Abstract
Objectives
MicroRNA (miRNA) profiles have been evaluated in several biospecimens in relation to common diseases for which diet may have a considerable impact. We aimed at characterising how specific diets are associated with the miRNome in stool of vegans, vegetarians and omnivores and how this is reflected in the gut microbial composition, as this is still poorly explored.
Design
We performed small RNA and shotgun metagenomic sequencing in faecal samples and dietary recording from 120 healthy volunteers, equally distributed for the different diets and matched for sex and age.
Results
We found 49 miRNAs differentially expressed among vegans, vegetarians and omnivores (adj. p <0.05) and confirmed trends of expression levels of such miRNAs in vegans and vegetarians compared with an independent cohort of 45 omnivores. Two miRNAs related to lipid metabolism, miR-636 and miR-4739, were inversely correlated to the non-omnivorous diet duration, independently of subject age. Seventeen miRNAs correlated (|rho|>0.22, adj. p <0.05) with the estimated intake of nutrients, particularly animal proteins, phosphorus and, interestingly, lipids. In omnivores, higher Prevotella and Roseburia and lower Bacteroides abundances than in vegans and vegetarians were observed. Lipid metabolism-related miR-425-3p and miR-638 expression levels were associated with increased abundances of microbial species, such as Roseburia sp. CAG 182 and Akkermansia muciniphila, specific of different diets. An integrated analysis identified 25 miRNAs, 25 taxa and 7 dietary nutrients that clearly discriminated (area under the receiver operating characteristic curve=0.89) the three diets.
Conclusion
Stool miRNA profiles are associated with specific diets and support the role of lipids as a driver of epigenetic changes and host-microbial molecular interactions in the gut.
Keywords: diet, molecular genetics, colonic microflora
Significance of this study.
What is already known on this subject?
The adoption of a vegetarian or vegan diet is associated with a lower intake of relevant risk factors for chronic diseases, such as cardiovascular diseases or cancer. However, it is still controversial whether such diets are always healthier and recommended for all.
Dietary components can modulate the composition and function of the gut microbiome and serve as a substrate for bacterial metabolism that could influence the host health.
A host-gut interplay may modulate the effects of the diet. MicroRNAs (miRNAs) found in stool may directly regulate specific bacterial gene expression and affect gut microbial growth, therefore being essential for maintaining normal gut microbiota.
What are the new findings?
Diet is associated with stool miRNome profile of omnivores, vegetarians and vegans, and several miRNAs characterise peculiar signatures of the different dietary regimes.
MiR-636 and miR-4739, having a significant role in lipid metabolism, show expression levels inversely related to the duration of the non-omnivorous dietary regime, independently of age. Furthermore, targets of stool miRNAs upregulated in vegetarian/vegan subjects are enriched in both lipid and folate metabolism.
Vegan and vegetarians are characterised by high stool levels of miR-425-3p, an miRNA related to lipid metabolism, whose levels in stool correlate with the relative abundance of specific microbial taxa, including Akkermansia muciniphila.
Stool miRNA profiles and microbiome composition, together with dietary nutrients, can accurately distinguish different diets and strengthen the evidence of host-gut microbiome crosstalk.
Significance of this study
How might it impact on clinical practice in the foreseeable future?
These findings constitute a significant step forward in the elucidation of the molecular mechanisms involving host and microbial activities associated with dietary component absorption and metabolism.
In monitoring adherence to specific dietary regimes, where the intake of nutrients should be carefully checked, a translational approach of a non-invasive stool miRNA profiling may be easily implemented.
An integrated analysis of gut microbiome and faecal miRNAs may provide insights for designing more accurate personalised dietary patterns and recommendations aimed at prevention/treatment of chronic diseases.
Introduction
Dietary habits are strongly related to lifestyle factors as well as social life and culture, and all are strictly connected with health. Diet and nutrition represent relevant risk factors for different metabolic and chronic diseases such as steatosis and non-alcoholic fatty liver disease, atherosclerosis and cancer.1 High-level consumption of red, processed meat and animal fats have been associated with an increase in cancer risk, including colorectal cancer (CRC),2 while the high intake of dietary fibres is considered a protective factor.3
An increasing number of studies showed that vegetarian (VT) and vegan (VN) dietary patterns are associated with significantly lower levels of the most relevant risk factors for chronic diseases (such as body mass index (BMI) or lipid and glucose levels) with a consequent reduced risk of incidence or mortality for ischaemic heart disease or cancers.4 However, it is difficult to discern whether the health advantages attributed to VNs could be generalised to VTs or even to moderate meat eaters following a healthy diet. Moreover, there is still the need to elucidate the precise molecular mechanisms of these associations. So far, only very few studies have performed rigorous experiments comparing omnivorous, VT and VN subjects as distinct experimental groups. The few available, investigating the molecular mechanisms involved with dietary components absorption, were generated from cell cultures and animal models.5
In the past few years, microRNAs (miRNAs), small non-coding RNAs that modulate gene expression at the post-transcriptional level, have been studied in detail as important regulators altered in human diseases, including cancer6 and neurodegenerative disorders.7 Nutrients and specific diet components have been implicated in the modulation of miRNA expression in in vitro/in vivo models and, as a consequence, in all the miRNA-mediated molecular mechanisms.8 Based on an miRNA candidate approach, we performed a pilot study on stool/plasma samples of healthy volunteers with different dietary habits and found differential miRNA expression levels among them.9 Different studies reported a relationship between the expression of faecal miRNAs and gastrointestinal disorders,10 including CRC.11 12 Then, stool samples can represent a proper non-invasive surrogate tissue to evaluate the transcriptional and epigenomic changes occurring on the adherence to a different dietary regimen. However, many issues remain unsolved regarding the quality and quantity of dietary components needed to regulate miRNA expression in humans.13 Recently, we clarified certain relationships between specific miRNA expression and the inclusion of specific nutrients in the diets of healthy individuals.14
Dietary components can modulate the composition and function of the gut microbiome15 16 and diets rich in fibres and poor in animal-based products (VN/VT regimes, Mediterranean diet) may drive a selection of specific taxa.17–19 In parallel, a growing number of studies reported the association of a gut microbiome dysbiosis with several gastrointestinal and non-gastrointestinal diseases, including diabetes,20 inflammatory bowel disease21 and cancer.22
Liu and colleagues23 have demonstrated that faecal miRNAs directly regulate specific bacterial gene expression and affect gut microbial growth, being therefore essential for the maintenance of normal gut microbiota. Moreover, an integrated analysis of the gut microbiome and small non-coding RNAs in the human stool may provide insights for designing more accurate tools for diagnostic purposes in CRC.12 Intestinal microbiota and miRNAs may interact to regulate host gene expression.24 However, the interplay existing between gut microbiome and miRNA expression profiles in the presence of a specific dietary regime is still unknown.
In the present study, the relationship between miRNA profiles and the gut microbiome according to different long-term dietary regimes was explored. Small non-coding RNA sequencing was performed in faecal and plasma samples from 120 healthy volunteers with omnivorous, VT and VN dietary habits. In addition, gut microbiome was analysed by shotgun metagenomics. The results from sequencing were integrated with detailed information on food and specific nutrient intake by validated questionnaires to elucidate the existence of molecular characterisation of a specific dietary regime.
By DESeq2 analysis, we found 49 miRNAs differentially expressed in stool of VNs and VTs in comparison with omnivores (Os) (adj. p<0.05). Interestingly, differentially expressed miRNAs among the dietary groups are involved in the regulation of lipid and folate metabolism, adipocyte differentiation and connected with obesity and obesity-connected diabetes. A combination of these miRNAs, microbial taxa and nutrients clearly distinguished subjects according to their diet (area under the receiver operating characteristic curve (AUC)=0.89).
Methods
Detailed methods are provided in the online supplemental materials of the manuscript.
gutjnl-2021-325168supp001.pdf (191.3KB, pdf)
Study population recruitment and dietary information
A cohort of 120 healthy individuals (72 women and 48 men), categorised as VTs, VNs and Os, was recruited on a voluntary basis in Turin (Italy) between May 2017 and July 2019. Eligible subjects were included in the study according to the following selection criteria: the associated dietary regime followed for more than 1 year, no consumption of antibiotics in the previous 3 months before sampling, no use of aspirin or other anti-inflammatory drugs in the month prior to the sampling, no evidence of intestinal and other pathologies, no pregnancy and lactation. Three subjects made use of antihypertensive drugs and only one made use of a cholesterol-lowering medication. The study cohort included the same proportion of VT, VN and O, matched for sex and age (figure 1A). Anthropometric data were collected for all subjects at the time of sampling. For an independent group of unmatched 45 omnivorous healthy individuals (31 women and 14 men), stool small RNA-Seq data (performed in the same laboratory and with the same protocol/pipeline of analysis) were also available for miRNA profiles validation.
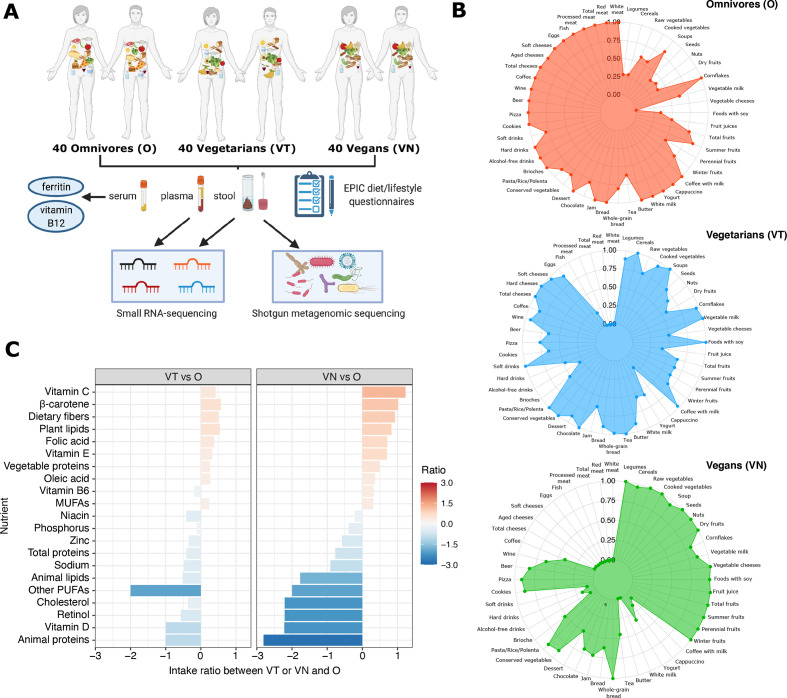
Figure 1.
(A) Workflow of the study; (B) Radar plots reporting the relative levels of weekly intake of specific food and drinks according to the dietary regimens as self-reported in questionnaires; (C) estimated nutrient intake ratio in vegetarians (VT, left panel) and vegans (VN, right panel) with respect to the omnivorous diet (O). MUFAs, monounsaturated fatty acids; PUFAs, polyunsaturated fatty acids.
All participants signed informed written consent. The design of the study, the informed consent and protocols were approved by the local Ethics Committee.
All recruited subjects filled in a validated self-administered Food Frequency Questionnaire (FFQ) assessing the usual diet, together with lifestyle and personal history data, in accordance with the European Prospective Investigation into Cancer and Nutrition EPIC study standards.25 The FFQ consisted of 248 questions concerning 188 different food items and included photos with two or three sample dishes of definite sizes or references to standard portion sizes. The composition in nutrients of individual food items was obtained from Italian food composition tables26 and the average daily intake of macronutrients and micronutrients for each volunteer was estimated.
Collection of biological samples
Naturally evacuated faecal samples were obtained from all volunteers previously instructed to self-collect the specimen at home. Stool were collected in nucleic acid collection and transport tubes with RNA/DNA stabilising solution (Norgen Biotek Corp) and stored at −80°C until RNA and DNA extraction.27
Blood samples were collected into EDTA and serum clot activator tubes, according to standard phlebotomy procedures, when volunteers brought the stool samples to the laboratory. EDTA tubes were centrifuged at ×1000 g for 10 min at room temperature and plasma was aliquoted and stored at −80°C. Serum tubes were centrifuged at ×4000 g for 10 min at room temperature, aliquoted and stored at −80°C. One aliquot of serum was used for the analyses of vitamin B12 and ferritin. The time between blood sample procurement and plasma/serum storage was less than 3 hours.
Extraction of total RNA from stool and plasma
Total RNA from stool was extracted using the Stool Total RNA Purification Kit (Norgen Biotek Corp) as described in Tarallo et al.12
Total RNA was extracted from 200 µL of plasma with the miRNeasy plasma/serum mini kit (Qiagen, Hilden, Germany) using the QiaCube extractor (Qiagen) following the manufacturer’s instructions and as described in the study by Ferrero et al 27 and Sabo et al.28
RNA concentration was quantified by Qubit microRNA Assay Kit (Invitrogen).
Extraction of total DNA from stool
The DNA extraction was performed with the QIAamp DNA stool MiniKit (QIAGEN) according to the instructions of the manufacturer and as described in the study by Tarallo et al 12 and Thomas et al.22 The DNA quantification was performed with Qubit fluorometer (Qubit DNA HS Assay Kit; Invitrogen).
Small RNA sequencing and bioinformatic analyses
Library preparation for small RNA (sRNA) transcripts was performed with the NEBNext Multiplex Small RNA Library Prep Set for Illumina (Protocol E7330, New England BioLabs, USA) as described in the study by Ferrero et al.27 For each sample, 250 ng of RNA was used as starting material to prepare libraries.
The obtained sequence libraries were subjected to the Illumina sequencing pipeline and 50 cycles sequencing-by-synthesis on the HiSeq 2000 (Illumina, USA) (in collaboration with Genecore Facility at EMBL, Heidelberg, Germany).
Small RNA sequencing analyses were performed using a previously described approach.12 The reads passing the quality control were trimmed from the adapter sequences using Cutadapt V.1.10. Trimmed reads were mapped against human miRNA sequences from miRBase V.2229 using BWA V.0.7.1230 with default settings. Summary statistics of the data and alignment are reported in online supplemental table 1A, B. Human miRNAs were annotated and quantified using two methods called the ‘knowledge-based’ and ‘position-based’ methods as described in Tarallo et al.12 miRNAs whose assigned arms were derived from the ‘position-based’ methodology were indicated in italic.
Differential expression analysis was performed with DESeq2 R package V.1.22.231 using the likelihood ratio test function including age and sex as covariates in the model. An miRNA was defined as differentially expressed (DEmiRNA) if associated with a Benjamini-Hochberg (BH)-adjusted p value<0.05 and supported by more than 15 reads in at least one of the dietary groups. The set of DEmiRNAs was overlapped with the annotation from Human microRNA Disease Database (HMDD) V.3.2,32 considering the annotations associated with each miRNA precursor and the database annotations related to gastrointestinal cancers, metabolic disorders, immune-related, or cardiovascular diseases.
Shotgun metagenomic and bioinformatic analyses
Libraries were prepared using the Nextera DNA Library Preparation kit (Illumina) and sequenced on an Illumina HiSeq platform as described in the study by Thomas et al.22
Host contamination was removed using the human sequence removal procedure from the Human Microbiome Project.33 Raw reads were quality-trimmed (Phred score <25) and reads shorter than 60 bp were discarded with the SolexaQA++ software.34 The number of reads obtained for each sample is reported in online supplemental table 1C. Taxonomic profiling was carried out by using MetaPhlAn 3.35 Functional profiles were obtained by HUMAnN 3.0.36
Integration between miRNA expression, nutrient intake and taxa
The R package mixOmics was employed for the integration of the three datasets (taxonomic profiles, miRNA expression profiles and dietary information), using the Data Integration Analysis for Biomarker discovery using Latent cOmponents (DIABLO) model.37 Datasets were integrated after normalisation (scale R function) and removal of near-zero variables (nearZeroVar function of R package caret).
Statistical analyses
All statistical analyses were performed using R software V.4.0.4. The analysis of the dietary and lifestyle covariates among subject groups was performed using the χ2 test for categorical variables. For continuous covariates, Kruskal-Wallis and Wilcoxon rank-Sum tests were performed.
Correlation analysis between DEmiRNA expression and nutrient intake was performed using the Spearman method. Multiple testing correction was performed using the BH method and correlations associated with an adjusted p value<0.05 were considered as statistically significant.
The Generalized Linear Regression Model (GLM) was computed using the glm R function to assess the relevance of subject covariates in explaining the observed stool DEmiRNA levels. DEmiRNA levels were considered as the dependent variable of the model while subjects age, sex, BMI, physical activity, waist circumference and season of sample collection were considered as the independent variables of the model. A covariate was considered related to DEmiRNA expression if associated with a p value<0.05. The relationship between DEmiRNA expression and the estimated intake of nutrients was evaluated computing a GLM adjusted for age, sex and BMI. The significance of the GLM was computed with respect to a null model using F-test implemented in the ANOVA R function. Models associated with a p value<0.05 were considered statistically significant. The contribution of a nutrient to the model was considered significant if the associated p value was lower than 0.05 and lower than those computed for the contribution of sex, age and BMI to the model.
Classification accuracy of the features identified by the DIABLO analysis was performed using a 10-fold cross-validation approach with the Random Forest classifier implemented in Weka V.3.8.5 (https://www.cs.waikato.ac.nz/ml/weka/). The classification efficiency was evaluated in terms of AUC.
MiRNA targets functional enrichment analysis
Functional enrichment analysis was performed using RBiomirGS V.0.2.1238 considering validated targets of: (1) DEmiRNAs coherently differentially expressed in VT and VN with respect to O; (2) miRNAs significantly correlated with cluster of nutrients or (3) taxa; (4) DEmiRNAs from the DIABLO analysis. The log2FC and adjusted p value computed between the VN and O diets were used as inputs for the tool. The gene sets characterised by an adjusted p value lower than 0.05 were considered as significantly enriched.
MiRNA expression and epigenetic state analysis in intestinal tissues
The analysis of miRNA expression in intestinal tissue samples was performed considering the data from the TissueAtlas,39 which includes miRNA expression levels measured in two colon and three small intestine samples. The epigenetic status of miRNA coding genes was evaluated using the chromatin status defined by the Roadmap Epigenomics Project.40 Specifically, the analysis was performed considering the 15 statuses (Core 15-state model) defined by integrating epigenetic data of duodenum, small intestine, colon, rectum mucosa, or colon smooth muscle samples. Only mature miRNAs with an univocal genomic locus were considered for the analysis.
Results
Study subjects and dietary characteristics
A total of 120 volunteers, divided into 40 Os, 40 VTs and 40 VNs, were included in the study. The characteristics of the subjects and the design of the study are described in table 1 and figure 1A, respectively. Subjects following an omnivorous diet displayed a slightly higher BMI (kg/m2) compared with the other groups (O: 23.6±3.9, VT: 21.9±2.6, VN: 21.8±2.9 kg/m2, in all comparisons p<0.05) and waist circumference (O: 83.8±11.6, VT: 78.1±8.6, VN:78.9±8.8 cm, in O vs VT, p<0.05) (table 1). Days of sample collection were differently distributed for VT and VN with respect to O individuals with respect to the different seasons of the year (p<0.05) (data not shown).
Table 1.
Study population characteristics
| Characteristics | Omnivores | Vegetarians | P value | Vegans | P value | |
| Sex | Females | 24 | 24 | – | 24 | – |
| Males | 16 | 16 | 16 | |||
| Age (years) | Mean±SD | 40.5±13.2 | 40.6±11.7 | 0.95 | 39.1±11.6 | 0.76 |
| Range | 20.6–64.1 | 21.1–67.4 | 18.5–60.6 | |||
| Years of vegetarian or vegan diet | Mean±SD | – | 9.7±9.0 | – | 4.8±2.5 | – |
| Range | – | 0.7–34.5 | 0.6–13.0 | |||
| BMI (kg/m2) | 23.6±3.9 | 21.9±2.6 | 0.05 | 21.8±2.9 | 0.03 | |
| BMI classes | Underweight (≤18.5) | 2 | 2 | 5 | ||
| Normal weight (18.5–24.9) | 26 | 33 | 0.13 | 30 | 0.13 | |
| Overweight (25–29.9) | 9 | 4 | 5 | |||
| Obese (≥30.0) | 3 | 0 | 0 | |||
| Missing | – | 1 | – | |||
| Waist circumference (cm) | 83.8±11.6 | 78.1±8.6 | 0.03 | 78.9±8.8 | 0.05 | |
| Highest level of education attained (n (%)) |
Primary or secondary school | 15 (37.5) | 14 (35.0) | 1.00 | 22 (55.0) | 0.18 |
| University or higher | 25 (62.5) | 26 (65.0) | 18 (45.0) | |||
| Physical Activity Total | Active | 2 | 4 | 0 | ||
| Moderately active | 16 | 6 | 0.02 | 18 | 0.09 | |
| Moderately inactive | 7 | 18 | 14 | |||
| Inactive | 15 | 12 | 8 | |||
| Smoking habit | Non-smoker | 21 | 23 | 26 | ||
| Former smoker | 10 | 15 | 0.06 | 10 | 0.29 | |
| Current smoker | 9 | 2 | 4 | |||
For continuous variables, the p value was computed using the Kruskal-Wallis method while χ2 test was used for analysing categorical variables.
Statistically significant values are in bold.
BMI, body mass index.
All the dietary and lifestyle characteristics are reported in detail in online supplemental table 2. In serum, the concentration of ferritin was significantly decreased in VT/VN with respect to O (p<0.01 and p<0.05, respectively), while vitamin B12 was lower in VT with respect to the other two groups. From the list of 188 items included in the EPIC questionnaires, the analysis was focused on the most representative nutrients characterising the differences in the dietary regimes (consumption of meat, processed meat, fish, cheese and dairy products, bread and pasta, total vegetables and fruits). The relative weekly intake of specific food and drinks as reported in the subject questionnaires is summarised in figure 1B and online supplemental figure 1A. A significantly higher intake of nutrients such as vitamin C, β-carotene and fibres was found in VN when compared with O. Conversely, in the same comparison, a significant decrease in vitamin D, animal proteins, retinol and cholesterol was observed in VN (figure 1C). In VT, the discrepancies with O were less marked but in line with those observed in VN: the only evident difference was in a decreased intake of polyunsaturated fatty acids in VT when compared with O (figure 1C). The distribution of the macronutrient and micronutrient intake among the three dietary groups is represented in online supplemental figure 1B.
gutjnl-2021-325168supp002.pdf (1.3MB, pdf)
Stool miRNA profiles according to different diets
After sequencing of stool RNA samples, an average of 64 681 reads (0.83%) was aligned to known human miRNA sequences and a median number of 253, 321 and 310 stool miRNAs were detected in O, VT and VN, respectively (online supplemental table 1A). Among these miRNAs, 145 (59.43%) were represented in all the three dietary groups (online supplemental table 3A).
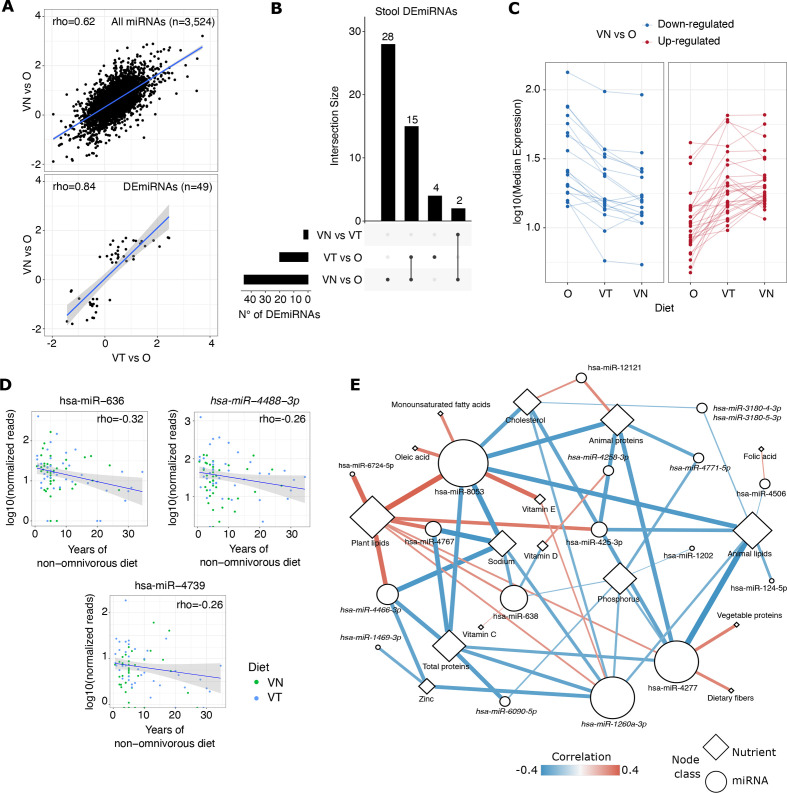
Fold changes (expressed as log2FC) of miRNA expression levels of VT and VN compared with O were significantly correlated (rho=0.62, p<0.0001), showing a coherent trend (figure 2A, upper panel).
Figure 2.
(A) Scatter plot reporting the correlation between miRNA expression log2FC resulting from the comparison between VT and O or VN and O. The correlation coefficient was computed using the Spearman method. In the upper panel are reported all miRNAs detected in the two comparisons; in the lower panel are reported only the DEmiRNAs. (B) Upset plot reporting the number of stool DEmiRNAs overlapped/specific among the three comparisons performed. (C) Line plot reporting the median expression of stool DEmiRNAs as distributed in the three dietary groups. MiRNAs were separated into downregulated and upregulated based on log2FC computed between VN and O. (D) Scatter plots reporting the normalised expression levels of stool DEmiRNAs whose expression decreased in relation with a non-omnivorous diet regime and its duration in time. (E) Network representation of the interactions between stool DEmiRNAs (circles) and nutrients (rhombus). The node size is proportional to the total node degree. Each edge represents a significant correlation (adjusted p<0.05) also supported by age-corrected, sex-corrected and BMI-corrected GLM analysis. Correlation coefficients are represented by the edge colours and width. O, omnivore; VN, vegan; VT, vegetarian
Forty-nine miRNAs were significantly differentially expressed among the three dietary groups by sex-adjusted and age-adjusted differential expression analysis (online supplemental table 3A). For these DEmiRNAs, fold changes from the comparisons of VT versus O and VN versus O were strongly correlated (rho=0.84, p<0.0001) (figure 2A, lower panel). Fifteen DEmiRNAs were differentially expressed in both VT and VN (figure 2B) and showed a coherent trend of expression: 11 upregulated and 4 downregulated in comparison with O. Considering the median expression of the 49 DEmiRNAs, a gradual change in the expression levels was observed from O to VN, with VT showing intermediate expression levels of these molecules in stool (figure 2C). A GLM adjusted for dietary group, sex, age, BMI, waist circumference, physical activity and the season of the sample collection showed that BMI, and age and season of sample collection contributed significantly in explaining the stool levels, respectively, of 11 and 8 DEmiRNAs out 49 (online supplemental table 3B). A limited number of DEmiRNAs was significantly affected by sex (one miRNA), physical activity (one miRNA) and waist circumference (three miRNAs). Correcting the DESeq2 analysis for the season of sample collection or BMI confirmed 38 and 30 DEmiRNAs out of the 49, respectively (online supplemental table 3C).
The expression levels of five DEmiRNAs significantly decreased with increasing years of non-omnivorous diet (p<0.05) (online supplemental table 3D; figure 2D). Among them, miR-636, miR-4488-3p and miR-4739 were also significantly downregulated in VT/VN in comparison with O (online supplemental figure 2A upper panel). In this respect, only the expression levels of miR-4488-3p were concomitantly inversely correlated with chronological age (online supplemental figure 2A, lower panel), strengthening the relationship between both miR-636 and miR-4739 down-regulation and the extent of a non-omnivorous dietary regime.
gutjnl-2021-325168supp003.pdf (824.8KB, pdf)
Clustering analysis of the correlation coefficients between estimated nutrient intake and the levels of all the identified stool miRNAs highlighted two main clusters composed of 30 and 8 miRNAs that were significantly correlated with nutrients enriched (eg, dietary fibres, plant lipids and vegetable proteins) or depleted (eg, animal lipids, animal proteins and sodium) in VT and VN, respectively (online supplemental figure 2B). Twenty-two miRNAs were among those identified as differentially expressed in VT/VN compared with O (online supplemental table 4A). To further explore the relationship between the nutrient estimated intake and the stool DEmiRNA levels, a GLM regression analysis adjusted for sex, age and BMI was performed revealing 56 miRNA-nutrient associations supported by both analyses (figure 2E, online supplemental table 4B). The nutrients characterised by the highest number of related DEmiRNAs were plant lipids (eight miRNAs), followed by animal lipids (seven miRNAs), and total/animal proteins and phosphorus (six miRNAs). Conversely, among all DEmiRNAs, miR-8053 levels were related to the highest number of nutrients (n=9), followed by miR-4277 and miR-1260a-3p (for both n=8). Out of 56 miRNA-nutrient associations, 28 associations involving nutrients not specific to a dietary regime like animal proteins/lipids were coherent also even considering the three dietary groups separately.
The analysis of HMDD annotations showed that 12 out of the 49 stool DEmiRNAs were characterised by precursor sequences that were related to a specific disease phenotype in different studies (n=191) (online supplemental figure 3A and online supplemental table 3E).
gutjnl-2021-325168supp004.pdf (576.2KB, pdf)
Validation in stool samples of an independent group of omnivores
The expression profiles of the 49 stool DEmiRNAs were also investigated in an independent cohort of 45 O analysed in another project (see Methods section). All DEmiRNAs were detected in stool of the validation cohort and their levels strongly correlated with those of the 40 O investigated in this study (rho=0.82, p<0.0001) (online supplemental figure 3B and online supplemental table 3F). Forty-one out of the 49 DEmiRNAs were validated as significantly differentially expressed in stool of these O subjects in comparison with the VT or VN (online supplemental figure 3C and online supplemental table 3F). Furthermore, the log2FC for these validated miRNAs were positively correlated between the two comparisons (rho=0.80 and 0.79, for VT vs O and VN vs O, respectively; p<0.0001). Among the eight non-significant miRNAs, seven were characterised by a coherent log2FC in at least one of the comparisons in both groups of Os.
Plasma miRNA profiles in relation to the diet
An average of 2 124 450 (30.70%) plasma small RNA sequencing reads were aligned to miRNA sequences (online supplemental table 1B). Plasma samples were characterised by a lower fraction of reads unmapped to human annotations (mean 2.36%) compared with stool (mean 60.75%). A median number of 382, 384 and 385 miRNAs were detected in plasma samples from O, VT and VN, respectively. Most of the miRNAs detected in plasma (n=252, 89%) were commonly detected in all three dietary groups.
Differently from miRNA profiles in stool samples, no DEmiRNAs were detected in plasma samples comparing the different dietary groups (online supplemental table 5A). Among the 49 DEmiRNAs obtained in the analysis of stool data, six (miR-181c-3p, miR-425-3p, miR-641-3p, miR-143-3p, miR-1260a-3p, miR-4508-3p) were also detected in plasma samples (online supplemental table 5B). Among them, only miR-143-3p was characterised by higher but still non-significantly different expression levels in VT compared with O and VN. None of the stool DEmiRNAs was significantly correlated with the expression levels of the same miRNA in plasma samples.
Among the other miRNAs detected in plasma samples, miR-362-3p showed the highest positive correlation comparing plasma and stool expression levels (rho=0.31, p<0.0001) while let-7a-1-3p was inversely correlated between the different biospecimens (rho=−0.34, p<0.0001) (online supplemental table 5C).
XenomiR expression levels in stool and plasma
The presence of food-derived non-human miRNAs (xenomiRs) was evaluated by analysing the fraction of human-unmapped reads. In stool samples, an average of 186 reads (0.005%) from the unmapped reads set was assigned to xenomiRs, while on plasma, the mapping rate was slightly higher (1290 reads on average, 0.87% of human-unmapped reads) (online supplemental table 1). Only two xenomiRs (gga-mir-1692 and gma-MIR6300) were detected above the expression threshold used in our analysis. Interestingly, the stool levels of gma-MIR6300, which is annotated in the soy genome (Glycine max), were significantly higher (p<0.05) in VT and VN with respect to O (online supplemental table 6). For this xenomiR, no sequence similarity was found among human miRNAs, while a homologous sequence was observed in the genome of other edible vegetables and fruits (eg, Solanum lycopersicum, Solanum tuberosum and Cucumis melo) (online supplemental table 6). Conversely, in plasma samples, 11 xenomiRs were detected, with chi-mir-30d (annotated to Capra hircus genome) characterised by the highest number of reads. All the 11 miRNAs were annotated to an animal species (C. hircus, Gallus gallus, Sus scrofa, Salmo salar or Gadus morhua). However, given the high homology of these miRNAs with their homologous sequence in humans, a real circulating level of these molecules is less reliable. The observed reads could be actually related to the sequencing of portions of the corresponding miRNA precursor in humans.
Gut microbiome composition and functions and relation to faecal miRNAs
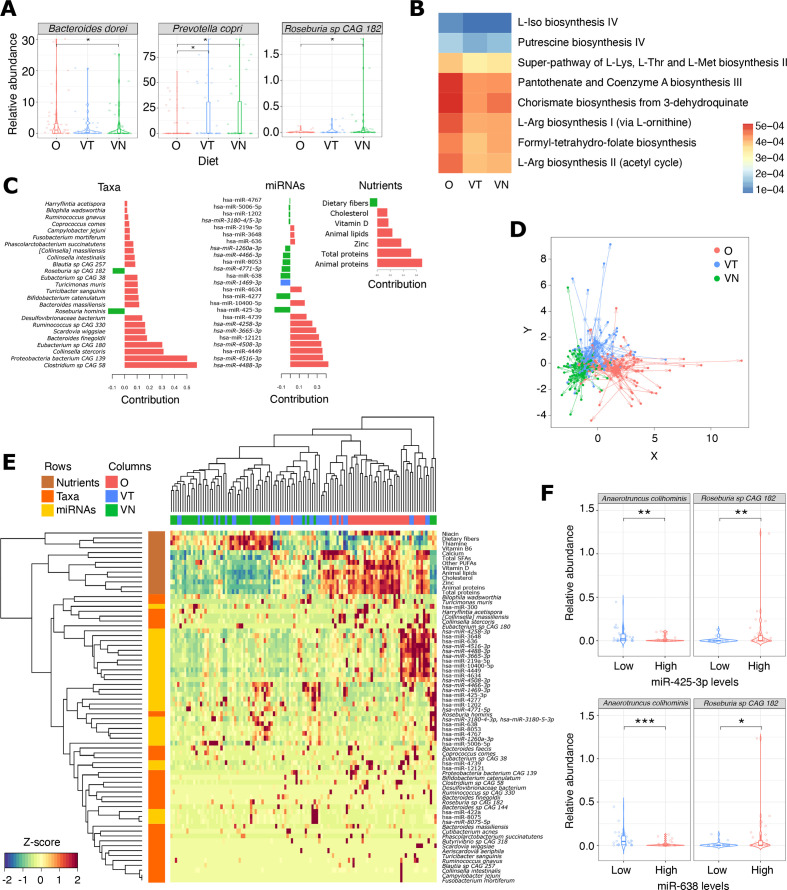
Gut microbiome taxonomic composition varied according to the different dietary patterns. Among differently abundant taxa, Prevotella copri and Roseburia sp. CAG 182 showed higher abundance in VT/VN compared with O, while Bacteroides dorei (currently Phocaeicola dorei) prevailed in O (p<0.05; figure 3A).
Figure 3.
(A) Box plots showing the relative abundance of three bacterial species differentially prevalent among the dietary groups. *Wilcoxon rank-sum p<0.05. (B) Heat map reporting the prevalence of metabolic pathways differentially represented among metagenomic profiles of the dietary groups. (C) Bar plot showing the loading weights of each DIABLO-selected variable on each component. The colour indicates the dietary group in which the variable has the maximum level. (D) Co-inertia analysis quantifying the co-variability between the three datasets (taxonomic composition, miRNA, dietary nutrients). Shapes represent the projected coordinates of each subject. The centroid for a given sample between all datasets is indicated by the start of the arrow and the location of the same sample in each dataset by the tips of the arrows. The length of the arrow is proportional to the divergence between data from different blocks. (E) Heat map showing the result of the hierarchical clustering analysis of the subjects based on discriminant variables identified by DIABLO. (F) Box plots reporting the relative abundances of two bacterial species whose levels are significantly different in stool of subjects characterised by higher or lower expression of both miR-425-3p (top) and miR-638 (bottom). O, omnivore; VN, vegan; VT, vegetarian.
A functional profiling of the gut metagenomes was performed with HUMAnN3. Different gut microbiome composition is reflected in differential potential activities. The microbiome of O showed increased abundance of several metabolic pathways, including pathways related to the biosynthesis of amino acids, biogenic amines or their precursors (L-isoleucine biosynthesis IV, L-arginine biosynthesis I and II, super-pathway of L-lysine, L-threonine and L-methionine biosynthesis II, putrescine biosynthesis IV, chorismate biosynthesis from 3-dehydroquinate), as well to the biosynthesis of vitamins (formyl-tetrahydrofolate biosynthesis, pantothenate and coenzyme A biosynthesis III) (FDR<0.05; figure 3B).
Clustering analysis of the correlation coefficient between bacterial abundance and stool miRNA expression levels resulted in less homogeneous clusters (online supplemental figure 2C). Despite two main clusters of bacterial species and miRNAs were identified, correlation coefficients were significant only on specific bacteria-miRNA pairs. The bacterial species characterised by the highest number of correlated miRNAs (p<0.05) were Ruminococcaceae bacterium D5 (11 correlated miRNAs), P. copri (n=8) and Phascolarctobacterium succinatutens (n=7). Among the miRNAs correlated with bacterial species, 10 were significantly differentially expressed in VT/VN compared with O (online supplemental table 4A).
Integration of miRNAs, microbiome and diet
The DIABLO model identified a set of 25 DEmiRNAs, 25 taxa and 7 dietary nutrients able, all together, to discriminate the three groups (figure 3C). As for stool DEmiRNAs, a GLM adjusted for age, sex, BMI, physical activity, waist circumference and season of sample collection was applied to evaluate the relationship between taxa and subject covariates. Only five species were associated with a significant contribution of at least one of these variables (online supplemental table 3B). Partial least squares discriminant analysis and the hierarchical clustering showed a separation of subjects according to their diet (figure 3D, E). To assess the robustness of the data reported in this study in discriminating the different dietary groups, a machine learning analysis using the combination of these features was applied. The analysis confirmed that the identified features were able to discriminate efficiently the different dietary groups (average AUC=0.89). As expected, higher accuracy was observed in classifying omnivorous (AUC=0.95) and VN (AUC=0.94) individuals than VT subjects (AUC=0.79).
Two DEmiRNAs, miR-425-3p and miR-638, attracted our interest since they were associated with a coherent higher expression in VT/VN compared with both the analysed O groups (among those not affected by BMI), they were also identified by DIABLO as discriminant features for the different dietary regimes, and they were significantly correlated with nutrients (figure 2E). Therefore, these DEmiRNAs were further investigated for their relationship with gut microbiome composition. Subjects within the highest quartile of miR-425-3p expression levels (regardless of the type of diet) showed a significantly higher abundance of Akkermansia muciniphila, Roseburia hominis, Roseburia sp. CAG 182, Oscillibacter sp. 57_20, Oscillibacter sp. CAG 241 and R. bacterium D5 compared with those in the lowest quartile (figure 3F and online supplemental figure 4), while Anaerotruncus colihominis and Flavonifractor plautii were more abundant in subjects with the lowest levels of the same miRNA. Consistently, higher levels of Roseburia sp. CAG 182 and R. bacterium D5, as well as a lower abundance of A. colihominis were found in subjects within the highest expression levels of miR-638.
gutjnl-2021-325168supp005.pdf (329.5KB, pdf)
Enrichment analysis
The miRNA target enrichment analysis was performed to explore whether independent sets of relevant stool miRNAs identified in this study (ie, all identified DEmiRNAs either upregulated or downregulated, miRNAs identified by DIABLO and miRNAs correlated with bacterial species or nutrient intake) were predicted to regulate common biological processes. A total of 927 functional terms were significantly enriched (adj. p<0.05) in target genes from each set of miRNAs considered (online supplemental table 7), with 68 terms enriched in at least three miRNA sets concomitantly (figure 4A). Out of them, 66 were enriched in miRNAs upregulated in VN/VT, suggesting an inhibition of the related process (negative coefficients in figure 4A). In particular, the terms that were redundantly enriched in the highest number of miRNA sets (five sets) were KEGG_One_carbon_pool_by_folate and GO_Regulation_of_gluconeogenesis_by_regulation_of_transcription_from_rna_polymerase_ii_promoter. Specifically, for KEGG metabolic-related terms, One_carbon_pool_by_folate and Glyoxylate_and_dicarboxylate_metabolism were the most represented in the analysis (figure 4B). These terms were enriched in the stool DEmiRNAs upregulated in VN/VT but also in miRNAs identified by DIABLO and those correlated with higher intake of nutrients observed in VN and VT. Finally, Arachidonic_acid_metabolism was the sole KEGG metabolic-related term enriched for DEmiRNAs downregulated in VN/VT.
Figure 4.
(A) Dot plot showing the statistical significance of the functional terms identified as enriched in the target of different groups of stool miRNAs differentially expressed among the dietary groups (DEmiRNAs), identified by DIABLO, correlated with nutrient groups or correlated with a group of bacterial species. The size of the dot is proportional to the significance while the colour code refers to the coefficient computed by RBiomirGS. Negative coefficients are related to process predicted to be downregulated based on the miRNA expression change. Only terms enriched in at least two miRNA groups are reported. (B) Table reporting the KEGG metabolic terms enriched in the target of different miRNA groups. The colour-coded bars represent the coefficient computed by RBiomirGS. Positive (red) or negative (blue) coefficients predicted based on the miRNA expression change.
Analysis of stool DEmiRNA expression and epigenetic state in intestinal tissue samples
Finally, we evaluated the expression levels of stool DEmiRNAs in intestinal tissues using the data from the TissueAtlas.39 The analysis confirmed that 19 DEmiRNAs out of 22 were detectable also in the intestine (online supplemental table 8A). In particular, miR-143-3p was the DEmiRNA characterised by the highest expression levels in colon samples (average Z-score=2.40), while others like miR-638, miR-425-3p and miR-636 were detected in colon/small intestine as well as in different tissue samples of the TissueAtlas repository.
Furthermore, since the epigenetic status of a gene locus can represent a reliable proxy of an active/repressive transcriptional state, the epigenetic state of DEmiRNA coding genes was evaluated in the Roadmap Epigenomics Project.40 The analysis confirmed a transcriptionally active epigenetic state in 41 out of the 49 stool DEmiRNAs in at least one of the sample types analysed (online supplemental table 8B). Specifically, colon mucosa was associated with the highest number of epigenetically active DEmiRNA loci (n=26), followed by duodenum mucosa (n=23), small intestine and colon smooth muscle tissues (n=18). The analysis confirmed, in colon smooth muscle samples, a chromatin state related to a strong transcription at miR-143 coding locus coherently with previous observations.41
Discussion
In the present study, we explored the faecal miRNome of 120 healthy subjects adhering to a VN, VT or omnivorous diet, reporting that miRNA expression levels are affected by diet, with a possible relationship with the gut microbiota composition and function. Forty-nine miRNAs were significantly differentially expressed among the three dietary groups and 15 of them showed a consistent trend of expression of upregulation or downregulation in both VT and VN in comparison with two independent groups of O. These findings suggest a gradual adaptation of the miRNome in response to a specific dietary regimen, probably as a consequence of the epigenetic and metagenomic changes occurring in the gut, which are reflected in stool. Studies on human and mouse models have already demonstrated that long-term dietary changes drive stable modifications in the host epigenome and gut microbiota.42 43 In this respect, in our study, two stool DEmiRNAs (miR-636 and miR-4739) showed a clear downregulation with the duration of the non-omnivorous diet, independently of VT/VN subject’s age. MiR-636, an intragenic miRNA coded from the SRSF2 locus, has been found downregulated in blood/serum and adipose tissue of obese subjects44 and, on the contrary, upregulated in young subjects with obesity and diabetic nephropathy.45 46 In general, this miRNA seems to be implicated in weight loss. Garcia-Lacarte and colleagues,47 for example, found miR-636 hypomethylated and overexpressed in patients with metabolic syndrome who were highly responsive to the weight loss regimen. Presumably, the downregulation of this miRNA observed in VT and VN may be associated with a low-fat diet, ultimately more protective to the risk of obesity and diabetes.
MiR-4739 has been reported as a hub in the regulatory network mediated by the long non-coding RNA NEAT1.48 NEAT1 works as an miRNA sponge and represses the proliferation and fibrosis in diabetic nephropathy.49 Therefore, the action of NEAT1 as a natural downregulator of miR-4739 may be imitated by a VT or VN diet. Moreover, miR-4739 has been associated with the induction of adipogenic differentiation of human bone marrow stromal cells by targeting low-density lipoprotein receptor-related protein 3 (LRP3).50 Several genes related to metabolism, including SHMT1 and MKNK2, have been annotated as having the highest number of validated interactions with miR-636 and miR-4739. SHMT1 codes for serine hydroxymethyltransferase 1, a key protein involved in one-carbon metabolism.51 Conversely, MKNK2, encoding for MAPK interacting serine/threonine kinase 2, was recently associated with the lipid metabolism in adipocytes.52 Despite the fact that more data are needed to support an miRNA expression modulation on a long-term plant-based diet, our data highlight that in stool it is possible to detect miRNAs modulating metabolic processes, such as those of lipids, largely affected by different dietary regimes.
Some of the other stool DEmiRNAs found in this study are dysregulated in different diseases, as retrieved from HMDD, a database of experimentally supported human miRNA disease associations. MiR-143 was the DEmiRNA associated with the highest number of annotations (101 studies), followed by miR-124 (44 studies), miR-425 (11 studies) and miR-638 (10 studies). Interestingly, in our study, all these miRNAs were upregulated in stool of both VT/VN when compared with O. MiR-143 is an evolutionarily conserved miRNA transcribed as bicistronic RNA in a cluster with miR-145. MiR-143/miR-145 are well-known tumour suppressor miRNAs dysregulated in several cancer types, including CRC.53 Consistently, low levels of miR-143-3p have been observed in stool of patients with CRC11 12 and their upregulation in stool of VT/VN supports the lower cancer risk observed in relation to these dietary habits.54 55 These two miRNAs target some fundamental tight junction proteins.56 Nonetheless, miR-143-3p is a regulator of adipocyte differentiation57 and decreased levels of this miRNA were observed in obese individuals compared with normal weight controls in two independent studies.58 59 MiR-143/miR-145 cluster has an essential role in intestinal epithelium regeneration by modulating the insulin growth factor signalling pathway.60 Interestingly, in a mouse model of colitis, the administration of two probiotics, Lactobacillus fermentum and L. salivarius, ameliorated colonic damage and, concomitantly, increased miR-143 expression in colonic tissue.61 Conversely, miR-124 is active in colon tissues and associated with the modulation of an inflammatory response by regulating the signal transducer and activator of transcription 3 and acetylcholinesterase.62 63 MiR-124-5p is frequently downregulated in CRC tissues.64 In stool of VT/VN, increased expression levels of such miRNAs regulating the inflammatory response and the maintenance of gut epithelium are coherent with the known anti-inflammatory effects of plant-based diets.65
In the past decade, metagenomics has deeply increased our knowledge of the composition and function of the gut microbiome and how it is shaped by external factors.66 However, to the best of our knowledge, only a few studies in humans have explored the role of host miRNAs as mediators or interactors of the gut microbiome.24 In 2019, Virtue and colleagues67 have shown that miR-181 is overexpressed in white adipocytes of obese mice and knockout mice were less susceptible to a high-fat diet. In the same study, the authors showed that the expression of this miRNA is influenced by variations in the mouse gut microbiota. MiR-181 was also observed upregulated in the postprandial period of a high-saturated fat meal in peripheral blood mononuclear cells of healthy individuals,68 supporting a link of this miRNA with lipid metabolism. Interestingly, in our study, miR-181c-3p stool levels progressively increased going from O to VN and were inversely correlated with the estimated animal lipid intake (Spearman rho=−0.22). A differential expression of various lipid-related miRNAs in stool of VT/VN suggests that the change in dietary lipid intake may drive effective epigenetic changes in the gut, as previously proposed.69 In low-lipid dietary intervention studies, the stool levels of these miRNAs could represent a valid non-invasive parameter to assess the patient adherence and response to the diet.
In the present study, the gut microbiome taxonomic composition varied according to the dietary patterns, with P. copri and Roseburia sp. CAG 182, having a higher abundance in VT/VN than O. Interestingly, A. muciniphila abundance was linked to both diet and miRNAs, in agreement with recent observations.24 A. muciniphila is a well-known beneficial microbe involved in mucin metabolism whose levels are decreased in several diseases, including ulcerative colitis and metabolic disorders.70 Previous evidence reported that human miRNAs could directly target genes of these bacteria. Specifically, Liu and colleagues71 showed that the oral administration of human faecal miR-30c in a mouse model of multiple sclerosis ameliorated the disease symptoms by directly targeting A. muciniphila β-galactosidase coding-gene, which in turn increased the bacterial abundance in the gut. Despite miR-30c-5p was associated with low stool levels in our study population, it was significantly more abundant in stool of VT/VN.
Of note, higher stool levels of miR-425-3p, an miRNA upregulated in VT/VN, were significantly related to the abundance of A. muciniphila in our study. This miRNA was observed as significantly downregulated in serum of hyperlipidemic subjects in comparison with healthy controls.72 Furthermore, the other mature miRNA derived from miR-425 locus, miR-425-5p, was shown to be able to respond to a lipid challenge in mice small intestine and to inhibit 3-hydroxy-3-methylglutaryl-CoA synthase coding (HMGCS2), a key enzyme of lipid metabolism.73 Finally, miR-425-5p overexpression in adipocytes was previously related to the regulation of lipogenesis and lipolysis.74 This data further strengthens our hypothesis on the role of the different dietary intake of lipids in VT/VN diets as a driver of consistent miRNA modulation in the gut. Higher stool levels of miR-425-3p and miR-638 were also significantly correlated with Roseburia and R. bacterium D5, which in our cohort were significantly more abundant in VT/VN, coherently with previous observation.18 75 This data could be a further evidence of the important host-microbial interactions mediated by diet and epigenetic factors such as miRNAs. In this sense, our results on miRNAs could reflect the gut microbiota shifts in response to dietary fat recently reviewed by Chadaideh and Carmody.76
In the present study, among the three investigated groups, the analysis of the dietary information recorded in the EPIC questionnaires confirmed the expected differences in weekly food consumption and estimated daily nutrient intake. Nutrients and specific diet components have been implicated in the modulation of miRNA expression in in vitro/in vivo models and, as a consequence, in all miRNA-mediated molecular mechanisms.13 In this study, vegetable lipids were the nutrients associated with the highest number of stool DEmiRNAs, together with animal lipids (namely miR-425-3p, miR-638, miR-8063 and miR-4277), but the latter showed an opposite trend. This result is consistent with previous evidence on the activity of lipids in modulating miRNA expression in different tissues, including gut,77 adipocytes,78 and plasma.79
A functional enrichment analysis was performed to identify the candidate downstream signalling and metabolic pathways that may be affected by an intestinal miRNA expression modulation, which is reflected in stool samples. This analysis confirmed a relationship between the identified stool DEmiRNAs and nutrient metabolism. Terms related to fatty acid metabolism (namely, Glyoxilate_and_dicarboxylate_metabolism and Phosphatidylglycerol_metabolic_process) and those related to folate metabolism (eg, One carbon pool by folate, Metabolism of folate and pterines and Tetrahydrofolate metabolic process) were frequently observed among the targets of miRNAs upregulated in stool of VT/VN. In particular, the gene MTHFD2 coding for the mitochondrial bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase was predicted as a target of two stool miRNAs upregulated in both VT/VN (miR-425-3p, miR-4277) (online supplemental table 7). The estimated dietary intake of folic acid was also correlated with the stool levels of these miRNAs in our analysis. The activity of miRNAs in regulating folate metabolism has been previously observed by others.80 We can hypothesise that miR-425-3p and miR-4277 may form a negative feedback loop on the increasing level of this nutrient in VT/VN. Interestingly, miR-425-3p was included in the list of miRNAs regulated by the activity of nuclear factor erythroid 2-related factor 2 (Nrf2),81 a transcription factor involved in the regulation of metabolism downstream of nutrient-sensing pathways, whose activity is pivotal for the gastrointestinal tract development and maintenance.82
MiRNAs are present in all eukaryotes: an intriguing aspect is that they could potentially ‘traverse’ from plants to animals through food via the gastrointestinal tract and access host cellular targets, where they could work as bioactive compounds and influence recipients’ physio-pathological conditions. However, this cross-kingdom interaction is still a topic of debate in the scientific community due to the lack of available data.83 Although some xenomiRs were detected in the present study, their low abundance in our data suggests a non-functional role of these molecules. Taking into consideration that xenomiR sequences may often be very similar to those of human miRNAs and the absence of evidence on the capacity of xenomiRs to resist food processing thus impedes a definitive investigation.
This study is the first to integrate stool miRNAs with gut microbiota profiles and estimated daily nutrient intake from dietary questionnaires. Despite significant differences in BMI in the study population, this covariate only narrowly influenced our results. Most of the stool identified DEmiRNAs, including miR-425-3p, miR-638, miR-636, miR-143-3p, miR-181c-3p and miR-124-5p, remained significantly differentially expressed after adjustment for this variable. Similarly, although individuals have different dietary habits over the course of the year, correcting the analysis by time of sample collection only marginally affected the significances of the identified DEmiRNAs.
We also investigated miRNA profiles in plasma samples from the same subjects. However, in this biofluid, no significant DEmiRNAs were found according to the different dietary habits. It is possible that stool may reflect epigenetic changes occurring in the intestinal cells as a consequence of a stable dietary habit, while plasma samples could better reflect miRNAs characterising transient cellular response to nutrition-related stimuli, including circulating nutrients, hormones and microbial metabolites. This is consistent with previous observations in all the three dietary groups between the estimated intake of particular nutrients and a set of plasma miRNAs.14 A rising number of available data support the modification of faecal miRNA levels as a proxy of alteration and response occurring in the gut epithelium.84 Indeed, the main source of faecal miRNAs is represented by intestinal epithelial cells, as described by Liu et al.23 Although, in the present study, we did not assess the cell of origins of the detected miRNAs, the analysis of the TissueAtlas and Roadmap Epigenomics data confirmed that most of the stool DEmiRNAs identified in the present study are expressed and active in the gut. Interestingly, the analysis also showed a chromatin state related to a strong transcription at miR-143 coding locus coherently with previous observations.41 Then, we cannot exclude the contribution of other cellular components of the gastrointestinal systems, including those from the stromal and immune compartments. This is particularly relevant when trying to understand the host-microbiome crosstalk within the complex intestinal microenvironment. However, novel co-culturing systems and in vitro models reproducing the gut system could be applied based on evidence of miRNA and bacteria dysregulation observed in faecal samples to improve our understanding of the molecular characteristics of this miRNA-mediated host-microbial interaction.85
In conclusion, we hereby present the first large-scale characterisation of faecal miRNAs in relation to different diets and to the gut microbial population with the state of the art of high-throughput technologies. We show a gradual modulation of the human miRNome as a consequence of different dietary regimens with important modifications in miRNAs involved in lipid and folate metabolism. Our data also provides novel evidence to improve our understanding of the role of small RNAs in the host-microbial crosstalk in physiological and pathological conditions. An integrated analysis of faecal miRNA and gut microbiome signatures might provide insights for designing more accurate personalised dietary patterns and recommendations aimed at the prevention/treatment of chronic diseases. In particular, in monitoring the adherence to specific dietary regimes, where the intake of nutrients should be carefully checked, a translational approach of a non-invasive stool miRNA profiling may be foreseen.
Acknowledgments
The authors are very grateful to all volunteers who participated with enthusiasm in the study. The authors are also thankful to Dr Vladimir Benes (Genecore Facility at EMBL, Heidelberg, Germany) for his kind collaboration and advice and to Mr Elton Jarvis Herman for his precious revision of the language of the text. The artwork for this study has been created with BioRender.com.
Footnotes
Twitter: @nsegata, @rupensa, @BarbaraPardini1
BP, DE and AN contributed equally.
Contributors: BP, ST and AN contributed to concept and design; BP, GF, RGP, SG, ST, VP and AN to collection and assembly of the data; BP, GF, ST, AF, FC and AN to small RNA-Seq data analyses and interpretation; DE, FDF, EP and NS to metagenomic data analyses and interpretation; ST, BP, GF, FC, DE, FDF and AN to manuscript writing; all authors gave final approval of the version.
Funding: This work was supported by Fondazione Umberto Veronesi postdoctoral fellowships 2017 and 2018 (recipients, ST and BP), Compagnia di San Paolo Torino, Italy (to BP, AN, ST), Lega Italiana per La Lotta contro i Tumori (to FC, BP and AN), Fondazione CRT (grant number 2019-0450 to RGP) the European H2020 research project Oncobiome (Grant number 825410), and the COST action TRANSCOLONCAN (CA17118). DE laboratory received funding from the JPI HDHL-INTIMIC - Knowledge Platform of Food, Diet, Intestinal Microbiomics and Human Health (ID 790) grant and the PRIN2017 (20174FHBWR_005) grant by the Italian Ministry of University and Research.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. Raw data are available upon request to the corresponding author.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Azienda Ospedaliera “SS. Antonio e Biagio e C. Arrigo” of Alessandria. ID: Colorectal_miRNA CEC2014.
References
- 1. Katz DL, Meller S. Can we say what diet is best for health? Annu Rev Public Health 2014;35:83–103. 10.1146/annurev-publhealth-032013-182351 [DOI] [PubMed] [Google Scholar]
- 2. Saliba W, Rennert HS, Gronich N, et al. Red meat and processed meat intake and risk of colorectal cancer: a population-based case-control study. Eur J Cancer Prev 2019;28:287–93. 10.1097/CEJ.0000000000000451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hullings AG, Sinha R, Liao LM, et al. Whole grain and dietary fiber intake and risk of colorectal cancer in the NIH-AARP diet and health study cohort. Am J Clin Nutr 2020;112:603–12. 10.1093/ajcn/nqaa161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dinu M, Abbate R, Gensini GF, et al. Vegetarian, vegan diets and multiple health outcomes: a systematic review with meta-analysis of observational studies. Crit Rev Food Sci Nutr 2017;57:3640–9. 10.1080/10408398.2016.1138447 [DOI] [PubMed] [Google Scholar]
- 5. Serrano JCE, Jove M, Gonzalo H, et al. Nutridynamics: mechanism(s) of action of bioactive compounds and their effects. Int J Food Sci Nutr 2015;66 Suppl 1:S22–30. 10.3109/09637486.2015.1035231 [DOI] [PubMed] [Google Scholar]
- 6. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther 2016;1:15004. 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharma S, Lu H-C. microRNAs in neurodegeneration: current findings and potential impacts. J Alzheimers Dis Parkinsonism 2018;8:420. 10.4172/2161-0460.1000420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gavrilas LI, Ionescu C, Tudoran O, et al. The role of bioactive dietary components in modulating miRNA expression in colorectal cancer. Nutrients 2016;8:590. 10.3390/nu8100590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tarallo S, Pardini B, Mancuso G, et al. Microrna expression in relation to different dietary habits: a comparison in stool and plasma samples. Mutagenesis 2014;29:385–91. 10.1093/mutage/geu028 [DOI] [PubMed] [Google Scholar]
- 10. Rashid H, Hossain B, Siddiqua T, et al. Fecal microRNAs as potential biomarkers for screening and diagnosis of intestinal diseases. Front Mol Biosci 2020;7:181. 10.3389/fmolb.2020.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Francavilla A, Turoczi S, Tarallo S, et al. Exosomal microRNAs and other non-coding RNAs as colorectal cancer biomarkers: a review. Mutagenesis 2020;35:243–60. 10.1093/mutage/gez038 [DOI] [PubMed] [Google Scholar]
- 12. Tarallo S, Ferrero G, Gallo G, et al. Altered fecal small RNA profiles in colorectal cancer reflect gut microbiome composition in stool samples. mSystems 2019;4:e00289–19. 10.1128/mSystems.00289-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. García-Segura L, Pérez-Andrade M, Miranda-Ríos J. The emerging role of microRNAs in the regulation of gene expression by nutrients. J Nutrigenet Nutrigenomics 2013;6:16–31. 10.1159/000345826 [DOI] [PubMed] [Google Scholar]
- 14. Ferrero G, Carpi S, Polini B, et al. Intake of natural compounds and circulating microRNA expression levels: their relationship investigated in healthy subjects with different dietary habits. Front Pharmacol 2020;11:619200. 10.3389/fphar.2020.619200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Angelis M, Ferrocino I, Calabrese FM, et al. Diet influences the functions of the human intestinal microbiome. Sci Rep 2020;10:4247. 10.1038/s41598-020-61192-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu GD, Compher C, Chen EZ, et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 2016;65:63–72. 10.1136/gutjnl-2014-308209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Filippis F, Pellegrini N, Vannini L, et al. High-Level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016;65:1812–21. 10.1136/gutjnl-2015-309957 [DOI] [PubMed] [Google Scholar]
- 19. Meslier V, Laiola M, Roager HM, et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020;69:1258–68. 10.1136/gutjnl-2019-320438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gross M. Does the gut microbiome hold clues to obesity and diabetes? Curr Biol 2013;23:R359–62. 10.1016/j.cub.2013.04.047 [DOI] [PubMed] [Google Scholar]
- 21. Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 2009;15:1183–9. 10.1002/ibd.20903 [DOI] [PubMed] [Google Scholar]
- 22. Thomas AM, Manghi P, Asnicar F, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 2019;25:667–78. 10.1038/s41591-019-0405-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu S, da Cunha AP, Rezende RM, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 2016;19:32–43. 10.1016/j.chom.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malmuthuge N, Guan LL. Noncoding RNAs: regulatory molecules of Host-microbiome crosstalk. Trends Microbiol 2021;29:713–24. 10.1016/j.tim.2020.12.003 [DOI] [PubMed] [Google Scholar]
- 25. Riboli E, Hunt KJ, Slimani N, et al. European prospective investigation into cancer and nutrition (EPIC): study populations and data collection. Public Health Nutr 2002;5:1113–24. 10.1079/PHN2002394 [DOI] [PubMed] [Google Scholar]
- 26. Salvini SPM, Gnagnarella P, Maisonneuve P, et al. Banca Dati di Composizione degli Alimenti per Studi Epidemiologici in Italia, 1998. [Google Scholar]
- 27. Ferrero G, Cordero F, Tarallo S, et al. Small non-coding RNA profiling in human biofluids and surrogate tissues from healthy individuals: description of the diverse and most represented species. Oncotarget 2018;9:3097–111. 10.18632/oncotarget.23203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sabo AA, Birolo G, Naccarati A, et al. Small non-coding RNA profiling in plasma extracellular vesicles of bladder cancer patients by next-generation sequencing: expression levels of miR-126-3p and piR-5936 increase with higher histologic grades. Cancers 2020;12. 10.3390/cancers12061507. [Epub ahead of print: 09 06 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47:D155–62. 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Z, Liu L, Gao Y, et al. Benchmark of computational methods for predicting microRNA-disease associations. Genome Biol 2019;20:202. 10.1186/s13059-019-1811-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007;449:804–10. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of illumina second-generation sequencing data. BMC Bioinformatics 2010;11:485. 10.1186/1471-2105-11-485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 2015;12:902–3. 10.1038/nmeth.3589 [DOI] [PubMed] [Google Scholar]
- 36. Franzosa EA, McIver LJ, Rahnavard G, et al. Species-Level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018;15:962–8. 10.1038/s41592-018-0176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh A, Shannon CP, Gautier B, et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 2019;35:3055–62. 10.1093/bioinformatics/bty1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J, Storey KB. RBiomirGS: an all-in-one miRNA gene set analysis solution featuring target mRNA mapping and expression profile integration. PeerJ 2018;6:e4262. 10.7717/peerj.4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res 2016;44:3865–77. 10.1093/nar/gkw116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kent OA, McCall MN, Cornish TC, et al. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res 2014;42:7528–38. 10.1093/nar/gku461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krautkramer KA, Kreznar JH, Romano KA, et al. Diet-Microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell 2016;64:982–92. 10.1016/j.molcel.2016.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dai Z, Ramesh V, Locasale JW. The evolving metabolic landscape of chromatin biology and epigenetics. Nat Rev Genet 2020;21:737–53. 10.1038/s41576-020-0270-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim NH, Ahn J, Choi YM, et al. Differential circulating and visceral fat microRNA expression of non-obese and obese subjects. Clin Nutr 2020;39:910–6. 10.1016/j.clnu.2019.03.033 [DOI] [PubMed] [Google Scholar]
- 45. Wang R, Hong J, Cao Y, et al. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur J Endocrinol 2015;172:291–300. 10.1530/EJE-14-0867 [DOI] [PubMed] [Google Scholar]
- 46. Wang L-P, Gao Y-Z, Song B, et al. Micrornas in the progress of diabetic nephropathy: a systematic review and meta-analysis. Evid Based Complement Alternat Med 2019;2019:3513179. 10.1155/2019/3513179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garcia-Lacarte M, Martinez JA, Zulet MA, et al. Implication of miR-612 and miR-1976 in the regulation of TP53 and CD40 and their relationship in the response to specific weight-loss diets. PLoS One 2018;13:e0201217. 10.1371/journal.pone.0201217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang Z, Winata WA, Zhang K, et al. Reconstruction of a lncRNA-Associated ceRNA network in endothelial cells under circumferential stress. Cardiol Res Pract 2020;2020:1481937. 10.1155/2020/1481937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang S, Xu Y, Ge X, et al. Long noncoding RNA NEAT1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating Akt/mTOR signaling pathway. J Cell Physiol 2019;234:11200–7. 10.1002/jcp.27770 [DOI] [PubMed] [Google Scholar]
- 50. Elsafadi M, Manikandan M, Alajez NM, et al. MicroRNA-4739 regulates osteogenic and adipocytic differentiation of immortalized human bone marrow stromal cells via targeting LRP3. Stem Cell Res 2017;20:94–104. 10.1016/j.scr.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 51. Ducker GS, Rabinowitz JD. One-Carbon metabolism in health and disease. Cell Metab 2017;25:27–42. 10.1016/j.cmet.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Merrett JE, Xie J, Psaltis PJ, et al. Mapk-Interacting kinase 2 (Mnk2) regulates adipocyte metabolism independently of its catalytic activity. Biochem J 2020;477:2735–54. 10.1042/BCJ20200433 [DOI] [PubMed] [Google Scholar]
- 53. Poli V, Seclì L, Avalle L. The microRNA-143/145 cluster in tumors: a matter of where and when. Cancers 2020;12:708. 10.3390/cancers12030708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Craig WJ. Health effects of vegan diets. Am J Clin Nutr 2009;89:1627S–33. 10.3945/ajcn.2009.26736N [DOI] [PubMed] [Google Scholar]
- 55. Appleby PN, Key TJ. The long-term health of vegetarians and vegans. Proc Nutr Soc 2016;75:287–93. 10.1017/S0029665115004334 [DOI] [PubMed] [Google Scholar]
- 56. Bi K, Zhang X, Chen W, et al. Micrornas regulate intestinal immunity and gut microbiota for gastrointestinal health: a comprehensive review. Genes 2020;11:1075. 10.3390/genes11091075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Esau C, Kang X, Peralta E, et al. Microrna-143 regulates adipocyte differentiation. J Biol Chem 2004;279:52361–5. 10.1074/jbc.C400438200 [DOI] [PubMed] [Google Scholar]
- 58. Can U, Buyukinan M, Yerlikaya FH. The investigation of circulating microRNAs associated with lipid metabolism in childhood obesity. Pediatr Obes 2016;11:228–34. 10.1111/ijpo.12050 [DOI] [PubMed] [Google Scholar]
- 59. Kilic ID, Dodurga Y, Uludag B, et al. Microrna -143 and -223 in obesity. Gene 2015;560:140–2. 10.1016/j.gene.2015.01.048 [DOI] [PubMed] [Google Scholar]
- 60. Chivukula RR, Shi G, Acharya A, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell 2014;157:1104–16. 10.1016/j.cell.2014.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rodríguez-Nogales A, Algieri F, Garrido-Mesa J, et al. Differential intestinal anti-inflammatory effects of Lactobacillus fermentum and Lactobacillus salivarius in DSS mouse colitis: impact on microRNAs expression and microbiota composition. Mol Nutr Food Res 2017;61:1700144. 10.1002/mnfr.201700144 [DOI] [PubMed] [Google Scholar]
- 62. Koukos G, Polytarchou C, Kaplan JL, et al. Microrna-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology 2013;145:842–52. 10.1053/j.gastro.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xiao Y-T, Wang J, Lu W, et al. Downregulated expression of microRNA-124 in pediatric intestinal failure patients modulates macrophages activation by inhibiting STAT3 and AChE. Cell Death Dis 2016;7:e2521. 10.1038/cddis.2016.426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xie X-Q, Wang M-J, Li Y, et al. miR-124 intensified oxaliplatin-based chemotherapy by targeting CAPN2 in colorectal cancer. Mol Ther Oncolytics 2020;17:320–31. 10.1016/j.omto.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Medawar E, Huhn S, Villringer A, et al. The effects of plant-based diets on the body and the brain: a systematic review. Transl Psychiatry 2019;9:226. 10.1038/s41398-019-0552-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scholz M, Ward DV, Pasolli E, et al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods 2016;13:435–8. 10.1038/nmeth.3802 [DOI] [PubMed] [Google Scholar]
- 67. Virtue AT, McCright SJ, Wright JM, et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci Transl Med 2019;11:eaav1892. 10.1126/scitranslmed.aav1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lopez S, Bermudez B, Montserrat-de la Paz S, et al. A microRNA expression signature of the postprandial state in response to a high-saturated-fat challenge. J Nutr Biochem 2018;57:45–55. 10.1016/j.jnutbio.2018.03.010 [DOI] [PubMed] [Google Scholar]
- 69. González-Becerra K, Ramos-Lopez O, Barrón-Cabrera E, et al. Fatty acids, epigenetic mechanisms and chronic diseases: a systematic review. Lipids Health Dis 2019;18:178. 10.1186/s12944-019-1120-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Macchione IG, Lopetuso LR, Ianiro G, et al. Akkermansia muciniphila: key player in metabolic and gastrointestinal disorders. Eur Rev Med Pharmacol Sci 2019;23:8075–83. 10.26355/eurrev_201909_19024 [DOI] [PubMed] [Google Scholar]
- 71. Liu S, Rezende RM, Moreira TG, et al. Oral administration of miR-30d from feces of MS patients suppresses MS-like symptoms in mice by expanding Akkermansia muciniphila. Cell Host Microbe 2019;26:779–94. 10.1016/j.chom.2019.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu S, Sagiv O, Rubin ML, et al. Validation study of the AJCC cancer staging manual, eighth edition, staging system for eyelid and periocular squamous cell carcinoma. JAMA Ophthalmol 2019;137:537–42. 10.1001/jamaophthalmol.2019.0238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ruiz-Roso MB, Gil-Zamorano J, López de Las Hazas MC, et al. Intestinal lipid metabolism genes regulated by miRNAs. Front Genet 2020;11:707. 10.3389/fgene.2020.00707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qi R, Wang J, Wang Q, et al. MicroRNA-425 controls lipogenesis and lipolysis in adipocytes. Biochim Biophys Acta Mol Cell Biol Lipids 2019;1864:744–55. 10.1016/j.bbalip.2019.02.007 [DOI] [PubMed] [Google Scholar]
- 75. Tomova A, Bukovsky I, Rembert E, et al. The effects of vegetarian and vegan diets on gut microbiota. Front Nutr 2019;6:47. 10.3389/fnut.2019.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chadaideh KS, Carmody RN. Host-Microbial interactions in the metabolism of different dietary fats. Cell Metab 2021;33:857–72. 10.1016/j.cmet.2021.04.011 [DOI] [PubMed] [Google Scholar]
- 77. Gil-Zamorano J, Tomé-Carneiro J, Lopez de Las Hazas M-C, et al. Intestinal miRNAs regulated in response to dietary lipids. Sci Rep 2020;10:18921. 10.1038/s41598-020-75751-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Carpi S, Scoditti E, Massaro M, et al. The extra-virgin olive oil polyphenols Oleocanthal and Oleacein counteract inflammation-related gene and miRNA expression in adipocytes by attenuating NF-κB activation. Nutrients 2019;11:2855. 10.3390/nu11122855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mantilla-Escalante DC, López de Las Hazas M-C, Gil-Zamorano J, et al. Postprandial circulating miRNAs in response to a dietary fat challenge. Nutrients 2019;11:1326. 10.3390/nu11061326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Beckett EL, Veysey M, Lucock M. Folate and microRNA: bidirectional interactions. Clin Chim Acta 2017;474:60–6. 10.1016/j.cca.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 81. Linna-Kuosmanen S, Tomas Bosch V, Moreau PR, et al. Nrf2 is a key regulator of endothelial microRNA expression under proatherogenic stimuli. Cardiovasc Res 2021;117:1339–57. 10.1093/cvr/cvaa219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Piotrowska M, Swierczynski M, Fichna J, et al. The Nrf2 in the pathophysiology of the intestine: molecular mechanisms and therapeutic implications for inflammatory bowel diseases. Pharmacol Res 2021;163:105243. 10.1016/j.phrs.2020.105243 [DOI] [PubMed] [Google Scholar]
- 83. Li Z, Xu R, Li N. Micrornas from plants to animals, do they define a new messenger for communication? Nutr Metab 2018;15:68. 10.1186/s12986-018-0305-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tili E, Michaille J-J, Piurowski V, et al. Micrornas in intestinal barrier function, inflammatory bowel disease and related cancers-their effects and therapeutic potentials. Curr Opin Pharmacol 2017;37:142–50. 10.1016/j.coph.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Udayasuryan B, Nguyen TTD, Slade DJ, et al. Harnessing tissue engineering tools to Interrogate Host-Microbiota crosstalk in cancer. iScience 2020;23:101878. 10.1016/j.isci.2020.101878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2021-325168supp001.pdf (191.3KB, pdf)
gutjnl-2021-325168supp002.pdf (1.3MB, pdf)
gutjnl-2021-325168supp003.pdf (824.8KB, pdf)
gutjnl-2021-325168supp004.pdf (576.2KB, pdf)
gutjnl-2021-325168supp005.pdf (329.5KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. Raw data are available upon request to the corresponding author.