Abstract
We used arrays of bioprinted renal epithelial cell spheroids for toxicity testing with cisplatin. The concentration-dependent cell death rate was determined using a lactate dehydrogenase assay. Bioprinted spheroids showed enhanced sensitivity to the treatment in comparison to monolayers of the same cell type. The measured dose-response curves revealed an inhibitory concentration of the spheroids of IC50 = 9 ± 3 μM in contrast to the monolayers with IC50 = 17 ± 2 μM. Fluorescent labeling of a nephrotoxicity biomarker, kidney injury molecule 1 indicated an accumulation of the molecule in the central lumen of the spheroids. Finally, we tested an approach for an automatic readout of toxicity based on microscopic images with deep learning. Therefore, we created a dataset comprising images of single spheroids, with corresponding labels of the determined cell death rates for training. The algorithm was able to distinguish between three classes of no, mild, and severe treatment effects with a balanced accuracy of 78.7%.
Keywords: Bioprinting, Spheroids, Kidney, Nephrotoxicity, Deep learning
1. Introduction
The development of novel three-dimensional (3D) cell culture models is motivated by their better accuracy in predicting the physiological response of a target organ in vitro[1]. This would be beneficial for a variety of applications, including preclinical drug testing for toxicity or personalized treatment optimizations. In this context, the kidney plays a crucial role. Many substances show nephrotoxic side effects in late stage clinical studies, which were not covered in preclinical screenings[2]. To model the kidney, specific cell types were isolated or reprogrammed to provide basic characteristics of the cells found in the functional units of the kidney, that is, the nephric tubules. These include the frequently used renal proximal tubule epithelial cells (RPTECs[3]), or the more sophisticated induced renal epithelial cells (iRECs[4]). The latter could be used prospectively to establish personalized testing, since they are derived by reprogramming fibroblasts, an accessible cell source. Besides the cell type, the structure of the cell models was found to significantly influence their functionality[1-3]. The simplest cell models are two-dimensional (2D) monolayers. On top of this, the structural complexity could be increased by embedding cells in artificial 3D scaffolds to mimic the natural extracellular matrix (ECM). In various tissue engineering studies, the embedded cells showed unique mechanisms of self-assembly and formed complex 3D structures over time, including hollow spheroids[4,5] and tubules[3], both of which recapitulated nephron tubule organization and functionality. Direct comparisons with 2D monolayers revealed an increased sensitivity to treatment with the known nephron-toxicant cisplatin, which is a common reference substance[3]. Bioprinting was established as an enabling technology for the biofabrication of 3D cell culture models by providing a high degree of automation and spatial resolution. Various bioprinting techniques were developed and applied to fabricate 3D cell models with defined size and shape by spatially controlling the cell distribution in an artificial ECM[6]. In this study, we used a drop-on-demand (DoD) bioprinting technology which was previously applied in the production, handling, and treatment of cell spheroids[7], attributed to its capability to precisely deposit low volumes of low viscous bioinks, such as spheroids or cells in suspensions. Compared to classical tissue engineering approaches, bioprinting provides increased sample reproducibility, which is one key requirement for systematic screening applications. In previous studies, we have described a scalable concept of DoD bioprinting and controlled cellular self-assembly to fabricate size-defined renal spheroids and tubules in a hydrogel scaffold[8]. These structures comprise of a hollow lumen surrounded by an organized epithelial cell layer, thereby closely mimicking the nephron tubule structure. Here, we applied this concept to fabricate 3D renal spheroids from iRECs for a head-to-head comparison with 2D monolayers of the same cell type. The sensitivity to the nephron-toxicant cisplatin was investigated with different readout methods. First, we determined the cell death rate Ψ using a lactate dehydrogenase (LDH) quantification assay. Next, we fluorescently labeled a nephrotoxicity biomarker for microscopic imaging. In the context of bioprinting, machine learning image processing could prospectively contribute to improve 3D cell model generation, fabrication, and readouts[9,10]. In the latter, microscope images of cell morphology could be used to assign biochemical values (e.g., viability) in an automated manner without requiring additional assays to be conducted for each experimental setting. This again addresses important aspects of prospective screening applications, such as automation and high throughput. Here, we present a feasibility study for an automated toxicity readout using deep learning image classification based on bioprinted renal spheroids[11,12]. We trained a convolutional neural network (CNN) through supervised learning to predict the Ψ of a spheroid from its microscopic image.
2. Materials and methods
2.1. Cell culture and hydrogels
For all sample preparations, we used iRECs[4]. A detailed description of the cell type and culture conditions can be found in previous studies[4]. The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, #41966029, Gibco), with additives of fetal bovine serum (10%) and penicillin/streptomycin (1%), for cell expansion. Matrigel (100%, #356231, Corning) was used as artificial ECM material. The 3D spheroid models were cultured in renal epithelial growth medium (REGM, #CC-3190, Lonza), without addition of additives.
2.2. DoD bioprinting
For bioprinting, we used a piezo-actuated DoD bioprinting technology (PipeJet®, Biofluidix GmbH). A detailed description of the printing process can be found in our previous study[6]. In brief, single droplets (10 nl) each containing about 150 cells were deposited onto a Matrigel substrate layer. The printed cell clusters were encapsulated with a second layer of Matrigel and incubated for subsequent incubation supporting the cellular self-assembly of one spheroid per cluster.
2.3. LDH toxicity assay
We used a colorimetric LDH Assay Kit (ab65393, Abcam) to quantify the cellular release of LDH enzyme caused by a treatment with different concentrations of cisplatin (ab141398, Abcam). The cell death rate Ψ was determined relative to a lysed control (Lysed Ctrl) obtained by sample treatments with Triton X (30 min, 37°C). The treatment was conducted by incubating the cells for 24 h and changing medium subsequently. To determine Ψ, the supernatant of treated samples was collected (10 μl) and compared to an un-lysed control (Neg. Ctrl). The measured absorbance was determined by measurements of the optical density at 450 nm wavelength (OD450). A normalized solution of LDH enzyme (0.25 μg/μl, LDH Ctrl) was used to assess the assay performance.
2.4. Fluorescent image acquisition
The treated samples were imaged using a fluorescence microscope (Axio Observer.Z1/7, Carl Zeiss) with a 20-fold magnification objective (EC Plan-Neofluar 20x/0.5 M27), LED excitation, and fluorescently labeled biomarkers for nephrotoxicity. The obtained images were correlated with the treatment dose and Ψ, which was chemically determined as the relative cytotoxicity with the released LDH amount. As primary biomarker of cytotoxicity, the integrity of the cell membranes was observed, which were labeled in the cell line (iRECs) with a stable expression of membrane-localized green fluorescent protein (GFP). The kidney injury molecule 1 (KIM-1) was labeled as a specifically expressed biomarker of nephrotoxic effects[13]. For this, the protein was fluorescently labeled post-treatment with a primary antibody (Invitrogen, #PA5-79345), and a secondary antibody (Abcam, #ab6939) for the microscopic detection within spheroids. For cultivation and microscopy, the spheroid arrays were fabricated in an 8-chamber microscopy slide (m-Slide 8 well; ibidi GmbH, #80826).
2.5. Deep learning
The code was written in Python 3.8.10 using Pytorch 1.8.1. Detailed information is listed in the Supplementary File. The dataset was made of 4974 spheroid images taken after 3 days of treatment with different concentrations of cisplatin. The composition of this dataset is described in the Table S1 (225.8KB, pdf) . The network architecture and hyperparameters were chosen by automatic hyperparameter optimization. The selected architecture was a VGG11 optimized with ADAM for 90 epochs. The full list of hyperparameters is provided in the Supplementary File. The concept of automated treatment effect readout was adapted from previous studies[11,12].
3. Results and discussion
3.1. Required 3D cell count for toxicity quantification
First, the required total cell count per 3D sample was defined, which enabled the quantification of toxicity with a chemical LDH assay readout. Therefore, cell cluster arrays were fabricated, ranging from 1 to 400 spheroids (Figure 1A). The cell clusters self-assembled to spheroids of uniform size within 4 days of incubation[8]. Then, the spheroids were lysed (Lysed Ctrl), and the amount of released LDH was compared to the Neg. Ctrl. (Figure 1B). The Lysed Ctrl samples showed an increasing amount of LDH in the supernatant in relation to an increasing number of spheroids per array, as expected. In contrast, the Neg. Ctrl samples showed a constant absorbance, independent of the cell count. The positive to negative signal ratio (SP/N = Lysed Ctrl/Neg. Ctrl) was calculated to determine the minimum spheroid count required to distinguish the Lysed Ctrl. from the Neg. Ctrl. The desired value of SP/N ≥3 was achieved for arrays with at least 100 spheroids. For these, an OD450_100 = 0.8 ± 0.6 was measured, showing a high coefficient of variation (75%). A number of 225 spheroids showed an absorbance of OD450_225 = 1.3 ± 0.4. In addition, the lowest coefficient of variation was found for this format (30%). In addition, the value was in the range of the LDH Ctrl, which was measured to OD450_LDH Ctrl = 1.6 ± 0.3, corresponding to 83 ± 25%. The largest array format (400 spheroids) showed an absorbance of 114 ± 47% relative to the LDH Ctrl, however, with an increased coefficient of variation. Based on these findings, spheroid arrays consisting of 225 spheroids, or 34×106 cells, were selected for the quantification of toxicity using an LDH assay.
Figure 1.
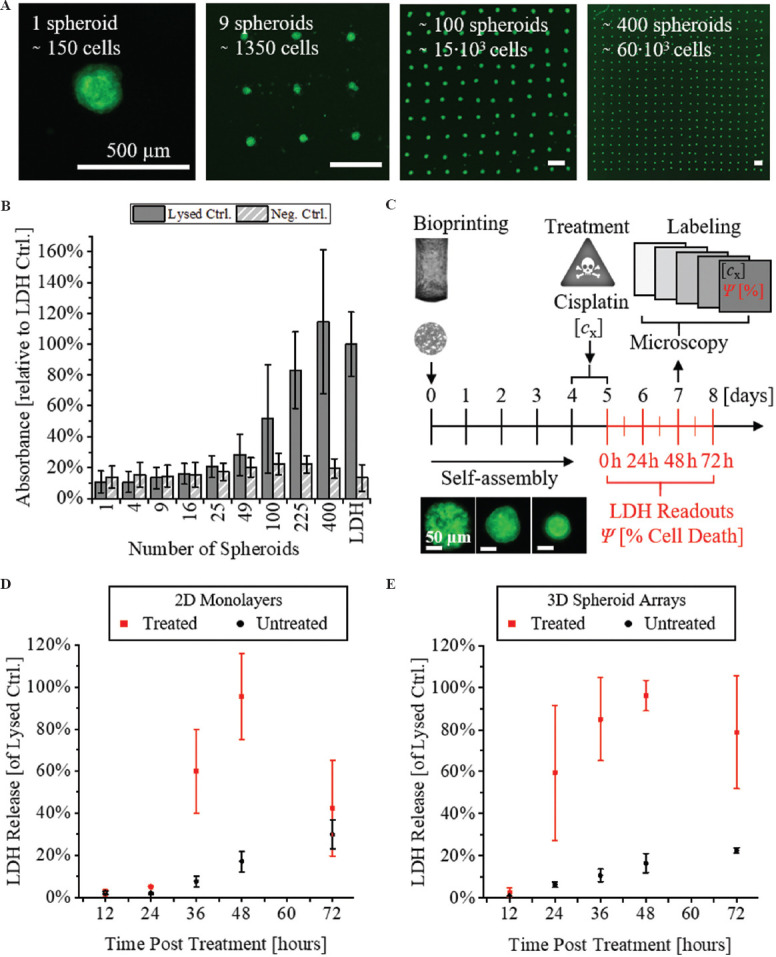
LDH toxicity readout with 3D renal spheroids. (A) Fluorescent microscope images of bioprinted spheroid arrays with different formats. Scale bars: 500 μm. (B) Quantification of total LDH release after lysis of different array formats. (C) Schematic timeline of sample preparation, treatment, and readout. (D and E) Time-dependent LDH release of 2D monolayers versus 3D spheroids at different time points post-treatment.
3.2. 3D response time to toxic treatment
Next, a suitable time-point for the toxicity readout was determined. A schematic timeline of the experimental procedure is shown in Figure 1C. All samples were prepared by bioprinting 4 days before a treatment with cisplatin, a common nephrotoxic substance[14]. Two types of samples were prepared, either classical monolayers (2D) or bioprinted arrays of 225 spheroids (3D Samples). On day 4 of incubation, the medium was replaced by a cell culture medium containing a dilution of cisplatin (cCisplatin = 100 μM). The treatment was conducted at 37°C for 24 h. Then, the cisplatin dilution was replaced by fresh medium. At this time point, we started the readout. During the subsequent incubation period, the supernatant was sampled at different time points (0, 12, 24, 36, 48, and 72 h post treatment), and the release of LDH was quantified. The measured LDH release curves for the 2 sample types are shown in Figure 1D and E. For 2D monolayer cultures, no significant LDH release was detected within the first 24 h (Figure 1D). After 36 h, the LDH release accounted for 60 ± 20% and increased to 96 ± 20% after 48 h, relative to the Lysed Ctrl. At 72 h post-treatment, the released LDH decreased to 42 ± 23%. The decrease in measured LDH could be related to the degradation of the enzyme in the culture supernatant. For the untreated control group (Neg. Ctrl), the release of LDH continuously increased over time, reaching a maximum of 10 ± 7% at 72 h. This could be explained by the fact that the 2D monolayer cultures were fully confluent. At this point, the cells in culture need to adapt to the limited growth area and nutrient deficiency, which could cause unspecific cell lysis and LDH release. The 3D spheroid arrays showed a distinct behavior compared to the 2D monolayers (Figure 1E). The first increase in LDH release occurred 24 h post-treatment, reaching 60 ± 32% of the Lysed Ctrl. After 48 h, the maximum was reached at 96 ± 7%. Afterward, the value declined to 79 ± 27% (72 h). Again, this decline could be explained by an impairment of the LDH activity due to the progressive incubation period. The untreated control shared a similar behavior with the 2D monolayers, with a slight and static linear increase of released LDH over time, reaching a maximum of 23 ± 1% at 72 h. This indicates again that within 3D spheroid cultures, unspecific cell lysis occurred, which was not related to a toxic treatment. This finding is in good accordance with the previously published data[15]. Taken together, from the experiments, the optimal time-point for the LDH-assay based readout was found to be at ttreatment = 48 h. The dynamic observation of LDH release after cisplatin treatment indicated significant differences between 2D and 3D cell models. The faster onset of toxicity in 3D models could be related to a more efficient uptake of the toxin. In the case of cisplatin, the uptake occurs through transport proteins located on the basal side of the cells. This side is facing toward the cisplatin containing culture medium in the case of hydrogel embedded 3D spheroids, while in 2D monolayers it is in contact to the solid culture vessel surface.
3.3. Dose-response curves
Next, dose-dependent treatment effects were quantified to derive dose-response curves for different cell culture models. For head-to-head comparison, three parameters were used, including the inhibitory concentration leading to 50% cell death (IC50), and the maximum and minimum response (Emax, Emin). As described in literature, clinically relevant cisplatin concentrations were in the range of 10 μM[16]. Based on this value, we prepared a dilution series (cCisplatin = 0.075, 0.5, 1, 2, 4, 8, 16, 32, 64, and 128 μM) for the treatment. Representative fluorescence microscope images of each sample type are shown in Figure 2A. For both sample types, the mean absorbance of three replicates for each treatment condition was calculated relative to the Lysed Ctrl. The dose-response curves were derived using a pharmacological dose-response fit function (see equation (1) in Supplementary File). The 2 sample types showed distinct curves and relations to the treatment concentration (Figure 2B). 2D monolayers showed an IC50 2D Ref. = 17 ± 2 μM, with a maximum response of Emax = 95 ± 7% and Emin = 15 ± 8% relative to the Lysed Ctrl. A significant lower IC50 was found for treated spheroid arrays compared to 2D monolayers, with an IC50 3D Spheroids = 9 ± 3 μM and similar values of Emin and Emax. The determined IC50 values of the 3D bioprinted models were in good accordance with the previously published data. In other studies, reported values were 5.72 μM[17], between 10 and 50 μM[3], or > 30 μM[18]. It should be noted that each of the reported data was based on experiments on different cell types, treatment protocols, and readout methods. Thus, a quantitative head-to-head comparison would not be conclusive. The reported values herein indicated that the fabricated 3D models showed similar sensitivities to cisplatin treatments as reported for other 3D models. In addition, a higher sensitivity compared to a 2D cell culture of the same cell type under identical treatment conditions were found, which is consistent with previous reports[3].
Figure 2.
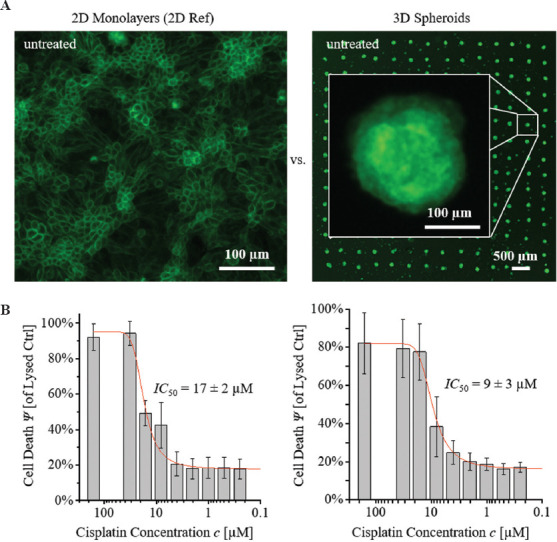
Head-to-head comparison. (A) 2D monolayers and 3D spheroid arrays, comprising of iREC cells. (B) Dose-response curves for a treatment with cisplatin.
3.4. Microscopic readout
In addition to the chemical readout, we investigated whether microscopic observations of the cell morphology could be used to assess toxicity effects in the 3D spheroids. With this, we aimed to achieve a higher sensitivity, compared to the chemical readout. Cytotoxic effects could become detectable before cell membrane lysis occurs, which leads to the release of LDH. Since the lysis of the cell membrane is considered the final step of cell death, this assumption is plausible. A cisplatin serial dilution was prepared in REGM, and the treatment was conducted according to the process and timeline described in Figure 1C. Three replicates of each treatment condition were used for microscopic observations. For the KIM-1 nephrotoxicity marker expression assessment, samples were fixed (PFA 4%, 1 h) before immunolabeling was performed. Representative false-color images of the observed spheroid morphologies for different treatment concentrations are shown in Figure 3. The observations showed a disintegration of the spheroids at treatment concentrations cCisplatin ≥ 32 μM. The outer boundary of the spheroids was found to be disrupted in the bright field and GFP channel, and no intact cell layer was outlining the spheroids. The KIM-1 signal was detected throughout the entire spheroid structures. For lower treatment concentrations (cCisplatin ≤ 16 μM), a distinct signal distribution was detected. The KIM-1 positive signal was exclusively located at the central lumen of the spheroids. Furthermore, we observed an intact cell layer surrounding the lumen without positive KIM-1 signal. This indicated that the KIM-1 proteins were transported out of the cells and accumulated in the lumen of the spheroids. This accumulation was later found to undergo a decline with decreasing treatment concentrations. The untreated control showed very low levels of KIM-1 positive signals. The KIM-1 unlabeled samples showed no detectable KIM-1 signals, confirming the specificity of the immunolabeling process. This observation was in good accordance with physiological observations in patients, where an increase in KIM-1 signal at the apical side of the renal tubules was detected as a consequence of kidney injury. This, in turn, led to an increase of KIM-1 concentrations in the urine, indicating an active transport and release of KIM-1 into the nephron lumen[19]. Thus, the accumulation of KIM-1 molecules found here hints at a basic physiological transport mechanism within the developed spheroid models.
Figure 3.
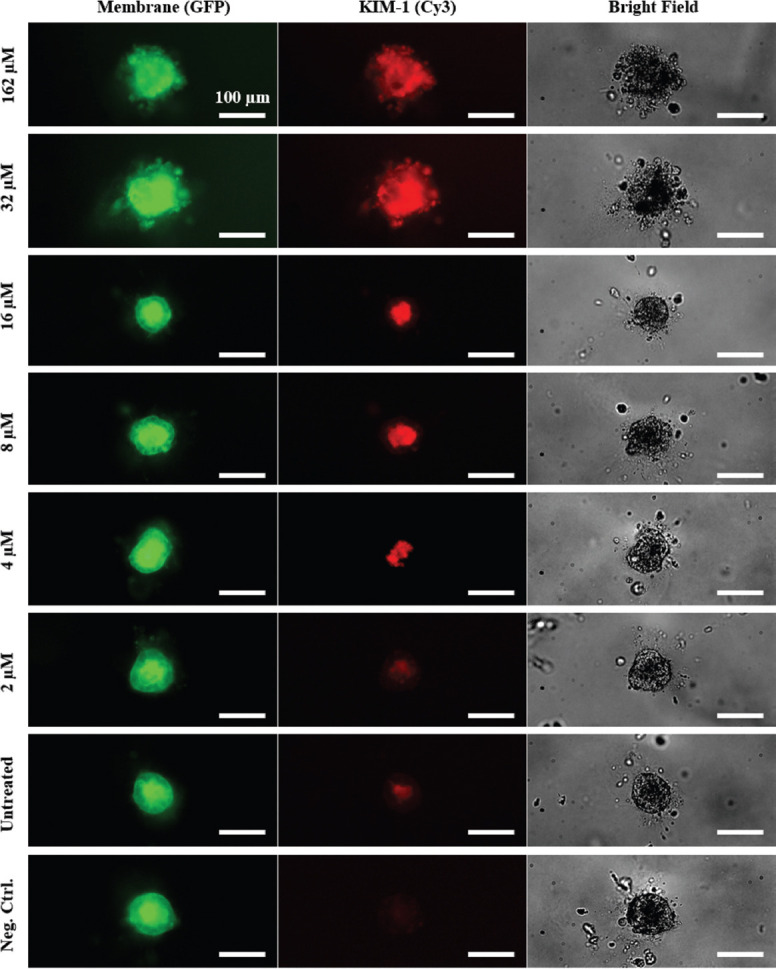
Fluorescence microscope images of cisplatin treated renal spheroids. The fluorescence channels show membrane-localized GFP (green) and nephrotoxicity biomarker KIM-1 (red). Scale bars: 100 μm.
3.5. Deep learning
Finally, we investigated the feasibility of automatic determination of Ψ of single spheroids from their morphological appearance in microscope images. A schematic process diagram is shown in Figure 4A. For this purpose, we developed a deep learning system based on CNN trained via supervised learning. We used fluorescence images from the GFP channel. The images were labeled with their Ψ values, as estimated by the dose-response curves (Figure 2). The network was trained as a regressor that takes an image of a single spheroid as input and a scalar corresponding to its estimated Ψ as output. The hyperparameters of the training pipeline were tuned via Bayesian Optimization[20], as implemented by (Bayesian Optimization with Hyperband [BOHB][20]). The hyperparameter search space and the optimum configuration found by BOHB are shown in the Table S2 (225.8KB, pdf) . The following results originate from this configuration. The dataset was split for training and testing in the proportions of 80 – 20%, respectively. The images were cropped to 450 × 450 px with the spheroid positioned at the center of the image. Online data augmentation was used. At each epoch, the images were randomly transformed within the ranges as shown in the Table S3 (225.8KB, pdf) . Training was conducted for 90 epochs, which resulted in a test mean squared error of 7.67×10-3. Figure 4B shows the predictions made by the network on the test dataset. It is possible to see that it performs well to differentiate spheroids with Ψ < 30% from those above 80%. Nevertheless, it fails to tell apart rates above 50%. Visual inspection of the dataset demonstrated that this was indeed a difficult task, as shown in Figure 4C. The accuracy of this network can be appreciated by framing the regression problem as a classification by binning the regressor outputs into discrete intervals.
Figure 4.
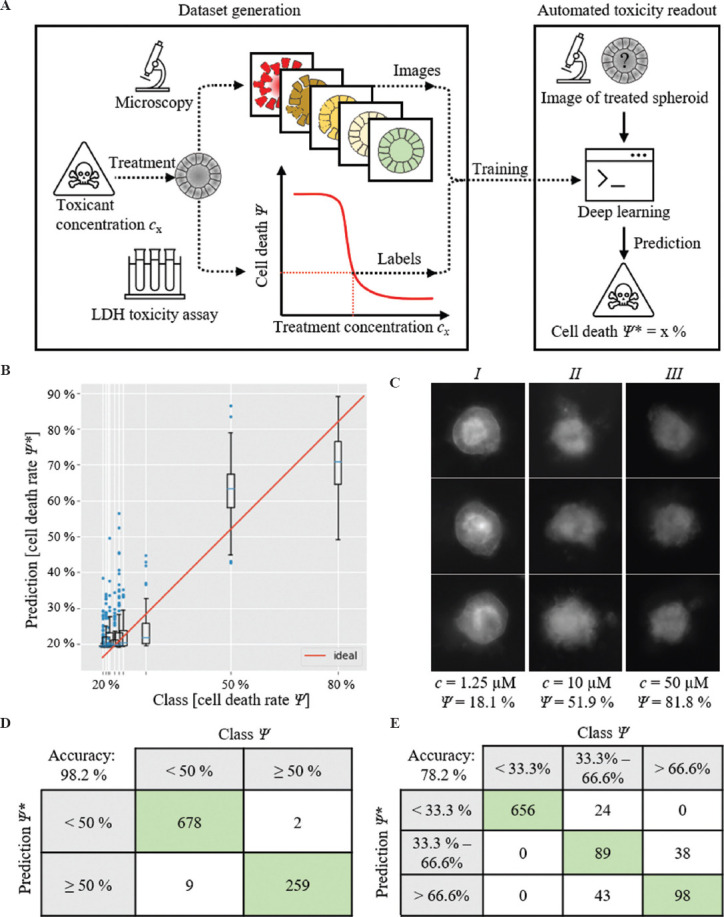
(A) Schematic of automatic toxicity readout. (B) Class vs. prediction box plot. (C) Fluorescent images of spheroids treated with different cisplatin concentrations c and corresponding cell death rates Ψ: Spheroids with low death rates (I) show obviously different appearances. Spheroids with intermediate (II) and high (III) death rates show similar appearances. (D and E) Confusion matrices when using the neural network as a classifier. The 2-class problem delivered a balanced accuracy: 98.2%. The 3-class problem delivered an accuracy of 78.7%.
We report the results of 2 classification formulations: first, a 3-class problem, where classes are divided based on death rate, that is, < 33.3%, between 33.3% and 66.6%, and more than 66.6%; second, a binary classification to distinguish 2 classes, either below or above the IC50. The results are shown as confusion matrices in Figure 4D and E. The 3-class problem achieved a balanced accuracy (mean per-class accuracy) of 78.7%. As expected, the main issue was to correctly differentiate spheroids with intermediary death rates from those with high death rates. On the other hand, the binary classification achieves a balanced accuracy of 98.2%, which confirms that the network can differentiate extreme values of death rate. These results are encouraging for a proof-of-concept study, but in the short term, small improvements should be attempted to further increase the network’s accuracy. The first step is to collect a dataset with good quality bright-field images. These images may contain vital information to improve the performance on the medium to high death rate range, where our network performed the worst. Second, the dataset should cover the intermediate death rate values better. The dataset used in this study has a uniform distribution of cisplatin concentrations (in the log scale), but due to the sigmoidal profile of the dose-response curve, this distribution translates to a sparse sampling of intermediate values of death rate. The current lack of training data for intermediate death rates is very likely to be reducing the performance of the deep learning network.
4. Conclusion and outlook
The presented results successfully demonstrated a concept for automated toxicity testing with renal spheroids. 3D bioprinting technology enabled scalable, reproducible, and automated fabrication of renal spheroids. In a head-to-head comparison, functional differences between 2D and 3D cell models were found. Toxic treatment effects varied with time and quantity, indicating increased sensitivity to the specific toxicant. This clearly demonstrated how 3D cell models could be of increasing relevance for future applications, such as toxicity studies. Deep learning image classification enabled an automated image-based readout. Although relatively high accuracies were achieved in this study, further improvements could be implemented in the future. To improve the performance, the present dataset could be augmented with additional images of the cell morphologies. Therefore, more biomarkers[13] and cell viability assays (e.g., MTT, PrestoBlue, etc.)[21] could be applied to generate ideally multi-channel and -dimensional fluorescence images, which could then be used to train the algorithm. With this additional image information, an increased performance could potentially be achieved, which would contribute to image-based toxicity readouts with higher precision in the future.
Acknowledgments
This work was supported by the Baden-Württemberg Stiftung (Grant No IAF-3). The authors acknowledge to S.S.L. and S.Z,. and the European Research Council to S.S.L. (grant agreement No 804474, DiRECT), and the Swiss National Science Foundation to S.S.L. (NCCR Kidney.CH).
Conflict of interest
The authors have no conflicts to declare.
Author contributions
K.T., G.M., S.Z., R.Z., S.L., R.P., and S.K. provided the conceptual design. S.L. and S.Z. were responsible for formal analysis. S.K., S.Z., R.Z., P.K., and S.L. acquired funding. K.T., G.M., L.R., R.P., and F.K. conducted experiments including cell culture, bioprinting, imaging, and investigation. F.K., K.T., R.P., L.R., P.K., and S.Z. provided resources. S.Z.; R.Z. supervised the work. K.T., G.M., and S.Z. were responsible for writing and visualization.
References
- 1.Breslin S, O'Driscoll L. Three-dimensional Cell Culture:The Missing Link in Drug Discovery. Drug Discov Today. 2013;18:240. doi: 10.1016/j.drudis.2012.10.003. https://doi.org/10.1016/j.drudis.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Homan KA, Kolesky DB, Skylar-Scott MA, et al. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Sci Rep. 2016;6:34845. doi: 10.1038/srep34845. https://doi.org/10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Secker PE, Luks L, Schlichenmaier N, et al. RPTEC/TERT1 Cells form Highly Differentiated Tubules when Cultured in a 3D Matrix. ALTEX. 2018;35:223. doi: 10.14573/altex.1710181. https://doi.org/10.14573/altex.1710181. [DOI] [PubMed] [Google Scholar]
- 4.Kaminski MM, Tosic J, Kresbach C, et al. Direct Reprogramming of Fibroblasts into Renal Tubular Epithelial Cells by Defined Transcription Factors. Nat Cell Biol. 2016;18:1269. doi: 10.1038/ncb3437. https://doi.org/10.1038/ncb3437. [DOI] [PubMed] [Google Scholar]
- 5.Buzhor E, Harari-Steinberg O, Omer D, et al. Kidney Spheroids Recapitulate Tubular Organoids Leading to Enhanced Tubulogenic Potency of Human Kidney-derived Cells. Tissue Eng Part A. 2011;17:2305. doi: 10.1089/ten.TEA.2010.0595. https://doi.org/10.1089/ten.TEA.2010.0595. [DOI] [PubMed] [Google Scholar]
- 6.Mota C, Camarero-Espinosa S, Baker MB, et al. Bioprinting:From Tissue and Organ Development to In Vitro Models. Chem Rev. 2020;120:10547. doi: 10.1021/acs.chemrev.9b00789. https://doi.org/10.1021/acs.chemrev.9b00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutzweiler L, Kartmann S, Troendle K, et al. Large Scale Production and Controlled Deposition of Single HUVEC Spheroids for Bioprinting Applications. Biofabrication. 2017;9:25027. doi: 10.1088/1758-5090/aa7218. https://doi.org/10.1088/1758-5090/aa7218. [DOI] [PubMed] [Google Scholar]
- 8.Tröndle K, Rizzo L, Pichler R, et al. Scalable Fabrication of Renal Spheroids and Nephron-like Tubules by Bioprinting and Controlled Self-assembly of Epithelial Cells. Biofabrication. 2021;13:185. doi: 10.1088/1758-5090/abe185. https://doi.org/10.1088/1758-5090/abe185. [DOI] [PubMed] [Google Scholar]
- 9.Ng WL, Chan A, Ong YS, et al. Deep Learning for Fabrication and Maturation of 3D Bioprinted Tissues and Organs. Virtual Phys Prototyp. 2020;15:340. https://doi.org/10.1080/17452759.2020.1771741. [Google Scholar]
- 10.Yu C, Jiang J. A Perspective on Using Machine Learning in 3D Bioprinting. Int J Bioprint. 2020;6:253. doi: 10.18063/ijb.v6i1.253. https://doi.org/10.18063/ijb.v6i1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benning L, Peintner A, Finkenzeller G, et al. Automated Spheroid Generation, Drug Application and Efficacy Screening Using a Deep Learning Classification:A Feasibility Study. Sci Rep. 2020;10:11071. doi: 10.1038/s41598-020-67960-0. https://doi.org/10.1038/s41598-020-67960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Greca A, Pérez N, Castañeda S, et al. Celldeath:A Tool for Detection of Cell Death in Transmitted Light Microscopy Images by Deep Learning-based Visual Recognition. PLoS One. 2020;16:e0253666. doi: 10.1371/journal.pone.0253666. https://doi.org/10.1101/2020.03.22.002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SY, Moon A. Drug-induced Nephrotoxicity and its Biomarkers. Biomol Ther. 2012;20:268. doi: 10.4062/biomolther.2012.20.3.268. https://doi.org/10.4062/biomolther.2012.20.3.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao X, Panichpisal K, Kurtzman N, et al. Cisplatin Nephrotoxicity:A Review. Am J Med Sci. 2007;334:115. doi: 10.1097/MAJ.0b013e31812dfe1e. https://doi.org/10.1097/MAJ.0b013e31812dfe1e. [DOI] [PubMed] [Google Scholar]
- 15.Tröndle K, Koch F, Finkenzeller G, et al. Bioprinting of High Cell Density Constructs Leads to Controlled Lumen Formation with Self-assembly of Endothelial Cells. J Tissue Eng Regen Med. 2019;13:1883–95. doi: 10.1002/term.2939. https://doi.org/10.1002/term.2939. [DOI] [PubMed] [Google Scholar]
- 16.Salas S, Mercier C, Ciccolini J, et al. Therapeutic Drug Monitoring for Dose Individualization of Cisplatin in Testicular Cancer Patients Based Upon Total Platinum Measurement in Plasma. Ther Drug Monit. 2006;28:532–9. doi: 10.1097/00007691-200608000-00008. https://doi.org/10.1097/00007691-200608000-00008. [DOI] [PubMed] [Google Scholar]
- 17.King SM, Higgins JW, Nino CR, et al. 3D Proximal Tubule Tissues Recapitulate Key Aspects of Renal Physiology to Enable Nephrotoxicity Testing. Front Physiol. 2017;8:123. doi: 10.3389/fphys.2017.00123. https://doi.org/10.3389/fphys.2017.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vormann MK, Gijzen L, Hutter S, et al. Nephrotoxicity and Kidney Transport Assessment on 3D Perfused Proximal Tubules. AAPS J. 2018;20:90. doi: 10.1208/s12248-018-0248-z. https://doi.org/10.1208/s12248-018-0248-z. [DOI] [PubMed] [Google Scholar]
- 19.Han WK, Bailly V, Abichandani R, et al. Kidney Injury Molecule-1 (KIM-1):A Novel Biomarker for Human Renal Proximal Tubule Injury. Kidney Int. 2002;62:237. doi: 10.1046/j.1523-1755.2002.00433.x. https://doi.org/10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- 20.Falkner S, Klein A, Hutter F. BOHB:Robust and Efficient Hyperparameter Optimization at Scale. 2018 International Conference on Machine Learning [Google Scholar]
- 21.Boncler M, Różalski M, Krajewska U, et al. Comparison of PrestoBlue and MTT Assays of Cellular Viability in the Assessment of Anti-proliferative Effects of Plant Extracts on Human Endothelial Cells. J Pharmacol Toxicol Methods. 2014;69:9. doi: 10.1016/j.vascn.2013.09.003. https://doi.org/10.1016/j.vascn.2013.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


