Abstract
Coronavirus disease 2019 (COVID-19) has caused significant global morbidity and mortality. The serology test that detects antibodies against the disease causative agent, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has often neglected value in supporting immunization policies and therapeutic decision-making. The ELISA-based antibody test is time-consuming and bulky. This work described a gold micro-interdigitated electrodes (IDE) biosensor for COVID antibody detection based on Electrochemical Impedance Spectroscopy (EIS) responses. The IDE architecture allows easy surface modification with the viral structure protein, Spike (S) protein, in the gap of the electrode digits to selectively capture anti-S antibodies in buffer solutions or human sera. Two strategies were employed to resolve the low sensitivity issue of non-faradic impedimetric sensors and the sensor fouling phenomenon when using the serum. One uses secondary antibody-gold nanoparticle (AuNP) conjugates to further distinguish anti-S antibodies from the non-specific binding and obtain a more significant impedance change. The second strategy consists of increasing the concentration of target antibodies in the gap of IDEs by inducing an AC electrokinetic effect such as dielectrophoresis (DEP). AuNP and DEP methods reached a limit of detection of 200 ng/mL and 2 μg/mL, respectively using purified antibodies in buffer, while the DEP method achieved a faster testing time of only 30 min. Both strategies could qualitatively distinguish COVID-19 antibody-positive and -negative sera. Our work, especially the impedimetric detection of COVID-19 antibodies under the assistance of the DEP force presents a promising path toward rapid, point-of-care solutions for COVID-19 serology tests.
Keywords: Gold interdigitated electrodes, Impedimetric biosensor, COVID-19 serology test, Gold nanoparticles, Dielectrophoresis force
1. Introduction
In December 2019, cluster cases of atypical pneumonia of unknown origin were reported in Wuhan, China and the disease was later named coronavirus disease 2019 (COVID-19). The agent causing COVID-19 was soon identified to be a novel betacoronavirus, designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (P. Zhou et al., 2020). The World Health Organization (WHO) declared COVID-19 a pandemic in March 2020 (Huang et al., 2020; Soares et al., 2022; Sun et al., 2020). This ongoing COVID-19 epidemic has caused global economic restraining due to multiple rounds of nationwide lockdowns in the effort of containing the spread of the disease. The disease has a wide clinical spectrum, ranging from asymptomatic infection to severe pneumonia resulting in multiorgan failure and death (Chen et al., 2020; Huang et al., 2020; Perez-Saez et al.; Richardson et al., 2020). Age and serious underlying health conditions are significant risk factors for COVID-19 patients to develop severe illnesses or suffer mortality (Eythorsson et al., 2020; Huang et al., 2020; Richardson et al., 2020b; F. Zhou et al., 2020).
In the early phases of an epidemic, timely diagnosis is essential for identifying and isolating infected individuals to reduce transmission (Kucharski et al., 2020). Given the high sensitivity and accuracy, the standard diagnosis is predominantly depending on reverse transcription and real-time quantitative polymerase chain reaction (RT-qPCR), which detects the viral nucleic acid extracted from nasopharyngeal/oropharyngeal swabs. Another diagnostic method is the direct detection of viruses or viral proteins (the antigen test) in clinical samples. Nevertheless, it is rarely applied in this pandemic because it could be up to 105 times less sensitive than RT-qPCR due to the lack of amplification steps of the viruses or viral proteins (Mak et al., 2020; Schildgen et al., 2020; Weitzel et al., 2020).
COVID-19 serology tests measure an individual's antibody (Ab) response to SARS-CoV-2 infection or vaccination. Although Ab detection is not the primary method of early diagnosis, it has many crucial roles in COVID-19 research, immunization policy formation and the development of therapeutics. The serology tests can be either qualitative or quantitative. The qualitative assay simply indicates whether the Ab is present or not based on a pre-determined threshold. Clinically, it may provide a fast and general indication of a current and/or past infection depending on when the sample is taken after exposure; hence is an excellent complement to the RT-qPCR test which can detect only active infections (Bortz et al., 2021). SARS-CoV-2 viruses have four structural proteins, Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N) proteins that encapsulate the viral RNA (Masters, 2006). The S protein contains two subunits, S1 and S2. Depending on target antigens of the assay, the qualitative assay can distinguish between immunity acquired from natural infection and vaccination. SARS-CoV-2 vaccines, including the BNT162b2 vaccine (Pfizer) and the mRNA-1273 vaccine (Moderna), only provoke Ab response against the S protein (Wheeler et al., 2021); while natural infections induce at least Abs against both the S and N proteins (Liu et al., 2020; K. K. K.-W. To et al., 2020). Moreover, qualitative tests can be carried out in time-course in convalescent patients and vaccinated individuals to know how long it takes before Abs decay and become non-detectable, aka, reaching seronegativity (Alejo et al., 2022; Iyer et al., 2020). On the other hand, the quantitative COVID-19 serology test measures the level of SARS-CoV-2 Abs using international reference materials as standards (Infantino et al., 2021). It helps to collect valuable data for answering fundamental clinical questions such as: What level of neutralizing Ab in an individual can ensure immunity to a future infection? What is the variation of Ab response among patients with different ages, genders, and underlying complications? Is there a correlation between the levels of Ab response and the severity of the disease (Röltgen et al., 2020)? Currently, enzyme-linked immunosorbent assay (ELISA) is one of the most common approaches for COVID-19 serology tests (Amanat et al., 2020; Stadlbauer et al., 2020); however, it has the shortcoming of being labor-intensive and requiring bulky equipments.
Biosensors employ various types of highly compact transducers consisting of nanomaterials or nanostructures that are functionalized with biorecognition elements to interact with the analytes. Alterations (e.g., physicochemical, optical, piezoelectric, electrochemical changes) resulted from the binding of analytes to the recognition elements can be transformed into quantitative and processable signals. Among these designs, electrochemical-based biosensors show high sensitivity, portability and the probability of being operated via smartphones (Zhao et al., 2021). Numerous groups have demonstrated prototypes of nanotechnology-enabled biosensors for direct COVID-19 virus/viral proteins detections (antigen tests) (Ahmed et al., 2018; Ahmed et al., 2017; Asif et al., 2020; Cabral-Miranda et al., 2018; Layqah and Eissa, 2019; Mahari et al., 2020; Roh and Jo, 2011; Seo et al., 2020; Yousefi et al., 2021). More specifically, the Electrochemical Impedance Spectroscopy (EIS)-based biosensors have been developed for SARS-CoV-2 detection and for screening of SARS-CoV-2 inhibitors (Kiew et al., 2021; Soares et al., 2022).
In comparison to the antigen test, relatively less attention was given to the development of point-of-care (POC) devices or biosensors for serology tests (Ab tests), especially one based on electrochemical detection (Elledge et al., 2021; Heggestad et al., 2021; Tan et al., 2020). Rashed et al. used electrochemical impedance-based detector for Ab detection, but in a 96-well format, analogous to that of the ELISA (Rashed et al., 2021). To date, lateral flow immunoassay (LFIA) based on colloidal gold is the only established POC serology test device available on the market. However, LFIAs suffer the disadvantage of non-quantitative, low sensitivity and inconsistent results.
In this article, we describe a biosensor employing transducers patterned with gold micro-interdigitated electrodes (IDE) to detect Abs against SARS-CoV-2 based on EIS responses. In-house recombinant viral S proteins or commercial recombinant S1 subunits are functionalized between the IDE digits to detect anti-S Abs (the Immunoglobulin G isotype). Using the in-house fabricated sensor chips, we realized EIS-based COIVD-19 antibody detection via two approaches: 1. probing the captured anti-S Ab using an AuNP-conjugated secondary Ab; 2. direct one-step detection with the assistance of dielectrophoretic (DEP) force. When using purified anti-S Abs, the AuNP strategy reached a Limit of Detection (LoD) at 200 ng/mL and the DEP approach have an LoD at 2 μg/mL. However, the DEP approach is simpler and faster, providing results in about 30min. Both methods were tested and verified using eight human serum samples (four positive and four negative, respectively) and can qualitatively separate COVID-19 Ab-positive or -negative sera. In summary, our sensor, especially with the assistance of the DEP force has the potential to meet the demand for a rapid POC device for COVID-19 serology testing at anytime and anywhere. When fully optimized, this sensor can help increase the capacity of serology tests, especially for remote communities.
2. Material and methods
2.1. Materials and chemicals
(3-Aminopropyl)triethoxysilane (APTES), N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide (SulfoNHS) ≥98%, glutaraldehyde 25% and bovine serum albumin 98% (BSA) were purchased from Sigma-Aldrich Canada Co. (Oakville, Ontario). Ultrapure water (18.2 MΩ/cm) was obtained from Millipore equipment (Mili-Q water).
The anti-human IgG F(ab')2 Fragment-15nm Gold Conjugate was purchased from Cytodiagnostics Inc (ON, Canada). The commercial His-tagged recombinant SARS-CoV-2 S1 (DAGC091) and human anti-SARS-CoV-2 S1 monoclonal antibody (IgG, clone BIB112) are procured from Creative Diagnostics (NY, USA). Horseradish Peroxidase (HRP)-conjugated anti-human IgG antibodies (catalogue no. CL2341HP) and Alexa Fluor® 488-conjugated goat anti-Human IgG antibodies (catalogue no.109-545-003) are obtained from Cedarlane (Burlington, Ontario). The Nunc MaxiSorp™ high protein-binding capacity 96 well ELISA plates (catalogue no. 44-2404-21) and 3, 3′, 5, 5;-tetramethylbenzidine (TMB) (catalogue no.002023) are from Thermo Fisher Scientific (MA, USA). Total IgGs from human serum (reagent grade) were purchased from Sigma-Aldrich (ON, Canada).
2.2. Fabrication of the IDE sensor
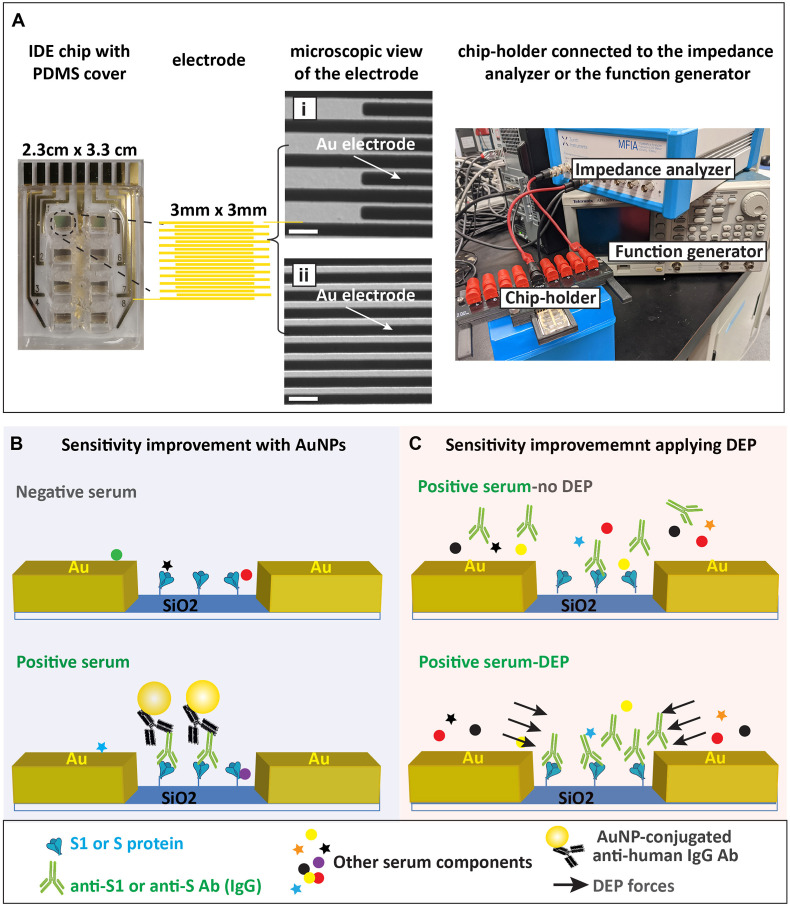
The IDE sensors were fabricated using the standard photolithographic procedure described in our previous work (Abdelrasoul et al., 2018, 2020; MacKay et al., 2017; Shoute et al., 2018). Each IDE comprises 168 digit pairs of gold electrodes with the width of 4 μm, thickness of 60 nm and 2 μm gap between electrodes. Each IDE is 3 mm × 3 mm in size and there are eight IDEs on each chip for conducting eight experiments in parallel (Fig. 1 A).
Fig. 1.
Experimental setup. (A) Basic components of the impedimetric sensing for COVID-19 serology test. From left to right are the IDE sensor chip with PDMS mask, detailed structure of each electrode and the chip-holder that connects the chip to the impedance analyzer or the functional generator. Differential interference contrast (DIC) microscopy images showing the edge of the electrode fingers (i) and the middle of the digit pattern (ii). Scale bar represents 10 μm. (B) Schematic representation of the COVID-19 Ab detection via the AuNP approach, which requires the immobilization of either the SARS-CoV-2 S1 subunit or the full-length S protein (S1/S) on the SiO2 substrate between the gold (Au) electrode digits. The anti-S1/S Abs in the positive serum sample will bind to the antigen (S1/S), resulting in the deposit of AuNPs when AuNP-conjugated anti-human IgG Abs are applied. The presence of AuNPs will cause measurable impedance changes. The negative serum may have some non-specific binding to S1/S, but no AuNP will be deposited under this scheme. (C) Schematic representation of the COVID-19 Ab detection via the DEP approach. Anti-S IgG Abs in the positive serum were selectively preconcentrated by the DEP force against other serum components. The binding of Anti-S IgG Abs (in the positive serum) to the sensor surface generates significant impedance change so that negative and positive sera can be separated.
2.3. Surface functionalization of the IDE sensor (the AuNP approach)
We hydroxylated the SiO2 surface by exposure of the chips to oxygen plasma (100 sccm O2, 150 mT pressure, 150 W RF) for 40 s using a bench-top reactive-ion etching (RIE) system (Trion Technology, USA). SARS-CoV-2 S1 or S proteins were immobilized to the SiO2 surface either by APTES-glutaraldehyde chemistries as described in our previous work (Abdelrasoul et al., 2018) or by APTES-EDC/sulfoNHS chemistries. For the APTES-EDC/sulfolNHS method, the cleaned chips were silanized by 5% APTES (prepared in 100% ethanol) for 3 h at 4 °C. After that, the chips were thoroughly washed sequentially with absolute ethanol, isopropanol, and Mili-Q water to remove the unreacted APTES. We then placed polydimethylsiloxane (PDMS) cover with eight wells on top of the chip to separate the analyte solutions (Fig. 1A). Meanwhile, 300 μg/mL of S protein or 144 μg/mL of S1 protein was activated on their carboxylic-acid groups by adding EDC and sulfo-NHS prepared in 0.1 M MES buffer (PH 4.7) at final concentrations of 0.65 mM and 2.5 mM, respectively (Dixit, Vashist, MacCraith and O'Kennedy, 2011). After 15 min of incubation at room temperature, 40 μL of activated S/S1 were added to each electrode for 1 h at 37 °C, allowing covalent bonding between carboxyl groups of the proteins and amine groups on the chip surface. Subsequently, the chips were rinsed two times with 10 mM PBS containing 0.2% tween (0.2% PBST), two times with 10 mM PBS and one time with H2O, followed by the incubation of analytes (either commercial anti-S1 Abs or human serum samples) with the chip for 1 h at 37 °C. After five washes, AuNP-conjugated anti-human IgG antibodies were allowed to react with the sensor surface for 3 h at room temperature.
2.4. Impedance measurement and DEP manipulation
A custom-made chip-holder based on pogo-pins (Mill-Max Corp.) was employed to electrically connect our biosensor to all equipment (Fig. 1A).
EIS measurements were performed using a high precision impedance analyzer (Zurich Instruments MFIA) controlled by the software LabOne® or the SP-200 potentiostat controlled by the EC-Lab® software package (Bio-Logic, TN, USA). We carried out EIS measurements after serum incubation and after AuNP binding with the presence of 50 μL of 10−6 M PBS inside the PDMS well. A sinusoidal voltage with an amplitude of 10 mV and zero DC bias was applied to the IDE sensor as an input, and the impedance spectrum was measured at the frequency range from 100 Hz to 1000 kHz.
During DEP experiments, sinusoidal signals were applied to the IDEs via the chip-holder by using a function generator (Tektronix AFG 3251) at various frequencies and AC voltages.
2.5. Imaging and field emission scanning electron microscopy
We acquired Differential Interference Contrast (DIC) images to show the detailed structure of the IDE digits (Fig. 1A) using a ZEISS LSM 710 confocal microscope system with a Plan-Apochromat 40x oil lens (NA:1.4).
Field Emission Scanning Electron Microscopy (FESEM) images were obtained using the ZEISS Sigma FESEM system with an in-lens secondary electron (SE) detector operating at 6 kV and an aperture of 20 m. The samples were examined at a magnification of 20,000×.
2.6. Surface roughness characterization via atomic force microscopy (AFM)
Atomic force microscope system (diMultiMode V SPM, Veeco Instruments Inc.) was used to characterize the surface topography of IDE. Samples were scanned with tapping mode at a scan rate of 0.1 Hz. AFM images were processed and analyzed through Nanoscope software.
3. Results and discussion
3.1. Experimental design and immunoassay scheme
Our IDE sensor assesses the Abs against SARS-CoV-2 via an on-chip immunoassay. The layout of the sensor chip and the device set up are demonstrated in Fig. 1A. The recombinant S protein or commercial S1 subunits (S1/S) was functionalized on the SiO2 substrate between the electrode digits to capture the analytes (either purified anti-S Abs or anti-S Abs in human serum samples). This work focused on detecting only the Immunoglobulin G (IgG) isotype of anti-S Abs since it is the most abundant Ab type in human serum and the long-lasting one after an infection (Gonzalez-Quintela et al., 2008). Two strategies were employed to enhance the sensitivity of the sensor: probing with AuNP-conjugated anti-human IgG Abs (Fig. 1B) and inducing DEP force to preconcentrate IgG Abs in the serum towards the gap of the electrode digits (which will be explained in detail in the later section) (Fig. 1C). In the AuNP approach, even if there were non-specific binding of other serum components to the S1/S protein, AuNP conjugated anti-human IgG Abs would not react to them; therefore, AuNPs will only be present with COVID-19 antibody-positive serum (Fig. 1B). Our previous work has established how the binding of AuNPs improves the sensitivity of the impedimetric IDE sensor(Abdelrasoul et al., 2018; MacKay et al., 2017).
Given that the AuNP approach is essentially similar to an indirect ELISA, we first validated the immunobinding scheme and the reagents via ELISA. During ELISA, HRP-conjugated anti-human IgG Abs were used instead of AuNP conjugates. The standard curves with various anti-S1 mAb concentrations were generated using the four-parameter logistic (4-PL) regression for two S1 coating concentrations in the wells (Fig. S1A). As a result, the R2 value for 250 ng and 150 ng of S1 coating were 0.999 and 0.996, respectively, indicating good fittings. Using 250 ng of S1 per well gave a standard curve with a steeper sloop and a lower EC50 at 130.3 ± 7.7 ng/mL, comparing to that of 150 ng of S1 coating (EC50 = 143.9 ± 43.7 ng/mL), meaning that a better resolution of detection was achieved. We then continued to validate the detection scheme on its clinical performance with serum samples using 250 ng of S1 per well. The ELISA reading of each sample was normalized to the reading of the blank sample to get the ELISA index, i.e. ELISA index = SampleO.D.(450nm)/blankO.D.(450nm). If we set the cutoff for a positive result at two standard deviations (SD) above the mean of all negative samples (Fig. S1B, black dotted line), all three positive sera had an ELISA index above the cutoff of 2.1 (Fig. S1B), confirming the fidelity of our assay design.
3.2. Validating the surface modification via fluorescent detection
Subsequently, we transferred this assay to our IDE sensors. To confirm a successful surface modification and the Ab binding on the chip, we first visualized the results using a fluorescent detection in which Alexa Fluor® 488-conjugated anti-human Abs was utilized instead of AuNP-conjugated anti-human Abs. We tried to immobilize S1 via the self-assembled monolayer (SAM) of amine functional groups on the chip using either APTES-glutaraldehyde or APTES-EDC/sulfoNHS chemistries. We then compared APES-glutaraldehyde and APTES-EDC/sulfoNHS methods for their efficiency in detecting the purified anti-S mAbs. Since the gold electrodes gave background fluorescent signal, we have included a blank sample (with 0 ng/mL of mAb) as a calibrator. The relative fluorescent intensity (RFI) for each sample was calculated at Samplefluorescent/Blankfluorescent. As a result, we found that APTES-EDC/sulfoNHS chemistry delivered higher RFI than the APTES-glutaraldehyde chemistry (p ≤ 0.05) (Figs. S2A–C). Therefore, in all the following experiments we used the APTES-EDC/sulfoNHS method for surface modification. Next, we again verified the success of surface modification using our in-house made S protein (antigen) with four positive and three negative serum samples. Like ELISA, the cutoff for a positive result was the mean of RFI for negative samples + 2SDs, which comes at 2.72. All four positive samples had the RFI above the cutoff (Figs. S2D–G), which confirmed the successful surface functionalization with the S protein and the specificity of our home-made S protein in binding the anti-S Ab in the serum.
3.3. Theories on impedimetric sensor data analysis and sensor sensitivity when using the AuNP approach
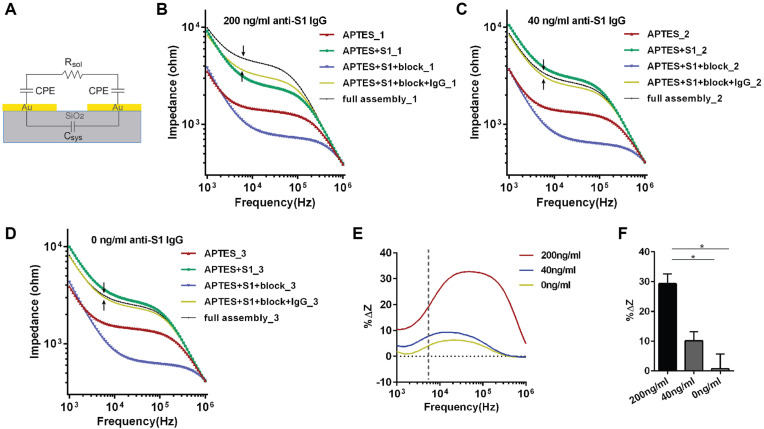
Electrochemical biosensors function by registering changes in the electrical properties caused by an analyte interacting with the biorecognition element on the sensing area (Grieshaber et al., 2008). EIS is a common technique for examining electrical properties of the sample, where a sinusoidal voltage or current perturbation is applied to the sample in an electrochemical cell to determine its impedance over a certain frequency range (Guiseppi-Elie and Lingerfelt, 2005; Mirsky et al., 1997). Impedance is typically measured with the electrode submerged in a buffer solution (in the presence of electrolytes). The EIS-based electrochemical biosensors can be categorized as faradic or non-faradic, depending on the presence or absence of redox species (Muñoz et al., 2017). The IDE biosensor described in this work is based on non-faradaic EIS with no redox species in either the electrode or electrolytes. The equivalent circuit of this sensor is modeled as a resistor (Rsol, representing the resistance of the buffer solution between the two electrodes), connected in series with two constant phase elements (CPEs) that depict the imperfect double layer capacitance at the electrode/electrolyte interface of the digits, which are connected in parallel with a capacitor modeling the system capacitance (Csys) (Fig. 2 A). Csys accounts for both the geometric capacitance (Cgeo) formed by the electrodes and the parasitic effects (Cpar) introduced by connection cables and SiO2 substrate. Since Cgeo and Cpar come with the IDE or the whole system itself and do not change significantly by surface modifications and analyte binding, we simplified them into one element: Csys.
Fig. 2.
Sensitivity of the sensor. (A) The equivalent circuit of the IDE sensor. Impedance spectra (magnitude) at the frequency range from 1k Hz – 1000 kHz measured after each step of surface modification when 200 ng/mL (B), 40 ng/mL (C) and 0 ng/mL of anti-S1 Ab (D) were tested. (B-D) IgG: anti-SARS-CoV-2 S1 IgG mAb; full assembly: APTES + S1 + block + anti-SARS-CoV-2 S1 IgG + anti-human IgG conjugated AuNPs. (E) Percentage change of impedance magnitude (%ΔZ) between the AuNPs binding step and the Ab binding step measured at frequency range from 1k Hz – 1000 kHz for different concentrations of anti-S mAb. %ΔZ=(ZAuNP-ZmAb)/ZmAb*100% where ZAuNP, and ZmAb represent the total impedance after AuNP and mAb step, respectively. (F) %ΔZ calculated at 5.76 kHz (pointed out by arrows in (B-D) and the dotted vertical line in (E)) for two experimental replicates. Error bars represent standard deviation. Statistics is determined by one-way ANOVA with Tukey's multiple comparisons test. *p<0.05.
We then used the commercial S1 protein (the biorecognition element) and the purified anti-S1 mAb (the analyte) to determine the IDE sensor's sensitivity. Typical impedance spectra (magnitude) after surface modification and each step of protein binding can be seen in Fig. 2B–D. First, the impedance increased substantially when EDC/NHS activated S1 protein was deposited (green curves in Fig. 2B–D) to the IDE that was just functionalized with APTES (red curve in Fig. 2B–D). More interestingly, although functionalization of the chip using EDC/NHS containing buffer alone without S1 protein give rise to non-specific impedance increases (blank in Fig. S3), it is significantly less than S1 deposition. Also, immobilizing a different protein at a higher concentration (1% BSA) resulted in an even greater impedance increase than S1 (Fig. S3). Therefore, we are confident that the increase in impedance is at least largely due to successful and specific immobilization of S1. Next, impedance declined after the blocking step (blue curve). Non-faradic mode did not have the sensitivity to indicate the presence or absence of anti-S1 mAb since 0 ng/mL of Ab (buffer alone) resulted in a comparable impedance rise as 200 ng/mL of Abs (Fig. 2C and D). It could be that our sensor, at this step, is sensitive to other interfering factors (e.g. residue salt from wash buffer settling on the electrode and in the gap), which masked the impedance changes caused specifically by Ab binding (also see section 3.5). Hence, commercial secondary Ab conjugated AuNPs were applied to resolve the issue.
For the AuNP approach, in the lower frequencies range (∼1 kHz–5.5 kHz), the impedance magnitude |Z| decreases as the frequency increases and the impedance is dominated by the CPE. Around the medium frequencies (∼5.5 kHz–150 kHz), impedance amplitude stays relatively stable and the impedance is dominated by Rsol. Lastly, the impedance amplitude decreases as the frequency increases again in the higher frequency range (∼150 kHz–1000 kHz) and this predominantly represents the Csys. The binding of AuNPs result in effective capacitance changes in CPEs and, to a certain extent, also affects the Rsol. And this combination of changes in CPEs and Rsol contributes to the total impedance change. Therefore, we believe that the effect of AuNP binding could be confidently picked up at the transition frequency between the low (where the CPE dominates) and the medium (where the Rsol dominates) frequency range (Fig. 2B–D, black arrows). For this reason, we obtain the assay result by calculating the percentage of impedance change (%ΔZ) between the AuNP binding step and its previous step (either the serum step or the mAb step, depending on the reagents used) at the transition frequency. Hence
| %ΔZ=(ZAuNP-Zserum)/Zserum*100% or %ΔZ=(ZAuNP-ZmAb)/ZmAb*100% | (1) |
where ZAuNP, Zserum or ZmAb represents the total impedance after reacting with AuNP, serum or mAb, respectively. Moreover, calculating %ΔZ at the transition frequency most likely best minimizes the variations among chips due to fabrication variations.
The %ΔZ for all frequencies measured (1 kHz–1000 kHz) was plotted in Fig. 2E. The %ΔZ for 0 ng/mL mAb represents the background impedance change and overall, the %ΔZ increases as the mAb concentration increases. When we calculated the %ΔZ at the transition frequency of 5.76 kHz (Fig. 2E, dotted vertical line) as explained previously, 0 ng/mL resulted on average 0.7% ± 5.0% of impedance change while 40 ng/mL and 200 ng/mL of mAb demonstrated 10.1% ± 3.0% and 29.3% ± 3.3% change, respectively (Fig. 2F). The difference between 0 ng/mL and 200 ng/mL is statistically significant (p < 0.05) (Fig. 2F). Therefore, we concluded that the sensor's LoD for anti-S1 mAbs is 200 ng/mL when using the method of secondary Ab-AuNP conjugates.
3.4. The clinical application of the AuNP approach using all in-house reagents
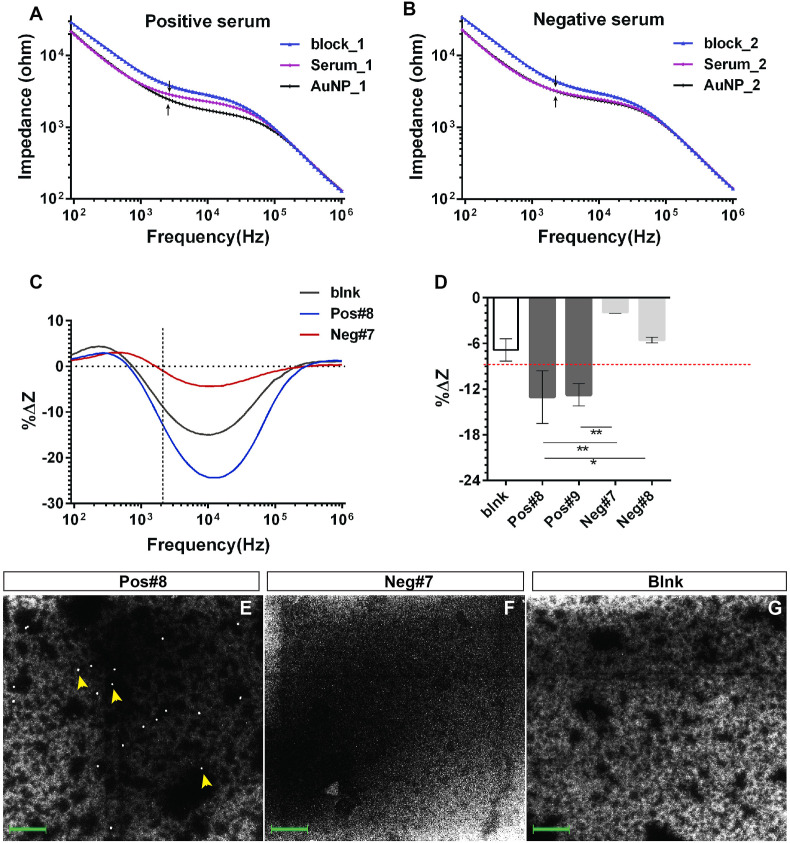
Next, we proceed to assess the sensor performance for real clinical applications using human serum samples. For this, we evaluated the AuNP approach using strictly our home-made reagents, which include the S antigen and in-house synthesized AuNPs conjugated with anti-human IgG Abs. Please refer to the Supplementary information for S protein expression and the characterization of AuNPs and Ab-AuNP conjugates (Fig. S4). Fig. 3 A–B showed impedance spectra (magnitude) after blocking, serum incubation and Ab-AuNPs binding step, respectively, for both the positive and negative serum samples. After blocking, negative serum binding again caused comparable impedance changes to positive serum binding, consistent with the result from purified Ab binding. Differences between positive and negative serum samples were only observed after applying secondary Ab conjugated AuNPs (Fig. 3A–B, black arrows). After the sensor reacted with our in-house Ab-AuNP conjugates, two negative serum samples caused −1.90% ± 0.0% and −5.6% ± 0.4% of impedance change, respectively; whereas the two positive serum samples experienced −13.1% ± 3.5% and −12.8% ± 1.5% of changes, respectively (Fig. 3D). Fig. 3E–G and Fig. S5 confirmed negligible AuNPs presence on the sensing surface for blank and negative samples and, in the meantime, showed observable AuNPs distribution in positive samples. In summary, these results indicate that a qualitatively discrimination between positive and negative samples can be achieved through our impedimetric method based on %ΔZ.
Fig. 3.
Evaluating the in-house made S protein and Ab-AuNP conjugates for sensor's clinical application. Impedance spectra (magnitude) at the frequency range from 1 kHz–1000 kHz measured after blocking step (blue), serum incubation step (red) and AuNPs binding step (black), respectively for a positive (A) and a negative serum (B). (C) %ΔZ between the AuNPs binding step and the serum step over all frequencies measured when using our in-house made AuNPs. %ΔZ=(ZAuNP-Zserum)/Zserum*100%. (D) %ΔZ calculated at 2.3 kHz from (C) for two positive and two negative sera. Error bars represent standard deviation. Statistics is determined by one-way ANOVA with Tukey's multiple comparisons test. *p<0.05. (E-G) FESEM images for positive (Pos#8), negative (Neg#7) and blank (blnk) samples after the AuNP step when using the in-house made AuNPs. Yellow arrowheads point to the individual AuNPs. Blank samples had no serum added. The green scale bar represents 300 nm.
Furthermore, we explored the sensor's potential for quantitative analysis of serum samples. The COVID-19 serum samples we received from the clinic were qualitatively determined and the concentration of anti-S Ab in the serum was unknown. For this reason, we generated serum samples whose relative anti-S Ab levels we at least know of by diluting one single positive serum. We also assumed that positive and negative serum have a similar matrix and their major difference is the presence or absence of anti-SARS-CoV-2 Abs. Based on this assumption, we diluted the one positive serum in a way that samples with the same matrix were obtained. In detail, one negative and one positive serum were firstly diluted 1 in 10 using the PBS-based buffer. Then 1 in 10 diluted positive serum is further diluted to achieve 1 in 50 and 1 in 100 dilutions using the 1 in 10 diluted negative serum. All samples hence have a matrix of 10% human serum. We tested the 1:10, 1:50 and 1:100 diluted positive samples via EIS and the 1:10 diluted negative serum served as a negative control. The results showed that when the positive serum was diluted 100 times, the %ΔZ was at the same level as that of the negative control, suggesting that the anti-S Ab level at this dilution dropped below the sensor's sensitivity (Fig. S6). In the meantime, %ΔZ (calculated at 5.6 kHz) differs nicely as levels of Abs change in the positive serum (Fig. S6), which indicates that our sensor has quantitative potential. These results suggested that we could potentially make the sensor semi-quantitative by the following two approaches: 1. use the dilution factor, in this case, 100 times (arbitrary unit) to indicate the relative Ab titer because following this dilution, the %ΔZ reached the same level as the negative control. 2. Using the %ΔZ value of negative control as the baseline and calculating the Area under Curve (see the triangle area in Fig. S6) to indicate Ab titer (Amanat et al., 2020).
In the future, we could further reduce the cost of our assay by optimizing the usage of S antigen for surface functionalization. Our preliminary study by ellipsometry suggested that a saturated surface functionalization could be reached with as low as 10 μg/mL of S protein by EDC/NHS conjugation. This is evidenced by Fig. S7 that for the concentration above 10 μg/mL, the substrate thickness after S protein functionalization remains almost constant at around 4 nm. However, when the concentrations of S protein fall below 10 μg/mL, the layer thickness decreases dramatically from 4 nm to 2.3 nm for 5 μg/mL, and to 0.6 nm for 1 μg/mL, which suggested that the optimal concentration with the lowest protein usage can be 10 μg/mL.
3.5. Using impedance measuring buffer at a higher conductivity improves the result consistency with trade-off of impedance signal robustness
Our sensor has several areas that could be improved for clinical applications: 1. Non-specific binding of irrelevant serum components was present, resulted in non-specific AuNP deposition even in negative serum (Fig. S5). 2. The blank sample where no serum was allowed to react with the S antigen resulted in a big impedance change after probing with AuNPs (Fig. 3D, blnk); this is not because of non-specific binding of AuNPs to the sensor since no AuNPs were observed in the blank sample via FESEM (Fig. 3G and Fig. S5, blnk). We believe that these impedance changes in blank samples are irrespective of the effective capacitance change in CPEs caused by AuNP binding on the sensing surface; rather are contributed solely by fluctuations in electrolyte (measuring buffer) concentrations during the two impedance measuring steps. The cause of fluctuation is that the immunoreaction and washing steps are performed in 10 mM PBS-based buffer (conductivity = 16 mS/cm), while following two washes with DI water, impedance measurements were carried out in 1 μM PBS (10,000 times diluted compared to the wash buffer and with a much higher conductivity of 3.35 μS/cm). Any residual salt from the 10 mM PBS-based wash buffer that was not thoroughly washed away from the IDE or PDMS wells during measurements would greatly affect the value of Rsol, thus the total impedance.
To overcome the above-mentioned shortcomings, we utilized a different wash buffer commonly used in ELISA (referred to as ELISA wash, conductivity = 2.84 mS/cm) after each step of immunobinding and then carried out impedance measurements in the same buffer. The recipe is listed in Supplementary Information. We expect that 1. it will more effectively wash away non-specific binding after both the serum and AuNP steps, and 2. washing and measuring in the same buffer will reduce the background Rsol change that is irrelevant to AuNP binding.
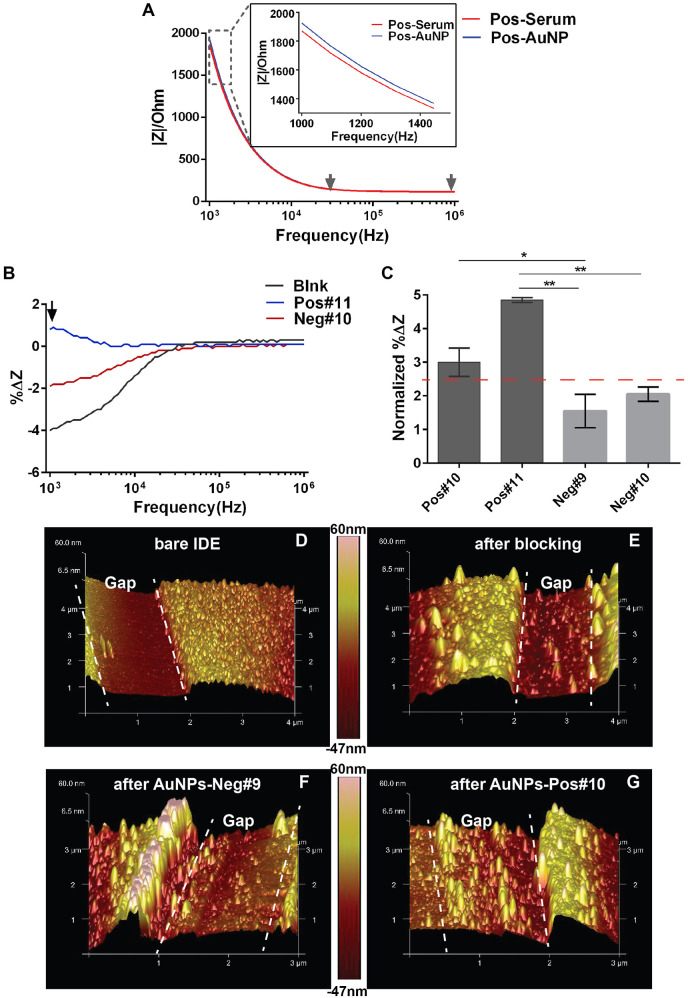
First of all, when using the ELISA wash for EIS measurement, impedance is dominated by Rsol at the frequency range from 20 kHz–100 kHz, evidenced from the constant impedance magnitude value in Fig. 4 A. We then calculated %ΔZ caused by AuNP binding across all frequencies that we tested and found that in the frequency range from 20 kHz−100 kHz, %ΔZ for blank, negative and positive serum were all centred around 0% (Fig. 4B). This result verified that there was indeed no change in Rsol after AuNP binding when EIS measuring was carried out in the same buffer as washing. In other words, the %ΔZ we observed in the lower frequency range (1 kHz–20 kHz) represents solely the effective capacitance change of CPEs caused by AuNP binding on the sensing surface (Fig. 4B). Next, we calculated the normalized %ΔZ (normalized %ΔZ = %ΔZsample-%ΔZblank) at 1 kHz, which is the frequency where maximal differences were observed between positive, negative serum and the blank sample (Fig. 4B, black arrow). The normalization essentially set the background impedance change of the blank samples to zero. It is clear in Fig. 4C that positive and negative serum could be significantly distinguished from each other after normalization.
Fig. 4.
The effect of the impedance measuring buffer. (A) The absolute impedance magnitude over frequencies from 1 kHz–1000 kHz of a typical EIS response before (Serum, red curve) and after reacting with secondary Ab-AuNP conjugates (AuNP, blue curve) for a COVID-19 positive serum sample. EIS was measured in the ELISA buffer. Gray arrows mark the frequency range where impedance is dominated by Rsol. (B) %ΔZ caused by the binding of Ab-AuNP conjugates over all frequencies tested when EIS responses were measured in the ELISA buffer. %ΔZ=(ZAuNP-Zserum)/Zserum*100%. (C) %ΔZ calculated at 1 kHz from (A) for two positive (Pos#9, Pos#10) and two negative (Neg#8, Neg#9) sera. Error bars represent standard deviation. Statistics is determined by one-way ANOVA with Tukey's multiple comparisons test. *p<0.05. **p<0.01. (D-G) AFM characterization of the cleaned gap surface (D), the sample before applying the serum (E), the samples after applying negative serum and Ab-AuNP conjugates (F), and the sample after applying positive serum and Ab-AuNP conjugates (G). The gap area between the electrode digits were outlined by white dotted line.
In addition, surface roughness characterization via atom force microscopy (AFM) confirmed that the non-specific binding from the negative serum was negligible. In more details, Fig. 4E revealed the status of the blocking sample, that is, before serum and AuNP incubation. Applying the negative serum and subsequently AuNPs did not substantially increase the surface roughness (Fig. 4F). On the contrary, positive serum surface showed more uniformly elevated morphology (Fig. 4G) compared to both the negative sample and the sample after blocking, confirming the binding of anti-S Abs and, in turn, anti-human IgG modified AuNPs to the sensing area. As a proof of concept, adjusting the protocol by employing ELISA buffer for both washing and EIS measurement resulted in a consistent and unambiguous distinction between the positive and negative serum (Fig. 4C). A cut-off for qualitative determination of positive and negative results can be calculated at the mean of normalized %ΔZ for all negative samples adding two SDs, which comes at 2.46% (Fig. 4C, dotted red line). In the future, with test data on a larger scale of clinical samples, a more accurate cut-off could be realized.
3.6. DEP facilitated one-step detection of COVID-19 antibodies
Human serum is a complex mixture of biological components which often cause the fouling phenomenon on the sensing surface due to the non-specific binding of irrelevant molecules. Therefore, we had to employ the secondary Abs conjugated AuNPs for further discrimination of target analytes (Abs against SARS-CoV-2 S protein). However, if we could achieve AuNP-free distinguishing of sera that are positive or negative for COVID-19 Abs, it will largely simplify and accelerate the detection process. Our pilot attempt was to use DEP force to selectively move and concentrate the target molecule to the sensing area (the gap of the IDE chip) against the irrelevant serum components (Fig. 1C). DEP is the movement of dielectric particles in the presence of an inhomogeneous electric field (for instance, by applying an AC voltage). The applied frequency and AC voltage is expressed as f and Vpp (peak to peak voltage), respectively. DEP has been used to directionally and selectively move nanomolecules like DNA and proteins according to the dielectric property of each molecule (Green et al., 1997; Kim et al., 2016; Mahshid et al., 2018).
A previous study reported the preconcentration of the amyloid beta 42 protein (4.5 kDa) and prostate specific antigen (34 kDa) to the gap area of the IDE sensor via DEP force by applying AC voltage between the IDE sensor's electrodes at 50 MHz (f), 0.5 Vpp and 50 MHz(f), 0.02 Vpp, respectively. In more detail, when the sample was incubated with the sensor, the suspended target protein was affected by the DEP force and moved towards the electrode gap to react more effectively with the biorecognition element in the gap. This manipulation has achieved a 2-fold increase of sensitivity for protein detection (Kim et al., 2016).
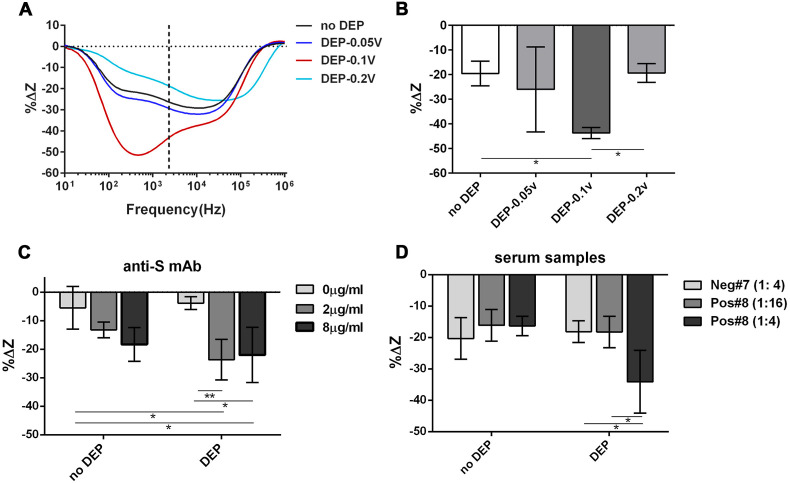
Similarly, we can apply DEP force to our target molecule, the IgG isotype of Abs, by simply connecting our chip to a function generator during serum incubation and configure it back to the detection mode by reconnecting to the impedance analyzer. In this way, we manipulated IgGs (with a molecular weight of 150 kD) by AC voltage at 50 MHz (f) and at 0.05, 0.1 and 0.2 Vpp, respectively. For proof of concept, the gap area was surface functionalized with total human IgGs and the anti-human IgG Abs (IgG isotype, at 1 μg/mL) were allowed to react for 30 min with or without the DEP force. After washing, the %ΔZ after the immunoreaction was computed at the transition frequency (Fig. 5 A, black dotted line). The binding of Abs resulted in a 19.5% ± 1.8% decrease of impedance without DEP force, while applying DEP at 50 MHz, 0.1Vpp gave twice as much of impedance change (%ΔZ = −43.7% ± 3.11%) (Fig. 5B). Hence, we concluded that 50 MHz, 0.1Vpp has realized preconcentration of the IgG isotype of Abs to the sensor gap.
Fig. 5.
Ab detection with the assistance of DEP. (A-B) optimization of DEP conditions by calculating %ΔZ between the Ab binding step and its previous step (blocking) over all frequencies measured. %ΔZ = (ZSerum-Zblocking)/Zblocking*100% where Zserum and Zblock represent the total impedance after the serum step and its previous step (blocking), respectively. Four DEP conditions were tested: no DEP, DEP by applying AC voltage at 50 MHZ (f) with 0.05Vpp, 0.1Vpp and 0.2Vpp, respectively. The immunoreaction between total human IgG and anti-human IgG Abs (at 1 μg/ml) was used as a test. (B) %ΔZ calculated at the transition frequency for all conditions in (A). Statistics is determined by one-way ANOVA with Tukey's multiple comparisons test. *p<0.05. (C-D) The effect of DEP force induced by AC voltage at 50 MHZ, 0.1Vpp on the detection of COVID-19 mAbs (C) or serum samples with different dilutions (D) by measuring %ΔZ at the transition frequency. The IDE was treated with S protein for COVID-19 Ab detection. One positive (Pos#8) and one negative (Neg#7) serum were used. (1:4): serum was diluted four times. (1:16): serum was diluted 16 times. Error bars represent the standard deviation of three replicates. Statistics is determined by two-way ANOVA with Tukey's multiple comparisons test. *p<0.05, **p<0.01.
All IgG molecules are very similar in structure and size, hence the COVID-19 Abs (IgG isotype) probably have the same dielectric properties as the anti-human IgG Abs (Green et al., 1997). We, therefore, tested the effect of DEP force on the detection of the anti-S1 mAb via applying the same AC voltage at 50 MHz, 0.1Vpp. In this case, the sensor is functionalized with S antigen. Anti-human IgG Abs at 2 μg/mL was used as a negative control, representing 0 ng/mL of anti-S1 mAb (the background change). Fig. 5C demonstrated that the impedance changes after incubation with 2 μg/mL or 8 μg/mL of the anti-S1 mAb had no significant difference from that of the background change without DEP force applied. When DEP force was applied, %ΔZ caused by 2 μg/mL of anti-S1 mAb rose significantly above the background level (p < 0.01) (Fig. 5C). However, the beneficial effect from DEP plateaued at 8 μg/mL of anti-S1 mAb since there was no obvious difference from 2 μg/mL under DEP conditions. This is probably because that the S antigen on the sensing surface was almost all occupied in these situations. Based on these results, we believe that DEP can indeed improve the performance of the sensor.
Lastly, we tested the effect of DEP on discrimination of human sera, positive or negative for COVID-19 Abs. It is clear from Fig. 5D that without DEP force, the positive and negative serum could not be distinguished from each other based on %ΔZ at the serum step. Moreover, positive serum diluted either 16 times or 4 times (1:16 vs 1:4) also had no significant difference (Fig. 5D). After applying DEP, however, the positive and negative serum could be separated based on %ΔZ when both of them are diluted four times (Fig. 5D, p < 0.05). This result shows the possibility that our sensor could differentiate positive and negative sera within half an hour.
4. Conclusion
In this work, an impedimetric biosensor device was proposed for qualitative determination of COVID-19 Abs in serum samples by using transducers with gold IDE patterned on the SiO2 substrate and with SARS-Cov-2 Spike (S) protein functionalized in between the electrode digits. This allows specific interaction between the S protein and anti-S Abs in the COVID-19 Ab-containing samples. The readout of results was based on impedance change via EIS analysis. We used two approaches to address the insensitivity of non-faradic impedimetric sensors and the fouling phenomenon when using serum samples. One approach used secondary Ab-AuNP conjugates to further distinguish anti-S Abs from the non-specific binding of other serum components and to obtain a more significant impedance change. The LoD for the AuNP strategy reached 200 ng/mL when using purified Abs in the buffer solution. The other strategy used DEP force to preconcentrate Abs to the gap area against non-relevant serum components. Despite having a poorer LoD than the AuNP strategy of only 2 μg/mL, this strategy achieved rapid one-step detection within half an hour. To date, only a couple of reports in the literature have described the POC devices for COVID-19 serology tests(Elledge et al., 2021; Heggestad et al., 2021; Tan et al., 2020) and our work seems to be the first report of using electrochemistry detection under the assistance of DEP force.
IgM and IgA are two other types of Abs that have diagnostic values for COVID-19, with IgM being the first Abs to emerge in response to pathogen contact (Isho et al., 2020; K. K. To et al., 2020). Our AuNP strategy can be quickly adapted for COVID-19 IgM and IgA detection by simply synthesizing anti-human IgM/IgA-AuNP conjugates using our established method. Alternatively, our sensor could measure all Ab isotypes without discrimination among IgG, IgM, and IgA by conjugating AuNPs with a specific in-house made anti-S monoclonal Ab to compete with anti-S Abs in the serum. However, for the DEP approach, fundamental studies need to be carried out on the suitable Vpp and frequency for moving the IgM or IgA Abs within the IDE sensor since IgM or IgA has significant differences from IgG in structure and size. Our work seems to open doors for many other COVID-19 related research topics.
Our ultimate goal is to quantitatively report SARS-CoV-2 Ab titers induced by vaccination or natural infections using our biosensor. To establish a real quantitative assay, we will need to obtain the WHO international standard for anti-SARS-CoV-2 immunoglobulin, which is pooled plasma obtained from eleven individuals recovered from the infection. This standard sample has an arbitrary unitage of 1000 binding antibody units (BAU)/mL(Kristiansen et al., 2021). We then can measure %ΔZ for the serial diluted standard samples. After normalizing readings to that of the blank (measuring buffer), we could establish an accurate calibration curve that correlates the normalized %ΔZ to Ab concentrations (expressed in BAU/mL). Moreover, we could scale up the testing capacity by having more IEDs per sensor chip, which could simply be achieved by using a new mask during nanofabrication. Testing many samples simultaneously can help to reduce the cost per sample. Lastly, engineering-wise, we are currently developing customized circuitries to replace the expensive signal generator and impedance analyzer so that they could be assembled with our sensor into a relatively low-cost POC device. Hence, our work, especially the DEP aspect, represents a novel avenue toward rapid, POC solutions for COVID-19 serology tests.
CRediT authorship contribution statement
Jie Zeng: Conceptualization, Methodology, Investigation, Data curation, Formal analysis, Visualization, Writing – original draft. Pedro A. Duarte: Conceptualization, Methodology, Formal analysis, Writing – review & editing. Yuhao Ma: Methodology, Investigation, Visualization. Oleksandra Savchenko: Methodology, Investigation. Lian Shoute: Methodology, Investigation. Yeganeh Khaniani: Methodology, Investigation, Visualization. Shawn Babiuk: Resources, Writing – review & editing. Ran Zhuo: Resources, Writing – review & editing. Gaser N. Abdelrasoul: Methodology, Investigation. Carmen Charlton: Resources. Jamil N. Kanji: Resources, Writing – review & editing. Lorne Babiuk: Resources. Cole Edward: Resources. Jie Chen: Funding acquisition, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The co-authors would like to thank the funding support from Alberta Innovates and the MITACS grant with Hidaca Ltd. Special thanks go to Dr. Shihong Xu and Shiau-Yin Wu for technical supports in FESEM imaging.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114476.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abdelrasoul G.N., Anwar A., MacKay S., Tamura M., Shah M.A., Khasa D.P.…Chen J. DNA aptamer-based non-faradaic impedance biosensor for detecting E. coli. Anal. Chim. Acta. 2020;1107:135–144. doi: 10.1016/j.aca.2020.02.004. [DOI] [PubMed] [Google Scholar]
- Abdelrasoul G.N., MacKay S., Salim S.Y., Ismond K.P., Tamura M., Khalifa C.…Khadaroo R.G. Non-invasive point-of-care device to diagnose acute mesenteric ischemia. ACS Sens. 2018;3(11):2296–2302. doi: 10.1021/acssensors.8b00558. [DOI] [PubMed] [Google Scholar]
- Ahmed S.R., Kang S.W., Oh S., Lee J., Neethirajan S. Chiral zirconium quantum dots: a new class of nanocrystals for optical detection of coronavirus. Heliyon. 2018;4(8) doi: 10.1016/j.heliyon.2018.e00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.R., Nagy É., Neethirajan S. Self-assembled star-shaped chiroplasmonic gold nanoparticles for an ultrasensitive chiro-immunosensor for viruses. RSC Adv. 2017;7(65):40849–40857. doi: 10.1039/C7RA07175B. [DOI] [Google Scholar]
- Alejo J.L., Mitchell J., Chang A., Chiang T.P.Y., Massie A.B., Segev D.L., Makary M.A. Prevalence and durability of SARS-CoV-2 antibodies among unvaccinated US adults by history of COVID-19. JAMA. 2022;327(11):1085–1087. doi: 10.1001/jama.2022.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M.…Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Ajmal M., Ashraf G., Muhammad N., Aziz A., Iftikhar T.…Liu H. The role of biosensors in COVID-19 outbreak. Curr. Opin. Electrochem. 2020 doi: 10.1016/j.coelec.2020.1008.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortz R.H., 3rd, Florez C., Laudermilch E., Wirchnianski A.S., Lasso G., Malonis R.J.…Chandran K. Single-dilution COVID-19 antibody test with qualitative and quantitative readouts. mSphere. 2021;6(2) doi: 10.1128/mSphere.00224-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral-Miranda G., Cardoso A.R., Ferreira L.C.S., Sales M.G.F., Bachmann M.F. Biosensor-based selective detection of Zika virus specific antibodies in infected individuals. Biosens. Bioelectron. 2018;113:101–107. doi: 10.1016/j.bios.2018.04.058. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y.…Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit C.K., Vashist S.K., MacCraith B.D., O'Kennedy R. Multisubstrate-compatible ELISA procedures for rapid and high-sensitivity immunoassays. Nat. Protoc. 2011;6(4):439–445. doi: 10.1038/nprot.2011.304. [DOI] [PubMed] [Google Scholar]
- Elledge S.K., Zhou X.X., Byrnes J.R., Martinko A.J., Lui I., Pance K.…Wells J.A. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 2021;39(8):928–935. doi: 10.1038/s41587-021-00878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eythorsson E., Helgason D., Ingvarsson R.F., Bjornsson H.K., Olafsdottir L.B., Bjarnadottir V.…Palsson R. Clinical spectrum of coronavirus disease 2019 in Iceland: population based cohort study. BMJ. 2020;371 doi: 10.1136/bmj.m4529. m4529-m4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Quintela A., Alende R., Gude F., Campos J., Rey J., Meijide L.M.…Vidal C. Serum levels of immunoglobulins (IgG, IgA, IgM) in a general adult population and their relationship with alcohol consumption, smoking and common metabolic abnormalities. Clin. Exp. Immunol. 2008;151(1):42–50. doi: 10.1111/j.1365-2249.2007.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N.G., Morgan H., Milner J.J. Manipulation and trapping of sub-micron bioparticles using dielectrophoresis. J. Biochem. Biophys. Methods. 1997;35(2):89–102. doi: 10.1016/S0165-022x(97)00033-X. [DOI] [PubMed] [Google Scholar]
- Grieshaber D., MacKenzie R., Vörös J., Reimhult E. Electrochemical biosensors - sensor principles and architectures. Sensors. 2008;8(3):1400–1458. doi: 10.3390/s80314000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiseppi-Elie A., Lingerfelt L. In: Immobilisation of DNA on Chips I. Wittmann C., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. Impedimetric detection of DNA hybridization: towards near-patient DNA diagnostics; pp. 161–186. [Google Scholar]
- Heggestad J.T., Kinnamon D.S., Olson L.B., Liu J., Kelly G., Wall S.A.…Chilkoti A. Multiplexed, quantitative serological profiling of COVID-19 from blood by a point-of-care test. Sci. Adv. 2021;7(26) doi: 10.1126/sciadv.abg4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.…Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infantino M., Pieri M., Nuccetelli M., Grossi V., Lari B., Tomassetti F.…Bernardini S. The WHO International Standard for COVID-19 serological tests: towards harmonization of anti-spike assays. Int. Immunopharm. 2021;100 doi: 10.1016/j.intimp.2021.108095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H.…Gingras A.C. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5(52) doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D.…Charles R.C. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020;5(52) doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiew L.V., Chang C.Y., Huang S.Y., Wang P.W., Heh C.H., Liu C.T.…Chang C.C. Development of flexible electrochemical impedance spectroscopy-based biosensing platform for rapid screening of SARS-CoV-2 inhibitors. Biosens. Bioelectron. 2021;183 doi: 10.1016/j.bios.2021.113213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim J., Yoo Y.K., Lee J.H., Park J.H., Hwang K.S. Sensitivity improvement of an electrical sensor achieved by control of biomolecules based on the negative dielectrophoretic force. Biosens. Bioelectron. 2016;85:977–985. doi: 10.1016/j.bios.2016.06.081. [DOI] [PubMed] [Google Scholar]
- Kristiansen P.A., Page M., Bernasconi V., Mattiuzzo G., Dull P., Makar K.…Knezevic I. WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet. 2021;397(10282):1347–1348. doi: 10.1016/S0140-6736(21)00527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski A.J., Klepac P., Conlan A.J.K., Kissler S.M., Tang M.L., Fry H.…Simons D. Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect. Dis. 2020;20(10):1151–1160. doi: 10.1016/S1473-3099(20)30457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186(4):224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W.…Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay S., Abdelrasoul G.N., Tamura M., Lin D.H., Yan Z.M., Chen J. Using impedance measurements to characterize surface modified with gold nanoparticles. Sensors. 2017;17(9) doi: 10.3390/s17092141. ARTN 2141, 10.3390/s17092141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahari S., Roberts A., Shahdeo D., Gandhi S. 2020. eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 Antigen, a Spike Protein Domain 1 of SARS-CoV-2. bioRxiv, 2020.2004.2024. [DOI] [Google Scholar]
- Mahshid S., Lu J., Abidi A.A., Sladek R., Reisner W.W., Ahamed M.J. Transverse dielectrophoretic-based DNA nanoscale confinement. Sci. Rep. 2018;8(1):5981. doi: 10.1038/s41598-018-24132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C.K., Cheng P.K.C., Lau S.S.Y., Wong K.K.Y., Lau C.S., Lam E.T.K.…Tsang D.N.C. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky V.M., Riepl M., Wolfbeis O.S. Capacitive monitoring of protein immobilization and antigen–antibody reactions on monomolecular alkylthiol films on gold electrodes. Biosens. Bioelectron. 1997;12(9):977–989. doi: 10.1016/S0956-5663(97)00053-5. [DOI] [PubMed] [Google Scholar]
- Muñoz J., Montes R., Baeza M. Trends in electrochemical impedance spectroscopy involving nanocomposite transducers: characterization, architecture surface and bio-sensing. TrAC, Trends Anal. Chem. 2017;97:201–215. doi: 10.1016/j.trac.2017.08.012. [DOI] [Google Scholar]
- Perez-Saez, J., Lauer, S. A., Kaiser, L., Regard, S., Delaporte, E., Guessous, I., . . . Zeballos Valle, A. Serology-informed estimates of SARS-CoV-2 infection fatality risk in Geneva, Switzerland. Lancet Infect. Dis. doi:10.1016/S1473-3099(20)30584-3. [DOI] [PMC free article] [PubMed]
- Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N.…Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112709. 112709-112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., the Northwell C.-R.C. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA. 2020;323(20):2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Consortium a. t.N.C.-R. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA. 2020;323(20):2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C., Jo S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011;86(12):1475–1479. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J.…Boyd S.D. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5(54) doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildgen V., Demuth S., Lüsebrink J., Schildgen O. 2020. Limits and Opportunities of SARS-CoV-2 Antigen Rapid Tests – an Experience Based Perspective. medRxiv, 2020.2009.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B.…Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shoute L.C.T., Anwar A., MacKay S., Abdelrasoul G.N., Lin D., Yan Z.…Li X.S. Immuno-impedimetric biosensor for onsite monitoring of ascospores and forecasting of sclerotinia stem rot of canola. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-30167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares J.C., Soares A.C., Angelim M.K.S.C., Proença-Modena J.L., Moraes-Vieira P.M., Mattoso L.H.C., Oliveira O.N., Jr. Diagnostics of SARS-CoV-2 infection using electrical impedance spectroscopy with an immunosensor to detect the spike protein. Talanta. 2022;239 doi: 10.1016/j.talanta.2021.123076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A.…Krammer F. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020;57(1) doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., He W.-T., Wang L., Lai A., Ji X., Zhai X.…Su S. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol. Med. 2020;26(5):483–495. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Krel M., Dolgov E., Park S., Li X., Wu W.…Fan X. Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C.…Yuen K.-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C.…Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel T., Legarraga P., Iruretagoyena M., Pizarro G., Vollrath V., Araos R.…Porte L. 2020. Head-to-head Comparison of Four Antigen-Based Rapid Detection Tests for the Diagnosis of SARS-CoV-2 in Respiratory Samples. bioRxiv, 2020.2005.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S.E., Shurin G.V., Yost M., Anderson A., Pinto L., Wells A., Shurin M.R. Differential antibody response to mRNA COVID-19 vaccines in healthy subjects. Microbiol. Spectr. 2021;9(1) doi: 10.1128/Spectrum.00341-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi H., Mahmud A., Chang D.R., Das J., Gomis S., Chen J.B.…Kelley S.O. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. 2021;143(4):1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L.…Li C.P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021:327. doi: 10.1016/j.snb.2020.128899. ARTN 128899, 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z.…Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W.…Shi Z.-L. Addendum: a pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;588(7836) doi: 10.1038/s41586-020-2951-z. E6-E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.