Abstract
Facioscapulohumeral Muscular Dystrophy (FSHD) is in the top three list of all dystrophies with an approximate 1:8000 incidence. It is not a life-threatening disease; however, the progression of the disease extends over being wheelchair bound. Despite some drug trials continuing, including DUX4 inhibition, TGF-ß inhibition and resokine which promote healthier muscle, there is not an applicable treatment option for FSHD today. Still, there is a need for new agents to heal, stop or at least slow down muscle wasting. Current FSHD studies involving nutraceuticals as vitamin C, vitamin E, coenzyme Q10, zinc, selenium, and phytochemicals as curcumin or genistein, daidzein flavonoids provide promising treatment strategies. In this review, we present the clinical and molecular nature of FSHD and focus on nutraceuticals and phytochemicals that may alleviate FSHD. In the light of the association of impaired pathophysiological FSHD pathways with nutraceuticals and phytochemicals according to the literature, we present both studied and novel approaches that can contribute to FSHD treatment.
Keywords: Facioscapulohumeral muscular dystrophy, FSHD, natural compound, phytochemical, nutraceutical, skeletal muscle, dystrophy
1. INTRODUCTION
Facioscapulohumeral Muscular Dystrophy (FSHD) is one of the most common dystrophies with 1:8000 incidence. The clinic of FSHD, as the name implies, is manifested by asymmetric progressive loss of power in the face, shoulder-scapular muscles and sometimes peroneal muscles [1, 2].
Although the time of onset of the disease is generally defined as the second and third decades [1], the age and clinical severity of the disease differ among individuals. While the prognosis of some patients is severe enough to start in the early 20s and leave them wheelchair-bound in the advanced ages, a mild clinical course can be observed in other patients with late-onset and limited muscle involvement. In a study involving large series of infantile cases, onset before the age of ten had been found to be related to de-novo mutation, while onset after the age of ten had been observed in familial cases [3].
Clinical Severity Score (CSS) is widely used in Europe and the United States of America, which examines the progression of the disease by dividing it into ten severity levels [4, 5]. There are other tools to standardize clinical severity. In the FSHD clinical score, Lamperti et al. described the strength and functionality of facial muscles as 0 to 2; scapular girdle muscles 0 to 3; upper limb muscles 0 to 2; leg muscles 0 to 2; pelvic girdle muscles 0 to 5; abdominal muscles 0 to 1; with total score ranges from 0 to 15 [6]. ReSolve is another study that brings together multiple evaluation strategies as Clinical Outcome Assessments (COA), electrical impedance myography, domain 1 of the motor function measure; reachable workspace; orofacial strength; muscle mass using Dual-Energy X-ray Absorptiometry (DEXA); quantitative myometry and manual muscle testing [7]. The outcomes of trials as antisense RNA mechanism of action for FSHD treatment may be followed with ReSolve [7].
Significant clinical differences between male and female patients have been identified in many studies, and clinical findings emerge at an earlier age and progress more severely in male patients diagnosed with FSHD [8]. This difference between men and women is not observed in infantile cases [9]. Although FSHD is known as muscle disease, eye, ear, bone, and tooth anomalies can accompany the disease; this condition is associated with the severity of the disease [2, 10, 11]. Other clinical findings that can be observed in FSHD are retinal vasculopathy, sensorineural hearing loss, restrictive lung disease, and right bundle branch block in the heart [12]. In addition, central nervous system involvements such as epilepsy and mental retardation can be observed in pediatric cases where FSHD progresses severely [13].
Magnetic resonance imaging studies reveal physical changes in FSHD. According to this, the most common findings are atrophy and fat infiltration [14]. Although no FSHD specific change which can enable it to be distinguished from other muscular dystrophies to date has been found, recently, a significant difference has been found in the proportion of trimethylamine/creatine studied as a metabolite in muscle groups with the Multivoxel Proton Magnetic Resonance Spectroscopy method compared to the control group [15]. EMG findings are found to be compatible with myopathic findings in FSHD, but no correlation has been found between electrophysiological findings and clinical features [16].
2. PATHOPHYSIOLOGY OF THE DISEASE
2.1. Genetic Background of FSHD
FSHD is a unique disease in which a contraction of macrosatellite repeat is present in contrast to other repeat diseases. FSHD is inherited in an autosomal dominant or a digenic manner. The relationship between FSHD and repeat units had been revealed by linkage studies [17-20]. These repeat units are located on the long arm of chromosome 4q35.2 region and named D4Z4 repeat units. In the normal population, D4Z4 macrosatellite on chromosome 4q35.2 consists of 11 to 100 repeat units, and each repeat unit consists of 3300 base pairs. On the other hand, 95% of individuals diagnosed with FSHD carry reduced [1-10] repeat unit numbers [20], and they are called FSHD1 cases. FSHD1 is inherited autosomal dominantly, and rarely, there are de novo cases [21].
There are many studies on gene expression levels in FSHD myoblast cell lines and patient biopsies [22-24]. Although there are many differences in results (which may result from experimental factors such as study conditions, sample heterogeneity), these studies show the change in expression of many genes in FSHD [22, 25-28]. In studies conducted, expression differences have been found to occur in many genes that have divergent functions such as apoptosis, embryological differentiation stages, when compared to healthy individuals. Although there are genes that show intersection in different studies, a clear responsible gene for FSHD disease has still not been identified except for Double Homeobox 4 (DUX4). However, there exist a considerable number of new studies indicating other responsible genes and pathways for the FSHD disease mechanism.
The only gene detected in the D4Z4 repeat sequence on 4q35 is the DUX4 retrogene. The DUX4 gene is located at the end of each repeat unit and encodes DUX4 transcription factor [29] containing two homeodomains [30, 31]. In healthy individuals, DUX4 is not expressed in muscle, and it is only expected to be expressed in testicular tissue [32]. In FSHD, DUX4 mRNA had been shown to be transcribed only from the last D4Z4 repeat unit in skeletal muscles [32, 33]. In addition to D4Z4 repeat deletion, one more genetic structure accompanies a single nucleotide polymorphism (SNP), which is called as qA allele. qA allele has been found in the normal population at a rate of 50% and settles close to the D4Z4 repeat [34, 35]. This qA allele creates a polyadenylation signal for the DUX4 mRNA and stabilizes that mRNA by protecting it from degradation.
The DUX4 gene has been found to be evolutionarily conserved [36], although it has a promoter and it does not have its own polyadenylation signal [37]; there is no FSHD patient who had completely deleted D4Z4 sequence [38]. Because of this, it is hypothesized that the main pathology in FSHD is the gain of funtion. Supporting this hypothesis, while nearly one of every 1000 FSHD myoblasts or nuclei has been found to be positive for the full length (fl) mRNA or protein of the DUX4, control group cultures have been found negative for DUX4 [32]. DUX4 with impaired expression is a transcription factor [29] indicating that DUX4 exhibits its effect with the target genes via binding to DNA rather than itself [39]. An example for this is FSHD Region Gene 2 (FRG2) gene which locates close to the D4Z4 area [40]. FRG2 was found to be upregulated in differentiating primary myoblasts of FSHD patients [41]. DUX4 can directly activate the FRG2 gene by binding target sequences on FRG2 promoter in myoblasts and fibroblasts derived from control individuals [42].
DUX4 is a double homeobox protein, especially expressed in embryological development [30, 43], and it is very similar to PAX protein family with their homeodomains. In one study, it was revealed that Paired Box 3 (PAX3) and PAX7 were the only members that could compete with DUX4, while other homeodomain proteins as PAX6, Paired Like Homeodomain 2 (PITX2c), Orthodenticle Homeobox 1 (OTX1), Retina and Anterior Neural Fold Homeobox (RAX), HESX Homeobox 1 (Hesx1), Mix Paired-Like (MIXL1) and T-Box Transcription Factor (TBX1) could not compete [44]. When PAX3 or PAX7 was upregulated, it was revealed that DUX4 was no longer toxic [45]. In addition, when PAX3 and PAX7 were overexpressed in FSHD cell lines, the level of DUX4 decreased, revealing a converse relationship between these proteins [46]. In a recent study, PAX7 target gene repression had been revealed as a biomarker in FSHD pathogenesis [47].
DUX4 related FSHD pathophysiology has not completely been enlightened. Still, there are many studies explaining the molecular action of DUX4. DUX4 is a pioneer transcription factor [48] that has a short life span. The main effect of pathologically re-expressed DUX4 is the formation of abnormal-shaped myotubes and cell death, leading to muscle atrophy and degeneration [45, 49, 50]. Its overexpression has been revealed to lead to apoptosis via increased activity of Caspase 3 and p53 pathway [33, 45, 51, 52]. Wnt/β-catenin signaling has also been shown to prevent apoptosis in FSHD1 and FSHD2 myotubes via suppressing DUX4 [53]. In 2015, Banerji et al. supported the central role of β-catenin, revealing its effect in the FSHD network [54].
2.2. Epigenetic Background of FSHD
The absence of D4Z4 deletion in approximately 5% of FSHD patients [55], the presence of asymptomatic cases with short repeat sequences in the D4Z4 on 4q35 region [56] and the emergence of FSHD symptoms in individuals with 11-12 repeats [57] have directed new studies for other gene/s that can be responsible for the FSHD. Results of these studies have shed light on a new area of FSHD pathophysiology, the epigenetic component of the disease.
Epigenetic component started with the discovery of Structural Maintenance of Chromosomes Flexible Hinge Domain Containing 1 (SMCHD1) mutation in a small proportion of FSHD patients (less than 5%) [43]. SMCHD1 gene is encoded from the p11.32 region of chromosome 18 and functions as a dimer, providing genomic DNA methylation. Later on, DNA Methyltransferase 3 Beta (DNMT3B) mutations were also detected in some of the FSHD patients [58]. DNMT3B is located in the q11.21 region on chromosome 20, and the product of this gene adds methyl groups to genomic DNA. When any of these two genes carry a mutation, general hypomethylation occurs at the DNA level. DNA hypomethylation occurring at D4Z4 repeats on the 4q35 region leads to DUX4 transcription [58] from contracted alleles. Just recently, in Ligand Dependent Nuclear Receptor Interacting Factor (LRIF1) gene, a homozygous mutation had been identified in a FSHD patient [59] leading to DUX4 transcription via chromatin relaxation in D4Z4 repeats. FSHD patients carrying mutations of SMCHD1, DNMT3B or LRIF1 genes were grouped as FSHD2. Clinically, FSHD2 has no different feature that had been described; because of that, FSHD1 and FSHD2 are undistinguishable in clinical presentation and can be distinguished only with molecular genetic analysis. However, for the clinical presentation of FSHD, the presence of a mutation in SMCHD1 or DNMT3B or LRIF1 genes is not sufficient. These patients also carry D4Z4 deletions between 8-20 repeats with qA allele. Because D4Z4 contraction exists together with SMCHD1, DNMT3B or LRIF1 gene mutations, FSHD2 has a digenic inheritance pattern. Still, challenges exist in the explanation of the interactions between SMCHD1, DNMT3B or LRIF1 proteins and D4Z4 repeat on 4q35 [60], and of the complexity of methylation status of D4Z4 region on FSHD clinical progression [61] directing FSHD to a complex disease more than a simply inherited Mendelian disease.
Epigenetic regulator complexes also have substantial roles in FSHD. Gabellini et al. showed that the genes located near the repeat sequence in FSHD are overexpressed with deregulation of repressor multiprotein complex (including YY1) via binding to 27 base pairs sequence called DNA Binding Element (DBE) at D4Z4 repeat on 4q35 [40]. Later on, they related this impact to a chromatin-associated long non-coding RNA named as DBE-T (D4Z4 binding element transcript), and they showed that DBE-T provided gene transcription by pulling Ash1L (ASH1 like Histone Lysine Methyltransferase) into the 4q35 region and driving histone H3 lysine 36 dimethylation [62]. The same group reported the role of D4Z4 repeat unit number and CpG composition in the repression of this area [63]. In another study, they also revealed that Protein Regulator of Cytokinesis 1 (PRC1) and Protein Regulator of Cytokinesis 2 (PRC2) complexes are enriched on D4Z4 repeats [64]. By inhibition of Smi1 (the core component of PRC1) and Suz12 (the core component of PRC2), they revealed that DUX4 de-repression was specifically related to PRC1 but not PRC2 [64].
In addition to lncRNA that is DBE-T, a wide variety of miRNAs have been found to be differentially expressed in FSHD muscle. Lim et al. reported that miRNAs play a role in the control of DUX4. Targeting the transcriptional starting region of DUX4 with siRNA decreases the DUX4 level. They supported this result by showing that the amount of DUX4 increases as a result of silencing DICER (Ribonuclease III) and Argonaute (AGO), which play a role in miRNA processing [65]. Compatible with these results, many sense and antisense transcripts from the D4Z4 unit have been identified in FSHD muscle cells [66]. A significant difference was detected in the expression level of miR-411 in both primary and immortalized FSHD myoblast cells [67]. In a study comparing FSHD and control primary myoblasts, a total of 29 different miRNAs consisting of 21 upregulated and 8 downregulated were detected in the FSHD group [68]. In the same study, the myogenic miRNAs miR-1, miR-133a, miR-133b, and miR-206 were found to be highly expressed in FSHD myoblasts suggesting that these cells escape differentiation via myogenic miRNA-induced repression. In another study which compared biopsies from FSHD1 patients and control group, miR-330, miR-331-5p, miR-34a, miR-380-3p, miR-516b, miR-582-5p, miR-517 and miR-625 were found to have expression differences [69]. In proliferation and differentiation stages, 6 different miRNAs specific to FSHD, miR-1268, 1268b, 1908, 4258, 4508 and 4516, were found to be changed [70]. Additionally, a change in different miRNA sets had been shown in FSHD1 and FSHD2 [71], indicating that divergent molecular pathways take place in two types of the disease. In the results of the aforementioned studies, similar to mRNA expression studies in FSHD, no overlapped certain one or two responsible miRNA(s) could be detected in independent studies, most probably because of a divergence in experimental designs and genetic heterogeneity of the disease.
D4Z4 repeat on chromosome 4 is very close to telomer (TTAGGG hexameric nucleotide repeats), which is an important functional unit in human chromosomes, located on the short and long arm of each chromosome. Having GC-rich sequences [20], D4Z4 repeat structure had been investigated, and it was found that guanine nucleotides in D4Z4 constructed G-quadruplex motifs in vitro [80, 81]. In a very recent study, it was also shown that by regulation of G-quadruplex motifs on D4Z4, DUX4 expression could be modulated.
These telomeric heterochromatin blocks cause reversible silencing in the subtelomeric genes causing an epigenetic regulation, which is called the Telomere Position Effect (TPE) [72]. TPE was first described in Drosophila melanogaster and shown in Saccharomyces cerevisiae [73]. The position effect of human telomeres was shown by Ofir et al., for the first time, with the addition of artificial telomeric sequences to the lymphoblastoid cells belonging to a case with 22q terminal deletion. In this study, it was shown that human telomeric sequences cause a delay in replication in the neighboring chromosomal regions [74]. On the other hand, every chromosome loses 30-200 bp telomere sequence in the 3’end of the single strand during replication of the discontinuous chain in each cell division due to the failure of DNA polymerase. While the average telomere length at birth is 8000, it goes down to approximately 1500 in old age [75]. In other words, age-related loss of telomeric sequence causes TPE to change as well. The shortening of the telomeric sequence leads to the expression of the repressed gene that is reproduced at an inappropriate time and place. In 2013, Stadler et al. revealed that compared with the samples of the unaffected individuals, myoblast subclones of FSHD patients containing the short telomeric sequence had increased DUX4 and DUX4 target genes’ (Zinc Finger and SCAN Domain Containing 4 -ZSCAN4 and KH Domain Containing 1 -KHDC1) expressions dramatically [76]. In parallel to this, Robin et al. proposed a model in which chromatin conformation and gene expression changes were a distinctive feature of FSHD muscle due to telomere shortening [77]. Another study that strengthened the role of TPE in FSHD revealed the difference in telomere length in semen samples and somatic samples that the sperm cell had strikingly shorter telomere length compared to somatic cell in Telomere Restriction Fragment (TRF) analysis [78]. In this study, re-elongation of these short telomeric sequences during early embryogenesis had been proposed. If the extension of these telomeric sequences does not occur, it may cause de-repression of adjacent genes in the allele with a short telomeric sequence, which may explain the heterogeneity of clinical findings even in the same family members having FSHD. Another study which might explain the relationship between TPE and FSHD is related to nuclear localization. Telomeric sequences show a peripheral location in the cell nucleus as they are poor in coding DNA sequences. The hypothesis is that the requirement of a threshold value is for the placement of D4Z4 sequence, thus TPE is epigenetically involved in silencing genes which are at the proximal subtelomeric location [79]. As a result, both shortened D4Z4 and shortened telomeres can disrupt the nuclear location of the 4q35 locus, causing de-repression of neighbor genes in FSHD disease.
2.3. Oxidative Stress and Inflammation
Oxidative stress is caused by free hydroxyl radicals (•OH), superoxide anions (O2•−) and hydrogen peroxide (H2O2) that are naturally produced as a result of enzymatic reactions in the cell from either mitochondria-related or other naturally occurring reactions. A healthy cell balances these free radicals by converting them into less harmful forms with the antioxidant enzymes, such as Superoxide Dismutase (SOD), Catalase, and Glutathione Peroxidase 3 (GPX3) [80]. Otherwise, these radicals can easily transfer their unpaired electron to nucleic acids, fat, protein or other compounds in the cell, leading to DNA damage, lipid peroxidation or damage to the protein structure.
Most recently, the disruption of oxidative stress has been identified as one of the components leading to muscle loss in FSHD patients. Laoudj-Chenivesse et al. revealed increased levels of Adenine Nucleotide Translocator (ANT1) protein corresponding to mitochondrial dysfunction together with a decrease in oxidative stress protection in FSHD muscle compared to control [81]. Supporting these results, Macaione et al. found an increased ANT1 expression and Nuclear Factor kappa B (NF-kB) DNA binding activity together with increased hydrogen peroxide and reduced peroxidase activity [82]. Winokur and colleagues identified the dysregulation of oxidative stress genes via global gene expression profiling in FSHD. They also observed that when they induced oxidative stress by using paraquat, an oxidative stressor, FSHD myoblasts were susceptible to oxidative stress, which was not observed in control myoblasts [83]. Another study revealed that DUX4 expression had been upregulated with oxidative stress [84]. With these aforementioned studies, it is obvious that oxidative stress has an important role in the pathology of FSHD. Another component observed in FSHD muscle tissue is histological inflammatory changes; mononuclear cell infiltrates in FSHD samples compared to controls which also enhance muscular damage found at higher amounts [85]. Pro-inflammatory cytokines have been reported to be increased in FSHD [86]; for example, Tumour Necrosis Factor alpha (TNF-α) had also been revealed as a perturbed pathway in FSHD network analysis [54]. DUX4 itself had also been shown to activate immune mediators [87], but not all dysregulated inflammatory genes are considered DUX4 targets. From a total of 118 immune genes that were expressed in FSHD biopsies, 80 were found to be DUX4 associated while 38 were not [88], which indicated an immune cell infiltration. These data render regulation of inflammation as another candidate for FSHD treatment.
2.4. Sex Steroids
Clinical variation between male and female FSHD patients indicated the effect of hormones on FSHD pathophysiology [8], which was found to be very similar to Limb-Girdle Muscle Dystrophy (LGMD) [89]. FSHD progresses mildly in females, and the disease progression is affected by pregnancy and birth in female patients [90-92]. An additional clue was the observation of intensified clinic findings with anti-estrogen therapy in FSHD cases with breast cancer [93]. These observations led to the investigation of sex steroids at both clinical and molecular levels. Mul et al. reported that lifetime estrogen exposure was not correlated with FSHD severity [94]; however, Hangul et al. reported a negative correlation between estradiol/total testosterone ratio and progesterone/total testosterone with the severity of FSHD, while there was a positive correlation found with total testosterone [95]. On molecular basis, Teveroni et al. treated differentiating FSHD cells with estradiol (E2) for 72 hours and detected that E2 antagonized DUX4-mediated impairment of myoblast differentiation [96]. Although DUX4 mRNA level did not change, E2 downregulated DUX4 target genes (e.g., TRIM43 and ZSCAN4) via epigenetic regulation of DUX4 target gene promoters [96]. Supporting the protective role of estrogen, Hangul et al. showed that DUX4 was downregulated at the protein level in myoblasts of one male FSHD patient [97].
In the previous part of this review, clinical features of FSHD, the studies on genetic and epigenetic components of the disease, the effect of oxidative stress, inflammation, and sex steroids have been summarized. In the following sections of this review, in addition to current clinical trials, the studies on natural compounds, which are promising candidate agents that can interfere with FSHD related genes and pathways, will be shared.
3. CLINICAL TRIALS FOR THE PHARMACOLOGICAL TREATMENT OF FSHD
Currently, there is no definitive specific medical treatment for FSHD other than symptomatic treatments for some complications of FSHD. Disease management may change in each patient and may include different therapies, such as physical therapy, speech therapy, surgery and respiratory support.
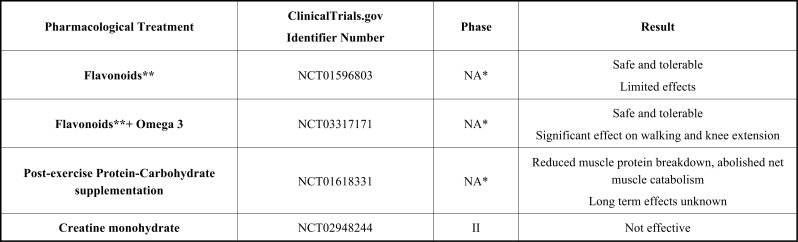
There are 43 clinical trials on the treatment of FSHD, of which 22 have been completed and 3 have been terminated. Of the ongoing 18 studies, 5 have not started recruitment yet, 9 are recruiting, and the status of 4 is unknown. These clinical trials are not only related to drugs but also diagnostic tests and devices for imaging technology. Only 18 of the 43 clinical trials are related to pharmacological treatment, 1 of which is about stem cell therapy and 3 of which are about dietary supplements. In this review, clinical trials on pharmacological treatment and stem cell therapy are discussed under section 3 (Table 1). Clinical trials on dietary supplements are discussed under section 4.
Table 1.
Clinical studies and summary of their results.
| Pharmacological Treatment |
ClinicalTrials.gov
Identifier Number |
Phase | Result |
|---|---|---|---|
| ACE-083 |
NCT02927080 NCT03943290 |
II | Safe and tolerable. Inconsistent clinical results. Studies terminated in Phase II |
| Albuterol (Salbutamol) |
NCT00004685 NCT00027391 |
NA* NA* |
Safe, tolerable with minor side effects. Only limited effect on muscle strength/mass |
| ATYR 1940 (Resolaris) |
NCT02603562 NCT02836418 NCT02579239 NCT02531217 NCT02239224 |
I/II | Safe and tolerable. Clinical improvement at weeks 12/24/36 |
| Losmapimod |
NCT04003974 NCT04264442 NCT04004000 |
I/II | Safe and tolerable. Ongoing Phase II trials |
| MYO-029 | NCT00104078 | I/II | Safe and tolerable. No clinical improvement but demonstrated bioactivity in limited subjects |
| Stem Cell | NCT02208713 | I | Ongoing |
|
Testosterone Enanthate and
Somatropin |
NCT03123913 | I | Recruiting |
*NA: Not applicable, means trials without FDA-defined phases.
3.1. Drugs
3.1.1. ACE-083
ACE-083 is a locally acting follistatin-based therapeutic that blocks proteins in the Transforming Growth Factor (TGF)-beta family, such as myostatin, which is responsible for reducing muscle strength and growth. ACE-083 had received orphan drug status from the U.S. Food and Drug Administration in 2018 for the treatment of FSHD. The efficacy and safety of ACE-083 were evaluated in a two-part Phase 2 clinical trial in patients with FSHD. Patients received either 150 mg or 200 mg doses of ACE-083 by local injection once every three weeks to the affected muscles for three months. Although the first part of the study showed that ACE-083 could increase patients’ muscle mass, the second part of the study failed to show an increase in muscle function with ACE-083 after six months [98]. As a result, the clinical trials of ACE-083 in FSHD were terminated by Acceleron Company.
3.1.2. ATYR 1940 (Resolaris)
Inflammation is involved in the pathophysiology of FSHD. Immunosuppression of the inflammation may slow down disease progression. Physiocrines are a novel group of naturally occurring proteins which are extracellular signaling regions of tRNA synthetases and modulate immune system pathways, thus being molecular targets for immunomodulatory therapies. ATYR1940 (Resolaris) is a physiocrine-based 506 amino acid recombinant protein that is a version of human histidyl-tRNA synthetase (HARS). Resolaris could potentially play a role in promoting skeletal muscle health by acting as an immunomodulator in skeletal muscle [99]. Resolaris has been shown to modulate immune responses in skeletal muscle in preclinical studies. Muscle strength of patients treated with Resolaris improved when compared to placebo. A two-part Phase I/II clinical trial, which was conducted by aTyr Pharma, showed that Resolaris 3,0 mg/kg led to an improvement of up to 25,5% in individualized neuromuscular quality of life (INQoL), and an increase of up to 3,8% in manual muscle testing (MMT) scores at week 12/14. Participants maintained or increased their MMT and INQoL scores at 24 and 36 weeks [100].
3.1.3. Albuterol (Salbutemol)
β2-adrenergic agonists have been shown to increase muscle strength and prevent muscle atrophy in experimental and clinical studies on healthy subjects. After albuterol, a β2-adrenergic agonist showed encouraging results in patients with FSHD in a small pilot study; randomized, double blind, placebo-controlled trials (RCT) were planned [101, 102]. The first RCT performed by Kissel et al. showed that both albuterol 8 and 16 mg did not improve global muscle strength and function but only exhibited some effect in increasing muscle mass. A later study conducted by Van der Kooi et al. showed that albuterol 8 mg exerted a limited positive effect on muscle strength and volume [103]. In a more recent study, albuterol 16 mg was inefficient in improving motor test scores [104]. Periodic use of salbutamol had been revealed to limit its side effects, like cramps, tremor and insomnia, in all studies [104].
3.1.4. MYO-029
Myostatin, a member of the transforming growth factor superfamily, is an endogenous inhibitor of muscle growth. MYO-029 is a recombinant human antibody that binds to myostatin and inhibits its activity. MYO-029 has been shown to increase muscle mass in immunodeficient mice by approximately 30% over 3 months [105]. A double-blind, placebo-controlled, multinational, randomized Phase I/II study evaluating MYO-029 in adult subjects with FSHD showed that MYO-029 exhibited good safety and tolerability with the exception of cutaneous hypersensitivity at 10 mg/kg and 30 mg/kg doses. The study failed to show improvement in muscle strength and function due to lack of power, but increased muscle size was demonstrated in a limited number of subjects by dual-energy radiographic absorptiometry and muscle biopsy showing MYO-029 bioactivity [106].
3.1.5. Losmapimod
DUX4 expression results in skeletal muscle loss and progressive motor disability. p38 inhibitors effectively suppress DUX4 expression in mouse xenograft models of human FSHD gene regulation [107]. Losmapimod is a selective small molecule inhibitor of p38α/β being developed by Fulcrum Therapeutics to reduce DUX4 expression. After pre-clinical studies demonstrated that losmapimod is a potent and highly selective inhibitor of p38 α/β resulting in the reduction of DUX4, a two-part Phase 1 clinical trial demonstrated that losmapimod 7,5 and 15 mg in capsule form was safe and well tolerated in healthy volunteers and patients with FSHD [108, 109]. A randomized controlled Phase IIb study is being conducted to evaluate drug efficacy [110].
3.1.6. Testosterone Enanthate and Somatropin
The safety and tolerability of combination therapy with Recombinant Human Growth Hormone (rHGH) 5.0 μg/kg SC and testosterone enanthate 140mg IM in adult male patients with FSHD are being investigated in the Phase 1 “Study of Testosterone and rHGH in FSHD (STARFISH)” study, which has started recently in 2020 (NCT03123913).
3.2. Stem Cell Treatment
A Phase-1 study on muscle-derived stem cells and adipose-derived mesenchymal stem cells in patients with FSHD is still going on. Patients were injected cell suspensions into biceps, triceps and trapezoids muscles and will be followed at 1, 2, 4, 6 and 12 months after cell injection (NCT02208713).
The pharmacological treatment, ClinicalTrials.gov Identifier number, clinical study phase, and a summary of the results, are shown in Table 1.
4. STUDIES INVOLVING NATURAL COMPOUNDS AND PHYTOCHEMICALS ON FSHD
Although there are plenty of studies on the genetic or epigenetic infrastructure of FSHD, there are a limited number of studies that have revealed the effect of natural compounds in vitro or in vivo. We have classified these few studies on natural compounds as DUX4 inhibition, telomere position effect, methylation, antioxidant interventions, phytoestrogen interventions and protein/creatin supplementation (Tables 2 and Table 3). Although classified basically, all these compounds (Fig. 1) have an effect on multiple pathways in a nested way.
Table 3.
Clinical studies with dietary supplements and summary of their results.
| Pharmacological Treatment |
ClinicalTrials.gov
Identifier Number |
Phase | Result |
|---|---|---|---|
| Flavonoids** | NCT01596803 | NA* | Safe and tolerable Limited effects |
| Flavonoids**+ Omega 3 | NCT03317171 | NA* | Safe and tolerable Significant effect on walking and knee extension |
| Post-exercise Protein-Carbohydrate supplementation | NCT01618331 | NA* | Reduced muscle protein breakdown, abolished net muscle catabolism Long term effects unknown |
| Creatine monohydrate | NCT02948244 | II | Not effective |
*NA: Not applicable; means trials without FDA-defined phases.
** Flavonoids refer to a variable combination of molecules like Vit C, Vit E, zinc, selenomethionine, coenzyme-Q10, amino acids, curcumin, acetyl-carnitin, etc. with various positive effects on free radicals, antioxidation, immune system, etc.
Fig. (1).
Chemical structures of phytochemicals studied in FSHD.
4.1. DUX4 Inhibition
Bosnakovski and colleagues completed a comprehensive study in which they tested 44.000 small compounds for DUX4 inhibition. From these compounds, they defined 52 compounds, and 60% of these inhibited DUX4 and protected cells from oxidative stress [111]. However, these compounds did not protect against Endoplasmic Reticulum (ER) stress, DNA damage and caspase activated cell death [111]. This study is valuable with its results, but they also implicated that inhibiting DUX4 is not enough to stop loss of muscle. Because of that, in addition to DUX4 inhibition, it is essential to target additional pathways that will block ER stress, DNA damage and caspase activated cell death related to FSHD pathophysiology.
Fisetin is a flavonol having antioxidant effect like other polyphenols. Sharma et al. found that protein Poly (ADP-ribose) Polymerase 1 (PARP1) was the top-ranked protein having interaction with DUX4 promoter in FSHD myoblasts but not in control myoblasts [112]. They proposed that the inhibition of PARP1 might affect DUX4 level and when they treated FSHD myoblasts with 0.5 mM fisetin, a polyphenolic PARP1 inhibitor, they identified the suppression of DUX4 expression as well as the suppression of DUX4 target gene ZSCAN4 [112].
Natural compounds that were used in two studies for DUX4 inhibition also exhibited antioxidant effects because of their chemical structure. Both of the studies aforementioned in this section indicate the effectiveness of natural compounds for DUX4 inhibition.
4.2. TPE Intervention
Another interesting study by Boussouar et al. revealed the effect of natural compounds on TPE in myoblasts. 72 hours treatment of two dietary flavones, acacetin and chyrisin, specifically alleviated TPE and led to the upregulation of FRG2 expression, a gene that has been found to be upregulated in FSHD, in the telomeric area [113]. A limitation of this study was that 72 hours time period did not reflect whether long-time effects would be in the same direction. The authors also tested other flavonoids, such as apigenin, luteolin, myricetin, quercetine dihydrate, and vitexin; however, similar effects were not observed [113]. Different effects of different polyphenols revealed that molecular effect is not a general property of polyphenolic compounds but the structure of the molecule. When the molecules in the study were carefully investigated, a right-left mirror difference was observed indicating the importance of the 3D structure of the molecule for future research. Even acacetin and chyrisin exhibited antioxidant and vascular relaxation effects; because of their upregulation effects on telomere-related genes, they are not good candidates for FSHD. However, if we examine this study up way down, we can conclude that polyphenols, such as apigenin, luteolin, myricetin, quercetine dihydrate, and vitexin, can be used in FSHD, as they would not significantly upregulate the telomeric genes as FRG2 and probably DUX4 which would be valuable to investigate. For instance, among these compounds, apigenin (a polyphenol found in plant-derived foods, including parsley, thyme, celery, and chamomile tea) might be a good candidate to study on FSHD cells. Apigenin has been reported to exert antioxidative effects by scavenging free radicals [114]. These effects are marked from the reduction of biomarkers of oxidative damage, such as reactive oxygen radicals (ROS) and lipid peroxidation (LPO), as well as pro-inflammatory cytokines [115], all of which have been shown to be disrupted in FSHD [81, 83, 84, 86]. To achieve a certain conclusion, detailed research with these compounds on FSHD cell lines is needed.
Being close to telomere and having GC rich sequences [20], D4Z4 repeat unit on chromosome 4 formed quadruplex complexes. Ciszewski et al. used berberine to modulate quadruplex structure. As a result, it was revealed that after berberine treatment, expression of DUX4 reduced, fibrosis was inhibited and an increase in muscle function had been observed [116].
4.2.1. Methylation Interventions
There is only one study on nutraceutical supplementation to manipulate methylation in FSHD [117]. Kooi and colleagues proposed that supplemental folic acid and methionine can reverse hypomethylation that is observed in FSHD. Because of that, they evaluated the effect of oral folic acid (5 mg per day) and methionine (1g three times per day) supplementation for 12 weeks on the methylation level of 4qter from peripheral blood lymphocytes of FSHD patients and the control group. The outcome of 4qter methylation was measured via CpG methylation-sensitive restriction sites (BsaAI and FseI) in the first (proximal) unit of the D4Z4 repeat array, which was still significantly hypomethylated compared to the control group. In addition, they performed total DNA methylation analysis to estimate CpG island methylation of the whole genome. As a result, they identified no significant methylation change, both in restriction and whole-genome molecular tests after supplementation. They reported 12 weeks folic acid and methionine supplementation to be safe [117]. Depending on these results, one can easily speculate the uselessness of these compounds for changing the methylation status of 4qter. However, there are multiple points that should be inferred and repeated. First, as Kooi and colleagues suggested, the folic acid dose might be insufficient. In addition, because the nutraceuticals have milder effects compared to drugs, it is important to use them for longer periods (more than 12 weeks) to reach a conclusion. Another thing to consider is the different responses of each CpG to folic acid and methionine supplementation. Recent studies on the methylation status of the first (proximal) unit of the D4Z4 repeat array have revealed complicated results. Jones et al. and Gaillard et al. reported that methylation in between 5’ and 3’ prime of D4Z4 area is divergent [118, 119]. They also found a correlation between 5’ prime methylation status and clinical status of FSHD [118, 119]. On the other hand, in a large cohort of genotype-phenotype study, Nicolic et al. reported that they did not find a certain correlation between the clinical progression and methylation status of this area [120]. In any way, it is clear that there are individual variances in the methylation of this area, including the same family members with the same repeat unit, in addition to the difference between FSHD1 and FSHD2 patients. Because of that, it is necessary to design new studies by considering the wide variability in the methylation status of the D4Z4 region.
4.2.2. Oxidative Stress Interventions-Antioxidants
Antioxidant molecules have two well-known groups as natural and synthetic antioxidants. Even though they are classified as antioxidants, some of them, on the other hand, such as α-tocopherol (vitamin E), vitamin C, and flavonoids, can become pro-oxidant when used at high concentrations [121]. Because of that, as a potential therapeutic option for FSHD [122], it is necessary to select the right antioxidant(s) in the right dose.
Bosnakovksi and colleagues revealed increased oxidative stress of DUX4-expressing myoblasts. They tested the effects of antioxidants via treating highly DUX4 exhibiting cells with β-mercaptoethanol, monothioglycerol, ascorbic acid, vitamin K2 and vitamin E. They observed these antioxidants enabled myoblasts even in the toxic levels of DUX4. They also analyzed Myogenic Differentiation 1 (MyoD) or Myogenic Factor 5 (Myf5) levels to reveal whether this effect was through these proteins. However, antioxidants had no effect on MyoD or Myf5, confirming that they exert effect via buffering the effect of DUX4 rather than inactivating it [45].
In a nutraceutical clinical study on FSHD, 500 mg vitamin C, 400 mg vitamin E, 25 mg zinc gluconate and 200 μg selenomethionine had been applied once a day for 17 weeks. 53 patients had been included in this study, and they were grouped as 26 treated and 27 matching placebos. The effect of this treatment had been tested via both clinical and molecular tests. The improvement in the two-minute walking test (2-MWT), maximal voluntary contraction, and endurance limit time of the dominant and nondominant quadriceps (MVCQD, MVCQND, TlimQD, and TlimQNDS) had been measured. Statistical significance had been observed only in MVCQ and TlimQ values, but not in 2-MWT. At the molecular level, lipid peroxidation had been found to be significantly different compared to the control group, indicating reduced oxidative stress [123]. These supplements were found to be safe and tolerable (Table 3); however, longer treatment periods than 17 weeks might have exhibited oxidant effects which might not be preferable in the treatment of FSHD; for clinical use in FSHD patients, time period should be clarified for these agents in future studies.
Another study tested the effect of Retinoic Acid (RA) on FSHD myoblasts from patient biopsies. RA reduced the cytotoxic damage and enhanced myoblast survival when applied before hydrogen peroxide, an oxidative stress inducer. The study also revealed that Glutathione Peroxidase 3 (GPX3) is an RA target in myoblasts, and RA executes antioxidant effect via upregulation of GPX3 [124].
Dmitriev et al. pointed out that DNA damage is caused by oxidative stress in FSHD. After treatment with strong antioxidants, tempol or N-Acetyl-L-cysteine (NAC), both DUX4-transfected myoblasts and FSHD myoblasts exhibited reduced levels of DNA damage confirming a DUX4 related oxidative stress in FSHD. Tempol also contributed to the healing of morphological defects in myotube formation [125].
Recently, results of a double-blind pilot trial in muscular dystrophies based on flavonoids and omega-3 have (FLAVOMEGA) been reported. 29 dystrophy patients (DMD, FSHD, and LGMD) were included in this study; 5 of them were FSHD patients. Placebo and treatment groups have been selected randomly. FLAVOMEGA supplementation had been tested for both safety and efficacy for 24 weeks of treatment. FLAVOMEGA composition consists of two phases as oily and powdered. In the oily phase, FLAVOMEGA contains Dokosaheksaenoik Acid (DHA), Eikosapentaenoic Acid (EPA), vitamin E, and lemon essential oil. In the powdered phase, it contains curcumin, acetyl L-carnitine, ascorbic acid, coenzymeQ10, skullcap (Scutellaria baicalensis Georgi) baicalin, and green tea. This mixture of nutraceuticals and phytochemicals is comprehensive as it targets multiple pathways essential for the treatment of dystrophies, such as disrupted mitochondria, oxidative stress and inflammatory pathways. In the FLAVOMEGA study, this supplement had been found to be well-tolerated without adverse effects (Table 3), and significant differences in 6-minute walk distance and isokinetic knee extension had been observed in the clinical outcomes of FSHD and LGMD patients [126].
In the light of these studies, targeting the oxidative stress pathway with nutraceuticals and phytochemicals is a promising approach for FSHD treatment.
4.2.3. Phytoestrogen Intervention
Banerji et al. investigated the altered myogenesis and identified suppression of mitochondrial biogenesis. This suppression was found to be related to Proliferator-Activated Receptor Gamma Coactivator 1-α (PGC1α) and Estrogen-Related Receptor α (ERRα). They performed knockdown of PGC1α in control myoblasts and observed hypertrophic myotubes, confirming their own results. Disturbed myogenesis was observed to be rescued by using food supplements as biochanin A, daidzein or genistein [127]. These results indicate specific targeting of phytoestradiols. Biochanin A, daidzein or genistein are isoflavones that can be taken from soy products. They are also grouped as phytoestrogens, because they can exhibit estrogenic activity. In addition to their estrogenic activity, belonging to the isoflavone class, these phytoestrogens exert antioxidant effects that make these natural compounds perfect candidates for the treatment of FSHD.
4.2.4. Protein and Creatine Supplementation
Protein supplementation after aerobic exercise has a stimulating effect on protein anabolism in healthy muscle. In a small group of patients with muscular dystrophy, post-exercise protein-carbohydrate supplementation within 3 hours led to reduced muscle protein breakdown resulting in an abolished net muscle catabolism, but the long-term effects of this treatment are unknown [128]. Creatine supplementation has been a target of interest in muscle diseases after it has been shown that the addition of creatine could attenuate weakness and metabolic disturbances in diseases characterized by atrophic conditions like in the muscle, bone or brain. Although efficient in some muscle disorders like dystrophinopathies, patients with FSHD did not benefit from creatine supplementation [129] (Table 3).
5. CANDIDATE NATURAL COMPOUNDS AND PHYTOCHEMICALS FOR FUTURE FSHD RESEARCH STUDIES
Due to few studies involving natural compounds focused on FSHD treatment, this area paves the way for a wide variety of novel studies.
Here, in this section, natural compounds related to some of the impaired FSHD pathways (Fig. 2) are discussed as promising possible candidates for future FSHD studies (Table 4). There are multiple nutraceuticals and phytochemicals that can contribute to impaired pathways. However, to have a powerful effect for slowing down FSHD, it is important to find out the right interaction of these natural compounds with the impaired molecular biology of the FSHD cell.
Fig. (2).
Chemical structures of candidate natural compounds and phytochemicals related to impaired FSHD molecular targets.
Table 4.
Candidate natural compounds and phytochemicals related to impaired FSHD molecular targets.
| Target Molecule | Natural Compound/Phytochemical | References |
|---|---|---|
| TNF-α IL-1B | Curcumin | [131] |
| NFκB | Curcumin, delphinidin | [134, 135, 143] |
| Smi1 (PRC2) | Curcumin | [136] |
| P300 | Curcumin, delphinidin, garcinol, anacardic acid, gallic acid and epigallocatechin-3-gallate (ECGC) | [137, 139-143] |
| STAT3 | Cryptotanshinone, curcumin, ursolic acid, cucurbitacin E, alantolactone, silibinin and piperlongumine | [142-147] |
| DNMTs | Resveratrol, schizandrol A (1 or 3 ug/ul) | [145, 146] |
| miR-206 PAX7 | TRF (25% alpha tocopherol and 75% tocotrienols) | [148] |
Curcumin, a phytopolyphenol isolated from Curcuma longa L., can interfere with multiple cellular targets; because of that, it is one of the promising candidates for muscle-wasting diseases [130]. It has been shown that curcumin contributes to the treatment of FSHD at the clinical level [126]. However, at the molecular level, the effects of curcumin have not been revealed yet; curcumin has been revealed to have a wide variety of molecular targets. In Mdx model of DMD, curcumin enhanced muscle strength by downregulating TNF-α and Interleukin 1 beta (IL-1β) levels [131]. In FSHD, the increased levels of reactive oxygen species related to TNF-α had been shown to lead to FSHD cell death [54]. However, it is not known yet whether curcumin has a similar regulating effect on TNF-α and IL-1β in FSHD muscle which would be interesting to investigate. Curcumin can also inhibit the NF-kB pathway [132, 133] which has also been revealed as another pathologically activated pathway in FSHD [82]. It had been revealed that treatment with 20 mg of curcuminoids daily for 6 weeks led to a significant lowering of NF-kB in the muscle of rats (134], and dietary curcumin supplementation alleviated NF-kB dependent muscle wasting [135]. Another target molecule of curcumin is Smi1; it can reduce Smi1 level [136]. Smi1 is a member of the PRC2 complex that is necessary for the epigenetic modulation of D4Z4 repeat on chromosome 4q35 [64]. Inhibition of a PRC2 complex member is an unwanted effect because when the activity of the PRC2 complex is reduced, the disruption of the epigenetic repression in the D4Z4 area might lead to an increased DUX4 expression. Until now, it has not been studied whether curcumin could exhibit its Smi1 reducing effect specifically on FSHD myoblasts/myotubes. The effects of curcumin on an FSHD cell need to be clarified. Another intriguing target of curcumin is P300, a histone acetyl transferase [137]. Bosnakovski et al. showed that P300 inhibitor reverses DUX4-mediated global histone H3 hyperacetylation, targets gene expression and cell death in FSHD myoblasts and FSHD animal model [138]. P300 can also be inhibited via other additional phytochemicals. Dietary natural compounds as Epigallocatechin-3-Gallate (EGCG), garcinol, anacardic acid and gallic acid can block P300 [139-142]. Delphinidin from Punica granatum L. recently had been revealed as a novel p300 inhibitor which interestingly had not affected other epigenetic enzymes as histone deacetylase and histone methyltransferase [143]. That specificity of delphinidin makes it a good candidate for FSHD treatment. Delphinidin additionally inhibits TNF-α stimulated NF-kB effect via blocking its target gene expression [143].
Epigenetic regulation of FSHD is one of the other major components that should be interpreted in treatment strategies. There is no natural compound in the literature that has been shown to affect SMCHD1 or LRIF1 proteins. However, in diseases other than FSHD, it has been revealed that DNMTs can be regulated by nutraceuticals and phytochemicals. DUX4 level has been revealed to be regulated by DNMT3B in a tissue-dependent manner [144]. These upregulating DNMT levels leading to hypermethylation in D4Z4 repeat may contribute to clinical severity via reducing DUX4 levels, especially in non-DNMT3B mutated FSHD1 patients or FSHD2 patients with the heterozygous mutation. Resveratrol via stilbene polyphenolic group restored oxidative stress condition by upregulating the decreased levels of DNMT1, DNMT3a, DNMT3b and Sirtuin 1 (SIRT1) in retinal pigment epithelium cells [145]. This upregulation of DNMTs increased the methylation levels of repeat sequence called Long Interspersed Nuclear element-1 (LINE-1) [145], which is a repeat sequence as D4Z4, making us curious to learn what the effect will be on FSHD. The treatment of another compound, schizandrol A, at 1 or 3 ug/ml dose increased mRNA expression of DNMT3A and DNMT3B on neuronal cell line, while 9 ug/ml exerted its effect in a completely opposite direction [146].
Another promising emergent research area in neuromuscular disorders is modulating miRNAs with nutraceuticals. Recently, miR-206 has been suggested as a circulating biomarker in LGMD patients since it has been found to be significantly elevated compared to the control group [147]. Similar to that, miR-206 has been reported to be expressed highly in FSHD myoblasts [68], and a possible issue of modulating miR-206 could be interesting. From FSHD literature, PAX7 has been known to be a key protein in FSHD pathophysiology [44, 46, 47]. A study in human myoblasts has shown miR-206 and PAX7 to be overlapping, which are two important molecules in FSHD pathophysiology. Razak et al. found out that 50 μg/mL tocotrienol-rich fraction (TRF; consists of 25% alpha tocopherol, 75% tocotrienols) treatment changed the expression of myomiRs in human skeletal muscle myoblasts [148]. TRF treatment promoted differentiation by modulating the expression of miR-206 which then resulted in the reduction of PAX7 expression [148]. Because TRF can target both miR-206 and PAX7, it is very interesting to search on FSHD at the molecular level. Supporting this, deprivation of vitamin E in diet resulted in muscle damage in rabbits, and this damage could be reversed with vitamin E treatment [149].
Signal Transducer and Activator of Transcription factors (STATs) are a key protein family playing a central role in multiple pathways. Although there is no study related to STATs in FSHD, we have recently reported a coincident FSHD-thrombocythemia case with Janus Kinase 2 (JAK2) mutation [150] which has recently aroused suspicion regarding the JAK/STAT pathway. In addition to this, convincing evidence exists regarding STAT3 playing a key role in muscle differentiation, regeneration and dystrophies. Tierney et al. revealed that STAT3 signaling controls satellite cell expansion and skeletal muscle repair; transient STAT3 inhibition promoted satellite cell expansion and enhanced tissue repair in dystrophic muscles [151]. However, continuous STAT3 inhibition leads to muscle wasting; because of that, transient STAT3 activation is thought to be beneficial for muscle regeneration [152]. Pharmaceutical STAT3 inhibitors are difficult to use in this two-edged situation in daily treatment because of their side effects. Here, natural STAT3 inhibitors might present a good alternative to use intermittently having lower side effects; on the other hand, it is worthy of notice that they can be inactivated easily, and they are not STAT3 specific. There are multiple natural compounds especially effective on STAT3; cryptotanshinone, curcumine, ursolic acid, cucurbitacin E, alantolactone, silibinin and piperlongumine [153-158] are some of these compounds.
To summarize, by matching natural compounds to impaired FSHD pathways (Fig. 3), promising treatment strategies might be developed. In this section, we have presented candidates targeting miR-206, DNMT3B, PRC2 (epigenetic modulation effect), p300, PAX7 (DUX4 modulation effect), Nf-kB, TGF-ß, (anti-inflammatory effect), and STAT3 for future research. These candidates can be expanded with the same approach via targeting several impaired pathways of FSHD. It is beneficial paying attention to some points while choosing the agent in order to achieve the desired success in the treatment. Natural compounds have multiple molecular targets in a cell. Because of that, it is necessary to test whether this compound affects the molecules that we want to target and whether it has no effect on other impaired pathways that we do not want to target in FSHD myoblast/myotube. Another point to be aware of is that natural compounds have milder effects compared to drugs which might lead to a wrong conclusion as they are ineffective. To overcome this, two strategies might be used: treatment for longer periods and combining more than one natural compound to get stronger effects.
Fig. (3).
Genetic and epigenetic background of FSHD with the indicated therapeutic interventions. (A higher resolution/colour version of this figure is available in the electronic copy of the article).
There are a few studies that have tested nutraceuticals and phytochemicals in cell lines. To obtain convincing evidence, it is necessary to perform an increased number of studies. Especially, evaluating right dosage and side effects in animal models might be useful. However, having different epigenetic states and translational differences, it is important to hold in mind that animal models might not provide complete reflection of treatment efficacy in humans in vivo. For this reason, after identifying dosage and side effects, studies on human cell lines and clinical trials need to be better performed for the clear reflection of efficacy. For the outcomes of clinical trials, evaluating some biomarker levels might be useful. Even though there is no FSHD specific severity, reflecting biomarkers as miyomiRs might provide information to follow the efficacy of potential treatment.
CONCLUSION
In this review, we have presented a variety of natural compounds which match up to each disrupted target in FSHD disease. As we put forward, natural compounds offer a wide variability of treatment options with matched molecular pathways observed in FSHD. Most of the natural compounds are safe when they are used in appropriate doses. As long as distinguished and combined in an appropriate way, nutraceuticals/phytochemicals can be suggested to ameliorate diseases; they can also be used as supplemental treatment options in addition to drugs for severely affected cases. In molecular studies, it would be useful to search for appropriate doses and combinations of right molecules on FSHD cell cultures and animal models, directing the results of these studies to clinical trials. With lower side effects and promising properties, these compounds deserve to take part in both clinical trials and molecular research studies.
KEY POINTS
Similar molecules in the same group of phytochemicals might have different effects on the same target. Example: Resveratrol via stilbene polyphenolic group restored oxidative stress condition by upregulating the decreased levels of DNMT1, DNMT3a, and DNMT3b [145]. EGCG, on the contrary, decreased DNMT3a and DNMT3b expression via catechin metabolite in the embryos of mice. Even though both stilbene and catechin are polyphenolic compounds, these counteracting results indicate the specificity of the effect for each compound and the importance of choosing the specifically correct metabolite for targeting wanted effect(s).
Table 2.
A summary of natural compounds and phytochemicals studied in FSHD.
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Tawil R. Facioscapulohumeral muscular dystrophy. Neurotherapeutics. 2008;5(4):601–606. doi: 10.1016/j.nurt.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzsimons R.B. Retinal vascular disease and the pathogenesis of facioscapulohumeral muscular dystrophy. A signalling message from Wnt? Neuromuscul. Disord. 2011;21(4):263–271. doi: 10.1016/j.nmd.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Nikolic A., Ricci G., Sera F., Bucci E., Govi M., Mele F., Rossi M., Ruggiero L., Vercelli L., Ravaglia S., Brisca G., Fiorillo C., Villa L., Maggi L., Cao M., D’Amico M.C., Siciliano G., Antonini G., Santoro L., Mongini T., Moggio M., Morandi L., Pegoraro E., Angelini C., Di Muzio A., Rodolico C., Tomelleri G., Grazia D’Angelo M., Bruno C., Berardinelli A., Tupler R. Clinical expression of facioscapulohumeral muscular dystrophy in carriers of 1-3 D4Z4 reduced alleles: Experience of the FSHD italian national registry. BMJ Open. 2016;6(1):e007798. doi: 10.1136/bmjopen-2015-007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricci E., Galluzzi G., Deidda G., Cacurri S., Colantoni L., Merico B., Piazzo N., Servidei S., Vigneti E., Pasceri V., Silvestri G., Mirabella M., Mangiola F., Tonali P., Felicetti L. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann. Neurol. 1999;45(6):751–757. doi: 10.1002/1531-8249(199906)45:6<751:AID-ANA9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.van Overveld P.G., Enthoven L., Ricci E., Rossi M., Felicetti L., Jeanpierre M., Winokur S.T., Frants R.R., Padberg G.W., van der Maarel S.M. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann. Neurol. 2005;58(4):569–576. doi: 10.1002/ana.20625. [DOI] [PubMed] [Google Scholar]
- 6.Lamperti C., Fabbri G., Vercelli L., D’Amico R., Frusciante R., Bonifazi E., Fiorillo C., Borsato C., Cao M., Servida M., Greco F., Di Leo R., Volpi L., Manzoli C., Cudia P., Pastorello E., Ricciardi L., Siciliano G., Galluzzi G., Rodolico C., Santoro L., Tomelleri G., Angelini C., Ricci E., Palmucci L., Moggio M., Tupler R. A standardized clinical evaluation of patients affected by facioscapulohumeral muscular dystrophy: The FSHD clinical score. Muscle Nerve. 2010;42(2):213–217. doi: 10.1002/mus.21671. [DOI] [PubMed] [Google Scholar]
- 7.LoRusso S., Johnson N.E., McDermott M.P., Eichinger K., Butterfield R.J., Carraro E., Higgs K., Lewis L., Mul K., Sacconi S., Sansone V.A., Shieh P., van Engelen B., Wagner K., Wang L., Statland J.M., Tawil R. Clinical trial readiness to solve barriers to drug development in FSHD (ReSolve): Protocol of a large, international, multi-center prospective study. BMC Neurol. 2019;19(1):224. doi: 10.1186/s12883-019-1452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin F., Wang Z.Q., Lin M.T., Murong S.X., Wang N. New insights into genotype-phenotype correlations in chinese facioscapulohumeral muscular dystrophy: A retrospective analysis of 178 Patients. Chin. Med. J. (Engl.) 2015;128(13):1707–1713. doi: 10.4103/0366-6999.159336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klinge L., Eagle M., Haggerty I.D., Roberts C.E., Straub V., Bushby K.M. Severe phenotype in infantile facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 2006;16(9-10):553–558. doi: 10.1016/j.nmd.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Matsuzaka T., Sakuragawa N., Terasawa K., Kuwabara H. Facioscapulohumeral dystrophy associated with mental retardation, hearing loss, and tortuosity of retinal arterioles. J. Child Neurol. 1986;1(3):218–223. doi: 10.1177/088307388600100308. [DOI] [PubMed] [Google Scholar]
- 11.Balatsouras D.G., Korres S., Manta P., Panousopoulou A., Vassilopoulos D. Cochlear function in facioscapulohumeral muscular dystrophy. Otol. Neurotol. 2007;28(1):7–10. doi: 10.1097/01.mao.0000244362.39696.c8. [DOI] [PubMed] [Google Scholar]
- 12.Tawil R., Kissel J.T., Heatwole C., Pandya S., Gronseth G., Benatar M. Guideline development, dissemination, and implementation subcommittee of the american academy of neurology; Practice issues review panel of the american association of neuromuscular & electrodiagnostic medicine. Evidence-based guideline summary: Evaluation, diagnosis, and management of facioscapulohumeral muscular dystrophy: Report of the guideline development, dissemination, and implementation subcommittee of the American academy of neurology and the practice issues review panel of the American association of neuromuscular & electrodiagnostic medicine. Neurology. 2015;85(4):357–364. doi: 10.1212/WNL.0000000000001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito Y., Miyashita S., Yokoyama A., Komaki H., Seki A., Maegaki Y., Ohno K. Facioscapulohumeral muscular dystrophy with severe mental retardation and epilepsy. Brain Dev. 2007;29(4):231–233. doi: 10.1016/j.braindev.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Iosa M., Mazzà C., Frusciante R., Zok M., Aprile I., Ricci E., Cappozzo A. Mobility assessment of patients with facioscapulohumeral dystrophy. Clin. Biomech. (Bristol, Avon) 2007;22(10):1074–1082. doi: 10.1016/j.clinbiomech.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 15.Leung D.G., Wang X., Barker P.B., Carrino J.A., Wagner K.R. Multivoxel proton magnetic resonance spectroscopy in facioscapulohumeral muscular dystrophy. Muscle Nerve. 2018;57(6):958–963. doi: 10.1002/mus.26048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorobek M., Szmidt-Sałkowska E., Rowińska-Marcińska K., Gaweł M., Hausmanowa-Petrusewicz I. Relationships between clinical data and quantitative EMG findings in facioscapulohumeral muscular dystrophy. Neurol. Neurochir. Pol. 2013;47(1):8–17. doi: 10.5114/ninp.2013.32936. [DOI] [PubMed] [Google Scholar]
- 17.Wijmenga C., Frants R.R., Brouwer O.F., Moerer P., Weber J.L., Padberg G.W. Location of facioscapulohumeral muscular dystrophy gene on chromosome 4. Lancet. 1990;336(8716):651–653. doi: 10.1016/0140-6736(90)92148-B. [DOI] [PubMed] [Google Scholar]
- 18.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2(1):26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 19.Wijmenga C., Sandkuijl L.A., Moerer P., van der Boorn N., Bodrug S.E., Ray P.N., Brouwer O.F., Murray J.C., van Ommen G.J., Padberg G.W. Genetic linkage map of facioscapulohumeral muscular dystrophy and five polymorphic loci on chromosome 4q35-qter. Am. J. Hum. Genet. 1992;51(2):411–415. [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt J.E., Lyle R., Clark L.N., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 1994;3(8):1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 21.van der Maarel S.M., Deidda G., Lemmers R.J., van Overveld P.G., van der Wielen M., Hewitt J.E., Sandkuijl L., Bakker B., van Ommen G.J., Padberg G.W., Frants R.R. De novo facioscapulohumeral muscular dystrophy: Frequent somatic mosaicism, sex-dependent phenotype, and the role of mitotic transchromosomal repeat interaction between chromosomes 4 and 10. Am. J. Hum. Genet. 2000;66(1):26–35. doi: 10.1086/302730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osborne R.J., Welle S., Venance S.L., Thornton C.A., Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68(8):569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- 23.Cheli S., François S., Bodega B., Ferrari F., Tenedini E., Roncaglia E., Ferrari S., Ginelli E., Meneveri R. Expression profiling of FSHD-1 and FSHD-2 cells during myogenic differentiation evidences common and distinctive gene dysregulation patterns. PLoS One. 2011;6(6):e20966. doi: 10.1371/journal.pone.0020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsumagari K., Chang S.C., Lacey M., Baribault C., Chittur S.V., Sowden J., Tawil R., Crawford G.E., Ehrlich M. Gene expression during normal and FSHD myogenesis. BMC Med. Genomics. 2011;4:67. doi: 10.1186/1755-8794-4-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tupler R., Perini G., Pellegrino M.A., Green M.R. Profound misregulation of muscle-specific gene expression in facioscapulohumeral muscular dystrophy. Proc. Natl. Acad. Sci. USA. 1999;96(22):12650–12654. doi: 10.1073/pnas.96.22.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winokur S.T., Chen Y.W., Masny P.S., Martin J.H., Ehmsen J.T., Tapscott S.J., van der Maarel S.M., Hayashi Y., Flanigan K.M. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12(22):2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 27.Celegato B., Capitanio D., Pescatori M., Romualdi C., Pacchioni B., Cagnin S., Viganò A., Colantoni L., Begum S., Ricci E., Wait R., Lanfranchi G., Gelfi C. Parallel protein and transcript profiles of FSHD patient muscles correlate to the d4z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics. 2006;6(19):5303–5321. doi: 10.1002/pmic.200600056. [DOI] [PubMed] [Google Scholar]
- 28.Arashiro P., Eisenberg I., Kho A.T., Cerqueira A.M., Canovas M., Silva H.C., Pavanello R.C., Verjovski-Almeida S., Kunkel L.M., Zatz M. Transcriptional regulation differs in affected facioscapulohumeral muscular dystrophy patients compared to asymptomatic related carriers. Proc. Natl. Acad. Sci. USA. 2009;106(15):6220–6225. doi: 10.1073/pnas.0901573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Mattéotti C., van Acker A.M., Leo O., Figlewicz D., Barro M., Laoudj-Chenivesse D., Belayew A., Coppée F., Chen Y.W. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA. 2007;104(46):18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyle R., Wright T.J., Clark L.N., Hewitt J.E. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28(3):389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- 31.Winokur S.T., Bengtsson U., Vargas J.C., Wasmuth J.J., Altherr M.R., Weiffenbach B., Jacobsen S.J. The evolutionary distribution and structural organization of the homeobox-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum. Mol. Genet. 1996;5(10):1567–1575. doi: 10.1093/hmg/5.10.1567. [DOI] [PubMed] [Google Scholar]
- 32.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., Miller D.G. Facioscapulohumeral dystrophy: Incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6(10):e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Mattéotti C., Arias C., Corona E.D., Nuñez N.G., Leo O., Wattiez R., Figlewicz D., Laoudj-Chenivesse D., Belayew A., Coppée F., Rosa A.L. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007;17(8):611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Lemmers R.J., Wohlgemuth M., van der Gaag K.J., van der Vliet P.J., van Teijlingen C.M., de Knijff P., Padberg G.W., Frants R.R., van der Maarel S.M. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81(5):884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spurlock G., Jim H.P., Upadhyaya M. Confirmation that the specific SSLP microsatellite allele 4qA161 segregates with fascioscapulohumeral muscular dystrophy (FSHD) in a cohort of multiplex and simplex FSHD families. Muscle Nerve. 2010;42(5):820–821. doi: 10.1002/mus.21766. [DOI] [PubMed] [Google Scholar]
- 36.Leidenroth A., Hewitt J.E. A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol. Biol. 2010;10:364. doi: 10.1186/1471-2148-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabriëls J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236(1):25–32. doi: 10.1016/S0378-1119(99)00267-X. [DOI] [PubMed] [Google Scholar]
- 38.Tupler R., Berardinelli A., Barbierato L., Frants R., Hewitt J.E., Lanzi G., Maraschio P., Tiepolo L. Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J. Med. Genet. 1996;33(5):366–370. doi: 10.1136/jmg.33.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsuhashi H., Mitsuhashi S., Lynn-Jones T., Kawahara G., Kunkel L.M. Expression of DUX4 in zebrafish development recapitulates facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 2013;22(3):568–577. doi: 10.1093/hmg/dds467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabellini D., Green M.R., Tupler R. Inappropriate gene activation in FSHD: A repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110(3):339–348. doi: 10.1016/S0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 41.Rijkers T., Deidda G., van Koningsbruggen S., van Geel M., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., van der Maarel S.M. FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD patients. J. Med. Genet. 2004;41(11):826–836. doi: 10.1136/jmg.2004.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thijssen P.E., Balog J., Yao Z., Pham T.P., Tawil R., Tapscott S.J., Van der Maarel S.M. DUX4 promotes transcription of FRG2 by directly activating its promoter in facioscapulohumeral muscular dystrophy. Skelet. Muscle. 2014;4:19. doi: 10.1186/2044-5040-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R., Krom Y.D., Klooster R., Sun Y., den Dunnen J.T., Helmer Q., Donlin-Smith C.M., Padberg G.W., van Engelen B.G., de Greef J.C., Aartsma-Rus A.M., Frants R.R., de Visser M., Desnuelle C., Sacconi S., Filippova G.N., Bakker B., Bamshad M.J., Tapscott S.J., Miller D.G., van der Maarel S.M. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012;44(12):1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosnakovski D., Toso E.A., Hartweck L.M., Magli A., Lee H.A., Thompson E.R., Dandapat A., Perlingeiro R.C.R., Kyba M. The DUX4 homeodomains mediate inhibition of myogenesis and are functionally exchangeable with the Pax7 homeodomain. J. Cell Sci. 2017;130(21):3685–3697. doi: 10.1242/jcs.205427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppée F., Belayew A., Perlingeiro R.R., Kyba M. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27(20):2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banerji C.R.S., Panamarova M., Hebaishi H., White R.B., Relaix F., Severini S., Zammit P.S. PAX7 target genes are globally repressed in facioscapulohumeral muscular dystrophy skeletal muscle. Nat. Commun. 2017;8(1):2152. doi: 10.1038/s41467-017-01200-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerji C.R.S., Zammit P.S. PAX7 target gene repression is a superior FSHD biomarker than DUX4 target gene activation, associating with pathological severity and identifying FSHD at the single-cell level. Hum. Mol. Genet. 2019;28(13):2224–2236. doi: 10.1093/hmg/ddz043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Himeda C.L., Jones P.L. The good, the bad, and the unexpected: Roles of DUX4 in Health and Disease. Dev. Cell. 2019;50(5):525–526. doi: 10.1016/j.devcel.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppée F., Belayew A. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One. 2011;6(10):e26820. doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rickard A.M., Petek L.M., Miller D.G. Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet. 2015;24(20):5901–5914. doi: 10.1093/hmg/ddv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandri M., El Meslemani A.H., Sandri C., Schjerling P., Vissing K., Andersen J.L., Rossini K., Carraro U., Angelini C. Caspase 3 expression correlates with skeletal muscle apoptosis in Duchenne and facioscapulo human muscular dystrophy. A potential target for pharmacological treatment? J. Neuropathol. Exp. Neurol. 2001;60(3):302–312. doi: 10.1093/jnen/60.3.302. [DOI] [PubMed] [Google Scholar]
- 52.Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2011;69(3):540–552. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Block G.J., Narayanan D., Amell A.M., Petek L.M., Davidson K.C., Bird T.D., Tawil R., Moon R.T., Miller D.G. Wnt/β-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet. 2013;22(23):4661–4672. doi: 10.1093/hmg/ddt314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banerji C.R., Knopp P., Moyle L.A., Severini S., Orrell R.W., Teschendorff A.E., Zammit P.S. β-Catenin is central to DUX4-driven network rewiring in facioscapulohumeral muscular dystrophy. J. R. Soc. Interface. 2015;12(102):20140797. doi: 10.1098/rsif.2014.0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35(4):315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 56.Nakagawa M., Matsuzaki T., Higuchi I., Fukunaga H., Inui T., Nagamitsu S., Yamada H., Arimura K., Osame M. Facioscapulohumeral muscular dystrophy: Clinical diversity and genetic abnormalities in Japanese patients. Intern. Med. 1997;36(5):333–339. doi: 10.2169/internalmedicine.36.333. [DOI] [PubMed] [Google Scholar]
- 57.Butz M., Koch M.C., Müller-Felber W., Lemmers R.J., van der Maarel S.M., Schreiber H. Facioscapulohumeral muscular dystrophy. Phenotype-genotype correlation in patients with borderline D4Z4 repeat numbers. J. Neurol. 2003;250(8):932–937. doi: 10.1007/s00415-003-1116-y. [DOI] [PubMed] [Google Scholar]
- 58.van den Boogaard M.L., Lemmers R.J.L.F., Balog J., Wohlgemuth M., Auranen M., Mitsuhashi S., van der Vliet P.J., Straasheijm K.R., van den Akker R.F.P., Kriek M., Laurense-Bik M.E.Y., Raz V., van Ostaijen-Ten Dam M.M., Hansson K.B.M., van der Kooi E.L., Kiuru-Enari S., Udd B., van Tol M.J.D., Nishino I., Tawil R., Tapscott S.J., van Engelen B.G.M., van der Maarel S.M. Mutations in dnmt3b modify epigenetic repression of the d4z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am. J. Hum. Genet. 2016;98(5):1020–1029. doi: 10.1016/j.ajhg.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamanaka K., Šikrová D., Mitsuhashi S., Masuda H., Sekiguchi Y., Sugiyama A., Shibuya K., Lemmers R.J.L.F., Goossens R., Ogawa M., Nagao K., Obuse C., Noguchi S., Hayashi Y.K., Kuwabara S., Balog J., Nishino I., van der Maarel S.M. Homozygous nonsense variant in LRIF1 associated with facioscapulohumeral muscular dystrophy. Neurology. 2020;94(23):e2441–e2447. doi: 10.1212/WNL.0000000000009617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greco A., Goossens R., van Engelen B., van der Maarel S.M. Consequences of epigenetic derepression in facioscapulohumeral muscular dystrophy. Clin. Genet. 2020;97(6):799–814. doi: 10.1111/cge.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salsi V., Magdinier F., Tupler R. Does DNA methylation matter in FSHD? Genes (Basel) 2020;11(3):E258. doi: 10.3390/genes11030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cabianca D.S., Casa V., Bodega B., Xynos A., Ginelli E., Tanaka Y., Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huichalaf C., Micheloni S., Ferri G., Caccia R., Gabellini D. DNA methylation analysis of the macrosatellite repeat associated with FSHD muscular dystrophy at single nucleotide level. PLoS One. 2014;9(12):e115278. doi: 10.1371/journal.pone.0115278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casa V., Runfola V., Micheloni S., Aziz A., Dilworth F.J., Gabellini D. Polycomb repressive complex 1 provides a molecular explanation for repeat copy number dependency in FSHD muscular dystrophy. Hum. Mol. Genet. 2017;26(4):753–767. doi: 10.1093/hmg/ddw426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim J.W., Snider L., Yao Z., Tawil R., Van Der Maarel S.M., Rigo F., Bennett C.F., Filippova G.N., Tapscott S.J. DICER/AGO-dependent epigenetic silencing of D4Z4 repeats enhanced by exogenous siRNA suggests mechanisms and therapies for FSHD. Hum. Mol. Genet. 2015;24(17):4817–4828. doi: 10.1093/hmg/ddv206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R., van der Maarel S.M., Filippova G.N., Tapscott S.J. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: New candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet. 2009;18(13):2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harafuji N., Schneiderat P., Walter M.C., Chen Y.W. miR-411 is up-regulated in FSHD myoblasts and suppresses myogenic factors. Orphanet J. Rare Dis. 2013;8:55. doi: 10.1186/1750-1172-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dmitriev P., Stankevicins L., Ansseau E., Petrov A., Barat A., Dessen P., Robert T., Turki A., Lazar V., Labourer E., Belayew A., Carnac G., Laoudj-Chenivesse D., Lipinski M., Vassetzky Y.S. Defective regulation of microRNA target genes in myoblasts from facioscapulohumeral dystrophy patients. J. Biol. Chem. 2013;288(49):34989–35002. doi: 10.1074/jbc.M113.504522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Portilho D.M., Alves M.R., Kratassiouk G., Roche S., Magdinier F., de Santana E.C., Polesskaya A., Harel-Bellan A., Mouly V., Savino W., Butler-Browne G., Dumonceaux J. miRNA expression in control and FSHD fetal human muscle biopsies. PLoS One. 2015;10(2):e0116853. doi: 10.1371/journal.pone.0116853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Colangelo V., François S., Soldà G., Picco R., Roma F., Ginelli E., Meneveri R. Next-generation sequencing analysis of miRNA expression in control and FSHD myogenesis. PLoS One. 2014;9(10):e108411. doi: 10.1371/journal.pone.0108411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lim J.W., Wong C.J., Yao Z., Tawil R., van der Maarel S.M., Miller D.G., Tapscott S.J., Filippova G.N. Small noncoding RNAs in FSHD2 muscle cells reveal both DUX4- and SMCHD1-specific signatures. Hum. Mol. Genet. 2018;27(15):2644–2657. doi: 10.1093/hmg/ddy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doheny J.G., Mottus R., Grigliatti T.A. Telomeric position effect--a third silencing mechanism in eukaryotes. PLoS One. 2008;3(12):e3864. doi: 10.1371/journal.pone.0003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63(4):751–762. doi: 10.1016/0092-8674(90)90141-Z. [DOI] [PubMed] [Google Scholar]
- 74.Ofir R., Wong A.C., McDermid H.E., Skorecki K.L., Selig S. Position effect of human telomeric repeats on replication timing. Proc. Natl. Acad. Sci. USA. 1999;96(20):11434–11439. doi: 10.1073/pnas.96.20.11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muraki K., Nyhan K., Han L., Murnane J.P. Mechanisms of telomere loss and their consequences for chromosome instability. Front. Oncol. 2012;2:135. doi: 10.3389/fonc.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stadler G., Rahimov F., King O.D., Chen J.C., Robin J.D., Wagner K.R., Shay J.W., Emerson C.P., Jr, Wright W.E. Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy. Nat. Struct. Mol. Biol. 2013;20(6):671–678. doi: 10.1038/nsmb.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robin J.D., Ludlow A.T., Batten K., Gaillard M.C., Stadler G., Magdinier F., Wright W.E., Shay J.W. SORBS2 transcription is activated by telomere position effect-over long distance upon telomere shortening in muscle cells from patients with facioscapulohumeral dystrophy. Genome Res. 2015;25(12):1781–1790. doi: 10.1101/gr.190660.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baird D.M., Britt-Compton B., Rowson J., Amso N.N., Gregory L., Kipling D. Telomere instability in the male germline. Hum. Mol. Genet. 2006;15(1):45–51. doi: 10.1093/hmg/ddi424. [DOI] [PubMed] [Google Scholar]
- 79.Ostlund C., Guan T., Figlewicz D.A., Hays A.P., Worman H.J., Gerace L., Schirmer E.C. Reduction of a 4q35-encoded nuclear envelope protein in muscle differentiation. Biochem. Biophys. Res. Commun. 2009;389(2):279–283. doi: 10.1016/j.bbrc.2009.08.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matés J.M., Pérez-Gómez C., De Castro I.N.J.C.b. Antioxidant enzymes and human diseases. Clin. Biochem. 1999;32(8):595–603. doi: 10.1016/S0009-9120(99)00075-2. [DOI] [PubMed] [Google Scholar]
- 81.Laoudj-Chenivesse D., Carnac G., Bisbal C., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J. Mol. Med. (Berl.) 2005;83(3):216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- 82.Macaione V., Aguennouz M., Rodolico C., Mazzeo A., Patti A., Cannistraci E., Colantone L., Di Giorgio R.M., De Luca G., Vita G. RAGE-NF-kappaB pathway activation in response to oxidative stress in facioscapulohumeral muscular dystrophy. Acta Neurol. Scand. 2007;115(2):115–121. doi: 10.1111/j.1600-0404.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 83.Winokur S.T., Barrett K., Martin J.H., Forrester J.R., Simon M., Tawil R., Chung S.A., Masny P.S., Figlewicz D.A. Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul. Disord. 2003;13(4):322–333. doi: 10.1016/S0960-8966(02)00284-5. [DOI] [PubMed] [Google Scholar]
- 84.Sasaki-Honda M., Jonouchi T., Arai M., Hotta A., Mitsuhashi S., Nishino I., Matsuda R., Sakurai H. A patient-derived iPSC model revealed oxidative stress increases facioscapulohumeral muscular dystrophy-causative DUX4. Hum. Mol. Genet. 2018;27(23):4024–4035. doi: 10.1093/hmg/ddy293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arahata K., Ishihara T., Fukunaga H., Orimo S., Lee J.H., Goto K., Nonaka I. Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): Immunocytochemical and genetic analyses. Muscle Nerve Suppl. 1995;(2):S56–S66. doi: 10.1002/mus.880181312. [DOI] [PubMed] [Google Scholar]
- 86.Turki A., Hayot M., Carnac G., Pillard F., Passerieux E., Bommart S., Raynaud de Mauverger E., Hugon G., Pincemail J., Pietri S., Lambert K., Belayew A., Vassetzky Y., Juntas M.R., Mercier J., Laoudj-Chenivesse D. Functional muscle impairment in facioscapulohumeral muscular dystrophy is correlated with oxidative stress and mitochondrial dysfunction. Free Radic. Biol. Med. 2012;53(5):1068–1079. doi: 10.1016/j.freeradbiomed.2012.06.041. [DOI] [PubMed] [Google Scholar]
- 87.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., Tapscott S.J. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012;22(1):38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yao Z., Snider L., Balog J., Lemmers R.J., Van Der Maarel S.M., Tawil R., Tapscott S.J. DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet. 2014;23(20):5342–5352. doi: 10.1093/hmg/ddu251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fanin M., Nascimbeni A.C., Angelini C. Gender difference in limb-girdle muscular dystrophy: A muscle fiber morphometric study in 101 patients. Clin. Neuropathol. 2014;33(3):179–185. doi: 10.5414/NP300728. [DOI] [PubMed] [Google Scholar]
- 90.Awater C., Zerres K., Rudnik-Schöneborn S. Pregnancy course and outcome in women with hereditary neuromuscular disorders: Comparison of obstetric risks in 178 patients. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012;162(2):153–159. doi: 10.1016/j.ejogrb.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 91.Ciafaloni E., Pressman E.K., Loi A.M., Smirnow A.M., Guntrum D.J., Dilek N., Tawil R. Pregnancy and birth outcomes in women with facioscapulohumeral muscular dystrophy. Neurology. 2006;67(10):1887–1889. doi: 10.1212/01.wnl.0000244471.05316.19. [DOI] [PubMed] [Google Scholar]
- 92.Rudnik-Schöneborn S., Glauner B., Röhrig D., Zerres K. Obstetric aspects in women with facioscapulohumeral muscular dystrophy, limb-girdle muscular dystrophy, and congenital myopathies. Arch. Neurol. 1997;54(7):888–894. doi: 10.1001/archneur.1997.00550190076017. [DOI] [PubMed] [Google Scholar]
- 93.Puma A., Garibaldi M., Teveroni E., Deidda G., Moretti F., Sacconi S.J.N.D. Estrogens as a potential disease modifier in FSHD: a retrospective clinical study. A Retrospective Clin. Study. 2017;27 doi: 10.1016/j.nmd.2017.06.386. [DOI] [Google Scholar]
- 94.Mul K., Horlings C.G.C., Voermans N.C., Schreuder T.H.A., van Engelen B.G.M. Lifetime endogenous estrogen exposure and disease severity in female patients with facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 2018;28(6):508–511. doi: 10.1016/j.nmd.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 95.Hangul C., Bozkurt S., Bilge U., Ozdem S., Altunbas H., Uysal H., Koc F., Karauzum S. The ratios of estradiol and progesterone to testosterone influence the severity of facioscapulohumeral muscular dystrophy. Neurol. Sci. Neurophysiol; 2020. [Google Scholar]
- 96.Teveroni E., Pellegrino M., Sacconi S., Calandra P., Cascino I., Farioli-Vecchioli S., Puma A., Garibaldi M., Morosetti R., Tasca G., Ricci E., Trevisan C.P., Galluzzi G., Pontecorvi A., Crescenzi M., Deidda G., Moretti F. Estrogens enhance myoblast differentiation in facioscapulohumeral muscular dystrophy by antagonizing DUX4 activity. J. Clin. Invest. 2017;127(4):1531–1545. doi: 10.1172/JCI89401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hangul C., Celik E., Kaya H., Eroglu O., Uysal H., Karauzum S. 2020.
- 98.Statland J., Bravver E., Karam C., Elman L., Johnson N., Joyce N., Kissel J., Shieh P., Korngut L., Weihl C. J. N. D. 2018.
- 99.Gershman A., Chiang K., Do M., Abbink E., Harbers V., Audebert C., Campana-Salort E., Monforte M., Iyadurai S., Carey L.J.N.D. A randomized, double-blinded, placebo-controlled, multiple ascending dose study to evaluate the safety, tolerability, pharmacokinetics, immunogenicity, and biological activity of ATYR1940 in adult patients with facioscapulohumeral muscular dystrophy. FSHD; 2016. p. 26. [Google Scholar]
- 100.Walker G., Butterfield R., Mathews K., Servais L., Day J., Gidaro T., Shukla S., Maggi L. J. N. D. 2017.
- 101.Kissel J.T., McDermott M.P., Natarajan R., Mendell J.R., Pandya S., King W.M., Griggs R.C., Tawil R. Pilot trial of albuterol in facioscapulohumeral muscular dystrophy. Neurology. 1998;50(5):1402–1406. doi: 10.1212/WNL.50.5.1402. [DOI] [PubMed] [Google Scholar]
- 102.Kissel J.T., McDermott M.P., Mendell J.R., King W.M., Pandya S., Griggs R.C., Tawil R. Randomized, double-blind, placebo-controlled trial of albuterol in facioscapulohumeral dystrophy. Neurology. 2001;57(8):1434–1440. doi: 10.1212/WNL.57.8.1434. [DOI] [PubMed] [Google Scholar]
- 103.van der Kooi E.L., Vogels O.J., van Asseldonk R.J., Lindeman E., Hendriks J.C., Wohlgemuth M., van der Maarel S.M., Padberg G.W. Strength training and albuterol in facioscapulohumeral muscular dystrophy. Neurology. 2004;63(4):702–708. doi: 10.1212/01.WNL.0000134660.30793.1F. [DOI] [PubMed] [Google Scholar]
- 104.Payan C.A., Hogrel J.Y., Hammouda E.H., Lacomblez L., Ollivier G., Doppler V., Eymard B., Attarian S., Pouget J., Desnuelle C., Laforêt P. Periodic salbutamol in facioscapulohumeral muscular dystrophy: A randomized controlled trial. Arch. Phys. Med. Rehabil. 2009;90(7):1094–1101. doi: 10.1016/j.apmr.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 105.Whittemore L.A., Song K., Li X., Aghajanian J., Davies M., Girgenrath S., Hill J.J., Jalenak M., Kelley P., Knight A., Maylor R., O’Hara D., Pearson A., Quazi A., Ryerson S., Tan X.Y., Tomkinson K.N., Veldman G.M., Widom A., Wright J.F., Wudyka S., Zhao L., Wolfman N.M. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem. Biophys. Res. Commun. 2003;300(4):965–971. doi: 10.1016/S0006-291X(02)02953-4. [DOI] [PubMed] [Google Scholar]
- 106.Wagner K.R., Fleckenstein J.L., Amato A.A., Barohn R.J., Bushby K., Escolar D.M., Flanigan K.M., Pestronk A., Tawil R., Wolfe G.I., Eagle M., Florence J.M., King W.M., Pandya S., Straub V., Juneau P., Meyers K., Csimma C., Araujo T., Allen R., Parsons S.A., Wozney J.M., Lavallie E.R., Mendell J.R. A phase I/IItrial of MYO-029 in adult subjects with muscular dystrophy. Ann. Neurol. 2008;63(5):561–571. doi: 10.1002/ana.21338. [DOI] [PubMed] [Google Scholar]
- 107.Oliva J., Galasinski S., Richey A., Campbell A.E., Meyers M.J., Modi N., Zhong J.W., Tawil R., Tapscott S.J., Sverdrup F.M. Clinically advanced p38 inhibitors suppress DUX4 expression in cellular and animal models of Facioscapulohumeral Muscular Dystrophy. J. Pharmacol. Exp. Ther. 2019;370(2):219–230. doi: 10.1124/jpet.119.259663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cadavid D., Mellion M., Wallace O., Ronco L., Thompson D., Rojas A., Hage M., Gould R. J. N. D. P. 2019.
- 109.Mellion M., Ronco L., Thompson L., Hage M., Brooks S., van Brummelen E., Pagan L., Badrising U., Raines S., Tracewell W. 2020. [Google Scholar]
- 110.Tawil A., Mellion M., Ronco L., Raines S., Tracewell W., Rahilly A., Rojas A., Hage M., Wagner K., Statland J. Design of a phase 2, randomized, double-blind, placebo-controlled, 24-week, parallel-group study of the efficacy and safety of losmapimod in treating subjects with facioscapulohumeral muscular dystrophy (fshd): redux4 (1592). AAN Enterprises; 2020. [Google Scholar]
- 111.Bosnakovski D., Choi S.H., Strasser J.M., Toso E.A., Walters M.A., Kyba M. High-throughput screening identifies inhibitors of DUX4-induced myoblast toxicity. Skelet. Muscle. 2014;4(1):4. doi: 10.1186/2044-5040-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sharma V., Pandey S.N., Khawaja H., Brown K.J., Hathout Y., Chen Y.W. PARP1 Differentially interacts with promoter region of DUX4 Gene in FSHD Myoblasts. J. Genet. Syndr. Gene Ther. 2016;7(4):303. doi: 10.4172/2157-7412.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boussouar A., Barette C., Nadon R., Saint-Léger A., Broucqsault N., Ottaviani A., Firozhoussen A., Lu Y., Lafanechère L., Gilson E., Magdinier F., Ye J. Acacetin and chrysin, two polyphenolic compounds, alleviate telomeric position effect in human cells. Mol. Ther. Nucleic Acids. 2013;2:e116. doi: 10.1038/mtna.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim M., Jung J., Jeong N.Y., Chung H.J. The natural plant flavonoid apigenin is a strong antioxidant that effectively delays peripheral neurodegenerative processes. Anat. Sci. Int. 2019;94(4):285–294. doi: 10.1007/s12565-019-00486-2. [DOI] [PubMed] [Google Scholar]
- 115.Oveissi V., Ram M., Bahramsoltani R., Ebrahimi F., Rahimi R., Naseri R., Belwal T., Devkota H.P., Abbasabadi Z., Farzaei M.H. Medicinal plants and their isolated phytochemicals for the management of chemotherapy-induced neuropathy: therapeutic targets and clinical perspective. Daru. 2019;27(1):389–406. doi: 10.1007/s40199-019-00255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ciszewski L., Lu-Nguyen N., Slater A., Brennan A., Williams H.E.L., Dickson G., Searle M.S., Popplewell L. G-quadruplex ligands mediate downregulation of DUX4 expression. Nucleic Acids Res. 2020;48(8):4179–4194. doi: 10.1093/nar/gkaa146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van der Kooi E.L., de Greef J.C., Wohlgemuth M., Frants R.R., van Asseldonk R.J., Blom H.J., van Engelen B.G., van der Maarel S.M., Padberg G.W. No effect of folic acid and methionine supplementation on D4Z4 methylation in patients with facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 2006;16(11):766–769. doi: 10.1016/j.nmd.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 118.Gaillard M.C., Roche S., Dion C., Tasmadjian A., Bouget G., Salort-Campana E., Vovan C., Chaix C., Broucqsault N., Morere J., Puppo F., Bartoli M., Levy N., Bernard R., Attarian S., Nguyen K., Magdinier F. Differential DNA methylation of the D4Z4 repeat in patients with FSHD and asymptomatic carriers. Neurology. 2014;83(8):733–742. doi: 10.1212/WNL.0000000000000708. [DOI] [PubMed] [Google Scholar]
- 119.Jones T.I., King O.D., Himeda C.L., Homma S., Chen J.C., Beermann M.L., Yan C., Emerson C.P., Jr, Miller J.B., Wagner K.R., Jones P.L. Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin. Epigenetics. 2015;7:37. doi: 10.1186/s13148-015-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nikolic A., Jones T.I., Govi M., Mele F., Maranda L., Sera F., Ricci G., Ruggiero L., Vercelli L., Portaro S., Villa L., Fiorillo C., Maggi L., Santoro L., Antonini G., Filosto M., Moggio M., Angelini C., Pegoraro E., Berardinelli A., Maioli M.A., D’Angelo G., Di Muzio A., Siciliano G., Tomelleri G., D’Esposito M., Della Ragione F., Brancaccio A., Piras R., Rodolico C., Mongini T., Magdinier F., Salsi V., Jones P.L., Tupler R. Interpretation of the epigenetic signature of facioscapulohumeral muscular dystrophy in light of genotype-phenotype studies. Int. J. Mol. Sci. 2020;21(7):E2635. doi: 10.3390/ijms21072635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bouayed J., Bohn T. Exogenous antioxidants--double-edged swords in cellular redox state: Health beneficial effects at physiologic doses versus deleterious effects at high doses. Oxid. Med. Cell. Longev. 2010;3(4):228–237. doi: 10.4161/oxim.3.4.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Denny A.P., Heather A.K. Are antioxidants a potential therapy for FSHD? A review of the literature. Oxid. Med. Cell. Longev. 2017;2017:7020295. doi: 10.1155/2017/7020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Passerieux E., Hayot M., Jaussent A., Carnac G., Gouzi F., Pillard F., Picot M.C., Böcker K., Hugon G., Pincemail J., Defraigne J.O., Verrips T., Mercier J., Laoudj-Chenivesse D. Effects of vitamin c, vitamin e, zinc gluconate, and selenomethionine supplementation on muscle function and oxidative stress biomarkers in patients with facioscapulohumeral dystrophy: a double-blind randomized controlled clinical trial. Free Radic. Biol. Med. 2015;81:158–169. doi: 10.1016/j.freeradbiomed.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 124.El Haddad M., Jean E., Turki A., Hugon G., Vernus B., Bonnieu A., Passerieux E., Hamade A., Mercier J., Laoudj-Chenivesse D., Carnac G. Glutathione peroxidase 3, a new retinoid target gene, is crucial for human skeletal muscle precursor cell survival. J. Cell Sci. 2012;125(Pt 24):6147–6156. doi: 10.1242/jcs.115220. [DOI] [PubMed] [Google Scholar]
- 125.Dmitriev P., Bou Saada Y., Dib C., Ansseau E., Barat A., Hamade A., Dessen P., Robert T., Lazar V., Louzada R.A.N., Dupuy C., Zakharova V., Carnac G., Lipinski M., Vassetzky Y.S. DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients. Free Radic. Biol. Med. 2016;99:244–258. doi: 10.1016/j.freeradbiomed.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 126.Sitzia C., Meregalli M., Belicchi M., Farini A., Arosio M., Bestetti D., Villa C., Valenti L., Brambilla P., Torrente Y. Preliminary evidences of safety and efficacy of flavonoids- and omega 3-based compound for muscular dystrophies treatment: a randomized double-blind placebo controlled pilot clinical trial. Front. Neurol. 2019;10:755. doi: 10.3389/fneur.2019.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Banerji C.R.S., Panamarova M., Pruller J., Figeac N., Hebaishi H., Fidanis E., Saxena A., Contet J., Sacconi S., Severini S., Zammit P.S. Dynamic transcriptomic analysis reveals suppression of pgc1α/errα drives perturbed myogenesis in facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 2019;28(8):1244–1259. doi: 10.1093/hmg/ddy405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Andersen G., Ørngreen M.C., Preisler N., Jeppesen T.D., Krag T.O., Hauerslev S., van Hall G., Vissing J. Protein-carbohydrate supplements improve muscle protein balance in muscular dystrophy patients after endurance exercise: a placebo-controlled crossover study. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015;308(2):R123–R130. doi: 10.1152/ajpregu.00321.2014. [DOI] [PubMed] [Google Scholar]
- 129.Walter M.C., Lochmüller H., Reilich P., Klopstock T., Huber R., Hartard M., Hennig M., Pongratz D., Müller-Felber W. Creatine monohydrate in muscular dystrophies: A double-blind, placebo-controlled clinical study. Neurology. 2000;54(9):1848–1850. doi: 10.1212/WNL.54.9.1848. [DOI] [PubMed] [Google Scholar]
- 130.Alamdari N., O’Neal P., Hasselgren P.O. Curcumin and muscle wasting: A new role for an old drug? Nutrition. 2009;25(2):125–129. doi: 10.1016/j.nut.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pan Y., Chen C., Shen Y., Zhu C.H., Wang G., Wang X.C., Chen H.Q., Zhu M.S. Curcumin alleviates dystrophic muscle pathology in mdx mice. Mol. Cells. 2008;25(4):531–537. [PubMed] [Google Scholar]
- 132.Singh S., Aggarwal B.B. Activation of transcription factor NF-kappa B is suppressed by curcumin (diferuloylmethane). J. Biol. Chem. 1995;270(42):24995–25000. doi: 10.1074/jbc.270.42.24995. [corrected]. [DOI] [PubMed] [Google Scholar]
- 133.Jobin C., Bradham C.A., Russo M.P., Juma B., Narula A.S., Brenner D.A., Sartor R.B. Curcumin blocks cytokine-mediated NF-kappa B activation and proinflammatory gene expression by inhibiting inhibitory factor I-kappa B kinase activity. J. Immunol. 1999;163(6):3474–3483. [PubMed] [Google Scholar]
- 134.Sahin K., Pala R., Tuzcu M., Ozdemir O., Orhan C., Sahin N., Juturu V. Curcumin prevents muscle damage by regulating NF-κB and Nrf2 pathways and improves performance: An in vivo model. J. Inflamm. Res. 2016;9:147–154. doi: 10.2147/JIR.S110873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He J., Xie H., Wu S. Dietary supplementation of curcumin alleviates NF-κB-dependent skeletal muscle wasting in Rat. Endocr. Metab. Immune Disord. Drug Targets. 2016;16(2):140–147. doi: 10.2174/1871530316666160613115221. [DOI] [PubMed] [Google Scholar]
- 136.Adeyeni T.A., Khatwani N., San K., Ezekiel U.R. BMI1 is downregulated by the natural compound curcumin, but not by bisdemethoxycurcumin and dimethoxycurcumin. Physiol. Rep. 2016;4(16):e12906. doi: 10.14814/phy2.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Balasubramanyam K., Varier R.A., Altaf M., Swaminathan V., Siddappa N.B., Ranga U., Kundu T.K. Curcumin, a novel p300/creb-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 2004;279(49):51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 138.Bosnakovski D., da Silva M.T., Sunny S.T., Ener E.T., Toso E.A., Yuan C., Cui Z., Walters M.A., Jadhav A., Kyba M. A novel P300 inhibitor reverses DUX4-mediated global histone H3 hyperacetylation, target gene expression, and cell death. Sci. Adv. 2019;5(9):eaaw7781. doi: 10.1126/sciadv.aaw7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Choi K.C., Jung M.G., Lee Y.H., Yoon J.C., Kwon S.H., Kang H.B., Kim M.J., Cha J.H., Kim Y.J., Jun W.J., Lee J.M., Yoon H.G. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69(2):583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 140.Balasubramanyam K., Altaf M., Varier R.A., Swaminathan V., Ravindran A., Sadhale P.P., Kundu T.K. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J. Biol. Chem. 2004;279(32):33716–33726. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 141.Sun Y., Jiang X., Chen S., Price B.D. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 2006;580(18):4353–4356. doi: 10.1016/j.febslet.2006.06.092. [DOI] [PubMed] [Google Scholar]
- 142.Choi K.C., Lee Y.H., Jung M.G., Kwon S.H., Kim M.J., Jun W.J., Lee J., Lee J.M., Yoon H.G. Gallic acid suppresses lipopolysaccharide-induced nuclear factor-kappaB signaling by preventing RelA acetylation in A549 lung cancer cells. Mol. Cancer Res. 2009;7(12):2011–2021. doi: 10.1158/1541-7786.MCR-09-0239. [DOI] [PubMed] [Google Scholar]
- 143.Seong A.R., Yoo J.Y., Choi K., Lee M.H., Lee Y.H., Lee J., Jun W., Kim S., Yoon H.G. Delphinidin, a specific inhibitor of histone acetyltransferase, suppresses inflammatory signaling via prevention of NF-κB acetylation in fibroblast-like synoviocyte MH7A cells. Biochem. Biophys. Res. Commun. 2011;410(3):581–586. doi: 10.1016/j.bbrc.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 144.Bouwman L. F., den Hamer B., Verveer E. P., Lerink L. J., Krom Y. D., van der Maarel S. M., de Greef J. C. J. S. M. 2020. [DOI] [PMC free article] [PubMed]
- 145.Maugeri A., Barchitta M., Mazzone M.G., Giuliano F., Basile G., Agodi A. Resveratrol modulates SIRT1 and DNMT functions and restores LINE-1 methylation levels in ARPE-19 cells under oxidative stress and inflammation. Int. J. Mol. Sci. 2018;19(7):E2118. doi: 10.3390/ijms19072118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang M., Huo D.S., Cai Z.P., Shao G., Wang H., Zhao Z.Y., Yang Z.J. The effect of schizandrol a-induced DNA methylation on SH-SY5YAB 1-40 altered neuronal cell line: a potential use in alzheimer’s disease. J. Toxicol. Environ. Health A. 2015;78(21-22):1321–1327. doi: 10.1080/15287394.2015.1085942. [DOI] [PubMed] [Google Scholar]
- 147.Pegoraro V., Angelini C. Circulating mir-206 as a biomarker for patients affected by severe limb girdle muscle dystrophies. Genes (Basel) 2021;12(1):85. doi: 10.3390/genes12010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Razak A.M., Khor S.C., Jaafar F., Karim N.A., Makpol S. Targeting myomiRs by tocotrienol-rich fraction to promote myoblast differentiation. Genes Nutr. 2018;13:31. doi: 10.1186/s12263-018-0618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Angelini C., Di Mauro S., Margreth A. Relationship of serum enzyme changes to muscle damage in vitamin E deficiency of the rabbit. Sperimentale. 1968;118(5):349–369. [PubMed] [Google Scholar]
- 150.Hangül C., Yücel O.K., Toylu A., Uysal H., Berker K.S. A Novel coincidence: essential thrombocythemia with facioscapulohumeral muscular dystrophy. Turk. J. Haematol. 2020;37(4):306–307. doi: 10.4274/tjh.galenos.2020.2020.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tierney M.T., Aydogdu T., Sala D., Malecova B., Gatto S., Puri P.L., Latella L., Sacco A. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat. Med. 2014;20(10):1182–1186. doi: 10.1038/nm.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guadagnin E., Mázala D., Chen Y.W. STAT3 in skeletal muscle function and disorders. Int. J. Mol. Sci. 2018;19(8):E2265. doi: 10.3390/ijms19082265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shin D.S., Kim H.N., Shin K.D., Yoon Y.J., Kim S.J., Han D.C., Kwon B.M. Cryptotanshinone inhibits constitutive signal transducer and activator of transcription 3 function through blocking the dimerization in DU145 prostate cancer cells. Cancer Res. 2009;69(1):193–202. doi: 10.1158/0008-5472.CAN-08-2575. [DOI] [PubMed] [Google Scholar]
- 154.Bharti A.C., Donato N., Aggarwal B.B. Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J. Immunol. 2003;171(7):3863–3871. doi: 10.4049/jimmunol.171.7.3863. [DOI] [PubMed] [Google Scholar]
- 155.Shanmugam M.K., Rajendran P., Li F., Nema T., Vali S., Abbasi T., Kapoor S., Sharma A., Kumar A.P., Ho P.C., Hui K.M., Sethi G. Ursolic acid inhibits multiple cell survival pathways leading to suppression of growth of prostate cancer xenograft in nude mice. J. Mol. Med. (Berl.) 2011;89(7):713–727. doi: 10.1007/s00109-011-0746-2. [DOI] [PubMed] [Google Scholar]
- 156.Sun C., Zhang M., Shan X., Zhou X., Yang J., Wang Y., Li-Ling J., Deng Y. Inhibitory effect of cucurbitacin E on pancreatic cancer cells growth via STAT3 signaling. J. Cancer Res. Clin. Oncol. 2010;136(4):603–610. doi: 10.1007/s00432-009-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bharadwaj U., Eckols T.K., Kolosov M., Kasembeli M.M., Adam A., Torres D., Zhang X., Dobrolecki L.E., Wei W., Lewis M.T., Dave B., Chang J.C., Landis M.D., Creighton C.J., Mancini M.A., Tweardy D.J. Drug-repositioning screening identified piperlongumine as a direct STAT3 inhibitor with potent activity against breast cancer. Oncogene. 2015;34(11):1341–1353. doi: 10.1038/onc.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bosch-Barrera J., Menendez J.A. Silibinin and STAT3: A natural way of targeting transcription factors for cancer therapy. Cancer Treat. Rev. 2015;41(6):540–546. doi: 10.1016/j.ctrv.2015.04.008. [DOI] [PubMed] [Google Scholar]