Abstract
Autophagy and phagocytosis are two important endogenous lysosomal dependent clearing systems in the organism. In some neurological disorders, excessive autophagy or dysfunctional phagocytosis has been shown to contribute to brain injury. Recent studies have revealed that there are underlying interactions between these two processes. However, different studies show inconsistent results for the contribution of autophagy to the phagocytic process in diverse phagocytes and relatively little is known about the link between them especially in the brain. It is critical to understand the role that autophagy plays in phagocytic process in order to promote the clearance of endogenous and exogenous detrimental materials. In this review, we highlight the studies focusing on phagocytosis and autophagy occurring in the brain and summarizing the possible regulatory roles of autophagy in the process of phagocytosis. Balancing the roles of autophagy and phagocytosis may be a promising therapeutic strategy for the treatment of some neurological diseases in the future.
Keywords: Phagocytosis, autophagy, cross-talk, interactions, brain, LC3 associated phagocytosis, microglia
1. INTRODUCTION
Autophagy and phagocytosis are two highly conserved endogenous lysosomal dependent clearing processes with similar morphological characteristics and functions. Both processes can form transient vesicular structures (autophagosomes and phagosomes, respectively) that engulf and deliver cargo to the lysosomes for digestion [1]. In addition, they are important in the maintenance of cellular and tissue homeostasis through degrading detrimental intra- and extra-cellular material [2]. Autophagy is an intracellular homeostatic mechanism whereby cytosolic constituents including aberrant organelles and proteins are delivered to the lysosomes for degradation. Unlike autophagy, phagocytosis comprises of the ingestion of extracellular agents, such as dying cells and pathogens to prevent the spillover of proinflammatory and neurotoxic molecules. However, excessive autophagy or dysfunctional phagocytosis can also exacerbate brain injury under certain pathological conditions [3-5]. Balancing the roles of autophagy and phagocytosis may be important for the treatment of some neurological disorders. Recently more attention has been given to the link between autophagy and phagocytosis [1, 2, 6-8]. Some research results have shown that autophagy can regulate phagocytosis via affecting the expression of target-recognizing receptors, phagosome maturation, and recycling of the phagocytic receptors. However, contrary findings have been reported in different studies about the relationship between these two processes. The nature of the interactions and underlying mechanisms between autophagy and phagocytosis remain unclear. In this review, we will discuss the role of autophagy and phagocytosis in the brain and summarize the recent advances that have been made in exploring the mechanisms underlying the cross-talk between autophagy and phagocytosis in diverse phagocytes. In summary, more experimental studies aiming to better understand the regulatory effect of autophagy on phagocytosis are needed to develop potential therapeutic avenues that can promote the phagocytic clearance of brain-derived pathological cargo in some neurological disorders.
2. PHAGOCYTOSIS IN THE BRAIN
2.1. Mechanism of Phagocytosis
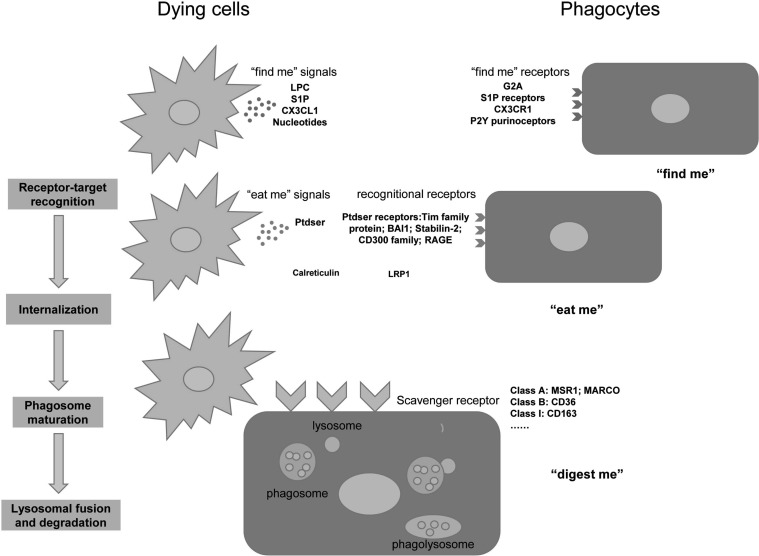
Phagocytosis is the process through which cells recognize, engulf, and digest large particles (>0.5um) [9, 10]. It is a receptor-mediated process consisting of three major steps: “find me”, “eat me”, and “digest me” that eventually results in the removal and elimination of particles including, but not limited to, bacteria, apoptotic cells, neoplastic cells, or cellular debris [10, 11]. Common phagocytes include monocytes, macrophages, microglia, dendritic cells, Langerhans cells, osteoclasts, and so on [12, 13]. Microglia have been regarded as the major phagocytes in the brain [14, 15]. During phagocytosis, self and non-self target particles can be recognized by specific receptors on the plasma membrane, and this recognition relies on coordinated specific engulfment signals [12]. We depict the engulfment of apoptotic cells as an example to elucidate the process of phagocytosis (Fig. 1).
Fig. (1).
The major steps of phagocytosis: “find me”, “eat me”, and “digest me”. This figure shows dying cells as an example to depict the process of phagocytosis. Dying cells release “find me” signals to attract the migration of phagocytes. The attracted phagocytes recognize and tether the target through “eat me” signals. Next, the phagocytes engulf and internalize the cells via scavenger receptors.Finally, the phagosome fuses with lysosome and the formed phagolysosome which degrades the target. LPC: lysophosphatidylcholine; G2A:LPC receptor, also termed G-protein–coupled receptor 132(GPR132); S1P:sphingosine-1-phosphate; CX3CL1:CX3C motif chemokine ligand 1; CX3CR1: CX3C chemokine receptor 1;PtdSer: phosphatidylserine; Tim: T-cell immunoglobulin and mucin domain-containing molecule; BAI1: Brain-specific angiogenesis inhibitor 1; RAGE: Receptor for advanced glycation end products; LRP1: Low density lipoprotein receptor-related protein 1; MSR1: Macrophage scavenger receptor 1; MARCO: macrophage receptor with collagenous structure. (A higher resolution/colour version of this figure is available in the electronic copy of the article).
First, the apoptotic cells release “find me” signals to attract phagocytes toward them [16]. Some of the most representative “find me” signals include lysophosphatidylcholine (LPC), sphingosine-1-phosphate (S1P), CX3C motif chemokine ligand 1 (CX3CL1), and nucleotides [17]. The signals bind with the corresponding “find me” receptors expressed on the phagocytes to facilitate the migration of macrophages to the apoptotic cells [12, 13, 16-18].
Next, when the macrophages are close enough to the target cells, a set of cell surface molecules expressed on the apoptotic cells can tag these cells as dead. These tags are the so-called “eat me” signals for phagocytosis [13]. Therefore, in the second step the dying cells expose “eat me” signals to be engulfed by macrophages. The most well-known and important “eat me” signal is phosphatidylserine (PtdSer). PtdSer is a type of phospholipid located on the inner membrane leaflet of the lipid bilayer in healthy cells, and when under apoptotic stress, PtdSer will be externalized on the outside cell surface [19]. The exposed PtdSer on apoptotic cell surface is recognized directly by PtdSer receptors (e.g. T cell immunoglobulin and mucin domain containing (Tim) family proteins, Brain-specific angiogenesis inhibitor 1(BAI1), Stabilin-2, CD300f and Receptor for advanced glycation end products (RAGE)) or indirectly by some bridging molecules (e.g. proteinS and C1q) [17]. In addition, Calreticulin (CRT), integrins, immunoglobulins (IgG superfamily), and complement proteins are other important “eat me” signals expressed on the apoptotic cell surface [17, 20]. As for healthy cells, “don’t eat me” signals on the cell surface such as CD47 and CD31 prevent live cells from being phagocytosed. CD47 was found to bind to signal regulatory protein-α(SIRPα) on macrophages to inhibit phagocytosis [13, 21]. It has been reported that there exists a repulsive signal CD31-CD31 homotypic interaction between viable neutrophils and phagocytes, which can mediate detachment of viable cells from phagocytes. However, this repulsive signal does not exist in apoptotic cells [22].
Finally, in the “digest me” stage, phagocytes engulf and completely degrade apoptotic cells in the lysosomal compartment [23]. After receptor-target recognition, complex signaling cascades are elicited leading to cytoskeletal rearrangement of actin filaments (F-actin). Actin polymerization serves as the force driving membrane extension and phagosome formation [12, 17]. The cargo is internalized into single-membrane phagosomes, and then these compartments fuse with lysosome to form the phagolysosome, promoting the cargo degradation [24].
2.2. Scavenger Receptors
Scavenger receptors(SRs) are transmembrane glycoproteins that typically bind numerous ligands and promote the clearance of non-self or altered-self targets [11]. They are important in the recognition and internalization of the targets. Based on the sequence similarity or shared structural features, scavenger receptors can be classified into different classes [25-28]. However, there is still a lack of a consensus standardized nomenclature system, which may affect communication and collaboration among investigators. The most studied classes of receptors include the class A, class B and class I scavenger receptors. SR-A1, also called macrophage scavenger receptor 1 (MSR1), is one of the most common class types of A scavenger receptors (SR-A) in macrophages, monocytes, microglia and dendritic cells. It has been found that SR-A1 can bind to β-amyloid (Aβ), heat shock proteins, surface molecules of bacteria, hepatitis C virus, and modified low density lipoprotein (acetylated LDL and oxidized LDL) [29]. The macrophage receptor with collagenous structure (MARCO) is another member of the class A SRs family. The expression level of MARCO in human brain, cultured human astrocytes and rodent microglia is very low. However, it is highly expressed in rodent astrocytes and macrophages [26]. CD36 is one of the most widely studied class B SRs expressed in monocytes, endothelial cells, and microglia. CD36 plays an important role in the recognition and endocytic uptake of erythrocytes, thrombospondin, collagen, lipids, fatty acids, apoptotic cells and amyloid proteins [30-32]. CD163 is the prototype class I scavenger receptor for haptoglobin-hemoglobin (Hp-Hb) complexes [25, 31]. In addition, high levels of CD163 is a feature of macrophages undergoing differentiation toward the “alternatively activated” M2 phenotype [27].
2.3. Detection of Phagocytosis
Quantifying phagocytosis is critical for understanding its contribution to pathophysiological process of various diseases. Multiple in vitro and in vivo techniques have been applied to study phagocytosis. Flow cytometry and microscopy are frequently used to assess the uptake by cells in vitro of fluorescently-labeled synthetic or physiological particles (e.g. latex, Aβ, myelin, zymosan or dextran) [10, 33, 34]. In one of the studies, the phagocytosis of erythrocytes by microglia was detected by 5(6)-carboxyfluorescein diacetate-labeled red blood cells (RBCs) applied to tag the target. The fluorescence intensity of the cell lysate from microglia containing engulfed RBCs can be referred to as phagocytosis index indicating the phagocytic efficacy of microglia [34]. However, the in vitro culture environment of phagocytes is different from the microenvironment in vivo. Furthermore, cultured cells are often used with synthetic phagocytic targets such as latex beads. Performing either in vitro or in vivo phagocytic assays with beads is artificial, as these particles do not release any chemoattractant and therefore cannot promote phagocytosis [35]. Thus in vitro assays may not accurately recapitulate physiological phagocytosis. It has been reported that alternatively activated M2-like microglia promote phagocytosis of RBCs and tissue debris in intracerebral hemorrhage (ICH) rodent model [36]. In addition, M2 microglia also play important roles in phagocytosis and toxicity clearance in other brain diseases, such as ischemic stroke [37, 38], traumatic brain injury (TBI) [39] and epilepsy [40]. In vivo studies have used the ratio of M2/M1 microglia (e.g. CD16, CD86 for M1 markers and CD206, Arginase 1 for M2 markers) to reflect phagocytosis indirectly [36, 41]. However, it might be misleading to use “markers” as a proxy for phagocytosis. Some studies have developed different methodological approaches to directly quantify phagocytosis in vivo, such as 3D electron microscopy reconstruction and live imaging using 2-photon microscopy [42-44]. In one study, researchers suggested that a novel magnetic resonance imaging(MRI) post-processing technique of gradient-recalledecho (GRE), the quantitative susceptibility mapping (QSM), is an accurate and noninvasive method for quantifying brain iron level, reflecting the hematoma clearance indirectly in an ICH Minipig Model [45].
2.4. Phagocytosis in Neurological Disorders
Microglia are the major phagocytes in the brain which can engulf and degrade microbes as well as various brain-derived cargo such as apoptotic cells, synapses, myelin and protein deposits such as Aβ and aggregated α-synuclein (α-syn) [12, 46, 47]. Whether microglial phagocytosis plays a beneficial or detrimental role in brain diseases remains controversial although most researchers now are inclined to the former [47]. Numerous studies show that microglia maintain homeostasis and possibly contribute to the neural network by assisting in synaptic remodeling and plasticity [48]. Synaptic pruning is mediated by microglia during neuronal circuit formation in the developing brain [49]. It has been found that efficient clearance of tissue debris is critical in the reconstruction and reorganization of neuronal networking after an injury in the brain [43, 50, 51]. Microglia phagocytosis promoted axon regeneration and microenvironment restoration during the recovery of an acute brain injury [47]. However, microglia have also been found to pathologically phagocytose synapses of neurons with tau pathology in Alzheimer’s disease (AD), which can indirectly lead to more tau-induced synapse loss [49], and the loss of synapses has emerged as a major correlate of cognitive decline in AD recently [52]. Apart from microglia, other phagocytes in the brain can also play a role in the removal of brain-derived cargo [53]. A study showed that monocyte-derived macrophages (MDMs), once localized to the site of injury, have a higher phagocytic capacity than microglia in cerebral ischemic stroke model [54]. In addition, astrocytes can also participate in the elimination of synapses and neuronal debris [10, 14, 15]. Recently, it has been found that phagocytosis by astrocytes can be actuated when microglial phagocytic activity is impaired. Astrocytes express TAM phagocytic receptors (MerTK and AXL),which are the main astrocytic phagocytic receptors for cell debris [55]. Moreover, infiltrated T cells also play an indirect role in regulation of phagocytosis. T cells extravasating into the brain can modulate the adaptive immune makeup in central nervous system (CNS)for not only the clearance of patho-proteins but can also promote the repair of damaged neurons by secreting neurotropic factors [56]. Furthermore, activated microglia enhanced the phagocytosis of degenerating myelin in the presence of infiltrated myelin-reactive T cells possibly through interleukin-2 (IL-2) and interleukin-1 beta (IL-1β) secretion [57, 58].
2.4.1. Phagocytosis and Neurodegenerative Diseases
Some human genetic and experimental studies showed a clear link between impaired microglial phagocytosis and neurodegenerative diseases [12, 59]. Triggering receptor expressed on myeloid cells 2(TREM2), a kind of pattern recognition receptor in the brain with various anionicligands (e.g. phosphatidylethanolamine (PE), phosphatidylserine (PS), cardiolipin (CL), phosphatidic acid (PA)), signals by forming a complex with the co-receptor DNAX-activation protein 12 (DAP12) [60, 61]. TREM2-DAP12 complex induces the activation of the spleen tyrosine kinase (Syk)/ phospholipase C-gamma (PLC-γ)/phosphoinositide 3-kinase (PI3K) signaling pathway, leading to the promotion of microglia phagocytosis [59]. Variants in TREM2 increase the risk for AD [59, 62], and have been associated with Parkinson’s disease (PD) [12]. For animal studies, TREM2-/- AD mice showed diffused amyloid plaques, with amyloid filaments extending outwards, and fewer compact amyloid plaques compared to control AD mice [59]. On the other hand, overexpression of TREM2 reduced amyloid burden and improved cognitive function in AD mice [63]. It has been reported that TREM2 can promote Aβ phagocytosis via activating CCAAT enhancer-binding protein alpha (C/EBPα)-dependent CD36 expression in microglia [64]. In addition, TREM2 has also been demonstrated to be involved in myelin phagocytosis in Multiple Sclerosis(MS) [10]. Furthermore, inhibition of TREM2 increased the severity of experimental autoimmune encephalomyelitis (EAE). TREM2 overexpression was found to be protective, partially caused by effects on clearance of myelin debris [65-67]. Recently, another famous member of TREM family, TREM1, has been reported to facilitate microglial phagocytosis of Aβ [68, 69]. Evidence showed that rs6910730G, an intronic variant of TREM1, reduced the ability of human monocytes for Aβ phagocytosis in a cohort human study [68]. Additionally, knockdown of TREM1 in the brains of APP/PSEN1 mice increased Aβ1-42 levels and total amyloid burden. While selective overexpression of TREM1 on microglia ameliorated Aβ neuropathology and rescued AD-related spatial cognitive dysfunction [68]. The TAM family receptors, MerTK and AXL, which bind PtdSer via the bridging molecules Protein S and Gas6, respectively, was shown to promote myelin phagocytosis in MS [70-72]. In the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced progressive model of PD, neurodegeneration was associated with decreased expression of scavenger receptor Mannose Receptor C-Type1 (MRC1) [46]. Scavenger receptors such as MSR1, CD36, and CD163 have been found to be involved in macrophage activation and increased phagocytosis of Aβ in AD [26, 73].
2.4.2. Phagocytosis and Acute Brain Disorders
Promoting microglia phagocytic clearance of hematoma has been found to be a promising therapeutic target to ameliorate inflammation and improve neurological outcomes after ICH and germinal matrix hemorrhage (GMH) [31, 74-76]. Endogenous scavenger receptors, such as CD36,MSR1, CD163 and low-density lipoprotein receptor related protein-1 (LRP1, also called CD91) have been reported to play important roles in hematoma clearance [77]. In adult hemorrhagic stroke models, activated transcription factors Nrf2 and peroxisome proliferator-activated receptor (PPAR-γ) could upregulate CD163 and CD36 expression, and consequently augmented microglia/macrophage phagocytic clearance of hematoma [77-79]. Hp-Hb complex can be recognized by CD163 receptors in microglia, promoting hemoglobin degradation and anti-inflammatory effects [80, 81]. Similarly, phagocytes expressing CD91 can endocytose the haem-haemopexin complex and dissociate it by lysosomal activity [82, 83]. In addition, the class A scavenger receptors MSR1 and MARCO play key roles in the internalization of damage-associated molecular patterns (DAMPs) by mononuclear phagocytes to resolve inflammation and prevent the exacerbation of ischemic stroke pathologies [84].
3. AUTOPHAGY IN THE BRAIN
3.1. Mechanism of Autophagy
Autophagy is a conserved cellular pathway involved in protein and organelle degradation that delivers cytoplasmic constituents to the lysosomes for degradation [85]. There are three different types of autophagy including macroautophagy, microautophagy, and chaperone-mediated autophagy [86-89]. Macroautophagy, the best characterized form of autophagy in mammalian cells, usually referred to simply as autophagy is mostly the focus in this review. Autophagy relies on the coordinated action of various members of the autophagy-related gene (ATG) protein family, which together underlie the induction and nucleation, elongation and maturation of autophagosome (AP), as well as the fusion of autophagosomes with lysosomes [90]. Autophagy can be induced by nutrient starvation through the inhibition of mammalian target of rapamycin (mTOR), resulting in translocation of mTOR substrate complex consisting of Unc-51-like kinase 1/2(ULK1/2), ATG13, FAK family interacting-protein of 200 kDa (FIP200) and ATG101 from the cytosol to certain domains of the endoplasmic reticulum or closely attached structures [3, 91]. Next, the recruited class III phosphatidylinositol-3-OH kinase (PI(3)K) complex (Beclin-1-Atg14L-Vps34) produces phosphatidylinositol-3-phosphate (PtdIns(3)P), which binds with effectors such as double FYVE-containing protein 1 (DFCP1) and WD-repeat domain phosphoinositide-interacting (WIPI) family proteins [89]. After the formation of phagophore, the membrane expands and envelopes the cargo to promote the maturation of the autophagosome. Next, the autophagosome delivers its cargo to the lysosome to form the autolysosomes for degradation [5, 89].
3.2. Microglia Autophagy in Neurological Disorders
Most of the literature have been published on the role of neuronal autophagy in the brain, and few studies have focused on glial autophagy, such as that performed by microglia and astrocytes. It has been reported that autophagy can regulate immune response of macrophages including inflammation and phagocytic functions [1, 91, 92]. However, there are controversial results whether microglia autophagy plays beneficial or detrimental functions under different pathogenic states in the brain. For example, moderate recruitment of autophagy is beneficial in abolishing brain hypoperfusion. However, its overactivity may be detrimental for cell survival [93]. The discrepant effects may depend on the timing and amount of autophagy activation, thus correctly fine tuning of the time and intensity will be important for clinical application [93].
3.2.1. Microglia Autophagy and Neurodegenerative Diseases
One of the pathological characteristics of PD is the presence of Lewy bodies, an eosinophilic cytoplasmic inclusion comprised largely of α-syn fibrils. It has been found that activation of autophagy in BV2 cells can lead to a reduction of α-syn induced pro-inflammation [94]. On the other hand, inhibition of autophagy in microglia caused PD-like symptoms in mice through activating NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome via phosphodiesterase 10A (PDE10A)–cyclic adenosine monophosphate (cAMP) signaling pathway [95]. In an Aβ-induced AD model, microglial autophagy was shown to participate in the degradation of extracellular Aβ fibrils, and the impairment of microglial autophagy resulted in increased inflammation via increasing IL-1β and activating inflammasome [96]. However, another study demonstrated positive correlation between Beclin-1, IL-1β, and Tumor necrosis factor alpha (TNF-α) in the cortex and/or hippocampus of AD mice, suggesting a relationship between inflammatory responses and autophagy [97].
3.2.2. Microglia Autophagy and Acute Brain Disorders
Microglia are activated in response to injury as one of the main drivers of inflammatory responses after neurotrauma [3, 98]. An increase in markers of autophagy has been reported in microglia after TBI, spinal cord injury(SCI), ischemic stroke and ICH in cellular and animal models [99-102]. However, whether autophagy has positive or negative effects on the inflammatory response after acute brain injury remains unclear. Some studies showed that autophagy exerted neurotoxic effects. It has been reported that cerebral ischemia induced-microglia autophagy closely accompanied ischemic neuroinflammation and injury in a permanent middle cerebral artery occlusion (pMCAO) model in mice [103]. And autophagy induced by toll-like receptor 4(TLR4) activation contributed to microglial activation and inflammatory injury in ICH model [104]. A study showed that the autophagy inhibitor 3-methyladenine (3-MA) attenuated white matter injury and improved working memory during chronic cerebral hypoperfusion in mice [105]. While some other studies have reported that autophagy inhibited neuroinflammatory damage, the autophagy activator rapamycin promoted a shift in microglia from M1 to the favorable M2 phenotype and produced anti-inflammatory effects in ischemic stroke and subarachnoid hemorrhage (SAH) models [106, 107].
4. LINK BETWEEN AUTOPHAGY AND PHAGOCYTOSIS
It has been reported that there potentially exists a link between autophagy and phagocytosis. Many studies have shown that phagocytosis can be regulated by autophagy (Table 1). The potential regulatory effects of autophagy over phagocytosis may occur at various steps of the phagocytic cascade, including cargo uptake, phagosomes maturation, fusion with lysosomes and recycling of phagocytic receptors [1].
Table1.
Current evidence of the cross-talk between autophagy and phagocytosis.
| - | Phagocyte | Disease/Model | Relationship | Mechanism | Refs. |
|---|---|---|---|---|---|
| Autophagy is negatively correlated with phagocytosis | In vitro studies | ||||
| Macrophages | Infecting macrophages with Mtb or BCG | Myeloid-specific Atg7-/- macrophages exhibited higher Mtb and BCG uptake | Accumulating SQSTM1 activated Nrf2, leading to upregulation of MSR1 and MARCO | [2] | |
| Dendritic cells | Irradiating live EG7 cells with γ-ray, and then coculturing with BMDCs | Atg5-deficient dendritic cells showed increased phagocytosis of apoptotic tumor cells | Increased expression of scavenger receptor CD36 | [6] | |
| Macrophages | Macrophages were exposed to CFDA-SE-labeled Escherichia coli K-12, CFDA-SE-labeled Lactobacillus reuteri ATCC PTA 6475, or Fluoresbrite YG microsphere |
Decreased phagocytosis was seen in Hrh2-/- macrophages with increased expression of autophagy genes Beclin-1 and ATG12 | Increased Beclin-1 and ATG12 expression was accompanied by decreased MSR1 expression and MSR1 surface abundance in Hrh2-/- macrophages | [11] | |
| Macrophages | Coculturing heat inactivated yeast particles or L. amazonensis stationary phase promastigotes with macrophages | Autophagy induced by physiological (starvation) and pharmacological (rapamycin) methods was shown to reduce the phagocytic capacity of murine macrophages | Inhibiting particle internalization | [126] | |
| Macrophages | Infecting RAW264.7 cells with ST239-MRSA | Autophagy inhibitor 3-MA promoted phagocytosis of macrophage | - | [127] | |
| In vivo studies | |||||
| Macrophages | Intranasally infecting mice with BCG | Atg7-/- mice showed increased bacterial loads | - | [2] | |
| Dendritic cells | Injecting apoptotic tumor cells into the footpads of the indicated mice | Atg5-deficient dendritic cells showed increased phagocytosis of apoptotic tumor cells | Increased expression of scavenger receptor CD36 | [6] | |
| Macrophages | Intranasally inserting ST239-MRSA to establish mouse pneumonia model | Autophagy inhibitor 3-MA protected mice from MRSA pneumonia | - | [127] | |
| Glia | Knocking down Draper (isoform I) in cortex glia in the brain of Drosophila melanogaster | Inactivating autophagy through TORC1 activation could rescue the accumulation of apoptotic neurons in the brain of Draper-/- deficient Drosophila melanogaster | - | [128] | |
| Microglia | AD patients carrying TREM2 risk variants; TREM2-deficient mice with AD-like pathology | Increased autophagy was detected in TREM2-deficient microglia and in AD patients carrying TREM2 variants |
TREM2 maintained microglia at high metabolic states through enhanced activation of the mTOR pathway | [130] | |
| Neutrophils | NLRP3-deficient (Nlrp3-/-) mice in polymicrobial sepsis model induced by CLP | HMGB1-/- peritoneal cells (primarily neutrophils) showed decreased autophagy and augmented phagocytosis | - | [131] | |
| Autophagy is positively correlated with phagocytosis | In vitro studies | ||||
| Microglia | LPS treated microglia was incubated with Aβ1−42 fibrils | TLR4 activation induced by LPS suppressed autophagy and impaired phagocytic capacity of microglia | - | [121] | |
| Macrophages | Macrophages were exposed to apoptotic lymphocytes or Jurkat T cells induced by cyclophosphamide | Autophagy inhibitor 3-MA decreased phagocytosis of the apoptotic cells by macrophages | - | [138] | |
| - | Phagocyte | Disease/Model | Relationship | Mechanism | Refs. |
| Microglia | Uptake of latex beads for Beclin-1 knockdown BV2 microglial cells |
Reducing microglial Beclin-1 levels significantly impaired phagocytosis, and phagocytosis was “rescued” by recovering Beclin-1 levels with a lentivirus encoding mouse Beclin-1 | Reduced Beclin-1 disrupted retromer-mediated recycling of phagocytic receptors CD36 and TREM2 | [139] | |
| Microglia | Incubation of DJ-1 deficient microglia with α-syn. | DJ-1 deficiency impaired autophagy and reduced uptake and clearance of α-syn in the phagocytosis in microglia | - | [144] | |
| In vivo studies | |||||
| Microglia | Injecting fibrillar Aβ into the frontal cortex of Beclin-1+/- mice | Reduced microglial Beclin-1 impaired Aβ phagocytosis | - | [139] | |
| Epithelial hyp7 cell | Apoptotic C. elegans apoptotic Q cells degraded by epithelial an hyp7 cell | Autophagy proteins LC3, ATG-18, EPG-5 act within the phagocyte to promote apoptotic cell degradation | ATG18 and EPG-5loss showed delayed recruitment of RAB-5, RAB-7, and lysosomal markers onto the apoptotic Q cell corpse | [143] | |
Abbreviations: Mtb: Mycobacterium tuberculosis; BCG: M. tuberculosis var. bovisBCG;MSR1: Macrophage scavenger receptor 1; MARCO: macrophage receptor with collagenous structure; BMDCs: Bone marrow-derived dendritic cells; Hrh2: histamine H2 receptor gene; MRSA: Methicillin-resistant Staphylococcus aureus; TORC1: Target of Rapamycin Complex 1; TREM2: Triggering receptor expressed on myeloid cells 2; mTOR: mammalian target of rapamycin; CLP: cecal ligation and puncture; LPS:lipopolysaccharide; Aβ:β-amyloid; TLR4: Toll-like receptor 4;α-syn: α -synuclein.
4.1. Microtubule-associated Protein 1 Light Chain 3 (LC3) Associated Phagocytosis
One recently discovered process called LC3-associated phagocytosis (LAP) bridges autophagy and phagocytosis in macrophages and retinal pigment epithelial (RPE) cells, since LC3 is involved in both processes. LAP is generally defined as a novel form of non-canonical autophagy, sometimes regarded as a special type of phagocytosis as well [46, 108, 109]. It is well-known that autophagosome formation involves the recruitment of LC3 from the cytosol to the limiting membrane of the phagophore where it provides a binding site for autophagy cargos and facilitates fusion with lysosomes [110]. LAP is a process activated by TLR signaling and NADPH oxidase (NOX) during phagocytosis of fungal and bacterial pathogens or apoptotic and necrotic cells, resulting in attachment of LC3 to the cytosolic side of the phagosome membrane where it facilitates phagosome maturation [108, 111]. LC3 is recruited to the single-membrane phagosome depending on the activity of autophagy enzymes Beclin-1, ATG5 and ATG7, but not the recruitment of ULK1 [112-114]. It has been reported that Rubicon and NOX2 are uniquely required by LAP, which can help distinguish between LAP and canonical autophagy [111].
The LC3-decorated phagosomes formed in this process, named LAPosomes, show enhanced fusion with lysosomes resulting in enhanced degradation of contained microbes [115]. In addition, LAP has been reported to be required for the daily clearance of ingested material in the RPE which is important for the photoreceptor outer segment (POS) renewal [116, 117].
Although most studies published so far have shown that LAP promotes phagosome maturation, a study in human macrophages observed quite the opposite-LC3 recruitment to zymosan-containing phagosomes was associated with delayed phagosome fusion with the lysosome [118]. Some researches suggest that whether LAP promotes or suppresses phagosome maturation depends on the foreign target and requires function of synaptosomal associated protein of 23 kDa (SNAP23). SNAP23 that is enriched on the phagosome membrane during LAP may be phosphorylated or dephosphorylated, thereby enhancing or inhibiting subsequent phagosome maturation, respectively [119]. Therefore, the influence of LC3 recruitment to phagosomes remains to be further studied.
In addition, it has been reported that LAP is not universally required for the fusion of phagosome with endosomes and lysosomes [24]. However, LC3-decorated phagosomes show enhanced lysosomal fusion, leading to a more effective degradation of their contents compared to LC3-lacking phagosomes [120]. Although few studies have focused on LAP in microglia, some researchers think this process exists in the brain [121, 122]. It has been reported that microglial autophagy associated phagocytosis is essential for the recovery of neuroinflammation in a murine model of MS [123]. Recently, Green DR et al. identified a related but distinct process of LC3-associated endocytosis (LANDO), which plays a protective role in the endocytosis of Aβ involved in AD [124]. It has been found that even in non-phagocytic cells, this process is required for the recycling of several internalized surface receptors to the plasma membrane. LANDO in microglia was shown to facilitate Aβ clearance and mitigate neurodegeneration in a murine model of AD [125].
In summary, LAP is a unique pathway that links signaling during phagocytosis with recruitment of some members of the autophagy machinery. However, the impact of LC3 recruitment to microglial phagocytosis remains to be tested.
4.2. Contribution of Autophagy to Phagocytic Efficiency
In addition to LAP, autophagy and phagocytosis may also be intimately reciprocally regulated in macrophages and microglia through other mechanisms. However, how autophagy modulates the efficiency of phagocytosis, such as promoting or inhibiting phagocytosis, is still unclear. Contrary results have been observed in different studies.
4.2.1. Autophagy is Negatively Correlated with Phagocytosis
Some studies showed that activation of autophagy suppressed phagocytosis [126], while inhibition or loss of autophagy could enhance phagocytosis [2, 127]. Lima et al. found that autophagy induction inhibited classical phagocytosis in murine macrophages via a mechanism that does not interfere with particle-receptor interaction [126]. Induction of autophagy did not affect the capacity of macrophages to recognize and bind to particles, even though particle internalization was suppressed [126]. Bonilla et al. demonstrated that the lack of ATG7 in peripheral macrophages increased phagocytic uptake of bacteria, possibly through enhanced expression of class A scavenger receptors MARCO and MSR1 in phagocyte cell surface [2]. Furthermore, an increase in scavenger receptors was observed with increased activity of the nuclear factor E2-related factor 2(Nrf2) resulting from the accumulation of sequestosome 1 (SQSTM1) in ATG7-/- macrophages [2]. In addition, the authors found that ATG3-/-, ATG5-/-, and ATG7-/- Mouse embryonic fibroblasts (MEFs) all had increased surface expression of MARCO and MSR1 and increased ability to internalize M. tuberculosis var. bovis BCG (BCG), suggesting that the observed changes in scavenger receptor expression and phagocytosis in ATG7-/- cells were autophagy dependent and not due to an autophagy-independent role of ATG7 [2]. A similar study recently found that lack of ATG5 in dendritic cells increased the phagocytic uptake of apoptotic tumor cells, possibly through the enhancement of CD36 expression [6]. It has been reported that the absence of phagocytic receptor Draper in glia can lead to a pronounced accumulation of apoptotic neurons in the brain of Drosophila melanogaster. Inactivating autophagy through Target of Rapamycin Complex 1(TORC1) activation or ATG1 inhibition in glia was sufficient to rescue apoptotic cells accumulation as well as neurodegeneration by regulating phagosome maturation [128]. Furthermore, increased Beclin-1 and ATG12 expression was accompanied by decreased MSR1 expression and MSR1 surface abundance in macrophages [11].
Recent studies showed that certain mediators are involved in the cross-talk between autophagy and phagocytosis. Among these factors, TREMs have been linked to the autophagy pathway [129]. One study showed that increased autophagy was detected in TREM2-deficient microglia and in AD patients carrying TREM2 variants, reflecting a negative correlation between phagocytic receptor TREM2 and autophagy [130]. Additionally, several recent studies indicate that the inflammasome NLRP3 and high-mobility group box protein 1 (HMGB1) may act as cross-talk mediators between autophagy and phagocytosis.After knockout of NLRP3 in peritoneal cells (primarily neutrophils), decreased autophagy, augmented phagocytosis and enhanced scavenger receptor MARCO and mannose binding leptin (MBL) expression was observed in a polymicrobial sepsis mice model [131]. Likewise, HMGB1 could inhibit microglia phagocytosis of Aβ in AD [132, 133]. HMGB1 regulates autophagy through interacting with autophagy related protein Beclin-1.The underlying mechanism might be that binding of HMGB1 in the cytoplasm with Beclin-1 promotes dissociation of Beclin-1 from the apoptosis inhibitor bcl-2, and facilitates binding of Beclin-1 and class III inositol 3 kinase (PI3K ClassIII)/ Vsp34 that activates autophagy [134-136]. However, no study to date has simultaneously evaluated the change in autophagy and phagocytosis under the action of HMGB1.
In summary, the papers discussed above indicate the inhibitory effect of autophagy on phagocytosis, and the mechanisms might be associated with the negative regulation of autophagy on the expression of scavenger receptors or maturation of phagosomes. Recently, some researchers proposed a new possible mechanism to explain the negative correlation effect. They think that since the formation of both phagolysosomes and autolysosomes requires involvement of membrane material from the plasma membrane, competitively to use the limited cellular membrane resources may be the reason for negative regulation between autophagy and phagocytosis [11].
4.2.2. Autophagy is Positively Correlated with Phagocytosis
On the other hand, some other research studies show a positive correlation effect of autophagy on the process of phagocytosis. It has been reported that inhibition of autophagy with autophagy inhibitor 3-MA led to reduced phagocytic efficiency in activated macrophages [137, 138]. Inhibiting Beclin-1disrupted phagocytic efficiency by impaired recycling of phagocytic receptor such as CD36 and TREM2 in AD model [139]. In addition, Beclin-1coordinated actin dynamics and membrane phospholipid synthesis to promote efficient apoptotic cell engulfment [140]. The C. elegans Q neuroblasts (Q cells) provide an appealing in vivo model system to understand the roles of autophagy genes in phagocytes for apoptotic cell removal, both apoptotic Q cells and the neighboring phagocyte hyp7 cell can be individually identified using cell type-specific promoters [141, 142]. Autophagy proteins LC3, ATG18 and Ectopic P-Granules Autophagy Protein 5 (EPG-5) acted within the phagocyte to promote apoptotic Q cells degradation, and the engulfment activity was decreased after the loss of autophagy genes ATG18 or EPG-5 [143]. As for the mechanism, ATG18 and EPG-5 loss showed delayed recruitment of lysosomal markers to the internalized apoptotic Q cell corpse. In addition, the recruitment of small guanosine triphosphatases RAB-7 and RAB-5 onto the phagosome was also delayed with ATG18 and EPG-5 loss, which affected phagolysosome formation [143].
In addition, some studies showed positive correlation between autophagy and phagocytosis. For example, TLR4 activation by lipopolysaccharide (LPS) in microglia could suppress autophagy and impair phagocytic capacity of microglia via inhibiting the activation of transcription factor Forkhead box O3 (FOXO3) [121]. Likewise, DJ-1 deficiency impaired autophagy and reduced uptake and clearance of α -syn in the phagocytosis by microglia, while the molecular mechanism remains unclear [144].
These studies indicate that autophagy may have a cytoprotective effect on phagocytosis. It should be noted that both 3-MA and Beclin-1 or LC3 reduction can also inhibit LAP. Of note, many of the pharmacological and gene approaches used in autophagy research may also affect LAP, which could complicate data interpretation [1],and should be taken into consideration when interpreting findings from the studies. Further research is essential to explore whether LAP plays a role in the phagocytosis process of macrophages and microglia.
CONCLUSION AND FUTURE DIRECTIONS
In this review, we focused on the emerging evidence and the roles of autophagy on phagocytosis. Published literature indicates that there are interactions between these two processes. The studies mostly showed that autophagy machinery can regulate engulfment activity of phagocytes through modulating the expression of the scavenger receptors. In this review we highlighted how autophagy can influence phagocytosis. Conversely, phagocytosis machinery might also affect autophagy. The activation of Class A scavenger receptor inhibited endoplasmic reticulum stress-induced autophagy in macrophage [145]. The phagocytic receptor TREM2 promoted the degradation oftype I transmembrane protein TMEM59, whose expression could facilitate autophagic flux through its carboxyl-terminus [146]. However, based on current studies it is unclear whether autophagy is positively or negatively correlated with phagocytosis. Contradictory results from different studies may be due to inconsistent spatial and temporal assessment of inflammation, phagocytic activity and outcomes in various models. In addition, the studies summarized in this review focused on diverse phagocytic cell types, which might also account for the discrepant results. More research is essential to determine whether autophagy can also regulate internalization process, phagosome maturation, fusion with lysosome and recycling of the phagocytic receptors. In addition, most of the studies to date were preliminary observations on how autophagy influences phagocytosis, while little is known about the molecular mechanisms that mediate the effects. Another challenge in the field has been the difficulty in detecting the in vivo phagocytosis process directly in different models. Although some new imaging techniques such as 3D electron microscopy reconstruction and live imaging with 2-photon microscopy have been proposed to observe the engulfment process, the cost effectiveness of these techniques needs may be limiting.
A better understanding of the role of autophagy in phagocytosis is critical, as promoting the phagocytic clearance of brain-derived cargo such as apoptotic cells, Aβ, myelin debris, and hematoma in the brain could be a beneficial therapeutic strategy for many neurological disorders. However, there are currently limited studies published on the interactions between autophagy and phagocytosis in the brain. Future research efforts are needed to study the cross-talk between autophagy and phagocytosis which may provide novel therapeutic avenues.
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This review was supported by NIH NS101284 to Jiping Tang.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Plaza-Zabala A., Sierra-Torre V., Sierra A. Autophagy and microglia: novel partners in neurodegeneration and aging. Int. J. Mol. Sci. 2017;18(3):18. doi: 10.3390/ijms18030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonilla D.L., Bhattacharya A., Sha Y., Xu Y., Xiang Q., Kan A., Jagannath C., Komatsu M., Eissa N.T. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity. 2013;39(3):537–547. doi: 10.1016/j.immuni.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H., Wu J., Shen H., Yao X., Liu C., Pianta S., Han J., Borlongan C.V., Chen G. Autophagy in hemorrhagic stroke: Mechanisms and clinical implications. Prog. Neurobiol. 2018;163-164:79–97. doi: 10.1016/j.pneurobio.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Scheiblich H., Bicker G. Regulation of microglial phagocytosis by RhoA/ROCK-inhibiting drugs. Cell. Mol. Neurobiol. 2017;37(3):461–473. doi: 10.1007/s10571-016-0379-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L., Bravo-San Pedro J.M., Blomgren K., Kroemer G. Autophagy in acute brain injury. Nat. Rev. Neurosci. 2016;17(8):467–484. doi: 10.1038/nrn.2016.51. [DOI] [PubMed] [Google Scholar]
- 6.Oh D.S., Lee H.K. Autophagy protein ATG5 regulates CD36 expression and anti-tumor MHC class II antigen presentation in dendritic cells. Autophagy. 2019;15(12):2091–2106. doi: 10.1080/15548627.2019.1596493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadwell K., Philips J.A. Autophagy meets phagocytosis. Immunity. 2013;39(3):425–427. doi: 10.1016/j.immuni.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 8.Oczypok E.A., Oury T.D., Chu C.T. It’s a cell-eat-cell world: autophagy and phagocytosis. Am. J. Pathol. 2013;182(3):612–622. doi: 10.1016/j.ajpath.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon S. Phagocytosis: An immunobiologic process. Immunity. 2016;44(3):463–475. doi: 10.1016/j.immuni.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Galloway D.A., Phillips A.E.M., Owen D.R.J., Moore C.S. Phagocytosis in the brain: Homeostasis and disease. Front. Immunol. 2019;10:790. doi: 10.3389/fimmu.2019.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fultz R., Engevik M.A., Shi Z., Hall A., Herrmann B., Ganesh B.P., Major A., Haag A., Mori-Akiyama Y., Versalovic J. Phagocytosis by macrophages depends on histamine H2 receptor signaling and scavenger receptor 1. MicrobiologyOpen. 2019;8(10):e908. doi: 10.1002/mbo3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tremblay M.E., Cookson M.R., Civiero L. Glial phagocytic clearance in Parkinson’s disease. Mol. Neurodegener. 2019;14(1):16. doi: 10.1186/s13024-019-0314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemke G. How macrophages deal with death. Nat. Rev. Immunol. 2019;19(9):539–549. doi: 10.1038/s41577-019-0167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung Y.J., Chung W.S. Phagocytic roles of glial cells in healthy and diseased brains. Biomol. Ther. (Seoul) 2018;26(4):350–357. doi: 10.4062/biomolther.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damisah E.C., Hill R.A., Rai A., Chen F., Rothlin C.V., Ghosh S., Grutzendler J. Astrocytes and microglia play orchestrated roles and respect phagocytic territories during neuronal corpse removal in vivo. Sci. Adv. 2020;6(26):eaba3239. doi: 10.1126/sciadv.aba3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz-Aparicio I., Beccari S., Abiega O., Sierra A. Clearing the corpses: Regulatory mechanisms, novel tools, and therapeutic potential of harnessing microglial phagocytosis in the diseased brain. Neural Regen. Res. 2016;11(10):1533–1539. doi: 10.4103/1673-5374.193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S.Y., Kim I.S. Engulfment signals and the phagocytic machinery for apoptotic cell clearance. Exp. Mol. Med. 2017;49(5):e331. doi: 10.1038/emm.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poon I.K., Lucas C.D., Rossi A.G., Ravichandran K.S. Apoptotic cell clearance: Basic biology and therapeutic potential. Nat. Rev. Immunol. 2014;14(3):166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fadok V.A., Voelker D.R., Campbell P.A., Cohen J.J., Bratton D.L., Henson P.M. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992;148(7):2207–2216. [PubMed] [Google Scholar]
- 20.Sierra A., Abiega O., Shahraz A., Neumann H. Janus-faced microglia: beneficial and detrimental consequences of microglial phagocytosis. Front. Cell. Neurosci. 2013;7:6. doi: 10.3389/fncel.2013.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao C., Keep R.F., Xi G., Hua Y. CD47 blocking antibody accelerates hematoma clearance after intracerebral hemorrhage in aged rats. Transl. Stroke Res. 2020;11(3):541–551. doi: 10.1007/s12975-019-00745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown S., Heinisch I., Ross E., Shaw K., Buckley C.D., Savill J. Apoptosis disables CD31-mediated cell detachment from phagocytes promoting binding and engulfment. Nature. 2002;418(6894):200–203. doi: 10.1038/nature00811. [DOI] [PubMed] [Google Scholar]
- 23.Arandjelovic S., Ravichandran K.S. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 2015;16(9):907–917. doi: 10.1038/ni.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cemma M., Grinstein S., Brumell J.H. Autophagy proteins are not universally required for phagosome maturation. Autophagy. 2016;12(9):1440–1446. doi: 10.1080/15548627.2016.1191724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.PrabhuDas M. R.; Baldwin, C. L.; Bollyky, P. L.; Bowdish, D.; Drickamer, K.; Febbraio, M.; Herz, J.; Kobzik, L.; Krieger, M.; Loike, J.; McVicker, B.; Means, T. K.; Moestrup, S. K.; Post, S. R.; Sawamura, T.; Silverstein, S.; Speth, R. C.; Telfer, J. C.; Thiele, G. M.; Wang, X. Y.; Wright, S. D.; El, K.J.A Consensus definitive classification of scavenger receptors and their roles in health and disease. J. Immunol. 2017;198:3775–3789. doi: 10.4049/jimmunol.1700373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y., Ye R.D. Microglial Aβ receptors in Alzheimer’s disease. Cell. Mol. Neurobiol. 2015;35(1):71–83. doi: 10.1007/s10571-014-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X., Guo C., Fisher P.B., Subjeck J.R., Wang X.Y. Scavenger receptors: Emerging roles in cancer biology and immunology. Adv. Cancer Res. 2015;128:309–364. doi: 10.1016/bs.acr.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Husemann J., Loike J.D., Anankov R., Febbraio M., Silverstein S.C. Scavenger receptors in neurobiology and neuropathology: their role on microglia and other cells of the nervous system. Glia. 2002;40(2):195–205. doi: 10.1002/glia.10148. [DOI] [PubMed] [Google Scholar]
- 29.Plüddemann A., Neyen C., Gordon S. Macrophage scavenger receptors and host-derived ligands. Methods. 2007;43(3):207–217. doi: 10.1016/j.ymeth.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Li Y. CD36 tango in cancer: signaling pathways and functions. Theranostics. 2019;9(17):4893–4908. doi: 10.7150/thno.36037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flores J.J., Klebe D., Tang J., Zhang J.H. A comprehensive review of therapeutic targets that induce microglia/macrophage-mediated hematoma resolution after germinal matrix hemorrhage. J. Neurosci. Res. 2020;98(1):121–128. doi: 10.1002/jnr.24388. [DOI] [PubMed] [Google Scholar]
- 32.Stewart C.R., Stuart L.M., Wilkinson K., van Gils J.M., Deng J., Halle A., Rayner K.J., Boyer L., Zhong R., Frazier W.A., Lacy-Hulbert A., El Khoury J., Golenbock D.T., Moore K.J. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 2010;11(2):155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lian H., Roy E., Zheng H. Microglial phagocytosis assay. Bio Protoc. 2016;6(21):6. doi: 10.21769/BioProtoc.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao X., Sun G., Zhang J., Ting S.M., Gonzales N., Aronowski J. Dimethyl fumarate protects brain from damage produced by intracerebral hemorrhage by mechanism involving Nrf2. Stroke. 2015;46(7):1923–1928. doi: 10.1161/STROKEAHA.115.009398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park D., Han C.Z., Elliott M.R., Kinchen J.M., Trampont P.C., Das S., Collins S., Lysiak J.J., Hoehn K.L., Ravichandran K.S. Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature. 2011;477(7363):220–224. doi: 10.1038/nature10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lan X., Han X., Li Q., Yang Q.W., Wang J. Modulators of microglial activation and polarization after intracerebral haemorrhage. Nat. Rev. Neurol. 2017;13(7):420–433. doi: 10.1038/nrneurol.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fumagalli S., Perego C., Pischiutta F., Zanier E.R., De Simoni M.G. The ischemic environment drives microglia and macrophage function. Front. Neurol. 2015;6:81. doi: 10.3389/fneur.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X., Wang H., Sun G., Zhang J., Edwards N.J., Aronowski J. Neuronal interleukin-4 as a modulator of microglial pathways and ischemic brain damage. J. Neurosci. 2015;35(32):11281–11291. doi: 10.1523/JNEUROSCI.1685-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanier E.R., Marchesi F., Ortolano F., Perego C., Arabian M., Zoerle T., Sammali E., Pischiutta F., De Simoni M.G. Fractalkine receptor deficiency is associated with early protection but late worsening of outcome following brain trauma in mice. J. Neurotrauma. 2016;33(11):1060–1072. doi: 10.1089/neu.2015.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson M.J., Manzanero S., Borges K. Complex alterations in microglial M1/M2 markers during the development of epilepsy in two mouse models. Epilepsia. 2015;56(6):895–905. doi: 10.1111/epi.12960. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Z.V., Lyu H., Lam S.Y.E., Lam P.K., Poon W.S., Wong G.K.C. The dynamics of microglial polarization reveal the resident neuroinflammatory responses after subarachnoid hemorrhage. Transl. Stroke Res. 2020;11(3):433–449. doi: 10.1007/s12975-019-00728-5. [DOI] [PubMed] [Google Scholar]
- 42.Savill J., Dransfield I., Gregory C., Haslett C. A blast from the past: Clearance of apoptotic cells regulates immune responses. Nat. Rev. Immunol. 2002;2(12):965–975. doi: 10.1038/nri957. [DOI] [PubMed] [Google Scholar]
- 43.Sierra A., Encinas J.M., Deudero J.J., Chancey J.H., Enikolopov G., Overstreet-Wadiche L.S., Tsirka S.E., Maletic-Savatic M. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell. 2010;7(4):483–495. doi: 10.1016/j.stem.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieger D., Moritz C., Ziegenhals T., Prykhozhij S., Peri F. Long-range Ca2+ waves transmit brain-damage signals to microglia. Dev. Cell. 2012;22(6):1138–1148. doi: 10.1016/j.devcel.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y., Zhang K., Yin X., Lei X., Chen X., Wang J., Quan Y., Yang L., Jia Z., Chen Q., Xian J., Lu Y., Huang Q., Zhang X., Feng H., Chen T. Quantitative iron neuroimaging can be used to assess the effects of minocycline in an intracerebral hemorrhage minipig model. Transl. Stroke Res. 2020;11(3):503–516. doi: 10.1007/s12975-019-00739-2. [DOI] [PubMed] [Google Scholar]
- 46.Janda E., Boi L., Carta A.R. Microglial phagocytosis and its regulation: a therapeutic target in Parkinson’s disease? Front. Mol. Neurosci. 2018;11:144. doi: 10.3389/fnmol.2018.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu R., Shen Q., Xu P., Luo J.J., Tang Y. Phagocytosis of microglia in the central nervous system diseases. Mol. Neurobiol. 2014;49(3):1422–1434. doi: 10.1007/s12035-013-8620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harry G.J. Microglia during development and aging. Pharmacol. Ther. 2013;139(3):313–326. doi: 10.1016/j.pharmthera.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogels T., Murgoci A.N., Hromádka T. Intersection of pathological tau and microglia at the synapse. Acta Neuropathol. Commun. 2019;7(1):109. doi: 10.1186/s40478-019-0754-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weldon D.T., Rogers S.D., Ghilardi J.R., Finke M.P., Cleary J.P., O’Hare E., Esler W.P., Maggio J.E., Mantyh P.W. Fibrillar beta-amyloid induces microglial phagocytosis, expression of inducible nitric oxide synthase, and loss of a select population of neurons in the rat CNS in vivo. J. Neurosci. 1998;18(6):2161–2173. doi: 10.1523/JNEUROSCI.18-06-02161.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito U., Nagasao J., Kawakami E., Oyanagi K. Fate of disseminated dead neurons in the cortical ischemic penumbra: ultrastructure indicating a novel scavenger mechanism of microglia and astrocytes. Stroke. 2007;38(9):2577–2583. doi: 10.1161/STROKEAHA.107.484394. [DOI] [PubMed] [Google Scholar]
- 52.Šišková Z., Tremblay M.È. Microglia and synapse: Interactions in health and neurodegeneration. Neural Plast. 2013;2013:425845. doi: 10.1155/2013/425845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei J., Wang M., Jing C., Keep R.F., Hua Y., Xi G. Multinucleated giant cells in experimental intracerebral hemorrhage. Transl. Stroke Res. 2020;11(5):1095–1102. doi: 10.1007/s12975-020-00790-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritzel R.M., Patel A.R., Grenier J.M., Crapser J., Verma R., Jellison E.R., McCullough L.D. Functional differences between microglia and monocytes after ischemic stroke. J. Neuroinflammation. 2015;12:106. doi: 10.1186/s12974-015-0329-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konishi H., Okamoto T., Hara Y., Komine O., Tamada H., Maeda M., Osako F., Kobayashi M., Nishiyama A., Kataoka Y., Takai T., Udagawa N., Jung S., Ozato K., Tamura T., Tsuda M., Yamanaka K., Ogi T., Sato K., Kiyama H. Astrocytic phagocytosis is a compensatory mechanism for microglial dysfunction. EMBO J. 2020;39(22):e104464. doi: 10.15252/embj.2020104464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das R., Chinnathambi S. Microglial priming of antigen presentation and adaptive stimulation in Alzheimer’s disease. Cell. Mol. Life Sci. 2019;76(19):3681–3694. doi: 10.1007/s00018-019-03132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nielsen H.H., Ladeby R., Fenger C., Toft-Hansen H., Babcock A.A., Owens T., Finsen B. Enhanced microglial clearance of myelin debris in T cell-infiltrated central nervous system. J. Neuropathol. Exp. Neurol. 2009;68(8):845–856. doi: 10.1097/NEN.0b013e3181ae0236. [DOI] [PubMed] [Google Scholar]
- 58.Grebing M., Nielsen H.H., Fenger C.D., Jensen T. K.; von Linstow, C.U.; Clausen, B.H.; Söderman, M.; Lambertsen, K.L.; Thomassen, M.; Kruse, T.A.; Finsen, B. Myelin-specific T cells induce interleukin-1beta expression in lesion-reactive microglial-like cells in zones of axonal degeneration. Glia. 2016;64(3):407–424. doi: 10.1002/glia.22937. [DOI] [PubMed] [Google Scholar]
- 59.Nizami S., Hall-Roberts H., Warrier S., Cowley S.A., Di Daniel E. Microglial inflammation and phagocytosis in Alzheimer’s disease: Potential therapeutic targets. Br. J. Pharmacol. 2019;176(18):3515–3532. doi: 10.1111/bph.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeh F.L., Hansen D.V., Sheng M. TREM2, microglia, and neurodegenerative diseases. Trends Mol. Med. 2017;23(6):512–533. doi: 10.1016/j.molmed.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 61.Kober D.L., Brett T.J. TREM2-ligand interactions in health and disease. J. Mol. Biol. 2017;429(11):1607–1629. doi: 10.1016/j.jmb.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gabandé-Rodríguez E., Keane L., Capasso M. Microglial phagocytosis in aging and Alzheimer’s disease. J. Neurosci. Res. 2020;98(2):284–298. doi: 10.1002/jnr.24419. [DOI] [PubMed] [Google Scholar]
- 63.Jiang T., Tan L., Zhu X.C., Zhang Q.Q., Cao L., Tan M.S., Gu L.Z., Wang H.F., Ding Z.Z., Zhang Y.D., Yu J.T. Upregulation of TREM2 ameliorates neuropathology and rescues spatial cognitive impairment in a transgenic mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2014;39(13):2949–2962. doi: 10.1038/npp.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S.M., Mun B.R., Lee S.J., Joh Y., Lee H.Y., Ji K.Y., Choi H.R., Lee E.H., Kim E.M., Jang J.H., Song H.W., Mook-Jung I., Choi W.S., Kang H.S. TREM2 promotes Aβ phagocytosis by upregulating C/EBPα-dependent CD36 expression in microglia. Sci. Rep. 2017;7(1):11118. doi: 10.1038/s41598-017-11634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piccio L., Buonsanti C., Mariani M., Cella M., Gilfillan S., Cross A.H., Colonna M., Panina-Bordignon P. Blockade of TREM-2 exacerbates experimental autoimmune encephalomyelitis. Eur. J. Immunol. 2007;37(5):1290–1301. doi: 10.1002/eji.200636837. [DOI] [PubMed] [Google Scholar]
- 66.Takahashi K., Prinz M., Stagi M., Chechneva O., Neumann H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 2007;4(4):e124. doi: 10.1371/journal.pmed.0040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poliani P.L., Wang Y., Fontana E., Robinette M.L., Yamanishi Y., Gilfillan S., Colonna M. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest. 2015;125(5):2161–2170. doi: 10.1172/JCI77983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang T., Zhang Y.D., Gao Q., Zhou J.S., Zhu X.C., Lu H., Shi J.Q., Tan L., Chen Q., Yu J.T. TREM1 facilitates microglial phagocytosis of amyloid beta. Acta Neuropathol. 2016;132(5):667–683. doi: 10.1007/s00401-016-1622-5. [DOI] [PubMed] [Google Scholar]
- 69.Saadipour K. TREM1: A potential therapeutic target for Alzheimer’s disease. Neurotox. Res. 2017;32(1):14–16. doi: 10.1007/s12640-017-9716-y. [DOI] [PubMed] [Google Scholar]
- 70.Weinger J.G., Brosnan C.F., Loudig O., Goldberg M.F., Macian F., Arnett H.A., Prieto A.L., Tsiperson V., Shafit-Zagardo B. Loss of the receptor tyrosine kinase Axl leads to enhanced inflammation in the CNS and delayed removal of myelin debris during experimental autoimmune encephalomyelitis. J. Neuroinflammation. 2011;8:49. doi: 10.1186/1742-2094-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ray A.K., DuBois J.C., Gruber R.C., Guzik H.M., Gulinello M.E., Perumal G., Raine C., Kozakiewicz L., Williamson J., Shafit-Zagardo B. Loss of Gas6 and Axl signaling results in extensive axonal damage, motor deficits, prolonged neuroinflammation, and less remyelination following cuprizone exposure. Glia. 2017;65(12):2051–2069. doi: 10.1002/glia.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Healy L.M., Jang J.H., Won S.Y., Lin Y.H., Touil H., Aljarallah S., Bar-Or A., Antel J.P. MerTK-mediated regulation of myelin phagocytosis by macrophages generated from patients with MS. Neurol. Neuroimmunol. Neuroinflamm. 2017;4(6):e402. doi: 10.1212/NXI.0000000000000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koronyo-Hamaoui M., Sheyn J., Hayden E.Y., Li S., Fuchs D., Regis G.C., Lopes D.H.J., Black K.L., Bernstein K.E., Teplow D.B., Fuchs S., Koronyo Y., Rentsendorj A. Peripherally derived angiotensin converting enzyme-enhanced macrophages alleviate Alzheimer-related disease. Brain. 2020;143(1):336–358. doi: 10.1093/brain/awz364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao X., Grotta J., Gonzales N., Aronowski J. Hematoma resolution as a therapeutic target: the role of microglia/macrophages. Stroke. 2009;40(3) Suppl.:S92–S94. doi: 10.1161/STROKEAHA.108.533158. [DOI] [PubMed] [Google Scholar]
- 75.Zhao X., Sun G., Ting S.M., Song S., Zhang J., Edwards N.J., Aronowski J. Cleaning up after ICH: the role of Nrf2 in modulating microglia function and hematoma clearance. J. Neurochem. 2015;133(1):144–152. doi: 10.1111/jnc.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu R., Li H., Hua Y., Keep R.F., Xiao J., Xi G., Huang Y. Early hemolysis within human intracerebral hematomas: an MRI study. Transl. Stroke Res. 2019;10(1):52–56. doi: 10.1007/s12975-018-0630-2. [DOI] [PubMed] [Google Scholar]
- 77.Wang G., Wang L., Sun X.G., Tang J. Haematoma scavenging in intracerebral haemorrhage: from mechanisms to the clinic. J. Cell. Mol. Med. 2018;22(2):768–777. doi: 10.1111/jcmm.13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deng S., Sherchan P., Jin P., Huang L., Travis Z., Zhang J.H., Gong Y., Tang J. Recombinant CCL17 enhances hematoma resolution and activation of CCR4/ERK/Nrf2/CD163 signaling pathway after intracerebral hemorrhage in mice. Neurotherapeutics. 2020;17(4):1940–1953. doi: 10.1007/s13311-020-00908-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y., Chen Q., Tan Q., Feng Z., He Z., Tang J., Feng H., Zhu G., Chen Z. Simvastatin accelerates hematoma resolution after intracerebral hemorrhage in a PPARγ-dependent manner. Neuropharmacology. 2018;128:244–254. doi: 10.1016/j.neuropharm.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 80.Moestrup S.K., Møller H.J. CD163: A regulated hemoglobin scavenger receptor with a role in the anti-inflammatory response. Ann. Med. 2004;36(5):347–354. doi: 10.1080/07853890410033171. [DOI] [PubMed] [Google Scholar]
- 81.Garton T., Keep R.F., Hua Y., Xi G. CD163, a hemoglobin/haptoglobin scavenger receptor, after intracerebral hemorrhage: functions in microglia/macrophages versus neurons. Transl. Stroke Res. 2017;8(6):612–616. doi: 10.1007/s12975-017-0535-5. [DOI] [PubMed] [Google Scholar]
- 82.Hvidberg V., Maniecki M.B., Jacobsen C., Højrup P., Møller H.J., Moestrup S.K. Identification of the receptor scavenging hemopexin-heme complexes. Blood. 2005;106(7):2572–2579. doi: 10.1182/blood-2005-03-1185. [DOI] [PubMed] [Google Scholar]
- 83.Wang G., Manaenko A., Shao A., Ou Y., Yang P., Budbazar E., Nowrangi D., Zhang J.H., Tang J. Low-density lipoprotein receptor-related protein-1 facilitates heme scavenging after intracerebral hemorrhage in mice. J. Cereb. Blood Flow Metab. 2017;37(4):1299–1310. doi: 10.1177/0271678X16654494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shichita T., Ito M., Morita R., Komai K., Noguchi Y., Ooboshi H., Koshida R., Takahashi S., Kodama T., Yoshimura A. MAFB prevents excess inflammation after ischemic stroke by accelerating clearance of damage signals through MSR1. Nat. Med. 2017;23(6):723–732. doi: 10.1038/nm.4312. [DOI] [PubMed] [Google Scholar]
- 85.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cuervo A.M., Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24(1):92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li W.W., Li J., Bao J.K. Microautophagy: lesser-known self-eating. Cell. Mol. Life Sci. 2012;69(7):1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levine B., Klionsky D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6(4):463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 89.Yamamoto A., Yue Z. Autophagy and its normal and pathogenic states in the brain. Annu. Rev. Neurosci. 2014;37:55–78. doi: 10.1146/annurev-neuro-071013-014149. [DOI] [PubMed] [Google Scholar]
- 90.Noda N.N., Inagaki F. Mechanisms of autophagy. Annu. Rev. Biophys. 2015;44:101–122. doi: 10.1146/annurev-biophys-060414-034248. [DOI] [PubMed] [Google Scholar]
- 91.Levine B., Mizushima N., Virgin H.W. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu M.Y., Lu J.H. Autophagy and macrophage functions: Inflammatory response and phagocytosis. Cells. 2019;9(1):70. doi: 10.3390/cells9010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ferrucci M., Biagioni F., Ryskalin L., Limanaqi F., Gambardella S., Frati A., Fornai F. Ambiguous effects of autophagy activation following hypoperfusion/ischemia. Int. J. Mol. Sci. 2018;19(9):19. doi: 10.3390/ijms19092756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bussi C., Peralta Ramos J.M., Arroyo D.S., Gaviglio E.A., Gallea J.I., Wang J.M., Celej M.S., Iribarren P. Autophagy down regulates pro-inflammatory mediators in BV2 microglial cells and rescues both LPS and alpha-synuclein induced neuronal cell death. Sci. Rep. 2017;7:43153. doi: 10.1038/srep43153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng J., Liao Y., Dong Y., Hu H., Yang N., Kong X., Li S., Li X., Guo J., Qin L., Yu J., Ma C., Li J., Li M., Tang B., Yuan Z. Microglial autophagy defect causes parkinson disease-like symptoms by accelerating inflammasome activation in mice. Autophagy. 2020;16(12):2193–2205. doi: 10.1080/15548627.2020.1719723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cho M.H., Cho K., Kang H.J., Jeon E.Y., Kim H.S., Kwon H.J., Kim H.M., Kim D.H., Yoon S.Y. Autophagy in microglia degrades extracellular β-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy. 2014;10(10):1761–1775. doi: 10.4161/auto.29647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.François A., Rioux Bilan A., Quellard N., Fernandez B., Janet T., Chassaing D., Paccalin M., Terro F., Page G. Longitudinal follow-up of autophagy and inflammation in brain of APPswePS1dE9 transgenic mice. J. Neuroinflammation. 2014;11:139. doi: 10.1186/s12974-014-0139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loane D.J., Byrnes K.R. Role of microglia in neurotrauma. Neurotherapeutics. 2010;7(4):366–377. doi: 10.1016/j.nurt.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strohm L., Behrends C. Glia-specific autophagy dysfunction in ALS. Semin. Cell Dev. Biol. 2020;99:172–182. doi: 10.1016/j.semcdb.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 100.Liu S., Sarkar C., Dinizo M., Faden A.I., Koh E.Y., Lipinski M.M., Wu J. Disrupted autophagy after spinal cord injury is associated with ER stress and neuronal cell death. Cell Death Dis. 2015;6:e1582. doi: 10.1038/cddis.2014.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarkar C., Zhao Z., Aungst S., Sabirzhanov B., Faden A.I., Lipinski M.M. Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy. 2014;10(12):2208–2222. doi: 10.4161/15548627.2014.981787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Datta A., Sarmah D., Mounica L., Kaur H., Kesharwani R., Verma G., Veeresh P., Kotian V., Kalia K., Borah A., Wang X., Dave K.R., Yavagal D.R., Bhattacharya P. Cell death pathways in ischemic stroke and targeted pharmacotherapy. Transl. Stroke Res. 2020;11(6):1185–1202. doi: 10.1007/s12975-020-00806-z. [DOI] [PubMed] [Google Scholar]
- 103.Yang Z., Zhong L., Zhong S., Xian R., Yuan B. Hypoxia induces microglia autophagy and neural inflammation injury in focal cerebral ischemia model. Exp. Mol. Pathol. 2015;98(2):219–224. doi: 10.1016/j.yexmp.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 104.Yang Z., Liu B., Zhong L., Shen H., Lin C., Lin L., Zhang N., Yuan B. Toll-like receptor-4-mediated autophagy contributes to microglial activation and inflammatory injury in mouse models of intracerebral haemorrhage. Neuropathol. Appl. Neurobiol. 2015;41(4):e95–e106. doi: 10.1111/nan.12177. [DOI] [PubMed] [Google Scholar]
- 105.Yang Z., Zhang N., Shen H., Lin C., Lin L., Yuan B. Microglial activation with reduction in autophagy limits white matter lesions and improves cognitive defects during cerebral hypoperfusion. Curr. Neurovasc. Res. 2014;11(3):223–229. doi: 10.2174/1567202611666140520124407. [DOI] [PubMed] [Google Scholar]
- 106.Li D., Wang C., Yao Y., Chen L., Liu G., Zhang R., Liu Q., Shi F.D., Hao J. mTORC1 pathway disruption ameliorates brain inflammation following stroke via a shift in microglia phenotype from M1 type to M2 type. FASEB J. 2016;30(10):3388–3399. doi: 10.1096/fj.201600495R. [DOI] [PubMed] [Google Scholar]
- 107.You W., Wang Z., Li H., Shen H., Xu X., Jia G., Chen G. Inhibition of mammalian target of rapamycin attenuates early brain injury through modulating microglial polarization after experimental subarachnoid hemorrhage in rats. J. Neurol. Sci. 2016;367:224–231. doi: 10.1016/j.jns.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 108.Fletcher K., Ulferts R., Jacquin E., Veith T., Gammoh N., Arasteh J.M., Mayer U., Carding S.R., Wileman T., Beale R., Florey O. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018;37(4):37. doi: 10.15252/embj.201797840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Galais M., Pradel B., Vergne I., Robert-Hebmann V., Espert L., Biard-Piechaczyk M. LAP (LC3-associated phagocytosis): Phagocytosis or autophagy? Med. Sci. (Paris) 2019;35(8-9):635–642. doi: 10.1051/medsci/2019129. [DOI] [PubMed] [Google Scholar]
- 110.Rai S., Arasteh M., Jefferson M., Pearson T., Wang Y., Zhang W., Bicsak B., Divekar D., Powell P.P., Naumann R., Beraza N., Carding S.R., Florey O., Mayer U., Wileman T. The ATG5-binding and coiled coil domains of ATG16L1 maintain autophagy and tissue homeostasis in mice independently of the WD domain required for LC3-associated phagocytosis. Autophagy. 2019;15(4):599–612. doi: 10.1080/15548627.2018.1534507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martinez J., Cunha L.D., Park S., Yang M., Lu Q., Orchard R., Li Q.Z., Yan M., Janke L., Guy C., Linkermann A., Virgin H.W., Green D.R. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533(7601):115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 112.Martinez J., Malireddi R.K.S., Lu Q., Cunha L.D., Pelletier S., Gingras S., Orchard R., Guan J.L., Tan H., Peng J., Kanneganti T.D., Virgin H.W., Green D.R. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 2015;17(7):893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 113.Martinez J., Almendinger J., Oberst A., Ness R., Dillon C.P., Fitzgerald P., Hengartner M.O., Green D.R. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA. 2011;108(42):17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., Green D.R. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450(7173):1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 115.Herb M., Gluschko A., Schramm M. LC3-associated phagocytosis - The highway to hell for phagocytosed microbes. Semin. Cell Dev. Biol. 2020;101:68–76. doi: 10.1016/j.semcdb.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 116.Frost L.S., Lopes V.S., Bragin A., Reyes-Reveles J., Brancato J., Cohen A., Mitchell C.H., Williams D.S., Boesze-Battaglia K. The contribution of melanoregulin to microtubule-associated protein 1 Light Chain 3 (LC3) associated phagocytosis in retinal pigment epithelium. Mol. Neurobiol. 2015;52(3):1135–1151. doi: 10.1007/s12035-014-8920-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ferguson T.A., Green D.R. Autophagy and phagocytosis converge for better vision. Autophagy. 2014;10(1):165–167. doi: 10.4161/auto.26735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Romao S., Gasser N., Becker A.C., Guhl B., Bajagic M., Vanoaica D., Ziegler U., Roesler J., Dengjel J., Reichenbach J., Münz C. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J. Cell Biol. 2013;203(5):757–766. doi: 10.1083/jcb.201308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hatsuzawa K., Sakurai C. Regulatory mechanism of SNAP23 in phagosome formation and maturation. Yonago Acta Med. 2020;63(3):135–145. doi: 10.33160/yam.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Birgisdottir Å.B., Johansen T. Autophagy and endocytosis - interconnections and interdependencies. J. Cell Sci. 2020;133(10):133. doi: 10.1242/jcs.228114. [DOI] [PubMed] [Google Scholar]
- 121.Lee J.W., Nam H., Kim L.E., Jeon Y., Min H., Ha S., Lee Y., Kim S.Y., Lee S.J., Kim E.K., Yu S.W. TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy. 2019;15(5):753–770. doi: 10.1080/15548627.2018.1556946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Green D.R., Oguin T.H., Martinez J. The clearance of dying cells: table for two. Cell Death Differ. 2016;23(6):915–926. doi: 10.1038/cdd.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Berglund R., Guerreiro-Cacais A.O., Adzemovic M.Z., Zeitelhofer M., Lund H., Ewing E., Ruhrmann S., Nutma E., Parsa R., Thessen-Hedreul M., Amor S., Harris R.A., Olsson T., Jagodic M. Microglial autophagy-associated phagocytosis is essential for recovery from neuroinflammation. Sci. Immunol. 2020;5(52):5. doi: 10.1126/sciimmunol.abb5077. [DOI] [PubMed] [Google Scholar]
- 124.Green D.R. Non-canonical functions of autophagy proteins. https://grantome.com/grant/NIH/R01-AI040646-22A1
- 125.Heckmann B.L., Teubner B.J.W., Tummers B., Boada-Romero E., Harris L., Yang M., Guy C.S., Zakharenko S.S., Green D.R. LC3-associated endocytosis facilitates β-Amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell. 2019;178(3):536–551.e14. doi: 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lima J.G., de Freitas Vinhas C., Gomes I.N., Azevedo C.M., dos Santos R.R., Vannier-Santos M.A., Veras P.S. Phagocytosis is inhibited by autophagic induction in murine macrophages. Biochem. Biophys. Res. Commun. 2011;405(4):604–609. doi: 10.1016/j.bbrc.2011.01.076. [DOI] [PubMed] [Google Scholar]
- 127.Zhu Y., Li H., Ding S., Wang Y. Autophagy inhibition promotes phagocytosis of macrophage and protects mice from methicillin-resistant staphylococcus aureus pneumonia. J. Cell. Biochem. 2018;119(6):4808–4814. doi: 10.1002/jcb.26677. [DOI] [PubMed] [Google Scholar]
- 128.Etchegaray J.I., Elguero E.J., Tran J.A., Sinatra V., Feany M.B., McCall K. Defective phagocytic corpse processing results in neurodegeneration and can be rescued by TORC1 activation. J. Neurosci. 2016;36(11):3170–3183. doi: 10.1523/JNEUROSCI.1912-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Natale G., Biagioni F., Busceti C.L., Gambardella S., Limanaqi F., Fornai F. TREM receptors connecting bowel inflammation to neurodegenerative disorders. Cells. 2019;8(10):8. doi: 10.3390/cells8101124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ulland T.K., Song W.M., Huang S.C., Ulrich J.D., Sergushichev A., Beatty W.L., Loboda A.A., Zhou Y., Cairns N.J., Kambal A., Loginicheva E., Gilfillan S., Cella M., Virgin H.W., Unanue E.R., Wang Y., Artyomov M.N., Holtzman D.M., Colonna M. TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell. 2017;170(4):649–663.e13. doi: 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jin L., Batra S., Jeyaseelan S. Deletion of Nlrp3 augments survival during polymicrobial sepsis by decreasing autophagy and enhancing phagocytosis. J. Immunol. 2017;198(3):1253–1262. doi: 10.4049/jimmunol.1601745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takata K., Takada T., Ito A., Asai M., Tawa M., Saito Y., Ashihara E., Tomimoto H., Kitamura Y., Shimohama S. Microglial Amyloid-β1-40 phagocytosis dysfunction is caused by high-mobility group box protein-1: implications for the pathological progression of Alzheimer’s disease. Int. J. Alzheimers Dis. 2012;2012:685739. doi: 10.1155/2012/685739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Takata K., Kitamura Y., Tsuchiya D., Kawasaki T., Taniguchi T., Shimohama S. High mobility group box protein-1 inhibits microglial Abeta clearance and enhances Abeta neurotoxicity. J. Neurosci. Res. 2004;78(6):880–891. doi: 10.1002/jnr.20340. [DOI] [PubMed] [Google Scholar]
- 134.Kang R., Livesey K.M., Zeh H.J., Loze M.T., Tang D. HMGB1: A novel Beclin 1-binding protein active in autophagy. Autophagy. 2010;6(8):1209–1211. doi: 10.4161/auto.6.8.13651. [DOI] [PubMed] [Google Scholar]
- 135.Qu L., Chen C., Chen Y., Li Y., Tang F., Huang H., He W., Zhang R., Shen L. High-Mobility Group Box 1 (HMGB1) and autophagy in acute lung injury (ALI): A review. Med. Sci. Monit. 2019;25:1828–1837. doi: 10.12659/MSM.912867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Angelopoulou E., Piperi C., Papavassiliou A.G. High-mobility group box 1 in Parkinson’s disease: from pathogenesis to therapeutic approaches. J. Neurochem. 2018;146(3):211–218. doi: 10.1111/jnc.14450. [DOI] [PubMed] [Google Scholar]
- 137.Song P., Wang Z., Zhang X., Fan J., Li Y., Chen Q., Wang S., Liu P., Luan J., Ye L., Ju D. The role of autophagy in asparaginase-induced immune suppression of macrophages. Cell Death Dis. 2017;8(3):e2721. doi: 10.1038/cddis.2017.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou P., Tan Y.Z., Wang H.J., Li T., He T., Yu Y., Zhang J., Zhang D. Cytoprotective effect of autophagy on phagocytosis of apoptotic cells by macrophages. Exp. Cell Res. 2016;348(2):165–176. doi: 10.1016/j.yexcr.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 139.Lucin K.M., O’Brien C.E., Bieri G., Czirr E., Mosher K.I., Abbey R.J., Mastroeni D.F., Rogers J., Spencer B., Masliah E., Wyss-Coray T. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer’s disease. Neuron. 2013;79(5):873–886. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Konishi A., Arakawa S., Yue Z., Shimizu S. Involvement of Beclin 1 in engulfment of apoptotic cells. J. Biol. Chem. 2012;287(17):13919–13929. doi: 10.1074/jbc.M112.348375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhou Z., Hartwieg E., Horvitz H.R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104(1):43–56. doi: 10.1016/S0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 142.Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A., McKay S., Okada H.M., Pan J., Schulz A.K., Tu D., Wong K., Zhao Z., Alexeyenko A., Burglin T., Sonnhammer E., Schnabel R., Jones S.J., Marra M.A., Baillie D.L., Moerman D.G. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5(9):e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li W., Zou W., Yang Y., Chai Y., Chen B., Cheng S., Tian D., Wang X., Vale R.D., Ou G. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J. Cell Biol. 2012;197(1):27–35. doi: 10.1083/jcb.201111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nash Y., Schmukler E., Trudler D., Pinkas-Kramarski R., Frenkel D. DJ-1 deficiency impairs autophagy and reduces alpha-synuclein phagocytosis by microglia. J. Neurochem. 2017;143(5):584–594. doi: 10.1111/jnc.14222. [DOI] [PubMed] [Google Scholar]
- 145.Huang H., Li X., Zhuang Y., Li N., Zhu X., Hu J., Ben J., Yang Q., Bai H., Chen Q. Class A scavenger receptor activation inhibits endoplasmic reticulum stress-induced autophagy in macrophage. J. Biomed. Res. 2014;28(3):213–221. doi: 10.7555/JBR.28.20130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu Z., Ning J., Zheng X., Meng J., Han L., Zheng H., Zhong L., Chen X.F., Zhang X., Luo H., Can D., Xu H., Zhang Y.W. TMEM59 interacts with TREM2 and modulates TREM2-dependent microglial activities. Cell Death Dis. 2020;11(8):678. doi: 10.1038/s41419-020-02874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]