Abstract
Extracellular vesicles, particularly exosomes, play a vital role via their cargoes. Their potential in pancreatic ductal adenocarcinoma (PDAC), one of the leading causes of cancer-related mortality worldwide is attracting interests. However, the roles and underlying mechanisms of exosomal circular RNAs (circRNAs) in the development of PDAC remain unclear yet. We aimed to illuminate the mechanisms of exosomal hsa_circ_0006790 (thereafter termed circ_6790) released by exosomes (Exo) derived from bone marrow mesenchymal stem cell (BM-MSC) during immune escape in PDAC in this study. BM-MSC-derived Exo inhibited growth, metastasis, and immune escape in PDAC. Exo enhanced circ_6790 expression in PDAC cells. Knockdown of circ_6790 in Exo significantly attenuated the anti-tumor effect of Exo. Circ_6790 facilitated the nuclear translocation of chromobox 7 (CBX7). CBX7 increased the DNA methylation of S100A11 by recruiting DNA methyltransferases to its promoter region, thereby inhibiting the transcription of S100A11. Inhibition of CBX7 or overexpression of S100A11 annulled the inhibitory effects of Exo on PDAC growth, metastasis, and immune escape. In conclusion, our results suggest that MSC-derived exosomal circ_6790 could downregulate S100A11 in PDAC cells and hamper immune escape via CBX7-catalyzed DNA hypermethylation.
Keywords: Bone marrow mesenchymal stem cell-derived exosome, circular RNA_0006790, CBX7, S100A11, pancreatic ductal adenocarcinoma, immunosuppression
Introduction
The pancreas is an organ linking the digestive system with the endocrine system, and pancreatic ductal adenocarcinoma (PDAC) holds accountable for the highest morbidity in both exocrine tissue-derived malignancies and pancreatic cancers [1]. PDAC is unique because there is remarkable desmoplasia, which leads to a large stromal compartment made up of immune cells, inflammatory cells, and fibroblasts, comprising the tumor microenvironment [2]. Immunotherapy has led to a substantial shift in the treatment of several advanced cancers, whereas its efficacy in PDAC is limited, possibly because of the immunosuppressive tumor microenvironment [3]. Therefore, identification of candidates that regulate the immune evasion in PDAC may improve patients’ prognosis and treatment effects.
Exosomes (Exo), cell-derived extracellular vesicles with a diameter between 30-150 nm, transport nucleic acids, proteins, and lipids for intercellular communication into target cells, thus participating in growth and metastasis of tumors by mediating the immune response [4]. Moreover, extracellular vesicles derived from mesenchymal stem cells (MSC) have been described to be superior to intact MSC as a medicinal product since they have a safer profile and can be stored without losing function [5]. Therefore, we chose MSC as our source cell type of Exo to study immune evasion in PDAC. As aforementioned, non-coding RNAs can be distributed among cancer cells and immune cells via extracellular vesicles, thereby influencing main cancer hallmarks, including evasion of the immune system and metastatic dissemination [6]. Circular RNAs (circRNAs), a group of conserved single-stranded RNAs derived from exonic or intronic sequences by precursor mRNA back-splicing, have emerged as a hotspot in the field of tumor pathology [7]. In this report, by comparing PDAC cells treated with MSC-derived Exo or phosphate-buffered saline (PBS), we identified hsa_circ_0006790 (herein termed circ-6790) as the most upregulated one in PDAC cells treated with Exo. RNA-binding proteins (RBPs) are a group of proteins playing major roles in the post-transcriptional regulation of RNAs [8]. For instance, circGOLPH3 could bind to chromobox 7 (CBX7) protein, thus regulating the proliferation and apoptosis of prostate cancer cells [9]. Here, our integrated bioinformatics tools also predicted the binding between CBX7 and circ-6790 in PDAC cells. However, the regulatory mechanism of circ-6790 delivered by Exo on CBX7 in PDAC has not been clearly established. We therefore proposed a hypothesis that circ-6790 incorporated by Exo may regulate the immune evasion in PDAC involving the interaction with CBX7. To address this hypothesis, we isolated Exo from bone marrow MSC (BM-MSC) and conducted co-culture experiments and gain- and loss-of-function assays.
Materials and methods
Cell culture
Human PDAC cells PANC-1 (CL-0184) and CFPAC-1 (CL-0059) were purchased Procell (Wuhan, Hubei, China). PDAC cells were grown in DMEM (PM150210B, Procell) containing 10% FBS and 1% penicillin/streptomycin at 37°C and 5% CO2. Human pancreatic ductal endothelial cells hTERT-HPNE (CRL-4023™) were from ATCC (Manassas, VA, USA) and cultured in DMEM supplemented with 5% FBS, 10 ng/mL human recombinant epidermal growth factor, 5.5 mM D-glucose, and 750 ng/mL puromycin at 37°C in 5% CO2. Human BM-MSC (HUXMF-01001) were acquired from Cyagen (Suzhou, Jiangsu, China) and cultured at 37°C in 5% CO2 using the BM-MSC Complete Medium (HUXMF-90011, Cyagen).
Exo concentration for cell co-culture was 50 μg/mL, and the duration was 24 h. The RNAi of circ_6790 and CBX7 (si-circ and si-CBX7) and negative control (si-Ctr) for cell transfection were acquired from Shanghai GenePharma Co., Ltd. (Shanghai, China). DNA overexpression plasmids of CBX7 and S100A11 (oe-CBX7 and oe-S100A11) and their negative control (oe-Ctr) were purchased from VectorBuilder (Guangzhou, Guangdong, China). Cell transfections were conducted using Lipofectamine 2000 as per the manufacturer’s protocol.
Characterization of BM-MSC and the derived Exo
Surface proteins of BM-MSC were identified by flow cytometry. Briefly, the cells were dissociated by trypsin, resuspended in PBS and counted. Cells (1 × 106) in eppendorf tubes were incubated with phycoerythrin-labeled monoclonal antibodies to CD44 (397503, BioLegend, San Diego, CA, USA), CD73 (344003, BioLegend), CD34 (343505, BioLegend), CD45 (304007, BioLegend) for 0.5 h at room temperature in the dark, and IgG was used as an isotype control (BioLegend). The cells were loaded onto a flow cytometrer (FACScalibur) and analyzed by FlowJo software. Osteogenic differentiation kit (HUXMX-90021, Cyagen) and adipogenic differentiation kit (HUXMX-90031, Cyagen) of BM-MSC were used to identify the differentiation of the cells.
The BM-MSC medium was refreshed with Exo-free medium, and the supernatant was harvested after 48 h, followed by differential centrifugation and filtration to isolate the Exo. In brief, the cell supernatant was centrifuged at 2,000 g for 20 min and at 10,000 g for 40 min, respectively. The supernatant was filtered through a 0.22-μm sterile filter and centrifuged at 100,000 g for 70 min, suspended in PBS, centrifuged at 100,000 g again for 70 min. The obtained Exo resuspended in PBS were subjected to concentration quantification using the BCA Protein Assay Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). The morphology of Exo was observed under TEM, and images were obtained at 120 kV using an HT7700 TEM (Hitachi, Tokyo, Japan). The particle size of Exo was determined by Malvern’s NanoSight NS300, and western blot was used to detect Exo-specific markers.
The knockdown efficiency was detected after 48-h transfection of si-circ and si-Ctr into BM-MSC by RT-qPCR. Subsequently, the medium of the transfected BM-MSC was replaced with Exo-free medium. The Exo were isolated from BM-MSC transfected with si-Ctr and si-circ as described above (named Exo-Ctr and Exo-KD, respectively). The successful establishment of Exo with knock down of circ_6790 was confirmed by RT-qPCR assay of circ_6790 expression in Exo.
Western blot
Total proteins were isolated from cells using RIPA lysis buffer (Beyotime, Shanghai, China), separated by SDS-PAGE, and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk for 60 min, followed by overnight incubation with the specific primary antibodies at 4°C. The membrane was re-probed with the secondary goat anti-rabbit antibody HRP-labeled IgG H&L (1:5,000, ab205718, Abcam) for 120 min at room temperature. Protein bands were observed using the enhanced chemiluminescent Substrate Kit (ab133409, Abcam) and quantified by Quantity One software as per the manufacturer’s protocol. The PARIS™ kit (Invitrogen Inc., Carlsbad, CA, USA) was used to extract cytosolic and cytoplasmic proteins, separately, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a cytoplasmic protein control and Lamin B1 as a nuclear protein control. Primary antibodies included were listed in Table 1.
Table 1.
Antibodies used for western blot
| Antibodies | Concentration | Catalog number | Manufacturer |
|---|---|---|---|
| CD63 | 1 μg/mL | GTX17441 | GeneTex |
| TSG101 | 2 μg/mL | GTX635396 | GeneTex |
| Calnexin | 0.3 μg/mL | GTX109669 | GeneTex |
| PD-L1 | 0.5 μg/mL | ab237726 | Abcam |
| CTLA-4 | 0.45 μg/mL | ab237712 | Abcam |
| CBX7 | 0.66 μg/mL | 26278-1-AP | ProteinTech |
| GAPDH | 0.1 µg/mL | #5174 | Cell Signaling Technologies |
| Lamin B1 | 0.1 µg/mL | ab16048 | Abcam |
| S100A11 | 0.5 µg/mL | ab169530 | Abcam |
| DNMT1 | 0.72µg/mL | ab92314 | Abcam |
| DNMT3A | 0.4 µg/mL | ab228691 | Abcam |
| DNMT3B | 0.4 µg/mL | ab227883 | Abcam |
Note: TSG101, Tumor susceptibility 101; PD-L1, Programmed cell death 1 ligand 1; CTLA-4, Cytotoxic T-lymphocyte protein 4; CBX7, Chromobox protein homolog 7; S100A11, Protein S100-A11; DNMT, DNA-methyltransferase.
Exo labeling and uptake
Exo were labeled using a PKH67 Green Fluorescent Cell Linker Kit (Sigma), and PDAC cells were co-cultured with 50 μg/mL PKH67-labeled Exo for 24 h. PBS was used to rinse off the un-uptaken Exo, followed by staining the nuclei with 5 μg/mL DAPI. Fluorescence images were captured by a confocal microscope to observe the uptake of Exo by PDAC cells.
CCK-8 assay
CCK-8 kits (Beyotime) were utilized to assess cell proliferation. Treated cells were plated at 2,000 cells/well in 96-well plates and cultured for the indicated time points (0, 24, 48, 72 h). Subsequently, the cells in each well were incubated with 10 μL CCK-8 solution at 37°C for 120 min. The OD value at 450 nm was read using a microplate reader.
Colony formation assay
Treated cells were plated at 800 cells/well in six-well plates and incubated at 37°C and 5% CO2 for 2 weeks. The cells were fixed in paraformaldehyde solution for 30 min and stained with crystal violet (Beijing Solarbio Life Sciences Co., Ltd., Beijing, China) for 15 min. The light microscopy was utilized to obtain images of each well, the number of colonies with more than 50 cells were counted.
Flow cytometry
Apoptosis was detected using the Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (Beyotime). Briefly, 1 × 105 resuspended cells were centrifuged at 1,000 g for 5 min and added with 195 μL Annexin V-FITC conjugate. After incubation with 5 μL Annexin V-FITC and 10 μL PI staining solution for 20 min at room temperature in the darkness, the staining was immediately assessed by flow cytometry (FACScalibur) and analyzed by FlowJo software.
Wound healing assay
The treated cells were plated onto 6-well plates (5 × 105 cells/well) and incubated to 80%~90% confluence. The cells were scratched using a 200-μL sterile pipette tip, rinsed with PBS, and then cultured in serum-free medium. Images were viewed under a light microscope (Olympus Optical Co., Ltd., Tokyo, Japan) at 0 and 48 h and photographed, and the migration rate was measured by calculating the total distance the cells migrated from the wound edge to the wound center using ImageJ.
Transwell assay
The invasive ability of the cells was tested using a Transwell insert (Costar Technologies, Inc., Coppell, TX, USA) pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA). The PDAC cells (1 × 105 in 200 µL) in medium without FBS was supplemented to the apical chamber, and 600 µL of medium plus 10% FBS (serum served as chemokine) was supplemented to the basolateral chamber. After 24 h of incubation, the cells on the upper surface of the insert were gently wiped off with a cotton swab, and the cells invading to the lower surface were fixed with 4% paraformaldehyde at room temperature for 15 min and stained with 2% crystal violet staining solution at room temperature for 10 min. The number of cells invaded through the Matrigel was counted under an inverted microscope, and the cells in five random fields of view were calculated using ImageJ software.
Immunosuppression assay
Peripheral blood mononuclear cells (PBMC) were extracted using ficoll-hypaque density gradient centrifugation from blood samples of healthy subjects. PBMC were cultured in RPMI-1640 medium, and T cells were isolated from PBMC by Dynabeads™ Untouched™ Human T Cell Kit (11344D, Thermo Fisher). Human T cells were activated and amplified by Dynabeads™ Human T-Activator CD3/CD28 (11161D, Thermo Fisher). Activated T cells were co-cultured with adherent PDAC cells at 3:1 for 24 h by Transwell inserts. The surviving tumor cells were fixed and stained with crystal violet solution. The crystal violet absorbed by the cells was extracted by adding 10% acetic acid, and the OD value at 595 nm was read on a microplate reader.
ELISA
The levels of the IFN-γ (E4825) and TNF-α (E4603) secreted by T cells co-cultured with tumor cells were determined by ELISA kits (both from BioVision, Inc., Exton, PA, USA) following the manufacturer’s protocols.
Animal assays
Animal manipulates were authorized by the Animal Research Ethics Committee of the China-Japan Union Hospital of Jilin University. All animals received humane care following the NIH guidelines. Sixty 4-5-week-old female BALB/c nude mice [10,11] were from Vital River (Beijing, China) and kept for at least 1 week before use. Nude mice were grouped into 6 groups of 10 mice each: the PBS, Exo, Exo + si-Ctr, Exo + si-CBX7, Exo + oe-Ctr, Exo + oe-S100A11 groups. A total of 1 × 106 CFPAC-1 cells after corresponding treatments suspended in 100 μL mixture of PBS and Matrigel were injected subcutaneously into five randomly selected nude mice. The change in volume of the xenograft tumor was measured weekly as volume = 0.5 × length × width2. The nude mice were euthanized by sodium pentobarbital (150 mg/kg) 4 weeks after transplantation, and tumor weights were harvested and measured.
For in vivo metastases of tumor cells, the remaining 5 nude mice in each group were injected with CFPAC-1 cells (1 × 106) after corresponding treatments via tail vein. After 6 weeks, the nude mice were euthanized by sodium pentobarbital (150 mg/kg), and their liver and lung tissues were collected. The area of metastatic tumor infiltrating in the liver and lung was detected by HE staining.
Thirty four- to five-week-old female C57BL/6 mice were from SLAC (Shanghai, China). The mice were grouped into 6 groups of 5 mice each: the PBS, Exo, Exo + si-Ctr, Exo + si-CBX7, Exo + oe-Ctr, Exo + oe-S100A11 groups. A total of 1 × 106 CFPAC-1 cells of the corresponding group suspended in 100 μL mixture of PBS and Matrigel were injected subcutaneously into C57BL/6 mice. CD8α neutralizing monoclonal antibody was administered intraperitoneally at 100 μg/mouse/3 days. The change in volume of the xenograft tumor was measured weekly as volume = 0.5 × length × width2. Nude mice were euthanized by an overdose of sodium pentobarbital (150 mg/kg) 4 weeks after transplantation, and the weight of harvested tumors was measured.
Immunohistochemistry
Xenograft tumors obtained from C57BL/6 mice were paraffin-embedded and sectioned (4 μm). The sections were dewaxed in xylene, dehydrated in graded ethanol, incubated in 3% H2O2 for 0.5 h, and then heated in citrate buffer (pH = 6.0) for 25 min at 95°C for antigen retrieval. The sections were sealed with 5% BSA at 37°C for 30 min and incubated overnight at 4°C with primary antibodies to CD8 (1:2000, ab209775, Abcam), PD-L1 (1:1.000, ab237726, Abcam), and CTLA-4 (1:500, ab237712, Abcam). Subsequently, the sections were incubated with secondary antibody IgG H&L (HRP) (1:5,000, ab205718, Abcam) for 120 min at room temperature, treated with diaminobenzidine and hematoxylin, and observed under a light microscope. Positive cell rates were quantified by ImageJ software in 5 randomly selected fields of view.
CircRNA microarray analysis
Sample preparation and microarray hybridization were conducted as per the standard protocol (Arraystar Inc., Rockville, MD, USA). In brief, total RNA was digested with RNase R (Epicentre Technologies, Madison, WI, USA) to remove linear RNA and enrich for circRNA. Total RNA was amplified and transcribed into fluorescent cRNA using an Arraystar Super RNA Labeling Kit with random primers. The labeled cRNA was hybridized to the Human Circular RNA Array (Catalog No: AS-S-CR-H-V2.0, 8x15K, Arraystar). After cleaning the slides, the arrays were scanned by an Axon GenePix 4000B microarray scanner. The scanned images were uploaded into GenePix Pro 6.0 software (Axon Instruments, Inc., Union City, CA, USA). When comparing the differences in profiles between the two groups, the “fold change” (i.e., the ratio of group means) between each circRNA group was calculated. |Fold change| ≥ 2 and p-value ≤ 0.05 was defined as having a significant difference in expression.
Fluorescence in situ hybridization (FISH) combined with immunofluorescence
For FISH, RiboBio (Guangzhou, Guangdong, China) was commissioned to synthesize digoxin-labeled probes specific for circ_6790. PDAC cells were fixed and pre-hybridized in a pre-hybridization solution (PBS containing 0.5% Triton X-100). Next, the cells were incubated overnight at 58°C with digoxin-binding probes in hybridization solutions (salmon sperm DNA, yeast tRNA, 40% formamide, 10% dextran sulfate, 1 × Denhardt’s solution, and 4 × sodium chloride-sodium citrate buffer). The cells were incubated overnight at 4°C with the primary antibody to CBX7 (MA5-31669, Thermo Fisher), followed by incubation with goat anti-Mouse IgG (H + L) (Alexa Fluor 488, A-11001, Thermo Fisher) for 120 min at room temperature. Cell nuclei were counter-stained with DAPI, and images were analyzed using a confocal fluorescence microscopy (Olympus).
RT-qPCR
Total RNA was extracted from the whole cell using Trizol (Invitrogen), while the PARIS™ kit (Invitrogen) was applied to isolate total RNA from the nucleus and cytoplasm. Comple-mentary DNA was synthesized using PrimeScript™ RT Master Mix (Takara, Kusatsu, Japan) as per the manufacturer’s instructions. PCR reactions were then performed by TB Green® Fast qPCR Mix (Takara) on an applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific). The relative transcription of the target gene was calculated using the 2-ΔΔCT method with GAPDH used as the internal reference. Table 2 shows the primers used.
Table 2.
Primer used for RT-qPCR
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| circ_6790 | ATGTTTGAACGCATGTCTCTGC | ATCGAGAGAGGGGCACATGA |
| MEMO1 | GAATGCACAGCTAGAAGGTTGGC | TAAGCATGGGCAGCACAAGACC |
| CBX7 | AGGAAGAGAGGTCCGAAACCCA | AGAAGCAGAGCTTCTCCTTGCC |
| S100A11 | CCAGAAGTATGCTGGAAAGGATG | CATCATGCGGTCAAGGACACCA |
| 18s | ACCCGTTGAACCCCATTCGTGA | GCCTCACTAAACCATCCAATCGG |
| U6 | CTCGCTTCGGCAGCACAT | TTTGCGTGTCATCCTTGCG |
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
Note: circRNAs, Circular RNAs; CBX7, Chromobox protein homolog 7; S100A11, Protein S100-A11; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
RNase R treatment
Total RNA (2 μg) after TRIzol reagent extraction was incubated with 3 U/μg RNase R or PBS at 37°C for 20 min, and the expression of MEMO1 or circ_6790 was detected by RT-qPCR.
RIP
The binding between circ_6790 and RBPs was examined using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) as per the instructions provided by the manufacturer. First, PDAC cells were lysed with RIP lysis buffer, and then cell lysates were incubated with RIP buffer containing antibodies to CBX7 (ab91431, Abcam), HNRNPC (PA5-22280, Thermo Fisher), IGF2BP2 (A303-316A, Thermo Fisher) coupled magnetic beads, with IgG as a control (Millipore). The products were digested with proteinase K buffer, and then RNA purification was performed. Finally, the enrichment of circ_6790 was determined by RT-qPCR.
Co-IP
Co-IP assays were carried out to determine if CBX7 interacted with DNMTs. PDAC cells were transfected using HA-tagged fusion protein pellet HA-CBX7, followed by preparation of cell lysates using RIPA buffer. Immunoprecipitation was conducted overnight at 4°C using Anti-HA antibody (1:200, ab9110, Abcam) and Anti-IgG (1:200, ab172730, Abcam). After the reaction with protein A/G agarose beads for 120 min, the beads were rinsed four times with Tris-NACL-EDT buffer. Then, the complexes were eluted by boiling in 2x lithium dodecyl sulfate loading buffer. Finally, western blot assay was carried out to measure the expression of DNMT1, DNMT3A, and DNMT3B.
Chromatin immunoprecipitation (ChIP)
The modification levels of CBX7 and DNMTs on the S100A11 promoter were detected using ChIP Assay Kit (Millipore). Briefly, the cells were cross-linked with 1% formaldehyde at 37°C for 10 min, and the chromatin was sonicated to an average size of 300-500 bp. Chromatin fragments were immunoprecipitated overnight at 4°C with antibodies to CBX7 (ab91431, Abcam), DNMT1 (ab92314, Abcam), DNMT3A (ab228691, Abcam), or DNMT3B (ab227883, Abcam), and IgG was used as a control (Millipore). Immunocomplexes were collected on protein A agarose beads, and the enrichment of the S100A11 promoter was analyzed by qPCR after elution and purification of DNA. The primer sequences used are were follows, S100A11 promoter forward primer: 5’-CTTACTCAGACAGCTTCCGC-3’; reverse primer: 5’-AGAGGGAAAGGAGGGAGAGT-3’.
Methylation-specific PCR (MSP)
MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) was utilized to predict the CpG island on the S100A11 promoter and to design MSP primers. Genomic DNA was extracted and purified from PADC cells by EasyPure® Genomic DNA Kit (TransGen Biotech, Beijing, China). Purified DNA was exposed to bisulfite using the DNA Bisulfite Transformation Kit (TianGen Biotech, Beijing, China) as per the manufacturer’s protocol. MSP of bisulfite-transformed DNA was performed using a nested two-stage PCR method. The amplified PCR products were separated and observed by agarose gel electrophoresis. Forward M primer: 5’-TTTTTTTTAATTTACGTTTTTTTCG-3’, reverse M primer: 5’-ACTCAATTCTATAATTAACGCCGAC-3’. Forward U primer: 5’-TTTTTTTAATTTATGTTTTTTTTGG-3’, reverse U primer: 5’-AAACTCAATTCTATAATTAACACCAAC-3’.
Statistics
Statistical analysis was done in GraphPad Prism 8 (GraphPad, San Diego, CA, USA). The measurement data were summarized as mean ± SD of three independent tests. All statistical comparisons between two groups were carried out using unpaired t-test unless stated otherwise. All statistical comparisons between three or more groups were done using one-way or two-way ANOVA, followed by Tukey’s multiple comparison tests. Statistical significance was defined as P < 0.05.
Results
Characterization of BM-MSC and the derived Exo
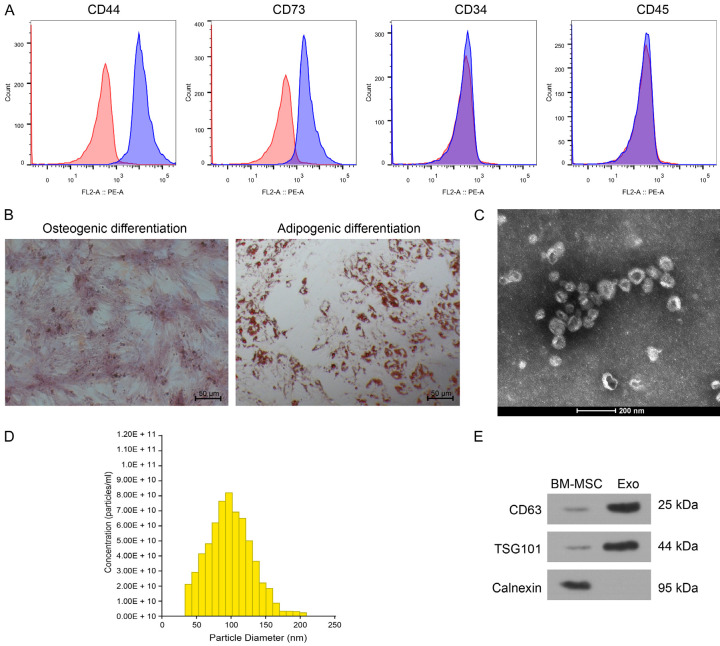
Phenotypic markers of purchased BM-MSC were detected by flow cytometry, and we found that CD44 and CD73 were positive; CD34 and CD45 were negative (Figure 1A). BM-MSC can differentiate into osteoblasts and adipocytes with good differentiation ability after induction culture (Figure 1B). The Exo isolated from BM-MSC showed a typical elliptical bilayer structure (Figure 1C), and the particle size was mainly distributed around 100 nm (Figure 1D). It was confirmed by western blot that the isolated Exo expressed the positive markers TSG101 and CD63, instead of the negative marker Calnexin (Figure 1E).
Figure 1.
Characterization of BM-MSC and derived Exo. A. Flow cytometry identification of surface markers of BM-MSC. B. Osteogenic differentiation of BM-MSC by alizarin red staining (left) and adipogenic differentiation of BM-MSC by oil red O staining (right). C. TEM observation of Exo structure. D. NTA analysis of Exo particle size. E. The expression of exosomal markers TSG101 and CD63 for derived Exo and BM-MSC was determined by Western blot. The data are a representative of 3 independent experiments.
Exo suppress the growth and metastasis of PDAC cells
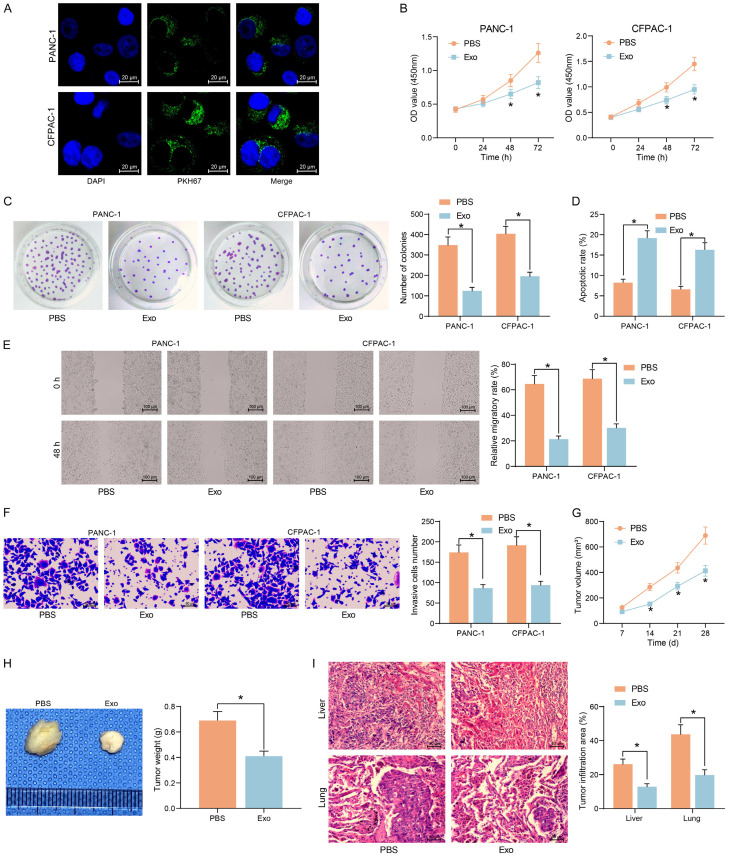
Using the internalization assay, we observed a significant uptake of Exo by PDAC cell lines after 24 h of Exo treatment (Figure 2A). Compared with PBS treatment, Exo treatment significantly inhibited the proliferative capacity (Figure 2B) and clonogenic capacity (Figure 2C) of PDAC cells. In contrast, the apoptosis of cells was significantly increased after Exo treatment (Figure 2D). The migration and invasion of cells were also significantly reduced upon Exo co-culture (Figure 2E, 2F).
Figure 2.
Exo repress the growth and migration of PDAC cells in vitro and in vivo. (A) Immunofluorescence detection of Exo uptake by PDAC cells. (B) The proliferation of cells measured by CCK8 assay. (C) The clonogenic ability of cells examined using colony formation assay. (D) Flow cytometry analysis of apoptosis rate of cells. (E) The cell migration of cells examined using wound healing assay. (F) Cell invasiveness examined using Transwell assays. (G) Volume changes of xenograft tumors in nude mice. (H) Weight of xenograft tumors. (I) The area of tumor infiltration in the liver and lungs of nude mice. *P < 0.05. Data are shown as mean ± SD of three technical replicates (n = 5). Unpaired t-test (H) and two-way ANOVA (B-G, I) were used for data comparison.
Subsequently, CFPAC-1 cells with a higher degree of malignancy were selected for in vivo experiments. A tumor xenograft model was established by subcutaneously injecting CFPAC-1 into nude mice, and the in vivo growth rate of Exo-treated tumor cells was significantly slowed down (Figure 2G), and the weight of harvested tumors were significantly decreased (Figure 2H). A tumor metastasis model was generated by tail vein injection of CFPAC-1 into nude mice. Using HE staining, we observed a significant decline in the area of infiltrating tumors in the lung and liver, indicating a significant decline in the metastatic capacity of Exo-treated tumor cells (Figure 2I).
Exo alleviate the immune escape and immunosuppressive microenvironment of PDAC
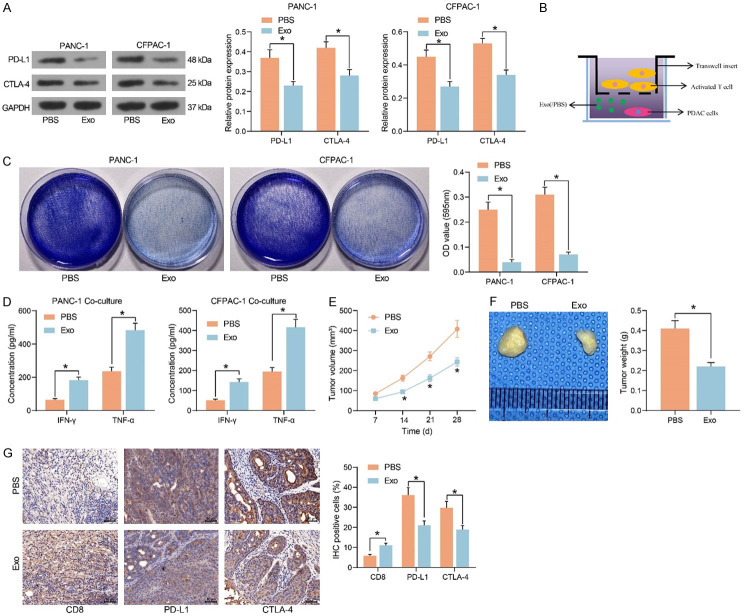
The impact of Exo treatment on the expression of tumor immune checkpoint PD-L1 and CTLA4 in PDAC cells was detected by western blot (Figure 3A). Exo significantly inhibited the expression of PD-L1 and CTLA-4 and hampered immune escape of tumor cells. Next, we co-cultured activated T cells with PDAC cells treated with Exo or not (Figure 3B). We found that Exo-treated PDAC cells were more sensitive to the killing effect of T cells by crystal violet staining (Figure 3C). Meanwhile, the levels of immune effectors IFN-γ and TNF-α secreted by T cells co-cultured with Exo-treated PADC cells were appreciably boosted, and the immunosuppressive effect of tumor cells was considerably diminished (Figure 3D).
Figure 3.
Exo repress immune escape and immunosuppression in PDAC. (A) The protein expression of PD-L1 and CTLA-4 in PDAC cells treated with PBS or Exo. (B) Co-culture Exo- or PBS-treated PDAC cells with activated T cells. (C) Surviving PDAC cells examined using crystal violet staining. (D) The levels of IFN-γ and TNF-α, the immune effectors released by T cells, examined by ELISA. (E) Volume changes of xenograft tumors. (F) Weight of xenograft tumors. (G) Immunohistochemical detection of CD8+ T cell infiltration and expression of PD-L1 and CTLA-4 in xenograft tumors. *P < 0.05. Data are shown as mean ± SD of three technical replicates (n = 5). Unpaired t-test (F) and two-way ANOVA (A, C-E, G) were used for data comparison.
CFPAC-1 cells with or without Exo treatment were injected into C57BL/6 mice, followed by the administration of CD8 mAB. We observed a significant decline in tumor growth rate (Figure 3E) and a decline in the weight of harvested xenograft tumors in the mice injected with cells co-cultured with Exo (Figure 3F). By immunohistochemical assay, we observed that CD8+ T cell infiltration was significantly enhanced in the tumor tissues, whereas the expression of PD-L1 and CTLA4 was significantly diminished in the mice injected with cells co-cultured with Exo (Figure 3G).
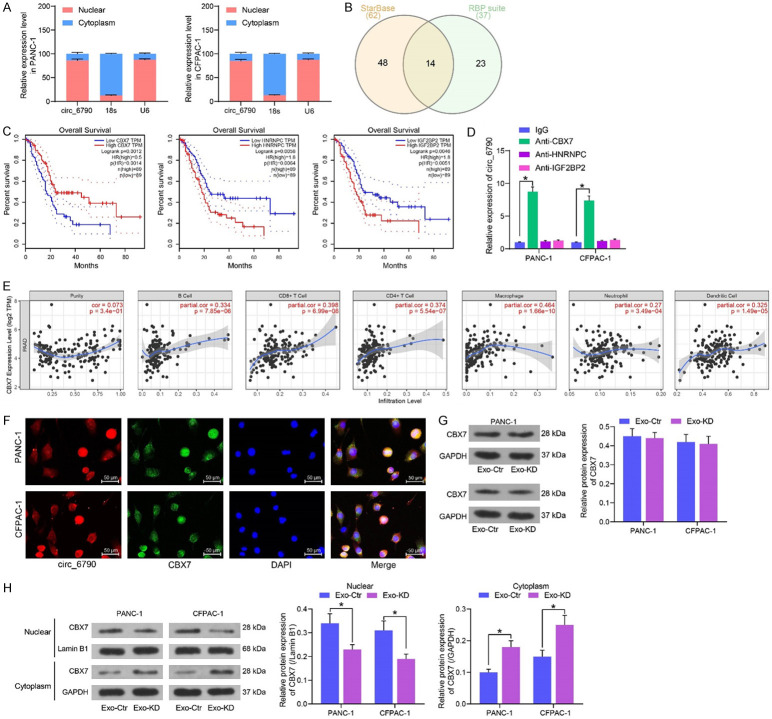
Exo treatment increases the expression of circ_6790 in PDAC cells
We examined the effect of Exo treatment on circRNA expression in CFPAC-1 cells by microarray analysis (Figure 4A). A substantial promotion in the hsa_circ_0006790 expression was observed in the cells after Exo treatment. In Circular RNA Interactome (https://circinteractome.irp.nia.nih.gov/index.html), hsa_circ_0006790 linear mRNA was MEMO1 with a spliced sequence length of 225 bp (Figure 4B). For further confirmation of the microarray results, we conducted RT-qPCR. The expression of circ_6790 was also significantly elevated in Exo-treated PANC-1 cells (Figure 4C). In exoRBase 2.0 (http://www.exorbase.org/exoRBaseV2/toIndex), we found that circ_6790 was present in exosomes from blood and urine samples (Figure 4D), but whether circ_6790 was present in exosomes from MSC samples has not been studied. We then measured the expression of circ_6790 in both BM-MSC and Exo, and the enrichment of circ_6790 was significantly higher in Exo (Figure 4E). In contrast, the expression of circ_6790 was much lower in PDAC cells than that in hTERT-HPNE cells (Figure 4F). RNase treatment significantly degraded linear mRNA MEMO1 in PDAC cells without significant effect on circ_6790 expression (Figure 4G).
Figure 4.
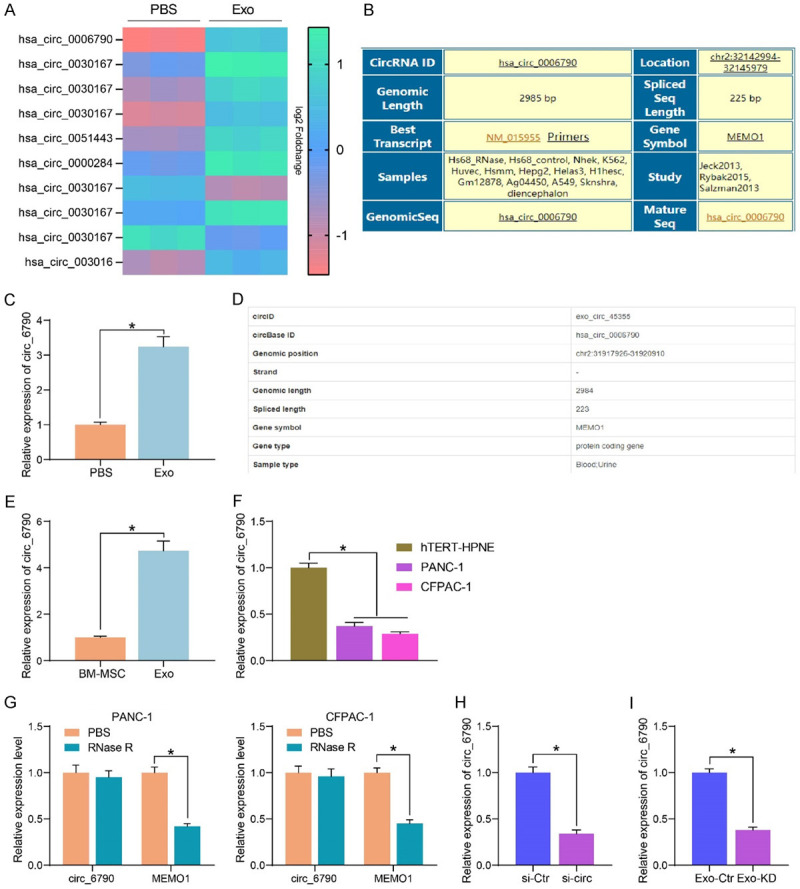
Identification of circRNA expression profiles in PDAC cells treated with Exo. (A) Microarray analysis of the effect of Exo treatment on circRNA expression in CFPAC-1 cells. (B) Sequence information of circ_6790. (C) circ_6790 expression in PANC-1 cells upon Exo treatment determined using RT-qPCR. (D) circ_6790 is present in exosomes. (E) The enrichment of circ_6790 in Exo and BM-MSC examined using RT-qPCR. (F) Detection of circ_6790 expression in PDAC cells by RT-qPCR. (G) The expression of circ_6790 and MEMO1 in PDAC cells upon RNase R treatment measured using RT-qPCR. (H) The expression profile of circ_6790 in BM-MSC upon si-circ treatment measured using RT-qPCR. (I) Detection of circ_6790 expression in Exo-Ctr and Exo-KD by RT-qPCR. *P < 0.05. Data are shown as mean ± SD of three technical replicates. Unpaired t-test (C, E, H, I) and one-way (F) or two-way ANOVA (G) were used for data comparison.
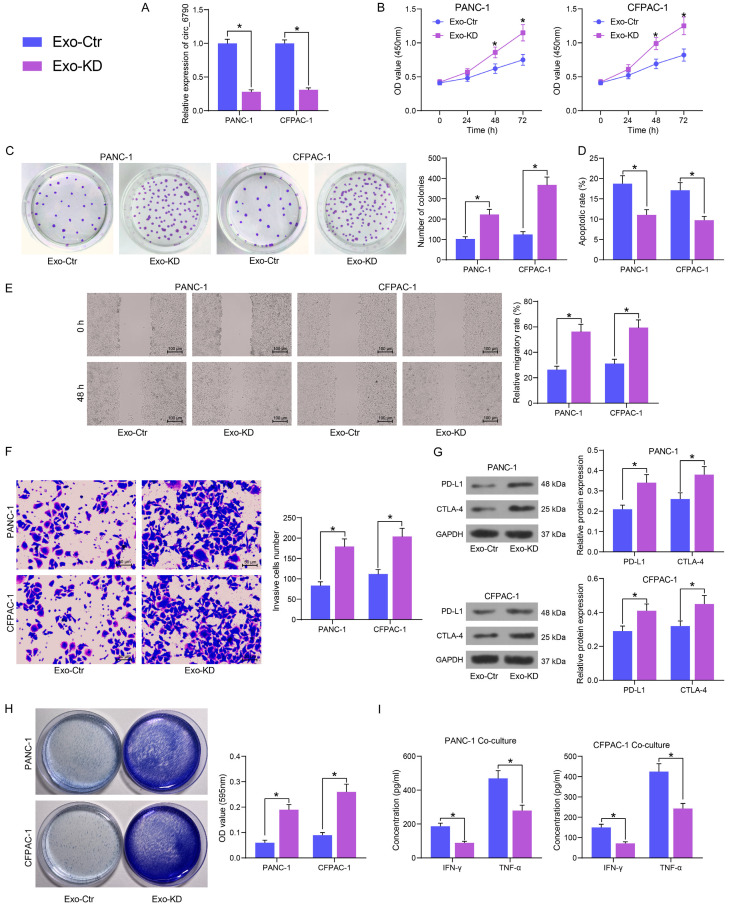
The expression of circ-6790 was inhibited by transfection of circ-6790 siRNA into BM-MSC (with those transfected with si-Ctr as control) (Figure 4H), and the Exo isolated from transfected BM-MSC were named Exo-Ctr and Exo-KD, respectively. The circ-6790 expression was much lower in Exo-KD than that in Exo-Ctr (Figure 4I).
Knockdown of circ_6790 reduces the anti-tumor effect of Exo in PDAC
PDAC cells were treated with both Exo-Ctr and Exo-KD, and the expression of circ_6790 was appreciably reduced in Exo-KD-treated PDAC cells compared to Exo-Ctr-treated ones (Figure 5A). By CCK8 and colony formation assays, we detected that depletion of circ_6790 considerably diminished the inhibitory ability of Exo on the proliferation and colony formation of PDAC cells (Figure 5B, 5C). Knockdown of circ_6790 also resulted in a significant attenuation of the pro-apoptotic effect of Exo on cell apoptosis and the repressive effect on cell migration and invasion (Figure 5D, 5F).
Figure 5.
Circ_6790 downregulation expedites the malignant phenotype and immune evasion of PDAC cells. A. Circ_6790 expression in PDAC cells after Exo treatment by RT-qPCR. B. The proliferation of cells measured by CCK8 assay. C. The clonogenic ability of cells examined using colony formation assay. D. The apoptosis rate of cells examined using flow cytometry. E. The cell migration of cells examined using wound healing assay. F. Cell invasiveness examined using Transwell assays. G. The PD-L1 and CTLA-4 protein expression in PDAC cells by Western blot. H. Surviving PDAC cells examined using crystal violet staining. I. The levels of IFN-γ and TNF-α, the immune effectors released by T cells, examined by ELISA. *P < 0.05. Data are shown as mean ± SD of three technical replicates. Two-way ANOVA was used for data comparison.
Knockdown of circ_6790 also reduced the inhibitory effect of Exo on PD-L1 and CTLA-4 expression in tumor cells (Figure 5G) and conferred the tumor cells resistance to the killing effect of T cells (Figure 5H). The secretion of IFN-γ and TNF-α by T cells was also significantly reduced following the downregulation of circ_6790 in Exo (Figure 5I).
Circ_6790 co-localizes with CBX7 in the nucleus of PDAC cells
To investigate the mechanism of action of circ_6790, we first determined the localization of circ_6790 by subcellular fractionation, and we observed that circ_6790 was primarily localized in the nucleus of PDAC cells (Figure 6A). We analyzed the RBPs that circ_6790 can bind to in StarBase (https://starbase.sysu.edu.cn/) and RBP suite (http://www.csbio.sjtu.edu.cn/bioinf/RBPsuite/), with a total of 14 intersections (Figure 6B). Subsequently, we analyzed the correlation between the expression of these factors and the survival of PDAC patients in GEPIA (http://gepia.cancer-pku.cn/index.html), where the expression of CBX7, HNRNPC, IGF2BP2 was significantly correlated with the survival of PDAC patients (Figure 6C). More specifically, survival rate was higher in patients with high CBX7 expression and worse in those with high HNRNPC or IGF2BP2 expression. We performed RIP experiments in PDAC cells. CBX7 significantly bound to circ-6790, whereas HNRNPC and IGF2BP2 had no significant enrichment effect on circ-6790 (Figure 6D). Meanwhile, TIMER website (https://cistrome.shinyapps.io/timer/) revealed that the expression of CBX7 was significantly and positively correlated with the level of immune infiltration in PDAC (Figure 6E).
Figure 6.
circ_6790 promotes CBX7 nuclear translocation. A. The localization of circ_6790 in PDAC cells examined using subcellular fractionation. B. The RBP of circ_6790 predicted using StarBase and RBP suite. C. Correlation of CBX7, HNRNPC, IGF2BP2 expression and survival of PDAC patients. D. The binding ability of CBX7, HNRNPC, and IGF2BP2 to circ_6790 assessed using RIP assay. E. Correlation of CBX7 expression with the level of immune infiltration of PDAC. F. Co-localization of circ_6790 and CBX7 detected by FISH combined with immunofluorescence. G. Western blot detection of CBX7 protein expression in PDAC cells co-cultured with Exo. H. Western blot detection of CBX7 protein expression in the cytoplasm and nucleus of PDAC cells co-cultured with Exo. *P < 0.05. Data are shown as mean ± SD of three technical replicates. Two-way ANOVA was used for data comparison.
By FISH combined with immunofluorescence assays, we found that circ_6790 and CBX7 were distributed in both the nucleus and cytoplasm (mainly in the nucleus), and co-localization of both occurred mainly in the nucleus (Figure 6F). There was no significant change in CBX7 expression in PDAC cells treated with Exo-KD or Exo-Ctr, indicating that circ_6790 expression did not affect the expression of CBX7 (Figure 6G). However, subcellular fractionation assays revealed that knockdown of circ_6790 significantly reduced CBX7 nuclear translocation (Figure 6H).
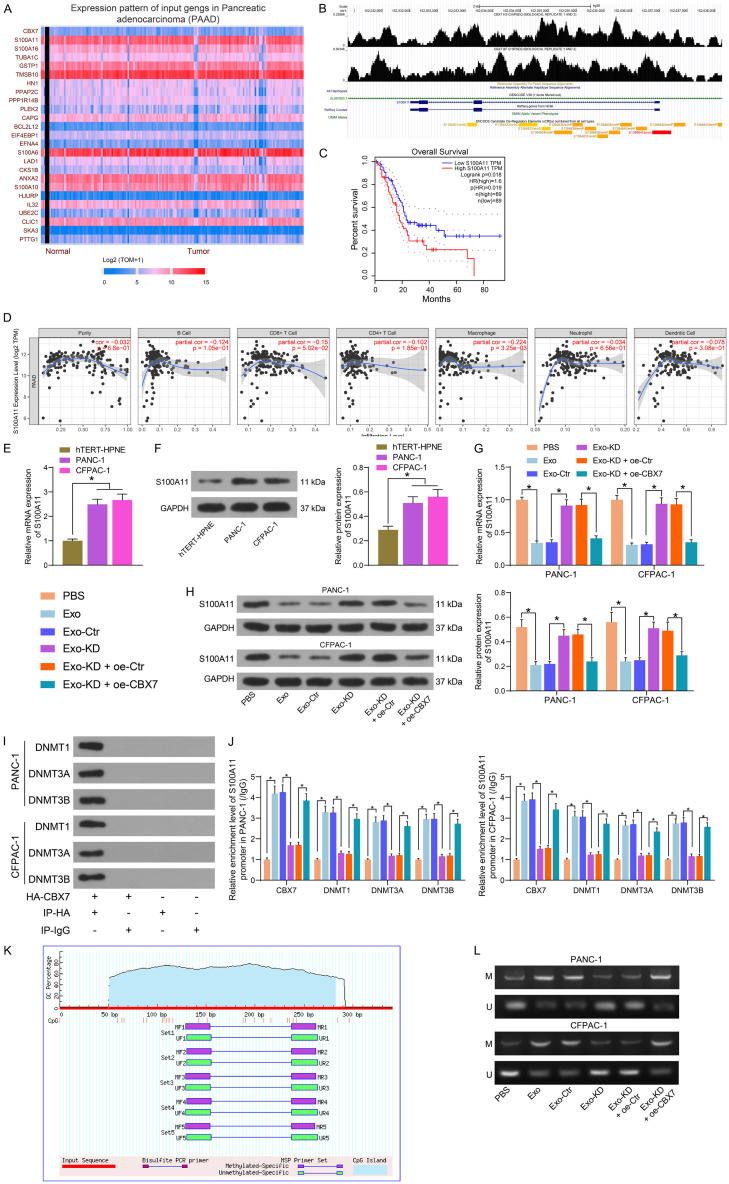
DNA methylation level of S100A11 is co-regulated by circ_6790 and CBX7
We first analyzed the genes negatively associated with CBX7 expression in PDAC in UALCAN (http://ualcan.path.uab.edu/index.html) (Figure 7A). Among them, S100A11 was the factor with the highest degree of correlation. In the ChIP-seq database Cistrome Data Browser (http://cistrome.org/db/#/), we observed a binding relationship between CBX7 and S100A11 promoter (Figure 7B). GEPIA database query revealed that PDAC patients with high S100A11 expression tended to have lower survival rates (Figure 7C). Additionally, a negative correlation between S100A11 expression and the level of immune infiltration in PDAC was observed in the TIMER website (Figure 7D). Through RT-qPCR and western blot, we detected a substantial elevation of S100A11 expression in PDAC cells (Figure 7E, 7F). We transfected CBX7 overexpression DNA plasmids into Exo-KD-treated PDAC cells. As expected, Exo treatment considerably diminished the expression profile of S100A11 in PDAC cells. The inhibitory effect of Exo on S100A11 expression in PDAC cells was significantly attenuated after knockdown of circ_6790 expression in Exo, while S100A11 expression was repressed again after overexpression of CBX7 (Figure 7G, 7H).
Figure 7.
circ_6790 regulates the expression of S100A11 through CBX7. (A) Differentially expressed genes in PDAC that were significantly and negatively associated with CBX7. (B) ChIP-seq database analysis of the binding of CBX7 to the S100A11 promoter. (C) Correlation between S100A11 expression and survival of PDAC patients. (D) Correlation between the expression of S100A11 and the level of immune cell infiltration in PDAC tissues. (E, F) Detection of S100A11 expression in PDAC cells and hTERT-HPNE cells by RT-qPCR (E) and western blot (F). (G, H) Detection of S100A11 expression in PDAC cells under different treatments by RT-qPCR (G) and western blot (H). (I) Co-IP detection of the binding relationship between CBX7 and DNMTs. (J) The enrichment ability of CBX7 and DNMTs for S100A11 promoter in each group of cells examined using ChIP-qPCR. (K) CpG site on S100A11 promoter. (L) The DNA methylation level of S100A11 promoter in each group of cells examined using MSP. *P < 0.05. Data are shown as mean ± SD of three technical replicates. One-way (E, F) or two-way ANOVA (G, H, J) were used for data comparison.
The enrichment ability of CBX7 on DNMTs was examined by Co-IP assays, and we detected a binding relationship between CBX7 and DNMTs in PDAC cells (Figure 7I). The levels of CBX7 and DNMTs on the S100A11 promoter were measured by ChIP-qPCR assay (Figure 7J). Exo treatment significantly increased CBX7, DNMT1, DNMT3A, and DNMT3B occupancy on the S100A11 promoter. Knockdown of circ_6790 significantly reduced the enrichment of relevant factors on the S100A11 promoter, which could be rescued by overexpression of CBX7. We predicted the CpG site on the S100A11 promoter in MethPrimer and designed MSP primers (Figure 7K). The DNA methylation levels of S100A11 promoter were detected in PADC cells by MSP assay (Figure 7L). We observed that Exo treatment drastically boosted the DNA methylation level of the S100A11 promoter. The promotion of DNA methylation level of S100A11 promoter by Exo was inhibited by knocking down circ_6790, which was increased again by overexpression of CBX7.
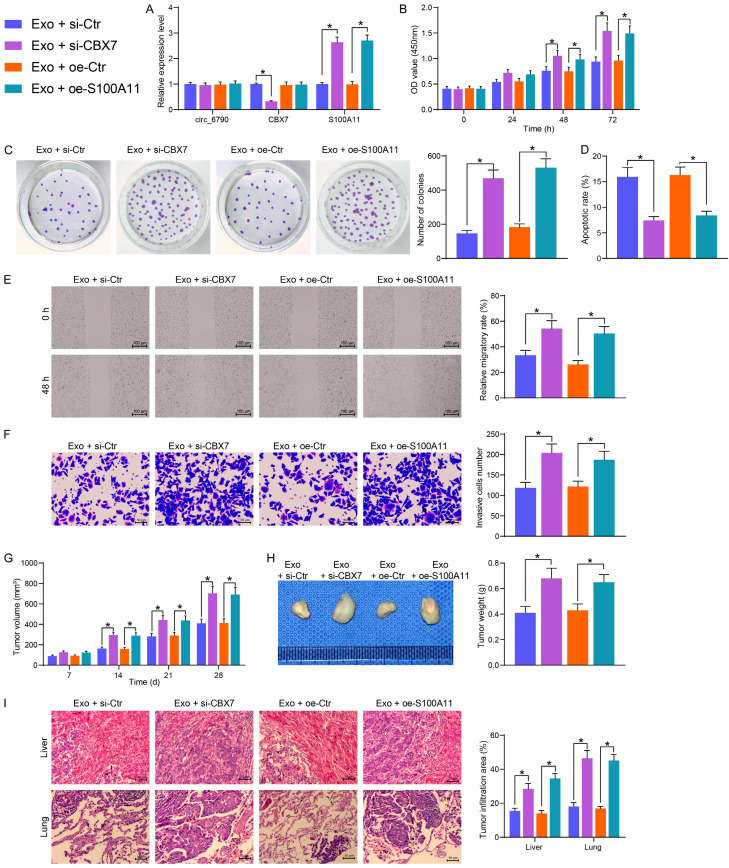
Circ_6790 hinders PDAC cell growth and metastasis by regulating S100A11 expression through binding to CBX7
Exo-treated PDAC cells were transfected with siRNA of CBX7 or overexpressed DNA plasmid of S100A11, and the expression of relevant factors was assessed using RT-qPCR (Figure 8A). No substantial changes were observed in the expression of circ_6790 in each group of cells. si-CBX7 inhibited the expression of CBX7 and elevated the expression of S100A11. Oe-S100A11 did not affect the expression of circ_6790 and CBX7, but it appreciably amplified the expression of S100A11 in the cells. Suppression of CBX7 or restoration of S100A11 mitigated the tumor-suppressive role of Exo, leading to the restoration of proliferation and clonogenic abilities of PDAC cells (Figure 8B, 8C). Moreover, a significant increase in the cell migration and invasion (Figure 8D, 8E), and a decrease in the level of apoptosis were observed in the PDAC cells transfected with si-CBX7 or oe-S100A11 (Figure 8F).
Figure 8.
The circ_6790/CBX7/S100A11 axis regulates the growth and metastases of PDAC. (A) circ_6790, CBX7, and S100A11 expression in PDAC cells after different treatments examined using RT-qPCR. (B) The proliferation of cells evaluated by CCK8 assay. (C) The clonogenic ability of cells examined using colony formation assay. (D) The apoptosis rate of cells by flow cytometry. (E) The cell migration of cells examined using wound healing assay. (F) Cell invasiveness examined using Transwell assays. (G) Volume changes of xenograft tumors. (H) Weight of xenograft tumors. (I) The area of tumor infiltration in the liver and lungs of nude mice. *P < 0.05. Data are shown as mean ± SD of three technical replicates (n = 5). One-way (C-F, H) or two-way ANOVA (A, B, G, I) were used for data comparison.
In in vivo experiments, inhibition of CBX7 or overexpression of S100A11 both significantly promoted tumor growth, resulting in heavier xenograft tumors (Figure 8G, 8H). Si-CBX7 or oe-S100A11 transfection in PDAC cells also led to increased infiltration of tumors into the lung and liver of nude mice (Figure 8I).
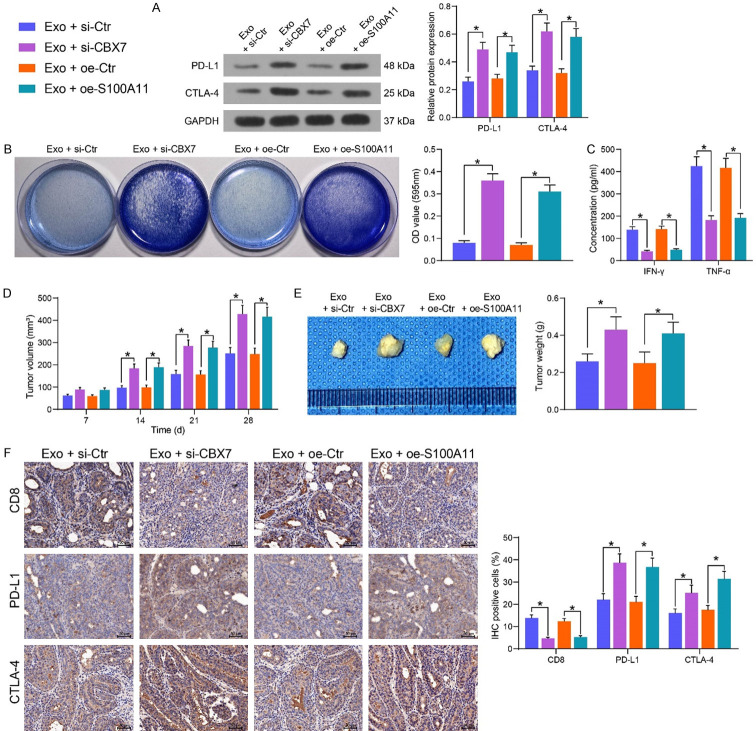
Circ_6790 affects immunosuppressive microenvironment of PDAC by regulating S100A11 expression through binding to CBX7
By western blot detection of PD-L1 and CTLA4 expression in CFPAC-1 cells transfected with si-CBX7 or oe-S100A11 in the presence of Exo, we observed that inhibition of CBX7 or upregulation of S100A11 considerably augmented the expression of both proteins in the cells (Figure 9A). The cells were co-cultured with activated T cells, followed by crystal violet staining. Inhibition of CBX7 or overexpression of S100A11 supported tumor cell evasion from the killing effects of T cells (Figure 9B). The levels of the immune effectors IFN-γ and TNF-α secreted by T cells were significantly reduced under the influence of si-CBX7 or oe-S100A11 (Figure 9C).
Figure 9.
The circ_6790/CBX7/S100A11 axis regulates immune escape of PDAC cells. (A) The protein expression of PD-L1 and CTLA-4 in PDAC cells. (B) Surviving PDAC cells examined using crystal violet staining. (C) The levels of IFN-γ and TNF-α, the immune effectors released by T cells, examined by ELISA. (D) Volume changes of xenograft tumors. (E) Weight of xenograft tumors. (F) Immunohistochemical detection of CD8+ T cell infiltration and expression of PD-L1 and CTLA-4 in xenograft tumors. *P < 0.05. Data are shown as mean ± SD of three technical replicates (n = 5). One-way (B, E) or two-way ANOVA (A, C, D, F) were used for data comparison.
Cells under different treatments were injected into C57BL/6 mice, followed by CD8 mAB administration. Tumor growth rate was accelerated upon inhibition of CBX7 or overexpression of S100A11 (Figure 9D), and we harvested heavier xenograft tumors (Figure 9E). Moreover, a significant decline in CD8+ T cell infiltration in tumor tissues was observed, which occurred concomitant with increased expression of PD-L1 and CTLA4 (Figure 9F).
Discussion
CircRNAs have attracted increasing attention over the past decade since they can mediate gene expression via different mechanisms and play versatile roles in various aspects of human cancer biology, and circRNAs are found enriched in Exo and the exosomal circRNAs play an important role in cancer biology [12]. Moreover, either mediation of the immune response, or targeting of epigenetic modifications alone or combined with chemotherapy, may open highly powerful therapeutic avenues in PDAC [13]. In the present study, we innovatively showed that BM-MSC deliver circ_6790 through Exo in PDAC cells, thereby downregulating S100A11 expression via CBX7-recurited DNMTs, and ultimately perturbs the immune escape of PDAC (Figure 10).
Figure 10.
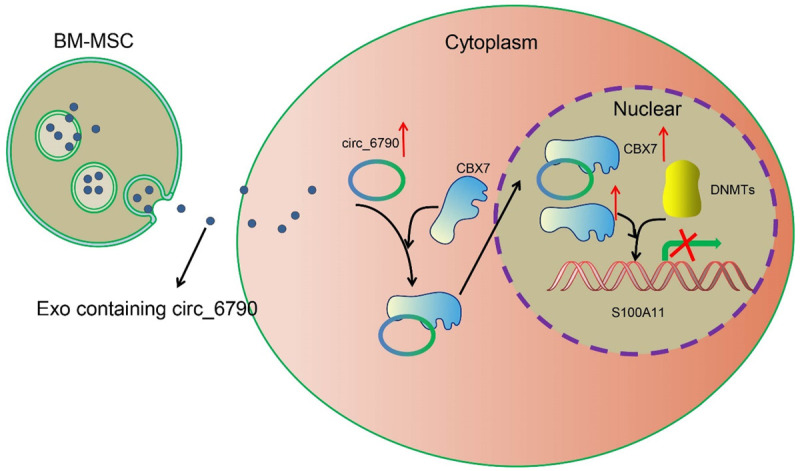
The mechanism diagram. BM-MSC-derived Exo repress the transcriptional expression of S100A11 by carrying circ_6790 that facilitates the CBX7 nuclear translocation, thereby ameliorating immune escape and immunosuppression in PDAC.
Exosomes can be supporters of tumor growth and invasion by benefitting the establishment of an immunosuppressive microenvironment and agents by promoting eradication of tumor cells via CD4+ and CD8+ T cells [14]. In the present study, we presented evidence not only showing the anti-tumor effects of Exo derived from BM-MSC in vitro and in vivo, but also demonstrating their supporting role in enhancing the killing effects of activated T cells. Inhibitory checkpoint molecules involving PD-L1 and CTLA-4 are upregulated during tumorigenesis, which are essential to the diminution of the immune system by blocking the activation of T cells [15]. We demonstrated here that Exo diminished the levels of PD-L1 and CTLA-4 in PDAC cells co-cultured with Exo and T cells in addition to reducing the secretion of IFN-γ and TNF-α. Gingival MSC-Exo have also been reported to reduce the release of inflammatory factors and support the polarization of pro-inflammatory macrophages into anti-inflammatory phenotype [16].
Circ_6790 (also known as circMEMO1) has been revealed subsequently as the cargo of MSC-Exo and has been found to be responsible for the anti-immunosuppressive effects of Exo in the present study. CircMEMO1 expression has been recently identified to be significantly reduced in hepatocellular carcinoma tissues, whereas circMEMO1 hampered cell proliferation, migration and invasion in hepatocellular carcinoma cells in vitro and in vivo [17]. However, its role regarding immunoregulation remains largely unclear. Here we observed the knockdown of circ_6790 in the source cell BM-MSC contributed to a decline in its expression in PDAC cells co-cultured with derived Exo and a decrease in the effects of Exo on immunoregulation. Even though most circRNA is located in cytoplasm (competing endogenous RNAs), some are existed in nucleus which can serve as transcriptional or splicing regulators to interfere with gene expression and involve in alternative splicing and transcription process [18]. RBPs are a group of proteins involved in gene transcription and translation, and interaction with RBPs is regarded as a significant part of circRNA function, including translation and transcriptional regulation of their targets [19]. Here, we determined the nuclear localization of circ_6790 in PDAC cells using both subcellular fractionation and FISH. To characterize the role of circ_6790 in PDAC cell immune evasion, we used two websites for the searching of RBP of circ_6790. After survival analysis and RIP assays, we identified CBX7 as the candidate, which also shares a positive relation with immune infiltration in PDAC. CBX7 has been summarized recently to play a dual role in cancers and can interact with different RNAs, including circRNAs in different cancer environments to affect the development of cancers [20]. Under the condition of PDAC, Karamitopoulou et al. suggested that depletion of CBX7 was correlated with a more aggressive phenotype [21]. In the present study, we determined the co-localization of CBX7 and circ_6790 in the nuclear of PDAC cells and proposed a possible mechanism that circ_6790 interacted with CBX7 and enhanced its nuclear translocation in PDAC cells.
Mohammad et al. reported the coimmunoprecipitation between DNMT1, DNMT3b, and CBX7 [22]. Therefore, we searched for genes with negative correlation with CBX7 in the UALCAN website, and S100A11 was the factor with the highest degree of correlation. We thus explored whether CBX7 bound to DNMTs to regulate S100A11 expression. Using ChIP-qPCR assays, we successfully confirmed that Exo treatment enhanced the levels of CBX7, DNMT1, DNMT3A, and DNMT3B in the promoter of S100A11, which was reduced by loss of circ_6790. However, CBX7 contributed to restoration of the enrichment of these factors in the S100A11 promoter region, preliminarily attest our hypothesis. Intriguingly, elevated S100A11 mRNA levels were associated with poor overall survival in glioblastoma patients, and S100A11 expression was negatively correlated with its corresponding methylation status [23]. This was largely in line with our MSP assay. Ohuchida et al. has revealed that S100A11 was overexpressed in the early stage of pancreatic carcinogenesis, which might serve as an effective tool for screening of patients with high-risk lesions that could develop into pancreatic cancer [24]. The latest report showed that S100A11 may impair the infiltration and cytolytic activity of CD8+ T cells in pancreatic cancer [25]. As regards to CBX7, more apoptotic cells were detected in CD4+ T cells with the CBX7 knockdown [26]. Our rescue experiments demonstrated that depletion of CBX7 or overexpression of S100A11 could mitigate the regulatory role of Exo on the malignant phenotype and immune evasion of PDAC cells.
In conclusion, the aforementioned findings demonstrated that Exo secreted by BM-MSC transfer circ_6790 into PDAC cells to suppress the invasion, migration, growth, and immune escape of PDAC cells through the downregulation of S100A11 via CBX7-recuited DNMTs. Therefore, Exo from BM-MSC with upregulation of circ_6790 could be a potential therapy for PDAC.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (82073299).
Disclosure of conflict of interest
None.
References
- 1.Uddin MH, Al-Hallak MN, Philip PA, Mohammad RM, Viola N, Wagner KU, Azmi AS. Exosomal microRNA in pancreatic cancer diagnosis, prognosis, and treatment: from bench to bedside. Cancers (Basel) 2021;13:2777. doi: 10.3390/cancers13112777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lafaro KJ, Melstrom LG. The paradoxical web of pancreatic cancer tumor microenvironment. Am J Pathol. 2019;189:44–57. doi: 10.1016/j.ajpath.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Timmer FEF, Geboers B, Nieuwenhuizen S, Dijkstra M, Schouten EAC, Puijk RS, de Vries JJJ, van den Tol MP, Bruynzeel AME, Streppel MM, Wilmink JW, van der Vliet HJ, Meijerink MR, Scheffer HJ, de Gruijl TD. Pancreatic cancer and immunotherapy: a clinical overview. Cancers (Basel) 2021;13:4138. doi: 10.3390/cancers13164138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, Fu W, Yi J, Wang J, Du G. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. 2021;11:2783–2797. doi: 10.1016/j.apsb.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rani S, Ryan AE, Griffin MD, Ritter T. Mesenchymal stem cell-derived extracellular vesicles: toward cell-free therapeutic applications. Mol Ther. 2015;23:812–823. doi: 10.1038/mt.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reese M, Dhayat SA. Small extracellular vesicle non-coding RNAs in pancreatic cancer: molecular mechanisms and clinical implications. J Hematol Oncol. 2021;14:141. doi: 10.1186/s13045-021-01149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. 2021;19:910–928. doi: 10.1016/j.csbj.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B, Li Z, Qin C, Li T, Wang Y, Cao H, Yang X, Wang W. Mobius strip in pancreatic cancer: biogenesis, function and clinical significance of circular RNAs. Cell Mol Life Sci. 2021;78:6201–6213. doi: 10.1007/s00018-021-03908-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong L, Tang Y, Jiang L, Tang W, Luo S. Regulation of circGOLPH3 and its binding protein CBX7 on the proliferation and apoptosis of prostate cancer cells. Biosci Rep. 2020;40:SR20200936. doi: 10.1042/BSR20200936. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Roca MS, Moccia T, Iannelli F, Testa C, Vitagliano C, Minopoli M, Camerlingo R, De Riso G, De Cecio R, Bruzzese F, Conte M, Altucci L, Di Gennaro E, Avallone A, Leone A, Budillon A. HDAC class I inhibitor domatinostat sensitizes pancreatic cancer to chemotherapy by targeting cancer stem cell compartment via FOXM1 modulation. J Exp Clin Cancer Res. 2022;41:83. doi: 10.1186/s13046-022-02295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Xu HX, Cho WCS, Cheuk W, Li Y, Huang QH, Yang W, Xian YF, Lin ZX. Brucein D augments the chemosensitivity of gemcitabine in pancreatic cancer via inhibiting the Nrf2 pathway. J Exp Clin Cancer Res. 2022;41:90. doi: 10.1186/s13046-022-02270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. circRNAs and exosomes: a mysterious frontier for human cancer. Mol Ther Nucleic Acids. 2020;19:384–392. doi: 10.1016/j.omtn.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hessmann E, Johnsen SA, Siveke JT, Ellenrieder V. Epigenetic treatment of pancreatic cancer: is there a therapeutic perspective on the horizon? Gut. 2017;66:168–179. doi: 10.1136/gutjnl-2016-312539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barros FM, Carneiro F, Machado JC, Melo SA. Exosomes and immune response in cancer: friends or foes? Front Immunol. 2018;9:730. doi: 10.3389/fimmu.2018.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X, Liu G, Li Y, Pan Y. Immune checkpoint: the novel target for antitumor therapy. Genes Dis. 2021;8:25–37. doi: 10.1016/j.gendis.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Wang Z, Shi B, Li Y, Wang R, Sun J, Hu Y, Yuan C, Xu Q. Effect of gingival mesenchymal stem cell-derived exosomes on inflammatory macrophages in a high-lipid microenvironment. Int Immunopharmacol. 2021;94:107455. doi: 10.1016/j.intimp.2021.107455. [DOI] [PubMed] [Google Scholar]
- 17.Dong ZR, Ke AW, Li T, Cai JB, Yang YF, Zhou W, Shi GM, Fan J. CircMEMO1 modulates the promoter methylation and expression of TCF21 to regulate hepatocellular carcinoma progression and sorafenib treatment sensitivity. Mol Cancer. 2021;20:75. doi: 10.1186/s12943-021-01361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Q, Hann SS. Biological roles and mechanisms of circular rna in human cancers. Onco Targets Ther. 2020;13:2067–2092. doi: 10.2147/OTT.S233672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Ouyang T, Li M, Hong T, Alriashy M, Meng W, Zhang N. CBX7 is dualistic in cancer progression based on its function and molecular interactions. Front Genet. 2021;12:740794. doi: 10.3389/fgene.2021.740794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karamitopoulou E, Pallante P, Zlobec I, Tornillo L, Carafa V, Schaffner T, Borner M, Diamantis I, Esposito F, Brunner T, Zimmermann A, Federico A, Terracciano L, Fusco A. Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreatic cancer. Eur J Cancer. 2010;46:1438–1444. doi: 10.1016/j.ejca.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Mohammad HP, Cai Y, McGarvey KM, Easwaran H, Van Neste L, Ohm JE, O’Hagan HM, Baylin SB. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 2009;69:6322–6330. doi: 10.1158/0008-5472.CAN-09-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y, Qing C, Wang J, Zeng Z. DNA methylation-based diagnostic and prognostic biomarkers for glioblastoma. Cell Transplant. 2020;29:963689720933241. doi: 10.1177/0963689720933241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohuchida K, Mizumoto K, Ohhashi S, Yamaguchi H, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. S100A11, a putative tumor suppressor gene, is overexpressed in pancreatic carcinogenesis. Clin Cancer Res. 2006;12:5417–5422. doi: 10.1158/1078-0432.CCR-06-0222. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang H, Chen X, Dong F, Zhang Z, Zhou Z, Ma Z, Huang S, Chen B, Zhang C, Hou B. Prognostic values and immune suppression of the S100A family in pancreatic cancer. J Cell Mol Med. 2021;25:3006–3018. doi: 10.1111/jcmm.16343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Li Y, Cao Y, Yuan M, Gao Z, Guo X, Zhu F, Wang Y, Xu J. Polycomb chromobox (Cbx) 7 modulates activation-induced CD4+ T cell apoptosis. Arch Biochem Biophys. 2014;564:184–188. doi: 10.1016/j.abb.2014.10.004. [DOI] [PubMed] [Google Scholar]