Abstract
To realize the goals of precision medicine in complex disease, discriminative clinical risk models are needed. One approach that has been proposed is polygenic risk scores (PRSs). PRSs incorporate information about inherited genetic risk for cancer, specifically those genetic variants that are common in the population. While PRSs are clearly associated with risk of cancer, there is an on-going debate on whether integrating PRSs into clinical practice have utility. Here, we present this important discussion to the cancer clinic. We argue that in cancer, the clinical utility of PRSs will depend on their actionability, or how such a score may guide clinical practice. In turn, the actionability depends on several factors. First, actionability depends on the discriminative power of the score, or how well it predicts who is at risk of the disease. Second, it depends on their comparative performance with respect to existing practice, as a score with good discriminative power will not be useful if there are better predictors used in the clinic. Finally, for a PRS to be useful there must also be available preventive actions. We discuss the strengths and challenges of utilizing a PRS in the context of each of these criteria, and provide insights on what is needed towards moving forward in translating PRSs into the cancer clinic. We further argue that in future studies, beyond predicting cancer risk, similarly developed PRS models may be of utility in predicting prognosis or treatment resistance.
Keywords: Polygenic risk, germline risk, cancer predisposition, cancer risk, polygenic risk score (PRS), genetic score, precision prevention, cancer screening
Introduction
To achieve precision cancer prevention across the lifespan, we need to predict personal risk for cancer. In risk prediction for complex diseases, there is a growing interest in utilizing polygenic risk scores (PRSs) both in clinical practice and population screening. Simply defined, a PRS is a sum of an individual’s germline genetic risk alleles for a trait, weighted by the allele’s corresponding effect sizes as estimated from a genome-wide association study (GWAS). It therefore provides a single score that summarizes an individual’s genetic risk of developing the disease. To construct a PRS, approaches vary from including only those variants that surpass stringent GWAS thresholds, to inclusion of all genome-wide variants. Several recent studies have utilized PRSs to identify high-risk individuals for complex disease (1-4), which have motivated calls for their integration into clinical practice (5,6). However, as others have described in detail, a strong statistical association between a PRS and disease does not mean that a score can easily discriminate between healthy individuals and those with the disease in question; thus translation of such scores into clinically relevant prediction models is a challenge (7,8).
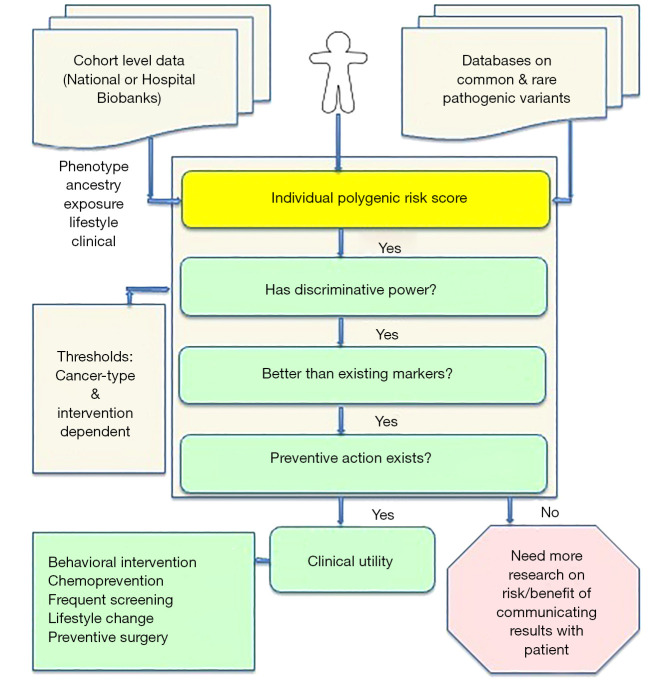
Here, we discuss the clinical utility of cancer PRSs for predicting personalized cancer risk. We argue that the clinical actions that can be taken in response to a PRS, or its actionability, determine its clinical utility. This actionability, in turn, depends on three aspects: (I) its discriminative power; (II) its performance in comparison to existing known risk factors; and (III) available preventive actions (see Figure 1). We describe the strengths and challenges in each of these aspects for utilizing a PRS, as well as what is still needed to integrate cancer PRSs to the clinic. Furthermore, we argue that the clinical utility of PRSs may not be limited to predicting risk in precision prevention for healthy individuals: there is also great potential for building and using PRSs in precision oncology for personal prediction of disease progression or drug resistance in individuals with cancer.
Figure 1.
Criteria for utilizing personal polygenic risk scores (PRSs) in the cancer clinic.
Discriminative power
In predicting germline genetic risk for complex disease, a PRS can only be useful if it adequately distinguishes between people at high and low risk. Just as numerous studies have shown that highly penetrant single germline rare pathogenic variants can lead to familial cancer syndromes that span cancers in multiple tissues, it is plausible to build PRSs that asses shared etiology across multiple cancer types (9-12). However, germline genetic factors that impact cancer are typically tissue-dependent (13) and current cancer PRSs are typically cancer-site specific. Thus, we consider here only site-specific cancer PRSs.
A PRS can bring added value to an existing cancer risk model in the clinic if it presents information independent of established clinical, environmental or lifestyle risk factors, and therefore improves the predictive power more than incrementally. Early reports on large-scale national biobanks support this. Data from the Finnish FinnGen biobank study has shown that PRSs provide additional value in clinical prediction of breast and prostate cancer risk (14). Similarly, in a study on germline genetic risk for 16 different cancers using UK BioBank data, PRSs have presented lower added value for cancers with strong modifiable risk factors (e.g., lung cancer risk in smokers), but higher in those without them (e.g., lung cancer risk in never-smokers) (15), with the greatest benefit for prostate, testicular and thyroid cancers, as well as leukemias and melanomas (15). It is also worth noting that a PRS can still be used if there is some overlap with existing factors. For example, the CanRisk model attenuates the PRS score in the context of a family history of breast cancer (16,17).
In reporting the predictive power of a cancer PRS, the standard epidemiological literature should be consulted to consider the relative merits of different reporting metrics, as these essentially have the same issues as for any predictor of disease or trait. Typically, a regression model is performed on the target sample, with the PRS as a predictor of the target trait or outcome, and covariates are included as appropriate. The metric that is most sensible to use is based on the context and the question being asked. To make quantifying the difference with other published studies more consistent, most studies use the incremental R2, where the effect of the PRS is separate from the effect of other covariates. However, if the model includes many covariates, then the incremental R2 may be higher because those other covariates will have explained a good fraction of the trait variance and thus some caution in this regard is warranted. For example, when a popular measure, the Net Reclassification Index (NRI) (18) is calculated on a large test dataset, it is likely to be positive even when the addition of the PRS to an existing model has no predictive information. A good discussion on measurements of PRS analysis results, plots, interpretation, predictive accuracy and power, as well as avoidance of over-fitting is provided in a recent study by Choi et al. [2020] (19).
In considering for which cancers a PRS may best discriminate germline genetic risk, the heritability of a cancer provides a natural upper limit to what can be solely achieved by a PRS. Twin studies may guide which cancers have sufficient genetic factors that a PRS could be of potential clinical utility (20,21). Note that this natural upper limit is higher than what can be estimated from current genome-wide association studies of cancer risk (22), as it includes yet to be resolved genetic associations such as the inclusion of rare variants. However, even for cancers where heritability is lower and modifiable risk factors are dominant, such as in lung cancer, a PRS can present clinical utility, as it could be used to modify screening guidelines. For instance, high PRS individuals could be added to screening protocols at younger ages than the standard recommendations, and screened more frequently, potentially saving lives (23).
One drawback to current PRSs is that their development is largely derived from individuals of European ancestry. This limits their applicability (24) and precision (25) for non-European populations and also exacerbates health inequalities. While genetic risk patterns are broadly similar across major population groups, and many common risk alleles discovered in one population group are detectable in others, allele frequencies and linkage disequilibrium (LD) may vary substantially. Consequently, how PRS information translates across various ancestries with different background genetic architecture—both in terms of allele frequencies, LD, and effect sizes—is critical (25,26). Genotyping will soon become widespread in high- and middle-income countries, further contributing to these health disparities. Including individuals of non-European ancestry in building PRSs not only reduces health disparities, but also generates better PRSs for individuals of European descent as well (27). It is notable that a recent paper on prostate cancer substantially improved risk prediction in non-European populations by including a modest fraction of non-Europeans (28). To better account for and utilize ancestry information, we need more diverse population allele frequency databases (29,30) disease association studies with other ancestral backgrounds, and new computational approaches (31).
Potential utility of PRS in practice
The practical clinical utility of a PRS that predicts personal cancer risk depends on how it compares to standard of care and existing risk prediction approaches rather than a null model of no risk prediction. We assume the PRS will be measured by the germline genetic analysis of a single blood draw. From a cost perspective, even if whole genome sequencing (WGS) of every patient is necessary for the cancer PRS, this will be performed only once in a person’s lifetime. As genetic sequencing becomes more common, these types of sequencing data will become available for screening for many different health outcomes. Hence, from the health system perspective, the cost will not be a major factor in utilizing PRSs at population level. We are already seeing efforts to that effect in research enterprises such as the Million Veteran Program (MVP) that plans to sequence a million veterans (32); national-level studies including the UK Biobank (33,34) and hospital-level efforts such as the BioMe Biobank of Icahn School of Medicine at Mount Sinai (35).
If the PRS were to be utilized to risk-stratify individuals for altered screening regimens, its utility would be guided by the relative expense and invasiveness of current screening modalities. For instance, in breast cancer, a PRS could theoretically be used to guide recommendations of frequency of mammography, which is a more involved procedure than a blood draw. In contrast, in prostate cancer screening, the existing standard of care when screening is desired is the prostate specific antigen (PSA) blood test. It has even been proposed that a single protein-based blood test at mid-life can be a predictor of clinically important prostate cancer in a man’s lifetime (36,37). Beyond comparison to existing screening and risk prediction approaches, care must be given to associations between elements of the PRS and other factors used in risk prediction. For instance, while a naïve PRS for lung cancer risk would include variants associated with smoking behavior (38), the performance of a PRS that includes such variants would need to be first carefully compared to those of existing clinical models that are based on asking individuals about their personal smoking history.
Available preventive actions
Finally, for a cancer risk PRS to be useful there must be some clinical action that can be taken in response to knowledge. In other words, it should lead to prevention (5). Similar to testing of single genes in monogenic cancer predisposition syndromes, a PRS may be able to identify a subset of high-risk individuals who would benefit from existing preventative actions. The thresholds of positive and negative predictive values for such a test will naturally depend on how invasive the preventive interventions being considered are. For example, currently in breast cancer prevention, the existence of a highly penetrant BRCA1/2 variant, together with other clinical factors, family history and patient preferences guide radical action such as double mastectomy. If a PRS is to be utilized for such radical preventive action with or without a BRCA1/2 variant, its accuracy thresholds need to be extremely stringent. However, less stringent thresholds may be considered for those individuals at high breast cancer risk but are instead only considering chemoprevention with a daily intake of selective estrogen receptor modulators (SERMs) or aromatase inhibitors. Furthermore, those at high-risk for colon cancer based on their PRS may simply undergo frequent screening and take a daily aspirin, affording even less stringent thresholds (39).
PRSs can also be useful in the context of modifiable environmental causes of cancer. For example, while smoking is a modifiable risk factor for lung cancer, most smokers are unable to quit. However, individuals are starting to access their complete genomic information (and risk) at ever-younger ages, which we anticipate will shift clinical utility from modifying established risky behaviors to prevention. Further research is needed in this area, on for example whether a high PRS for lung cancer will impact young never-smokers’ future smoking decisions, shifting focus from supporting smoking cessation to smoking prevention on high PRS individuals before they even start smoking.
Research is also needed on the impact of cancer PRS information on possibly altering the modifiable behaviors of individuals. There is already research in this direction in other complex diseases. For example, in coronary heart disease research, a smart phone app that calculates people’s PRSs from their combined genetic profile, mobile health and questionnaire data is being used to monitor changes in their health-related behaviors afterwards (mygenerank.scripps.edu). Similar studies and user-friendly technologies in the cancer domain are needed to better understand individual responses to high cancer risk PRS information.
So far, studies suggest that when individuals are confronted with their own genetic risk data, their preventive behavioral responses are impacted by multilevel sociocultural and policy-related factors (40). While limited, studies on communicating DNA-based disease risk estimates have reported little or no effect on smoking or physical activity, and a small effect on self-reported diet and on intentions to change behavior in the short term (41,42). However, users of direct-to-consumer of genetic testing services (who tend to self-select for individuals of higher educational levels), were reported to adopt long-term changes into healthier diets (43). In addition, a recent personalized genetically informed risk tool showed to promote progress towards smoking cessation (44). Furthermore, a recent melanoma clinical trial reported that while personal genomic risk information did not influence sun exposure patterns, it did improve some skin cancer prevention and early detection behaviors (45). Finally, multiple studies have reported that personal risk information is potentially useful for shared and informed decision-making in the clinic (46), and that individuals at high cancer risk tend to take preventive action. Of note, women’s health behavior tended to change after receiving breast cancer risk estimates with tailored screening and prevention recommendations, highlighting the importance of effective risk communication (47).
In the absence of concrete actions that can be taken, there is a debate as to whether knowledge of individual risk is useful. While some individuals still prefer to know their risk for lethal disease to prepare for treatment and to understand the potential impact of disease on their future (48), just having the knowledge without available actions can be harmful in certain cases (49). We need to develop guidelines on how best to educate individuals and the medical community on what a PRS is: that a high score does not mean an individual will definitely develop cancer, and a low score does not mean freedom from disease; and that all individuals should adopt healthy habits such as eating less, exercising more, or quitting smoking, regardless of their personal PRS. We will need considerable education of both the public and medical community on the interpretation of PRS results, and to integrate the patients into the discussions, whenever a risk score is to be given to an individual.
Are we ready?
Though PRSs show much promise, several challenges remain. First, building a PRS requires accessible data on a large number of individuals that exhibit the phenotype. For most cancers, germline genetic risk is shared across thousands of mostly common variants with individually modest effects on population risk. The largest current resources for building a PRS include national-level biobanks such as UK Biobank (though individuals in this databank are healthier than average) (33); all of Us Research Program (50); Million Veteran Program (32); NHLBI The Trans-Omics for Precision Medicine (TOPMed) program (51), as well as individual hospital system biobanks integrated with electronic health record (EHR) data (35). Thus, PRSs built for common cancers with GWAS data in large cohorts are likely to be more ready for the clinic. However, as these cohorts have relatively few individuals with specific rare cancers compared to dedicated case-control studies, PRSs for such rare cancers will need to wait for larger genetic epidemiology studies, though the low frequency of these cancers means the benefit of population-level PRS screening would be reduced as there would be less cases to detect. For example, so far there has been a limited number of genetic epidemiology studies solely devoted to understanding the risk for the relatively rare small cell lung cancers. This problem is exacerbated by the use of ICD billing codes to determine diagnosis in biobanks. For instance, an ICD code will distinguish between a right or a left lung, yet, it will not distinguish between squamous cell carcinoma, adenocarcinoma, or small cell carcinoma of the lung. Heterogeneity between the cohorts is another barrier to pooling data among studies. Similarly, as discussed above, to be broadly applicable, PRSs need to be developed using cohorts of diverse ancestry; the focus of many of the large studies on individuals of European ancestry remains a problem for developing generally applicable PRSs.
Second, current cancer PRSs typically only consider common genetic variants; however, to improve predictive accuracy, we need to integrate variants over the continuous spectrum of risk alleles, across a range of frequencies and effect sizes observed within a population. Notably, rare deleterious variants (RDVs) can play key roles in risk, as they tend to have high penetrance. Multiple studies have identified RDVs in DNA damage repair genes with large or moderate effect sizes in cancer risk (52,53) (e.g., Fanconi Anemia genes in lung squamous cancers; ATM gene in lung adenocarcinomas). Furthermore, variants that impact outcome independent of risk may also improve clinical risk models of lethal cancers. For example, RDVs in DNA repair genes have been shown to increase risk for aggressive prostate cancers (54,55). To incorporate RDVs into PRSs, large population-based sequencing studies for gene detection (56) are needed for their systematic and unbiased functional assessment, in addition to a better understanding of their complex interplay with polygenic background. For example, individuals at highest risk by PRS can yield the equivalent risk of RDVs (5). The polygenic background itself can also impact RDV penetrance (57). For example, it has been shown that individuals with moderately penetrant CHEK2 RDVs can be better advised about their risk in light of their PRSs (58). Thus, PRSs that extend into including RDVs will add value to clinical risk models.
Third, genetic risk often depends on clinical, lifestyle and environmental risk factors, which should be carefully integrated with demographic and clinical factors. For example, biological females clearly will not develop prostate cancer, no matter how high their PRS, and risk for all cancers increases with age. Clearly, there is a benefit for measuring PRS earlier in life, so as to understand modifiable factors to decrease disease risk. This is where additional clinical data from EHR (age, gender, smoking and alcohol history, viral infections, blood tests, etc.) will come into play. More research is needed to understand how best to integrate PRS with existing risk models (17). In addition, benefits of measuring PRS early in life must be carefully balanced against the potential risks of stigmatization and discrimination, as well as implications for parents having this information for their children (59).
Fourth, any meaningful translation of independent research studies into clinical practice will require standardization of all aspects of PRS measurement. PRSs are generally developed from panels of whole-genome SNP genotyping or sequencing which are then processed using statistical imputation approaches to fill in missing data. How such statistical approaches will fit into the regulatory frameworks guiding clinical tests remains an open challenge. Similarly, it is important to report PRS with standardized nomenclature so that there is no confusion as to the identity of the SNPs used in the PRS, direction of effect for each allele, and specific algorithm for summing across the SNPs (i.e., how to deal with negative weights on some alleles). The recent review from the Clinical Genome Resource (ClinGen) Complex Disease Working Group and the Polygenic Score (PGS) Catalog is an important step towards such standardization (60).
Fifth, the clinical success of PRSs will depend on their uptake—both adoption by clinicians and acceptance by patients. To that end, a recent clinical trial on a prostate cancer PRS is informative (61). This study aimed to recruit men, measure them for a prostate cancer PRS, and perform intensive screening (MRI and biopsy) in those men with a PRS in the top decile. Interestingly, only 26% of men invited to participate by letter chose to participate, suggesting that uptake of PRS-based screening may be low. Further studies on this issue will be needed, however, as there is a difference between being invited to participate in a research study by letter and a screening recommendation made by your primary care physician in person.
Finally, with the advent of new technologies, it remains to be seen whether we can improve PRSs based on the incorporation of additional genetic factors, such as those associated with methylation-based “age acceleration”, clonal hematopoiesis and/or telomere length (62), as well as germline human leukocyte antigen (HLA) typing in cancers where these factors play important roles.
Beyond risk prediction—PRS in prognosis and treatment response
Beyond their immediate use in risk prediction and cancer prevention, it is possible that germline genetic information could also be utilized to build PRSs that predict clinically significant end-points such as cancer metastasis, death or treatment response. For example, in prostate cancer, while various PRSs were developed for the endpoint of incident prostate cancer (9,28,63), this is not the endpoint of most clinical interest because many incident prostate cancers are indolent. Thus, a PRS that predicts incident prostate cancer will suffer from the same problem of over-diagnosis as existing prostate cancer screening approaches. In fact, SNPs associated with prostate cancer risk do not appear to correlate with survival or outcome (64,65). In contrast, separate SNPs have been identified that are associated with prostate cancer survival (66). Thus, it would be worth exploring if PRSs could be designed to predict the most clinically significant endpoint in cancers such as prostate cancer where risk-stratifying cases is as important, if not more important, than predicting risk of incident cancer.
In cardiology, studies show that a PRS can have diagnostic utility in identifying individuals who can benefit most from a treatment (67). However, in the precision oncology domain, research is still needed to understand the utility of PRSs in better predicting personal resistance or response to treatment. For this purpose, we would need to build PRSs to match drugs to individuals who are most likely to benefit from them. Briefly, germline genotype data can be pooled from multiple medical centers on participants of a clinical trial for maximum statistical power to build PRSs and to test their predictive ability in stratifying patients that are most likely to resist or respond to a specific treatment. If successful, such approaches would have high impact on clinical trials by focusing on individuals with highest response PRSs to a specific treatment, or by removing those with high resistance PRSs. These can be further combined with additional information such as those from mining EHR databases, to test whether even more powerful treatment response models can be developed. In support of the idea that germline genetic variation can influence treatment response, we note that PARP inhibitors are approved for several cancer indications on the basis of the presence of either a germline variant or a tumor mutation in DNA repair genes.
Conclusions
In summary, PRSs have great potential to play a key role in cancer screening, prevention, early detection, as well as in precision oncology. Their eventual clinical use will depend on the cancer type, other existing risk markers, clinical parameters, and available preventive actions. Towards this end, beyond continuing to genotype large numbers of individuals at population or hospital-scale for all cancer types and ancestries, we need to focus research on considering where PRS may be most discriminative and impactful at the population level, which will greatly aid in the development of new methods to overcome the challenges described here.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors thank Paul O’Reilly and Kenan Onel for careful reviews of the manuscript.
Funding: This work was supported by the LUNGevity foundation and the National Institutes of Health (No. R33 CA263705 to Z.H.G and No. R01 CA244948 to R.J.K).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-698/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-698/coif). RJK reports that this work was supported by a grant from the National Institutes of Health (No. R01 CA244948). ZHG reports that this work was supported by a grant from the LUNGevity foundation (No. R33 CA263705). The authors have no other conflicts of interest to declare.
References
- 1.Aragam KG, Chaffin M, Hindy G, et al. Clinical characteristics and treatment patterns of patients at high polygenic risk for coronary artery disease. J Am Coll Cardiol 2019. [Epub ahead of print]. doi: . 10.1016/S0735-1097(19)33681-2 [DOI] [Google Scholar]
- 2.Yengo L, Sidorenko J, Kemper KE, et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641-9. 10.1093/hmg/ddy271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inouye M, Abraham G, Nelson CP, et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J Am Coll Cardiol 2018;72:1883-93. 10.1016/j.jacc.2018.07.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219-24. 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 2018;19:581-90. 10.1038/s41576-018-0018-x [DOI] [PubMed] [Google Scholar]
- 6.Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet 2019;28:R133-42. 10.1093/hmg/ddz187 [DOI] [PubMed] [Google Scholar]
- 7.Ala-Korpela M, Holmes MV. Polygenic risk scores and the prediction of common diseases. Int J Epidemiol 2020;49:1-3. 10.1093/ije/dyz254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sud A, Turnbull C, Houlston R. Will polygenic risk scores for cancer ever be clinically useful? NPJ Precis Oncol 2021;5:40. 10.1038/s41698-021-00176-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graff RE, Cavazos TB, Thai KK, et al. Cross-cancer evaluation of polygenic risk scores for 16 cancer types in two large cohorts. Nat Commun 2021;12:970. 10.1038/s41467-021-21288-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashkin SR, Graff RE, Kachuri L, et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat Commun 2020;11:4423. 10.1038/s41467-020-18246-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X, Finucane HK, Schumacher FR, et al. Shared heritability and functional enrichment across six solid cancers. Nat Commun 2019;10:431. 10.1038/s41467-018-08054-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sampson JN, Wheeler WA, Yeager M, et al. Analysis of Heritability and Shared Heritability Based on Genome-Wide Association Studies for Thirteen Cancer Types. J Natl Cancer Inst 2015;107:djv279. 10.1093/jnci/djv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.GTEx Consortium . The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020;369:1318-30. 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mars N, Koskela JT, Ripatti P, et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med 2020;26:549-57. 10.1038/s41591-020-0800-0 [DOI] [PubMed] [Google Scholar]
- 15.Kachuri L, Graff RE, Smith-Byrne K, et al. Pan-cancer analysis demonstrates that integrating polygenic risk scores with modifiable risk factors improves risk prediction. Nat Commun 2020;11:6084. 10.1038/s41467-020-19600-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carver T, Hartley S, Lee A, et al. CanRisk Tool-A Web Interface for the Prediction of Breast and Ovarian Cancer Risk and the Likelihood of Carrying Genetic Pathogenic Variants. Cancer Epidemiol Biomarkers Prev 2021;30:469-73. 10.1158/1055-9965.EPI-20-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med 2019;21:1708-18. 10.1038/s41436-018-0406-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pepe MS, Fan J, Feng Z, et al. The Net Reclassification Index (NRI): a Misleading Measure of Prediction Improvement Even with Independent Test Data Sets. Stat Biosci 2015;7:282-95. 10.1007/s12561-014-9118-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi SW, Mak TS, O'Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc 2020;15:2759-72. 10.1038/s41596-020-0353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 2000;343:78-85. 10.1056/NEJM200007133430201 [DOI] [PubMed] [Google Scholar]
- 21.Mucci LA, Hjelmborg JB, Harris JR, et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA 2016;315:68-76. 10.1001/jama.2015.17703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang YD, Hurson AN, Zhang H, et al. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat Commun 2020;11:3353. 10.1038/s41467-020-16483-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung RJ, Warkentin MT, Brhane Y, et al. Assessing Lung Cancer Absolute Risk Trajectory Based on a Polygenic Risk Model. Cancer Res 2021;81:1607-15. 10.1158/0008-5472.CAN-20-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landry LG, Rehm HL. Association of Racial/Ethnic Categories With the Ability of Genetic Tests to Detect a Cause of Cardiomyopathy. JAMA Cardiol 2018;3:341-5. 10.1001/jamacardio.2017.5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin AR, Kanai M, Kamatani Y, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 2019;51:584-91. 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin AR, Gignoux CR, Walters RK, et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet 2017;100:635-49. 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gettler K, Levantovsky R, Moscati A, et al. Common and Rare Variant Prediction and Penetrance of IBD in a Large, Multi-ethnic, Health System-based Biobank Cohort. Gastroenterology 2021;160:1546-57. 10.1053/j.gastro.2020.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti DV, Darst BF, Moss LC, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet 2021;53:65-75. 10.1038/s41588-020-00748-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434-43. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GenomeAsia100K Consortium. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 2019;576:106-11. 10.1038/s41586-019-1793-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Márquez-Luna C, Loh PR; South Asian Type 2 Diabetes (SAT2D) Consortium, et al. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet Epidemiol 2017;41:811-23. 10.1002/gepi.22083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016;70:214-23. 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed] [Google Scholar]
- 33.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203-9. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abul-Husn NS, Manickam K, Jones LK, et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science 2016;354:aaf7000. 10.1126/science.aaf7000 [DOI] [PubMed] [Google Scholar]
- 36.Sjoberg DD, Vickers AJ, Assel M, et al. Twenty-year Risk of Prostate Cancer Death by Midlife Prostate-specific Antigen and a Panel of Four Kallikrein Markers in a Large Population-based Cohort of Healthy Men. Eur Urol 2018;73:941-8. 10.1016/j.eururo.2018.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vertosick EA, Häggström C, Sjoberg DD, et al. Prespecified 4-Kallikrein Marker Model at Age 50 or 60 for Early Detection of Lethal Prostate Cancer in a Large Population Based Cohort of Asymptomatic Men Followed for 20 Years. J Urol 2020;204:281-8. 10.1097/JU.0000000000001007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen LS, Hung RJ, Baker T, et al. CHRNA5 risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis--a meta-analysis. J Natl Cancer Inst 2015;107:djv100. 10.1093/jnci/djv100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsioulias GJ, Go MF, Rigas B. NSAIDs and Colorectal Cancer Control: Promise and Challenges. Curr Pharmacol Rep 2015;1:295-301. 10.1007/s40495-015-0042-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein WMP, O'Connell ME, Bloch MH, et al. Behavioral Research in Cancer Prevention and Control: Emerging Challenges and Opportunities. J Natl Cancer Inst 2022;114:179-86. 10.1093/jnci/djab139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marteau TM, French DP, Griffin SJ, et al. Effects of communicating DNA-based disease risk estimates on risk-reducing behaviours. Cochrane Database Syst Rev 2010;(10):CD007275. 10.1002/14651858.CD007275.pub2 [DOI] [PubMed] [Google Scholar]
- 42.Hollands GJ, French DP, Griffin SJ, et al. The impact of communicating genetic risks of disease on risk-reducing health behaviour: systematic review with meta-analysis. BMJ 2016;352:i1102. 10.1136/bmj.i1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliveri S, Cincidda C, Ongaro G, et al. What people really change after genetic testing (GT) performed in private labs: results from an Italian study. Eur J Hum Genet 2022;30:62-72. 10.1038/s41431-021-00879-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey AT, Bourdon JL, Bray M, et al. Proof of Concept of a Personalized Genetic Risk Tool to Promote Smoking Cessation: High Acceptability and Reduced Cigarette Smoking. Cancer Prev Res (Phila) 2021;14:253-62. 10.1158/1940-6207.CAPR-20-0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smit AK, Allen M, Beswick B, et al. Impact of personal genomic risk information on melanoma prevention behaviors and psychological outcomes: a randomized controlled trial. Genet Med 2021;23:2394-403. 10.1038/s41436-021-01292-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards AG, Naik G, Ahmed H, et al. Personalised risk communication for informed decision making about taking screening tests. Cochrane Database Syst Rev 2013;(2):CD001865. 10.1002/14651858.CD001865.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rainey L, van der Waal D, Donnelly LS, et al. Women's health behaviour change after receiving breast cancer risk estimates with tailored screening and prevention recommendations. BMC Cancer 2022;22:69. 10.1186/s12885-022-09174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tibben A. Predictive testing for Huntington’s disease. Brain Research Bulletin 2007;72:165-71. 10.1016/j.brainresbull.2006.10.023 [DOI] [PubMed] [Google Scholar]
- 49.Lerman C, Gold K, Audrain J, et al. Incorporating biomarkers of exposure and genetic susceptibility into smoking cessation treatment: effects on smoking-related cognitions, emotions, and behavior change. Health Psychol 1997;16:87-99. 10.1037/0278-6133.16.1.87 [DOI] [PubMed] [Google Scholar]
- 50.All of Us Research Program Investigators , Denny JC, Rutter JL, et al. The "All of Us" Research Program. N Engl J Med 2019;381:668-76. 10.1056/NEJMsr1809937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taliun D, Harris DN, Kessler MD, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 2021;590:290-9. 10.1038/s41586-021-03205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esai Selvan M, Klein RJ, Gümüş ZH. Rare, Pathogenic Germline Variants in Fanconi Anemia Genes Increase Risk for Squamous Lung Cancer. Clin Cancer Res 2019;25:1517-25. 10.1158/1078-0432.CCR-18-2660 [DOI] [PubMed] [Google Scholar]
- 53.Esai Selvan M, Zauderer MG, Rudin CM, et al. Inherited Rare, Deleterious Variants in ATM Increase Lung Adenocarcinoma Risk. J Thorac Oncol 2020;15:1871-9. 10.1016/j.jtho.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallagher DJ, Gaudet MM, Pal P, et al. Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res 2010;16:2115-21. 10.1158/1078-0432.CCR-09-2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016;375:443-53. 10.1056/NEJMoa1603144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Dhindsa RS, Carss K, et al. Rare variant contribution to human disease in 281,104 UK Biobank exomes. Nature 2021;597:527-32. 10.1038/s41586-021-03855-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fahed AC, Wang M, Homburger JR, et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun 2020;11:3635. 10.1038/s41467-020-17374-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borde J, Ernst C, Wappenschmidt B, et al. Performance of Breast Cancer Polygenic Risk Scores in 760 Female CHEK2 Germline Mutation Carriers. J Natl Cancer Inst 2021;113:893-9. 10.1093/jnci/djaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polygenic Risk Score Task Force of the International Common Disease Alliance . Responsible use of polygenic risk scores in the clinic: potential benefits, risks and gaps. Nat Med 2021;27:1876-84. 10.1038/s41591-021-01549-6 [DOI] [PubMed] [Google Scholar]
- 60.Wand H, Lambert SA, Tamburro C, et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature 2021;591:211-9. 10.1038/s41586-021-03243-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eeles RA, Ni Raghallaigh H, The BARCODE1 Study Group. BARCODE 1: A pilot study investigating the use of genetic profiling to identify men in the general population with the highest risk of prostate cancer to invite for targeted screening. J Clin Oncol 2020;38:abstr 1505.
- 62.Dugué PA, Bassett JK, Wong EM, et al. Biological Aging Measures Based on Blood DNA Methylation and Risk of Cancer: A Prospective Study. JNCI Cancer Spectr 2020;5:pkaa109. 10.1093/jncics/pkaa109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fritsche LG, Patil S, Beesley LJ, et al. Cancer PRSweb: An Online Repository with Polygenic Risk Scores for Major Cancer Traits and Their Evaluation in Two Independent Biobanks. Am J Hum Genet 2020;107:815-36. 10.1016/j.ajhg.2020.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan J, Kopp R, Stratton K, et al. An analysis of the association between prostate cancer risk loci, PSA levels, disease aggressiveness and disease-specific mortality. Br J Cancer 2015;113:166-72. 10.1038/bjc.2015.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gallagher DJ, Vijai J, Cronin AM, et al. Susceptibility loci associated with prostate cancer progression and mortality. Clin Cancer Res 2010;16:2819-32. 10.1158/1078-0432.CCR-10-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li W, Middha M, Bicak M, et al. Genome-wide Scan Identifies Role for AOX1 in Prostate Cancer Survival. Eur Urol 2018;74:710-9. 10.1016/j.eururo.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mega JL, Stitziel NO, Smith JG, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet 2015;385:2264-71. 10.1016/S0140-6736(14)61730-X [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as