Abstract
The interspecific transfer of two giant linear plasmids was investigated in sterile soil microcosms. Plasmids pRJ3L (322 kb) and pRJ28 (330 kb), both encoding mercury resistance, were successfully transferred in amended soil microcosms from their streptomycete hosts, the isolates CHR3 and CHR28, respectively, to a plasmidless and mercury-sensitive strain, Streptomyces lividans TK24. Transconjugants of S. lividans TK24 were first observed after 2 to 3 days of incubation at 30°C, which corresponded to the time taken for the formation of mycelia in soil. Transfer frequencies were 4.8 × 10−4 and 3.6 × 10−5 CFU/donor genome for pRJ3L and pRJ28, respectively. Transconjugants were analyzed by pulsed-field gel electrophoresis for the presence of plasmids, and plasmid identity was confirmed by restriction digests. Total genomic DNA digests confirmed that transconjugants were S. lividans TK24. The mercury resistance genes were shown to be on the plasmid in the transconjugants by hybridization analysis and were still functional. This is the first demonstration of transfer of giant linear plasmids in sterile soil microcosms. Giant linear plasmids were detected in many Streptomyces spp. isolated from mercury-contaminated sediments from Boston Harbor (United States), Townsville Harbor (Australia), and the Sali River (Tucuman, Argentina). Mercury resistance genes were shown to be present on some of these plasmids. Our findings that giant linear plasmids can be transferred between Streptomyces spp. and are common in environmental Streptomyces isolates suggest that these plasmids are important in gene transfer between streptomycetes in the environment.
Streptomycetes are common inhabitants of the soil environment and are regarded as the most numerous actinomycetes isolated from soil (31). Predominantly found as spores, streptomycetes can germinate and grow into a mycelial state for brief periods of time when nutrients become available (19). The growth of streptomycetes in soil is discontinuous in space and time, and survival between brief periods of vegetative growth is by production of spores resistant to starvation and desiccation (30, 31). Streptomyces species can survive for several weeks in sterile and nonsterile soil as spores after a short mycelial growth phase of 2 to 3 days (28). Under these conditions, the covalently closed circular plasmid pIJ673 was transferred intra- or interspecifically in sterile and nonsterile soil (28). The detection of transconjugants correlated with the mycelial stage of both the donor and recipient (28).
Plasmid transfer between streptomycetes in soil has been well studied (2, 22, 27–29), and a mathematical model for the transfer of covalently closed circular plasmids in soil microcosms has been deduced, taking into account the reduction in the exponential growth rate due to depletion of nutrients and/or moisture from soil microcosms, important factors influencing the development of streptomycetes in soil (5). However, there are no reports of transfer of Streptomyces giant linear plasmids in soil microcosms. Linear plasmids were first described in Streptomyces spp. by Hayakawa et al. (9), and studies by Kinashi and coworkers (16, 17) have demonstrated the presence of giant linear plasmids (>100 kb) in several antibiotic-producing Streptomyces spp. Linear plasmids ranging in size from 12 kb to 1 Mb have now been reported in more than 10 Streptomyces spp. (4, 8, 10, 32, 34), and they have also been found in other actinomycete genera such as Nocardia, Rhodococcus, and Mycobacterium (6, 12, 13, 21). Giant linear plasmids confer advantageous phenotypes and have been shown to carry genes encoding antibiotic biosynthesis, resistance to heavy metals, and ability to break down xenobiotics (for a review, see reference 20). At the molecular level, some linear plasmids have been shown to integrate into the chromosome, existing as an integrated or a free form (18). Recently, we demonstrated that mercury resistance genes were located on transmissible giant linear plasmids in two Chesapeake Bay Streptomyces isolates (24). This finding has important ecological implications, as plasmid transfer might be responsible for the spread of mercury resistance genes in polluted environments. Transfer functions of linear plasmids are still unknown. However, Zotchev and Schrempf (33) have cloned a 5.74-kb region of pBL1, a 43-kb Streptomyces linear plasmid, containing at least five open reading frames ORFs necessary for plasmid transfer and regulation.
Using two well-characterized Streptomyces strains (23, 24), we studied the transfer of two giant linear plasmids encoding mercury resistance, pRJ3L (322 kb) and pRJ28 (330 kb), between their respective hosts, Streptomyces strains CHR3 and CHR28 and a mercury-sensitive laboratory strain, S. lividans TK24, in amended sterile soil microcosms. We report the first example of transfer of giant linear plasmids in soil microcosms and compare transfer frequencies and time scales of initial transfer with those found with the covalently closed circular plasmid pIJ673 (28). In addition, we show that large Streptomyces plasmids encoding mercury resistance are widespread in mercury-contaminated sediments.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains used in plasmid transfer experiments are described in Table 1. All strains were maintained as spores prepared on solid agar ISP Medium 2 (YME) (Difco Laboratories, Detroit, Mich.) incubated at 30°C, as described by Hopwood et al. (11). Streptomyces strains were isolated from mercury-contaminated sediments in Boston Harbor (3), Townsville Harbor, Australia (7), and the Sali River, Tucuman, Argentina (1), by our previously described isolation procedure (23).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Phenotypea | Linear plasmid(s) | Source or reference |
|---|---|---|---|
| Streptomyces sp. strain CHR3 | Hgr Clms | pRJ3H, pRJ3L | 23 |
| Streptomyces sp. strain CHR28 | Hgr Clms | pRJ28 | 23 |
| S. lividans CHR3T3 | Hgr Clmr | pRJ3LT3 | This study |
| S. lividans CHR3T7 | Hgr Clmr | pRJ3T7 | This study |
| S. lividans CHR3T14 | Hgr Clmr | pRJ3T14 | This study |
| S. lividans CHR28T3 | Hgr Clmr | pRJ28T3 | This study |
| S. lividans CHR28T7 | Hgr Clmr | pRJ28T7 | This study |
| S. lividans CHR28T14 | Hgr Clmr | pRJ28T14 | This study |
| S. lividans TK24 | Hgs Clmr | 11 | |
| S. lividans 66 | Hgr Clms | SLP2 | 11 |
Hg, mercury; Clm, chloramphenicol.
Soil microcosm experiments.
The soil used in this study was taken from a local wheat field site at Cryfield, University of Warwick, Coventry, United Kingdom. Soil was prepared as previously described (28). Soil subsamples (10 g in 50 ml-Universal glass vials) were amended with 1% (wt/wt) soluble starch (Sigma Chemical Co., St. Louis, Mo.) and 1% (wt/wt) crabshell chitin (Sigma) prior to sterilization at 121°C for 15 min. Separate duplicate microcosms were prepared for sacrificial sampling at each time point. Spores were counted directly by microscopy and by plating in triplicate onto YME plus chloramphenicol (25 μg/ml) or mercuric chloride (0.05 mM). Inocula consisted of a mixture of donor and recipient spores in sufficient distilled water to give a final moisture content of 15% (wt/wt) in the microcosms. The donor-to-recipient ratio was 1:5 (105 spores of donor) in all experiments. Microcosms were incubated at 30°C. Spores and mycelia were extracted with one-fourth-strength Ringer solution (0.6 mM NaHCO3, 39 mM NaCl, 1.4 mM KCl, 1.6 mM CaCl2) by shaking 1 g of soil in 2 ml of solution on a Griffin flask shaker for 10 min. Appropriate dilutions prepared in one-fourth-strength Ringer solution were plated onto three selective media and incubated at 30°C: (i) YME agar containing 0.01 mM HgCl2 (selection of donor); (ii) YME agar containing 25 μg of chloramphenicol (selection of recipient) per ml; and (iii) YME agar containing 0.01 mM HgCl2 and 25 μg of chloramphenicol (selection of transconjugants) per ml. All experiments were carried out in duplicate, and samples taken at each time point were plated in triplicate.
PFGE.
DNA plug preparation, plasmid restriction digests, and pulsed-field gel electrophoresis (PFGE) analysis were performed as described previously (24). Ramping times are indicated in the legends of Fig. 2 to 4. Ladders of λ DNA concatamers and Saccharomyces cerevisiae YNN 295 chromosomes (Bio-Rad Laboratories) were used as molecular weight standards. DNA was stained with SYBR Green I (Molecular Probes, Inc., Eugene, Oreg.) prior to photography with 302-nm UV light illumination by using a SYBR Green I gel stain photographic filter (Molecular Probes, Inc.). The gels were digitized by using a FluorImager 573 (Molecular Dynamics, Sunnyvale, Calif.).
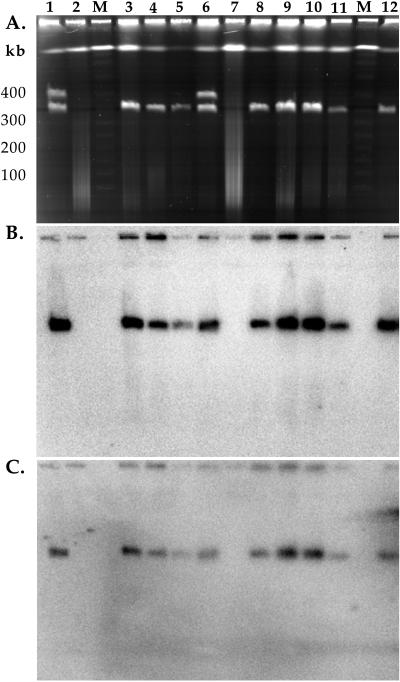
FIG. 2.
PFGE analysis of transconjugants (A) and Southern hybridization analysis of transconjugants with probe MER-A (B) and MER-B (C). Lane 1, Streptomyces sp. strain CHR3 at day 14; lane 2, S. lividans TK24 at day 14; lanes 3 to 5 S. lividans CHR3T3, CHR3T7, and CHR3T14; lane 6, Streptomyces sp. strain CHR3; lane 7, S. lividans TK24; lane 8, Streptomyces sp. strain CHR28; lanes 9 to 11, S. lividans CHR28T3, CHR28T7, and CHR28T14; lane 12, Streptomyces sp. strain CHR28 at day 14. The pulse time was 30 s for 24 h (6 V/cm at 14°C). Lanes designated M are λ DNA concatamer molecular weight markers.
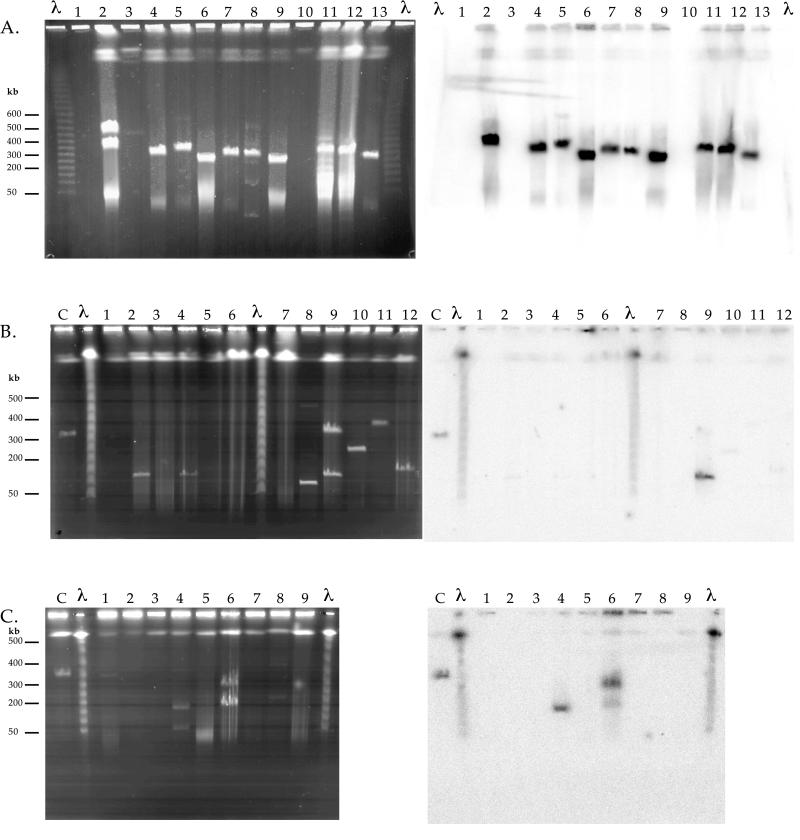
FIG. 4.
PFGE and Southern hybridization with probe MER-A of large plasmids in Streptomyces strains isolated from sediments as follows: (A) Boston Harbor (United States), isolates in lanes 1 to 13; (B) Townsville Harbor (Australia), isolates in lanes 1 to 12; lanes C, Streptomyces sp. strain CHR28; and (C) the Sali River (Tucuman, Argentina) isolates in lanes 1 to 9; lanes C, Streptomyces sp. strain CHR28. Pulse time was 30 to 50 s for 22 h (6 V/cm at 14°C). Molecular weight markers in the lanes designated λ were λ DNA concatamers.
DNA labeling and Southern hybridization.
An 816-bp PvuII fragment (MER-A) of the S. lividans 66 mercuric reductase gene merA and a 716-bp SalI-EcoRV fragment (MER-B) of the organomercurial lyase gene merB were prepared from plasmid pJOE851.2 (25), kindly donated by J. Altenbuchner, Stuttgart, Germany. Probes were labeled and Southern hybridizations were performed as described previously (23).
Determination of mercury resistance by agar diffusion assay.
Sensitivity of strains to mercury was tested by agar diffusion assay (26), as described previously (24).
Plasmid curing by growth at elevated temperature.
Streptomyces strains CHR3 and CHR28 were cultured in YME and incubated with shaking (300 rpm) at 30, 42, and 45°C for 48 h. Mycelia were collected and PFGE plugs were prepared and digested with restriction enzymes AseI as described previously (24).
RESULTS AND DISCUSSION
Streptomyces species isolated from the marine environment and carrying plasmid-borne mercury resistance were found to survive in soil and transfer mercury resistance. Transfer of two giant linear plasmids pRJ3L and pRJ28 (322 and 330 kb in size, respectively) was observed. Both plasmids are well characterized (24) and were used to estimate the extent of giant linear plasmid transfer in soil and to determine the time scale of the initial transfer in soil microcosms.
Transfer frequencies and growth curves.
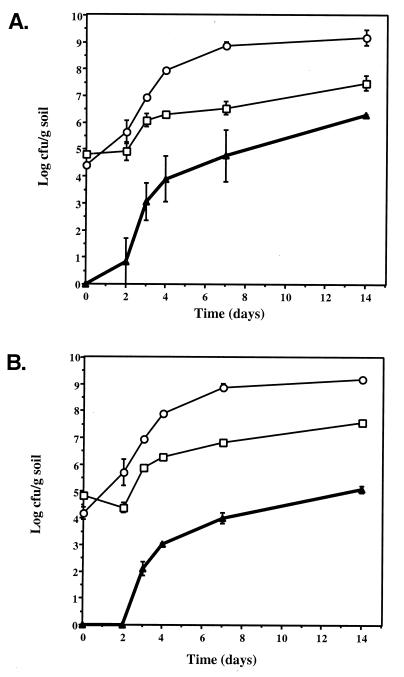
Spores and mycelia were extracted from soil microcosms, and appropriate dilutions were plated on three selective media for enumeration of donors (CHR3 or CHR28), recipient S. lividans TK24, and transconjugants. Plate counts are shown in Fig. 1. Transconjugants were detectable at days 2 and 3 for pRJ3L and pRJ28, respectively. The numbers of transconjugants then increased exponentially until day 4, followed by a slower rate of increase from day 7 to 14 (1.9 × 106 CFU/g for CHR3T and 1.2 × 105 CFU/g for CHR28T after 14 days) (Fig. 1). This profile is in agreement with earlier observations made with a small covalently closed circular plasmid pIJ673 for transfer in soil microcosms (28) and described by Clewlow et al. (5) in a mathematical model of plasmid transfer between strains of streptomycetes in soil microcosms. In the model, donors and recipient grew exponentially up to ca. day 7, after which competition for nutrients and/or decreasing moisture levels began to result in a reduced growth rate. Transconjugants formed during a period of plasmid transfer from day 0 to day 2, suggesting that mycelial growth and contact were necessary for plasmid transfer. After day 2, transconjugants grew exponentially until day 7 where growth rate reduction occurred, as for the donor and recipient populations. Successful plasmid transfer is unlikely to occur after 7 days under these poor growth conditions. Rate parameters for the model were obtained from experiments done with Warwick soil, and similar methods were applied in this study. In general, the fit of this model was good for plasmid pIJ673, and similar trends were observed in the present study. Nutrient and moisture depletion are likely to have had a similar effect on giant linear plasmid transfer.
FIG. 1.
Growth and recovery of donors and recipients from amended sterile soil microcosms inoculated with recipient strain S. lividans TK24 and donor Streptomyces sp. strains CHR3 (A) and CHR28 (B). Donor Streptomyces sp. strains CHR3 or CHR28 (○), recipient S. lividans TK24 (□), and transconjugants (▴) are as marked. Plate counts are the results from duplicate microcosms (triplicate plating), and the standard errors are indicated.
Frequencies of transfer at day 3 were 4.8 × 10−4 and 3.6 × 10−5 CFU/donor genome for CHR3 and CHR28, respectively. These frequencies of transfer were similar to those found with the covalently closed circular plasmid pIJ673, which transferred interspecifically with an efficiency of 7.4 × 10−4 CFU/donor genome (28). We previously found transfer frequencies of 1.5 × 10−2 and 1.8 × 10−3 transconjugants per donor genome for CHR3 and CHR28, respectively, in plasmid transfer experiments on agar plates (24). Mechanisms of transfer of giant linear plasmids are not well understood. The similarities in rate and frequency of transfer of covalently closed circular and giant linear plasmids in sterile soil suggest that a similar mechanism may be present in both cases. For covalently closed circular plasmid transfer, an initial mycelial contact was necessary, followed by mycelial fusion upon which transfer occurs. This was followed by spreading of plasmids into the mycelial mass (14, 15). At present, the tra genes on giant linear plasmids are not known, and it will be interesting to determine whether they are homologous to the plasmid transfer genes in covalently closed circular plasmids (14, 15).
Confirmation of transconjugants.
Colonies growing on YME containing 0.01 mM HgCl2 and chloramphenicol (25 μg/ml) were considered as transconjugants and were transferred individually onto fresh YME plates containing the same concentration of mercuric chloride and chloramphenicol to confirm their phenotypic traits. All transconjugants tested maintained growth after subculture on YME containing HgCl2 and chloramphenicol. Moreover, transconjugants recovered from the different crosses showed the characteristic gray spore color of S. lividans TK24, easily distinguishable from the white spores of Streptomyces sp. strain CHR3 and the dark green spores of Streptomyces sp. strain CHR28.
In order to compare the resistance of transconjugants and donors to mercury, one transconjugant of each cross from time points of 3, 7, and 14 days was analyzed by agar diffusion disk assay for HgCl2 and phenylmercuric acetate resistance (results not shown). No differences in mercury resistance profiles were observed between the original donors (CHR3 and CHR28) and the three transconjugants tested for each donor.
To confirm the transfer of the plasmids as nonintegrative elements, transconjugants from microcosms sampled at days 3, 7, and 14 were analyzed by PFGE (Fig. 2A). A plasmid of identical size to pRJ28 (330 kb) was present in each transconjugant. Crosses between CHR3 and S. lividans TK24 gave transconjugants that contained only one plasmid of identical size to pRJ3L (322 kb). No plasmid bands corresponding in size to pRJ3H were observed in transconjugants. pRJ3H does not encode mercury resistance (24), and therefore there would be no selection for transconjugants containing pRJ3H under these experimental conditions (i.e., on mercury-containing media). Transfer of pRJ3H is therefore unlikely to be detectable in these experiments and may or may not have occurred. The original donor strains, sampled at day 14, retained their original plasmids (pRJ28 in strain CHR28, pRJ3H and pRJ3L in strain CHR3). No plasmids were present in mercury-sensitive S. lividans TK24 after 14 days in soil microcosms. Hybridization with probes MER-A (Fig. 2B) and MER-B (Fig. 2C) confirmed the presence of the mercury resistance genes on the plasmids present in transconjugants. Also, mercury resistance genes were located on plasmids pRJ3L and pRJ28 in the two original donor strains (CHR3 and CHR28, respectively) and in the donor strains sampled at 14 days. Both probes hybridized with the plasmids in each transconjugant but not with the chromosomal DNA of transconjugants (Fig. 2B and C).
The plasmids present in each transconjugant were identical to the original plasmids pRJ3L and pRJ28, since no differences were observed between their restriction digest pattern with restriction endonucleases HindIII and XbaI (results not shown). In addition, transconjugants were confirmed to be derived from S. lividans TK24 by comparing total genomic DNA digest patterns of the transconjugants and the original donors and recipient. The restriction pattern of total genomic DNA from each transconjugant was identical to the pattern of S. lividans TK24 and different from CHR3 and CHR28 restriction patterns (results not shown).
Interspecific transfer and curing experiments.
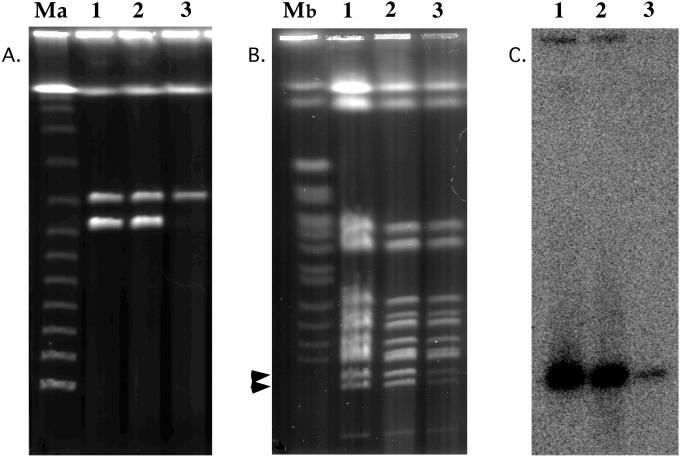
The transfer described here between Streptomyces sp. strains CHR3 or CHR28 and S. lividans TK24 is interspecific. 16S rRNA sequencing (23) clearly showed that both Streptomyces CHR3 and CHR28 are closely related to, although not necessarily the same species as, S. rochei, whereas the recipient in this experiment was S. lividans. No plasmidless strains of CHR3 or CHR28 were available. Multiple rounds (thirty) of sporulation/ germination on rich media were unsuccessful in curing the plasmids, as reported previously (24). In another attempt to cure plasmids pRJ3L and pRJ28, CHR3 and CHR28 were cultured at elevated temperatures (42 and 45°C). The plasmid band intensity of pRJ3L was dramatically reduced when Streptomyces sp. strain CHR3 was grown at 45°C (Fig. 3A). However, plasmid pRJ3H band intensity remained similar at 30, 42, and 45°C (Fig. 3A). Digestion of total genomic DNA with AseI, which cuts pRJ3L once, also showed reduced intensity of both pRJ3L restriction fragments indicated by the arrows in Fig. 3B. Growth at 42°C did not change band intensity of pRJ3L (Fig. 3A and B) or pRJ28 (data not shown). Streptomyces sp. strain CHR28 did not grow at 45°C, and CHR3 did not grow at temperatures higher than 45°C. Hybridization with probe MER-A confirmed the presence of pRJ3L in CHR3 grown at 45°C, and this probe did not hybridize to any chromosomal bands, indicating that pRJ3L had not integrated into the chromosome (Fig. 3C). After plating of the culture grown at 45°C on YME plates without mercury selection, subculturing of 300 individual clones gave colonies that were all mercury resistant. We were unsuccessful in curing pRJ28 or pRJ3L, making it impossible to examine the intraspecific transfer of linear plasmids in soil microcosms. The fact that all colonies tested after culture at elevated temperature retained mercury resistance suggests that there was an overall reduction in copy number rather than persistence of plasmid copies in only part of the population.
FIG. 3.
PFGE analysis of total genomic DNA (A), total genomic DNA digested with restriction enzyme AseI (B), and Southern hybridization (C) with probe MER-A of Streptomyces sp. strain CHR3 grown at 30°C (lane 1), 42°C (lane 2), and 45°C (lane 3). Pulse time was 30 s for 20 h (A) and 60 to 180 s for 26 h (B) (6 V/cm at 14°C). Molecular weight markers in lanes Ma (λ DNA concatamers) and Mb (S. cerevisiae YNN 295 chromosomes) are as indicated. Arrows indicate the two AseI restriction fragments of pRJ3L.
Widespread occurrence of Streptomyces giant linear plasmids encoding mercury resistance.
Twenty-nine Streptomyces strains isolated from highly mercury contaminated Boston Harbor sediment samples (3) were screened for the presence of giant linear plasmids. Overall, 20 of these strains contained one or more giant linear plasmids, ranging in size between ca. 50 and 600 kb. Five strains contained two plasmids. Hybridization with probe MER-A confirmed that 18 of the total of 25 giant linear plasmids encoded mercury resistance. A set of the giant linear plasmids detected in Boston Harbor Streptomyces isolates is shown in Fig. 4A. Of 12 Streptomyces isolates from the Sali River, Argentina, 7 contained giant linear plasmids ranging in size from ca. 60 to 400 kb, 1 of which encoded mercury resistance as indicated by hybridization with the MER-A gene probe (Fig. 4B). Three of nine Streptomyces strains from Townsville Harbor, Australia, contained giant linear plasmids ranging from ca. 50 to 300 kb, two of which showed homology to merA (Fig. 4C).
It is apparent from these data that large plasmids are common in environmental Streptomyces isolates and may be important in mercury resistance in the natural environment. This raises the intriguing possibility that transfer of giant linear plasmids such as we have demonstrated in sterile soil may be a widespread occurrence under natural conditions.
We have demonstrated that giant linear plasmids can be transferred in soil microcosms with efficiencies comparable to those of covalently closed circular plasmids. We also showed that giant linear plasmids are structurally stable upon transfer, and multiple rounds of sporulation did not cure the transconjugants of the newly acquired plasmid. Taking into account the stability of the plasmid in the transconjugants, their large size, and the type of information potentially transferable (e.g., biosynthetic pathways, biodegradative genes, and metal resistance genes [20]), these plasmids could confer numerous phenotypic advantages to the transconjugants in microcosms and in natural communities. In some cases, giant linear plasmids may also be stably integrated into the chromosome (18), although this has not been found with pRJ3L or pRJ28 (reference 24 and Fig. 2). Recombination events can occur between similar DNA sequences, such as genes of secondary metabolite biosynthetic pathways, and create new metabolites. Transfer of giant linear plasmids between different genera of actinomycetes has not yet been demonstrated, but the common presence of linear plasmids in Nocardia, Rhodococcus, and Mycobacterium (6, 12, 13, 21) raises the possibility of intergeneric transfer. Our finding that giant linear plasmids can be transferred between Streptomyces species gives these plasmids a potentially important role in gene transfer between streptomycetes in the environment.
ACKNOWLEDGMENTS
This study was supported by the Schering-Plough Research Institute.
We thank Ann Horan for her support and Frank Robb for comments on the manuscript. We thank Michael Bothner and Maria Amoroso for supplying sediments from Boston Harbor and from the Sali River, Argentina, respectively.
Footnotes
Contribution no. 500 from the Center of Marine Biotechnology.
REFERENCES
- 1.Amoroso M J, Castro G R, Carlino F J, Romero N C, Hill R T, Oliver G. Screening of heavy metal-tolerant actinomycetes isolated from the Sali River. J Gen Appl Microbiol. 1998;44:129–132. doi: 10.2323/jgam.44.129. [DOI] [PubMed] [Google Scholar]
- 2.Bleakey B H, Crawford D L. The effects of varying moisture and nutrient levels on the transfer of a conjugative plasmid between Streptomyces species in soil. Can J Microbiol. 1989;35:544–549. [Google Scholar]
- 3.Bothner M H, Buchholtz ten Brink M, Manheim F T. Metal concentrations in surface sediments of Boston Harbor—changes with time. Mar Environ Res. 1998;45:127–155. [Google Scholar]
- 4.Chen C W, Yu T-W, Lin Y-S, Kieser H M, Hopwood D A. The conjugative plasmid SLP2 of Streptomyces lividans is a 50 kb linear molecule. Mol Microbiol. 1993;7:925–932. doi: 10.1111/j.1365-2958.1993.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 5.Clewlow L J, Cresswell N, Wellington E M H. Mathematical model of plasmid transfer between strains of streptomycetes in soil microcosms. Appl Environ Microbiol. 1990;56:3139–3145. doi: 10.1128/aem.56.10.3139-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabrock B, Kesseler M, Averhoff B, Gottschalk G. Identification and characterization of a transmissible linear plasmid from Rhodococcus erythropolis BD2 that encodes isopropylbenzene and trichloroethane catabolism. Appl Environ Microbiol. 1994;60:853–860. doi: 10.1128/aem.60.3.853-860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbs R J. Metals of the bottom muds in Townsville Harbor, Australia. Environ Pollut. 1993;81:297–300. doi: 10.1016/0269-7491(93)90212-7. [DOI] [PubMed] [Google Scholar]
- 8.Gravius B, Glocker D, Pigac J, Pandza K, Hranueli D, Cullum J. The 387 kb linear plasmid pPZG101 of Streptomyces rimosus and its interactions with the chromosome. Microbiology. 1994;140:2271–2277. doi: 10.1099/13500872-140-9-2271. [DOI] [PubMed] [Google Scholar]
- 9.Hayakawa T, Tanaka T, Sakaguchi K, Otake N, Yonehara H. A linear plasmid-like DNA in Streptomyces sp. producing lankacidin group antibiotics. J Gen Appl Microbiol. 1979;25:255–260. [Google Scholar]
- 10.Hirochika H, Nakamura K, Sakaguchi K. A linear DNA plasmid from Streptomyces rochei with an inverted repetition of 614 base pairs. EMBO J. 1984;3:761–766. doi: 10.1002/j.1460-2075.1984.tb01881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D T, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 12.Kalkus J, Dörrie C, Fisher D, Reh M, Schegel H G. The giant plasmid pHG207 from Rhodococcus sp. encoding hydrogen autotrophy: characterization of the plasmid and its termini. J Gen Microbiol. 1993;139:2055–2060. doi: 10.1099/00221287-139-9-2055. [DOI] [PubMed] [Google Scholar]
- 13.Kalkus J, Reh M, Schegel H G. Hydrogen autotrophy of Nocardia opaca strains is encoded by linear megaplasmids. J Gen Microbiol. 1990;136:1145–1151. doi: 10.1099/00221287-136-6-1145. [DOI] [PubMed] [Google Scholar]
- 14.Kendall K J, Cohen S N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988;170:4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendall K J, Stein D S, Cohen S N. Transfer functions, promoters and sequence analysis of the Streptomyces plasmid pIJ101. In: Okami Y, Beppu T, Ogawara H, editors. Biology of actinomycetes '88. Tokyo: Japan Scientific Societies Press; 1988. pp. 52–57. [Google Scholar]
- 16.Kinashi H, Shimaji M. Detection of giant linear plasmids in antibiotic producing strains of Streptomyces by the OFAGE technique. J Antibiotics. 1987;40:913–916. doi: 10.7164/antibiotics.40.913. [DOI] [PubMed] [Google Scholar]
- 17.Kinashi H, Shimaji M, Sakai A. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. Nature (London) 1987;328:454–456. doi: 10.1038/328454a0. [DOI] [PubMed] [Google Scholar]
- 18.Kinashi H, Shimaji-Murayama M, Hanafusa T. Integration of SCP1, a giant linear plasmid, into the Streptomyces coelicolor chromosome. Gene. 1992;115:35–41. doi: 10.1016/0378-1119(92)90537-y. [DOI] [PubMed] [Google Scholar]
- 19.Mayfield C I, Williams S T, Ruddick S M, Hatfield H L. Studies on the ecology of actinomycetes in soil. IV. Observations on the form and growth of streptomycetes in soil. Soil Biol Biochem. 1972;4:79–91. [Google Scholar]
- 20.Meinhardt F, Schaffrath R, Larsen M. Microbial linear plasmids. Appl Microbiol Biotechnol. 1997;47:329–336. doi: 10.1007/s002530050936. [DOI] [PubMed] [Google Scholar]
- 21.Picardeau M, Vincent V. Characterization of large linear plasmids in mycobacteria. J Bacteriol. 1997;179:2753–2756. doi: 10.1128/jb.179.8.2753-2756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rafii F, Crawford D L. Transfer of conjugative plasmids and mobilization of a nonconjugative plasmid between Streptomyces strains on agar and in soil. Appl Environ Microbiol. 1988;54:1334–1340. doi: 10.1128/aem.54.6.1334-1340.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravel J, Amoroso M J, Colwell R R, Hill R T. Mercury-resistant actinomycetes from the Chesapeake Bay. FEMS Microbiol Lett. 1998;162:177–184. doi: 10.1111/j.1574-6968.1998.tb12996.x. [DOI] [PubMed] [Google Scholar]
- 24.Ravel J, Schrempf H, Hill R T. Mercury resistance is encoded by transferable giant linear plasmids in two Chesapeake Bay Streptomyces strains. Appl Environ Microbiol. 1998;64:3383–3388. doi: 10.1128/aem.64.9.3383-3388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedlmeier R, Altenbuchner J. Cloning and DNA sequence analysis of the mercury resistance genes of Streptomyces lividans. Mol Gen Genet. 1992;236:76–85. doi: 10.1007/BF00279645. [DOI] [PubMed] [Google Scholar]
- 26.Weiss A A, Murphy S D, Silver S. Mercury and organomercurial resistances determined by plasmids in Staphylococcus aureus. J Bacteriol. 1977;132:197–208. doi: 10.1128/jb.132.1.197-208.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wellington E M H, Cresswell N, Herron P R, Clewlow L J, Saunders V A, Wipat A. Gene transfer between streptomycetes in soil. In: Fry J C, Day M J, editors. Bacterial genetics in natural environments. London, England: Chapman & Hall; 1990. pp. 216–230. [Google Scholar]
- 28.Wellington E M H, Cresswell N, Saunders V A. Growth and survival of streptomycete inoculants and extent of plasmid transfer in sterile and nonsterile soil. Appl Environ Microbiol. 1990;56:1413–1419. doi: 10.1128/aem.56.5.1413-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellington E M H, Saunders V A, Cresswell N, Wipat A. Plasmid transfer between streptomycetes in soil. In: Okami Y, Beppu T, Ogawara H, editors. Biology of actinomycetes '88. Tokyo: Japan Scientific Societies Press; 1988. pp. 300–305. [Google Scholar]
- 30.Williams S T. Streptomycetes in the soil ecosystem. In: Mordarski M, Kurylowicz W, Jeljaszewicz J, editors. Nocardia and Streptomyces. New York, N.Y: Fisher Verlag; 1978. pp. 137–144. [Google Scholar]
- 31.Williams S T, Lanning S, Wellington E M H. Ecology of actinomycetes. In: Goodfellow M, Mordarski M, Williams S T, editors. The biology of the actinomycetes. London, England: Academic Press; 1984. pp. 481–528. [Google Scholar]
- 32.Wu X, Roy K L. Complete nucleotide sequence of a linear plasmid from Streptomyces clavuligerus and characterization of its RNA transcripts. J Bacteriol. 1993;175:37–52. doi: 10.1128/jb.175.1.37-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zotchev S B, Schrempf H. The linear Streptomyces plasmid pBL1: analyses of transfer functions. Mol Gen Genet. 1994;242:374–382. doi: 10.1007/BF00281786. [DOI] [PubMed] [Google Scholar]
- 34.Zotchev S B, Soldatova L I, Orekhov A V, Schrempf H. Characterization of a linear extrachromosomal DNA element (pBL1) isolated after interspecific mating between Streptomyces bambergiensis and S. lividans. Res Microbiol. 1992;143:839–845. doi: 10.1016/0923-2508(92)90071-u. [DOI] [PubMed] [Google Scholar]