Abstract
Purpose
Risk of advanced age-related macular degeneration (AAMD) is associated with rare genetic variants in the gene encoding complement factor I (CFI), which is associated with lower circulating CFI protein levels, but the nature of the relationship is unclear. Can genetic factors be used to infer whether low circulating CFI is associated with AAMD risk?
Design
Two-sample inverse variance-weighted Mendelian randomization (MR) was used to evaluate evidence for a relationship between CFI levels and AAMD risk, comparing CFI levels from genetically predefined subsets in AAMD and control cohorts.
Participants
We derived genetic instruments for systemic CFI level in 3301 healthy INTERVAL study participants. To evaluate a genetic causal odds ratio (OR) for the effect of CFI levels on AAMD risk, results from an AAMD genome-wide association study from the International AMD Genomics Consortium, were combined with CFI levels from SCOPE and SIGHT AAMD patients.
Methods
Published genetic and proteomic data was combined with data from cohorts of patients with geographic atrophy (GA) in a series of MR analyses.
Main Outcome Measures
Establishing a causal relationship for CFI on AAMD.
Results
One common CFI variant, rs7439493, was associated strongly with low CFI level, explaining 4.8% of phenotypic variance. Using rs7439493, MR estimates for AAMD odds increased per standard deviation (SD) CFI decrease in were 1.47 (95% confidence interval [CI], 1.30–1.65; P = 2.1 × 10–10). MR using rare variant (rs141853578 encoding p.Gly119Arg) indicated 1-SD decrease in CFI led to increased AAMD OR of 1.79 (95% CI, 1.46–2.19; P = 1.9 × 10–8). The rare variant rs141853578 explained a further 1.7% of phenotypic variance. To benchmark the effect of low CFI levels on AAMD ORs using a CFI-specific proteomic assay, we estimated the effect using CFI levels from 24 rs141853578 positive GA patients; each 1-SD reduction (3.5 μg/mL) in CFI associated with a 1.67-fold increased odds of AAMD (95% CI, 1.40–2.00; P = 1.85 × 10–8).
Conclusions
Concordance in MR calculations provide good genetic evidence for a potentially causal role of lower CFI level increasing AAMD risk.
Keywords: Advanced age-related macular degeneration, Biomarkers, CFI, Factor I, Genetics
Abbreviations and Acronyms: AMD, age-related macular degeneration; AAMD, advanced age-related macular degeneration; AP, alternative pathway; ARMS2, age-related maculopathy susceptibility 2; BP, base pair; CFB, complement factor B; CFH, complement factor H; CFI, complement factor I; CI, confidence interval; CS, complement system; C3, complement component 3; GA, geographic atrophy; GWAS, genome-wide association study; HTRA1, high-temperature requirement A serine peptidase 1; IAMDGC, International AMD Genomics Consortium; IRB, institutional review board; LD, linkage disequilibrium; MAF, minor allele frequency; MNV, macular neovascularization; MR, Mendelian randomization; OR, odds ratio; PQTL, protein quantitative trait loci; SD, standard deviation; SNP, single nucleotide polymorphism
Age-related macular degeneration (AMD) is the leading cause of irreversible central vision loss among elderly Western populations.1, 2, 3 Age-related macular degeneration is a progressive retinal disease in which the early stages are characterized by drusen and pigmentary changes, causing mild visual impairment. Many patients progress to advanced AMD (AAMD), which has 2 subtypes: exudative AMD, involving angiogenesis in the choroid and macular neovascularization (MNV), and nonexudative AMD, involving degeneration or geographic atrophy (GA) of the retinal pigment epithelium.4 The prevalence of AAMD in European individuals 65 to 69 years of age is approximately 0.5%, rising to 9.8% in those 85 years of age or older.5
Antiangiogenic agents are effective at controlling MNV6; however, treated patients still have residual visual disability because of varying levels of retinal tissue disruption and atrophy, and monthly intravitreal injections are invasive and costly, with increased risk of intraocular infection.7 Antioxidant and mineral supplementation can reduce the progression of nonexudative AMD to exudative AMD8; however, to date, no effective treatment exists for nonexudative AMD.9
The natural history of AMD has been studied extensively,4,10 and despite much effort invested in the identification of different biomarkers to identify high-risk individuals to guide screening, monitoring, and treatment options and outcomes, these remain confined to research settings and have not yet moved into clinical practice.11 Advances in retinal imaging techniques have improved diagnosis and guide disease management, and efforts continue to refine disease characterization and selection of anatomic features that may be used as end points in future clinical trials to improve chances of success.12
Understanding the cause of nonexudative AMD has been limited partly because of lack of relevant animal disease models.13 Age-related macular degeneration development is influenced by advancing age, lifestyle factors like smoking and a high body mass index, and a positive family history of the disease.14,15 Compared with other complex traits, AMD has a strong genetic influence: heritability is estimated to be approximately 46% for disease development and for 71% disease severity.16 Study of human genetic variation can help to improve our biological understanding of causal pathways for disease, and this in turn has the potential to translate into improved clinical diagnostics and drug target selection.17 For AMD, several hypotheses for disease development and progression have focused on the underlying pathogenic pathways related to genetic predisposition.18,19
Genome-wide association studies (GWASs) have identified common genetic variants that increase risk of AAMD, with the two strongest risk factors mapping to the region surrounding complement factor H (CFH) on chromosome 1q32, and the age-related maculopathy susceptibility 2 (ARMS2)/ high-temperature requirement A serine peptidase 1 (HTRA1) region on chromosome 10q26. CFH codes for the CFH protein, implicating the complement system (CS) in AAMD disease pathogenesis.20,21 Less is understood about how the association at ARMS2/HTRA1 contributes functionally to disease,22,23 but carriers of this risk factor are reported to have more severe disease and a MNV-like phenotype compared with carriers of CFH risk alleles.24,25 Other AAMD GWAS risk factors also mapped to genes coding for proteins in the alternative pathway (AP) of the CS; these include the CFI, complement factor B (CFB), and complement component (C3) genes.19,26 Additionally, AAMD GWASs also identified genetic risk factors mapping to genes in other biological pathways, such as the high-density lipoprotein cholesterol, collagen synthesis, receptor-mediated endocytosis, extracellular matrix organization and assembly, and angiogenesis pathways.19
Detailed analyses of underlying biological pathways identified by GWAS can help to identify important regulators or modifiers that may be targeted therapeutically. The CS is a promising target pathway for intervention indicated by genetic evidence, immunohistochemistry, and protein biomarker studies. The prolonged overactivity of the CS is a main driver of AMD development and progression,27,28 and the link to disease is supported by the observations of a number of CS proteins, activators, and regulatory proteins being identified as molecular constituents of drusen, the hallmark extracellular deposits associated with AMD that are found alongside retinal pigment epithelium cells at the site of disease.29, 30, 31 The CS is a critical feature of the innate immune response, comprising complex pathways of multiple cascading proteins with tightly controlled enzymatic functions, and it is this complexity that makes it challenging to determine the optimal position to intervene.32
Intravitreal injection of different inhibitors against various complement proteins has been trialed in AMD with limited success33, 34, 35; for example, lampalizumab (anti–complement factor D; Genentech, Inc),33,36 LFG316 (anti–C5 antibody; Novartis AG), eculizumab (anti–C5 antibody; Alexion Pharmaceuticals, Inc), and CLG561 (antiproperdin; Novartis AG). A phase 2/3 trial of an intravitreal pegylated RNA aptamer that inhibits complement factor C5 (avacincaptad pegol; Iveric Bio) did meet its primary end point of slowing GA lesion area growth35 and is undergoing further evaluation. Another strategy overcoming the requirement for regular intravitreal injections is using adeno-associated virus gene therapy to deliver the human complement factor I (CFI) gene to the retinal pigment epithelium to drive expression of endogenous CFI protein to slow atrophic disease.37 The rationale for ocular CFI supplementation is to enable normalization of complement control at the level of the retina and to restore homeostasis to slow the disease process and retinal degeneration. However, it remains unclear what the most appropriate target patient population is and what role CFI plays as a biomarker in clinical development of new therapies. Compared with the common CFI AAMD-associated risk variant identified by GWAS, individuals carrying rare genetic variants in CFI are at an even greater risk of developing AAMD.19,21,26,38, 39, 40, 41 Rare CFI variants have been described in familial AMD, and sporadic patients with AMD carrying rare CFI variants are more likely to report a positive family history and a younger age at symptom onset.42,43 The CFI protein is produced systemically by the liver44 and locally in the eye by the retinal pigment epithelium tissues.45 Complement factor I is a critical inhibitor of the AP and is a key regulator of all 3 complement activating pathways including the AP, by irreversibly cleaving C3b or C4b and halting further complement activation.46 Complement factor I-mediated cleavage of C3b represents a critical step in regulation of the AP, which is under a finely tuned positive feedback loop controlling complement deposition.47,48
Normal variation in systemic CFI protein level is influenced by age,49 immunologic processes,50 and genetic background. Approximately 4% to 7% of patients with AAMD carry a rare genetic variant in CFI,51,52 and a reported 36% have low CFI protein levels in the blood serum.51 Low CFI protein level is considered the functional consequence of some underlying rare CFI variant genotype, which ultimately fails to produce a secreted CFI protein.40,51,53, 54, 55
Levels of intraocular CFI are correlated positively with systemic CFI levels in healthy individuals and those with AMD.56 Lower ocular CFI level is hypothesized to contribute to uncontrolled C3b accumulation, resulting in an imbalance in the AP, which over time leads to deposition of complement at the site of disease, driving macular degeneration.27,28,57
Mendelian randomization (MR) is a statistical approach that can be applied to investigate the causal relationships between risk factors and outcomes via the use of genetic instruments (in this context, genetic variants58,59). Mendelian randomization uses genetic association data with the risk factor (systemic CFI protein levels) and genetic association data with disease outcome (risk of development of AAMD) to assess whether the risk factor is likely to be associated causally with disease. Because genetic instruments are distributed randomly at conception, the genetically predicted CFI levels are unlikely to be related to confounders of AMD risks or to be influenced consequentially by AMD disease status through reverse causality.58
In this study, we investigated whether common and rare genetic factors play a part in driving variation in circulating CFI. These data then were used to infer the causal relationship between CFI on AAMD risk; showing a causal relationship would validate CFI further as an important prognostic biomarker and would have therapeutic implications for disease prevention and clinical development of treatments for AAMD.
Methods
Overview of Methods
We conducted a series of 2-sample MR analyses to evaluate the association between genetically predicted circulating CFI level and AAMD risk using genetic instruments identified in 3,301 healthy European participants in the INTERVAL study60 considering both directly genotyped common variants and a selected set of previously reported rare variants that were assessed via genotype imputation (Supplemental Fig 1). To evaluate a genetic causal odds ratio (OR) for the effect of CFI levels on AAMD risk, we used inverse variance-weighted MR analysis, using results from an AAMD meta-GWAS.19 To benchmark the effect of low CFI levels on AAMD odds, we used CFI-specific proteomic measurements from patients with GA carrying a rare CFI variant, rs141853578, who have taken part in two natural history studies, SCOPE (ClinicalTrials.gov identifier, NCT03894020) and SIGHT.61
Genomic and Proteomic Biomarker Datasets
We used genetic and proteomics data from Sun et al60 that links germline genotypes to plasma protein levels, allowing determination of particular genetic markers acting in an allele-specific manner. After quality control, data were available for use in protein quantitative trait loci analysis from 3283 proteins, including CFI.
We also used blood serum CFI and genetic data from 24 pseudoanonymized patients with GA who harbored the rs141853578 variant, of whom 14 were recruited into SCOPE, and a further 10 recruited from the SIGHT study.61 Serum CFI levels were compared with 329 CFI rare variant-negative patients with GA from SIGHT. Geographic atrophy was determined in both studies using fundus autofluorescence, and patients were entered based on the presence of unilateral or bilateral GA and a reading performance of ≥ 40 letters by best-corrected visual acuity, but not MNV or diabetic retinopathy, as determined by a retinal specialist.
For SCOPE patients, saliva DNA was screened for CFI rare variants using targeted next-generation sequencing conducted by Molecular Vision Laboratory. A customized capture panel was designed using Agilent SureSelect Target Enrichment kit to amplify the CFI coding region. Paired-end reads were sequenced using Illumina Miseq version 2 platform, using acceptance thresholds of more than 30 times coverage over more than 98% of the target region. Sequencing reads were aligned to the human reference genome (National Center for Biotechnology Information build GRCh37 version 3) using NextGENe software version 2.4.2.3, and genotypes were called using the Genetics Assistant version 1.4.7 program. SIGHT patient blood DNA was screened for CFI variants using Sanger sequencing, described elsewhere.61
SCOPE and SIGHT patient serum samples were collected according to standard protocols and were stored at –80 °C. Serum CFI protein concentrations were measured using a validated sandwich enzyme-linked immunosorbent assay (Hycult Biotech), by Eurofins BioPharma Service. Serum CFI levels were compared with those generated in 125 normal control participants (BioIVT, UK).
Ethics Statement
For the SCOPE study, written consent was obtained from all participants in accordance with the tenets of the Declaration of Helsinki after explanation of the nature and implications of the study, and all methods were carried out in accordance with the relevant guidelines and regulations of local or country-specific ethics committees. For SCOPE, institutional review board (IRB)/ ethics committe/ regulatory authority approval was obtained on a local or country-specific basis; United Kingdom (Medicines and Healthcare products Regulatory Agency, London; North of Scotland Research Ethics Committee), United States of America (Oregon Health & Science University, Portland; The Johns Hopkins Medicine IRB, Baltimore; Wills Eye Hospital IRB, Philadelphia; Columbia University IRB, New York; Advarra), Australia (Therapeutic Goods Administration, Woden; Bellberry Human Research Ethics Committee, Eastwood), Poland (Komisja Bioetyczna przy Bydgoskiej Izbie Lekarskej; Komisja Bioetyczna przy Okregowej Izbie Lekarskiej w Lodzi; Komisja Bioetyczna Slaskiej Izby Lekarskiej w Katowicach; Office for Registration of Medicinal Products, Medical Devices and Biocidal Products, Warszawa), Germany (Ethikkommission der Medizinischen Fakultät der Eberhard Karls Universität und am Universitätsklinikum Tübingen), France (Comité de protection des Personnes Sud Méditerrannée I - Marseille), The Netherlands (CMO Regio Arnhem-Nijmegen UMC St. Radboud), and Spain (CEIC-Hospital Clinico San Carlos, Madrid), SIGHT study IRB/ethics approval, registration and regulation were described previously.61
Advanced Age-Related Macular Degeneration Data
Age-related macular degeneration GWAS data were used to characterize the relationship between genetic markers and AAMD risk. The dataset comprised 12 711 patients with AAMD and 14 590 control participants of European descent from the International AMD Genomics Consortium (IAMDGC).19 Data were handled as described previously,62 and the resultant GWAS summary statistics were taken forward for analysis.
Statistical Analysis
For the proteomics dataset, Sun et al60 provided protein quantitative trait loci GWAS summary statistics. Methods of the protein GWAS were described previously. Briefly, the residuals of protein abundance levels from linear regression were rank-inverse normalized, meaning the effect sizes for single nucleotide polymorphisms (SNPs) represent per standard deviation (SD) change in protein abundances.
To allow assessment of genetic polymorphisms not presented in the original article, we performed genome-wide imputation on individual-level genotyping array data. Although the original manuscript presented some imputed SNPs, many rare SNPs of interest were filtered out (SNPs with minor allele count of < 8, minor allele frequency [MAF] of approximately 0.1%) and are not presented. In particular, we were interested in several rare variants recently reported in the literature (rs141853578, rs1017242313, rs752163277, and rs587779635).51,54 We imputed these variants in the Michigan Imputation Server based on approximately 600 K high-quality SNPs with MAF of more than 0.01 that overlapped with approximately 800 K UK Biobank array SNPs.63 The association between the imputed CFI rare variants and CFI level was assessed in a linear regression model in R software (R Foundation for Statistical Computing, Vienna, Austria).
The linkage disequilibrium (LD) score regression method was used to estimate the genetic correlation between the biomarker traits and AAMD using GWAS summary statistics (sample size and genotyping limitations precluded this being undertaken in SIGHT and SCOPE data). An MR approach was used to investigate the potential causal relationship between biomarker traits and AAMD. In MR analysis, we obtained genetic instruments for biomarker traits from the above biomarker GWAS summary statistics. We focused in the first instance on individual genetic markers with large effects on the trait (this contrasts with the situation with LD score regression, where a large number of genetic markers of typically small effect are used). For our primary analysis, we selected lead independent genome-wide significant variants (significance set at 2-tailed P < 5 × 10−8 and LD between single nucleotide variants r2 < 0.001).
The inverse-variance weighted regression method was used as our primary analysis. The MR analysis was conducted in R packages MendelianRandomization and TwoSampleMR.64 Because in some scenarios relatively few genetic markers are available that exceed the genome-wide significance threshold in a GWAS, we conducted a secondary analysis considering more genetic markers. We used a less strict P value threshold to include more SNPs, with independent SNPs attaining P < 5 × 10–6 included in the model. Using this larger set of SNPs, we generated results that should be more reliable in the face of deviations from the MR assumptions using alternative MR estimators. These alternative estimators were the weighted median and MR-Egger regression methods. Analyses were performed with R software version 3.4.1.
To examine the relationship between rare variation and complement protein levels, we used linear regression models. The results from the linear regressions were then taken forward for use in causal inference.64 The variance explained in CFI levels by genetic variants was computed using the formula 2 × f × (1 – f) × β2, where f is the allele frequency of the variant and β is the estimated effect on CFI levels (in SD units).
The LD was determined using LDlink, using data from European populations.65 Variants were annotated with non-Finnish European MAF from GnomAD version 2.1.1,66 function was annotated with using HaploReg version 4.1,67 clinical variant was annotated with ClinVar,68 and prior trait associations were annotated using Open Targets Genetics Portal.69
Results
Genetic Determinants of Normal Variation in Systemic Complement Factor I Level
The systemic CFI GWAS focused on common variants and identified one genome-wide significant SNP, rs7439493, located near the CFI gene loci (Supplementary Fig 2). Rs7439493 explained 4.8% of the variance in circulating CFI levels, indicating that it confers a relatively strong cis effect on CFI levels.
Investigating the Genetic Correlation between Complement Factor I and Advanced Age-Related Macular Degeneration Risk
The LD score regression first was used to explore the genome-wide genetic correlation between the CFI level and risk of different AMD subtypes (Supplemental Table 1); however, this failed to detect any significant correlations. This indicates that beyond the genomic region around CFI (where a strong association exists between CFI variants and AMD), no clear evidence exists that genes distant from the target gene are important determinants of CFI level.
Association Between Complement Factor I and the Risk of Advanced Age-Related Macular Degeneration from Mendelian Randomization Analysis
The lead CFI SNP rs7439493 was taken forward as an instrument variant for MR analysis using the single SNP Wald ratio method. The results show that CFI level was associated negatively with risk of AAMD (Table 1), where a 1-SD decrease in CFI level leads to an OR increase of 1.47 (47% increased risk) of AAMD (95% confidence interval [CI] 1.30–1.65; P = 2.1 × 10–10).
Table 1.
Common Genetic Instrument Variable for Circulating Complement Factor I level
| Single-Nucleotide Polymorphism | Chromosome | Base Pair Position | Effect Allele | Noneffect Allele | β | P Value | Age-Related Macular Degeneration Type | Odds Ratio (95% Confidence Interval) | P Value |
|---|---|---|---|---|---|---|---|---|---|
| rs7439493 | 4 | 110656730 | A | G | 0.31 | 8.9 × 10–37 | Advanced | 1.47 (1.30–1.65) | 2.1 × 10–10 |
| Intermediate | 1.24 (1.07–1.44) | 0.004 | |||||||
| GA | 1.54 (1.26–1.88) | 2.1 × 10–5 | |||||||
| MNV | 1.44 (1.26–1.64) | 7.4 × 10–8 |
GA = geographic atrophy; MNV = macular neovascularization.
The effect estimate was most pronounced in the advanced age-related macular degeneration subgroups compared with the intermediate age-related macular degeneration subgroup. Mendelian randomization estimates of odds ratios and 95% confidence intervals are based on a 1-standard deviation decrease in complement factor I protein levels based on the Wald ratio method.
To test the sensitivity of this MR estimate to our modelling assumptions, we applied alternative MR methods using a less strict P value threshold for SNP selection (P < 5 × 10–6). We applied MR Egger, weighted mean, and inverse variance-weighted methods, and their estimates broadly were consistent in effect size and direction to the estimate from the single SNP method (Fig 1). The MR-Egger intercepts showed no evidence of directional pleiotropy effects (intercepts were approximately 0; P > 0.05). All three MR methods showed increased risk of AAMD from lower levels of CFI, showing that our results are robust with respect to the SNP instrument used.
Figure 1.
Forest plot showing the Mendelian randomization (MR) estimates of the causal effects of circulating complement factor I (CFI) level on advanced age-related macular degeneration (AMD). The x-axis is the estimated odds ratio (OR) for AMD subtypes per 1-standard deviation increase in genetically predicted CFI level for the common rs7439493 variant evaluated. The vertical dashed line is the reference at OR = 1. The y-axis lists the different AMD subtypes. Different MR methods are displayed with different line types: solid line, 1 single nucleotide polymorphism (SNP) MR inverse variance-weighted (IVW) analysis; dashed line, MR IVW analysis; dotted line, MR-Egger analysis; larger dashed line, weighted mean. The line on either side of the point estimates represent the 95% confidence interval (CI). CNV = choroidal neovascularization; GA = geographic atrophy.
We then evaluated the relationship between systemic CFI levels and different AMD subtypes. The effect estimate was consistent between GA and MNV (OR, 1.54 [95% CI, 1.26–1.88; P = 2.1 × 10–5] and 1.44 [95% CI, 1.26–1.64; P = 7.8 × 10–8], respectively), but was weaker in intermediate AMD (OR, 1.24; 95% CI, 1.07–1.44; P = 0.004). These findings suggest that CFI may be involved comparably in progression to all forms of AAMD.
Rare Variant Analysis for Systemic Complement Factor I Level on Advanced Age-Related Macular Degeneration Risk
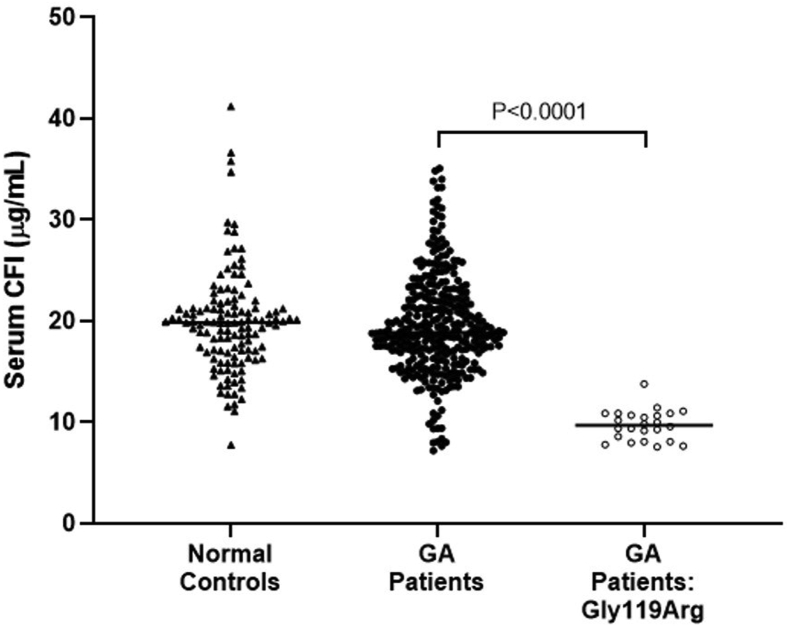
We also conducted an analysis based on a rare CFI variant, rs141853578, encoding p.Gly119Arg linked to AAMD risk.19,42 We used serum CFI protein levels from 24 individuals with GA who carried a heterozygous p.Gly119Arg variant. The CFI levels were compared with those measured in 125 normal control participants. The mean CFI level in p.Gly119Arg-positive patients with GA was 9.735 μg/ml (SD, 1.457 μg/ml) compared with the mean CFI level of 19.140 μg/ml (SD, 3.5 μg/ml) observed in normal control participants (Fig 2). The mean difference (9.404 μg/ml) then was used to compute an estimate of the causal effect of serum CFI protein on AAMD risk. The rare variant MR estimate revealed that a 1-SD (3.5 μg/ml) reduction in CFI protein level led to a 1.67 OR (67%) increased risk of AAMD (95% CI, 1.40–2.00; P = 1.85 × 10–8; Table 2).
Figure 2.
Scatterplot showing serum complement factor I (CFI) levels measured in normal control participants (n = 125), patients with geographic atrophy (GA; n = 329), and patients with GA carrying a CFI p.Gly119Arg variant (n = 24). Serum CFI data are shown as individual results, and the line represents the median. A Mann–Whitney U test was used for statistical analysis.
Table 2.
Rare Genetic Instrument Variable for Circulating Complement Factor I Level Based on SCOPE and SIGHT Proteomic Data
| Single Nucleotide Polymorphism | Chromosome | Base Pair Position | Effect Allele | Noneffect Allele | β | P Value | Age-Related Macular Degeneration Type | Odds Ratio (95% Confidence Interval) | P Value |
|---|---|---|---|---|---|---|---|---|---|
| rs141853578 | 4 | 110685820 | T | C | –9.41 | 1.5 × 10–25 | Advanced | 1.67 (1.40–2.00) | 1.85 × 10–8 |
| Intermediate | 1.47 (1.19–1.82) | 3.80 × 10–4 | |||||||
| GA | 1.98 (1.59–2.46) | 7.04 × 10–10 | |||||||
| MNV | 1.53 (1.25–1.88) | 3.73 × 10–5 |
GA = geographic atrophy; MNV = macular neovascularization.
The effect estimate was most pronounced in the GA subgroup compared with the MNV subgroup and intermediate age-related macular degeneration subgroup. Mendelian randomization estimates of odds ratios and 95% confidence intervals are based on 1-standard deviation (3.5 μg/ml) decrease in complement factor I protein levels based on the Wald ratio method.
To verify this finding, we used the imputed genetic data for p.Gly119Arg in the Sun et al60 dataset, achieving a good imputation quality score (0.8). The frequency of the minor allele T was 0.15%, and the effect of the rare variant on CFI levels was statistically significant (P = 5.58 × 10–14, β = 2.37, implying that this SNP explains 1.7% of the variance in CFI levels in this population). Mendelian randomization analysis then was conducted using the imputed p.Gly119Arg variant and variant association data from the IAMDGC GWAS. The MR estimate for the causal effect of CFI on AAMD risk for a 1-SD decrease in CFI protein level was calculated to confer an increased OR of 1.79 (95% CI, 1.46–2.19; P = 5.58 × 10–14; Table 3). The effect estimate was similar in direction and magnitude to our previous findings using direct measurement of CFI levels in p.Gly119Arg variant carriers and control participants, building confidence in the accuracy of our prediction.
Table 3.
Rare Genetic Instrument Variable for Circulating Complement Factor I Level Based on Sun et al60 Proteomic Data
| Single Nucleotide Polymorphism | Chromosome | Base Pair Position | Ancestral Allele | Effect Allele | Imputation Quality Score | β | P Value | Age-Related Macular Degeneration Type | Odds Ratio (95% Confidence Interval) | P Value |
|---|---|---|---|---|---|---|---|---|---|---|
| rs141853578 | 4 | 110685820 | T | C | 0.80 | –2.37 | 5.58 × 10–14 | Advanced | 1.79 (1.46–2.19) | 1.85 × 10–8 |
| Intermediate | 1.55 (1.22–1.97) | 3.80 × 10–4 | ||||||||
| GA | 2.17 (1.69–2.77) | 7.04 × 10–10 | ||||||||
| MNV | 1.62 (1.29–2.04) | 3.73 × 10–5 |
GA = geographic atrophy; MNV = macular neovascularization.
The effect estimate was most pronounced in the advanced age-related macular degeneration and GA subgroup compared with the MNV and intermediate age-related macular degeneration subgroups. Mendelian randomization estimates of odds ratios and 95% confidence intervals are based on 1-standard deviation (3.5 μg/ml) decrease in complement factor I protein levels based on the Wald ratio method.
Discussion
In this study, we conducted comprehensive MR analysis using common and rare variation at CFI to demonstrate a causal relationship between lower (genetically predicted) CFI levels and increased risk of AAMD and different AMD subtypes. We separately estimated the causal effect of low serum CFI level on AAMD risk by selecting first a common CFI variant and second the rare p.Gly119Arg CFI variant to use as genetic instruments in a 2-sample MR analysis. The concordance in direction and effect size from both calculations provide good genetic evidence for a potentially causal role that lower CFI level increases AAMD risk. The MR estimates from the common and rare variant calculations showed that a 1-SD decrease in CFI level led to increased odds of AAMD developing of between 47% and 67%, respectively. Using the SD determined in serum CFI level from normal control participants, we estimate that an 18.3% (3.5 μg/ml) reduction in CFI levels from the mean causes an approximate 50% increased odds of AAMD (Supplemental Fig 3).
Our study confirmed that a proportion of normal variation in CFI level is explained by genetic factors located at the CFI gene. The variant rs7439493 is common in the European population (MAF, 41.3%) and accounted for 4.8% of normal variation in CFI levels, representing a substantial contribution from a single region. In contrast, the rare p.Gly119Arg variant (annotated as a variant with “conflicting interpretations of pathogenicity”) conferred a greater magnitude of effect on CFI levels, but given the rarer allele frequency (MAF, 0.08%), this resulted in a smaller overall contribution to normal phenotypic CFI variance of 1.7%. When present on an AMD background, p.Gly119Arg accounted for 3.3% of variance in CFI level.
The p.Gly119Arg variant results in a protein secretion failure leading to lower CFI and increased complement activation in vitro.26 The functional mechanism explaining why the common rs7439493 variant is linked to CFI levels is not known. Rs7439493 is in LD with the lead AAMD risk factor previously identified at CFI, rs10033900 (r2 = 0.38),19,38 suggesting that both variants tag the same functional effect on AAMD risk and variation in macular thickness,70 but have no impact on risk of other diseases.69 Gene expression studies in healthy systemic tissues link rs7439493 to expression levels of CFI.71,72 A correlation was not detectable in retinal tissues, but this is likely because of sample size limitations and lack of testing individuals carrying rare CFI variants.51,73,74
Systemic CFI levels have been shown to be associated with intraocular CFI levels in both normal eyes as well as in eyes with AMD.56 Because ocular CFI level studies involve small sample sizes that are not sufficiently powered to detect genetic associations, and given that intraocular CFI levels are correlated strongly with systemic CFI levels, we used systemic CFI levels in this study as a surrogate for local CFI levels in the eye. Evidence from genetics, immunohistochemistry, and biomarker studies confirm a critical role for complement dysregulation and low CFI at the site of disease being a main driver for macular degeneration.27,28,57 Our study suggested that levels of the master regulator of the alternative pathway, CFI, is under relatively tight local genetic control. A one-time administration approach using adeno-associated virus to enable cellular transduction and to induce sustained expression of CFI after subretinal injection currently is being tested in patients with GA in phase 1 and 2 clinical trials (ClinicalTrials.gov identifiers, NCT03846193, NCT04437368, and NCT04566445).
Recent phase 2 studies suggesting intraocular inhibition of complement factor C3 cleavage using an intravitreal cyclic peptide-bound polyethylene glycol polymer (pegcetacoplan; Apellis Pharmaceuticals, Waltham, MA) may slow GA growth significantly,34 adding further weight to controlling complement dysregulation at the site of disease being a viable approach.37 This is despite a lack of success reported in earlier phase 3 trials targeting complement factor D using an intravitreal monoclonal antibody approach (lampalizumab; Genentech, Inc., San Francisco, CA).36 After detection of a positive signal using retrospective subgroup analysis using a CFI genetic variant on the phase 2 data,33 the lampalizumab phase 3 trial studies used the same CFI genetic variant as a biomarker to select a patient subpopulation for investigation, not the same core clinical trial design as the phase 2 study, which may be one reason for trial failure.
Our study adds support that CFI is an optimal target for therapeutic intervention in the CS pathway, that enhancing ocular CFI activity may be beneficial for treating AAMD,37 and that serum CFI level is a useful biomarker for understanding ocular CFI level. Moreover, CFI levels are driven by genetic determinants, and genetic stratification can be used to identify patients at a higher risk of developing AAMD, as well as being used as a surrogate marker for predicting CFI levels.
Study Limitations
Our approach used studies conducted in different populations of undefined ethnicity and combined proteomic datasets generated using different CFI assays. Our findings warrant replication in independent AAMD cohorts, correlating CFI levels with genetic and environmental factors.
Conclusions
Genetically predicted lower CFI levels were associated with increased risk of all AMD subtypes. The causal estimates derived using the rare and common CFI variants were strongly concordant, which provides confidence in a potential causal role for low levels on development of AMD. This furthers our understanding of the underlying pathologic mechanism of AMD, where CFI levels can be used to identify high-risk individuals.
Acknowledgments
The authors thank all the participants in the SCOPE and SIGHT studies, the IAMDGC, and the INTERVAL study.
Manuscript no. XOPS-D-21-00118
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s): A.V.J.: Employee and Equity owner – Gyroscope Therapeutics Ltd.
S.M.: Advisory board and Equity owner – StratifEYE, Ltd.
J.F.: Employee – Gyroscope Therapeutics Ltd.
C.H.: Employee – Gyroscope Therapeutics Ltd; Consultant – Q32 Bio Inc, Chinook Therapeutics, Biocryst Pharmaceuticals; Financial support – Ra Pharmaceuticals; Royalties – Hycult Biotech; Equity owner – Gyroscope Therapeutics Ltd.
D.K.: Consultant – Gyroscope Therapeutics Ltd, Alexion Pharmaceuticals, Novartis, Apellis Pharmaceuticals, Silence Therapeutics, Sarepta Therapeutics; Lecturer – Alexion Pharmaceuticals, Novartis; Patents – Gyroscope Therapeutics Ltd; Data Safety Monitoring Board or Advisory Board – Sarepta Therapeutics; Equity owner – Gyroscope Therapeutics Ltd.
A.L.: Consultant, Financial support, Equity owner – Gyroscope Therapeutics Ltd.
N.W.: Employee – Gyroscope Therapeutics Ltd.; Consultant – Apellis, Nidek, Boehringer Ingelheim; Financial support – Carl Zeiss Meditec, Topcon, Regeneron, Heidelberg, Nidek, Optovue; Equity owner – Ocudyne
Gyroscope Therapeutics Ltd., London, United Kingdom, provided funding for the design and conduct of the SCOPE study; approved the design, conduct, analysis, and interpretation of the data; preparation of the manuscript; and made the decision to submit the manuscript for publication. Supported by the Australian National Health and Medical Research Fellowship (grant nos.: 1150144 and 1116360 [S.M.]); the University of Queensland Research Training Scholarship (X. H.); and the QIMR Berghofer PhD Top Up Scholarship (X. H.).
Drs Jones, Han, and MacGregor had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. For the AMD datasets, all contributing sites and additional funding information are acknowledged in this publication: Fritsche et al. Nat Genet. 2016;48(134–143). The International AMD Genomics consortium’s web page is: http://eaglep.case.edu/iamdgc_web/, and additional information is available at: http://csg.sph.umich.edu/abecasis/public/amd2015/. Contributory IAMDGC research studies were funded in Australia by the National Health and Medical Research Council (1023911, 974159, 211069, 457349, 512423, 302010, 571013, 358702, 632909), the Ophthalmic Research Institute of Australia, the BrightFocus Foundation, a Ramaciotti Establishment Grant and the Victorian Government. In the US, contributory IAMDGC research studies were funded by grants or contracts awarded by the National Institute of Health or National Eye Institute (R01 EY09859, EY06594, AG019085, EY023164, EY022310, EY012118, AG044089, T32 EY007157, EY021532, EY0105712, 1U54HG006542-01, UL1TR000427, 1U01HG006389, EY023194, P30-EY005722, ZO1 EY000475, HHS-NOI-EY-0-2127, HHS-N-260-2005-00007-C, EY0022005, HG006513, HG007022, EY016862, EY021900, EY017362, EY13824, EY009611, CA87969, CA49449, HL35464, EY014428, EY018660, P30EY022589, 1R01EY018660-01A10, R01-EY013435, P30-EY019007), Arnold and Mabel Beckman Initiative for Macular Research, Research to Prevent Blindness Inc, EyeSight Foundation of Alabama, the Marshfield Clinic Research Foundation, 863 Program (2014AA021604), the ALSAM Foundation, VA Merit Review, Foundation Fighting Blindness, and the International Retinal Research Foundation. In the UK, contributory IAMDGC research studies were funded by grants from the Medical Research Council (G0000067), the Guide Dogs for the Blind Association and the Department of Health's NIHR Biomedical Research Centre for Ophthalmology at Moorfields Eye Hospital and UCL Institute of Ophthalmology, the Wellcome Trust (076169/A), American Health Assistance Foundation (M2007110), Macula Vision Research Foundation, TFC Frost Charitable Trust, Brian Mercer Charitable Trust, Macular Society, Hobart Trust, and the Gift of Sight appeal. The contributory Scottish AMD study was funded by the Chief scientists Office (Scotland, CZB/4/79). In the European Union, contributory IAMDGC research studies were funded by the EUGENDA-Cologne group was supported by a grant from the Retinovit foundation, the French Ministère de l'Enseignement Supèrieur et de la Recherche, Foundation Fighting Blindness Clinical Research Institute, BMBF-01ER1206, EFKS 2012_A147 , BMBF-01GP1308, the Deutsche Forschungsgemeinschaft (1259/19-1 and WE1259/19-2, BHFW), the Alcon Research Institute, ZoNMW (170885606), MDfonds, Landelijke Stichting voor Blinden en Slechtzienden, Gelderse Blindenstichting, and Algemene Vereniging ter Voorkoming van Blindheid, Stichting Nederlands Oogheelkundig Onderzoek, Oogfonds. The Jerusalem study was supported by grants from the Israel Science fund and the Israeli Ministry of Health. In China, contributory IAMDGC research studies were funded by National Natural Science Foundation of China (81170883 and 81430008). In South Korea, contributory IAMDGC research studies were funded by the National Research Foundation of Korea, funded by the Ministry of Education, Science, and Technology (NRF-2009-0072603 and NRF-2012R1A1A2008943). NRF-2012R1A1A2008943). For the INTERVAL dataset, all contributing sites and additional funding information are acknowledged in this publication: Sun et al. Nature. 2018;558(7708):73–79. The INTERVAL research study was funded by the NHS Blood and Transplant, National Institute for Health Research, UK Medical Research Council, and British Heart Foundation. The SomaLogic plasma protein GWAS summary statistics web page containing instructions for access is: Proteins—Cardiovascular Epidemiology Unit (cam.ac.uk). The SIGHT study was funded by Gyroscope Therapeutics Ltd., and additional information is provided here: Khan et al. Hum Mutat. 2021;42(9):1139-1152.
SCOPE Study Group: Dr Catherine Creuzot-Garcher, CHU Dijon—Hopital Mitterrand, Cedex, France; Dr Salvatore Grisanti, University Hospital Schleswig-Holstein Campus, Luebeck, Germany; Dr Martin Spitzer, Universitaetsklinikum Hamburg-Eppendorf, Hamburg, Germany; Dr Reiner Schlingerman, Amsterdam UMC—Locatie AMC, Amsterdam, The Netherlands; Dr Tsveta Ivanova, Manchester Royal Eye Hospital, Manchester, United Kingdom; Prof. David Steele, Sunderland Eye Infirmary, Sunderland, United Kingdom; Dr Jared Nielsen, Wolfe Eye Clinic, West Des Moines, Iowa; Dr Raj Maturi, Midwest Eye Institute Northside, Indianapolis, Indiana; Dr Jeff Heier, Ophthalamic Consultants of Boston (OCB), Boston, Massachusetts; Dr Tongalp Tezel, Columbia University Medical Center, New York, New York; Dr Ghassan Ghorayeb, West Virginia University, Morgantown, West Virginia; and Dr David Eichenbaum, Retina Vitreous Associates of Florida, St. Petersburg, Florida.
HUMAN SUBJECTS: Human subjects were included in this study. All methods were carried out in accordance with the relevant guidelines and regulations of local or country-specific Ethics Committees. All research adhered to the tenets of the Declaration of Helsinki. All participants provided informed consent.
No animal subjects were included in this study.
Author Contributions:
Conception and design: Jones, MacGregor, Han, Kavanagh, Lotery, Waheed
Analysis and interpretation: Jones, MacGregor, Han, Francis, Harris, Kavanagh, Lotery, Waheed
Data collection: Jones, MacGregor, Han, Francis, Kavanagh, Lotery, Waheed
Obtained funding: N/A; Study was performed as part of the authors' regular employment duties. No additional funding was provided.
Overall responsibility: Jones, MacGregor, Han, Francis, Harris, Kavanagh, Lotery, Waheed
Supplementary Data
References
- 1.Friedman D.S., O’Colmain B.J., Muñoz B., Tomany S.C. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122(4):564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 2.Wong W.L., Su X., Li X., et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Global Heal. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392(10153):1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 4.Lim L.S., Mitchell P., Seddon J.M., et al. Age-related macular degeneration. Lancet. 2012;379(9827):1728–1738. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 5.Colijn J.M., Buitendijk G.H.S., Prokofyeva E., et al. Prevalence of age-related macular degeneration in Europe: the past and the future. Ophthalmology. 2017;124(12):1753–1763. doi: 10.1016/j.ophtha.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenfeld P.J., Brown D.M., Heier J.S., et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355(14):1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 7.Raftery J., Clegg A., Jones J., et al. Ranibizumab (Lucentis) versus bevacizumab (Avastin): modelling cost effectiveness. Br J Ophthalmol. 2007;91(9):1244–1246. doi: 10.1136/bjo.2007.116616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunness J.S., Gonzalez-Baron J., Applegate C.A., et al. Enlargement of atrophy and visual acuity loss in the geographic atrophy form of age-related macular degeneration. Ophthalmology. 1999;106(9):1768–1779. doi: 10.1016/S0161-6420(99)90340-8. [DOI] [PubMed] [Google Scholar]
- 9.Maguire M.G., Martin D.F., Ying G.S., et al. Five-year outcomes with anti–vascular endothelial growth factor treatment of neovascular age-related macular degeneration. Ophthalmology. 2016;123(8):1751–1761. doi: 10.1016/j.ophtha.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleckenstein M., Mitchell P., Freund K.B., et al. The progression of geographic atrophy secondary to age-related macular degeneration. Ophthalmology. 2018;125(3):369–390. doi: 10.1016/j.ophtha.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 11.Lambert N.G., ElShelmani H., Singh M.K., et al. Risk factors and biomarkers of age-related macular degeneration. Prog Retin Eye Res. 2016;54:64–102. doi: 10.1016/j.preteyeres.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadda S.R., Chakravarthy U., Birch D.G., et al. Clinical endpoints for the study of geographic atrophy secondary to age-related macular degeneration. Retina. 2016;36(10):1806–1822. doi: 10.1097/IAE.0000000000001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennesi M.E., Neuringer M., Courtney R.J. Animal models of age related macular degeneration. Mol Aspects Med. 2012;33(4):487–509. doi: 10.1016/j.mam.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seddon J.M., Reynolds R., Maller J., et al. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Opthalmol Vis Sci. 2009;50(5):2044–2053. doi: 10.1167/iovs.08-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buitendijk G.H.S., Rochtchina E., Myers C., et al. Prediction of age-related macular degeneration in the general population the Three Continent AMD Consortium. Ophthalmology. 2013;120(12):2644–2655. doi: 10.1016/j.ophtha.2013.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seddon J.M., Cote J., Page W.F., et al. The US Twin Study of Age-Related Macular Degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123(3):321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 17.Nelson M.R., Tipney H., Painter J.L., et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47(8):856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 18.Kauppinen A., Paterno J.J., Blasiak J., et al. Inflammation and its role in age-related macular degeneration. Cell Mol Life Sci. 2016;73(9):1765–1786. doi: 10.1007/s00018-016-2147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritsche L.G., Igl W., Bailey J.N.C., et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards A.O., Ritter R., Abel K.J., et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 21.Fritsche L.G., Chen W., Schu M., et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grassmann F., Heid I.M., Weber B.H.F., International AMD Genomics Consortium (IAMDGC) Recombinant haplotypes narrow the ARMS2/HTRA1 association signal for age-related macular degeneration. Genetics. 2017;205(2):919–924. doi: 10.1534/genetics.116.195966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams B.L., Seager N.A., Gardiner J.D., et al. Chromosome 10q26–driven age-related macular degeneration is associated with reduced levels of HTRA1 in human retinal pigment epithelium. Proc Natl Acad Sci U S A. 2021;118(30) doi: 10.1073/pnas.2103617118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitz-Valckenberg S., Fleckenstein M., Zouache M.A., et al. Progression of age-related macular degeneration among individuals homozygous for risk alleles on chromosome 1 (CFH-CFHR5) or chromosome 10 (ARMS2/HTRA1) or both. JAMA Ophthalmol. 2022;140(3):252–260. doi: 10.1001/jamaophthalmol.2021.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobrin L., Reynolds R., Yu Y., et al. ARMS2/HTRA1 locus can confer differential susceptibility to the advanced subtypes of age-related macular degeneration. Am J Ophthalmol. 2011;151(2):345–352. doi: 10.1016/j.ajo.2010.08.015. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van de Ven J.P.H., Nilsson S.C., Tan P.L., et al. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat Genet. 2013;45(7):813–817. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 27.Clark S.J., Bishop P.N. The eye as a complement dysregulation hotspot. Semin Immunopathol. 2018;40(1):65–74. doi: 10.1007/s00281-017-0649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schramm E.C., Clark S.J., Triebwasser M.P., et al. Genetic variants in the complement system predisposing to age-related macular degeneration: a review. Mol Immunol. 2014;61(2):118–125. doi: 10.1016/j.molimm.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson D.H., Radeke M.J., Gallo N.B., et al. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29(2):95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson D.H., Mullins R.F., Hageman G.S., Johnson L.V. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134(3):411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 31.Hageman G.S., Luthert P.J., Chong N.H.V., et al. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20(6):705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 32.Clark S.J., Bishop P.N. The eye as a complement dysregulation hotspot. Semin Immunopathol. 2018;40(1):65–74. doi: 10.1007/s00281-017-0649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaspan B.L., Williams D.F., Holz F.G., et al. Targeting factor D of the alternative complement pathway reduces geographic atrophy progression secondary to age-related macular degeneration. Sci Transl Med. 2017;9(395) doi: 10.1126/scitranslmed.aaf1443. [DOI] [PubMed] [Google Scholar]
- 34.Wykoff C.C., Rosenfeld P.J., Waheed N.K., et al. Characterizing new-onset exudation in the randomized phase 2 FILLY trial of complement inhibitor pegcetacoplan for geographic atrophy. Ophthalmology. 2021;128(9):1325–1336. doi: 10.1016/j.ophtha.2021.02.025. [DOI] [PubMed] [Google Scholar]
- 35.Jaffe G.J., Westby K., Csaky K.G., et al. C5 inhibitor avacincaptad pegol for geographic atrophy due to age-related macular degeneration: a randomized pivotal phase 2/3 trial. Ophthalmology. 2021;128(4):576–586. doi: 10.1016/j.ophtha.2020.08.027. [DOI] [PubMed] [Google Scholar]
- 36.Holz F.G., Sadda S.R., Busbee B., et al. Efficacy and safety of lampalizumab for geographic atrophy due to age-related macular degeneration: Chroma and Spectri phase 3 randomized clinical trials. JAMA Ophthalmol. 2018;136(6):666. doi: 10.1001/jamaophthalmol.2018.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dreismann A.K., McClements M.E., Barnard A.R., et al. Functional expression of complement factor I following AAV-mediated gene delivery in the retina of mice and human cells. Gene Ther. 2021;28(5):265–276. doi: 10.1038/s41434-021-00239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fagerness J.A., Maller J.B., Neale B.M., et al. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17(1):100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexander P., Gibson J., Cree A.J., et al. Complement factor I and age-related macular degeneration. Mol Vis. 2014;20:1253–1257. [PMC free article] [PubMed] [Google Scholar]
- 40.Seddon J.M., Yu Y., Miller E.C., et al. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet. 2013;45(11):1366–1370. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hageman G.S., Anderson D.H., Johnson L.V., et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102(20):7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saksens N.T.M., Geerlings M.J., Bakker B., et al. Rare genetic variants associated with development of age-related macular degeneration. JAMA Ophthalmol. 2016;134(3):287–293. doi: 10.1001/jamaophthalmol.2015.5592. [DOI] [PubMed] [Google Scholar]
- 43.Shoshany N., Weiner C., Safir M., et al. Rare genetic variants in Jewish patients suffering from age-related macular degeneration. Genes (Basel) 2019;10(10):825. doi: 10.3390/genes10100825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris K.M., Aden D.P., Knowles B.B., Colten H.R. Complement biosynthesis by the human hepatoma-derived cell line HepG2. J Clin Invest. 1982;70:906–913. doi: 10.1172/JCI110687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schäfer N., Wolf H.N., Enzbrenner A., et al. Properdin modulates complement component production in stressed human primary retinal pigment epithelium cells. Antioxidants. 2020;9(9):793. doi: 10.3390/antiox9090793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gitlin J., Rosen F., Lacchman P. The mechanism of action of the C3b inactivator (conglutinogen- activating factor) on its naturally occurring substrate, the major fragment of the third component of complement (C3b) J Exp Med. 1975;141(5):1221–1226. doi: 10.1084/jem.141.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medof M.E., Iida K., Mold C., Nussenzweig V. Unique role of the complement receptor CR1 in the degradation of C3b associated with immune complexes. J Exp Med. 1982;156:1739–1754. doi: 10.1084/jem.156.6.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pangburn M.K., Schreiber R.D., Muller H.J. Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein β1H for cleavage of C3b and C4b in solution. J Exp Med. 1977;146:257–270. doi: 10.1084/jem.146.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hecker L.A., Edwards A.O., Ryu E., et al. Genetic control of the alternative pathway of complement in humans and age-related macular degeneration. Hum Mol Genet. 2010;19(1):209–215. doi: 10.1093/hmg/ddp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nilsson S.C., Sim R.B., Lea S.M., et al. Complement factor I in health and disease. Mol Immunol. 2011;48(14):1611–1620. doi: 10.1016/j.molimm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Hallam T.M., Marchbank K.J., Harris C.L., et al. Rare genetic variants in complement factor I lead to low FI plasma levels resulting in increased risk of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2020;61(6) doi: 10.1167/iovs.61.6.18. 18–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Java A., Baciu P., Widjajahakim R., et al. Functional analysis of rare genetic variants in complement factor I (CFI) using a serum-based assay in advanced age-related macular degeneration. Transl Vis Sci Technology. 2020;9(9) doi: 10.1167/tvst.9.9.37. 37–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kavanagh D., Yu Y., Schramm E.C., et al. Rare genetic variants in the CFI gene are associated with advanced age-related macular degeneration and commonly result in reduced serum factor I levels. Hum Mol Genet. 2015;24(13):3861–3870. doi: 10.1093/hmg/ddv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Jong S., Volokhina E.B., de Breuk A., et al. Effect of rare coding variants in the CFI gene on factor I expression levels. Hum Mol Genet. 2020;29(14):2313–2324. doi: 10.1093/hmg/ddaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geerlings M.J., de Jong E.K., den Hollander A.I. The complement system in age-related macular degeneration: a review of rare genetic variants and implications for personalized treatment. Mol Immunol. 2017;84:65–76. doi: 10.1016/j.molimm.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schick T., Steinhauer M., Aslanidis A., et al. Local complement activation in aqueous humor in patients with age-related macular degeneration. Eye. 2017;31(5):810–813. doi: 10.1038/eye.2016.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan X., Fujiu K., Manabe I., et al. Choroidal neovascularization is inhibited via an intraocular decrease of inflammatory cells in mice lacking complement component C3. Sci Rep. 2015;5(1):15702. doi: 10.1038/srep15702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawlor D.A., Harbord R.M., Sterne J.A.C., et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;15(27):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 59.Roberts R. Mendelian randomization studies promise to shorten the journey to FDA approval. JACC Basic Transl Sci. 2018;3(5):690–703. doi: 10.1016/j.jacbts.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun B.B., Maranville J.C., Peters J.E., et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan A.H., Sutton J., Cree A.J., et al. Prevalence and phenotype associations of complement factor I mutations in geographic atrophy. Hum Mutat. 2021;42(9):1139–1152. doi: 10.1002/humu.24242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han X., Ong J.S., Hewitt A.W., et al. The effects of eight serum lipid biomarkers on age-related macular degeneration risk: a Mendelian randomization study. Int J Epidemiol. 2021;50(1):325–336. doi: 10.1093/ije/dyaa178. [DOI] [PubMed] [Google Scholar]
- 63.Das S., Forer L., Schönherr S., et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yavorska O.O., Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6) doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karczewski K.J., Francioli L.C., Tiao G., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Landrum M.J., Lee J.M., Benson M., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2017;46(database issue) doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mountjoy E., Schmidt E.M., Carmona M., et al. Open Targets Genetics: an open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat Genet. 2021;53(11):1527–1533. doi: 10.1038/s41588-021-00945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaye R.A., Patasova K., Patel P.J., et al. Macular thickness varies with age-related macular degeneration genetic risk variants in the UK Biobank cohort. Sci Rep. 2021;11(1):23255. doi: 10.1038/s41598-021-02631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lonsdale J., Thomas J., Salvatore M., et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strunz T., Lauwen S., Kiel C., et al. A transcriptome-wide association study based on 27 tissues identifies 106 genes potentially relevant for disease pathology in age-related macular degeneration. Sci Rep. 2020;10(1):1584. doi: 10.1038/s41598-020-58510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ratnapriya R., Sosina O.A., Starostik M.R., et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genet. 2019;51(4):606–610. doi: 10.1038/s41588-019-0351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orozco L.D., Chen H.H., Cox C., et al. Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep. 2020;30(4):1246–1259. doi: 10.1016/j.celrep.2019.12.082. e6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.