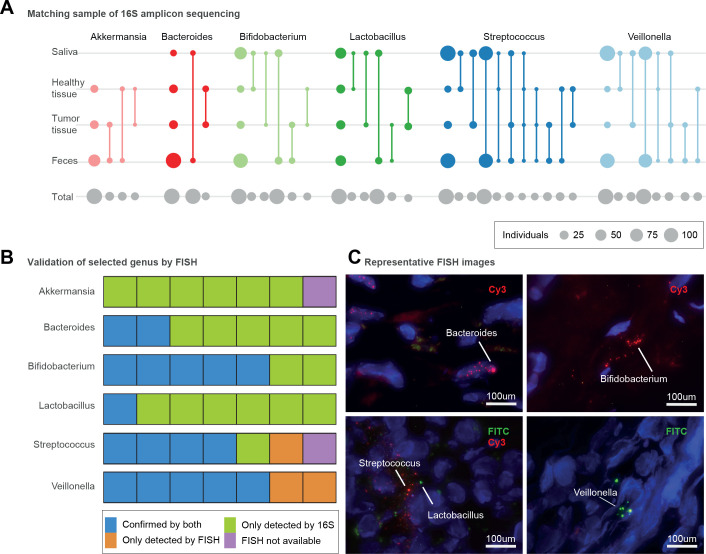
Figure 4.
Presence of microbiomes in different sections of the pancreas with different conditions. (A) Presence of different genera in four different body sites including faecal, saliva, pancreatic tumour and healthy tissue samples, as inferred by 16S amplicon data. Circle size corresponds to the total number of subjects available for each comparison (grey, bottom row) or with intra-individually matched amplicon sequence variants (coloured); matched sample types are connected by lines. The first column shows the total number of samples per site in which the genus was detected. (B) Seven selected pancreatic tissue samples (five tumour and two non-tumour) to show bacterial presence/absence with both 16S amplicon and fluorescence in situ hybridisation (FISH) methods. Validation of bacterial presence with both 16S amplicon sequencing and FISH is shown in blue. Samples showing bacterial presence according to 16S only are displayed in green. Bacterial presence validated only by FISH is shown in orange, and samples not subjected to FISH validation owing to lack of tissue material are shown in purple. (C) Representative microscopy images for Bacteroides (intranuclear, tumour tissue), Bifidobacterium (extranuclear, tumour tissue), Lactobacillus (extranuclear, non-tumour tissue), Streptococcus (extranuclear, non-tumour tissue), Veillonella (extranuclear, tumour tissue). Fluorescein isothiocyanate (FITC) and Cy3 fluorescent dyes were used as indicated, and DAPI (4',6,-diamidino-2-phenylindole; blue) was used to label the nucleus.