Abstract
Objective
Tissue-resident memory T cells (TRM) are vital immune sentinels that provide protective immunity. While hepatic CD8+ TRM have been well described, little is known about the location, phenotype and function of CD4+ TRM.
Design
We used multiparametric flow cytometry, histological assessment and novel human tissue coculture systems to interrogate the ex vivo phenotype, function and generation of the intrahepatic CD4+ T-cell compartment. We also used leukocytes isolated from human leukocyte antigen (HLA)-disparate liver allografts to assess long-term retention.
Results
Hepatic CD4+ T cells were delineated into three distinct populations based on CD69 expression: CD69−, CD69INT and CD69HI. CD69HICD4+ cells were identified as tissue-resident CD4+ T cells on the basis of their exclusion from the circulation, phenotypical profile (CXCR6+CD49a+S1PR1−PD-1+) and long-term persistence within the pool of donor-derived leukcoocytes in HLA-disparate liver allografts. CD69HICD4+ T cells produced robust type 1 polyfunctional cytokine responses on stimulation. Conversely, CD69INTCD4+ T cells represented a more heterogenous population containing cells with a more activated phenotype, a distinct chemokine receptor profile (CX3CR1+CXCR3+CXCR1+) and a bias towards interleukin-4 production. While CD69INTCD4+ T cells could be found in the circulation and lymph nodes, these cells also formed part of the long-term resident pool, persisting in HLA-mismatched allografts. Notably, frequencies of CD69INTCD4+ T cells correlated with necroinflammatory scores in chronic hepatitis B infection. Finally, we demonstrated that interaction with hepatic epithelia was sufficient to generate CD69INTCD4+ T cells, while additional signals from the liver microenvironment were required to generate liver-resident CD69HICD4+ T cells.
Conclusions
High and intermediate CD69 expressions mark human hepatic CD4+ TRM and a novel functionally distinct recirculating population, respectively, both shaped by the liver microenvironment to achieve diverse immunosurveillance.
Keywords: T lymphocytes, hepatitis B, immunology, liver immunology, cellular immunity
Significance of this study.
What is already known on this subject?
Tissue-resident memory CD4+ and CD8+ T cells are important front-line immune sentinels in many human tissues.
The human liver has been shown to contain long-lived tissue-resident CD8+ T cells that are capable of rapid effector function.
Liver-resident CD4+ T cells remain uncharacterised, and their contribution to health and disease has not yet been studied.
What are the new findings?
CD69 expression identifies three phenotypically and functionally distinct intrahepatic CD4+ T-cell populations: CD69−CD4+, CD69INTCD4+ and CD69HICD4+.
CD69HICD4+ T cells represent a long-lived liver-resident population that expresses classical retention markers, occupies sinusoidal and periportal niches, and is maintained in a resting and restrained state.
CD69INT marks a population containing both resident and recirculating T cells with differential chemokine and activation profiles.
CD69HICD4+ T cells produce robust TH1 cytokine responses, while CD69INTCD4+T-cells cells favour the production of interleukin-4 on short-term T-cell receptor engagement.
The frequency of CD69INTCD4+ T cells correlates with necroinflammatory scores in patients with chronic hepatitis B infection.
Novel autologous liver slice coculture models promote the differentiation of both CD69INTCD4+ and CD69HICD4+ cells from blood, but hepatic epithelia were sufficient to induce the CD69INTCD4+ phenotype.
Significance of this study.
How might it impact on clinical practice in the foreseeable future?
Our study identifies distinct intrahepatic CD4+ T cells not detectable in the blood, underscoring the need for continued sampling of the liver.
An understanding of the differential functionality of CD69HICD4+ and CD69INTCD4+ T cells compartmentalised at the site of pathology has important implications for current intensive efforts to develop immunotherapies for liver diseases.
The capacity of liver-derived signals to allow in vitro recapitulation of tissue-resident CD4+ T cells could be exploited for therapeutic targeting.
Introduction
Tissue-resident memory T cells (TRM) are a non-recirculating population that are critical in front-line adaptive immunity. Strategically positioned within tissues, these cells react to pathogen re-exposure more efficiently than circulating memory subsets.1 This function is mediated directly and by employing an innate-like ‘sensing and alarm’ strategy to enable recruitment and activation of other effector cells.2 3 Human TRM have now been identified in many organs1 4 5 and differ substantially from their circulating counterparts in phenotype,6 function,7 8 metabolism,9 10 maintenance requirements11 and responsiveness to stimuli.12 Expression of tissue retention molecules CD69, CD103 and CD49a and a lack of tissue egress markers including CCR7 and sphingosine-1-phosphate receptor 1 (S1PR1) define TRM.13 Of these, CD69 is particularly important as a marker preserved on CD4+ and CD8+ TRM in all tissues,14 and separation through expression of this molecule alone has recently been used to define a human TRM transcriptome with strong fidelity to more established murine TRM profiles.13 15
Recently, Pallett et al identified intrahepatic CD8+ TRM (CD69+CD103+CXCR6+CXCR3+PD-1+ (PD-1 – programmed cell death protein-1)), capable of robust interleukin (IL)-2 production, associated with viral control in the liver of HBV-infected individuals.5 However, little is known about CD4+ TRM and how the liver shapes their biology. In one study, Wong et al outlined distinct activation, differentiation and homing receptor profiles of liver perfusate CD4+ T cells as part of a multiorgan mapping study,16 supporting the possibility of a liver-resident CD4+ T-cell population.
Here, we provide the first comprehensive phenotypical and functional analysis of intrahepatic CD4+ TRM in the human liver. We identified two distinct populations of CD69-expressing intrahepatic CD4+ T cells: CD69HI and CD69INT. CD69HICD4+ T cells within the human liver had prototypical hallmarks of tissue residency, including high expression of retention markers, exclusion from the circulation and rapid multifunctional type 1 cytokine production on stimulation. We also report a novel population of intrahepatic CD69INTCD4+ T cells characterised by a unique chemokine receptor profile (CD69INTCX3CR1+CXCR3+CXCR1+). CD69INTCD4+ cells retained the ability to recirculate and on stimulation produced the TH2 cytokine IL-4. The frequency of these CD69INTCD4+ T cells also correlated with necroinflammatory scores in patients with chronic hepatitis B. Finally, we demonstrated that contact with hepatic epithelia drives the CD69INTCD4+ phenotype, while CD69HICD4+ cells required additional signals from the liver microenvironment.
Materials and methods
Patient samples and immune cell isolation
Blood, liver and lymph node (LN) samples were obtained from centre A, the Queen Elizabeth Hospital, Birmingham (references 06/Q2702/61 and 06/Q2708/11). Blood, liver (resections, biopsies, fine needle aspirates, HLA-mismatched explants), gut, spleen and LN samples from centre B were obtained from either the Royal Free Hospital, London (references 16/WA/0289, 11/WA/0077, 11/H0720/4 (RIPCOLT clinical trial number 8191) or 11/LO/0421) or Royal London Hospital, Barts Health NHS Trust (references P/01/023, 16/LO/1699 or 17/LO0266). Immune cells were isolated from tissues/blood through tissue digestion and density centrifugation (see online supplemental experimental methods). See online supplemental table 1 for full patient details.
gutjnl-2020-323771supp011.pdf (182.2KB, pdf)
Flow cytometry
For surface staining, cells were incubated with fluorescence-conjugated antibodies on ice for 20–30 min. For intracellular staining, cells were either fixed with 1% formaldehyde (Sigma-Aldrich) for 15 min, permeabilised with 0.1% Saponin (Sigma-Aldrich) and stained with relevant antibodies in 0.1% saponin (30 min, 20°C), or fixed and permeabilised with Cytofix/Cytoperm (BD Bioscience) or FoxP3 Buffer Set (BD Bioscience) according to the manufacturer’s instructions, and stained in 0.1% saponin. Dead intrahepatic lymphocytes (IHLs) were identified and excluded using either a fixable live/dead dye (Thermo Fisher) for all centre B samples or zombie dyes (Biolegend) for all cultured centre A samples. Samples were analysed on an ADP CyAn flow cytometer running Summit software (Beckmann Coulter, centre A) or LSRII or X20 flow cytometers running FACSDiva software (BD Bioscience) for samples from centre B (see online supplemental table 2 for the list of antibodies used and online supplemental figure 1 for gating strategies). CD69− and CD69INT populations were distinguished using isotype-matched controls, in combination with peripheral blood staining to determine CD69INT versus CD69HI gate positions.
gutjnl-2020-323771supp001.pdf (1.1MB, pdf)
Immunofluorescence
Formalin-fixed paraffin-embedded 3 µm liver sections were deparaffinised with xylene, rehydrated with 99% industrial denatured alcohol and underwent antigen retrieval by microwaving in Tris-based antigen-unmasking solution (Vector Labs). Slides were washed with TBS+0.1% Tween (TBST) and 2× casein solution (Vector labs) was added for 10 min, before 1-hour incubation with primary antibodies diluted in TBST. For antibodies used, see online supplemental experimental procedures. Following three washes with TBST, secondary antibodies were applied for 1 hour in TBST; autofluorescence was quenched with the TrueVIEW autofluorescence quenching kit (Vector Labs); and tissues were mounted with VECTASHIELD Vibrance Antifade Mounting Medium with DAPI (4′,6-diamidino-2-phenylindole, Vector Labs). Tissues were imaged using the Zeiss LSM 880 confocal microscope (Carl Zeiss) equipped with a ×63 water immersion objective.
T-cell stimulation for assessment of cytokine production
Peripheral blood mononuclear cells (PBMCs) and IHLs were first stained for surface antigens then cultured alone, with 1:1 ratio of anti-CD3/CD28 beads (Dynabeads, ThermoFisher), or 50 ng/mL phorbol 12-myristate 13-acetate (PMA) and 1 µM ionomycin (both Sigma Aldrich, UK), all with 10 µg/mL Brefeldin A (Sigma Aldrich). For culture and media details, see online supplemental experimental procedures.
CD4+ T-cell isolation and cell culture
CD4+ T cells were isolated from PBMCs with the EasySep human CD4+ T-cell enrichment kit, or EasySep naïve/memory human CD4+ T-cell enrichment kits (all StemCell Technologies). T cells/PBMCs were cultured with hepatic epithelial cell lines (Huh-7, HepG2 and Hep3B), hepatic stellate cell line LX-2, primary hepatic sinusoidal endothelial cells (HSECs) and primary biliary epithelial cells (BECs). Primary BEC and HSEC were isolated in-house as previously described.17 18 For media details, see online supplemental experimental procedures. 1×106 PBMCs/T-cells were added per well and cultured for up to 7 days. For transwell separation experiments, T cells were added to the top of the 0.4 µm pore transwell insert, separated from hepatic cells at the bottom of the 24-well plate.
Liver slice cultures
Precision-cut liver slices of 2 mm were prepared using a TruSlice tissue slicer (CellPath) and were cultured in complete Dulbecco's Modified Eagle Medium (DMEM) with 2% foetal bovine serum (FBS) in 48-well plates. Autologous total PBMCs were added in T-cell media (1×106/well), and plates were cultured for 5 hours at 37°C before PBMC harvest and were used in downstream assays.
Data analysis and statistics
All flow cytometry data were analysed using FlowJo V.9–10 (FlowJo LLC). Statistical testing was applied in Prism V.8 (GraphPad). Median average values and and non-parametric testing were used throughout.
Results
CD69 expression distinguishes three intrahepatic CD4+ T-cell populations with differential homing potentials
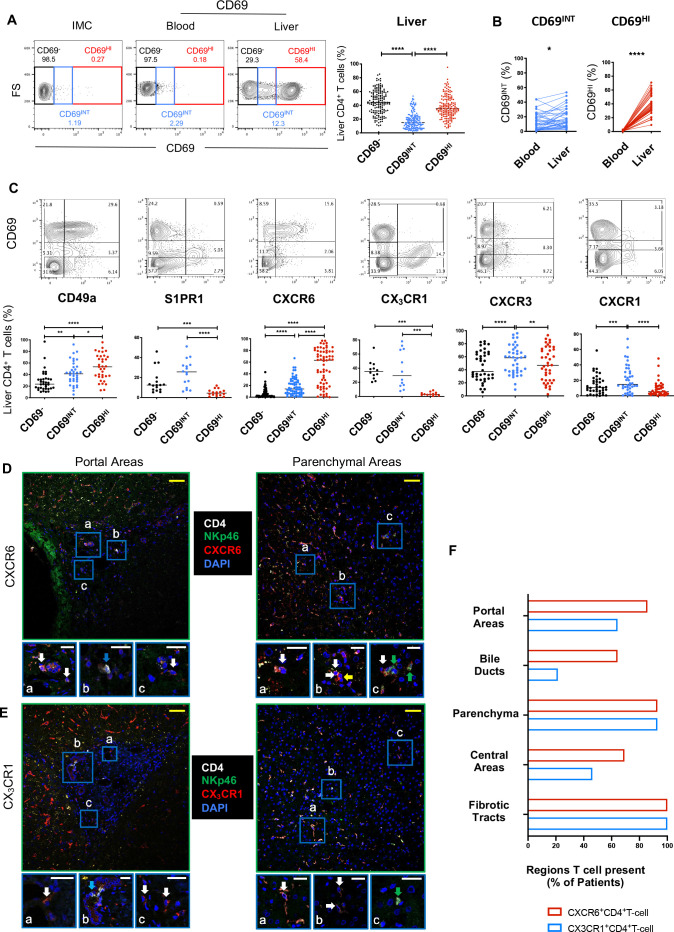
To identify intrahepatic CD4+ TRM, we analysed CD69 expression in over 160 liver samples from two research centres. Three intrahepatic CD4+ T-cell phenotypes were identified: CD69−, CD69INT and CD69HI (figure 1A). CD69HICD4+ cells were negligible in blood, while CD69INTCD4+ T cells were detected in both intrahepatic and peripheral pools (figure 1B). In the liver, CD69HICD4+ T cells displayed striking concordance with a residency-associated profile (CD49a+CXCR6+S1PR1−CX3CR1−) (figure 1C). By contrast, CD69INTCD4+ T cells retained expression of the tissue egress marker S1PR1 and fractalkine receptor CX3CR1, which is associated with migratory T cells,13 19 20 as well as the strongest expression of parenchymal homing receptors CXCR3 and CXCR1.21 Hepatic CD69INTCD4+ T cells expressed less CD49a and CXCR6 than CD69HICD4+ T cells, although these residence markers were all expressed to a higher extent than on the CD69−CD4+ T cells.
Figure 1.
CD69 expression distinguishes three intrahepatic CD4+ T-cell populations with differential homing potentials. (A) Gating strategy showing CD69−, CD69INT and CD69HI populations. Representative flow cytometry plot for CD4+ T-cell distribution in blood and liver, and summary data showing % CD4+ T cells in IHL from two independent centres (n=162). Isotype-matched controls were used to set CD69− gates. (B) % CD69-expressing T-cell populations in paired blood and liver (n=39). (C) Expression of key homing and retention markers on CD69-expressing CD4+ T cells (% of total CD4+ T cells). Images depicting localisation of CXCR6+ CD4+ T cells (D) or CX3CR1+ CD4+ T cells (E) in portal and parenchymal areas of human livers (representative of n=14 livers (5 control, 4 patients with HBV and 5 patients with PBC)). Sections stained for CD4, NKp46, DAPI and chemokine receptor indicated. Cells of interest expressed both the chemokine receptor and CD4 and lacked NK cell marker NKp46. Areas of interest (A–C) shown at higher magnification below each main image. White arrows: cell of interest, green arrows: NKp46+ cell, yellow arrow: chemokine receptor+ CD4− cell, blue arrows: CD4+ NKp46- CXCR6- cells (D) or CD4+NKp46-CX3CR1- cells (E). Yellow scale bars: 50 µm, white scale bars: 20 µm. (F) Cumulative scoring of the presence of each cell of interest within different liver regions. Cells of interest were scored as present in specific areas if at least three cells were present within each region. Plot shows the % of each region that contained cells of interest (n=14, as above; fibrotic tracts in non-control livers only, n=9). Cells were classed as present in portal regions and central regions if they were identified within 50 µm of their respective vasculature. Association with bile ducts was scored if cells were making direct contact. Statistical comparisons by Freidman tests with Dunn’s multiple tests (A, C); Wilcoxon matched-pair, signed-rank tests (B). p < 0.05 (*), < 0.01 (**), < 0.001 (***), < 0.0001 (****) FS, forward scatter; IHL, intrahepatic lymphocyte; IMC, isotype-matched control; NK, natural killer; PBC, primary biliary cholangitis.
In keeping with their association with residence, fine-needle aspirate (FNA) samples (that sample more blood-derived than interstitial T cells compared with biopsies22) showed a more marked reduction in the frequency of CD69HICD4+ than CD69INTCD4+ T cells compared with matched liver tissue obtained by biopsy (online supplemental figure 2A). Similarly, of the three populations, only CD69HICD4+ T cells were enriched for an effector memory phenotype—a prerequisite for TRM cells (online supplemental figure 2B). Interestingly, more CD69INTCD4+ T cells expressed a gut homing signature (CCR9, integrin α4β7) than the other two CD69-expressing populations, suggestive of a potential wider enteric surveillance role (online supplemental figure 2C). Additional profiling revealed increased CCR5 expression on CD69HICD4+ T cells, higher expression of CCR6 on both CD69-expressing populations than CD69−CD4+ T cells and no differential CCR10 expression. The retention marker CD103 was expressed most on CD69HICD4+ T cells, although this frequency was low, as reported for other human resident CD4+ T-cell subsets (online supplemental figure 2C).4 Together, our data reveal two distinct CD69-expressing CD4+ T-cell populations in the human liver: CD69HICD4+ T cells with the strongest TRM profile and CD69INTCD4+ T cells with differential homing potential.
gutjnl-2020-323771supp002.pdf (612.1KB, pdf)
Next, we assessed whether differential expression of CXCR6 and CX3CR1 between the different intrahepatic CD4+ T-cell populations affected their hepatic distribution (online supplemental figure 3A). CD4+ T cells expressing either chemokine receptor were found throughout the liver—in both portal and central areas, in fibrotic tracts and throughout the parenchyma where they likely play a crucial role in the immunosurveillance of hepatocytes (figure 1D, E, and online supplemental figure 3A–C). CXCR6+CD4+ T cells (enriched for the CD69HICD4+ T-cell population) were found more frequently in association with bile ducts (figure 1F), in keeping with the role of this receptor in biliary homing.23
gutjnl-2020-323771supp003.pdf (711.8KB, pdf)
High CD69 expression marks a CD4+ T-cell population capable of long-term residence within the liver
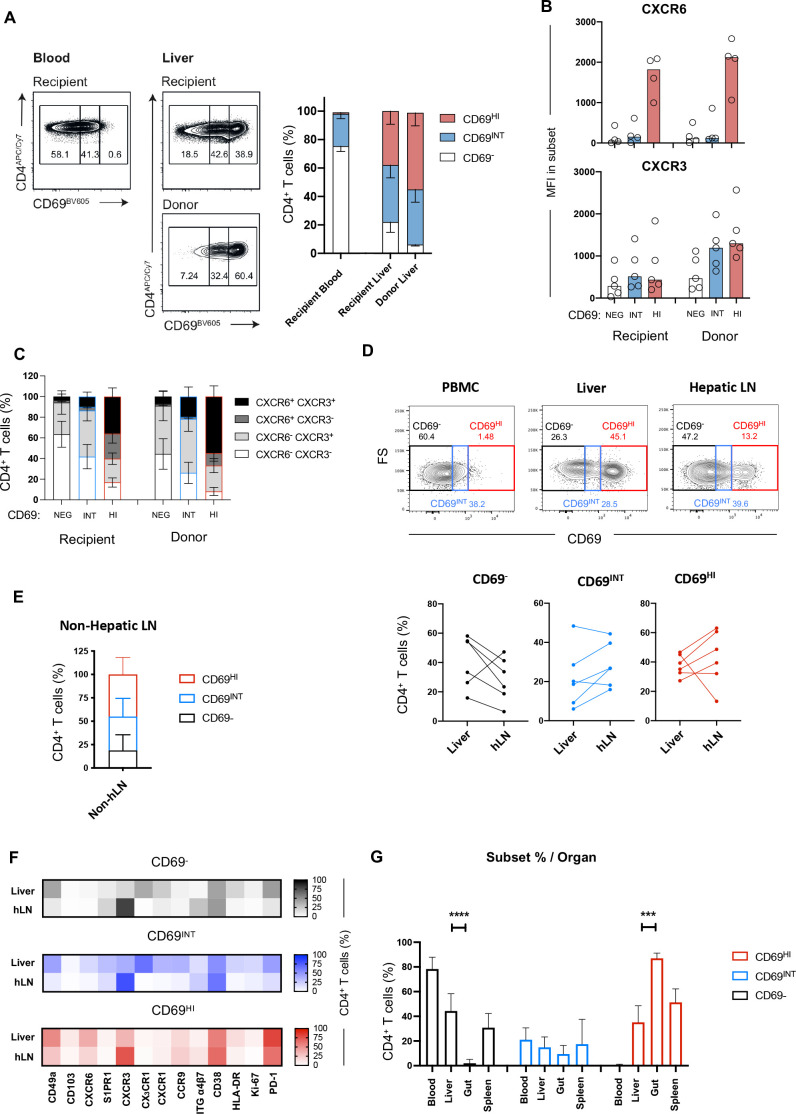
To ascertain which population was strictly resident in the human liver, we examined HLA-mismatched allograft samples explanted up to a decade after initial transplantation.24 In our recent study, we showed that in all cases, a small pool of long-lived, donor-derived CD4+ T cells were detected by staining with HLA-specific antibodies.24 No donor-derived CD4+ T cells were detected in the blood, confirming that donor-derived cells and their progeny were maintained locally in the liver allograft.24 Re-examining the CD4+ T-cell fraction from donor and recipient pools, we observed that CD69HICD4+ T cells were significantly enriched in the persisting, donor-derived fraction, establishing these cells as TRM (figure 2A). By contrast, CD69−CD4+ T cells comprised a negligible fraction of the long-lived donor-derived T cells. Interestingly, however, a population of CD69INTCD4+ T cells was detected in the donor-derived compartment in all cases, suggestive of the long-term retention of some of these cells. Examining the recipient-derived CD4+ T cells infiltrating the allograft, we found these to be capable of acquiring both a CD69HICXCR6HI and a CD69INTCXCR6LO phenotype, suggesting that infiltrating T cells are shaped by the hepatic microenvironment (figure 2B). By contrast, recipient CD4+ T cells within the allograft only showed a subtle increase in CXCR3 compared with their circulating counterparts, expressing much less than the donor-derived CD69HICD4+ and CD69INTCD4+ T cells, suggesting CXCR3 is less easily imprinted on liver infiltration (figure 2B). Donor-derived CD69HICD4+ T cells contained a greater representation of dual CXCR6+CXCR3+-expressing cells than their recipient-derived counterparts, while the CXCR6−CXCR3+ was most enriched on the donor-derived CD69INTCD4+ T-cell population (figure 2C). This suggests expression of both markers is important in long-term retention of CD69HICD4+ T cells, with CD69INTCD4+ T cells more reliant on CXCR3 alone.
Figure 2.
High CD69 expression marks a CD4+ T-cell population capable of long-term residence within the liver. (A) HLA-mismatched allograft sampling allows assessment of resident T cells. Donor-derived T cells are distinguished from recipient-derived T cells through HLA staining. Example distributions of CD69−, CD69INT and CD69HI cells in recipient and donor pools of liver and blood samples, and combined data across five patient samples. (B) MFI of CXCR6 and CXCR3 expressions in the three populations in donor and recipient pools. (C) Breakdown of CXCR3/CXCR6 coexpression patterns in different donor and recipient subpopulations (n=4). (D) Staining and combined data showing population distribution from liver (n=6), hepatic LNs (n=6) and non-hepatic (mesenteric) LNs (n=6). Example plots show hLN, PBMC and liver as gating controls. (E) Subset breakdown in distal non-hLNs. (F) Heatmap of % marker expression in CD69− (top), CD69INT (middle) and CD69HI (bottom) from matched liver and hLN samples. CD103, n=6; CD49a, n=5; CXCR6 and HLA-DR, n=4; CXCR3, CXCR1, PD-1 and CD38, n=3; S1PR1, CCR9, integrin α4β7 and Ki-67, n=2; CX3CR1, n=1. (G) Frequency of CD69−, CD69INT and CD69HI CD4+ T cells in blood (n=103), liver (n=118), gut (n=6) and spleen (n=4) samples. Statistical comparisons on paired populations by Wilcoxon matched-pair, signed-rank tests (A–D, F), and Kruskal-Wallis tests with duns post hoc tests on liver, gut and spleen samples within each CD4+ T-cell subset (G). HI, high; hLN, hepatic lymph node; INT, intermediate; LN, lymph node; MFI, Median fluorescence intensity, NEG, negative.
Reasoning that expression of tissue egress markers S1PR1 and CX3CR1 would imbue CD69INTCD4+ T cells with the ability to recirculate through lymphatics, we assessed the make-up of matched liver and liver-draining hepatic hilar LN (hepatic lymph node (hLN), figure 2D) and distal non-hLN (figure 2E). Both CD69INTCD4+ and CD69HICD4+ T cells were present in LNs, supporting this possibility. While donor-matched liver and hLN CD69HICD4+ T cells were phenotypically similar, reflective of a common residency signature, CD69INTCD4+ and CD69−CD4+ cells in hLNs differed substantially from their liver equivalents in their chemokine receptor profile (subsets in hLNs enriched for CXCR3, but depleted for CX3CR1, CXCR1 and CCR9 expression; figure 2F). Furthermore, CD69HICD4+ and CD69INTCD4+ were detectable not only in the liver and LNs but also in the gut, where CD69HICD4+ predominates, and in spleen samples (figure 2G).
Thus, while the properties of long-lived tissue enrichment and absence from the peripheral circulation mean CD69HICD4+ T cells comply with a tissue-resident definition, CD69INTCD4+ T cells may represent a population with a context-dependent capacity for liver occupancy and egress.
CD69HICD4+ TRM demonstrates a restrained, resting phenotype, while CD69INTCD4+ T cells exhibit features of activation
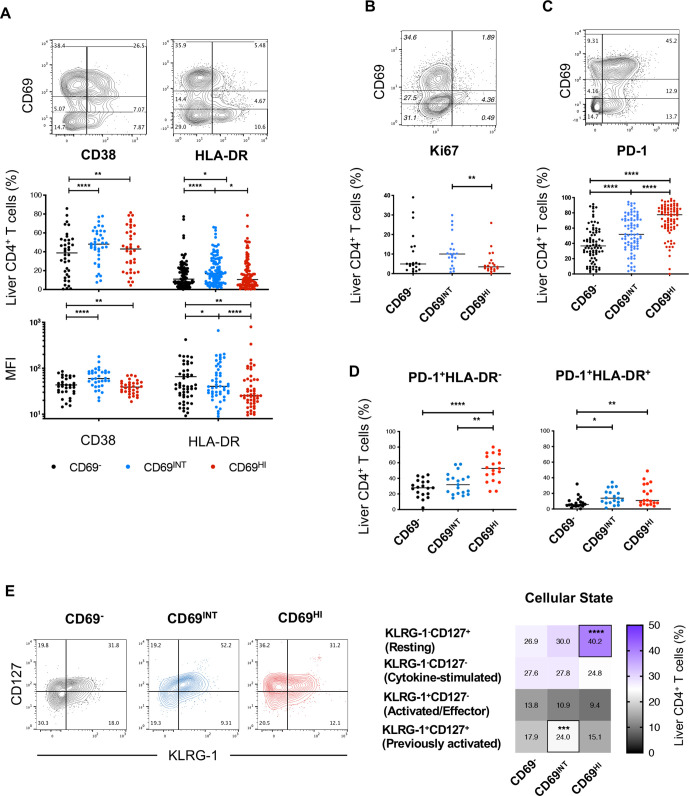
Alongside a tissue-residence marker, CD69 has been used as an indicator of early lymphocyte activation.25 Therefore, we examined the activation status of the three hepatic CD4+ T-cell populations. Intriguingly, the extent of cellular activation did not correlate with levels of CD69 expression; CD69INTCD4+ T cells were enriched for activation markers CD38 and HLA-DR, expressing more CD38 than CD69−CD4+ T cells, and more HLA-DR than CD69HICD4+ T cells (figure 3A). Consistent with recent activation in vivo, the CD69INTCD4+ T-cell population also expressed more Ki67 than their CD69HICD4+ T-cell counterparts (figure 3B). Regulatory T cells (TREG, CD4+CD25HICD127LO) were not significantly enriched in any intrahepatic CD4+ population, irrespective of CD69 expression, and TREG functional markers cytotoxic T lymphocyte-associated protein-4 (CTLA4) and CD39 were similarly expressed by both CD69-expressing populations when compared with CD69−CD4+ T cells (online supplemental figure 4).
Figure 3.
CD69HICD4+ TRM demonstrate a restrained, resting phenotype, while CD69INTCD4+ T cells exhibit features of activation. (A) % and MFI expression of CD38 and HLA-DR. % expression of (B) Ki-67 and (C) PD-1 expressions among the three CD4+ T-cell populations. (D) Subset representation among PD-1+HLA-DR− and PD-1+HLA-DR+ designations (n=19). (E) Analysis of the four differentiation/cellular states based on KLRG-1 and CD127 expressions (n=41). Heatmap shows % expression of each designation. Freidman’s tests with Dunn’s multiple tests were used for statistical analysis (A–E) p < 0.05 (*), < 0.01 (**), < 0.001 (***), < 0.0001 (****). MFI, mean fluorescence intensity.
gutjnl-2020-323771supp004.pdf (1MB, pdf)
Another hallmark of human TRM is the adoption of a self-restrained, resting state necessary to prevent inflammatory damage to residing tissues.5 13 15 Correspondingly, CD69HICD4+ T cells were enriched for PD-1, with CD69−CD4+ T cells displaying the lowest frequency (figure 3C). As PD-1 can also denote activation,26 we assessed the coexpression of PD-1 and HLA-DR. The percentage of PD-1+HLA-DR− cells were enriched within the CD69HICD4+ T-cell population, suggesting PD-1 upregulation in CD69HICD4+ T cells was not simply an activation phenomenon (figure 3D). To investigate cellular activation states in more detail, we also analysed coexpression patterns of killer cell lectin-like receptor-G1 (KLRG-1, a marker of antigen experience) and CD127, an indicator of common γ-chain cytokine sensitivity.27 28 As in human TRM studies,29 CD69HICD4+ T cells contained the most resting (KLRG-1−CD127+) cells, whereas CD69INTCD4+ T cells were enriched for the previously activated (KLRG-1+CD127+) population (figure 3E).
These data illustrate differences in activation states between CD69HICD4+ and CD69INTCD4+ T cells, with the former exhibiting a resting/restrained phenotype in keeping with their profile as liver TRM, while the latter population displayed features consistent with recent activation.
Liver CD69HICD4+ and CD69INTCD4+ T cells are skewed towards TH1 and TH2 functional profiles, respectively
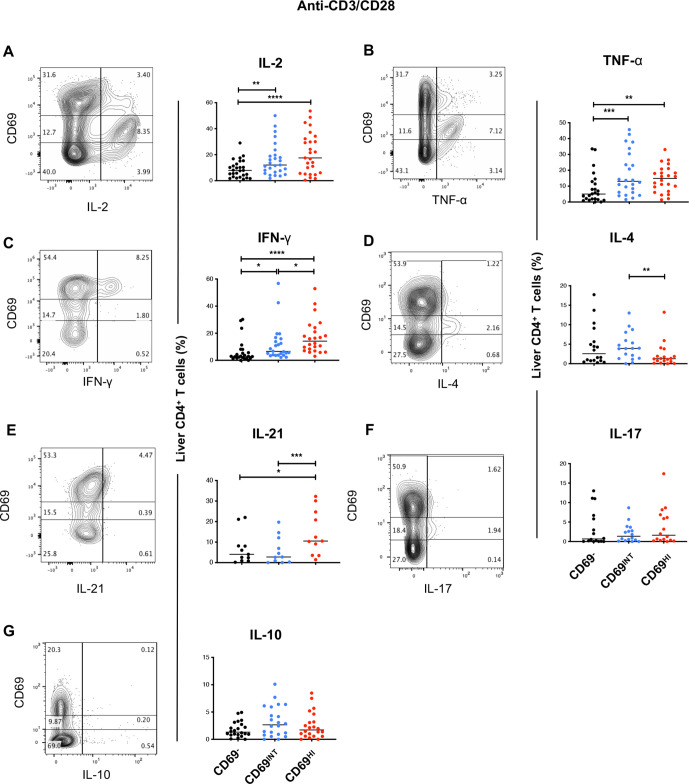
CD4+ TRM cells have a superior functional capacity to circulating T cells and mediate protection against a number of viral infections in multiple organs.1 15 To assess the functional potential of CD69HICD4+ TRM and CD69INTCD4+ T cells, intrahepatic leukocytes from a subset of livers were first prestained for CD69 expression to rule out stimulation-induced changes to CD69 expression and then were stimulated to assess their capacity for cytokine production. Following T-cell receptor (TCR) ligation with 5-hour anti-CD3/CD28 stimulation, more CD69HICD4+ T cells produced interferon gamma (IFN-γ) and IL-21 than any other population, and both CD69HI and CD69INTCD4+ T cells were enriched for IL-2, and tumour necrosis factor alpha (TNF-α) compared with their CD69−CD4+ counterparts (figure 4). Among the two CD69-expressing populations, CD69INTCD4+ T cells expressed more IL-4, with no differential expression patterns noted for IL-17 or IL-10.
Figure 4.
Liver CD69HICD4+ and CD69INTCD4+ T cells are skewed towards TH1 and TH2 functional profiles, respectively. Each sample was stained with CD69 prior to stimulation to exclude effects of altered CD69 levels due to cellular activation. Representative flow plots and combined data of % expression of six prototypical TH cytokines after 5 hours of stimulation of IHL with anti-CD3/CD28: (A) IL-2 (n=27), (B) TNF-α (n=24), (C) IFN-γ (n=24), (D) IL-4 (n=18), (E) IL-21 (n=11), (F) IL-17 (n=16), (G) IL-10 (n=22). See online supplemental table 4 for disease breakdowns. Freidman’s tests with Dunn’s multiple tests were used for statistical analysis (A–F). IFN-γ, interferon gamma; IHL, intrahepatic lymphocyte; IL, interleukin; TNF-α, tumour necrosis factor alpha.
We also assessed the maximum functional capacity of these cells following stimulation with mitogens PMA and ionomycin, and the direct ex vivo cytokine levels produced without exogenous stimulation. Similar to anti-CD3/CD28 stimulation, IFN-γ and IL-21 were also highest in CD69HICD4+ T cells in PMA/ionomycin-stimulated conditions, and IL-4 was similarly enriched in CD69INT versus CD69HICD4+ T cells (online supplemental figure 5A). This stimulation also demonstrated CD69HICD4+ T cells possessed the greatest potential to produce IL-2 and TNF-α and a higher potential than CD69−CD4+ T cells to produce IL-17. Importantly, in the absence of an exogenous stimulation, CD69INTCD4+ T cells produced IL-4, and CD69HICD4+ T cells showed a small enrichment for IFN-γ and IL-21 production compared with CD69−CD4+T-cells (online supplemental figure 5B), perhaps reflecting recent in vivo stimulation. IL-10 was not enriched in any subset irrespective of stimulation, in line with the lack of an over-representation of a TREG phenotype in either CD4+ T-cell subset (online supplemental figure 4).
gutjnl-2020-323771supp005.pdf (223.2KB, pdf)
Human TRM from other organs are often polyfunctional, producing the cytokines IFN-γ, TNF-α and IL-2 simultaneously, a property that equips these T cells for better pathogen control.30–34 Likewise, we observed an increase in IL-2+TNF-α+IFN-γ+ type 1 multifunctional cells in CD69HICD4+ T cells compared with CD69−CD4+ T cells, a feature not shared by CD69INTCD4+ T cells (online supplemental figure 5C). Finally, assessment of CD4+ T-cell transcription factors revealed an increase in T-bet in CD69HICD4+ T cells in line with an enrichment for IFN-γ production, but we were unable to detect TH2 transcription factor GATA-3 in hepatic CD4+ T cells (online supplemental figure 6). RORγt and FoxP3 were not differentially enriched in either CD69-expressing subset.
gutjnl-2020-323771supp006.pdf (991.4KB, pdf)
Taken together, CD69HICD4+ and CD69INTCD4+ T cells are functionally distinct, with CD69HICD4+ T cells favouring IFN-γ and type 1 multifunctional responses, while CD69INT expression predisposed CD4+ T cells to enhanced IL-4 production.
Increased CD69INTCD4+ T-cell frequencies correlate with necroinflammation in chronic HBV infection (CHB)
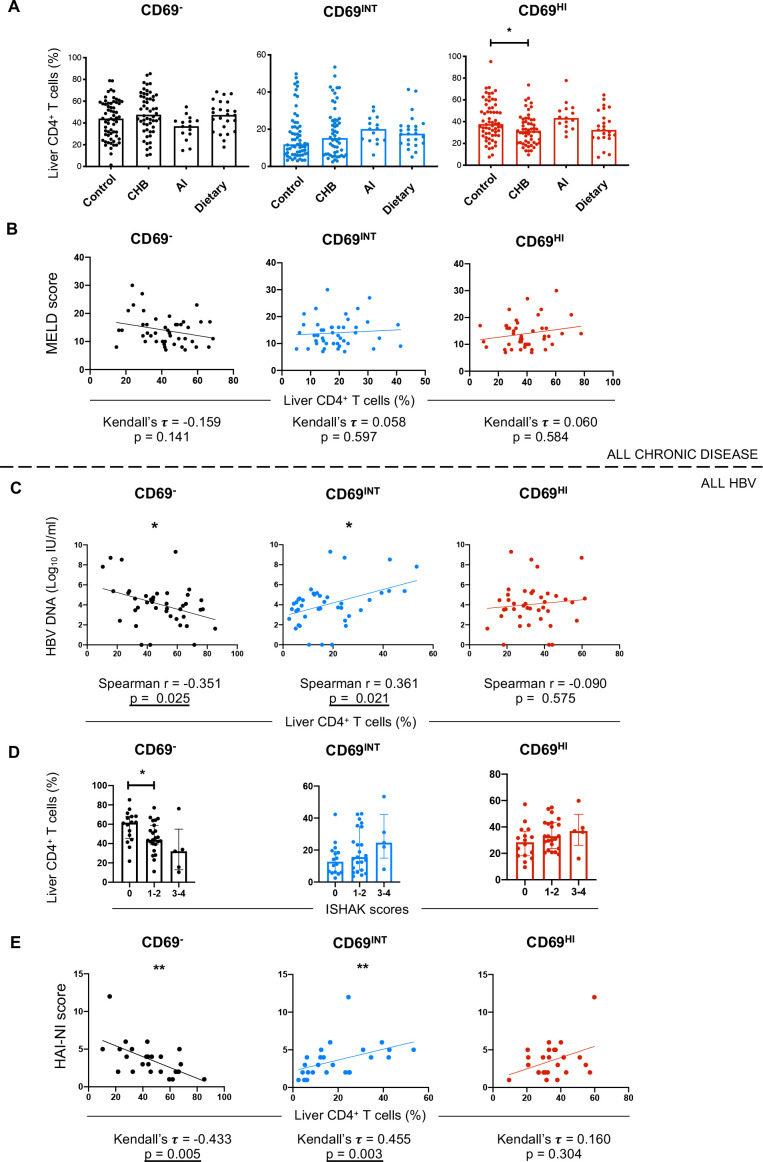
Next, we stratified liver samples into CHB, autoimmune (autoimmune hepatitis, primary biliary cholangitis and primary sclerosing cholangitis), dietary-induced liver disease (alcoholic liver disease and non-alcoholic steatohepatitis) and control (healthy preimplant, healthy transplant rejections, non-tumour-associated colorectal cancer and hepatocellular carcinoma margins and cyst-free margins of polycystic liver disease tissue) groups to test for population enrichment across diseases. CD69HICD4+ T-cell frequencies showed a modest yet consistent reduction in patients with CHB compared with control livers, but no other disease-specific differences were observed (figure 5A). To test if activated CD69INTCD4+ T cells correlated with disease progression, we analysed model for end-stage liver disease (MELD) scores (a commonly used metric to assess severity of non-viral chronic liver disease35) in explanted livers from patients with chronic hepatitis. There was no correlation with either CD4+T-cell population and MELD score, irrespective of liver disease aetiology, potentially reflective of a putative role for these cells in both health and disease (figure 5B).
Figure 5.
Increased CD69INTCD4+ T-cell frequencies correlate with necroinflammation in CHB. (A) Representation of each population in control livers (n=62 (11 donor explant transplant rejections, 5 healthy tissue biopsies, 36 colorectal cancer margin liver explants, 8 HCC margin liver explants and 2 cyst-free areas of PLD explants)), patients with chronic HBV (CHB, n=54), autoimmune liver disease (n=15 (6 PBC, 8 PSC and 1 AIH)), and dietary liver disease (n=24 (16 ALD and 8 NASH)). (B) Correlation analysis of patient MELD scores versus % of each of the three subsets for all donors with end-stage liver disease from centre A. (C) HBV DNA, Ishak scoring (D) and HAI-NI scoring (E) plotted against % of each T-cell population in the HBV cohort. Correlation and p values reported for each plot. Statistical testing used: Kruskal-Wallis tests with Dunn’s multiple post hoc tests (A, C), Kendall’s tau rank correlation tests (B, E), Spearman’s rank order correlation (C). AI, autoimmune; AIH, autoimmune hepatitis; ALD, alcoholic liver disease; CHB, chronic HBV infection; HAI-NI, Histology Activity Index-Necroinflammatory; HCC, hepatocellular carcinoma; NASH, non-alcoholic steatohepatitis; PBC, primary biliary cholangitis; PLD, polycystic liver disease; PSC, primary sclerosing cholangitis.
Progression to advanced fibrosis in CHB is highly heterogeneous, with the duration of infection and phase of disease contributing to this process.36 When analysing patients with CHB by hepatitis B ‘e’ antigen (HBeAg) seropositivity, viraemia, or the extent of liver inflammation using serum alanine aminotransferase (ALT) concentrations,36 we noted that CD69INTCD4+ T-cell frequencies correlated weakly with serum HBV DNA (figure 5C). The presence of HBeAg or serum ALT did not correlate with any CD4+ T-cell population (online supplemental figure 7A–B). Combined analysis of these three metrics into the distinct clinical phases also revealed no subpopulation-linked association (online supplemental figure 7C).
gutjnl-2020-323771supp007.pdf (318.5KB, pdf)
In order to further assess any links between populations and degree of fibrosis, and ongoing necroinflammatory activity, we subcategorised the patients using the validated Ishak and Histology Activity Index-Necroinflammatory scoring systems.37 Frequencies of CD69INTCD4+ T cells and CD69HICD4+ TRM cells did not correlate with the extent of fibrosis by Ishak scoring (figure 5D). However, CD69INTCD4+ T cells were more frequently observed in patients with a higher intrahepatic necroinflammatory score (figure 5E). Together, these data suggest that activated CD69INT cells may play a role in inflammatory processes of CHB.38
CD69INTCD4+and CD69HICD4+ T-cells are differentially induced by the liver microenvironment
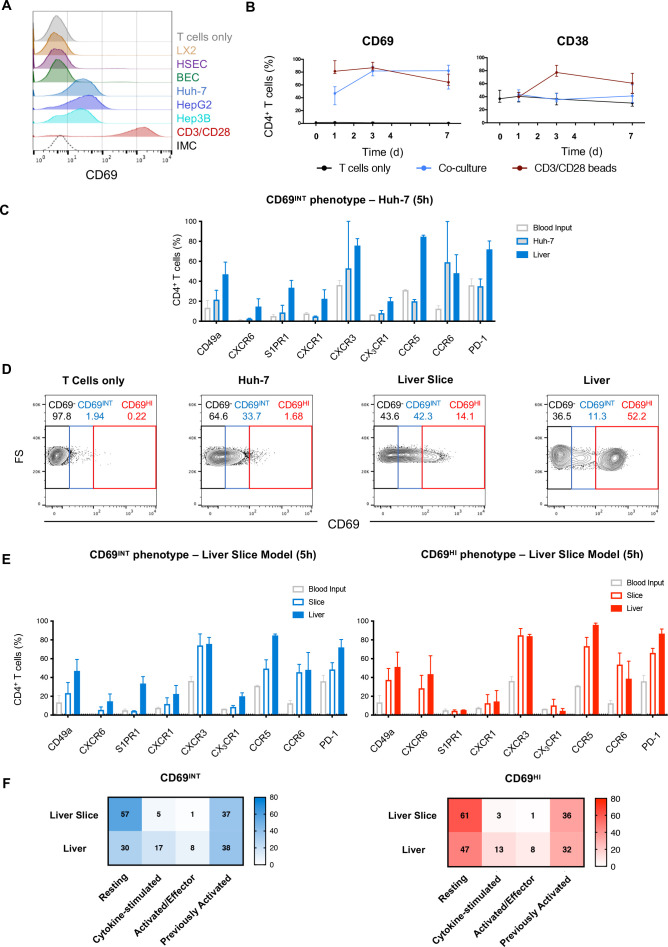
Finally, we sought to determine the origin of these distinct liver CD4+ T-cell populations by deconstructing the contribution of different hepatic cell types in vitro. To investigate the role of hepatic epithelia, we first cultured PBMC-derived CD4+ T cells with different hepatocyte cell lines (Huh-7, HepG2 and Hep3B); within 16 hours, we observed a clear induction of intermediate CD69 expression (figure 6A). By contrast, neither primary HSEC, primary BEC nor the hepatic stellate cell line, LX-2, was able to induce CD69 in the same time frame. However, primary BECs were able to strongly promote the increase in CD69INTCD4+ T cells from 72 hours, suggesting that sustained interaction with liver epithelia may be necessary for the generation of this population (online supplemental figure 8A–B). Hepatic epithelial-induced CD69 upregulation to an intermediate level was not simply a feature of their activation, as no concomitant upregulation of prototypical T-cell activation marker CD38 was seen, and conventional T cell activation with anti-CD3/CD28 led only to high CD69 expression (figure 6A–B). Mechanistically, this intermediate CD69 induction required direct T cell–epithelial cell contact, and was most efficient in memory CD4+ T cells (online supplemental figure 8C–D). In vitro-generated CD69INTCD4+ T cells partially recapitulated the phenotypical signature of intrahepatic CD69INTCD4+ T cells observed ex vivo after just 5 hours in culture, with upregulation of CXCR3 and CD49a, but not S1PR1, CXCR1 or CX3CR1 (figure 6C). CD69INTCD4+ T cells generated in vitro were also capable of producing IL-4 on stimulation, again resembling intrahepatic CD69INTCD4+ T cells (online supplemental figures 4 and 9). Thus, short-term contact with hepatic epithelia can induce a population of CD69INTCD4+ T cells in vitro.
Figure 6.
CD69INTCD4+ and CD69HI CD4+ T cells are differentially induced by the liver microenvironment. (A) % CD69 expression on PBMC-derived CD4+ T cells cultured for 16 hours with primary HSEC, primary BEC; hepatic stellate cell line LX-2; hepatocyte cell lines HuH-7, HepG2 or Hep3B; with anti-CD3/CD28; or alone. Histogram displays representative CD69 expression levels in each condition. (B) % CD69 and CD38 expressions on blood CD4+ T cells over a 7-day culture period with HuH-7 (n=8–10/timepoint). (C) Comparison of key phenotypical markers in Huh-7-generated CD69INT cells from PBMC following 5-hour culture, matched patient IHL CD69INT cells and blood T cells alone (n=2). (D) Representative flow plots showing degree of CD69INT and CD69HI generation within PBMC after 5 hours of culture: alone, with Huh-7 cells, with precision-cut donor-matched liver slices; or from directly isolated IHLs from matched human liver. (E) Comparison of CD69INT cells (left) and CD69HI cells (right) generated from donor-matched PBMCs in a precision-cut liver slice model, with matched donor-derived liver populations, and input blood CD4+ T cells alone (n=2). (F) Activation/differentiation statuses of CD69INT and CD69HI cells in the different conditions as assessed by KLRG-1/CD127 costaining patterns (as in figure 3E). Colour intensity and displayed numbers represent median % in each KLRG-1/CD127 designation. BEC, biliary epithelial cell; FS, forward scatter; HL, intrahepatic lymphocyte; HSEC, hepatic sinusoidal endothelial cell; IMC, isotype-matched control.
gutjnl-2020-323771supp008.pdf (1.2MB, pdf)
gutjnl-2020-323771supp009.pdf (552.6KB, pdf)
We further considered whether additional signals from the liver microenvironment were required to generate CD69HICD4+ T cells. To investigate this, we used a coculture model of patient-derived PBMCs with autologous precision-cut liver slices to allow full retention of the native liver microenvironment (figure 6D). Coculture of autologous PBMC for 5 hours with matched liver slices led to an increase in T-cell expression of both intermediate and high levels of CD69, not seen with hepatic epithelia coculture. Remarkably, short-term slice-culture-generated CD69HICD4+ T cells phenotypically resembled ex vivo intrahepatic CD69HICD4+ T cells, with high expression of CXCR6, CD49a, CCR5 and PD-1, low expression of S1PR1 and a largely resting (KLRG-1−CD127+) phenotype (figure 6E–F). Correspondingly, CD69INTCD4+ T cells also generated through hepatic slice culture acquired many of the phenotypical characteristics of their ex vivo counterparts, notably expression of CXCR3, and partial acquisition of the residency markers CD49a and CXCR6. Together these results suggest that CD4+ T cell contact with hepatic epithelia promotes their differentiation to a CD69INTCD4+ phenotype in the liver, whereas the generation of CD69HICD4+ TRM requires additional signals from the liver microenvironment.
Discussion
In this study, we characterised two distinct CD69-expressing CD4+ T-cell populations in the human liver — a long-lived CD69HICD4+ TRM subset, with a prototypical tissue-retention signature, a resting restrained phenotype and the ability to instigate type-1 multifunctional responses on stimulation; and a novel population of CD69INTCD4+ T cells with a CXCR3+CXCR1+CX3CR1+ phenotype that are more activated, recirculation-competent and skewed towards TH2 responses on stimulation. We show that these two populations possess different generation requirements and are equipped to play differential roles in liver disease.
In agreement with other human CD4+ TRM studies, liver CD69HICD4+ T cells expressed TRM-associated retention molecules CD49a and CXCR6,8 16 39 have low expression of the homing receptors S1PR1 and CX3CR1,13 a resting and restrained phenotype including high PD-1 expression,13 39 and the ability to produce TH1 cytokines.33 34 39 CXCR6 is of particular importance as a key liver retention molecule that is required for residence of multiple lymphocyte subpopulations in the liver,40–42 and our data demonstrate that the liver microenvironment is able to rapidly induce a CXCR6+ signature in newly formed resident CD4+ T cells.
We recently described human liver CD8+ TRM that share some of these key features (CXCR6+, PD-1+ and rapid functionality).5 Intriguingly, intrahepatic CD8+ TRM in both mouse and humans are thought to uniquely reside within the liver vasculature.5 43 44 Our data suggest that liver CD69HICD4+ TRM can also be found throughout the parenchyma, including within sinusoids. Candidate molecules for maintaining the CD69HITRM in this niche include CXCR6 through interactions with its ligand CXCL16, expressed on the sinusoidal lumen,23 40 or integrin αLβ2-ICAM interactions.44 Furthermore, our findings suggest CD69HICD4+TRM can be found in portal regions, likely directed specifically to portal vasculature by CCR5 ligands.45 The strategic positioning of CD4+ TRM in both vascular sites could allow efficient targeted immunosurveillance and opportunities to interact with other key immune cells within the liver. Finally, we demonstrated our CD69HICD4+TRM are most likely enriched for IL-21+TFH-like cells, in keeping with the emerging overlap between TFH and TRM phenotypes.46 47
Hepatic CD69INTCD4+ T cells have not been previously described. Functionally, CD69INTCD4+ T cells were most able to produce the TH2 cytokine IL-4. Key distinguishing features of hepatic CD69INTCD4+ T cells were expression of CXCR3 and CXCR1 required for hepatocyte homing,21 and importantly, CX3CR1. CD69INTCD4+ T cells were also found in hLNs, consistent with a wide-ranging immune surveillance role, analogous to the CX3CR1INT ‘peripheral memory’ CD8+ T cells that survey peripheral tissues in both humans and mice.19 20 Further, a small population of non-resident CD69INTCD8+ T cells in mice has been previously reported.48 The ‘migratory memory’ CD4+ T cells described by Watanabe et al showed variable CD69 positivity and recirculated through the skin slower than conventional TCM.49 Interestingly, activated, recirculating CD69INTCD4+ T cells, rather than resident CD69HICD4+ T cells correlated with necroinflammatory scores in CHB. This suggests CD69INTCD4+ T-cell involvement in proinflammatory processes in chronic viral disease, but whether this contributes to the cause, or is a consequence of the inflammation, remains to be determined.
Although CD69INTCD4+ T cells expressed a tissue egress signature (S1PR1 and CX3CR1) and were found in the blood, these cells may also contain a population capable of long-term residence. While this resident pool could conceivably derive from resident CD69HICD4+ T cells, the clear disparity in chemokine receptor expression suggests this outcome is unlikely. Resident CD69HICD4+ T cells may use CXCR6 for long-term retention more than CD69INTCD4+ T cells, allowing the possibility of distinct liver immunosurveillance roles. Alternatively, short-term resident populations may exist within the CD69INTCD4+ pool, as has been described for murine tissue CD4+ T cells,50 possibly with the potential to transdifferentiate into long-term ‘conventional’ CD69HITRM or ‘alternative’ CD69INTTRM. Future studies using single-cell sequencing approaches or TCR-repertoire analysis to further dissect the CD69INTCD4+ T-cell compartment are necessary to address these hypotheses.
Our data revealed insights into the mechanisms behind the generation of both CD69INTCD4+ and CD69HICD4+ T cells. CD69INTCD4+-like cells were generated following short-term direct contact with hepatic epithelial cell lines, and primary BEC. Although the molecular mechanism for this remains undefined, in situ hepatocyte contact may promote CD4+ T-cell CD69 upregulation to an intermediate level, increasing liver dwell time and allowing more efficient immunosurveillance. Conversely, CD69HICD4+ T cells required additional signals from the liver microenvironment, given that cells with this phenotype could only be formed from blood-derived CD4+ T cells when cocultured with autologous liver slices. Interestingly, robust initiation of a residency transcriptional programme can happen within 2 days in mice.48 Cytokines such as IL-15 and TGF-β can induce a CD8+ T-cell tissue residency programme,11 and we previously demonstrated that combinations of both cytokines were sufficient to generate cells with a CD8+ TRM phenotype.5 This raises the possibility that these cytokines provide the same additional signals for CD69HICD4+TRM formation. Unfortunately, extension of liver-slice coculture beyond 5 hours was not technically possible. Mouse models could be used to better test the longevity of these phenotypes.
In conclusion, this study provides a phenotypical and functional framework for understanding liver CD4+ TRM biology and characterises a novel heterogeneous CD69INTCD4+ T-cell population that is shaped by the liver microenvironment. We suggest that for at least some peripheral tissues, binary expression of CD69 alone is not sufficient to define tissue-resident CD4+ T cells. This work will facilitate the understanding of the role of liver CD4+ T cells in hepatic immune homeostasis, with implications for the development of novel immunotherapeutic strategies for chronic liver diseases.
gutjnl-2020-323771supp010.pdf (950.2KB, pdf)
Acknowledgments
We are grateful to all patients and donors at the Queen Elizabeth Hospital Birmingham, and the Royal Free Hospital, London. We thank the whole liver transplantation unit and all the clinical pathology team at the Queen Elizabeth Hospital for sample allocation; and all clinical staff who helped with patient recruitment across both centres including the Tissue Access for Patient Benefit project at The Royal Free Hospital. Thanks to Anthony Bolan (National Health Service, London, UK), and staff at the MRC Weatherall Institute of Molecular Medicine Sequencing Facility (Oxford, UK) for their help with HLA-haplotyping. Our thanks also to Loraine Brown and Bridget Gunson for their help with access and management of patient information and to the support staff at the UCL Infection and Immunity Flow Cytometry Core Facility.
Footnotes
Twitter: @BenWiggins_093, @pallett_lab, @druppygill, @arzii_x, @maini_lab, @zaniastamataki
Contributors: BGW, LJP, MKM and ZS conceived the project, designed experiments, critically evaluated the data and edited the manuscript; BGW, LJP, XL, SPD, OEA, USG, AMP, SK, KA, YSL and GMR generated data; BGW, LJP, XL and SPD analysed data; USG, GMR, GH and PTFK provided access to patient material; BGW wrote the manuscript and carried out statistical comparisons; BGW, LJP, SPD and ZS constructed figures; YH, MKM and ZS acquired funding and supervised the project; all authors reviewed the manuscript. BGW, LJP and XL are joint first authors. YH, MKM and ZS are joint last authors.
Funding: BGW and SPD were funded by PhD studentships from the Medical Research Council Centre for Immune Regulation, University of Birmingham. SPD was funded by an NC3R training fellowship. MKM and LJP are supported by Wellcome Trust Investigator award 214191/Z/18/Z and CRUK Immunology grant 26603. XL was funded by a Guangzhou Municipal Government (GMG 2016201604030021) award to YH and ZS. KA is funded by a Dr Falk studentship to GH. YSL was funded by an MRC Confidence in Concept Award to ZS. ZS was funded by the MRC, Wellcome Trust, GMG, The Birmingham Children’s Hospital Research Foundation the Royal Society and an intermediate career fellowship from the Medical Research Foundation. USG was funded by A Wellcome Trust Clinical Research Training Fellowship (107389/Z/15/Z), NIHR Academic Clinical Lectureship (018/064/A), Academy of Medical Sciences Starter Grant (SGL021/1030) and Seedcorn funding from Rosetrees/Stoneygate Trust (A2903). PTFK is supported by Barts Charity Project Grants (723/1795 and MGU/0406) and an NIHR Research for patient benefit award (PB-PG-0614-34087).
Competing interests: BW collaborated with and received funding from Bioniz. LJP sat on advisory boards/provided consultancy for Gilead Sciences and SQZ Biotech. KA was funded by a studentship with Dr Falk. MKM received research funding from Gilead Sciences, Hoffmann La Roche and Immunocore. MKM sat on advisory boards/provided consultancy for Gilead, Hoffmann La Roche, Immunocore, VIR, Galapagos NV, GSK, Abbvie and Freeline. ZS collaborated with Bioniz and AstraZeneca and consulted for Boehringer Ingelheim. All other authors declare no conflict of interest.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
This study was approved by local research ethics committees in Birmingham and London. All samples were obtained with written informed patient consent. All study protocols complied with the 1975 Declaration of Helsinki.
References
- 1. Clark RA. Resident memory T cells in human health and disease. Sci Transl Med 2015;7:269rv1. 10.1126/scitranslmed.3010641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schenkel JM, Fraser KA, Vezys V, et al. Sensing and alarm function of resident memory CD8⁺ T cells. Nat Immunol 2013;14:509–13. 10.1038/ni.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beura LK, Fares-Frederickson NJ, Steinert EM, et al. CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J Exp Med 2019;216:1214–29. 10.1084/jem.20181365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park CO, Kupper TS. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat Med 2015;21:688–97. 10.1038/nm.3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pallett LJ, Davies J, Colbeck EJ, et al. IL-2 high tissue-resident T cells in the human liver: Sentinels for hepatotropic infection. J Exp Med 2017;214:1567–80. 10.1084/jem.20162115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mueller SN, Mackay LK. Tissue-Resident memory T cells: local specialists in immune defence. Nat Rev Immunol 2016;16:79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- 7. Jiang X, Clark RA, Liu L, et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature 2012;483:227–31. 10.1038/nature10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Teijaro JR, Turner D, Pham Q, et al. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol 2011;187:5510–4. 10.4049/jimmunol.1102243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pan Y, Tian T, Park CO, et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 2017;543:252–6. 10.1038/nature21379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pallett LJ, Schmidt N, Schurich A. T cell metabolism in chronic viral infection. Clin Exp Immunol 2019;197:143–52. 10.1111/cei.13308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mackay LK, Wynne-Jones E, Freestone D, et al. T-Box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 2015;43:1101–11. 10.1016/j.immuni.2015.11.008 [DOI] [PubMed] [Google Scholar]
- 12. McMaster SR, Wilson JJ, Wang H, et al. Airway-Resident memory CD8 T cells provide antigen-specific protection against respiratory virus challenge through rapid IFN-γ production. J Immunol 2015;195:203–9. 10.4049/jimmunol.1402975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumar BV, Ma W, Miron M, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep 2017;20:2921–34. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turner DL, Farber DL. Mucosal resident memory CD4 T cells in protection and immunopathology. Front Immunol 2014;5:331. 10.3389/fimmu.2014.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szabo PA, Miron M, Farber DL. Location, location, location: tissue resident memory T cells in mice and humans. Sci Immunol 2019;4. 10.1126/sciimmunol.aas9673. [Epub ahead of print: 05 04 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong MT, Ong DEH, Lim FSH, et al. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 2016;45:442–56. 10.1016/j.immuni.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 17. Morland CM, Fear J, McNab G, et al. Promotion of leukocyte transendothelial cell migration by chemokines derived from human biliary epithelial cells in vitro. Proc Assoc Am Physicians 1997;109:372–82. [PubMed] [Google Scholar]
- 18. Lalor PF, Adams DH. The liver: a model of organ-specific lymphocyte recruitment. Expert Rev Mol Med 2002;4:1–15. 10.1017/S1462399402004155 [DOI] [PubMed] [Google Scholar]
- 19. Gerlach C, Moseman EA, Loughhead SM, et al. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity 2016;45:1270–84. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gordon CL, Lee LN, Swadling L, et al. Induction and Maintenance of CX3CR1-Intermediate Peripheral Memory CD8+ T Cells by Persistent Viruses and Vaccines. Cell Rep 2018;23:768–82. 10.1016/j.celrep.2018.03.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Adams DH, Ju C, Ramaiah SK. Mechanisms of immune-mediated liver injury. Toxicological 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gill US, Pallett LJ, Thomas N, et al. Fine needle aspirates comprehensively sample intrahepatic immunity. Gut 2019;68:1493–503. 10.1136/gutjnl-2018-317071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heydtmann M, Lalor PF, Eksteen JA, et al. Cxc chemokine ligand 16 promotes integrin-mediated adhesion of liver-infiltrating lymphocytes to cholangiocytes and hepatocytes within the inflamed human liver. J Immunol 2005;174:1055–62. 10.4049/jimmunol.174.2.1055 [DOI] [PubMed] [Google Scholar]
- 24. Pallett LJ, Burton AR, Amin OE, et al. Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J Exp Med 2020;217. 10.1084/jem.20200050. [Epub ahead of print: 07 09 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Testi R, D'Ambrosio D, De Maria R, et al. The CD69 receptor: a multipurpose cell-surface trigger for hematopoietic cells. Immunol Today 1994;15:479–83. 10.1016/0167-5699(94)90193-7 [DOI] [PubMed] [Google Scholar]
- 26. Agata Y, Kawasaki A, Nishimura H, et al. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol 1996;8:765–72. 10.1093/intimm/8.5.765 [DOI] [PubMed] [Google Scholar]
- 27. Henson SM, Akbar AN. KLRG1--more than a marker for T cell senescence. Age 2009;31:285–91. 10.1007/s11357-009-9100-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang H-Y, Luther SA. Expression and function of interleukin-7 in secondary and tertiary lymphoid organs. Semin Immunol 2012;24:175–89. 10.1016/j.smim.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 29. Thome JJC, Yudanin N, Ohmura Y, et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 2014;159:814–28. 10.1016/j.cell.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kannanganat S, Ibegbu C, Chennareddi L, et al. Multiple-cytokine-producing antiviral CD4 T cells are functionally superior to single-cytokine-producing cells. J Virol 2007;81:8468–76. 10.1128/JVI.00228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seder RA, Darrah PA, Roederer M. T-Cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 2008;8:247–58. 10.1038/nri2274 [DOI] [PubMed] [Google Scholar]
- 32. Sathaliyawala T, Kubota M, Yudanin N, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 2013;38:187–97. 10.1016/j.immuni.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Purwar R, Campbell J, Murphy G, et al. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One 2011;6:e16245. 10.1371/journal.pone.0016245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Booth JS, Toapanta FR, Salerno-Goncalves R, et al. Characterization and functional properties of gastric tissue-resident memory T cells from children, adults, and the elderly. Front Immunol 2014;5:294. 10.3389/fimmu.2014.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singal AK, Kamath PS. Model for end-stage liver disease. J Clin Exp Hepatol 2013;3:50–60. 10.1016/j.jceh.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suk-Fong Lok A. Hepatitis B treatment: what we know now and what remains to be Researched. Hepatol Commun 2019;3:8–19. 10.1002/hep4.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goodman ZD. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J Hepatol 2007;47:598–607. 10.1016/j.jhep.2007.07.006 [DOI] [PubMed] [Google Scholar]
- 38. Maini MK, Boni C, Lee CK, et al. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J Exp Med 2000;191:1269–80. 10.1084/jem.191.8.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oja AE, Piet B, Helbig C, et al. Trigger-happy resident memory CD4+ T cells inhabit the human lungs. Mucosal Immunol 2018;11:654–67. 10.1038/mi.2017.94 [DOI] [PubMed] [Google Scholar]
- 40. Geissmann F, Cameron TO, Sidobre S, et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol 2005;3:e113. 10.1371/journal.pbio.0030113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stegmann KA, Robertson F, Hansi N, et al. Cxcr6 marks a novel subset of T-betloEomeshi natural killer cells residing in human liver. Sci Rep 2016;6:26157. 10.1038/srep26157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tse S-W, Radtke AJ, Espinosa DA, et al. The chemokine receptor CXCR6 is required for the maintenance of liver memory CD8⁺ T cells specific for infectious pathogens. J Infect Dis 2014;210:1508–16. 10.1093/infdis/jiu281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fernandez-Ruiz D, Ng WY, Holz LE, et al. Liver-Resident Memory CD8+ T Cells Form a Front-Line Defense against Malaria Liver-Stage Infection. Immunity 2016;45:889–902. 10.1016/j.immuni.2016.08.011 [DOI] [PubMed] [Google Scholar]
- 44. McNamara HA, Cai Y, Wagle MV, et al. Up-Regulation of LFA-1 allows liver-resident memory T cells to patrol and remain in the hepatic sinusoids. Sci Immunol 2017;2. 10.1126/sciimmunol.aaj1996. [Epub ahead of print: 17 03 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oo YH, Shetty S, Adams DH. The role of chemokines in the recruitment of lymphocytes to the liver. Dig Dis 2010;28:31–44. 10.1159/000282062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Künzli M, Schreiner D, Pereboom TC, et al. Long-Lived T follicular helper cells retain plasticity and help sustain humoral immunity. Sci Immunol 2020;5. 10.1126/sciimmunol.aay5552. [Epub ahead of print: 06 03 2020]. [DOI] [PubMed] [Google Scholar]
- 47. Chen M-M, Xiao X, Lao X-M, et al. Polarization of tissue-resident TFH-Like cells in human hepatoma bridges innate monocyte inflammation and M2b macrophage polarization. Cancer Discov 2016;6:1182–95. 10.1158/2159-8290.CD-16-0329 [DOI] [PubMed] [Google Scholar]
- 48. Skon CN, Lee J-Y, Anderson KG, et al. Transcriptional downregulation of S1PR1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol 2013;14:1285–93. 10.1038/ni.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Watanabe R, Gehad A, Yang C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med 2015;7:279ra39. 10.1126/scitranslmed.3010302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Collins N, Jiang X, Zaid A, et al. Skin CD4(+) memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat Commun 2016;7:11514. 10.1038/ncomms11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-323771supp011.pdf (182.2KB, pdf)
gutjnl-2020-323771supp001.pdf (1.1MB, pdf)
gutjnl-2020-323771supp002.pdf (612.1KB, pdf)
gutjnl-2020-323771supp003.pdf (711.8KB, pdf)
gutjnl-2020-323771supp004.pdf (1MB, pdf)
gutjnl-2020-323771supp005.pdf (223.2KB, pdf)
gutjnl-2020-323771supp006.pdf (991.4KB, pdf)
gutjnl-2020-323771supp007.pdf (318.5KB, pdf)
gutjnl-2020-323771supp008.pdf (1.2MB, pdf)
gutjnl-2020-323771supp009.pdf (552.6KB, pdf)
gutjnl-2020-323771supp010.pdf (950.2KB, pdf)
Data Availability Statement
Data are available upon reasonable request.