Abstract
Peptide nucleic acid (PNA)-mediated PCR clamping (H. Ørum, P. E. Nielsen, M. Egholm, R. H. Berg, O. Buchardt, and C. Stanley, Nucleic Acids Res. 21:5332–5336, 1993) was introduced as a novel procedure to selectively amplify ribosomal DNAs (rDNAs) which are not frequently found in clone libraries generated by standard PCR from complex microbial consortia. Three different PNA molecules were used; two of these molecules (PNA-ALF and PNA-EUB353) overlapped with one of the amplification primers, whereas PNA-1114F hybridized to the middle of the amplified region. Thus, PCR clamping was achieved either by competitive binding between the PNA molecules and the forward or reverse primers (competitive clamping) or by hindering polymerase readthrough (elongation arrest). Gene libraries generated from mixed rDNA templates by using PCR clamping are enriched for clones that do not contain sequences homologous to the appropriate PNA oligomer. This effect of PCR clamping was exploited in the following two ways: (i) analysis of gene libraries generated by PCR clamping with PNA-ALF together with standard libraries reduced the number of clones which had to be analyzed to detect all of the different sequences present in an artificial rDNA mixture; and (ii) PCR clamping with PNA-EUB353 and PNA-1114F was used to selectively recover rDNA sequences which represented recently described phylogenetic groups (NKB19, TM6, cluster related to green nonsulfur bacteria) from an anaerobic, dechlorinating consortium described previously. We concluded that PCR clamping might be a useful supplement to standard PCR amplification in rDNA-based studies of microbial diversity and could be used to selectively recover members of undescribed phylogenetic clusters from complex microbial communities.
Culture-independent analysis of PCR-amplified 16S rRNA gene (rDNA) libraries is a powerful approach for determining the diversity of complex microbial environments (7, 8). By using PCR-primers that target conserved regions of the 16S rDNA sequences can be retrieved both from well-known bacterial or archaeal phyla and from phylogenetic groups represented exclusively by uncultured microorganisms (10).
Here we describe peptide nucleic acid (PNA)-mediated PCR clamping (18) as a novel approach for generating rDNA clone libraries from environmental samples. PNA-mediated PCR clamping relies on the following two unique properties of PNA oligomers: (i) PNA-DNA duplexes generally have greater thermal stability than the corresponding DNA-DNA duplexes (17); and (ii) PNA oligomers are not recognized by DNA polymerases and consequently cannot serve as primers during PCR amplification (18). Bound PNA does not inhibit PCR completely but reduces amplification efficiency. PCR clamping of mixed DNA templates (e.g., total rDNA of a microbial community) inhibits amplification of sequences which are perfectly homologous to the respective PNA oligomer, which results in preferential amplification of sequences with mismatches to the PNA. Thus, PCR clamping introduces a preferential bias to selectively enrich nontarget sequences of a mixed template. By using both an artificial rDNA mixture and natural community rDNA we found that this ability of PCR clamping can supplement standard PCR amplification in rDNA-based studies of microbial diversity.
MATERIALS AND METHODS
Bacterial strains and environmental rDNA templates.
The bacterial strains and cloned rDNAs from an environmental source used as references in PCR clamping experiments are listed in Table 1. Reamplified rDNA inserts were generated from a 16S rDNA library as described previously (23). Full-length 16S rRNA genes of bacterial reference strains were PCR amplified by using primers TPU1 and RTU8 and the following hot-start protocol: initial denaturation at 98°C for 30 s and at 93°C for 2 min, addition of AmpliTaq polymerase (Perkin-Elmer, Weiterstadt, Germany), 25 cycles consisting of denaturation at 93°C for 1 min, primer annealing at 53°C for 1 min, and elongation at 72°C for 2 min, and a final elongation step consisting of 72°C for 7 min. Amplification products were cloned as described below and were reamplified by using vector-specific primers M13(−40)F and M13(−24)R. For reamplification we used the conditions described above except that the hot start was omitted and the initial denaturation step consisted of denaturation at 95°C for 5 min. The resulting PCR amplicons were purified with a silica suspension (5) and Geneclean spin filters (Dianova, Hamburg, Germany). For PCR clamping experiments performed with PNA-EUB353 and PNA-1114F amplified community 16S rDNA was used as the template. The DNA concentrations in all of the rDNA controls were determined spectrophotometrically by using an Ultrospec III photometer (Pharmacia, Freiburg, Germany).
TABLE 1.
Bacterial strains and environmental rDNA clones that were used as controls in PCR clamping experiments
| Organism or clonea | No. of mismatches with PNA oligomer:
|
Accession no.b | ||
|---|---|---|---|---|
| EUB353 | 1114F | ALF | ||
| Verrucomicrobium spinosum DSM 4136T | 2 | 1 | X90515 | |
| Carnobacterium gallinarum DSM 4847T | 2 | AJ387905 | ||
| Pseudomonas sp. strain OLB-1 | 1 | 2 | AJ387904 | |
| Pseudomonas aeruginosa ATCC 25330 | 1 | 2 | M34133 | |
| rDNA clone SJA-22 | 1 | 2 | AJ009456 | |
| rDNA clone SJA-43 | 3 | 2 | AJ009463 | |
| rDNA clone SJA-131 | 2 | 2 | 3 | AJ009492 |
| rDNA clone SJA-4 | 3 | 3 | AJ009448 | |
| rDNA clone SJA-9 | AJ009451 | |||
| rDNA clone SJA-105 | AJ009482 | |||
| rDNA clone SJA-53 | 1 | AJ009467 | ||
| rDNA clone SJA-186 | 2 | AJ009507 | ||
Pseudomonas sp. strain OLB-1 was isolated from an anaerobic, trichlorobenzene-dechlorinating consortium (24), whereas SJA rDNA clones were retrieved from the same community by direct PCR cloning as described previously (23).
For V. spinosum DSM 4136T the sequence corresponding to E. coli 16S rRNA positions 8 to 30 was determined (5′ AGAGUUUGAUCCUGGCUCAGAAC 3′) as this region was not included in the previously published sequence.
Synthesis of PNA.
PNA were synthesized with a model 8900 Expedite nucleic acid synthesizer (PerSeptive Biosystems, Framingham, Mass.) by using an amide resin, Fmoc-protected monomers (Perkin-Elmer), and the protocol recommended by the manufacturer (PNA II method; PerSeptive). The resin was washed with dichloromethane (Merck, Darmstadt, Germany), and the products were deprotected with 80% trifluoroacetic acid (TFA)–20% m-cresol (Merck) for 90 min at room temperature (21°C). The product was precipitated with 6 volumes of ether (Merck), incubated for 10 min at −18°C, and pelleted by centrifugation. The pellet was washed with ether, dried with a Speed Vac apparatus, resuspended in 50 μl of 0.1% TFA, resolved completely by adding 200 μl of acetic acid, and precipitated again as described above. The dried pellet was resolved with 500 μl of 0.1% TFA and was incubated for 1 h at 60°C. The PNA was purified by reversed-phase high-performance liquid chromatography by using a pepRPC HR5/5 column (Amersham-Pharmacia, Freiburg, Germany) with aqueous 0.1% TFA and 0.08% TFA in acetonitrile (PROLIGO, Hamburg, Germany). The fractions that were collected were lyophilized and dissolved in 0.1% TFA. The yield was determined by measuring absorbance at 260 nm. The purity of the PNA oligomers was confirmed by mass spectroscopy (PerSeptive).
PNA-mediated PCR clamping.
The DNA and PNA oligomers used in this study are shown in Table 2. All PCRs were performed by using a total volume of 25 μl and a Trioblock thermocycler (Biometra, Göttingen, Germany). Each reaction mixture contained 1× PCR buffer (Gibco; Roche Diagnostics, Mannheim, Germany), 7.5 pmol of primer, 1.5 mM MgCl2, each deoxynucleoside triphosphate (Roche Diagnostics) at a concentration of 25 μM, 9 pmol of PNA (omitted in the control PCR), and 0.5 or 1.25 U of AmpliTaq polymerase (Perkin-Elmer). The template concentrations used in most reaction mixtures were 16 pg/μl for reference rDNA and 80 pg/μl for community rDNA; the only exception was the experiment in which PCR clamping with PNA-ALF was used, in which a mixture of different templates was used, as described below. The cycling conditions used were similar in all of the PCR clamping experiments (variations are indicated in parentheses), as follows: initial denaturation at 95°C for 5 min, followed by 20 cycles consisting of denaturation at 93°C for 1 min (92°C for PNA-EUB353), PNA annealing at 70°C for 1 min, primer annealing at 53°C for 1 min (61°C for PNA-EUB353), and elongation at 72°C for 1 min (90 s for PNA-1114F) and a final elongation step consisting of 72°C for 7 min (only for PNA-EUB353). Amplification products were separated by agarose gel electrophoresis and were visualized by staining the gels with ethidium bromide (100 μg/liter).
TABLE 2.
Sequences of DNA and PNA oligomers used in this study
| Oligomer | Sequence (5′→3′)a | Target
|
Reference | |
|---|---|---|---|---|
| Orientationb | Position | |||
| TPU1 | AGAGTTTGATCMTGGCTCAG | F | 8-27 | 13 |
| 365R-2 | ATTCYYGACTGCAGYCAC | R | 348-365 | This study |
| 365R-5 | ATTCYYGACTGCTGYCAC | R | 348-365 | This study |
| 365R-6 | ATTCYYTACTGCTGYCAC | R | 348-365 | This study |
| 365R-7 | ATTCYYCACTGCTGYCTC | R | 348-365 | This study |
| 365R-9 | ATTCYYTACTGCTGYCTC | R | 348-365 | This study |
| 365R | Equimolar mixture of 365R-2, -5, -6, -7, and -9 | R | 348-365 | This study |
| EUB353 | TGCCTCCCGTAGGAGT | R | 338-353 | 1c |
| RTU2 | TGCCTCCCGTAGGAGTYTGG | R | 334-53 | 13 |
| RTU8 | AAGGAGGTGATCCANCCRCA | R | 1522-41 | 13 |
| PNA-EUB353 | NH2-TGCCTCCCGTAGGAGT-CONH2 | R | 338-353 | 1c |
| PNA-ALF | NH2-TGGCTCAGAGCGAAC-CONH2 | F | 20-34 | 15d |
| PNA-1114F | NH2-GCAACGAGCGCAACCC-CONH2 | F | 1099-1114 | 13 |
Analysis of amplicons generated by PCR clamping. (i) PCR cloning.
PCR amplicons were directly ligated into vector pCR2.1 of a TA cloning kit by following the instructions of the manufacturer (Invitrogen, de Schelp, The Netherlands). Amplicons obtained from PCR clamping with PNA-1114F were purified by preparative gel electrophoresis by using a QiaQuick gel extraction kit (Qiagen, Hilden, Germany). To compensate for the loss of DNA during purification, excised agarose bands from three PCR were combined and eluted in 50 μl of the buffer supplied.
(ii) Dot blot hybridization.
For dot blot hybridization 16S rDNA inserts of randomly selected recombinant Escherichia coli clones were PCR amplified by using M13 primers as described above. Alternatively, plasmid DNA was isolated by using a GFX plasmid preparation kit (Amersham-Pharmacia, Freiburg, Germany) as recommended by the manufacturer. The presence of inserts of the expected size was determined by digesting plasmid DNA with EcoRI. Plasmid DNA or PCR amplicons were heat denatured (95°C, 4 min) and immediately cooled on ice. Aliquots (1.5 μl) were spotted onto a positively charged nylon membrane (Roche Diagnostics) and fixed by UV cross-linking. Prior to hybridization with FAM-labeled oligonucleotide probe EUB353, membranes were prehybridized with 25 ml of standard hybridization buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium lauroylsarcosine, 0.02% sodium dodecyl sulfate, and 1% blocking reagent (Roche Diagnostics) at 42°C for 30 min. Hybridization was performed with 25 ml of standard hybridization buffer containing 60 pmol of the probe at 42°C for 2 h and this was followed by two stringency washes at 54°C with 40 ml of 6× SSC–0.1% sodium dodecyl sulfate for 15 min. Digoxigenin (DIG)-labeled polynucleotide probes were generated and hybridization was performed as described previously (23), except that two probes were used simultaneously. Hybrids were detected on X-ray film by chemoluminescence by using the reagents of a DIG luminescence detection kit (Roche Diagnostics). To detect the FAM-labeled oligonucleotide, the anti-DIG conjugate was replaced by an alkaline phosphatase coupled to anti-FAM immunoglobulin G antibodies (TIB MOLBIOL, Berlin, Germany).
(iii) Sequencing and phylogenetic analyses.
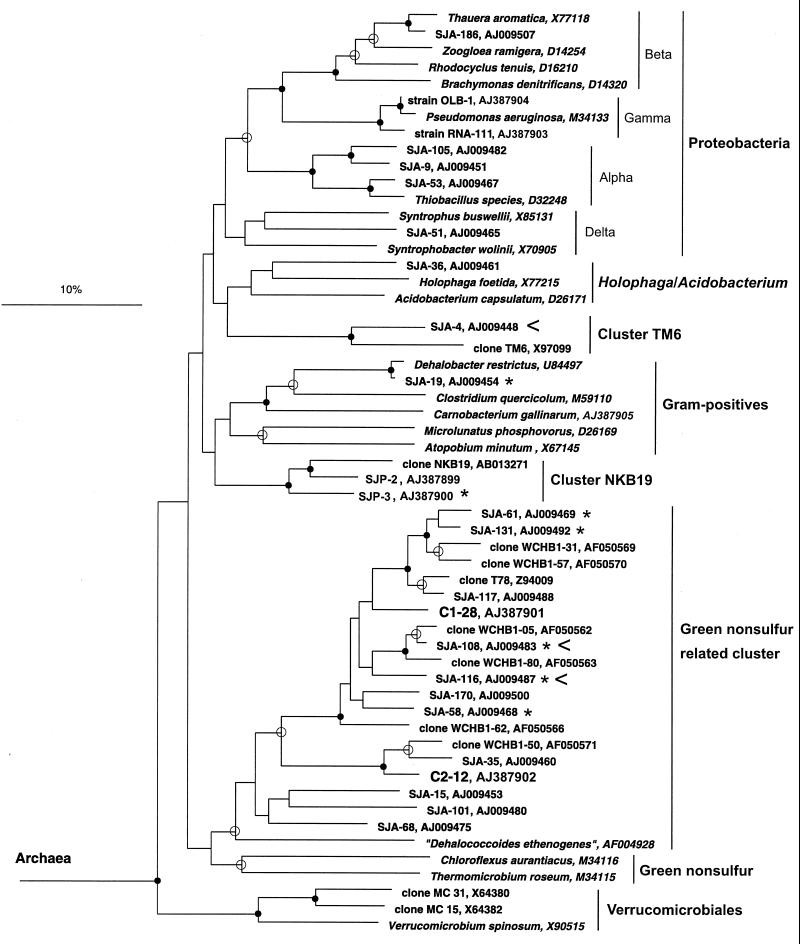
Plasmid DNA or silica-purified PCR products were used as templates for cycle sequencing in which we used a Thermo Sequenase fluorescently labeled primer cycle sequencing kit (Amersham-Pharmacia) and fluorescently labeled universal M13 primers and 16S rDNA-specific primers. Sequencing reactions were analyzed by using an automated model LICOR DNA4000L sequencer (MWG-BIOTECH, Ebersberg, Germany). Phylogenetic analyses were performed by using the ARB software package (http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps). A phylogenetic tree (see Fig. 5) was constructed by using the Jukes-Cantor correction (11) and the neighbor-joining method (20) with 1,000 bootstrap resamplings. Sequences were analyzed for possible chimeric structures by the fractional treeing method as described previously (23).
FIG. 5.
Phylogenetic dendrogram showing clusters mentioned in this study. For sequences marked with an asterisk a corresponding rDNA clone (level of sequence similarity, >99%) was recovered by competitive clamping with PNA-EUB353 (Table 5). For sequences marked with a less-than sign the corresponding rDNA clone was obtained by elongation arrest with PNA-1114F. PNA-1114F nontarget clones C1-28 and C2-12 could not be affiliated with any of the previously described SJA clone families in the cluster related to GNS bacteria and were included in the dendrogram. 16S rDNA clone sequences SJP-2 and SJP-3 (∼1,300 bp) were amplified from total community rDNA by a seminested PCR performed with forward primer NKB19F (5′ GCTGCAAGGCGTCGCCG 3′) derived from NKB19-related clones B1-9, B1-11, and B3-6 and universal reverse primer RTU8. Branch points supported by bootstrap values of >74% are indicated by solid circles. Open circles indicate branch points supported by bootstrap values between 50 and 74%. Branch points without circles were not resolved (bootstrap values, <50%). Scale bar = 10% sequence divergence.
Nucleotide sequence accession numbers.
Nineteen nucleotide sequences of rDNA clones and reference strains have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. AJ387887 to AJ387905.
RESULTS
PCR clamping of an artificial rDNA mixture.
We designed a competitive clamping experiment in which primer TPU1 and the PNA-ALF oligomer, which had an eight-base overlap, were used (Table 2). First, the specificity was tested by using different reference rDNA templates. As shown in Fig. 1, PCR amplification of completely complementary rDNAs (clones SJA-9 and SJA-105) was suppressed by PNA-ALF, whereas rDNA of clone SJA-53 or Verrucomicrobium spinosum (one mismatch each) and rDNA of clone SJA-186 (two mismatches) were amplified.
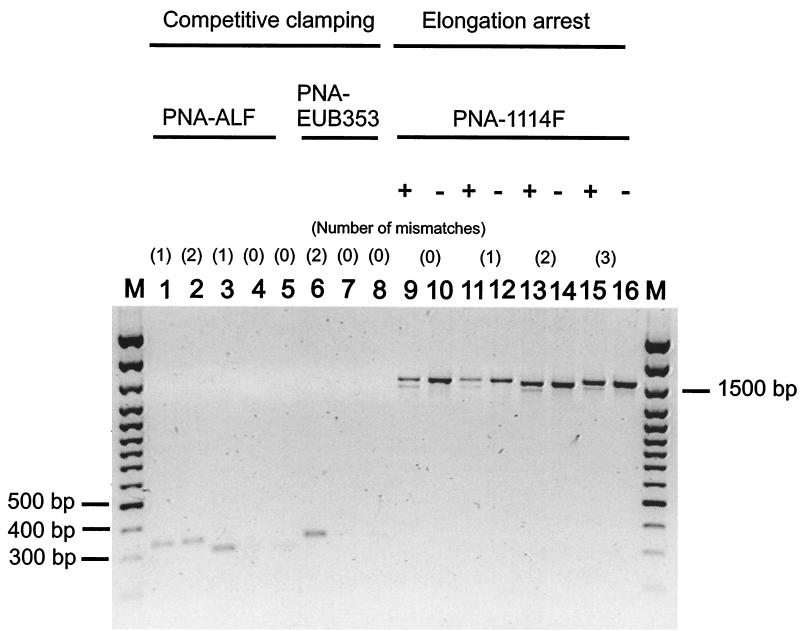
FIG. 1.
Agarose gel electrophoresis analysis of products of various PCR clamping reactions. Lanes 1 through 5, PCR performed with PNA-ALF and reference rDNAs (V. spinosum [one mismatch], SJA-186 [two mismatches], SJA-53 [one mismatch], SJA-9 [no mismatch], SJA-105 [no mismatch]); lanes 6 through 8, PCR performed with PNA-EUB353 and reference rDNAs (V. spinosum [two mismatches], strain OLB-1 [no mismatch], C. gallinarum [no mismatch]); lanes 9 through 16, duplicate PCR performed with (+) or without (−) PNA-1114F and reference rDNAs (V. spinosum [no mismatch], strain OLB-1 [one mismatch], SJA-131 [two mismatches], SJA-4 [three mismatches]); lanes M, length standard (100 bp plus; MBI-Fermentas, St.-Leon Roth, Germany). The sizes of relevant bands are shown on the left and right.
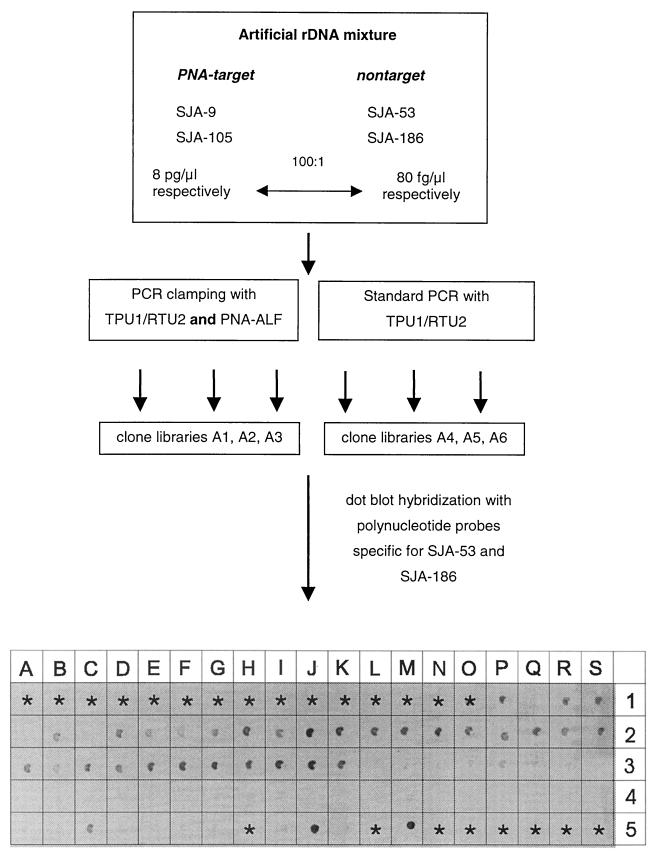
Second, an artificial rDNA mixture was analyzed. The sample used contained a 100-fold excess of target rDNAs (SJA-9 and SJA-105) compared with nontarget rDNAs (SJA-186 and SJA-53). As controls, PCR without PNA-ALF were performed in parallel. All PCR and cloning reactions were performed in triplicate, which resulted in three independent clone libraries each for both clamped PCR (clone libraries A1 to A3) and control PCR (clone libraries A4 to A6) (Fig. 2). A total of 68 randomly selected clones (34 clones from each set) were analyzed. As shown in Fig. 2, simultaneous dot blot hybridization performed with DIG-labeled polynucleotide probes specific for SJA-186 and SJA-53 revealed that 28 of the 34 clones (82.4%) in the libraries derived from PCR clamping were positive, while only 1 positive clone (2.9%) was detected among the 34 clones derived from standard PCR.
FIG. 2.
Analysis of an artificial rDNA mixture by competitive clamping: schematic diagram of PCR clamping performed with PNA-ALF and results of dot blot hybridizations with polynucleotide probes specific for SJA-53 and SJA-186. Positions P1 to K3, selected clones from libraries A1 to A3 (PCR clamping); positions L3 to G5, selected clones from libraries A4 to A6 (control PCR). The following 16S rDNAs were used as controls: SJA-9 (position I5), SJA-53 (position J5), SJA-105 (position K5), and SJA-186 (position M5). Asterisks indicate positions where no samples were applied.
PCR clamping of total community rDNA.
To demonstrate the ability of PCR clamping to selectively recover rDNA sequences of recently described phylogenetic groups from total community rDNA, we used the rDNA amplicon of an anaerobic dechlorinating consortium as the template; it had been shown previously that this consortium included several representatives of recently described phylogenetic groups (23). The following two strategies were used; (i) competitive clamping with primers TPU1 and 365R and PNA-EUB353, which had a binding site that overlapped with the binding site of the reverse primer 365R; and (ii) elongation arrest performed with primers TPU1 and RTU8 and PNA-1114F, which bound to the middle of the PCR target region (Fig. 3B). The specificities of the two PCR clamping approaches were tested with individual reference rDNA templates before complex target rDNAs were used.
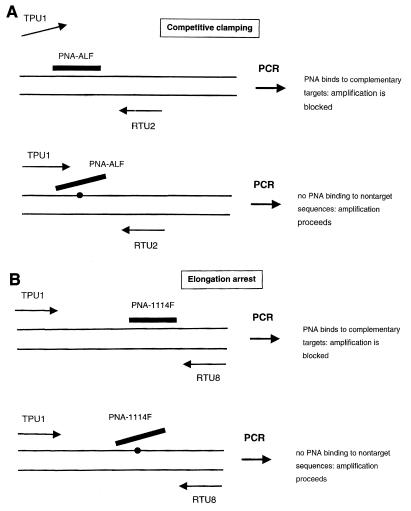
FIG. 3.
Schematic diagram of PNA-mediated PCR clamping of 16S rDNA amplification. (A) Competitive clamping: inhibition of PCR amplification by PNA-ALF-mediated exclusion of primer TPU1. (B) Elongation arrest: inhibition of PCR amplification by binding of PNA-1114F to an internal target sequence, which prevents readthrough by the Taq polymerase. In both cases amplification proceeds only if one or more base substitutions in the binding sites of appropriate PNA molecules are present.
While PCR amplification of complementary sequences of strain OLB-1 and Carnobacterium gallinarum was specifically inhibited by PNA-EUB353, template rDNA derived from V. spinosum (two mismatches) was readily amplified (Fig. 1). Compared to the competitive PCR clamping approach, which resulted in complete suppression of PCR amplification, PNA-1114F-mediated elongation arrest led to reduced amplification of target rDNAs but not to complete amplification arrest (Fig. 1).
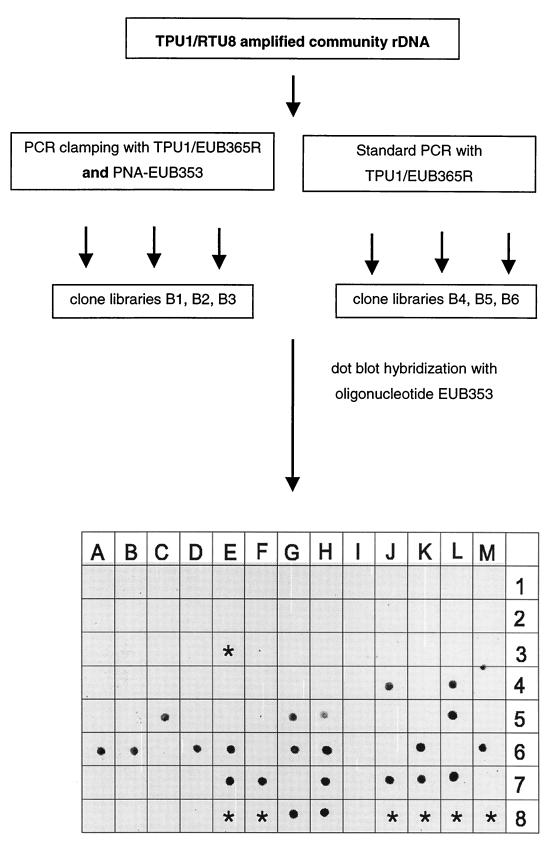
The efficiency of competitive PCR clamping by PNA-EUB353 when total community rDNA was used was analyzed as shown in Fig. 4. A standard PCR (without a PNA oligomer) served as the control. All PCR and cloning reactions were performed in triplicate, which resulted in clone libraries B1 to B3 (PCR clamping) and B4 to B6 (control PCR). A total of 46 clones in libraries B1 to B3 and 45 clones in libraries B4 to B6 were randomly chosen and used for further analysis. Dot blot hybridization with FAM-labeled oligonucleotide probe EUB353 revealed no positive clones among the 46 clones in libraries derived from PCR clamping, whereas 20 (44.4%) of the 45 clones obtained from the control PCR were hybridization positive (Fig. 4). To verify the hybridization results and to perform an additional phylogenetic analysis, 17 EUB353-negative clones in libraries B1 to B3 and 9 positive clones were sequenced. Sequencing of the oligonucleotide EUB353 target site confirmed the specificity of the dot blot hybridization procedure and revealed that all of the EUB353-negative clones had several base substitutions (Table 3). A subsequent phylogenetic analysis showed that 16 of 17 EUB353-negative clones in libraries B1 to B3 either were affiliated with a phylogenetic cluster close to the green nonsulfur (GNS) bacteria (10) or were related to rDNA sequence NKB19 (14), which was not closely related to any of the phylogenetic groups described previously (Table 4). In contrast, seven of nine EUB353-positive clones either were representatives of previously described proteobacterial SJA clone families or were identical to the 16S rDNA sequence of Pseudomonas sp. strain RNA-111, which had been isolated from the same community previously (24).
FIG. 4.
Analysis of community rDNA by competitive clamping: schematic diagram of PCR clamping performed with PNA-EUB353 and results of dot blot hybridizations with oligonucleotide probe EUB353. Positions A1 to H4, selected clones from libraries B1 to B3 (PCR clamping); positions I4 to A8, selected clones from libraries B4 to B6 (standard PCR). The following 16S rDNAs were used as controls: SJA-22 (position B8), SJA-43 (position C8), SJA-131 (position D8), Pseudomonas aeruginosa ATCC 25330 (position G8), C. gallinarum DSM 4847T (position H8), and V. spinosum DSM 4136T (position I8). Asterisks indicate positions where no samples were applied.
TABLE 3.
Competitive clamping of total community rDNA: analysis of PNA-EUB353 nontarget sequences (E. coli positions 336 to 355) of clones belonging to gene libraries B1 to B3 generated by PCR clamping and of selected reference sequences
| Group | Organism or clonea | Target sequence (5′→3′)b |
|---|---|---|
| Gene libraries B1 to B3 | rDNA clone B1-7 | AGACACCTACGGGTGACAGC |
| rDNA clone B1-10 | ATACACCTACGGGTGGCAGC | |
| rDNA clone B1-12 | ATACACCTACGGGTGGCAGC | |
| rDNA clone B1-13 | ATACACCTACGGGTGGCAGC | |
| rDNA clone B1-11 | AGACACCTACGGGTGGCAGC | |
| rDNA clone B1-9 | AGACACCTACGGGTGACAGC | |
| Reference sequences | Carnobacterium gallinarum DSM 4847T | AGACTCCTACGGGAGGCAGC |
| Verrucomicrobium spinosum DSM 4136I | AGACACCTACGGGTGGCAGC | |
| rDNA clone SJA-58 | ATACACCTACGGGTGGCAGC | |
| rDNA clone SJA-61 | ATACACCTACGGGTGGCAGC | |
| rDNA clone SJA-116 | AGACACCTACGGGTGGCAGC | |
| rDNA clone SJA-170 | ATACACCTACGGGTGGCAGC | |
| rDNA clone WCHB1-50 | AGACGCCTACGGGTGGCAGC | |
| rDNA clone NKB19 | AGACACCTACGGGTGGCAGC |
The sources of the reference sequences were as follows: SJA rDNA clones, von Wintzingerode et al. (23); rDNA clone WCHB1-50, Dojka et al. (6); and rDNA clone NKB19, Li et al. (14).
The target region of PNA-EUB353 is underlined. The boldface letters indicate nucleotides that differed from nucleotides in sequences found in the 16S rDNA of C. gallinarum and most other members of the domain Bacteria.
TABLE 4.
Phylogenetic affiliations of 17 EUB353-negative clones and 9 EUB353-positive clones selected from 16S rDNA clone libraries generated by competitive clamping (clone libraries B1 to B3) and standard PCR of total community rDNA, respectively
| Phylogenetic groupa | Clone | Closest phylogenetic neighbor in databaseb | % Sequence similarityc | Accession no. |
|---|---|---|---|---|
| Cluster related to GNS bacteria | B1-7 | rDNA clone SJA-116 | 99.7 | AJ009487 |
| B1-10 | rDNA clone SJA-170 | 86.2 | AJ009500 | |
| B1-12 | rDNA clone SJA-170 | 88.4 | AJ009500 | |
| B1-13 | rDNA clone SJA-131 | 99.4 | AJ009492 | |
| B1-14 | rDNA clone SJA-170 | 85.9 | AJ009500 | |
| B1-15 | rDNA clone WCHB1-50 | 95.6 | AF050566 | |
| B1-16 | rDNA clone SJA-116 | 99.4 | AJ009487 | |
| B1-18 | rDNA clone SJA-61 | 99.4 | AJ009469 | |
| B2-3 | rDNA clone SJA-108 | 100 | AJ009483 | |
| B2-8 | rDNA clone SJA-170 | 88.7 | AJ009500 | |
| B2-10 | rDNA clone SJA-61 | 99.7 | AJ009469 | |
| B3-4 | rDNA clone SJA-116 | 99.7 | AJ009487 | |
| B3-8 | rDNA clone SJA-58 | 99.4 | AJ009468 | |
| NKB19 | B1-9 | rDNA clone NKB19 | 84.1 | AB013271 |
| B1-11 | rDNA clone NKB19 | 83.8 | AB013271 | |
| B3-6 | rDNA clone NKB19 | 84.1 | AB013271 | |
| Low-G+C-content gram-positive bacteria | B3-14 | rDNA clone SJA-19 | 99.4 | AJ009454 |
| α-Proteobacteria | B4-53 | rDNA clone SBR1100 | 96.0 | X84528 |
| B4-62 | rDNA clone SJA-53 | 98.0 | AJ009467 | |
| β-Proteobacteria | B4-68 | rDNA clone SJA-186 | 99.7 | AJ009507 |
| B5-11 | rDNA clone SJA-186 | 99.7 | AJ009507 | |
| B6-52 | rDNA clone SJA-186 | 99.4 | AJ009507 | |
| B6-60 | rDNA clone SJA-186 | 99.4 | AJ009507 | |
| γ-Proteobacteria | B4-69 | Pseudomonas sp. strain RNA-111 | 100 | AJ387903 |
| δ-Proteobacteria | B5-4 | rDNA clone SJA-51 | 96.9 | AJ009465 |
| Low-G+C-content gram-positive bacteria | B6-59 | Clostridium quercicolum | 90.2 | M59110 |
α-, β-, γ-, and δ-Proteobacteria, α, β, γ, and δ subdivisions of the Proteobacteria, respectively.
References were as follows: SJA rDNA clones, reference 23; rDNA clone WCHB1-50, reference 6; rDNA clone NKB19, reference 14; rDNA clone SBR100, reference 4; and strain RNA-111, reference 24.
Based on comparisons with E. coli positions 7 to 347.
PCR clamping performed by the elongation arrest strategy with PNA-1114F and primers TPU1 and RTU8 was done in triplicate and resulted in three clone libraries, which were designated C1 to C3. The effect of PCR clamping was measured by using clone library SJA as the control; this clone library had been generated previously by standard PCR from total community DNA by using the same broad-spectrum primers (23). Twenty-four randomly chosen clones in libraries C1 to C3 were sequenced (∼800 to 1,500 bp) in order to determine both their phylogenetic affiliation and the sequence of the PNA-1114F target site. This analysis revealed that 11 of the 24 clones (45.8%) were PNA-1114F negative and had up to three base substitutions in the PNA-1114F target site (Table 5). Thus, when PNA-1114F-mediated elongation arrest was used, an approximately sixfold relative increase (6.9% PNA-1114F-negative clones among SJA clones) in nontarget sequences was observed compared to the SJA clone library generated by standard PCR alone. Sequences of nontarget clones either were nearly identical (>99%) to the sequence of clone SJA-4 in the TM6 cluster (19) or were affiliated with the phylogenetic cluster related to GNS bacteria (Fig. 5). One clone was identified as a sequence chimera.
TABLE 5.
PCR clamping by elongation arrest: analysis of PNA-1114F target sequences (E. coli positions 1097 to 1116) of selected clones belonging to gene libraries generated by PCR clamping and reference sequences
| Group | Organism or clonea | Target sequence (5′→3′)b |
|---|---|---|
| Gene libraries generated by PCR clamping | rDNA clone C1-28 | CGCTAACGAGCGCAACCCCT |
| rDNA clone C2-12 | CCCTAACGAGCGCAACCCTC | |
| rDNA clone C1-3 (>99% similarity to rDNA clone SJA-4) | CTCTAACGAGCGCAACTCTT | |
| Reference sequences | rDNA clone SJA-4 | CTCTAACGAGCGCAACTCTT |
| Verrucomicrobium spinosum DSM 4136T | CCGCAACGAGCGCAACCCCT | |
| Pseudomonas sp. strain OLB-1 | CCGTAACGAGCGCAACCCTT | |
| rDNA clone SJA-108 | CGCTAACGAGCGCAACCCTC | |
| rDNA clone SJA-116 | CGCTAACGAGCGCAACCCTC | |
| rDNA clone WCHB1-50 | CCCTAACGAGCGCAACCCTC | |
| rDNA clone TM6 | CTTTAACGAGCGCAACTCCT |
The sources of the reference sequences were as follows: Pseudomonas sp. strain OLB-1, von Wintzingerode (24); SJA rDNA clones, von Wintzingerode et al. (23); rDNA clone WCHB1-50, Dojka et al. (6); and rDNA clone TM6, Rheims et al. (19).
The target region of PNA-1114F is underlined. The boldface letters indicate nucleotides that differed from nucleotides in sequences found in the 16S rDNA of V. spinosum and most other members of the domain Bacteria.
DISCUSSION
Culture-independent analysis of PCR-amplified 16S rRNA gene libraries for studying the diversity of complex microbiota has become an important tool in microbial ecology. This method includes analysis of both cultured and uncultured microorganisms (7, 8). Various investigations of environmental and man-made microbial habitats not only have increased our knowledge concerning the phylogenetic diversity of well-known bacterial or archaeal phyla but also have revealed the presence of novel phylogenetic groups represented exclusively by uncultured microorganisms (for a review, see reference 10). However, the laborious and time-consuming steps involved in analysis of gene libraries present a serious problem since usually a large number of clones have to be screened to detect not only the dominant sequences but also the less abundant clones in a rDNA gene library. Here, we used PNA-mediated PCR clamping, which has been used to suppress certain variants in order to identify clinical relevant polymorphisms (3, 16, 18, 21), to reduce amplification of abundant sequences from a standard clone library and to enrich rare sequences, which were less homologous to the PNA oligomer used. PNA-mediated PCR clamping might be a useful supplement to standard PCR when it is included in the following strategy: (i) generation of a rDNA clone library by standard PCR and determination of the abundant sequence types; (ii) design of a PNA oligomer(s) that targets the abundant sequences; and (iii) analysis of an additional rDNA clone library generated by PNA-mediated PCR clamping. In this study we used PCR clamping for both possible strategies, competitive clamping that attacked either the forward primer (PNA-ALF) or the reverse primer (PNA-EUB353) and the elongation arrest approach (PNA-1114F).
PCR clamping of an artificial rDNA mixture.
PCR clamping to selectively enrich nontarget sequences was first used with an artificial mixture containing 1% SJA-53 rDNA and 1% SJA-186 rDNA. Only one of these rDNAs was detected in 34 clones belonging to gene libraries A4 to A6 generated by standard PCR. This changed, however, if PNA-ALF, which specifically binds to SJA-9 and SJA-105 rDNAs, was added to the PCR mixture. In this case, the initially less numerous SJA-53 and SJA-186 rDNAs clearly dominated, resulting in clone libraries A1 to A3 (28 of 34 clones). This example demonstrated the potential of PCR clamping as a supplement for standard PCR in rDNA-based diversity studies. When both PCR approaches were used, analysis of a few clones was sufficient to detect all of the different sequences in the artificial mixture, and consequently screening a large number of standard PCR clones was not necessary.
PCR clamping of natural community rDNA.
In a recent phylogenetic analysis of an anaerobic, trichlorobenzene-dechlorinating microbial consortium (23), it was shown that several rDNA clones (SJA sequences) were affiliated with recently described phylogenetic clusters (clusters OP10 [9], WS1 [6], and TM6 [19], and a cluster related to the GNS bacteria [10]) and were closely related to rDNA sequences of uncultivated bacteria in another anaerobic, dechlorinating community (6). In most of these SJA sequences there were variations in highly conserved rDNA signature sites for the domain Bacteria (EUB353 site [1] and 1114F site [13]), and consequently the sequences were designated EUB353/1114F-negative sequences. Future studies of anaerobic, dechlorinating microbial consortia should focus on EUB353/1114F-negative sequences since they might represent indicator organisms for the dechlorination process (23). However, for such studies analysis of standard rDNA clone libraries alone is not reasonable since EUB353/1114F-negative sequences accounted for only about 19% of all of the SJA sequences analyzed; consequently, screening a large number of clones will be necessary to detect these sequences. Therefore, PCR clamping with PNA-EUB353 and PNA-1114F was tested to determine its ability to selectively recover EUB353/1114F-negative sequences from previously studied total community rDNA.
Indeed, PCR clamping with PNA-EUB353 and bacterium-specific primers specifically amplified rDNA sequences belonging to the cluster related to the GNS bacteria and sequences affiliated with rDNA sequence NKB19, which had not been detected among the SJA clones previously (23).
Compared to competitive clamping with PNA-EUB353, clamping by elongation arrest with PNA-1114F was less effective since it resulted in less enrichment of rDNA sequences affiliated with the GNS bacterium-related cluster and with cluster TM6. This might have been due to interactions among the three reaction molecules (PNA, DNA, and Taq polymerase), which were obviously more complex than the interactions between PNA and DNA in competitive clamping. For example, our efforts to use PNA-EUB353 (16-mer) or an extended PNA (19-mer) for PCR clamping by elongation arrest consistently failed. This finding is consistent with previously described problems with this PCR clamping type (18; O. Landt, unpublished data). However, compared to competitive clamping, clamping by elongation arrest offers greater flexibility in chosing target sites for PNA oligomers and primers. Our results for both sensitivity and specificity of PCR clamping confirm the previous findings of Behn and Schuermann (2) that PCR clamping selectively amplifies nontarget sequences in the presence of a high background level of target sequences. In the study of these authors PCR clamping followed by SSCP analysis allowed detection of p53 gene mutations even in samples with a 200-fold excess of wild-type genes. Similar to our data, most PCR clamping protocols result in specific binding of PNA oligomers at 68 or 70°C (2, 3, 12, 16, 21). The fact that binding of PNA oligomers is less sensitive to base composition and length of the oligomer definitely simplifies the design of PCR clamping experiments. We are aware that biases introduced by standard PCR amplification and subsequent cloning (for a review, see reference 22) are not compensated for with this method. Furthermore, less abundant sequences that are fully complementary to the PNA oligomer are not selectively amplified by PCR clamping. However, as shown in this study, PCR clamping might be a useful supplement for standard PCR. Further work with different PNA oligomers and other complex microbiota is urgently needed to fully explore the potential of PNA-mediated PCR clamping in rDNA-based studies of microbial diversity.
ACKNOWLEDGMENTS
We thank Heidemarie Hans (Forschungsinstitut für Molekulare Pharmakologie, Berlin im Forschungsverbund Berlin e.V.) for excellent technical assistance and Claudia Bergmüller, Klaus Heuner, and Michael Rohrbach for helpful comments.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 193; Biological Treatment of Industrial Wastewaters) to U.B.G.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behn M, Schuermann M. Sensitive detection of p53 gene mutations by a 'mutant enriched' PCR-SSCP technique. Nucleic Acids Res. 1998;26:1356–1358. doi: 10.1093/nar/26.5.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behn M, Schuermann M. Simple and reliable factor V genotyping by PNA-mediated PCR-clamping. Thromb Haemostasis. 1998;79:773–777. [PubMed] [Google Scholar]
- 4.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle J S, Lew A M. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 1995;11:8. doi: 10.1016/s0168-9525(00)88977-5. [DOI] [PubMed] [Google Scholar]
- 6.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Göbel U B. Phylogenetic amplification for the detection of uncultured bacteria and the analysis of complex microbiota. J Microbiol Methods. 1995;23:117–128. [Google Scholar]
- 8.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 9.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 12.Kyger E M, Krevolin M D, Powell M J. Detection of the hereditary hemochromatosis gene mutation by real-time fluorescence polymerase chain reaction and peptide nucleic acid clamping. Anal Biochem. 1998;260:142–148. doi: 10.1006/abio.1998.2687. [DOI] [PubMed] [Google Scholar]
- 13.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley and Sons Ltd.; 1991. pp. 115–173. [Google Scholar]
- 14.Li L, Guenzennec J, Nichols P, Henry P, Yanagibayashi M, Kato C. Microbial diversity in Nankai trough sediments at a depth of 3,843 m. J Oceanogr. 1999;55:635–642. [Google Scholar]
- 15.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 16.Mrozikiewicz P M, Landt O, Cascorbi I, Roots I. Peptide nucleic acid-mediated polymerase chain reaction clamping allows allelic allocation of CYP1A1 mutations. Anal Biochem. 1997;250:256–257. doi: 10.1006/abio.1997.2246. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen P E. Applications of peptide nucleic acids. Curr Opin Biotechnol. 1999;10:71–75. doi: 10.1016/s0958-1669(99)80013-5. [DOI] [PubMed] [Google Scholar]
- 18.Ørum H, Nielsen P E, Egholm M, Berg R H, Buchardt O, Stanley C. Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res. 1993;21:5332–5336. doi: 10.1093/nar/21.23.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rheims H, Rainey F A, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17:159–169. [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Thiede C, Bayerdörffer E, Blasczyk R, Wittig B, Neubauer A. Simple and sensitive detection of mutations in the ras proto-oncogenes using PNA-mediated PCR clamping. Nucleic Acids Res. 1996;24:983–984. doi: 10.1093/nar/24.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 23.von Wintzingerode F, Selent B, Hegemann W, Göbel U B. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl Environ Microbiol. 1999;65:283–286. doi: 10.1128/aem.65.1.283-286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Wintzingerode F. Ph.D. thesis. Berlin, Germany: Humboldt-Universität zu Berlin; 1999. [Google Scholar]