Abstract
Viruses are the causing agents for many relevant diseases, including influenza, Ebola, HIV/AIDS, and COVID-19. Its rapid replication and high transmissibility can lead to serious consequences not only to the individual but also to collective health, causing deep economic impacts. In this scenario, diagnosis tools are of significant importance, allowing the rapid, precise, and low-cost testing of a substantial number of individuals. Currently, PCR-based techniques are the gold standard for the diagnosis of viral diseases. Although these allow the diagnosis of different illnesses with high precision, they still present significant drawbacks. Their main disadvantages include long periods for obtaining results and the need for specialized professionals and equipment, requiring the tests to be performed in research centers. In this scenario, biosensors have been presented as promising alternatives for the rapid, precise, low-cost, and on-site diagnosis of viral diseases. This critical review article describes the advancements achieved in the last five years regarding electrochemical biosensors for the diagnosis of viral infections. First, genosensors and aptasensors for the detection of virus and the diagnosis of viral diseases are presented in detail regarding probe immobilization approaches, detection methods (label-free and sandwich), and amplification strategies. Following, immunosensors are highlighted, including many different construction strategies such as label-free, sandwich, competitive, and lateral-flow assays. Then, biosensors for the detection of viral-diseases-related biomarkers are presented and discussed, as well as point of care systems and their advantages when compared to traditional techniques. Last, the difficulties of commercializing electrochemical devices are critically discussed in conjunction with future trends such as lab-on-a-chip and flexible sensors.
Keywords: Biosensors, Viral diseases, Genosensor, Electrochemistry, Immunosensor, Biomarkers
Graphical abstract
1. Introduction
Viruses are responsible for causing many different diseases which lead to hundreds of thousands of deaths every year [1], including influenza [2], Ebola [3], MERS (Middle East respiratory syndrome) [4], HIV/AIDS (human immunodeficiency virus/Acquired immunodeficiency syndrome) [5] and, more recently, COVID-19 (coronavirus disease 2019) [6]. These are one of the smallest infectious agents known to men, presenting only a few numbers of genes usually encased in a proteic capsid and a lipidic envelope [7]. The translation of such genes occurs only by the machinery of the host cell, which is used by the intercellular parasite to synthesize millions of new viral particles [8] which will act similarly, causing harsh consequences quickly [9]. In this scenario, the rapid and precise diagnosis of viral diseases is of extreme importance to improve clinical outcomes, allowing doctors to take adequate measures.
Recently, the deep global health and economic impact caused by the pandemic of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) has also raised great attention to the need for low-cost diagnosis of viral diseases and the preparation for future outbreaks. Besides being able to mutate quickly and to recombine genetic material [10], viruses can present excellent resistance and high transmissibility [11], increasing the chance of a pandemic, especially in a globalized world. Future pandemic events caused by viruses are not only possible but also probable, requiring preparation from the scientific community, governments, and the general population [12]. In this context, the rapid diagnosis of novel viral infections (e.g. SARS-CoV-2) plays an essential role in its adequate control, allowing the first isolation of patients and preventing its incontrollable spreading.
Currently, the gold-standard techniques applied for the diagnosis of viral diseases are PCR (polymerase chain reaction) and ELISA (enzyme-linked immunosorbent assay). These present adequate limit of detection (LOD) and precision but require specialized reagents, personnel, and facilities to be performed, presenting low accessibility and delayed results. Therefore, the development of new and improved strategies able to perform the rapid, simple, and in situ diagnosis of viral diseases with low-costs is of great interest. Such tools would allow results to be obtained in a few minutes in a decentralized fashion, possibly dramatically increasing the number of tests that could be performed. Furthermore, even regions with low-resources and lacking specialized personnel would be able to perform the tests along the hospital beds.
This review article critically discusses new and improved electrochemical biosensors for efficient virus detection and the diagnosis of viral diseases published in the last five years. The precise diagnosis of COVID-19 (coronavirus disease 2019) is highlighted in a specialized section. Diverse construction techniques for building immunosensors and genosensors are covered, as well as the advantages of point of care (POC) devices when compared to traditional techniques. Furthermore, the challenges of commercializing electrochemical biosensors are discussed along with future tendencies on the field, including lab-on-a-chip and flexible devices. Other recent review papers on detection of viruses with extremely valuable content can be found in the literature, including the ones published by Ribeiro et al. [13], Mokhtarzadeh et al. [14], and Ji et al. [15]. This is, however, to the best of our knowledge, the most detailed review regarding current electrochemical biosensors for viral detection published in recent years.
2. Genosensors or DNA/RNA sensors for viral detection
Genosensors or DNA/RNA sensors are compact analytical devices frequently used to identify pathogens, such as bacteria and viruses, diseases, and/or predisposition for diseases through genetic material analysis. Genosensors use immobilized DNA/RNA probes as a recognition element, allowing specific hybridization reactions to occur usually by DNA–DNA or DNA–RNA molecular recognition [16,17]. These specific bindings provide high specificity and allow the direct analysis of complex samples [16,18]. Among the several advantages attributed to genosensors are the simplicity of construction and the fast results acquisition. Furthermore, these usually involve fewer measurement steps and use lower reagent amounts if compared to traditional techniques [19,20]. The many different genosensor types mainly differ regarding the transduction method, which is typically optical [21,22], piezoelectric [23,24], or electrochemical [25,26]. Electrochemical genosensors, in special, are one of the most commonly found in the literature. Some of the main strengths of these devices are their great miniaturization potential, high sensitivity, extremely low LODs, and usually not requiring complex sample pretreatment [27]. On the other hand, its main disadvantages include higher costs and greater instrumental complexity, especially if compared to similar colorimetric devices. It is important to emphasize, however, that unlike electrochemical biosensors, colorimetric devices usually present the limited potential for performing precise target quantification, which is important for indicating the severity of an infection, for example [28,29].
In recent years, advances in electrochemical genosensors can be categorized into three main topics: probe immobilization method, detection design, and signal amplification strategy. Thus, each of these points will be better explored next.
2.1. Methods for DNA immobilization on electrodes
In general, the development of electrochemical sensors is based on the premise that the proposed electrode presents one or more interesting physical-chemical properties, such as high electrical conductivity, high sensitivity, and selectivity or specificity to the desired analyte [30,31]. In this way, usually, the surface of an electric transducer is modified by the incorporation of synthetic or natural materials that promote the required properties, being called modified electrodes [32,33]. Regarding genosensors, it is not different. Indeed, the main concern is the probe immobilization, which should allow efficient hybridization while maintaining the desired electrical properties of the transducer [34]. The amount of probe immobilized on the genosensor surface (i.e., probe density) is directly related to the availability of analyte binding sites [35]. However, excessive probe density can lead to the saturation of the sensor surface and steric hindrance, significantly decreasing hybridization efficiency [36,37]. Therefore, the conditions and methods applied for probe immobilization are crucial to ensure the good performance of genosensors.
Among many different strategies, adsorption is the simplest way to incorporate genetic material into the transducer surface. Carbon nanomaterials are commonly employed for this purpose, such as graphene or carbon nanotubes (CNT), acting mainly through van der Waals interactions [25,38,39]. Although being the simplest strategy and not presenting limitations regarding the nucleic acids to be applied, this strategy is not widely used [16]. There are a few publications that use the adsorption strategy in biosensors development [[40], [41], [42], [43]] but, usually, strong and oriented interactions are preferred for probe immobilization [44], such as covalent bonds [[45], [46], [47]] and crosslinking [42,48,49] as these are commonly able to provide devices with a greater availability of analyte biding sites and more stability.
Regarding the use of covalent bonds for probe immobilization, thiol-Au interactions are one of the most used approaches [[50], [51], [52]]. Manzano et al. [52] have applied this technique to construct a genosensor for the sensitive detection of hepatitis A virus cDNA. Specific thiolated DNA probes were immobilized in the disposable screen-printed gold electrodes (SPAuE) by thiol-Au bonds. Upon hybridization with the target, changes in the oxidation potential of the indicator tripropylamine were observed due to the blocking effects for charge transfer and diffusion. The device was easy to use, provided rapid results, and was capable of achieving a limit of detection (LOD) of 6.94 fg μL−1 for viral cDNA, which is similar to the values obtained using nRT-PCR (Nested-reverse transcription-polymerase chain reaction) (6.4 fg μL–1). However, no evidence was provided regarding its direct application to real samples, which is essential to assure the biosensors functioning in a complex environment.
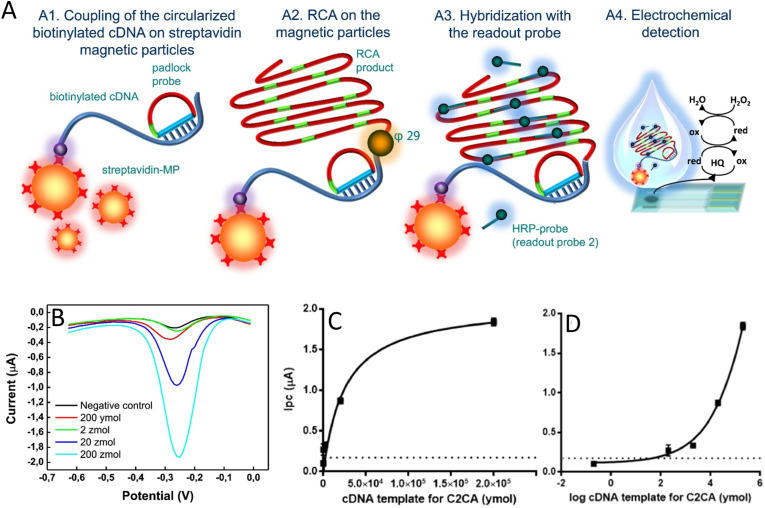
Probably, the interaction between avidin (or streptavidin) and biotin is one of the most used probe immobilization strategies, being characterized as a cooperative hydrogen bond (crosslinking) [34,48,53,54]. These interactions are highly strong and very close, promoting great immobilization stability [55]. Carinelli and coauthors used this strategy for the efficient Ebola virus cDNA quantification [56]. The detection strategy was based on the use of magnetic particles modified with streptavidin to bind to cDNA biotinylated probes. These, in turn, were amplified by rolling-circle-amplification (RCA). HRP (horseradish peroxidase) probes (readout probe) were then added to the system and hybridized to the amplified portion of the cDNA, allowing the electrochemical detection using the H2O2 voltammetric reduction by square-wave voltammetry (SWV). A step-by-step of the applied method is shown in Fig. 1 A. The high stability of the bond between streptavidin and biotin allowed the target amplification and the incorporation of several HRP-probes, significantly increasing the sensitivity of the developed device. The further reamplification of the target using the circle to circle amplification (C2CA) enabled the designed system to detect amounts of the target as low as 200 ymol (ca. 120 cDNA molecules) in less than 2.5 h (Fig. 1 B, C and D). Although the low LOD achieved by the developed strategy presents the potential to detect the disease in its early-stage (106 to 1010 RNA copies/mL) [57], no evidence of its application in real samples was provided due to the challenges involved in using Ebola-virus contaminated fluids. C2CA application in real samples was already proven possible by previous studies [58], but the complexity involved in the designed system brings important drawbacks for its practical use.
Fig. 1.
Basic device operation and results obtained by Carinelli et al. using RCA and C2CA combined with electrochemistry for the precise detection of Ebola virus cDNA. (A) Step-by-step schematic representation of the proposed genosensor. Step A1 - Coupling of the circularized biotinylated cDNA to streptavidin-magnetic particles; step A2 - amplification by RCA using ϕ29 DNA polymerase; step A3 - Hybridization step with a probe labeled with HRP and; step A4 - Electrochemical determination on SPCEs by SWV with an enzymatic reaction in the presence of hydroquinone as a mediator. (B) Square wave voltammograms for different concentrations of cDNA templates for C2CA (C) calibration curve obtained by one-site binding fitting and by (D) four-parameter logistic fitting. (Reprinted with permission from Carinelli et al. Yoctomole electrochemical genosensing of Ebola virus cDNA by rolling circle and circle to circle amplification. Biosens. and Bioelectron. 93, 65–71, © 2017 Elsevier BV [56]).
Some common crosslinkers are also widely explored to perform efficient and stable probe immobilization, including glutaraldehyde (GA), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS) [26,37,59]. For example, Farzin et al. [37] reported a device modified with an amine-ionic liquid functionalized reduced graphene oxide (rGO) for the detection of human papillomavirus (HPV)-16 cDNA (complementary DNA). The synthesized material was deposited on top of electrodes modified with multiwalled carbon nanotubes (MWCNTs), which was used for the covalent binding of aminated DNA probes with the use of GA. The authors then used the nanostructured platform for detecting a fragment of HPV-16 cDNA by differential pulse voltammetry (DPV). The response mechanism is label-free, using the anthraquinone-2-sulfonic acid monohydrate sodium salt as a signal probe. In this case, the salt was only incorporated into dsDNA (double-stranded DNA) after the hybridization of the probe with HPV-16 cDNA. The applied strategy provided the genosensor with high stability due to the existence of a strong and stable bond between the DNA fragment and the amine-ionic liquid functionalized rGO. With this strategy, the authors have obtained a LOD of 1.3 nmol L−1 for HPV-16 cDNA. To evaluate the applicability of the device to real samples, HN5 head and neck squamous carcinoma cell lines were spiked with target DNA (0–5.0 μM), showing recoveries ranging from 94 to 103%. It is important to notice, however, that the target strand is a small portion of the viral genetic material, being considerably shorter (17 pb) than the ones typically found in real, untreated samples.
2.2. Label-free and sandwich genosensors
DNA/RNA target determinations can be easily performed by the incorporation of a label sequence that presents a specific response, by using optical [21,60], fluorimetric [61,62], or electrochemical [63,64] detection systems. Regarding electrochemical genosensors, labels can be added to the capture probe or, more commonly, to a reporter probe. When the capture probe itself is labeled, the analytical response is usually based on the proximity of the label and the electrode, which varies in the presence of the analyte [65]. When a labeled reporter probe is used, on the other hand, a sandwich-like structure is formed with the capture probe and the desired target, and then the analytical response relies on the presence and on the amount of label itself [66,67].
Commonly, the use of labels can bring great advantages to the analytical performance of genosensors, including higher sensitivity and significantly lower LODs and LOQs. Furthermore, different labels can be applied for the efficient simultaneous or differential detection of analytes even in a complex biological matrix. The main challenge, in this case, is to improve the label immobilization on the genetic probe while still maintaining the accessibility to efficient hybridization [68]. The methods depend on the affinity of the label species for the biomolecule and must present great stability. Disadvantages of using sandwich-like strategies are usually related to higher analysis costs, more complex designs, and delayed results [68].
Recently, approaches based on simple construction strategies or that are capable of producing devices with lower financial resources have been attracting great attention from the scientific community. In this context, electrochemical viral determination using label-free devices has been widely explored [69,70], with a greater number of examples being found in the literature, if compared to sandwich-based (labeled) designs. Such determinations are usually based on the difference of charge transfer resistance when the genetic material is in its single- or double-stranded form. Although usually simple to use, label-free biosensors might present higher LODs than the similar labeled ones.
The label-free strategy was employed by Srisomwat et al. [71] in the development of a pop-up origami-based genosensor for label-free electrochemical detection of hepatitis B virus DNA. For that, the authors screen-printed a working electrode using graphene ink on one side of the paper device and modified the backside with specific pyrrolidinyl peptide nucleic acid by covalent binding. While unhybridized, the probe allows efficient electron transfer. After target hybridization, however, electron transfer resistance increases significantly, allowing its precise determination through current monitoring using DPV in the presence of an electrochemical probe (potassium hexacyanoferrate (III)/(II)). The simple methodology allowed the development of a low cost and sensitive device for the precise detection of hepatitis B virus DNA with a LOD of 1.45 pmol L−1. Although the applicability of the device in real scenarios was tested using samples prepared from plasmid constructs in concentrations typically found in serum of HBV patients (102 to 105 copies/μL), it is unclear if the device was tested in plasma itself – which may bring substantial challenges for the practical use of the biosensor.
Similarly, the decrease in electronic transfer capacity due to DNA target hybridization can be monitored through the direct measure of charge transfer resistance, which can be easily obtained using electrochemical impedance spectroscopy (EIS). This technique is probably the most common one for the construction of label-free genosensors [50,51,[72], [73], [74], [75]]. For example, Faria and Zucolotto [50] have constructed an impedimetric genosensor for the label-free detection of the Zika virus. For that, the authors used gold-based three-electrode disposable strips, and the determination of the target was performed by EIS. The SH-probe DNA oligonucleotide was immobilized by self-assembled monolayers (SAMs) on gold working electrodes and, afterward, incubated in the sample. The measurements were based on a decrease of charge transfer resistance after hybridization in the presence of a widely applied electrochemical probe (potassium hexacyanoferrate (III)/(II)). The proposed device displayed a great selectivity for Zika virus cDNA sequences in the presence of the non-complementary cDNA sequence for the dengue virus, and a LOD of 25 nmol L−1. Although being extremely simple to produce and presenting adequate analytical performance, the device was only tested in samples produced after PCR amplification, considerably restricting its point-of-care and rapid application. Furthermore, no real samples were used, and further studies must be performed in order to prove its clinical applicability.
Many other types of arrangements can be used in genosensors development [[76], [77], [78], [79]]. The use of nanomaterials or nanostructures, for example, can provide greater sensitivity to the developed devices [[80], [81], [82]]. In this case, the unique properties of nanomaterials can be extremely useful, which include their high electron mobility and specific surface area, and their electrical and thermal properties [83]. Furthermore, the possibility of tailoring nanostructures using different materials (e.g., Au, Ag, C, and Si), in different sizes and shapes enables the design of novel and enhanced biosensors. The improvement of the analytical performance of genosensors using nanomaterials is usually achieved by the modification of electrodes for obtaining high electrical conductivity, and to increase biomolecules loading, or through the use of labels that provide greater sensitivity [84].
Shariati et al. [82] developed an impedimetric genosensor for the simple detection of HPV DNA. A conventional three-electrode array was employed, with the working electrode being composed of gold nanotubes/nanoporous polycarbonate templates. Specific thiolated ssDNA was covalently immobilized into the pores of the working electrode. The identification of HPV was then performed through monitoring the charge transfer resistance before and after the hybridization of target sequences. The electrode structure allowed an extremely low LOD of 1.0 fmol L−1, which is considerably lower if compared to a similarly constructed device that does not contain nanostructures (LOD 25 nmol L−1) [50]. Besides, the genosensor was able to distinguish the target DNA from non-complementary oligonucleotides, including a sequence containing a single mismatch, showing the great potential of the genosensor to be applied in complex samples. No real samples, however, were used in the study, and a short DNA sequence (25 pb) was used for testing, which suggests the need for substancial studies prior to direct application in clinical samples.
In another example, Hu et al. [85] applied nanomaterials in the development of an electrochemical genosensor for a conserved fragment of the pol gene sequence of human immunodeficiency virus 1 (HIV-1) detection. The authors used a glassy carbon electrode modified with graphene sheets and decorated with “green”-synthesized gold nanoparticles. The specific DNA probes, on its turn, were immobilized on the modified working electrode through electrostatic interactions. The HIV-1 detection was performed with the EIS technique in the presence of potassium hexacyanoferrate (III)/(II). In this case, an increase in charge transfer resistance values was observed after the hybridization with target DNA and a LOD of 34 fmol/L−1 was obtained. Once more, the electrode modification with nanomaterials has led to an improvement in the genosensor performance with great simplicity even in label-free systems. As in previously mentioned examples, the device was not tested in any real samples, bringing doubts regarding its practical applications.
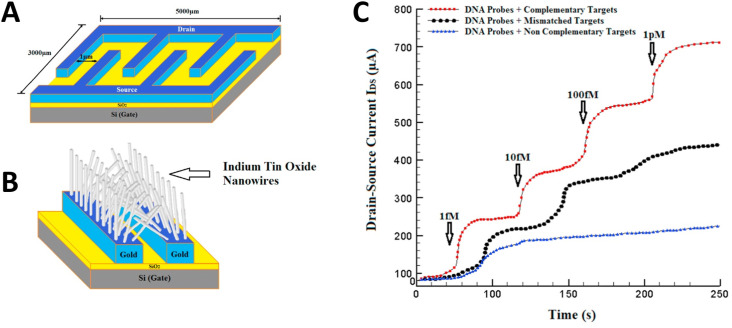
A strategy for determining hepatitis B virus using electrochemical genosensors has also been proposed by Shariati [68]. In this work, a field-effect transistor (FET) was constructed for the precise determination of the hepatitis B virus. The system was based on the synthesis of Au-decorated ITO nanowires on the FET system that allowed the simple attachment of the desired specific thiolated oligonucleotides (Fig. 2 A and B). The hepatitis B virus DNA determination was then performed by electrical transfer shifts caused by the negatively-charged complementary DNA (Fig. 2C). The proposed system using an electrochemical transducer presented a LOD of less than 1.0 fmol L−1 and displayed the ability to differentiate between target and mismatch sequences, which would be practically impossible to achieve using a colorimetric device, for example. Comparing this developed strategy with the one described by Srisomwat et al. [71] for the detection of the same virus (previously discussed), it is clear the device created by Shariati presents a more complex construction procedure, including E-beam lithography and the use of tubular furnaces, and relies on more expensive substrates, such as gold and ITO. However, the analytical performance of the biosensor described by Shariati is superior to the one achieved by Srisomwat, with a LOD 1000x lower. Therefore, this protocol may hold potential to improve clinical diagnosis, with further testing with biological samples being required to prove its practical use.
Fig. 2.
Strategy developed by Shariati et al. for the sensitive determination of hepatitis B virus. (A) Schematics showing the electrodes and Fet pattern; (B) Growth of ITO nanowires over Au patterns. C) Systems response upon the addition of different concentrations of complementary (red), mismatch (black) and non-complementary (blue) sequences. (Reprinted with permission from Shariati, M. et al. The field-effect transistor DNA biosensor based on ITO nanowires in label-free hepatitis B virus detecting compatible with CMOS technology. Biosens. and Bioelectron. 105, 58–64, © 2018 Elsevier BV [68]). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As previously stated, the use of labels in sandwich-like designs has the potential to improve the genosensors analytical performance, if compared to label-free similar assembles. An example was published by Alzate et al. [60], who constructed a genosensor for the selective detection of the Zika virus concomitantly in the presence of dengue and chikungunya homologous arboviruses. The detection of the genetic target was achieved using a biotin-labeled capture probe and the signal probe with the HPR label. A SPAuE was used as a transductor electrode for chronoamperometric measurements. The use of an enzymatic label, although adding complexity and increasing the costs of the tests, allows LODs in a pmol L−1 range to be achieved. In this case, a 1000 lower LOD was observed than a previously described label-free approach for the detection of the same virus (25 nmol L−1) [50]. The device was also tested in conditions close to clinical applications, with the biosensor being able to adequately detect and differenciate Zika virus synthetic genetic material from other arboviruses in spiked serum, urine, and saliva samples without the need of PCR amplification (0.5 nmol L−1). The device was also tested in cDNA and RNA from patients infected with Zika virus, being able to differenciate between control and positive samples.
In a different example, Cajugas and collegues [66] used the AuNP/DNA conjugate as a label for the ultrasensitive electrochemical detection of Zika virus genetic material. In this case, a capture probe specifically designed to interact with Zika virus DNA was immobilized at the surface of screen-printed electrodes either based on gold (SPAuE) or carbon (SPCE). Upon the contact of the genosensor with a positive sample, the hybridization occurs and the nanobioconjugate label (AuNP/DNA) becomes attached to the platform through the target sequence. The application of DPV measurements with Ru3+ as an electrochemical reporter allows the precise determination of Zika virus, with a linear signal being obtained in concentrations ranging from 10 to 600 fM of target for SPAuEs and from 500 fM to 10 pM for SPCEs. Genosensors based on SPAuEs achieved LODs as low as 0.2 fM, one of the lowest values reported in this review, and more than ten million times lower than the ones reported by a similar label free assay [50]. Furthermore, the device presented great potential for the simple diagnosis of the disease in situ, being able to detect Zika virus genetic material in real serum samples with no need for sample extraction or PCR amplification. Therefore, although being based on an analytical mechanism with additional elements and higher complexity (if compared to label-free systems), the developed device eliminates the need for amplification steps, significantly reducing the total analysis time and allowing the full diagnosis to be performed using a single equipment.
Chen et al. [67], on its turn, have developed a dual-probe sandwich-like electrochemical genosensor for the B/C genotyping of hepatitis B virus. In this case, the use of sequential hybridization steps in different temperatures was used to differentiate the virus genotypes while the use of reporter probes conjugated with HRP allowed the amperometric detection of both of the targets. This strategy, therefore, demands performing several hybridization steps and measurements for obtaining the final results, significantly increasing total analysis time and reducing its practical applications. To achieve fast responses, reporter probes labeled with nanomaterials presenting different properties (e.g., oxidation potentials) could be applied [86].
2.3. Signal amplification method
Although some electrochemical genosensors are currently able to detect viral genetic material in the same range of concentrations found in vivo [52,66,87], others still lack sensitivity for the direct analysis of biological fluids. In this regard, signal amplification strategies can be employed, aiming at amplifying the target itself or the signal magnitude generated by the label species [88].
PCR analysis and its variations are the most common amplification strategies for virus determination [[89], [90], [91]], are extremely sensitive and present high specificity [[92], [93], [94]]. These, however, require specialized instrumentation and trained personnel to be performed, which increases their cost and restricts the applications. Alternative methodologies that present higher simplicity and lower costs while exhibiting good sensitivity have been proposed in the literature [56,95]. An example can be found in the paper published by Ciftci et al. [95]. The authors developed a rapid and portable genosensor for the determination of the Ebola virus using magnetic beads and RCA as a signal amplification system. The use of an isothermal amplification strategy significantly increases the portability potential of the tests, allowing its execution in low-resource areas. The Ebola virus cDNA was amplificated by RCA on magnetic beads and further labeled with glucose oxidase enzyme (GOx). After GOx labeling, a chronoamperometric determination was performed using a Prussian blue (PB) modified screen-printed electrode (SPE) for hydrogen peroxide monitoring. The methodology proposed in this paper has proved to be fast and sensitive, allowing the Ebola virus cDNA determination in up to 1.0 pmol L−1 (approximately 6 × 108 targets/mL), which is in the concentration range for diagnosing the disease on its early-stage [57]. Furthermore, the device was tested using clinical samples with high viral loads and was able to differenciate between negative and Malaria-contaminated samples. Therefore, despite its increased complexity, this could be an excellent alternative for traditional methods in determining the pathogen in clinical samples, with important future steps being the validation of the assay using PCR and its comparison with the standard technique.
Another exciting strategy for genetic material amplification is the loop-mediated isothermal amplification (LAMP) technique [96,97]. LAMP uses four to six primers and the enzyme Bst DNA polymerase, which besides synthesis activity, allows the opening of the dsDNA [[96], [97], [98]]. This strategy allows the isothermal amplification of genetic fragments with high specificity, sensitivity, speed, and reduced cost, being of great interest for the detection of pathogens [[99], [100], [101], [102]]. An interesting combination of LAMP and electrochemical genosensors was described by Bartosik et al. [103] for the precise determination of HPV-16 and HPV-18 DNA fragments. In this paper, LAMP was applied to generate target amplicons containing digoxigenin-11-dUTP (DIG-dUTP), which were captured by magnetic particles conjugated to the desired specific DNA probes. Then, anti-DIG-HRP were applied to label the target and a magnetic pull-down was used to concentrate the particles on the surface of carbon-based working electrodes. Last, amperometric measurements were applied to precisely detect and quantify the desired target. The LAMP reaction required shorter amplification times than conventional PCR (∼2.5 h), and the LOD for electrochemical determination using LAMP amplification (0.1 ng) was seven times smaller than the one obtained with PCR (0.7 ng). It is important to mention that the developed technique displayed great performance for clinical applications, being validated with DNA isolated from cervical smears, not relying on thermocycling, and presenting the potential for the simultaneous analysis of different samples.
Table 1 shows a summary of genosensors for viral detection, including the immobilization and detection technique applied, sample type, application, and LOD.
Table 1.
Genosensors for virus detection.
| Viruses | Sensor | Sample type | Application | Immobilization | Label | Amplification | Technique | LOD | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Chikungunya virus | PDNA/Fe3O4@Au/ePAD | CHIK PDNA | Serum sample | Electrostatic interaction | Methylene blue | Nanomaterial | DPV | 0.1 nmol L−1 | [267] |
| Ebola | Au-SPE | EBOV DNA in PBS | – | crosslinking (streptavidin–biotin) | Label-free | – | DPV | 4.7 nmol L−1 | [268] |
| Hepatitis B | NPGE/MPA | HBV DNA | Blood sample | coordinate bond (thiol-gold) | Ferrocene | PCR | DPV | 0.8 μmol L−1 | [269] |
| Hepatitis B | ITO NWs | HBV DNA | – | – | Label-free | – | FET | 1 fmol L−1 | [68] |
| HIV1 | AuIL/PTCA/graphene/GCE | HIV DNA | – | coordination bonds | Label-free | – | EIS | 3.4 × 10−14 mol L−1 | [85] |
| HPV | AuNTs-PC electrode | HPV DNA16 | – | coordinate bond (thiol-gold) | Label-free | Electric field | EIS | 1 fmol L−1 | [82] |
| Zika | SPAuE | RNA (NS5 protein) | – | coordinate bond (thiol-gold) | Label-free | PCR | EIS | 25.0 ± 1.7 nmol L−1 | [50] |
| Hepatitis B | 3D microfluidic paper-based | HBV acpcPNA | – | Hydrogen bond | Label-Free | – | DPV | 1.45 pmol L−1 | [71] |
| Hepatitis B | sandwich-type DNA | HBV-DNA in human serum | Serum sample | host-guest (coordination bonds) | HP5–Au/CoS-aDNA | Nanocomposite material | Chronoamperometry | 0.32 fmol L−1 | [270] |
| HPV | GCE/MWCNT/NH2-IL-rGO | HPV DNA16 | HN5 head and neck squamous carcinoma cell line | coordination bonds | Label-free | Nanocomposite material | DPV | 1.3 nmol L−1 | [37] |
| Haemophilus influenza | NPs/Ag-DPA-GQDs/GCE | Haemophilus influenza genome | Human plasma samples | Covalent bond | TB | PCR | SWV | 1 zmol L−1 | [47] |
| Ebola | SPCE | EBOV cDNA in PBS | – | crosslinking (streptavidin–biotin) | HRP | RCA | SWV | 33 cDNA molecules | [56] |
| Ebola | SPE/PB | EBOV cDNA in PBS | Clinical samples | crosslinking (streptavidin–biotin) | GOx | RCA | Chronoamperometry | 100 fmol L−1 | [95] |
| Zika | SPAuEs | ZIKV – R2 | Serum, urine, and saliva samples | crosslinking (streptavidin–biotin) | HRP | RT-PCR | Chronoamperometry | 0.7 pmol L−1 | [60] |
| HPV | SPCE | HPV16 and HPV18 DNA | Clinical samples | crosslinking (streptavidin–biotin) | HRP | LAMP | Chronoamperometry | 0.1 ng | [103] |
PDNA/Fe3O4@Au/ePAD: gold nanoparticles associated with magnetic nanoparticles and CHIK PDNA paper analytical devices; CHIK PDNA:20-mer oligonucleotide sequence for chikungunya capture probe; ITO NWs: indium tin oxide nanowires electrode; FET: field effect transistor; H1N1: influenza A; mini-HA-GA-APTES-ITO: indium tin oxide-based electrode modified with aptamers, glutaraldehyde and mini-hemagglutinin; Au-SPE: gold screen-printed electrode; Cu3(PO4)2-BSA-GO-AuNP: Nanoflowers based on copper(II) sulfate pentahydrate, bovine serum albumin, graphite oxide and gold nanoparticles; NPGE/MPA: 3-mercaptopropionic acid self-assembled monolayer nanoporous gold electrode; GOx: Glucose Oxidase; RCA: Rolling Circle Amplification; SPE/PB: screen-printed electrode with Prussian blue; SPAuEs: screen-printed gold electrodes; HRP: horseradish peroxidase; HPV: human papillomaviruses; SPCE: screen-printed carbon electrode; LAMP: Loop mediated isothermal amplification; GCE/MWCNT/NH2-IL-rGO: glassy carbon electrode modified with multiwalled carbon nanotubes with reduced graphene oxide and an amine-ionic liquid functionalized; NPs/Ag-DPA-GQDs/GCE: glassy carbon electrode modified with silver nanoparticle-d-penicillamine functionalized graphene quantum dots; TB: toluidine blue; SWV: square-wave voltammetry; acpcPNA: Pyrrodinyl peptide nucleic acid; EIS: electrochemical impedance spectroscopy; AuNTs-PC: AuNTs decorated nanoporous polycarbonate; HIV1: human immunodeficiency virus 1; AuIL/PTCA/graphene/GCE: glassy carbon electrode modified with graphene sheets functionalized with 3,4,9,10-perylene tetracarboxylic acid with amine-terminated ionic liquid protected gold nanoparticles.
3. Aptamer-based biosensors for viral diseases diagnosis
Aptamers are single-stranded artificial nucleotides (DNA or RNA) capable of specifically binding to a target (e.g., drugs, proteins, viruses, whole cells). Usually, these are composed of a few dozen to hundreds of nucleotides and are produced by a technique named Systematic Evolution of Ligands by Exponential Enrichment (SELEX), performed by iterative cycles of binding, washing, and amplification [104]. In recent years, aptamers have drawn significant attention due to their low synthesis cost, easy modification (e.g., thiol, enzymatic labels), and high stability, being adequate bioreceptors for the construction of biosensors and increasing its commercial potential.
Currently, diverse important aptamer-based biosensors can be found in the literature for the detection of viruses. Considerations regarding its adequate construction methods and parameters can be considered to be parallel to the ones made for genosensors. Therefore, methods previously discussed in the genosensors section can be used for aptamer immobilization and labeling, for example. To illustrate, aptamer-based biosensors for aiding in the diagnosis of viral diseases will be presented and discussed.
Bhardwaj et al. [105] applied aptamers to develop a label-free electrochemical biosensor able to differentiate between subtypes of influenza A H1N1 virus. Specific influenza A mini-hemagglutinin protein was used for the production of aptamers with high binding affinities using SELEX, allowing the detection of the desired virus subtypes (seasonal and 2009 pandemic H1N1). The immobilization of these aptamers onto an ITO electrode allowed the detection of the desired targets with a LOD of 3.7 plaque-forming units mL−1. It is important to mention that the method used for immobilizing the aptamers on the surface of the electrodes was only based on electrostatic interactions, possibly limiting its use in complex samples and its clinical applications.
Bai et al. [106], on its turn, published an EIS aptamer-based biosensor for the detection of inactivated H1N1 virus. First, SELEX technique was applied to generate high binding affinity aptamers to the inactivated viruses. Then, the candidates were applied in a direct enzyme-linked oligonucleotide assay (ELONA), with the best ones being selected for a sandwich ELONA and the construction of the electrochemical aptasensor. While the sandwich-ELONA achieved a LOD of 0.3 ng μL−1, EIS was able to lower these values 300 × (0.9 pg μL−1) while presenting also great selectivity. Furthermore, the analytical performance of the aptasensor was assessed in low and high probe densities. While the best selectivity was displayed at the low probe density, extremely higher sensitivity was achieved at the high probe density. Regarding clinical analysis, it is important to mention that the developed method presents the advantage of not requiring sample pretreatment and shows greater safety as inactivated samples are applied in the analysis. However, no tests with real samples were performed in this work.
Lee et al. [107], on its turn, have introduced a multi-functional bioprobe, the DNA 3-way-junction (3 WJ), to detect the avian influenza virus (H5N1) in a rapid and simple manner. The DNA 3 WJ is composed of three independent, but connected sequences. The first one is an aptamer responsible for the recognition of H5N1 HA protein, while the second and third ones are an HRP-mimicked DNAzyme for generating the EC signal and a thiol group for the efficient immobilization of the structure, respectively. The 3 WJ was then immobilized into porous AuNPs present on the top of the working electrode and cyclic voltammetry was applied to detect the virus in buffer and chicken serum samples. The technique stands out for being both label-free and redox reporter-free, which simplifies its practical use. It is also important to mention that the developed technique can be extended for the detection of different targets and, with the use of different labels, a multiplexed system can be achieved. This is of great interest for practical applications, allowing the easy and quick differenciation between viral targets.
4. Immunosensors for viral diseases diagnosis
Immunosensors have been widely used for detecting different types of viruses, including coronaviruses, hepatitis B and C, influenza, dengue, chikungunya, Zika, Japanese encephalitis, measles, Newcastle disease, porcine epidemic diarrhea, rotavirus, enterovirus 71 and HIV, as summarized in Tables 2 and 3 . Immunosensors have been considered as a complementary method to the traditional reverse-transcription polymerase chain reaction (RT-PCR) protocols due to their high sensitivity and specificity. Also, the rapid diagnosis of viruses can be performed without the sample preparation steps required by the PCR-based tests, which may bring significant clinical advantages.
Table 2.
Label-free electrochemical immunosensor for virus detection.
| Virus | Target | Application | Electrode | Modifier | Biorecognition element | Immobilization | Technique | Redox probe | LOD | Linear range | Incubation time | Response time | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chikungunya virus | CHIKV-nsP3 antigena | Human serum samples | Au | SAM-MUA, MH | anti-CHIKV-nsP3 | Covalent, EDC/NHS coupling | EIS | [Fe(CN)6]3-/4- | 8 ng mL−1 | 25 ng m−1-1 μg mL−1 | 90 min | – | [144] |
| Dengue virus | DENV-NS1 antigena | Human serum samples | SPCE | BSA | anti-DENV-NS1 | Covalent, EDC/NHS coupling | EIS | [Fe(CN)6]3-/4- | 0.3 ng mL−1 | 1–200 ng mL−1 | 60 min | – | [271] |
| DENV-NS1 antigena | Human serum samples | ITO | SP:TMAP:PPD/AuNPs/PPD | anti-DENV-NS1 | Covalent, EDC/NHS coupling | DPV | [Fe(CN)6]3-/4- | 5 ng mL−1 | 5–4000 ng mL−1 | 45 min | – | [272] | |
| anti-DENV-NS1b | Bovine blood plasma samples | GCE | pp-NHS/CNT | DENV2- NS1 | Covalent, NHS coupling | EIS | [Fe(CN)6]3-/4- | 10−12 g mL−1 in PBS and plasma | 10−13-10−5 g mL−1 in PBS, 10−11-10−5 in plasma | 30 min | – | [273] | |
| DENV2-NS1 antigena | – | Au | SAM-MUA | DGV BP1 | Covalent, EDC/NHS coupling | SWV | [Fe(CN)6]3-/4- | 1.49 μg mL−1 | – | 60 min | – | [147] | |
| DENV-NS1 antigena | Human serum samples | SPCE | PS/DA | NS1 protein MIP | Molecular imprinting | EIS | [Fe(CN)6]3-/4- | 0.3 ng mL−1 | (1–200 ng mL−1 | – | – | [141] | |
| DENV-NS1 antigena | Human sera samples | ITO | MoS2/AuNP/ITO | anti-DENV-NS1 | Covalent, EDC/NHS coupling | EIS | [Fe(CN)6]3-/4- | 1.67 ng mL−1 in PBS, 1.19 ng mL−1 in human sera | 102–108 ng mL−1 | 25 min | – | [274] | |
| anti-DENV-NS1b | Clinical human serum samples | SPE | polymeric films from 4-APA | DENV-NS1 protein antigen | Adsorption | EIS | [Fe(CN)6]3-/4- | – | – | 20 min | – | [275] | |
| DENV-NS1 antigena | Commercial human serum | Au | SAM-PEG-thiol-11Fc | anti-DENV-NS1 | Covalent, EDC/NHS coupling | ECS/EIA | Ferrocene | ECS: 340 pg mL−1 in PBS and 1200 pg mL−1 in diluted serum EIA: 220 pg mL−1 in PBS and 611 pg mL−1 in diluted serum |
ECS: 1–5000 ng mL−1 in PBS and 5–1000 ng mL−1 in diluted serum EIA: 1–5000 ng mL−1 in PBS and 1–1000 ng mL−1 in diluted serum |
30 min | 4 min (ECS) <4s (EIA) |
[117] | |
| anti-DENV-NS1b | Commercial human serum | Au | SAM-PEG-thiol-11Fc | DENV-NS1 protein antigen | Covalent, EDC/NHS coupling | ECS/EIA | Ferrocene | ECS: 231 pg mL−1 in PBS and 6100 pg mL−1 in diluted serum EIA: 393 pg mL−1 in PBS and 9500 pg mL−1 in diluted serum |
ECS and EIA: 1-1000 mL−1 in PBS and, 10–1000 ng mL−1 in diluted serum | 30 min | 4 min (ECS) <4s (EIA) |
[117] | |
| DENV-NS1 antigena | PBS and neat serum biological samples | Au | SAM-16-MDHDA-11-FcC | anti-DENV-NS1 | Covalent, EDC/NHS coupling | ECS | Ferrocene | 0.2 ng mL−1 in PBS and 0.5 ng mL−1 in neat serum | 5–1000 ng mL−1 in PBS and neat serum | 30 min | – | [222] | |
| DENV-NS1 antigena | PBS and neat serum biological samples | Au | SAM-MUA, 6COH | anti-DENV-NS1 | Covalent, EDC/NHS coupling | EIS | [Fe(CN)6]3-/4- | 3 ng mL−1 in PBS and 30 ng mL−1 in neat serum | 10–2000 ng mL−1 in PBS and 10–1000 ng mL−1 in neat serum | 30 min | – | [222] | |
| DENV-NS1 antigena | Human serum samples | SPCE | GO-Ru(II)/CS | anti-DENV-NS1 | Protein-G affinity interaction | Amperometry | Ru(II) | 0.38 ng mL−1 | 1–10 ng mL−1 | 20 min | <1 min | [152] | |
| Hepatitis B virus | HBsAga | – | SPCE | BSA | anti-HBs | Covalent, EDC/NHS coupling | EIS | [Fe(CN)6]3-/4- | 2.1 ng mL−1 | 5–3000 ng mL−1 | 45 min | – | [142] |
| anti-HBcb | – | GCE | HAc–NH2-CNT | HBcAg | Covalent, EDC/NHS coupling | EIS | [Fe(CN)6]3-/4- | 0.03 ng mL−1 | 1–6 ng mL−1 | 20 min | – | [131] | |
| HBsAga | Human serum samples | Au | GO/Fc-CS/AuNPs | anti-HBs antibodies | Adsorption | DPV | Fc-CS | 0.1 ng mL−1 | 0.1 ng mL−1-350 ng mL−1 | 15 min | – | [153] | |
| HBsAga | Human serum samples | SPE | GO/Fe3O4/PB@AuNPs | anti-HBs antibodies | Adsorption | DPV | PB | 0.00016 ng mL−1 | 0.5 pg mL−1-200 ng mL−1 | 40 min | – | [151] | |
| anti-HBcb | Human serum samples | Au | PTy–COOH–CNT | HBcAg | Covalent, EDC/NHS coupling | SWV | PTy | 0.89 ng mL−1 | 1.0–5.0 ng mL−1 | 15 min | – | [155] | |
| Hepatitis C virus | HCV antigena | – | Glass substrate, WE and CE: Au; RE: Ag | AuNP/ZnONR | anti-HCV | Covalent, GA coupling | CV | [Fe(CN)6]3-/4- | 0.25 μg μL−1 | – | – | – | [143] |
| HCV antigena | Human serum samples | GCE | AgNPs/GQD-SH | anti-HCV | Adsorption | DPV | Riboflavin | 3 fg mL−1 | 0.05 pg mL−1-60 ng mL mL-1 | 30 min | – | [148] | |
| HCV antigena | Human serum samples | GCE | V2O5 nanobelts | anti-HCV | Adsorption | DPV | Riboflavin | 1.3 fg mL−1 | 10 fg mL−1-100 ng mL−1 | 35 min | – | [149] | |
| Human immune deficiency virus | HIV-p24a | Human serum samples | GCE | MIPs/MWCNTs | HIV-p24 MIP | Covalent, GA coupling | EIS | [Fe(CN)6]3-/4- | 0.083 pg cm−3 | 1.0 × 10−4-2 ng cm−3 | 10 min | – | [276] |
| Influenza A virus | H1N1 antigenc | Saliva samples | ITO/glass | TrGO | anti-H1N1 | Covalent, PBSE linker | EIS | [Fe(CN)6]3-/4- | 26.04 PFU mL−1 in PBS and 33.11 PFU mL−1 in saliva | – | – | – | [139] |
| HA proteins of H5N1 and H1N1 antigensa | – | Dual SPCE | GO-MB/CS | anti-H5N1-HA and anti-H1N1-HA | Protein-A affinity interaction | DPV | MB | 9.4 pmol L−1 H1 N1, 8.3 pmol L−1 H5N1 | 25–500 pmol L−1 | 30 min | <1 min | [150] | |
| H1N1 virus antigenc | – | Au microelectrode | CA/rGO | anti-H1N1 | Covalent, EDC/NHS coupling | Amperometry | [Fe(CN)6]3-/4- | 0.5 PFU mL−1 | 1-104 PFU mL−1 | Flow | Flow | [277] | |
| Japanese encephalitis virus | JEV antigenc | Human serum samples | SPCE | CNPs | anti-JEV | Covalent, EDC/NHS coupling | SWV | [Fe(CN)6]3-/4- | 2 ng mL−1 | 5–20 ng mL−1 | 20 min | – | [278] |
| JEV antigenc | Healthy mouse serum | Pt microelectrodes | PAni/MWCNTs | anti-JEV | Covalent, EDC/NHS coupling | Amperometry | PAni | 2 ng mL−1 | 2–250 ng mL−1 | 45 min | 13 s | [154] | |
| Measles virus | Anti-measles virusb | – | GCE | Unmodified GCE | measles virus antigen | Adsorption | LSV | [Fe(CN)6]3-/4- | 1.5 × 10−5 IU mL−−1 | 0.3–3 × 10−4 IU mL−1 | 20 min | [140] | |
| Anti-measles virusb | – | GCE | Functionalized GCE | Measles virus antigen | Covalent, GA coupling | LSV | [Fe(CN)6]3-/4- | 9.3 × 10–6 IU mL--1 | 0.3–3 × 10−5 IU mL−1 | 10 min | – | [140] | |
| Newcastle Disease Virus | NDV antigenc | – | Au | Protein A | IgY | Covalent, GA coupling | CV | [Fe(CN)6]3-/4- | 100.95 EID50 mL−1 | 106-102 EID50 mL−1 | 60 min | – | [145] |
| NDV antigenc | – | Au | SAM-TGA | IgY | Covalent, DDC/NHS coupling | CV | [Fe(CN)6]3-/4- | 100.67 EID50 mL−1 | 106-102 EID50 mL−1 | 60 min | – | [145] | |
| Porcine Epidemic Diarrhea Virus | PEDV antigenc | Pig manure samples | GCE | AuNP/MoS2/rGO | PEDV-2C11 antibodies | Adsorption | EIS | [Fe(CN)6]3-/4- | – | 82.5–1.65 × 104 TCID50 mL−1 | 140 min | – | [138] |
| Rotaviruses | rotaviruses (RVs), rotavirus antigenc | – | GCE | AuNPs/SAM-CA | Rotavirus antibodies | Covalent, GA coupling | EIS | [Fe(CN)6]3-/4- | 2.3 PFU mL−1 | 4.6–4.6 × 105 PFU mL−1 | 60 min | 55 min | [279] |
| Zika virus | ZIKV-NS1 antigena | Human urine samples | Au (WE, PCB) | ZnO nanostructures | anti-ZIKV-NS1 | Adsorption | CV | [Fe(CN)6]3-/4- | 1.00 pg mL−1 | 0.1–100 ng mL−1 | 60 min | – | [146] |
| ZIKV-envelop protein antigena | – | IDE-Au | SAM-DTSP | Zev-Abs | Adsorption | EIS | [Fe(CN)6]3-/4- | 10 pmol L−1 | 10 pmol L−1-1 nmol L−1 | 30 min | 40 min | [280] |
Type of target:a viral protein, b antibody, c whole virus. CHIKV-nsP3 antigen: chikungunya virus non-structural protein antigen, anti-CHIKV-nsP3: anti chikungunya virus non-structural protein antibody. SAM: self-assembled monolayer, MUA: 11-mercaptoundecanoic acid, MH: 6-mercapto-1-hexanol, EDC: 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, NHS: N-Hydroxysuccinimide, EIS: electrochemical impedance spectroscopy, DENV-NS1 antigen: Dengue virus non-structural 1 protein antigen, anti-DENV-NS1: anti Dengue virus non-structural 1 protein monoclonal antibody, SPCE: screen-printed carbon electrode, BSA: bovine serum albumin, ITO: indium tin oxide, SP: 4-sulfophenyl, TMAP: 4-trimethylammoniophenyl, PPD:1,4-phenylenediamine, AuNPs: gold nanoparticles, DPV: differential pulse voltammetry, DENV2- NS1: recombinant protein NS1 Dengue Virus Type-2, GCE: glassy carbono electrode, pp-NHS (N-hydroxysuccinimido 11-(pyrrol-1-yl) undecanoate), CNT (carbono nanotube), DGV BP1: promising recognition peptide for DENV-2 NS1, SWV: square wave voltammetry, PS: polysulfone nanofibers, DA: dopamine, MIP: molecularly imprinted polymers, 4-APA: 4-aminophenylacetic acid, PEG-thiol: HS-C11-EG3-OCH2-COOH, EG = ethylene glycol, 11Fc: 11-(ferrocenyl)-undecanethiol, ECS: Electrochemical capacitance spectroscopy, EIA: Electrochemical immittance analysis, 16-MHDA: 16-mercaptohexadecanoic acid, 6COH: 6-mercapto-1-hexanol, HBsAg: hepatitis B surface antigen, anti-HBs: anti hepatitis B surface antibody, anti-HBc: anti hepatitis B core protein antibody, HBcAg: hepatitis B core protein antigen, HAc: hyaluronic acid, NH2-CNT: amino multi-walled carbon nanotubes, PB: Prussian blue, PTy: polytyramine, COOH-CNT: carboxylated multi-walled carbon nanotubes, GA: glutaraldheide, HCV antigen: hepatitis C virus antigen, anti-HCV: anti hepatitis C virus antibody, ZnONR: ZnO nanorods, GQD-SH: thiol graphene quantum dots, HIV-p24 antigen: human immunodeficiency virus p24, MWCNTs: multi-walled carbon nanotubes, H1N1 antigen: influenza virus H1N1 antigen, anti-H1N1: anti influenza virus H1N1 antibody, TrGO: thermally-decomposed rGO, PBSE: 1-pyrenebutanoic acid succinimidyl ester, HA proteins of H5N1 and H1N1 antigens: recombinant hemagglutinin influenza A virus H5N1 and H1N1, anti-H5N1-HA and anti-H1N1-HA: anti influenza-A hemagglutinin monoclonal H5N1 and H1N1 antibodies, GO: graphene oxide, MB: methylene blue, CS: chiosan, CA: cystamine, rGO: reduced graphene oxide, JEV antigen, anti-JEV:, CNPs: carbono nanoparticles, PAni: polyaniline, TGA: thioglycolic acid, IgY: chicken egg yolk antibodies against Newcastle disease virus, PEDV antigen: porcine epidemic diarrhea virus antigen, anti-PEDV: anti porcine epidemic diarrhea virus antibody, IDE-Au: interdigitated micro-electrode of gold, ZIKV-NS1: Zika virus NS1 protein antigen, anti-ZIKV-NS1: anti-Zika virus NS1 protein antbody, PCB: Printed Circuit Board (PCB), DTSP: dithiobis(succinimidyl propionate), ZIKV-envelop protein antigen: Zika-virus envelop protein antigen, Zev-Abs: ZIKV-envelop protein antibody.
Table 3.
Sandwich-type electrochemical immunosensor for virus detection.
| Virus | Target | Application | Electrode | Modifier | Biorecognition element | Immobilization | Label | Technique | Redox probe | LOD | Linear range | Incubation time | Response time | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avian Leukosis Virus | ALV-J antigenc | Avian serum samples | GCE | GR-PTCA | Ab1: anti-ALV-J | Covalent, EDC/NHS coupling | Fc-AuNP-β-CD-Ab2 | DPV | Ferrocene | 101.93 TCID50 mL−1 | 102.0104.0 TCID50 mL−1 | 30 min | – | [156] |
| ALV-J antigenc | Avian serum samples | GCE | rGO-TA-Fe3O4 | Ab1: anti-ALV-J | Covalent, cis-diols in TA with saccharides on Ab1 | eZIF-Ab2-HRP | DPV | HRP catalyzed reduction of H2O2, HQ as redox mediator | 140 TCID50 mL−1 | 152-10000 TCID50 mL−1 | 60 min | – | [166] | |
| ALV-J antigenc | Avian serum samples | GCE | mpg-C3N4 | Ab1: anti-ALV-J | Covalent, GA coupling | Th-mpg-C3N4/Ab2 | DPV | Thionine | 120 TCID50 mL−1 | 102.08-104.0 TCID50 mL−1 | 40 min | – | [157] | |
| ALV-J antigenc | – | GCE | GR-PTCA | Ab1: anti-ALV-J | Covalent, EDC/NHS coupling | NC-Au-Ab2-ALP | DPV | ALP-catalyzed silver deposition | 101.98 TCID50 mL−1 | 102.08-104.0 TCID50 mL−1 | 40 min | – | [164] | |
| Enterovirus 71 | EV71c | Clinical samples | ITO | AuNPs/MA | mAb: anti-EV71 | Covalent, EDC/NHS coupling | MBs-mAb-HPR | Amperometry | HRP catalyzed reduction of H2O2, TBM as redox mediator | 0.1 ng mL−1 | 0.1–6000 ng mL−1 | 30 min | – | [165] |
| Hepatitis B virus | HBeAga | Human serum samples | GCE | pGO@Au | Ab1: anti-HBeAg | Adsorption | Ab2-Au@Pd/MoS2@MWCNTs | Amperometry | Catalytic reduction of H2O2 | 26 fg mL−1 | 0.1 fg mL−1-500 pg mL−1 | 30 min | – | [169] |
| HbsAga | Human serum samples | GCE | GO-CS | Ab1: Anti-HBsAg | Covalent, GA coupling | Ab2/Fe3O4-AuNPs- DNAzyme/MB | DPV | DNAzyme catalyzed reduction of H2O2, MB as redox mediator | 60 fg mL−1 | 0.1–300 pg mL−1 | 30 min | – | [167] | |
| HbsAga | Human serum samples | GCE | GO-CS | Ab1: Anti-HBsAg | Covalent, GA coupling | Ab2-DNAzyme-H-amino-rGO/Au | DPV | DNAzyme catalyzed reduction of H2O2, MB as redox mediator | 10 fg mL−1 | 0.1–1000 pg mL−1 | 30 min | – | [167] | |
| HbsAga | Human serum samples | Au | Fe3O4 | Ab1: Anti-HBsAg | Covalent, EDC/NHS coupling | Ab2-MB-Au-DNAzyme | SWV | DNAzyme catalyzed reduction of H2O2, MB as redox mediator | 0.19 pg mL−1 | 0.3–1000 pg mL−1 | 30 min | – | [168] | |
| HbsAga | Human serum samples | GCE | pGO/Au | Ab1: Anti-HBsAg | Adsorption | MoS2@Cu2O–Pt/Ab2 | Amperometry | Catalytic reduction of H2O2 | 0.15 pg mL−1 | 0.5 pg mL−1-200 ng mL−1 | 60 min | – | [170] | |
| Hepatitis C virus | HCV antigena | Human serum samples | SPCE | Gr-IL-Fu/RhNPs | Ab1: anti-HCV | Adsorption | CB/Ab2/Nafion@TiO2 | DPV | CB | 25 fg mL−1 | 0.1–250 pg mL−1 | 35 min | – | [158] |
| Influenza A | H5N1 antigenc | Chicken swab sample | Au | DTSP-ProteinA-anti-H5N1 | Anti-H5N1 | Protein-A affinity interaction | Con A-MNPs | CV | ECC of Fe3O4 to PB | 0.0022 HAU | 0.0025–0.16 HAU | 60 min | – | [171] |
| H7N9 antigenc | – | Au | AuNP-G | mAb: anti- H7N9 | C | pAb-AgNP-G | LSV | AgNPs | 1.6 pg mL−1 | 1.6 × 10−3 -16 ng mL−1 | 30 min | – | [160] | |
| H7N9 antigenc | Chicken serum and liver samples | AuMA | AuNPs/mAb | mAb: anti-H7N9 | Covalent, EDC coupling | pAb-bi-FMNs-ALP | LSV | ALP-catalyzed silver deposition | 7.8 fg mL−1 | 0.01–1.5 pg mL−1 | 30 min | – | [163] | |
| H9N2 antigenc | – | ITO | pAb-Au-MnO2/rGO | pAb: anti-H9N2 | Covalent, COOH of FMNCs with amino groups of mAb | mAb-FMCNs-ALP | LSV | ALP-catalyzed silver deposition | 10 pg mL−1 | 0.1–1.000 ng mL−1 | 30 min | – | [162] | |
| H9N2 antigena | Allantoic cavity of a 10-days old pathogen free embryonated chicken eggs | SPCE | – | anti- H9N2 -M2 | Covalent, EDC coupling | MNP-anti-M2-H9N9–AuNP | Amperometry | Catalytic HER by AuNPs | 16 HAU | – | 60 min | 160 s | [281] | |
| Influenza virusc | Human serum samples | GCE | AuNPs | Ab1: anti- Influenza | Adsorption | Ab2/ALP/Pt-pZnO-hemin | DPV | ALP-catalyzed 1-naphthol production | 0.76 pg mL−1 | 0.001–60 ng mL−1 | 30 min | – | [161] | |
| Tick-Borne Encephalitis Virus | anti-TBEVb | Biological fluids | GCCE | Cysteamine-GA-TBEV | TBEV antigens and antibodies | Covalent, GA coupling | Ab@AgNPs | LSV | AgNPs | 50 IU mL−1 | 50–1600 IU mL−1 | 60 min | – | [159] |
Type of target:a viral protein, b antibody, c whole virus. Ab1: primary antibody, Ab2: secondary antibory, ALV-J antigen: avian leukosis virus subgroup J antigen, anti-ALV-J: anti avian leukosis virus subgroup J antibody, GCE: glassy carbono electrode, GR-PTCA graphene-perylene-3,4,9,10-tetracarboxylic acid nanocomposite, EDC: 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, NHS: N-Hydroxysuccinimide, Fc: ferrocene, AuNP: gold nanoparticle, β-CD: β-cyclodextrin, DPV: differential pulse voltammetry, rGO: reduced graphene oxide, TA: tannic acid, eZIF: zeolitic imidazolate frameworks, HRP: horseradish peroxidase, mpg-C3N4: mesoporous graphitic carbon nitride, GA: glutaraldheide, Th: thionine, NC-Au: nanocellulose-Au nanocomposites, ALP: alkaline phosphatase, EV71 antigen: human enterovirus 71 antigen, ITO: indium tin oxide electrode, MA:, mAb: monoclonal antibody, pAb: polyclonal antibody, mAb: anti-EV71: human anti enterovirus 71 monoclonal antibody, MBs: magnetic nanobeads, HBeAg: hepatitis B e antigen, anti-HBeAg: anti hepatitis B e antibody, HBsAg: hepatitis B surface antigen, anti-HBs: anti hepatitis B surface antibody, anti-HBc: anti hepatitis B core protein antibody, pGO@Au: Au nanoparticles functionalized porous graphene oxide (pGO@Au), GO: graphene oxide, CS: chiosan, DNAzyme: hemin/G-quadruplex DNAzyme, MB: methylene blue, HCV antigen: hepatitis C virus antigen, anti-HCV: anti hepatitis C virus antibody, Gr-IL-Fu: graphene/ionic liquid/fullerene nanocomposite, RhNPs: Rh nanoparticles, CB: celestine blue, H5N1 antigen: influenza virus H5N1 antigen, anti-H5N1: anti influenza virus H5N1 hemagglutinin antibody, DTSP: dithiobis(succinimidyl propionate), Com A: concanavalin A, MNPs: magnetic nanoparticles, ECC: electrochemical conversion, H7N9 antigen: influenza virus H7N9 antigen, anti-H7N9: anti influenza virus H7N9 hemagglutinin antibody, AuNP-G: gold nanoparticle-graphene nanocomposite, AgNP-G: silver nanoparticle-graphene nanocomposite, bi-FMNs: bifunctional fluorescence magnetic nanospheres, Au-MA: Au microelectrode array, H9N2 antigen: influenza virus H9N2 antigen, anti-H9N2: anti influenza virus H9N2 hemagglutinin antibody, FMCNs: fluorescent-magnetic-catalytic nanospheres, anti-H9N2-M2: anti influenza virus H9N2 M2 protein antibody, MNP: magetic nanoparticles, Pt-pZnO-hemin: nanohybrid of Pt-porous ZnO spheres-hemin.
The principle of operation of immunosensors consists of converting the information associated with an immunochemical reaction into a measurable signal proportional to the concentration of the analyte. These biosensing devices employ an antibody or antigen as a biorecognition element, which is usually immobilized on a transducer surface [108,109]. Immunosensors can be classified as electrochemical (alteration in electrical signal), optical (variation in phase, polarization speed, or frequency of input light), or piezoelectric (mass change), depending on the transducer employed [110]. Although optical and piezoelectric immunosensors will not be focused on the present report, notable achievements in the utilization of these devices for the detection of viruses were recently reviewed [[111], [112], [113]]. For example, a digital immunoassay method, based on fluorescence images, was used for simultaneous detection of the inactivated H9N2, H1N1, and H7N9 avian influenza viruses at the single virus level [114]. Zuo et al. [115] have proposed a reproducible and stable piezoelectric immunosensor for the detection of the inactivated SARS-CoV in sputum with an analysis time of ca. 2 min.
Electrochemical immunosensors for diagnosis of viral diseases have attracted considerable attention due to the possibility of miniaturization, cost-effectivity, operational simplicity, fast response, high sensitivity, and specificity meeting the requirements for the fabrication of point of care devices [116]. As can be seen in Table 2, Table 3, different types of electrochemical immunosensors for the detection of viruses have been reported, such as voltammetric, amperometric, impedimetric, and potentiometric. To decrease the analysis time, Santos et al. [117] have proposed the utilization of a redox capacitive transducer for the detection of clinically relevant concentrations of dengue non-structural protein 1 (NS1) antigen and IgG antibody (anti-NS1) in a few seconds. In the proposed method, data can be acquired in an optimized frequency, without equivalent circuits fitting, offering advantages over the traditional impedimetric approaches. The simple operation of the device combined to the quick response of biosensors is a key feature for aiding in the practical diagnosis of viral diseases. Although its application in diluted serum (20%) shows an increase in LOD (0.611 ng mL−1) if compared to the values obtained in PBS (0.220 ng mL−1), the clinically relevant concentration range of NS1 is still greatly appraised (1–5000 ng mL−1) [118].
The combination of immunoassays and commercially available electrochemical biosensors presents an excellent potential for virus detection in remote areas. In this sense, Taebi et al. [119] have proposed a method for the diagnosis of hepatitis B using a personal glucose meter. For the immunoassay, the authors immobilized glucoamylase in a bioconjugate containing different concentrations of two subtypes of the hepatitis B surface antigens (HBsAg), which is composed of three structurally related envelope proteins of hepatitis B virus [120]. After the immunoreaction, in the presence of starch, glucose was produced, and a glucometer measured its concentration. Therefore, it was possible to establish a linear relationship between the concentration of the target and the glucometer signal. The proposed method has a high potential for diagnosis of hepatitis B, exhibiting LODs of 0.3–0.4 ng mL−1 for the subtypes “ad” and “ay”, respectively, which are acceptable based on the LOD of 0.5 ng mL−1 presented by Food and Drug Administration of USA (FDA). It is noteworthy to mention that this work presents an important advantage regarding its commerciability, as it makes use of a widely available and inexpensive equipment for achieving a sensitive analysis – the glucometer.
The biorecognition elements of electrochemical immunosensors for virus detection can be antigens or antibodies. The antigens usually are glycoproteins present on the surface of viruses responsible for binding to the host cell receptor. For example, coronavirus entry into the host cell is mediated by the spike (S) glycoprotein [121], which was recently employed as an antigen in immunosensor for diagnosis of COVID-19 [122]. The antibodies, on its turn, are specific glycoproteins produced as a response to the antigens in the host cell after a few days [110]. As antibody production can persist for several years, its presence in blood might indicate that the patient is currently or was previously ill, or that he received a vaccine against that specific virus [123,124]. The concentration of a specific antibody varies in acute or long term production, aiding in the timely diagnosis of infections.
4.1. Immobilization of the biorecognition element
The performance of an electrochemical immunosensor is intimately dependent on the immobilization method of biorecognition elements, which should ensure the stability of the antibodies or antigens on the transducer surface, maintaining their specificity and biological activity [125,126].
Strategies for the immobilization of antibodies were recently reviewed [[126], [127], [128], [129], [130], [131]]. Among the possibilities, adsorption methods including electrostatic, hydrophobic, and van der Waals interactions are attractive due to their simplicity. However, these result in randomly oriented antibodies, reducing antigen-binding capacity, and desorption processes can seriously compromise the stability and reproducibility of the immunosensor, especially when applying real samples. Therefore, this strategy is not commonly applied.
Another approach is the covalent immobilization based on the interactions of the functionalized transducer surface with the amine, carboxyl, carbohydrate, and thiol moieties of the antibodies. An example was proposed by Cabral et al. [131], who developed a label-free immunosensor to detect anti hepatitis B virus core protein antibodies (anti-HBc). In this case, the authors deposited a nanohybrid film of amino-functionalized carbon nanotubes recovered by hyaluronic acid onto a glassy carbon electrode. The hyaluronic acid was anchored to the amino group on carbon nanotubes by covalent binding (amide bond) and the antigen (HBc) was covalently linked to the carbon nanotubes by amine bonds. The proposed immunosensor was able to detect anti-HBc with high stability by square wave voltammetry (SWV), with a LOD of 0.03 ng mL−1. This stability, even after 20 voltammetric cycles at 100 mV s−1, was attributed to the strong covalent bonds between the electrode and the hybrid film-HBc antigen. Therefore, the presence of functional groups allowed not only the efficient immobilization of the biological material but also the incorporation of a high conductive material on the base electrode, significantly contributing to its sensitivity. The linear range achieved by the biosensor is in accordance to the ones required in clinical applications, but full analytical assessments using real samples should be performed before accessing its potential for practical blood bank analysis.
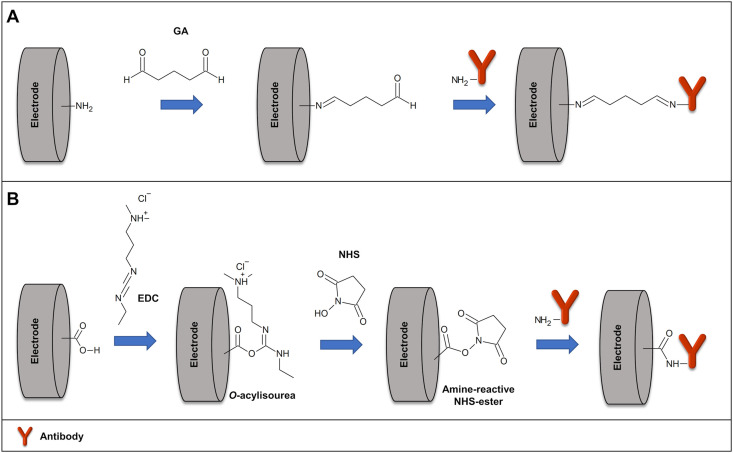
One of the most conventional methods for covalent immobilization of antibodies is the use of crosslinkers. GA, for example, may be used to activate the amino groups on the immunosensor, producing reactive aldehyde groups on its surface [132,133]. The activated surface reacts with the amino groups of the antibodies to form imine bonds, as can be seen in Fig. 3 A. Moreover, EDC is usually employed for carboxy-to-amine linking. Thus, a carboxyl-functionalized surface can be activated with EDC to generate a highly reactive O-acylisourea active intermediate, allowing the formation of amide bonds with the amino groups of the antibodies. The same method can be used to bind the carboxyl of the antibodies to an amine-functionalized surface. To improve the efficiency of the covalent immobilization, the combination of EDC with NHS has been employed, resulting in a more stable active intermediate, Fig. 3B [134].
Fig. 3.
Schematic illustration of antibody covalent immobilization of (A) a carboxyl-functionalized surface activated with EDC/NHS, and (B) an amine-functionalized surface activated with glutaraldehyde.
An example of the use of this strategy can be observed in the work reported by Devarakonda et al. [133], which describes the development of a paper-based immunosensor for the label-free detection of influenza virus H1N1. A stencil-printed carbon electrode was first modified with single-walled carbon nanotubes and chitosan. Anti-influenza A antibodies were then immobilized on the electrode by highly stable glutaraldehyde cross-linking. The proposed device was effective for quantifying influenza virus using DPV with a LOD of 113 PFU mL−1. Among the several advantages presented by the biosensor, it is noteworthy to highlight its high temporal stability, which displayed a relative standard deviation of only 4.4% after 15 days. This was attributed, in part, to the presence of strong and stable covalent bonds between the electrode and the receptors.
Although the covalent immobilization via glutaraldehyde or EDC/NHS can improve the stability of the immunosensor, it usually results in random orientation, due to the abundance of amino and carboxyl groups well-distributed on the antibody surface. On the other hand, stable covalent immobilization can be performed from the selective reduction of the disulfide bonds present in specific regions of the antibodies [135]. This allows reactive thiol groups to be generated, allowing the oriented arrangement of the antibodies [135]. Another possibility is the formation of reactive aldehyde groups from the oxidation of hydroxyl groups of carbohydrates moieties of antibodies. However, this activation process may involuntarily oxidize amino acids present on the antibodies, affecting the site-selectivity and antigen-binding capacity. Oriented immobilization of the antibodies may also be achieved via specific affinities interaction of proteins A and G with the Fc (fragment crystallizable) domain of the antibodies. Although recent examples of the use of this immobilization method for the electrochemical detection of viruses were hardly found, it is worth mentioning that an interesting study was published by Huy et al., in 2011 [44]. In this paper, the authors have reported different techniques to immobilize anti-japanese encephalitis virus antibodies in a silanized surface, including direct covalent bonding, crosslinking, anti-IgG, and protein A. The use of protein A presented the best analytical performance, causing a two-to three-fold improvement in the electrochemical signal of the antigen.
The immobilization of the biorecognition element is usually followed by the incubation of the immunosensor with blocking agents, such as bovine serum albumin (BSA), casein, and surfactants. The utilization of these compounds can avoid non-specific adsorption of interfering species that decreases the sensitivity of the immunosensor, which is essential for its application in complex samples.
4.2. Immunoassay designs
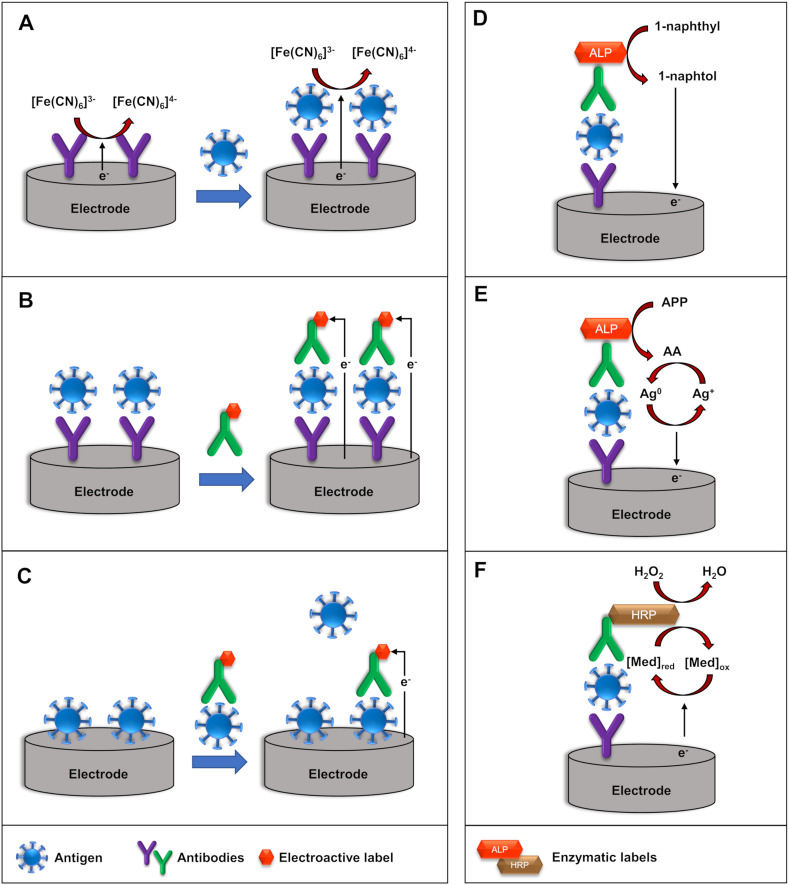
Electrochemical immunosensors can be categorized according to the immunoassay format, into a label-free, sandwich (non-competitive) and competitive immunosensors, as presented in Fig. 4 (left).
Fig. 4.
Schematic representation of (A) label-free, (B) sandwich, and (C) competitive electrochemical immunosensors (Left) and schematic representation of the mechanism using enzymatic-labels (right): (D) ALP enzymatic-label for production of electrochemically active 1-naphthol, (E) ALP-catalyzed deposition of electroactive silver, and (F) HRP-catalyzed oxidation of the redox mediator.
4.2.1. Label-free electrochemical immunosensors
Label-free electrochemical immunosensors are widely used for virus detection due to their simplicity, fast response, and the possibility of real-time monitoring (Table 2). Antibodies and antigens are not commonly electrochemically active, thus a redox probe is introduced into the electrolyte solution. The formation of the antibody-antigen immunocomplex blocks the electron transfer between the transducer and the redox probe, changing the analytical signal [136,137]. In voltammetric transducers, the peak currents typically decrease with the increase of the concentration of the analyte. In impedimetric immunosensors, the charge transfer resistance typically increases upon the addition of the analyte. Ferricyanide/ferrocyanide, [Fe(CN)6]3–/4–, is extensively used as a redox probe in label-free immunosensors for virus detection [[138], [139], [140], [141], [142], [143], [144], [145], [146], [147]] due to its well-defined redox processes, reversible heterogeneous kinetics and stability in aqueous solution. Moreover, the use of redox probes with low oxidation potential can minimize or avoid interfering species, which is especially important for real samples. For example, Valipour and Roushani have employed riboflavin as a redox probe in a label-free immunosensor for detection of hepatitis C core antigen protein (HCcAg), due to its negative oxidation potential [148,149]. Nevertheless, other species may also undergo electrochemical processes at −0.45 V vs. Ag/AgCl, potential value at which riboflavin was detected, and the reduced solubility of riboflavin in water may be a drawback to be overcome.
Since the electrochemical processes of the redox probe in solution are diffusion-limited, strategies for its immobilization on the transducer surface can benefit the performance of the immunosensor. In this regard, Veerapandian et al. [150] have used methylene blue (MB) as a redox probe for the development of a voltammetric immunosensor for simultaneous detection of influenza A H1N1 and H5N1 viruses. For multiplexed detection, the authors have used SPE with two working electrodes modified with graphene oxide, MB, and specific antibodies against the hemagglutinin proteins of H1N1 or H5N1. Upon the addition of the analytes, an insulating layer of the immune complex is formed, creating a barrier toward the electron-transfer process at the electrode interface. Therefore, the anodic current associated with the oxidation of MB decreases with the concentrations of the analytes. Limits of detection at the picomolar level were obtained, 9.4 and 8.3 pmol L−1 for H1N1 and H5N1 antigenic hemagglutinin proteins, respectively. The authors also found that chronoamperometry may be applied for the quick detection of targets, which significantly increases the potential of the biosensor to be applied in practical scenarios. Other electroactive compounds, such as PB [151], ruthenium complexes [152], ferrocene [117,153], polyaniline [154], and polytryamine [155], have also been used as redox mediators in label-free immunosensors for the detection of viruses. However, the redox probe [Fe(CN)6]3–/4– has been preferred due to its high water solubility, well known electrochemical behavior on different electrodes, and low cost.
4.2.2. Sandwich-type electrochemical immunosensors
In sandwich-type format (Fig. 4B), after the immunochemical reaction between the biorecognition element (primary antibody) and the analyte (antigen), a labeled secondary antibody is added. Thus, the formation of the sandwich complex results in an electrochemical signal proportional to the concentration of the analyte. Several labels, such as electroactive compounds, enzymes, and electrocatalysts have been employed for the fabrication of sandwich-type immunosensors for virus detection, as presented in Table 3. The use of such labels commonly increases the biosensors complexity and associated costs but also improves its analytical performance when compared to a similar label-free design.
Among the electroactive compounds, ferrocene [156], thionine [157], and celestine blue [158] were used in voltammetric immunosensors. To improve the performance of the immunosensor, the electroactive compounds are often combined with carbon-based materials or metallic nanoparticles. Khristunova et al. [159] employed electroactive silver nanoparticles (AgNPs) as a label for the fabrication of an immunosensor for the detection of antibodies against tick-borne encephalitis virus. Huang et al. [160] have used a label of AgNPs/graphene nanocomposites for the detection of picomolar concentrations of inactivated avian influenza virus H7N9. In this case, the AgNPs act as a redox probe, while graphene was used for signal amplification due to its high electrical conductivity, surface-to-volume ratio, and electron transfer rate. The sensitivity achieved by the developed method, however, was not superior to the traditional ELISA method, presenting low practical advantages.
Enzymatic labels can be used in two different approaches for the generation of the analytical signal. In the first one, enzymes are employed for the production of electrochemically active compounds. For example, alkaline phosphatase (ALP) enzyme catalyzes the conversion of 1-naphthyl phosphate into electroactive 1-naphthol (1-NP), Fig. 4D. Therefore, the electrochemical signal generated from the oxidation of 1-NP can be employed to determine the concentration of the analyte. A signal amplification strategy for influenza antigen detection was reported by Yan et al. [161]. The authors have applied an ALP label containing porous zinc oxide functionalized with platinum nanoparticles and hemin, which catalyzes the oxidation of 1-NP in the presence of H2O2. ALP enzyme is also employed for the catalytic deposition of electroactive metallic silver, as shown in Fig. 4E. In the presence of ALP, ʟ-ascorbic acid-2-phosphate (AAP) or p-aminophenyl phosphate (APP) is converted into ascorbic acid (AA) or p-aminophenol (AP), respectively. Then, AA or AP can reduce AgNO3, producing metallic silver on the surface of the immunosensor [[162], [163], [164]]. The device presented a sub-picomolar LOD (0.76 pg mL−1) and adequate performance in a 10% human serum samples. Furthermore, at least 90% of its original electrochemical signal was maintained after 25 days of storage at 4 °C, which is essential for commercialization purposes.
In the second approach, enzymatic labels, such as HRP, are used in the presence of a redox mediator. As can be seen in Fig. 4F, HRP catalyzes the oxidation of redox mediator in the presence of H2O2. Then, the analytical signal, associated with the reduction of the oxidized mediator, increases with the concentration of the analyte. The redox mediators TMB (3,3′,5,5′-tetramethylbenzidine) and hydroquinone were utilized in immunosensors for the detection of inactive human enterovirus 71 [165] and the whole avian leukosis virus subgroup J [166] in avian serum samples, respectively.
Since peroxidase mimetics are more stable and present lower costs if compared to HPR, their use as labels in immunosensors is attractive. Among the possibilities, hemin/G-quadruplex HRP-mimicking DNAzyme has been used in electrochemical immunosensors, employing methylene blue as a redox mediator. These immunosensors have shown satisfactory stability, reproducibility, and selectivity for hepatitis B virus surface antigen. Limits of detection in the femtomolar range can be achieved and the biosensors can be applied for the determination of HBsAg in spiked human serum samples [167,168].
Additionally, labels of electrocatalysts materials are promising alternatives to HRP labels due to the possibility of direct electron transfer to the transducer surface. Thus, the immunoassay can be performed without redox mediators [169]. For example, Li et al. [170] have proposed a nanohybrid label of molybdenum disulfide, cuprous oxide, and platinum nanoparticles (MoS2@Cu2O–Pt), due to their electrocatalytic activity towards the reduction of H2O2 (Fig. 5 A). The complex was then applied for the precise quantification of the hepatitis B surface antigen through a sandwich-like structure with glassy carbon electrodes (GCE) modified with porous graphene oxide, Au composites, and specific antibodies (Fig. 5B, C, and D). A LOD of 0.15 pg mL−1 was obtained, and the immunosensor was effectively applied for quantitative detection of the antigen in human serum samples. This biosensor presents considerably lower LOD than the one produced by Taebi et al. [119] (previously discussed) for the same virus (0.3 ng mL−1 0.4 ng mL−1 for subtypes “ay” and “ad”, respectively). Both biosensors, however, present acceptable LODs based on the value defined by the Food and Drug Administration of USA (FDA) (0.5 ng mL−1), with the biosensor developed by Taebi et al. [119] presenting advantages due to its simple application and for using a widely available instrument for the measurements, a glucometer.
Fig. 5.
Detailed construction of sandwich immunosensor using (A) MoS2@Cu2O–Pt/Ab2 and (B) GCE/GO-Au/Ab1. (C) Amperometric response obtained with different concentrations of hepatitis B surface antigen, and (D) its corresponding calibration curve. (Reprinted with permission from Li, F. et al. Facile synthesis of MoS2@Cu2O–Pt nanohybrid as enzyme-mimetic label for the detection of the Hepatitis B surface antigen. Biosens. and Bioelectron. 100, 512–518, © 2018 Elsevier BV [170]).
Another strategy for the generation of electrochemical signals in immunosensors is based on the use of self-sacrificial labels. An example was reported by Zhang et al. [171], who proposed an electrochemical conversion (ECC) method for the production of Prussian white (PW) from a label of Fe3O4 magnetic NPs. The cathodic current associated with the reduction process of PB to PW was adopted as the analytical signal and shown to be proportional to the concentration of influenza virus H5N1 [172]. The developed method exploits an interesting mass transport mode, promoting surface generation-confinement in with greater production efficiency than conventional approaches. The biosensor also presented good sensitivity and a LOD of 0.0022 HAU, which is one of the lowest ones presented in this work for influenza virus. It is important to notice that such approaches brings opportunities for the development of new and improved similar electrochemical approaches in the near future.
4.2.3. Competitive electrochemical immunosensors
In competitive electrochemical immunosensors, labeled and free biomolecules compete for a limited number of binding sites, which are usually immobilized on a transducer surface. Due to the problems associated with the random orientation of the antibodies, immobilization of the antigen is the most frequently used strategy. In this case, the immobilized antigen usually reacts with the labeled antibody in competition with the free antigens (Fig. 4C) with the analytical signal decreasing upon the increase in the concentration of the free antigen in the sample. It is important to notice that the competitive approach is especially interesting for the detection of small molecules that do not support sandwich assays or do not produce relevant signal changes in label-free strategies.
Regarding their application in the diagnosis of viral diseases, Mandli et al. [173] have proposed a competitive amperometric immunosensor for hepatitis A virus antigen detection obtained from AVAXIM vaccine, which contains the inactivated virus [174]. After the competition between the immobilized antigen and the free antigen for the primary antibodies, secondary antibodies labeled with HRP were added. Hydroquinone was employed as a redox mediator for the generation of the analytical signal. Therefore, in the presence of H2O2, the amperometric signal decreases with the increase in the concentration of the virus. Although presenting slightly higher complexity, the developed sensor presented lower LODs than indirect ELISA. However, the detection of hepatitis A virus antigen in real samples was only reported in spiked tap water, creating doubts regarding its clinical application.
Recently, a competitive voltammetric immunosensor was described for the determination of coronaviruses (CoV), with an analysis time of 20 min [175]. Since the proposed immunosensor array chip contains eight working electrodes, multiplex detection of different types of CoV can be performed. The immunosensor was applied in the determination of the spike protein S1 of the Middle East respiratory syndrome coronavirus (MERS-CoV) and the nucleocapsid protein N of human coronavirus (HCoV). The free antigen in the sample competes with the immobilized antigenic proteins of CoV for a fixed concentration of the antibody (anti-MERS-CoV or anti-HCoV) added to the sample. Then, in the presence of the redox probe [Fe(CN)6]3–/4–, the cathodic peak current decreased with the increase in the concentration of the HCoV and MERS-CoV antigen. Limits of detection of 0.4 and 1.0 pg mL−1 were obtained for HCoV and MERS-CoV antigens, respectively. The proposed immunosensor was successfully applied in spiked nasal samples, confirming its high potential for a rapid and multiplex detection of coronaviruses. It is also important to highlight that its multiple working electrodes allow great practical applicability, allowing the simple and quick differenciation between viral strains.
4.2.4. Lateral-flow assays
Lateral flow immunoassays (LFIs) are one of the most used point of care diagnostic technology, presenting simple construction and manipulation, low operational costs, versatility, portability, small sample volume requirements, long-term stability, and quick response as advantages when compared to traditional assays [[176], [177], [178]]. LFIs have been widely used for the detection of biomarkers for diagnosing infectious diseases and for the detection of viruses, such as dengue [[179], [180], [181], [182]], Zika [183], Ebola [184,185], HIV [186], HPV [187], pseudo-rabies [188], hepatitis [[189], [190], [191]], infectious bronchitis and respiratory syncytial virus [192,193] (Table 4 ). In recent years, several works have also reported the determination of different types of avian influenza A by LFI [[194], [195], [196], [197], [198]].
Table 4.
Summary of the Lateral Flow Immunoassays (LFI) reported for the detection of different viruses.
| Viruses | Assay | Sample type | Detection (Time) | LOD | Ref. |
|---|---|---|---|---|---|
| Dengue | Recombinase polymerase amplification combined with lateral flow dipstick | DENV-1, -2, -3, -4 RNA serotypes in human serum from infected patients | Colorimetric | 10 copies/RNA molecules | [179] |
| LFI combined with near-infrared fluorescent | Anti-DENV-1 IgG antibodiesb in human serum from infected patients | Near-infrared fluorescence | – | [180] | |
| Electrochemical lateral-flow immunoassay | dengue NS1 proteina in buffer | Electrochemical | 0.5 ng mL−1 | [182] | |
| Electrochemical lateral-flow immunoassay | dengue NS1 proteina in human serum samples | Electrochemical | 50 ng mL−1 | [181] | |
| Zika | LFI based on quantum dot microspheres | ZIKV NS1 proteina in spiked human serum | Fluorescence (20 min) | 0.15 ng mL−1 | [183] |
| Ebola | LFI based on RT-PCR | EBOV glycoprotein and nucleoproteina in human plasma and oral swab | Colorimetric | – | [184] |
| LFI based on Fe3O4 magnetic nanoparticles | EBOV antigena in human serum samples | Colorimetric (30 min) | 1.0 ng mL−1 | [185] | |
| HIV | Commercial Daina Screen Combo and Architect Combo assays | HIV-1/2 antibody and HIV-1 p24 antigena in human blood | Chromatography (18.8–19.3 days) | 200 copies mL−1 | [186] |
| HPV | Magnetic immunochromatographic strip with PDZ ligands | HPV E6 antigena in cervical swab | Colorimetric | – | [187] |
| Pseudo-rabies | LFI based on SERS | PRV antigenc in pig tissues and gE-deleted vaccine | Raman spectroscopy (15 min) | 5 ng mL−1 | [188] |
| Hepatitis B | Luminescent quantum dot-beads sandwich | HBsAga antigen in human serum | Luminescence | 75 pg mL−1 | [189] |
| Hepatitis C | LFI based on up-converting nanoparticles | Anti-HCV antibodiesb in human serum | Colorimetric | – | [191] |
| Bronchitis and Newcastle disease virus | Multiple reverse-transcription LAMP and lateral flow dipstick | Simultaneous IBV and NDS RNA in tracheas and lungs tissues, oral and cloacal swabs | Electrophoresis | 100.8 and 100.7 RNA copies/reaction (IBV and NDV) | [192] |
| Respiratory syncytial | Recombinase polymerase amplification combined with lateral flow dipstick | RSV-A and RSV-B RNA in nasopharyngeal aspirates from healthy and sick people (respiratory diseases) | Colorimetry | 10 copies RNA/assay | [193] |
| Avian influenza A | LFI based on near-infrared (NIR)-to-NIR upconversion gold nanoparticles | AIV nucleoproteinsa in avian stool | Photoluminescence (20 min) | 102 and 103.5 EID50 mL−1 (H5N2 and H5N6) | [194] |
| LFI based on sandwich-type aptamers using graphene-oxide | AIV H5N2 antigenc in ducks feces | Visual | 1.27 × 105 EID50 mL−1 | [195] | |
| LFI based on label-free quantum dots | AIV antigenc in serum samples from infected chicken | Fluorescence | 0.09 ng mL−1 | [196] | |
| RT-RPA with lateral flow dipsticks | H1 and H3 hemagglutinin genes in throat swabs | Colorimetric | 677.1 and 112.2 copies/reaction (H1 and H3) | [197] | |
| LFI based on SERS | AIV H7N9 hemagglutinina in poultry organs, cloacal and throat swabs | Raman spectroscopy (20 min) | 0.0018 HAU | [198] |
Type of target: aviral protein, bantibody,cwhole virus.
LFIs are typically based on a sandwich strategy and are composed of four individual zones (sample pad, conjugate pad, detection pad, and absorbent pad) overlaid and assembled on an inert platform (Fig. 6 ). The LFI principle is based on the ability of the sample to flow through the capillary strip and on the specific antigen-antibody interaction promoted by the biorecognition element immobilized in the detection pad. For this, the sample is dripped on the sample pad and allowed to flow freely to the conjugate pad. The conjugate pad contains a labeled biomolecule conjugate (antigen or antibody, depending on the target), allowing the virus immunocapture. The complex then flows to the detection pad, which is usually composed of a nitrocellulose membrane with specific antigens (for antibody detection) or antibodies (to virus/protein detection) immobilized on its surface. The continuous sample flow is guaranteed by the presence of an absorbent pad at the end of the strip. Cellulosic, glass fiber and paper-based sample pads have been reported as porous membranes for virus detection.
Fig. 6.
Detailed mechanism of a sandwich LFI (Reprinted with permission from Liu, B. et al. Paper-based electrochemical biosensors: from test strips to paper-based microfluidics. Electroanalysis. 26, 1214–1223, © 2014 WILEY-VCH [199]).
The detection of the virus in a sample can usually be performed by the naked eye due to the application of colorful labels. Commercial colorimetric LFIs are currently common and appraise the direct analysis of saliva, urine, and other biological fluids for assessing pregnancy or diagnosing viral diseases, such as HIV and COVID-19. AuNPs are the most common label used due to their high stability, biocompatibility, excellent optical properties, and the red color easily observed in the final results [200,201]. However, latex beads [202], gold colloids [184,197], quantum dots [183,189,196], gold-silver core-shell NPs [188,198], europium [203], and AgNPs [204] are also explored in the literature. These species can be used not only as labels but also to improve the biological material immobilization or interaction as well as the detection sensitivity and reproducibility [176]. Also, quantum dots present unusual fluorescence/luminescence characteristics, allowing their easy detection. LFI devices can also have their advantages enhanced by coupling to different detection methods, such as chromatography [186], near-infrared fluorescence [180], indirect fluorescence/luminescence [189,196], surface-enhanced Raman scattering (SERS) [188,198], and electrochemical detection [163,181,182,205].
Regarding electrochemical detection, Sinawang et al. [182] developed an interesting LFI coupled to SPAuE for the detection and quantification of the dengue NS1 protein. Gold nanoparticles with anti-NS1 antibodies conjugated to ferrocene were applied to electrochemically detect the desired target. A different electrochemical LFI was developed by Sinawang et al., in 2018 [181]. In this case, a miniaturized LFI based on 4-amino-2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPO)-tagged AuNPs for an amperometric point-of-care dengue NS1 protein detection was described. The authors used thiolated polyethylene glycol (PEG-thiol) to stabilize the AuNPs and as an anchor for the conjugation of antibodies and redox species. The assay provided rapid results (less than 30 min) and was able to capable quantify dengue NS1 protein. In this case, the low sample volume may be an advantage if compared to traditional assays, especially regarding low resource settings.
Bhardwaj et al. [205], in this turn, developed an innovative vertical LFI for the detection of influenza virus that combined both optical and electrochemical methods. In this work, a different pore size membrane was applied as a sample pad, allowing uniform flows to be achieved for the efficient HRP-labeled anti-HA antibodies interaction with the target. The detection was subsequently performed through the use of EIS (for electrochemical detection) and a scanner (for colorimetric detection). The immunosensor allowed the rapid (6 min) and sensitive electrochemical and colorimetric detection of influenza H1N1 virus in saliva samples. Both colorimetric and electrochemical techniques presented similar LODs in saliva (1.34 and 3.3 PFU mL–1, respectively), and adequate performance for the direct analysis of saliva and nasal secretions - as the infectious dose of H1N1 in these secretions range from 103 to 105 TCID50 mL–1 [206].
It is important to mention that the typical analytical parameters currently achieved by the electrochemical LFIs for viral detection are similar to the ones obtained by optical detection. Optical biosensors, however, commonly rely on simpler instrumentation (e.g., smartphones or the naked eye) and are easier to use. Advantages regarding the use of electrochemical LFIs are mainly related to greater reproducibility in a variety of conditions, such as lighting, and greater quantification precision, which is important to determine the extension of an infection, for example. New and improved electrode fabrication and modification techniques, and labeling methods hold great promises for such devices soon.
5. Biomarkers for viral diseases diagnosis
Besides the identification of the pathogen itself or the antibodies related to it, the detection of specific biomarkers is a valuable tool to assist in the diagnosis of viral diseases and their complications. According to the Biomarkers Definition Working Group, biomarkers correspond to “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” [207]. It can be related to biochemical, cellular, molecular alterations in biological systems, and includes tools and technologies for understanding the prediction, diagnosis, and other aspects of a disease treatment [208,209]. In this section, we will focus on specific biomolecules that present altered concentrations due to the occurrence of a viral disease.
One of the first steps to follow when a patient presents an infection is to determine its origin, allowing adequate treatment. In this sense, biomarkers for different diseases are already known, including C-reactive protein (CRP), procalcitonin (PCT), and proadrenomedullin [210,211], and they are essential to distinguish between bacterial and viral infections. A study performed by Yusa and coworkers [212] have shown the set of interleukins (IL) IL-4, IL-8, IL-10, IL-12 and tumor necrosis factor-alpha (TNF-α) might be suitable biomarkers for infections and that IL-2, IL-8 or IL-10 present the potential to distinguish between bacterial and viral infections, being great tools for assisting in precise diagnosis, increasing the chances of treatments success.
Sharma et al. [213] have described the development of a label-free electrochemical biosensor for the detection of human IL-8 in full serum. To perform the specific detection of the target, a small, robust, and synthetic capture protein based on a cystatin scaffold was immobilized onto gold electrodes using a monothiol-alkane-poly(ethylene glycol) (PEG) acid self-assembly monolayer (SAM) and EDC/NHS. Then, changes in the phase were measured using EIS, allowing the sensitive detection of IL-8. Such devices, therefore, could significantly aid in the differentiation of bacterial and viral infections. It is important to mention that, although different studies show the potential of IL-8 as a biomarker for various diseases, large-scale studies that validate its accuracy and outcome are needed. Currently, IL-6 is more commonly applied in clinical diagnosis, acting as a marker of immune system activation [214].
Another biomarker that can aid in the distinction between bacterial and viral infections is the C-reactive protein (CRP). According to a study developed by Sasaki et al. [215] the protein usually displays significantly altered levels during bacterial infections, remaining in normal concentrations during viral ones. Currently, CRP is commonly used in clinical diagnosis both to detect inflammation due to acute conditions or to monitor chronic diseases [216]. Dong et al. [217] recently developed a new electrochemical immunosensor based on an ionic liquid and ZnO/porous carbon matrix composite for the detection of CRP in human serum samples. DPV technique was used for the precise quantification of CRP.
Hafaiedh et al. [218] also developed an electrochemical biosensor for the detection of CRP. In this case, two different immobilization methods for anti-CRP antibodies were proposed, being one based on the simple physisorption of the antibody on a gold electrode surface, and the other based on oriented anti-CRP antibody immobilization using protein G as an intermediate layer. The authors performed the detection of the protein directly in human plasma, using EIS, validating the developed device for possible clinical use. In comparison to the previous work, Hafaiedh and collaborators presented a more sensitive immunosensor probably due to the organization of the receptors.
Regarding dengue fever, NS1 (non-structural protein 1) is one of the most applied biomarkers for its diagnosis due to its specificity and sensitivity. When the dengue virus infects patients, NS1 is secreted into the bloodstream in the first days of infection, allowing its simple identification using serum samples during the acute phase of the disease [219]. Several publications in the literature relate the suitability of this biomarker [[220], [221], [222]] and, although PCR-based methods are currently the most common techniques applied for the clinical diagnosis of the disease, NS1-based rapid tests are also used [223].
A significant number of biosensors for the rapid and straightforward determination of NS1 can be found in the literature. For example, Lim et al. [221] have developed a label-free electrochemical biosensor for the quantification of NS1. To identify affinity peptides that can bind to the biomarker, a polyvalent phage display was applied. Afterward, cyclic voltammetry (CV) and EIS were applied to quantify the protein. The developed sensor presented specificity towards NS1 when compared to BSA. The LOD presented by the device (1.49 μg mL−1), however, did not cover the full range of concentration of NS1 found in primary (0.04–2.00 μg mL−1) and secondary (0.01–2.00 μg mL−1) infections [118], limiting its clinical applicability.
Figueiredo et al. [219] have also described a label-free electrical NS1 biosensor for the early diagnosis of dengue. For that, the authors used, for the first time, egg yolk immunoglobulin as a biorecognition element. Comparing the proposed system to the one previously presented [221], there are advantages regarding specific clinical applications. The first one is related to the simple electrical instrumentation required for the measurements, which can be miniaturized and transported to perform the diagnosis in situ, allowing quick medical decisions. Furthermore, the LOD achieved by the biosensor is about 16 times lower than the one presented by the similar electrochemical system [221], covering a greater portion of the usual concentrations of NS1 found in plasma. Lower LODs however, must be achieved to provide tests with low false-negative rates. This could be performed with the use of signal amplification techniques or by approaching the antibody recognition sites from the electrodes.
Considered by the World Health Organization (WHO) as a global epidemy, AIDS/HIV has already affected 76 million people and caused 33 million deaths [224]. As HIV infects the cells of the immune system, primarily CD4 lymphocytes, the decrease of its plasma levels is an important clinical parameter for AIDS diagnosis and for assessing the stage of the disease. Carinelli et al. [225] developed a magneto electrochemical immunosensor for the rapid lymphocyte T CD4+ count in the blood. Two different antibodies were added to the sample: an anti-CD3 antibody, conjugated to magnetic particles, and a biotinylated anti-CD4 antibody. When in the presence of the desired lymphocyte, both antibodies were attached to the cell surface. The addition of a streptavidin-HRP reporter and the pull-down of the cells to the surface of the working electrode was then performed, allowing the precise electrochemical quantification of the target. The developed device presents great potential to be used in clinical diagnosis as it can be applied directly to whole blood, besides being able to detect the whole range of clinical interest for AIDS diagnosis and follow up (89–912 cells μL−1, with an LOD of 44 cells μL−1). Combining its outstanding analytical performance with the possibility to perform measurements using simple instrumentation, the technique offers great advantages over the gold standard flow cytometry method. It still requires, however, the performance of several steps by a specialized worker for its adequate functioning.
Still regarding the quantification of CD4+ T cells, Kim et al. [226] have developed an immunosensor based on a two-step O2 plasma treatment of single-wall carbon nanotube (SWCNT) modified electrodes and covalent immobilization of anti-CD4 monoclonal antibodies. With CD4+ T cells being detected by SWV, the immunosensor presented a LOD (100 cells mL−1) considerably lower than the one reported in the previous work [225] (44 cells μL−1). However, the device was tested using single cells obtained from mice spleens dispersed in the buffer and there is no evidence of its performance in whole blood. Furthermore, the two steps of O2 plasma treatment need to be performed 35 days apart to avoid the physical destruction of SWCNTs, making the construction of this immunosensor extremely time demanding.
Biomarkers can also be useful for differentiating mild from severe cases of viral diseases. Dengue virus infection, for example, can present a vast spectrum of clinical manifestations, ranging from asymptomatic conditions to dengue hemorrhagic fever and dengue shock syndrome, which can lead to death. Therefore, the determination of the initial stage of dengue hemorrhagic fever is of great importance to determine the adequate therapies and significantly improves the chances of recovery of patients. John et al. [227] published a review on this topic and stated that severe dengue biomarkers include, among others, T and B cell activation and apoptosis and cytokine storms.
Baraket and coworkers [228] reported a fully integrated electrochemical platform for multiple cytokines detection (IL-10 and IL-1b). First, anti-human IL-1b and anti-human IL-10 monoclonal antibodies were attached to 4-carboxymethyl aryl diazonium (CMA) through the use of EDC and NHS crosslinkers. Then, the diazotization of CMA was performed, allowing its immobilization onto the gold working electrodes by the application of repetitive CV cycles. The detection of ILs was then performed using EIS. The platform displayed high sensitivity and specificity, presenting no interference by other cytokines, and was able to detect the targets in a 1–15 pg mL−1 range. The described device can significantly lower the total analysis time and can be used by physicians to identify an increase in IL secretion, aiding in the identification of cytokine storms and, therefore, in severe dengue cases, for example. As previously cited, although different IL are indicated as biomarkers by recent studies, IL-6 is the only one currently being used for the clinical diagnosis of diseases [214].
The above-cited devices exemplify how biomarkers-based detection can significantly aid in the diagnosis and treatment of viral diseases. Its rapid, low-cost, and simple responses can dramatically increase the accessibility and response rates, improving the clinical treatment of the affected patients and allowing quick practical decision making.
6. Methods and devices for COVID-19 diagnosis
The understanding of the structural and genomic features of the SARS-CoV-2 has a crucial role in the investigation of its origin [229], mechanism of receptor recognition [230], mutations, and genetic recombination [231,232]. Furthermore, the knowledge concerning the SARS-CoV-2 virology and the pathogenesis of COVID-19 can guide therapeutic strategies, the development of vaccines [233,234], and the identification of biomarkers, opening up the possibilities of diagnostic tests.
The SARS-CoV-2 is a spherical virus with a diameter ranging from 60 to 140 nm [235], containing positive-sense and single strands of RNA, enveloped with a crown-like structure of spike (S) glycoproteins. Besides the S proteins, the virus is also structurally composed of the envelope (E), membrane (M), and nucleocapsid (N) proteins, as shown in Fig. 7 .
Fig. 7.
Structure and genomic organization of SARS-CoV-2. The open reading frame 1 ab gene (ORF1ab, in yellow) encodes non-structural proteins, including the RNA-dependent RNA polymerase (RdRp). The structural genes (in green) encode the structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N). The genes highlighted in gray encode the accessory proteins (AP). (Adapted from Alanagreh, L. et al. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens 9, 5, 331 © 2020 MDPI [236]). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Once the viral genome of SARS-CoV-2 is sequenced, the ORF1ab, RNA-dependent RNA polymerase (RdRp), E, N, and S genes can be employed as targets in the PCR-based diagnostic kits for COVID-19. Currently, different PCR-based kits are commercially available for use and their evaluation is essential not only to compare each product but also to further improve their performance. Based on this, comparative studies have evaluated the performance of commercially available RT-PCR tests for COVID-19 [237,238]. For example, Nalla et al. [237] have compared seven different RT-PCR kits and reported superior sensitivities of the assays proposed by Corman et al. [237] and the CDC (Centers for Disease Control and Prevention) [239], which target the E-gene and the N2 region of the N-gene of SARS-Cov-2, respectively. The E-gene primer-probe sets can detect both SARS-CoV and SARS-CoV-2. On the other hand, the commercial kits based on the ORF1ab, RdRp, N, and S genes targets do not exhibit cross-reactivity towards other respiratory coronaviruses, such as human coronavirus (HCoV) NL63, HCoV-OC43, HCoV-229E, SARS-CoV-1, and MERS. Also, a PCR efficiency above 96% was found for all different commercial assays [238].
The genetic material of the virus can also be detected by genosensors, as presented in section 2. For the SARS-CoV-2, Qiu et al. [240] have reported the use of the RdRp, ORF1ab, and E genes as target analytes in plasmonic photothermal genosensors. The analytical signals result from the hybridization process between the genes and their complementary cDNA sequences. Since the signals were proportional to the physical length and molecular weight of the genes, the strongest and the lowest responses were obtained for the sensors of the ORF1ab and the E sequences, respectively. The sensor of the RdRp gene presented a LOD of 0.22 pmol L−1 and could discriminate similar biomarkers from SARS-CoV-2 and SARS-CoV. The complex instrumentation required for adequate measurements, however, is a major drawback for its practical applications.
In contrast to the RNA-based methods, the ELISA tests, point of care LFI, and immunosensors employ the proteins of the virus as biomarkers. In this sense, the diagnosis of COVID-19 can be performed by measuring the antibodies, IgM and IgG, produced in the host as an immune response to viral proteins. Another possibility is the utilization of these antibodies as biorecognition elements in biosensors. Therefore, the virus can be directly detected by the specific interaction of its proteins with the antibodies immobilized on the transducer surface.
Among the proteins of SARS-CoV-2, the N and S glycoproteins are abundantly expressed during the infection and highly immunogenic, which makes them suitable for utilization as biomarkers in the diagnostic tests. Several commercial immuno-based assays have received emergency authorizations due to the COVID-19 pandemic situation. However, caution is advised regarding the applicability and validation of such tests. For example, the use of some lateral flow assays has been questioned due to the lack of validation related to its sensitivity and specificity [241].
Liu et al. [242] have compared the performance of ELISA kits based on N and S proteins for the detection of IgM and IgG antibodies. The study was performed with 214 COVID-19 patients. The positive rates of the N-based and S-based tests for the detection of antibodies were 80.4% and 82.2%, respectively. The higher sensitivity of S-based tests can be associated with the earlier response to the S antigen than the N antigen in patients with COVID-19 [243]. One of the drawbacks associated with the N-based ELISA tests is the possibility of false-positive results because the nucleocapsid proteins of the human coronaviruses are highly homologous, e.g., the SARS-CoV-2 N protein is 90% similar to SARS-CoV N protein [244].
On the other hand, the S protein, which is responsible for recognizing host cell receptors [245], shares ca. 76% amino acid identity with the SARS-CoV. Since the S protein contains two subunits, S1 and S2, different S-based ELISA tests can be performed. In this regard, Okba et al. [246] have compared the use of the S protein and the S1 subunit as antigens in ELISA tests. Among them, the S1-ELISA presented higher specificity to the SARS-CoV-2 antibodies. Possibly, this finding can be associated with higher conservation of the S2 subunit of the SARS-CoV-2, resulting in cross-reaction with the S antibodies of the MERS-CoV. Furthermore, the S1 subunit mediates the cell attachment and contains four domains, including the receptor-binding domain (RBD), which is the most variable part of the coronavirus genome [229] and the principal target of the antibodies that neutralize SARS-CoV-2 [247].
Regarding electrochemical detection, Vadlamani et al. [248] reported an electrochemical biosensor for the detection of SARS-CoV-2 through the spike RBD of the virus. For this, cobalt-functionalized TiO2 nanotubes (Co-TNT) were obtained by electrochemical anodization and wet ion exchange, being applied as the working electrode. Although, in principle, the sensor was able to detect the RBD protein with the use of amperometric techniques it is far from being adequate for clinical use. Its main drawback is the lack of a specific biorreceptor for the target, making the detection in complex biological samples impracticable.
An impressive work that employed electrochemistry for aiding in COVID-19 diagnosis was developed by Zhao et al. [87]. The authors developed a diagnostic system for the detection of SARS-COV-2 RNA without the need for nucleic acid amplification using only a smartphone. For that, a super sandwich genosensor was applied, with a capture probe being anchored by thiol bonds to Au@Fe3O4 nanoparticles and a label probe being anchored to AuNPs present in a p-sulfocalix [8]arene functionalized graphene containing toluidine blue (TB) as an electrochemical reporter. A third sequence, named auxiliary probe, can hybridize with two different label probe areas, producing long concatamers and high sensitivity. The developed genosensor was tested in RNA extracts from SARS-CoV-2 confirmed patients, presenting detection rates higher than the ones presented by RT-qPCR in the same samples (between 85.5 and 46.2% for the developed biosensor and between 56.5 and 7.7% for RT-qPCR). Furthermore, the LOD achieved by the device is the lowest reported (200 copies mL−1) among the SARS-CoV-2 genosensors published to date. The device presents itself as a great tool for aiding in the diagnosis of SARS-CoV-2 especially in low resource settings, as it relies on simple instrumentation and does not require any amplification methods to be performed.
Since the diagnosis testing for COVID-19 is based on the presence of the virus or its antibodies, the results vary over time [249]. In this sense, He et al. [250] have investigated the viral load dynamics of SARS-CoV-2 in throat swabs from patients with COVID-19. According to the authors, the viral load peak is reached at the time of symptom onset and gradually decreases towards the LOD of RT-PCR at about day 21. Thus, although the PCR-based tests allow the selective detection of SARS-CoV-2, false-negative results can be observed during the incubation time, estimated to be 5.1 days [251]. The sensibility of the PCR assays also depends on the sampling technique. For example, Wang et al. reported higher positive rates in bronchoalveolar lavage fluid specimens compared to the nasal or pharyngeal swabs [252] due to the slower decline of the viral RNA in these samples. While the viral load decreases over the second week, the IgG and IgM antibodies concentrations increase from day ten after symptom onset [243]. Therefore, the use of serological tests is not indicated for the early diagnosis of COVID-19 and is more suitable during the convalescent phase of the disease.
Rashed et al. [253] demonstrated the feasibility of using the EIS technique for rapid and accurate detection of SARS-COV-2 antibodies. A 16-well plate containing gold electrodes was pre-coated with RBD. The plate interface independently addressed each well to acquire single frequency measurements (10 kHz) every few seconds. The association and disassociation of the protein RBD and the SARS-COV-2 antibodies provoked rapid changes in the total impedance, indicating if the sample is positive, or negative for SARS-CoV-2 antibodies. Fast (5 min) and reliable results were obtained for the screening of serum samples, with the immunosensor being able to differentiate all positive clinical samples from negative controls. The technique is label-free, relies on commercially available equipment, and requires simple sample handling, being a great candidate for clinical applications.
Outstanding work by Seo et al. [122] recently reported a FET-based immunosensor for rapid diagnosis of COVID-19, which can operate without any sample preparation and with a processing time lower than 1 min. The proposed immunosensor successfully detected the SARS-CoV-2 spike protein at fg mL−1 concentrations. Moreover, it was able to detect the whole SARS-CoV-2 virus in culture medium and in clinical samples, collected from the nasopharyngeal swab of COVID-19 patients. The limit of detection of 2.42 × 102 copies mL−1, for the clinical samples, was comparable to the values reported for the current molecular diagnostic tests of COVID-19 [254].
Variations in the virus dynamics and the host immune response have been observed in mild and severe cases of COVID-19. For example, patients with severe COVID-19 can present viral load 60 times higher than that one associated with mild cases [255]. Shen et al. [256] reported delayed specific IgM antibody responses in patients with severe COVID-19 patients. According to this study, the positive rate of IgM, at ≤3 days after symptom onset, was 38% for the mild group against 0% for the severe group. To avoid misinterpretation of the COVID-19 diagnostics, the serological tests can complement the PCR-based assays. Furthermore, the diagnosis or even the treatment of COVID-19 patients can be guided by the identification of other biomarkers. In this regard, a systematic review [257] explored the role of the biomarkers in the diagnosis of COVID-19, including CRP, serum amyloid A, interleukin-6, lactate dehydrogenase, neutrophil-to-lymphocyte ratio, D-dimer, cardiac troponin, renal biomarkers, lymphocytes, and platelet count.
Recently, Guan et al. [258] have described interesting biomarkers for COVID-19, including CRP, procalcitonin, and lactate dehydrogenase (LDH). Besides CRP, procalcitonin, and LDH presenting elevated concentrations in the patients, the concentrations of the biomarkers were even further increased in severely ill patients when compared to mild cases. IL-6 was also reported as an essential biomarker for COVID-19 in a study published by Chen et al. [259]. The authors stated that 52% of the tested COVID-19 patients presented elevated IL-6 levels at admission.
7. Concluding remarks and future perspectives
The recent advances highlighted in this review make clear that biosensors display high applicability towards the rapid, low-cost, and straightforward diagnosis of viral diseases. These devices show similar analytical performance to traditional virus-detection techniques, such as PCR and ELISA, while presenting improvements in many different aspects. One of the main advantages presented by them is the ability of miniaturization, which allows the construction of portable, versatile, and low-sample consumption biosensors. Furthermore, such small dimensions allow the integration of diverse complex bench-top protocols for viral analysis to be performed in a single hand-held device – named lab-on-a-chip. Its ability to perform genetic material extraction, isolation, amplification, and detection in a rapid, portable and automated manner brings great promises to the area, including low-cost analysis being performed by untrained users even in low-resource settings. The recent miniaturization and flexibility regarding electrical and electrochemical instrumentation also contribute greatly to the use of biosensors in situ, providing rapid medical responses and no sample transportation costs, presenting great potential for improving clinical diagnosis [260,261].
Furthermore, the use of new materials for building microfluidic devices and electrodes brings novel applications to the table. The use of flexible substrates, for example, allows the construction of compact and mechanically robust platforms with great adherence to the skin or soft organs [262]. This strategy, in its turn, makes direct and continuous sample collection possible, allowing rapid responses in the advent of infections, improving the patients’ outcome, and possibly reducing treatment-associated costs [263,264]. The use of paper for building state-of-the-art biosensors has also been increasing due to its ability to store active biomolecules for long periods, providing energy-free fluid flux, presenting low-cost and great commercial availability.
New and improved electrodes building strategies also brings great promises to the area. The use of 3D-printing and laser inscription allows the customized construction of low-cost electrodes for specific applications – greatly increasing, therefore, the versatility of construction and analytical performance of devices while lowering its costs [265]. Emerging technologies regarding improvements in bioreceptors, such as DNA origami, can also provide biosensors with a low limit of detection and great sensitivity. In this regard, DNA programmability has been applied to create supramolecular structures in desired shapes and sizes with great reproducibility, allowing bioanalytical chemistry to advance towards single-molecule detection [266]. Last, the association of biosensors with recently emerged molecular biology techniques, such as RCA and LAMP, is an excellent alternative for the construction of devices able to perform analysis with great sensitivity and specificity without the need of thermocyclers or complex equipment.
Despite their advantages, it is noteworthy to mention that currently electrochemical biosensors are not commercially available - except for very specific examples such as the glucometer – making it worthwile to hightlight some of the challenges for its routine use. Different aspects possibly contribute to this, including: 1) The lack of stability of the biorecognition element layer. Biosensors commonly make use of biomolecules structured in a specific manner in the biorecognition layer, with the stability of the biomolecule itself and also the layer being essential for the adequate functioning of the device. It is still a great challenge to keep the adequate functionality of the biorecognition layer for long periods, under different temperatures and storage conditions. This instability significantly hampers the lifetime of the devices, bringing great challenges to its commercialization. 2) The complex fabrication and the use of high-cost materials. Many biosensors, in order to improve their analytical parameters (sensitivity, limit of detection, reproducibility), are based on a highly complex design and/or make use of materials that present high costs (nanoparticles, rare elements, liquid cristals). While complex construction methods might not be adequate for batch construction, high-cost materials inviabilize the competition with current techniques. 3) The barrier for market entrance. The industrial interest in the commercialization of electrochemical biosensors depends on a clear market demand, as there are many costs associated with new technologies. First, regulatory agencies must approve the product. Then, all machinery and protocol must be adequate to batch produce the devices at low costs and with reproducibility. 4) Last but not least, the adequate preparation of real samples. Some devices, although presenting great analytical performance under controlled environments, are not able to perform proper readouts in raw real samples. The reasons for that may vary, including the presence of interfering species, biofouling, and the analyte nature (the size of a target DNA or the formation of complexes with the analyte, for example). These are still great challenges to be addressed in order to make general electrochemical biosensors become commercially available.
It is noteworthy to mention that different advancements in electrochemical sensing offer promises for addressing these commercial challenges. These include, for example, the use of 3-D printed electrodes (for lower costs and higher accessibility), the use of new assembles and labels (for a longer lifetime and more stability, sensitivity, and reliability of the devices), the use of innovative bioreceptors (biomimetic enzymes, molecularly imprinted membranes, DNA origami), and the development of flexible devices (improving the range of application of the sensors). Furthermore, the development of new and improved electronics and the recent advancements on the “Internet of Things” hold great promises for future commercial devices.
In conclusion, the determination of whole viruses, parts of them, or even biomarkers related to specific illness can significantly aid in the differentiation and diagnosis of diseases with similar clinical symptoms, greatly assisting physicians in daily clinical practice. The recent advances in biosensors for viral disease diagnosis have allowed rapid in situ monitoring to be performed at low costs even in the absence of complex equipment or specialized workers. Although the manufacture and application of these biosensors still present many challenges to be overcome, these devices hold great promises for the real-time and continuous monitoring of specific pathogens. Furthermore, these are extremely valuable tools in endemic and pandemic scenarios, allowing the sensitive and specific detection of pathogens to be performed in an uncentralized and low-cost manner with minimal resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful to São Paulo Research Foundation - FAPESP (Grant# 2013/22127-2; 2017/21097-3; 2018/19750-3; 2019/00473-2; 2019/01844-4), Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (Grant # 307271/2017-0; 303338/2019-9), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) (Finance code 001; grant # 88887.504861/2020-00, and grant # 3007/2014), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais - FAPEMIG (Grant# APQ-03141-18) for financial support. We would also like to thank the INCTBio grants (FAPESP #2014/40867-3 and CNPq #465389/2014–7).
Biographies

Laís Canniatti Brazaca received her M.S. and Ph.D. degrees from São Carlos Institute of Physics, University of São Paulo (IFSC/USP), Brazil, in 2015 and 2019, respectively. She is currently a postdoctoral researcher at São Carlos Institute of Chemistry, University of São Paulo (IQSC/USP). Her specialties include biosensors for medical diagnosis, microfabrication, electrodes modification, nanomaterials for sensing applications and paper-based devices.

Pãmyla L. dos Santos is graduated in Chemistry from the Federal University of Lavras (2012), and received her master’s (2015) and Ph.D (2019) degree in Inorganic Chemistry from the University of Campinas, with an internship period at the Manchester Metropolitan University, UK (2018). She currently works as a post-doctoral research fellow at the Federal University of Santa Catarina. Her research interests are focused on synthesis and characterization of graphene composites, and development of 3D-printed electrodes for electroanalysis and energy conversion applications.

Paulo Roberto de Oliveira received his Ph.D. in Analytical Chemistry from Federal University of Paraná (UFPR), Curitiba-PR, Brazil, in 2016. He is now a Post-doc research in the Federal University of São Carlos (UFSCar), Araras-SP, Brazil. His current interest is the development of electrochemical sensors and biosensors for environmental applications.

Diego Pessoa Rocha is graduated in Chemistry from the Federal University of Uberlândia in 2011, and achieved his master (2015) and Ph.D. degree (2020) at the same University in the Analytical Chemistry area with an internship period at the Manchester Metropolitan University, UK (2018–2019). He is currently a postdoctoral researcher at Federal University of Uberlândia. He has experience on modified electrodes with carbon nanomaterials, such as carbon nanotubes and graphene oxide, and their application in hydrodynamic systems, such as batch-injection and flow-injection analysis.

Jéssica Santos Stefano is graduated in Chemistry from the Federal University of Uberlândia in 2014, and achieved her master (2016) and Ph.D. degree (2020) at the same University in the Analytical Chemistry area with an internship period at the Ruhr-Universität Bochum, DE (2018–2019). She is currently a postdoctoral researcher at Federal University of Uberlândia. She has experience on modified electrodes with carbon nanomaterials, such as carbon nanotubes, and their application in hydrodynamic systems, such as batch-injection and flow-injection analysis, as well as with bipolar electrochemistry.

Cristiane Kalinke is graduated in Chemistry from the Federal Technological University of Paraná, Brazil (2013), and received her master’s (2015) and Ph.D. degree (2019) in Analytical Chemistry from Federal University of Paraná, Brazil. She is now a postdoctoral research at the University of Campinas. Her research interest is the development of biochar-based and 3D-printed electrochemical sensors and biosensors for biological applications.

Rodrigo A. A. Munoz is graduated in Chemistry from the University of Sao Paulo, Brazil (2002), and received his Ph.D. in Analytical Chemistry from the same university in 2006, an internship period at the Oxford University, UK (2005). He completed a postdoctoral research at the Arizona State University (USA) during 2006–2007 and a postdoctoral research at the University of Sao Paulo during 2007–2008. He is currently Associate Professor of Chemistry at the Federal University of Uberlandia, Brazil. His current research interests focus on the development of analytical methods and electrochemical (bio)sensors associated with flow-injection and batch-injection analyses using the 3D-printing technology with applications on forensics, food, biological and fuels.

Juliano Alves Bonacin received his Ph.D. degree in 2007 (University of São Paulo, Brazil) and was Postdoctoral Fellow between 2007 and 2009 (University of São Paulo, Brazil). He founded one of the first companies in the Nanotechnology field in Brazil and nowadays has a permanent position as Associate Professor at Institute of Chemistry of University of Campinas (UNICAMP). His research interests are energy conversion, water oxidation, electrocatalysis, and 3D-printing technology.

Bruno Campos Janegitz received Ph.D. degree from Federal University of S ∼ ao Carlos, in 2012. He was a postdoctoral researcher at University of São Paulo between 2012 and 2014. At present, he is Professor at Federal University of São Carlos. His research interests include electroanalytical chemistry, nanostructured electrode materials and modified electrode surfaces, electrochemical sensors and biosensors for medical and environmental analysis.

Emanuel Carrilho is a chemist (1987) with a Master of Science in Analytical Chemistry from the University of São Paulo (USP) at São Carlos, Brazil (1990). He obtained his Ph.D. at the Northeastern University, Boston, Ma (1997), and was a visiting scholar at Harvard University in Whitesides’ group (2007–2009). Carrilho is full professor at the São Carlos Institute of Chemistry, USP since 2013. Carrilho’s group has been working on the development of new bioanalytical methods covering the broad aspects of genomics, proteomics, metabolomics for human health and applied microbiology in the search for cancer biomarkers and neglected tropical diseases. Recently, BioMicS Group is developing microfluidic applications for low-cost diagnostics for developing countries using paper-based analytical devices (μPADs) and developing new ultra-sensitive contactless conductivity detection.
References
- 1.Saylan Y., Erdem Ö., Ünal S., Denizli A. An alternative medical diagnosis method: biosensors for virus detection. Biosensors. 2019;9:65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56 doi: 10.1128/mr.56.1.152-179.1992. http://mmbr.asm.org/content/56/1/152.abstract 152 LP – 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldmann H., Geisbert T.W. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellors J.W., Rinaldo C.R., Gupta P., White R.M., Todd J.A., Kingsley L.A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science (80-. ) 1996;272 doi: 10.1126/science.272.5265.1167. 1167 LP – 1170. [DOI] [PubMed] [Google Scholar]
- 6.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. U.K. HLH across Speciality Collaboration, COVID-19: Consider Cytokine Storm Syndromes and Immunosuppression. Lancet (London, England) 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryu W.-S. W.-S.B.T.-M.V. of H.P.V. Ryu. Academic Press; Boston: 2017. Chapter 2-virus structure; pp. 21–29. [DOI] [Google Scholar]
- 8.Cohen F.S. How viruses invade cells. Biophys. J. 2016;110:1028–1032. doi: 10.1016/j.bpj.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryu W.-S. W.-S.B.T.-M.V. of H.P.V. Ryu. Academic Press; Boston: 2017. Chapter 3-virus life cycle; pp. 31–45. [DOI] [Google Scholar]
- 10.Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 11.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross A.G.P., Crowe S.M., Tyndall M.W. Planning for the next global pandemic. Int. J. Infect. Dis. 2015;38:89–94. doi: 10.1016/j.ijid.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribeiro B.V., Cordeiro T.A.R., Oliveira e Freitas G.R., Ferreira L.F., Franco D.L. Biosensors for the detection of respiratory viruses: a review. Talanta Open. 2020;2:100007. doi: 10.1016/j.talo.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mokhtarzadeh A., Eivazzadeh-Keihan R., Pashazadeh P., Hejazi M., Gharaatifar N., Hasanzadeh M., Baradaran B., de la Guardia M. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal. Chem. 2017;97:445–457. doi: 10.1016/j.trac.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji T., Liu Z., Wang G., Guo X., Akbar khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Detection of COVID-19: a review of the current literature and future perspectives, Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pividori M.I., Merkoçi A., Alegret S. Electrochemical genosensor design: immobilisation of oligonucleotides onto transducer surfaces and detection methods. Biosens. Bioelectron. 2000;15:291–303. doi: 10.1016/s0956-5663(00)00071-3. [DOI] [PubMed] [Google Scholar]
- 17.Liu S.-J., Nie H.-G., Jiang J.-H., Shen G.-L., Yu R.-Q. Electrochemical sensor for mercury(II) based on conformational switch mediated by interstrand cooperative coordination. Anal. Chem. 2009;81:5724–5730. doi: 10.1021/ac900527f. [DOI] [PubMed] [Google Scholar]
- 18.Mikkelsen S.R. Electrochecmical biosensors for DNA sequence detection. Electroanalysis. 1996;8:15–19. doi: 10.1002/elan.1140080104. [DOI] [Google Scholar]
- 19.Nolasco G., Sequeira Z., Soares C., Mansinho A., Bailey A.M., Niblett C.L. Asymmetric PCR ELISA: increased sensitivity and reduced costs for the detection of plant viruses. Eur. J. Plant Pathol. 2002;108:293–298. doi: 10.1023/A:1015649429160. [DOI] [Google Scholar]
- 20.Kara P., Kerman K., Ozkan D., Meric B., Erdem A., Ozkan Z., Ozsoz M. Electrochemical genosensor for the detection of interaction between methylene blue and DNA. Electrochem. Commun. 2002;4:705–709. doi: 10.1016/S1388-2481(02)00428-9. [DOI] [Google Scholar]
- 21.Mazlan N.-F., Tan L.L., Karim N.H.A., Heng L.Y., Jamaluddin N.D., Yusof N.Y.M., Quay D.H.X., Khalid B. Acrylic-based genosensor utilizing metal salphen labeling approach for reflectometric dengue virus detection. Talanta. 2019;198:358–370. doi: 10.1016/j.talanta.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira D.A., V Silva J., Flauzino J.M.R., Castro A.C.H., Moço A.C.R., Soares M.M.C.N., Madurro J.M., Brito-Madurro A.G. Application of nanomaterials for the electrical and optical detection of the hepatitis B virus. Anal. Biochem. 2018;549:157–163. doi: 10.1016/j.ab.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 23.Abdul Rasheed P., Sandhyarani N. Quartz crystal microbalance genosensor for sequence specific detection of attomolar DNA targets. Anal. Chim. Acta. 2016;905:134–139. doi: 10.1016/j.aca.2015.11.033. [DOI] [PubMed] [Google Scholar]
- 24.Guliy O.I., Zaitsev B.D., Larionova O.S., Borodina I.A. Virus detection methods and biosensor technologies. Biophysics. 2019;64:890–897. doi: 10.1134/S0006350919060095. [DOI] [Google Scholar]
- 25.Rasheed P.A., Sandhyarani N. Carbon nanostructures as immobilization platform for DNA: a review on current progress in electrochemical DNA sensors. Biosens. Bioelectron. 2017;97:226–237. doi: 10.1016/j.bios.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 26.da Fonseca Alves R., Franco D.L., Cordeiro M.T., de Oliveira E.M., Fireman Dutra R.A., Del Pilar Taboada Sotomayor M. Novel electrochemical genosensor for Zika virus based on a poly-(3-amino-4-hydroxybenzoic acid)-modified pencil carbon graphite electrode. Sens. Actuator. B Chem. 2019;296:126681. doi: 10.1016/j.snb.2019.126681. [DOI] [Google Scholar]
- 27.Dungchai W., Chailapakul O., Henry C.S. Electrochemical detection for paper-based microfluidics. Anal. Chem. 2009;81:5821–5826. doi: 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- 28.Koczula K.M., Gallotta A. Lateral flow assays. Essays Biochem. 2016;60:111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma S., Zapatero-Rodriguez J., Estrela P., O’Kennedy R. Point-of-care diagnostics in low resource settings: present status and future role of microfluidics. Biosensors. 2015;5:577–601. doi: 10.3390/bios5030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akanda M.R., Sohail M., Aziz M.A., Kawde A.-N. Recent advances in nanomaterial-modified pencil graphite electrodes for electroanalysis. Electroanalysis. 2016;28:408–424. doi: 10.1002/elan.201500374. [DOI] [Google Scholar]
- 31.Zhao G., Wang H., Liu G. Recent advances in chemically modified electrodes, microfabricated devices and injection systems for the electrochemical detection of heavy metals: a review. Int. J. Electrochem. Sci. 2017;12:8622–8641. [Google Scholar]
- 32.Sajid M., Nazal M.K., Mansha M., Alsharaa A., Jillani S.M.S., Basheer C. Chemically modified electrodes for electrochemical detection of dopamine in the presence of uric acid and ascorbic acid: a review. TrAC Trends Anal. Chem. 2016;76:15–29. doi: 10.1016/j.trac.2015.09.006. [DOI] [Google Scholar]
- 33.Redepenning J.G. Chemically modified electrodes: a general overview. TrAC Trends Anal. Chem. 1987;6:18–22. doi: 10.1016/0165-9936(87)85014-8. [DOI] [Google Scholar]
- 34.González-López A., Fernández Abedul M.T. Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. Lab. Methods Dyn. Electroanal. 2020:213–220. doi: 10.1016/B978-0-12-815932-3.00021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Movilli J., Rozzi A., Ricciardi R., Corradini R., Huskens J. Control of probe density at DNA biosensor surfaces using poly(l-lysine) with appended reactive groups. Bioconjug. Chem. 2018;29:4110–4118. doi: 10.1021/acs.bioconjchem.8b00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Idili A., Amodio A., Vidonis M., Feinberg-Somerson J., Castronovo M., Ricci F., Folding-Upon-Binding, Electrochemical Signal-On. DNA sensor with high affinity and specificity. Anal. Chem. 2014;86:9013–9019. doi: 10.1021/ac501418g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farzin L., Sadjadi S., Shamsipur M., Sheibani S. Electrochemical genosensor based on carbon nanotube/amine-ionic liquid functionalized reduced graphene oxide nanoplatform for detection of human papillomavirus (HPV16)-related head and neck cancer. J. Pharm. Biomed. Anal. 2020;179:112989. doi: 10.1016/j.jpba.2019.112989. [DOI] [PubMed] [Google Scholar]
- 38.Janegitz B.C., Silva T.A., Wong A., Ribovski L., Vicentini F.C., del M., Taboada Sotomayor P., Fatibello-Filho O. The application of graphene for in vitro and in vivo electrochemical biosensing, Biosens. Bioelectron. 2017;89:224–233. doi: 10.1016/j.bios.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 39.Silva T.A., Moraes F.C., Janegitz B.C., Fatibello-Filho O. Electrochemical biosensors based on nanostructured carbon black: a review. J. Nanomater. 2017;2017:4571614. doi: 10.1155/2017/4571614. [DOI] [Google Scholar]
- 40.Wang J., Cai X., Rivas G., Shiraishi H., Dontha N. Nucleic-acid immobilization, recognition and detection at chronopotentiometric DNA chips. Biosens. Bioelectron. 1997;12:587–599. doi: 10.1016/S0956-5663(96)00076-0. [DOI] [PubMed] [Google Scholar]
- 41.Wang J., Cai X., Fernandes J.R., Grant D.H., Ozsoz M. Electrochemical measurements of oligonucleotides in the presence of chromosomal DNA using membrane-covered carbon electrodes. Anal. Chem. 1997;69:4056–4059. doi: 10.1021/ac9704319. [DOI] [PubMed] [Google Scholar]
- 42.Marrazza G., Chianella I., Mascini M. vol. 14. 1999. Disposable DNA Electrochemical Sensor for Hybridization detection1; pp. 43–51. (This Paper Was Presented at the Fifth World Congress on Biosensors, Berlin, Germany, 3–5 June 1998.1, Biosens. Bioelectron). [DOI] [PubMed] [Google Scholar]
- 43.Erdem A., Pividori M.I., del Valle M., Alegret S. Rigid carbon composites: a new transducing material for label-free electrochemical genosensing. J. Electroanal. Chem. 2004;567:29–37. doi: 10.1016/j.jelechem.2003.10.049. [DOI] [Google Scholar]
- 44.Huy T.Q., Hanh N.T.H., Van Chung P., Anh D.D., Nga P.T., Tuan M.A. Characterization of immobilization methods of antiviral antibodies in serum for electrochemical biosensors. Appl. Surf. Sci. 2011;257:7090–7095. doi: 10.1016/j.apsusc.2011.03.051. [DOI] [Google Scholar]
- 45.Zhang Y., Wang J., Xu M. A sensitive DNA biosensor fabricated with gold nanoparticles/ploy (p-aminobenzoic acid)/carbon nanotubes modified electrode. Colloids Surf. B Biointerfaces. 2010;75:179–185. doi: 10.1016/j.colsurfb.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 46.Balasubramanian K., Burghard M. Chemically functionalized carbon nanotubes. Small. 2005;1:180–192. doi: 10.1002/smll.200400118. [DOI] [PubMed] [Google Scholar]
- 47.Saadati A., Hassanpour S., Hasanzadeh M., Shadjou N. Binding of pDNA with cDNA using hybridization strategy towards monitoring of Haemophilus influenza genome in human plasma samples. Int. J. Biol. Macromol. 2020;150:218–227. doi: 10.1016/j.ijbiomac.2020.02.062. [DOI] [PubMed] [Google Scholar]
- 48.dos C., Riccardi S., Dahmouche K., Santilli C.V., da Costa P.I., Yamanaka H. Immobilization of streptavidin in sol–gel films: application on the diagnosis of hepatitis C virus. Talanta. 2006;70:637–643. doi: 10.1016/j.talanta.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 49.Chung D.-J., Kim K.-C., Choi S.-H. Electrochemical DNA biosensor based on avidin–biotin conjugation for influenza virus (type A) detection. Appl. Surf. Sci. 2011;257:9390–9396. doi: 10.1016/j.apsusc.2011.06.015. [DOI] [Google Scholar]
- 50.Faria H.A.M., Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019;131:149–155. doi: 10.1016/j.bios.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 51.Brazaca L.C., Bramorski C.B., Cancino-Bernardi J., Janegitz B.C., Zucolotto V. A genosensor for sickle cell anemia trait determination. Electroanalysis. 2016;29:773–777. doi: 10.1002/elan.201600573. [DOI] [Google Scholar]
- 52.Manzano M., Viezzi S., Mazerat S., Marks R.S., Vidic J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018;100:89–95. doi: 10.1016/j.bios.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 53.Wu B., Yang T., Zou D., Jin L., Liang X., Li T., Huang G., Zhang J. Nuclear magnetic resonance biosensor based on streptavidin–biotin system and poly-l-lysine macromolecular targeted gadolinium probe for rapid detection of Salmonella in milk. Int. Dairy J. 2020;102:104594. doi: 10.1016/j.idairyj.2019.104594. [DOI] [Google Scholar]
- 54.Jimenez-Falcao S., Parra-Nieto J., Pérez-Cuadrado H., Martínez-Máñez R., Martínez-Ruiz P., Villalonga R. Avidin-gated mesoporous silica nanoparticles for signal amplification in electrochemical biosensor. Electrochem. Commun. 2019;108:106556. doi: 10.1016/j.elecom.2019.106556. [DOI] [Google Scholar]
- 55.Wong J., Chilkoti A., Moy V.T. Direct force measurements of the streptavidin–biotin interaction. Biomol. Eng. 1999;16:45–55. doi: 10.1016/S1050-3862(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 56.Carinelli S., Kühnemund M., Nilsson M., Pividori M.I. Yoctomole electrochemical genosensing of Ebola virus cDNA by rolling circle and circle to circle amplification. Biosens. Bioelectron. 2017;93:65–71. doi: 10.1016/j.bios.2016.09.099. [DOI] [PubMed] [Google Scholar]
- 57.Gire S.K., Goba A., Andersen K.G., Sealfon R.S.G., Park D.J., Kanneh L., Jalloh S., Momoh M., Fullah M., Dudas G., Wohl S., Moses L.M., Yozwiak N.L., Winnicki S., Matranga C.B., Malboeuf C.M., Qu J., Gladden A.D., Schaffner S.F., Yang X., Jiang P.-P., Nekoui M., Colubri A., Coomber M.R., Fonnie M., Moigboi A., Gbakie M., Kamara F.K., Tucker V., Konuwa E., Saffa S., Sellu J., Jalloh A.A., Kovoma A., Koninga J., Mustapha I., Kargbo K., Foday M., Yillah M., Kanneh F., Robert W., Massally J.L.B., Chapman S.B., Bochicchio J., Murphy C., Nusbaum C., Young S., Birren B.W., Grant D.S., Scheiffelin J.S., Lander E.S., Happi C., Gevao S.M., Gnirke A., Rambaut A., Garry R.F., Khan S.H., Sabeti P.C. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science (80-. ) 2014;345 doi: 10.1126/science.1259657. 1369 LP – 1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mezger A., Ohrmalm C., Herthnek D., Blomberg J., Nilsson M. Detection of rotavirus using padlock probes and rolling circle amplification. PloS One. 2014;9 doi: 10.1371/journal.pone.0111874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin X., Lian X., Luo B., Huang X.-C. A highly sensitive and stable electrochemical HBV DNA biosensor based on ErGO-supported Cu-MOF. Inorg. Chem. Commun. 2020;119:108095. doi: 10.1016/j.inoche.2020.108095. [DOI] [Google Scholar]
- 60.Alzate D., Cajigas S., Robledo S., Muskus C., Orozco J. Genosensors for differential detection of Zika virus. Talanta. 2020;210:120648. doi: 10.1016/j.talanta.2019.120648. [DOI] [PubMed] [Google Scholar]
- 61.Ziółkowski R., Oszwałdowski S., Kopyra K.K., Zacharczuk K., Zasada A.A., Malinowska E. Toward fluorimetric-paired-emitter-detector-diode test for Bacillus anthracis DNA based on graphene oxide. Microchem. J. 2020;154:104592. doi: 10.1016/j.microc.2019.104592. [DOI] [Google Scholar]
- 62.El-Yazbi A.F., Loppnow G.R. Probing DNA damage induced by common antiviral agents using multiple analytical techniques. J. Pharmaceut. Biomed. Anal. 2018;157:226–234. doi: 10.1016/j.jpba.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 63.Singhal C., Pundir C.S., Narang J. A genosensor for detection of consensus DNA sequence of Dengue virus using ZnO/Pt-Pd nanocomposites. Biosens. Bioelectron. 2017;97:75–82. doi: 10.1016/j.bios.2017.05.047. [DOI] [PubMed] [Google Scholar]
- 64.Malecka K., Stachyra A., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Radecka H., Radecki J. Electrochemical genosensor based on disc and screen printed gold electrodes for detection of specific DNA and RNA sequences derived from Avian Influenza Virus H5N1. Sens. Actuator. B Chem. 2016;224:290–297. doi: 10.1016/j.snb.2015.10.044. [DOI] [Google Scholar]
- 65.Teengam P., Siangproh W., Tuantranont A., Henry C.S., Vilaivan T., Chailapakul O. Electrochemical paper-based peptide nucleic acid biosensor for detecting human papillomavirus. Anal. Chim. Acta. 2017;952:32–40. doi: 10.1016/j.aca.2016.11.071. [DOI] [PubMed] [Google Scholar]
- 66.Cajigas S., Alzate D., Orozco J. Gold nanoparticle/DNA-based nanobioconjugate for electrochemical detection of Zika virus. Microchim. Acta. 2020;187:594. doi: 10.1007/s00604-020-04568-1. [DOI] [PubMed] [Google Scholar]
- 67.Chen J., Liu Z., Zheng Y., Lin Z., Sun Z., Liu A., Chen W., Lin X. B/C genotyping of hepatitis B virus based on dual-probe electrochemical biosensor. J. Electroanal. Chem. 2017;785:75–79. doi: 10.1016/j.jelechem.2016.12.017. [DOI] [Google Scholar]
- 68.Shariati M. The field effect transistor DNA biosensor based on ITO nanowires in label-free hepatitis B virus detecting compatible with CMOS technology. Biosens. Bioelectron. 2018;105:58–64. doi: 10.1016/j.bios.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 69.Steinmetz M., Lima D., Viana A.G., Fujiwara S.T., Pessôa C.A., Etto R.M., Wohnrath K. A sensitive label-free impedimetric DNA biosensor based on silsesquioxane-functionalized gold nanoparticles for Zika Virus detection. Biosens. Bioelectron. 2019;141:111351. doi: 10.1016/j.bios.2019.111351. [DOI] [PubMed] [Google Scholar]
- 70.Du M., Yang T., Li X., Jiao K. Fabrication of DNA/graphene/polyaniline nanocomplex for label-free voltammetric detection of DNA hybridization. Talanta. 2012;88:439–444. doi: 10.1016/j.talanta.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 71.Srisomwat C., Teengam P., Chuaypen N., Tangkijvanich P., Vilaivan T., Chailapakul O. Pop-up paper electrochemical device for label-free hepatitis B virus DNA detection. Sens. Actuator. B Chem. 2020;316:128077. doi: 10.1016/j.snb.2020.128077. [DOI] [Google Scholar]
- 72.Mashhadizadeh M.H., Talemi R.P. Synergistic effect of magnetite and gold nanoparticles onto the response of a label-free impedimetric hepatitis B virus DNA biosensor. Mater. Sci. Eng. C. 2016;59:773–781. doi: 10.1016/j.msec.2015.10.082. [DOI] [PubMed] [Google Scholar]
- 73.Wang S., Liu Q., Li H., Li Y., Hao N., Qian J., Zhu W., Wang K. Fabrication of label-free electrochemical impedimetric DNA biosensor for detection of genetically modified soybean by recognizing CaMV 35S promoter. J. Electroanal. Chem. 2016;782:19–25. doi: 10.1016/j.jelechem.2016.09.052. [DOI] [Google Scholar]
- 74.Avelino K.Y.P.S., Oliveira L.S., Lucena-Silva N., de Melo C.P., Andrade C.A.S., Oliveira M.D.L. Metal-polymer hybrid nanomaterial for impedimetric detection of human papillomavirus in cervical specimens. J. Pharm. Biomed. Anal. 2020;185:113249. doi: 10.1016/j.jpba.2020.113249. [DOI] [PubMed] [Google Scholar]
- 75.Ribovski L., Zucolotto V., Janegitz B.C. A label-free electrochemical DNA sensor to identify breast cancer susceptibility. Microchem. J. 2017;133:37–42. doi: 10.1016/j.microc.2017.03.011. [DOI] [Google Scholar]
- 76.Civit L., Fragoso A., Hölters S., Dürst M., O’Sullivan C.K. Electrochemical genosensor array for the simultaneous detection of multiple high-risk human papillomavirus sequences in clinical samples. Anal. Chim. Acta. 2012;715:93–98. doi: 10.1016/j.aca.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 77.Manzanares-Palenzuela C.L., Martín-Fernández B., Sánchez-Paniagua López M., López-Ruiz B. Electrochemical genosensors as innovative tools for detection of genetically modified organisms. TrAC Trends Anal. Chem. 2015;66:19–31. doi: 10.1016/j.trac.2014.10.006. [DOI] [Google Scholar]
- 78.Berti F., Laschi S., Palchetti I., Rossier J.S., Reymond F., Mascini M., Marrazza G. Microfluidic-based electrochemical genosensor coupled to magnetic beads for hybridization detection. Talanta. 2009;77:971–978. doi: 10.1016/j.talanta.2008.07.064. [DOI] [PubMed] [Google Scholar]
- 79.Vo-Dinh T. Development of a DNA biochip: principle and applications1The submitted manuscript has been authored by a contractor of the US Government under contract No. DE-AC05-96OR22464. Accordingly, the US Government retains a nonexclusive royalty-free license to pu. Sens. Actuator. B Chem. 1998;51:52–59. doi: 10.1016/S0925-4005(98)00182-8. [DOI] [Google Scholar]
- 80.Li Y., Wang Z., Sun L., Liu L., Xu C., Kuang H. Nanoparticle-based sensors for food contaminants. TrAC Trends Anal. Chem. 2019;113:74–83. doi: 10.1016/j.trac.2019.01.012. [DOI] [Google Scholar]
- 81.Saleh T.A., Fadillah G., Saputra O.A. Nanoparticles as components of electrochemical sensing platforms for the detection of petroleum pollutants: a review. TrAC Trends Anal. Chem. 2019;118:194–206. doi: 10.1016/j.trac.2019.05.045. [DOI] [Google Scholar]
- 82.Shariati M., Ghorbani M., Sasanpour P., Karimizefreh A. An ultrasensitive label free human papilloma virus DNA biosensor using gold nanotubes based on nanoporous polycarbonate in electrical alignment. Anal. Chim. Acta. 2019;1048:31–41. doi: 10.1016/j.aca.2018.09.062. [DOI] [PubMed] [Google Scholar]
- 83.Brazaca L.C., Ribovski L., Janegitz B.C., Zucolotto V. Elsevier, Woodhead Publishing; 2016. Nanostructured Materials and Nanoparticles for Point of Care (POC) Medical Biosensors, in: Med. Biosens. Point Care Appl; pp. 229–254. [Google Scholar]
- 84.Bonanni A., del Valle M. Use of nanomaterials for impedimetric DNA sensors: a review. Anal. Chim. Acta. 2010;678:7–17. doi: 10.1016/j.aca.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 85.Hu Y., Hua S., Li F., Jiang Y., Bai X., Li D., Niu L. Green-synthesized gold nanoparticles decorated graphene sheets for label-free electrochemical impedance DNA hybridization biosensing. Biosens. Bioelectron. 2011;26:4355–4361. doi: 10.1016/j.bios.2011.04.037. [DOI] [PubMed] [Google Scholar]
- 86.Merkoçi A., Aldavert M., Marín S., Alegret S. New materials for electrochemical sensing V: nanoparticles for DNA labeling. TrAC Trends Anal. Chem. 2005;24:341–349. doi: 10.1016/j.trac.2004.11.007. [DOI] [Google Scholar]
- 87.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jevšnik M., Lusa L., Uršič T., Glinšek Biškup U., Petrovec M. Detection of herpes simplex and varicella-zoster virus from skin lesions: comparison of RT-PCR and isothermal amplification for rapid identification. Diagn. Microbiol. Infect. Dis. 2020:115015. doi: 10.1016/j.diagmicrobio.2020.115015. [DOI] [PubMed] [Google Scholar]
- 89.Steininger C., Kundi M., Aberle S.W., Aberle J.H., Popow-Kraupp T. Effectiveness of reverse transcription-PCR, virus isolation, and enzyme-linked immunosorbent assay for diagnosis of influenza A virus infection in different age groups. J. Clin. Microbiol. 2002;40 doi: 10.1128/JCM.40.6.2051-2056.2002. 2051 LP – 2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watzinger F., Ebner K., Lion T. Detection and monitoring of virus infections by real-time PCR. Mol. Aspect. Med. 2006;27:254–298. doi: 10.1016/j.mam.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kwit E., Osiński Z., Rzeżutka A. Detection of viral DNA of myxoma virus using a validated PCR method with an internal amplification control. J. Virol. Methods. 2019;272:113709. doi: 10.1016/j.jviromet.2019.113709. [DOI] [PubMed] [Google Scholar]
- 92.Zhang S.-Q., Tan B., Li P., Wang F.-X., Guo L., Yang Y., Sun N., Zhu H.-W., Wen Y.-J., Cheng S.-P. Comparison of conventional RT-PCR, reverse-transcription loop-mediated isothermal amplification, and SYBR green I-based real-time RT-PCR in the rapid detection of bovine viral diarrhea virus nucleotide in contaminated commercial bovine sera batches. J. Virol. Methods. 2014;207:204–209. doi: 10.1016/j.jviromet.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 93.Madi M., Hamilton A., Squirrell D., Mioulet V., Evans P., Lee M., King D.P. Rapid detection of foot-and-mouth disease virus using a field-portable nucleic acid extraction and real-time PCR amplification platform. Vet. J. 2012;193:67–72. doi: 10.1016/j.tvjl.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 94.Olmos A., Bertolini E., Cambra M. Simultaneous and co-operational amplification (Co-PCR): a new concept for detection of plant viruses. J. Virol. Methods. 2002;106:51–59. doi: 10.1016/S0166-0934(02)00132-5. [DOI] [PubMed] [Google Scholar]
- 95.Ciftci S., Cánovas R., Neumann F., Paulraj T., Nilsson M., Crespo G.A., Madaboosi N. The sweet detection of rolling circle amplification: glucose-based electrochemical genosensor for the detection of viral nucleic acid, Biosens. Bioelectron. 2020;151:112002. doi: 10.1016/j.bios.2019.112002. [DOI] [PubMed] [Google Scholar]
- 96.Singh M., Pal D., Sood P., Randhawa G. Loop-mediated isothermal amplification assays: rapid and efficient diagnostics for genetically modified crops. Food Contr. 2019;106:106759. doi: 10.1016/j.foodcont.2019.106759. [DOI] [Google Scholar]
- 97.Parida M., Sannarangaiah S., Dash P.K., Rao P.V.L., Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008;18:407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Techathuvanan C., Draughon F.A., D’Souza D.H. Loop-mediated isothermal amplification (LAMP) for the rapid and sensitive detection of Salmonella typhimurium from pork. J. Food Sci. 2010;75:M165–M172. doi: 10.1111/j.1750-3841.2010.01554.x. [DOI] [PubMed] [Google Scholar]
- 99.Kashir J., Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses. 2020;141:109786. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang D., Yu J., Wang Y., Zhang M., Li P., Liu M., Liu Y. Development of a real-time loop-mediated isothermal amplification (LAMP) assay and visual LAMP assay for detection of African swine fever virus (ASFV) J. Virol. Methods. 2020;276:113775. doi: 10.1016/j.jviromet.2019.113775. [DOI] [PubMed] [Google Scholar]
- 101.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ji J., Xu X., Wu Q., Wang X., Li W., Yao L., Kan Y., Yuan L., Bi Y., Xie Q. Simple and visible detection of duck hepatitis B virus in ducks and geese using loop-mediated isothermal amplification. Poult. Sci. 2020;99:791–796. doi: 10.1016/j.psj.2019.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bartosik M., Jirakova L., Anton M., Vojtesek B., Hrstka R. Genomagnetic LAMP-based electrochemical test for determination of high-risk HPV16 and HPV18 in clinical samples. Anal. Chim. Acta. 2018;1042:37–43. doi: 10.1016/j.aca.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 104.Zou X., Wu J., Gu J., Shen L., Mao L. Application of aptamers in virus detection and antiviral Therapy. Front. Microbiol. 2019;10:1462. doi: 10.3389/fmicb.2019.01462. https://www.frontiersin.org/article/10.3389/fmicb.2019.01462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhardwaj J., Chaudhary N., Kim H., Jang J. Subtyping of influenza A H1N1 virus using a label-free electrochemical biosensor based on the DNA aptamer targeting the stem region of HA protein. Anal. Chim. Acta. 2019;1064:94–103. doi: 10.1016/j.aca.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 106.Bai C., Lu Z., Jiang H., Yang Z., Liu X., Ding H., Li H., Dong J., Huang A., Fang T., Jiang Y., Zhu L., Lou X., Li S., Shao N. Aptamer selection and application in multivalent binding-based electrical impedance detection of inactivated H1N1 virus. Biosens. Bioelectron. 2018;110:162–167. doi: 10.1016/j.bios.2018.03.047. [DOI] [PubMed] [Google Scholar]
- 107.Lee T., Park S.Y., Jang H., Kim G.-H., Lee Y., Park C., Mohammadniaei M., Lee M.-H., Min J. Fabrication of electrochemical biosensor consisted of multi-functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater. Sci. Eng. C. 2019;99:511–519. doi: 10.1016/j.msec.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 108.Luppa P.B., Sokoll L.J., Chan D.W. Immunosensors - principles and applications to clinical chemistry. Clin. Chim. Acta. 2001;314:1–26. doi: 10.1016/s0009-8981(01)00629-5. [DOI] [PubMed] [Google Scholar]
- 109.Thévenot D.R., Toth K., Durst R.A., Wilson G.S. Electrochemical biosensors: recommended definitions and classification1International union of pure and applied chemistry: physical chemistry division, commission I.7 (biophysical chemistry); analytical chemistry division. Commission V.5 (Electroanalytical, Biosens. Bioelectron. 2001;16:121–131. doi: 10.1016/s0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 110.Lim S.A., Ahmed M.U. Immunosensors, the Royal Society of Chemistry. 2019. CHAPTER 1 introduction to immunosensors; pp. 1–20. [DOI] [Google Scholar]
- 111.Hassanpour S., Baradaran B., de la Guardia M., Baghbanzadeh A., Mosafer J., Hejazi M., Mokhtarzadeh A., Hasanzadeh M. Diagnosis of hepatitis via nanomaterial-based electrochemical, optical or piezoelectrical biosensors: a review on recent advancements. Microchim. Acta. 2018;185:568. doi: 10.1007/s00604-018-3088-8. [DOI] [PubMed] [Google Scholar]
- 112.Moulick A., Richtera L., Milosavljevic V., Cernei N., Haddad Y., Zitka O., Kopel P., Heger Z., Adam V. Advanced nanotechnologies in avian influenza: current status and future trends – a review. Anal. Chim. Acta. 2017;983:42–53. doi: 10.1016/j.aca.2017.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hassanpour S., Baradaran B., Hejazi M., Hasanzadeh M., Mokhtarzadeh A., de la Guardia M. Recent trends in rapid detection of influenza infections by bio and nanobiosensor. TrAC Trends Anal. Chem. 2018;98:201–215. doi: 10.1016/j.trac.2017.11.012. [DOI] [Google Scholar]
- 114.Wu Z., Zeng T., Guo W.-J., Bai Y.-Y., Pang D.-W., Zhang Z.-L. Digital single virus immunoassay for ultrasensitive multiplex avian influenza virus detection based on fluorescent magnetic multifunctional nanospheres. ACS Appl. Mater. Interfaces. 2019;11:5762–5770. doi: 10.1021/acsami.8b18898. [DOI] [PubMed] [Google Scholar]
- 115.Zuo B., Li S., Guo Z., Zhang J., Chen C. Piezoelectric immunosensor for SARS-associated coronavirus in sputum. Anal. Chem. 2004;76:3536–3540. doi: 10.1021/ac035367b. [DOI] [PubMed] [Google Scholar]
- 116.da Silva E.T.S.G., Souto D.E.P., Barragan J.T.C., de J., Giarola F., de Moraes A.C.M., Kubota L.T. Electrochemical biosensors in point-of-care devices: recent advances and future trends. ChemElectroChem. 2017;4:778–794. doi: 10.1002/celc.201600758. [DOI] [Google Scholar]
- 117.Santos A., Bueno P.R., Davis J.J. A dual marker label free electrochemical assay for Flavivirus dengue diagnosis. Biosens. Bioelectron. 2018;100:519–525. doi: 10.1016/j.bios.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 118.Alcon S., Talarmin A., Debruyne M., Falconar A., Deubel V., Flamand M. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 2002;40:376–381. doi: 10.1128/jcm.40.02.376-381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taebi S., Keyhanfar M., Noorbakhsh A. A novel method for sensitive, low-cost and portable detection of hepatitis B surface antigen using a personal glucose meter. J. Immunol. Methods. 2018;458:26–32. doi: 10.1016/j.jim.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 120.Shouval D. Hepatitis B vaccines. J. Hepatol. 2003;39:70–76. doi: 10.1016/s0168-8278(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 121.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Il Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 123.Poovorawan Y., Chongsrisawat V., Theamboonlers A., Crasta P.D., Messier M., Hardt K. Long-term anti-HBs antibody persistence following infant vaccination against hepatitis B and evaluation of anamnestic response. Hum. Vaccines Immunother. 2013;9:1679–1684. doi: 10.4161/hv.24844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Theeten H., Van Herck K., Van Der Meeren O., Crasta P., Van Damme P., Hens N. Long-term antibody persistence after vaccination with a 2-dose HavrixTM (inactivated hepatitis A vaccine): 20 years of observed data, and long-term model-based predictions. Vaccine. 2015;33:5723–5727. doi: 10.1016/j.vaccine.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 125.Trilling A.K., Beekwilder J., Zuilhof H. Antibody orientation on biosensor surfaces: a minireview. Analyst. 2013;138:1619–1627. doi: 10.1039/C2AN36787D. [DOI] [PubMed] [Google Scholar]
- 126.Sharma S., Byrne H., O’Kennedy R.J. Antibodies and antibody-derived analytical biosensors. Essays Biochem. 2016;60:9–18. doi: 10.1042/EBC20150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mollarasouli F., Kurbanoglu S., Ozkan S.A. The role of electrochemical immunosensors in clinical analysis. Biosensors. 2019;9:86. doi: 10.3390/bios9030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shen M., Rusling J.F., Dixit C.K. Site-selective orientated immobilization of antibodies and conjugates for immunodiagnostics development. Methods. 2017;116:95–111. doi: 10.1016/j.ymeth.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Welch N.G., Scoble J.A., Muir B.W., Pigram P.J. Orientation and characterization of immobilized antibodies for improved immunoassays (Review) Biointerphases. 2017;12 doi: 10.1116/1.4978435. 02D301. [DOI] [PubMed] [Google Scholar]
- 130.Vashist S.K., Luong J.H.T. S.K. Vashist, J.H.T.B.T.-H. Of I.T. Luong. Academic Press; 2018. Chapter 2-Antibody immobilization and surface functionalization chemistries for immunodiagnostics; pp. 19–46. [DOI] [Google Scholar]
- 131.Cabral D.G.A., Lima E.C.S., Moura P., Dutra R.F. A label-free electrochemical immunosensor for hepatitis B based on hyaluronic acid–carbon nanotube hybrid film. Talanta. 2016;148:209–215. doi: 10.1016/j.talanta.2015.10.083. [DOI] [PubMed] [Google Scholar]
- 132.Migneault I., Dartiguenave C., Bertrand M.J., Waldron K.C. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques. 2004;37:790–802. doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 133.Devarakonda S., Singh R., Bhardwaj J., Jang J. Cost-effective and handmade paper-based immunosensing device for electrochemical detection of influenza virus. Sensors. 2017;17:2597. doi: 10.3390/s17112597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grabarek Z., Gergely J. Zero-length crosslinking procedure with the use of active esters. Anal. Biochem. 1990;185:131–135. doi: 10.1016/0003-2697(90)90267-D. [DOI] [PubMed] [Google Scholar]
- 135.Karyakin A.A., V Presnova G., Rubtsova M.Y., Egorov A.M. Oriented immobilization of antibodies onto the gold surfaces via their native thiol groups. Anal. Chem. 2000;72:3805–3811. doi: 10.1021/ac9907890. [DOI] [PubMed] [Google Scholar]
- 136.Brazaca L.C., Bramorski C.B., Cancino-Bernardi J., da Silveira Cruz-Machado S., Markus R.P., Janegitz B.C., Zucolotto V. An antibody-based platform for melatonin quantification. Colloids Surf. B Biointerfaces. 2018;171:94–100. doi: 10.1016/j.colsurfb.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 137.de Oliveira G.C.M., de J.H., Carvalho S., Brazaca L.C., Vieira N.C.S., Janegitz B.C. Flexible platinum electrodes as electrochemical sensor and immunosensor for Parkinson’s disease biomarkers. Biosens. Bioelectron. 2020;152:112016. doi: 10.1016/j.bios.2020.112016. [DOI] [PubMed] [Google Scholar]
- 138.Li X., Wang Y., Zhang X., Gao Y., Sun C., Ding Y., Feng F., Jin W., Yang G. An impedimetric immunosensor for determination of porcine epidemic diarrhea virus based on the nanocomposite consisting of molybdenum disulfide/reduced graphene oxide decorated with gold nanoparticles. Microchim. Acta. 2020;187:217. doi: 10.1007/s00604-020-4166-2. [DOI] [PubMed] [Google Scholar]
- 139.Joshi S.R., Sharma A., Kim G.-H., Jang J. Low cost synthesis of reduced graphene oxide using biopolymer for influenza virus sensor. Mater. Sci. Eng. C. 2020;108:110465. doi: 10.1016/j.msec.2019.110465. [DOI] [PubMed] [Google Scholar]
- 140.Svalova T.S., Malysheva N.N., Bubekova A.K., Saigushkina A.A., V Medvedeva M., Kozitsina A.N. Effect of the method for immobilizing receptor layer on the analytical characteristics of a label-free electrochemical immunosensor for the determination of measles antibodies. J. Anal. Chem. 2020;75:254–261. doi: 10.1134/S106193482002015X. [DOI] [Google Scholar]
- 141.Arshad R., Rhouati A., Hayat A., Nawaz M.H., Yameen M.A., Mujahid A., Latif U. MIP-based impedimetric sensor for detecting dengue fever biomarker. Appl. Biochem. Biotechnol. 2020 doi: 10.1007/s12010-020-03285-y. [DOI] [PubMed] [Google Scholar]
- 142.Akkapinyo C., Khownarumit P., Waraho-Zhmayev D., Poo-arporn R.P. Development of a multiplex immunochromatographic strip test and ultrasensitive electrochemical immunosensor for hepatitis B virus screening. Anal. Chim. Acta. 2020;1095:162–171. doi: 10.1016/j.aca.2019.10.016. [DOI] [PubMed] [Google Scholar]
- 143.Biasotto G., Costa J.P.C., Costa P.I., Zaghete M.A. ZnO nanorods-gold nanoparticle-based biosensor for detecting hepatitis C. Appl. Phys. A. 2019;125:821. doi: 10.1007/s00339-019-3128-1. [DOI] [Google Scholar]
- 144.George A., Amrutha M.S., Srivastava P., Sai V.V.R., Sunil S., Srinivasan R. Label-free detection of chikungunya non-structural protein 3 using electrochemical impedance spectroscopy. J. Electrochem. Soc. 2019;166:B1356–B1363. doi: 10.1149/2.1081914jes. [DOI] [Google Scholar]
- 145.Tran L.T., Tran T.Q., Ho H.P., Chu X.T., Mai T.A. Simple label-free electrochemical immunosensor in a microchamber for detecting Newcastle disease virus. J. Nanomater. 2019;2019:3835609. [Google Scholar]
- 146.Faria A.M., Mazon T. Early diagnosis of Zika infection using a ZnO nanostructures-based rapid electrochemical biosensor. Talanta. 2019;203:153–160. doi: 10.1016/j.talanta.2019.04.080. [DOI] [PubMed] [Google Scholar]
- 147.Kim J.H., Cho C.H., Ryu M.Y., Kim J.-G., Lee S.-J., Park T.J., Park J.P. Development of peptide biosensor for the detection of dengue fever biomarker, nonstructural 1. PloS One. 2019;14:e0222144. doi: 10.1371/journal.pone.0222144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Valipour A., Roushani M. Using silver nanoparticle and thiol graphene quantum dots nanocomposite as a substratum to load antibody for detection of hepatitis C virus core antigen: electrochemical oxidation of riboflavin was used as redox probe. Biosens. Bioelectron. 2017;89:946–951. doi: 10.1016/j.bios.2016.09.086. [DOI] [PubMed] [Google Scholar]
- 149.Valipour A., Roushani M. A glassy carbon immunoelectrode modified with vanadium oxide nanobelts for ultrasensitive voltammetric determination of the core antigen of hepatitis C virus. Microchim. Acta. 2017;184:4477–4483. doi: 10.1007/s00604-017-2449-z. [DOI] [Google Scholar]
- 150.Veerapandian M., Hunter R., Neethirajan S. Dual immunosensor based on methylene blue-electroadsorbed graphene oxide for rapid detection of the influenza A virus antigen. Talanta. 2016;155:250–257. doi: 10.1016/j.talanta.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 151.Wei S., Xiao H., Cao L., Chen Z. A label-free immunosensor based on graphene oxide/Fe3O4/prussian blue nanocomposites for the electrochemical determination of HBsAg. Biosensors. 2020;10:24. doi: 10.3390/bios10030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kanagavalli P., Veerapandian M. Opto-electrochemical functionality of Ru(II)-reinforced graphene oxide nanosheets for immunosensing of dengue virus non-structural 1 protein. Biosens. Bioelectron. 2020;150:111878. doi: 10.1016/j.bios.2019.111878. [DOI] [PubMed] [Google Scholar]
- 153.Zhao F., Cao L., Liang Y., Wu Z., Chen Z., Zeng R. Label-free amperometric immunosensor based on graphene oxide and ferrocene-chitosan nanocomposites for detection of Hepatitis B virus antigen. J. Biomed. Nanotechnol. 2017;13:1300–1308. [Google Scholar]
- 154.Hien H.T., Giang H.T., Trung T., Van Tuan C. Enhancement of biosensing performance using a polyaniline/multiwalled carbon nanotubes nanocomposite. J. Mater. Sci. 2017;52:1694–1703. doi: 10.1007/s10853-016-0461-z. [DOI] [Google Scholar]
- 155.Trindade E.K.G., Dutra R.F. A label-free and reagentless immunoelectrode for antibodies against hepatitis B core antigen (anti-HBc) detection. Colloids Surf. B Biointerfaces. 2018;172:272–279. doi: 10.1016/j.colsurfb.2018.08.050. [DOI] [PubMed] [Google Scholar]
- 156.Ning S., Zhou M., Liu C., Waterhouse G.I.N., Dong J., Ai S. Ultrasensitive electrochemical immunosensor for avian leukosis virus detection based on a β-cyclodextrin-nanogold-ferrocene host-guest label for signal amplification. Anal. Chim. Acta. 2019;1062:87–93. doi: 10.1016/j.aca.2019.02.041. [DOI] [PubMed] [Google Scholar]
- 157.Zhou D., Wang M., Dong J., Ai S. A novel electrochemical immunosensor based on mesoporous graphitic carbon nitride for detection of subgroup J of avian leukosis viruses. Electrochim. Acta. 2016;205:95–101. doi: 10.1016/j.electacta.2016.04.101. [DOI] [Google Scholar]
- 158.Valipour A., Roushani M. TiO2 nanoparticles doped with Celestine Blue as a label in a sandwich immunoassay for the hepatitis C virus core antigen using a screen printed electrode. Microchim. Acta. 2017;184:2015–2022. doi: 10.1007/s00604-017-2190-7. [DOI] [Google Scholar]
- 159.Khristunova Y., Korotkova E., Kratochvil B., Barek J., Dorozhko E., Vyskocil V., Plotnikov E., Voronova O., Sidelnikov V. Preparation and investigation of silver nanoparticle-antibody bioconjugates for electrochemical immunoassay of tick-borne encephalitis. Sensors. 2019;19:2103. doi: 10.3390/s19092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Huang J., Xie Z., Xie Z., Luo S., Xie L., Huang L., Fan Q., Zhang Y., Wang S., Zeng T. Silver nanoparticles coated graphene electrochemical sensor for the ultrasensitive analysis of avian influenza virus H7. Anal. Chim. Acta. 2016;913:121–127. doi: 10.1016/j.aca.2016.01.050. [DOI] [PubMed] [Google Scholar]
- 161.Yang Z.-H., Zhuo Y., Yuan R., Chai Y.-Q. A nanohybrid of platinum nanoparticles-porous ZnO–hemin with electrocatalytic activity to construct an amplified immunosensor for detection of influenza. Biosens. Bioelectron. 2016;78:321–327. doi: 10.1016/j.bios.2015.10.073. [DOI] [PubMed] [Google Scholar]
- 162.Peng X., Luo G., Wu Z., Wen W., Zhang X., Wang S. Fluorescent-magnetic-catalytic nanospheres for dual-modality detection of H9N2 avian influenza virus. ACS Appl. Mater. Interfaces. 2019;11:41148–41156. doi: 10.1021/acsami.9b16718. [DOI] [PubMed] [Google Scholar]
- 163.Wu Z., Guo W.-J., Bai Y.-Y., Zhang L., Hu J., Pang D.-W., Zhang Z.-L. Digital single virus electrochemical enzyme-linked immunoassay for ultrasensitive H7N9 avian influenza virus counting. Anal. Chem. 2018;90:1683–1690. doi: 10.1021/acs.analchem.7b03281. [DOI] [PubMed] [Google Scholar]
- 164.Liu C., Dong J., Waterhouse G.I.N., Cheng Z., Ai S. Electrochemical immunosensor with nanocellulose-Au composite assisted multiple signal amplification for detection of avian leukosis virus subgroup. J. Biosens. Bioelectron. 2018;101:110–115. doi: 10.1016/j.bios.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 165.Hou Y.-H., Wang J.-J., Jiang Y.-Z., Lv C., Xia L., Hong S.-L., Lin M., Lin Y., Zhang Z.-L., Pang D.-W. A colorimetric and electrochemical immunosensor for point-of-care detection of enterovirus 71. Biosens. Bioelectron. 2018;99:186–192. doi: 10.1016/j.bios.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 166.Liu C., Dong J., Ning S., Hou J., Waterhouse G.I.N., Cheng Z., Ai S. An electrochemical immunosensor based on an etched zeolitic imidazolate framework for detection of avian leukosis virus subgroup. J. Microchim. Acta. 2018;185:423. doi: 10.1007/s00604-018-2930-3. [DOI] [PubMed] [Google Scholar]
- 167.Alizadeh N., Hallaj R., Salimi A. Dual amplified electrochemical immunosensor for hepatitis B virus surface antigen detection using Hemin/G-quadruplex immobilized onto Fe3O4-AuNPs or (Hemin-amino-rGO-Au) nanohybrid. Electroanalysis. 2018;30:402–414. doi: 10.1002/elan.201700727. [DOI] [Google Scholar]
- 168.Alizadeh N., Hallaj R., Salimi A. A highly sensitive electrochemical immunosensor for hepatitis B virus surface antigen detection based on Hemin/G-quadruplex horseradish peroxidase-mimicking DNAzyme-signal amplification. Biosens. Bioelectron. 2017;94:184–192. doi: 10.1016/j.bios.2017.02.039. [DOI] [PubMed] [Google Scholar]
- 169.Gao Z., Li Y., Zhang X., Feng J., Kong L., Wang P., Chen Z., Dong Y., Wei Q. Ultrasensitive electrochemical immunosensor for quantitative detection of HBeAg using Au@Pd/MoS2@MWCNTs nanocomposite as enzyme-mimetic labels. Biosens. Bioelectron. 2018;102:189–195. doi: 10.1016/j.bios.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 170.Li F., Li Y., Feng J., Gao Z., Lv H., Ren X., Wei Q. Facile synthesis of MoS2@Cu2O-Pt nanohybrid as enzyme-mimetic label for the detection of the Hepatitis B surface antigen. Biosens. Bioelectron. 2018;100:512–518. doi: 10.1016/j.bios.2017.09.048. [DOI] [PubMed] [Google Scholar]
- 171.Zhang Q., Li L., Qiao Z., Lei C., Fu Y., Xie Q., Yao S., Li Y., Ying Y. Electrochemical conversion of Fe3O4 magnetic nanoparticles to electroactive prussian blue analogues for self-sacrificial label biosensing of avian influenza virus H5N1. Anal. Chem. 2017;89:12145–12151. doi: 10.1021/acs.analchem.7b02784. [DOI] [PubMed] [Google Scholar]
- 172.Amano Y., Cheng Q. Detection of influenza virus: traditional approaches and development of biosensors. Anal. Bioanal. Chem. 2005;381:156–164. doi: 10.1007/s00216-004-2927-0. [DOI] [PubMed] [Google Scholar]
- 173.Mandli J., Attar A., Ennaji M.M., Amine A. Indirect competitive electrochemical immunosensor for hepatitis A virus antigen detection. J. Electroanal. Chem. 2017;799:213–221. doi: 10.1016/j.jelechem.2017.05.047. [DOI] [Google Scholar]
- 174.Vidor E., Fritzell B., Plotkin S. Clinical develpment of a new inactivated hepatitis A vaccine. Infection. 1996;24:447–458. doi: 10.1007/BF01713047. [DOI] [PubMed] [Google Scholar]
- 175.Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sajid M., Kawde A.-N., Daud M. Designs, formats and applications of lateral flow assay: a literature review. J. Saudi Chem. Soc. 2015;19:689–705. doi: 10.1016/j.jscs.2014.09.001. [DOI] [Google Scholar]
- 177.Eltzov E., Guttel S., Low Yuen Kei A., Sinawang P.D., Ionescu R.E., Marks R.S. Lateral flow immunoassays – from paper strip to smartphone technology. Electroanalysis. 2015;27:2116–2130. doi: 10.1002/elan.201500237. [DOI] [Google Scholar]
- 178.Brazaca L.C., Moreto J.R., Martín A., Tehrani F., Wang J., Zucolotto V. Colorimetric paper-based immunosensor for simultaneous determination of fetuin B and clusterin toward early Alzheimer’s diagnosis. ACS Nano. 2019 doi: 10.1021/acsnano.9b06571. [DOI] [PubMed] [Google Scholar]
- 179.Xi Y., Xu C.-Z., Xie Z.-Z., Zhu D.-L., Dong J.-M. Rapid and visual detection of dengue virus using recombinase polymerase amplification method combined with lateral flow dipstick. Mol. Cell. Probes. 2019;46:101413. doi: 10.1016/j.mcp.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 180.Chen L., Wang H., Guo T., Xiao C., Liu L., Zhang X., Liu B., Li P., Liu A., Li B., Li B., Mao Y. A rapid point-of-care test for dengue virus-1 based on a lateral flow assay with a near-infrared fluorescent dye. J. Immunol. Methods. 2018;456:23–27. doi: 10.1016/j.jim.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 181.Sinawang P.D., Fajs L., Elouarzaki K., Nugraha J., Marks R.S. TEMPO-based immuno-lateral flow quantitative detection of dengue NS1 protein. Sens. Actuators B Chem. 2018;259:354–363. doi: 10.1016/j.snb.2017.12.043. [DOI] [Google Scholar]
- 182.Sinawang P.D., Rai V., Ionescu R.E., Marks R.S. Electrochemical lateral flow immunosensor for detection and quantification of dengue NS1 protein. Biosens. Bioelectron. 2016;77:400–408. doi: 10.1016/j.bios.2015.09.048. [DOI] [PubMed] [Google Scholar]
- 183.Rong Z., Wang Q., Sun N., Jia X., Wang K., Xiao R., Wang S. Smartphone-based fluorescent lateral flow immunoassay platform for highly sensitive point-of-care detection of Zika virus nonstructural protein 1. Anal. Chim. Acta. 2019;1055:140–147. doi: 10.1016/j.aca.2018.12.043. [DOI] [PubMed] [Google Scholar]
- 184.Phan J.C., Pettitt J., George J.S., Fakoli L.S., III, Taweh F.M., Bateman S.L., Bennett R.S., Norris S.L., Spinnler D.A., Pimentel G., Sahr P.K., Bolay F.K., Schoepp R.J. Lateral flow immunoassays for Ebola virus disease detection in Liberia. J. Infect. Dis. 2016;214:S222–S228. doi: 10.1093/infdis/jiw251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Duan D., Fan K., Zhang D., Tan S., Liang M., Liu Y., Zhang J., Zhang P., Liu W., Qiu X., Kobinger G.P., Fu Gao G., Yan X. Nanozyme-strip for rapid local diagnosis of Ebola. Biosens. Bioelectron. 2015;74:134–141. doi: 10.1016/j.bios.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 186.Nakagiri I., Tasaka T., Okai M., Nakai F., Bunya R., Nagai S., Yoshida T., Tokunaga H., Kondo E., Wada H. Screening for human immunodeficiency virus using a newly developed fourth generation lateral flow immunochromatography assay. J. Virol. Methods. 2019;274:113746. doi: 10.1016/j.jviromet.2019.113746. [DOI] [PubMed] [Google Scholar]
- 187.Peck R.B., Schweizer J., Weigl B.H., Somoza C., Silver J., Sellors J.W., Lu P.S. A magnetic immunochromatographic strip test for detection of human papillomavirus 16 E6. Clin. Chem. 2006;52:2170–2172. doi: 10.1373/clinchem.2006.072884. [DOI] [PubMed] [Google Scholar]
- 188.Shen H., Xie K., Huang L., Wang L., Ye J., Xiao M., Ma L., Jia A., Tang Y. A novel SERS-based lateral flow assay for differential diagnosis of wild-type pseudorabies virus and gE-deleted vaccine. Sens. Actuators B Chem. 2019;282:152–157. doi: 10.1016/j.snb.2018.11.065. [DOI] [Google Scholar]
- 189.Shen J., Zhou Y., Fu F., Xu H., Lv J., Xiong Y., Wang A. Immunochromatographic assay for quantitative and sensitive detection of hepatitis B virus surface antigen using highly luminescent quantum dot-beads. Talanta. 2015;142:145–149. doi: 10.1016/j.talanta.2015.04.058. [DOI] [PubMed] [Google Scholar]
- 190.Martiskainen I., Talha S.M., Pettersson K. Sensitive lateral flow immunoassay for the detection of hepatitis B virus surface antigen. Clin. Chim. Acta. 2019;493:S656–S657. doi: 10.1016/j.cca.2019.03.1384. [DOI] [Google Scholar]
- 191.Bayoumy S., Juntunen E., Pettersson K., Talha S.M. Detection of antibodies to the hepatitis C virus using up-converting nanoparticles − Based lateral flow immunoassay. Clin. Chim. Acta. 2019;493:S653–S654. doi: 10.1016/j.cca.2019.03.1378. [DOI] [Google Scholar]
- 192.Wu X., Song Z., Zhai X., Zuo L., Mei X., Xiang R., Kang Z., Zhou L., Wang H. Simultaneous and visual detection of infectious bronchitis virus and Newcastle disease virus by multiple LAMP and lateral flow dipstick. Poult. Sci. 2019;98:5401–5411. doi: 10.3382/ps/pez372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xu Y., Fang D., Chen F., Zhao Q., Cai C., Cheng M. Utilization of recombinase polymerase amplification method combined with lateral flow dipstick for visual detection of respiratory syncytial virus. Mol. Cell. Probes. 2020;49:101473. doi: 10.1016/j.mcp.2019.101473. [DOI] [PubMed] [Google Scholar]
- 194.Kim J., Kwon J.H., Jang J., Lee H., Kim S., Hahn Y.K., Kim S.K., Lee K.H., Lee S., Pyo H., Song C.-S., Lee J. Rapid and background-free detection of avian influenza virus in opaque sample using NIR-to-NIR upconversion nanoparticle-based lateral flow immunoassay platform. Biosens. Bioelectron. 2018;112:209–215. doi: 10.1016/j.bios.2018.04.047. [DOI] [PubMed] [Google Scholar]
- 195.Kim S.H., Lee J., Lee B.H., Song C.-S., Gu M.B. Specific detection of avian influenza H5N2 whole virus particles on lateral flow strips using a pair of sandwich-type aptamers. Biosens. Bioelectron. 2019;134:123–129. doi: 10.1016/j.bios.2019.03.061. [DOI] [PubMed] [Google Scholar]
- 196.Li X., Lu D., Sheng Z., Chen K., Guo X., Jin M., Han H. A fast and sensitive immunoassay of avian influenza virus based on label-free quantum dot probe and lateral flow test strip. Talanta. 2012;100:1–6. doi: 10.1016/j.talanta.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 197.Sun N., Wang W., Wang J., Yao X., Chen F., Li X., Yinglei Y., Chen B. Reverse transcription recombinase polymerase amplification with lateral flow dipsticks for detection of influenza A virus and subtyping of H1 and H3. Mol. Cell. Probes. 2018;42:25–31. doi: 10.1016/j.mcp.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Xiao M., Xie K., Dong X., Wang L., Huang C., Xu F., Xiao W., Jin M., Huang B., Tang Y. Ultrasensitive detection of avian influenza A (H7N9) virus using surface-enhanced Raman scattering-based lateral flow immunoassay strips. Anal. Chim. Acta. 2019;1053:139–147. doi: 10.1016/j.aca.2018.11.056. [DOI] [PubMed] [Google Scholar]
- 199.Liu B., Du D., Hua X., Yu X.-Y.Y., Lin Y. Paper-based electrochemical biosensors: from test strips to paper-based microfluidics. Electroanalysis. 2014;26:1214–1223. doi: 10.1002/elan.201400036. [DOI] [Google Scholar]
- 200.Reid R., Chatterjee B., Das S.J., Ghosh S., Sharma T.K. Application of aptamers as molecular recognition elements in lateral flow assays. Anal. Biochem. 2020;593:113574. doi: 10.1016/j.ab.2020.113574. [DOI] [PubMed] [Google Scholar]
- 201.Yu L., Song Z., Peng J., Yang M., Zhi H., He H. Progress of gold nanomaterials for colorimetric sensing based on different strategies. TrAC Trends Anal. Chem. 2020;127:115880. doi: 10.1016/j.trac.2020.115880. [DOI] [Google Scholar]
- 202.Mao X., Wang W., Du T.E. Dry-reagent nucleic acid biosensor based on blue dye doped latex beads and lateral flow strip. Talanta. 2013;114:248–253. doi: 10.1016/j.talanta.2013.04.044. [DOI] [PubMed] [Google Scholar]
- 203.Juntunen E., Myyryläinen T., Salminen T., Soukka T., Pettersson K. Performance of fluorescent europium(III) nanoparticles and colloidal gold reporters in lateral flow bioaffinity assay. Anal. Biochem. 2012;428:31–38. doi: 10.1016/j.ab.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 204.Wu Y., Zhou Y., Leng Y., Lai W., Huang X., Xiong Y. Emerging design strategies for constructing multiplex lateral flow test strip sensors. Biosens. Bioelectron. 2020;157:112168. doi: 10.1016/j.bios.2020.112168. [DOI] [PubMed] [Google Scholar]
- 205.Bhardwaj J., Sharma A., Jang J. Vertical flow-based paper immunosensor for rapid electrochemical and colorimetric detection of influenza virus using a different pore size sample pad. Biosens. Bioelectron. 2019;126:36–43. doi: 10.1016/j.bios.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 206.Yezli S., Otter J.A. Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ. Virol. 2011;3:1–30. doi: 10.1007/s12560-011-9056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Biomarkers Definition Working Group Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 208.Sadana A., Sadana N. Elsevier Inc.; Amsterdam: 2014. Biomarkers and Biosensors: Detection and Binding to Biosensor Surfaces and Biomarkers Applications. [Google Scholar]
- 209.Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Havelka A., Sejersen K., Venge P., Pauksens K., Larsson A. Calprotectin, a new biomarker for diagnosis of acute respiratory infections. Sci. Rep. 2020;10:4208. doi: 10.1038/s41598-020-61094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Riedel S. Predicting bacterial versus viral infection, or none of the above: current and future prospects of biomarkers. Clin. Lab. Med. 2019;39:453–472. doi: 10.1016/j.cll.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 212.Yusa T., Tateda K., Ohara A., Miyazaki S. New possible biomarkers for diagnosis of infections and diagnostic distinction between bacterial and viral infections in children. J. Infect. Chemother. 2017;23:96–100. doi: 10.1016/j.jiac.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 213.Sharma R., Deacon S.E., Nowak D., George S.E., Szymonik M.P., Tang A.A.S., Tomlinson D.C., Davies A.G., McPherson M.J., Wälti C. Label-free electrochemical impedance biosensor to detect human interleukin-8 in serum with sub-pg/ml sensitivity. Biosens. Bioelectron. 2016;80:607–613. doi: 10.1016/j.bios.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lab tests online, Interleukin-6. 2020. https://labtestsonline.org/tests/interleukin-6
- 215.Sasaki K., Fujita I., Hamasaki Y., Miyazaki S. Differentiating between bacterial and viral infection by measuring both C-reactive protein and 2′-5′-oligoadenylate synthetase as inflammatory markers. J. Infect. Chemother. 2002;8:76–80. doi: 10.1007/s101560200010. [DOI] [PubMed] [Google Scholar]
- 216.Lab tests online C-reactive protein (CRP) 2020. https://labtestsonline.org/tests/c-reactive-protein-crp
- 217.Dong S., Zhang D., Cui H., Huang T. ZnO/porous carbon composite from a mixed-ligand MOF for ultrasensitive electrochemical immunosensing of C-reactive protein. Sens. Actuator. B Chem. 2019;284:354–361. doi: 10.1016/j.snb.2018.12.150. [DOI] [Google Scholar]
- 218.Hafaiedh I., Chammem H., Abdelghani A., Ait E., Feldman L., Meilhac O., Mora L. Supported protein G on gold electrode: characterization and immunosensor application. Talanta. 2013;116:84–90. doi: 10.1016/j.talanta.2013.04.059. [DOI] [PubMed] [Google Scholar]
- 219.Figueiredo A., Vieira N.C.S., dos Santos J.F., Janegitz B.C., Aoki S.M., Junior P.P., Lovato R.L., Nogueira M.L., Zucolotto V., Guimarães F.E.G. Electrical detection of dengue biomarker using egg yolk immunoglobulin as the biological recognition element. Sci. Rep. 2015;5:7865. doi: 10.1038/srep07865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wasik D., Mulchandani A., Yates M.V. Salivary detection of dengue virus NS1 protein with a label-free immunosensor for early dengue diagnosis. Sensors. 2018;18:2641. doi: 10.3390/s18082641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lim J.M., Kim J.H., Ryu M.Y., Cho C.H., Park T.J., Park J.P. An electrochemical peptide sensor for detection of dengue fever biomarker NS1. Anal. Chim. Acta. 2018;1026:109–116. doi: 10.1016/j.aca.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 222.Cecchetto J., Fernandes F.C.B., Lopes R., Bueno P.R. The capacitive sensing of NS1 Flavivirus biomarker. Biosens. Bioelectron. 2017;87:949–956. doi: 10.1016/j.bios.2016.08.097. [DOI] [PubMed] [Google Scholar]
- 223.BVS Atenção primária em Saúde, Qual a especificidade e sensibilidade do teste rápido da dengue e que tipos existem? 2020. https://aps.bvs.br/aps/qual-a-especificidade-e-sensibilidade-do-teste-rapido-da-dengue-e-que-tipos-existem/
- 224.World Health Organization, Estimated number of people (all ages) living with HIV. 2020. https://www.who.int/data/gho/data/indicators/indicator-details/GHO/estimated-number-of-people-(all-ages)-living-with-hiv
- 225.Carinelli S., Xufré Ballesteros C., Martí M., Alegret S., Pividori M.I. Electrochemical magneto-actuated biosensor for CD4 count in AIDS diagnosis and monitoring. Biosens. Bioelectron. 2015;74:974–980. doi: 10.1016/j.bios.2015.07.053. [DOI] [PubMed] [Google Scholar]
- 226.Kim J., Park G., Lee S., Hwang S.-W., Min N., Lee K.-M. Single wall carbon nanotube electrode system capable of quantitative detection of CD4+ T cells. Biosens. Bioelectron. 2017;90:238–244. doi: 10.1016/j.bios.2016.11.055. [DOI] [PubMed] [Google Scholar]
- 227.John D.V., Lin Y.-S., Perng G.C. Biomarkers of severe dengue disease - a review. J. Biomed. Sci. 2015;22:83. doi: 10.1186/s12929-015-0191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Baraket A., Lee M., Zine N., Sigaud M., Bausells J., Errachid A. A fully integrated electrochemical biosensor platform fabrication process for cytokines detection. Biosens. Bioelectron. 2017;93:170–175. doi: 10.1016/j.bios.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 229.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Liu C., Zhou Q., Li Y., V Garner L., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 235.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Alanagreh L., Alzoughool F., Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9:331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00557-20. e00557–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.U.S. Department of Health and Human Services 2019-Novel coronavirus (2019-nCoV) real-time rRT-PCR panel primers and probes. 2020. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf
- 240.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 241.Krammer F., Simon V. Serology assays to manage COVID-19. Science (80) 2020;368 doi: 10.1126/science.abc1227. 1060 LP – 1061. [DOI] [PubMed] [Google Scholar]
- 242.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. e00461–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., Lau D.P.-L., Choi C.Y.-C., Chen L.-L., Chan W.-M., Chan K.-H., Ip J.D., Ng A.C.-K., Poon R.W.-S., Luo C.-T., Cheng V.C.-C., Chan J.F.-W., Hung I.F.-N., Chen Z., Chen H., Yuen K.-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 246.Okba N.M.A., Muller M.A., Li W., Wang C., GeurtvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Hingrat Q.L., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Vadlamani B.S., Uppal T., Verma S.C., Misra M. Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors. 2020;20:5871. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. J. Am. Med. Assoc. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 250.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 251.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Mostafa H.H., Hardick J., Morehead E., Miller J.-A., Gaydos C.A., Manabe Y.C. Comparison of the analytical sensitivity of seven commonly used commercial SARS-CoV-2 automated molecular assays. J. Clin. Virol. 2020;130:104578. doi: 10.1016/j.jcv.2020.104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Liu Y., Yan L.-M., Wan L., Xiang T.-X., Le A., Liu J.-M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Shen L., Wang C., Zhao J., Tang X., Shen Y., Lu M., Ding Z., Huang C., Zhang J., Li S., Lan J., Wong G., Zhu Y. Delayed specific IgM antibody responses observed among COVID-19 patients with severe progression. Emerg. Microb. Infect. 2020;9:1096–1101. doi: 10.1080/22221751.2020.1766382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kermali M., Khalsa R.K., Pillai K., Ismail Z., Harky A. The role of biomarkers in diagnosis of COVID-19 – a systematic review. Life Sci. 2020;254:117788. doi: 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Adams S.D., Doeven E.H., Quayle K., Kouzani A.Z. MiniStat: development and evaluation of a mini-Potentiostat for electrochemical measurements. IEEE Access. 2019;7:31903–31912. doi: 10.1109/ACCESS.2019.2902575. [DOI] [Google Scholar]
- 261.Hoilett O.S., Walker J.F., Balash B.M., Jaras N.J., Boppana S., Linnes J.C. KickStat: a Coin-Sized Potentiostat for high-Resolution electrochemical analysis. Sensors. 2020;20:2407. doi: 10.3390/s20082407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Sempionatto J.R., Brazaca L.C., García-Carmona L., Bolat G., Campbell A.S., Martin A., Tang G., Shah R., Mishra R.K., Kim J., Zucolotto V., Escarpa A., Wang J. Eyeglasses-based tear biosensing system: non-invasive detection of alcohol, vitamins and glucose. Biosens. Bioelectron. 2019;137:161–170. doi: 10.1016/j.bios.2019.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Inan H., Poyraz M., Inci F., Lifson M.A., Baday M., Cunningham B.T., Demirci U. Photonic crystals: emerging biosensors and their promise for point-of-care applications. Chem. Soc. Rev. 2017;46:366–388. doi: 10.1039/C6CS00206D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yang B., Fang X., Kong J. Situ sampling and monitoring cell-free DNA of the Epstein–Barr virus from dermal interstitial fluid using wearable microneedle patches. ACS Appl. Mater. Interfaces. 2019;11:38448–38458. doi: 10.1021/acsami.9b12244. [DOI] [PubMed] [Google Scholar]
- 265.Silva V.A.O.P., Fernandes-Junior W.S., Rocha D.P., Stefano J.S., Munoz R.A.A., Bonacin J.A., Janegitz B.C. 3D-printed reduced graphene oxide/polylactic acid electrodes: a new prototyped platform for sensing and biosensing applications, Biosens. Bioelectron. 2020;170:112684. doi: 10.1016/j.bios.2020.112684. [DOI] [PubMed] [Google Scholar]
- 266.Kogikoski S., Paschoalino W.J., Kubota L.T. Supramolecular DNA origami nanostructures for use in bioanalytical applications. TrAC Trends Anal. Chem. 2018;108:88–97. doi: 10.1016/j.trac.2018.08.019. [DOI] [Google Scholar]
- 267.Singhal C., Dubey A., Mathur A., Pundir C.S., Narang J. Paper based DNA biosensor for detection of chikungunya virus using gold shells coated magnetic nanocubes. Process Biochem. 2018;74:35–42. doi: 10.1016/j.procbio.2018.08.020. [DOI] [Google Scholar]
- 268.Ilkhani H., Farhad S. A novel electrochemical DNA biosensor for Ebola virus detection. Anal. Biochem. 2018;557:151–155. doi: 10.1016/j.ab.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 269.Ahangar L.E., Mehrgardi M.A. Amplified detection of hepatitis B virus using an electrochemical DNA biosensor on a nanoporous gold platform. Bioelectrochemistry. 2017;117:83–88. doi: 10.1016/j.bioelechem.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 270.Qian X., Tan S., Li Z., Qu Q., Li L., Yang L. A robust host-guest interaction controlled probe immobilization strategy for the ultrasensitive detection of HBV DNA using hollow HP5–Au/CoS nanobox as biosensing platform. Biosens. Bioelectron. 2020;153:112051. doi: 10.1016/j.bios.2020.112051. [DOI] [PubMed] [Google Scholar]
- 271.Nawaz M.H., Hayat A., Catanante G., Latif U., Marty J.L. Development of a portable and disposable NS1 based electrochemical immunosensor for early diagnosis of dengue virus. Anal. Chim. Acta. 2018;1026:1–7. doi: 10.1016/j.aca.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 272.Darwish N.T., Alrawi A.H., Sekaran S.D., Alias Y., Khor S.M. Electrochemical immunosensor based on antibody-nanoparticle hybrid for specific detection of the dengue virus NS1 biomarker. J. Electrochem. Soc. 2015;163:B19–B25. doi: 10.1149/2.0471603jes. [DOI] [Google Scholar]
- 273.Palomar Q., Gondran C., Marks R., Cosnier S., Holzinger M. Impedimetric quantification of anti-dengue antibodies using functional carbon nanotube deposits validated with blood plasma assays. Electrochim. Acta. 2018;274:84–90. doi: 10.1016/j.electacta.2018.04.099. [DOI] [Google Scholar]
- 274.Solanki S., Soni A., Pandey M.K., Biradar A., Sumana G. Langmuir–Blodgett nanoassemblies of the MoS2–Au composite at the air–water interface for dengue detection. ACS Appl. Mater. Interfaces. 2018;10:3020–3028. doi: 10.1021/acsami.7b14391. [DOI] [PubMed] [Google Scholar]
- 275.Pimenta T.C., da C., Santos C., Thomasini R.L., Ferreira L.F. Impedimetric immunosensor for dengue diagnosis using graphite screen-printed electrodes coated with poly(4-aminophenylacetic acid) Biomed. Microdevices. 2018;20:78. doi: 10.1007/s10544-018-0324-2. [DOI] [PubMed] [Google Scholar]
- 276.Ma Y., Shen X.-L., Zeng Q., Wang H.-S., Wang L.-S. A multi-walled carbon nanotubes based molecularly imprinted polymers electrochemical sensor for the sensitive determination of HIV-p24. Talanta. 2017;164:121–127. doi: 10.1016/j.talanta.2016.11.043. [DOI] [PubMed] [Google Scholar]
- 277.Singh R., Hong S., Jang J. Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 2017;7:42771. doi: 10.1038/srep42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Chin S.F., Lim L.S., Pang S.C., Sum M.S.H., Perera D. Carbon nanoparticle modified screen printed carbon electrode as a disposable electrochemical immunosensor strip for the detection of Japanese encephalitis virus. Microchim. Acta. 2017;184:491–497. doi: 10.1007/s00604-016-2029-7. [DOI] [Google Scholar]
- 279.Attar A., Mandli J., Ennaji M.M., Amine A. Label-free electrochemical impedance detection of rotavirus based on immobilized antibodies on gold sononanoparticles. Electroanalysis. 2016;28:1839–1846. doi: 10.1002/elan.201600179. [DOI] [Google Scholar]
- 280.Kaushik A., Yndart A., Kumar S., Jayant R.D., Vashist A., Brown A.N., Li C.-Z., Nair M. A sensitive electrochemical immunosensor for label-free detection of Zika-virus protein. Sci. Rep. 2018;8:9700. doi: 10.1038/s41598-018-28035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sayhi M., Ouerghi O., Belgacem K., Arbi M., Tepeli Y., Ghram A., Anik Ü., Österlund L., Laouini D., Diouani M.F. Electrochemical detection of influenza virus H9N2 based on both immunomagnetic extraction and gold catalysis using an immobilization-free screen printed carbon microelectrode. Biosens. Bioelectron. 2018;107:170–177. doi: 10.1016/j.bios.2018.02.018. [DOI] [PubMed] [Google Scholar]