Here, Kundu et al. investigated the role of the microRNA-183/96/182 cluster (m96cl) in lung cancer and used a novel conditional m96cl mouse to establish that loss of m96cl accelerated the growth of K-Ras mutant autochthonous lung adenocarcinomas. Overall, the authors identified a novel mechanistic role of the m96cl in the suppression of lung cancer growth and metastasis by inducing an IL2-mediated systemic CD8+ CTL immune response.
Keywords: CD8+ T lymphocytes, CTL, tumor immune microenvironment, microRNA-183/96/182, antitumor immune response, interleukin-2, EMT, Zeb1, Foxf2, lung cancer
Abstract
One of the mechanisms by which cancer cells acquire hyperinvasive and migratory properties with progressive loss of epithelial markers is the epithelial-to-mesenchymal transition (EMT). We have previously reported that in different cancer types, including nonsmall cell lung cancer (NSCLC), the microRNA-183/96/182 cluster (m96cl) is highly repressed in cells that have undergone EMT. In the present study, we used a novel conditional m96cl mouse to establish that loss of m96cl accelerated the growth of Kras mutant autochthonous lung adenocarcinomas. In contrast, ectopic expression of the m96cl in NSCLC cells results in a robust suppression of migration and invasion in vitro, and tumor growth and metastasis in vivo. Detailed immune profiling of the tumors revealed a significant enrichment of activated CD8+ cytotoxic T lymphocytes (CD8+ CTLs) in m96cl-expressing tumors, and m96cl-mediated suppression of tumor growth and metastasis was CD8+ CTL-dependent. Using coculture assays with naïve immune cells, we show that m96cl expression drives paracrine stimulation of CD8+ CTL proliferation and function. Using tumor microenvironment-associated gene expression profiling, we identified that m96cl elevates the interleukin-2 (IL2) signaling pathway and results in increased IL2-mediated paracrine stimulation of CD8+ CTLs. Furthermore, we identified that the m96cl modulates the expression of IL2 in cancer cells by regulating the expression of transcriptional repressors Foxf2 and Zeb1, and thereby alters the levels of secreted IL2 in the tumor microenvironment. Last, we show that in vivo depletion of IL2 abrogates m96cl-mediated activation of CD8+ CTLs and results in loss of metastatic suppression. Therefore, we have identified a novel mechanistic role of the m96cl in the suppression of lung cancer growth and metastasis by inducing an IL2-mediated systemic CD8+ CTL immune response.
The development of new therapeutic strategies for the treatment of metastatic lung cancers necessitates the thorough understanding of the underlying basic biological mechanisms of metastasis. For this, the genetically engineered KrasLA1/+; p53R172HΔG/+ (KP) (Zheng et al. 2007; Gibbons et al. 2009) mouse model is used, as it closely recapitulates the human disease and produces spontaneous multifocal lung adenocarcinomas with local invasion and a 30% incidence rate for fulminant metastases. From these murine models, syngeneic immunocompetent cell line models have been derived, which are comprised of a panel of tumor cell lines isolated from primary lung tumors or metastases from the KP mice. These cell lines vary in their metastatic potentials when injected in syngeneic wild-type animals (Gibbons et al. 2009; Ahn et al. 2012; Yang et al. 2014). Multiple groups have shown that in these models and in many other tumor types, the microRNA-200 family (miR-200) and the ZEB family of transcriptional repressors modulate EMT by regulating each other through a double-negative feedback loop (Bracken et al. 2008; Burk et al. 2008; Gregory et al. 2008; Gibbons et al. 2009). MicroRNA-200 expression was strongly correlated with expression of microRNA-183/96/182 (m96cl) in different epithelial tumor types, including NSCLC, whereas both these microRNA groups showed significant negative correlation with EMT status (Kundu et al. 2016). It was also shown that these microRNA families convergently target the EMT transcriptional repressors Zeb1 and Foxf2, which can potently regulate EMT, invasion, and metastasis in lung cancers by transcriptional repression of E-Cadherin and miR-200. However, it has not been determined whether m96cl has an independent function in regulating tumorigenesis or metastasis in lung cancers.
An adaptive immune response to a growing tumor is a hallmark of cancer and plays a dynamic role in the progression and outcome of the disease (Hanahan and Weinberg 2011; Hanahan and Coussens 2012). One of the crucial antitumor immune cell populations in the tumor microenvironment is the tumor-infiltrating effector CD8+ cytotoxic T lymphocytes (CTLs) (Restifo et al. 2012; Farhood et al. 2019; van der Leun et al. 2020). Upon primary stimulation of CD8+ T lymphocytes by dendritic cell-mediated tumor antigen presentation through MHC-I, the effector CD4+ T cells costimulate the CD8+ T lymphocytes into an effector cytolytic state through release of a milieu of inflammatory cytokines (Chen et al. 2018b; Farhood et al. 2019). Among these cytokines, interleukin-2 (IL2) is of primary importance, as at lower doses it is critical to drive the proliferation and sustained maintenance of the activated CD8+ CTLs in an effector cytolytic state. In contrast, high levels of IL2 induce proliferation of CD4+ Treg cells, which in combination with other immune-suppressive cells (myeloid-derived suppressor cells and macrophages) results in switching of effector CD8+ CTLs to a nonfunctional exhausted state (Kalia et al. 2010; Malek and Castro 2010; Wherry 2011; Josefowicz et al. 2012; Spolski et al. 2018). Therefore, a fine balance of IL2 in the tumor microenvironment is critical for the cytolytic activity of antitumor CD8+ CTLs. Cancer types that show increased abundance of these CD8+ CTLs in the tumor microenvironment show an elevated and sustainable response toward targeted immunotherapies (Restifo et al. 2012; Spolski et al. 2018). The underlying mechanisms influencing the activation, recruitment, and infiltration of CD8+ CTLs in the tumor microenvironment is not fully understood and is a topic of great interest to the scientific community. Our previous work has established that in lung and breast cancer models, EMT is a major factor that controls the infiltration of CD8+ CTLs in the tumors and also determines metastatic outcomes (Chen et al. 2014b; Lou et al. 2016; Joseph et al. 2021).
In the present study, we used a novel conditional m96cl mouse to show that m96cl functions as an important tumor and metastasis suppressor in lung adenocarcinomas. We further elucidated a mechanism whereby elevated expression of m96cl in epithelial tumors results in an IL2-mediated paracrine activation of CD8+ CTLs to elicit an antitumor immune response.
Results
Loss of the microRNA-183/96/182 cluster results in accelerated tumor growth and decreased survival of Kras-driven lung cancers in a novel GEMM
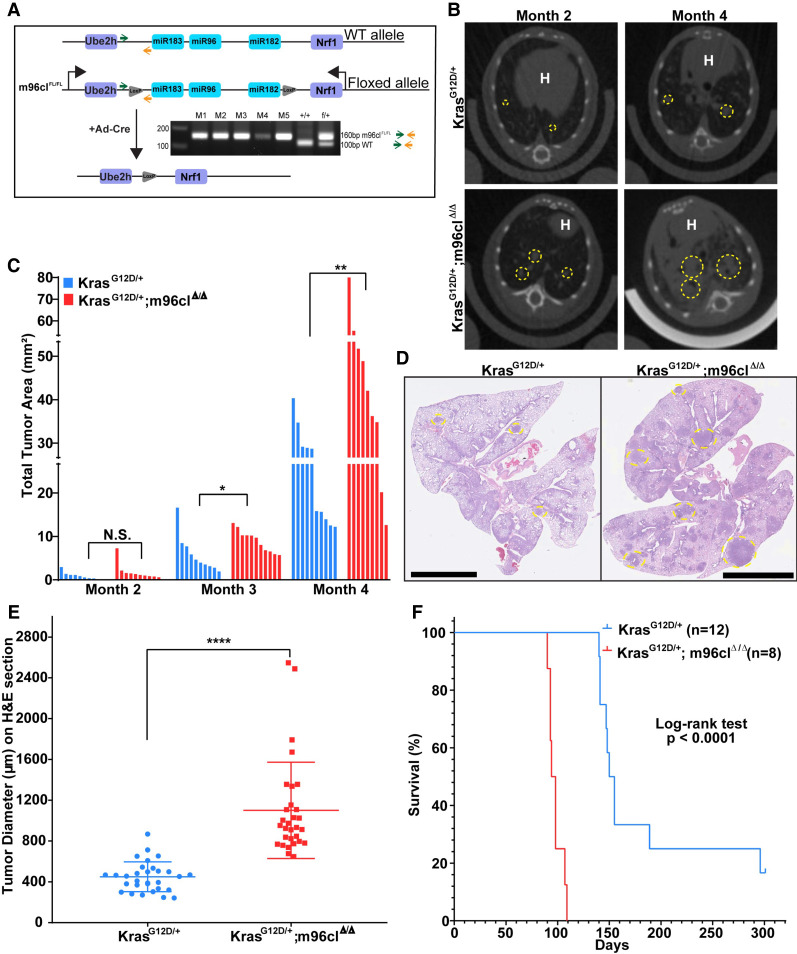
Previous work from our group has shown the combined role of the microRNA-183/96/182 cluster (m96cl) and the miR-200 family in repressing lung cancer metastasis (Kundu et al. 2016). Therefore, to determine whether m96cl has an independent role in driving tumorigenesis and metastasis, we generated the novel genetically engineered mouse, m96clFL, where Cre-recombinase-mediated deletion of the m96cl could be induced by introduction of loxP sites flanking the microRNA-183/96/182 cluster stem–loop (Supplemental Fig. S1A, panel i). Allelic integration was validated by Southern blotting (Supplemental Fig. S1E–G). We crossed m96clFL mice with KrasLSL-G12D mice to generate m96clFL/FL; KrasLSL-G12D mice. Mice expressing the targeted alleles were validated by PCR amplification of the stem–loop loxP site (Fig. 1A; Supplemental Fig. S1A, panel ii). Intratracheal delivery of adenoviral-Cre produced autochthonous lung adenocarcinomas with an activated KrasG12D and homozygous loss of the microRNA-183/96/182 cluster (KrasG12D/+; m96clΔ/Δ). Furthermore, we verified by qPCR analysis that m96cl expression was lost in the autochthonous lung tumors from the KrasG12D/+; m96clΔ/Δ mice, as compared with the KrasG12D/+ mice (Supplemental Fig. S1B). Micro-CT image analysis performed monthly after induction revealed that loss of m96cl caused a significant acceleration in growth of lung adenocarcinomas from as early as month 3 (Fig. 1B,C; Supplemental Fig. S1C). To ascertain the increased tumor burden upon m96cl loss, we quantified the tumor diameters in the H&E-stained lung sections upon sacrifice of the animals. We observed that the average size of the tumor nodules in the KrasG12D/+; m96clΔ/Δ mice was significantly larger than those from the KrasG12D animals (Fig. 1D,E; Supplemental Fig. S1D). Additionally, we found that the KrasG12D/+; m96clΔ/Δ mice had a median survival of 96 d, which was significantly shorter than the KrasG12D/+ mice, with a median survival of 152 d based on a log rank test (Fig. 1F). Overall, these data establish that loss of the microRNA-183/96/182 cluster increases tumor growth and has a consequential role in regulating lung tumorigenesis and progression.
Figure 1.
Loss of microRNA-183/96/182 cluster results in accelerated tumor growth and decreased survival of Kras-driven lung cancers in a novel GEMM. (A) Schematic showing deletion of m96cl in the m96clFL mice upon Ad-Cre infection. PCR gel showing screening for mice with the engineered allele to show the presence of the LoxP site. (B) Representative micro-CT images of KrasG12D/+ and KrasG12D/+; m96clΔ/Δ lungs at months 2 and 4. Yellow dashed circles outline target lesions. “H” indicates the mouse heart. (C) Total tumor area as assessed by micro-CT imaging of mouse lungs at months 2, 3, and 4. n = 9–12. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (*) P < 0.05, (**) P < 0.01. (D) Representative H&E images of mouse lungs from C at month 4. Yellow dashed circles outline lung tumors. Scale bar, 6 mm. (E) Quantification of tumor diameter on H&E-stained lungs from KrasG12D/+ and KrasG12D/+; m96clΔ/Δ mice. (****) P < 0.0001. (F) Kaplan–Meier survival curve of KrasG12D/+ and KrasG12D/+; m96clΔ/Δ mice. n = 8–12. Statistical difference determined using log rank test.
Induced expression of the microRNA-183/96/182 cluster suppresses cancer cell invasion and metastasis
We have reported that transient coexpression of miR-183, miR-96, and miR-182 in NSCLC cells resulted in reduced cellular migration and invasion (Kundu et al. 2016). To understand the functional implication of stable overexpression of m96cl on cancer cell growth, invasion, and metastasis, we generated a doxycycline-inducible construct expressing the complete microRNA-183/96/182 genomic cluster. This construct was used to produce stable inducible m96cl-expressing cell lines using the mesenchymal-like, metastatic (KrasLA1/+; TP53R172HΔG/+ [KP]) 344SQ and 344LN murine lung cancer cells (Zheng et al. 2007; Gibbons et al. 2009). We confirmed overexpression of all members of the m96cl as early as 24 h after Dox induction (Fig. 2A; Supplemental Fig. S2A). Consistent with our previously published data, the induced expression of miR96cl promoted a mesenchymal-to-epithelial transition in the mesenchymal-like 344SQ and 344LN cells. This was evidenced phenotypically, as the cells showed a rounded and clustered epithelial morphology in 2D (Fig. 2B), as well as molecularly by elevated E-Cadherin expression and suppression of Zeb1 and vimentin expression as shown by qPCR analysis. Foxf2 and Adcy6 are known targets of the m96cl and were included as controls (Supplemental Fig. S2B,D). Overexpression of m96cl suppressed invasiveness of the 344SQ and 344LN cells in 2D transwell assays (Fig. 2C; Supplemental Fig. S2C,E). Additionally, using a 1.5 mg/mL collagen/Matrigel matrix, which recapitulates the tumor extracellular matrix (ECM), and quantifying the structures with invasive protrusions on days 4, 6, and 9, we showed that m96cl represses invasion in 3D cultures (Fig. 2D; Supplemental Fig. S2F). Next, to test whether m96cl expression altered the in vivo metastatic potency, the syngeneic mouse lung cancer cell line 344SQ with inducible expression of m96cl or control was injected subcutaneously in syngeneic 129/Sv WT mice and monitored for tumor growth with or without doxycycline induction. In the doxycycline-induced cohorts, the m96cl-induced tumors showed a severe decrease in final tumor volume as compared with the control tumors (Fig. 2E). These results were analogous to the phenotype observed in vitro, where m96cl expression resulted in significantly repressed cellular growth (Supplemental Fig. 2G). Interestingly, in the doxycycline group, the m96cl-induced tumors showed robust repression of lung metastases when compared with the vector control tumors, which was evident from the quantification of the macrometastatic nodules in the lung, as well as by the H&E-stained lung sections (Fig. 2F). The doxycycline-untreated cohorts showed no difference in either tumor volumes or metastasis between the m96cl and vector controls. Furthermore, to ascertain that the reduced metastatic potential of the m96cl tumors is not a consequence of reduced tumor volume, we included an additional cohort in which the m96cl-induced tumors were allowed to grow longer in order to become size-matched with the vector controls. We also observed in this group that the tumors showed a significant reduction in metastasis when compared with the control group (Fig. 2E,F), establishing that m96cl could significantly suppress metastasis independent of primary tumor size.
Figure 2.
Induced expression of the microRNA-183/96/182 cluster suppresses cancer cell invasion and metastasis. (A) qPCR analysis for relative expression of miR-96, miR-182, and miR-183 in 344SQ Vec and 344SQ m96cl at 24, 48, or 72 h of doxycycline induction. Statistical significance is as indicated. (**) P < 0.01, (***) P < 0.001. (B) Bright-field microscopy images showing morphology of 344SQ Vec or 344SQ m96cl cells induced with doxycycline for 48 h. Scale bar, 100 µm. (C) Quantitation of cells for invasion (red) and migration (blue) of 344SQ Vec and 344SQ m96cl cells at 16 h after seeding. n = 3. (*) P < 0.05. (D) 344SQ Vec or 344SQ m96cl cells were plated in triplicate wells in 1:1 Matrigel:collagen-I matrix. Images were taken at days 4, 6, and 9. Representative images from day 9 are shown at the left, and quantification of invasive structures are shown at the right. Statistical significance is as indicated. (**) P < 0.01, (***) P < 0.001. (E) 344SQ Vec or 344SQ m96cl cells were implanted subcutaneously into 129/sv mice. Doxycycline-containing chow or normal chow was placed as indicated. Mice were sacrificed after 8 wk, except for the primary tumor size-matched cohort of 344SQ m96cl(+Dox), which was grown for 12 wk. Tumor volume was quantified and analyzed using unpaired Student's t-test. (N.S.) Not significant, (**) P < 0.01. (F) Quantitation of lung metastatic nodule formation from mice corresponding to E. (Top) Metastatic nodules counted from lung surface. (Bottom) Whole-lung tissue H&E. Scale bar, 6 mm. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (***) P < 0.001.
Next, we wanted to determine whether loss of m96cl in vivo could drive metastasis. For this, we generated cell lines from autochthonous lung tumors, which were formed by delivery of Ad-CRE in conditional GEMMs with different genotypes. We generated the KrasLSL-G12D; P53FL/FL ; m96clFL/FL mice and used the KrasLSL-G12D; p53FL/FL mice as controls. Autochthonous lung tumors were harvested, and cells lines were generated with the respective genotypes. We first validated that in the KPM96 cells (derived from KrasG12D/−; p53−/−; m96cl−/− tumors), m96cl expression was completely lost as compared with the KP cells (derived from KrasG12D/−; p53−/− tumors) (Supplemental Fig. S2H). We also observed that the KPM96 cell lines were more migratory and invasive in vitro versus the control KP lines (Supplemental Fig. S2I). Furthermore, when we implanted these cells in syngeneic hosts, we observed that in comparison with the KP cells, the KPM96 cell lines formed significantly larger tumors and significantly greater numbers of lung metastases (Supplemental Fig. S2J,K). The KPM96 cells also formed distant metastasis in other organs (the kidney, liver, and heart), whereas the KP cells did not show observable organ metastases (data not shown). Therefore, our data strongly indicate that in addition to controlling tumor cell proliferation, m96cl also alters other cellular processes to regulate in vivo metastasis.
The microRNA-183/96/182 cluster stimulates an antitumor CD8+ CTL response
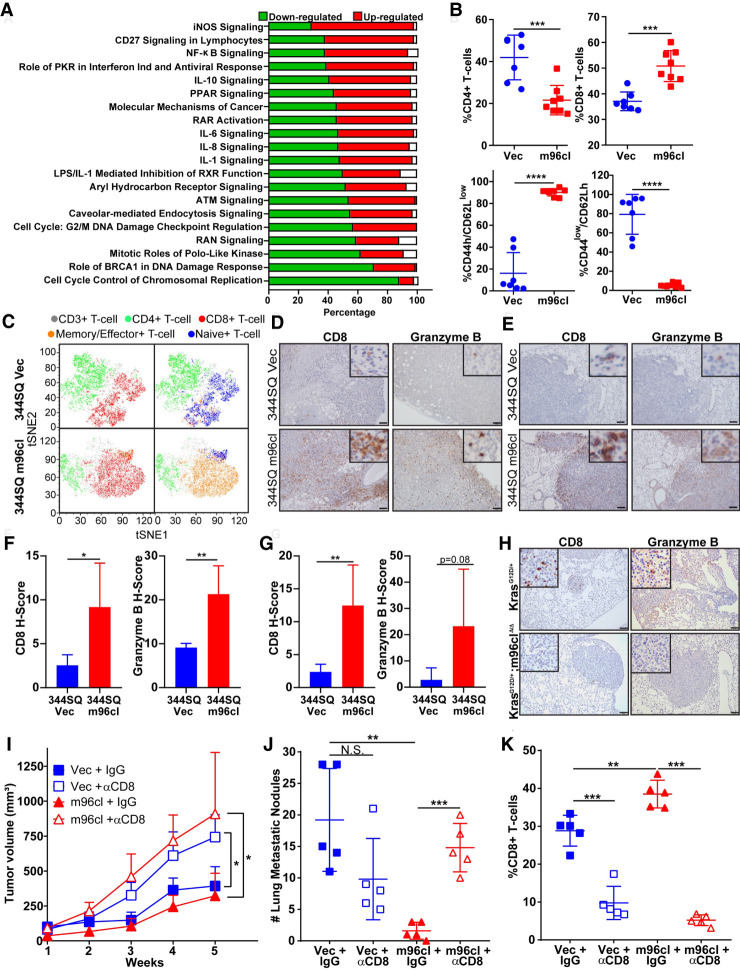
To identify downstream targets and pathways altered by m96cl, we performed an mRNA microarray analysis using the 344SQ cells expressing the m96cl or vector control. Upon analyzing the data set using Ingenuity pathway analysis, we observed that multiple gene and signaling networks were differentially altered upon induction of m96cl (Supplemental Fig. S3A–E). Of the top 20 most altered pathways, we noted that canonical mitotic and cell cycle pathways underwent m96cl-mediated down-regulation (Fig. 3A). This is consistent with our data showing that decreased proliferation was manifested as reduced tumor (Fig. 2E) and cellular (Supplemental Fig. S2G) growth upon m96cl overexpression. Interestingly, we observed that a host of immune pathways was up-regulated upon m96cl induction, which could elucidate the tumor growth-independent role of m96cl as a metastasis suppressor (Fig. 3A). The canonical pathways that were up-regulated upon m96cl overexpression include iNOS signaling, NFκB signaling, and several interleukin signaling processes. Thus, to elucidate the specific changes in the tumor immune microenvironment associated with m96cl tumor expression, we performed immune profiling by flow cytometry using the early primary syngeneic tumors formed by 344SQ cells expressing the m96cl or vector as controls 3 wk after injection. We observed that m96cl induction results in a prominent and significant increase in total CD8+ CTLs with a decrease in the total CD4+ T cells. Upon further analysis, we detected that within the CD8+ CTLs, the effector/memory CD8+ CTL subpopulation (CD44h/CD62Llow) was substantially elevated, with a concurrent reduction in the naive CD8+ CTL subpopulation (CD44low/CD62Lh) (Fig. 3B). Upon tSNE analysis of representative samples, it was revealed that in tumors with m96cl overexpression, the majority of the CD8+ CTLs were in an effector/memory state, whereas in the vector control tumors, the CD8+ CTLs were in a naïve state (Fig. 3C). Analysis for other immune cells revealed no detectable differences in percentages of immunosuppressive Treg cells, ICOS CD4+ T cells or exhausted CD8+ CTLs (Supplemental Fig. S3F) in the tumors with induced m96cl expression compared with the control tumors. We also observed a significant decrease in MDSCs in the m96cl cohort but no discernable difference in the antigen-presenting cell (APC) populations (Supplemental Fig. S3G) between the two groups. We observed a similar significant increase in CD8+ CTL response in m96cl-induced tumors collected 6 wk after implantation (i.e., the stage of macroscopic metastatic outgrowth), confirming that this is a durable long-term effect (Supplemental Fig. S3H). Furthermore, we confirmed similar effects in additional lung adenocarcinoma syngeneic models, including the 344P and LLC-JSP, that showed an up-regulation of total and effector/memory CD8+ CTLs upon induced expression of m96cl in the tumor cells (Supplemental Fig. S3I,J). To assess the cytolytic functions of the elevated CD8+ T cells in the tumors with induced m96cl expression, we probed for CD8 and granzyme B in sections of the primary tumors and lung tissue with metastatic nodules. IHC staining and subsequent quantification revealed a significant increase in both CD8+ CTLs and granzyme B-positive cells in both the primary and metastatic tumors in the m96cl-induced tumor cohort as compared with the control (Fig. 3D–G; Supplemental Fig. S3K). Conversely, we observed a decrease in the abundance of CD8+ CTLs and granzyme B+ cells in the autochthonous lung tumors from the KrasG12D/+; m96clΔ/Δ mice, as compared with KrasG12D/+ mice (Fig. 3H). Furthermore, to ascertain the role of elevated CD8+ CTL response as a mechanism of m96cl-mediated suppression of tumor growth and metastasis, we let tumors from 344SQ cells with induced expression of m96cl or vector control grow in syngeneic mice that were continually depleted for CD8+ CTLs by biweekly injections with anti-CD8 antibodies. We observed that with near-complete depletion of CD8+ CTLs (Fig. 3K; Supplemental Fig. S3M) there was a significant increase in tumor volumes (Fig. 3I; Supplemental Fig. S3L) in both the vector control and m96cl cohorts. More strikingly, CD8+ CTL depletion completely abrogated the metastatic suppression due to m96cl expression in tumors (Fig. 3J). Furthermore, to ascertain whether m96cl expression could alter MHC-I expression, we assayed the cell surface expression of MHC-I in either cells with induced expression of m96cl or tumors formed using these cells. We observed that m96cl did not significantly alter baseline MHC-I expression on the tumor cells or their ability to increase MHC-I in response to IFNγ. We also assayed for MHC-I expression in a panel of KP cell lines and did not observe a discernable contribution of the EMT status on the expression of MHC-I (Supplemental Fig. S3N–P). These results indicate that m96cl expression in tumors induces a suppression of tumor growth and metastasis by stimulating the activation and function of CD8+ CTLs.
Figure 3.
The microRNA-183/96/182 cluster stimulates an antitumor CD8+ CTL response. (A) The top canonical pathways affected by m96cl induction were identified using IPA analysis, quantifying fold change between 344SQ Vec and 344SQ m96cl cells at 72 h after induction. Bar chart representing the percentage of genes from the data set that are found in each canonical pathway. Genes up-regulated are shown in red, genes down-regulated are shown in green, and genes that did not change in the data set are shown in white. (B) Percentage of tumor-infiltrating CD4+ T cells (top left), CD8+ T cells (top right), memory/effector CD8+ T cells (bottom left), and naïve CD8+ T cells (bottom right) from 344SQ Vec and 344SQ m96cl tumors at 3 wk after implantation. (C) Representative tSNE plots from B for 344SQ Vec or 344SQ m96cl tumors. (Green) CD3+ T cells (gray), CD4+ T cells, (red) CD8+ T cells, (orange) memory/effector CD8+ T cells, (blue) naïve CD8+ T cells. (D) Representative images of CD8 and granzyme B IHC staining in primary tumors formed by subcutaneous implantation of 344SQ Vec or 344SQ m96cl after week 8. Scale bar, 100 µm. (E) Representative images of CD8 and granzyme B IHC staining in lung metastasis formed in the mice as described in D. Scale bar, 100 µm. (F) CD8 H-score (left) and granzyme B H-score (right) from images as in D. n = 3 mice per group. Data were analyzed using unpaired Student's t-test. (*) P < 0.05, (**) P < 0.01. (G) CD8 H-score (left) and granzyme B H-score (right) from images as in E. n = 3 mice per group. Data were analyzed using unpaired Student's t-test. (**) P < 0.01. (H) Representative images of CD8 and granzyme B IHC staining in lungs from KrasG12D/+ and KrasG12D/+; m96clΔ/Δ mice 3 mo after Ad-Cre infection. Scale bar, 100 µm. (I) Tumor volumes for 344SQ Vec and 344SQ m96cl tumor-bearing mice treated with IgG or α-CD8 for 5 wk after implantation. Week 5 data were analyzed using unpaired Student's t-test. (*) P < 0.05. (J) Quantitation of lung metastatic nodule formation in mice corresponding to I. Metastatic nodules counted from lung surface. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (**) P < 0.01, (***) P < 0.001. (K) Percentage of tumor-infiltrating CD8+ T cells from different treatment cohorts as indicated, at week 5. Data were analyzed using unpaired Student's t-test. (**) P < 0.01, (***) P < 0.001.
MicroRNA-183/96/182 cluster expression in tumor cells induces paracrine activation of CD8+ CTL proliferation and function
Activation of CD8+ CTLs is dependent on antigenic TCR stimulation by two independent signals and costimulation by inflammatory cytokines. Sustained functionality and proliferation are also dependent on the availability of cytokine-mediated stimulation. Therefore, we next sought to examine the mechanism by which m96cl expression in tumor cells influences CD8+ CTL expansion and functionality. First, we wanted to determine whether direct cell–cell contact between the tumor cells and immune cells is necessary for this process. For this, we seeded 344SQ cells with induced expression of m96cl or vector control with a suspension of naïve immune cells isolated from syngeneic WT mouse spleens for a period of 96 h in the presence of CD3 and CD28 stimulatory antibodies. We then performed flow cytometric analysis to determine changes in the immune subpopulations (Fig. 4A). Results from the coculture assays recapitulated the in vivo phenotype. When in coculture with m96cl-expressing tumor cells, compared with control cells, we observed a significant elevation in the abundance of total and proliferating CD8+ CTLs and the effector/memory CD8+ CTL subpopulation, with no significant changes to the proliferating naïve population (Fig. 4B). We further collected conditioned media from the 344SQ cells with inducible expression of m96cl or vector control to culture the naïve immune cells (Fig. 4A). Upon immune profiling of the naïve immune cells after culture, we observed that the results from the conditioned media assay closely mimicked what we noted in the coculture assay (Fig. 4C). We also confirmed these immune response phenotypes using additional KP tumor models (the 344P and 531LN3) with inducible expression of m96cl (Supplemental Fig. S4A–D). To further assess the functionality of these CD8+ CTLs, we analyzed the CD8+ CTLs that had been cultured in conditioned media from 344SQ vector or m96cl cells for IFNγ response. We observed that the CD8+ CTLs stimulated in the conditioned media of m96cl-induced cancer cells showed a significant increase in IFNγ, indicating greater functional activation (Fig. 4D). To further validate the functional properties of these stimulated CD8+ CTLs, we performed a cytotoxicity assay. To do this, we enriched stimulated CD8+ CTLs from naïve immune cells that were cultured in the conditioned media from 344SQ cells with induced expression of vector or m96cl. These primed CD8+ CTLs were incubated at varying ratios with the respective cells as target and cell death was measured using an LDH release assay. We observed that the CD8+ CTLs primed and stimulated in the conditioned media from m96cl-expressing cells were significantly more cytotoxic to the respective target cancer cells as compared with the controls (Fig. 4E). These results further established that cancer cells expressing miR-96c could induce paracrine stimulation of effector CD8+ CTLs.
Figure 4.
MicroRNA-183/96/182 cluster expression in tumor cells induces paracrine activation of CD8+ CTL proliferation and function. (A) Schema representing the methodology of culturing naïve immune cells with cancer cell lines (coculture, left) or culturing them in conditioned media (right) collected from cancer cells grown in vitro for 72–96 h with or without Dox induction. (B) Coculture of naïve immune cells with 344SQ Vec or 344SQ m96cl cells while induced for 96 h with Dox. After coculturing, immune cells were then analyzed by flow cytometry. (Panel i) Percentage total CD8+ T cells. (Panel ii) Percentage effector/memory CD8+ T cells (CD44hCD62Llow). (Panel iii) Percentage Ki-67+ effector/memory (CD44hCD62Llow) CD8+ T cells. (Panel iv) Percentage naïve CD8+ T cells (CD44low/CD62Lh). (Panel v) Percentage Ki-67+ naive (CD44lowCD62Lh) CD8+ T cells. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (***) P < 0.001. (C) Conditioned media (CM) from 344SQ Vec or 344SQ m96cl cells induced for 72 h was used to culture naïve immune cells for 72–96 h. Immune cells were then analyzed by flow cytometry as described above in B. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (*) P < 0.05, (***) P < 0.001. (D) Quantitation of IFN-γ-expressing CD8+ T cells in the immune cell population, cultured in conditioned media from 344SQ Vec or 344SQ m96cl cells that were either uninduced or induced with Dox for 96 h. Data were analyzed using unpaired Student's t-test. (***) P < 0.001. (E) Percentage cytotoxicity of 344SQ Vec or 344SQ m96cl cells when cultured for 24 h with CD8+ T cells that were precultured for 6 d in conditioned media from 344SQ Vec or 344SQ m96cl cells. A ratio of 1:0–1:40 of tumor cell line:CD8+ T cells was used. Data were analyzed using unpaired Student's t-test. (**) P < 0.01.
Last, we evaluated whether m96cl expression in cancer cells could affect the relative numbers of immunosuppressive Th17 cells within the CD4+ T cells. We observed no detectable difference in the stimulation of Th17 cells between the m96cl and vector control group (Supplemental Fig. S4E,F). Thus, our data indicate that m96cl expression in cancer cells results in paracrine stimulation of CD8+ CTLs, thereby inducing their differentiation into an effector/memory state primed for killing target cancer cells.
MicroRNA-183/96/182 cluster expression induces interleukin-2 family signaling in cancer cells
To identify immune-associated genes and pathways that are induced upon m96cl expression and result in stimulation of CD8+ CTL response, we performed a qRT-PCR analysis to study a panel of cytokines, chemokines, and transcription factors that regulate common immune pathways in cancers. From the qPCR analysis, we identified several cytokines (IL1, IL2, IL4, and IL23) and chemokines (CXCL9, CXCL11, CCL2, and CCL20) that were up-regulated in the m96cl-expressing cells (Fig. 5A; Supplemental Fig. S5A). We validated several of these hits in the KP murine 531LN3 cell line and in the human H157 cells with induced expression of m96cl (Supplemental Fig. S5B,C). In parallel, to study a broader panel of tumor-immune microenvironment-associated genes and pathways in cancer, we performed NanoString nSolver advanced gene expression analysis on either cell lines with induced expression of m96cl or vector as control grown in vitro or syngeneic tumors formed by these cell lines in vivo. The NanoString nSolver analysis from the cells and tumors revealed significant alteration in several genes and pathways of the tumor-immune microenvironment upon m96cl expression (Fig. 5B,C; Supplemental Fig. S5D–I). We observed from the three independent analyses that the interleukin-2 (IL2) gene/signaling pathway was the only one commonly up-regulated because of m96cl expression in all conditions (Fig. 5D). Next, we wanted to functionally validate whether m96cl-mediated increased expression and secretion of IL2 from the cancer cells was sufficient to induce the CD8+ T-cell stimulation. For this, we individually depleted IL2 or the other cytokines and chemokines that were observed to be elevated in the m96cl-expressing cells from conditioned media collected from culturing either the vector control or miR-96-induced cells using specific depletion antibodies or IgG as control. The depleted conditioned media was then used to culture naïve immune cells over a period of 4 d, followed by flow analysis to quantify total or the effector/memory subpopulation of CD8+ CTLs. We observed that depletion of only IL2 or CCL2 was sufficient to revert the stimulation of CD8+ CTLs by the conditioned media (Fig. 5E; Supplemental Fig. S5J). We also observed that IL2 protein expression was significantly elevated in the m96cl-induced cells both in an unstimulated state or when stimulated by ionomycin and PMA (Fig. 5F; Supplemental Figs. S5K–L, S8H). However, we did not observe any such elevation in protein levels for CCL2 (Supplemental Fig. S5M) in the m96cl-induced cells. Therefore, we inferred that IL2 was the cytokine that was elevated in the m96cl-expressing cancer cells, which when secreted could articulate a paracrine stimulation of the CD8+ CTLs. We further validated this by performing an IFNγ response assay and a cytotoxicity assay as described in Figure 4 using the IL2-depleted conditioned media. We observed that IL2 depletion from the conditioned media of the m96cl-induced cells significantly abrogated the IFNγ expression and targeted cytotoxicity of the cultured CD8+ T cells (Fig. 5G–H). To further ascertain the role of IL2 as the downstream effector of m96cl expression, we generated CRISPR-mediated mIL-2 knockout clones in 344SQ m96cl and 344SQ Vec cell lines. Genomic sequences were verified for loss of the 136-bp region covering IL2_codon1 (Supplemental Fig. S8J). Conditioned media from these lines was used for coculture assays using naïve immune cells (as described in the Materials and Methods). Reassuringly, we observed that upon loss of IL2 expression, the 344SQ m96cl_IL2 KO cells completely lost the ability to induce the proliferation and activation of total and effector CD8+ T cells as compared with the control 344SQ Vec_IL2 KO cells. Concurrently, we observed that the m96cl_IL2 KO cells could induce a significant increase in CD4+ T cells and naïve CD8+ T cells as compared with the controls (Supplemental Fig. S8K). These results establish that increased IL2 expression and secretion because of elevated m96cl expression in tumor cells are the driving mechanisms for the m96cl-induced paracrine stimulation of CD8+ CTL function.
Figure 5.
MicroRNA-183/96/182 cluster expression induces interleukin-2 family signaling in cancer cells. (A) Heat map of qRT-PCR expression analysis for cytokines, chemokines, and immune TFs in 344SQ Vec or 344SQ m96cl cells after 72 h of Dox induction. (B) Heat map of pathway scores from nSolver Nanostring advanced analysis. 531LN3 Vec or m96cl and 344SQ Vec or m96cl cells were induced for 72 or 96 h in vitro. The top 10 pathways are shown. (C) Heat map of pathway scores from nSolver Nanostring advanced analysis done on RNA extracted from tumors grown from subcutaneous implantation of 344SQ Vec or 344SQ m96cl cells that were harvested after 3 wk of Dox induction in vivo. Each number represents a distinct mouse. The top 10 pathways are shown. (D) Venn diagram depicting overlap between qRT-PCR, in vitro Nanostring pathway scores, and in vivo Nanostring pathway scores described in A–C. (E) CD8+ T cells (panel i) or effector/memory CD8+ T cells (panel ii) in immune cell population that were cultured in conditioned media from 344SQ Vec or 344SQ m96cl cells (induced for 72 h with Dox), which were depleted of IL2 or IgG (as control). Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (*) P < 0.05, (**) P < 0.01. (F) Percentage of IL2-expressing 344SQ Vec or 344SQ m96cl cells (induced for 72 h with Dox) with and without ionomycin/PMA stimulation for 24 h. Data were analyzed using unpaired Student's t-test. (*) P < 0.05, (***) P < 0.001. (G) Percentage IFN-γ-expressing CD8+ T cells in immune cells cultured in conditioned media from 344SQ Vec or 344SQ m96cl cells (induced for 72 h with Dox), which were depleted of IL2 or IgG (as control). Data were analyzed using unpaired Student's t-test. (*) P < 0.05, (**) P < 0.01. (H) Percentage cytotoxicity of 344SQ Vec or 344SQ m96cl cells when cultured for 24 h with CD8+ T cells that were precultured for 6 d in conditioned media from 344SQ Vec or 344SQ m96cl cells (Dox-induced), which were depleted of IL2 or IgG (as control). A ratio of tumor cells:CD8+ T cells of 1:10 was used. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (**) P < 0.01.
The microRNA-183/96/182 cluster modulates IL2 expression by repressing Zeb1 and Foxf2
To understand the molecular mechanism underlying the regulation of IL2 expression downstream from m96cl, we studied the promoter of IL2 to identify possible binding sites for transcriptional repressors that are known targets of m96cl. JASPAR analysis revealed several binding motifs with varying affinities for ZEB1 and Foxf2, which we have previously identified as bona fide transcription factor targets of the m96cl and miR-200 microRNA family (Fig. 6A; Supplemental Fig. S6A,S6D; Kundu et al. 2016). To ascertain the transcriptional regulation of IL2 by Foxf2 and ZEB1, we performed a luciferase reporter assay. We observed that inducible expression of m96cl and miR-200 could elevate IL2 promoter activity in different cell lines, whereas inducible expression of Foxf2 and ZEB1 significantly inhibited IL2 promoter activity (Fig. 6B). We further performed ChIP analysis and identified that both ZEB1 and Foxf2 bind to the IL2 promoter at the motifs that were predicted for the higher affinity by JASPAR (Fig. 6C; Supplemental Fig. S6D). To elucidate the regulation of the IL2 signaling by Foxf2 and ZEB1, we performed a qRT-PCR analysis looking at the expression of members of the IL2 signaling pathways as annotated by the NanoString nSolver analysis. As expected, induced expression of m96cl in murine and human cells resulted in a significant up-regulation of the genes of the IL2 signaling pathway, but Foxf2 and ZEB1 expression completely reversed the expression profiles of these genes (Fig. 6D). We also confirmed that IL2 protein expression was significantly repressed by Foxf2 and ZEB1 expression in the same cells (Supplemental Fig. S6C). To address the functional outcome of Foxf2 and ZEB1 expression on CD8+ CTL activation, we collected conditioned media from cultures of cells expressing the control vectors m96cl, miR-200, Foxf2, and ZEB1. We cultured naïve immune cells in the respective conditioned media in the presence of activation signals (anti-CD3 and anti-CD28) and assessed the effect on CD8+ CTL expansion. We noted that while conditioned media from m96cl- and miR-200-expressing cells significantly boosted the total and effector/memory CD8+ CTL population, this stimulation was strongly inhibited when conditioned media from Foxf2- and ZEB1-expressing cells were used (Fig. 6E; Supplemental Fig. S6B). We further validated this by performing an IFN-γ response assay and a cytotoxicity assay as described in Figure 4 using the different conditioned media as described above. We observed that conditioned media from the m96cl- and miR-200-induced cells was able to stimulate IFN-γ expression and targeted cytotoxicity of the CD8+ CTLs, whereas the conditioned media from Foxf2- and ZEB1-expressing cells significantly reversed these effects (Fig. 6F,G). To further ascertain the role of Zeb1 and Foxf2 downstream from m96cl, we performed siRNA-mediated knockdown of either Zeb1, Foxf2, or both in 344SQ m96cl and 344SQ Vec cells. qPCR analysis was performed to confirm specific knockdown of the target genes (Supplemental Fig. S6E). We collected conditioned media from cells transfected with the different targeting siRNAs as indicated. Naïve immune cells were cultured in the different pools of conditioned media, and immune profiling was performed by flow cytometry. We observed that, only for 344SQ Vec cells that were treated with siRNA targeting Zeb1, Foxf2, or both, there was a significant elevation in the abundance of total and proliferating CD8+ CTLs and effector/memory CD8+ CTL subpopulation, with no significant changes to the naïve population or exhausted CD8+ T cells. These changes were not observed in the cohorts of the 344SQ m96cl cells transfected with the siRNAs (Supplemental Fig. S6F, panels i–iv). These results confirm that only in the 344SQ Vec control cells when the high levels of Zeb1/Foxf2 were knocked down were the proliferation and activation increased in CD8+ T cells. In case of the m96cl-expressing cells in which the levels of Zeb1/Foxf2 were already low, further knockdown of their expression did not have any effect on the CD8+ T cell subpopulations.
Figure 6.
The microRNA-183/96/182 cluster modulates IL2 expression by repressing Zeb1 and Foxf2. (A) Schematic representation of potential E-boxes (black) located on the IL2 promoter. Arrows indicate primer binding sites used to determine binding of Zeb1 (green) and Foxf2 (purple) to the IL2 promoter after ChIP of the respective proteins. (B) Luciferase reporter assay using the dual-luciferase reporter system (Firefly [GL3] and Renilla [RL] as control) for IL2 promoter activity upon expression of either Vec, m96cl, miR-200, Zeb1, or Foxf2 in 344SQ (blue), 531LN3 (red), and 393P (green) cells as indicated. Data were analyzed using unpaired Student's t-test. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. (C) Chromatin immunoprecipitation for Zeb1 and Foxf2 binding to IL2 promoter. GFP only, GFP-Zeb1, or GFP-Foxf2 from inducible 393P cells were chromatin-immunoprecipitated using an anti-GFP antibody, and qPCR was performed to assay for binding of these proteins to the predicted respective high-affinity E-boxes on the IL2 promoter. Control ChIP was performed with rabbit IgG. Data were analyzed using unpaired Student's t-test. (*) P < 0.05, (***) P < 0.001. (D) Heat map of qRT-PCR expression analysis for IL2 pathway-associated genes in 344SQ Vec or m96cl; H157 Vec or m96cl; and 393P Vec, Zeb1, or Foxf2 cells after 72 h of Dox induction. (E) Conditioned media from 344SQ Vec or m96cl; H157 Vec or m96cl; and 393P Vec, Zeb1, or Foxf2 cells after 72 h of Dox induction was used to culture naïve immune cells for 96 h. A percentage CD8+ T cells was quantified in the cultured immune cell population. (F) Quantitation of IFN-γ-expressing CD8+ T cells in the immune cell population, cultured for 96 h in conditioned media from either Vec-, m96cl-, miR-200-, Zeb1-, or Foxf2-expressing 344SQ (blue), 531LN3 (red), and 393P (green) cells that were induced for 72 h with Dox. Data were analyzed using unpaired Student's t-test. (***) P < 0.001, (****) P < 0.0001. (G) Percentage cytotoxicity of Vec-, m96cl-, miR-200-, Zeb1-, or Foxf2-expressing 344SQ (blue), 531LN3 (red), and 393P (green) cells (Dox-induced) when cultured for 24 h with CD8+ T cells that were precultured for 6 d in conditioned media collected from culturing the respective (Dox-induced) cell lines. A ratio of tumor cells:CD8+ T cells of 1:10 was used. Data were analyzed using unpaired Student's t-test. (*) P < 0.05, (***) P < 0.001.
These results show that m96cl modulates the expression of IL2 in cancer cells by regulating the expression of the transcriptional repressors Foxf2 and ZEB1, thereby altering the levels of secreted IL2 in the tumor microenvironment.
IL2 depletion abrogates microRNA-183/96/182 cluster-mediated metastasis suppression in vivo
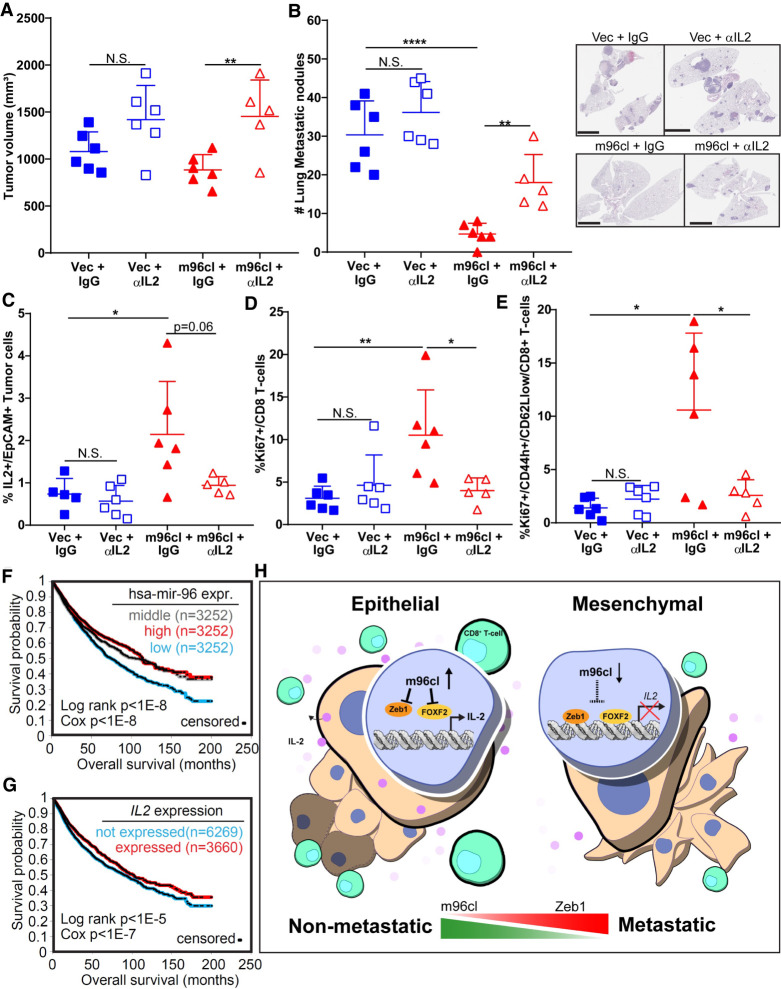
We have shown that lung cancer cells expressing m96cl could inhibit metastasis by stimulating IL2-mediated functional activation and expansion of CD8+ CTLs. To extend these analyses, we performed an in vivo metastasis study with antibody-mediated IL2 depletion. The murine 344SQ cells with inducible expression of vector or m96cl were implanted into syngeneic mice. These mice were fed with doxycycline-containing feed. Half of the mice from either vector or m96cl cell-injected cohorts were injected with depleting antibodies targeting IL2 (200 μg/mouse biweekly, IP), and the remaining half were dosed with IgG as control. After 6 wk of treatment, we noted that there was a significant increase in the volume of the primary tumors of the IL2-depleted cohorts when compared with the respective IgG control groups (Fig. 7A; Supplemental Fig. S7A). Upon quantifying the metastasis, we observed that the IL2-depleted 344SQ m96cl cohort showed a significantly higher number of lung and other organ metastasis when compared with the IgG-treated 344SQ m96cl group. Similar high lung and other organ metastasis was also observed in the 344SQ vector groups (Fig. 7B; Supplemental Fig. S7B,C). Flow cytometric analysis revealed a significant drop in the IL2+ tumor cells in the IL2-depleted tumors as compared with the IgG-treated 344SQ m96cl tumors (Fig. 7C), validating the effective depletion of systemic IL2. We also observed a significant drop in the counts of the proliferating (Ki67+) total and effector/memory CD8+ CTLs in the 344SQ m96cl tumors that were depleted for IL2 as compared with the IgG-treated tumors (Fig. 7D,E). We did not observe a significant difference in the overall counts of the CD8+ CTL subpopulations (Supplemental Fig. S7D) between the respective cohorts. To substantiate the role of tumor cell-expressed IL2 in regulating CD8+ T-cell response, we generated IL2-overexpressing cell line using the parental 344SQ cells. IL2 expression was confirmed using qPCR, Western blotting, and ELISA (Supplemental Fig. S8E–G). These cell lines were implanted in syngeneic mice, and tumors were processed for immune profiling. We observed that tumors with IL2 expression showed significantly elevated total and proliferating CD8+ T cells and effector memory T cells, with significantly lower naïve T cells, CD4+ T cells, and T regulatory cells (Supplemental Fig. S8I), as was observed in the m96cl-expressing tumors. Next, we wanted to assess the clinical relevance of these findings to determine the status of m96cl and IL2 expression in human pan-cancer data sets and whether their expression profile could be a prognostic indicator of disease outcome. We observed in pan-cancer data sets that patients with higher expression of either IL2, hsa-miR-96, or hsa-miR-182 showed significantly better overall survival (Fig. 7F,G; Supplemental Fig. S7G). We also observed that in the TCGA lung adenocarcinoma-specific data set, high IL2 expression was significantly associated with better overall survival (Supplemental Fig. S7E,F).
Figure 7.
IL2 depletion abrogates microRNA-183/96/182 cluster-mediated metastasis suppression in vivo. (A) Tumor volumes for 344SQ Vec and 344SQ m96cl tumor-bearing mice treated with IgG or anti-IL2 antibodies for 6 wk after implantation. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (**) P < 0.01. (B) Quantitation of lung metastatic nodule formation in mice corresponding to A. (Left) Metastatic nodules counted from lung surface. (Right) H&E-stained sections of representative whole lungs. Scale bar, 6 mm. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (**) P < 0.01, (****) P < 0.0001. (C) Percentage of IL2+ EpCAM+ (IL2+EpCAM+CD31−CD45−) tumor cells from 344SQ Vec and 344SQ m96cl tumors from A. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (*) P < 0.05. (D) Percentage of tumor-infiltrating proliferating Ki67+ CD8+ T cells from 344SQ Vec and 344SQ m96cl tumors from A. Data were analyzed using unpaired Student's t-test. (N.S.) Not significant, (*) P < 0.05, (**) P < 0.01. (E) Percentage of tumor-infiltrating proliferating effector/memory CD8+ T cells (Ki-67+/CD44h/CD62Llow/CD8+) from 344SQ Vec and 344SQ m96cl tumors from A. Data were analyzed using unpaired Student's t-test. (*) P < 0.05. (F) Kaplan–Meier survival curves for TCGA lung adenocarcinoma patients with m96cl high (red) versus m96cl low (blue) expression. Statistical difference was determined using log rank Cox test. (G) Kaplan–Meier survival curves for TCGA lung adenocarcinoma patients with IL-2 high (red) versus IL-2 low (blue) expression. Statistical difference was determined using log rank Cox test. (H) Working model: High m96cl expression in epithelial tumors results in reduced expression of EMT TFs like Zeb1 and Foxf2, which leads to increased IL2 expression and secretion in the tumor microenvironment. This increased local intratumoral IL2 gradient results in a paracrine activation and proliferation of CD8+ CTLs, which consequently primes an elevated antitumor immune response and increased immune clearance of metastatic tumors, whereas repressed m96cl expression in mesenchymal tumors suppresses this mechanism and results in increased tumor growth and metastasis.
Based on our findings, we propose a model in which elevated levels of m96cl in primary epithelial lung cancers lead to an increased suppression of the transcriptional repressors ZEB1 and Foxf2, which results in an elevated expression and secretion of IL2. Increased IL2 in the tumor microenvironment of epithelial tumors induces a paracrine stimulation of tumor-infiltrating CD8+ CTLs, resulting in their expansion and increased functionality. These elevated levels of cytotoxic CD8+ CTLs result in enhanced tumor clearance and suppression of metastatic outgrowth. In more advanced mesenchymal states, the tumors have reduced levels of m96cl and other epithelial microRNAs like miR-200, resulting in inhibition of the IL2-mediated CD8+ T-cell activation and loss of effective immune surveillance, with increased tumor growth, invasion, and distant metastasis (Fig. 7H).
Discussion
Detailed elucidation of the various underlying molecular mechanisms of lung cancer progression and metastasis is of primary concern to the cancer research community. One of the established models for initiation and progression of metastasis is EMT, where epithelial cancer cells progressively lose cell–cell contacts, induce stem cell-like properties, and elevate motility and invasion. This is brought about by orchestrated molecular changes, including up-regulation of Zeb1, Foxf2, and several other transcriptional repressor proteins with coordinated repression of epithelial microRNAs like miR-200 and the m96cl (Gibbons et al. 2009; Liu et al. 2012; Pacurari et al. 2013; Tsai and Yang 2013; Feng et al. 2014; Kundu et al. 2016; Krebs et al. 2017; Pastushenko and Blanpain 2019). EMT, and specifically the role of the Zeb1/miR-200 double-negative feedback in modulating the reversible alteration between a growth-promoting epithelial state and an invasive mesenchymal state, have been studied by us and others in the context of metastasis in lung and multiple other cancer types (Burk et al. 2008; Korpal et al. 2008; Gibbons et al. 2009; Ahn et al. 2012). Our previous observations on the functional role of m96cl as an inhibitor of cellular invasion (Kundu et al. 2016) laid the foundation for us to query for an independent role of m96cl in lung tumorigenesis and metastasis. The accelerated tumor growth and severe reduction of overall survival of the new conditional m96cl knockout GEMMs (KrasG12D/+; m96clΔ/Δ) establishes m96cl as a prominent tumor suppressor in lung adenocarcinoma. The lack of metastasis in these animals could potentially be attributed to wild-type p53 levels, as the loss of m96cl with Kras mutation only is possibly an insufficient genetic driver to initiate metastasis.
Although the advent and success of immune checkpoint blockade (ICB) therapies in various cancers has altered the treatment landscape, a majority of patients still do not receive durable benefit from this therapeutic modality (Fridman et al. 2012; Larkin et al. 2015a,b). It has been documented that tumors that have a higher abundance of infiltrated CD8+ T cells have a higher predisposition for better response to immunotherapy. Therefore, insights into the mechanisms that regulate the recruitment or activation of CTLs in the tumor microenvironment are of utmost importance (Hildner et al. 2008; Fridman et al. 2012; Spranger and Gajewski 2018; Garner and de Visser 2020). Inactivation of major tumor suppressors like p53, LKB1, and PTEN or activation of oncogenic signaling like WNT/b-Catenin and MYC has been shown to diminish the infiltration of activated CTLs within the tumor microenvironment and also lead to their elevated exhaustion (Xue et al. 2007; Spranger et al. 2015; Casey et al. 2016; Koyama et al. 2016; Peng et al. 2016; Topalian et al. 2016; Skoulidis et al. 2018; Spranger and Gajewski 2018; Sitthideatphaiboon et al. 2021). EMT has also been implicated as an immunosuppressive mechanism in different cancers by elevating inflammation and induction of exhaustion of the CD8+ CTLs at the tumor microenvironment (Derynck and Weinberg 2019). We have previously shown in NSCLC that epithelial tumor cells with high miR-200 could directly target expression of the immunosuppressive ligand PD-L1 on tumor cells, resulting in decreased exhaustion and increased infiltration of CTLs in the tumor microenvironment (Chen et al. 2014a). Here we elucidate another novel mechanism in which elevated m96cl, an epithelial microRNA whose expression is tightly correlated to miR-200, could induce the paracrine activation and proliferation of CTLs in the tumors by increased IL2 secretion. These results are exciting, as they define a parallel function for m96cl to epigenetically regulate a tumor cell-nonautonomous immune response to inhibit metastasis.
Materials and methods
Cell culture
Human and murine lung cancer cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS). KrasLA1-G12D (KP) murine cell lines were derived and cultured as previously described (Gibbons et al. 2009). The Lewis lung adenocarcinoma LLC-JSP was obtained from ATCC. Cells were cultured in a 37°C humidified incubator with 5% CO2. Cells were mycoplasma-tested using LookOut mycoplasma PCR detection kit (Sigma-Aldrich). For doxycycline (Sigma-Aldrich) induction, a final concentration of 2 μg/mL was added for the indicated time.
Plasmid constructs and lentivirus generation
For generating the human and mouse m96cl-inducible lentiviral constructs, miR-183-96 genomic fragment was PCR-amplified from genomic DNA isolated from H157 cells as a XmaI–KpnI fragment. (Human and mouse mature miR-183 and miR-96 have identical core sequences, so the same genomic fragment was used for both human and mouse m96cl constructs.) Human or mouse genomic fragments for miR-182 were individually PCR-amplified as KpnI–SalI fragments from genomic DNA isolated from H157 or 344SQ cells, respectively. The miR-183-96 fragment and respective miR-182 fragment were ligated in the Dox-inducible pTRIPZ-GFP vector (Kundu et al. 2016, 2018) digested with AgeI–XhoI (pTRIPZ-GFP digested with AgeI + XhoI excised out GFP). The resulting constructs were sequenced for verification. Lentiviruses generated using these constructs were used to generate human or mouse stable lines after infection, followed by puromycin selection. These stable inducible cell lines expressed mature miR-183, miR-96, and miR-182 when induced with doxycycline. All primers used are listed in Supplemental Table S1.
For generating the mouse IL2 promoter reporter construct, an ∼1.2-kb fragment of mouse IL2 promoter (starting from the TSS) was PCR-amplified as a KpnI–HindIII fragment from genomic DNA isolated from murine 344SQ cells. This was ligated in pGL3 (Promega) vector digested with the same restriction enzymes. All primers used are listed in Supplemental Table S1.
Lentiviruses were generated by transfecting pTRIPZ construct with psPAX2/pMD2.G into HEK 293T cells using Lipofectamine 2000 (Thermo Fisher). Fresh media was added after 24 h and incubated for 48 h. Virus was collected and syringe-filtered through 0.45-μm filter. Virus and polybrene (8 μg/mL; Millipore) were added to cell lines for 48 h. The transduced cells were then selected for using puromycin.
Generation of m96clFL mice
The m96clFL mice were generated in a manner similar to miR182FL mice (Sachdeva et al. 2016), except that the loxP sites flank the entire miR183-96-182 cluster. A targeting construct was generated in E. coli from a 129Sv BAC by recombineering (Zhang et al. 1998). A mobile pSIM vector recombineering system (Datta et al. 2006) was introduced into BAC clone bMQ340c02 (obtained from Source BioScience LifeSciences) containing 120 kb of mouse chromosome 6 harboring miR-183-96-182. LoxP sites and a neomycin cassette flanked by frt sites were recombineered into the BAC, followed by recombineering out into the targeting vector (Supplemental Fig. S1A, panel i). A linearized targeting vector was electroporated into 129Sv ES cells, and homologous recombination events were detected by PCR (Nagy et al. 1993). After verification of correct rearrangement by Southern blot analysis, clones were injected into C57BL/6 blastocysts following standard procedures. Chimeric males were mated to WT C57BL/6 females to generate heterozygote miR-m96clFL mice (Supplemental Fig. S1A, panel i).
Animal experiments
Animal experiments were approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center (protocol 1271).
The m96clFl/FL mice on a mixed 129Sv and C57BL/6 background were bred with 129sv mice and backcrossed for at least five generations to obtain a background of ∼98%. The m96clFL/FL mice were then crossed with the KrasLSL-G12D mice to generate the KrasLSL-G12D; m96clFL/FL mouse model. Adeno-cre-inducible mice of 129sv background were infected with virus at 3 mo of age by intratracheal intubation. Formation of lung tumors was assessed monthly with cross-sectional micro-CT imaging for tumor formation and tumor area measurement. Mice were genotyped to determine mutational status with ear snips at 3 wk of age. Only mice showing expected mutations were used for further assays. Mice were sacrificed upon visible morbidity or completion of the experimental period.
For experiments with syngeneic tumor assays, cells (unless otherwise noted, 1 × 106 cells in 100 μL of DPBS) were implanted subcutaneously into the right flanks of male or female 129sv mice of 3–6 mo of age. Tumor size as measured by digital caliper was calculated based on formula 1/2(length × width2) at the time point indicated. Mice were placed on doxycycline-containing chow (625 mg/kg doxycycline; Envigo) 2–3 d after tumor implantation. Doxycycline feed was replaced weekly. After euthanasia, metastatic nodules on the lung surface were counted. Lungs were perfused, fixed in 10% formalin, and processed for sectioning.
Antibody-mediated cell depletion was performed using anti-CD8 antibody (clone 2.34, BioXcell) at 400 μg/mouse (intraperitoneally) 1 wk before tumor cell implantation. A maintenance dose of 200 μg/mouse twice weekly was started on day 1 after subcutaneous cancer cell implantation.
IL2 depletion in vivo was performed by administration (intraperitoneally) of 200 μg/mouse twice weekly of anti-IL2 (JES6-1A12; BioXcell BE0043) or rat IgG2a,κ (2A3; BioXcell BE0089) as control.
Invasion and migration assays
Transwell migration of 8 μM inserts (BD Biosciences) and invasion (BD Biosciences) assays was performed for 16 h as previously described (Kundu et al. 2016, 2018). Inserts were imaged using crystal violet solution, and migratory or invasive cells were analyzed on an Olympus IX73 microscope and counted using ImageJ software (National Institutes of Health). For 3D invasion assays, single cells were seeded on a matrix of Matrigel (BD-Biosciences) or a 1.5 mg/mL Matrigel/collagen mixture (BD-Biosciences). Media was replenished every 2 d. Structures were imaged on days 4, 6, and 9 using an Olympus IX73 microscope and counted.
RNA isolation and qPCR analysis
Total RNA was isolated using TRIzol reagent (Thermo Fisher) following the manufacturer's recommendations. cDNA was synthesized using qScript cDNA supermix (Quantabio). Real-time PCR was performed using SYBR Green PCR master mix (Life Technologies) with primers listed in Supplemental Table S1. L32 was used to normalize expression. TaqMan assays (Life Technologies) for miRNA qPCR were normalized to miR-16. The 7500 Fast real-time PCR system (Applied Biosystems) were used for qPCR reactions. Low-abundance cytokines and chemokines were amplified via qPCR by using the PerfeCTa PreAmp supermix (Quantabio). cDNA was synthesized using qScript XLT cDNA supermix (Quantabio). Primers were pooled, and preamplification reactions with PerfeCta PreAmp supermix were performed following the manufacturer's recommendations. Real-time PCR was performed using iTaq universal SYBR Green supermix (Bio-Rad) with primers listed in Supplemental Table S1. L32 was used to normalize expression. The CFX384 real-time PCR detection system (Bio-Rad) was used.
Microarray and IPA analysis
mRNA was isolated from 344SQ Vec or 344SQ m96cl cells that were induced for 24, 48, or 72 h with 2 μg/mL doxycycline and checked for quality before processing for microarray analysis using in-house custom-made mouse MTA1.0 chips and an Affymetrix platform as described earlier (Kundu et al. 2016). Differential expression was determined using two-sided tests on log2 transformed data. SigTerms (Creighton et al. 2008) was used to identify enriched GO terms.
Pathway analysis was evaluated using Qiagen Ingenuity pathway analysis (IPA). IPA upstream analysis, canonical pathway analysis, regulator effects analysis, and networks were performed and visualized following the manufacturer's recommendations. A cutoff at FDR adjusted P < 0.05 and fold change 0.5 were used to identify significantly differentially expressed genes. Enriched canonical pathway analysis of the differentially expressed genes from 344SQ Vec and 344SQ m96cl induced for 72 h was performed. The top 20 significant canonical pathways were identified. Statistical analysis of canonical pathways was determined using right-tailed Fisher's exact test as listed in Supplemental Table S2. The genes down-regulated, up-regulated, with no change, and with no overlap with the data set for each canonical pathway are listed in Supplemental Table S2.
Flow cytometry
Tumors were processed following the MACS (Miltenyi Biotec, Inc.) mouse tumor dissociation kit. Tumors were chopped with a sterile scalpel until 2–3 mm, and then placed in digestion media containing 0.05% collagenase I (Sigma), 30 U/mL DNase type IV (Sigma), and 0.01% hyaluronidase type V (Sigma). Mechanical dissociation using the gentleMACS dissociator (Miltenyi Biotec, Inc.) was performed followed by 40-min incubation at 37°C. Samples were further mechanically dissociated and passed through a 70-μm filter. RBC lysis (BioLegend) was performed following the manufacturer's recommendation. Tumor cells were stained for 1 h with cell surface antibodies as listed in Supplemental Table S1. Cells were fixed in 2% PFA for 15 min, followed by two washes with permeabilization buffer (BioLegend). Cells were further stained with intracellular antibodies as listed in Supplemental Table S1 and washed with FACS buffer. Cells were acquired using BD LSR Fortessa (BD Biosciences) and analyzed using FlowJo software (10.6.2). The gating scheme is shown in Supplemental Figure S8. T cell gating strategy was as follows: CD8+ were gated from parent CD3+ CD45+ cells, which were gated from live cells > from singlets. Antigen-presenting populations were gated from parent CD45+ live cells. All antibodies used for flow cytometry are listed in Supplemental Table S1. tSNE analysis was performed using FlowJo software (10.6.2), and manual gates were applied that excluded doublets, debris, and dead cells. DownSampling gate tool (plugin) was used for 50,000 events. All populations from down-sampled gate were then concatenated, and tSNE parameters were applied.
Immunohistochemistry
Paraffin-embedded tissue sections were processed as described earlier (Peng et al. 2019). H&E sections were examined to count and measure metastatic lung lesions. For IHC staining, mouse CD8 (antibody clone D4W2Z; 1:80; Cell Signaling Technology 98941) and mouse granzyme B (1:800; Abcam ab4059) were detected using the Leica detection kit (Leica Microsystems). The stained slides were digitally scanned using the Aperio ScanScope Turbo slide scanner (Leica Microsystems).
IHC quantitation
Hematoxylin and eosin (H&E)- and immunohistochemistry (IHC)-stained slides were scanned at 20×, using an Aperio AT2 scanner (Leica Biosystem). We performed digital image analysis using Halo (Indica Lab) image analysis software to assess the expression of Ki67 in tumor cells and the presence of CD8+ cells and granzyme B+ cells in the tumor microenvironment. The analysis was supervised by a pathologist with training in digital image analysis. For all biomarkers, we selected tumor area and excluded necrosis, and processed or stained artifacts using Halo annotation tools. For Ki67 assessment, we used the “cytonuclear v2.0.5” algorithm to detect the number of tumor cells with positive expression at “weak,” “moderate,” and “strong” levels of intensity. We reported the percentage of Ki67+ tumor cells and the H-score (0–300), and the tissue area analyzed. For the assessment of CD8 and granzyme B, we used the “cytonuclear v2.0.5” algorithm to detect the number of positive immune cells at any intensity level. We exported the results as total number of positive cells and tumor area analyzed, and calculated densities of CD8- and granzyme B-positive cells (n/mm2, where n is the number of CD8/granzyme B-positive cells per square millimeter of the sample section). In a subset of cases, we assessed CD8 and granzyme B in tumor metastasis and adjacent normal lung tissue area.
In vitro coculture/conditioned media culture
Cancer cells were seeded in a 24-well plate, and naïve immune cells processed from spleens of wild-type 129sv mice were added at 1:6–1:8 ratio. All cocultures were supplemented with 5 μg/mL anti-CD3 and anti-CD28 (BioLegend or BioXcell) antibodies and 50 μM β-mercaptoethanol. Cells were cocultured and induced with doxycycline for 96 h at 37°C with 5% CO2. For conditioned media assays, conditioned media was collected after 72 h of doxycycline induction of cancer cells, centrifuged at 1500 rpm to remove potential detached cancer cells, and then added to naïve immune cells and cultured for 72–96 h at 37°C with 5% CO2.
Cytotoxicity assay
Naïve immune cells were precultured for 6 d in conditioned media collected from either doxycycline-induced 344SQ Vec or 344SQ m96cl cells. CD8+ T lymphocytes were enriched using the mouse CD8a+ T-cell isolation kit (Miltenyi Biotec 130-104-075). Enrichment of CD8+ T cells was confirmed via flow cytometry, and purity was found at >98%. Enriched CD8+ T cells that were precultured in 344SQ Vec or 344SQ m96cl conditioned media were then added to induced 344SQ Vec or 344SQ m96cl cells, respectively, for 24 h at the indicated ratios. Cytotoxicity was determined using the LDH cytotoxicity assay (Thermo Fisher) following the manufacturer's instructions. For cytotoxicity experiments after cytokine/chemokine depletion, the naïve immune cells were precultured in cytokine/chemokine-depleted conditioned media from respective cells and used as described above.
Nanostring analysis
NanoString nCounter expression assay was performed on 344SQ Vec, 344SQ m96cl, 531LN3 Vec, or 531LN3 m96cl cells that were induced for 72 h with doxycycline. RNA was collected from cell lines using TRIzol reagent (Thermo Fisher) following the manufacturer's recommendations. 129/sv mice implanted with 344SQ Vec or 344SQ m96cl cells were on doxycycline feed for 3 wk. Tumors were harvested and flash-frozen, and total RNA was isolated using the mirVana isolation kit (Thermo Fisher). Gene expression analysis was done using a custom mouse tumor microenvironment panel. A total of 100 ng of total RNA in a final volume of 5 μL was mixed with capture probes and reporter probes tagged with a fluorescent barcode from the custom gene expression code set. Probes and targets were hybridized for 12–24 h at 65°C. Hybridized samples were run on the NanoString nCounter preparation station following the manufacturer's recommendations. The samples were run on maximum scan resolution on the nCounter digital analyzer. Data were analyzed using nSolver analysis software. Additional statistical analysis on NanoString nCounter data was conducted as described (Chen et al. 2018a) and suggested by the manufacturer using R version 3.4.2 (R Core Team 2016, https://www.r-project.org). Samples were scaled by the geometric mean of the positive spike-in RNA hybridization controls, as described in the nCounter Expression Data Analysis Guide (NanoString Technologies, Inc.). The expression of each endogenous gene was then tested against the detected expression of all negative control genes using one-sided, two-sample t-tests. Genes showing greater expression than the negative control genes with P < 0.001 were included in further analysis, while others were omitted. Housekeeping gene normalization was applied using the same geometric mean scaling, with a housekeeping gene set identified as most stable in a larger data set by the method of Vandesompele et al. (2002). This set consisted of Alas1, Abcf1, Tbp, Ppia, and Tubb5. Differential expression analysis was conducted on the data after log2 transformation, comparing 344SQ Vec versus 344SQ m96cl using the Empirical Bayes method in LIMMA (Smyth 2004; Ritchie et al. 2015). Differential expression (DE) values for all immune genes as assayed and analyzed by NanoString in in vitro and in vivo samples are listed in Supplemental Table S3.
Cytokine/chemokine depletion experiments
Conditioned media from 344SQ Vec or m96cl cells was collected after 72 h of doxycycline induction. The conditioned media was selectively depleted of individual cytokine/chemokine or IgG (control) by incubating with specific antibodies as listed in Supplemental Table S1 and Protein G beads for 6 h. Beads bound to antibody-protein complexes were centrifuged, and the cytokine/chemokine-depleted supernatant conditioned media was used to culture the naïve immune cells for 72–96 h before analysis by flow cytometry for different immune subpopulations.
Cytokine stimulation
KP cells were seeded at 100,000 cells per well in a 24-well plate. Dox was added to the treated groups (+Dox) only. After 72 h, cells in both +Dox and −Dox cohorts were stimulated for cytokine production using PMA and ionomycin for 24 h. Four hours prior to completion, Brefeldin-A (Golgi-Plug, BD Bioscience) was added as per the manufacturer's recommended concentration for maximal intracellular retention of proteins.
Prediction of binding sites
To generate the predicted transcription factor (TF) binding sites on the IL2 promoter, the promoter sequence for the mouse IL2 gene was used for the matrix profile search on the JASPAR web portal (Fornes et al. 2020). For the search, the vertebrate database selecting the different TF binding motifs was selected to search through the promoter sequence of IL2 with a relative profile threshold score cutoff of ≥80%. The resulting sites were ranked accordingly, and the highest-scoring motifs were annotated on the promoter segment.
Luciferase reporter assay
For reporter assays, respective cells were cotransfected with 500 ng of the reporter construct (GL3) with 10 ng of control (RL) constructs for 16 h with Lipofectamine LTX (Thermo Fisher Scientific) and then assayed for luciferase activity after 48 h of doxycycline induction using the dual-luciferase reporter assay system (Promega E1980).
ChIP assay
ChIP was performed in 393P Vec (GFP), 393P-GFP-Zeb1 (Manshouri et al. 2019), and 393P-GFP-Foxf2 (Kundu et al. 2016) using the Zymo-Spin ChIP kit (Zymo Research D5210) according to the manufacturer's protocol. An anti-GFP antibody (Thermo Fisher Scientific A-11122) or isotype IgG (BioXcell) was used to perform ChIP for GFP only or GFP-tagged Zeb1 or Foxf2. IL2 promoter segment enrichment was analyzed by qPCR using primers as listed under “ChIP Primers” in Supplemental Table S1.
Clinical analysis
For pan-cancer expression analyses, we collected RNA-seq and miRNA-seq data from The Cancer Genome Atlas (TCGA) representing tumors of various histological subtypes as previously described (PMID: 29440175). RNA-seq and miRNA-seq data were obtained from The Broad Institute Firehose pipeline (http://gdac.broadinstitute.org). For miRNA-seq, we previously corrected for batch effects between data platforms (Illumina GAIIx or HiSeq 2000; PMID: 29440175). Patient survival data from TCGA were current as of March 31, 2016.
Statistics
All statistical analyses were performed using GraphPad Prism version 8.0.0. Unless otherwise noted, a one-way ANOVA test was used for multigroup comparisons, while unpaired Student's t-test (two-tailed) was performed for two-group comparisons. A P-value of <0.05 was considered statistically different. Figures denote statistical differences of P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****). Survival experiments used log rank test for survival analysis. Animal experiments were repeated twice, and all in vitro experiments were repeated at least three times with comparable results.
Supplementary Material
Acknowledgments
This study used the resources of the Research Histology, Pathology, and Imaging Core at the University of Texas MD Anderson Cancer Center (MDACC), supported by P30 CA16672 from the Department of Health and Human Services (DHHS)/National Cancer Institute (NCI) Cancer Center Support Grant (CCSG), and The Advanced Cytometry and Sorting Facility (ACSF) Core at MDACC, supported by CCSG NCI P30 CA16672. The m96clFL mice were generated by the Duke Transgenic Core, supported by P30 CA014236 from the DHHS/NCI CCSG and D.L.G. This study was supported by the National Institutes of Health (NIH) CA125123 to C.J.C., Cancer Prevention and Research Institute of Texas (CPRIT) Research Training Grant RP170067 to R.B., NIH R35 CA197616 to D.G.K., and NIH R37CA214609-01A1, CPRIT-Multi-Investigator Research Award (MIRA) RP160652-P3, and Rexanna's Foundation for Fighting Lung Cancer to D.L.G. The work was also supported by generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shots Program.
Author contributions: S.T.K, B.L.R., and D.L.G. conceived, designed, and implemented the study. M.S. and D.G.K. designed and developed the m96clFL mice. S.T.K., B.L.R., L.A.G., A.N.W., M.G.P., R.B., J.J.F., C.A.C, L.M.S., and F.R.R.A. acquired and analyzed the data. S.T.K., B.L.R., R.B., A.N.W., M.G.P., L.D., F.C., J.W., C.J.C., and D.L.G. performed the statistical analysis and interpreted and represented the data. S.T.K., B.L.R., R.B. wrote and critically revised the manuscript, and prepared the figures and tables. S.T.K. and D.L.G. provided overall supervision and execution of the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349321.121.
Competing interest statement
D.L.G. has received research grant funding from AstraZeneca, Janssen, Takeda, Astellas, Ribon Therapeutics, and NGM Biopharmaceuticals. D.G.K. is a cofounder of and stockholder in XRAD Therapeutics and NGM Biopharmaceuticals, which is developing radiosensitizers. D.G.K. is a member of the scientific advisory board for and owns stock in Lumicell, Inc., a company commercializing intraoperative imaging technology. D.G.K. is a coinventor on a patent for a handheld imaging device and is a coinventor on a patent for radiosensitizers. XRAD Therapeutics, Merck, Bristol Myers Squibb, and Varian Medical Systems provide research support to D.G.K. All other authors declare no competing interests.
References
- Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, Pertsemlidis A, Gregory PA, Wright JA, Goodall GJ, et al. 2012. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest 122: 3170–3183. 10.1172/JCI63608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition. Cancer Res 68: 7846–7854. 10.1158/0008-5472.CAN-08-1942 [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. 2008. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 9: 582–589. 10.1038/embor.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M, et al. 2016. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352: 227–231. 10.1126/science.aac9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al. 2014a. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 5: 5241. 10.1038/ncomms6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al. 2014b. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 5: 5241. 10.1038/ncomms6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Diao L, Yang Y, Yi X, Rodriguez BL, Li Y, Villalobos PA, Cascone T, Liu X, Tan L, et al. 2018a. CD38-Mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov 8: 1156–1175. 10.1158/2159-8290.CD-17-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zander R, Khatun A, Schauder DM, Cui W. 2018b. Transcriptional and epigenetic regulation of effector and memory CD8 T cell differentiation. Front Immunol 9: 2826. 10.3389/fimmu.2018.02826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. 2008. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA 14: 2290–2296. 10.1261/rna.1188208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Costantino N, Court DL. 2006. A set of recombineering plasmids for Gram-negative bacteria. Gene 379: 109–115. 10.1016/j.gene.2006.04.018 [DOI] [PubMed] [Google Scholar]
- Derynck R, Weinberg RA. 2019. EMT and cancer: more than meets the eye. Dev Cell 49: 313–316. 10.1016/j.devcel.2019.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhood B, Najafi M, Mortezaee K. 2019. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: a review. J Cell Physiol 234: 8509–8521. 10.1002/jcp.27782 [DOI] [PubMed] [Google Scholar]
- Feng X, Wang Z, Fillmore R, Xi Y. 2014. MiR-200, a new star miRNA in human cancer. Cancer Lett 344: 166–173. 10.1016/j.canlet.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranasic D, et al. 2020. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 48: D87–D92. 10.1093/nar/gkz1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman WH, Pagès F, Sautès-Fridman C, Galon J. 2012. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 12: 298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- Garner H, de Visser KE. 2020. Immune crosstalk in cancer progression and metastatic spread: a complex conversation. Nat Rev Immunol 20: 483–497. 10.1038/s41577-019-0271-z [DOI] [PubMed] [Google Scholar]
- Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, et al. 2009. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 23: 2140–2151. 10.1101/gad.1820209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601. 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Coussens LM. 2012. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21: 309–322. 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. 2008. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322: 1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30: 531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Soundararajan R, Vasaikar S, Yang F, Allton KL, Tian L, den Hollander P, Isgandarova S, Haemmerle M, Mino B, et al. 2021. CD8+ t cells inhibit metastasis and CXCL4 regulates its function. Br J Cancer 125: 176–189. 10.1038/s41416-021-01338-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. 2010. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32: 91–103. 10.1016/j.immuni.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283: 14910–14914. 10.1074/jbc.C800074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, Buczkowski KA, Liu Y, Awad MM, Denning WL, et al. 2016. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res 76: 999–1008. 10.1158/0008-5472.CAN-15-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P, et al. 2017. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 19: 518–529. 10.1038/ncb3513 [DOI] [PubMed] [Google Scholar]
- Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, Tong P, Creighton CJ, Gibbons DL. 2016. The miR-200 family and the miR-183∼96∼182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene 35: 173–186. 10.1038/onc.2015.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu ST, Grzeskowiak CL, Fradette JJ, Gibson LA, Rodriguez LB, Creighton CJ, Scott KL, Gibbons DL. 2018. TMEM106B drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Nat Commun 9: 2731. 10.1038/s41467-018-05013-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. 2015a. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 373: 23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Hodi FS, Wolchok JD. 2015b. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 373: 1270–1271. 10.1056/NEJMc1509660 [DOI] [PubMed] [Google Scholar]
- Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK. 2012. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Medical oncology 29: 618–626. 10.1007/s12032-011-9923-y [DOI] [PubMed] [Google Scholar]
- Lou Y, Diao L, Cuentas ER, Denning WL, Chen L, Fan YH, Byers LA, Wang J, Papadimitrakopoulou VA, Behrens C, et al. 2016. Epithelial–mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res 22: 3630–3642. 10.1158/1078-0432.CCR-15-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek TR, Castro I. 2010. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33: 153–165. 10.1016/j.immuni.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manshouri R, Coyaud E, Kundu ST, Peng DH, Stratton SA, Alton K, Bajaj R, Fradette JJ, Minelli R, Peoples MD, et al. 2019. ZEB1/NuRD complex suppresses TBC1D2b to stimulate E-cadherin internalization and promote metastasis in lung cancer. Nat Commun 10: 5125. 10.1038/s41467-019-12832-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci 90: 8424–8428. 10.1073/pnas.90.18.8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacurari M, Addison JB, Bondalapati N, Wan YW, Luo D, Qian Y, Castranova V, Ivanov AV, Guo NL. 2013. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells. Int J Oncol 43: 548–560. 10.3892/ijo.2013.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastushenko I, Blanpain C. 2019. EMT transition states during tumor progression and metastasis. Trends Cell Biol 29: 212–226. 10.1016/j.tcb.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. 2016. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov 6: 202–216. 10.1158/2159-8290.CD-15-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng DH, Kundu ST, Fradette JJ, Diao L, Tong P, Byers LA, Wang J, Canales JR, Villalobos PA, Mino B, et al. 2019. ZEB1 suppression sensitizes KRAS mutant cancers to MEK inhibition by an IL17RD-dependent mechanism. Sci Transl Med 11: eaaq1238. 10.1126/scitranslmed.aaq1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restifo NP, Dudley ME, Rosenberg SA. 2012. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol 12: 269–281. 10.1038/nri3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Mito JK, Lee CL, Zhang M, Li Z, Dodd RD, Cason D, Luo L, Ma Y, Van Mater D, et al. 2016. MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. J Clin Invest 126: 1606. 10.1172/JCI86573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitthideatphaiboon P, Galan-Cobo A, Negrao MV, Qu X, Poteete A, Zhang F, Liu DD, Lewis WE, Kemp HN, Lewis J, et al. 2021. STK11/LKB1 mutations in NSCLC are associated with KEAP1/NRF2-dependent radiotherapy resistance targetable by glutaminase inhibition. Clin Cancer Res 27: 1720–1733. 10.1158/1078-0432.CCR-20-2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, Schrock AB, Hartmaier RJ, Trabucco SE, Gay L, et al. 2018. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov 8: 822–835. 10.1158/2159-8290.CD-18-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: 1–25. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Spolski R, Li P, Leonard WJ. 2018. Biology and regulation of IL-2: from molecular mechanisms to human therapy. Nat Rev Immunol 18: 648–659. 10.1038/s41577-018-0046-y [DOI] [PubMed] [Google Scholar]
- Spranger S, Gajewski TF. 2018. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer 18: 139–147. 10.1038/nrc.2017.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S, Bao R, Gajewski TF. 2015. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523: 231–235. 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- Topalian SL, Taube JM, Anders RA, Pardoll DM. 2016. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 16: 275–287. 10.1038/nrc.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Yang J. 2013. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 27: 2192–2206. 10.1101/gad.225334.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Leun AM, Thommen DS, Schumacher TN. 2020. CD8+ t cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer 20: 218–232. 10.1038/s41568-019-0235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.1. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ. 2011. T cell exhaustion. Nat Immunol 12: 492–499. 10.1038/ni.2035 [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660. 10.1038/nature05529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ahn YH, Chen Y, Tan X, Guo L, Gibbons DL, Ungewiss C, Peng DH, Liu X, Lin SH, et al. 2014. ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. J Clin Invest 124: 2696–2708. 10.1172/JCI72171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JP, Stewart AF. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet 20: 123–128. 10.1038/2417 [DOI] [PubMed] [Google Scholar]
- Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. 2007. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene 26: 6896–6904. 10.1038/sj.onc.1210493 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.