Abstract
Viridans group streptococci are major constituents of the normal human oral flora and are also identified as the predominant pathogenic bacteria in native valve infective endocarditis. Little information is available regarding the regulation of gene expression in viridans group streptococci, either in response to changes in the oral environment or during development of endocarditis. We therefore constructed a set of broad-host-range vectors for the isolation of promoters from viridans group streptococci that are activated by specific environmental stimuli in vitro or in vivo. A genomic library of Streptococcus gordonii strain CH1 was constructed in one of the new vectors, and this library was introduced into a homologous bacterium by using an optimized electroporation protocol for viridans group streptococci. Because viridans group streptococci entering the bloodstream from the oral cavity encounter an increase in pH, we selected promoters upregulated by this specific stimulus. One of the selected promoter sequences showed homology to the promoter region of the hydA gene from Clostridium acetobutylicum, the expression of which is known to be regulated by the environmental pH. The isolation of this pH-regulated promoter shows that S. gordonii can sense an increase in the environmental pH, which serves as a signal for bacterial gene activation. Furthermore, this demonstrates the usefulness of these new selection vectors in research on adaptive gene expression of viridans group streptococci and possibly also of other gram-positive bacteria.
Viridans group streptococci (VS) are major constituents of the human commensal oral flora (15). They constantly have to adapt to the rapid changes in their natural habitat (4). Adaptation is characterized firstly by sensing of environmental changes, followed by signal transduction, which can result in the expression of genes whose products are involved in the adaptive process (17, 30, 43). One of the environmental changes VS encounter is variation in the extracellular pH, which drops rapidly after carbohydrate consumption by the host. Streptococcus mutans responds to such a decrease in pH by rapid upregulation of the expression of several regulatory genes and of genes involved in stress responses, including hcrA, grpE, and dnaK (23). However, knowledge on gene expression in other VS induced by pH or other environmental stimuli is scarce.
VS, including Streptococcus sanguis, Streptococcus oralis, and Streptococcus gordonii, are the most frequently encountered bacterial causes of native valve infective endocarditis (IE) (10, 40). This disease is caused by the rapid growth and persistence of bacteria embedded in a platelet-fibrin thrombus (a vegetation) present on damaged endocardium or heart valves (12). Studies on virulence factors of VS in the pathogenesis of IE have mainly focused on components involved in bacterial adherence to the vegetation. These include exopolysaccharides (7, 31) and adhesins for connective-tissue proteins, for adhesive macromolecules present in plasma, and for blood platelets (3, 19, 25, 47). Little information is available on the regulation of these and other possible virulence factors of VS in the host.
Therefore, we have developed a plasmid-based selection system for the isolation of inducible VS promoters. The system is designed partly in analogy to the in vivo expression technology (IVET) system (18, 28), since IVET has been shown to be a promising tool in the study of adaptive gene expression of VS in an experimental rabbit model of IE [20; A. O. Kiliç, M. C. Herzberg, X. Zhao, M. W. Meyer, and L. Tao, Abstr. ASM Conf. Streptococcal Genet. (Genet. Streptococci, Enterococci, Lactococci), abstr. LB-03, p. 41, 1998]. As VS experience an increase in the environmental pH from slightly acidic to neutral levels when entering the blood from the oral plaque (33), we used this selection system to identify genes whose expression was influenced by this specific stimulus. A pH-regulated promoter of S. gordonii strain CH1 was isolated and characterized, showing that VS indeed recognize an increase in pH as a signal for adaptive gene expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Escherichia coli strains BHB2600 and DH5α were cultured in Luria-Bertani broth and on Luria-Bertani agar at 37°C. For maintenance of plasmids, ampicillin (50 μg/ml) or erythromycin (150 μg/ml for strain BHB2600 and 300 μg/ml for strain DH5α) was added to the growth medium. VS were cultured in Todd-Hewitt (TH) broth and on TH agar (Difco Laboratories, Detroit, Mich.) at 37°C in a 5% CO2 atmosphere or anaerobically. When required, TH broth and TH agar were supplemented with erythromycin (5 μg/ml).
TABLE 1.
Bacterial strains used in this study
| Strain | Source or reference |
|---|---|
| Escherichia coli DH5α | Gibco-BRL (Breda, The Netherlands) |
| Escherichia coli BHB2600 | 21 |
| Streptococcus gordonii CH1 | 53 |
| Streptococcus sanguis U108 | 8 |
| Streptococcus sanguis S221 | Our culture collection |
| Streptococcus sanguis S263 | Our culture collection |
| Streptococcus bovis S225 | Our culture collection |
| Streptococcus bovis S228 | Our culture collection |
| Streptococcus oralis S195 | Our culture collection |
| Streptococcus mutans S39 | Our culture collection |
| Streptococcus mutans V403 | 31 |
| Streptococcus oralis J30 | 8 |
| Streptococcus oralis S223 | Our culture collection |
| Streptococcus oralis S235 | Our culture collection |
| Streptococcus oralis S245 | Our culture collection |
| Streptococcus salivarius S304 | Our culture collection |
| Streptococcus salivarius S310 | Our culture collection |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant features | Reference |
|---|---|---|
| pEC2A | E. coli plasmid containing the trp′-′lacZ fusion gene; 7.5 kb; Emr | 5 |
| pIC20R | E. coli plasmid; 2.7 kb; Apr | 29 |
| pLITMUS28 | E. coli plasmid; 2.8 kb; Apr | 14 |
| pMG36 | Lactococcal expression vector; 3.7 kb; Kmr | 49 |
| pMG36e | Lactococcal expression vector; 3.6 kb; Emr | 49 |
| pGKV210 | Lactococcal shuttle vector; 4.4 kb; Emr | 50 |
| pORI19S | Lactococcal shuttle vector; 3.9 kb; Spr | Smith, personal communication |
| pSU31 | pUC19 containing the transcriptional terminator from S. equisimilis H46A; 6.2 kb; Apr | 45 |
| pMM201 | pMG36e containing the EcoRI-BamHI part of pIC20R MCS and the trp′-′lacZ gene from pEC2A; 6.6 kb; Emr | This study |
| pMM206 | Derivative of pGKV210 with a deletion of the EcoRI-BamHI fragment; 4.4 kb; Emr | This study |
| pMM210 | pLITMUS28 containing the 3.4-kb EcoRI-KpnI fragment of pMM201; 6.2 kb; Apr | This study |
| pMM211 | pMM210 with self-annealed primer AV7 cloned into the KpnI site; 6.2 kb; Apr | This study |
| pMM214 | pMM211 with the promoterless aad(9) gene of pORI19S cloned into the BamHI site; 6.9kb; Apr | This study |
| pMM218 | pMM214 with the SpeI-EcoRI transcription terminator fragment from the pSU31; 7.0 kb; Apr | This study |
| pMM219 | pMM218 with the promoterless aphIII gene of pMG36 replacing the promoterless aad(9) gene; 7.1 kb; Apr | This study |
| pMM223 | pMM206 with the 4.3-kb SpeI fragment from pMM218 in the XbaI site; 8.1 kb; Emr | This study |
| pMM225 | pMM206 with the 4.4-kb SpeI fragment from pMM219 in the XbaI site; 8.2 kb; Emr | This study |
| pMM239 | pMM223 with the streptococcal rgg-gftG promoter fragment from S. gordonii CH1 in the EcoRI-SalI site; 9.2 kb; Em | This study |
| pMM240 | pMM223 with the Sau3A fragment containing a constitutive promoter from S. gordonii CH1 in the BglII site; 9.0 kb; Emr | This study |
| pMM243 | pMM223 with the Sau3A fragment containing the neutral-pH-inducible promoter from S. gordonii CH1 in the BglII site; 8.6 kb; Emr | This study |
MIC and MBC determination.
The MICs and minimal bactericidal concentrations (MBCs) of spectinomycin for VS were determined by broth microdilution assays (32). Dilution series of spectinomycin or kanamycin, ranging from 25 to 1,000 μg/ml, in TH broth supplemented with 5% horse blood were prepared in microtiter plates. To each well 100 μl of TH broth containing 106 bacteria was added, and the plates were incubated overnight at 37°C in a 5% CO2 atmosphere. The MIC was defined as the lowest antibiotic concentration at which no growth was visible. To determine the MBC, 1 μl from each well without visible bacterial growth was cultured on blood agar plates. The MBC was defined as the lowest antibiotic concentration that reduced the bacterial inoculum at least 1,000-fold.
DNA isolation.
Plasmid DNA was isolated from E. coli using Qiagen GmbH (Hilden, Germany) plasmid DNA isolation kits and was isolated from VS as described previously (52). Chromosomal DNA from VS was isolated using the Puregene chromosomal DNA isolation kit for gram-positive bacteria and yeast (Gentra Systems Inc., Minneapolis, Minn.).
Molecular cloning and DNA sequence determination.
DNA manipulations were done according to standard techniques (39). Plasmid DNA was introduced into E. coli by electroporation using the method of Dower et al. (11). DNA sequencing was performed with the PCR-mediated Taq Dye Deoxy Terminator Cycle sequencing kit (Perkin-Elmer, Foster City, Calif.) using an Applied Biosystems model 373 DNA sequencer.
Construction of selection vectors pMM223 and pMM225.
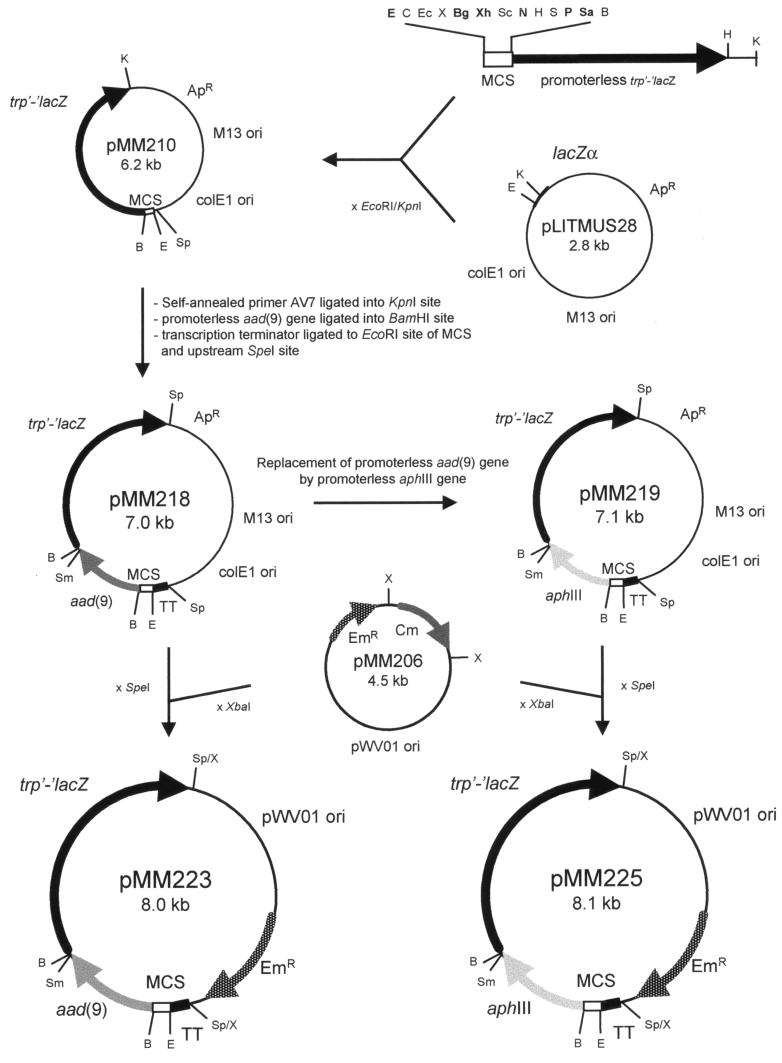
Plasmids used for the construction of the selection vectors pMM223 and pMM225 are listed in Table 2. The 3.3-kb BamHI-HindIII fragment of pEC2A (5), containing the promoterless trp′-′lacZ fusion gene (9), was ligated to the EcoRI-BamHI part of the pIC20R multiple cloning site (MCS) (29) and the product was introduced into the EcoRI- and HindIII-digested shuttle vector pMG36e (49), resulting in pMM201 (data not shown). To achieve higher cloning efficiencies, the MCS-trp′-′lacZ fragment was excised from pMM201 as an EcoRI-KpnI fragment and ligated to EcoRI- and KpnI-digested E. coli plasmid pLITMUS28 (14), creating pMM210 (Fig. 1). Self-annealed primer AV7 (Table 3) was cloned into the KpnI site of pMM210, replacing this site by an SpeI site (pMM211). The gene aad(9), originating from Enterococcus faecalis and conferring resistance to spectinomycin (27), was amplified as a promoterless gene from plasmid pORI19S (H. E. Smith, personal communication) using primers AV3 and AV4 (Table 3). The 795-bp PCR fragment was digested with BamHI and cloned into the BamHI site of pMM211, resulting in pMM214. A bidirectional transcriptional terminator of Streptococcus equisimilis H46A present on a 922-bp PstI-HindIII fragment of plasmid pSU31 (45) was amplified using primers AV8 and AV9 (Table 2). The terminator fragment was digested with SpeI and EcoRI and ligated in the reversed orientation in front of the selection cassette of pMM214 to generate plasmid pMM218 (Fig. 1). For the construction of the second selection cassette, the aphIII gene originating from E. faecalis plasmid pJH1 and conferring resistance to kanamycin (48) was selected. It was amplified from plasmid pMG36 (49) as a promoterless gene by using primers AV1 and AV2 (Table 3). The 855-bp product was digested with BamHI and used to replace the BamHI fragment of pMM218 comprising the promoterless aad(9) gene. The resulting plasmid was designated pMM219. The selection cassettes were excised as SpeI fragments from pMM218 and pMM219 and ligated to XbaI-digested pMM206, a derivative of the lactococcal shuttle vector pGKV210 (50), to yield the vectors pMM223 [aad(9) Sp] and pMM225 (aphIII Km), respectively (Fig. 1).
FIG. 1.
Schematic representation of the construction of selection vectors pMM223 and pMM225. Details of the construction are given in the text. TT, streptococcal transcription terminator; ApR, ampicillin resistance gene; EmR, erythromycin resistance gene; Cm, promoterless chloramphenicol acetyltransferase gene; B, BamHI; Bg, BglII; C, ClaI; E, EcoRI; Ec, EcoRV; H, HindIII; K, KpnI; N, NruI; P, PstI; Sc, SacI; Sa, SalI; Sm, SmaI; S, SphI; Sp, SpeI; X, XbaI; Xh, XhoI. Endonucleases printed in bold represent unique sites in the MCS.
TABLE 3.
Synthetic oligonucleotides used in this studya
| Primer | Sequence (5′-3′) | Restriction site |
|---|---|---|
| AV1 | AACAGGATCCGGGGTATCTTTAAATACTGTAG | BamHI |
| AV2 | AACAGGATCCCGGGCTAGGTACTAAAACAATTCATCC | BamHI/SmaI |
| AV3 | AATTGGATCCCTAATCAAAATAGTGAGGAGG | BamHI |
| AV4 | AATTGGATCCCGGGTTTTTTTATAATTTTTTTAATCTG | BamHI/SmaI |
| AV7 | ATCATACTAGTATGATGTAC | SpeI |
| AV8 | AACAGAATTCACGGTCTTCTAAAACGATG | EcoRI |
| AV9 | ATGTCACTAGTCTCTACAAC | SpeI |
| AV13 | GATAAGATCTTGACGGAGATTAGCAAAAAG | BglII |
| AV16 | GATACTGCAGCCTTCTGAAAATAGTATAAAG | PstI |
| AV19 | CCTCCTCACTATTTTGATTAG |
Oligonucleotides were obtained from Perkin-Elmer Nederland B.V. (Nieuwerkerk aan de Ijssel, The Netherlands). Primers AV1-AV2, AV3-AV4, and AV8-AV9 were used to amplify the promoterless aad(9) gene, the promoterless aphIII gene, and the streptococcal transcriptional terminator, respectively. Oligonucleotide AV7 was self-annealed and used in the construction of the cassettes (Fig. 1). Underlined and bold sequences are recognition sites for the indicated endonucleases. AV9 and AV19 were used for sequencing of the cloned chromosomal fragments.
Electrotransformation of VS.
Electrocompetent VS cells were prepared according to the method of Smith et al. (44), with minor modifications. Briefly, bacteria harvested from mid-log cultures in TH broth were washed three times in ice-cold, sterile distilled water and three times in ice-cold 0.3 M sucrose–10% glycerol. Bacteria were resuspended in 1/500 volume of the latter solution, and 50 μl of this suspension was used for electroporation with a Gene-Pulser and Pulse Controller (Bio-Rad Laboratories B.V., Veenendaal, The Netherlands). Immediately after electroporation, 0.95 ml of TH broth supplemented with 0.3 M sucrose was added, and the bacteria were incubated for 2 h at 37°C and subsequently plated on the appropriate selective agar medium. Colonies of VS transformants were visible after 24 to 48 h of incubation at 37°C in a 5% CO2 atmosphere. To obtain maximal transformation frequencies, cuvettes with different electrode gap sizes (0.1 or 0.2 cm) were tested, and resistance setting (100 or 200 Ω), field strength (10 to 25 kV/cm), and type of electrical pulse (decayed pulse or squared pulse) were varied. In addition, the influence of procedures affecting VS cell wall integrity on electroporation efficiencies was tested. dl-Threonine (40 mM), 1% (vol/vol) glycine, or a sub-MIC concentration of penicillin G was added to the growth medium used for the preparation of competent VS cells, or competent VS were enzymatically treated with lysozyme (0.5 mg/ml; Sigma Chemical Co., St. Louis, Mo.) and mutanolysin (2.5 U/ml; Sigma) for 1 h.
Natural transformation of S. gordonii CH1.
S. gordonii CH1 cells were made competent for natural transformation according to the method of Jenkinson (24). Briefly, an overnight culture in brain heart yeast (brain heart infusion medium [Difco Laboratories] with 5 g of yeast extract [Difco] per liter) was diluted 100-fold in fresh brain heart yeast supplemented with 5% horse serum and 1% glucose and then incubated for 3 h at 37°C. This culture was diluted 100-fold in fresh medium and incubated for 1 h at 37°C. From this culture 1-ml aliquots were taken, 1.5 μg of plasmid DNA was added, and incubation was continued for another 3 h at 37°C. Aliquots of 10 μl were plated onto selective TH agar and incubated for 24 to 48 h at 37°C in a 5% CO2 atmosphere to select for plasmid-containing transformants.
β-Galactosidase activity assay.
β-Galactosidase activities of the VS clones were determined using the fluorescent substrate fluorescein di-β-galactopyranoside (FDG; Molecular Probes Europe BV, Leiden, The Netherlands). Bacteria cultured overnight in TH broth containing erythromycin (5 μg/ml) were harvested by centrifugation, washed, and resuspended in STES buffer (10 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 20% sucrose) to an optical density at 620 nm (A620) of 1.0. Of this suspension 1.5 ml was centrifuged, the pellet was resuspended in 1 ml of STES buffer supplemented with 50 mg of lysozyme per ml and 200 U of mutanolysin per ml, and this suspension was incubated at 37°C. After 2 h, 50 μl of 1% sodium dodecyl sulfate and 50 μl of chloroform were added and the samples were mixed for 10 s and then were left standing for 15 min at room temperature. Six replicate samples of 50 μl of this suspension were transferred to wells of microtiter plates, 150 μl of Z buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 10 mM KCl, 1 mM MgSO4, 40 mM β-mercaptoethanol [pH 7.0]) containing 33 μM FDG was added to each replicate, and microtiter plates were incubated at 37°C. The emission was measured at different times at 530 nm (band-pass wavelength = 30 nm) after excitation at 485 nm (band-pass wavelength = 20 nm) using a Cytofluor II fluorescence multiwell plate reader (PerSeptive Biosystems, Inc., Framingham, Mass.). The β-galactosidase activity was plotted as arbitrary fluorescence units over time, and results are the averages of six reactions.
Construction of an S. gordonii CH1 genomic library.
A genomic library of S. gordonii CH1 was constructed using the selection vector pMM223. Vector DNA was digested with BglII and dephosphorylated using calf intestine alkaline phosphatase (Boehringer Mannheim GmbH, Mannheim, Germany). Genomic DNA isolated from S. gordonii CH1 was digested to completion with Sau3A. Vector and chromosomal fragments were ligated, and the ligation mixture was introduced into E. coli BHB2600 by electroporation. Plasmid DNA was isolated from erythromycin-resistant transformants constituting the genomic library and introduced into the homologous streptococcal strain CH1 by electroporation.
Selection of pH-regulated promoters.
The streptococcal genomic bank was plated onto TH agar of pH 7.3 supplemented with erythromycin (5 μg/ml) for plasmid maintenance and spectinomycin (500 μg/ml) for selection of active streptococcal promoters. After anaerobic incubation at 37°C for 36 h, colonies resistant to erythromycin as well as spectinomycin were replated onto TH agar of pH 6.2 supplemented with erythromycin and spectinomycin. As a control for the viability of the isolated S. gordonii clones, these were also restreaked onto TH agar plates of pH 6.2 supplemented with erythromycin only and onto plates of pH 7.3 supplemented with erythromycin and spectinomycin. Plasmids were isolated from clones that failed to grow on the pH 6.2 agar but that did grow on the agar of pH 7.3 in the presence of spectinomycin. The fragments cloned in these plasmids were amplified by PCR using primers AV4 and AV9 (Table 3), and the PCR products were sequenced using primer AV19 (Table 3). The obtained sequences were analyzed using the BLAST program (2).
Measurement of in vitro growth rate.
To determine the relative activities of promoters at different pH values, the growth rates of selected clones were determined in TH broth of pH 6.2 and pH 7.3, in the presence and absence of spectinomycin. A single colony of each clone was cultured at 37°C in TH broth supplemented with erythromycin for plasmid maintenance. Overnight cultures were diluted 100-fold in fresh TH broth containing both erythromycin (5 μg/ml) and spectinomycin (500 μg/ml) or containing erythromycin alone. Growth was monitored by measuring the A620 over time, and the mid-log-phase doubling time (t½) was determined. Relative promoter activity at pH 6.2 and 7.3 was expressed as the ratio of growth in the presence and absence of spectinomycin at each pH [t½ (+spec)/t½(−spec)].
Statistical evaluation.
The significance of the differences between the growth rate ratios at either pH was calculated with Student's t test.
Nucleotide sequence accession numbers.
The sequences of the vectors pMM223 and pMM225 have been assigned GenBank accession numbers AF076212 and AF076213, respectively. The sequence of the isolated pH-regulated promoter fragment from S. gordonii CH1 pMM243 has been assigned GenBank accession number AF127175.
RESULTS
New broad-host-range promoter selection vectors pMM223 and pMM225.
We constructed a set of self-replicating broad-host-range promoter selection vectors for gram-positive bacteria (Fig. 1). Firstly, selection cassettes were constructed in E. coli plasmid pLITMUS28 (14). Each selection cassette contains two promoterless genes. The first gene is either a promoterless aphIII gene (48), conferring resistance to kanamycin, or an aad(9) (27) gene, conferring resistance to spectinomycin, for the selection of active promoters. The second gene is the trp′-′lacZ fusion gene, encoding β-galactosidase (9), to be used for discrimination between constitutive and induced promoter activities. In front of the two promoterless genes, the MCS from pIC20R (29) was introduced for insertion of DNA fragments with possible promoter activity. A bidirectional transcriptional terminator from S. equisimilis strain H46A (45) was cloned in front of the MCS to prevent possible readthrough into the promoterless cassette. The terminator was inserted in its reversed orientation, which has the highest termination activity in E. coli TG1 (45). The resulting selection cassettes have a total size of approximately 4.3 kb, and all components of the cassettes can be replaced or removed separately or in combination, using common restriction endonucleases (Fig. 1).
The cassettes were cloned as SpeI fragments into XbaI-digested plasmid pMM206, a derivative of the lactococcal shuttle vector pGKV210 (50), replacing the promoterless chloramphenicol acetyltransferase gene of pMM206. Plasmid pMM206 had been constructed by digestion of pGKV210 with EcoRI and BamHI, filling in of the ends with Klenow large-fragment DNA polymerase I, and ligation of the blunt ends. This vector contains an erythromycin resistance gene for plasmid maintenance and the broad-host-range origin of replication (ori) of plasmid pWV01 (26). The sequences of the new vectors were assembled from published sequences of the different fragments used and sequences obtained after sequence determination of the borders of these fragments after ligation.
Transformation of VS.
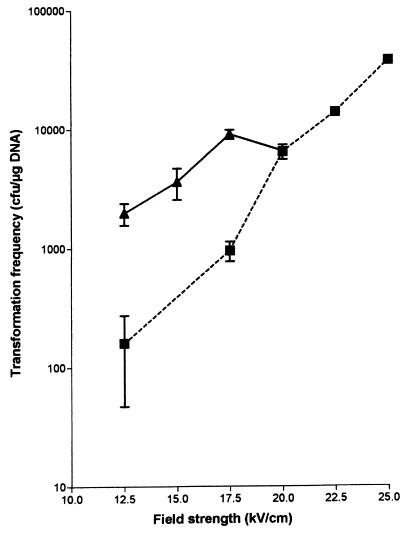
We optimized the electroporation procedure using our standard S. sanguis test strain U108 and shuttle vector pGKV210. Maximal efficiencies were obtained using cuvettes with a 0.1-cm electrode gap at a resistance setting of 100 Ω, a capacitance of 25 μF, and a field strength of 25 kV/cm (Fig. 2). At these settings time constants ranged from 2.0 to 2.5 ms. The number of transformants increased linearly with the plasmid DNA concentration over a range from 5 to 500 ng (data not shown). Despite the high field strength, survival rates of S. sanguis U108 generally were 70% or higher. Colonies of the U108 transformants were often variable in size, but the introduced plasmid pGKV210 could be isolated from the transformants in all cases.
FIG. 2.
Electroporation frequency of S. sanguis strain U108 as function of field strength. The lactococcal shuttle vector pGKV210 was used as the donor DNA. Electroporation cuvettes with an electrode gap of 0.1 cm were used at a resistance setting of 100 Ω (squares) or 200 Ω (triangles) and a capacitance of 25 μF.
Either culturing S. sanguis U108 in the presence of 1% (vol/vol) glycine (22) or 40 mM dl-threonine (6) or treatment of the bacteria with lysozyme (36, 42) and mutanolysin reduced transformation frequencies at least fivefold. Addition of a sub-MIC concentration of penicillin G to the medium used to prepare electrocompetent cells (35) or the use of a squared instead of a decayed pulse (34) did not improve electroporation.
To determine the transformability of other VS strains, several laboratory strains and clinical endocarditis isolates were electrotransformed with pGKV210. In these experiments, the field strength was reduced to 20 kV/cm, since at 25 kV/cm arcing regularly occurred. S. gordonii CH1 and S. sanguis U108 had the highest transformation frequencies (2 × 104 and 7 × 103 CFU/μg of DNA, respectively), while for three other strains (S. oralis J30, S. mutans S39, and S. sanguis S263) reasonable numbers of transformants were obtained (4 × 103, 1 × 103, and 4 × 102 CFU/μg of DNA, respectively). At 25 kV/cm, transformation of S. gordonii CH1 was over 100-fold more efficient than that of S. sanguis U108 (Table 4).
TABLE 4.
Efficiencies of transformation of pGKV210 and derived vectors into S. sanguis U108 and S. gordonii CH1
| Vector | Frequency in S. sanguis U108 of electroporation | Frequency in S. gordonii CH1 of:
|
|
|---|---|---|---|
| Electroporation | Natural transformation | ||
| pGKV210 | 4 × 104 | 5 × 106 | 2 × 105 |
| pMM223 | <102 | 7 × 103 | 3 × 102 |
| pMM225 | <102 | 5 × 103 | <102 |
Subsequently, electroporation efficiencies of pGKV210, pMM223, and pMM225 were determined for both S. sanguis U108 and S. gordonii CH1. The highest frequencies were obtained with S. gordonii CH1 for all vectors tested (Table 4). As this strain is known to be naturally transformable, efficiencies of the optimized electroporation procedure and of natural transformation were compared. The optimized electroporation procedure proved to be superior, especially for the newly constructed vectors (Table 4).
Testing of expression vectors.
To determine the spectinomycin level required for the selection of active promoters, plasmid pMM239 was constructed. A 1.1-kb rgg-gtfG promoter fragment (46) amplified from the genomic DNA of S. gordonii CH1 using primers AV13 and AV16 was digested with BglII and PstI and then cloned into the MCS of pMM223. The MICs and MBCs for S. gordonii CH1 and S. sanguis U108 harboring either plasmid pMM223 or pMM239 were assessed. The presence of the active gtfG promoter in pMM239 was well detectable, since without plasmid or with pMM223 both strains were susceptible to spectinomycin concentrations of <25 μg/ml, whereas S. gordonii CH1 and S. sanguis U108 harboring pMM239 were resistant to >1,000 and >800 μg of spectinomycin per ml, respectively. The MICs of kanamycin for our test strains, S. gordonii CH1 and S. sanguis U108 with or without pMM225, were relatively high, 250 μg/ml, but the presence of an active promoter in pMM225 increased the level of resistance to >1,000 mg of kanamycin per ml. Thus, vectors pMM223 and pMM225 allowed selection of active promoters.
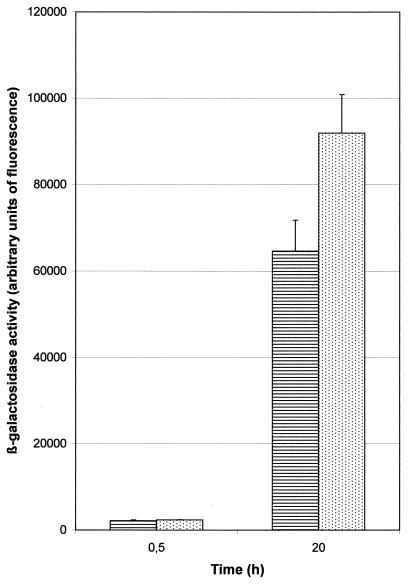
Subsequently, we evaluated the functionality of the promoterless lacZ reporter gene. In E. coli, promoter activity could easily be detected by plating the organism on agar plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) per ml as a substrate. In S. gordonii CH1, the presence of an active promoter could not be discriminated using either agar plates or agar overlays containing up to 500 μg of X-Gal per ml (1). All strains, carrying vectors with or without active promoters, produced blue colonies after 2 to 3 days of incubation. In liquid assays using FDG the presence of the rgg-gtfG promoter on plasmid pMM239 could be detected in S. gordonii CH1 after 20 h of incubation, despite the endogenous β-galactosidase activity (Fig. 3). Although addition of 2% glucose to the growth medium reduced this endogenous activity, it did not improve the ability to detect an active promoter (data not shown).
FIG. 3.
Assessment of β-galactosidase activity in S. gordonii CH1 harboring either plasmid pMM223 (no promoter [hatched bars]) or pMM239 (active rgg-gtfG promoter [dotted bars]) using the fluorescent substrate FDG.
Selection of pH-regulated promoters of S. gordonii CH1.
A genomic library of S. gordonii CH1 was constructed in pMM223 by using E. coli as an intermediate host, since direct transformation of ligation mixtures to S. gordonii CH1 resulted in low numbers of recombinant clones. We used pMM223 because the background resistance of S. gordonii CH1 to spectinomycin was very low (see above). The final streptococcal genomic library contained approximately 105 independent clones, with an average insert size of approximately 500 bp. Statistically, this library represents the entire genome of S. gordonii CH1 (39).
VS causing IE translocate from the oral plaque to the blood, and the bacteria are exposed to a pH shift from pH 6.0 to 6.5 (33) to pH 7.3 to 7.4. Therefore, we selected promoters which were upregulated at pH 7.3. Of 143 individual clones that grew on TH agar of pH 7.3 supplemented with 500 μg of spectinomycin per ml, 5 clones were identified which hardly grew on TH agar of pH 6.2 when spectinomycin was present. The pH of the medium influenced the activity of β-galactosidase (data not shown), hampering assessment of relative promoter activity. Therefore, the promoter activities of one of these clones [CH1(pMM243] at pH 6.2 and 7.3 were further tested by determination of the in vitro growth rate in the presence or absence of spectinomycin. The growth rate was significantly higher at pH 7.3 than at pH 6.2 (0.94 ± 0.01 and 0.69 ± 0.04, respectively [P = 0.0005] as compared to a clone [CH1(pMM240)] that harbors a constitutive streptococcal promoter (0.98 ± 0.01 and 0.95 ± 0.01 at pH 7.3 and pH 6.2, respectively), demonstrating the upregulation of the pH-regulated promoter at pH 7.3.
Sequencing of the pH-regulated promoter fragment and comparison to GenBank sequences revealed homology to the promoter region of the hydA gene of Clostridium acetobutylicum (Fig. 4). The −35 and −10 regions were identified at almost identical positions in the two sequences. The extended −10 sequence present in the clostridial hydA promoter region (5′-AatATga-3′) (lowercase letters represent nucleotides that were not conserved in the extended −10 region)(54) was found at the same position upstream of the −10 box of the streptococcal promoter. The inverted repeat sequences present upstream and downstream of the Clostridium promoter stretch, involved in catabolite repression (13, 16) and repression of the transcription of hydA (16, 51), respectively, were, however, not identified in the streptococcal sequence. A possible Shine-Dalgarno sequence and translation start were found (Fig. 4). The cloned fragment contained only the sequence encoding the first 24 amino acids of the putative coding region following this translation start. This sequence did not show homology to any entry in the GenBank database or to the partly sequenced genomes of S. mutans, Streptococcus pneumoniae, and Streptococcus pyogenes.
FIG. 4.
Nucleotide sequence of the isolated pH-regulated promoter fragment from S. gordonii CH1(pMM243) and homology to the promoter region of hydA of C. acetobutylicum ATCC 824. The −35 and −10 boxes are underlined, and the extended −10 region, as described for C. acetobutylicum (54), is printed in italics. The inverted repeat sequences in the C. acetobutylicum sequence upstream and downstream of the promoter region are indicated by asterisks. A putative Shine-Dalgarno (SD) sequence and translational start site in the streptococcal sequence are printed in bold. The translated amino acid sequence of the putative open reading frame is shown.
DISCUSSION
Apart from being important colonizers of the oral cavity and upper pharyngeal tract in humans, VS are recognized as the most common bacterial agents causing native valve IE (10, 40). Little information is available regarding the adaptive potential of this group of microorganisms, either in their natural habitat or during the pathogenesis of IE. Of techniques to study adaptive gene expression (38), the IVET system is most suited for selection of promoters of genes induced under specific in vitro conditions or in the complex in vivo environment (28). In its original design, the IVET system used integration of a nonreplicative vector into genomic DNA via a single crossover. This approach requires high frequencies of transformation, which are difficult to achieve in many gram-positive bacteria. In addition, although the IVET system was originally developed to not disrupt any bacterial genes (28), integration of a nonreplicative vector containing a fragment without a promoter may still cause mutations. We therefore developed an IVET-based selection-reporter system using self-replicating plasmids to study inducible genes in VS. Promoterless genes conferring resistance to kanamycin (aphIII) and spectinomycin [aad(9)] were chosen for the selection of active promoters, as these antibiotics are bactericidal for our VS (data not shown). The use of bacteriostatic antibiotics and associated resistance genes [20; Kiliç et al., Abstr. ASM Conf. Streptococcal Genet. (Genet. Streptococci, Enterococci, Lactococci)] may result in erroneous selection of surviving bacteria that do not have a cloned active promoter. Plasmids pMM223 and pMM225 carry the replication origin (ori) of the lactococcal plasmid pWV01 (three to five copies per cell in lactococci, streptococci, and Bacillus subtilis) (26). Because of their low copy number, these selection vectors are expected not to interfere significantly with regulation of gene expression in these gram-positive bacteria.
VS generally are refractory to electrotransformation. Weakening of cell walls by various methods, which increased transformation frequencies for several gram-positive bacteria (6, 22, 35, 36, 42), did not improve the transformation frequencies of our VS strains. With the optimized electroporation protocol, transformation frequencies appeared to increase almost linearly with increasing field strength and were maximal at 25 kV/cm (Fig. 2). At this maximum attainable field strength, only approximately 30% of the competent VS were killed. For E. coli, maximal transformation frequencies are obtained when 50 to 75% of the cells are killed due to the electrical discharge (11). Thus, even higher transformation frequencies might be possible with equipment capable of generating higher field strengths. S. gordonii CH1 and S. sanguis U108 had the highest transformation frequencies of the 15 strains tested. Plasmid size and possibly the plasmid-borne genes appeared to influence transformation efficiencies, as pMM223 and pMM225 had lower frequencies than pGKV210 in both S. sanguis U108 and S. gordonii CH1. Electroporation was superior to natural transformation of S. gordonii CH1 (Table 4), and frequencies were sufficiently high to construct a representative genomic bank.
Active promoters of S. gordonii CH1 could be detected using the promoterless spectinomycin resistance gene of pMM223 and the kanamycin resistance gene of pMM225. Detection of the induced expression of the promoterless trp′-′lacZ in these plasmids proved to be more difficult in our test strains, although this reporter gene has successfully been used in the closely related S. pneumoniae (5). S. gordonii CH1 harboring pMM239, which carries the active rgg-gtfG promoter fragment (46), could not be discriminated from S. gordonii CH1 carrying pMM223 without a promoter, either on X-Gal-containing plates or in agar overlays (1). Using the fluorescent substrate FDG, elevated β-galactosidase levels in lysates of bacterial clones harboring pMM239 could be recorded after 20 h of incubation (Fig. 3). Although the β-galactosidase expression of S. gordonii CH1 was subject to catabolite repression, as demonstrated by reduction of expression in the presence of 2% glucose, glucose did not improve discrimination between the presence and absence of an active promoter. The use of a streptococcal mutant with lower endogenous β-galactosidase activity, as described for S. pneumoniae (1), could be a suitable alternative. Secondly, the E. coli lacZ gene might be replaced by the recently described β-galactosidase gene of Bacillus stearothermophilus [Poyart and Trieu-Cuot, Abstr. ASM Conf. Streptococcal Genet. (Genet. Streptococci, Enterococci, Lactococci)].
From an S. gordonii CH1 genomic library constructed in vector pMM223 a promoter fragment was isolated whose activity was upregulated by a shift from oral plaque pH (6.2) to blood pH (7.3). Part of this fragment showed homology to the promoter region of the hydA gene of C. acetobutylicum ATCC 824 (16), which encodes a putative hydrogenase with strong homology to the [Fe] hydrogenases from Desulfovibrio and other Clostridium species. The expression of C. acetobutylicum hydA is downregulated at the transcriptional level by a decrease in environmental pH (16). A more detailed study on the putative streptococcal promoter would be required to confirm its localization on the isolated fragment and determine the mode of transcription regulation. However, the activity of the promoter on the isolated fragment was clearly regulated by the environmental pH, showing that our selection method allows isolation of promoters responding to environmental changes.
In their study using an IVET chromosomal integration system, Kiliç and coworkers selected 13 inducible promoters from S. gordonii V288 in a rabbit model of IE. The genes controlled by the isolated promoters encode proteins involved in different cellular functions, including rapid bacterial growth and resistance to host defense [20; Kiliç et al., Abstr. ASM Conf. Streptococcal Genet. (Genet. Streptococci, Enterococci, Lactococci)]. A hydA promoter homolog as identified in our study was not among these selected promoters. The environmental stimuli responsible for induction of the identified inducible genes are still unknown.
To gain a more complete understanding of the regulation of expression of in vivo-induced genes, and of other virulence genes, it is crucial to identify the stimuli responsible for their induction. To our knowledge this report is the first one on the isolation of a VS promoter sequence whose activity is directly influenced by a shift from oral to blood pH. Such a stimulus might also be an important signal for gene induction in other bacterial species. We are presently studying whether the expression of more VS genes is influenced in response to this pH change and whether this response involves a common regulatory mechanism. These insights will possibly also contribute to a better understanding of the pathogenesis of IE.
ACKNOWLEDGMENTS
We thank Hilde Smith (ID-DLO, Lelystad, The Netherlands) for plasmid pORI19S, Elizabeth Campbell (Rockefeller University, New York, N.Y.) for plasmid pEC2A, and Horst Malke (Friedlich-Schiller-Universität, Jena, Germany) for providing the bidirectional transcriptional terminator from S. equisimilis H46A. We also thank Hilde Dijstelbloem for statistical analyses and Martine van Vugt for critically reading the manuscript.
REFERENCES
- 1.Alloing G, Granadel C, Morrison D A, Claverys J-P. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol Microbiol. 1996;21:471–478. doi: 10.1111/j.1365-2958.1996.tb02556.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E F, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Baddour L M. Virulence factors among gram-positive bacteria in experimental endocarditis. Infect Immun. 1994;62:2143–2148. doi: 10.1128/iai.62.6.2143-2148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden G H, Hamilton I R. Survival of oral bacteria. Crit Rev Oral Biol Med. 1998;9:54–85. doi: 10.1177/10454411980090010401. [DOI] [PubMed] [Google Scholar]
- 5.Campbell E A, Choi S Y, Masure H R. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol. 1998;27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 6.Chassy B M. A gentle method for the lysis of oral streptococci. Biochem Biophys Res Commun. 1976;68:603–608. doi: 10.1016/0006-291x(76)91188-8. [DOI] [PubMed] [Google Scholar]
- 7.Dall L H, Herndon B L. Association of cell-adherent glycocalyx and endocarditis production by viridans group streptococci. J Clin Microbiol. 1990;28:1698–1700. doi: 10.1128/jcm.28.8.1698-1700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dankert J, van der Werff J, Zaat S A J, Joldersma W, Klein D, Hess J. Involvement of bactericidal factors from thrombin-stimulated platelets in clearance of adherent viridans streptococci in experimental infective endocarditis. Infect Immun. 1995;63:663–671. doi: 10.1128/iai.63.2.663-671.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas C W I, Heath J, Hampton K K, Preston F E. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol. 1993;39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 11.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durack D T, Beeson P B. Experimental bacterial endocarditis. I. Colonization of a sterile vegetation. Br J Exp Pathol. 1972;53:44–49. [PMC free article] [PubMed] [Google Scholar]
- 13.Ebright R H, Kolb A, Buc H, Kunkel T A, Krakow J S, Beckwith J. Role of glutamic acid-181 in DNA-sequence recognition by the catabolic gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc Natl Acad Sci USA. 1987;84:6083–6087. doi: 10.1073/pnas.84.17.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans P D, Cook C N, Riggs P D, Noren C J. LITMUS; multipurpose cloning vectors with a novel system for bidirectional in vitro transcription. BioTechniques. 1995;19:130–135. [PubMed] [Google Scholar]
- 15.Frandsen E V G, Pedrazzoli V, Kilian M. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol. 1991;6:129–133. doi: 10.1111/j.1399-302x.1991.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 16.Gorwa M-F, Croux C, Soucaille P. Molecular characterization and transcriptional analysis of the putative hydrogenase gene of Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:2668–2675. doi: 10.1128/jb.178.9.2668-2675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross R. Signal transduction and virulence regulation in human and animal pathogens. FEMS Microbiol Rev. 1993;10:301–326. doi: 10.1111/j.1574-6968.1993.tb05873.x. [DOI] [PubMed] [Google Scholar]
- 18.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herzberg M C. Platelet-streptococcal interactions in endocarditis. Crit Rev Oral Biol Med. 1996;7:222–236. doi: 10.1177/10454411960070030201. [DOI] [PubMed] [Google Scholar]
- 20.Herzberg M C, Meyer M W, Kiliç A O, Tao L. Host-pathogen interactions in bacterial endocarditis: streptococcal virulence in the host. Adv Dent Res. 1997;11:69–74. doi: 10.1177/08959374970110011901. [DOI] [PubMed] [Google Scholar]
- 21.Hohn B. In vitro packaging of λ and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- 22.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayaraman G C, Penders J E, Burne R A. Transcriptional analysis of the Streptococcus mutans hcrA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x. [DOI] [PubMed] [Google Scholar]
- 24.Jenkinson H F. Novobiocin-resistant mutants of Streptococcus sanguis with reduced cell hydrophobicity and defective in coaggregation. J Gen Microbiol. 1987;133:1909–1918. doi: 10.1099/00221287-133-7-1909. [DOI] [PubMed] [Google Scholar]
- 25.Johnson C M. Adherence events in the pathogenesis of infective endocarditis. Infect Dis Clin N Am. 1993;7:21–36. [PubMed] [Google Scholar]
- 26.Kok J, van der Vossen J M B M, Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984;48:726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeBlanc D J, Lee L N, Inamine J M. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 29.Marsh J L, Erfle M, Wykes E J. The pIC plasmids and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 30.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munro C L, Macrina F L. Sucrose-derived exopolysaccharides of Streptococcus mutans V403 contribute to infectivity in endocarditis. Mol Microbiol. 1993;8:133–142. doi: 10.1111/j.1365-2958.1993.tb01210.x. [DOI] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved standard M7-A4. 4th edition. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 33.Nolte W A. Defense mechanisms of the mouth. In: Nolte W A, editor. Oral microbiology. St. Louis, Mo: The C. V. Mosby Company; 1982. pp. 245–260. [Google Scholar]
- 34.Ohse M, Takahashi K, Kadowaki Y, Kusaoke H. Effects of plasmid DNA sizes and several other factors on transformation of Bacillus subtilis ISW1214 with plasmid DNA by electroporation. Biosci Biotechnol Biochem. 1995;59:1433–1437. doi: 10.1271/bbb.59.1433. [DOI] [PubMed] [Google Scholar]
- 35.Park S F, Stewart G S. High efficiency transformation of Listeria monocytogenes by electroporation of penicillin treated cells. Gene. 1990;94:129–132. doi: 10.1016/0378-1119(90)90479-b. [DOI] [PubMed] [Google Scholar]
- 36.Powell I B, Achen M G, Hillier A J, Davidson B E. A simple and rapid method for genetic transformation of lactic streptococci by electroporation. Appl Environ Microbiol. 1988;54:655–660. doi: 10.1128/aem.54.3.655-660.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pozzi G, Musmanno R A, Lievens P M-J, Oggioni M R, Plevani P, Manganelli R. Method and parameters for genetic transformation of Streptococcus sanguis Challis. Res Microbiol. 1990;141:659–670. doi: 10.1016/0923-2508(90)90060-4. [DOI] [PubMed] [Google Scholar]
- 38.Quinn F D, Newman G W, King C H. In search of virulence factors of human bacterial disease. Trends Microbiol. 1997;5:20–26. doi: 10.1016/S0966-842X(97)81770-2. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sandre R M, Shafran S D. Infective endocarditis: review of 135 cases over 9 years. Clin Infect Dis. 1996;22:276–286. doi: 10.1093/clinids/22.2.276. [DOI] [PubMed] [Google Scholar]
- 41.Scheld W M, Valone J A, Sande M A. Bacterial adherence in the pathogenesis of endocarditis. Interaction of bacterial dextran, platelets, and fibrin. J Clin Investig. 1978;61:1394–1404. doi: 10.1172/JCI109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott P T, Rood J I. Electroporation-mediated transformation of lystaphin-treated Clostridium perfringens. Gene. 1989;82:327–333. doi: 10.1016/0378-1119(89)90059-0. [DOI] [PubMed] [Google Scholar]
- 43.Smith H. What happens in vivo to bacterial pathogens. Ann N Y Acad Sci. 1996;797:77–92. doi: 10.1111/j.1749-6632.1996.tb52951.x. [DOI] [PubMed] [Google Scholar]
- 44.Smith H E, Wisselink H J, Vecht U, Gielkens A L J, Smits M A. High-efficiency transformation and gene inactivation in Streptococcus suis type 2. Microbiology. 1995;141:181–188. doi: 10.1099/00221287-141-1-181. [DOI] [PubMed] [Google Scholar]
- 45.Steiner K, Malke H. Transcription termination of the streptokinase gene of Streptococcus equisimilis H46A: bidirectionality and efficiency in homologous and heterologous hosts. Mol Gen Genet. 1995;246:374–380. doi: 10.1007/BF00288611. [DOI] [PubMed] [Google Scholar]
- 46.Sulavik M C, Tardif G, Clewell D B. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J Bacteriol. 1992;174:3577–3586. doi: 10.1128/jb.174.11.3577-3586.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullam P M. Host-pathogen interactions in the development of bacterial endocarditis. Curr Opin Infect Dis. 1994;7:304–309. [Google Scholar]
- 48.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 49.Van de Guchte M, van der Vossen J M B M, Kok J, Venema G. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:224–228. doi: 10.1128/aem.55.1.224-228.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Vossen J M B M, Kok J, Venema G. Construction of cloning, promoter-screening, and terminator-screening shuttle vectors for Bacillus subtilis and Streptococcus lactis. Appl Environ Microbiol. 1985;50:540–542. doi: 10.1128/aem.50.2.540-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voordouw G, Brenner S. Nucleotide sequence of the gene encoding the hydrogenase from Desulfovibrio vulgaris (Hildenborough) Eur J Biochem. 1985;148:515–520. doi: 10.1111/j.1432-1033.1985.tb08869.x. [DOI] [PubMed] [Google Scholar]
- 52.Vriesema A J M, Zaat S A J, Dankert J. A simple procedure for isolation of cloning vectors and endogenous plasmids from viridans group streptococci and Staphylococcus aureus. Appl Environ Microbiol. 1996;62:3527–3529. doi: 10.1128/aem.62.9.3527-3529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wells V D, Munro C L, Sulavik M C, Clewell D B, Macrina F L. Infectivity of a glucan synthesis-defective mutant of Streptococcus gordonii (Challis) in a rat endocarditis model. FEMS Microbiol Lett. 1993;112:301–306. doi: 10.1111/j.1574-6968.1993.tb06466.x. [DOI] [PubMed] [Google Scholar]
- 54.Young M, Minton N P, Staudenbauer W L. Recent advances in the genetics of clostridia. FEMS Microbiol Rev. 1989;63:301–326. doi: 10.1111/j.1574-6968.1989.tb03402.x. [DOI] [PubMed] [Google Scholar]