Abstract
Background
This study aimed to summarize the available data on the association between the severity of (COVID-19) and routine blood indicators, inflammatory, biochemical parameters and coagulation parameter.
Methods
A literature search was conducted of PubMed, EMBASE, and Web of Sciences, CNKI, WanFang database providing relevant data. Random-effects meta-analysis was used to pool effect sizes.
Results
In patients with severe symptoms, interleukin-6, [IL-6; standardized mean difference (SMD) =1.15, 95% confidence interval (95% CI): 1.01, 1.29, P<0.001, n=1,121], interleukin-10 (IL-10; SMD =0.92, 95% CI: 0.75, 1.08, P<0.001, n=782), interleukin-4 (IL-4; SMD =0.2, 95% CI: 0.01, 0.39, P=0.04, n=500), procalcitonin (PCT; SMD =1.16, 95% CI: 0.99, 1.33, P<0.001, n=734), C-reactive protein (CRP; SMD =1.42, 95% CI: 1.27, 1.57, P<0.001, n=1,286), serum amyloid A (SAA; SMD =2.82, 95% CI: 2.53, 3.11, P<0.001, n=502) neutrophil count (SMD =0.63, 95% CI: 0.44, 0.82, P<0.001, n=558), alanine aminotransferase (ALT; SMD =2.72, 95% CI: 2.43, 3.02, P<0.001, n=538), aspartate aminotransferase (AST; SMD =2.75, 95% CI: 2.37, 3.12, P<0.001, n=313), lactate dehydrogenase (LDH; SMD =4.01, 95% CI: 3.79, 4.24, P<0.001, n=1,055), creatine kinase (CK; SMD =2.62, 95% CI: 2.2, 3.03, P<0.001, n=230;), CK-MB isoenzyme (CK-MB; SMD =3.07, 95% CI: 2.81, 3.34, P<0.001, n=600, activated partial thromboplastin time (APTT; SMD =0.63, 95% CI: 0.39, 0.87, P<0.001, n=351), and prothrombin time (P-T; SMD =1.83, 95% CI: 1.55, 2.11, P<0.001, n=351) were significantly higher than in patients with mild symptoms. On the contrary, lymphocyte count (SMD =−1.04, 95% CI: −1.21, −0.86, P<0.001, n=805) platelets (SMD =−1.47, 95% CI: −1.7, −1.24, P<0.001, n=653), monocyte count (SMD =−0.56, 95% CI: −0.8, −0.32, P<0.001, n=403), and albumin (SMD =−2.95, 95% CI: −3.21, −2.7, P<0.001, n=637) was significantly lower in patients with severe symptoms than in patients with mild symptoms. IL-6 (SMD =2.62, 95% CI: 2.15, 3.09, P<0.001, n=185), PCT (SMD =0.2, 95% CI: 0.16, 0.23, P<0.001, n=156), creatinine (SMD =2.29, 95% CI: 1.87, 2.7, P<0.001, n=213), and neutrophil counts (SMD =2.77, 95% CI: 2.38, 3.16, P<0.001, n=260) in patients with COVID-19 in the death group were significantly higher than that in patients in the survival group, while the lymphocyte count was significantly lower.
Conclusions
In summary, current evidence show that those laboratory indicators are associated with the severity of COVID-19 and thus could be used as prognostic risk stratification of patients with COVID-19.
Keywords: Coronavirus disease 2019 (COVID-19), laboratory biomarker, prognosis, meta-analysis
Introduction
Coronavirus disease 2019 (COVID-19) is a self-limiting illness in approximately 80% of patients. However, some patients will develop more severe symptoms, including dyspnea, multifunctional organ failure, and even death (1). With a mortality rate of 1–2%, COVID-19 requires intensive care in 5% of patients (2).
Currently, there is no systemic treatment for COVID-19. Although some drugs, such as hydroxychloroquine, have been included in treatment guidelines or are being developed in interventional studies, treatment of COVID-19 is mainly supportive (3). Aberrant biomarkers help to better understand the biological characteristics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus causing COVID-19, and contribute to the development of targeted drugs (4). In the context of out stretched heath care systems and limited resources, risk stratification is important to identify patients who the most need in-hospital and in-depth management. laboratory parameters along with some clinical factors might help to predict disadvantage outcomes among COVID-19 patients. The combination of biomarkers and other clinical parameters could help predict disease progression in COVID-19 patients. These parameters might help in prognostic risk layering of patients with COVID-19. We present the following article in accordance with the MOOSE reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-345/rc).
Methods
Inclusion criteria
The inclusion criteria were as follows: (I) The inclusion criteria strictly following PICOS criteria (II) study subjects were patients diagnosed with COVID-19; (III) grouping included a mild symptom group and severe symptom group. The mild group had mild clinical symptoms, accompanied by fever and respiratory symptoms, and pneumonia findings were indicated by imaging examination. The clinical symptoms of patients with severe symptoms were shortness of breath, respiratory rate ≥30 beats/minute, oxygen saturation ≤93% at resting state, and arterial partial pressure of oxygen (PaO2)/inspired oxygen concentration (FiO2) ≤300 mmHg. The severity of COVID-19 was defined in accordance with international guidelines for community-acquired pneumonia (5).
Literature search
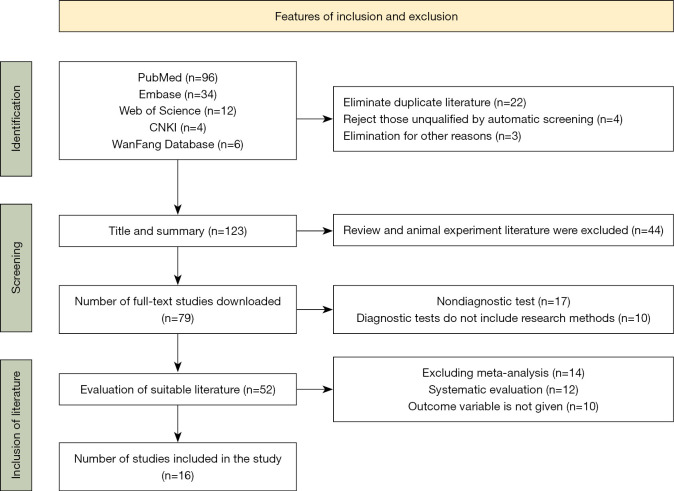
A total of 152 relevant studies published between April 2020 and October 2021 were identified using the keywords ‘COVID-19 Coronary Pneumonia Laboratory Indicators, such as biochemical parameters, coagulation indicators, inflammatory factors’ such as IL-6, IL-4, PCT to search PubMed, EMBASE, Web of Science, and CNKI, WanFang database. Finally, 16 articles were included in the study. A flow chart summarizing the inclusion and exclusion of articles is shown in Figure 1.
Figure 1.
Inclusion and exclusion summary of literature.
Study selection
Two independent investigators (B Cao and C Wang) assessed the titles and abstracts of the articles obtained from the above databases, and included all literature that met the study criteria. The included data was analyzed using Review Manager 5.3 software for meta-analysis.
Data extraction and statistical analysis
Data from the included articles were extracted in the form of word tables. The extracted contents included the name of the first author of the article, sample size, comorbidities of COVID-19 patients, and standardized mean difference (SMD) values.
The data were saved in Microsoft excel 2007 and then imported into Review Manager 5.3 software to analyze the SMD values of laboratory parameters of patients with mild symptoms and patients with severe symptoms. In cases where studies did not report the SMD of patient biomarkers, the SMD of each mean was calculated using the first quartile (Q1), fourth quartile (Q4), the median, and the number of cases before combining the effect sizes, as recommended in the Cochrane Handbook for Systematic Reviews of Interventions (6). For each study, we calculated their SMD and the corresponding 95% CI, the pooled SMD with 95% CI was summarized to represent the total effect. The presence and amount of heterogeneity were assessed using the χ2 test and I2 test, with I2 of 25%, 50%, and 75% representing low, moderate, and high heterogeneity, respectively (7,8). A two-sided P value of less than 0.05 was considered statistically significant for all analyses. I2>50%, the heterogeneity is considered high. Considering the age difference, stratified analysis was performed between the mild group patients and the severe group patients with the average age younger than 55 years and elder than 55 years Table 1. Publication bias was assessed using funnel plots and Egger’s test (P<0.10) (9). The characteristics of the included literature are shown in Table 2.
Table 1. Stratified analysis of standardized mean difference of biological indicators in meta-analysis for patients with severe and mild disease.
| Anomaly indicator | Age (years) | SMD (95% CI) | P value | Heterogeneity I2 (%) | Sample size for mild | Sample size for severe | Included literature |
|---|---|---|---|---|---|---|---|
| Lymphocyte count | >55 | −0.42 (−0.5, −0.34) | <0.001 | 45 | 219 | 171 | 3 |
| <55 | −0.27 (−0.31, 0.24) | <0.001 | 48 | 301 | 71 | 3 | |
| IL-6 | >55 | 1.54 (1.29, 1.79) | <0.001 | 72 | 201 | 150 | 4 |
| <55 | 3.49 (2.99, 3.98) | <0.001 | 0 | 48 | 160 | 3 | |
| CRP | >55 | 0.48 (0.08, 0.89) | 0.02 | 38 | 175 | 159 | 3 |
| <55 | 3.53 (2.97, 4.08) | <0.001 | 51 | 65 | 325 | 3 | |
| D-D | >55 | 1.07 (0.79, 1.35) | <0.001 | 31 | 71 | 270 | 3 |
| <55 | 2.7 (2.29, 3.11) | <0.001 | 37 | 58 | 174 | 3 | |
| Neutrophil count | >55 | 0.44 (0.24, 0.65) | <0.001 | 80 | 191 | 270 | 4 |
| <55 | 1.79 (1.28, 2.3) | <0.001 | 0 | 29 | 68 | 2 |
SMD, standardized mean difference; 95% CI, confidence interval; IL-6, interleukin 6; CRP, C-reactive protein; D-D, D-dimer.
Table 2. Characteristics of the included literature.
| Author | Research type | Group | Number of people | Age (years) | Complications |
|---|---|---|---|---|---|
| Qian GQ (10) | Multicenter retrospective study | Mild symptom | 82 | 49 | Not reported |
| Severe symptom | 9 | 66 | Not reported | ||
| Xie Y (11) | Single retrospective study | Mild symptom | 22 | 58 | Not reported |
| Severe symptom | 7 | 69 | Not reported | ||
| Zhu Z (12) | Single retrospective study | Mild symptom | 111 | 49.9±15 | High blood pressure 50%, heart disease 12.5%, cancer 6.25%, COPD 12.5% |
| Severe symptom | 16 | 57.5±11 | Diabetes 9.01%, hypertension 20.72%, heart disease 3.6%, cancer 3.6%, chronic obstructive pulmonary disease 3.6% | ||
| Zheng F (13) | Single retrospective study | Mild symptom | 131 | 40 | Diabetes 3.8%, hypertension 7.6%, heart disease 1.5%, chronic obstructive pulmonary disease 3.1%, cerebrovascular disease 2.3%, chronic liver disease 3.1% |
| Severe symptom | 30 | 57 | Diabetes 6.7%, hypertension 40%, heart disease 6.7%, chronic obstructive pulmonary disease 6.7%, cerebrovascular disease 3.3% | ||
| Lv Z (14) | Multicenter retrospective study | Mild symptom | 115 | 62 | Diabetes 7.83%, hypertension 20%, coronary heart disease 4.35%, chronic obstructive pulmonary disease 1.74% |
| Severe symptom | 155 | 61 | Diabetes 11.6%, hypertension 21.29%, heart disease 4.52%, chronic obstructive pulmonary disease 1.94% | ||
| Zhang G (15) | Single retrospective study | Mild symptom | 166 | 51 | Diabetes 9%, hypertension 16.9%, heart disease 5.4%, chronic obstructive pulmonary disease 1.2%, chronic kidney disease 0.6%, cerebrovascular disease 2.4%, chronic liver disease 1.8%, tumor 1.2%, |
| Severe symptom | 55 | 62 | Diabetes 12.7%, hypertension 47.3%, heart disease 23.6%, chronic obstructive pulmonary disease 7.3%, chronic kidney disease 9.1%, cerebrovascular disease 20%, chronic liver disease 7.3%, tumor 7.8%, immunosuppression 1.8% | ||
| Zhang H (16) | Multicenter retrospective study | Mild symptom | 9 | 47±11.7 | Not reported |
| Severe symptom | 4 | 55.2±6 | Not reported | ||
| Fu J (17) | Single retrospective study | Mild symptom | 22 | 40.7±9 | Hyperlipidemia 4.5% |
| Severe symptom | 13 | 60±15.5 | Diabetes 23.1%, hypertension 38.5%, chronic obstructive pulmonary disease 7.7%, cerebrovascular | ||
| Liu SL (18) | Single retrospective study | Mild symptom | 194 | 43 | Not reported |
| Severe symptom | 31 | 64 | Not reported | ||
| Liu Q (19) | Multicenter retrospective study | Mild symptom | 59 | 49 | Diabetes 3.4%, chronic kidney disease 4%, cerebrovascular disease 1.7%, chronic obstructive pulmonary disease 3.4% |
| Mild symptom | 25 | 52 | Diabetes 12%, chronic kidney disease 4%, heart disease 8%, cancer 4%, COPD 12% | ||
| Xu B (20) | Single retrospective study | Mild symptom | 80 | 56 | Not reported |
| Severe symptom | 45 | 60 | Not reported | ||
| Burgos-Blasco B (21) | Single retrospective study | Mild symptom | 35 | 68.5 | Cardiovascular disease 31%, chronic kidney disease 6%, chronic liver disease 6%, tumor 6% |
| Severe symptom | 27 | 71 | Cardiovascular disease 41%, chronic kidney disease 19%, chronic liver disease 4%, tumor 15% | ||
| Vultaggio A (22) | Single retrospective study | Mild symptom | 63 | 72±13 | Cardiovascular disease 41%, chronic kidney disease 19%, chronic liver disease 4%, tumor 15% |
| Severe symptom | 145 | 63±15 | Diabetes 27%, hypertension 58.7%, chronic lung disease 17.5%, cardiovascular disease 11.1% | ||
| Gao Y (23) | Single retrospective study | Mild symptom | 28 | 42.96±14 | Diabetes 3.57%, hypertension 25%, heart disease 8% |
| Severe symptom | 15 | 45±7.68 | Diabetes 40%, hypertension 40%, heart disease 6% | ||
| Zeng Z (24) | Single retrospective study | Mild symptom | 93 | 59 | Diabetes 18.3%, hypertension 43%, chronic kidney disease 3.2%, tumor 3.2%, chronic respiratory disease 4.3%, heart disease 10.8% |
| Severe symptom | 167 | 62 | Diabetes 18%, hypertension 36.5%, chronic kidney disease 0.6%, tumor 1.2%, chronic respiratory disease 5.9%, heart disease 10.8%, 8.4% | ||
| Liu J (25) | Single retrospective study | Mild symptom | 13 | 59.7±10.1 | Diabetes 30.8%, hypertension 38.5%, pituitary adenoma 7.7% |
| Severe symptom | 27 | 43.2±12.3 | Diabetes 7.4%, hypertension 3.7%, pituitary adenoma 3.7% |
COPD, chronic obstructive pulmonary disease.
Results
Abnormal routine blood indicators
In patients with severe COVID-19, lymphocyte count [SMD =−1.04, 95% confidence interval (95% CI): −1.21, −0.86, P<0.001, number of patients n=805], monocyte count (SMD =−0.56, 95% CI: −0.8, −0.32, P<0.001, n=403), and platelets (SMD =−1.47, 95% CI: −1.7, −1.24, P<0.001, n=653) were significantly lower compared to patients with mild symptoms. In contrast, the number of neutrophils in patients with severe symptoms (SMD =0.63, 95% CI: 0.44, 0.82, P<0.001, n=558) was higher than that in patients with mild symptoms (Table 3). The number of neutrophils in cases of death (SMD =2.77, 95% CI: 2.38, 3.16, P<0.001, n=260) was significantly higher than that in survival cases. However, the number of lymphocytes in the death cases (SMD =−2.1, 95% CI: −2.46, −1.75, P<0.001, n=260) was significantly lower than that in survival cases (Table 4).
Table 3. Summary results of standardized mean difference of biological indicators in meta-analysis for patients with severe and mild disease.
| Anomaly indicator | SMD (95% CI) | P value | I2 (%) | Number of studies | Patients with severe symptoms | Patients with mild symptoms |
|---|---|---|---|---|---|---|
| Inflammatory factors | ||||||
| IL-6 | 1.15 (1.01, 1.29) | <0.001 | 95 | 10 | 492 | 629 |
| IL-4 | 0.2 (0.01, 0.39) | 0.04 | 0 | 6 | 233 | 267 |
| IL-10 | 0.92 (0.75, 1.08) | <0.001 | 98 | 3 | 383 | 399 |
| SAA | 2.82 (2.53, 3.11) | <0.001 | 88 | 5 | 101 | 401 |
| CRP | 1.42 (1.27, 1.57) | <0.001 | 98 | 11 | 368 | 918 |
| Procalcitonin | 1.16 (0.99, 1.33) | <0.001 | 98 | 5 | 396 | 338 |
| Blood coagulation factors | ||||||
| D-D | 1.18 (0.96, 1.4) | <0.001 | 93 | 6 | 129 | 444 |
| APTT | 0.63 (0.39, 0.87) | <0.001 | 83 | 4 | 97 | 254 |
| P-T | 1.83 (1.55, 2.11) | <0.001 | 89 | 4 | 97 | 254 |
| Routine blood indexes | ||||||
| Lymphocyte count | −1.04 (−1.21, −0.86) | <0.001 | 96 | 7 | 257 | 548 |
| Neutrophil count | 0.63 (0.44, 0.82) | <0.001 | 87 | 6 | 220 | 338 |
| Monocyte count | −0.56 (−0.8, −0.32) | <0.001 | 47 | 5 | 96 | 307 |
| Platelets | −1.47 (−1.7, −1.24) | <0.001 | 97 | 6 | 117 | 536 |
| Biochemical indicators | ||||||
| AST | 2.75 (2.37, 3.12) | <0.001 | 91 | 4 | 61 | 252 |
| ALT | 2.72 (2.43, 3.02) | <0.001 | 96 | 4 | 92 | 446 |
| ALB | −2.95 (−3.21, −2.7) | <0.001 | 93 | 4 | 229 | 408 |
| LDH | 4.01 (3.79, 4.24) | <0.001 | 84 | 6 | 351 | 704 |
| Myocardial indexes | ||||||
| CK | 2.62 (2.2, 3.03) | <0.001 | 87 | 4 | 50 | 180 |
| CK-MB | 3.07 (2.81, 3.34) | <0.001 | 92 | 4 | 138 | 462 |
SMD, standardized mean difference; 95% CI, confidence interval; I2 (%), heterogeneity; IL-6, interleukin 6; IL-4, interleukin 4; IL-10, interleukin 10; SAA, serum amyloid A; CRP, C-reactive protein; D-D, D-dimer; APTT, activated partial thromboplastin time; P-T, prothrombin time; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALB, albumin; LDH, lactate dehydrogenase; CK, creatine kinase; CK-MB, creatine kinase MB isoenzyme.
Table 4. Meta-analysis summary of standardization of laboratory indicators of surviving cases and death cases.
| Anomaly indicator | SMD (95% CI) | P value | I2 (%) | Included literature | Death cases | Surviving cases |
|---|---|---|---|---|---|---|
| Inflammatory factors | ||||||
| IL-6 | 2.62 (2.15, 3.09) | <0.001 | 78 | 2 | 33 | 152 |
| PCT | 0.2 (0.16, 0.23) | <0.001 | 39 | 2 | 29 | 127 |
| Routine blood indicators | ||||||
| neutrophils | 2.77 (2.38, 3.16) | <0.001 | 55 | 3 | 54 | 206 |
| Lymphocyte count | −2.1 (−2.46, −1.75) | <0.001 | 77 | 3 | 54 | 206 |
| Biochemical indicator | ||||||
| Creatinine | 2.29 (1.87, 2.7) | <0.001 | 96 | 2 | 42 | 171 |
SMD, standardized mean difference; 95% CI, 95% confidence interval; I2 (%), heterogeneity; IL-6, interleukin 6; PCT, procalcitonin.
Lymphocytes in the patients who older than 55 years (SMD =−0.42,95% CI: −0.5, −0.34, P<0.001, n=390) was higher than that in patients who younger than 55years (Table 1).
Inflammatory factors
In patients with severe COVID-19, levels of serum interleukin-6 (IL-6; SMD =1.15, 95% CI: 1.01, 1.29, P<0.001, n=1,121), interleukin-4 (IL-4; SMD =0.2, 95% CI: 0.01, 0.39, P=0.04, n=500), interleukin-10 (IL-10; SMD =0.92, 95% CI: 0.75, 1.08, P<0.001, n=782), procalcitonin (PCT; SMD =1.16, 95% CI: 0.99, 1.33, P<0.001, n=734), serum amyloid A (SAA; SMD =2.82, 95% CI: 2.53, 3.11, P<0.001, n=502), and C-reactive protein (CRP; SMD =1.42, 95% CI: 1.27, 1.57, P<0.001, n=1,286) were significantly higher than in the milder COVID-19 patients (Table 3). In cases resulting in death, IL-6 (SMD =2.62, 95% CI: 2.15, 3.09, P<0.001, n=185) and PCT (SMD =0.2, 95% CI: 0.16, 0.23, P<0.001, n=156) were significantly higher than that in survival cases (Table 4).
Biochemical abnormalities
Aspartate aminotransferase (AST; SMD =2.75, 95% CI: 2.37, 3.12, P<0.001, n=313), alanine aminotransferase (ALT; SMD =2.72, 95% CI: 2.43, 3.02, P<0.001, n=538), and lactate dehydrogenase (LDH; SMD =4.01, 95% CI: 3.79, 4.24, P<0.001, n=1,055) in patients with severe symptoms were significantly higher than in patients with mild symptoms (Table 3), while albumin (SMD =−2.95, 95% CI: −3.21, −2.7, P<0.001, n=637) was significantly lower in patients with severe symptoms. The cardiac indexes creatine kinase (CK; SMD =2.62, 95% CI: 2.2, 3.03, P<0.001, n=230) and creatine kinase MB isoenzyme (CK-MB; SMD =3.07, 95% CI: 2.81, 3.34, P<0.001, n=600) in patients with severe symptoms were significantly higher in patients with non-severe symptoms (Table 3), while creatinine (SMD =2.29, 95% CI: 1.87, 2.7, P<0.001, n=213) in the survival cases was significantly lower than in patients who had died (Table 4).
Coagulation parameters
Patients with severe COVID-19 disease had higher levels of D-dimer (SMD =1.18, 95% CI: 0.96, 1.4, P<0.001, n=573), activated partial thromboplastin time (SMD =0.63, 95% CI: 0.39, 0.87, P<0.001, n=351), and prothrombin time (SMD =1.83, 95% CI: 1.55, 2.11, P<0.001, n=351) compared with patients with mild symptoms (Table 3).
Discussion
The results showed that IL-6, IL-4, IL-10, PCT, SAA, CRP, LDH, AST, ALT, D-dimer, P-T, APTT, CK, and neutrophil count of patients with severe COVID-19 were higher than that of patients with mild symptoms. However, the number of lymphocytes, platelets, and monocytes was low in patients with severe symptoms.
Clinical features and inflammatory factors
The most typical clinical features of patients with COVID-19 infection include fever, cough, dyspnea, muscle pain, fatigue, and symptoms of pneumonia (10) and some patients have sputum (11). Studies have shown that older patients and patients with underlying diseases will be more severely ill (12,13). Patients co-infected with COVID-19 and other respiratory pathogens had higher white blood cell count, neutrophil count, D-dimer, CRP, IL-6, and PCT than patients infected with COVID-19 alone (14). Unlike SARS-CoV, the SARS-CoV-2 exhibit a highly contagious even during the asymptomatic period (15). Studies have shown that serum levels of CRP and SAA were higher in patients with severe symptoms than in patients with mild symptoms, and that SAA better predicts disease severity than CRP (16,17). However, increased SAA levels were not able to indicate the cause or distinguish COVID-19 patients from non-infected patients (18). Therefore, detection of combined laboratory parameters is required to better indicate the development of COVID-19. In addition, these indicators can guide clinicians in assessing and treating patients, as well as assist physicians to better distinguish COVID-19 from other infections (19).
COVID-19 disease can cause multifunctional organ failure in the body accompanied by an increase in systemic blood inflammatory factors (1), with IL-6 one of the key mediators regulating the inflammatory response. Furthermore, SARS-CoV-2 is highly pathogenic, associated with rapid viral replication, and has a tendency to infect the lower respiratory tract, which could cause a severe respiratory distress response caused by IL-6 (20). Effective inhibition of the cytokine storm may contribute to further deterioration of the condition in COVID-9 patients (21). Vultaggio et al. reported that in patients with COVID-19 infection, IL-6 can be used as a biomarker for early prediction of disease progression (22). Our results suggested that increased IL-6 levels are important for assessing disease progression. Gao et al. reported that a concentration of IL-6 higher than 24.3 pg/mL could indicate the severity of COVID-19 viral pneumonia, with a sensitivity and specificity of 73.3% and 89.3%, respectively (23). The level of various inflammatory factors and liver markers, such as sIL-2R, IL-10, TNF-a, hsCRP, LDH, CK-MB, ALT, were higher in severe patients than in mild patients. (24,25). Infection with COVID-19 is associated with a ‘cytokine storm’ caused by a systemic immune response in vivo (26). After activation by infectious agents such as SARS-CoV-2, macrophages, endothelial cells, and T cells produce IL-6 and activate tissue cells, including endothelial cells and parenchymal cells, to produce inflammatory effector molecules (27). Our study indicated that elevated serum concentrations of IL-6 correlated with disease severity. An increased level of IL-6 is common in patients with respiratory dysfunction (27), which implied that the possible mechanism of cytokine-guided lung injury was caused by COVID-9 viral infection. IL-10, mainly produced by monocytes and T cells, is the most effective cytokine for reducing the inflammatory process and thus limiting tissue damage caused by inflammation (28,29). In this study, the SMD of IL-10 indicated the progression of the disease course from mild disease to severe disease. Huang et al. and Zeng et al. reported that interleukin-1 receptor antagonist (IL-1RA), interleukin-1 alpha (IL-1a), IL-2, IL-4, IL-6, IL-8, IL-7, IL-10, IL-12, IL-17, interferon gamma-induced protein 10 (IP-10), platelet-derived growth factor (PDGF), tumor necrosis factor α (TNF-a), interferon gamma (IFN-γ), granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (G-CSF), and hepatocyte growth factor (HGF) were associated with lung injury and disease severity (24,30).
Abnormal myocardial indexes and biochemical indexes
COVID-19 can affect the heart and cause fatal damage to other organs. COVID-19 infection causes chronic myocardial damage and severe cardiovascular system damage (31). Han et al. reported that high concentrations of biomarkers such as myoglobin, CK isoenzyme, amino-terminal pro-brain natriuretic peptide, and troponin I were associated with disease severity and death (32). Our analysis showed that the serum myocardial injury indexes CK and CK-MB in patients with severe COVID-19 were significantly higher than those in patients with mild COVID-19. Elevated myocardial indexes correlate with the progression of the patient's disease.
In order to better explain the link between COVID-19 and cardiovascular disease, it is necessary to understand the underlying pathological mechanisms of COVID-19 infection, SARS-CoV-2 virus, and cellular transmembrane proteins. Angiotensin-converting enzyme-2 (ACE2), the ACE homologous protein, binds to enter type II alveolar epithelial cells, macrophages, and other types of cells. This process requires cellular serine protease (TMPRSS2.12) to initiate the priming of the SARS-CoV-2 virus S protein. Therefore, the infection of SARS-CoV-2 requires the co-expression of ACE2 and TMPRSS2 in the same cell type. As a cleavage protein, the viral S protein is a necessary special protein when COVID-19 binds to ACE2 (33). In addition to type II alveolar epithelial cells, ACE2 is highly expressed in pericytes, which may contribute to microvascular dysfunction and explain a greater predisposition to acute coronary syndrome (ACS) (34).
Our results showed that ALT, AST, and LDH were significantly increased in the serum of patients with severe symptoms compared with the mild symptom group. Therefore, in addition to the lungs in patients with severe symptoms, there were other areas of organ dysfunction, including in the liver, kidney, and heart. Significantly lower albumin in patients with severe symptoms suggested that malnutrition is a common feature of COVID-19 patients.
Abnormal routine blood indicators
Alterations in several biological parameters, especially the reduction of lymphocyte counts and neutrophils, are associated with disease progression from severe disease to death (25,35). Moreover, low lymphocyte counts have been reported in other viral respiratory diseases, such as respiratory syncytial virus infection (36). The immune response marked by severe lymphopenia appears to suggest delayed complications following the early massive release of cytokines during SRAS-Cov-2 lung injury (37-39).
Another key finding of this study was thrombocytopenia in severe COVID-19 patients. It was reported that a COVID-19 thrombocytopenia-related mechanism has been proposed. COVID-19 can infect bone marrow cells, thereby reducing platelet production (40). The elevated PCT and neutrophil counts may be associated with disease severity and associated bacterial infection, which may indicate severe progression in patients.
Indicators of abnormal blood coagulation
Disseminated intravascular coagulation (DIC) and pulmonary embolism generally increase D-dimer concentrations and fibrin degradation products in COVID-19, and DIC was observed in 71.4% of patients who died from COVID-19. Currently, massive pulmonary embolism has been reported (41), although the early appearance of DIC features is usually evident. It is notable that the studies from China showed that D-dimer could increase a high degree of prediction of poor COVID-19 outcomes.
The retrospective study reported by Zhou et al. indicated that elevated D-dimer levels (>1 g/L) were strongly associated with in-hospital mortality [odds ratio (OR) 18.4, 95% CI: 2.6, 28.6, P=0.003] (42). In addition, it was reported that more discrete changes in D-dimer levels could be observed earlier in rapidly developing disease stages.
There were some limitations in our study. Firstly, due to the limited number of studies, Egger's test and funnel plots were chosen to assess publication bias, rather than forest plots and publication bias was confirmed that (P<0.05). Secondly, although some of the included studies were from outside of China, most were based in China and regional and ethnic differences are not universal.
Conclusions
Some inflammatory factors (IL-6, IL-10, IL-4, SAA, PCT, and CRP), biochemical indicators (CK-MB, D-dimer, AST, ALT, LDH, and creatinine), and routine blood indicators (lymphocyte count, neutrophil count, and platelets) were significantly associated with the progression of COVID-19 disease. These biomarkers may be useful for predicting the classification of patients at risk for COVID-19 and for further treatment and prognostic assessment.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was financially supported by the Baotou Medical College Scientific Research Fund Project (Project No. BYJJ-XG 202006).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Reporting Checklist: The authors have completed the MOOSE reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-345/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-345/coif). The authors have no conflicts of interest to declare.
References
- 1.Gandhi RT, Lynch JB, Del Rio C. Mild or Moderate Covid-19. N Engl J Med 2020;383:1757-66. 10.1056/NEJMcp2009249 [DOI] [PubMed] [Google Scholar]
- 2.Yang P, Ding Y, Xu Z, et al. Epidemiological and clinical features of COVID-19 patients with and without pneumonia in Beijing, China. bioRxiv 2020. doi: . 10.1101/2020.02.28.20028068 [DOI]
- 3.Zhou M, Qi J, Li X, et al. The proportion of patients with thrombocytopenia in three human-susceptible coronavirus infections: a systematic review and meta-analysis. Br J Haematol 2020;189:438-41. 10.1111/bjh.16655 [DOI] [PubMed] [Google Scholar]
- 4.Taghiloo S, Soltanshahi M, Aliyali M, et al. Cytokine profiling in Iranian patients with COVID-19; association with clinical severity. Iran J Immunol 2021;18:54-64. [DOI] [PubMed] [Google Scholar]
- 5.Metlay JP, Waterer GW, Long AC, et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 2019;200:e45-e67. 10.1164/rccm.201908-1581ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins J, Thomas J, Chandler J, et al. Cochrane Handbook for Systematic Reviews of Interventions (Internet). Available online: www.training.cochrane.org/handbook
- 7.Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, et al. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods 2006;11:193-206. 10.1037/1082-989X.11.2.193 [DOI] [PubMed] [Google Scholar]
- 8.Cochran WG. The combination of estimates from different experiments. Biometrics 1954;10:101-29. 10.2307/3001666 [DOI] [Google Scholar]
- 9.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian GQ, Yang NB, Ding F, et al. Epidemiologic and clinical characteristics of 91 hospitalized patients with COVID-19 in Zhejiang, China: a retrospective, multi-centre case series. QJM 2020;113:474-81. 10.1093/qjmed/hcaa089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y, You Q, Wu C, et al. Impact of Cardiovascular Disease on Clinical Characteristics and Outcomes of Coronavirus Disease 2019 (COVID-19). Circ J 2020;84:1277-83. 10.1253/circj.CJ-20-0348 [DOI] [PubMed] [Google Scholar]
- 12.Zhu Z, Cai T, Fan L, et al. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int J Infect Dis 2020;95:332-9. 10.1016/j.ijid.2020.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng F, Tang W, Li H, et al. Clinical characteristics of 161 cases of corona virus disease 2019 (COVID-19) in Changsha. Eur Rev Med Pharmacol Sci 2020;24:3404-10. [DOI] [PubMed] [Google Scholar]
- 14.Lv Z, Cheng S, Le J, et al. Clinical characteristics and co-infections of 354 hospitalized patients with COVID-19 in Wuhan, China: a retrospective cohort study. Microbes Infect 2020;22:195-9. 10.1016/j.micinf.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, Hu C, Luo L, et al. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J Clin Virol 2020;127:104364. 10.1016/j.jcv.2020.104364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Du F, Cao XJ, et al. Clinical characteristics of coronavirus disease 2019 (COVID-19) in patients out of Wuhan from China: a case control study. BMC Infect Dis 2021;21:207. 10.1186/s12879-021-05897-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu J, Huang PP, Zhang S, et al. The value of serum amyloid A for predicting the severity and recovery of COVID-19. Exp Ther Med 2020;20:3571-7. 10.3892/etm.2020.9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu SL, Wang SY, Sun YF, et al. Expressions of SAA, CRP, and FERR in different severities of COVID-19. Eur Rev Med Pharmacol Sci 2020;24:11386-94. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Dai Y, Feng M, et al. Associations between serum amyloid A, interleukin-6, and COVID-19: A cross-sectional study. J Clin Lab Anal 2020;34:e23527. 10.1002/jcla.23527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu B, Fan CY, Wang AL, et al. Suppressed T cell-mediated immunity in patients with COVID-19: A clinical retrospective study in Wuhan, China. J Infect 2020;81:e51-60. 10.1016/j.jinf.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgos-Blasco B, Güemes-Villahoz N, Santiago JL, et al. Hypercytokinemia in COVID-19: Tear cytokine profile in hospitalized COVID-19 patients. Exp Eye Res 2020;200:108253. 10.1016/j.exer.2020.108253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vultaggio A, Vivarelli E, Virgili G, et al. Prompt Predicting of Early Clinical Deterioration of Moderate-to-Severe COVID-19 Patients: Usefulness of a Combined Score Using IL-6 in a Preliminary Study. J Allergy Clin Immunol Pract 2020;8:2575-2581.e2. 10.1016/j.jaip.2020.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol 2020;92:791-6. 10.1002/jmv.25770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng Z, Yu H, Chen H, et al. Longitudinal changes of inflammatory parameters and their correlation with disease severity and outcomes in patients with COVID-19 from Wuhan, China. Crit Care 2020;24:525. 10.1186/s13054-020-03255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Li S, Liu J, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020;55:102763. 10.1016/j.ebiom.2020.102763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y, Yang H, Ji W, et al. Virology, Epidemiology, Pathogenesis, and Control of COVID-19. Viruses 2020;12:372. 10.3390/v12040372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Luo S, Shen Y, et al. Multiple enzyme release, inflammation storm and hypercoagulability are prominent indicators for disease progression in COVID-19: a multi-centered, correlation study with CT imaging score. SSRN 2020. doi: . 10.2139/ssrn.3544837 [DOI]
- 28.Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol 2010;10:170-81. 10.1038/nri2711 [DOI] [PubMed] [Google Scholar]
- 29.Iyer SS, Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol 2012;32:23-63. 10.1615/CritRevImmunol.v32.i1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lippi G, Lavie CJ, Sanchis-Gomar F. Cardiac troponin I in patients with coronavirus disease 2019 (COVID-19): Evidence from a meta-analysis. Prog Cardiovasc Dis 2020;63:390-1. 10.1016/j.pcad.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han H, Xie L, Liu R, et al. Analysis of heart injury laboratory parameters in 273 COVID-19 patients in one hospital in Wuhan, China. J Med Virol 2020;92:819-23. 10.1002/jmv.25809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020;181:271-280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Li X, Chen M, et al. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc Res 2020;116:1097-100. 10.1093/cvr/cvaa078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danwang C, Endomba FT, Nkeck JR, et al. A meta-analysis of potential biomarkers associated with severity of coronavirus disease 2019 (COVID-19). Biomark Res 2020;8:37. 10.1186/s40364-020-00217-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leroy EM, Baize S, Debre P, et al. Early immune responses accompanying human asymptomatic Ebola infections. Clin Exp Immunol 2001;124:453-60. 10.1046/j.1365-2249.2001.01517.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Liu L, Zhang D, et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet 2020;395:1517-20. 10.1016/S0140-6736(20)30920-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol 2020;92:424-32. 10.1002/jmv.25685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y, Wang Y, Shao C, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ 2020;27:1451-4. 10.1038/s41418-020-0530-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chng WJ, Lai HC, Earnest A, et al. Haematological parameters in severe acute respiratory syndrome. Clin Lab Haematol 2005;27:15-20. 10.1111/j.1365-2257.2004.00652.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang N, Li D, Wang X, et al. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 2020;18:844-7. 10.1111/jth.14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou F, Yu T, Du R, et al. Clinical course and risk actors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054-62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as