Abstract
Bacterial community composition, enzymatic activities, and carbon dynamics were examined during diatom blooms in four 200-liter laboratory seawater mesocosms. The objective was to determine whether the dramatic shifts in growth rates and ectoenzyme activities, which are commonly observed during the course of phytoplankton blooms and their subsequent demise, could result from shifts in bacterial community composition. Nutrient enrichment of metazoan-free seawater resulted in diatom blooms dominated by a Thalassiosira sp., which peaked 9 days after enrichment (≈24 μg of chlorophyll a liter−1). At this time bacterial abundance abruptly decreased from 2.8 × 106 to 0.75 × 106 ml−1, and an analysis of bacterial community composition, by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA gene fragments, revealed the disappearance of three dominant phylotypes. Increased viral and flagellate abundances suggested that both lysis and grazing could have played a role in the observed phylotype-specific mortality. Subsequently, new phylotypes appeared and bacterial production, abundance, and enzyme activities shifted from being predominantly associated with the <1.0-μm size fraction towards the >1.0-μm size fraction, indicating a pronounced microbial colonization of particles. Sequencing of DGGE bands suggested that the observed rapid and extensive colonization of particulate matter was mainly by specialized α-Proteobacteria- and Cytophagales-related phylotypes. These particle-associated bacteria had high growth rates as well as high cell-specific aminopeptidase, β-glucosidase, and lipase activities. Rate measurements as well as bacterial population dynamics were almost identical among the mesocosms indicating that the observed bacterial community dynamics were systematic and repeatable responses to the manipulated conditions.
During phytoplankton blooms in the ocean, primary productivity and its processing by the food web create a heterogeneous environment of particulate, colloidal, and dissolved organic matter in a continuum of size classes and concentrations (2, 4, 35, 76). Major changes in organic matter concentration and composition are expected to occur at different stages of the bloom. The variations in the organic matter regime are typically accompanied by pronounced changes in bacterial abundance, productivity, ectohydrolase activities, and colonization of particles (48, 68). In one recent mesocosm experiment (68), growth rates, colonization, and enzyme activities generally increased following the peak of a diatom bloom but different enzymes were found to peak at different times. In principle, these changes could have occurred without major shifts in the phylogenetic composition of the bacterial community. Shifts in activity and surface attachment would represent plasticity in the bacterial phenotypes, with enzyme expression and growth being regulated in response to the available organic substrates. Alternatively, the observed changes could have resulted from community succession, with bacteria with inherently different metabolic capabilities predominating at different stages of the bloom. Species successions could then affect as well as reflect bacteria-organic matter coupling and the biogeochemical fate of the bloom. For instance, a dominance of bacteria specialized in colonizing and hydrolyzing phytoplankton detritus would greatly influence the biogeochemical transformation of the detritus. At the same time, free-living species adapted for efficiently utilizing dissolved organic matter could prevent large-scale dissolved organic matter accumulation despite its high production rates typical in the blooms (35, 68).
Consistent with the latter interpretation, Martinez et al. (41) observed that different isolates of marine bacteria grown under the same conditions can vary dramatically in the absolute and relative levels of their various ectoenzymes. The enzyme profiles of 44 isolates indicated specialization among marine heterotrophic bacteria for different polymeric substrates and suggested that community composition is an important factor influencing bulk measurements of biochemical activity in seawater (41). Other studies have revealed phylogenetic differences among free-living and attached bacteria (1, 5, 10), which also suggest that community composition is likely to change under postbloom conditions when the fraction of attached bacteria can be high (43, 68). Some recent studies have demonstrated bacterial succession both seasonally in the field (44) and in response to organic enrichment in a continuous-flow system (74). However, studies that simultaneously measure changes in the bulk biochemistry and phylogenetic composition of the bacterial community are just beginning to be done (12).
In the present study we sought to determine whether the colonization and hydrolysis of decaying phytoplankton would be accompanied by significant changes in bacterial community composition. We found that the pronounced increases in enzymatic activities and bacterial growth during the course of a mesocosm diatom bloom were concomitant with an extensive colonization of the decaying diatoms and major changes in total bacterial community composition. These data suggest that colonization was accomplished by fast-growing, highly hydrolytic “particle specialist” bacteria. These were primarily related to Cytophagales and the “marine alpha group” of the class Proteobacteria.
MATERIALS AND METHODS
Experimental design.
Diatom blooms were generated in the absence of metazoan grazers in four laboratory mesocosms. Surface seawater (from 0.5-m depth) was collected off Scripps Pier (San Diego, Calif.) on 2 December 1997. Filtered surface water (<33-μm mesh; Nitex) was partitioned between four 200-liter cylindrical polyethylene tanks (Nalgene) and transferred to an 18 ± 1°C climate room within 30 min. All four mesocosms were amended with NaH2PO4 (1.0 μM) and Na2SiO3 (10.8 μM), and different nitrogen sources were added to pairs of tanks in an attempt to induce dominance by different phytoplankton species (NaNO3: tank 1, 6.9 μM; tank 2, 10.3 μM; NH4Cl: tanks 3 and 4, 10.1 μM). The tanks were exposed to artificial light at a surface irradiance of ≈200 μE m−2 s−1 (12 h of light and 12 h of darkness). After each sampling, the light sources were lowered to maintain them at a constant distance from the water surface. Mixing was provided by bubbling from the bottom center of each tank. Bubbling rates were estimated by capturing the bubbles in an inverted, water-filled graduated cylinder held with the opening just below the surface of the water and recording the rate of water displacement. Air flow in each tank was adjusted to ca. 800 ± 40 ml of air min−1. Just prior to sampling, the tanks were gently stirred with a polyvinyl chloride pipe to resuspend any particulates accumulating around the bottom edges of the tanks. Sampling was accomplished by siphoning 4 to 7 liters of water from the center of each tank with silicone tubing into polycarbonate carboys, starting ≈12 h after filling the tanks. Except for filtration for DNA, all variables were monitored every day in tanks 1 and 3. Aliquots for microbial counts were fixed immediately after sampling, and rate measurement was initiated thereafter. During incubations for the rate measurements (and within 2 h of sampling) samples for nutrients, DNA extraction, particulate organic carbon (POC), and chlorophyll a were processed and then immediately frozen for later analysis. Only total bacterial abundance, chlorophyll a levels, and bacterial community composition were monitored in tanks 2 and 4. After the experiment it was confirmed that no macrozooplankton were present in the tanks by filtering 10 liters from each tank onto a 33-μm Nitex mesh and examining the mesh microscopically.
DNA filtration and extraction.
Bacterial DNA was obtained every second day by filtering 2 to 3 liters of water through 0.22-μm-pore-size Sterivex capsule filters (diameter, 1.7 cm; length, 6.7 cm; Millipore) via a peristaltic pump (≈100 ml min−1). Filters were frozen at −80°C until extraction. DNA was extracted from the filters essentially by the method of Somerville et al. (69) with slight modifications. Lysis was accomplished within the Sterivex filter housing in 1.8 ml of SET buffer (20% sucrose, 50 mM EDTA, 50 mM Tris-HCl, pH 8.0) containing freshly made lysozyme (5 mg ml−1, final concentration) for 1 h at 37°C. Proteinase K (2 mg ml−1, final concentration) and sodium dodecyl sulfate (0.5%, final concentration) were added, and the mixture was incubated at 60°C for 2 h. Samples were heated just to the boiling point (10 to 20 s) in a microwave oven and then the lysate was drawn into a syringe. The filter was washed with 1 ml of TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0), which was then pooled with the lysate. After treatments with RNase (Sigma; 0.1 mg ml−1; 10 min at room temperature) and with 0.5 volume of 7.5 M ammonium acetate (15 min, room temperature), the lysates were briefly centrifuged (5 min, 14,500 × g, room temperature). The supernatants were precipitated with 2 volumes of ethanol, and the resulting pellets were resuspended in 400 μl of TE buffer. The solutions were extracted twice with 500 μl of phenol-chloroform-isoamyl alcohol (25:24:1) and once with 500 μl of chloroform-isoamyl alcohol (24:1) and were precipitated with ammonium acetate-ethanol (59). Precipitated DNA was resuspended in TE and quantified fluorometrically (PicoGreen; Molecular Probes).
PCR amplification.
A 16S rRNA gene fragment (169 to 194 bp in length) was amplified by PCR using a universal primer complementary to position 517 to 534 (5′-ATTACCGCGGCTGCTGG-3′) and a bacterial primer complementary to position 341 to 358 with a 40-bp GC clamp (underlined) (5′-CGCCCGCCGCG CGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGC AG-3′ [45]) following the protocol described in detail by Riemann et al. (57). In brief, after an initial denaturation for 5 min at 94°C, samples were amplified for 30 cycles by touchdown PCR (11). For each cycle, denaturation was at 94°C for 1 min. Annealing was for 1 min at a temperature which decreased 1°C every two cycles from an initial 65°C to a final of 50°C. Primer extension was at 72°C for 3 min in each cycle, with a final 7-min extension following the last cycle. A negative control, in which the template was replaced with an equal volume of sterile water, was included in each batch of PCRs.
DGGE and cloning.
PCR products were precipitated with ethanol, resuspended in TE buffer, and quantified (PicoGreen; Molecular Probes). Five hundred nanograms of PCR product was loaded on 8% polyacrylamide gels (acrylamide–N,N′-methylenebisacrylamide [37:1]) containing denaturant gradients of 30 to 46% from top to bottom (where 100% is defined as 7 M urea and 40% [vol/vol] formamide). Electrophoresis was performed with a hot-bath denaturing gradient gel electrophoresis (DGGE) unit (CBS Scientific, Del Mar, Calif.) using 0.5× TAE running buffer (20 mM Tris, 10 mM acetate, 0.5 mM Na2-EDTA, pH 8.2) at 60°C for 5.5 h at 200 V. Gels were stained for 30 min in SYBR Green I nucleic acid stain (Molecular Probes), destained for 10 min in 0.5× TAE, and photographed with UV transillumination.
DGGE bands were excised using a sterile razor blade, and DNA was eluted overnight at 37°C in 400 μl of 1× SSC buffer (0.6 M NaCl, 60 mM trisodium citrate, pH 7). The eluant was centrifuged briefly to pellet acrylamide fragments. The supernatant was precipitated with 1/10 volume of LiCl and 3 volumes of ethanol, resuspended in 10 μl of TE and cloned using the Original TA cloning kit or the TOPO TA cloning kit (Invitrogen). Insert-containing clones were identified by agarose gel electrophoresis of PCR amplification products of the inserts using M13F and M13R primers.
DGGE profiles of reamplified, cloned DNA were used to check for heteroduplexes (13) and to confirm the positions of cloned bands relative to the original sample as described previously (57).
Sequencing and phylogenetic analysis.
Bidirectional sequencing was performed with the ABI PRISM sequencing kit (Perkin-Elmer) using M13 primers and an automated ABI DNA sequencer. Sequences were aligned to known sequences using BLAST (basic local alignment search tool) (3). All sequences were analyzed by the program CHECK_CHIMERA from the Ribosomal Database Project (RDP) (39). Phylogenetic relationships were inferred by the maximum-likelihood, maximum-parsimony, and neighbor joining methods, which all gave similar results. Sequences were aligned using RDP, version 7.0 (39), and the tree was constructed using the program package ARB (Department of Microbiology, Technical University of Munich, Munich, Germany [http://isar.mpi-bremen.de/arb/carb.html]).
Sampling of bacteria.
The discrimination between abundance, carbon production, and enzyme activities of free-living versus particle-attached bacteria was operationally defined by size fractionation. Subsamples (100 to 200 ml) were gravity filtered through 47-mm-diameter, 1.0-μm-pore-size polycarbonate filters (Nuclepore). The abundance and activity of free-living bacteria were obtained from measurements on the filtrate. The abundance and activity of attached bacteria were calculated as the difference between measurements on unfiltered water and water filtered through the polycarbonate filters.
Bacterial abundance.
For routine counts, aliquots (1 to 2 ml) of unfiltered and gravity-filtered (1.0-μm pore size) water were fixed with filtered (0.2-μm pore size) electron microscopy (EM) grade glutaraldehyde (1%, final concentration). After the addition of Tween 80 (Sigma; 10 μg ml−1), the samples were sonicated for 30 s, stained with 4′, 6′-diamidino-2-phenylindole (DAPI; 1 μg ml−1) for 10 min (77) and filtered onto black 0.2-μm-pore-size polycarbonate filters (Poretics). Within a few hours of sampling >200 bacteria filter−1 (or >20 fields filter−1) were counted at ×1,250 using epifluorescence microscopy (Olympus BH-2). To estimate variability among filters, duplicate sets of filters were made on days 5, 9, and 13.
Additional counts by a different procedure were done to test for bias in our operational definition of free-living and particle-attached bacteria. Unfiltered, unsonicated samples from tanks 1 and 3 on days 13, 14, and 15 were stained with DAPI and then filtered through 1.0-μm-pore-size black polycarbonate filters. Bacteria on the filters which were not associated with visible particles were counted and expressed as a percentage of the total bacterial count obtained by the routine procedure.
Heterotrophic flagellate abundance and size.
Aliquots (10 to 40 ml) were fixed with filtered (0.2-μm pore size) EM grade glutaraldehyde (1%, final concentration), stained with DAPI (1 μg ml−1, final concentration), and filtered through 0.8-μm-pore-size black polycarbonate filters (Nuclepore). One filter diameter on each filter was counted at ×1,250 using epifluorescence microscopy. Heterotrophic flagellates were defined as flagellate-sized cells without visible chlorophyll. To estimate variability among filters, duplicate sets of filters were made on days 14 and 15. The sizes of individual flagellates were measured using a calibrated ocular grid.
Viral abundance.
Aliquots (0.5 to 1.0 ml) were fixed with filtered (0.2-μm pore size) EM grade glutaraldehyde (1%, final concentration), filtered through 0.02-μm-pore-size Anodisc filters (Whatman), stained with SYBR Green (Molecular Probes), and mounted in glycerol as described by Noble and Fuhrman (47). Filters were counted at ×1,250 using epifluorescence microscopy and a charge-coupled device camera connected to a monitor (system IFG-300; MTI). To estimate variability among filters, duplicate sets of filters were made on days 7, 8, and 13.
Bacterial production.
Bacterial production was measured by [3H]leucine incorporation (33), as modified for microcentrifugation by Smith and Azam (66). Triplicate 1.7-ml aliquots of unfiltered and filtered (1.0-μm pore size) samples were incubated with l-[4,5-3H]leucine (20 nM, final concentration; DuPont NEN) in sterile 2.0-ml polypropylene tubes for ca. 1 h at 18 ± 1°C. Samples with 5% trichloracetic acid added prior to the addition of [3H]leucine served as blanks. Bacterial carbon production was calculated as described by Simon and Azam (64). A carbon-to-cell ratio of 20 fg of C bacterium−1 (37) was used for free-living bacteria, and 53 fg of C bacterium−1 was used for attached bacteria (average of attached bacterial cells on diatom flocs) (65). Cell-specific growth rates were calculated assuming exponential growth.
Chlorophyll a levels, phytoplankton cell counts, and POC.
Samples (0.2 to 1.0 liter) were filtered onto duplicate glass fiber filters (GF/F; Whatman) and frozen. Chlorophyll a was extracted in 96% ethanol and measured spectrophotometrically as described by Jespersen and Christoffersen (30). For phytoplankton enumeration, bulk samples of 50 ml were fixed with Lugol's solution and cells were enumerated with an inverted microscope using sedimentation chambers. For POC analysis, duplicate samples (50 to 200 ml) were filtered onto precombusted GF/F filters and frozen. Filters were dried, acidified, and analyzed on a Perkin-Elmer model 2400 CHN analyzer.
Hydrolytic ectoenzyme activities.
Triplicate unfiltered and gravity-filtered (1.0- and 0.2-μm pore size) samples (4 ml) were incubated with fluorogenic substrates (methylumbelliferyl [MUF] and aminomethyl coumarin [AMC] derivatives [29]) to determine potential hydrolysis rates in the three operationally defined fractions: attached (unfiltered minus 1.0-μm filtrate), free (1-μm filtrate minus 0.2-μm filtrate), and dissolved (0.2-μm filtrate). The substrates used and enzymes assayed were as follows: l-leucine–AMC, aminopeptidase; MUF-β-d-glucoside, β-glucosidase; MUF-α-d-glucoside, α-glucosidase; MUF-phosphate, alkaline phosphatase; MUF-oleate, lipase. Substrate hydrolysis rates were measured with a Hoefer TKO-100 fluorometer (356-nm excitation; 460-nm emission) using heat-killed samples as controls. The fluorometer was calibrated with standard solutions of MUF and AMC, and potential activities at 100 μM substrate concentration were measured.
Nutrients.
Four 12-ml samples were filtered through glass fiber filters (GF/F; Whatman) into 15-ml polypropylene tubes, stored frozen (−20°C), and analyzed within 4 months for nitrate, ammonia, silicate, and phosphate as described by Parsons et al. (49).
Nucleotide sequence accession numbers.
The following DGGE band sequences have been deposited in GenBank under the indicated accession numbers (from band 1 to band 16, in order): MBE1, AF191752; MBE2, AF191753; MBE3, AF191754; MBE4, AF191755; MBE5, AF191756; MBE6, AF191757; MBE7, AF191758; MBE8, AF191759; MBE9, AF191760; MBE10, AF191761; MBE11, AF191762; MBE12, AF191763; MBE13, AF191764; MBE14, AF191765; MBE15, AF191766; MBE16, AF191767. (MBE stands for marine bloom experiment.)
RESULTS
Chlorophyll a and microbial abundances.
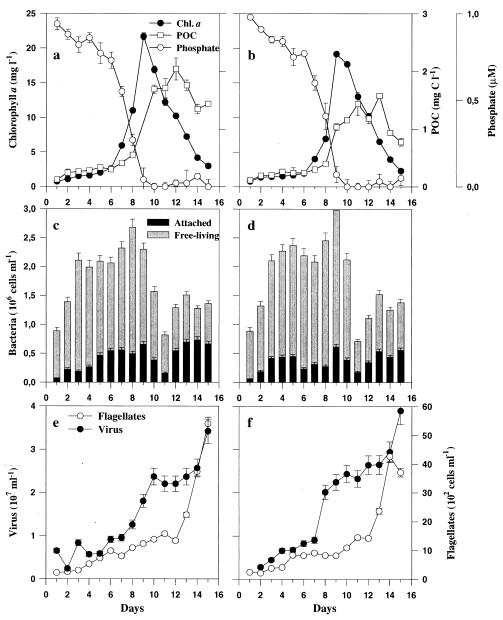
The addition of different nitrogen sources had no significant effect on phytoplankton or other microbial compositions or activities. Chlorophyll a levels (means ± standard deviations [SD] for the four tanks) increased from 2.3 ± 0.3 μg of chlorophyll a liter−1 on day 6 to 21.7 ± 2.2 μg of chlorophyll a liter−1 on day 9, on which day the nitrogen (data not shown) and phosphate concentrations were less than 0.1 μM (Fig. 1a and b). Chlorophyll a level then decreased to 2.6 ± 0.5 μg liter−1 by day 15 (Fig. 1a and b). The phytoplankton community was dominated by Thalassiosira sp., which reached cell densities of 15 × 103 to 18 × 103 cells ml−1 at days 9 to 10 (data not shown). All other diatom species and genera combined (Pseudo-nitzschia spp., Nitzschia spp., Thalassionema frauenfeldii) represented <10% of the total phytoplankton cell count. POC values increased up to days 12 to 13 (1.6 ± 0.4 mg of C liter−1) and then started to decline (Fig. 1a and b).
FIG. 1.
(a and b) Chlorophyll a (Chl. a), phosphate, and POC in tank 1 (a) and tank 3 (b), which were amended with NO3− and NH4+, respectively. (c and d) Abundances of free-living and attached bacteria in tank 1 (c) and tank 3 (d) are presented as stacked bars, which sum to total bacterial abundance. (e and f) Abundances of virus and flagellates in tank 1 (e) and tank 3 (f). Error bars indicate SD.
Bacterial abundances were very similar in the four tanks, except that the abundance in tank 2 was higher prior to the bloom (data not shown). Bacterial abundance increased to a peak value of (2.8 ± 0.2) × 106 ml−1 at days 8 to 9 but then decreased dramatically to (0.7 ± 0.1) × 106 ml−1 at day 11 (Fig. 1c and d). Bacterial abundance increased again towards the end of the experiment. Whereas free-living bacteria dominated bacterial abundance (73 to 93% of the total) prior to the peak of the bloom, attached bacteria increased from 7 to 27% of the total count prior to the peak of the bloom to 20 to 58% of the total count in the late postbloom phase (days 12 to 15; Fig. 1c and d). On days 13 to 15, the number of bacteria retained on a 1.0-μm-pore-size filter which were not associated with visible particulate material ranged from 15 to 26% of the total count (data not shown). Virus abundance increased steadily from day 7 to day 15 (9 × 106 to 37 × 106 ml−1), except for a stagnation during the time of low bacterial abundance (Fig. 1e and f). Flagellate abundance increased throughout the experiment and reached values of 4 × 103 to 5 × 103 ml−1 at the end of the experiment (Fig. 1e and f).
Bacterial production and specific growth rates.
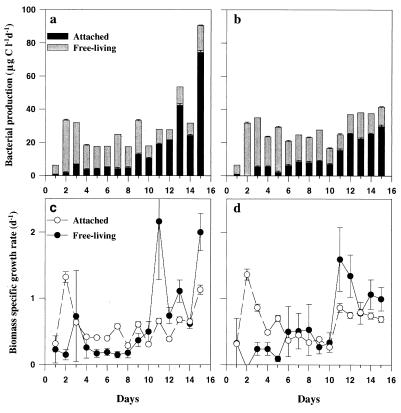
Secondary productivity and specific growth rates of the free-living bacteria peaked within the first 2 days of the experiment. Between days 1 and 2, total production showed the largest relative increase (ca. fivefold), which was attributed solely to the free-living bacteria. Total bacterial production reached maximum values of 41 to 90 μg of C liter−1 day−1 after the peak of the bloom (Fig. 2a and b). While the <1.0-μm filtrate accounted for most (65 to 100%) of the bacterial carbon production before the peak of the bloom (days 1 to 8), attached bacteria accounted for most (43 to 82%) of the production in the postbloom phase (days 10 to 15; Fig. 2a and b). Throughout the experiment, the free-living bacteria had low specific growth rates (μ = 0.6 ± 0.3 day−1; range = 0.3 to 1.4 day−1) while attached bacteria had their highest specific growth rates after the peak of the bloom (μ = 1.2 ± 0.6 day−1; range = 0.3 to 2.2 day−1; Fig. 2c and d).
FIG. 2.
(a and b) Stacked bar graphs showing carbon production of free-living and attached bacteria over time in tank 1 (a) and tank 3 (b). (c and d) Biomass-specific growth rates of free-living and attached bacteria versus time in tank 1 (c) and tank 3 (d). Error bars indicate SD.
Enzyme activities.
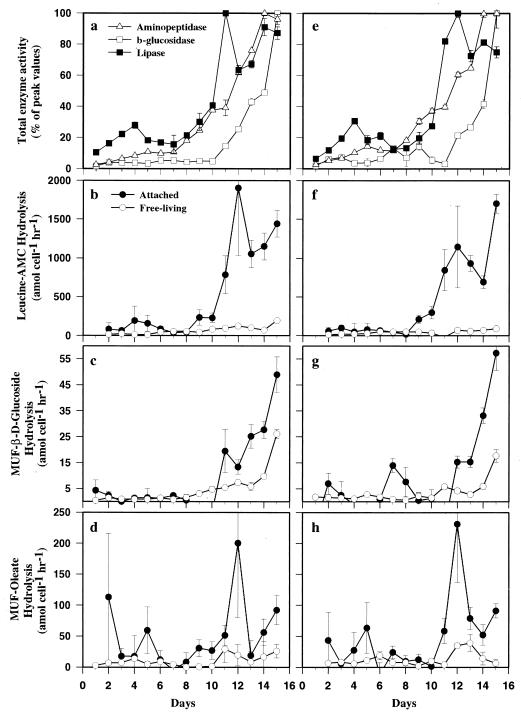
Enzyme activities for all five measured enzymes peaked during the postbloom phase, and activities in tanks 1 and 3 were very similar (Fig. 3a and e; only data for aminopeptidase, β-glucosidase, and lipase are presented). During the experiment aminopeptidase activity increased from 198 ± 39 to 1,065 ± 192 nmol liter−1 h−1, with the majority of the total activity associated with the attached bacteria. Cell-specific hydrolysis rates in the attached bacterial fraction were typically higher than those in the free-living fraction, often by more than an order of magnitude (Fig. 3b and f). Total β-glucosidase activity increased throughout the experiment to 50 ± 3 nmol liter−1 h−1 on day 15. Most activity was associated with the >1.0-μm fraction, but some was also associated with the free-living bacteria (Fig. 3c and g). Total α-glucosidase activity was also highest on day 15 (21 ± 1 nmol liter−1 h−1) and was mainly associated with the free-living bacteria. Phosphatase and lipase activities were mainly found in the dissolved (<0.2-μm) fraction and in the attached fractions, reaching maximum activities 2 to 3 days after the peak of the bloom (87 to 106 and 137 to 180 nmol liter−1 h−1, respectively). Distinct peaks in cell-specific lipase activity of attached bacteria were observed on day 11, whereas free-living bacteria had very low activities throughout the experiment (Fig. 3d and h).
FIG. 3.
Time course of potential ectoenzyme activities in tank 1 (a to d) and tank 3 (e to h). (a and b) Total potential enzyme activities presented as percentages of their peak values. (c to h) Cell-specific hydrolysis rates for attached and free-living bacteria. Error bars indicate SD.
DGGE analysis.
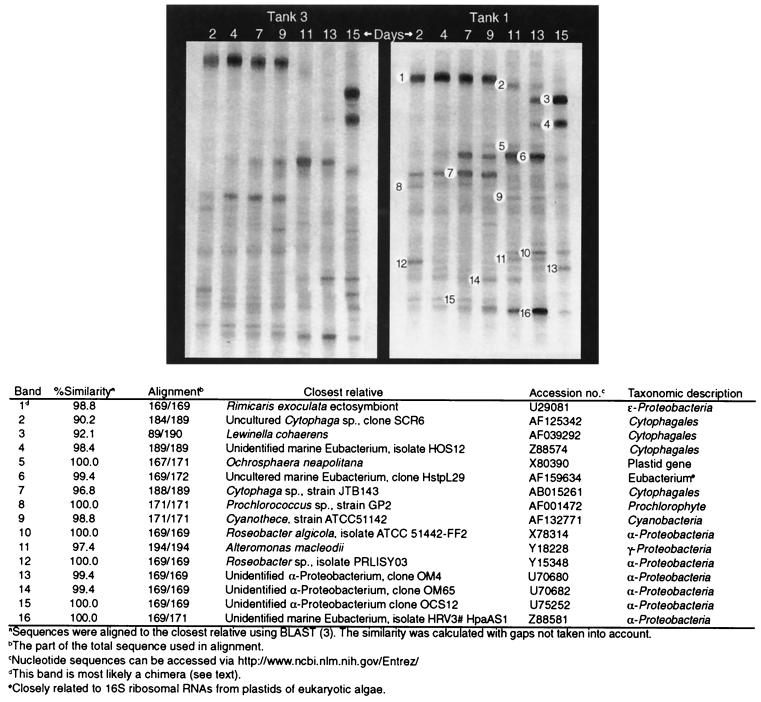
Pronounced changes in relative brightness of DGGE bands of PCR-amplified community DNA were observed during the course of the experiment, especially after the peak of the bloom (day 9) in association with the rapid decline in bacterial abundance (Fig. 1c and d and 4). Bands 1, 7, and 15 disappeared, whereas bands 2, 6, 10, 11, and 16 increased in brightness. Bands 3 and 4 became very bright at day 15, while band 16 disappeared. DGGE profiles for tank 1 and tank 3 were very similar in appearance, despite the use of two different gels (Fig. 4). DGGE profiles from tank 1 and tank 3 were compared with those for the duplicate tanks (tank 2 and tank 4) on days 2, 9, and 15 (data not shown). Only minor differences comparable to those between tank 1 and tank 3 were seen.
FIG. 4.
DGGE profiles of the bacterial community composition over time in tanks 1 and 3. Separate gels (each with 30 to 46% denaturant gradients) were used for the two tanks. Each excised, cloned, and sequenced band is numbered on the left. The relationships of excised band sequences to other sequences in the GenBank database are indicated in the table under the gels.
The PCR amplicons consisted of 15 to 20 resolvable phylotypes. Sixteen bands from different vertical positions in the tank 1 profile were excised, cloned, and sequenced. Bands from different days with identical vertical positions in a gel were assumed to have identical sequences, as shown previously (57). No heteroduplexes were found among the excised bands. The majority of the amplicons were related to α-Proteobacteria (38%), Cytophagales (25%), or Cyanobacteria (13%) (Fig. 4 and 5). Except for two amplicons related to Cytophagales, the amplicons were closely related to known sequences in the databases (96.8 to 100% similarity).
FIG. 5.
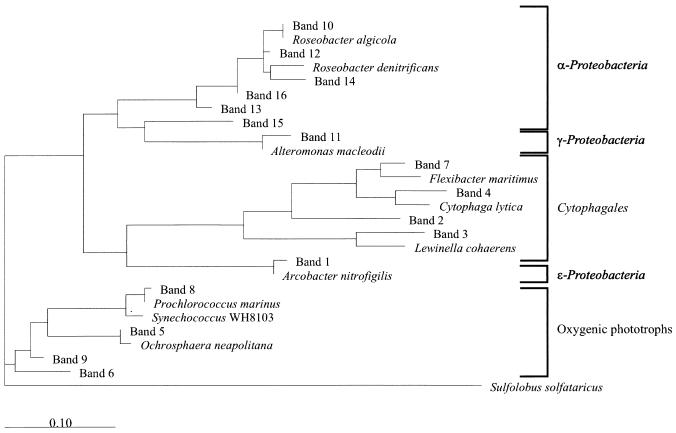
Phylogenetic tree showing relationships of the sequences found in our samples to representative bacterial 16S rRNA genes. Sequences of the cultured bacteria were retrieved from GenBank. The tree was inferred by the maximum-likelihood method using full-length genes for the cultured bacteria and the entire length of the sequenced bands (169 to 194 bp) beginning at the equivalent to base 341 and going towards the 3′ end of the 16S rRNA molecule (Escherichia coli numbers). An Archaea (Sulfolobus solfataricus) was used as an outgroup. GenBank accession numbers of the cultured bacteria are as follows: R. algicola, X78314; R. denitrificans, M96746; A. macleodii, Y18228; F. maritimus, M64629; C. lytica, M62796; L. cohaerens, AF039292; A. nitrofigilis, L14627; P. marinus, X63140; Synechococcus sp. strain WH8103, AF001479; O. neapolitana, X80390. The scale bar indicates substitutions per nucleotide position.
Band 1 was found to be a possible chimera composed of sequences from a Campylobacter sp. (ɛ-Proteobacteria) and Arcobacter nitrofigilis (ɛ-Proteobacteria), which is a NaCl-requiring, nitrogen-fixing bacterium isolated from plant roots (42). Each part composing this purported chimera had a 100% match to database sequences, which indicates a very high probability of it being a chimera (58). Thus, band 1 may be an artifact produced by the PCR. Four bands (bands 2, 3, 4, and 7) were related to Cytophagales. Band 7 was seen as a very dominant band until day 9, while bands 2, 3, and 4 were observed only in the postbloom phase. Bands 3 and 4 appeared on day 13 and became the most dominant bands on day 15.
Another four bands (bands 5, 6, 8, and 9) were related to photosynthetic organisms. Of these, two were closely related to phytoplankton plastid DNA, band 5 being most closely related to the plastid of Ochrosphaera neapolitana (Haptophyceae) and band 6 being most closely related to that of an uncultured marine bacterium showing a high degree of similarity to 16S ribosomal RNAs from plastids of eukaryotic algae. Band 6 appeared as a bright band from days 7 to 13. Two bands were related to Cyanobacteria sequences. The band 8 sequence was identical to those of several Prochlorococcus strains, and the band was seen throughout the experiment, while band 9 was related to a Cyanothece species and was most conspicuous on days 11 and 13.
Six bands were related to two different clusters of α-Proteobacteria. Bands 10, 12, 13, 14, and 16 clustered close to or inside the marine alpha group (20), while band 15 was identical to that of the environmental clone OCS12 (14) within the “SAR11 cluster”. Bands 10 and 12 were 100 and 98% identical to those of Roseobacter algicola and a Roseobacter sp., respectively, both isolated from dinoflagellates (36, 52). Bands 13 and 14 were closely related to two environmental clones recently found by Rappé et al. (54) off Cape Hatteras, N.C. Of these bands, 12 and 15 appeared in the beginning of the experiment, while bands 10, 13, 14, and 16 were most prominent towards the end of the experiment.
Band 11 falls among environmental clones isolated near a deep-sea hydrothermal vent (53), which are related to the polymer-secreting Alteromonas macleodii (γ-Proteobacteria). This band was seen on days 11 and 13, only.
DISCUSSION
The majority (62%) of phylotypes identified in this study fell into two major phylogenetic groups (α-Proteobacteria and Cytophagales), which have previously been associated with surface-attached growth. Clones clustering within or close to the α-Proteobacteria have been obtained from a number of marine environments, for example the Sargasso Sea (17), the Oregon coast (70), the Georgia coast (20), and the Antarctic (6), and members of this group have also been found to be numerically dominant in coastal waters (20). Several members of the marine alpha group (including R. algicola) have been found on marine snow (55), and some have been shown to be culturable on plates (23, 70). In addition, Roseobacter species (bands 10 and 12) have been isolated from the phycosphere of dinoflagellates (36, 52). In this study, the predominance of DGGE bands related to the marine alpha group during the time of high particle colonization in the postbloom phase also suggests that at least certain members of this group are adapted for growth on particles.
Cytophaga-related species are an abundant group in marine systems (19) and have previously been observed to be dominant on marine macroaggregates (10, 55), on sludge flocs (40), and in freshwater mesocosms (28, 73). This group is known to be involved in the degradation of complex macromolecules (60), and recently Cytophaga-related species were found to have cell-specific growth rates up to five times faster than the total community turnover in a protein-enriched mesocosm (51). Although no generalization on substrate preference can be made for Cytophagales, it is notable that bands 2, 3, and 4 appear at days 11 and 13 coincident with pronounced increases in cell-specific growth rates (Fig. 2c and d) as well as cell-specific enzyme activities among particle-attached bacteria (Fig. 3b to d and f to h). In contrast, band 7 is seen as a bright band prior to the peak of the bloom and is likely to represent a predominantly free-living phylotype. Similarly, by DGGE analyses Fandino et al. (12) found Cytophaga-related species to be members of the free-living bacterial assemblages during a dinoflagellate bloom in the southern California bight. Thus, it appears that phylotypes within Cytophagales, in addition to their predominance on particles, can also be major components of free-living marine bacterial communities under nutrient-rich conditions (12).
Like most studies which discriminate free-living and attached bacteria, ours relied on size fractionation. This can result in overestimation of the contribution of attached bacteria due to trapping of some free-living bacteria on the filters. We estimated the filtration bias due to trapping using samples from the postbloom phase as a “worst-case scenario” since the average size of free-living bacteria was expected to increase during the experiment. We consider the result (15 to 26%) to be an overestimation, since not all particulate matter can be observed using DAPI staining (e.g., transparent exopolymer particles [2], Coomassie-stained particles [38]). However, it is also conceivable that some bacteria attached to very small or very fragile particles could pass the filter. Therefore, we emphasize that the distinction between free-living and attached bacteria is strictly operational. Some degree of bias in defining these categories is inevitable, but the distinction can still be useful if the caveats are kept in mind.
The colonization of particulate matter during the postbloom phase was measured as increased particle-associated microbial abundance and activity. The predominance of DGGE bands related to Cytophagales and marine α-Proteobacteria during this period is interpreted as an indication of a marked role for these groups in the colonization and hydrolysis of particles. Thus, we assume that DGGE reflects at least the major variations in the relative abundances of PCR-amplifiable phylotypes (57). It is well known that there are several potential biases which may affect DGGE analysis of PCR amplicons (possible biases introduced by the PCR and tests of the reproducibility and sensitivity of DGGE were discussed in more detail by Riemann et al. [57]). However, replicate PCRs and DGGE gels confirmed that DGGE patterns were highly reproducible, so we assume that a possible amplification bias for or against a certain sequence is constant. Thus, changes in relative band intensity with time should reflect actual changes in the relative abundance of a phylotype. While we believe this assumption to be valid, we emphasize that DGGE does not provide a complete or quantitative view of the bacterial community composition, but rather a somewhat simplified “fingerprint.” Further, since this approach is based on analyses of 16S rRNA genes, the microbial diversity may be underestimated due to the conserved nature of this gene (16, 75).
The idea that the present phylotypes related to Cytophagales and the marine alpha group could be particle specialist bacteria responsible for the high growth rates and hydrolytic activities associated with particles is indirectly supported by a number of studies. These indicate that attached bacteria are phylogenetically (1, 5, 10) and functionally different from free-living bacteria. Attached bacteria have indeed been found to have higher cell-specific enzyme activities (25, 68) and sometimes higher growth rates (24, 68). They are also generally larger (43, 63) and have higher specific uptake rates for some substrates (48, 72). While particle-attached bacteria normally account for <10 to 15% of the total bacterial production and abundance (24, 71), they can account for >50% of total bacterial biomass and/or hydrolytic activity during the senescence of algal blooms (43, 68). Therefore, we propose a scenario where bacteria specialized in particle colonization and hydrolysis proliferated during the detritus-dominated death phase of the diatom bloom leading to the predominance of DGGE bands related to Cytophagales and the marine alpha group.
Dominance of phylotypes related to Cytophaga and the marine alpha group during the late stage of the diatom bloom was concomitant with pronounced increases in potential enzymatic activities and changes in relative enzyme activities, indicating that bacterial community composition strongly influences biochemistry vis-à-vis bacterial cycling of particulate and polymeric organic matter. Similar observations were made for a protein-enriched mesocosm (51) and during an intense dinoflagellate bloom off the southern California coast (L. B. Fandino, unpublished observations). Both groups found a correlation between the level of enzymatic activity and the composition of the bacterial community. Although our data are insufficient to demonstrate a causal link between the shifts in community composition and enzyme activities, we hypothesize that substrate and niche availability may drive bacterial population successions and consequently the variations in enzyme activities. For example, since lipase activity is a prominent phenotype among some marine bacterial isolates (41) and is produced mainly by bacteria (18), lipid compounds released from lysed diatoms (56) and from flagellates (46) may have selected for bacterial populations expressing high levels of lipase activity in the postbloom phase. Recently, van Hannen et al. (74) showed that the origin of detritus can indeed affect the structure of the bacterial community.
Except for phosphatase, which is produced by bacteria as well as by phytoplankton (9, 41), we speculate that the distribution and the levels of enzyme activities among the analyzed size fractions reflect specializations for the hydrolysis of different polymers by free-living and attached bacteria. An example is the high total and cell-specific aminopeptidase activities of attached bacteria, which were severalfold higher than those of free-living bacteria and significantly higher than the glucosidase activities. This has often been observed (67, 68) and can be interpreted as the potential for a faster solubilization of proteins relative to carbohydrates, especially on particles. However, in general the comparison of relative hydrolysis rates for different substrates must be interpreted with caution since actual in situ hydrolysis rates will depend on the quality and concentration of the naturally occurring substrates (25). Additionally, the role of protozoa in ectohydrolase production is unclear but is potentially significant (32).
During the experiment several bands decreased in brightness and/or disappeared, e.g., bands 1, 2, 7, 12, 14, and 15 (Fig. 4). The most dramatic change was the disappearance of three dominant bands (bands 1, 7, and 15) associated with a marked decrease in bacterial abundance on days 8 to 11, indicating pronounced mortality of the associated three phylotypes. Grazing and viral infection rates were not explicitly measured, so the fate of the bacteria is uncertain. However, we assume that the apparently extensive phylotype-specific mortality from days 9 to 11 was caused mainly by flagellate grazing and/or viral lysis.
Viruses are considered important in regulating and maintaining the diversity of microbial communities (15), and a high concentration of a susceptible host can lead to rapid viral propagation and lysis of a particular host population (see, e.g., references 7 and 27). Given the relative host specificity of viruses, they may be expected to cause phylotype-specific losses such as those we observed. However, a number of studies have also demonstrated that flagellate grazing has the potential to affect the species composition of a mixed bacterial community. The extent of flagellate grazing of bacteria is affected by bacterial size and motility (22, 61), and protozoa consume and digest various bacterial species with variable efficiencies (21). Further, Caron (8) showed a strong specialization among flagellate species in the ability to graze free-living and attached bacteria and Šimek et al. (62) observed a shift in the numerical dominance of different subdivisions of the class Proteobacteria as a consequence of heavy flagellate grazing.
To estimate the impact of grazing, we applied a grazing model which considers mean measured flagellate size d (4.35 ± 0.69 μm; n = 60), bacterial and flagellate abundances, and temperature (reference 50, equation 4). The model yielded a grazing rate estimate of 0.28 × 106 bacteria ml−1 day−1 on day 10 (tank 1), which corresponds to 31% of the bacterial production on day 10 or 36% of the observed decrease in bacterial abundance from day 10 to day 11. These estimates suggest that flagellates could have had a significant impact on community composition.
Another possible effect of extensive flagellate grazing (up to >4 × 103 flagellates ml−1, Fig. 1e and f) was observed towards the end of the experiment where large chains (up to >100 μm or longer) of filamentous bacterial morphotypes increased from 0 (day 12) to 38 to 52% (day 15) of total bacterial abundance (data not shown). These bacteria, presumably resistant to grazing (31), were mainly associated with particles and may have resulted from increased growth rates of species already present on particles (26) and/or the introduction of new dominant phylotypes derived from the free-living populations (bands 3 and 4; Fig. 4).
Summary and conclusions.
This study shows for the first time that the colonization of particles during the postbloom phase of a diatom bloom is accompanied by major changes in bacterial community composition. These changes happened on a very short time-scale (1 to 2 days) and were concomitant with pronounced increases in the abundance, growth rate, and hydrolytic activity of the attached bacteria. The measured variables from the four mesocosms were very similar, which indicates that the observed bacterial dynamics represent a robust and systematic community response to the environmental conditions during this experimental diatom bloom. Our data suggest that colonization of decaying diatoms was accomplished by fast-growing, highly hydrolytic particle-specialist bacteria. These were primarily related to Cytophagales and the marine alpha group of the class Proteobacteria. The present study shows the highly dynamic nature of bacterial community composition and strongly suggests that nutrient-induced changes in natural phytoplankton communities lead to significant effects on the structure and functioning of bacterial assemblages as well as on the nature and the rates of bacterially mediated organic matter cycling. Hence, this study emphasizes the need to incorporate community composition into our conceptual thinking of the biogeochemical activities of marine microbial assemblages.
ACKNOWLEDGMENTS
This work was funded by NSF grants OCE98-19603, OPP95-30851, and OPP96-17045 to F.A.
We thank Morten Søndergaard and Mathias Middelboe for useful criticism of an earlier version of the manuscript, Thomas Leser for assistance in generating the phylogenetic tree, and Laura B. Fandino for sharing unpublished data.
REFERENCES
- 1.Acinas S G, Antón J, Rodríguez-Valera F. Diversity of free-living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alldredge A L, Passow U, Logan B E. The abundance of a class of large, transparent organic particles in the ocean. Deep-Sea Res. 1993;40:1131–1140. [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azam F. Microbial control of oceanic carbon flux: the plot thickens. Science. 1998;280:694–696. [Google Scholar]
- 5.Bidle K, Fletcher M. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl Environ Microbiol. 1995;61:944–952. doi: 10.1128/aem.61.3.944-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman J P, McCammon S A, Brown M V, Nichols D S, McMeekin T. Diversity and association of psychrophilic bacteria in Antarctic Sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bratbak G, Heldal M, Norland S, Thingstad T F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990;56:1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caron D A. Grazing of attached bacteria by heterotrophic microflagellates. Microb Ecol. 1987;13:203–218. doi: 10.1007/BF02024998. [DOI] [PubMed] [Google Scholar]
- 9.Chróst R J, Overbeck J. Kinetics of alkaline phosphatase activity and phosphorus availability for phytoplankton and bacterioplankton in Lake Plußsee (north German eutrophic lake) Microb Ecol. 1987;13:229–248. doi: 10.1007/BF02025000. [DOI] [PubMed] [Google Scholar]
- 10.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 11.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fandino L B, Riemann L, Steward G F, Long R A, Azam F. Bacterial succession during a dinoflagellate bloom analyzed by DGGE and rDNA sequencing. Suppl EOS Trans Am Geophys Union. 1998;79:OS63. [Google Scholar]
- 13.Fernandez E, Bienvenu T, Arramond F D, Beldjord K, Kaplan J C, Beldjord C. Use of chemical clamps in denaturing gradient gel electrophoresis: application in the detection of the most frequent Mediterranean β-thalassemic mutations. PCR Methods Applic. 1993;3:122–124. doi: 10.1101/gr.3.2.122. [DOI] [PubMed] [Google Scholar]
- 14.Field K G, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrman J A. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 16.Fuhrman J A, Campbell L. Microbial microdiversity. Nature. 1998;393:410–411. [Google Scholar]
- 17.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gajewski A J, Chróst R J, Siuda W. Bacterial lipolytic activity in a eutrophic lake. Arch Hydrobiol. 1993;128:107–126. [Google Scholar]
- 19.Glöckner F O, Fuchs B M, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González J M, Iriberri J, Egea L, Barcina A. Differential rates and digestion of bacteria by freshwater and marine phagotrophic protozoa. Appl Environ Microbiol. 1990;56:1851–1857. doi: 10.1128/aem.56.6.1851-1857.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González J M, Sherr E B, Sherr B F. Differential feeding by marine flagellates on growing versus starving, and on motile versus nonmotile, bacterial prey. Mar Ecol Prog Ser. 1993;102:257–267. [Google Scholar]
- 23.González J M, Kiene R P, Moran M A. Transformations of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith P, Shiah F-K, Gloersen K, Ducklow H W, Fletcher M. Activity and distribution of attached bacteria in Chesapeake Bay. Mar Ecol Prog Ser. 1994;108:1–10. [Google Scholar]
- 25.Grossart H-P, Simon M. Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow) Aquat Microb Ecol. 1998;15:127–140. [Google Scholar]
- 26.Hahn M W, Moore E R B, Höfle M G. Bacterial filament formation, a defense against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl Environ Microbiol. 1999;65:25–35. doi: 10.1128/aem.65.1.25-35.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennes K P, Suttle C A, Chan A M. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl Environ Microbiol. 1995;61:3623–3627. doi: 10.1128/aem.61.10.3623-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Höfle M G. Bacterioplankton community structure and dynamics after large-scale release of nonindigenous bacteria as revealed by low-molecular-weight analysis. Appl Environ Microbiol. 1992;58:3387–3394. doi: 10.1128/aem.58.10.3387-3394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoppe H-G. Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylumbelliferyl-substrates. Mar Ecol Prog Ser. 1983;11:299–308. [Google Scholar]
- 30.Jespersen A-M, Christoffersen K. Measurements of chlorophyll-a from phytoplankton using ethanol as extraction solvent. Arch Hydrobiol. 1987;109:445–454. [Google Scholar]
- 31.Jürgens K, Güde H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser. 1994;112:169–188. [Google Scholar]
- 32.Karner M, Ferrier-Pagés C, Rassoulzadegan F. Phagotrophic nanoflagellates contribute to occurrence of α-glucosidase and aminopeptidase in marine environments. Mar Ecol Prog Ser. 1994;114:237–244. [Google Scholar]
- 33.Kirchman D, Nees E K, Hodson R. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol. 1985;49:599–607. doi: 10.1128/aem.49.3.599-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirchman D L, Suzuki Y, Garside C, Ducklow H W. High turnover rates of dissolved organic carbon during a spring phytoplankton bloom. Nature. 1991;352:612–614. [Google Scholar]
- 35.Koike I, Hara S, Terauchi K, Kogure K. Role of sub-micrometre particles in the ocean. Nature. 1990;345:242–244. [Google Scholar]
- 36.Lafay B, Ruimy R, Trauenberg C R, Breittmayer V, Gauthier M J, Christen R. Roseobacter algicola sp nov, a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int J Syst Bacteriol. 1995;45:290–296. doi: 10.1099/00207713-45-2-290. [DOI] [PubMed] [Google Scholar]
- 37.Lee S, Fuhrman J A. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Microbiol. 1987;53:1298–1303. doi: 10.1128/aem.53.6.1298-1303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long R A, Azam F. Abundant protein-containing particles in the sea. Aquat Microb Ecol. 1996;10:213–221. [Google Scholar]
- 39.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manz W, Amann R, Ludwig W, Vancanney M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 41.Martinez J, Smith D C, Steward G F, Azam F. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat Microb Ecol. 1996;10:223–230. [Google Scholar]
- 42.McClung C R, Patriquin D G, Davis R E. Campylobacter nitrofigilis sp nov, a nitrogen-fixing bacterium associated with roots of Spartina alterniflora Loisel. Int J Syst Bacteriol. 1983;33:605–612. [Google Scholar]
- 43.Middelboe M, Søndergaard M, Letarte Y, Borch N H. Attached and free-living bacteria: production and polymer hydrolysis during a diatom bloom. Microb Ecol. 1995;29:231–248. doi: 10.1007/BF00164887. [DOI] [PubMed] [Google Scholar]
- 44.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagata T, Kirchman D L. Release of macromolecular organic complexes by heterotrophic marine flagellates. Mar Ecol Prog Ser. 1992;83:233–240. [Google Scholar]
- 47.Noble R T, Fuhrman J A. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- 48.Palumbo A V, Ferguson R L, Rublee P A. Size of suspended bacterial cells and association of heterotrophic activity with size fractions of particles in estuarine and coastal waters. Appl Environ Microbiol. 1984;48:157–164. doi: 10.1128/aem.48.1.157-164.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsons T R, Maita Y, Lalli C M. A manual of chemical and biological methods for seawater analysis. Elmsford, N.Y: Pergamon Press; 1984. [Google Scholar]
- 50.Peters F. Prediction of planktonic protistan grazing rates. Limnol Oceanogr. 1994;39:195–206. [Google Scholar]
- 51.Pinhassi J, Azam F, Hempälä J, Long R A, Martinez J, Zweifel U L, Hagström Å. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat Microb Ecol. 1999;17:13–26. [Google Scholar]
- 52.Prokic I, Bruemer F, Brigge T, Goertz H D, Gerdts G, Schuett C, Elbraechter M, Mueller W E G. Bacteria of the genus Roseobacter associated with the toxic dinoflagellate Prorocentrum lima. Protist. 1999;149:347–357. doi: 10.1016/S1434-4610(98)70041-0. [DOI] [PubMed] [Google Scholar]
- 53.Raguenes G, Pignet P, Gauthier G, Peres A, Christen R, Rougeaux H, Barbier G, Guezennec J. Description of a new polymer-secreting bacterium from a deep-sea hydrothermal vent, Alteromonas macleodii subsp. fijiensis, and preliminary characterization of the polymer. Appl Environ Microbiol. 1996;62:67–73. doi: 10.1128/aem.62.1.67-73.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 55.Rath J, Wu K Y, Herndl G J, DeLong E F. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:262–269. [Google Scholar]
- 56.Reynolds C S. The ecology of freshwater phytoplankton. Cambridge, United Kingdom: Cambridge University Press; 1984. [Google Scholar]
- 57.Riemann L, Steward G F, Fandino L B, Campbell L, Landry M R, Azam F. Bacterial community composition during two consecutive NE monsoon periods in the Arabian Sea studied by denaturing gradient gel electrophoresis (DGGE) of rRNA genes. Deep Sea Res II. 1999;46:1791–1811. [Google Scholar]
- 58.Robison-Cox J F, Bateson M M, Ward D M. Evaluation of nearest-neighbor methods for detection of chimeric small-subunit rRNA sequences. Appl Environ Microbiol. 1995;61:1240–1245. doi: 10.1128/aem.61.4.1240-1245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 60.Shewan J M, McMeekin T A. Taxonomy (and ecology) of Flavobacterium and related genera. Annu Rev Microbiol. 1983;37:233–252. doi: 10.1146/annurev.mi.37.100183.001313. [DOI] [PubMed] [Google Scholar]
- 61.Šimek K, Chrzanowski T H. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl Environ Microbiol. 1992;58:3715–3720. doi: 10.1128/aem.58.11.3715-3720.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Šimek K, Vrba J, Pernthaler J, Posch T, Hartman P, Nedoma J, Psenner R. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl Environ Microbiol. 1997;63:587–595. doi: 10.1128/aem.63.2.587-595.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simon M. Biomass and production of small and large free-living and attached bacteria in Lake Constance. Limnol Oceanogr. 1987;32:591–607. [Google Scholar]
- 64.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 65.Simon M, Alldredge A L, Azam F. Bacterial carbon dynamics on marine snow. Mar Ecol Prog Ser. 1990;65:205–211. [Google Scholar]
- 66.Smith D C, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs. 1992;6:107–114. [Google Scholar]
- 67.Smith D C, Simon M, Alldredge A L, Azam F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature. 1992;359:139–142. [Google Scholar]
- 68.Smith D C, Steward G F, Long R A, Azam F. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep Sea Res II. 1995;42:75–97. [Google Scholar]
- 69.Somerville C C, Knight I T, Straube W L, Colwell R T. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989;55:548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Strobel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turley C M, Mackie P J. Biogeochemical significance of attached and free-living bacteria and the flux of particles in the NE Atlantic Ocean. Mar Ecol Prog Ser. 1994;115:191–203. [Google Scholar]
- 72.Unanue M, Ayo B, Azúa I, Barcina I, Iriberri J. Temporal variability of attached and free-living bacteria in coastal waters. Microb Ecol. 1992;23:27–39. doi: 10.1007/BF00165905. [DOI] [PubMed] [Google Scholar]
- 73.van Hannen E J, Zwart G, van Agterveld M P, Gons H J, Ebert J, Laanbroek H J. Changes in bacterial and eukaryotic community structure after mass lysis of filamentous Cyanobacteria associated with viruses. Appl Environ Microbiol. 1999;65:795–801. doi: 10.1128/aem.65.2.795-801.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Hannen E J, Mooij W, van Agterveld M P, Gons H J, Laanbroek H J. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2478–2484. doi: 10.1128/aem.65.6.2478-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ward D M, Ferris M J, Nold S C, Bateson M M. A natural view of microbial biodiversity within hot spring Cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wells M L, Goldberg E D. Occurrence of small colloids in sea water. Nature. 1991;353:342–344. [Google Scholar]
- 77.Yoon W B, Rosson R A. Improved method for enumeration of attached bacteria for study of fluctuation in the abundance of attached and free-living bacteria in response to diel variation in seawater turbidity. Appl Environ Microbiol. 1990;56:595–600. doi: 10.1128/aem.56.3.595-600.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]