In this study, Jones and Kraus integrated data from metabolomic, proteomic, and transcriptomic assays to characterize how alterations in NAD+ metabolism during the differentiation of mouse ESCs lead to alteration of the PARP1-mediated ADP-ribosylated (ADPRylated) proteome and mRNA isoform specialization. Their findings demonstrate a role for the NAD+–PARP1 axis in the maintenance of the mESC state, specifically in the splicing program during differentiation.
Keywords: ADP-ribosylation, de novo pathway, embryonic stem cells (ESCs), metabolomics, nicotinamide adenine dinucleotide (NAD+), NAD+ biosynthesis, PARP1, Preiss–Handler pathway, proteomics, salvage pathway, spliceosome, splicing, U2AF35, U2AF65
Abstract
The differentiation of embryonic stem cells (ESCs) into a lineage-committed state is a dynamic process involving changes in cellular metabolism, epigenetic modifications, post-translational modifications, gene expression, and RNA processing. Here we integrated data from metabolomic, proteomic, and transcriptomic assays to characterize how alterations in NAD+ metabolism during the differentiation of mouse ESCs lead to alteration of the PARP1-mediated ADP-ribosylated (ADPRylated) proteome and mRNA isoform specialization. Our metabolomic analyses indicate that mESCs use distinct NAD+ biosynthetic pathways in different cell states: the de novo pathway in the pluripotent state, and the salvage and Preiss–Handler pathways as differentiation progresses. We observed a dramatic induction of PARP1 catalytic activity driven by enhanced nuclear NAD+ biosynthesis during the early stages of mESC differentiation (e.g., within 12 h of LIF removal). PARP1-modified proteins in mESCs are enriched for biological processes related to stem cell maintenance, transcriptional regulation, and RNA processing. The PARP1 substrates include core spliceosome components, such as U2AF35 and U2AF65, whose splicing functions are modulated by PARP1-mediated site-specific ADP-ribosylation. Finally, we observed that splicing is dysregulated genome-wide in Parp1 knockout mESCs. Together, these results demonstrate a role for the NAD+–PARP1 axis in the maintenance of mESC state, specifically in the splicing program during differentiation.
PARP1 is an abundant nuclear protein that controls a variety of cellular processes, including DNA repair, histone modification, chromatin remodeling, and transcription regulation (Gibson and Kraus 2012; Gupte et al. 2017). Growing evidence has suggested a role for PARP1 in RNA biology, including RNA processing and splicing (Kim et al. 2020). PARP1 is an enzyme with a C-terminal catalytic domain, which allows it to use oxidized nicotinamide adenine dinucleotide (NAD+) as a substrate for the covalent attachment of chains of ADP-ribose (ADPR) [poly(ADP-ribose) (PAR)] on substrate proteins through a process known as PARylation (Gibson and Kraus 2012; Gupte et al. 2017). Great strides have been made over the past decade in the identification of specific sites of (ADP-ribosyl)ation (ADPRylation) by mass spectrometry and the functional characterization of those sites (Daniels et al. 2015; Hendriks et al. 2019).
NAD+ is a metabolic cofactor with critical roles in a variety of physiological and pathological processes (Cambronne and Kraus 2020). In mammals, NAD+ can be synthesized through three distinct pathways that start from different metabolites: the de novo, salvage, and Preiss–Handler pathways (Houtkooper et al. 2010). During PARylation, NAD+ is cleaved into ADPR and nicotinamide, with the latter feeding into the salvage pathway. Recent studies have demonstrated the importance of cellular context in pathway usage; however, how the pathways are used in different ESC states remains unknown (Folmes et al. 2012; Minhas et al. 2019; Zhang et al. 2019).
The nicotinamide mononucleotide adenylyltransferases (NMNAT-1, NMNAT-2, and NMNAT-3) are required for all three NAD+ biosynthetic pathways (Fortunato et al. 2021). The restricted subcellular localization of the NMNATs (i.e., NMNAT-1 in the nucleus and NMNAT-2 in the cytosol) underlies the compartmentalized biosynthesis of NAD+. The NMNATs and their respective NAD+ pools are nonredundant, as mice genetically lacking either cytosolic or nuclear NMNAT are nonviable (Conforti et al. 2011; Hicks et al. 2012). We have shown that NAD+ is generally confined to the compartment of its production and regulates the catalytic activity of PARP family members within that compartment (Ryu et al. 2018; Challa et al. 2021). PARP1 is a major consumer of NAD+ and its catalytic activity is modulated by NAD+ availability, although little is known about the impact of PARP1 on NAD+ biosynthesis and its role in the differentiation of ESCs.
The transition of ESCs from the pluripotent state to a lineage-committed state is known to involve many dynamic cellular processes, including modifications to the epigenome, transcriptome, and metabolome (Young 2011; Carey et al. 2015; Sperber et al. 2015). Recent work has implicated PARP1 and its enzymatic activity in the regulation of ESC self-renewal and lineage commitment. PARP1 promotes the maintenance of ground-state pluripotency and inhibits the differentiation of ESCs into trophoblast derivatives (Hemberger et al. 2003; Roper et al. 2014). Adding to this complexity, PARP1 regulates the transcriptional program of ESCs in both catalytic-independent and catalytic-dependent manners, with distinct outcomes (Doege et al. 2012; Chiou et al. 2013; Roper et al. 2014; Liu and Kraus 2017). Inhibition of PARP1 catalytic activity causes a reduced propensity to differentiate and, in the process of reprogramming, a diminished capacity to return to pluripotency (Ji and Tulin 2012; Weber et al. 2013).
Here, we used both wild-type and Parp1−/− murine ESCs (mESCs), as well as a suite of “omics” assays (i.e., metabolomic, proteomic, and genomic), to explore (1) NAD+ biosynthetic pathway utilization and compartmentalization during the differentiation of mESCs, (2) the connection between NAD+ availability and PARP1 catalytic activity in mESCs, and (3) PARP1-mediated PARylation of RNA binding proteins that regulates splicing to control the differentiation of mESCs.
Results
In this study, we examined potential alterations in NAD+ biosynthetic pathways, PARP1 activity, PARP1 substrates, and molecular outcomes during the differentiation of mESCs upon removal of leukemia inhibitory factor (LIF) and a shift to suspension in low-attachment plates for embryoid formation. As described below, we executed and integrated a suite of “omics” assays (i.e., metabolomics, proteomics, and genomics) to connect the NAD+–PARP1 axis to site-specific ADPRylation of splicing factors and alterations in mRNA isoforms that determine mESC state.
The levels and localization of NAD+ biosynthesis change during the differentiation of mESCs
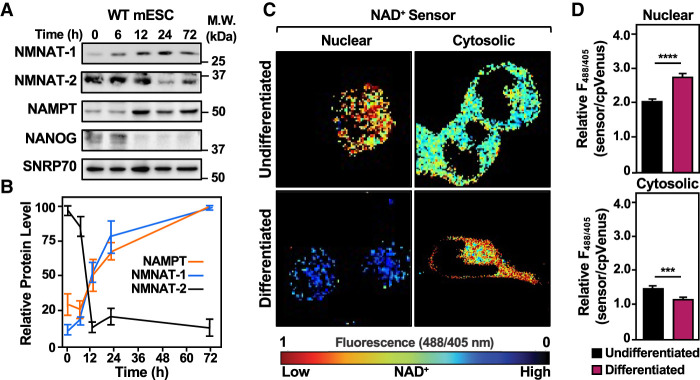
To explore how NAD+ levels, localization, and biosynthesis pathways change during the differentiation of mESCs, we began by investigating the levels of enzymes in the salvage pathway during a time course of mESC differentiation by Western blotting. We observed that NMNAT-1 and NMNAT-2 protein levels change in an inverse manner; NMNAT-1 levels were low in the pluripotent state and increased during differentiation, while NMNAT-2 levels were high in the pluripotent state and decreased during differentiation (Fig. 1A,B). Interestingly, we found that NAMPT levels mirror NMNAT-1 levels, increasing during differentiation.
Figure 1.
Dynamic shifts in the levels of enzymes in the NAD+ salvage pathway and compartmentalized biosynthesis lead to increased nuclear NAD+ levels during mESC differentiation. (A,B) The levels of enzymes in the NAD+ salvage pathway are dynamic during mESC differentiation. (A) Western blot analysis of whole-cell extracts prepared from wild-type (WT) mESCs for NMNAT-1, NMNAT-2, and NAMPT during a time course of differentiation upon LIF removal. SNRP70 and NANOG were used as markers for loading and differentiation, respectively. (B) Quantification of multiple Western blots, like those shown in A. Each point represents the mean ± SEM; n = 4. (C,D) Compartment-specific NAD+ levels in WT mESCs undifferentiated and 12-h differentiated by LIF removal. The fluorescence images in C were generated using nuclear and cytosolic NAD+ sensors. The scale bar shows the inverse relationship between fluorescent signal and NAD+ levels. The bars in the graphs in D represent the mean ± SEM of the relative NAD+ concentrations calculated using sensor(488/405 nm)/control(488/405 nm) fluorescence ratios determined by live-cell imaging (n = 3, ANOVA). (∗∗∗) P < 0.001, (∗∗∗∗) P < 0.0001. (Top) Nuclear. (Bottom) Cytosolic.
To determine how the changes in NMNAT-1, NMNAT-2, and NAMPT affect subcellular NAD+ levels, we used genetically encoded nuclear and cytosolic NAD+ sensors coupled with live-cell imaging (Cambronne et al. 2016; Ryu et al. 2018; Challa et al. 2021). Nuclear NAD+ levels were low in the pluripotent state and elevated in the differentiated state, following the pattern of NMNAT-1 and NAMPT protein levels during differentiation (Fig. 1C,D; Supplemental Fig. S1A,B). Cytosolic NAD+ levels were comparatively high in the pluripotent state and decreased in the differentiated state, following the pattern of NMNAT-2 protein levels during differentiation (Fig. 1C,D; Supplemental Fig. S1A,B). Together, these results show that compartment-specific NAD+ synthesis is driven by changes in the levels of enzymes in the NAD+ salvage pathway during differentiation.
The pathways used for NAD+ biosynthesis change during the differentiation of mESCs
The biological programs that maintain pluripotency in ESCs have metabolite requirements distinct from those that maintain terminal differentiation in cells (Carey et al. 2015; Sperber et al. 2015; Dahan et al. 2019). Biosynthesis of NAD+ occurs through three pathways: the Preiss–Handler, de novo, and salvage pathways (Fig. 2A; Houtkooper et al. 2010; Cambronne and Kraus 2020). Each pathway uses a distinct starting metabolite, but all use NMNATs to convert either nicotinamide mononucleotide (NMN) or nicotinic acid mononucleotide (NAMN) into NAD+ or NAAD, respectively (Fig. 2A). In most tissue types, the main source of NAD+ is thought to be the salvage pathway, but recent work has shown that cellular context can impact pathway usage (Folmes et al. 2012; Zhang et al. 2019).
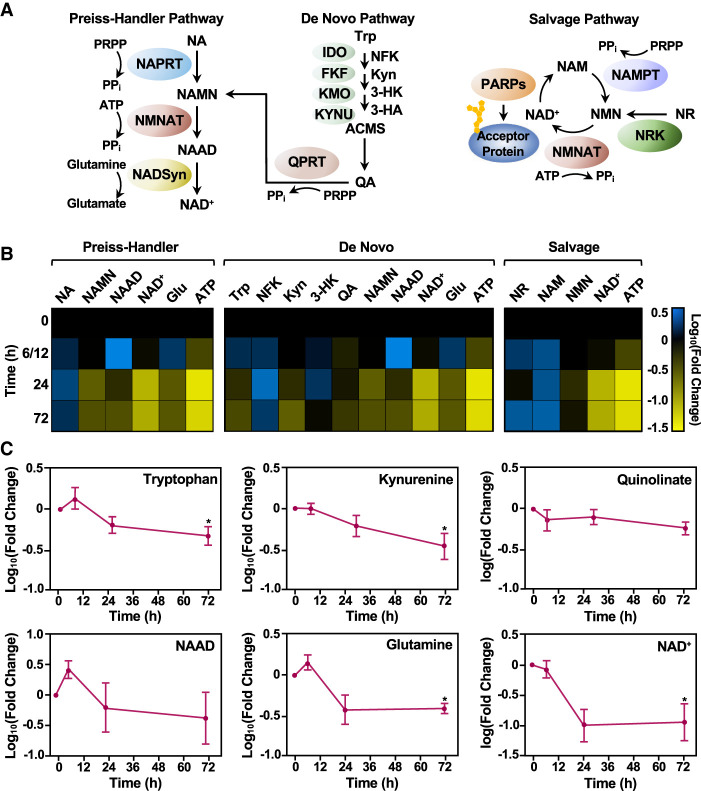
Figure 2.
Different NAD+ biosynthetic pathways are used in distinct mESC states. (A) Schematic representation of the three major NAD+ biosynthesis pathways (Preiss–Handler, de novo, and salvage), including the metabolites in the pathway and the enzymes that catalyze each step. Enzymes are represented as colored ovals. (NA) Nicotinic acid, (NAMN) nicotinic acid mononucleotide, (NAAD) nicotinic acid adenine dinucleotide, (Trp) tryptophan, (NFK) N-formyl-kynurenine, (Kyn) kynurenine, (3-HK) 3-hydroxykynurenine, (3-HA) 3-hydroxyanthranilate, (QA) quinolinate, (NAM) nicotinamide, (NR) nicotinamide riboside, (NMN) nicotinamide mononucleotide, (NAD+) nicotinamide adenine dinucleotide, (PRPP) phosphoribosyl pyrophosphate, (Glu) glutamine, (ATP) adenosine triphosphate. (B) Heat map representation of the relative levels of metabolites in the NAD+ biosynthetic pathways in WT mESCs during a time course of differentiation as determined by mass spectrometry. Each column represents a single metabolite throughout time course, and each row represents the time points (the data for the 6- and 12-h time points were combined). Metabolites that increased relative to the 0-h time point are represented in blue (log10 fold change [FC]), and metabolites that decreased in abundance are indicated in yellow. Metabolites are grouped and ordered according to the NAD+ metabolic pathways (Preiss–Handler, de novo, and salvage), as indicated. (C) Line plot representation of average relative levels (log10FC) of key metabolites in the de novo pathway (n = 7; ANOVA). (∗) P < 0.05.
To investigate in more detail NAD+ biosynthesis in mESCs in the pluripotent state and during differentiation, we performed targeted steady-state metabolomics using mass spectrometry. We hypothesized that the pathways feeding NMNAT-1 and NMNAT-2 would show predictable metabolite levels corresponding to the changing levels of NMNAT-1, NMNAT-2, and NAMPT during differentiation, as shown in Figure 1, A and B (e.g., high metabolite levels in the pluripotent state that decrease during differentiation for the pathway feeding NMNAT-2). We collected mESCs at various times after differentiation initiated by LIF removal (0, 6, 12, 24, and 72 h). Cell extracts were subjected to LC-MS/MS analysis to quantify nearly all metabolites in the three NAD+ biosynthetic pathways (Fig. 2A,B; Supplemental Table S1). Given the variability in the early phases of differentiation, we combined the results from the 6- and 12-h time points.
Changes in the levels of metabolites in the NAD+ biosynthetic pathways were evident. For example, we observed reductions in the levels of metabolites in the de novo pathway during differentiation (Fig. 2B [middle, blue-to-yellow shift], C). These included significant depletions of tryptophan (Trp) and kynurenine (Kyn; log10 fold changes of −0.342 and −0.46 [fold changes of 0.46 and 0.35], respectively), both of which are unique to the de novo pathway. In addition, we observed reductions in quinolinate (QA), nicotinic acid mononucleotide (NAMN), and nicotinic acid adenine dinucleotide (NAAD), although they did not reach significance (Fig. 2B [middle], C). Glutamine, a donor metabolite for NAD+ synthase (NADSyn), which participates in both the Preiss–Handler and de novo pathways, also decreased significantly during differentiation (log10 fold change of −0.49 [fold change of 0.39]) (Fig. 2B [middle], C).
We observed that not all early de novo metabolites followed this trend; however, kynurenine and its intermediates are either active themselves in various biological processes (e.g., aryl hydrocarbon receptor activation, NMDA receptor agonism, and melatonin production) or serve as starting materials for other metabolic pathways that do not funnel into de novo NAD+ synthesis (Wogulis et al. 2008). In fact, Trp, 3-HK, and Kyn all can be metabolized by non-NAD+-related enzymes and form products not within the NAD+ synthesis pathway (Gostner et al. 2020). In contrast, we observed increases in the levels of metabolites in the salvage pathway during differentiation (Fig. 2B, right, black-to-blue shift). For example, nicotinamide (NAM) and nicotinamide riboside (NR), which are unique to the salvage pathway, increased during the time course of differentiation (log10 fold changes of 0.292 and 0.27 [fold changes of 2.0 and 1.9], respectively) (Fig. 2B, right). Nicotinamide mononucleotide (NMN) levels, however, remained relatively stable during differentiation (Fig. 2B, right). Similar results were observed for nicotinic acid (NA) in the Preiss–Handler pathway (Fig. 2B, left). Together, these data suggest that mESCs differentially use NAD+ biosynthetic pathways in different cell states: the de novo pathway in the pluripotent state, and the salvage and Preiss–Handler pathways as differentiation progresses.
To further investigate the effects of NAD+ biosynthetic pathway usage on embryonic stem cell state, we knocked down mRNAs encoding key enzymes from the de novo and salvage pathways using siRNAs, focusing on Qprt and Nampt, respectively (Supplemental Fig. S2A). Upon knockdown of Qprt in the undifferentiated state, we observed a decrease in pluripotency marker gene expression and an increase in some differentiation marker gene expression. However, knockdown of Qprt during the course differentiation did not alter the expression of these genes, highlighting the key role of the de novo pathway in the maintenance of ESC pluripotency (Supplemental Fig. S2B,C). In contrast, upon knockdown of Nampt, we observed increases in the expression of pluripotency marker genes, except for Sox2, which remained unchanged. Notably, this alteration was present when Nampt was knocked down during differentiation—when its protein levels are normally elevated, and the salvage pathway metabolites are elevated (Supplemental Fig. S2B,C). Together, these results indicate that NAD+ biosynthesis pathway availability can play a role in controlling cell state. Specifically, inhibiting de novo biosynthesis of NAD+ increases the propensity to differentiate, and inhibiting the salvage pathway supports pluripotency maintenance.
Knockout of Parp1 during differentiation in mESCs alters compartmentalized NAD+ biosynthesis and cell state
We have previously shown that changes in NAD+ levels within specific cellular compartments control the activity of PARPs within those compartments (Ryu et al. 2018; Challa et al. 2021). To determine how compartmentalized NAD+ biosynthesis in mESCs might affect PARP1 activity in the nucleus, we examined changes in PARylation (specifically, PARP1 automodification) by Western blotting throughout a time course of differentiation (Fig. 3A). Interestingly, although PARP1 protein levels were unchanged during differentiation, PARP1 catalytic activity increased significantly within 12 h of LIF removal (Fig. 3A; Supplemental Fig. S3A). These results suggested a link between increased levels of nuclear NAD+ synthesis and increased PARP1 activity during differentiation.
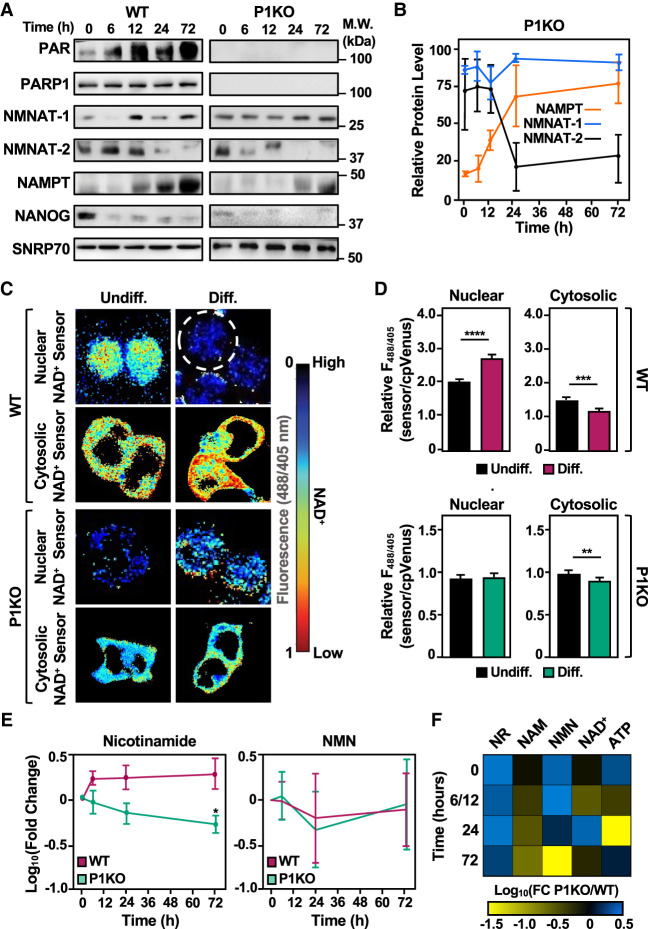
Figure 3.
Genetic deletion of Parp1 alters NAD+ biosynthesis and the levels of nuclear and cytosolic NAD+. (A,B) PARP1 catalytic activity, but not PARP1 protein levels, increases during the differentiation of wild-type (WT) mESCs. Automodification of PARP1 monitored by Western blotting with a PAR detection reagent was used to assess PARP1 activity. (A) Western blot analysis of whole-cell extracts prepared from WT and Parp1−/− (P1KO) mESCs as indicated. NANOG and SNRP70 were used as markers for differentiation and loading, respectively. (B) Quantification of Western blots for NAMPT, NMNAT-1, and NMNAT-2 assessing their levels over a time course of differentiation in P1KO mESCs (cf. Fig. 1B). (C,D) Compartment-specific NAD+ levels in wild-type (WT) and Parp1−/− (P1KO) mESCs undifferentiated (Undiff.) and 12-h differentiated by LIF removal (Diff.). The fluorescence images in C were generated using nuclear and cytosolic NAD+ sensors as indicated. The scale bar shows the inverse relationship between fluorescent signal and NAD+ levels. The bars in the graphs in D represent the mean ± SEM of the relative NAD+ concentrations calculated using sensor(488/405 nm)/control(488/405 nm) fluorescence ratios determined by live-cell imaging (n = 3, ANOVA). (∗∗) P < 0.01, (∗∗∗) P < 0.001, (∗∗∗∗) P < 0.0001. (Top) WT. (Bottom) P1KO. (Left) Nuclear. (Right) Cytosolic. (E) Line plot representation of average relative levels (log10FC) of NAM and NMN in the salvage pathway (n = 7; ANOVA). (∗) P < 0.05. (F) Heat map representation of the relative ratios of metabolites in the salvage pathway (nicotinamide riboside [NR], nicotinamide [NAM], nicotinamide mononucleotide [NMN], nicotinamide adenine dinucleotide [NAD+], and adenine triphosphate [ATP]) in P1KO mESCs versus WT mESCs during a time course of differentiation as determined by mass spectrometry. Each column represents a single metabolite throughout the time course, and the rows represent the time points (the data for the 6- and 12-h time points were combined). Metabolites that increased in P1KO mESCs versus WT mESCs at the same time point of differentiation are represented in blue (log10FC), and metabolites that decreased in abundance are indicated in yellow.
To explore the connections between PARP1 and NAD+ biosynthesis in mESCs in more detail, we used wild-type (WT) and Parp1−/− (P1KO) mESCs. We have shown that genetic depletion of PARP1 creates a metastable pluripotent state in which the expression of some of the core transcription factors (e.g., Sox2 and Oct4) is paired with increased expression of differentiation markers (Liu and Kraus 2017). We confirmed these results here, including a significant reduction in alkaline phosphatase (AP) activity, a marker of pluripotency, in P1KO cells compared with the WT cells (Supplemental Fig. S3B–E). Furthermore, we found that depletion of NMNAT-1, the nuclear NAD synthase supplying PARP1 catalytic activity, affected these key genes in a manner similar to that of P1KO (Supplemental Fig. S4). It is important to note that although they exist in a metastable pluripotent state, P1KO mESCs differentiate to a similar extent and with patterns similar to that of WT mESCs (Supplemental Fig. S5).
These effects were accompanied by an elevation in the levels of NMNAT-1, the nuclear NAD+ synthase, in the undifferentiated state in P1KO cells compared with the WT cells (Figs. 1A,B, 3A,B). These results suggest that elevated NMNAT-1 levels represent another marker of the metastable pluripotent state upon PARP1 depletion. In assays of compartmentalized NAD+ biosynthesis using the sensors described above, genetic depletion of PARP1, a major consumer of NAD+, eliminated the differentiation-induced increase in nuclear NAD+ observed in WT cells without affecting the differentiation-induced decrease in cytosolic NAD+ (Fig. 3C,D; Supplemental Fig. S6). The results reflect the steady levels of NMNAT-1 in the P1KO cells. Treatment of WT mESCs with PARP inhibitor during differentiation mimicked the patterns, both in enzyme level and NAD+ compartmentalization, that were observed in P1KO ESCs (cf. Fig. 3A–D and Supplemental Fig. S7).
The effect of PARP1 depletion on NAD+ metabolism was also observed in targeted metabolic pathway analysis. Nicotinamide (NAM), a product released when PARP1 cleaves NAD+ to ADP-ribose (ADPR), was significantly decreased in P1KO cells compared with WT cells during the time course of differentiation, as expected due to the loss of PARP1 activity (Fig. 3E,F). Strikingly, P1KO cells exhibited elevated levels of nicotinamide riboside (NR) compared with WT cells at all time points (Fig. 3F); NR synthesis bypasses the recycling of NAM by NAMPT. The levels of other metabolites in the salvage pathway were similar between P1KO and WT mESCs (Fig. 3E,F). Additionally, metabolite levels in the Preiss–Handler and de novo pathways, with the exception of NAMN, were similar in WT and P1KO mESCs (Supplemental Fig. S8). Together, these results highlight the “PARP1–NAD+ axis” during the differentiation of mESCs, as illustrated by the reciprocal effects between PARP1 and NAD+ biosynthetic enzymes, such as NMNAT-1.
PARP1 ADPRylates RNA binding factors and core spliceosome components, including U2AF, in embryonic stem cells
Our previous (Ryu et al. 2018) and current results point to the coordination of nuclear NAD+ biosynthesis and PARP1 activity during cellular differentiation programs. A logical consequence of this “NAD+–PARP1 axis” is the NAD+-dependent ADPRylation of substrate proteins by PARP1, presumably to achieve specific regulatory outcomes. In this regard, we sought to identify site-specific PARylation events on PARP1 substrates that could support the differentiation of mESCs.
To this end, we used an NAD+ analog-sensitive PARP1 (asPARP1) approach that we developed previously (Supplemental Fig. S9A,B; Gibson et al. 2016; Gibson and Kraus 2017) to identify the PARP1-mediated Asp/Glu ADPRylated proteome in mESCs. In this approach, an asPARP1 mutant (L877A), but not wild-type PARP1, can ADPRylate substrates in cell extracts using the NAD+ analog 8-Bu(3-yne)T-NAD+ (Supplemental Fig. S9C–E; Gibson et al. 2016; Gibson and Kraus 2017). The substrate proteins and the specific sites of PARylation on those proteins are then identified by mass spectrometry. Using this approach, we identified 472 high-confidence, unique PARP1-mediated Asp/Glu ADPRylated protein substrates, as well as 293 specific sites of ADPRylation (Fig. 4A; Supplemental Fig. S9F; Supplemental Table S2). Gene ontology analysis of these substrates identified roles in well-studied cellular processes known to be regulated by PARP1 activity, such as transcription, DNA damage repair, and chromatin modification, as well as biologically relevant processes, such as stem cell maintenance and differentiation (Fig. 4B). Interestingly, the GO analyses also identified terms associated with RNA processing and splicing (Fig. 4B). We focused on the latter.
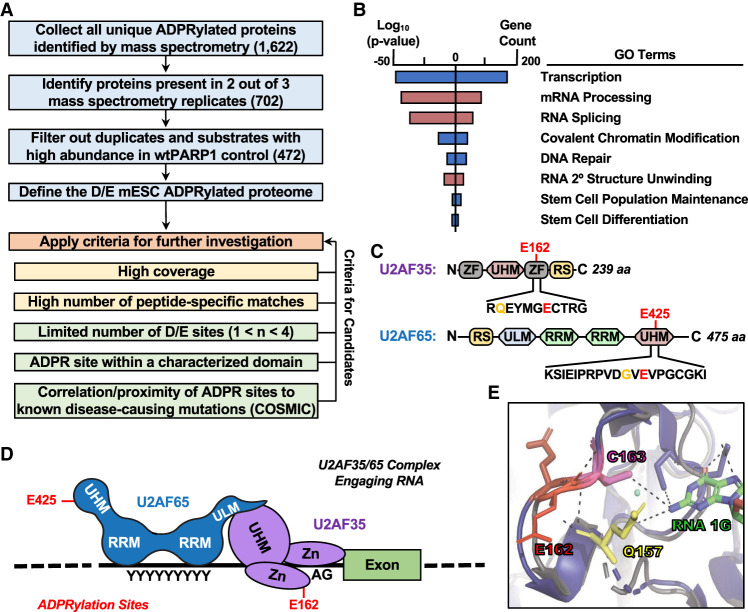
Figure 4.
Identification of PARP1 substrates and specific sites of ADPRylation in differentiating mESCs. (A) Flowchart illustrating the cutoffs and criteria used for defining the ADP-ribosylated proteome and selecting candidates for further investigation using mass spectrometry data from an asPARP1 approach. (B) Gene ontology terms enriched for the protein substrates of PARP1 performed using the DAVID tool. (C) Schematic diagrams of U2AF35 and U2AF65 showing key functional domains and the PARP1-mediated ADPRylation sites (Glu162 and Glu425 in U2AF35 and U2AF65, respectively), determined by LC-MS/MS coupled with an asPARP1 approach. Hotspot mutation sites are highlighted in yellow (Gln157 and Gly423 in U2AF35 and U2AF65, respectively), determined by COSMIC. (D) Model of the U2AF35/65 complex associated with its cognate RNA sequences. The ADPRylation sites (Glu162 and Glu425 in U2AF35 and U2AF65, respectively) are shown. (E) Close-up view of the X-ray crystal structure of yeast U2AF1 complexed with RNA overlaid with an illustration of mU2AF35 (AlphaFold) highlighting the region around Glu162, a site of PARP1-mediated ADPRylation. Resolved and potential hydrogen bonds between amino acid side chains and RNA are shown as dashed lines.
To facilitate subsequent analyses, we applied a set of criteria to ensure selection of proteins with biologically relevant ADPRylation events that could be readily and unambiguously interrogated in a range of assays (Fig. 4A). Application of these criteria yielded U2 snRNP auxiliary factor 35-kDa and 65-kDa subunits (U2AF35 and U2AF65, respectively; collectively, U2AF). Single sites of Glu ADPRylation were identified on U2AF35 and U2AF65 (E162 and E425, respectively) (Fig. 4C), although additional sites on other residues may be possible. Both sites are near known hotspot mutations identified in multiple cancer types in the COSMIC database (Q157 in U2AF35, and G423 in U2AF65). Together, U2AF35 and U2AF65 form the U2AF complex, a core component of the earliest intron/exon-defined spliceosome, the E complex, and define the U2-type form of intron removal (Sharp and Burge 1997; Will and Luhrmann 2011). U2AF35 interacts with the 3′ splice site (3′SS) AG dinucleotide, while U2AF65 binds the polypyrimidine tract (PPT) (Fig. 4D).
In U2AF35, E162 resides at the surface of the second zinc finger domain and is predicted to hydrogen-bond with Q157 (the hotspot mutation site), Y158, and T164 (Fig. 4E; Jumper et al. 2021). The crystal structure of the yeast homolog of U2AF35 (U2AF1) with RNA shows that both Q157 and C163 form hydrogen bonds with the N1 atom of guanine (1G) in the 3′ splice site RNA (Fig. 4E; Yoshida et al. 2020). The hydrogen bonding between E162 and T164 is thought to aid the proper conformation of C163, which contributes to U2AF35 selectivity for guanine at the 3′ splice site, rather than adenine. In U2AF65, E425 resides within a noncanonical RNA recognition motif (UHM; U2AF homology motif) (Fig. 4D). In contrast to the first two RRMs of U2AF65, which bind directly to pre-mRNA, UHMs have been shown to bind tryptophan-containing linear peptide motifs (ULMs) (Kielkopf et al. 2004). The interaction between the UHM and ULM is known to enhance binding of U2AF65 to the PPT (Berglund et al. 1998; Rudner et al. 1998). These structural analyses suggest plausible molecular effects of PARP1-mediated site-specific PARylation on U2AF function, which we explore in more detail below.
Functional interplay among U2AF, ADPRylation, and RNA binding
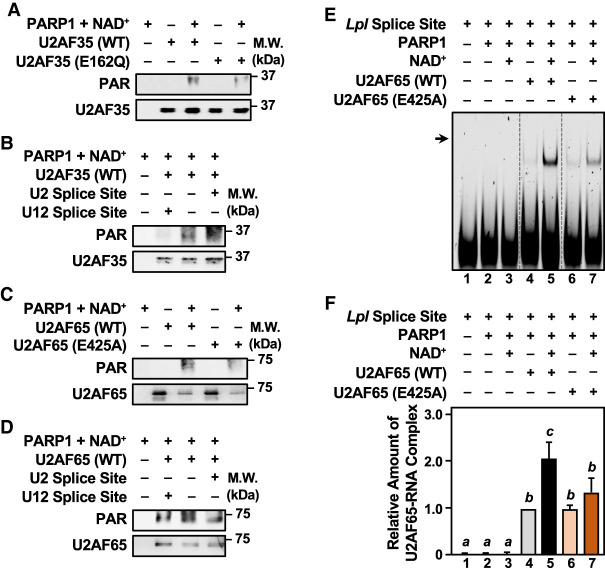
To elucidate how PARP1-mediated PARylation of U2AF might regulate RNA binding activity, we generated ADPRylation site mutants of U2AF35 and U2AF65 (E162Q and E425A, respectively). We expressed and purified these mutants (Supplemental Fig. S10) and then used them with recombinant wild-type PARP1 (Supplemental Fig. S9C) and NAD+ in PARylation and RNA recognition/binding assays with RNA containing either (1) a strong U2-type 3′ splice site acceptor sequence (U2 splice site), the preferred substrate for U2AF, or (2) a U12-type 3′ splice site (U12 splice site) that should not be recognized by U2AF. As expected, mutation of E162 reduced PARP1-mediated PARylation of U2AF35 (Fig. 5A; Supplemental Fig. S11A). These results were confirmed in mESCs (Supplemental Fig. S11B). Incubation of U2AF35 with its cognate RNA (U2 splice site), but not the U12 splice site RNA, enhanced PARP1-mediated PARylation of U2AF35 by about twofold versus no RNA (Fig. 5B; Supplemental Fig. S11C). In fact, PARP1-mediated PARylation of U2AF35 was inhibited by the U12 splice site RNA (Fig. 5B; Supplemental Fig. S11C). Thus, PARP1-mediated PARylation of U2AF35 is enhanced by binding to its specific RNA substrate.
Figure 5.
Interplay between splice site RNA binding by the U2AF complex and PARP1-mdiated ADP-ribosylation of U2AF35 and U2AF65. (A,B) In vitro PARylation assays with WT or ADPRylation site mutant (E162Q) U2AF35 in the presence of PARP1 and NAD+. Western blots showing the levels of PARylated U2AF35 and total U2AF35. Size markers in kilodaltons are shown. Representative images from n ≥ 4. (A) Assay with U2AF35 WT versus E162Q. (B) Assay with RNA oligonucleotides containing either a strong U2 splice site acceptor or a U12 class splice site. (C,D) In vitro PARylation assays with WT or ADPRylation site mutant (E425A) U2AF65 in the presence of PARP1 and NAD+. Western blots showing the levels of PARylated U2AF65 and total U2AF65. Size markers in kilodaltons are shown. Representative images from n ≥ 4. (C) Assay with U2AF65 WT versus E425A. (D) Assay with RNA oligonucleotides containing either a strong U2 splice site acceptor sequence or a U12 class splice site. (E,F) RNA electrophoretic mobility shift assays (EMSAs) with WT or ADPRylation site mutant (E425A) U2AF65 following in vitro PARylation by PARP1. Cy5-labeled Lpl RNA was used as a splice site in the binding reactions. The binding reactions were run on a native 6% polyacrylamide gel and then imaged directly using a ChemiDoc MP system. (E) Comparison of the ability of WT or ADPR site mutant (E425A) U2AF65 to bind to the Lpl RNA after being subjected to in vitro PARylation with PARP1 and NAD+ as indicated. The vertical dashed lines indicate lanes from the same experiment that were moved to maintain a consistent experimental order, without changing the exposure or intensity. (F) Quantification of Lpl RNA binding in multiple experiments like those shown in E (n = 5). Bars marked with different letters are significantly different (one-way ANOVA; Sidak).
Likewise, as expected, mutation of E425 reduced PARP1-mediated PARylation of U2AF65 (Fig. 5C; Supplemental Fig. S11D). These results were confirmed in mESCs (Supplemental Fig. S11E). In contrast to U2AF35, PARP1-mediated PARylation of U2AF65 was independent of interaction with its cognate RNA (U2 splice site) (Fig. 5D; Supplemental Fig. S11F). We considered, however, that PARylation of U2AF65 might affect its binding to RNA. We performed RNA electrophoretic mobility shift assays (EMSAs) using a Cy5-labeled strong U2-type 3′ splice site acceptor from the lipoprotein lipase (Lpl) gene shown previously to be bound by U2AF65 (Fu et al. 2011). Both wild-type (WT) and E425A U2AF65 bound to the Cy5-labeled Lpl splice site, but not to a Cy5-labeled U12-type 3′ splice site from Scn8a (Supplemental Fig. S11G). Interestingly, PARP1-mediated PARylation enhanced the binding of U2AF65 to the Cy5-labeled Lpl splice site (Fig. 5E,F; Supplemental Fig. S11H).
Together, these results demonstrate the functional interplay among U2AF, ADPRylation, and RNA binding. PARP1-mediated PARylation of U2AF35 is enhanced by binding to cognate RNA, whereas PARP1-mediated PARylation of U2AF65 enhances its binding to cognate RNA.
Splicing is dysregulated in P1KO mESCs genome-wide
RNA isoform switching occurs during mESC differentiation, with broad isoform diversity in the pluripotent state and increasing specificity of isoform usage as cells progress to a lineage-committed cell type (Trapnell et al. 2010). We found many components of the spliceosome in the mESC PARP1-mediated ADPRylated proteome (Supplemental Table S2). Thus, we asked whether loss of PARP1 would have an impact on alternative splicing events (ASEs) in mESCs during differentiation. The mRNA transcribed from the Ctnnd1 (catenin δ one) gene exhibits distinct isoform usage dependent on the pluripotent state of stem cells (Supplemental Fig. S12A; Salomonis et al. 2010). Using a semiquantitative PCR assay, we observed that genetic depletion of PARP1 alters the splicing pattern of Ctnnd1 during the differentiation of mESCs (Supplemental Fig. S12A,B).
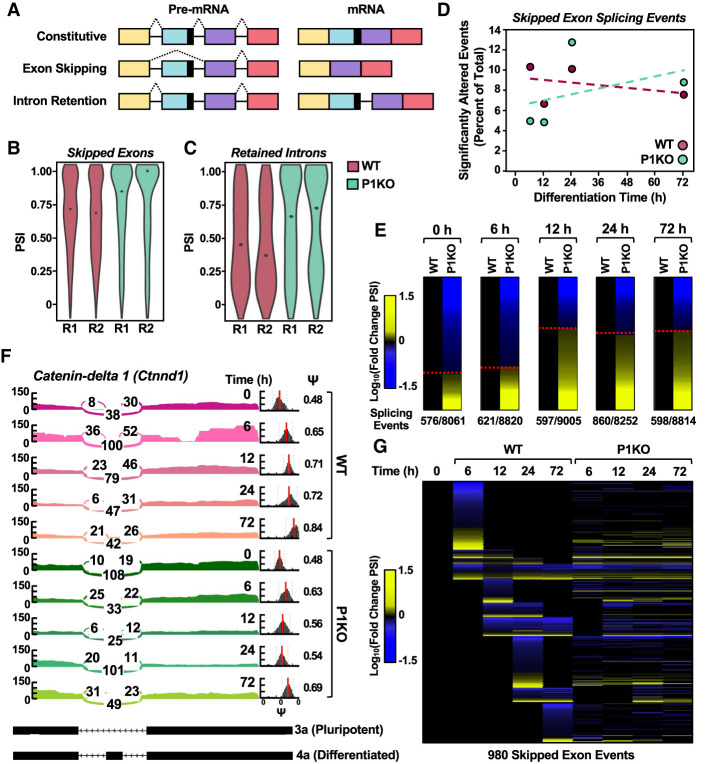
To investigate the role of PARP1 in splicing globally, we performed paired-end RNA sequencing on WT and P1KO mESCs during a time course of differentiation and analyzed the sequencing results using both MISO (primarily for visualization) and rMATs (primarily for quantitative analyses) software (Supplemental Tables S3, S4; Katz et al. 2010; Shen et al. 2014). In our analysis, we focused on skipped exon (SE) or retained intron (RI) events (Fig. 6A) because altered recognition of 3′ splice site intron–exon boundaries by U2AF during constitutive splicing would most likely affect these event classes (Reed and Maniatis 1986). In agreement with previous studies, genetic depletion of PARP1 promoted ASEs in mESCs at time points during differentiation when PARP1 has minimal catalytic activity, suggesting catalytic-independent functions of PARP1 in splicing (Matveeva et al. 2016; Matveeva et al. 2019). However, at 12 h of differentiation, we observed a marked increase in splicing inclusion (quantified as “percent spliced in” [PSI]) in the significantly altered SE and RI events in P1KO mESCs (Fig. 6B,C, respectively). Additionally, the distribution and frequency of ASEs among the splicing event classes were altered in the P1KO cells at 12 h of differentiation compared with the WT cells (Supplemental Fig. S12C).
Figure 6.
Genetic deletion of Parp1 alters splicing patterns in mESCs. (A) Diagram of splicing events thought to be most affected by disruption of U2AF complex function. The pre-mRNA and mature mRNA products resulting from the different types of splicing events are shown. (B,C) Violin plots tallying significantly altered splicing events (ASEs) between WT and P1KO mESCs at 12 h of differentiation from the skipped exon (597-event; B) and retained intron (148-event; C) splicing classes. Significantly altered splicing events were defined as a ΔPSI >0.1 and FDR =0.05. (D) Line plot representation showing significantly altered skipped exon splicing events as a percent of total during a time course of differentiation in WT and P1KO mESCs. The splicing events are expressed relative to the undifferentiated state (0 h) in either WT or P1KO mESCs. Percentages of significantly altered splicing events (ASEs) were calculated by dividing statistically significant events (ΔPSI >0.2 and Bayes factor >5) by the total number of splicing events for that event class and multiplying by 100. (E) Heat map representation showing the number of significantly altered skipped exon splicing events in mESCs over a time course of differentiation comparing WT and P1KO mESCs. Each row represents a single splicing event, and columns represent WT versus P1KO at each time point. Events that increased in P1KO cells relative to WT are indicated in yellow (log10FC), and decreased splicing events are indicated in blue. The ASEs are ordered by increasing exon incorporation from top to bottom. (F) Sashimi plots with accompanying PSI (ψ) histograms generated by MISO showing an example of an ASE in Ctnnd1 mediated by PARP1 and PARylation in mESCs over a time course of differentiation. The splicing pattern in WT mESCs is shown in pink and the pattern in P1KO mESCs is shown in green. RNA-seq reads contributing to the included or excluded exon species are represented numerically, as well as visually by line thickness. The red bar in the PSI histogram indicates the median value, with the spread of the histogram indicating the confidence interval. A shift of 0.2 in PSI value is considered significant. (G) Heat map representation showing the number of significantly altered skipped exon splicing events in mESCs over a time course of differentiation comparing WT and P1KO mESCs. Each row represents a single splicing event. Events that increased relative to the initial (0-h) time are indicated in yellow (log10FC), and decreased splicing events are indicated in blue. The ASEs are ordered by increasing exon incorporation from top to bottom in each cluster.
To understand how splicing events are differentially altered over time in WT and P1KO mESCs, we compared significant ASEs between the two cell types. Although we did not observe a significant change in the percentage of SE events in P1KO cells compared with WT cells during differentiation (Fig. 6D), we did observe a significant increase in the percentage of significantly altered RI splicing events during the same time course (Supplemental Fig. S12D). However, when we analyzed SE events at individual time points, hundreds of splicing events were included or excluded upon loss of PARP1, as denoted by changes in PSI (Fig. 6E). Strikingly, at 12 h of differentiation, when PARP1 first reaches maximal catalytic activity during differentiation, we observed a marked increase in the inclusion isoform event (Fig. 6E). Interestingly, this increase in inclusion isoform events was not seen in all ASE classes (Supplemental Fig. S12E). Using Ctnnd1 as an example, we observed over differentiation a dysregulation of splicing and a significant decrease in PSI in the P1KO cells versus the WT cells (Fig. 6F). Similar results were observed globally (Fig. 6G).
When examining the contribution of site-specific ADPRylation to the ASEs, we observed perturbation in selected inclusion and exclusion of events in the presence of the ADPR site mutant U2AF35 or U2AF65 compared with wild type (Supplemental Fig. S13). Specifically, for Ctnnd1, we observed elevated levels of the 3a isoform during differentiation in mESCs expressing the ADPRylation-deficient U2AF mutants (Supplemental Fig. S13A), similar to what is seen in P1KO mESCs (cf, Supplemental Fig. S12A,B). Likewise, for Irf9, we observed a modest but reproducible elevation in the levels of the included isoform relative to the excluded isoform in mESCs expressing the ADPRylation-deficient U2AF mutants (Supplemental Fig. S13B). In contrast, we did not observe similar effects with Rbm5 (Supplemental Fig. S13C), suggesting that ADPRylation of other factors may play a role. Together, these results clearly demonstrate a role for PARP1 in the maintenance of the splicing program in mESCs during differentiation.
Inhibition of PARP1 catalytic activity increases U2AF-mediated splicing in mESCs
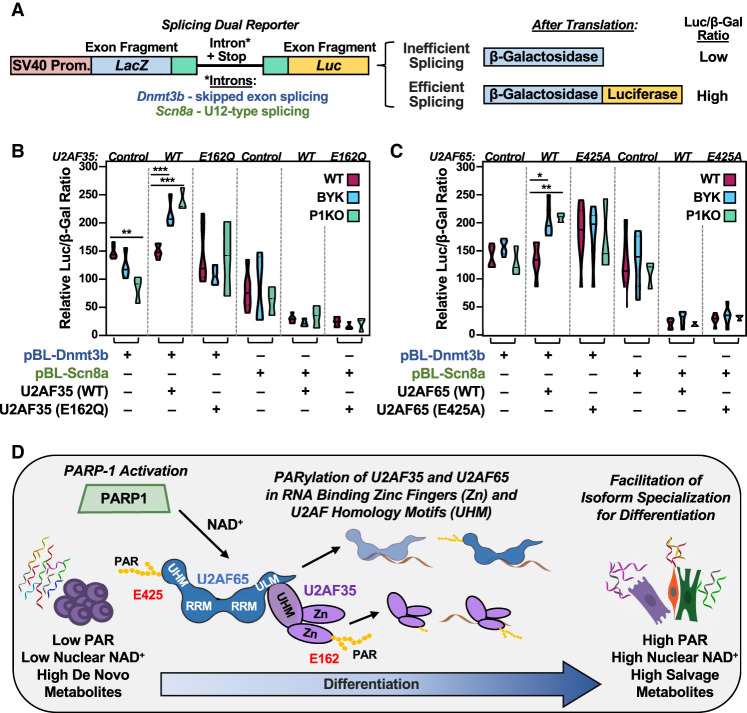
The results presented above point to a regulatory effect of PARP1-mediated PARylation on the functions of U2AF35 and U2AF65 in splicing. To further investigate the function of U2AF PARylation on splicing in mESCs, we used a cell-based dual-reporter splicing assay (Fig. 7A; Kollmus et al. 1996; Nasim and Eperon 2006). When proper in-frame splicing of the intron occurs, the reporter expresses both β-galactosidase and luciferase. If splicing does not occur, or is out of frame, then only β-galactosidase is expressed (Fig. 7A). Thus, efficient splicing can be shown as a ratio between β-galactosidase and luciferase expression/activity (Fig. 7A). Alternatively, splicing in this assay can be detected by PCR, and we found that these results mirrored those of the luminescence-based approach (Supplemental Fig. S14).
Figure 7.
Inhibition of PARP1 catalytic activity affects U2AF splicing functions during the differentiation of mESCs. (A) Diagram of a pBPLUGA in vivo dual-splicing reporter containing fragments of splice sites of the Dnmt3b (U2-type splicing) and Scn8a (U12-type splicing) genes. The reporter contains two reporter genes (LacZ encoding β-galactosidase and Luc encoding luciferase). Efficient splicing removes in-frame stop codons in the intron, allowing for expression of a LacZ + Luc fusion. (B,C) Relative luciferase/β-galactosidase activity ratios from assays in WT and P1KO mESCs using the Dnmt3b and Scn8a pBPLUGA constructs in the presence of WT or ADPRylation site mutant U2AF (E162Q U2AF35 and E425A U2AF65). The reporter and U2AF35 or U2AF65 expression plasmids were electroporated into mESCs. The mESCs were then differentiated for 12 h by LIF removal. In some cases, as indicated, the cells were treated with the PARP inhibitor BYK204165 at 10 µM for 2 h prior to collection. Summary of results from multiple experiments displayed as violin plots (n ≥ 3, statistical tests were performed using two-way ANOVA and multiple comparisons within each group of three conditions [WT, BYK, and P1KO]; Sidak). (∗) P < 0.05, (∗∗) P < 0.01, (∗∗∗) P < 0.001. (D) Model showing the regulation of embryonic stem cell state by the NAD+–PARP1 axis through site-specific PARylation of the U2AF complex proteins U2AF35 and U2AF65, resulting in altered splicing of mRNAs encoding key embryonic stem cell state proteins.
We inserted the exon 1–intron–exon 2 boundary from Dnmt3b between the two reporter genes; Dnmt3b mRNA exhibited a significantly altered SE splicing event in our RNA-seq data and has been shown by others to be differentially spliced in association with U2AF35 mutations (Ilagan et al. 2015). As a negative control, we inserted the exon 2–intron–exon 3 boundary from Scn8a between the two reporter genes; Scn8a mRNA contains a U12-type splice site. These two constructs, which are denoted as pBL-Dnmt3b and pBL-Scn8a, respectively, were electroporated into WT and P1KO mESCs with expression constructs for ectopic expression of wild-type or ADPRylation site mutants U2AF35 and U2AF65. After electroporation, the cells were differentiated by LIF removal and then briefly treated with BYK204165 (BYK), a PARP1-selective PARP inhibitor (PARPi).
In splicing assays with pBL-Dnmt3b, expression of wild-type U2AF35 showed no difference in splicing compared with vector alone. However, when WT or P1KO mESCs ectopically expressing wild-type U2AF35 were treated with BYK, they exhibited increased splicing of the reporter compared with untreated cells (Fig. 7B). These effects were abrogated with the U2AF35 E162Q ADPRylation site mutant, as well as the pBL-Scn8a reporter (Fig. 7B). Similar results were observed with U2AF65; that is, when WT or P1KO mESCs ectopically expressing the pBL-Dnmt3b reporter and wild-type U2AF65 were treated with BYK, they exhibited increased splicing of the reporter compared with untreated cells (Fig. 7C). Again, these effects were abrogated with the U2AF65 E425A ADPRylation site mutant, as well as the pBL-Scn8a reporter (Fig. 7C). Relatedly, we found that depletion of NMNAT-1, which reduces the nuclear NAD+ pool used by PARP1, produced results consistent with those of P1KO and BYK treatment (Supplemental Fig. S15).
Discussion
The differentiation of ESCs from a pluripotent state is tightly regulated. We have characterized a regulatory pathway that integrates the levels, subcellular location, and pathways of NAD+ biosynthesis with the activity of PARP1, the site-specific ADPRylation of U2AF, the downstream regulation of splicing, and specification of mRNA isoforms in differentiating mESCs (Fig. 7D). Specifically, our studies demonstrate a transition from elevated de novo metabolites to increased salvage pathway metabolites, leading to increased NAD+ levels in the nucleus that coincide with the catalytic activation of PARP1. The enhanced catalytic activity is directed toward the site-specific ADPRylation of RNA processing proteins and core components of the spliceosome complex, including U2AF35 and U2AF65. Moreover, PARylation of these splicing proteins regulates their molecular functions and contributions to global changes in mRNA isoform specification. Collectively, our metabolomic, proteomic, genomic, computational, and biochemical studies demonstrate the intricate interplay between nuclear NAD+ levels and PARP1 activity during differentiation, highlighting the importance of site-specific ADPRylation of splicing factors in the maintenance of a canonical splicing program in mESCs. These results point to the role and regulation of PARP1 catalytic activity in the control of embryonic stem cell state.
Nuclear PARP1 activity is driven by pathway-specific, compartmentalized NAD+ biosynthesis and mESC state
Historically, the major pathway for the biosynthesis of NAD+ within most cell types, excluding the liver, is the salvage pathway (Liu et al. 2018; Xie et al. 2020). Recent studies, however, have begun to challenge this assumption by characterizing the impact of cellular state on the usage of the different NAD+ biosynthetic pathways. For example, immune cells switch from the salvage pathway to the de novo pathway for inflammatory responses (Minhas et al. 2019; Zhang et al. 2019). Additionally, some cancer subtypes use the Preiss–Handler pathway to sustain their unique metabolic landscape (Piacente et al. 2017; Chowdhry et al. 2019). Our results quantifying the (1) steady-state intermediate metabolite levels during the course of ESC differentiation and (2) impact of depletion of key pathway-specific enzymes on cell state provide insights into the pathway usage that occurs with differentiation. However, additional investigation of pathway flux is needed to ascertain the cause and extent of metabolite dynamics. Taken together, the previous studies and our current work demonstrate the importance of cellular context in the regulation of NAD+ biosynthetic pathway selection.
In addition, we also examined the role of a major NAD+ consumer, PARP1, on NAD+ metabolism in mESCs. Other work found that loss of NMNAT-2 supports mESC differentiation, mimicking the physiological decrease we observed (Feng et al. 2016). These results, coupled with prior studies demonstrating the impact of key NAD+ biosynthetic enzymes (e.g., NMNAT-1) on PARP1 activity, highlight the importance of compartmentalized NAD+ pools (Ryu et al. 2018). However, the effect of PARP1 on the levels of NAD+ biosynthetic enzymes has not been fully explored. In this regard, we observed that loss of PARP1 and its catalytic activity alters the levels of NMNAT-1, the amount of NAD+ in different cellular compartments, and the production of NAD+ intermediate metabolites. Our results examining the role of PARP1 and PARylation in ESC biology fit well with the growing recognition of the functional relationships between PARP1 and the nuclear NAD+ pool. Interestingly, alterations in nuclear PAR and NAD+ levels during differentiation do not necessarily proceed in the same manner in all cell types (e.g., preadipocytes → high to low [Ryu et al. 2018] and mESCs → low to high [this study]), suggesting specific contributions to biological needs of that system.
PARP1-mediated, site-specific ADPRylation is required for the differentiation of mESCs
PARP1 is a multifaceted protein that functions both as a nucleic acid-binding protein and an enzyme mediating PARylation of substrates (Gibson and Kraus 2012; Gupte et al. 2017; Kim et al. 2020). Previous studies have demonstrated the importance of PARP1 in the regulation of ESC state or reprogramming by genetic deletion of Parp1 or through inhibition of its catalytic activity (Gao et al. 2009; Doege et al. 2012; Ji and Tulin 2012; Chiou et al. 2013; Weber et al. 2013; Roper et al. 2014; Jiang et al. 2015; Liu and Kraus 2017). Prior work has demonstrated a catalytic-independent function of PARP1 in mESC state regulation and found that PARP1 binds to intractable genomic loci, acting as a “pioneer factor” for pluripotency factor Sox2 (Liu and Kraus 2017). Others have observed the onset of PARP1 catalytic activity in mESC differentiation and demonstrated a role for PARP1 using PARP inhibitors (Ji and Tulin 2012), while others have demonstrated a role for PARP1 in reprogramming using RNA-mediated knockdown of Parp1 mRNA and pharmacological inhibition of PARP1 activity (Doege et al. 2012; Chiou et al. 2013; Weber et al. 2013; Jiang et al. 2015). In spite of these studies, which have focused primarily on the role of PARP1 in the regulation of chromatin and transcription, the mechanisms by which PARP1-mediated PARylation regulates the differentiation program in ESCs, including potential effects on RNA splicing, remain poorly characterized.
In this regard, our asPARP1 approach coupled with mass spectrometry identified ADPRylated proteins with functions in the maintenance of stem cell pluripotency, indicating that the substrates that we identified are biologically relevant to ESCs. In addition, the substrates that we identified were enriched in biological processes known to be regulated by PARP1, including transcription and DNA repair, as well as processes less well connected to PARP1 function, such as RNA processing and splicing. The latter, however, represents an emerging area of research. In this regard, Matveeva et al. (2016, 2019) have demonstrated a role for PARP1 in cotranscriptional alternate splicing, suggesting a need for a broader view of PARP1's cellular functions.
Site-specific ADPRylation of U2AF35 and U2AF65 by PARP1
mRNA isoform diversity is highest in undifferentiated ESCs and decreases with differentiation (Trapnell et al. 2010; Wu et al. 2010). Two unique spliceosomes that mediate splicing exist in the nucleus: the U2- dependent spliceosome, which catalyzes the removal of U2-type introns, and the less abundant U12-dependent spliceosome, which catalyzes U12-type intron splicing (Will and Luhrmann 2011). The recognition and recruitment of the U2-dependent spliceosome to GT and AG dinucleotides at the 5′ and 3′ intron boundaries by the U2AF complex is a defining feature of this class of intron removal (Sharp and Burge 1997; Shao et al. 2014). The activity of spliceosome proteins and related factors is regulated by expression, as well as post-translational modifications (Chen and Manley 2009).
We found that both components of U2AF—U2AF35 and U2AF65—are PARylated at sites within functional domains that facilitate their interaction with recognition of the 3′ splice site (i.e., the zinc finger in U2AF35, and the U2AF homology motif in U2AF65). Mutation of these sites reduced PARylation of U2AF35 and U2AF65 in vitro and in mESCs, as well as blocked the stimulation of specific splicing events by a PARP inhibitor in reporter assays. Interestingly, PARylation of U2AF35, but not U2AF65, increased upon interaction with its cognate splice site RNA. Our results suggest that U2AF35 recognition and binding of a 3′ splice site may occur before PARylation, while U2AF65 may be PARylated prior to interactions with RNA. Differences between U2AF35 and U2AF65 with respect to the impact of PARylation may be due to the distinct domains PARylated by PARP1, the different binding sites within RNA, or distinct roles of PARylation; cumulatively, we found that U2AF function is regulated by the PARylation status of its individual subunits.
Global patterns of splicing in mESCs are controlled by PARP1
A previous study demonstrated the capacity of PARP1 to regulate cotranscriptional splicing events in a catalytic-independent manner by chromatin occupancy at intron–exon boundaries and reducing RNA polymerase II progression (Matveeva et al. 2019). In this regard, in our studies, we observed substantial differences in splicing inclusion events in PARP1 knockout mESCs versus wild-type cells, even at time points when PARP1 was shown to be catalytically inactive. Taken together, these results point to a role for PARP1 in splicing independent of its catalytic activity. These same studies, however, also demonstrated that alternative splicing events are affected by inhibition of PARP1 catalytic activity (Matveeva et al. 2016, 2019), demonstrating distinct roles of PARP1 protein and its catalytic activity in splicing regulation. ADPRylation has been implicated in splicing regulation in other studies as well. RNA splicing proteins hnRNP A1 and A2 interact with PAR chains, which alters their functions (Ji and Tulin 2009). These effects, however, were not in the context of covalent attachment of PAR to the hnRNP proteins, and the PAR chains were not attributed to a specific PARP family member (Ji and Tulin 2009).
By examining mESCs during a time course of differentiation and cataloging splicing trends globally, we were able to identify alterations in splicing that occurred after enhancement of PARP1 catalytic activity. Using this approach, we observed a shift from splicing exclusion to splicing inclusion at 12 h of differentiation, the same time point of catalytic activation. Our genome-wide approach investigating the relationship between PARP1 and splicing regulation complement existing knowledge to highlight the role of PARP1 in regulating splicing activity. Further studies using PARP1 DNA/RNA binding or catalytic activity-deficient mutants in a Parp1-null background will be useful to further elucidate the catalytic-dependent and catalytic-independent roles of PARP1 in splicing in mESCs.
Materials and methods
Additional details on the Materials and Methods are in the Supplemental Material.
Antibodies
A detailed list of the antibodies used is in the Supplemental Material.
Cell culture
Parp1+/+ (wild-type [WT]) and Parp1−/− (knockout [P1KO]) mESCs (Yang et al. 2004; Gao et al. 2009) were maintained on a feeder layer of CF6Neo mouse embryonic fibroblasts in Dulbecco's modified Eagle medium containing 15% (v/v) fetal bovine serum and other additives as described in the Supplemental Material. Feeder cells were depleted for experimental assays by two passages on 0.2% gelatin-coated plates. mESCs were differentiated in ultralow-attachment plates (Thermo Fisher 05-539-101) coupled with LIF removal, unless noted. HEK-293T cells were cultured in DMEM containing 10% fetal bovine serum. For both cell lines, fresh cell stocks were regularly replenished from the original stocks, verified for cell type identity using the GenePrint 24 system (Promega) or by unique genetic identifiers (e.g., Parp1−/−), and confirmed as mycoplasma-free every 3 mo using a commercial testing kit.
Cell treatments
mESCs were differentiated by LIF removal and by growth in ultralow-attachment plates. For treatment with 10 µM BYK204165 (Santa Cruz Biotechnology), the cells were exposed for 2 h prior to the end of the differentiation time point before collection.
Molecular cloning and expression plasmid generation
cpVenus-based NAD+ sensor constructs
Expression vectors for cpVenus-based nuclear and cytoplasmic NAD+ sensors and their corresponding cpVenus-only controls were kindly provided by Dr. Michael Cohen and Dr. Richard Goodman, Vollum Institute, Oregon Health and Science University (Cambronne et al. 2016). DNA coding for the sensors or controls was amplified using the primers listed in the Supplemental Material and then cloned into the pCDH-EF1α expression vector.
Splicing factor expression vectors
cDNAs coding for N-terminally FLAG-tagged U2AF35 or U2AF65 were amplified from cDNA-containing vectors using primers listed in the Supplemental Material. The amplified products were then cloned into pCDH-EF1α and pFASTBAC. Similar vectors containing cDNAs for ADPRylation site mutants of U2AF35 (E162Q) and U2AF65 (E425A) were generated by site-directed mutagenesis with primers listed in the Supplemental Material.
β-Galactosidase and luciferase dual-reporter splicing assay
DNA covering the relevant intron/exon boundaries of Dnmt3b and Scn8a were amplified from genomic DNA with primers listed in the Supplemental Material. The amplified products were then cloned into pBPLUGA (Kollmus et al. 1996).
Generation of mESCs with ectopic expression of proteins
Stable ectopic protein expression in WT mESCs
mESCs were infected with lentiviruses generated from the pCDH vectors described and were treated with 250 µg/mL neomycin to select cells expressing wild type and E162Q U2AF35 or 1 μg/mL puromycin to select cells expressing wild type and E425A U2AF65.
Transient ectopic protein expression in WT and PARP1 KO mESCs
For fluorescent NAD+ determinations, mESCs were transfected with the cpVenus NAD+ sensors or corresponding control plasmids. All experiments were performed 48 h after transfection. For splicing assays, mESCs were electroporated with the pBPLUGA-Dntm3b or Scn8a vectors alone or with the pCDH-EF1α-U2AF35(WT)/U2AF65(WT) or U2AF35(E162Q)/U2AF65(E425A) vectors. All experiments were performed within 72 h of electroporation.
Preparation of cell extracts and Western blotting
Whole-cell extracts were prepared from mESCs in lysis buffer (details are in the Supplemental Material). For Western blotting, volumes of cell extract containing equivalent amounts of total protein, or individual in vitro PARylation reactions (see below), were run on polyacrylamide–SDS gels, transferred to a nylon-backed nitrocellulose membrane, and blotted using a variety of antibodies as noted. Western blot signals were detected using an ECL detection reagent (Thermo Fisher) and a ChemiDoc imaging system (Bio-Rad Laboratories).
Determination of nuclear and cytoplasmic NAD+ levels using cpVenus-based sensors
mESCs expressing nuclear or cytoplasmic NAD+ sensors or their corresponding cpVenus-only controls (Cambronne et al. 2016) were used to determine changes in subcellular NAD+ levels during differentiation as described previously (Ryu et al. 2018). The cells were differentiated solely by LIF removal as indicated before imaging. The ratios of fluorescence intensities were determined at 488 nm to 405 nm of the sensors and the cpVenus controls for three independent biological replicates for each condition. Average ratiometric values for the undifferentiated cpVenus transfected mESCs were set to 1, and the rest of the data were normalized accordingly.
Quantification of NAD+-related metabolites using mass spectrometry
Metabolite-containing supernatants were prepared from mESCs and stored at −80°C until analyzed by mass spectrometry. For targeted metabolomic analysis, the metabolite-containing supernatants were analyzed using a SCIEX QTRAP 5500 liquid chromatograph/triple quadrupole mass spectrometer. Most metabolites were analyzed using a Nexera ultrahigh-performance liquid chromatograph system (Shimadzu Corporation) with a SeQuant ZIC-pHILIC HPLC column at 35°C. For nicotinate, we used a Phenomenex Synergi Polar-RP HPLC column at 35°C. The mass spectrometer was used with an electrospray ionization (ESI) source in multiple reaction monitoring (MRM) mode. The peak values assigned to each metabolite in each sample per replicate were normalized to the cell count for that replicate. The values from the 6-h and 12-h time points were averaged together to reduce the noise from these temporally related collections, which exhibited the greatest biological variability. Statistical significance was calculated using a two-way ANOVA/Tukey's multiple comparison test.
Expression and purification of recombinant proteins
We used Sf9 insect cells to express and purify PARP1, asPARP1, and wild-type and mutant U2AF35 and U2AF65 as described previously (Gibson et al. 2016, 2017). The eluted proteins were aliquoted and flash-frozen in liquid nitrogen. Quality assessment and determination of the concentrations of the purified proteins were performed by SDS-PAGE analysis versus BSA protein standards followed by staining with Coomassie Brilliant Blue.
Identification of PARP1 substrates using an NAD+ analog-sensitive PARP1 (asPARP1) approach
We used an NAD+ analog-sensitive PARP1 (asPARP) approach coupled with protein mass spectrometry as described previously (Gibson et al. 2016, 2017) to identify protein substrates of PARP1 catalytic activity, as well as the specific amino acid residues modified by PARP1 in those substrates. First, we verified the system using in vitro enzyme assays and in-gel fluorescence assays. Then, we used the asPARP1 approach with cell extract from WT mESCs at 12 h of differentiation, followed by mass spectrometry to identify the ADPRylated proteins (trypsin digestion elution) and the specific sites of modification (hydroxylamine elution) as described previously (Gibson et al. 2016, 2017). Peptides identified from samples prepared with wild-type PARP1 (wtPARP1) were treated as nonspecific background.
Analysis of U2AF35 structure
The crystal structure of the yeast homolog of U2AF1 (U2AF35) bound to 3′ splice site RNA was downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) using PDB ID 7c06.6 (Yoshida et al. 2020). The predicted structure of mouse U2AF35was downloaded from AlphaFold Protein Structure Database using ID AF-Q9D883-F1 (Jumper et al. 2021). Both structures were uploaded, overlaid, positioned, and colorized using PyMOL.
RNA isolation and PCR-based gene expression and splicing assays
Total RNA was isolated from mESCs using an RNeasy Plus mini kit (Qiagen). Total RNA was reverse-transcribed using oligo(dT) primers or random hexamers (for downstream use in semiquantitative PCR) plus MMLV reverse transcriptase (Promega) to generate cDNA pools. For reverse transcription quantitative PCR (RT-qPCR), the cDNA pools were subjected to qPCR using the gene-specific primers listed in the Supplemental Material as described previously (Liu and Kraus 2017). For reverse transcription semiquantitative PCR (RT-sqPCR), the cDNA samples were subjected to PCR using the isoform-specific primers listed in the Supplemental Material as described previously (Harvey et al. 2021).
RNA sequencing (RNA-seq)
Total RNA was isolated from mESCs and then enriched for polyA+ RNA using Dynabeads oligo(dT)25 (Invitrogen). The polyA+ RNA was then used to generate paired-end strand-specific RNA-seq libraries as described previously (Zhong et al. 2011). The RNA-seq libraries were subjected to QC analyses (i.e., number of PCR cycles required to amplify each library, the final library yield, and the size distribution of final library DNA fragments) and sequenced using an Illumina HiSeq 2000. The raw data were subjected to QC analyses using the FastQC tool (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were trimmed to remove adapter sequences and shorter reads and then mapped to mouse genome mm10 using the spliced reader aligner TopHat version 2.1.1 (Kim et al. 2013). The aligned reads were sorted and indexed using SamTools (Li et al. 2009) to format properly for downstream analysis.
Differential alternative splicing events were detected by integrating both replicates with rMATS version 4.1.1 (Shen et al. 2014). Additionally, alternative splicing events and isoform usage were detected and filtered using MISO version 0.5.3 (Katz et al. 2010) with each replicate handled separately. Detected events were visualized using the Sashimi program (Katz et al. 2010). Heat maps visualizing changes in PSI value were generated using Morpehus (https://software.broadinstitute.org/morpheus/).
Gene ontology (GO) analyses
Gene ontology analyses were performed using DAVID (Database for Annotation, Visualization, and Integrated Discovery) (Huang et al. 2009a,b). Inputs for the GO analyses included the results from the proteomic studies filtered for high-confidence proteomic hits.
In vitro ADPRylation assays
In vitro PARylation assays were performed as described previously (Zhang et al. 2012; Lin et al. 2018). The reactions contained 0.1 μM purified PARP1, NAD+, a purified protein substrate, and polypyrimidine tract RNA. U2AF35 and U2AF65 proteins were purified as described above and were included at a concentration of 1 µM and 0.5 µM, respectively, per reaction. Sheared salmon sperm DNA (100 ng/μL) was used as an allosteric activator of PARP1 enzymatic activity. The reaction products were then resolved on an SDS-PAGE gel, transferred to a nylon-backed nitrocellulose membrane, and blotted with an anti-PAR detection reagent (EMD Millipore).
RNA electrophoretic mobility assays with U2AF65
Interactions between U2AF65 and various splice site RNAs were analyzed by RNA electrophoretic mobility assays (EMSAs). Cy5-labeled RNAs containing splice sites from the mouse Lpl (Fu et al. 2011) or Scn8a (Sharp and Burge 1997) genes were used in binding reactions as described previously (Fu et al. 2011). The reactions were run on native 6% polyacrylamide gels in 1× TBE. The gel was then imaged directly using a ChemiDoc MP system (Bio-Rad).
Immunoprecipitation and interaction assays
Immunoprecipitation from mESCs
Whole-cell extracts were prepared from mESCs expressing FLAG-tagged wild-type or mutant (E162Q) U2AF35, FLAG-tagged wild-type or mutant (E425A) U2AF65, or parental mESCs. The extracts were incubated with anti-FLAG M2 agarose resin, and the immunoprecipitated FLAG-tagged U2AF proteins were recovered from the beads by either eluting with 0.5 mg/mL 3xFLAG peptide or by heating for 5 min at 100°C in 2× SDS-PAGE loading buffer. The immunoprecipitated material was subjected to Western blotting as described above.
Cell-based dual-reporter splicing assays
mESCs were electroporated with the pBPLUGA constructs containing the exon–intron–exon boundary of Dnmt3b or Scn8a alone or in combination with pCDH-U2AF35(WT), pCDH-U2AF35(E162Q), pCDH-U2AF65(WT), or pCDH-U2AF65(E425A). After electroporation, the cells were seeded on gelatin-coated 12-well plates and grown overnight. Differentiation was performed by LIF removal for 12 h. The dual-reporter splicing assays were performed as reported previously (Nasim and Eperon 2006). The luciferase luminescence intensities were quantified immediately, followed by β-galactosidase luminescence intensities, using a plate reader (CLARIOstar BMG Labtech) for both.
Data availability
The metabolomics mass spectrometry data can be accessed from MassIVE (https://massive.ucsd.edu) using accession number MSV000088612. Processed data identifying specific NAD+ metabolites and precursors are in Supplemental Table S1. The asPARP1/ADPRylated proteome mass spectrometry data can be accessed from MassIVE (https://massive.ucsd.edu) using accession number MSV000088580. Processed data identifying ADPRylated peptides and specific sites of ADPRylation are in Supplemental Table S2. The RNA-seq data sets generated for this study can be accessed from the NCBI's Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo) using accession number GSE192380. Processed data quantifying specific splicing events are in Supplemental Tables S3 and S4.
Supplementary Material
Acknowledgments
We acknowledge and thank Ziying Liu for her initial work on PARP1 in mESCs in the Kraus laboratory; Tim Hou, Sridevi Challa, Ralph DeBerardinis, Laura Banaszynski, Nicholas Conrad, and MiKayla Stokes for providing critical feedback on this work and the manuscript; Anusha Nagari and Tulip Nandu for assistance with the analysis of the sequencing data; Aishwarya Sundaresan for assistance with analysis of the mass spectrometry data; Sridevi Challa for help with microscopy and luciferase assays; and Luka Zrnic for help with PCR assays. We also thank Zhao-Qi Wang for the WT and P1KO mESCs, Richard Goodman and Michael Cohen for the cpVenus-based NAD+ sensors, and John McCarthy for the dual-reporter pBPLUGA vector. We also acknowledge and thank the University of Texas Southwestern Medical Center Live-Cell Imaging Core (Katherine Luby-Phelps), Next-Generation Sequencing Core (Vanessa Schmid), CRI Metabolomics Facility (Ralph DeBerardinis and Feng Cai), and Proteomics Core (Andrew Lemoff). This work was supported by a grant from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (NIH/NIDDK; R01 DK058110) and funds from the Cecil H. and Ida Green Center for Reproductive Biology Sciences Endowment to W.L.K., and by a National Science Foundation Graduate Research fellowship to A.J.
Author contributions: A.J. and W.L.K conceived this project, and A.J. developed the ideas further in consultation with W.L.K. A.J. and W.L.K. designed the experiments and oversaw their execution. A.J. performed all of the wet laboratory experiments and computational analyses. A.J. prepared the initial drafts of the figures and text, which were edited and finalized by W.L.K. W.L.K. secured funding to support this project and provided intellectual support for all aspects of the work.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349335.121.
Competing interest statement
W.L.K. is a founder for Ribon Therapeutics, Inc.; a founder, consultant, and Science Advisory Board member for ARase Therapeutics, Inc.; and a consultant and Science Advisory Board member for Alphina Therapeutics, Inc. He is also a holder of U.S. patent number 9,599,606, covering the ADP-ribose detection reagent used here, which has been licensed to and is sold by EMD Millipore.
References
- Berglund JA, Abovich N, Rosbash M. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev 12: 858–867. 10.1101/gad.12.6.858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne XA, Kraus WL. 2020. Location, location, location: compartmentalization of NAD+ synthesis and functions in mammalian cells. Trends Biochem Sci 45: 858–873. 10.1016/j.tibs.2020.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne XA, Stewart ML, Kim D, Jones-Brunette AM, Morgan RK, Farrens DL, Cohen MS, Goodman RH. 2016. Biosensor reveals multiple sources for mitochondrial NAD+. Science 352: 1474–1477. 10.1126/science.aad5168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. 2015. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518: 413–416. 10.1038/nature13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa S, Khulpateea BR, Nandu T, Camacho CV, Ryu KW, Chen H, Peng Y, Lea JS, Kraus WL. 2021. Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers. Cell 184: 4531–4546.e26. 10.1016/j.cell.2021.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754. 10.1038/nrm2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou SH, Jiang BH, Yu YL, Chou SJ, Tsai PH, Chang WC, Chen LK, Chen LH, Chien Y, Chiou GY. 2013. Poly(ADP-ribose) polymerase 1 regulates nuclear reprogramming and promotes iPSC generation without c-Myc. J Exp Med 210: 85–98. 10.1084/jem.20121044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhry S, Zanca C, Rajkumar U, Koga T, Diao Y, Raviram R, Liu F, Turner K, Yang H, Brunk E, et al. 2019. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 569: 570–575. 10.1038/s41586-019-1150-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti L, Janeckova L, Wagner D, Mazzola F, Cialabrini L, Di Stefano M, Orsomando G, Magni G, Bendotti C, Smyth N, et al. 2011. Reducing expression of NAD+ synthesizing enzyme NMNAT1 does not affect the rate of Wallerian degeneration. FEBS J 278: 2666–2679. 10.1111/j.1742-4658.2011.08193.x [DOI] [PubMed] [Google Scholar]
- Dahan P, Lu V, Nguyen RMT, Kennedy SAL, Teitell MA. 2019. Metabolism in pluripotency: both driver and passenger? J Biol Chem 294: 5420–5429. 10.1074/jbc.TM117.000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CM, Ong SE, Leung AK. 2015. The promise of proteomics for the study of ADP-ribosylation. Mol Cell 58: 911–924. 10.1016/j.molcel.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. 2012. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488: 652–655. 10.1038/nature11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Tong M, Xia B, Luo GZ, Wang M, Xie D, Wan H, Zhang Y, Zhou Q, Wang XJ. 2016. Ubiquitously expressed genes participate in cell-specific functions via alternative promoter usage. EMBO Rep 17: 1304–1313. 10.15252/embr.201541476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Dzeja PP, Nelson TJ, Terzic A. 2012. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11: 596–606. 10.1016/j.stem.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato C, Mazzola F, Raffaelli N. 2021. The key role of the NAD biosynthetic enzyme nicotinamide mononucleotide adenylyltransferase in regulating cell functions. IUBMB Life 10.1002/iub.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Masuda A, Ito M, Shinmi J, Ohno K. 2011. AG-dependent 3′-splice sites are predisposed to aberrant splicing due to a mutation at the first nucleotide of an exon. Nucleic Acids Res 39: 4396–4404. 10.1093/nar/gkr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Kwon SW, Zhao Y, Jin Y. 2009. PARP1 poly(ADP-ribosyl)ates Sox2 to control Sox2 protein levels and FGF4 expression during embryonic stem cell differentiation. J Biol Chem 284: 22263–22273. 10.1074/jbc.M109.033118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411–424. 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. 2017. Identification of protein substrates of specific PARP enzymes using analog-sensitive PARP mutants and a ‘clickable’ NAD+ analog. Methods Mol Biol 1608: 111–135. 10.1007/978-1-4939-6993-7_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, Schwede F, Yu Y, Kraus WL. 2016. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353: 45–50. 10.1126/science.aaf7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Conrad LB, Huang D, Kraus WL. 2017. Generation and characterization of recombinant antibody-like ADP-ribose binding proteins. Biochemistry 56: 6305–6316. 10.1021/acs.biochem.7b00670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostner JM, Geisler S, Stonig M, Mair L, Sperner-Unterweger B, Fuchs D. 2020. Tryptophan metabolism and related pathways in psychoneuroimmunology: the impact of nutrition and lifestyle. Neuropsychobiology 79: 89–99. 10.1159/000496293 [DOI] [PubMed] [Google Scholar]
- Gupte R, Liu Z, Kraus WL. 2017. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 31: 101–126. 10.1101/gad.291518.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SE, Lyu J, Cheng C. 2021. Methods for characterization of alternative RNA splicing. Methods Mol Biol 2372: 209–222. 10.1007/978-1-0716-1697-0_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Nozaki T, Winterhager E, Yamamoto H, Nakagama H, Kamada N, Suzuki H, Ohta T, Ohki M, Masutani M, et al. 2003. Parp1-deficiency induces differentiation of ES cells into trophoblast derivatives. Dev Biol 257: 371–381. 10.1016/S0012-1606(03)00097-6 [DOI] [PubMed] [Google Scholar]
- Hendriks IA, Larsen SC, Nielsen ML. 2019. An advanced strategy for comprehensive profiling of ADP-ribosylation sites using mass spectrometry-based proteomics. Mol Cell Proteomics 18: 1010–1026. 10.1074/mcp.TIR119.001315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks AN, Lorenzetti D, Gilley J, Lu B, Andersson KE, Miligan C, Overbeek PA, Oppenheim R, Bishop CE. 2012. Nicotinamide mononucleotide adenylyltransferase 2 (Nmnat2) regulates axon integrity in the mouse embryo. PLoS One 7: e47869. 10.1371/journal.pone.0047869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. 2010. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31: 194–223. 10.1210/er.2009-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, Bradley RK. 2015. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res 25: 14–26. 10.1101/gr.181016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. 2009. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res 37: 3501–3513. 10.1093/nar/gkp218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. 2012. Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte localization. Nat Commun 3: 760. 10.1038/ncomms1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BH, Chen WY, Li HY, Chien Y, Chang WC, Hsieh PC, Wu P, Chen CY, Song HY, Chien CS, et al. 2015. CHD1L regulated PARP1-driven pluripotency and chromatin remodeling during the early-stage cell reprogramming. Stem Cells 33: 2961–2972. 10.1002/stem.2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Lücke S, Green MR. 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev 18: 1513–1526. 10.1101/gad.1206204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Challa S, Jones A, Kraus WL. 2020. PARPs and ADP-ribosylation in RNA biology: from RNA expression and processing to protein translation and proteostasis. Genes Dev 34: 302–320. 10.1101/gad.334433.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus H, Flohe L, McCarthy JE. 1996. Analysis of eukaryotic mRNA structures directing cotranslational incorporation of selenocysteine. Nucleic Acids Res 24: 1195–1201. 10.1093/nar/24.7.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KY, Huang D, Kraus WL. 2018. Generating protein-linked and protein-free mono-, oligo-, and poly(ADP-ribose) in vitro. Methods Mol Biol 1813: 91–108. 10.1007/978-1-4939-8588-3_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Kraus WL. 2017. Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol Cell 65: 589–603.e9. 10.1016/j.molcel.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Su X, Quinn WJ, Hui S, Krukenberg K, Frederick DW, Redpath P, Zhan L, Chellappa K, White E, et al. 2018. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab 27: 1067–1080.e5. 10.1016/j.cmet.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva E, Maiorano J, Zhang Q, Eteleeb AM, Convertini P, Chen J, Infantino V, Stamm S, Wang J, Rouchka EC, et al. 2016. Involvement of PARP1 in the regulation of alternative splicing. Cell Discov 2: 15046. 10.1038/celldisc.2015.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, Al-Tinawi QMH, Rouchka EC, Fondufe-Mittendorf YN. 2019. Coupling of PARP1-mediated chromatin structural changes to transcriptional RNA polymerase II elongation and cotranscriptional splicing. Epigenetics Chromatin 12: 15. 10.1186/s13072-019-0261-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minhas PS, Liu L, Moon PK, Joshi AU, Dove C, Mhatre S, Contrepois K, Wang Q, Lee BA, Coronado M, et al. 2019. Macrophage de novo NAD+ synthesis specifies immune function in aging and inflammation. Nat Immunol 20: 50–63. 10.1038/s41590-018-0255-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim MT, Eperon IC. 2006. A double-reporter splicing assay for determining splicing efficiency in mammalian cells. Nat Protoc 1: 1022–1028. 10.1038/nprot.2006.148 [DOI] [PubMed] [Google Scholar]
- Piacente F, Caffa I, Ravera S, Sociali G, Passalacqua M, Vellone VG, Becherini P, Reverberi D, Monacelli F, Ballestrero A, et al. 2017. Nicotinic acid phosphoribosyltransferase regulates cancer cell metabolism, susceptibility to NAMPT inhibitors, and DNA repair. Cancer Res 77: 3857–3869. 10.1158/0008-5472.CAN-16-3079 [DOI] [PubMed] [Google Scholar]
- Reed R, Maniatis T. 1986. A role for exon sequences and splice-site proximity in splice-site selection. Cell 46: 681–690. 10.1016/0092-8674(86)90343-0 [DOI] [PubMed] [Google Scholar]
- Roper SJ, Chrysanthou S, Senner CE, Sienerth A, Gnan S, Murray A, Masutani M, Latos P, Hemberger M. 2014. ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells. Nucleic Acids Res 42: 8914–8927. 10.1093/nar/gku591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC. 1998. Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol Cell Biol 18: 1765–1773. 10.1128/MCB.18.4.1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, Kraus WL. 2018. Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360: eaan5780. 10.1126/science.aan5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, et al. 2010. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci 107: 10514–10519. 10.1073/pnas.0912260107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, Li H, et al. 2014. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol 21: 997–1005. 10.1038/nsmb.2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA, Burge CB. 1997. Classification of introns: U2-type or U12-type. Cell 91: 875–879. 10.1016/S0092-8674(00)80479-1 [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci 111: E5593–E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, et al. 2015. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol 17: 1523–1535. 10.1038/ncb3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber FA, Bartolomei G, Hottiger MO, Cinelli P. 2013. Artd1/Parp1 regulates reprogramming by transcriptional regulation of Fgf4 via Sox2 ADP-ribosylation. Stem Cells 31: 2364–2373. 10.1002/stem.1507 [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wogulis M, Chew ER, Donohoue PD, Wilson DK. 2008. Identification of formyl kynurenine formamidase and kynurenine aminotransferase from Saccharomyces cerevisiae using crystallographic, bioinformatic and biochemical evidence. Biochemistry 47: 1608–1621. 10.1021/bi701172v [DOI] [PubMed] [Google Scholar]
- Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, et al. 2010. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci 107: 5254–5259. 10.1073/pnas.0914114107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie N, Zhang L, Gao W, Huang C, Huber PE, Zhou X, Li C, Shen G, Zou B. 2020. NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential. Signal Transduct Target Ther 5: 227. 10.1038/s41392-020-00311-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. 2004. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23: 3872–3882. 10.1038/sj.onc.1207491 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Park SY, Sakashita G, Nariai Y, Kuwasako K, Muto Y, Urano T, Obayashi E. 2020. Elucidation of the aberrant 3′ splice site selection by cancer-associated mutations on the U2AF1. Nat Commun 11: 4744. 10.1038/s41467-020-18559-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. 2011. Control of the embryonic stem cell state. Cell 144: 940–954. 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Yao J, DuMond ME, Krishnakumar R, Ruhl DD, Ryu KW, Gamble MJ, Kraus WL. 2012. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J Biol Chem 287: 12405–12416. 10.1074/jbc.M111.304469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tao J, Ling Y, Li F, Zhu X, Xu L, Wang M, Zhang S, McCall CE, Liu TF. 2019. Switch of NAD salvage to de novo biosynthesis sustains SIRT1-RelB-dependent inflammatory tolerance. Front Immunol 10: 2358. 10.3389/fimmu.2019.02358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. 2011. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: 940–949. 10.1101/pdb.prot5652 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolomics mass spectrometry data can be accessed from MassIVE (https://massive.ucsd.edu) using accession number MSV000088612. Processed data identifying specific NAD+ metabolites and precursors are in Supplemental Table S1. The asPARP1/ADPRylated proteome mass spectrometry data can be accessed from MassIVE (https://massive.ucsd.edu) using accession number MSV000088580. Processed data identifying ADPRylated peptides and specific sites of ADPRylation are in Supplemental Table S2. The RNA-seq data sets generated for this study can be accessed from the NCBI's Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo) using accession number GSE192380. Processed data quantifying specific splicing events are in Supplemental Tables S3 and S4.