Abstract

Developmental self-assembly of DNA nanostructures provides an ideal platform for studying the power and programmability of kinetically controlled structural growth in engineered molecular systems. Triggered initiation and designated sequencing of assembly and disassembly steps have been demonstrated in structures with branches and loops. Here we introduce a new strategy for selectively activating distinct subroutines in a developmental self-assembly program, allowing structures with distinct properties to be created in response to various molecular signals. We demonstrate this strategy in triggered self-assembly of a DNA ring, the size and growth direction of which are responsive to a key molecule. We articulate that reversible assembly steps with slow kinetics at appropriate locations in a reaction pathway could enable multiple populations of structures with stimulus-responsive properties to be simultaneously created in one developmental program. These results open up a broad design space for the self-assembly of molecules with adaptive behaviors toward advanced control in synthetic materials and molecular motors.
Introduction
Molecular self-assembly is key to the functionality of living cells, allowing lipids, nucleic acids, and proteins to organize themselves into structures with desired shapes and properties. Understanding the principles of self-assembly in engineered molecular systems is fundamentally important to control the behavior of biomolecules for technological advances. DNA self-assembly is one of the most well-studied areas of engineered molecular self-assembly.1 Complex shapes with up to gigadalton sizes have been created with nanometer precision.2−5 Moreover, a self-assembly process could be designed to carry out complex computation and algorithms.6,7
Most DNA self-assembly processes investigated so far take place spontaneously during thermal annealing, but some exhibit isothermal behavior in response to a triggering signal.8−10 Triggered self-assembly processes allow desired structures to grow at desired times, while the isothermal property allows for applications where temperature changes are undesired, for example, in a biological environment.11,12 Similar to how the kinetics of growth in multicellular development is orchestrated by genetic programs, the kinetic pathway of triggered self-assembly can be controlled by molecular programs encoded in DNA; this type of behavior has been referred to as developmental self-assembly.13 The kinetic control was achieved by toehold-mediated DNA strand displacement,14 where the reaction of an initiator strand with a hairpin motif by toehold binding and branch migration reveals a previously sequestered toehold for subsequent reactions. Using this mechanism, dendritic structures9 and a tetrahedron13 were created with prescribed sequences for every self-assembly step.
The prior investigations raised an important challenge regarding the design space of triggered self-assembly. As seen in biological systems, development can be influenced by changing environmental conditions throughout the entire growth process rather than just within the initiation step. What new design principles can be established to enable the self-assembly of DNA nanostructures with stimuli-responsiveness more deeply embedded within the growth process? To begin answering this question, here we show that distinct signal molecules can be designed to selectively activate a subroutine (e.g., a subset of steps) in a developmental self-assembly program, resulting in the assembly of structures of varying sizes or growth directions from the same set of building blocks. In these examples, the signal molecules encode both the start and stop conditions of a growth process, paving the way for future explorations involving more complex conditions.
Various strategies for growing DNA nanostructures with programmable sizes have been developed. For example, increased numbers of unique strands were used to create DNA tubes with increased circumferences,15 and distinct connector strands with a specific offset were used to enforce how wide a sheet must grow before it rolled up into a tube.16 Due to the nature of spontaneous self-assembly, these strategies lead to the immediate growth of partial target structures once the DNA strands are mixed together (Figure 1a). Seeded growth is possible in tile self-assembly systems, which allow the width of DNA ribbons to be controlled using a DNA origami structure as an information-bearing seed.17 In that case, nucleation only occurs when the seed is present. However, once the growth begins, DNA tiles will spontaneously bind to each other, as by default they are all activated. By contrast, developmental self-assembly utilizes hairpin motifs that are activated one at a time. This unique property makes it possible to design a system where the entire growth process is inhibited through kinetic traps, parts of which can be selectively activated upon specific signals. These signals could then alter the outcome of self-assembly (Figure 1b).
Figure 1.
Concept of (a) spontaneous and (b) developmental self-assembly, which create DNA rings with distinct sizes.
Results and Discussion
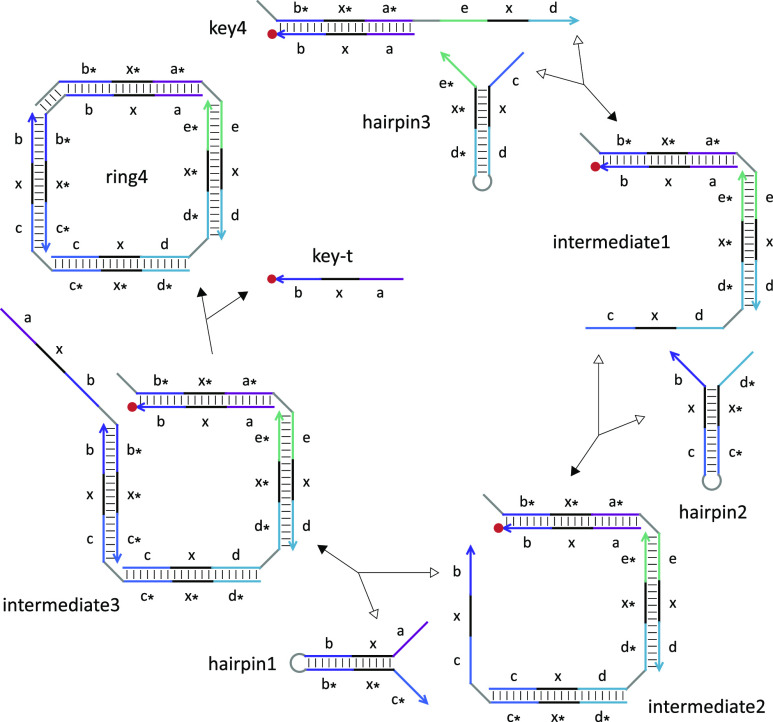
To demonstrate the concept of developmental self-assembly with stimulus-responsive properties, we designed a set of hairpins that could be triggered to form DNA rings with varying sizes depending on the identity of a key that functioned as both an initiator and a terminator (Figure 2a). As shown in the abstract reaction graph (notation explained in the figure caption), a unique toehold composing the output port in each of the hairpins is initially sequestered, preventing the hairpins from interacting with each other when all keys are absent. A key reacts specifically with one of the hairpins, activating its output port for the next assembly step. A cascade of assembly reactions occurs until the activation of a hairpin whose output port matches the input port on the key. A disassembly reaction then takes place to release a strand from the key and close the ring, completing the self-assembly pathway. As shown in Figure 2b, each of the the seven unique hairpins consists of two exposed toeholds (input ports), a common branch migration domain (colored in black), and a sequestered toehold (output port). Each of the three unique key molecules consists of two strands, one of which opens up the first hairpin and the other of which will be released by the last hairpin in the designated self-assembly pathway. The released strand is labeled with a fluorophore to detect pathway completion.
Figure 2.
Design of a DNA ring with a stimulus-responsive size. (a) Abstract reaction graph. Each hairpin is represented as a node with three ports, and each key is represented as a node with two ports. Triangles and circles indicate input and output ports, respectively, while their open or filled representation corresponds to an exposed or sequestered toehold. Solid and dashed black arrows indicate assembly and disassembly reactions, respectively. Gray arrows indicate possible reactions that are not used in the designed ring formation. (b) Domain-level strand diagrams. Each unique toehold and branch migration domain is labeled with a distinct letter. Asterisks indicate sequence complementary. The three keys that trigger the self-assembly of DNA rings with four, six, and eight strands are labeled as key4, key6, and key8, respectively.
Each assembly step that opens up a hairpin is a reversible strand-displacement reaction (Figure 3). The forward reaction is driven by additional base pairs in the toehold domain, while the backward reaction is driven by the entropic gain of one free molecule. Toehold sequences were designed in NUPACK18 to ensure approximately equal forward and backward reaction rates based on the equilibrium constant estimated with 100 nM reactant concentrations. The final disassembly step that closes the ring is an irreversible strand-displacement reaction driven forward by entropy and possibly additional base pairs. A short loop domain in each hairpin provides the desired structural flexibility for ring formation. The loop domain in the last assembled hairpin (hairpin1) also provides an option for controlling the kinetics of ring closure by serving as a toehold to initiate the displacement of the fluorophore-labeled strand (key-t) in a key complex. Once key-t is released, no open toehold will be available for further reactions and thus the self-assembly pathway will be complete.
Figure 3.
Reaction pathway of the formation of a four-stranded ring. Forward and backward reactions are indicated by filled and open arrowheads, respectively. Each toehold, branch migration, and loop domain has seven, six, and four nucleotides, respectively.
We first characterized the ring formation by gel electrophoresis. For control purposes, we designed a set of nonhairpin DNA strands that spontaneously self-assembled into a ring or a linear structure upon thermal annealing, with sizes similar to either each of the triggered rings (Figure 4a) or the largest linear structures before ring closure. When the seven hairpins were mixed together without any keys (Figure 4b, lane 11), no products of larger sizes were observed. When key4, key6, or key8 was present, products with increasing sizes were formed (lanes 12–14, respectively), comparable to the four-stranded, six-stranded, and eight-stranded annealed ring controls (lanes 6, 8, and 10, respectively). In addition to SYBR Gold staining, the same gel was also imaged with ATTO590 fluorescence (Figure 4c). All keys were consumed (comparing with lane 3), and the fluorophore-labeled key-t strand was released (compared with lane 4) in all lanes that contained hairpins and a key, indicating successful ring formation. Unlike the annealed controls of linear structures (lanes 5, 7, and 9), no key-t strands were observed in the products (lanes 12–14), confirming the differences between intermediates and products.
Figure 4.
The formation of DNA rings. (a) Rings of three distinct sizes. Gel electrophoresis performed with (b) SYBR Gold staining and (c) ATTO590 fluorescence. Lanes containing DNA ladders, control structures, and hairpins with or without a key are labeled in blue, black, and orange, respectively. Dashed black and orange lines indicate a comparison between controls and reactants or intermediates, and between controls and products, respectively. Hairpins, keys, and control structures were at concentrations of 150, 100, and 100 nM, respectively. Samples of hairpins and a key were incubated at room temperature for roughly 1 h before they were loaded onto the gel.
Intermediate steps of ring formation were also studied on a gel (Figure S6). When key8 was mixed with an increased number of hairpins 7–2, various sized products with an increasingly wider spectrum were observed (lanes 6–11). These products appeared both with SYBR Gold staining and with ATTO590 fluorescence, indicating that they contained the key-t strand. The spectra of the products confirmed that each assembly step was reversible, while the darkest band in each lane represented the most common product at equilibrium. Only when all seven hairpins were present (lane 12), a product band appeared at a position similar to that of the eight-stranded annealed ring control, while all products with smaller sizes largely disappeared. The disappearance of smaller-sized products confirmed that the final disassembly step was largely irreversible.
Interestingly, the gels also revealed the growth of polymers larger than the target ring size as well as double-sized rings. This is because a linear polymer (e.g., intermediate3 in Figure 3) in competition with ring closure could also react with a second copy of the key molecule (e.g., key4) or any other linear polymers containing a key (e.g., intermediates 1 through 3) to form a longer polymer. A linear polymer with twice as many strands in the target ring could then close to form a double-sized ring. The ring closure is an unimolecular reaction, and the growth of a longer polymer is a bimolecular reaction; thus, the competition should favor the formation of the desired ring at a sufficiently low concentration. Indeed, insignificant amounts of larger polymers and double-sized rings were observed at 100 nM (Figure S4) compared to 1 μM (Figure S5), agreeing with simulation predictions (Figure S7).
Next, to quantitatively understand the reaction kinetics and the completion of the ring formation, we performed fluorescence kinetics experiments at 100 nM. A reporter molecule was used to detect the amount of key-t strand released (Figure 5b). The change of fluorescence in bulk was normalized to concentration based on control experiments (Figure S8). To investigate the range of kinetics that could be controlled by a toehold on the key (Figure 5a), we varied the toehold length in a set of experiments on four-stranded ring formation (Figure 5c). A simple model was developed to simulate the expected system behavior (Supplementary Note S2). The forward rate of each assembly step was estimated using the effective strand displacement rate quantified in previous studies.19,20 The backward rate was calculated using the equilibrium constant from the NUPACK analysis (Figure S2a). Comparing simulation with experimental data, we then estimated the unimolecular rate constant (k) of strand displacement occuring in the disassembly step that leads to ring closure, assuming that the rate was inversely proportional to the ring size. Kinetics over a range of approximately four orders of magnitude was observed with 0–7-nt toeholds (Figure 5c), where the lower bound was comparable to the rate of strand displacement with a remote toehold and a double-stranded spacer.21
Figure 5.
Kinetics of ring formation. (a) Toehold design for controlling the kinetics of the disassembly step that leads to ring closure. (b) Reporting mechanism that detects the amount of the key-t strand released. Simulation (solid trajectories) and fluorescence kinetics data (dotted trajectories) for (c) the formation of four-stranded rings with various toehold sizes and (d) ring formation with various keys. Initial concentrations of keys, hairpins, and the reporter were 100, 150, and 150 nM, respectively.
To investigate the impact of ring size on the overall assembly kinetics, we used the same toehold on three distinct keys, which were separately added to a mixture of seven hairpins. The formations of four-, six-, and eight-stranded rings all approached near completion within 1 h, while the fluorescence change without a key was hardly detectable (Figure 5d). Much longer experiments over 20 h allowed the rate of leak between a key and the reporter to be estimated (Figure S9a); this rate was comparable to the strand displacement rate without a toehold.20 Similarly, the leak between hairpin1 and a key was also estimated (Figure S9b). In this case, because the loop domain in haripin1 was used as a toehold to displace the key-t strand while closing the ring (Figure 5a), a four-way strand displacement leak reaction was possible when the hairpin was closed. With a 4-nt loop, the leak was not measurable. With a 7-nt loop, the leak was still lower than the four-way strand displacement rate with a single exposed toehold.22
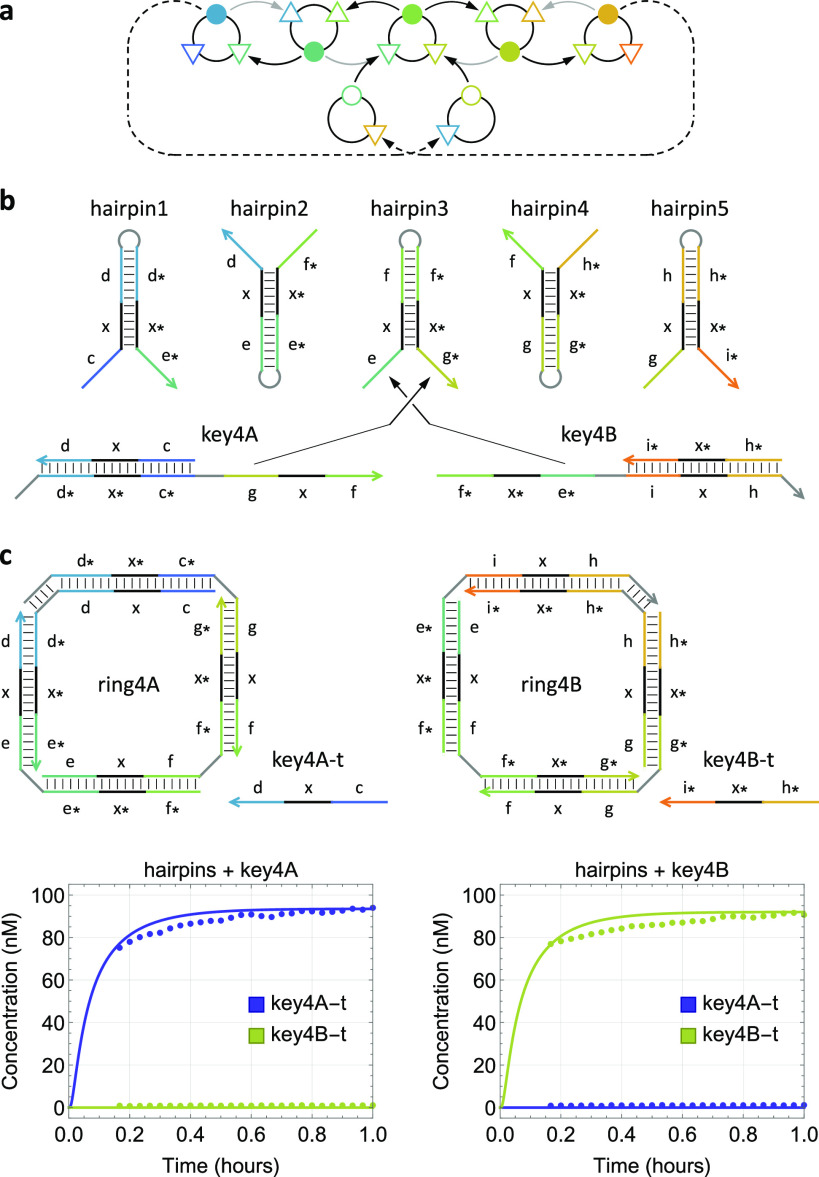
Finally, we demonstrated a second type of stimulus-responsive growth, where two unique keys were designed to control the self-assembly in opposite directions of a reaction graph (Figure 6a). This is possible because each hairpin has two input ports (i.e., two exposed toeholds), enabling subsequent assembly steps involving different hairpins (Figure 6b). Fluorescence data suggested that each of the two desired rings formed when the corresponding key was present (Figures 6c and S11b). Additionally, gel electrophoresis confirmed that both rings were of the correct size (Figure S11a).
Figure 6.
Stimulus-responsive growth direction in developmental self-assembly. (a) Abstract reaction graph. (b) Domain-level strand diagrams. The toehold for ring closure is 4-nt long in each of the two keys, complementary to the loop domains in hairpin1 and hairpin5. (c) Simulation and fluorescence kinetics data for the formation of rings with identical sizes but opposite growth directions controlled by two distinct keys. Initial concentrations of keys and hairpins were 100 and 150 nM, respectively. The reporting mechanism is shown in Figure S10.
Conclusions
To summarize, we have shown that the information on both start and stop conditions of a developmental self-assembly pathway can be encoded in a key molecule, allowing structures with distinct properties to grow in response to stimuli. The same strategy could be generalized to self-assembled structures with stimulus-responsive shapes. For example, linear, ring, and tree structures could be created as distinct subroutines of the same developmental program by choosing appropriate start and stop conditions within one self-assembly reaction graph. We have also demonstrated that an overall self-assembly process can be composed of reversible and irreversible steps, where the kinetics of a certain step can be tuned to be substantially slower than others. Exploiting this strategy, populations of structures with responsive growth properties could be created. For example, when the disassembly step of ring closure was removed, linear structures with various sizes were created. In more complex reaction graphs with reversible steps, multiple slow assembly steps could be employed at desired locations to drive the growth of multiple populations of structures with distinct properties.
In general, developmental self-assembly is relevant not only for growing static structures but also important for controlling the kinetics of dynamical structures that function as motors.9,23 Understanding the full range of behavior that can be programmed in developmental self-assembly is critical for embedding sophisticated control within materials such as hydrogels24−26 and engineered molecular devices such as synthetic membrane channels,27,28 enabling them to exhibit adaptive properties in response to a changing molecular environment.
Acknowledgments
A.T.G. and S.R.D. were supported by Caltech internal funds for BE/CS 196, a course on the design and construction of programmable molecular systems. S.R.D. was also supported by a NIH/BLP training grant (5 T32 GM 112592-5). L.Q. was supported by a NSF award (1813550).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c03853.
Supporting methods, notes, and DNA sequences (PDF)
Author Contributions
¶ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Seeman N. C.; Sleiman H. F. DNA nanotechnology. Nat. Rev. Mater. 2018, 3, 17068. 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- Rothemund P. W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Ong L. L.; Hanikel N.; Yaghi O. K.; Grun C.; Strauss M. T.; Bron P.; Lai-Kee-Him J.; Schueder F.; Wang B.; Wang P.; et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 2017, 552, 72–77. 10.1038/nature24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenbauer K. F.; Sigl C.; Dietz H. Gigadalton-scale shape-programmable DNA assemblies. Nature 2017, 552, 78–83. 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- Tikhomirov G.; Petersen P.; Qian L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 2017, 552, 67–71. 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W.; Papadakis N.; Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2004, 2, e424. 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.; Doty D.; Myhrvold C.; Hui J.; Zhou F.; Yin P.; Winfree E. Diverse and robust molecular algorithms using reprogrammable DNA self-assembly. Nature 2019, 567, 366–372. 10.1038/s41586-019-1014-9. [DOI] [PubMed] [Google Scholar]
- Dirks R. M.; Pierce N. A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 15275–15278. 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P.; Choi H. M.; Calvert C. R.; Pierce N. A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y.; Hariadi R. F.; Choi H. M. T.; Winfree E. Integrating DNA strand-displacement circuitry with DNA tile self-assembly. Nat. Commun. 2013, 4, 1965. 10.1038/ncomms2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. M.; Chang J. Y.; Trinh L. A.; Padilla J. E.; Fraser S. E.; Pierce N. A. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010, 28, 1208–1212. 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delebecque C. J.; Lindner A. B.; Silver P. A.; Aldaye F. A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 2011, 333, 470–474. 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- Sadowski J. P.; Calvert C. R.; Zhang D. Y.; Pierce N. A.; Yin P. Developmental self-assembly of a DNA tetrahedron. ACS Nano 2014, 8, 3251–3259. 10.1021/nn4038223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurke B.; Turberfield A. J.; Mills A. P.; Simmel F. C.; Neumann J. L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- Yin P.; Hariadi R. F.; Sahu S.; Choi H. M.; Park S. H.; LaBean T. H.; Reif J. H. Programming DNA tube circumferences. Science 2008, 321, 824–826. 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Chen X.; Kang G.; Peng R.; Pan V.; Sundaresan R.; Wang P.; Ke Y. Programming DNA tube circumference by tile offset connection. J. Am. Chem. Soc. 2019, 141, 19529–19532. 10.1021/jacs.9b08921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish R. D.; Schulman R.; Rothemund P. W.; Winfree E. An information-bearing seed for nucleating algorithmic self-assembly. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 6054–6059. 10.1073/pnas.0808736106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadeh J. N.; Steenberg C. D.; Bois J. S.; Wolfe B. R.; Pierce M. B.; Khan A. R.; Dirks R. M.; Pierce N. A. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y.; Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009, 131, 17303–17314. 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- Srinivas N.; Ouldridge T. E.; Šulc P.; Schaeffer J. M.; Yurke B.; Louis A. A.; Doye J. P.; Winfree E. On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res. 2013, 41, 10641–10658. 10.1093/nar/gkt801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genot A. J.; Zhang D. Y.; Bath J.; Turberfield A. J. Remote toehold: a mechanism for flexible control of DNA hybridization kinetics. J. Am. Chem. Soc. 2011, 133, 2177–2182. 10.1021/ja1073239. [DOI] [PubMed] [Google Scholar]
- Dabby N. L.The kinetics of toehold-mediated four-way branch migration in synthetic molecular machines for active self-assembly: Prototype algorithms, designs, and experimental study. Ph.D. thesis, California Institute of Technology, Pasadena, CA, 2013. [Google Scholar]
- Venkataraman S.; Dirks R. M.; Rothemund P. W.; Winfree E.; Pierce N. A. An autonomous polymerization motor powered by DNA hybridization. Nat. Nanotechnol. 2007, 2, 490–494. 10.1038/nnano.2007.225. [DOI] [PubMed] [Google Scholar]
- Wang J.; Chao J.; Liu H.; Su S.; Wang L.; Huang W.; Willner I.; Fan C. Clamped hybridization chain reactions for the self-assembly of patterned DNA hydrogels. Angew. Chem., Int. Ed. 2017, 56, 2171–2175. 10.1002/anie.201610125. [DOI] [PubMed] [Google Scholar]
- Cangialosi A.; Yoon C.; Liu J.; Huang Q.; Guo J.; Nguyen T. D.; Gracias D. H.; Schulman R. DNA sequence-directed shape change of photopatterned hydrogels via high-degree swelling. Science 2017, 357, 1126–1130. 10.1126/science.aan3925. [DOI] [PubMed] [Google Scholar]
- Fern J.; Schulman R. Modular DNA strand-displacement controllers for directing material expansion. Nat. Commun. 2018, 9, 3766. 10.1038/s41467-018-06218-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langecker M.; Arnaut V.; Martin T. G.; List J.; Renner S.; Mayer M.; Dietz H.; Simmel F. C. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012, 338, 932–936. 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. R.; Seifert A.; Fertig N.; Howorka S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016, 11, 152–156. 10.1038/nnano.2015.279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.