Abstract
Circular RNAs (circRNAs) have been reported to play crucial roles in the progression of various cancers, including esophageal squamous cell carcinoma (ESCC). However, the function of circRNAs in ESCC stemness has not been reported. This study aimed to identify novel circRNAs that regulate ESCC stemness and explore their internal mechanisms in ESCC. We found that hsa_circ_0001741 was upregulated in ESCC tissues and was positively related to lymphatic metastasis, higher TNM stage, and poor prognosis. Functionally, hsa_circ_0001741 promoted ESCC cell stemness, invasion, and migration in vitro. Mechanistically, analysis of the relationship between hsa_circ_0001741 and tumor suppressor miR-491-5p revealed that hsa_circ_0001741 functioned as a miR-491-5p sponge. Specifically, hsa_circ_0001741 bound to miR-491-5p to prevent the microRNA from binding to the 3’-UTR of NOTCH3 mRNA and suppressing NOTCH3 expression. Moreover, the ablation of hsa_circ_0001741 significantly inhibited the tumorigenicity in vivo. In conclusion, hsa_circ_0001741 promotes ESCC stemness, invasion, and migration by sponging tumor suppressor miR-491-5p to upregulate NOTCH3 expression. Our findings identify a novel therapeutic target for ESCC patients and the expression level of hsa_circ_0001741 has the potential to serve as a prognostic biomarker for ESCC.
Keywords: Circular RNA, hsa_circ_0001741, esophageal squamous cell carcinoma, cancer stemness
Introduction
Esophageal cancer is a crucial global health problem. According to reported Global Cancer Incidence, Mortality, and Prevalence (GLOBOCAN) statistics, 604,100 new cases of esophageal cancer and 544,076 esophageal cancer deaths occurred in 2020 (https://gco.iarc.fr/). Esophageal cancers, which are classified as carcinomas, fall into two main histological subtypes: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). On a global scale, more than 85% of esophageal cancers belong to the ESCC subtype [1]. Despite recent advances in medical care, the 5-year survival rate of ESCC is less than 20% which reflects enhanced ESCC cell abilities to engage in both local invasions of tissues and distant metastasis [2,3]. Ultimately, the tumor metastasis led to multiple complications, and death in patients. Therefore, it is particularly important to study mechanisms underlying ESCC progression in order to identify molecules with key roles in this process. In turn, such studies should facilitate the discovery of potential therapeutic targets for the development of ESCC treatments.
Mounting evidence suggests that cancer cells can exhibit stem cell-like features of self-renewal and multilineage differentiation, which are often referred to collectively as “cancer stemness” [4]. Cancer stemness drives tumorigenesis and metastasis by generating cells with enhanced abilities to evolve, diversify, and adapt, qualities that tend to be associated with tumor cell invasion and metastatic capabilities [5]. However, the detailed molecular mechanisms by which cancer stemness promotes tumor development remain largely undefined for many cancers, including ESCC.
Circular RNAs (circRNAs) belong to a novel class of endogenous RNAs and are characterized by covalently closed-loop structures that lack both 5’-3’ polarity and polyadenylated tails and possess greater stability compared with corresponding linear RNAs [6]. The progress of deep RNA sequencing has made it possible to identify a large number of exon and intron circRNAs which are expressed in tissue-specific patterns and functionally associated with various cancers. [6,7]. Meanwhile, recent studies have illustrated that the expression of circRNAs could be dysregulated in cancer cells, thus suggesting that circRNAs could serve as potentially valuable diagnostic biomarkers and therapeutic targets [8]. Competing endogenous RNA (ceRNA) is the most classical mechanism for circrRNA to function and has been reported extensively in various cancers [9-13]. Nowadays, multiple circRNAs have been reported to be associated with cancer stemness and metastasis; for example, circ-Cdyll acts as a miRNA sponge to promote hepatocellular carcinoma stemness and tumor growth [14], and miRNA sponge hsa_circ_0003222 promotes stemness and progression of lung cancer [15]. Notably, although several circRNAs have been found to be associated with ESCC progression, circRNAs that influence cancer stemness in ESCC have not yet been reported.
In this study, the use of circRNA microarray profiling enabled us to determine that hsa_circ_0001741 was upregulated in ESCC patient tissues, with higher hsa_circ_0001741 expression levels in tissues found to be associated with enhanced lymphatic metastasis, higher TNM (tumor, node, metastasis) stage, and poor prognosis. Additional experiments revealed that hsa_circ_0001741 regulated NOTCH3 expression by sponging miR-491-5p to ultimately promote ESCC stemness, invasion, and migration.
Materials and methods
Patient specimens
ESCC and adjacent normal tissues were collected from 109 patients undergoing surgery at Sun Yat-sen Memorial Hospital. The tissue samples were surgically taken and stored in liquid nitrogen until assayed. None of the patients received preoperative chemoradiotherapy. The research program was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital. Written informed consent was obtained from all patients before their inclusion in the study.
Microarray analysis
The circRNA microarray (GSE131969) uploaded to the GEO database was used to detect differentially expressed circRNA between ESCC and normal tissues (http://www.ncbi.nlm.nih.gov/geo/). CircRNAs with “group RAW” values of > 250 were retained to exclude circRNAs with low expression levels.
Cell culture
Four human esophageal squamous epithelial cell lines (TE1, KYSE150, KYSE180, and ECA109) and a human normal esophageal epithelial cell line (HEEC) that were used in this study were obtained from the Shanghai Cell Bank. All cells were grown in 1640 (Gibco) medium containing 10% fetal bovine serum (Gibco) and 100 U/mL streptomycins and penicillin (HyClone).
Total RNA extraction and qRT-PCR
The TRIzol (Invitrogen) was used to obtain total RNA following the standard protocol. A NanoDrop system was used to measure the purity and concentration of total RNA samples. The PrimeScript RT Master Mix (TaKaRa) was used to perform cDNA synthesis. MiRNAs reverse transcription was performed according to instructions provided with the miRNA First Strand cDNA Synthesis Kits (Accurate Biotechnology). A LightCycler® 480 System (Roche) was used to perform qRT-PCR. We calculated the relative changes in target RNA expression levels based on the 2-ΔΔCt method.
Tumorsphere formation assay
After digestion with trypsin, ESCC cells were resuspended in a serum-free medium after being washed twice with PBS and then counted. We added 1500 cells/well in 1000 μL Serum-free Tumorsphere Media (R&D StemXVivo) to 24-well CORNING ultra-low attachment plates then incubated in a 5% CO2 humidified incubator at 37°C. Spheres with diameters of > 70 µm were counted under magnification using a microscope after 2 weeks.
Cell transfection
ESCC cells were inoculated in 6-well plates until cell growth density was approximately 60%. Next, cells were transfected with shRNA, siRNA, miRNA mimics, miRNA inhibitors and overexpression plasmids using lipo3000 and P3000 transfection reagents (Invitrogen) following the protocol provided by the manufacturer.
Transwell assay
Invasion and migration abilities of ESCC cells were tested by transwell assays using transwell inserts (CORNING) precoated with or without Matrigel. 500 μL of RPMI 1640 complete medium was placed in the bottom chambers. 100 μL serum-free RPMI 1640 which contained 20,000 ESCC cells was placed in the upper chambers. After 1-2 days, 4% paraformaldehyde was added to cells bound to membranes to fix the cells, followed by crystal violet staining. Afterward, the stained cells were observed and photographed under a light microscope (Leica).
RNA fluorescence in situ hybridization (FISH)
The hsa_circ_0001741 specific probe targeting the back-splice junction region was cy3-labeled while the miR-491-5p specific probe was FAM-labeled. Both probes were synthesized by GenePharma. A FISH Kit (RiboBio) was used to carry out the RNA FISH assay following the protocol. A confocal microscope (Zeiss) equipped with a digital camera was used to acquire multi-channel fluorescence Images.
RNA-binding protein immunoprecipitation (RIP) and RNA pull-down assay
A Millipore Magna RIP Kit was used to perform the RIP assays. We performed RNA pull-down assays as previously described [30]. Biotinylated hsa_circ_0001741 or oligo probe was synthesized by Kidan Biosciences (Guangzhou, China). Meanwhile, the hsa_circ_0001741 probe was designed to target the back-splice junction region sequence. Pull-down assays were conducted following a standard protocol. Briefly, we collected ESCC cells followed by cross-linking, lysis, and sonication, and then the hybridization was performed by incubating cell lysates with hsa_circ_0001741 or a control probe at 37°C for 12 h. Afterward, to generate circRNA-probe-bead complexes, precleaned Streptavidin Magnetic Beads (Thermo Fisher) were added to the lysates then the mixtures were incubated at 37°C for 1 h. After we clean the beads with wash buffer, the RNA was eluted then TRIzol (Invitrogen) was used to extract the total RNA followed by cDNA synthesis and qRT-PCR.
Western blot (WB) analysis
The RIPA buffer (CWBIO, China) with 1% phosphatase and protease inhibitors was used to lysis ESCC cells and extract proteins. After proteins were electrophoresed and separated via SDS-PAGE, we transferred the proteins to the PVDF membranes (Millipore, USA). Next, a 5% BSA solution was used to block the membranes at room temperature for 1 h. Thereafter, blocked membranes were incubated with primary antibodies (1:1000) specific for NOTCH3 (CST, USA), SOX2 (ABclonal, China), Nanog (Proteintech, China), E-cadherin (ImmunoWay, China), N-cadherin (ImmunoWay, China), or glyceraldehyde dehydrogenase (GAPDH) (ABclonal, China) at 4°C overnight. After that, the membranes were incubated with goat anti-mouse or goat anti-rabbit secondary antibody (Proteintech) for 1 hour. Signals corresponding to protein bands bound to antibody probes were detected after adding an enhanced chemoluminescent (ECL) reagent (Thermo Fisher Scientific).
Luciferase reporter assay
The wild-type (hsa_circ_0001741-WT or NOTCH3 3’UTR-WT) or mutated (hsa_circ_0001741-Mut or NOTCH3 3’UTR-Mut) sequences containing miR-491-5p binding sites were constructed and inserted into luciferase-report vectors separately. TE1, ECA109, or HEK-293T cells were cultured in 96-well plates then cells were co-transfected with miR-491-5p or control mimics and recombinant luciferase reporter plasmids for 48 h. Luciferase activity was measured and normalized to Renilla luciferase activity by using a dual-luciferase reporter assay kit (Vazyme).
In vivo limiting-dilution assay
The 4 weeks-old BALB/c nude mice (Charles River, China) were raised in the SPF animal facility of the Institute of Biological and Medical Engineering, Guangdong Academy of Sciences. Prior approval was acquired from the ethics committee of this research institute before experiments were conducted. For in vivo limiting-dilution assays, different numbers of TE1 cells with or without knockdown of hsa_circ_0001741 expression were injected into flanks of nude mice after being mixed with an equal volume of Matrigel. Tumor volume was calculated from external measurements (Volume = W2 × L/2, W stand for width, L stand for length) and was monitored for 7 weeks. We used the software (http://bioinf.wehi.edu.au/software/elda/) to conduct an Extreme Limiting Dilution Analysis. After the mice were sacrificed, tumors were subjected to further immunohistochemical analysis using primary antibodies against SOX2 (Proteintech, China), Nanog (Proteintech, China), N-cadherin (Immunoway, China), E-cadherin (Immunoway, China) and NOTCH3 (Proteintech, China).
Statistical analysis
Statistical analyses were performed using PRISM 8 and SPSS 25.0 statistical software. In this paper, the Chi-square test was used to analyze the relationship between hsa_circ_0001741 expression and the characteristics of patient. Kaplan-Meier analysis was used to assess overall survival (OS). ROC curve was applied to evaluate the diagnostic value of hsa_circ_0001741. P < 0.05 was considered statistically significant.
Results
ESCC tissues exhibit upregulated hsa_circ_0001741 expression
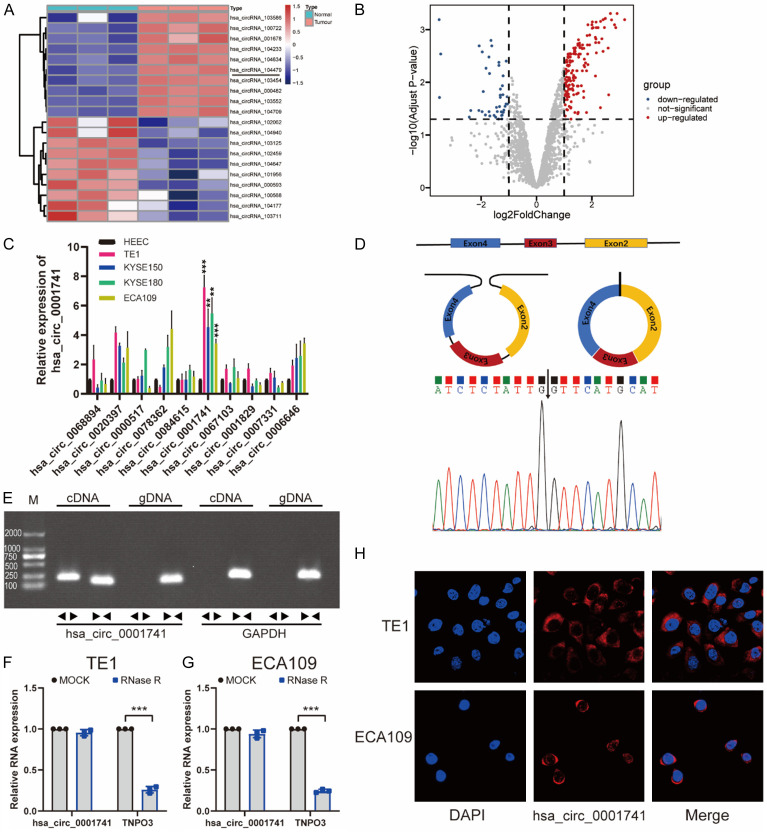
Volcano plot-based analysis of circRNA expression profiles of three paired ESCC tissue samples that we had previously uploaded in the GEO (Gene Expression Omnibus) database revealed variations of circRNA expression as log-fold changes of > 1 (P < 0.05) (Figure 1B). After the top 10 upregulated or downregulated circRNAs were screened out via hierarchical clustering (Figure 1A), we examined the 10 upregulated circRNAs expression changes between 4 ESCC cell lines and a normal epithelial cell line of esophageal using qRT-PCR. The results revealed that hsa_circ_0001741 was significantly upregulated in all ESCC cell lines as compared with corresponding expression levels in HEECs (Figure 1C), thus prompting us to focus on hsa_circ_0001741 thereafter.
Figure 1.
Screening and identification of hsa_circ_0001741 in ESCC cells. A. Clustered heatmap showing the top 10 differentially expressed circRNAs between ESCC tissue and adjacent normal tissue. B. Volcano plot showing screened circRNAs with significant differential expression between ESCC tissue and adjacent normal tissue (adjust P < 0.05, |Log FC| > 1). The red dots represent significantly upregulated circRNAs. C. Graph showing hsa_circ_0001741 was significantly upregulated in ESCC cell lines. D. Schematic illustration demonstrates hsa_circ_0001741 formation via three exons circularization of the TNPO3 gene. Sanger sequencing result obtained for hsa_circ_0001741 showing the back-spliced junction region. E. The presence of hsa_circ_0001741 was confirmed by PCR and gel electrophoresis using convergent and divergent primers. PCR result showing hsa_circ_0001741 could only be amplified from cDNA. F, G. Stability of hsa_circ_0001741 and linear TNPO3 as assessed via RNaseR treatment followed by qRT-PCR. H. Immunofluorescence detection was combined with RNA FISH assay of hsa_circ_0001741 and the majority of hsa_circ_0001741 was found within the cytoplasm. **P < 0.01, ***P < 0.001.
According to information in the circBase database (http://circbase.org), hsa_circ_0001741 is formed via back-splicing of three exons (exons 2-4) of the TNPO3 gene (Figure 1D). This information was confirmed by Sanger sequencing, which showed that the hsa_circ_0001741 sequence was identical to the sequence described in the circBase database annotation (Figure 1D). Given that back-splice junctions can be caused by either the genomic rearrangements or reverse splicing of cDNA, two sets of primers (divergent and convergent primers) were designed to determine which mechanism was responsible for generating hsa_circ_0001741 in our samples. We used divergent primers to detect the hsa_circ_0001741 circular form while using convergent primers to amplify the linear transcripts of TNPO3 mRNA. Next, both cDNA and gDNA prepared from our sample were used to amplify circular and linear transcripts of TNPO3 deriving from the two sets of primers. The results indicated that hsa_circ_0001741 was amplified by divergent primers only from cDNA but not from gDNA (Figure 1E). RNase R endoribonuclease was used to treat RNA extracted from TE1 and ECA109 cell lines followed by qRT-PCR analysis. The results showed a sharp decrease in the level of linear TNPO3 RNAs after RNase R treatment, whereas hsa_circ_0001741 molecules were resistant to RNase R treatment (Figure 1F). Moreover, we determined the cellular location of the circRNA using FISH assays based on the binding of a cy3-labeled fluorescent probe targeting the hsa_circ_0001741 head-to-tail spliced junction region and hsa_circ_0001741 was mainly located in the cytoplasm as the results demonstrated (Figure 1H).
Overexpression of hsa_circ_0001741 was correlated with poor survival
To analyze the clinical significance of hsa_circ_0001741 expression in ESCC, we used qRT-PCR to measure relative expression levels of hsa_circ_0001741 in 109 pairs of ESCC tissues as compared with adjacent normal tissues and hsa_circ_0001741 was found overexpressed in tissues of 70.6% (77 of 109) of ESCC patients (Figure 2A). Notably, compared with that in adjacent normal tissues, hsa_circ_0001741 expression levels were significantly higher in ESCC tissues (Figure 2C). Moreover, high hsa_circ_0001741 expression levels were significantly associated with more extensive lymph node metastasis and higher ESCC TNM stage (Table 1) (Figure 2E). Further investigation of the relationship between the expression level of hsa_circ_0001741 and patient prognosis revealed an inverse association between hsa_circ_0001741 expression level and overall survival time by using Kaplan-Meier analysis (Figure 2B). This result prompted us to further analyze receiver operating characteristic (ROC) curves of hsa_circ_0001741 expression to investigate the diagnostic value of this circRNA by analyzing the area under the curve (AUC) values. The results indicated that hsa_circ_0001741 level could be used to predict the incidence of lymph node metastasis based on an AUC value of 0.72 (Figure 2D).
Figure 2.
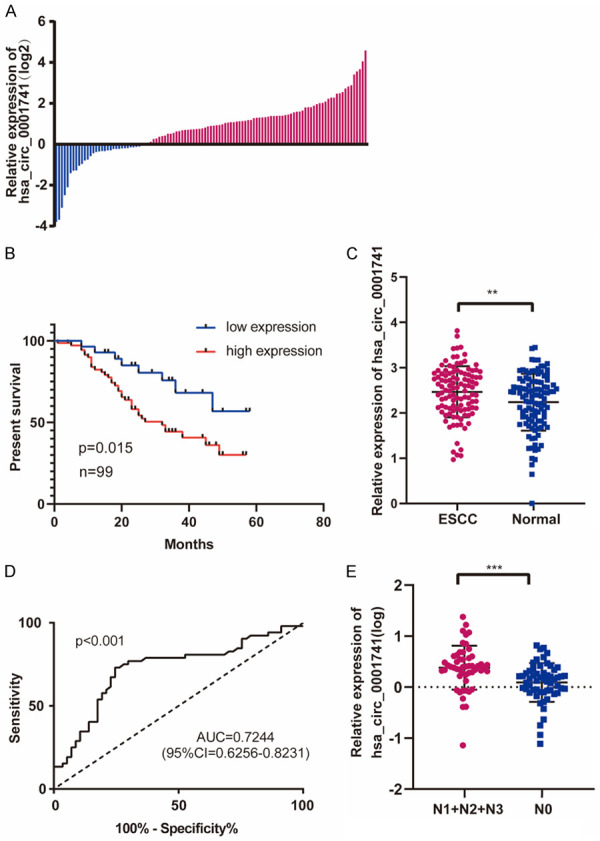
hsa_circ_0001741 was upregulated in ESCC and correlated with poor patient prognosis. A. Relative expression levels of hsa_circ_0001741 in 109 paired ESCC patients. Blue: down-regulation, 29.3% (32/109); Red: up-regulation, 70.6% (77/109). B. Kaplan-Meier’s analyses of the correlations between the hsa_circ_0001741 expression and OS (overall survival) in 99 ESCC patients (patients lost to follow-up were excluded). C. Relative expression levels of hsa_circ_0001741 in ESCC and adjacent normal tissues were analyzed by qRT-PCR. D. The ROC curve for the diagnosis of lymph node metastasis in ESCC. E. Graph showing that expression levels of hsa_circ_0001741 are significantly higher in patients with lymph node metastasis than in patients without lymph node metastasis. **P < 0.01, ***P < 0.001.
Table 1.
Relationship between clinicopathological characteristics and hsa_circ_0001741 expression in ESCC patients
| Clinicopathological parameters | Number of cases | hsa_circ_0001741 expression level | ||
|---|---|---|---|---|
|
| ||||
| Low | High | P-value | ||
| Total | 109 | 32 | 77 | |
| Gender | 0.996 | |||
| Male | 92 | 27 | 65 | |
| Female | 17 | 5 | 12 | |
| Age | 0.772 | |||
| < 60 | 42 | 13 | 29 | |
| ≥ 60 | 67 | 19 | 48 | |
| Histologic grade | 0.148 | |||
| Well | 10 | 4 | 6 | |
| Moderate | 60 | 13 | 47 | |
| Poor | 39 | 15 | 24 | |
| T stage1 | ||||
| T1 + T2 | 35 | 13 | 22 | 0.22 |
| T3 + T4 | 74 | 19 | 55 | |
| N stage1 | 0.027* | |||
| N0 | 57 | 22 | 35 | |
| N1 + N2 + N3 | 52 | 10 | 42 | |
| TNM stage1 | 0.022* | |||
| I-II | 56 | 22 | 34 | |
| III-IV | 53 | 10 | 43 | |
Tumor, Node, Metastasis stage (TNM stage);
P < 0.05.
hsa_circ_0001741 promotes ESCC cell stemness, invasion, and migration
To investigate the biological function of hsa_circ_0001741, we used two short hairpin RNAs (shRNA1 and shRNA2) targeted to the back-spliced junction sequence to knockdown hsa_circ_0001741 expression in TE1. The results revealed a successful knockdown of hsa_circ_0001741 expression that did not significantly affect expression of its linear TNPO3 mRNA counterpart (Figure 3E). However, shRNA1 was more effective than shRNA2 in knocking down hsa_circ_0001741 expression. Meanwhile, due to the fact that only low-level upregulation of hsa_circ_0001741 was observed in ECA109 cells, we constructed a plasmid to enable overexpression of hsa_circ_0001741 in ECA109 cells and found again that overexpression of hsa_circ_0001741 expression did not significantly affect expression of its linear TNPO3 mRNA counterpart (Figure 3F).
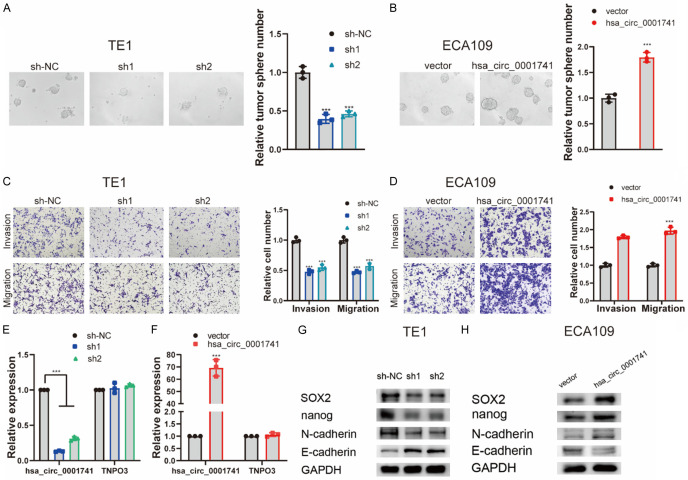
Figure 3.
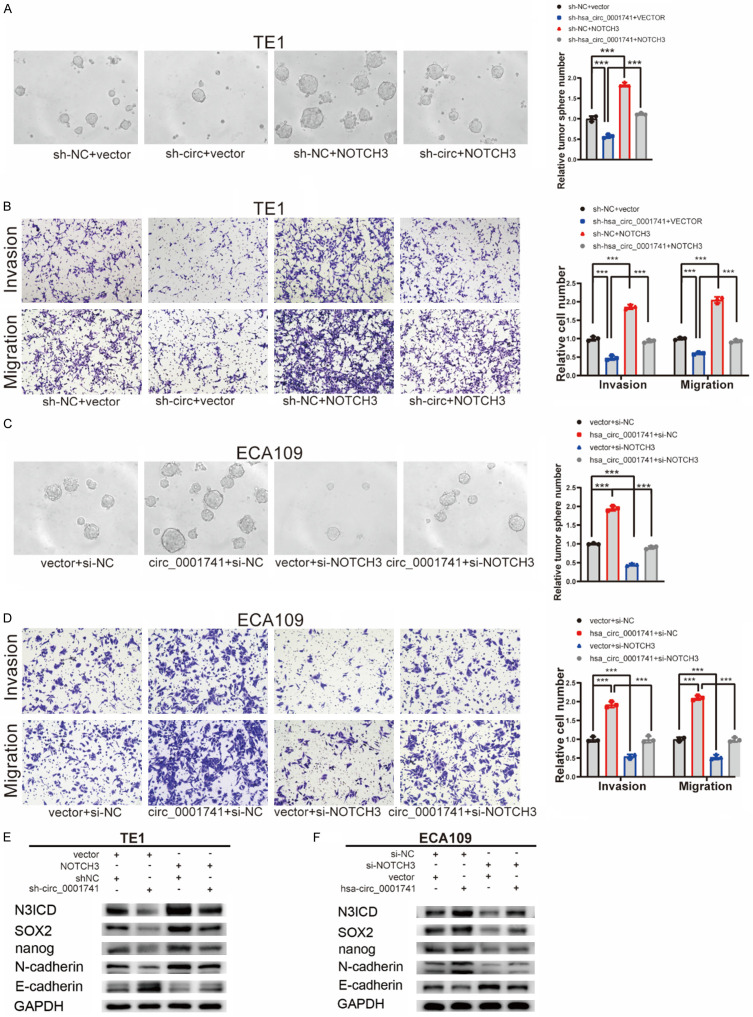
hsa_circ_0001741 promotes ESCC cell stemness, invasion, and migration. (A, B) Representative image showing tumorsphere formation (A) after knockdown hsa_circ_0001741 in TE-1 cells and (B) after overexpression of hsa_circ_0001741 in ECA109 cells. (C, D) The effect of hsa_circ_0001741 on ESCC cells invasion and migration abilities using transwell assay (C) after knockdown of hsa_circ_0001741 in TE1 cells and (D) after overexpression of hsa_circ_0001741 in ECA109 cells. (E) The expression of hsa_circ_0001741 in TE1 cell line transfected with siRNAs or si-NC measured by qRT-PCR. (F) The overexpression plasmid of hsa_circ_0001741 or the control vector was transfected into ECA109 cell line, and the expression level of hsa_circ_0001741 was measured by qRT-PCR. (G, H) Cancer stemness and EMT marker protein levels in ESCC cells (G) after hsa_circ_0001741 knockdown or (H) after hsa_circ_0001741 overexpression was detected by Western blot. **P < 0.01, ***P < 0.001.
Next, we conducted tumorsphere assays to assess the cancer stemness of ESCC cells. The results of tumorsphere assays showed that downregulation of hsa_circ_0001741 expression was associated with significantly reduced tumorsphere number, while upregulation of hsa_circ_0001741 expression markedly promoted tumorsphere formation (Figure 3A, 3B). Similarly, transwell assay results showed that hsa_circ_0001741 downregulation inhibited ESCC cell invasive and migratory activities, while hsa_circ_0001741 overexpression promoted both of these activities (Figure 3C, 3D). Thus, results of the abovementioned assays showed that downregulation of hsa_circ_0001741 expression decreased ESCC cell cancer stemness, invasion, and migration, while hsa_circ_0001741 overexpression enhanced these activities in ESCC cells.
There is a close relationship between the cancer stemness and the epithelial-mesenchymal transition that prompted us to investigate cancer stemness and its association with EMT-related markers using Western blot (WB) analysis. The results revealed that knockdown of hsa_circ_0001741 expression led to downregulated expression of SOX2 and Nanog along with downregulated expression of N-cadherin which is a mesenchymal cell marker and upregulated expression E-cadherin which is an epithelial marker. By contrast, the expression level of E-cadherin was decreased while those of N-cadherin, SOX2, and Nanog were increased in ESCC cells with hsa_circ_0001741 overexpressed (Figure 3G, 3H). These results demonstrated that hsa_circ_0001741 promoted ESCC cancer stemness and EMT.
hsa_circ_0001741 acts as a miR-491-5p sponge in ESCC cells
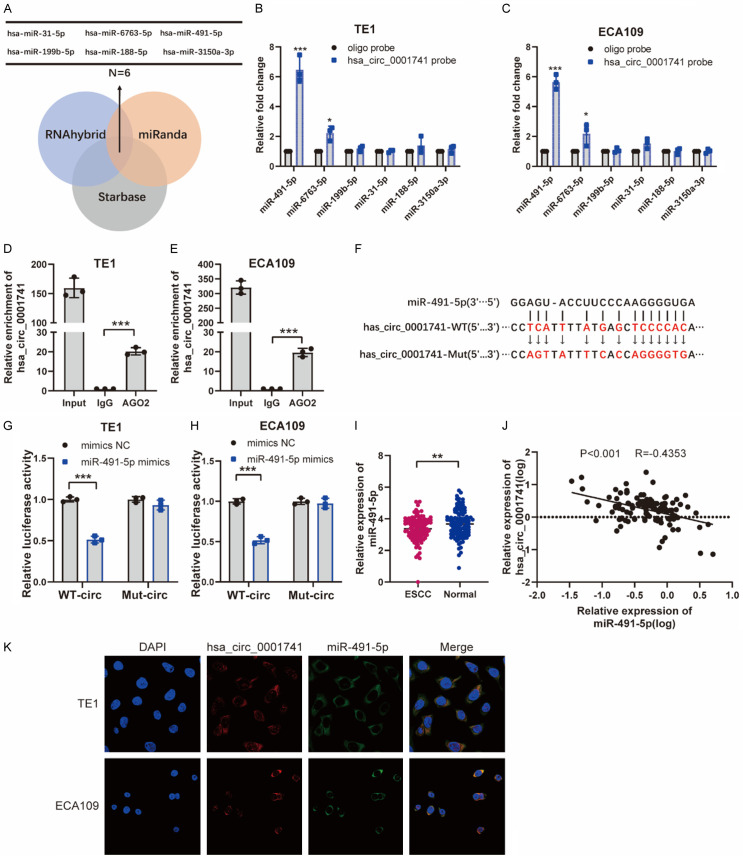
It has been extensively reported that circRNAs could regulate the transcriptional activity of target genes by acting as miRNA sponges. Thus, we searched three different databases (RNAhybrid, miRanda, and Starbase) in order to identify potential miRNAs that bind to hsa_circ_0001741 and subsequently detected 6 miRNAs that were common to all three databases (Figure 4A). To verify whether any of the six candidate miRNAs interacted with hsa_circ_0001741, a specific biotin-labeled hsa_circ_0001741 probe was used to conduct an RNA pull-down assay. Among the 6 candidate miRNAs, miR-491-5p was most significantly pulled down by the hsa_circ_0001741 probe in TE1 and ECA109 cell lines according to the qRT-PCR results (Figure 4B, 4C). Meanwhile, the results of AGO2 RNA immunoprecipitation (RIP) assays that were performed in these cell lines revealed that AGO2 pulled down hsa_circ_0001741. Moreover, qRT-PCR results showed that hsa_circ_0001741 circRNA was obviously enriched in AGO2-containing beads as compared with beads containing control IgG-immunoprecipitated antigens (Figure 4D, 4E). To determine whether miR-491-5p could bind to hsa_circ_0001741, we conducted a dual-luciferase reporter assay. Our results revealed a significant reduction (~50%) in luciferase reporter activity only when the wild-type hsa_circ_0001741 and miR-491-5p mimics were co-transfected while co-transfection of miR-491-5p mimics and mutant hsa_circ_0001741 not (Figure 4F-H). In addition, we used qRT-PCR to validate the expression of miR-491-5p and found that miR-491-5p in adjacent normal tissues were significantly higher than those in ESCC tissues (Figure 4I). Furthermore, the expression of hsa_circ_0001741 was negatively correlated with miR-491-5p based on the qRT-PCR results (Figure 4J). Taken together, these results revealed a direct interaction occurred between miR-491-5p and hsa_circ_0001741, which prompted us to perform FISH assays in ESCC cell lines to identify subcellular locations of hsa_circ_0001741 and miR-491-5p. The results showed that hsa_circ_0001741 and miR-491-5P were located at the same intracytoplasmic locations (Figure 4K), thus suggesting that hsa_circ_0001741 acts as a miR-491-5p sponge in ESCC cells by functioning as a competitive endogenous RNA.
Figure 4.
hsa_circ_0001741 acts as a sponge of miR-491-5p in ESCC. A. Diagram showing identities of six predicted potential target miRNAs of hsa_circ_0001741 that were detected via searches of RNAhybrid, miRanda, and Starbase databases. B, C. RNA pull-down was performed in TE1 and ECA109 cell lines using hsa_circ_0001741 and oligo control probes. D, E. RIP assays were carried out using an AGO2 antibody. F. Putative complementary binding sites between hsa_circ_0001741 and miR-491-5p, as predicated using miRanda. G, H. Luciferase reporter assay of TE1 and ECA109 cells after cells were transformed with reporter plasmids Luc-hsa_circ_0001741-WT or Luc-hsa_circ_0001741-Mut and co-transfected with mimic miR-491-5p or NC. I. The relative expression of miR-491-5p in ESCC and adjacent normal tissues were was analyzed by qRT-PCR. J. The expression of hsa_circ_0001741 and miR-491-5p was negatively correlated in 109 ESCC patients. K. hsa_circ_0001741 and miR-491-5p were detected by the FISH assay. **P < 0.01, ***P < 0.001.
Hsa_circ_0001741 upregulates NOTCH3 by sponging miR-491-5p
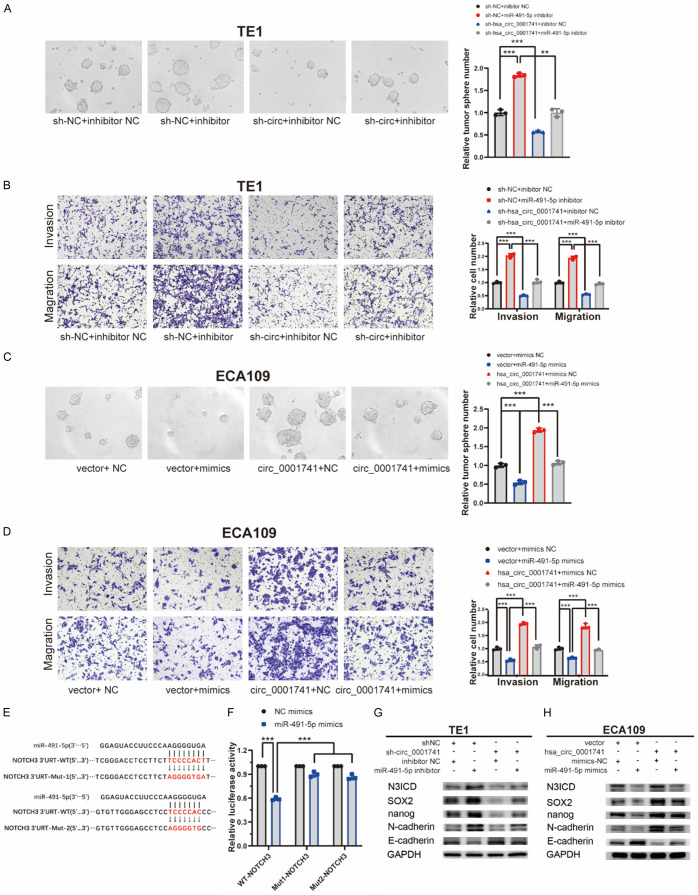
To determine whether miR-491-5p could reverse the function of hsa_circ_0001741 in ESCC cells, we next constructed a stable hsa_circ_0001741 knockdown TE1 cell line and its control cell line. Concurrently, a vector that enabled stable overexpression of hsa_circ_0001741 or an empty vector was transfected into the ECA109 cell line (Figure S1). Thereafter, we performed rescue experiments to verify the functional relationship between miR-491-5p and hsa_circ_0001741. The results revealed that tumorsphere formation, invasion, and migration of control TE1 cells transfected with negative control (NC) inhibitor were all significantly lower than corresponding activities observed for cells transfected with miR-491-5p inhibitor, suggesting that miR-491-5p significantly inhibited tumorsphere formation, invasion, and migration. Additionally, the knockdown of hsa_circ_0001741 led to decreased tumorsphere formation, invasion, and migration, while miR-491-5p inhibition led to increases in all three activities (Figure 5A, 5B). Moreover, hsa_circ_0001741 overexpression in cells promoted tumorsphere formation, invasion, and migration, while transfection of cells with miR-491-5p mimics led to decreases in all three activities (Figure 5C, 5D).
Figure 5.
hsa_circ_0001741 upregulates NOTCH3 by sponging miR-491-5p. (A, B) The cancer stemness (A) and invasion and migration (B) abilities in TE1 cells inhibited by hsa_circ_0001741 knockdown were reversed after transfection with miR-491-5p inhibitor using tumorsphere formation and transwell assay. (C, D) The cancer stemness (C) and invasion and migration (D) abilities promoted by hsa_circ_0001741 overexpression in ECA109 cells were reversed after transfection with miR-491-5p mimics using tumorsphere formation and transwell assay. (E) Predicted putative miR-491-5p binding sites in 3’UTR of NOTCH3 gene. (F) Luciferase reporter assay showing luciferase activity of NOTCH3-3’UTR WT or NOTCH3-3’UTR mutant in cells co-transfected with miRNA mimics. (G) The expression of NOTCH3 (N3ICD), SOX2, Nanog, N-cadherin protein levels in TE1 cells downregulated by hsa_circ_0001741 knockdown were reversed after transfection with miR-491-5p inhibitor and detected by western blotting. (H) The expression of NOTCH3 NICD, SOX2, Nanog, N-cadherin protein levels in ECA109 cells upregulated by hsa_circ_0001741 overexpression were reversed after transfection with miR-491-5p mimics and detected by Western blotting. **P < 0.01, ***P < 0.001.
NOTCH3 is a gene that has been found to promote cancer stemness and progression. In previous studies, NOTCH3 was found to be directly targeted by miR-491-5p, whereby NOTCH3 activation resulted from inhibition of miR-491-5p that led to cancer progression. Notably, bioinformatics analysis conducted here revealed that miR-491-5p binds to two potential binding sites within the 3’UTR of NOTCH3 mRNA, with their binding interactions confirmed by luciferase assay (Figure 5E, 5F).
In order to evaluate the effects of hsa_circ_0001741 and miR-491-5p expressions in cancer stemness, the expression levels of key proteins were examined by conducting the Western blotting assay. The results indicated that knockdown of hsa_circ_0001741 could induce downregulation of NOTCH3 NICD (N3ICD), SOX2, Nanog, and N-cadherin, while increasing E-cadherin. Additionally, the results mentioned above were partially replicated in hsa_circ_0001741-overexpressing ESCC cell line. In addition, the results of Western blotting further indicated that hsa_circ_0001741 could function as a miR-491-5p sponge (Figure 5G, 5H). In summary, these findings suggest that hsa_circ_0001741 promotes cancer stemness and EMT by acting as a miR-491-5p sponge to regulate the expression of NOTCH3.
Silencing of NOTHC3 reverses the tumorigenic effect induced by hsa_circ_0006948 overexpression
To further verify whether hsa_circ_0001741 promotes ESCC cancer stemness and EMT by allowing NOTCH3 expression, we conducted a rescue experiment by transfecting ESCC cells with a NOTCH3 overexpression plasmid or a NOTCH3 siRNA. In TE1 cell lines, downregulation of hsa_circ_0001741 inhibited tumorsphere formation, invasion, and migration, while overexpression of NOTCH3 reversed this effect (Figure 6A, 6B). Furthermore, overexpression of NOTCH3 reversed hsa_circ_0001741 knockdown-induced effects (i.e., downregulation of expressions of NOTCH3 NICD, SOX2, Nanog, and N-cadherin; upregulation of E-cadherin expression) (Figure 6E). Moreover, knockdown of NOTCH3 expression abrogated the increased tumorigenic effect induced by hsa_circ_0001741 overexpression in ECA109 cells (Figure 6C, 6D, 6F).
Figure 6.
NOTCH3 reverses the function of hsa_circ_0001741 in ESCC cells. (A, B) The cancer stemness (A) and invasion and migration (B) abilities in TE1 cells inhibited by hsa_circ_0001741 knockdown were reversed after transfection with NOTCH3 overexpression plasmid using tumorsphere formation and transwell assay. (C, D) The cancer stemness (C) and invasion and migration (D) abilities promoted by hsa_circ_0001741 overexpression in ECA109 cells were reversed after transfection with miR-491-5p mimics using tumorsphere formation and transwell assay. (E) The downregulation of NOTCH3, SOX2, Nanog, N-cadherin, and the upregulation of E-cadherin induced by hsa_circ_0001741 knockdown in TE-1 was reversed by overexpression of NOTCH3 as detected by Western blotting. (F) The upregulation of NOTCH3, SOX2, Nanog, N-cadherin, and the downregulation of E-cadherin induced by hsa_circ_0001741 overexpression in ECA109 were abolished by knockdown NOTCH3 and detected by Western blotting. **P < 0.01, ***P < 0.001.
hsa_circ_0001741 influences tumorigenicity of ESCC in vivo
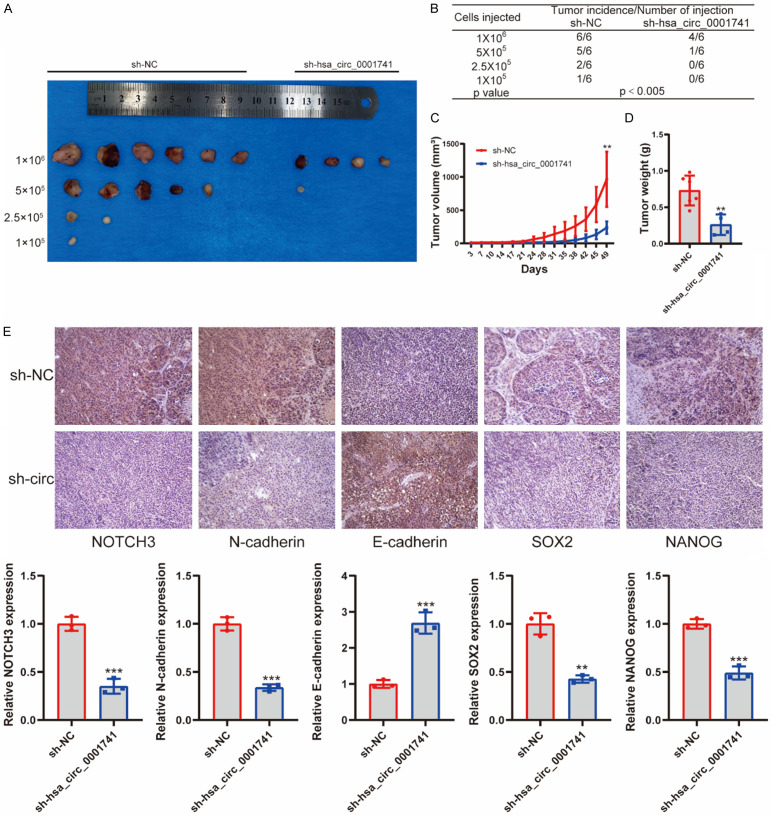
In order to explore tumorigenic effects of hsa_circ_0001741 expression in vivo, TE1 cells (with or without hsa_circ_0001741 knockdown) were grafted onto flanks of BALB/c nude mice followed by extreme limiting dilution analysis (ELDA). Knockdown of hsa_circ_0001741 in TE1 cells was found to reduce tumor incidence as compared with the control TE1 cells, as assessed by a limiting dilution xenograft analysis. These results argued that hsa_circ_0001741 overexpression led to increased tumor-initiating capacity. By contrast, grafting of TE1 cells after hsa_circ_0001741 knockdown led to development of fewer tumors and tumors were detected in only 5 of 24 mice engrafted with TE1 cells with hsa_circ_0001741 knocked down as compared with 14 of 24 mice engrafted with TE1 control cells (Figure 7A, 7B). Moreover, hsa_circ_0001741 knockdown led to significantly reduced xenograft tumor growth (Figure 7C, 7D).
Figure 7.
Ablation of hsa_circ_0001741 inhibits tumorigenicity of ESCC cells. (A) In vivo limiting dilution analysis of TE1 cells with or without hsa_circ_0001741 knockdown (n = 6). (B) The frequency of tumor formation at each cell dose injected was shown. The data were analyzed using ELDA software. (C, D) Tumor volumes (C) were measured with a caliper over a 7-week time period, and tumor weights (D) were also measured at the end of the study, with both graphs generated using data from mice receiving 1 × 106 injected tumor cells. (E) Expression levels of NOTCH3, SOX2, Nanog, N-cadherin E-cadherin and in tumors were analyzed by IHC staining. **P < 0.01, ***P < 0.001.
Finally, the abundances of NOTCH3, SOX2, Nanog, N-cadherin, and E-cadherin proteins in ESCC tissues were assessed by immunohistochemical staining (IHC). The results consistently demonstrated that knockdown of hsa_circ_0001741 led to downregulation of expression of SOX2, Nanog, and N-cadherin and upregulation of E-cadherin. Taken together, our findings demonstrated that hsa_circ_0001741 enhanced ESCC tumorigenic activities in vivo (Figure 7E).
Discussion
In recent years, accumulating evidence demonstrates that circRNAs are critical for various biological processes, especially those related to tumorigenesis, invasion, and metastasis. Previous studies had revealed that expression profiles of circRNAs are cell type-specific and tissue-type specific such that circRNAs could serve as potential biomarkers even though the functions of most circRNAs remain unclear [8,16]. In this study, we found that hsa_circ_0001741 was significantly upregulated in ESCC tissues and that this characteristic was associated with higher lymph node metastasis, TNM stage, and poorer patient survival. Therefore, hsa_circ_0001741 holds promise as a biomarker for ESCC prognosis as well as a potential therapeutic target.
Growing evidence indicates that cancer stemness plays a key role in tumor development, progression and metastasis [4], with stemness of some cancers observed to be regulated by several circRNAs. In this study, we screened for upregulated circRNAs in ESCC cells using microarray analysis and determined that hsa_circ_0001741 promoted cancer stemness in vitro and in vivo. Importantly, induction of EMT programs in a broad range of cancer types has been shown to be correlated with the activation of cancer stem cell properties [17]. In the current study, expression levels of key factors associated with both cancer stemness and EMT induction were significantly increased, as shown via WB analysis. Notably, this is the first study to show that circRNA regulates cancer stemness in ESCC.
The most frequently reported function of exonic circRNAs is acting as “miRNA sponges,” whereby they bind to and inhibit miRNA functions by protecting miRNA-targeted mRNAs from miRNA-mediated degradation [18]. One such circRNA, hsa_circ_0001741, which is back-spliced from three exons of TNPO3, is predominantly located at the cytoplasm and has been confirmed to contain miRNA binding sites as previously predicted via bioinformatics analysis. Based on the characteristics mentioned above, we speculated that hsa_circ_0001741 might function as a miRNA sponge in ESCC. Subsequently, results we obtained using luciferase reporter assays, RIP assays, and pull-down assays showed that hsa_circ_0001741 could directly bind to miR-491-5p, while results of “rescue” assays showed that hsa_circ_0001741 acted as a miR-491-5p sponge in ESCC cells. Interestingly, it has been reported that miR-491-5p plays tumor suppressor roles in multiple cancers [19-24]. In this study, we found that miR-491-5p overexpression could inhibit cancer stemness, invasion, and migration in ESCC cells, suggesting that miR-491-5p plays a tumor suppressor role in ESCC as well.
Notch signaling has been found to profoundly affect cancer progression, as aberrantly high expression levels of NOTCH3 have been found in multiple tumor types, such as hepatocellular carcinoma [25], nasopharyngeal carcinoma [26], ovarian cancer [27] and gastric cancer [21]. Moreover, activation of NOTCH3 signaling has been reported to be closely related to cancer stemness [28], metastasis, and poor prognosis [29]. Importantly, the results of a previous study have shown that NOTCH3 was negatively regulated by miR-491-5p [21]. In this study, downregulation of hsa_circ_0001741 inhibited tumorsphere formation, invasion and migration, while also reducing expression of NOTCH3, with all of these effects found to be reversed in the presence of miR-491-5p inhibitor. Furthermore, knockdown of NOTCH3 expression could reverse hsa_circ_0001741-induced effects on expression of SOX2, Nanog, N-cadherin, and E-cadherin. Taken together, our findings demonstrate that hsa_circ_0001741 regulates cancer stemness and EMT by preventing miR-491-5p from suppressing NOTCH3 expression.
In summary, hsa_circ_0001741 was upregulated in ESCC tissues, which is associated with more lymph node metastasis, higher TNM stage and poor prognosis. Mechanistically, hsa_circ_0001741 expression promoted cancer stemness and induced EMT of ESCC cells by acting as a miRNA sponge by binding to miR-491-5p and preventing it from suppressing the expression of its target gene NOTCH3, thus leading to upregulated NOTCH3 expression. Therefore, our results provide potential biomarkers and therapeutic targets for application in the diagnosis and treatment of ESCC.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81871886, 81672415, 82073121), the Natural Science Foundation of Guangdong Province (No. 2017A030313474), Guangdong Science and Technology Department (No. 2017B030314026), Guangdong Basic and Applied Basic Research Foundation (No. 2020A1515010041), Guangzhou Science and Technology Project (No. 202103000063) and the Fundamental Research Funds for the Central Universities (No. 19ykpy110).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 2.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 3.Abnet CC, Arnold M, Wei WQ. Epidemiology of Esophageal squamous cell carcinoma. Gastroenterology. 2018;154:360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miranda A, Hamilton PT, Zhang AW, Pattnaik S, Becht E, Mezheyeuski A, Bruun J, Micke P, de Reynies A, Nelson BH. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci U S A. 2019;116:9020–9029. doi: 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13:669–680. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, Li X, Sun Z. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19:14. doi: 10.1186/s12943-019-1125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 9.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 10.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kuhn R, Rosenmund C, Birchmeier C, Rajewsky N. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 11.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, Li B, Qin M, Yang C, He J, Zuo Q, Wang S, Liu J, Ye W, Lv Y, Zhao F, Yao M, Jiang L, Qin W. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, Shao B, Sun Z, Ji Z. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021;20:93. doi: 10.1186/s12943-021-01372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, Zhang H, Yang P, Cui X, Ren Y, Xin X, Li H, Wang R, Wang W, Jiang F, Liu S, Ding J, Zhang B, Li L, Wang H. A noncoding regulatory rnas network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2020;71:130–147. doi: 10.1002/hep.30795. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Zhang J, Yang X, Hu C, Chu T, Zhong R, Shen Y, Hu F, Pan F, Xu J, Lu J, Zheng X, Zhang H, Nie W, Han B, Zhang X. hsa_circ_0003222 accelerates stemness and progression of non-small cell lung cancer by sponging miR-527. Cell Death Dis. 2021;12:807. doi: 10.1038/s41419-021-04095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson MM, Weinberg RA, Lees JA, Guen VJ. Emerging mechanisms by which EMT programs control stemness. Trends Cancer. 2020;6:775–780. doi: 10.1016/j.trecan.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 19.Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, Wang M, Gu C, Zhang X, Jiang G. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39:1696–1709. doi: 10.1038/s41388-019-1092-z. [DOI] [PubMed] [Google Scholar]
- 20.Sun R, Liu Z, Tong D, Yang Y, Guo B, Wang X, Zhao L, Huang C. miR-491-5p, mediated by Foxi1, functions as a tumor suppressor by targeting Wnt3a/beta-catenin signaling in the development of gastric cancer. Cell Death Dis. 2017;8:e2714. doi: 10.1038/cddis.2017.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang W, Zhang J, Huang T, Zhou Y, Wong CC, Chan RCK, Dong Y, Wu F, Zhang B, Wu WKK, Chan MWY, Cheng ASL, Yu J, Wong N, Lo KW, To KF. NOTCH3, a crucial target of miR-491-5p/miR-875-5p, promotes gastric carcinogenesis by upregulating PHLDB2 expression and activating Akt pathway. Oncogene. 2021;40:1578–1594. doi: 10.1038/s41388-020-01579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denoyelle C, Lambert B, Meryet-Figuiere M, Vigneron N, Brotin E, Lecerf C, Abeilard E, Giffard F, Louis MH, Gauduchon P, Juin P, Poulain L. miR-491-5p-induced apoptosis in ovarian carcinoma depends on the direct inhibition of both BCL-XL and EGFR leading to BIM activation. Cell Death Dis. 2014;5:e1445. doi: 10.1038/cddis.2014.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Liu Y, Granberg KJ, Wang Q, Moore LM, Ji P, Gumin J, Sulman EP, Calin GA, Haapasalo H, Nykter M, Shmulevich I, Fuller GN, Lang FF, Zhang W. Two mature products of MIR-491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene. 2015;34:1619–1628. doi: 10.1038/onc.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang WC, Chan SH, Jang TH, Chang JW, Ko YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, Juang JL, Wang HC, Cheng AJ, Lu YC, Wang LH. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74:751–764. doi: 10.1158/0008-5472.CAN-13-1297. [DOI] [PubMed] [Google Scholar]
- 25.Giovannini C, Gramantieri L, Chieco P, Minguzzi M, Lago F, Pianetti S, Ramazzotti E, Marcu KB, Bolondi L. Selective ablation of Notch3 in HCC enhances doxorubicin’s death promoting effect by a p53 dependent mechanism. J Hepatol. 2009;50:969–979. doi: 10.1016/j.jhep.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 26.Man CH, Wei-Man Lun S, Wai-Ying Hui J, To KF, Choy KW, Wing-Hung Chan A, Chow C, Tin-Yun Chung G, Tsao SW, Tak-Chun Yip T, Busson P, Lo KW. Inhibition of NOTCH3 signalling significantly enhances sensitivity to cisplatin in EBV-associated nasopharyngeal carcinoma. J Pathol. 2012;226:471–481. doi: 10.1002/path.2997. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Yun R, Yu X, Hu H, Huang G, Tan B, Chen T. Overexpression of Notch3 and pS6 is associated with poor prognosis in human ovarian epithelial cancer. Mediators Inflamm. 2016;2016:5953498. doi: 10.1155/2016/5953498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiu M, Wang Y, Li B, Wang X, Xiao F, Chen S, Zhang L, Zhou B, Hua F. The role of Notch3 signaling in cancer stemness and chemoresistance: molecular mechanisms and targeting strategies. Front Mol Biosci. 2021;8:694141. doi: 10.3389/fmolb.2021.694141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inder S, O’Rourke S, McDermott N, Manecksha R, Finn S, Lynch T, Marignol L. The Notch-3 receptor: a molecular switch to tumorigenesis? Cancer Treat Rev. 2017;60:69–76. doi: 10.1016/j.ctrv.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 30.He J, Huang Z, He M, Liao J, Zhang Q, Wang S, Xie L, Ouyang L, Koeffler HP, Yin D, Liu A. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol Cancer. 2020;19:17. doi: 10.1186/s12943-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.