Abstract
In Lactococcus lactis, which is widely used as a starter in the cheese industry, the first step of aromatic and branched-chain amino acid degradation is a transamination which is catalyzed by two major aminotransferases. We have previously purified and characterized biochemically and genetically the aromatic aminotransferase, AraT. In the present study, we purified and studied the second enzyme, the branched-chain aminotransferase, BcaT. We cloned and sequenced the corresponding gene and used a mutant, along with the luciferase gene as the reporter, to study the role of the enzyme in amino acid metabolism and to reveal the regulation of gene transcription. BcaT catalyzes transamination of the three branched-chain amino acids and methionine and belongs to class IV of the pyridoxal 5′-phosphate-dependent aminotransferases. In contrast to most of the previously described bacterial BcaTs, which are hexameric, this enzyme is homodimeric. It is responsible for 90% of the total isoleucine and valine aminotransferase activity of the cell and for 50 and 40% of the activity towards leucine and methionine, respectively. The original role of BcaT was probably biosynthetic since expression of its gene was repressed by free amino acids and especially by isoleucine. However, in dairy strains, which are auxotrophic for branched-chain amino acids, BcaT functions only as a catabolic enzyme that initiates the conversion of major aroma precursors. Since this enzyme is still active under cheese-ripening conditions, it certainly plays a major role in cheese flavor development.
Lactococcus lactis is becoming one of the best-characterized gram-positive bacteria, probably because it is widely used as a starter in the dairy industry. Besides its essential role in milk acidification, this organism plays a major role in proteolysis (46) and also has enzymatic potential to transform amino acids to aroma compounds (1, 7, 16, 18). In particular, L. lactis can transform aromatic amino acids (ArAAs), branched-chain amino acids (BcAAs), and methionine to potent aroma compounds that have been identified as major aroma components of cheese flavors (9, 14, 17, 19, 35, 36). Previously, we demonstrated that the first step in degradation of ArAAs and BcAAs is a transamination (42, 50; S. Thirouin, L. Rijnen, J.-C. Gripon, and M. Yvon, Club des bactéries lactiques—7ème Colloque, abstr. M4, 1995), and we identified two major aminotransferases in L. lactis subsp. cremoris NCDO 763 that are responsible for the transamination (50). The first aminotransferase is an aromatic aminotransferase (AraT) that we purified and characterized previously (50). This enzyme is active with the three ArAAs and also with leucine and methionine. It is produced constitutively and plays a dual role in biosynthesis and catabolism of ArAAs (41). While AraT plays a major role in ArAA catabolism, degradation of leucine and methionine can also be initiated by a second aminotransferase which is also active with isoleucine and valine. The aminotransferase responsible for BcAA transamination is very interesting since its substrates are precursors of major aroma compounds of cheese, such as isobutyrate, isovalerate, 3-methylbutanal, 2-methylbutanal, and 3-methylpropanal (9, 14, 17, 30, 35, 36). Biochemical and genetic characterization of this enzyme could make it possible to control its action during cheese ripening.
In contrast to AraTs and aspartate aminotransferases, branched-chain aminotransferases (BcaTs) (EC 2.6.1.42) have not been extensively studied. While 25 gene sequences are available in gene banks, only a few bacterial BcaTs have been well characterized (10, 27, 29, 32, 37, 48, 49). All of these enzymes belong to class IV of the pyridoxal phosphate-dependent aminotransferases (3, 24). In Escherichia coli or Salmonella enterica sevovar Typhimurium, the ilvE gene encoding BcaT is part of the BcAA biosynthetic operon, which is regulated by multivalent repression by the three BcAAs, while in Pseudomonas aeruginosa or Pseudomonas putida, BcaT apparently is synthesized constitutively. In L. lactis, the gene encoding BcaT is not part of the leu-ilv cluster, which contains all of the other structural genes for BcAA biosynthesis; transcription of this gene cluster is controlled mainly by a repression mechanism regulated only by isoleucine (8, 22).
The aims of the present work were to characterize biochemically and genetically the BcaT of L. lactis and to determine whether the corresponding gene is regulated or not regulated and then to evaluate the role and importance of the enzyme in amino acid metabolism. To do this, we used a mutant strain with a disrupted bcaT gene.
MATERIALS AND METHODS
Chemicals.
Amino acids, keto acids, inhibitors, pyridoxal 5′-phosphate (PLP), EDTA, streptomycin sulfate, erythromycin, and lysozyme were obtained from Sigma Chemical Co. (St. Louis, Mo.). Q-Sepharose Fast Flow gels and Mono-Q HR 10/10 and Superose 12 HR 10/30 columns were purchased from Pharmacia Biotech (Uppsala, Sweden). Radiolabeled amino acids were obtained from Isotopchim (Peyruis, France).
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. L. lactis strains were grown at 30°C either in M17 medium supplemented with 0.5% (wt/vol) glucose (45) or in modified or unmodified chemically defined medium (CDM) (44). The casein used in modified CDM was prepared by precipitating at pH 4.6 milk reconstituted from NILAC low-heat spray powder (NIZO, Ede, The Netherlands). E. coli was grown aerobically in Luria-Bertani medium (43) at 37°C. When needed, erythromycin (5 μg · ml−1 for L. lactis and 150 μg · ml−1 for E. coli) or ampicillin (50 μg · ml−1 for E. coli) was added to the culture medium.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| L. lactis subsp. cremoris | ||
| NCDO 763 | Wild type | National Collection of Food Bacteria, Shinfield, Reading, England |
| TIL 46 | NCDO 763 cured of its 2-kb plasmid | |
| TIL 354 | TIL 46 bcaT mutant with luxAB downstream of the bcaT promoter | This study |
| E. coli TG1 | 20 | |
| Plasmids | ||
| pGEM-T easy | Cloning vector (T overhangs), lacZ f1 ori Ampr, 3,018 bp | Promega Corp., Madison, Wis. |
| pTIL250 | 693-bp fragment of bcaT in pGEM-T | This study |
| pTIL252 | 2.5-kb TIL 46 DNA fragment containing bcaT in pGEM-T | This study |
| pJIM2374 | Eryr, integrative transcriptional fusion vector with the luxAB genes | 12 |
| pTIL253 | pTIL250 in SalI site of pJIM2374 | This study |
Enzyme purification.
Cells and cellular extract were prepared from a 5-liter culture of L. lactis NCDO 763 in CDM as previously described (50). The enzyme was then purified by a three-step procedure, as follows.
(i) Step 1.
The dialyzed cellular extract was loaded onto a Q-Sepharose Fast Flow column (gel bed volume, 83 ml) equilibrated with 50 mM potassium phosphate buffer (pH 7.5) containing 2 mM β-mercaptoethanol, 2 mM EDTA, and 0.1 mM PLP. The retained proteins were eluted at a rate of 3 ml/min with a 150-min linear 0.1- to 0.5-mol/liter NaCl gradient in the same buffer. Fractions containing isoleucine aminotransferase (Ile-AT) activity, which eluted at NaCl concentrations between 0.13 and 0.27 mol/liter, were pooled and dialyzed against 25 mM Tris-HCl buffer (pH 8.8) (Tris buffer).
(ii) Step 2.
The dialyzed fraction was loaded onto a Mono-Q HR 10/10 column equilibrated with Tris buffer, and the enzyme was eluted with a 100-min linear 0.25- to 0.45-mol/liter sodium acetate gradient in the same buffer at a rate of 3 ml/min. The eluent was collected in 3-ml fractions.
(iii) Step 3.
Each of the two most active fractions (which eluted at sodium acetate concentrations around 0.35 mol/liter) was concentrated by using Ultrafree-MC 10,000 NMWL filter units (Millipore Corp., Bedford, Mass.) to a volume of 0.2 ml and separately injected onto a Superose 12 HR 10/30 column. Elution was performed at a rate of 0.2 ml/min with 25 mM Tris-HCl buffer (pH 8) containing 0.15 M NaCl. The purity of active fractions was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the pure fractions were pooled. PLP was added to a final concentration of 0.05 mM, and the final preparation was stored at 6°C until it was used for characterization studies.
Enzyme characterization. (i) Molecular mass determination.
The molecular mass of the enzyme was estimated after gel filtration with the Superose 12 HR 10/30 column (purification step 3). The column had been calibrated previously under similar conditions with a mixture of marker proteins that included tyroglobulin (molecular mass, 670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa) (Bio-Rad Laboratories, Hercules, Calif.). The subunit molecular mass was also determined by SDS-PAGE.
(ii) SDS-PAGE.
A running gel was prepared with 12.5% acrylamide, and a stacking gel was prepared with 6% acrylamide. Low-range SDS-PAGE standards (Bio-Rad) were used as molecular weight references. Proteins were visualized by silver staining as described by Blum et al. (5).
(iii) Specificity.
Specificity for different amino acids and α-keto acids was determined under the conditions described below for determining aminotransferase activity.
(iv) Dependence on pH and temperature.
The pH dependence was investigated at pHs ranging from 4.5 to 9.5 as previously described (50), and the temperature dependence was determined at the optimum pH (7.5).
(v) Effects of inhibitors and bivalent cations.
The effects of inhibitors and bivalent cations were established after preincubation of pure enzyme preparations for 5 min with different inhibitors at final concentrations of 1 and 10 mM.
(vi) Amino acid sequencing.
The NH2-terminal amino acid sequence of purified BcaT was determined by using a pulse liquid sequencer (model 494A; Applied Biosystems). The enzyme was electroblotted from an SDS-PAGE gel onto a polyvinylidene difluoride membrane, as described by Matsudaira (34). The membrane was stained with Coomassie blue and then directly used in the sequencer.
Gene cloning, sequencing, and inactivation. (i) DNA techniques.
All DNA manipulations were performed as described by Sambrook et al. (43). L. lactis and E. coli electrocompetent cells were prepared and transformed by using standard techniques (26, 43). E. coli plasmid DNA was prepared with a plasmid purification kit obtained from Qiagen Inc. (Chatsworth, Calif.), and L. lactis plasmid DNA was prepared by the method of O'Sullivan and Klaenhammer (38). L. lactis chromosomal DNA was prepared as previously described (33, 43). For Southern blot analysis, a 1-kb fragment of the gene was used to prepare the DNA probe with an ECL kit (Amersham, Buckinghamshire, United Kingdom), and hybridization was performed as described by the supplier.
(ii) Northern blot analysis.
Total RNA was prepared as previously described for Bacillus subtilis (21). After extraction and treatment with phenol-chloroform, RNA was precipitated with ethanol; 50 μg of glyoxalated RNA was electrophoresed through a 1% agarose gel. Hybridization was performed as described above for Southern blot analysis by using the same DNA probe.
(iii) PCR, cloning, and sequencing.
PCR amplification was performed by using a Perkin-Elmer model 480 or 2400 DNA thermal cycler and Taq DNA polymerase (Appligene, Illkirch, France); 30 cycles were performed. A 2.5-kb DNA fragment containing bcaT was amplified from the total DNA of strain NCDO 763 by PCR by using two oligonucleotides (5′-TAT CAG CGA CTA AAT CTC-3′ and 5′-AAT TTG GGC AAT GAA GCC-3′) obtained from sequences of lactococcal DNA fragments homologous to the ilvE gene of Haemophilus influenzae (A. Sorokin and A. Bolotin, personal communication). The fragment was cloned into the pGEM-T vector to obtain pTIL252, in which the direction of the insert was opposite that of lacZ.
pTIL252 was used as a sequencing template. The nucleotide sequence was determined at least twice for both strands with a model 370A automatic DNA sequencer (Applied Biosystems). The samples used for sequencing were prepared with a PRISM Ready Reaction Dye Deoxy terminator cycle sequencing kit (Applied Biosystems, Inc.).
The DNA and protein sequences were analyzed with the GCG program (Genetics Computer Group, Inc., Madison, Wis.). Protein homology searches were carried out by using the BLAST network service (2). ProDom database 99.1 (11) was used to search for homologous domains, and Prosite (3) was used to search for characteristic motifs.
(iv) Gene inactivation.
A bcaT mutant with the luxAB genes under the control of the bcaT promoter was constructed in L. lactis subsp. cremoris TIL 46. To do this, a 693-bp internal fragment of the bcaT gene was generated by PCR by using nucleotides 5′-GCA ATT AAT TTA GAC TGG-3′ and 5′-GCT GTA ATT CCA AAG AAA-3′, and this fragment was cloned into the pGEM-T vector to obtain pTIL250. The latter was then inserted into the SalI site of the integrative transcription fusion vector p-orinewlux (p-JIM 2374) to create pTIL253, and the pGEM-T vector was eliminated by double digestion with BstXI and ApaI. After blunt ends were created with T4 DNA polymerase, the DNA fragment was religated in a dilute solution (500 ng of DNA · ml−1). Homologous integration of the fragment into the chromosome left bcaT disrupted and the luxAB genes under the control of the bcaT promoter in strain TIL 354. The final construction was similar to the construction described previously for araT (41). Interruption of bcaT and the integration site were verified by PCR and DNA sequencing.
Luciferase assay.
Luciferase activity was monitored throughout growth in different media, as previously described (15, 40). Briefly, 1 ml of culture was mixed with 5 μl of nonaldehyde, and light emission was measured immediately with a Berthold luminometer. Simultaneously, the optical density at 600 nm (OD600) of an identical sample was measured. The values given below for this experiment are the values read on a curve (lux versus OD600) at an OD600 of 0.6 and then expressed as kilolux per OD600 unit. The data reported below are means based on at least three replicates.
Aminotransferase activity assay.
During enzyme purification, aminotransferase activity was monitored as previously described for AraT (50), except that the amino acid substrate used was isoleucine instead of phenylalanine.
To determine specific activities, specificity for amino acids and α-keto acids, dependence on pH and temperature, and inhibitors of the pure enzyme, the same protocol was used, but the level of l-glutamic acid (or the amino acid corresponding to the α-keto acid used as the amino group acceptor) was measured by performing an amino acid analysis with a model LC3000 automatic analyzer (Biotronick, Maintal, Germany) as previously described (50).
The aminotransferase activities in extracts of cells grown in different media were determined as described by Rijnen et al. (41). Cell extracts were prepared as described previously (41) and were diluted in such a way that after 15 min of reaction no more than 10% of the substrate was used. The data reported below are means of the results obtained with triplicate cultures.
Amino acid catabolism.
We examined the catabolism of amino acids by whole cells of L. lactis subsp. cremoris TIL 46 and its bcaT mutant by using radiolabeled amino acids as tracers and the protocol described previously (41, 50). Briefly, each reaction mixture contained 100 mM Tris-HCl buffer (pH 8), 2 mM unlabeled amino acid, 0.05 μM tritiated amino acid, and 10 mM α-ketoglutarate. Cells grown in CDM to an OD480 of 10 were added to 500 μl of the reaction mixture. After incubation at 37°C for 0, 10, 20, and 40 h, the reaction mixtures were analyzed by reverse-phase high-performance liquid chromatography with both UV detection at 214 nm and radioactivity detection. The data reported below are means of the results obtained with duplicate reaction mixtures.
Protein determination.
Protein concentrations were determined by the micromethod of Bradford (6) by using bovine serum albumin as the standard, as recommended by the manufacturer (Pierce Chemical Co., Rockford, Ill.).
Nucleotide sequence accession number.
The nucleotide sequence described in this paper has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AF164204.
RESULTS
Purification and characterization of the enzyme.
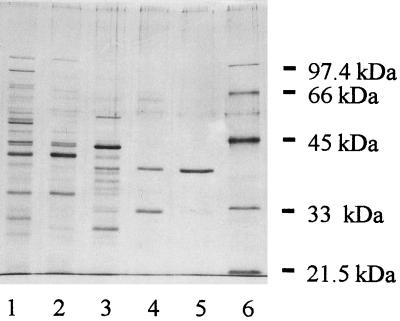
Most of the Ile-AT activity was recovered in the intracellular extract, and in all three chromatographic steps the Ile-AT activity was recovered as one major peak, indicating that the purified aminotransferase accounted for most (more than 90%) of the Ile-AT activity of the cell extract. Finally, the enzyme was purified about 421-fold by using a three-step procedure, and the level of recovery was less than 0.1%. The final fraction was considered pure as determined by SDS-PAGE with silver staining (Fig. 1). The pure fraction, which had a specific activity of 94 U/mg of protein with isoleucine and α-ketoglutarate as the substrates, was used for characterization.
FIG. 1.
SDS-PAGE (10% polyacrylamide) with silver staining, showing the different steps used for purification of Ile-AT activity. Lane 1, intracellular extract; lanes 2 and 3, pooled active fractions from Q-Sepharose and Mono-Q columns; lane 4, the most active fraction from the Mono-Q column; lane 5, purest fraction from the Superose 12 column; lane 6, Bio-Rad low-molecular-mass markers.
The molecular mass of the enzyme was about 64 kDa as determined by Superose 12 gel filtration and 38 kDa as determined by SDS-PAGE, which suggested that the enzyme is homodimeric.
The enzyme catalyzed transamination of only l-amino acids. The substrate specificity is shown in Table 2. The best amino acid substrates were branched-chain amino acids, especially isoleucine, but the enzyme also exhibited low activity with methionine and very slight activity with phenylalanine and cysteine. All other common amino acids were not substrates. The enzyme could use all of the α-keto acids corresponding to its amino acid substrates and also could use pyruvate weakly as an amino group acceptor.
TABLE 2.
Substrate specificity of the aminotransferase isolated from L. lactis subsp. cremoris NCDO 763
| Substrate | Relative activity (%)a |
|---|---|
| Amino donorsb | |
| l-Isoleucine | 100 |
| l-Leucine | 65 |
| l-Valine | 56 |
| l-Methionine | 6.4 |
| l-Cysteine | 1.6 |
| l-Phenylalanine | 0.5 |
| l-Alanine | NDd |
| Amino acceptorsc | |
| α-Ketoglutarate (Glu) | 100 |
| α-Ketoisocaproate (Leu) | 73.3 |
| α-Ketoisovalerate (Val) | 69.1 |
| 4-Methylthio-2-oxobutyrate (Met) | 32.3 |
| β-Phenylpyruvate (Phe) | 14 |
| Pyruvate (Ala) | 2.5 |
| Oxaloacetate (Asp) | ND |
Determined by production of the amino acid corresponding to the amino group receptor. Values are means of two determinations.
α-Ketoglutarate (10 mM) was used as the amino group acceptor. The concentration of each amino acid was 3 mM.
Isoleucine (3 mM) was used as the amino group donor. All keto acids were assayed at a concentration of 10 mM. The amino acid corresponding to the amino group acceptor is given in parentheses.
ND, not detected (less than 0.5%).
The optimum pH for activity was around pH 7.5, and the optimum temperature was between 35 and 40°C. At pH 5.5 the activity was about 20% of the activity at the optimum pH, and the activity at 10°C was about 10% of the activity at the optimum temperature.
The pure enzyme was not stable. Storage of the purest fraction at −20 or 0°C resulted in a very rapid loss of all activity. However, the enzyme was fairly stable at 6°C for 1 week. It was partially inactivated by heating at 50°C (40% inactivation after 30 min) and was inactivated more by heating at 60°C (more than 80% inactivation after 30 min).
As expected, the enzyme was strongly inhibited by carbonyl reagents, such as hydroxylamine and phenylhydrazine, which are known inhibitors of PLP-dependent enzymes (Table 3). It was also sensitive to sulfhydryl reagents, such as iodoacetamide and iodoacetic acid, suggesting that thiol groups are involved in the enzyme activity. The enzyme was not metal ion dependent, since the chelating agent EDTA did not have an inhibitory effect and no bivalent cation stimulated activity. In contrast, Cu2+ and Co2+ had inhibitory effects, and the inhibitory effect of Cu2+ was greater. Finally, the activity of the enzyme was not affected by Ca2+ or by 0.4% NaCl and was only slightly reduced (20%) by 4% NaCl. Therefore, the enzyme could still be active under cheese-ripening conditions.
TABLE 3.
Effects of protein labeling reagents, transaminase inhibitors, and metal ions on the activity of BcaT from L. lactis subsp. cremoris NCDO 763a
| Compound | Concn | % Inhibition |
|---|---|---|
| Iodoacetic acid | 1 mmol/liter | 56 |
| 10 mmol/liter | 97 | |
| Iodoacetamide | 1 mmol/liter | 57 |
| 10 mmol/liter | 65 | |
| Hydroxylamine | 1 mmol/liter | 100 |
| Phenylhydrazine | 1 mmol/liter | 100 |
| EDTA | 10 mmol/liter | 10 |
| NaCl | 0.4% | 0 |
| 4% | 18 | |
| CaCl2 | 10 mmol/liter | 21 |
| CuCl2 | 1 mmol/liter | 54 |
| 10 mmol/liter | 100 | |
| CoCl2 | 10 mmol/liter | 63 |
| ZnCl2 | 10 mmol/liter | 23 |
Purified enzyme was incubated with the inhibitors for 5 min at pH 8 before we assayed for Ile-AT activity; α-ketoglutarate was the amino group acceptor used. NaCl, EDTA, CaCl2, CuCl2, CoCl2, and ZnCl2 were added to the assay buffer.
The N-terminal sequence was identified as A-I-N-L-D-W-E-N-L-G-F-S-Y and exhibited 65% identity with the N-terminal sequence deduced from ilvE of H. influenzae.
Genetic characterization of BcaT.
The analysis of the DNA sequence revealed an open reading frame that encodes a 340-amino-acid protein with a calculated molecular mass of 36,944 Da, which is in excellent agreement with the molecular mass determined by SDS-PAGE (38 kDa). We found a putative ribosome binding site (GGAGG) starting 10 bp upstream of the ATG start codon and two potential promoters (TTGTTT-TATAAT and TTCTTG-TAATAT) 20 and 23 bp upstream of the ribosome binding site. Finally, we also observed a putative ρ-independent transcriptional terminator 8 bp downstream of the TAA stop codon. Southern blot analyses performed under different DNA digestion conditions (XmnI, XbaI, SpeI, NcoI, HpaI, HindIII, EcoRI, BstI, and AAtII) revealed that the probe (gene fragment) hybridized with a unique fragment of DNA from each preparation, indicating that a single copy of the gene was present in the chromosome. An RNA analysis performed by the Northern blot method showed that bcaT was expressed in a 1.1-kb transcript (from the potential promoter to the putative terminator).
Homologies.
The bcaT sequence of L. lactis NCDO 763 is 98.5% identical to the bcaT sequence of L. lactis NCDO 1403 (A. Sorokin and A. Bolotin, personal communication). The amino acid sequence deduced from the nucleotide sequence exhibited significant similarity (levels of identity, 30 to 68%) with the amino acid sequences of BcaTs from mammals, yeast, bacteria, and plants. In particular, it exhibited 68 and 60% homology with the amino acid sequences deduced from the nucleotide sequences of ilvE of H. influenzae and ilvE of Helicobacter pylori, respectively. The PLP binding site of L. lactis BcaT is most likely Lys-184 in the conserved sequence K-x-G-x-N-Y found in all BcaT sequences (3). The bcaT sequence of L. lactis NCDO 763 also exhibited the aminotransferase class IV signature (Prosite accession no. PS00770) (3), which locates 36 residues at the C-terminal side of the PLP lysine. We constructed a dendrogram from a multiple alignment of the deduced amino acid sequences of BcaTs that belong to aminotransferase class IV, including our BcaT sequence (data not shown). Bacterial BcaTs were segregated in three subclasses. The first subclass contained hexameric BcaTs, and transaminase B of E. coli was the best-characterized enzyme in this subclass. The second subclass contained bacterial BcaTs homologous to B. subtilis ILVE, which were similar to yeast and mammal (vertebrate) BcaTs. Most of the enzymes in this subclass that have been characterized are homodimeric. Finally, our L. lactis BcaT was classified in the third subclass along with H. influenzae ILVE and H. pylori ILVE, both of which had a homologous N-terminal domain that was made up of 55 amino acid residues and is called domain 38216 in the ProDom 99.1 database (11).
Role of BcaT in degradation of amino acids to aroma compounds.
The aminotransferase activities of the bcaT mutant were compared with those of the wild-type strain (Table 4). Inactivation of bcaT reduced the Ile-AT and valine aminotransferase activities by more than 90% and the leucine aminotransferase activity by approximately 50%. It also decreased, to a lesser extent (around 40%), the methionine aminotransferase activity, while it did not alter the activity with ArAAs.
TABLE 4.
Aminotransferase activities of the wild-type strain and the bcaT mutant
| Substrate | Aminotransferase activity ofa:
|
|
|---|---|---|
| Wild-type strain | bcaT mutant | |
| Phenylalanine | 115 ± 11 | 107 ± 7 |
| Tyrosine | 113 ± 12 | 103 ± 6 |
| Tryptophan | 78 ± 10 | 69 ± 6 |
| Leucine | 303 ± 31 | 126 ± 48 |
| Isoleucine | 272 ± 20 | 25 ± 12 |
| Valine | 122 ± 5 | 14 ± 1 |
| Methionine | 42 ± 2 | 26 ± 2 |
Activities were determined by using cellular extracts and are expressed in nanomoles of glutamate produced from α-ketoglutarate per minute per milligram of protein. The data are means ± standard deviations based on two determinations.
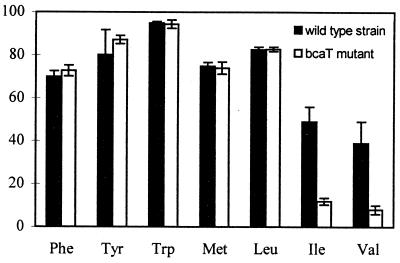
The role of BcaT in amino acid catabolism was studied by comparing amino acid degradation and metabolite formation by whole cells of both strains for 40 h in reaction mixtures that contained or did not contain α-ketoglutarate as the α-keto acid acceptor. In medium lacking α-ketoglutarate, no degradation occurred either with the wild-type strain or with the mutant strain (results not shown). In contrast, in medium containing α-ketoglutarate (Fig. 2), the wild-type strain degraded all amino acids, and bcaT inactivation strongly inhibited degradation of isoleucine and valine while it did not affect degradation of leucine, methionine, and ArAAs.
FIG. 2.
Percentages of amino acid degradation by the wild-type strain and the bcaT mutant after incubation of whole cells for 20 h in reaction medium containing radiolabeled amino acids as tracers and α-ketoglutarate under conditions described in the text. The amino acids whose degradation was studied are phenylalanine (Phe), tyrosine (Tyr), tryptophan (Trp), methionine (Met), leucine (Leu), isoleucine (Ile), and valine (Val). The results are means and standard deviations based on two determinations.
Regulation of bcaT transcription.
A gene fusion with luxAB genes as reporter genes was introduced downstream of the promoter in the bcaT mutant, and luciferase activity was measured in different media (Table 5). The transcriptional activity of the bcaT promoter was around 48 klx/OD600 unit in CDM and did not change when we omitted phenylalanine or all three ArAAs in the medium. In contrast, the activity increased two- to threefold when we reduced the BcAA concentration or the isoleucine concentration in CDM or when we decreased the methionine concentration, while it decreased twofold when we increased the BcAA concentration fivefold. In fact, we could not completely remove BcAAs or methionine from the medium since our strain was auxotrophic for these amino acids (39). Furthermore, gene transcription increased significantly (more than 10-fold) when free amino acids were replaced by casein in CDM, and a large part of the increase was repressed when free amino acids were added to the medium containing casein.
TABLE 5.
Bioluminescence in growing cultures of TIL 354 (bcaT mutant) in different media (complete CDM or modified CDM)
| Culture mediumb | Luciferase activity (klx/OD600 unit)a |
|---|---|
| CDM | 48 ± 6 |
| CDM without aromatic amino acids | 60 ± 8 |
| CDM with reduced ILV concentration (1/10) | 141 ± 8 |
| CDM with reduced M concentration (1/50) | 124 ± 2 |
| CDM with reduced I concentration (1/100) | 112 ± 6 |
| CDM with higher ILV concentration (×5) | 30 ± 5 |
| CDM with casein instead of free amino acids | 606 ± 47 |
| CDM with casein and all free amino acids | 194 ± 37 |
Data are means ± standard deviations based on at least three determinations.
I, L, V, and M refer to isoleucine, leucine, valine, and methionine, respectively. 1/10, 1/50, 1/100, and ×5 indicate the dilution or the increase in amino acid concentration in CDM.
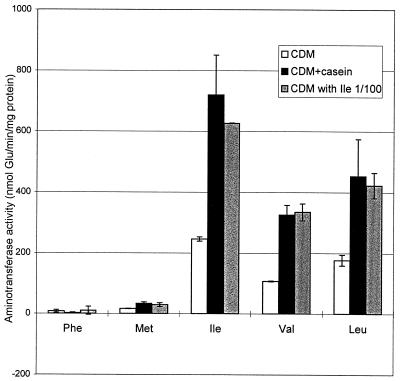
The aminotransferase activities of the wild-type strain and the mutant were also determined by using cells grown in certain media in which increases in bcaT transcription were observed. In contrast to the activities of the wild-type strain, the activities of the mutant were not affected by the growth medium (results not shown). Figure 3 shows the aminotransferase activity due to BcaT, which was calculated by subtracting the activity of the mutant from the activity of the wild-type strain. The BcaT activity increased 2.5- to 3-fold when the free amino acids in CDM were replaced by casein or when the isoleucine concentration in CDM was reduced 100-fold (to 2 mg/liter).
FIG. 3.
Aminotransferase activity of BcaT (activity of the wild-type strain minus activity of the bcaT mutant) in cells grown in CDM, in CDM containing casein instead of free amino acids (CDM+casein), and in CDM containing a 100-fold lower concentration of isoleucine (CDM with Ile 1/100). Aminotransferase activity was measured with phenylalanine (Phe), methionine (Met), isoleucine (Ile), valine (Val), and leucine (Leu) as substrates and α-ketoglutarate as the cosubstrate. Results are means and standard deviations based on at least two determinations.
DISCUSSION
Because transamination is the last step in BcAA biosynthesis and the first step in catabolism of BcAAs, BcaTs occur widely in animals, yeast, and bacteria. We isolated the BcaT responsible for most of the Ile-AT activity of L. lactis subsp. cremoris NCDO 763. In general, bacterial BcaTs are quite different from the mammalian and yeast enzymes, but the lactococcal enzyme also appears to be different from bacterial BcaTs which have been characterized previously; it seems to belong to a new class of BcaTs. Indeed, lactococcal BcaT differs from previously described bacterial BcaTs by its much lower molecular weight (4, 27, 31, 32, 37, 47, 49) and by not being active or being only very slightly active with ArAAs (29, 31, 37, 48, 49). In contrast, the lactococcal enzyme has a molecular weight similar to the molecular weights of mammalian BcaTs, but mammalian BcaTs are often specific for only one (or two) BcAAs (25, 28) while the lactococcal enzyme exhibits little preference among these amino acids. The gene encoding BcaT in L. lactis also differs from most previously described genes encoding BcaTs in mammals, yeast, or bacteria, but it is similar to the ilvE genes of H. influenzae and H. pylori, whose products have not been characterized yet. The differences between groups of enzymes are probably related to their physiological roles. In mammals the BcAAs are essential, and therefore the BcaTs are not biosynthetic, while in bacteria BcaTs, especially the hexameric enzymes, are biosynthetic and are often part of the BcAA biosynthetic operon (4). However, since lactococcal BcaT is different from all of the previously described BcaTs, there is a question concerning its physiological role.
In dairy L. lactis strains, such as L. lactis NCDO 763, BcaT cannot play a biosynthetic role since all BcAAs are essential (39). However, in these dairy strains, as in nondairy strains which are prototrophic for BcAAs, all of the genes required for BcAA synthesis are present. Auxotrophy in dairy strains seems to be the result of several mutations and deletions in these genes, which might be a consequence of adaptation of the organisms to milk and dairy products (23). Therefore, originally, BcaT may have been the last enzyme in the BcAA biosynthesis pathway. The original function of BcaT in BcAA biosynthesis is reinforced by the fact that bcaT transcription seems to be repressed by free amino acids, especially isoleucine, which is also the regulator of the BcAA biosynthetic operon (8, 22). However, the regulation mechanisms remain to be determined since other BcAAs and perhaps also methionine seem to be involved in regulation. Dias and Weimer (13) also observed that the methionine aminotransferase activity of L. lactis was repressed when the methionine concentration in the growth medium was increased. The variation in methionine aminotransferase activity is probably due to BcaT since the activity of AraT, which also catalyzes methionine transamination, seems to be constitutive (41).
Therefore, in dairy strains, BcaT is mainly involved in BcAA degradation and especially in conversion of BcAAs to aroma compounds. Indeed, inactivation of bcaT resulted in a significant decrease in degradation of Ile and Val, which are precursors of aroma compounds. The low level of degradation observed with the mutant (less than 10% of the initial degradation level) was the result of residual aminotransferase activity since it occurred only in the presence of an α-keto acid acceptor. This residual aminotransferase activity did not appear to be due to AraT since we did not detect such an activity with the purified enzyme (50). This suggests that another aminotransferase may be responsible for the low level of residual activity. This will have to be verified by using a double mutant with both inactivated bcaT and inactivated araT. Although inactivation of bcaT clearly reduced the leucine aminotransferase and methionine aminotransferase activities of the strain, it did not affect degradation in liquid medium, probably because the residual activities, which were mainly due to AraT (41), were sufficient to result in total degradation of Leu and Met under the experimental conditions used. However, in the wild-type strain, BcaT certainly participates in degradation of leucine and methionine to α-keto acids which are also direct precursors of aroma compounds, such as isovalerate, methanethiol, and other sulfur compounds (19, 51).
Since BcaT plays a major role in isoleucine and valine degradation and also contributes to leucine and methionine transamination and since the enzyme appeared to be active under cheese-ripening conditions (pH, salt concentration, and temperature), it is certainly involved in the development of cheese flavor. However, the role of BcaT should be verified by performing cheese-making trials. Additional studies on the regulation mechanism are in progress, and these studies should provide tools which can be used to control amino acid transamination during cheese ripening.
ACKNOWLEDGMENTS
This work was supported by contract FAIR CT 97-3173 from the Commission of the European Communities.
We thank J. C. Huet for N-terminal sequencing, M. Nardi for providing technical advice concerning molecular biology, A. Gaudin and V. Schrepfer for providing technical assistance, and J.-C. Gripon for critically reading the manuscript. We are indebted to K. Lynch (I.N.R.A. Translation Unit, Jouy-en-Josas, France) for revising the English.
REFERENCES
- 1.Alting A C, Engels W J M, van Schalkwijk S, Exterkate F A. Purification and characterization of cystathionine β-lyase from Lactococcus lactis subsp. cremoris B78 and its possible role in flavor development in cheese. Appl Environ Microbiol. 1995;61:4037–4042. doi: 10.1128/aem.61.11.4037-4042.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zang J, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A, Bucher P, Hofmann K. The Prosite database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg C M, Liu L, Vartak N B, Whalen W A, Wang B. The branched-chain amino acid transaminase genes and their products in Escherichia coli. In: Barak Z, Chipman D M, Schloss J V, editors. Biosynthesis of branched-chain amino acids—1990. Weinheim, Germany: VCH Publishers, Inc.; 1990. pp. 131–162. [Google Scholar]
- 5.Blum H, Beier H, Gross J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Bruinenberg P G, de Roo G, Limsowtin G K. Purification and characterization of cystathionine γ-lyase from Lactococcus lactis subsp. cremoris SK11: possible role in flavor compound formation during cheese maturation. Appl Environ Microbiol. 1997;63:561–566. doi: 10.1128/aem.63.2.561-566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopin A. Organization and regulation of amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:21–38. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 9.Christensen K R, Reineccius G A. Aroma extract dilution analysis of aged Cheddar cheese. J Food Sci. 1995;60:218–220. [Google Scholar]
- 10.Coleman M S, Armstrong F B. Branched-chain amino acid aminotransferase of Salmonella typhimurium. Biochim Biophys Acta. 1970;227:56–66. doi: 10.1016/0005-2744(71)90167-7. [DOI] [PubMed] [Google Scholar]
- 11.Corpet F, Grouzy J, Kahn D. The ProDom database of protein families. Nucleic Acids Res. 1998;26:323–326. doi: 10.1093/nar/26.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delorme C, Ehrlich D S, Renault P. Regulation of expression of the Lactococcus lactis histidine operon. J Bacteriol. 1999;181:2026–2037. doi: 10.1128/jb.181.7.2026-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias B, Weimer B. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl Environ Microbiol. 1998;64:3320–3326. doi: 10.1128/aem.64.9.3320-3326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn H C, Lindsay R C. Evaluation of the role of microbial Strecker-derived aroma compounds in unclean-type flavors of Cheddar cheese. J Dairy Sci. 1985;68:2859–2874. [Google Scholar]
- 15.Eaton T J, Shearman C A, Gasson M J. The use of bacterial luciferase genes as reporter genes in Lactococcus: regulation of the Lactococcus lactis subsp. lactis lactose genes. J Gen Microbiol. 1993;139:1495–1501. doi: 10.1099/00221287-139-7-1495. [DOI] [PubMed] [Google Scholar]
- 16.Engels W J M, Visser S. Development of cheese flavour from peptides and amino acids by cell-free extracts of Lactococcus lactis subsp. cremoris B78 in a model system. Neth Milk Dairy J. 1996;50:3–17. [Google Scholar]
- 17.Engels W J M, Dekker R, de Jong C, Neeter R, Visser S. A comparative study of volatile compounds in the water-soluble fraction of various types of ripened cheese. Int Dairy J. 1997;7:255–263. [Google Scholar]
- 18.Gao S, Oh D H, Broadbent J R, Johnson M E, Weimer B C, Steele J L. Aromatic amino acid catabolism by lactococci. Lait. 1997;77:371–381. [Google Scholar]
- 19.Gao S, Mooberry E S, Steele J L. Use of 13C nuclear magnetic resonance and gas chromatography to examine methionine catabolism by lactococci. Appl Environ Microbiol. 1998;64:4670–4675. doi: 10.1128/aem.64.12.4670-4675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson T J. Studies on the Eppstein Barr virus genome. Ph.D. thesis. Cambridge, England: University of Cambridge; 1984. [Google Scholar]
- 21.Glatron M F, Papoport G. Biosynthesis of the parasporal reclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie. 1972;54:1291–1301. doi: 10.1016/s0300-9084(72)80070-1. [DOI] [PubMed] [Google Scholar]
- 22.Godon J-J, Chopin M-C, Ehrlich S D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992;174:6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godon J-J, Delorme C, Bardowski J, Chopin M-C, Ehrlich S D, Renault P. Gene inactivation in Lactococcus lactis: branched-chain amino acid biosynthesis. J Bacteriol. 1993;175:4383–4390. doi: 10.1128/jb.175.14.4383-4390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green J M, Merkel W K, Nichols B P. Characterization and sequence of Escherichia coli pabC, the gene encoding aminodeoxychorismate lyase, a pyridoxal phosphate-containing enzyme. J Bacteriol. 1992;174:5317–5323. doi: 10.1128/jb.174.16.5317-5323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall T R, Wallin R, Reinhart G D, Hutson S M. Branched chain aminotransferase isoenzymes. Purification and characterization of the rat brain isoenzyme. J Biol Chem. 1993;266:3092–3093. [PubMed] [Google Scholar]
- 26.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue K, Kuramitsu S, Aki K, Watanabe Y, Takagi T, Nishigai M, Ikai A, Kagamiyama H. Branched-chain amino acid aminotransferase of Escherichia coli: overproduction and properties. J Biochem. 1988;104:777–784. doi: 10.1093/oxfordjournals.jbchem.a122549. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins W T, Taylor R T. Branched-chain amino acid aminotransferase (pig heart soluble) Methods Enzymol. 1970;17:802–807. [Google Scholar]
- 29.Kanda M, Hori K, Kurotsu T, Ohgishi K, Hanawa T. Purification and properties of branched chain amino acid aminotransferase from gramicidin S-producing Bacillus brevis. J Nutr Sci Vitaminol. 1995;41:51–60. doi: 10.3177/jnsv.41.51. [DOI] [PubMed] [Google Scholar]
- 30.Kubickovà J, Grosch W. Evaluation of potent odorants of Camembert cheese by dilution and concentration techniques. Int Dairy J. 1997;7:65–70. [Google Scholar]
- 31.Lee-peng F-G, Hermodson M A, Kohlhaw G B. Transaminase B from Escherichia coli: quaternary structure, amino-terminal sequence, substrate specificity, and absence of a separate valine-α-ketoglutarate activity. J Bacteriol. 1979;139:339–345. doi: 10.1128/jb.139.2.339-345.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipscomb E L, Horton H R, Armstrong F B. Molecular weight, subunit structure, and amino acid composition of the branched-chain amino acid aminotransferase of Salmonella typhimurium. Biochemistry. 1974;13:2070–2077. doi: 10.1021/bi00707a011. [DOI] [PubMed] [Google Scholar]
- 33.Loureiro dos Santos A L, Chopin A. Shotgun cloning in Streptococcus lactis. FEMS Microbiol Lett. 1987;42:209–212. [Google Scholar]
- 34.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 35.Milo C, Reineccius G A. Identification and quantification of potent odorants in regular-fat and low-fat mild Cheddar cheese. J Agric Food Chem. 1997;45:3590–3594. [Google Scholar]
- 36.Morgan M E, Lindsay R C, Libbey L M. Identity of additional aroma constituents in milk cultures of Streptococcus lactis var. maltigenes. J Dairy Sci. 1966;49:15–18. doi: 10.3168/jds.S0022-0302(66)87777-9. [DOI] [PubMed] [Google Scholar]
- 37.Norton J E, Sokatch J R. Purification and partial characterization of the branched chain amino acid transaminase of Pseudomonas aeruginosa. Biochim Biophys Acta. 1970;206:261–269. doi: 10.1016/0005-2744(70)90109-9. [DOI] [PubMed] [Google Scholar]
- 38.O'Sullivan D J, Klaenhammer T R. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol. 1993;59:2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiter J, Oram J D. Nutritional studies on cheese starters. I. Vitamin and amino acid requirements of single strain starters. J Dairy Res. 1962;29:63–77. [Google Scholar]
- 40.Renault P, Corthier G, Goupil N, Delorme C, Ehrlich S D. Plasmid vectors for gram-positive bacteria switching from high to low copy number. Gene. 1996;183:175–182. doi: 10.1016/s0378-1119(96)00554-9. [DOI] [PubMed] [Google Scholar]
- 41.Rijnen L, Bonneau S, Yvon M. Genetic characterization of the major lactococcal aromatic aminotransferase and its involvement in conversion of amino acids to aroma compounds. Appl Environ Microbiol. 1999;65:4873–4880. doi: 10.1128/aem.65.11.4873-4880.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roudot-Algaron F, Yvon M. Le catabolisme des acides aminés aromatiques et des acides aminés à chaîne ramifiée chez Lactococcus lactis. Lait. 1998;78:23–30. [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Smid E J, Konings W N. Relationship between utilization of proline and proline-containing peptides and growth of Lactococcus lactis. J Bacteriol. 1990;172:5286–5292. doi: 10.1128/jb.172.9.5286-5292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visser S. Proteolytic enzymes and their relation to cheese ripening and flavor: an overview. J Dairy Sci. 1993;76:329–350. [Google Scholar]
- 47.Whitaker R J, Gaines C G, Jensen R A. A multispecific quintet of aromatic aminotransferases that overlap different biochemical pathways in Pseudomonas aeruginosa. J Biol Chem. 1982;257:13550–13556. [PubMed] [Google Scholar]
- 48.Wong H C, Lessie T G. Branched chain amino acid aminotransferase isoenzymes of Pseudomonas cepacia. Arch Microbiol. 1979;120:223–229. doi: 10.1007/BF00423069. [DOI] [PubMed] [Google Scholar]
- 49.Xing R, Whitman W B. Characterization of amino acid aminotransferase of Methanococcus aeolicus. J Bacteriol. 1992;174:541–548. doi: 10.1128/jb.174.2.541-548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yvon M, Thirouin S, Rijnen L, Fromentier D, Gripon J-C. An aminotransferase from Lactococcus lactis initiates conversion of aromatic amino acids to flavor compounds. Appl Environ Microbiol. 1997;63:414–419. doi: 10.1128/aem.63.2.414-419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yvon M, Berthelot S, Gripon J-C. Adding α-ketoglutarate to semi-hard cheese curd highly enhances the conversion of amino acids to aroma compounds. Int Dairy J. 1999;8:889–898. [Google Scholar]