick WC, Zheng S, Shi ZZ. OLA1 regulates protein synthesis and integrated stress response by inhibiting eIF2 ternary complex formation. Sci Rep. 2015;5:13241. doi: 10.1038/srep13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mazor KM, Stipanuk MH. GCN2- and eIF2alpha-phosphorylation-independent, but ATF4-dependent, induction of CARE-containing genes in methionine-deficient cells. Amino Acids. 2016;48:2831–2842. doi: 10.1007/s00726-016-2318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wanders D, Stone KP, Forney LA, Cortez CC, Dille KN, Simon J, Xu M, Hotard EC, Nikonorova IA, Pettit AP, Anthony TG, Gettys TW. Role of GCN2-independent signaling through a noncanonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes. 2016;65:1499–1510. doi: 10.2337/db15-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pettit AP, Jonsson WO, Bargoud AR, Mirek ET, Peelor FF 3rd, Wang Y, Gettys TW, Kimball SR, Miller BF, Hamilton KL, Wek RC, Anthony TG. Dietary methionine restriction regulates liver protein synthesis and gene expression independently of eukaryotic initiation factor 2 phosphorylation in mice. J Nutr. 2017;147:1031–1040. doi: 10.3945/jn.116.246710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ehren JL, Maher P. Concurrent regulation of the transcription factors Nrf2 and ATF4 mediates the enhancement of glutathione levels by the flavonoid fisetin. Biochem Pharmacol. 2013;85:1816–1826. doi: 10.1016/j.bcp.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 100.Huang HC, Nguyen T, Pickett CB. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc Natl Acad Sci U S A. 2000;97:12475–12480. doi: 10.1073/pnas.220418997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ye J, Palm W, Peng M, King B, Lindsten T, Li MO, Koumenis C, Thompson CB. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 2015;29:2331–2336. doi: 10.1101/gad.269324.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K, Cecconi F, Choi AMK, Choi ME, Chu CT, Codogno P, Colombo MI, Cuervo AM, Deretic V, Dikic I, Elazar Z, Eskelinen EL, Fimia GM, Gewirtz DA, Green DR, Hansen M, Jaattela M, Johansen T, Juhasz G, Karantza V, Kraft C, Kroemer G, Ktistakis NT, Kumar S, Lopez-Otin C, Macleod KF, Madeo F, Martinez J, Melendez A, Mizushima N, Munz C, Penninger JM, Perera RM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Sadoshima J, Santambrogio L, Scorrano L, Simon HU, Simon AK, Simonsen A, Stolz A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Galluzzi L, Pietrocola F. Autophagy in major human diseases. EMBO J. 2021;40:e108863. doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perrone CE, Malloy VL, Orentreich DS, Orentreich N. Metabolic adaptations to methionine restriction that benefit health and lifespan in rodents. Exp Gerontol. 2013;48:654–660. doi: 10.1016/j.exger.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 104.Ables GP, Hens JR, Nichenametla SN. Methionine restriction beyond life-span extension. Ann N Y Acad Sci. 2016;1363:68–79. doi: 10.1111/nyas.13014. [DOI] [PubMed] [Google Scholar]
- 105.Ruckenstuhl C, Netzberger C, Entfellner I, Carmona-Gutierrez D, Kickenweiz T, Stekovic S, Gleixner C, Schmid C, Klug L, Sorgo AG, Eisenberg T, Buttner S, Marino G, Koziel R, Jansen-Durr P, Frohlich KU, Kroemer G, Madeo F. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 2014;10:e1004347. doi: 10.1371/journal.pgen.1004347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zou K, Ouyang Q, Li H, Zheng J. A global characterization of the translational and transcriptional programs induced by methionine restriction through ribosome profiling and RNA-seq. BMC Genomics. 2017;18:189. doi: 10.1186/s12864-017-3483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Plummer JD, Johnson JE. Extension of cellular lifespan by methionine restriction involves alterations in central carbon metabolism and is mitophagy-dependent. Front Cell Dev Biol. 2019;7:301. doi: 10.3389/fcell.2019.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu WC, Saha A, Yan W, Garrison K, Lamb C, Pandey R, Irani S, Lodi A, Lu X, Tiziani S, Zhang YJ, Georgiou G, DiGiovanni J, Stone E. Enzyme-mediated depletion of serum l-Met abrogates prostate cancer growth via multiple mechanisms without evidence of systemic toxicity. Proc Natl Acad Sci U S A. 2020;117:13000–13011. doi: 10.1073/pnas.1917362117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu H, Zhang W, Wang K, Wang X, Yin F, Li C, Wang C, Zhao B, Zhong C, Zhang J, Peng F, Bi Y, Shen C, Hou X, Zhang D, Liu Y, Ai J, Zhao S. Methionine and cystine double deprivation stress suppresses glioma proliferation via inducing ROS/autophagy. Toxicol Lett. 2015;232:349–355. doi: 10.1016/j.toxlet.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 110.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 111.Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502:489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- 112.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deroo LA, Bolick SC, Xu Z, Umbach DM, Shore D, Weinberg CR, Sandler DP, Taylor JA. Global DNA methylation and one-carbon metabolism gene polymorphisms and the risk of breast cancer in the Sister Study. Carcinogenesis. 2014;35:333–338. doi: 10.1093/carcin/bgt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, Voravud N, Sriuranpong V, Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 116.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, Polyak K. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 117.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23:853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu L, Wylie RC, Andrews LG, Tollefsbol TO. Aging, cancer and nutrition: the DNA methylation connection. Mech Ageing Dev. 2003;124:989–998. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 119.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 120.Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, Tajara EH, Issa JP. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim JH, Dhanasekaran SM, Prensner JR, Cao X, Robinson D, Kalyana-Sundaram S, Huang C, Shankar S, Jing X, Iyer M, Hu M, Sam L, Grasso C, Maher CA, Palanisamy N, Mehra R, Kominsky HD, Siddiqui J, Yu J, Qin ZS, Chinnaiyan AM. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res. 2011;21:1028–1041. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, Vaurs-Barriere C, Bignon YJ, Ramus S, Benitez J, Caldes T, Akiyama Y, Yuasa Y, Launonen V, Canal MJ, Rodriguez R, Capella G, Peinado MA, Borg A, Aaltonen LA, Ponder BA, Baylin SB, Herman JG. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 123.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S, Stauffer KM, Makohon-Moore A, Zhong Y, Wu H, Wellen KE, Locasale JW, Iacobuzio-Donahue CA, Feinberg AP. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49:367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, Irizarry RA, Feinberg AP. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Etchegaray JP, Mostoslavsky R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell. 2016;62:695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sproul D, Kitchen RR, Nestor CE, Dixon JM, Sims AH, Harrison DJ, Ramsahoye BH, Meehan RR. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol. 2012;13:R84. doi: 10.1186/gb-2012-13-10-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- 129.Kaelin WG Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kera Y, Katoh Y, Ohta M, Matsumoto M, Takano-Yamamoto T, Igarashi K. Methionine adenosyltransferase II-dependent histone H3K9 methylation at the COX-2 gene locus. J Biol Chem. 2013;288:13592–13601. doi: 10.1074/jbc.M112.429738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Haws SA, Yu D, Ye C, Wille CK, Nguyen LC, Krautkramer KA, Tomasiewicz JL, Yang SE, Miller BR, Liu WH, Igarashi K, Sridharan R, Tu BP, Cryns VL, Lamming DW, Denu JM. Methyl-metabolite depletion elicits adaptive responses to support heterochromatin stability and epigenetic persistence. Mol Cell. 2020;78:210–223. e218. doi: 10.1016/j.molcel.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Loenen W. S-adenosylmethionine: jack of all trades and master of everything? Biochem Soc Trans. 2006;34:330–333. doi: 10.1042/BST20060330. [DOI] [PubMed] [Google Scholar]
- 136.Roundtree IA, He C. RNA epigenetics--chemical messages for posttranscriptional gene regulation. Curr Opin Chem Biol. 2016;30:46–51. doi: 10.1016/j.cbpa.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell. 2013;154:416–429. doi: 10.1016/j.cell.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cowling VH. Myc up-regulates formation of the mRNA methyl cap. Biochem Soc Trans. 2010;38:1598–1601. doi: 10.1042/BST0381598. [DOI] [PubMed] [Google Scholar]
- 139.Shima H, Matsumoto M, Ishigami Y, Ebina M, Muto A, Sato Y, Kumagai S, Ochiai K, Suzuki T, Igarashi K. S-adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 2017;21:3354–3363. doi: 10.1016/j.celrep.2017.11.092. [DOI] [PubMed] [Google Scholar]
- 140.Gomez A, Gomez J, Lopez Torres M, Naudi A, Mota-Martorell N, Pamplona R, Barja G. Cysteine dietary supplementation reverses the decrease in mitochondrial ROS production at complex I induced by methionine restriction. J Bioenerg Biomembr. 2015;47:199–208. doi: 10.1007/s10863-015-9608-x. [DOI] [PubMed] [Google Scholar]
- 141.Altundag O, Canpinar H, Celebi-Saltik B. Methionine affects the expression of pluripotency genes and protein levels associated with methionine metabolism in adult, fetal, and cancer stem cells. J Cell Biochem. 2022;123:406–416. doi: 10.1002/jcb.30180. [DOI] [PubMed] [Google Scholar]
- 142.Luo M, Clouthier SG, Deol Y, Liu S, Nagrath S, Azizi E, Wicha MS. Breast cancer stem cells: current advances and clinical implications. Methods Mol Biol. 2015;1293:1–49. doi: 10.1007/978-1-4939-2519-3_1. [DOI] [PubMed] [Google Scholar]
- 143.Luo M, Wicha MS. Targeting cancer stem cell redox metabolism to enhance therapy responses. Semin Radiat Oncol. 2019;29:42–54. doi: 10.1016/j.semradonc.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Luo M, Shang L, Brooks MD, Jiagge E, Zhu Y, Buschhaus JM, Conley S, Fath MA, Davis A, Gheordunescu E, Wang Y, Harouaka R, Lozier A, Triner D, McDermott S, Merajver SD, Luker GD, Spitz DR, Wicha MS. Targeting breast cancer stem cell state equilibrium through modulation of redox signaling. Cell Metab. 2018;28:69–86. e66. doi: 10.1016/j.cmet.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR, Brown JM, Weissman IL, Clarke MF. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jeselsohn R, Cornwell M, Pun M, Buchwalter G, Nguyen M, Bango C, Huang Y, Kuang Y, Paweletz C, Fu X, Nardone A, De Angelis C, Detre S, Dodson A, Mohammed H, Carroll JS, Bowden M, Rao P, Long HW, Li F, Dowsett M, Schiff R, Brown M. Embryonic transcription factor SOX9 drives breast cancer endocrine resistance. Proc Natl Acad Sci U S A. 2017;114:E4482–E4491. doi: 10.1073/pnas.1620993114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sufrin JR, Coulter A, Talalay P. Structural and conformational analogues of L-methionine as inhibitors of the enzymatic synthesis of S-adenosyl-L-methionine. IV. Further mono-, bi-and tricyclic amino acids. Mol Pharmacol. 1979;15:661–677. [PubMed] [Google Scholar]
- 149.Zhang W, Sviripa V, Chen X, Shi J, Yu T, Hamza A, Ward ND, Kril LM, Vander Kooi CW, Zhan CG, Evers BM, Watt DS, Liu C. Fluorinated N,N-dialkylaminostilbenes repress colon cancer by targeting methionine S-adenosyltransferase 2A. ACS Chem Biol. 2013;8:796–803. doi: 10.1021/cb3005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Strekalova E, Malin D, Good DM, Cryns VL. Methionine deprivation induces a targetable vulnerability in triple-negative breast cancer cells by enhancing TRAIL receptor-2 expression. Clin Cancer Res. 2015;21:2780–2791. doi: 10.1158/1078-0432.CCR-14-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yamamoto J, Miyake K, Han Q, Tan Y, Inubushi S, Sugisawa N, Higuchi T, Tashiro Y, Nishino H, Homma Y, Matsuyama R, Chawla SP, Bouvet M, Singh SR, Endo I, Hoffman RM. Oral recombinant methioninase increases TRAIL receptor-2 expression to regress pancreatic cancer in combination with agonist tigatuzumab in an orthotopic mouse model. Cancer Lett. 2020;492:174–184. doi: 10.1016/j.canlet.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 152.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835. e814. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Plummer R, Attard G, Pacey S, Li L, Razak A, Perrett R, Barrett M, Judson I, Kaye S, Fox NL, Halpern W, Corey A, Calvert H, de Bono J. Phase 1 and pharmacokinetic study of lexatumumab in patients with advanced cancers. Clin Cancer Res. 2007;13:6187–6194. doi: 10.1158/1078-0432.CCR-07-0950. [DOI] [PubMed] [Google Scholar]
- 154.Tolcher AW, Mita M, Meropol NJ, von Mehren M, Patnaik A, Padavic K, Hill M, Mays T, McCoy T, Fox NL, Halpern W, Corey A, Cohen RB. Phase I pharmacokinetic and biologic correlative study of mapatumumab, a fully human monoclonal antibody with agonist activity to tumor necrosis factor-related apoptosis-inducing ligand receptor-1. J. Clin. Oncol. 2007;25:1390–1395. doi: 10.1200/JCO.2006.08.8898. [DOI] [PubMed] [Google Scholar]
- 155.Wakelee HA, Patnaik A, Sikic BI, Mita M, Fox NL, Miceli R, Ullrich SJ, Fisher GA, Tolcher AW. Phase I and pharmacokinetic study of lexatumumab (HGS-ETR2) given every 2 weeks in patients with advanced solid tumors. Ann Oncol. 2010;21:376–381. doi: 10.1093/annonc/mdp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Merchant MS, Geller JI, Baird K, Chou AJ, Galli S, Charles A, Amaoko M, Rhee EH, Price A, Wexler LH, Meyers PA, Widemann BC, Tsokos M, Mackall CL. Phase I trial and pharmacokinetic study of lexatumumab in pediatric patients with solid tumors. J. Clin. Oncol. 2012;30:4141–4147. doi: 10.1200/JCO.2012.44.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Greco FA, Bonomi P, Crawford J, Kelly K, Oh Y, Halpern W, Lo L, Gallant G, Klein J. Phase 2 study of mapatumumab, a fully human agonistic monoclonal antibody which targets and activates the TRAIL receptor-1, in patients with advanced non-small cell lung cancer. Lung Cancer. 2008;61:82–90. doi: 10.1016/j.lungcan.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 158.Ross RB, Noll CI, Ross WC, Nadkarni MV, Morrison BH Jr, Bond HW. Cycloaliphatic amino acids in cancer chemotherapy. J Med Pharm Chem. 1961;3:1–23. doi: 10.1021/jm50014a001. [DOI] [PubMed] [Google Scholar]
- 159.Johnson RO. Preliminary phase II trials with 1-aminocyclopentane carboxylic acid (Nsc-1026) Cancer Chemother Rep. 1963;32:67–71. [PubMed] [Google Scholar]
- 160.Zhang J, Li X, Han X, Liu R, Fang J. Targeting the thioredoxin system for cancer therapy. Trends Pharmacol Sci. 2017;38:794–808. doi: 10.1016/j.tips.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 161.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 162.Fath MA, Ahmad IM, Smith CJ, Spence J, Spitz DR. Enhancement of carboplatin-mediated lung cancer cell killing by simultaneous disruption of glutathione and thioredoxin metabolism. Clin Cancer Res. 2011;17:6206–6217. doi: 10.1158/1078-0432.CCR-11-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA, Elia A, Berger T, Cescon DW, Adeoye A, Brustle A, Molyneux SD, Mason JM, Li WY, Yamamoto K, Wakeham A, Berman HK, Khokha R, Done SJ, Kavanagh TJ, Lam CW, Mak TW. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 164.Mandal PK, Schneider M, Kolle P, Kuhlencordt P, Forster H, Beck H, Bornkamm GW, Conrad M. Loss of thioredoxin reductase 1 renders tumors highly susceptible to pharmacologic glutathione deprivation. Cancer Res. 2010;70:9505–9514. doi: 10.1158/0008-5472.CAN-10-1509. [DOI] [PubMed] [Google Scholar]
- 165.Roder C, Thomson MJ. Auranofin: repurposing an old drug for a golden new age. Drugs R D. 2015;15:13–20. doi: 10.1007/s40268-015-0083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hoffman RM. Methionine dependence in cancer cells - a review. In Vitro. 1982;18:421–428. doi: 10.1007/BF02796468. [DOI] [PubMed] [Google Scholar]
- 167.Lin DW, Chung BP, Kaiser P. S-adenosylmethionine limitation induces p38 mitogen-activated protein kinase and triggers cell cycle arrest in G1. J Cell Sci. 2014;127:50–59. doi: 10.1242/jcs.127811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Higuchi T, Han Q, Miyake K, Oshiro H, Sugisawa N, Tan Y, Yamamoto N, Hayashi K, Kimura H, Miwa S, Igarashi K, Bouvet M, Singh SR, Tsuchiya H, Hoffman RM. Combination of oral recombinant methioninase and decitabine arrests a chemotherapy-resistant undifferentiated soft-tissue sarcoma patient-derived orthotopic xenograft mouse model. Biochem Biophys Res Commun. 2020;523:135–139. doi: 10.1016/j.bbrc.2019.12.024. [DOI] [PubMed] [Google Scholar]
- 169.Higuchi T, Sugisawa N, Yamamoto J, Oshiro H, Han Q, Yamamoto N, Hayashi K, Kimura H, Miwa S, Igarashi K, Tan Y, Kuchipudi S, Bouvet M, Singh SR, Tsuchiya H, Hoffman RM. The combination of oral-recombinant methioninase and azacitidine arrests a chemotherapy-resistant osteosarcoma patient-derived orthotopic xenograft mouse model. Cancer Chemother Pharmacol. 2020;85:285–291. doi: 10.1007/s00280-019-03986-0. [DOI] [PubMed] [Google Scholar]
- 170.Nagahama T, Goseki N, Endo M. Doxorubicin and vincristine with methionine depletion contributed to survival in the Yoshida sarcoma bearing rats. Anticancer Res. 1998;18:25–31. [PubMed] [Google Scholar]
- 171.Kawaguchi K, Miyake K, Han Q, Li S, Tan Y, Igarashi K, Lwin TM, Higuchi T, Kiyuna T, Miyake M, Oshiro H, Bouvet M, Unno M, Hoffman RM. Targeting altered cancer methionine metabolism with recombinant methioninase (rMETase) overcomes partial gemcitabine-resistance and regresses a patient-derived orthotopic xenograft (PDOX) nude mouse model of pancreatic cancer. Cell Cycle. 2018;17:868–873. doi: 10.1080/15384101.2018.1445907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lu S, Chen GL, Ren C, Kwabi-Addo B, Epner DE. Methionine restriction selectively targets thymidylate synthase in prostate cancer cells. Biochem Pharmacol. 2003;66:791–800. doi: 10.1016/s0006-2952(03)00406-4. [DOI] [PubMed] [Google Scholar]
- 173.Ham MS, Lee JK, Kim KC. S-adenosyl methionine specifically protects the anticancer effect of 5-FU via DNMTs expression in human A549 lung cancer cells. Mol Clin Oncol. 2013;1:373–378. doi: 10.3892/mco.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Visentin M, Zhao R, Goldman ID. The antifolates. Hematol Oncol Clin North Am. 2012;26:629–648. ix. doi: 10.1016/j.hoc.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang YC, Chiang EP. Low-dose methotrexate inhibits methionine S-adenosyltransferase in vitro and in vivo. Mol Med. 2012;18:423–432. doi: 10.2119/molmed.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Conklin KA. Chemotherapy-associated oxidative stress: impact on chemotherapeutic effectiveness. Integr Cancer Ther. 2004;3:294–300. doi: 10.1177/1534735404270335. [DOI] [PubMed] [Google Scholar]
- 178.Kery M, Papandreou I. Emerging strategies to target cancer metabolism and improve radiation therapy outcomes. Br J Radiol. 2020;93:20200067. doi: 10.1259/bjr.20200067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Esen H, Erdi F, Kaya B, Feyzioglu B, Keskin F, Demir LS. Tissue thioredoxin reductase-1 expression in astrocytomas of different grades. J Neurooncol. 2015;121:451–458. doi: 10.1007/s11060-014-1661-5. [DOI] [PubMed] [Google Scholar]
- 180.Cadenas C, Franckenstein D, Schmidt M, Gehrmann M, Hermes M, Geppert B, Schormann W, Maccoux LJ, Schug M, Schumann A, Wilhelm C, Freis E, Ickstadt K, Rahnenfuhrer J, Baumbach JI, Sickmann A, Hengstler JG. Role of thioredoxin reductase 1 and thioredoxin interacting protein in prognosis of breast cancer. Breast Cancer Res. 2010;12:R44. doi: 10.1186/bcr2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fu B, Meng W, Zeng X, Zhao H, Liu W, Zhang T. TXNRD1 is an unfavorable prognostic factor for patients with hepatocellular carcinoma. Biomed Res Int. 2017;2017:4698167. doi: 10.1155/2017/4698167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Epner DE, Morrow S, Wilcox M, Houghton JL. Nutrient intake and nutritional indexes in adults with metastatic cancer on a phase I clinical trial of dietary methionine restriction. Nutr Cancer. 2002;42:158–166. doi: 10.1207/S15327914NC422_2. [DOI] [PubMed] [Google Scholar]
- 183.Plaisance EP, Greenway FL, Boudreau A, Hill KL, Johnson WD, Krajcik RA, Perrone CE, Orentreich N, Cefalu WT, Gettys TW. Dietary methionine restriction increases fat oxidation in obese adults with metabolic syndrome. J Clin Endocrinol Metab. 2011;96:E836–840. doi: 10.1210/jc.2010-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Orgeron ML, Stone KP, Wanders D, Cortez CC, Van NT, Gettys TW. The impact of dietary methionine restriction on biomarkers of metabolic health. Prog Mol Biol Transl Sci. 2014;121:351–376. doi: 10.1016/B978-0-12-800101-1.00011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fang H, Stone KP, Forney LA, Sims LC, Gutierrez GC, Ghosh S, Gettys TW. Implementation of dietary methionine restriction using casein after selective, oxidative deletion of methionine. iScience. 2021;24:102470. doi: 10.1016/j.isci.2021.102470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Goseki N, Endo M. Thiol depletion and chemosensitization on nimustine hydrochloride by methionine-depleting total parenteral nutrition. Tohoku J Exp Med. 1990;161:227–239. doi: 10.1620/tjem.161.227. [DOI] [PubMed] [Google Scholar]
- 187.Durando X, Thivat E, Farges MC, Cellarier E, D’Incan M, Demidem A, Vasson MP, Barthomeuf C, Chollet P. Optimal methionine-free diet duration for nitrourea treatment: a phase I clinical trial. Nutr Cancer. 2008;60:23–30. doi: 10.1080/01635580701525877. [DOI] [PubMed] [Google Scholar]
- 188.Thivat E, Farges MC, Bacin F, D’Incan M, Mouret-Reynier MA, Cellarier E, Madelmont JC, Vasson MP, Chollet P, Durando X. Phase II trial of the association of a methionine-free diet with cystemustine therapy in melanoma and glioma. Anticancer Res. 2009;29:5235–5240. [PubMed] [Google Scholar]
- 189.Kokkinakis DM, von Wronski MA, Vuong TH, Brent TP, Schold SC Jr. Regulation of 06-methylguanine-DNA methyltransferase by methionine in human tumour cells. Br J Cancer. 1997;75:779–788. doi: 10.1038/bjc.1997.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kokkinakis DM, Hoffman RM, Frenkel EP, Wick JB, Han Q, Xu M, Tan Y, Schold SC. Synergy between methionine stress and chemotherapy in the treatment of brain tumor xenografts in athymic mice. Cancer Res. 2001;61:4017–4023. [PubMed] [Google Scholar]
- 191.Thivat E, Durando X, Demidem A, Farges MC, Rapp M, Cellarier E, Guenin S, D’Incan M, Vasson MP, Chollet P. A methionine-free diet associated with nitrosourea treatment down-regulates methylguanine-DNA methyl transferase activity in patients with metastatic cancer. Anticancer Res. 2007;27:2779–2783. [PubMed] [Google Scholar]
- 192.Hoffman RM, Stern PH. Dietary methionine restriction-based cancer chemotherapy in rodents. Methionine Dependence of Cancer and Aging: Springer; 2019. pp. 83–94. [DOI] [PubMed] [Google Scholar]
- 193.Kokkinakis DM, Liu X, Neuner RD. Modulation of cell cycle and gene expression in pancreatic tumor cell lines by methionine deprivation (methionine stress): implications to the therapy of pancreatic adenocarcinoma. Mol Cancer Ther. 2005;4:1338–1348. doi: 10.1158/1535-7163.MCT-05-0141. [DOI] [PubMed] [Google Scholar]
- 194.Durando X, Farges MC, Buc E, Abrial C, Petorin-Lesens C, Gillet B, Vasson MP, Pezet D, Chollet P, Thivat E. Dietary methionine restriction with FOLFOX regimen as first line therapy of metastatic colorectal cancer: a feasibility study. Oncology. 2010;78:205–209. doi: 10.1159/000313700. [DOI] [PubMed] [Google Scholar]
- 195.Goseki N, Endo M, Onodera T, Kosaki G. Anti-tumor effect of L-methionine-deprived total parenteral nutrition with 5-fluorouracil administration on Yoshida sarcoma-bearing rats. Ann Surg. 1991;214:83. doi: 10.1097/00000658-199107000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Goseki N, Yamazaki S, Shimojyu K, Kando F, Maruyama M, Endo M, Koike M, Takahashi H. Synergistic effect of methionine-depleting total parenteral nutrition with 5-fluorouracil on human gastric cancer: a randomized, prospective clinical trial. Jpn J Cancer Res. 1995;86:484–489. doi: 10.1111/j.1349-7006.1995.tb03082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Goseki N, Maruyama M, Nagai K, Kando F, Endo M, Shimoju K, Wada Y. Clinical evaluation of anticancer effect of methionine-depleting total parenteral nutrition with 5-fluorouracil and/or mitomycin C. Gan To Kagaku Ryoho. 1995;22:1028–35. [PubMed] [Google Scholar]
- 198.Xiao HB, Cao WX, Yin HR, Lin YZ, Ye SH. Influence of L-methionine-deprived total parenteral nutrition with 5-fluorouracil on gastric cancer and host metabolism. World J Gastroenterol. 2001;7:698. doi: 10.3748/wjg.v7.i5.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cao WX, Ou JM, Fei XF, Zhu ZG, Yin HR, Yan M, Lin YZ. Methionine-dependence and combination chemotherapy on human gastric cancer cells in vitro. World J Gastroenterol. 2002;8:230. doi: 10.3748/wjg.v8.i2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cao WX, Cheng QM, Fei XF, Li SF, Yin HR, Lin YZ. A study of preoperative methionine-depleting parenteral nutrition plus chemotherapy in gastric cancer patients. World J Gastroenterol. 2000;6:255. doi: 10.3748/wjg.v6.i2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.US National Library of Medicine. ONC201 with and without methionine-restricted diet in patients with metastatic triple negative breast cancer. ClinicalTrials.gov 2018-2020; https://ClinicalTrials.gov/show/ NCT03733119.
- 202.US National Library of Medicine. Methionine-restricted diet to potentiate the effects of radiation therapy. ClinicalTrials.gov 2018-2020; https://ClinicalTrials.gov/show/ NCT03574194.
- 203.US National Library of Medicine. A window of opportunity study of methionine deprivation in triple negative breast cancer. ClinicalTrials.gov 2017-2020; https://ClinicalTrials.gov/show/ NCT03186937.
- 204.US National Library of Medicine. Dietary methionine restriction plus temozolomide for recurrent GBM. ClinicalTrials.gov 2007-2012; https://clinicaltrials.gov/ct2/show/ NCT00508456.
- 205.US National Library of Medicine. Methionine-restriction diet (MRD) in obese adults with metabolic syndrome. ClinicalTrials.gov 2008-2015; https://clinicaltrials.gov/ct2/show/ NCT00640757.
- 206.US National Library of Medicine. Dietary methionine and cysteine restriction in healthy adults. ClinicalTrials.gov 2014-2020; https://clinicaltrials.gov/ct2/show/ NCT02192437.
- 207.US National Library of Medicine. Methionine and cysteine restriction in humans. ClinicalTrials.gov 2018-2020; https://clinicaltrials.gov/ct2/show/ NCT03629392.
- 208.US National Library of Medicine. Sulfur amino acids, energy metabolism and obesity (STAY) ClinicalTrials.gov 2021; https://clinicaltrials.gov/ct2/show/ NCT04701346.
- 209.Roy DG, Chen J, Mamane V, Ma EH, Muhire BM, Sheldon RD, Shorstova T, Koning R, Johnson RM, Esaulova E, Williams KS, Hayes S, Steadman M, Samborska B, Swain A, Daigneault A, Chubukov V, Roddy TP, Foulkes W, Pospisilik JA, Bourgeois-Daigneault MC, Artyomov MN, Witcher M, Krawczyk CM, Larochelle C, Jones RG. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab. 2020;31:250–266. e259. doi: 10.1016/j.cmet.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 210.Tan Y, Sun X, Xu M, Tan X, Sasson A, Rashidi B, Han Q, Tan X, Wang X, An Z, Sun FX, Hoffman RM. Efficacy of recombinant methioninase in combination with cisplatin on human colon tumors in nude mice. Clin Cancer Res. 1999;5:2157–2163. [PubMed] [Google Scholar]
- 211.Tan Y, Xu M, Hoffman RM. Broad selective efficacy of recombinant methioninase and polyethylene glycol-modified recombinant methioninase on cancer cells in vitro. Anticancer Res. 2010;30:1041–1046. [PubMed] [Google Scholar]
- 212.Hoffman RM. Methioninase: a therapeutic for diseases related to altered methionine metabolism and transmethylation: cancer, heart disease, obesity, aging, and Parkinson’s disease. Hum Cell. 1997;10:69–80. [PubMed] [Google Scholar]
- 213.Yang Z, Wang J, Yoshioka T, Li B, Lu Q, Li S, Sun X, Tan Y, Yagi S, Frenkel EP. Pharmacokinetics, methionine depletion, and antigenicity of recombinant methioninase in primates. Clin Cancer Res. 2004;10:2131–2138. doi: 10.1158/1078-0432.ccr-03-0068. [DOI] [PubMed] [Google Scholar]
- 214.Tan Y, Zavala J, Xu M, Zavala J, Hoffman RM. Serum methionine depletion witout side effects by methioninase in metastatic breast cancer patients. Anticancer Res. 1996;16:3937–3942. [PubMed] [Google Scholar]
- 215.Lim HI, Hamada K, Yamamoto J, Han Q, Tan Y, Choi HJ, Nam SJ, Bouvet M, Hoffman RM. Oral methioninase inhibits recurrence in a PDOX mouse model of aggressive triple-negative breast cancer. In Vivo. 2020;34:2281–2286. doi: 10.21873/invivo.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kawaguchi K, Han Q, Li S, Tan Y, Igarashi K, Kiyuna T, Miyake K, Miyake M, Chmielowski B, Nelson SD, Russell TA, Dry SM, Li Y, Singh AS, Eckardt MA, Unno M, Eilber FC, Hoffman RM. Targeting methionine with oral recombinant methioninase (o-rMETase) arrests a patient-derived orthotopic xenograft (PDOX) model of BRAF-V600E mutant melanoma: implications for chronic clinical cancer therapy and prevention. Cell Cycle. 2018;17:356–361. doi: 10.1080/15384101.2017.1405195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sugisawa N, Hamada K, Han Q, Yamamoto J, Sun YU, Nishino H, Kawaguchi K, Bouvet M, Unno M, Hoffman RM. Adjuvant oral recombinant methioninase inhibits lung metastasis in a surgical breast-cancer orthotopic syngeneic model. Anticancer Res. 2020;40:4869–4874. doi: 10.21873/anticanres.14489. [DOI] [PubMed] [Google Scholar]
- 218.Park JH, Zhao M, Han Q, Sun Y, Higuchi T, Sugisawa N, Yamamoto J, Singh SR, Clary B, Bouvet M, Hoffman RM. Efficacy of oral recombinant methioninase combined with oxaliplatinum and 5-fluorouracil on primary colon cancer in a patient-derived orthotopic xenograft mouse model. Biochem Biophys Res Commun. 2019;518:306–310. doi: 10.1016/j.bbrc.2019.08.051. [DOI] [PubMed] [Google Scholar]
- 219.Oshiro H, Tome Y, Kiyuna T, Yoon SN, Lwin TM, Han Q, Tan Y, Miyake K, Higuchi T, Sugisawa N. Oral recombinant methioninase overcomes colorectal-cancer liver metastasis resistance to the combination of 5-fluorouracil and oxaliplatinum in a patient-derived orthotopic xenograft mouse model. Anticancer Res. 2019;39:4667–4671. doi: 10.21873/anticanres.13648. [DOI] [PubMed] [Google Scholar]
- 220.Han Q, Hoffman RM. Lowering and stabilizing PSA levels in advanced-prostate cancer patients with oral methioninase. Anticancer Res. 2021;41:1921–1926. doi: 10.21873/anticanres.14958. [DOI] [PubMed] [Google Scholar]
- 221.Han Q, Hoffman RM. Chronic treatment of an advanced prostate-cancer patient with oral methioninase resulted in long-term stabilization of rapidly rising PSA levels. In Vivo. 2021;35:2171–2176. doi: 10.21873/invivo.12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Machover D, Rossi L, Hamelin J, Desterke C, Goldschmidt E, Chadefaux-Vekemans B, Bonnarme P, Briozzo P, Kopecny D, Pierige F, Magnani M, Mollicone R, Haghighi-Rad F, Gaston-Mathe Y, Dairou J, Boucheix C, Saffroy R. Effects in cancer cells of the recombinant l-methionine gamma-lyase from brevibacterium aurantiacum. Encapsulation in human erythrocytes for sustained l-methionine elimination. J Pharmacol Exp Ther. 2019;369:489–502. doi: 10.1124/jpet.119.256537. [DOI] [PubMed] [Google Scholar]
- 223.Gay F, Aguera K, Senechal K, Tainturier A, Berlier W, Maucort-Boulch D, Honnorat J, Horand F, Godfrin Y, Bourgeaux V. Methionine tumor starvation by erythrocyte-encapsulated methionine gamma-lyase activity controlled with per os vitamin B6. Cancer Med. 2017;6:1437–1452. doi: 10.1002/cam4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Stone E, Paley O, Hu J, Ekerdt B, Cheung NK, Georgiou G. De novo engineering of a human cystathionine-gamma-lyase for systemic (L)-Methionine depletion cancer therapy. ACS Chem Biol. 2012;7:1822–1829. doi: 10.1021/cb300335j. [DOI] [PMC free article] [PubMed] [Google Scholar]



