Abstract
Neurodegenerative diseases (ND), as a group of central nervous system (CNS) disorders, are among the most prominent medical problems of the 21st century. They are often associated with considerable disability, motor dysfunction and dementia and are more common in the aged population. ND imposes a psychologic, economic and social burden on the patients and their families. Currently, there is no effective treatment for ND. Since many ND result from the gain of function of a mutant allele, small interference RNA (siRNA) can be a potential therapeutic agent for ND management. Based on the RNA interference (RNAi) approach, siRNA is a powerful tool for modulating gene expression through gene silencing. However, there are some obstacles in the clinical application of siRNA, including unfavorable immune response, off-target effects, instability of naked siRNA, nuclease susceptibility and a need to develop a suitable delivery system. Since there are some issues related to siRNA delivery routes, in this review, we focus on the application of siRNA in the management of ND treatment from 2000 to 2020.
Keywords: Central nervous system, neurodegenerative disorders, siRNA, RNAi, delivery system, antisense technology
1. INTRODUCTION
According to the World Health Organization (WHO) reports, the central nervous system (CNS) related diseases are the leading medical problem in the 21st century and have a high prevalence worldwide. Neurodegenerative diseases (ND) are a large group of CNS disorders. They are often associated with disability, motor dysfunction and dementia (the weakness of mental functions that could affect the different intellectual process, including language, learning, thinking, calculation, behavior, and memory) due to the progressive deterioration and death of the neurons [1-3]. ND consist of various disorders such as Alzheimer's disease, Huntington's disease, Parkinson's disease, Multiple Sclerosis and spinal cord injury [4].
Currently, the treatment of these diseases is a big challenge for clinicians and researchers. The currently available medications can only relieve some of the symptoms, but they cannot stop the progression of these diseases [5]. However, the characterization of the genes and the molecular pathway involved in the pathogenesis of ND, as well as the advancement in the gene therapy methods, have made some advances towards finding an effective and satisfactory treatment approach for the management of these disorders [6]. One of the most potent strategies to fight ND is antisense technology due to its high ability to target mutant genes. This technique includes various methods, such as antisense oligonucleotides (ASO), RNAi technology (siRNA, miRNA, shRNA), ribozyme, DNAzyme and aptamer. Many studies based on antisense technology in pre-clinical and clinical phases are currently under research to find a suitable answer for the ND challenge. For example, RO7234292 or Tominersen, an investigational drug from ASO class, is undergoing clinical trials at phase 3 (NCT03761849) to treat patients with Huntington's disease. WVE-120102 is another ASO that is currently under investigation at Phase 1b/2a clinical study (NCT03225846) to the same condition [7, 8]. Some of them have even received FDA approval. Spinraza™ (Nusinersen) is the first FDA-approved antisense drug based on a CNS disease, spinal muscular atrophy (SMA), that recovers the expression of Survival motor neuron protein through splicing correction [9, 10].
One of the gene targeting procedures is RNAi technology that has also been used to treat ND in recent years. siRNA, a valuable class of RNAi mechanism, has been a significant achievement in the world of biology in the last two decades [11]. Theoretically, siRNA can target any mRNA that is translated into a protein [12]. Hence, siRNA is a powerful mean for drug discovery in medical research [13]. siRNA has some advantages over other common therapeutic approaches such as antibodies, small molecules, and proteins. siRNA does not require a particular target on the cell membrane surface or a druggable target [14]. Compared to other typical drugs, siRNA can be designed efficiently since it includes a small number of nucleotides (21-23) and follows the Watson–Crick base pairing rules [15]. The siRNA can work in lower concentrations than other antisense oligonucleotides and ribozymes, suggesting siRNA has high fidelity and efficacy [12]. siRNA also has some advantages over ASO; for instance, finding a potent siRNA is comparably easier than ASO because ASOs must have chemical modifications to function appropriately. For this reason, siRNAs are preferred for in vitro experiments [16].
Considering the highlighted benefits of siRNA technology, it is not surprising many investigators used this method to find a solution for the treatment of ND. This review focuses on the application of the therapeutic potential of siRNA in the treatment of ND based on the existing evidence.
2. sIRNA-MEDIATED RNAI PATHWAY
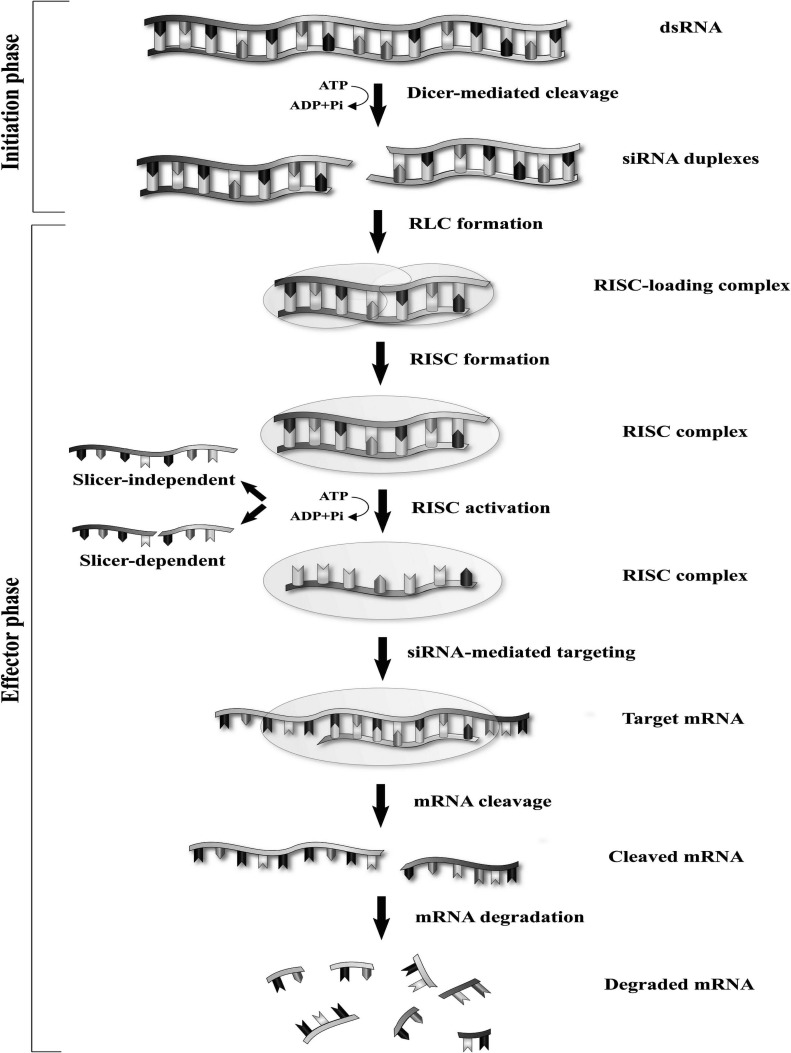
The RNAi process is started when a double-strand (ds) RNA is introduced into the cell [17]. It comprises an initial stage followed by an effector stage. In the initiation stage, an endoribonuclease enzyme, Dicer, cleaves the dsRNA and produces a shorter fragment (21-23 base pair), called siRNA. Dicer belongs to the RNase III family and is described as the “molecular ruler” (Fig. 1). The 3’ end of new siRNA has two nucleotides overhangs necessary for its specific function, whereas the 5’ end consists of a monophosphate group [18-22]. In the second (effector) stage, the siRNA molecule is loaded into a multiprotein complex called the RISC (RNA induced silencing complex). The rest of the steps, such as completing the siRNA processing, target recognition and digestion, are facilitated with this complex [23]. After the siRNA-RISC formation, one of the two-strands with the more stable 5’ end, namely the guide or antisense strand, remains connected with the RISC. While the other strand, the so-called passenger or sense strand, is digested and is discharged from the complex by the argonaute protein 2 (AGO2), which is an integral part of the RISC [24-28]. AGO is the major player and the critical effector molecule in RNAi associated silencing. It is a family with four members (AGO 1-4) in which only the AGO2 has the catalytic function in the mammalian cells [26, 29]. It is thought that following the release of the passenger strand, the RISC is activated, and then the guide strand can bind to the target mRNA. An impressive gene silencing will be accessible only if the guide strand of siRNA and mRNA transcript are paired completely (unlike miRNA), which leads to the cleavage of the target mRNA by the AGO2 part of RISC [26, 30]. Subsequently, the cleaved mRNA is degraded by the cellular nuclease [31].
Fig. (1).
Mechanism of RNAi by dicer. (A higher resolution/colour version of this figure is available in the electronic copy of the article).
3. sIRNA DELIVERY SYSTEMS
The development of an effective and safe approach for the siRNA delivery to the target cells is the major impediment for siRNA's clinical use [32-33]. There are various reasons for these challenges with siRNA, including the large size of siRNA (13 kD), its polyanionic nature and its inability to pass from the cell membrane because of the negative charge [13, 34]. A suitable delivery approach should have some features such as no or low toxicity, improve the cellular uptake of the siRNA, siRNA protection from the serum nuclease attack, lowering the rate of siRNA renal filtration, ability to extravasate from the blood to the target site (after intravenous injection administration) and preservation of siRNA from the phagocytosis [33, 35-38].
There are two major types of delivery systems for the siRNA transfer: viral vectors and non-viral vectors. The hallmark of viral vectors is their high efficiency, but some safety issues limit their clinical application. The most common viral vectors consist of adenoviruses (AVs), adeno-associated viruses (AAVs) and lentiviruses (LVs) [39]. The non-viral vectors are more preferred than the viral vectors due to their safety profile, although their efficiency is not very high. They can be divided into different types [40] including lipid base (e.g. liposome)[38, 41], non-lipid inorganic-based (e.g. golden nanoparticles [42] and superparamagnetic iron oxide nanoparticles (SPIONs)) [43] and non-lipid organic-based (e.g. chitosan, PEI, polyplexes) [38, 41]. Moreover, siRNA can be modified to increase its stability [44-46].
4. sIRNA AND NEURODEGENERATIVE DISORDERS
4.1. Alzheimer’s Disease
Alzheimer’s disease (AD) is a progressive, devastating and the most prevalent neurodegenerative disorder [47]. The clinical symptoms of the disease include ongoing deterioration of memory, learning, cognition and consequently personality and behavioral changes [47, 48]. AD is an age-dependent disease and the most common reason for dementia (>80%) in the aging population. It is predicted that by 2060, the number of peoples affected by AD in the U.S. will increase to 9.3 million [49].
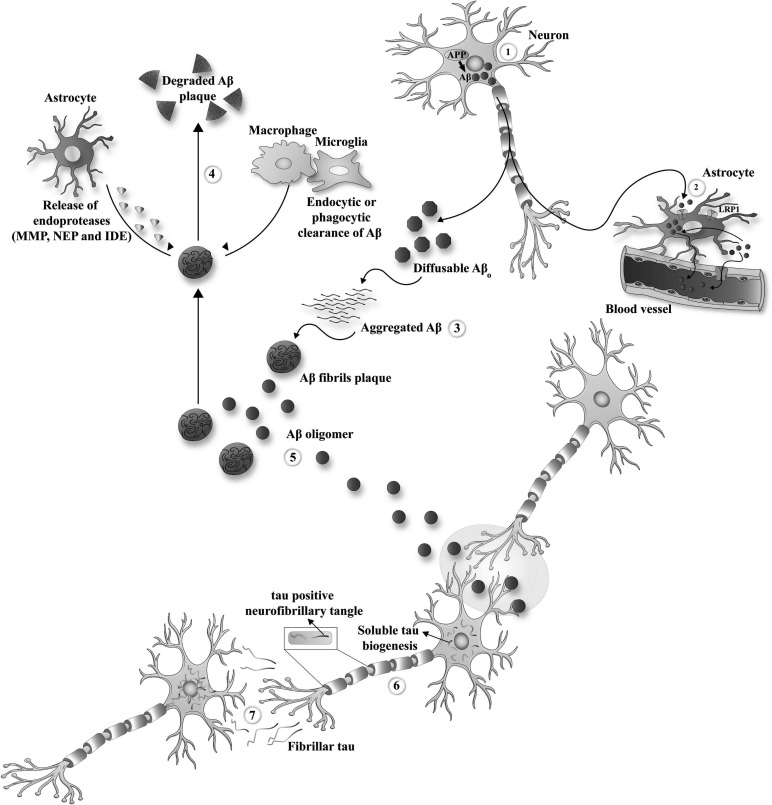
AD is highlighted by two major forms of pathological protein aggregates, namely extracellular amyloid plaques, which are an accumulation of β-amyloid (Aβ) and intracellular neurofibrillary tangles (NFTs), which are an aggregation of abnormally phosphorylated tau (Fig. 2) [50]. The precise process of AD is not elucidated yet. Many factors could promote the development of AD, but it is not easy to ascertain the exact role of each in the development of AD [51].
Fig. (2).
Mechanism of Tau formation and aggregation in Alzheimer. (A higher resolution/colour version of this figure is available in the electronic copy of the article).
There is no effective treatment for AD yet [52], however, a few drugs are prescribed to alleviate some of the symptoms of patients suffering from AD. As mentioned above, the siRNA is a powerful technique to suppress the expression of specific genes [53]. Inhibition of AD-related genes by the siRNA approach could be an excellent therapeutic option for AD’s treatment (Table 1).
Table 1.
siRNA therapeutic applications in Alzheimer’s disease.
| Target gene(s) | Delivery approach | Model(s) | Effect(s) | References |
|---|---|---|---|---|
| BACE1-AS | lentiviral vector |
In vivo: SAMP8 mice |
-Improvement of memory and learning behaviors | [52]* |
| APP | Naked siRNA |
In vitro: human neuroblastoma cell line (SH-SY5Y) |
-Improvement in synaptic activity and mitochondrial function | [68]* |
| Tau | ||||
| VDAC1 | ||||
| BACE1 | Lentiviral vectors |
In vivo: mouse model of Alzheimer disease |
-Decreasing amyloid plaque rate -improvement in neuropathological and behavioral signs |
[71]* |
| presenilin1 (PS1) | Naked siRNA |
In vitro: IMR-32 (human neuroblastoma cells) |
-Reducing the level of Aβ 42 | [75]* |
| ROCK-II | PEG–PEI co-polymer |
In vitro: C17.2 (neural stem cells) |
-Promoting axonal regeneration | [157] |
| mutant presenilin1 (L392V PS-1) | Lentiviral vector and synthetic chemically modified siRNA |
In vivo: rat model In vitro: dividing and neural stem cells |
-Decreasing the level of amyloid plaque | [158] |
| BACE1 | ||||
| I2 PP-2A | lentiviral vector |
In vivo: TG2576 mice |
-Decreasing the level of Aβ and APP and phosphorylated tau -Improvement of memory and learning ability |
[159] |
| ACAT-1 | chemically synthesized siRNA |
In vitro: human APP751 (H4APP751) |
-Reducing the enzymatic process of APP -Enhancing the level of free cholesterol |
[160] |
| BACE1 | PEGylated magnetite nanoparticles |
In vitro: HFF-1 cells |
-Significant suppression of BACE1 expression | [161] |
| BACE1 | Fusion protein TARBP-BTP |
In vivo: AbPP-PS1 mice |
-Reduction of plaque load in the cerebral cortex and hippocampus | [162] |
| Nogo receptor | poly - lysine starch nanoparticle |
In vivo: Male SD mice |
- Promoting the regeneration and repair of cholinergic neurons | [163] |
| BACE1 | PEG-PDMAEMA nanocomplex |
In vivo: APP/PS1 transgenic mice In vitro: bEnd.3 |
- Increasing the level of synaptophysin - Rescued memory loss |
[164] |
*Explained in the text.
One of the most recognized hypotheses regarding the development of AD is the amyloid cascade theory. This hypothesis suggests that the aggregations of Aβ activate a harmful cascade in the brain, which leads to the degeneration of the neurons, progressive deterioration of cognition and development of dementia [54-56]. Aβ is a peptide that is typically produced from the amyloid precursor protein (APP) cleavage. The APP is a membrane protein that takes part in cell signaling. Alternative splicing of its pre-mRNA can produce different isoforms of the APP [50, 57]. In normal cell processing, the APP is cut by the α-secretase(s) and then is cleaved by the γ-secretase. The result of this enzymatic process is a remarkably soluble and non-pathological product, a p3/p3-like portion [54, 58-59]. In opposition to this, the APP could be cut first by the β-secretase, then the different left-over membrane linked fragments are cleaved by the γ-secretase [60-61]. The resultant fragment consists of 99 residues from the C-terminal of APP. Hereafter a distinctive γ-secretase cleaves this fragment at position 40 (Aβ 1-40) or 42 of the Aβ region (Aβ 1-42). Notably, these forms of Aβ could pass from the presynaptic end to the ECM (extracellular matrix). Consequently, the insoluble fibrillary Aβ plaques are formed in the outer space of the neurons [62-65].
Besides Aβ, tau is another major player in AD pathology. In AD, hyperphosphorylated tau protein can aggregate and form intracellular bodies known as NFT. Tau is a microtubule-related protein with an essential role in both axonal and dendritic functions. It has been revealed that tau protein could mediate Aβ toxicity through dendritic function regulation [66].
APP and tau could be promising target for RNAi therapy because of their critical role in both familial and sporadic forms of AD [54, 67]. Accordingly, in an in vitro study using SHSY5Y cells (human neuroblastoma cell line), the expression of three AD-related genes, APP, tau and VDAC1, were silenced by a specific siRNA. Following the reduction of APP, tau and VDAC1 mRNAs and their proteins, the synaptic activity and mitochondrial function were improved in the transfected SHSY5Y cells. Based on these findings, the reduction of expression of these three genes could have a protective role in AD [68].
The siRNA could target the APP gene and the APP-related pathways to decrease the Aβ plaque formation. For example, BACE1 is a β-secretase involved in the cleavage processing of APP, a rate-limiting step in Aβ formation [69-70]. Hence, it is not surprising that this gene quickly became an attractive therapeutic target for the researchers. For instance, in 2005, a group of investigators used a transgenic mouse model of AD to assess the effect of reducing the BACE1 level on the improvement of Alzheimer-like symptoms in AD’s models. They utilized a lentiviral vector that expressed siRNA against the BACE1. Their experiment showed that the BACE1 suppression specifically diminished amyloid plaque rate in vivo, and the neuropathological and behavioral signs of mouse models got better [71].
Interestingly, BACE1 has a positive regulator known as BACE1-antisense transcript (BACE1-AS). It is a long noncoding RNA (lncRNA) transcribed from the reverse strand of the BACE1 gene. The BACE1-AS enhances the stability of BACE1 by forming the RNA duplex. It has been demonstrated that the concentrations of BACE1-AS are increased in patients with AD and also in the transgenic model mouse of AD. Also, changes in the BACE1-AS level could alter the amount of Aβ1–40 and Aβ 1–42 products [72]. Consistently, in a recent study, BACE1-AS expression was inhibited by administering siRNA lentivirus to bilateral hippocampi of SAMP8 mice (an AD mouse model). The main result of BACE1-AS knockdown was the amelioration of learning problem and memory loss in mice models, probably because of the improvement in neuronal growth in the hippocampus, BACE1 suppression, blocking of Aβ accumulation and decreasing of the phosphorylated tau protein [52].
As mentioned earlier, γ-secretase has an essential function in the cleavage of APP and producing the Aβ peptide. Additionally, it has been proven that presenilins (PS1 and PS2) are a critical unite of the γ-secretase complex and are needed for the γ-secretase cleavage action. On the other hand, some mutations in the PS1 gene are seen in many inherited AD [73, 74]. Hence, some researchers studied the siRNA technology in the IMR-32 neuronal cell line to determine the role of the PS1 in Aβ42 formation. Their results showed that the transfected IMR-32 cells with anti-PS1 siRNA reduced the level of Aβ42. Therefore, PS1 also could be a potential therapeutic target for gene therapy of AD [75].
4.2. Parkinson’s Disease
Parkinson’s disease (PD) is the most common movement-related disorder and the second most prevalent neurodegenerative disease following AD [76]. PD is an extremely disabling, finally fatal, and until now, an incurable disease [77]. The frequency of PD has grown up during the past two decades [76]. PD has some common symptoms, including rigidity, resting tremor, bradykinesia and posture instability [78]. The psychological problems may also appear in later stages. Two main processes lead to the progression of PD, including the formation of intracellular bodies, Lewy bodies (LB), which consist of filamentous α-synuclein aggregations in the brain of patients and the destruction of the dopaminergic neurons [79].
The current accessible therapeutic approach for PD is limited to some medications. None of them can cure the symptoms of disease entirely. They only can decelerate the progression of the disease and also have unfavorable side effects [80]. PD is a multifactorial disorder with a combination of both genetics and environmental factors [81]. Based on this rationale, the siRNA dependent approach suggests a novel treatment strategy for the management of PD.
Many studies that used the siRNA technology to treat PD focused on the α-Synuclein (α-syn) gene because of its critical role in PD pathology. α-syn is a small peripheral membrane protein that is expressed in the axonal end of the neurons [82]. The main role of this protein is to process the neurotransmitters in the presynaptic region. In this region, the α-syn interacts with the presynaptic membrane proteins, and synapsis derived vesicles. The other functions of α-syn include proteasome processing and mitochondrial function [83-87]. The first evidence demonstrating the vital role of α-syn in PD pathology was obtained from identifying a missense mutation (A53T) in the α-syn in four unrelated families with inherited PD. The high susceptibility of people with duplicated α-syn to the PD is another confirmation for the critical role of α-syn [82, 88-89].
Several lines of experiments using different methods to target the α-syn by siRNA were used to assess the potential of this approach in the treatment of PD (Table 2). For instance, the effect of the naked siRNA against the SNCA (the gene of α-syn) was evaluated, both in vivo and in vitro, and the ability of this siRNA to decrease the expression of SNCA was demonstrated [90]. In another study for the first time, the anti-SNCA siRNA was administrated to the brain (substantia nigra) of a monkey model. There was a reduction in the level of α-syn mRNA and protein. Also, no tissue-specific or systematic toxicity was reported in these monkeys. These results showed the feasibility and safety of using siRNA in the primates [91]. The efficacy of naked siRNA is very low for the reasons mentioned before. Hence, a research group used a viral vector (AAV vector) containing α-syn siRNA in a mice model. This vector was tolerated well in the mouse models of PD and successfully reduced α-syn mRNA and protein [92]. In another study, an anomalous RNAi by siRNA, namely “expression-control RNAi” (ExCont-RNAi), was developed. This method was designed to regulate the level of overexpressed SNCA. In this study, the PD model flies were exposed to the ExCont-RNAi. They showed motor function recovery following the reduced level of the SNCA. There was a positive association between the grade of motor dysfunction and the level of SNCA in the PD flies [93].
Table 2.
siRNA therapeutic applications in Parkinson’s disease.
| Target gene | Delivery system | Model(s) | Effect(s) | Reference |
|---|---|---|---|---|
|
α-synuclein (SNCA) |
Anionic liposomes decorated with a rabies virus glycoprotein -derived peptide |
In vitro: neuronal cell from P0 newborn C57BL/6J mice |
-Reducing the level of SNCA | [77] |
| Naked siRNA |
In vitro: human neuroblastoma cells (BE(2)-M17) In vivo: wild-type C57BL6 female mice |
-Reducing the level of SNCA | [90]* | |
| Naked siRNA |
In vivo: Primate Substantia Nigra |
Reducing the level of SNCA and the first evidence of successful anti-α-syn uclein intervention in the primate |
[91]* | |
| Viral vector (AAV vectors)) |
In vivo: Thy1-hSNCA mice |
-Decreased hSNCA expression -Rescue of hSNCA-mediated behavioral deficits |
[92]* | |
| ExCont-RNAi |
In vitro: Drosophila S2 cells and human fibroblasts In vivo: flies model of PD |
-Reducing the level of SNCA - Improvement in motor dysfunction |
[93]* | |
| Nanoparticle (LDH) |
In vitro: human neuroblastoma cell line (SH-SY5Y) |
-Reducing the level of SNCA | [165] | |
| PEG-PEI |
In vitro: PC12 cells |
-Protect cells from death via apoptosis | [166] | |
| PEI F25- LMW |
In vitro: human neuroblastoma cell line (SH-SY5Y) In vivo: Thy1-aSyn mice |
-Reducing the level of SNCA | [167] | |
| Peptide mediated delivery |
In vivo:
transgenic mouse model of PD |
-Reducing the accumulation of α-syn -Amelioration of inflammatory pathology |
[168] |
*Explained in the text
4.3. Huntington’s Disease
Huntington’s disease (HD) is an inherited disorder with an autosomal dominant pattern. The genetic cause of HD is trinucleotide expansion (CAG: glutamine codon) in the exon 1 of the Htt gene [94-95]. The product of this gene is the huntingtin protein, a 348-kDa protein that is present in various cells, especially in the neurons of the brain [96]. This protein plays a crucial role in a wide range of functions, including endocytosis, regulation of transcription, transport in synapsis and axonal transport [97]. The normal alleles of the Htt gene have <36 repeats of the CAG. But if these repeats increase to 36 and more, the mutant alleles are formed at the HD locus [98]. Cognitive impairment, motor dysfunction, dementia and neuronal death result from this gain of function mutation in patients with HD [99].
Among all the neurodegenerative diseases, HD is one of the best ones to be targeted by siRNA since this treatment is a suitable therapy for autosomal dominant disorders [100-104]. The effect of the Htt gene silencing by the siRNA method was assessed through different in vitro and in vivo experiments (Table 3). As a first step towards developing effective siRNAs as a therapeutic tool for HD, three different siRNAs against Htt was tested in the cell culture. The results showed that one of them, which was specific for an upstream region of CAG repeated, successfully suppressed the expression of Htt [105].
Table 3.
siRNA therapeutic applications in Huntington’s disease.
| Target gene(s) | Delivery approach | Model(s) | Effect(s) | References |
|---|---|---|---|---|
|
Htt |
Naked siRNA |
In vivo: HD transgenic mouse model, R6/2 |
-Inhibition of the Htt expression - reduction of size and number of NIIs |
[101]* |
| cholesterol-conjugated (cc) siRNA |
In vivo: viral transgenic mouse model of HD |
-Inhibition of the Htt expression - Improvement of some movement problem -Survival of neurons |
[104]* | |
|
Naked siRNA |
In vitro: -COS-7 (African green monkey fibroblasts); -SH-SY5Y (human neuroblastoma); -Neuro-2A (mouse neuroblastoma). |
- Inhibition of the Htt expression | [105]* | |
| Chitosan-based nanoparticle |
In vivo: transgenic YAC128 mouse |
- Decreasing the level of mutant htt protein | [169] |
*Explained in the text
In a study of the anti-Htt siRNA in HD, R6/2, a transgenic mouse model of HD was used. These animals expressed the mutant alleles of Huntingtin and had unusual behavior. They also formed the aggregations of polyglutamine in their neurons, namely neuronal intranuclear inclusions (NIIs). Intraventricular injection of anti-Htt siRNA showed promising results, including inhibition of the Htt in transgenic mice and reduced size and number of NIIs [101]. In a modified study, a “cholesterol-conjugated (cc) siRNA” was used to target the Htt gene. This was used since it has been demonstrated that in vitro conjugation of cholesterol and bioactive molecules could improve the uptake process [106]. This conjugation could also increase siRNA uptake [107]. Besides, the LDL receptors are present in the brain cells [108]. Their results also showed the knockdown of the Htt gene, extended survival of neurons, diminished NIIs and improvement of movement with the cholesterol-conjugated (cc) siRNA [104].
4.4. Spinal Cord Injury
Spinal cord injury (SCI) is a serious clinical issue worldwide because of the irreversible impairment of the neurons and secondary problems [109]. SCI has a heavy economic and social burden on the affected people, family, and health services [110]. SCI results in transitional or constant damage in the sensory, motor and autonomous function of the spinal cord [111]. It is regarded as a permanent disability since the CNS cannot regenerate its neuronal axons [112]. So far, significant progress has been made in the diagnosis and recovery and has increased SCI's survival rate, although there is a long way to develop an effective treatment.
Since some genetic aspects of SCI were established in the last years, using the siRNA technology to silence the involving genes has been considered an alternative therapeutic approach (Table 4). For example, ephrinB3 (ephB3) is a useful target since it has been proven that this gene is involved in the inhibition of axonal growth and decreasing the recovery rate after the CNS injury [113]. Accordingly, the effects of a lentiviral vector expressing the anti-ephB3 siRNA were tested in a rat model. This experiment revealed that the spinal cord administration of anti-ephB3 siRNA and consequently reducing the expression of the ephB3 gene lead to the recovery of the axonal regeneration and the motor function after SCI. It could also enhance the Basso-Beattie-Bresnahan (BBB) score [114].
Table 4.
siRNA therapeutic applications in spinal cord injury.
| Gene target(s) | Delivery system | Mode(s) | Effect(s) | References |
|---|---|---|---|---|
| GFAP | adenovirus vectors |
In vitro: C6 glioma cells In vivo: SCI model rat |
-Improvement of urinary function | [112]* |
| Vimentin | ||||
| EphB3 | Lentiviral vector |
In vivo: female Wistar rats |
-Improvement in axonal regeneration and the motor function | [114]* |
| iNOS | chitosan |
In vivo: Female BALB/c mice1 |
-Improvement of the secondary damage following SCI | [121]* |
| Nischarin | PEI-ALG |
In vivo: SCI model rat |
-Improvement of motor function | [124]* |
| RhoA | 2’O-methylated siRNA |
In vivo: female Sprague- Dawley rats |
-Improvement in walking -declining of allodynia |
[170] |
*Explained in the text
One of the pathological features of SCI is the accumulation of reactive astrocytes in the damaged region. The regeneration process of the neurons is disrupted, and the permanent disability is the inescapable result of such events. Reactive astrocytes are characterized by up-regulation of the intermediate filament (IF) proteins such as glial fibrillary acidic protein (GFAP) and vimentin [115]. In a study using siRNA technology, the expression of GFAP and vimentin were down-regulated in a rat model. For the assessment of its efficacy, the improvement in bladder function was tested. An improvement was evident in bladder function, demonstrating the efficacy of siRNA [112].
Another pathological condition in the SCI is neuroinflammation, where M1 macrophages have a critical role [116-118]. M1 macrophages produce a large number of inducible nitric oxide synthase (iNOS) and its product, nitric oxide (NO), which following SCI can lead to axon degeneration and demyelination [119, 120]. Hence, in the acute stage of SCI, iNOS can be a suitable target. Recently a siRNA-chitosan-antibody nanoparticle complex was used to suppress the iNOS expression in vitro and in vivo. This antibody complex helped the M1 macrophages to phagocytosis the nanoparticle by the Fc-receptor. There was a successful reduction of the iNOS expression by this complex. The results demonstrate promising evidence for improving the secondary damage following the SCI [121].
Recently a newly discovered protein with specific expression in neurons, Nischarin (Nis), was used as a target for siRNA therapy in the SCI. Nischarin can suppress neurite outgrowth as well as neurons regeneration [122-123]. For silencing of the Nis, a nano complex consisting of Nis-siRNA and PEI-ALG was developed and then administrated to a rat model with SCI. The improvement of motor function in the rat models confirmed the therapeutic potency of this method [124].
4.5. Multiple Sclerosis
Multiple sclerosis (MS) is the most common non-traumatic debilitating disorder that affects a young person [125]. It is a chronic, demyelinating, neurodegenerative and inflammatory disorder of the CNS [126]. Although this disease's precise aetiology is not clear, it is evident that MS is a heterogeneous, multifactorial complex disease developed by genetic susceptibility and environmental factors [126-127].
The focal plaques made of demyelinating lesions are the generic hallmark of all MS subtypes. They appear over the post-capillary venules in the grey and white matter of the spinal cord as well as the brain of the patients [126, 128-129]. MS is also defined as an autoimmune disorder in which both autoantibody and autoreactive T cells can destroy the myelin sheath [130]. It has an early inflammation stage and a delayed neurodegeneration stage related to, respectively, relapsing-remitting form and non-relapsing forms such as the primary and secondary progressive MS [131, 132].
The existing MS treatments are limited to the immunomodulatory or immunosuppressant agents, meaning they have to continuously take treatment. Moreover, these medications do not improve the patient’s quality of life [133, 134]. It can be said that MS is a more convenient target for the treatment by siRNA than other neurodegenerative diseases. Firstly, MS has an immunological basis so that the target cells can be triggered easily through systemic administration. Secondly, usually in MS, BBB has been broken; hence getting the siRNA to the target lesion is simpler [135].
Different genes and molecular pathways can be triggered by this method (Table 5). It has been revealed that T-bet is an essential regulator of the IFN-γ gene in Th1 (major T cell in MS pathogenesis), but not TH2. IFN-y is also a major mediator in the signaling pathway that leads to the naive T cell differentiation into the T helper cells [136-138]. The investigation through siRNA against T-bet had exciting results in both prevention and treatment. Normally, the transfection of myelin derived antigens into the mice could induce MS, namely the EAE model. But if treated T cells with both specific myelin antigen and anti-T-bet siRNA transfer to the naïve mice, the EAE induction process would fail [139]. However, if anti-T-bet siRNA was injected intravenously during the EAE induction, it will block the disease's development [139].
Table 5.
siRNA therapeutic applications in multiple sclerosis.
| Target | Delivery System | Model(s) | Effect(s) | References |
|---|---|---|---|---|
| T-bet | Naked siRNA |
In vivo: EAE mice (mouse model of MS) |
- Specifically regulate IFN - Prevented the onset of disease |
[139]* |
| Notch1 | pIRES2‐EGFP vector |
In vivo: Mouse model of acute demyelination |
- Promotion of the remyelination - Improve OL differentiation -Increase mature OL |
[142]* |
| LINGO-1 | Chitosan nanoparticles |
In vivo: Male Wistar rats |
- Better motor function -Repair in histopathological sections |
[143]* |
| NR4A2 | hemagglutinating Virus of Japan envelope (HVJ-E) vector kit |
In vivo: EAE mice (mouse model of MS) |
-Inhibiting the pathogenic potentials of IFN and IL-17 | [171] |
| TRIF | Liposome |
In vivo: EAE mice (mouse model of MS) |
-Alleviating the severity of EAE via the inhibition of interleukin and cytokine release | [172] |
| caspase-2 | Naked siRNA |
In vivo: EAE mice (mouse model of MS) |
-Significant inhibition of nerve cell loss -Decreasing in RNFL thickness - Increased survival of RGC after ON |
[173] |
| CaMKII | Naked siRNA |
In vivo: EAE mice (mouse model of MS) |
-Reduced mechanical and thermal hypersensitivity - Essential role of CaMKII_ in inducing and maintaining the evoked and non-evoked pain in EAE. |
[174] |
*Explained in the text.
There is a close association between the potency of remyelination and the level of oligodendrocyte progenitors in MS [140]. It has been revealed that in the animal models, the noch1 signaling pathway plays a role in the inadequate and impaired remyelination process [141]. More confirmation was obtained by a study in which the Notch1 specific siRNA was injected into the MS mice models. Improvement in the potency of oligodendrocyte differentiation and promotion of remyelination were the major results of this study [142]. It has also been demonstrated that LINGO-1 protein could suppress myelination and oligodendrocyte differentiation. Accordingly, in a recent study, a chitosan-based nanoparticle was loaded with siRNA against LINGO-1, and it was administrated intranasally to the rat model of demyelination. The results in the treatment group were promising. In the molecular sight, the downregulation of LINGO-1 leads to a higher level of myelin basic protein (MBP) and a lower level of caspase-3. The motor function in the remyelination treated group was also improved, indicating the neuroprotective effect of LINGO-1 silencing via siRNA [143].
5. OVERCOMING BARRIERS FOR SIRNA THERAPEUTICS
Although several studies have shown the significant potential of siRNA therapeutics in-vitro, systemic siRNA therapy faces several extracellular and intracellular barriers for translation of siRNA therapeutics from bench to bedside. Several documents have been published to comprehensively address this topic, particularly on siRNA therapeutics applications [144, 145]. However, there is an urgent need for a thorough investigation of barriers reported in the scientific literature. Also, several studies describe the methods and techniques in overcoming these barriers for the translation of siRNA therapeutics to the clinics. Briefly, the major extracellular barriers, enzymatic degradation of siRNAs by serum endonucleases and RNAases, rapid renal clearance of siRNA delivery system, impermeability of siRNA to the biological membranes, activation of the immune system, plasma protein sequestration, and capillary endothelium crossing. In contrast, the intracellular obstacles to siRNA action are the endosomal trap, arrival at the correct intracellular site of action (cytosol), and off-target effects [146].
A recent review delineates in detail the use of bioinformatics tools, software packages, and protocols in helping to design precise siRNA sequences for silencing desired genes and avoiding off-target effects [147].
Several approaches have been investigated to solve these challenges. For instance, chemical modification of the dsRNA has shown in numerous studies to attenuate immune response, permit resistance to endogenous endonucleases and exonucleases [148], improve sequence selectivity to reduce off-target RNAi activity, enhanced cellular permeability, and improved antisense strand selectivity by the RISC complex [149-153]. Another important factor in improving the effectiveness of siRNA therapy is sequence selection. The antisense strand in dsRNA is the guide strand for the activation of RISC binding to the target mRNA. The sequences of the antisense strand are the single most important determinant of siRNA effectiveness. The sequence selection is not only important on-target potency, but also it can profoundly minimize the off-target RNAi activity [154]. Avoiding siRNA's endosomal trap and enhancing its escape for its cytoplasmic activity is a major challenge in RNAi-based therapy [155]. The siRNA, along with the carriers, enters the cells via endocytosis by interacting with anionic proteoglycans to form endocytic vesicles. After entry into the cell, the siRNA is entrapped in the endosomal vesicle. In case the siRNA delivery system stays trapped in the vesicle; the late endosomal vesicle becomes acidified by membrane proton pump ATPase. It is relocated to the lysosome, where it is further acidified, resulting in degradation of the siRNA. Several carriers have been investigated to enhance the endosomal escape and increased transfection efficiency [156]. Keeping in view the progress made so far in addressing the barriers to siRNA therapy, numerous ongoing clinical trials in different stages, improved protocols for the systemic administration of siRNA treatment for targeted therapy, more mature clinical research and development, and the Good Manufacturing Practices, it is hoped that numerous siRNA based therapies will get approval from FDA in the years to come. Beyond these novel techniques and modifications in siRNA delivery systems for enabling siRNA therapeutics' success, it is much likely that RNAi therapy has several paths for impactful innovations in challenging diseases over the next decade [146].
CONCLUSION
Finding an optimal treatment for ND is still a tremendous medical challenge, maybe due to the specific conditions of these diseases, such as their complex nature, incompletely understood etiology or the physiological barrier such as BBB (blood-brain barrier), which make them difficult for drug delivery. As an alternative strategy, with its features to specific gene silencing, siRNA is a potential therapeutic option for the treatment of ND. In vivo experiments, continuous development has resulted in successful new ways of designing, identifying, and delivering siRNAs. Proof-of-principle studies in vivo have clearly demonstrated that both viral and non-viral delivery methods can provide selective and potent target gene suppression without any apparent toxic effects. There are also persistent problems with off-target effects, competition with cellular RNAi components, and effective delivery in vivo. Although recent researches and trials from a large number of animal model studies have confirmed that most off-target effects are not dangerous, other important issues need to be addressed before RNAi-based drugs are ready for clinical use. Currently, RNAi may be harnessed as a new therapeutic modality for brain diseases. Finally, there are already several RNAi-based human clinical trials in progress. It is hoped that this technology will also have effective applications in ND.
ACKNOWLEDGEMENTS
Declared none.
LIST OF ABBREVIATIONS
- AAVs
Adeno-associated Viruses
- Aβ
β-amyloid
- ACAT-1
Acetyl-CoA Acetyltransferase 1
- AD
Alzheimer’s disease
- AGO
Argonaute protein
- APP
Amyloid precursor protein
- AVs
Adenoviruses
- BACE
Beta-Secretase
- BACE1-AS
BACE1-antisense transcript
- BBB
Basso-Beattie-Bresnahan
- CaMKII
Calcium/calmodulin dependent protein kinase II
- CC
Cholesterol-conjugated
- CNS
Central Nervous System
- EAE
Experimental Autoimmune Encephalomyelitis
- ECM
Extracellular Matrix
- EphB3
EphrinB3
- ExCont-RNAi
Expression-control RNAi
- GFAP
Glial Fibrillary Acidic Protein
- HD
Huntington’s Disease
- HVJ-E
Hemagglutinating Virus of Japan Envelope
- I2 PP-2A
Inhibitor 2 of protein phosphatase 2A
- IFN-γ
Interferon gamma
- iNOS
Inducible nitric oxide synthase
- IL-17
Interleukin 17
- IF
Intermediate filament
- LDL
Low-density lipoprotein
- LVs
Lentiviruses
- LBs
Lewy bodies
- lncRNA
Long noncoding RNA
- MS
Multiple sclerosis
- ND
Neurodegenerative diseases
- NFTs
Neurofibrillary tangles
- NIIs
Neuronal intranuclear inclusions
- Nis
Nischarin
- NO
nitric oxide
- Notch1
Notch homolog 1 translocation-associated (Drosophila)
- NR4A2
Nuclear receptor subfamily 4 group A member 2
- ON
Optic neuritis
- OLs
Oligodendrocytes
- PD
Parkinson’s disease
- PEI
Polyethylenimine
- PEI-ALG
Polyethyleneimine-alginate
- PS
Presenilin
- RGC
Retinal ganglion cells
- RhoA
Ras homolog family member A
- RISC
RNA induced silencing complex
- RNAi
RNA interference
- RNFL
Retinal nerve fibre layer
- ROCK
Rho-associated protein kinase
- TRIF
TIR-domain-containing adapter-inducing interferon-β
- SAMP8
Senescence accelerated mouse-prone 8
- SCI
Spinal cord injury
- siRNA
Small Interference RNA
- SPIONs
Superparamagnetic iron oxide nanoparticles
- T-bet
T-box transcription factor
- VDAC1
Voltage-dependent anion-selective channel 1
- WHO
World Health Organization
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Gomes M.J., Martins S., Sarmento B. siRNA as a tool to improve the treatment of brain diseases: Mechanism, targets and delivery. Ageing Res. Rev. 2015;21:43–54. doi: 10.1016/j.arr.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Chekani F., Bali V., Aparasu R.R. Quality of life of patients with Parkinson’s disease and neurodegenerative dementia: A nationally representative study. Res. Social Adm. Pharm. 2016;12(4):604–613. doi: 10.1016/j.sapharm.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Batista P., Pereira A. Quality of life in patients with neurodegenerative diseases. J. Neurol. Neurosci. 2016;7(1) doi: 10.21767/2171-6625.100074. [DOI] [Google Scholar]
- 4.Paulsen J.S., Nance M., Kim J-I., Carlozzi N.E., Panegyres P.K., Erwin C., Goh A., McCusker E., Williams J.K. A review of quality of life after predictive testing for and earlier identification of neurodegenerative diseases. Prog. Neurobiol. 2013;110:2–28. doi: 10.1016/j.pneurobio.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahir-Jouzdani F., Mottaghitalab F., Dinarvand M., Atyabi F. siRNA delivery for treatment of degenerative diseases, new hopes and challenges. J. Drug Deliv. Sci. Technol. 2018;45:428–441. doi: 10.1016/j.jddst.2018.04.001. [DOI] [Google Scholar]
- 6.Brown R.H., Al-Chalabi A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2017;377(2):162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 7.Coutinho M.F., Matos L., Santos J.I., Alves S. The mRNA Metabolism in Human Disease. Springer; 2019. RNA therapeutics: how far have we gone? pp. 133–177. [DOI] [PubMed] [Google Scholar]
- 8.Sharad S. 2019. [Google Scholar]
- 9.Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E., Norris D.A., Xia S., Bennett C.F., Bishop K.M. Treatment of infantile-onset spinal muscular atrophy with nusinersen: A phase 2, open-label, dose-escalation study. Lancet. 2016;388(10063):3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 10.Li Q. Nusinersen as a therapeutic agent for spinal muscular atrophy. Yonsei Med. J. 2020;61(4):273–283. doi: 10.3349/ymj.2020.61.4.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koutsilieri E., Rethwilm A., Scheller C. Neuropsychiatric Disorders An Integrative Approach. Springer; 2007. The therapeutic potential of siRNA in gene therapy of neurodegenerative disorders. pp. 43–49. [DOI] [PubMed] [Google Scholar]
- 12.Cho K.J., Kim G.W. RNA Interference. IntechOpen; 2016. RNAi Therapeutic potentials and prospects in CNS disease. [Google Scholar]
- 13.de Fougerolles A., Vornlocher H-P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6(6):443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 15.Seyhan A.A. RNAi: a potential new class of therapeutic for human genetic disease. Hum. Genet. 2011;130(5):583–605. doi: 10.1007/s00439-011-0995-8. [DOI] [PubMed] [Google Scholar]
- 16.Watts J.K., Corey D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012;226(2):365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Paula D., Bentley M.V.L., Mahato R.I. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA. 2007;13(4):431–456. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 20.Nykänen A., Haley B., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107(3):309–321. doi: 10.1016/S0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 21.Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20(23):6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M., Doudna J. A. 2008.
- 23.Chernikov I.V., Vlassov V.V., Chernolovskaya E.L. Current development of siRNA bioconjugates: from research to the clinic. Front. Pharmacol. 2019;10:444. doi: 10.3389/fphar.2019.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J-J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305(5689):1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 25.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J-J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 27.Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 28.Rand T.A., Petersen S., Du F., Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123(4):621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 29.Hutvagner G., Simard M.J. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 30.Martinez J., Patkaniowska A., Urlaub H., Lührmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110(5):563–574. doi: 10.1016/S0092-8674(02)00908-X. [DOI] [PubMed] [Google Scholar]
- 31.Orban T.I., Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11(4):459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burnett J.C., Rossi J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012;19(1):60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shajari N., Mansoori B., Davudian S., Mohammadi A., Baradaran B. Overcoming the challenges of siRNA delivery: Nanoparticle strategies. Curr. Drug Deliv. 2017;14(1):36–46. doi: 10.2174/1567201813666160816105408. [DOI] [PubMed] [Google Scholar]
- 35.Jain R.K., Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 2010;7(11):653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumari A., Kumar V., Yadav S.K. Nanocarriers: a tool to overcome biological barriers in siRNA delivery. Expert Opin. Biol. Ther. 2011;11(10):1327–1339. doi: 10.1517/14712598.2011.587801. [DOI] [PubMed] [Google Scholar]
- 37.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16(9):543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H.J., Kim A., Miyata K., Kataoka K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv. Drug Deliv. Rev. 2016;104:61–77. doi: 10.1016/j.addr.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Wang Y., Zhu Y., Oupický D. Recent advances in delivery of drug-nucleic acid combinations for cancer treatment. J. Control. Release. 2013;172(2):589–600. doi: 10.1016/j.jconrel.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marquez A. R., Madu C. O., Lu Y. 2018.
- 41.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44(14):6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y., Huo S., Hardie J., Liang X-J., Rotello V.M. Progress and perspective of inorganic nanoparticle-based siRNA delivery systems. Expert Opin. Drug Deliv. 2016;13(4):547–559. doi: 10.1517/17425247.2016.1134486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arami H., Khandhar A., Liggitt D., Krishnan K.M. In vivo delivery, pharmacokinetics, biodistribution and toxicity of iron oxide nanoparticles. Chem. Soc. Rev. 2015;44(23):8576–8607. doi: 10.1039/C5CS00541H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choung S., Kim Y.J., Kim S., Park H-O., Choi Y-C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342(3):919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 45.Kenski D.M., Butora G., Willingham A.T., Cooper A.J., Fu W., Qi N., Soriano F., Davies I.W., Flanagan W.M. siRNA-optimized modifications for enhanced in vivo activity. Mol. Ther. Nucleic Acids. 2012;1:e5. doi: 10.1038/mtna.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terrazas M., Kool E.T. RNA major groove modifications improve siRNA stability and biological activity. Nucleic Acids Res. 2009;37(2):346–353. doi: 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie C., Smailagic N., Noel‐Storr A.H., Ukoumunne O., Ladds E.C., Martin S. CSF tau and the CSF tau/ABeta ratio for the diagnosis of Alzheimer’s disease dementia and other dementias in people with mild cognitive impairment (MCI). Cochrane Database Syst. Rev. 2017;3(3):CD010803. doi: 10.1002/14651858.CD010803.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selkoe D.J. Alzheimer’s disease: genes, proteins, and therapy. Physiol. Rev. 2001;81(2):741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 49.Brookmeyer R., Abdalla N., Kawas C.H., Corrada M.M. Forecasting the prevalence of preclinical and clinical Alzheimer’s disease in the United States. Alzheimers Dement. 2018;14(2):121–129. doi: 10.1016/j.jalz.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh S.K., Srivastav S., Yadav A.K., Srikrishna S., Perry G. Overview of Alzheimer’s disease and some therapeutic approaches targeting Aβ by using several synthetic and herbal compounds. Oxid. Med. Cell. Longev. 2016;2016:7361613. doi: 10.1155/2016/7361613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen S., Ge X., Chen Y., Lv N., Liu Z., Yuan W. Advances with RNA interference in Alzheimer’s disease research. Drug Des. Devel. Ther. 2013;7:117–125. doi: 10.2147/DDDT.S40229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W., Zhao H., Wu Q., Xu W., Xia M. Knockdown of BACE1-AS by siRNA improves memory and learning behaviors in Alzheimer’s disease animal model. Exp. Ther. Med. 2018;16(3):2080–2086. doi: 10.3892/etm.2018.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Alegre P., Paulson H.L. Technology insight: Therapeutic RNA interference--how far from the neurology clinic? Nat. Clin. Pract. Neurol. 2007;3(7):394–404. doi: 10.1038/ncpneuro0551. [DOI] [PubMed] [Google Scholar]
- 54.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 55.Hardy J. Alzheimer’s disease: the amyloid cascade hypothesis: an update and reappraisal. J. Alzheimers Dis. 2006;9(3) Suppl.:151–153. doi: 10.3233/JAD-2006-9S317. [DOI] [PubMed] [Google Scholar]
- 56.Golde T.E., Petrucelli L., Lewis J. Targeting Abeta and tau in Alzheimer’s disease, an early interim report. Exp. Neurol. 2010;223(2):252–266. doi: 10.1016/j.expneurol.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson G.V., Bailey C.D. The p38 MAP kinase signaling pathway in Alzheimer’s disease. Exp. Neurol. 2003;183(2):263–268. doi: 10.1016/S0014-4886(03)00268-1. [DOI] [PubMed] [Google Scholar]
- 58.Selkoe D.J. Amyloid β-peptide is produced by cultured cells during normal metabolism: A reprise. J. Alzheimers Dis. 2006;9(3) Suppl.:163–168. doi: 10.3233/JAD-2006-9S319. [DOI] [PubMed] [Google Scholar]
- 59.Selkoe D.J. Alzheimer’s disease results from the cerebral accumulation and cytotoxicity of amyloid beta-protein. J. Alzheimers Dis. 2001;3(1):75–80. doi: 10.3233/JAD-2001-3111. [DOI] [PubMed] [Google Scholar]
- 60.Eggert S., Paliga K., Soba P., Evin G., Masters C.L., Weidemann A., Beyreuther K. The proteolytic processing of the amyloid precursor protein gene family members APLP-1 and APLP-2 involves α-, β-, γ-, and ϵ-like cleavages: Modulation of APLP-1 processing by n-glycosylation. J. Biol. Chem. 2004;279(18):18146–18156. doi: 10.1074/jbc.M311601200. [DOI] [PubMed] [Google Scholar]
- 61.Kaether C., Haass C., Steiner H. Assembly, trafficking and function of γ-secretase. Neurodegener. Dis. 2006;3(4-5):275–283. doi: 10.1159/000095267. [DOI] [PubMed] [Google Scholar]
- 62.Turner R.S. Alzheimer’s disease. Semin. Neurol. 2006;26(5):499–506. doi: 10.1055/s-2006-951622. [DOI] [PubMed] [Google Scholar]
- 63.Ohyagi Y., Tabira T. Intracellular amyloid beta-protein and its associated molecules in the pathogenesis of Alzheimer’s disease. Mini Rev. Med. Chem. 2006;6(10):1075–1080. doi: 10.2174/138955706778560175. [DOI] [PubMed] [Google Scholar]
- 64.Newman M., Musgrave I.F., Lardelli M. Alzheimer disease: Amyloidogenesis, the presenilins and animal models. Biochim. Biophys. Acta. 2007;1772(3):285–297. doi: 10.1016/j.bbadis.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Multhaup G. Amyloid precursor protein and BACE function as oligomers. Neurodegener. Dis. 2006;3(4-5):270–274. doi: 10.1159/000095266. [DOI] [PubMed] [Google Scholar]
- 66.Ittner L.M., Ke Y.D., Delerue F., Bi M., Gladbach A., van Eersel J., Wölfing H., Chieng B.C., Christie M.J., Napier I.A., Eckert A., Staufenbiel M., Hardeman E., Götz J. Dendritic function of tau mediates amyloid-β toxicity in Alzheimer’s disease mouse models. Cell. 2010;142(3):387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- 67.Lee V.M., Goedert M., Trojanowski J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24(1):1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 68.Manczak M., Reddy P.H. RNA silencing of genes involved in Alzheimer’s disease enhances mitochondrial function and synaptic activity. Biochim. Biophys. Acta. 2013;1832(12):2368–2378. doi: 10.1016/j.bbadis.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munro K.M., Nash A., Pigoni M., Lichtenthaler S.F., Gunnersen J.M. Functions of the Alzheimer’s disease protease BACE1 at the synapse in the central nervous system. J. Mol. Neurosci. 2016;60(3):305–315. doi: 10.1007/s12031-016-0800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haniu M., Denis P., Young Y., Mendiaz E.A., Fuller J., Hui J.O., Bennett B.D., Kahn S., Ross S., Burgess T., Katta V., Rogers G., Vassar R., Citron M. Characterization of Alzheimer’s β -secretase protein BACE. A pepsin family member with unusual properties. J. Biol. Chem. 2000;275(28):21099–21106. doi: 10.1074/jbc.M002095200. [DOI] [PubMed] [Google Scholar]
- 71.Singer O., Marr R.A., Rockenstein E., Crews L., Coufal N.G., Gage F.H., Verma I.M., Masliah E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005;8(10):1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 72.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G., III, Kenny P.J., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oikawa N., Walter J. Presenilins and γ-Secretase in membrane proteostasis. Cells. 2019;8(3):E209. doi: 10.3390/cells8030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X., Li Y., Xu H., Zhang Y.W. The γ-secretase complex: from structure to function. Front. Cell. Neurosci. 2014;8:427. doi: 10.3389/fncel.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kandimalla R.J., Wani W.Y., Binukumar B.K., Gill K.D. siRNA against presenilin 1 (PS1) down regulates amyloid β42 production in IMR-32 cells. J. Biomed. Sci. 2012;19(1):2. doi: 10.1186/1423-0127-19-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tysnes O-B., Storstein A. Epidemiology of Parkinson’s disease. J. Neural Transm. (Vienna) 2017;124(8):901–905. doi: 10.1007/s00702-017-1686-y. [DOI] [PubMed] [Google Scholar]
- 77.Schlich M., Longhena F., Faustini G., O’Driscoll C.M., Sinico C., Fadda A.M., Bellucci A., Lai F. Anionic liposomes for small interfering ribonucleic acid (siRNA) delivery to primary neuronal cells: evaluation of alpha-synuclein knockdown efficacy. Nano Res. 2017;10(10):3496–3508. doi: 10.1007/s12274-017-1561-z. [DOI] [Google Scholar]
- 78.Chen S-Y., Tsai S-T. The epidemiology of Parkinson’s disease. Tzu-Chi Med. J. 2010;22(2):73–81. doi: 10.1016/S1016-3190(10)60044-4. [DOI] [Google Scholar]
- 79.Lees A.J., Hardy J., Revesz T. Parkinson’s disease. Lancet. 2009;373(9680):2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 80.Savitt J.M., Dawson V.L., Dawson T.M. Diagnosis and treatment of Parkinson disease: Molecules to medicine. J. Clin. Invest. 2006;116(7):1744–1754. doi: 10.1172/JCI29178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Del Rey N.L-G., Quiroga-Varela A., Garbayo E., Carballo-Carbajal I., Fernández-Santiago R., Monje M.H.G., Trigo-Damas I., Blanco-Prieto M.J., Blesa J. Advances in Parkinson’s disease: 200 years later. Front. Neuroanat. 2018;12:113. doi: 10.3389/fnana.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stefanis L. α-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012;2(2):a009399. doi: 10.1101/cshperspect.a009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nemani V.M., Lu W., Berge V., Nakamura K., Onoa B., Lee M.K., Chaudhry F.A., Nicoll R.A., Edwards R.H. Increased expression of α-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65(1):66–79. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanaka Y., Engelender S., Igarashi S., Rao R.K., Wanner T., Tanzi R.E., Sawa A.L., Dawson V., Dawson T.M., Ross C.A. Inducible expression of mutant α-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum. Mol. Genet. 2001;10(9):919–926. doi: 10.1093/hmg/10.9.919. [DOI] [PubMed] [Google Scholar]
- 85.Lashuel H.A., Overk C.R., Oueslati A., Masliah E. The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013;14(1):38–48. doi: 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bellucci A., Mercuri N.B., Venneri A., Faustini G., Longhena F., Pizzi M., Missale C., Spano P. Review: Parkinson’s disease: From synaptic loss to connectome dysfunction. Neuropathol. Appl. Neurobiol. 2016;42(1):77–94. doi: 10.1111/nan.12297. [DOI] [PubMed] [Google Scholar]
- 87.Bellucci A., Zaltieri M., Navarria L., Grigoletto J., Missale C., Spano P. From α-synuclein to synaptic dysfunctions: new insights into the pathophysiology of Parkinson’s disease. Brain Res. 2012;1476:183–202. doi: 10.1016/j.brainres.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 88.Chartier-Harlin M-C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., Waucquier N., Defebvre L., Amouyel P., Farrer M., Destée A. α-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364(9440):1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 89.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M.R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. α-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302(5646):841–841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 90.Lewis J., Melrose H., Bumcrot D., Hope A., Zehr C., Lincoln S., Braithwaite A., He Z., Ogholikhan S., Hinkle K., Kent C., Toudjarska I., Charisse K., Braich R., Pandey R.K., Heckman M., Maraganore D.M., Crook J., Farrer M.J. In vivo silencing of alpha-synuclein using naked siRNA. Mol. Neurodegener. 2008;3(1):19. doi: 10.1186/1750-1326-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McCormack A.L., Mak S.K., Henderson J.M., Bumcrot D., Farrer M.J., Di Monte D.A. α-synuclein suppression by targeted small interfering RNA in the primate substantia nigra. PLoS One. 2010;5(8):e12122. doi: 10.1371/journal.pone.0012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim Y.C., Miller A., Lins L.C., Han S.W., Keiser M.S., Boudreau R.L., Davidson B.L., Narayanan N.S. RNA Interference of human α-Synuclein in mouse. Front. Neurol. 2017;8:13. doi: 10.3389/fneur.2017.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takahashi M., Suzuki M., Fukuoka M., Fujikake N., Watanabe S., Murata M., Wada K., Nagai Y., Hohjoh H. Normalization of overexpressed α-Synuclein causing Parkinson’s disease by a moderate gene silencing with RNA Interference. Mol. Ther. Nucleic Acids. 2015;4:e241. doi: 10.1038/mtna.2015.14. [DOI] [PubMed] [Google Scholar]
- 94.La Spada A.R., Taylor J.P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11(4):247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Budworth H., McMurray C.T. Trinucleotide Repeat Protocols. Springer; 2013. A brief history of triplet repeat diseases. pp. 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saudou F., Humbert S. The biology of huntingtin. Neuron. 2016;89(5):910–926. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 97.Imarisio S., Carmichael J., Korolchuk V., Chen C-W., Saiki S., Rose C., Krishna G., Davies J.E., Ttofi E., Underwood B.R., Rubinsztein D.C. Huntington’s disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412(2):191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 98.Lee J-M., Ramos E.M., Lee J-H., Gillis T., Mysore J.S., Hayden M.R., Warby S.C., Morrison P., Nance M., Ross C.A., Margolis R.L., Squitieri F., Orobello S., Di Donato S., Gomez-Tortosa E., Ayuso C., Suchowersky O., Trent R.J., McCusker E., Novelletto A., Frontali M., Jones R., Ashizawa T., Frank S., Saint-Hilaire M.H., Hersch S.M., Rosas H.D., Lucente D., Harrison M.B., Zanko A., Abramson R.K., Marder K., Sequeiros J., Paulsen J.S., Landwehrmeyer G.B., Myers R.H., MacDonald M.E., Gusella J.F. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012;78(10):690–695. doi: 10.1212/WNL.0b013e318249f683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamamoto A., Lucas J.J., Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101(1):57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 100.Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T., Paulson H.L., Yang L., Kotin R.M., Davidson B.L. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10(8):816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 101.Wang Y-L., Liu W., Wada E., Murata M., Wada K., Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington’s disease by siRNA. Neurosci. Res. 2005;53(3):241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 102.Xia H., Mao Q., Paulson H.L., Davidson B.L. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002;20(10):1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 103.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. USA. 2005;102(16):5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G., Manoharan M., Sah D.W., Zamore P.D., Aronin N. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. USA. 2007;104(43):17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu W., Goto J., Wang Y-L., Murata M., Wada K., Kanazawa I. Specific inhibition of Huntington’s disease gene expression by siRNAs in cultured cells. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2003;79(10):293–298. doi: 10.2183/pjab.79B.293. [DOI] [Google Scholar]
- 106.Cheng K., Ye Z., Guntaka R.V., Mahato R.I. Enhanced hepatic uptake and bioactivity of type alpha1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J. Pharmacol. Exp. Ther. 2006;317(2):797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- 107.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., John M., Kesavan V., Lavine G., Pandey R.K., Racie T., Rajeev K.G., Röhl I., Toudjarska I., Wang G., Wuschko S., Bumcrot D., Koteliansky V., Limmer S., Manoharan M., Vornlocher H.P. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 108.Hofmann S.L., Russell D.W., Goldstein J.L., Brown M.S. mRNA for low density lipoprotein receptor in brain and spinal cord of immature and mature rabbits. Proc. Natl. Acad. Sci. USA. 1987;84(17):6312–6316. doi: 10.1073/pnas.84.17.6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kose E. A., Bakar B., Ayva S. K., Kilinc K., Apan A. 2012. [PubMed]
- 110.Hachem L.D., Ahuja C.S., Fehlings M.G. Assessment and management of acute spinal cord injury: From point of injury to rehabilitation. J. Spinal Cord Med. 2017;40(6):665–675. doi: 10.1080/10790268.2017.1329076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Krassioukov A. Autonomic function following cervical spinal cord injury. Respir. Physiol. Neurobiol. 2009;169(2):157–164. doi: 10.1016/j.resp.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 112.Toyooka T., Nawashiro H., Shinomiya N., Shima K. Down-regulation of glial fibrillary acidic protein and vimentin by RNA interference improves acute urinary dysfunction associated with spinal cord injury in rats. J. Neurotrauma. 2011;28(4):607–618. doi: 10.1089/neu.2010.1520. [DOI] [PubMed] [Google Scholar]
- 113.Duffy P., Wang X., Siegel C.S., Tu N., Henkemeyer M., Cafferty W.B., Strittmatter S.M. Myelin-derived ephrinB3 restricts axonal regeneration and recovery after adult CNS injury. Proc. Natl. Acad. Sci. USA. 2012;109(13):5063–5068. doi: 10.1073/pnas.1113953109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qu Y., Zhao J., Wang Y., Gao Z. Silencing ephrinB3 improves functional recovery following spinal cord injury. Mol. Med. Rep. 2014;9(5):1761–1766. doi: 10.3892/mmr.2014.2019. [DOI] [PubMed] [Google Scholar]
- 115.Pekny M., Pekna M. Astrocyte intermediate filaments in CNS pathologies and regeneration. J. Pathol. 2004;204(4):428–437. doi: 10.1002/path.1645. [DOI] [PubMed] [Google Scholar]
- 116.Kigerl K.A., Gensel J.C., Ankeny D.P., Alexander J.K., Donnelly D.J., Popovich P.G. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J. Neurosci. 2009;29(43):13435–13444. doi: 10.1523/JNEUROSCI.3257-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.David S., Kroner A. Repertoire of microglial and macrophage responses after spinal cord injury. Nat. Rev. Neurosci. 2011;12(7):388–399. doi: 10.1038/nrn3053. [DOI] [PubMed] [Google Scholar]
- 118.Kuo H-S., Tsai M-J., Huang M-C., Chiu C-W., Tsai C-Y., Lee M-J., Huang W-C., Lin Y-L., Kuo W-C., Cheng H. Acid fibroblast growth factor and peripheral nerve grafts regulate Th2 cytokine expression, macrophage activation, polyamine synthesis, and neurotrophin expression in transected rat spinal cords. J. Neurosci. 2011;31(11):4137–4147. doi: 10.1523/JNEUROSCI.2592-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Greenhalgh A.D., David S. Differences in the phagocytic response of microglia and peripheral macrophages after spinal cord injury and its effects on cell death. J. Neurosci. 2014;34(18):6316–6322. doi: 10.1523/JNEUROSCI.4912-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.David S. Mechanisms underlying macrophage polarization in spinal cord injury-detrimental and beneficial influences on recovery. FASEB. 2015;29(1):210. [Google Scholar]
- 121.Gao W., Li J. Targeted siRNA delivery reduces nitric oxide mediated cell death after spinal cord injury. J. Nanobiotechnology. 2017;15(1):38. doi: 10.1186/s12951-017-0272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ding Y., Li Y., Lu L., Zhang R., Zeng L., Wang L., Zhang X. Inhibition of nischarin expression promotes neurite outgrowth through regulation of PAK activity. PLoS One. 2015;10(12):e0144948. doi: 10.1371/journal.pone.0144948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alahari S.K., Lee J.W., Juliano R.L. Nischarin, a novel protein that interacts with the integrin α5 subunit and inhibits cell migration. J. Cell Biol. 2000;151(6):1141–1154. doi: 10.1083/jcb.151.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ding Y.M., Li Y.Y., Wang C., Huang H., Zheng C.C., Huang S.H., Xuan Y., Sun X.Y., Zhang X. Nischarin-siRNA delivered by polyethylenimine-alginate nanoparticles accelerates motor function recovery after spinal cord injury. Neural Regen. Res. 2017;12(10):1687–1694. doi: 10.4103/1673-5374.217348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kobelt G., Thompson A., Berg J., Gannedahl M., Eriksson J., Group M.S., Platform E.M.S. New insights into the burden and costs of multiple sclerosis in Europe. Mult. Scler. 2017;23(8):1123–1136. doi: 10.1177/1352458517694432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Filippi M., Bar-Or A., Piehl F., Preziosa P., Solari A., Vukusic S., Rocca M.A. Multiple sclerosis. Nat. Rev. Dis. Primers. 2018;4(1):43. doi: 10.1038/s41572-018-0041-4. [DOI] [PubMed] [Google Scholar]
- 127.Ascherio A. Environmental factors in multiple sclerosis. Expert Rev. Neurother. 2013;13(12) Suppl.:3–9. doi: 10.1586/14737175.2013.865866. [DOI] [PubMed] [Google Scholar]
- 128.Minagar A., Alexander J.S. Blood-brain barrier disruption in multiple sclerosis. Mult. Scler. 2003;9(6):540–549. doi: 10.1191/1352458503ms965oa. [DOI] [PubMed] [Google Scholar]
- 129.Ortiz G.G., Pacheco-Moisés F.P., Macías-Islas M.Á., Flores-Alvarado L.J., Mireles-Ramírez M.A., González-Renovato E.D., Hernández-Navarro V.E., Sánchez-López A.L., Alatorre-Jiménez M.A. Role of the blood-brain barrier in multiple sclerosis. Arch. Med. Res. 2014;45(8):687–697. doi: 10.1016/j.arcmed.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 130.Ma X., Jiang Y., Wu A., Chen X., Pi R., Liu M., Liu Y. Berberine attenuates experimental autoimmune encephalomyelitis in C57 BL/6 mice. PLoS One. 2010;5(10):e13489. doi: 10.1371/journal.pone.0013489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Leray E., Yaouanq J., Le Page E., Coustans M., Laplaud D., Oger J., Edan G. Evidence for a two-stage disability progression in multiple sclerosis. Brain. 2010;133(Pt 7):1900–1913. doi: 10.1093/brain/awq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Coles A.J., Cox A., Le Page E., Jones J., Trip S.A., Deans J., Seaman S., Miller D.H., Hale G., Waldmann H., Compston D.A. The window of therapeutic opportunity in multiple sclerosis: evidence from monoclonal antibody therapy. J. Neurol. 2006;253(1):98–108. doi: 10.1007/s00415-005-0934-5. [DOI] [PubMed] [Google Scholar]
- 133.Ransohoff R.M., Hafler D.A., Lucchinetti C.F. Multiple sclerosis-a quiet revolution. Nat. Rev. Neurol. 2015;11(3):134–142. doi: 10.1038/nrneurol.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dobson R., Giovannoni G. Multiple sclerosis - a review. Eur. J. Neurol. 2019;26(1):27–40. doi: 10.1111/ene.13819. [DOI] [PubMed] [Google Scholar]
- 135.Lovett-Racke A.E., Cravens P.D., Gocke A.R., Racke M.K., Stüve O. Therapeutic potential of small interfering RNA for central nervous system diseases. Arch. Neurol. 2005;62(12):1810–1813. doi: 10.1001/archneur.62.12.1810. [DOI] [PubMed] [Google Scholar]
- 136.Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., Glimcher L.H. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. doi: 10.1016/S0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 137.Afkarian M., Sedy J.R., Yang J., Jacobson N.G., Cereb N., Yang S.Y., Murphy T.L., Murphy K.M. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat. Immunol. 2002;3(6):549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 138.Shier P., Hofstra C.L., Ma X-J., Wu Y., Ngo K., Fung-Leung W-P. Tbt-1, a new T-box transcription factor induced in activated Th1 and CD8+ T cells. Immunogenetics. 2000;51(10):771–778. doi: 10.1007/s002510000212. [DOI] [PubMed] [Google Scholar]
- 139.Lovett-Racke A.E., Rocchini A.E., Choy J., Northrop S.C., Hussain R.Z., Ratts R.B., Sikder D., Racke M.K. Silencing T-bet defines a critical role in the differentiation of autoreactive T lymphocytes. Immunity. 2004;21(5):719–731. doi: 10.1016/j.immuni.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 140.Zhang Y., Zhang J., Navrazhina K., Argaw A.T., Zameer A., Gurfein B.T., Brosnan C.F., John G.R. TGFbeta1 induces Jagged1 expression in astrocytes via ALK5 and Smad3 and regulates the balance between oligodendrocyte progenitor proliferation and differentiation. Glia. 2010;58(8):964–974. doi: 10.1002/glia.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hanafy K.A., Sloane J.A. Regulation of remyelination in multiple sclerosis. FEBS Lett. 2011;585(23):3821–3828. doi: 10.1016/j.febslet.2011.03.048. [DOI] [PubMed] [Google Scholar]
- 142.Fan H., Zhao J.G., Yan J.Q., Du G.Q., Fu Q.Z., Shi J., Yang Y.H., Du X.W., Bai X.L. Effect of Notch1 gene on remyelination in multiple sclerosis in mouse models of acute demyelination. J. Cell. Biochem. 2018;119(11):9284–9294. doi: 10.1002/jcb.27197. [DOI] [PubMed] [Google Scholar]
- 143.Youssef A.E.H., Dief A.E., El Azhary N.M., Abdelmonsif D.A., El-Fetiany O.S. LINGO-1 siRNA nanoparticles promote central remyelination in ethidium bromide-induced demyelination in rats. J. Physiol. Biochem. 2019;75(1):89–99. doi: 10.1007/s13105-018-00660-6. [DOI] [PubMed] [Google Scholar]
- 144.Zhang P., An K., Duan X., Xu H., Li F., Xu F. Recent advances in siRNA delivery for cancer therapy using smart nanocarriers. Drug Discov. Today. 2018;23(4):900–911. doi: 10.1016/j.drudis.2018.01.042. [DOI] [PubMed] [Google Scholar]
- 145.Setten R.L., Rossi J.J., Han S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18(6):421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 146.Sajid M.I., Moazzam M., Kato S., Yeseom Cho K., Tiwari R.K. Overcoming barriers for siRNA therapeutics: From bench to bedside. Pharmaceuticals (Basel) 2020;13(10):E294. doi: 10.3390/ph13100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fakhr E., Zare F., Teimoori-Toolabi L. Precise and efficient siRNA design: A key point in competent gene silencing. Cancer Gene Ther. 2016;23(4):73–82. doi: 10.1038/cgt.2016.4. [DOI] [PubMed] [Google Scholar]
- 148.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 149.Robbins M., Judge A., Liang L., McClintock K., Yaworski E., MacLachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 2007;15(9):1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- 150.Snead N.M., Escamilla-Powers J.R., Rossi J.J., McCaffrey A.P. 5′ Unlocked nucleic acid modification improves siRNA targeting. Mol. Ther. Nucleic Acids. 2013;2:e103. doi: 10.1038/mtna.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shum K., Rossi J. SiRNA delivery methods. 2016.
- 152.Janas M.M., Schlegel M.K., Harbison C.E., Yilmaz V.O., Jiang Y., Parmar R., Zlatev I., Castoreno A., Xu H., Shulga-Morskaya S., Rajeev K.G., Manoharan M., Keirstead N.D., Maier M.A., Jadhav V. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018;9(1):723. doi: 10.1038/s41467-018-02989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bramsen J.B., Laursen M.B., Nielsen A.F., Hansen T.B., Bus C., Langkjaer N., Babu B.R., Højland T., Abramov M., Van Aerschot A., Odadzic D., Smicius R., Haas J., Andree C., Barman J., Wenska M., Srivastava P., Zhou C., Honcharenko D., Hess S., Müller E., Bobkov G.V., Mikhailov S.N., Fava E., Meyer T.F., Chattopadhyaya J., Zerial M., Engels J.W., Herdewijn P., Wengel J., Kjems J. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37(9):2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 155.Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35(3):222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 156.Subhan M.A., Torchilin V.P. siRNA based drug design, quality, delivery and clinical translation. Nanomedicine (Lond.) 2020;29:102239. doi: 10.1016/j.nano.2020.102239. [DOI] [PubMed] [Google Scholar]
- 157.Liu Y., Liu Z., Wang Y., Liang Y-R., Wen X., Hu J., Yang X., Liu J., Xiao S., Cheng D. Investigation of the performance of PEG-PEI/ROCK-II-siRNA complexes for Alzheimer’s disease in vitro. Brain Res. 2013;1490:43–51. doi: 10.1016/j.brainres.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 158.Sierant M., Kubiak K., Kazmierczak-Baranska J., Paduszynska A., Kuwabara T., Warashina M., Nacmias B., Sorbi S., Nawrot B. Nucleic Acids Symposium Series. Oxford: University Press; 2008. In RNA interference in silencing of genes of Alzheimer’s disease in cellular and rat brain models. pp. 41–42. [DOI] [PubMed] [Google Scholar]
- 159.Wei W. Effect of targeting i2pp-2a with sirnas on alzheimer-like pathology in tg2576 mice. Alzheimers Dement. 2009;5(4):43. doi: 10.1016/j.jalz.2009.05.652. [DOI] [Google Scholar]
- 160.Huttunen H.J., Greco C., Kovacs D.M. Knockdown of ACAT-1 reduces amyloidogenic processing of APP. FEBS Lett. 2007;581(8):1688–1692. doi: 10.1016/j.febslet.2007.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lopez-Barbosa N., Garcia J.G., Cifuentes J., Castro L.M., Vargas F., Ostos C., Cardona-Gomez G.P., Hernandez A.M., Cruz J.C. Multifunctional magnetite nanoparticles to enable delivery of siRNA for the potential treatment of Alzheimer’s. Drug Deliv. 2020;27(1):864–875. doi: 10.1080/10717544.2020.1775724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Haroon M.M., Saba K., Boddedda V.H., Kumar J.M., Patel A.B., Gopal V. Delivery of BACE1 siRNA mediated by TARBP-BTP fusion protein reduces β-amyloid deposits in a transgenic mouse model of Alzheimer’s disease. J. Biosci. 2019;44(1):1. doi: 10.1007/s12038-018-9822-x. [DOI] [PubMed] [Google Scholar]
- 163.Wang Z., Cheng Y., Zhao D., Pliss A., Liu J., Luan P. Synergic treatment of Alzheimer’s disease with brain targeted nanoparticles incorporating NgR-siRNA and brain derived neurotrophic factor. Smart Materials in Medicine. 2020;1:125–130. doi: 10.1016/j.smaim.2020.08.001. [DOI] [Google Scholar]
- 164.Guo Q., Zheng X., Yang P., Pang X., Qian K., Wang P., Xu S., Sheng D., Wang L., Cao J., Lu W., Zhang Q., Jiang X. Small interfering RNA delivery to the neurons near the amyloid plaques for improved treatment of Alzheimer׳s disease. Acta Pharm. Sin. B. 2019;9(3):590–603. doi: 10.1016/j.apsb.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Acharya R., Chakraborty M., Chakraborty J. Prospective treatment of Parkinson’s disease by a siRNA-LDH nanoconjugate. MedChemComm. 2019;10(2):227–233. doi: 10.1039/C8MD00501J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu Y.Y., Yang X.Y., Li Z., Liu Z.L., Cheng D., Wang Y., Wen X.J., Hu J.Y., Liu J., Wang L.M., Wang H.J. Characterization of polyethylene glycol-polyethyleneimine as a vector for alpha-synuclein siRNA delivery to PC12 cells for Parkinson’s disease. CNS Neurosci. Ther. 2014;20(1):76–85. doi: 10.1111/cns.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Helmschrodt C., Höbel S., Schöniger S., Bauer A., Bonicelli J., Gringmuth M., Fietz S.A., Aigner A., Richter A., Richter F. Polyethylenimine nanoparticle-mediated siRNA delivery to reduce α-Synuclein expression in a model of Parkinson’s disease. Mol. Ther. Nucleic Acids. 2017;9:57–68. doi: 10.1016/j.omtn.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Spencer B., Trinh I., Rockenstein E., Mante M., Florio J., Adame A., El-Agnaf O.M.A., Kim C., Masliah E., Rissman R.A. Systemic peptide mediated delivery of an siRNA targeting α-syn in the CNS ameliorates the neurodegenerative process in a transgenic model of Lewy body disease. Neurobiol. Dis. 2019;127:163–177. doi: 10.1016/j.nbd.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sava V., Fihurka O., Khvorova A., Sanchez-Ramos J. Enriched chitosan nanoparticles loaded with siRNA are effective in lowering Huntington’s disease gene expression following intranasal administration. Nanomedicine (Lond.) 2020;24:102119. doi: 10.1016/j.nano.2019.102119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Otsuka S., Adamson C., Sankar V., Gibbs K.M., Kane-Goldsmith N., Ayer J., Babiarz J., Kalinski H., Ashush H., Alpert E., Lahav R., Feinstein E., Grumet M. Delayed intrathecal delivery of RhoA siRNA to the contused spinal cord inhibits allodynia, preserves white matter, and increases serotonergic fiber growth. J. Neurotrauma. 2011;28(6):1063–1076. doi: 10.1089/neu.2010.1568. [DOI] [PubMed] [Google Scholar]
- 171.Doi Y., Oki S., Ozawa T., Hohjoh H., Miyake S., Yamamura T. Orphan nuclear receptor NR4A2 expressed in T cells from multiple sclerosis mediates production of inflammatory cytokines. Proc. Natl. Acad. Sci. USA. 2008;105(24):8381–8386. doi: 10.1073/pnas.0803454105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang X., Zheng X., Ma C., Zhao L. Role of TRIF small interference RNA (siRNA) in chronic Experimental Allergic Encephalomyelitis (EAE). Med. Sci. Monit. 2015;21:2583–2587. doi: 10.12659/MSM.894564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lidster K., Jackson S.J., Ahmed Z., Munro P., Coffey P., Giovannoni G., Baker M.D., Baker D. Neuroprotection in a novel mouse model of multiple sclerosis. PLoS One. 2013;8(11):e79188. doi: 10.1371/journal.pone.0079188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hu X., Huang F., Wang Z.J. CaMKIIα mediates the effect of IL-17 to promote ongoing spontaneous and evoked pain in multiple sclerosis. J. Neurosci. 2018;38(1):232–244. doi: 10.1523/JNEUROSCI.2666-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]