Graphical abstract
Keywords: SARS-CoV-2, RdRp, Insilico molecular docking analysis, Molecular dynamics simulation, ADMET analysis, Fungal metabolites
Abstract
Various protein/receptor targets have been discovered through in-silico research. They are expanding rapidly due to their extensive advantage of delivering new drug candidates more quickly, efficiently, and at a lower cost. The automation of organic synthesis and biochemical screening will lead to a revolution in the entire research arena in drug discovery. In this research article, a few fungal metabolites were examined through an in-silico approach which involves major steps such as (a) Molecular Docking Analysis, (b) Drug likeness and ADMET studies, and (c) Molecular Dynamics Simulation. Fungal metabolites were taken from Antibiotic Database which showed antiviral effects on severe viral diseases such as HIV. Docking, Lipinski's, and ADMET analyses investigated the binding affinity and toxicity of five metabolites: Chromophilone I, iso; F13459; Stachyflin, acetyl; A-108836; Integracide A (A-108835). Chromophilone I, iso was subjected to additional analysis, including a 50 ns MD simulation of the protein to assess the occurring alterations. This molecule's docking data shows that it had the highest binding affinity. ADMET research revealed that the ligand might be employed as an oral medication. MD simulation revealed that the ligand–protein interaction was stable. Finally, this ligand can be exploited to develop SARS-CoV-2 therapeutic options. Fungal metabolites that have been studied could be a potential source for future lead candidates. Further study of these molecules may result in creating an antiviral drug to battle the SARS-CoV-2 virus.
1. Introduction
SARS-CoV-2 is an RNA virus with a positive strand. It has a genome about 30 kb in size and has 14 open reading frames (ORFs). The 50-ORF-1a/1b gene encodes polyprotein 1a (pp1a) and polyprotein 1 ab (pp1ab) precursor polyproteins, which are subsequently cleaved into 16 nonstructural proteins (nsp1-16) in the SARS-CoV-2 genome (Wu et al., 2020, Gordon et al., 2020). In addition to papain-like protease (nsp3), RdRp (nsp12), chymotrypsin-like main protease (3CL protease, nsp5), exoribonuclease (nsp14) and helicase (nsp13), all of these nonstructural proteins play an important role in the development of viruses (Thoms et al., 2020). The host immune system and replication of SARS-CoV-2 are regulated by other NSP proteins. Nsp 1 binds to the ribosome and suppresses the host translation machinery (Yuan et al., 2020, Littler et al., 2020). Nsp9 is implicated in the replication and pathogenicity of viral genomic RNA (Pillon et al., 2020). Nsp15 is an endoribonuclease that degrades viral RNA to avoid detection by the host defense system (Rosas-Lemus et al., 2020). Nsp16, a methyltransferase that works in tandem with nsp10, can cap viral mRNA transcripts for effective translation and immune evasion (Subissi et al., 2014). The four structural proteins, the spike (S), envelope (E), membrane (M), and nucleocapsid (N) comprise approximately 33% of the virus genome at the C-terminus and play a critical role in viral structure integrity, such as spike protein, which allowed SARS-CoV-2 entry into the host. There are nine potential ORFs for auxiliary factors at the 30 ends of the genome (Thoms et al., 2020).
RdRp's general structure is like that of SARS-CoV RdRp (Kirchdoerfer and Ward, 2019). In Europe, an RdRp mutation 14408C > T was recently discovered, which is linked to a higher mutation rate in viral genomes from Asia via an unknown mechanism (Pachetti et al., 2020). The resultant mutation P323L is found in RdRp's interface domain, which is far from the active catalytic site, and may exercise its effects by altering interactions with other replication-transcription complex components or the RNA template (Eskier et al., 2020).
RdRp is a possible therapeutic target (Mohapatra et al., 2022, Mohapatra et al., 2021a, Mohapatra et al., 2021b) for COVID-19 treatment since it is required for viral RNA replication. To begin with, RdRp, like other SARS-CoV-2 proteins, lacks closed-related host cell equivalents. As a result, addressing RdRp may help to avoid off-target consequences. Second, the active catalytic motifs of the RdRp are different from those of the spike protein and other virus surface proteins. Remdesivir inhibits RdRp via a mechanism. (A) Remdesivir's chemical structure and cellular metabolic route (Godoy et al., 2017, Gerlach et al., 2015, Campagnola et al., 2011). (B) favipiravir binding mechanism at the active catalytic site of the SARS-CoV-2 RdRp (PDB code: 7AAP). Among RNA viruses RdRp is highly conserved making it a promising antiviral therapeutic target for a wide range of viruses. Many nucleoside analog inhibitors were inhibitory to a wide range of RNA viruses (Shrikanth et al., 2021). Finally, repurposing RdRp-targeted medicines for COVID-19 treatment remains a potential technique.
This study examines fungal metabolites from different fungi in silico to identify potential SARS-CoV-2 RdRp inhibitors, one of the important target proteins in SARS-CoV2. To evaluate SARS-CoV-2 protease inhibition capability, a docking study and pharmacokinetics tests were conducted to estimate binding energies and evaluate site interactions.
2. Materials and methods
2.1. Preparation of receptor molecule for docking
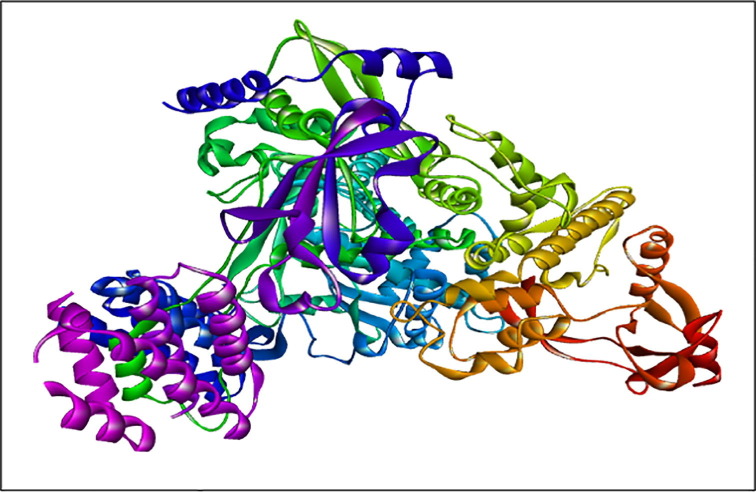
The Fig. 1 shows RDRP- RNA Dependent RNA Polymerase protein of SARS CoV-2 was selected as a target receptor in this in silico research. The 3D structure of the protein which in complex with favipiravir drug was identified as 7AAP of a resolution of 2.50 Å, the protein-bound hetero atoms were removed and the Polar Hydrogen atoms and Kollman Charges (Abdalla et al., 2021) were included for proper optimization using AutoDock Vina MGL tools (Boufissiou et al., 2022). The active sites of the 7AAP were considered as the bound site of favipiravir.
Fig. 1.
RDRP (7AAP) protein structure.
2.2. Preparation of lead molecules
Lead molecules were fungal metabolites that were taken from a database of an antibiotic to extract fungal metabolites which are antiviral, and the structures were obtained from the PubChem database in 3D SDF format. After searching their structures in PubChem, got the structures of 31 metabolites. The selected 31 metabolites were subjected to molecular docking. The docking results in binding affinity were spread between −5.1 to −8.7. Here we fixed the threshold value as −7, and in this, 18 metabolites with the binding affinity above −7 were selected and studied further.
The detailed structures and properties of the studied 18 fungal metabolites are described in Table 1 . (“Novel Antibiotics Database. Available at: https://www.antibiotics.or.jp/journal/database/database-top.htm).
Table 1.
List of selected metabolites.
| Fungi |
Compounds | PubChem-CID's | Mol. Wt. (g/mol) | Log P (<5) | Binding Affinity | |
|---|---|---|---|---|---|---|
| Genus | Species | |||||
| Chrysosporium | merdarium | Semicochliodinol A | 474302 | 438.5 | 5.548 | −8.7 |
| Chrysosporium | merdarium | Semicochliodinol B | 474303 | 438.5 | 4.545 | −8.6 |
| Stachybotrys | SQ-02-S-L2 | 11028093 | 556.7 | 5.65 | −7.8 | |
| Penicillium | Chromophilone I, iso | 6438449 | 416.9 | 5.54 | −7.8 | |
| Hypoxylon | fragiforme | L-696,474 | 6473821 | 477.6 | 3.16 | −7.8 |
| Penicillium | F13459 | 10119161 | 528.5 | 4.62 | −7.7 | |
| Stachybotrys | SQ-02-S-V1 | 11103392 | 527.6 | 1.25 | −7.6 | |
| Stachybotrys | Stachyflin | 493326 | 385.5 | 4.62 | −7.6 | |
| Hypoxylon | fragiforme | L-696,475 | 6474904 | 461.6 | 4.40 | −7.6 |
| Hypoxylon | fragiforme | L-697,318 | 101629194 | 477.6 | 3.22 | −7.5 |
| Stachybotrys | Stachyflin, acetyl | 493325 | 427.5 | 4.93 | −7.5 | |
| Fusarium | compactum | A-108836 | 10506876 | 550.8 | 1.88 | −7.4 |
| Fusarium | compactum | Integracide A (A-108835) |
460025 | 594.8 | 4.11 | −7.3 |
| Trichoderma | harzianum | Fleephilone | 10575150 | 441.5 | 4.75 | −7.2 |
| Trichothecium | roseum | Trichothecin | 12444502 | 332.4 | 4.62 | −7.2 |
| phycomyces blake skeanus | Phycomysterol A | 11794191 | 380.6 | 3.54 | −7.1 | |
| Suillus granulatus | Flazin | 5377686 | 308.29 | 4.62 | −7.1 | |
| Arthrinium | Terpestacin | 6443294 | 402.6 | 6.53 | −7 | |
Semicochliodinol A and B: Produced by the Fungus Chrysosporium merdarium. They were shown Inhibition of HIV-1 Protease and EGF-R Protein Tyrosine Kinase Related to Asterriquinones and specifically Semicochliodinol B also shown therapeutic contender for Marburg virus's VP35 and VP40 proteins (Hasan et al., 2022). IsoChromophilone I was produced from Penicillium, which was found to inhibit HIV Proliferation (Omura et al., 1993, Matsuzaki et al., 1995). L-696,475, L-696,474 & L-697,318 were isolated from Hypoxylon fragiforme fungal extracts. Which were shown inactive against the HIV – 1 protease (Lingham et al., 1992). Stachyflin and its derivatives acetyl Stachyflin, SQ-02-S-L2, SQ-02-S-V1 showed a potent anti-influenza virus activity which was extracted from Stachybotrys fungal species (Shaikh et al., 2022a, Shaikh et al., 2022b, Shaikh et al., 2022c, Qian-Cutrone et al., 1996). F13459 was shown to inhibit the effect of synthesis and trafficking of virus glycoprotein isolated from Penicillium species (Minagawa et al., 2002). Flazin isolated from the fruiting bodies of Suillus granulatus was found to have weak anti-HIV activity (Tang et al., 2008). Fleephilone was shown to inhibit HIV by binding to REV/RRE, produced by Trichoderma harzianum. It was discovered that A-108836 was extracted from Fusarium compactum species, which inhibited Rhinovirus 3C protease (Koshino et al., 2001), and Integracide (A-108835) was isolated from the fermentation broth of a Fusarium species. It was found that the sulfated ester Integracide A significantly inhibited strand transfer reactions of HIV-1 integrase.
2.3. Docking analysis
To perform molecular docking lead molecules energies were minimized using PyRx software. After that docking was done with RDRP (7AAP) structured protein as a macromolecule. The configuration file for the grid parameters was generated using DSV from that site of favipiravir which was already present in the structure, then is x, y, and z coordinates were noted down, and In PyRx grid box was set according to obtained values for a specific active site for docking to be carried out.
2.4. ADMET prediction
Swiss ADME and pKCSM were used to determine the drug-like nature of the molecules (Brill et al., 1996). An effective drug must interact with pharmacokinetics, toxicity (Pires et al., 2015, Chandramohan et al., 2015) and potency. Drug pharmacokinetic profiles provide detailed information about absorption, distribution, metabolism, and excretion (ADME) characteristics. A toxicity study was carried out using the webserver pkCSM to predict the drug-likeness of metabolites (Pires et al., 2015).
2.5. MD simulation
MD Simulation helps identify changes and fluctuations taking place in Protein. The interaction stability confirmations are done by analyzing simulation trajectories and calculating the root mean square deviation, root means square fluctuations, interactions between residues of the Protein and ligands (Priya et al., 2019, Wang and Zhu, 2016), changes occurring within the protein backbone, and changes occurring within ligand atoms, for a time period of 50 ns (Hollingsworth and Dror, 2018).
3. Results and discussion
3.1. Drug likeness, and oral bioavailability based on Swiss ADME predictions
A total of 18 lead molecules (fungal metabolites) were taken for docking analysis, which had a binding affinity above −7. The docked studies have shown promising results against RdRp of SARS CoV-2.
Compared to remdesivir which is indicated for Covid users (Chandramohan et al., 2015). Remdesivir was stated to have slowed the progression of severe respiratory disease and had a faster recovery time (Shanmuga Priya et al., 2022). Other medications have exhibited binding energy against docking of Protein RDRP as compared to ligands of oseltamivir, ritonavir, remdesivir, ribavirin, favipiravir, chloroquine, and hydroxychloroquine, they calculated −4.7, −7.3, −6.5, −5.6, −5.4, −5.1, and hydroxychloroquine was found to be – Lipinski's rule of five is a broad rule that describes a molecule's drug ability. This rule can help you figure out if a biologically active molecule has the chemical and physical qualities to be taken orally (Hosseini et al., 2021a, Hosseini et al., 2021b, Beigel et al., 2020) as shown in Table 2 .
Table 2.
Properties of fungal Metabolites related to Drug ability Violations.
| SL.no | Metabolites | Lipinski violations | Ghose violations | Veber violations | Egan violations | Muegge violations | Bioavailability Score | No. of H-bond donor (5) |
No. of H-bond acceptor (<10) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Semicochliodinol A | 0 | 0 | 0 | 0 | 1 | 0.56 | 4 | 4 |
| 2 | Semicochliodinol B | 0 | 0 | 0 | 0 | 1 | 0.56 | 0 | 5 |
| 3 | SQ-02-S-L2 | 1 | 3 | 0 | 1 | 0 | 0.55 | 1 | 3 |
| 4 | Chromophilone I, iso | 0 | 0 | 0 | 0 | 0 | 0.55 | 4 | 4 |
| 5 | L-696,474 | 0 | 2 | 0 | 0 | 0 | 0.55 | 3 | 4 |
| 6 | F13459 | 2 | 2 | 1 | 1 | 2 | 0.17 | 2 | 4 |
| 7 | SQ-02-S-V1 | 1 | 3 | 0 | 0 | 1 | 0.56 | 2 | 8 |
| 8 | Stachyflin | 0 | 0 | 0 | 0 | 0 | 0.55 | 2 | 4 |
| 9 | L-696,475 | 1 | 2 | 0 | 0 | 1 | 0.55 | 3 | 8 |
| 10 | L-697,318 | 0 | 2 | 0 | 0 | 0 | 0.55 | 4 | 11 |
| 11 | Stachyflin, acetyl | 0 | 0 | 0 | 0 | 0 | 0.55 | 2 | 7 |
| 12 | A-108836 | 2 | 4 | 0 | 1 | 1 | 0.56 | 3 | 5 |
| 13 | Integracide A (A-108835) | 1 | 4 | 0 | 2 | 1 | 0.56 | 2 | 5 |
| 14 | Fleephilone | 0 | 0 | 0 | 0 | 0 | 0.55 | 1 | 4 |
| 15 | Trichothecin | 0 | 0 | 0 | 0 | 0 | 0.55 | 1 | 4 |
| 16 | Phycomysterol A | 1 | 1 | 0 | 1 | 2 | 0.55 | 3 | 4 |
| 17 | Flazin | 0 | 0 | 0 | 0 | 0 | 0.56 | 1 | 4 |
| 18 | Terpestacin | 0 | 0 | 0 | 0 | 0 | 0.55 | 2 | 6 |
The cost of a drug's failure goes up the longer it stays in the development pipeline. So, if scientists want it to fail in the end, they should let it fail early, preferably before in vivo tests. In vitro ADMET assay, many of which are automated or have a high throughput, are used more and more in the early stages of drug discovery to predict how a drug will likely act when it is given to a person. This helps get rid of drugs with bad properties while letting promising leads move forward.
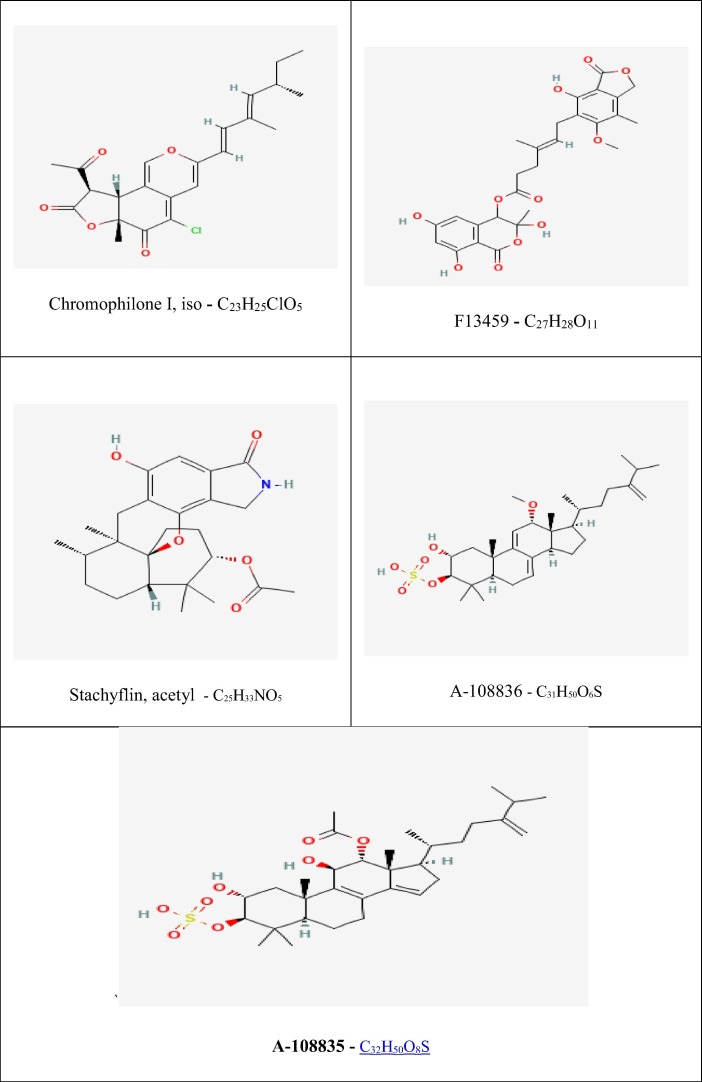
The predicted toxicity profile of the 18 fungal metabolites, with their prophecy probability, is shown in Table 3 . Understanding the above results regarding toxicity, out of 18 metabolites, 5 namely Chromophilone I, iso; F13459; Stachyflin, acetyl; A-108836; Integracide A (A-108835) have shown no AMES toxicity properties of the molecules and indicates that they are non-mutagenic. They were found to be weak or non-inhibitors of the hERGI & II potassium ion channel, which is primarily studied for its role in cardiac electrical activity that coordinates the heart's beating. Also, the metabolites showed acceptable Oral Rat Acute Toxicity (LD50), which indicates that metabolites will not be harmful at lower concentrations (mol/kg). Hepatotoxicity indicates that the drug molecules are likely to disrupt the liver's normal function. They tested five metabolites that did not exhibit hepatotoxicity and skin allergy. Hence based on analysis of all the properties, these 5 metabolites shown in Fig. 2 , assure safety and are said to be non-toxic and non-carcinogenic.
Table 3.
Toxicity profile of selected metabolites.
| Sl. No | Metabolites |
Toxicity predictors |
||||||
|---|---|---|---|---|---|---|---|---|
| AMES toxicity |
hERG I inhibitor |
hERG II inhibitor |
Oral Rat Acute Toxicity (LD50) |
Oral Rat Chronic Toxicity (LOAEL) |
Hepatotoxicity |
Skin Sensitisation |
||
| Unit | Categorical (Yes/No) | Categorical (Yes/No) | Categorical (Yes/No) | Numeric (mol/kg) | Numeric (log mg/kg) | Categorical (Yes/No) | Categorical (Yes/No) | |
| 1 | Semicochliodinol A | NO | NO | YES | 2.58 | 2.832 | YES | NO |
| 2 | Semicochliodinol B | NO | NO | YES | 2.644 | 2.74 | YES | NO |
| 3 | SQ-02-S-L2 | NO | NO | NO | 2.38 | 2.238 | YES | NO |
| 4 | Chromophilone I, iso | NO | NO | NO | 2.197 | 0.711 | NO | NO |
| 5 | L-696,474 | NO | NO | NO | 2.972 | 0.631 | YES | NO |
| 6 | F13459 | NO | NO | NO | 2.436 | 3.145 | NO | NO |
| 7 | SQ-02-S-V1 | NO | NO | NO | 2.945 | 2.447 | YES | NO |
| 8 | Stachyflin | NO | NO | NO | 2.219 | 0.063 | YES | NO |
| 9 | L-696,475 | |||||||
| 10 | L-697,318 | NO | NO | NO | 3.07 | 0.633 | YES | NO |
| 11 | Stachyflin, acetyl | NO | NO | NO | 2.129 | 1.782 | NO | NO |
| 12 | A-108836 | NO | NO | NO | 2.323 | 2.532 | NO | NO |
| 13 | Integracide A (A-108835) | NO | NO | NO | 2.409 | 2.423 | NO | NO |
| 14 | Fleephilone | YES | NO | NO | 2.53 | 1.777 | NO | NO |
| 15 | Trichothecin | NO | NO | NO | 2.487 | 1.519 | NO | NO |
| 16 | Phycomysterol A | NO | NO | YES | 2.332 | 0.845 | NO | NO |
| 17 | Flazin | NO | NO | NO | 2.696 | 1.321 | YES | NO |
| 18 | Terpestacin | NO | NO | NO | 2.437 | 1.785 | NO | NO |
Fig. 2.
Structures of selected 5 fungal metabolites obtained from the PubChem database in 3D SDF format.
3.2. Molecular interaction profile
Each ligand successfully docked with the target proteins, indicating significant binding. The protein-metabolite interaction was visualized in Discovery Studio Visualizer and depicted below.
-
i.
Chromophilone I, iso
- ii.
-
iii.
Stachyflin, acetyl
-
iv.
A-108836
-
v.
Integracide A (A-108835)
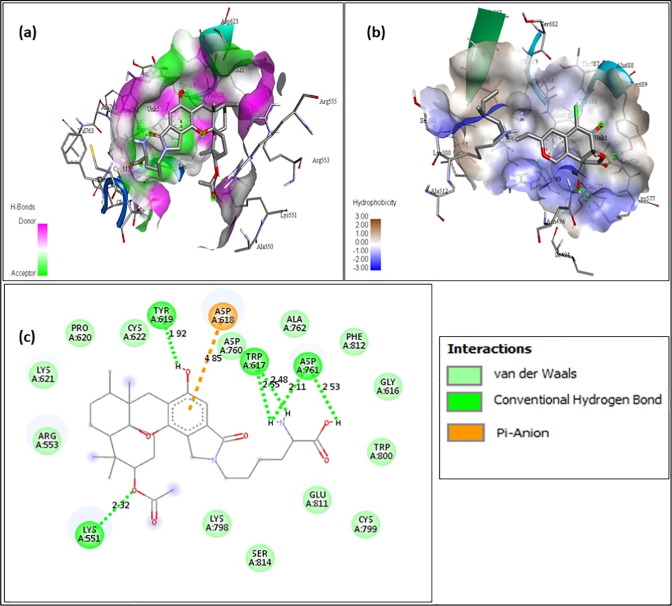
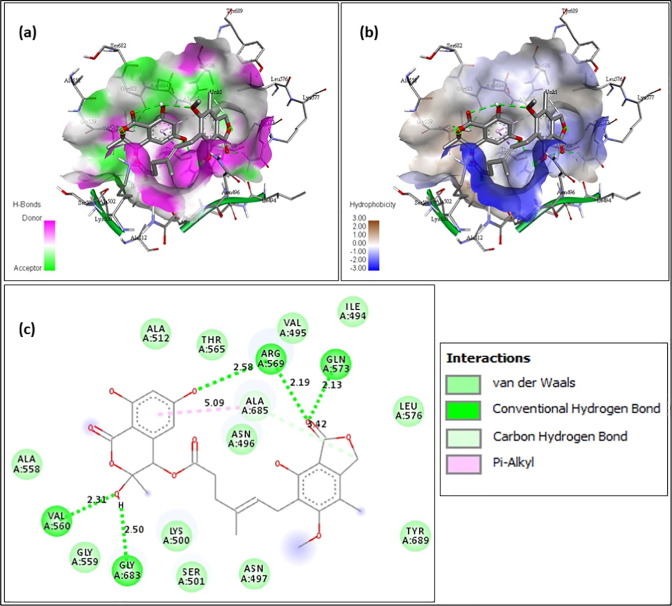
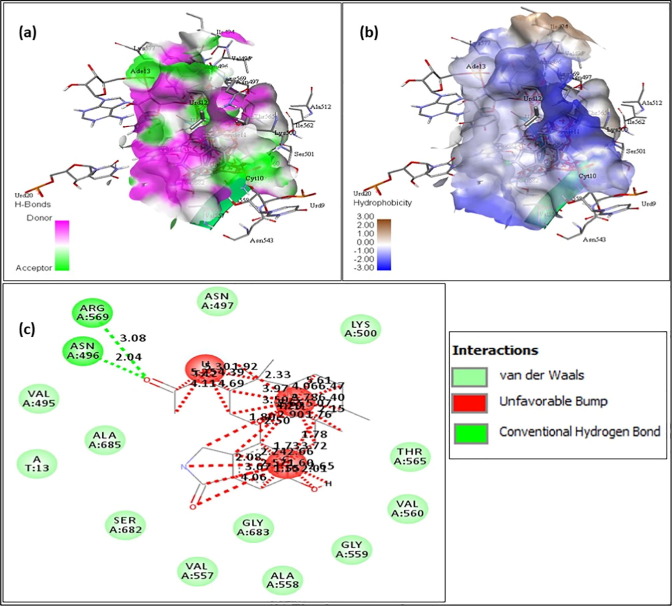
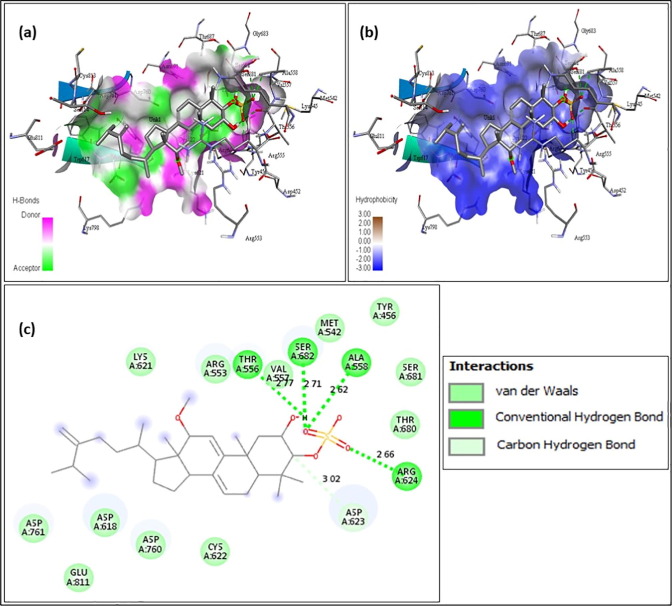
The above figures contain the interaction plots of (a) Hydrogen bond, (b) Hydrophobicity and (c) 2D interaction. In Fig. 3 , the compound Chromophilone I, iso lead molecule shows a binding affinity of −7.8 and the amino acids interacting with the protein RdRp are Tyr619 interacting at a distance of 1.92 Å, Trp617 at 2.55 Å and 2.48 Å, Asp761 at 2.11 and 2.53 Å, Lys551 at 2.32 Å with conventional hydrogen bond and the amino acid Asp618 was interacting at 4.85 Å distance with pi-Anion bond. In Fig. 4 F13459 which shows an interaction score of −7.7 and amino acids interacting were, Gln573 had a distance of 2.13 Å, Arg569 at 2.19 Å and 2.58 Å, Gly683 at 2.50 Å and Val560 at 2.31 Å by making interaction with hydrogen bond and at 5.09 Å, Ala685 had formed a pi-alkyl bond. From Fig. 5 Stachyflin, acetyl has shown a binding affinity of −7.5 and amino acids Asn496 was interacting with conventional hydrogen bonds and Arg569 was interacting with Vander Waals bonds at a distance of 2.04 Å and 3.08 Å respectively. In Fig. 6 the lead molecule A-108836 showed binding energy of −7.4,the amino acids were Ala558 at a distance of 2.62 Å, Arg 624 at 2.66 Å, Ser682 at 2.71 Å and Thr556 at 2.77 Å interacting with conventional hydrogen bond and Asp623 at 3.02 Å interacting with carbon hydrogen bond.
Fig. 3.
Chromophilone I, iso - (a) Hydrogen bond interaction; (b) Hydrophobicity interaction; and (c) 2D interaction.
Fig. 4.
F13459- (a) Hydrogen bond interaction; (b) Hydrophobicity interaction; and (c) 2D interaction.
Fig. 5.
Stachyflin, acetyl- (a) Hydrogen bond interaction; (b) Hydrophobicity interaction; and (c) 2D interaction.
Fig. 6.
A-108836- (a) Hydrogen bond interaction; (b) Hydrophobicity interaction; and (c) 2D interaction.
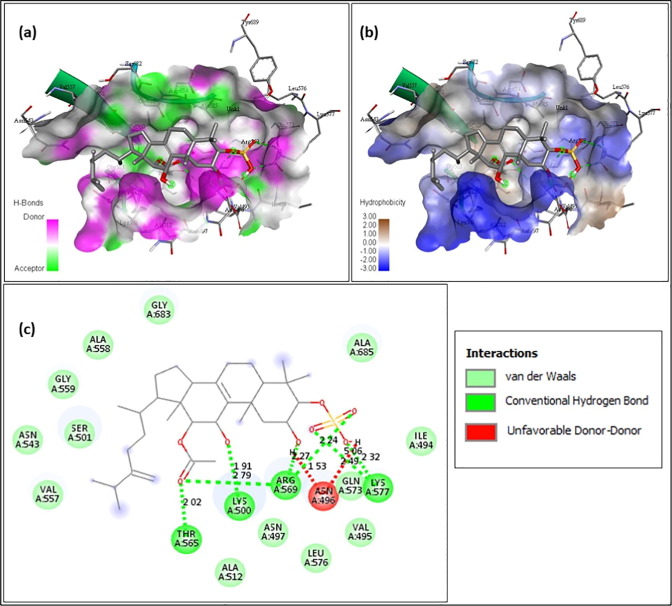
In Fig. 7 the molecule Integracide A (A-108835) showed binding energy of −7.3 and the amino acids interacting were Lys500 at 1.91 Å, Thr565 at 2.02 Å, Arg569 at 2.27 Å, at 2.24 Å and at 2.79 Å, Lys577 interacting at a distance of 2.32 Å and at 2.49 Å were shown to interact with conventional hydrogen bond and Asn496 interacting at 1.53 Å and 5.06 Å by making unfavorable donor bond.
Fig. 7.
Integracide A (A-108835)- (a) Hydrogen bond interaction; (b) Hydrophobicity interaction; and (c) 2D interaction.
3.3. Molecular dynamics of computer simulation with Desmond
The binding affinity and structure of the ligand–protein complex (Wang and Zhu, 2016) can be predicted via molecular docking (Wong and Lightstone, 2011; “Novel Antibiotics Database. Available at: https://www.antibiotics.or.jp/english/data-base/data-base-top/ (Accessed: January 30, (2022)). To assess the stability of the protein–ligand complex, MD simulates the dynamic behavior of the molecular system because docking is merely a static view of the molecules arranged in the active site of the protein (Choudhary et al., 2020). Simulations of molecular dynamics estimate how each atom in a protein or other molecular system will move over time (Hollingsworth and Dror, 2018, Muddapur et al., 2022, Muhsinah et al., 2022) which was accessed using Schrodinger Desmond (Alam, 2021, Duong et al., 2020) simulation. In silico molecular docking is used to identify important structural features for binding as well as for in silico screening efforts to find suitable binding partners (Al-Wasidi et al., 2020a, Al-Wasidi et al., 2020b, Iqbal et al., 2021, Shaikh et al., 2022a, Shaikh et al., 2022b, Shaikh et al., 2022c). Molecular docking is extensively utilized in the computer-assisted drug design process. It can be used to: (1) anticipate the binding mode of previously recognized ligands; (2) identify novel and potent ligands; and (3) predict binding affinity at various stages of the drug development process (Zhou et al., 2020). The data thus obtained will guide researchers to find new leads for developing novel potential strategies for the management of malignancies, diabetes and other chronic metabolic disorders (Hosseini et al., 2021a, Hosseini et al., 2021b, Kumar et al., 2021, Liu et al., 2020, Al-Wasidi et al., 2020a, Al-Wasidi et al., 2020b).
For simulation studies, the ligand Chromophilone I, iso was selected and performed simulation study since it shows negative for toxicity studies.
A simulation was performed to access the structural conformation of protein RDRP and Ligand Chromophilone I, iso. Simulations were subjected for 50 ns, during the complete simulation time the protein–ligand tends to be stable and are in contact with each other and it was observed that there is no considerable fluctuation found during the simulation run time since the stability of a protein is relative to its conformation, as depicted in Fig. S1 (supplementary file), which infers the RMSD evolution of a protein (left Y-axis) and ligand (right Y-axis).
In addition, the RMSF of the protein was also obtained with fluctuations, i.e. in the range of 1–5. These changes suggest that the ligand has a significant impact on silencing the receptor and, hence decreasing its function, as shown in Fig. S2 (supplementary file).
The timeline of all the hydrogen bonds, hydrophobic contacts, ionic contacts, and water bridges formed during PL interactions and the amino acids involved in the interaction has been depicted in Fig. S3 (Supplementary file). Two types of interaction graphs are displayed here: the top panel displays the total number of individual connections the protein makes with the ligand during the length of the trajectory simulation, and the bottom panel depicts a good contact.
In each trajectory frame, the second section of the bottom panel illustrates how 24 residues interact with the ligand. According to the scale to the right of the plot, some residues have multiple specific contacts with the ligand, which is shown by darker orange shading, namely Arg349, Phe396 and Val675 interacting constantly when compared to other amino acids.
PL interactions (or 'contacts') are usually categorized into four types: Hydrogen Bonds, Hydrophobic, Ionic and Water Bridges which is shown in histogram in Fig. S4 (Supplementary file). The stacked bar of three amino acids Arg349, Phe396 and Val675 have greater than or equal to 60% of simulation time the specific interaction is maintained with water bridge formation. While other amino acids interacts by making hydrophobic bonds with 0–28% of simulation contacts.
The ligand RMSF displays the ligand's fluctuations by atom, based on the 2D structure's top panel as depicted in Fig. S5 (Supplementary file). It could reveal how ligand fragments interact with proteins and serve an entropic function in the binding process. The fit ligand to protein line in the bottom panel shows fluctuations of the ligand concerning the protein at a rate of 0.6 to 1.5. The PL complex is thus aligned on the protein backbone first, and then the ligand RMSF on the ligand heavy atoms is calculated. Overall, the docked complex and the dynamic interactions involved appear to be very stable.
4. Conclusions
In the search for a cure for COVID-19, the exploration of natural resources is necessary. Antiviral effects of fungal metabolites were investigated in this work, which focused on the protein RdRp, a vital enzyme for the life cycle of RNA viruses. Five metabolites namely Semicochliodinol A; Semicochliodinol B; SQ-02-S-L2; Chromophilone I, iso; L-696,474 were examined using docking, Lipinski's, and ADMET analysis to determine binding affinity and toxicity. Chromophilone I, iso – C23H25ClO5 was subjected to further study, such as a 50 ns MD simulation of the protein to determine the alterations taking place. The docking data for this molecule revealed that it has the optimum binding affinity. Research on ADMET showed that the ligand could be used as an oral drug. MD simulation showed stable ligand–protein interaction. Lastly, this ligand can be used to make treatment options that fight SARS-CoV-2. Fungal metabolites that have been tested may be an excellent source to make future lead candidates. Further research into this chemical could lead to the development of an antiviral medication to combat the SARS-CoV-2 virus.
Funding
The authors are grateful to the Deanship of Scientific Research, Najran University, Najran, Saudi Arabia, for funding this research through grant research code NU/RC/MRC/11/1; this work was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R199), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia; was supported by AlMaarefa University researchers supporting program (grant number: MA-006), AlMaarefa University, Riyadh, Saudi Arabia;
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to Najran University, Najran, KSA; and KLE Technological University, BVB Campus, Hubballi-580031, Karnataka, India; and the authors deeply acknowledge the Researchers Supporting Program, Princess Nourah bint Abdulrahman University Researchers Supporting Project, Riyadh, Saudi Arabia; and AlMaarefa University, Riyadh, Saudi Arabia for supporting steps of this work.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102147.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abdalla M., Mohapatra R.K., Sarangi A.K., Mohapatra P.K., Eltayb W.A., Alam M., El-Arabey A.A., Azam M., Al-Resayes S.I., Seidel V., Dhama K. In silico studies on phytochemicals to combat the emerging COVID-19 infection. J. Saudi Chem. Soc. 2021;25(12) doi: 10.1016/j.jscs.2021.101367. [DOI] [Google Scholar]
- Alam M. Multispectroscopic and molecular modeling strategy to explore the interaction of cholest-5-en-7-one with human serum albumin: DFT and Hirshfeld surface analysis. J. King Saud Univ. Sci. 2021;33(8) doi: 10.1016/j.jksus.2021.101661. [DOI] [Google Scholar]
- Al-Wasidi, A. S., Al-Jafshar, N.M., Al-Anazi, A.M., Refat, M.S., El-Metwaly, N.M., Ibrahim, H.K., El-Fattah, W.A., Naglah, A.M., Al-Omar, M.A., Kalmouch, A., 2020. In methanolic solvent synthesis of New MnII, CoII, NiII and CuII schiff base of aromatic _ amino acids: Spectroscopic, thermal, molecular docking and antimicrobial studies. Sci. Adv. Mater 12.
- Al-Wasidi A.S., Naglah A.M., Al-Omar M.A., Al-Obaid A.-R.-M., Alosaimi E.H., El-Metwaly N.M., Refat M.S., Ahmed A.S., El-Deen I.M., Soliman A.H., Emam A. Manganese (II), ferric (III), cobalt (II) and copper (II) thiosemicarbazone Schiff base complexes: Synthesis, spectroscopic, molecular docking and biological discussions. Mater. Express. 2020;10:290–300. doi: 10.1166/mex.2020.1636. [DOI] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.-D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boufissiou A., Abdalla M., Sharaf M., Al-Resayes S.I., Imededdine K., Alam M., Yagi S., Azam M., Yousfi M. In-silico investigation of phenolic compounds from leaves of Phillyrea angustifolia L. as a potential inhibitor against the SARS-CoV-2 main protease (Mpro PDB ID:5R83) using a virtual screening method. J. Saudi Chem. Soc. 2022;26(3) doi: 10.1016/j.jscs.2022.101473. [DOI] [Google Scholar]
- Brill G.M., Kati W.M., Montgomery D., Karwowski J.P., Humphrey P.E., Jackson M., Clement J.J., Kadam S., Chen R.H., Mcalpine J.B. Novel triterpene sulfates from Fusarium compactum using a Rhinovirus 3C protease inhibitor screen. J. Antibiot. (Tokyo) 1996;49:541–546. doi: 10.7164/antibiotics.49.541. [DOI] [PubMed] [Google Scholar]
- Campagnola G., Gong P., Peersen O.B. High-throughput screening identification of poliovirus RNA-dependent RNA polymerase inhibitors. Antiviral Res. 2011;91:241–251. doi: 10.1016/j.antiviral.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramohan V., Nagaraju N., Rathod S., Kaphle A., Muddapur U. Identification of deleterious SNPs and their effects on structural level in CHRNA3 gene. Biochem. Genet. 2015;53:159–168. doi: 10.1007/s10528-015-9676-y. [DOI] [PubMed] [Google Scholar]
- Choudhary M.I., Shaikh M., tul-Wahab A., ur-Rahman A., Salahub D. In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation. PLoS One. 2020;15(7) doi: 10.1371/journal.pone.0235030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T.-H., Paramita Devi A., Tran N.-M.-A., Phan H.-V.-T., Huynh N.-V., Sichaem J., Tran H.-D., Alam M., Nguyen T.-P., Nguyen H.-H., Chavasiri W., Nguyen T.-C. Synthesis, α-glucosidase inhibition, and molecular docking studies of novel N-substituted hydrazide derivatives of atranorin as antidiabetic agents. Bioorg. Med. Chem. Lett. 2020;30(17) doi: 10.1016/j.bmcl.2020.127359. [DOI] [PubMed] [Google Scholar]
- Eskier D., Karakülah G., Suner A., Oktay Y. RdRp mutations are associated with SARS-CoV-2 genome evolution. PeerJ. 2020;8:e9587. doi: 10.7717/peerj.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach P., Malet H., Cusack S., Reguera J. Structural insights into Bunyavirus replication and its regulation by the vRNA promoter. Cell. 2015;161:1267–1279. doi: 10.1016/j.cell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy A.S., Lima G.M.A., Oliveira K.I.Z., Torres N.U., Maluf F.V., Guido R.V.C., Oliva G. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 2017;8 doi: 10.1038/ncomms14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M., Mia M.M., Islam M.M., Hasan Saraf M.S., Islam M.S. A computerized pharmaceutical repurposing approach reveals Semicochliodinol B synthesized from Chrysosporium merdarium as a viable therapeutic contender for Marburg virus's VP35 and VP40 proteins. Inform. Med. Unlocked. 2022;28 doi: 10.1016/j.imu.2021.100821. [DOI] [Google Scholar]
- Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini M., Chen W., Xiao D., Wang C. Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs. Precis. Clin. Med. 2021;4:1–16. doi: 10.1093/pcmedi/pbab001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini, M., Chen, W., Xiao, D., Wang, C., 2021. Computational molecular docking and virtual screening revealed promising SARS-CoV-2drugs. Precis. Clin. Med. 4. [DOI] [PMC free article] [PubMed]
- Iqbal N., Iqubal S.M.S., Khan A.A., Mohammed T., Alshabi A.M., Aazam E.S., Rafiquee M.Z.A. Effect of CTABr (surfactant) on the kinetics of formation of silver nanoparticles by Amla extract. J. Mol. Liq. 2021;329 doi: 10.1016/j.molliq.2021.115537. [DOI] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshino H., Muroi M., Tajika T., Kimura Y., Takatsuki A. F13459, a new derivative of mycophenolic acid. II. Physico-chemical properties and structural elucidation. J. Antibiot. (Tokyo) 2001;54:494–500. doi: 10.7164/antibiotics.54.494. [DOI] [PubMed] [Google Scholar]
- Kumar, S.R., Alshabi, A.M., Shaikh, I.A., Almehizia, A.A., Kulkarni, V.H., Joshi, S.D., 2021. Synthesis, in silico molecular docking and antimicrobial study of some New 3-(Substituted-quinolin-3-yl)- 1-[4-(1H-pyrrol-1-yl) phenyl] prop-2-en-1-one derivatives. Indian J. Heterocycl. Chem 31.
- Lingham R.B., Hsu A., Silverman K.C., Bills G.F., Dombrowski A., Goldman M.E., Darke P.L., Huang L., Koch G., Ondeyka J.G., Goetz M.A. L-696,474, a novel cytochalasin as an inhibitor of HIV-1 protease. III. Biological activity. J. Antibiot. (Tokyo) 1992;45:686–691. doi: 10.7164/antibiotics.45.686. [DOI] [PubMed] [Google Scholar]
- Littler, D.R., Gully, B.S., Colson, R.N., Rossjohn, J., 2020. Crystal structure of the SARS-CoV-2 nonstructural protein 9, Nsp9. iScience 23, 101258. https://doi.org/10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed]
- Liu L., Wang Q., Chen Z., Duan B., Jin L., Zhang D. A synthetic peptide AWRK6 combined with epigallocatechin gallate alleviates type 2 diabetes in mice. Sci. Adv. Mater. 2020;12:740–745. doi: 10.1166/sam.2020.3719. [DOI] [Google Scholar]
- Matsuzaki K., Ikeda H., Masuma R., Tanaka H., Omura S. Isochromophilones I and II, novel inhibitors against gpl20-CD4 binding produced by Penicillium multicolor FO-2338. I. screening, taxonomy, fermentation, isolation and biological activity. J. Antibiot. (Tokyo) 1995;48:703–707. doi: 10.7164/antibiotics.48.703. [DOI] [PubMed] [Google Scholar]
- Minagawa K., Kouzuki S., Kamigauchi T. Stachyflin and acetylstachyflin, novel anti-influenza A virus substances, produced by Stachybotrys sp. RF-7260. II. Synthesis and preliminary structure-activity relationships of stachyflin derivatives. J. Antibiot. (Tokyo) 2002;55:165–171. doi: 10.7164/antibiotics.55.165. [DOI] [PubMed] [Google Scholar]
- Mohapatra R.K., Dhama K., El–Arabey A.A., Sarangi A.K., Tiwari R., Emran T.B., Azam M., Al-Resayes S.I., Raval M.K., Seidel V., Abdalla M. Repurposing benzimidazole and benzothiazole derivatives as potential inhibitors of SARS-CoV-2: DFT, QSAR, molecular docking, molecular dynamics simulation, and in-silico pharmacokinetic and toxicity studies. J. King Saud Univ. Sci. 2021;33(8) doi: 10.1016/j.jksus.2021.101637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra, R.K., Perekhoda, L., Azam, M., Suleiman, M., Sarangi, A.K., Semenets, A., Pintilie, L., Al-Resayes, S.I., 2021b. Computational investigations of three main drugs and their comparison with synthesized compounds as potent inhibitors of SARS-CoV-2 main protease (Mpro): DFT, QSAR, molecular docking, and in silico toxicity analysis. J. King Saud Univ. Sci. 33, 101315. https://doi.org/10.1016/j.jksus.2020.101315 [DOI] [PMC free article] [PubMed]
- Mohapatra R.K., Sarangi A.K., Kandi V., Azam M., Tiwari R., Dhama K. Omicron (B.1.1.529 variant of SARS-CoV-2); an emerging threat: Current global scenario. J. Med. Virol. 2022;94:1780–1783. doi: 10.1002/jmv.27561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddapur U.M., Alshehri S., Ghoneim M.M., Mahnashi M.H., Alshahrani M.A., Khan A.A., Iqubal S.M.S., Bahafi A., More S.S., Shaikh I.A., Mannasaheb B.A., Othman N., Maqbul M.S., Ahmad M.Z. Plant-based synthesis of gold nanoparticles and theranostic applications: A review. Molecules. 2022;27(4):1391. doi: 10.3390/molecules27041391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhsinah A.B., Maqbul M.S., Mahnashi M.H., Jalal M.M., Altayar M.A., Saeedi N.H., Alshehri O.M., Shaikh I.A., Khan A.A.L., Shakeel Iqubal S.M., Khan K.A., Dawoud A., Mannasaheb B.A., Azzouz S., Mohammed T. Antibacterial activity of Illicium verum essential oil against MRSA clinical isolates and determination of its phyto-chemical components. J. King Saud Univ. Sci. 2022;34(2) doi: 10.1016/j.jksus.2021.101800. [DOI] [Google Scholar]
- Omura S., Tanaka H., Matsuzaki K., Ikeda H., Masuma R. Isochromophilones I and II, novel inhibitors against gp120-CD4 binding from Penicillium sp. J. Antibiot. (Tokyo) 1993;46:1908–1911. doi: 10.7164/antibiotics.46.1908. [DOI] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18 doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon, M.C., Frazier, M.N., Dillard, L.B., Williams, J.G., Kocaman, S., Krahn, J.M., Perera, L., 2020. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15, BioRxiv Prepr. BioRxiv Prepr. Serv. Biol. [DOI] [PMC free article] [PubMed]
- Pires D.E.V., Blundell T.L., Ascher D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graphbased signatures. J. Med. Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya V.G., Swaminathan P., Muddapur U.M., Fandilolu P.M., Parulekar R.S., Sonawane K.D. Peptide similarity search based and virtual screening based strategies to identify small molecules to inhibit CarD-RNAP interaction in M. tuberculosis. Int. J. Peptide Res. Therap. 2019;25(2):697–709. [Google Scholar]
- Qian-Cutrone J., Huang S., Chang L.P., Pirnik D.M., Klohr S.E., Dalterio R.A., Hugill R., Lowe S., Alam M., Kadow K.F. Harziphilone and fleephilone, two new HIV REV/RRE binding inhibitors produced by Trichoderma harzianum. J. Antibiot. (Tokyo) 1996;49:990–997. doi: 10.7164/antibiotics.49.990. [DOI] [PubMed] [Google Scholar]
- Rosas-Lemus, M., Minasov, G., Shuvalova, L., Inniss, N.L., Kiryukhina, O., Wiersum, G., Kim, Y., Jedrzejczak, R., Maltseva, N.I., Endres, M., Jaroszewski, L., Godzik, A., Joachimiak, A., Satchell, K.J.F., 2020. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. bioRxivorg. https://doi.org/10.1101/2020.04.17.047498
- Shaikh I.A., Muddapur U.M., C K., Badiger S., Kulkarni M., Mahnashi M.H., Alshamrani S.A., Huneif M.A., More S.S., Khan A.A., Iqubal S.M.S. In silico molecular docking and simulation studies of protein HBx involved in the pathogenesis of hepatitis B virus-HBV. Molecules. 2022;27(5):1513. doi: 10.3390/molecules27051513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh I.A., Muddapur U.M., Kulkarni M., Badiger S., Krithika C., Mahnashi M.H., Mannasaheb B.A., Alsaikhan F., Bahafi A., Iqubal S.M.S., Khan A.A., Shaikh M.A.K., Maqbul M.S. in-Silico analysis of VP4 protein causing pathogenesis in Rotavirus and its interaction studies. Sci. Adv. Mater. 2022;14:67–79. doi: 10.1166/sam.2022.4193. [DOI] [Google Scholar]
- Shaikh, I.A., Muddapur, U.M., Srikanth, B., Madhura Ashwin Kulkarni, C., Mater, H., 2022c. Basheerahmed Abdulaziz Mannasaheb. Silico approach in targeting SARS-CoV-2 virus protein mpro using fungal metabolites 14, 43–54. https://doi.org/10.1166/sam.2022.4185
- Shanmuga Priya V.G., Bhandare V., Muddapur U.M., Swaminathan P., Fandilolu P.M., Sonawane K.D. Molecular modeling approach to identify inhibitors of Rv2004c (rough morphology and virulent strain gene), a DosR (dormancy survival regulator) regulon protein from Mycobacterium tuberculosis. J. Biomol. Struct. Dyn. 2022;40:3242–3257. doi: 10.1080/07391102.2020.1846620. [DOI] [PubMed] [Google Scholar]
- Shrikanth U.M., Shrinivas L., Vasanad S., Mathapati V., Biradar P., Swaminathan V.G. Exploring phytochemicals of Ficus religiosa against coronavirus protein using molecular modeling techniques. Res. Rev. 2021;8:18–26. [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111 doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.-G., Wang Y.-H., Wang R.-R., Dong Z.-J., Yang L.-M., Zheng Y.-T., Liu J.-K. Synthesis of analogues of flazin, in particular, flazinamide, as promising anti-HIV agents. Chem. Biodivers. 2008;5:447–460. doi: 10.1002/cbdv.200890044. [DOI] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Stürzel C.M., Fröhlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhu W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Fut. Med. Chem. 2016;8:1707–1710. doi: 10.4155/fmc-2016-0143. [DOI] [PubMed] [Google Scholar]
- Wong S.E., Lightstone F.C. Accounting for water molecules in drug design. Expert Opin. Drug Discov. 2011;6:65–74. doi: 10.1517/17460441.2011.534452. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B., Shen Q., Wu S., Chen S., Lomakin I.B., Xiong Y. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell. 2020;80:1055–1066.e6. doi: 10.1016/j.molcel.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu K., Sun Z., Wang T. Synthesis of novel biomaterial for the treatment of sepsis in rats. Sci. Adv. Mater. 2020;12:1175–1183. doi: 10.1166/sam.2020.3783. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.