Abstract
For biodetection of mutagenic pollution of marine environments, an organism naturally occurring in these habitats should be used. We found that marine bacterium Vibrio harveyi may be an appropriate bioindicator of mutagenic pollution. For positive selection of mutants, we developed a simple method for isolation of V. harveyi mutants resistant to neomycin. We constructed genetically modified V. harveyi strains that produce significantly more neomycin-resistant mutants upon treatment with low concentrations of mutagens than the wild-type counterpart. The sensitivity of the mutagenicity test with the V. harveyi strains is at least comparable to (if not higher than) that of the commonly used Ames test, which uses Salmonella enterica serovar Typhimurium strains. Therefore, we consider that the V. harveyi strains described in this report could be used as potential bioindicators of mutagenic pollution of marine environments.
Mutagenic pollution of natural environments seems to be a general and serious problem (see, for instance, information provided by the U.S. Environmental Protection Agency [6]) that has been extensively investigated (the Environmental Mutagen Information Center database contains over 20,000 citations to literature on agents that have been tested for mutagenic activity; see http://www.nlm.nih.gov/pubs/factsheets/emicfs.html). This problem also concerns marine habitats. Therefore, detection of the presence of mutagens in the environment is important. This is not an easy procedure, as mutagenic components usually occur in natural habitats at low concentrations. Moreover, mutagens are only a fraction of contaminating chemicals in natural environments. Thus, biological mutagenicity tests seem to be more sensitive and accurate than chemical analyses.
The most commonly used mutagenicity test is that described by Ames (1) and subsequently modified by Ames and coworkers (2, 13). In this test, a series of genetically modified Salmonella enterica serovar Typhimurium strains are used. The presence of mutations in his genes allows positive selection of his+ revertants on minimal agar plates. A deletion of the uvrB gene in most of these tester strains ensures a higher efficiency of mutagenesis due to inactivation of one of the bacterial DNA repair systems (2). Moreover, these bacteria bear the rfa mutation, which causes a partial loss of the lipopolysaccharide barrier that coats the surface of the bacteria and increases permeability to large molecules (including some mutagens) that do not penetrate the normal cell wall (2). Some of these strains harbor, in addition, plasmid pKM101, which contains the mucA and mucB genes, responsible for the enhancement of an error-prone DNA repair system (14, 22, 25).
Although the Ames test is very useful for detecting mutagens under laboratory conditions, we considered that for monitoring of marine environments, an organism that naturally lives in these habitats should be used. Vibrio harveyi is a free-living bacterium found in diverse marine environments (18, 19). Moreover, it is easily cultivated under laboratory conditions and completely safe to work with as a nonpathogenic microbe. Therefore, we have chosen V. harveyi as an organism that could serve as a bioindicator of mutagenic pollution of marine environments. We aimed to genetically modify this bacterium to obtain a highly mutagenic strain that would allow the detection of low concentrations of mutagens. On the other hand, since we wanted to construct a bacterium that could be used as a potential bioindicator in marine habitats, we wanted to introduce as few genetic changes as possible to avoid obtaining bacteria unable to survive under natural environmental conditions.
MATERIALS AND METHODS
Bacterial strains, bacteriophage, and plasmids.
Bacterial strains are listed in Table 1. A thermoinducible bacteriophage, P1CMcrl100, has already been described (17). Plasmid pSUPTn5pMCS (12) bears a Tn5-derived transposon carrying a trimethoprim resistance gene. For construction of plasmid pAB91273, the SspI-ScaI fragment (containing the mucA and mucB genes) of plasmid pGW1700 (16) was inserted into the EcoRI site of plasmid pFF1 (7). The genetic engineering procedures used for the construction of the plasmid described above were performed as described by Sambrook et al. (20).
TABLE 1.
Bacterial strains
| Strain | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Escherichia coli | ||
| MG1655 | Wild type | 8 |
| S17-1 | pro hsdR recA, with integrated plasmid RP4 (Tc::Mu-Km::Tn7) | 12 |
| MC1061 | hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi | 15 |
| Salmonella enterica serovar Typhimurium | ||
| LT2 | Wild type | 21 |
| TA98 | hisD3052 rfa Δ(chl bio uvrB), bearing plasmid pKM101 | 13 |
| Vibrio harveyi | ||
| BB7 | Wild type | 3 |
| BB7X | As BB7, but bearing Tn5Tp inserted into the chromosome; UV sensitive | This work |
| BB7M | As BB7, but bearing plasmid pAB91273 | This work |
| BB7XM | As BB7X, but bearing plasmid pAB91273 | This work |
Culture media.
Luria-Bertani (LB) and BOSS media have already been described (10, 20). Minimal medium 3 (26) was used, but for V. harveyi cultivation, the concentration of NaCl was 3%. RGMC medium was described previously (23), but NaCl was added to a final concentration of 3%. Antibiotics were added (when necessary) to the following concentrations: amplicillin up to 50 μg/ml, chloramphenicol up to 35 μg/ml, trimethoprim up to 200 μg/ml, rifampin up to 50 μg/ml, and neomycin up to 50 μg/ml.
Bacterial conjugation.
Conjugation between Escherichia coli (donor) and V. harveyi (recipient) strains was performed by a previously described method (23).
UV sensitivity assays.
Bacteria were grown in LB (E. coli) or BOSS (V. harveyi) medium at 37°C (E. coli) or 30°C (V. harveyi) to an optical density at 575 nm (OD575) of 0.3. The culture was centrifuged (2,000 × g, 10 min), and the bacterial pellet was suspended in an equal volume of minimal medium salts (27) (E. coli) or the same salts but containing 3% NaCl (V. harveyi). Five milliliters of the suspension was transferred to a petri dish and UV irradiated. Bacteria were titrated on LB (E. coli) or BOSS (V. harveyi) agar plates at 37°C (E. coli) or 30°C (V. harveyi), and the fractions of survivors were calculated. For the agar plate test, bacteria were streaked across the LB (E. coli) or BOSS (V. harveyi) plate, and sectors of the plate were irradiated with different UV doses. The plate was incubated overnight at 37°C (E. coli) or 30°C (V. harveyi), and growth inhibition was estimated.
Crystal violet sensitivity assays.
Serial dilutions of the bacterial culture grown in LB (Salmonella serovar Typhimurium) or BOSS (V. harveyi) medium (to an OD575 of 0.3) were spread on LB (Salmonella serovar Typhimurium) or BOSS (V. harveyi) plates containing no crystal violet or different concentrations of this reagent. The plates were incubated overnight, and the fractions of survivors were calculated.
Transposon mutagenesis of V. harveyi.
E. coli S17-1 bearing plasmid pSUPTn5pMCS was lysogenized with phage P1CMcrl100 as described previously (17). The lysogenic strain was cultivated for 6 days in LB medium at 30°C, with 1:100 dilution into fresh medium every day. Following thermal induction of the prophage (17), the phage lysate was used for lysogenization of E. coli MC1061 with selection for trimethoprim resistance (200 μg/ml). A strain containing the P1CMcrl100Tn5Tp prophage was then used for propagation of the phage after thermal induction. The phage lysate was used for the transduction of V. harveyi BB7 (with selection for trimethoprim resistance as described above) by a previously described procedure (17) but without the addition of CaCl2, as we found that this reagent caused problems with V. harveyi growth. Since phage P1 can adsorb to V. harveyi cells but is not able to replicate in this bacterium, we considered that most of the trimethoprim-resistant cells contained Tn5Tp integrated into the host chromosome due to its transposition from phage DNA. From each independent experiment, only one V. harveyi mutant was chosen for further analysis.
Mutagenicity tests.
For the plate tests, 4 × 106 V. harveyi cells grown in BOSS medium to mid-log phase (OD575, 0.2) were spread on BOSS plates containing various amounts of mutagens. Plates were incubated for 48 h at 30°C, and colonies were counted. For the liquid medium tests, bacteria were grown in BOSS medium (short-term test) or minimal medium 3 containing 3% NaCl (long-term test) in the presence of different amounts of mutagens for various times (the experiments were started when bacterial cultures reached an OD575 of 0.1). Bacteria were titrated on BOSS plates and BOSS plates with neomycin (final concentration, 50 μg/ml), and the fraction of neomycin-resistant mutants was calculated.
RESULTS
UV sensitivity of V. harveyi.
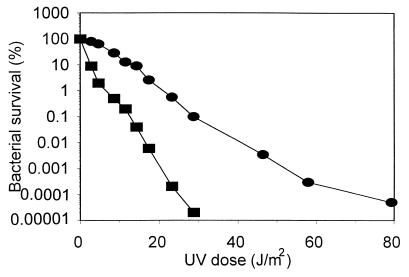
Organisms used as bioindicators of mutagenic pollution should be relatively sensitive to mutagens. Therefore, we tested the UV sensitivity of wild-type V. harveyi BB7. We found that this bacterium is significantly more sensitive to UV irradiation than wild-type E. coli MG1655 (Fig. 1). Therefore, we considered that V. harveyi BB7 can be used for further work on the construction of the bioindicator strains.
FIG. 1.
UV sensitivity of E. coli MG1655 (circles) and V. harveyi BB7 (squares).
Sensitivity of V. harveyi to crystal violet.
Some mutagens cannot significantly enhance a number of bacterial mutants, as they are not able to enter the cell through the envelope of certain bacteria, including Salmonella serovar Typhimurium (2). In the Ames test, this problem has been overcome by using rfa mutants of Salmonella serovar Typhimurium, which have changes in lipopolysaccharide that allow the penetration of many mutagens into the cells (2, 13).
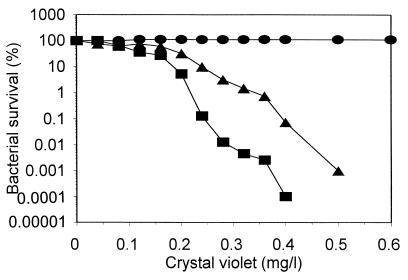
Crystal violet is one of the reagents inhibiting bacterial grow only when it can enter cells efficiently. Therefore, crystal violet can be used in tests for bacterial cell envelope permeability by simply measuring the sensitivity of cells to it (2, 13). Wild-type Salmonella serovar Typhimurium is resistant to crystal violet (Fig. 2) because this reagent cannot penetrate the cells (13), while the survival of rfa mutants in the presence of crystal violet is considerably lower (Fig. 2) because the mutation permits large molecules (such as crystal violet) to enter and kill the bacteria (13). We found that the sensitivity of V. harveyi BB7 to crystal violet is even higher than that of the Salmonella serovar Typhimurium rfa mutant used in the Ames test (Fig. 2). These results indicate that for using V. harveyi in mutagenicity tests, there is no need for isolation or construction of rfa or other mutations affecting lipopolysaccharide structure.
FIG. 2.
Sensitivity to crystal violet of Salmonella serovar Typhimurium LT2 (circles) and TA98 (triangles) and V. harveyi BB7 (squares).
Isolation of V. harveyi mutants very sensitive to UV irradiation.
The results presented above indicated that V. harveyi may be a suitable organism for the bioindication of mutagens. Therefore, we aimed to construct a modified strain of this bacterium which would possess even more features useful in biological tests for the presence of mutagens in marine environments. The first step was to isolate a mutant very sensitive to UV irradiation. We performed transposon mutagenesis and isolated over 80 independent mutants (only 1 mutant was taken from each mutagenesis experiment). Among these mutants, six were very sensitive to UV irradiation, as tested by the plate assay (growth of the wild-type V. harveyi strain was still observed at a UV dose of 15 J/m2, whereas complete inhibition of growth of the six mutants was observed upon UV irradiation at a dose as low as 7 J/m2). However, we found that of these six mutants, five had simultaneously lost the ability to luminesce. Because of this additional phenotype, for further studies we chose the mutant that was luminescent and very sensitive to UV irradiation. We named this strain BB7X.
Construction of V. harveyi strains bearing a plasmid expressing the mucA and mucB genes.
Apart from isolating appropriate mutants, enhanced mutagenesis in bacteria may be achieved by introducing mucA and mucB genes, originally present in plasmid R46 and in its derivative, pKM101 (16). Expression of these genes causes an increase in the efficiency of an error-prone DNA repair system. We have constructed plasmid pAB91273, bearing the origin of replication (oriV) from plasmid RK2, the oriT region from RK2, the mucA and mucB genes, and ampicillin resistance and chloramphenicol resistance genes. Following transformation of E. coli, this plasmid was introduced into V. harveyi strains by conjugation and was found to be stably maintained in these strains (data not shown). We named strains BB7 and BB7X bearing plasmid pAB91273 BB7M and BB7XM, respectively.
Positive selection for V. harveyi mutants.
In order to use V. harveyi strains as bioindicators of mutagens, we had to develop a system for the positive selection of mutants. V. harveyi is sensitive to neomycin, but we found that neomycin-resistant mutants appear spontaneously at a low frequency (about 10−5 to 10−4). This frequency is nevertheless higher than that of the appearance of specific substitution mutants (see, for instance, reference 13). This fact is due to the nature of neomycin action. Neomycin is an aminoglycoside antibiotic that interferes with decoding at site A at the ribosome during translation (5). Resistance to this antibiotic occurs as a result of various rRNA modifications in the decoding site (5).
The addition of a mutagen to V. harveyi cultures resulted in the appearance of a considerably increased number of neomycin-resistant mutants relative to those obtained under normal growth conditions (see below). Therefore, we decided to use this assay in our further tests.
Mutagenicity tests using V. harveyi strains.
The wild-type V. harveyi strain (BB7), its UV-sensitive mutant derivative (BB7X), and strains bearing plasmid pAB91273 (BB7M and BB7XM) were assayed in mutagenicity tests. We used several mutagens previously used in classical mutagenicity tests (13).
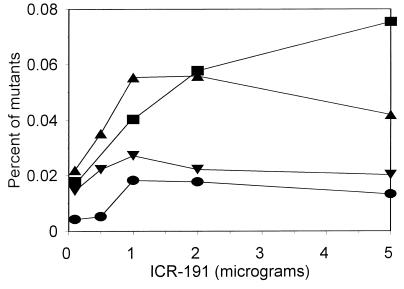
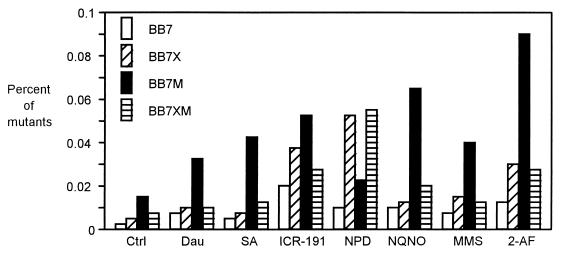
In the first type of experiment, we followed the procedure used in the Ames test (13); i.e., dilutions of a bacterial culture were spread on an agar plate containing a mutagen, and after 48 h of incubation the number and percentage of neomycin-resistant mutants were determined. An example of the dose-response effects of mutagens is presented in Fig. 3, and the results are summarized in Fig. 4. We found that even wild-type V. harveyi produced an increased number of neomycin-resistant mutants on plates with tested mutagens. Generally, more mutants appeared when strain BB7X was tested. However, the best results were obtained with strains BB7M and BB7XM; BB7M responded especially effectively to practically all tested mutagens. Different numbers of spontaneous neomycin-resistant mutants appeared (the lowest number for BB7 and the highest number for BB7M), but the numbers of mutagen-induced neomycin-resistant colonies were significantly higher.
FIG. 3.
Dose-response effects of 2-methoxy-6-chloro-9-(3-(2-chloroethyl)aminopropylamino)acridine · 2HCl (ICR-191) on V. harveyi BB7 (circles), BB7X (squares), BB7M (triangles), and BB7XM (inverted triangles).
FIG. 4.
Agar plate mutagenicity test with V. harveyi BB7, BB7X, BB7M, and BB7XM. V. harveyi cells (4 × 106) grown in BOSS medium were spread on BOSS plates containing mutagens; after incubation for 48 h at 30°C, the number of neomycin-resistant mutants was counted and the percentage of mutants was calculated. Ctrl, control experiments (without any mutagen). The following amounts of mutagens were added per plate: daunomycin (Dau), 6 μg/ml; sodium azide (SA), 1.5 μg/ml; 2-methoxy-6-chloro-9-(3-(2-chloroethyl)aminopropylamino)acridine · 2HCl (ICR-191), 1 μg/ml; 4-nitro-o-phenylenediamine (NPD), 20 μg/ml; 4-nitroquinolone-N-oxide (NQNO), 0.5 μg/ml; methyl methanesulfonate (MMS), 1 μg/ml; and 2-aminofluorene (2-AF), 10 μg/ml. The results presented are mean values from two independent experiments.
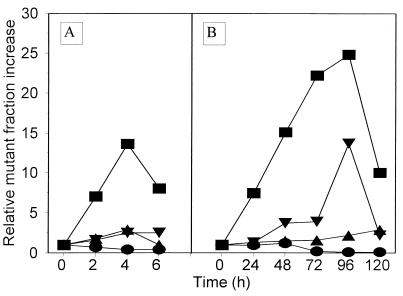
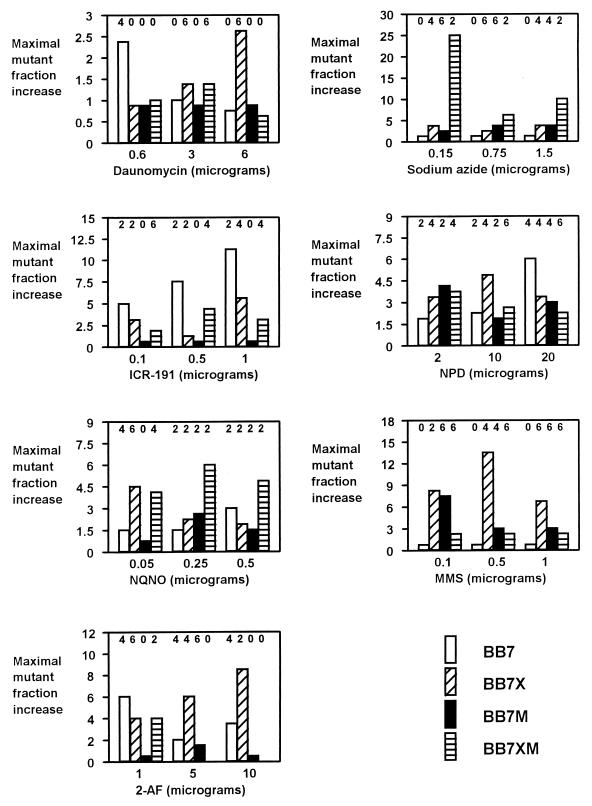
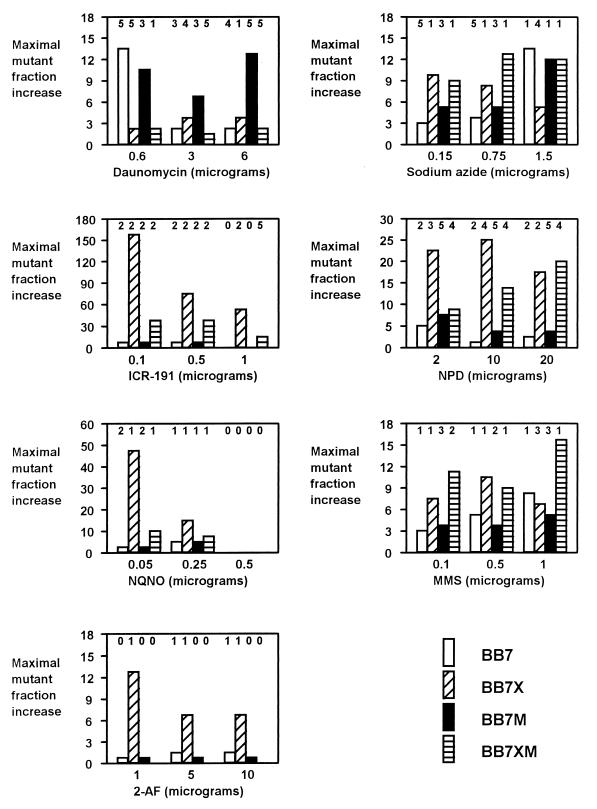
The above-described tests indicated that V. harveyi strains may be useful test strains for detecting mutagens. However, for using these bacteria as bioindicators in natural habitats, a significant number of mutants should also appear upon cultivation of the test strains in liquid media containing small amounts of mutagens. Therefore, we cultivated bacteria in rich medium for several hours or in minimal medium for several days in the presence of mutagens and determined the fractions of neomycin-resistant mutants among all living cells at different times of incubation. Since bacteria were growing continuously under these conditions (except with concentrations of certain mutagens that were toxic for bacteria), an increase in the fraction of mutants rather than the percentage of mutants (calculated on the basis of the actual number of mutants) was considered, contrary to the method used in the agar plate mutagenicity test. The kinetics of increase of the mutant fraction during cultivation of bacteria in the presence of mutagens are shown in Fig. 5, and the results are summarized in Fig. 6 and 7. The results show that practically each tested mutagen at each tested amount caused a significant increase in the mutant fraction (at least severalfold) of at least one of the test strains. The results obtained with minimal medium (Fig. 7) are important, as the test conditions resemble the conditions found in natural environments. In fact, the response of the test strains to mutagens in minimal medium (Fig. 7) was generally more pronounced than that in rich medium (Fig. 6).
FIG. 5.
Increase in the neomycin-resistant mutant fraction during cultivation of V. harveyi BB7 (circles), BB7X (squares), BB7M (triangles), and BB7XM (inverted triangles) in BOSS medium (A) and in minimal medium 3 containing 3% NaCl (B) in the presence of 0.5 μg of methyl methanesulfonate (A) and 10 μg of 4-nitro-o-phenylenediamine (B).
FIG. 6.
Liquid rich medium mutagenicity test with V. harveyi BB7, BB7X, BB7M, and BB7XM. BOSS medium was used. Abbreviations: ICR-191, 2-methoxy-6-chloro-9-(3-(2-chloroethyl)aminopropylamino)acridine · 2HCl; NPD, 4-nitro-o-phenylenediamine; NQNO, 4-nitroquinolone-N-oxide; MMS, methyl methanesulfonate; 2-AF, 2-aminofluorene. Amounts of mutagens were per 25 ml of liquid culture. The fraction of neomycin-resistant mutants was calculated by dividing the number of neomycin-resistant cells by the number of all living cells per milliliter of culture. An increase in the fraction of neomycin-resistant mutants was calculated by dividing the value of the fraction of neomycin-resistant mutants at a particular time after the addition of a mutagen by the value of the fraction of neomycin-resistant mutants before the addition of the mutagen. The values of the fraction of neomycin-resistant mutants before the addition of a mutagen ranged from 10−5 to 10−4. Times after the addition of mutagens (in hours) at which maximal increases in the fractions of neomycin-resistant mutants were observed are indicated above columns (this item concerns only experiments in which the increase in values was greater than 1); 0 denotes no increase in the fraction of neomycin-resistant mutants due to either a lack of strain response or toxicity of the mutagen for the strain at a particular concentration.
FIG. 7.
Liquid minimal medium mutagenicity test with V. harveyi BB7, BB7X, BB7M, and BB7XM. Minimal medium 3 containing 3% NaCl was used. Abbreviations: ICR-191, 2-methoxy-6-chloro-9-(3-(2-chloroethyl)aminopropylamino)acridine · 2HCl; NPD, 4-nitro-o-phenylenediamine; NQNO, 4-nitroquinolone-N-oxide; MMS, methyl methanesulfonate; 2-AF, 2-aminofluorene. Amounts of mutagens were per 25 ml of liquid culture. The fraction of neomycin-resistant mutants was calculated by dividing the number of neomycin-resistant cells by the number of all living cells per milliliter of culture. An increase in the fraction of neomycin-resistant mutants was calculated by dividing the value of the fraction of neomycin-resistant mutants at a particular time after the addition of a mutagen by the value of the fraction of neomycin-resistant mutants before the addition of the mutagen. The values of the fraction of neomycin-resistant mutants before the addition of a mutagen ranged from 10−5 to 10−4. Times after the addition of mutagens (in days) at which maximal increases in the fractions of neomycin-resistant mutants were observed are indicated above columns (this item concerns only experiments in which the increase in values was greater than 1); 0 denotes no increase in the fraction of neomycin-resistant mutants due to either a lack of strain response or toxicity of the mutagen for the strain at a particular concentration.
DISCUSSION
We found that V. harveyi possesses several features making it a potential bioindicator of mutagenic pollution of marine environments. These features are as follows: (i) it is a free-living bacterium found in diverse marine environments (18, 19); (ii) it can be easily cultivated under laboratory conditions; (iii) it is a nonpathogenic microbe and thus is completely safe to work with; and (iv) neomycin-resistant mutants can be easily isolated, and their frequency increases in the presence of mutagens (Fig. 3 to 7). Genetic modifications of the wild-type V. harveyi strain by isolation of a transposon mutant very sensitive to UV irradiation and introduction of a plasmid bearing mucA and mucB genes led to the construction of a series of strains that can be used in mutagenicity tests.
Different strains produced the largest numbers of mutants in response to various kinds of mutagens (Fig. 4, 6, and 7); thus, we recommend the use of a set of strains for the detection of an unknown mutagen rather than only one strain, as in the Ames test (13). Nevertheless, strain BB7M responded to all tested mutagens in the plate mutagenicity assay (Fig. 4) and seems to be the most general test strain, since all previously described Salmonella serovar Typhimurium strains responded to a significantly more narrow spectrum of mutagens (13). Like strain BB7M in the plate assay, strain BB7X responded to all tested mutagens in the liquid medium assay (Fig. 6 and 7); thus, it may also be considered a general test strain.
It is intriguing that strains BB7M and BB7X often showed a higher percentage of mutants (Fig. 4) or a more significant increase in the fraction of mutants (Fig. 6 and 7) than strain BB7XM. One might expect that BB7XM should be more mutagenic than other test strains used in this work because it bears both a mutation causing high UV sensitivity and a plasmid expressing mucA and mucB genes. On the other hand, the very high mutagenicity of BB7XM may cause the appearance of lethal mutations at a rate significantly higher than that occurring in other strains. Thus, the viability of BB7XM in the presence of mutagens (especially at higher concentrations) is considerably decreased, and many potential neomycin-resistant mutants may not be detected due to the simultaneous appearance of lethal mutations in the same cells. According to this prediction, the best results with strain BB7XM were often observed at lower mutagen concentrations (Fig. 6 and 7). Therefore, it seems that BB7XM may be particularly useful in tests where very small amounts of mutagen(s) occur.
The sensitivity of the mutagenicity tests described in this report is at least comparable to (if not higher than) that obtained in the Ames test, which uses Salmonella serovar Typhimurium mutant strains (compare Fig. 4 in this article with Table 4 in reference (13)). However, note that in the Ames test, 1 × 108 to 2 × 108 bacteria are used per plate (13); in our test, we spread 4 × 106 cells per plate and obtained similar numbers of mutants (compare Fig. 4 in this article with Table 4 in reference (13)). A relatively high fraction of V. harveyi mutants appearing in the presence of mutagens was confirmed in the second type of mutagenicity test, in which bacteria were cultivated in liquid media (Fig. 6 and 7). The possibility of using a relatively small number of bacterial cells should be important for the bioindication of mutagenic pollution of marine environments, as it might be difficult to obtain a very high density of test bacteria in natural habitats, even assuming their immobilization on a carrier surface or trapping with a vessel permeable for mutagenic molecules but not for bacterial cells.
The use of bioluminescent bacteria for the detection of toxic chemicals was described (4), and V. harveyi was used as a test organism in toxicity studies (11, 24). In those studies, the toxicity of different chemicals was determined by a relatively simple method based on measuring changes in bioluminescence. However, toxicity assays should be distinguished from mutagenicity tests. Toxic agents are not always mutagens, and mutagenic chemicals are often toxic only at relatively high concentrations. Therefore, although the previously described bioluminescence assays (4, 11, 24) are simple, they can be useful for the detection of toxic substances rather than mutagens that occur in natural habitats in concentrations too low to provoke serious toxic effects in bacterial cells. Recently, a system for the detection of mutagenic repair in E. coli, based on a fusion plasmid containing the E. coli umuC gene and V. harveyi luxAB genes, has been developed (9). However, like Salmonella serovar Typhimurium used in the Ames test, E. coli is not a marine bacterium; thus, it may be successfully used in laboratory conditions rather than in mutagenicity assays for natural marine environments.
In conclusion, here we describe the use of V. harveyi strains as bioindicators of the presence of mutagens at low concentrations. The assays proposed in this report are very simple, as they are based on positive selection of bacterial mutants (neomycin-resistant mutants) on agar plates. The most important advantages of the mutagenicity tests described in this report are that (i) low concentrations of mutagens can be detected; (ii) V. harveyi is a marine bacterium, so that the detection of mutagens present in natural marine environments is possible; and (iii) unlike Salmonella serovar Typhimurium, V. harveyi is a nonpathogenic bacterium and completely safe to work with. The last item could also be valuable for classification schemes for biohazardous organisms.
ACKNOWLEDGMENT
This work was supported by the Polish State Committee for Scientific Research (KBN; project 6 P04C 100 12).
REFERENCES
- 1.Ames B N. The detection of chemical mutagens with enteric bacteria. In: Hollaender A, editor. Chemical mutagens, principles and methods for their detection. New York, N.Y: Plenum Press; 1971. pp. 267–282. [Google Scholar]
- 2.Ames B N, Lee F D, Durston W E. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc Natl Acad Sci USA. 1973;70:782–786. doi: 10.1073/pnas.70.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belas R, Mileham A, Cohn D, Hilmen M, Simon M, Silverman M. Bacterial luminescence: isolation and expression of the luciferase genes from Vibrio harveyi. Science. 1982;218:791–793. doi: 10.1126/science.10636771. [DOI] [PubMed] [Google Scholar]
- 4.Bulich A A, Isenberg D L. Use of the luminescent bacterial system for the rapid assessment of aquatic toxicity. ISA Trans. 1981;20:29–33. [PubMed] [Google Scholar]
- 5.Dahlberg A E. The functional role of ribosomal RNA in protein synthsis. Cell. 1989;57:525–529. doi: 10.1016/0092-8674(89)90122-0. [DOI] [PubMed] [Google Scholar]
- 6.Davey K. EPA's strategy for priority persistent, bioaccumulative, and toxic (PBT) pollutants. In: McDonald G, Sheridan D, Wigginton M, editors. Chemicals in our community. Vol. 2. Washington, D.C.: Office of Pollution Prevention and Toxics, U.S. Environmental Protection Agency; 1999. p. 9. [Google Scholar]
- 7.Durland R H, Toukdarian A, Fang F, Helinski D R. Mutations in the trfA replication gene of the broad-host-range plasmid RK2 result in elevated plasmid copy number. J Bacteriol. 1990;172:3859–3867. doi: 10.1128/jb.172.7.3859-3867.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen K F. The Escherichia coli “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Justus T, Thomas S M. Construction of a umuC′-luxAB plasmid for the detection of mutagenic DNA repair via luminescence. Mutat Res. 1998;398:131–141. doi: 10.1016/s0027-5107(97)00215-7. [DOI] [PubMed] [Google Scholar]
- 10.Klein G, Żmijewski M, Krzewska J, Czeczatka M, Lipińska B. Cloning and characterization of the dnaK heat shock operon of the marine bacterium Vibrio harveyi. Mol Gen Genet. 1998;259:179–189. doi: 10.1007/s004380050803. [DOI] [PubMed] [Google Scholar]
- 11.Lange J H, Thomulka K W. Use of the Vibrio harveyi toxicity test for evaluating mixture interactions of nitrobenzene and dinitrobenzene. Ecotoxicol Environ Safety. 1997;38:2–12. doi: 10.1006/eesa.1997.1546. [DOI] [PubMed] [Google Scholar]
- 12.MacKenzie C, Chidambaram M, Sodergren E J, Kaplan S, Weinstock G M. DNA repair mutants of Rhodobacter sphaeroides. J Bacteriol. 1995;177:3027–3035. doi: 10.1128/jb.177.11.3027-3035.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maron D M, Ames B N. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 14.McCann J, Springarn N E, Kobori J, Ames B N. Detection of carcinogens and mutagens: bacterial tester strains with R factor plasmids. Proc Natl Acad Sci USA. 1975;72:979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meissner P S, Sisk W P, Berman M L. Bacteriophage λ cloning system for the construction of directional cDNA libraries. Proc Natl Acad Sci USA. 1987;84:4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry K L, Walker G C. Identification of plasmid (pKM101)-coded proteins involved in mutagenesis and UV resistance. Nature. 1982;300:278–281. doi: 10.1038/300278a0. [DOI] [PubMed] [Google Scholar]
- 17.Rosner J L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972;49:679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- 18.Ruby E G, Morin J G. Luminous enteric bacteria of marine fishes: a study of their distribution, densities, and dispersion. Appl Environ Microbiol. 1979;38:406–411. doi: 10.1128/aem.38.3.406-411.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruby E G, Greenberg E P, Hastings J W. Planktonic marine luminous bacteria: species distribution in the water column. Appl Environ Microbiol. 1980;39:302–306. doi: 10.1128/aem.39.2.302-306.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Sanderson K E, Hessel A, Stocker B A D. Strains of Salmonella typhimurium and other Salmonella species used in genetic analysis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 2496–2503. [Google Scholar]
- 22.Shanabruch W G, Walker G C. Localization of the plasmid (pKM101) gene(s) involved in recA+ lexA+-dependent mutagenesis. Mol Gen Genet. 1980;179:289–297. doi: 10.1007/BF00425456. [DOI] [PubMed] [Google Scholar]
- 23.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 24.Thomulka K W, Lange J H. A mixture toxicity study employing combinations of tributyltin chloride, dibutyltin dichloride, and tin chloride using the marine bacterium Vibrio harveyi as the test organism. Ecotoxicol Environ Safety. 1996;34:76–84. doi: 10.1006/eesa.1996.0047. [DOI] [PubMed] [Google Scholar]
- 25.Walker G C, Dobson P P. Mutagenesis and repair deficiencies of Escherichia coli umuC mutants are suppressed by the plasmid pKM101. Mol Gen Genet. 1979;172:17–24. doi: 10.1007/BF00276210. [DOI] [PubMed] [Google Scholar]
- 26.Węgrzyn G, Taylor K. Inheritance of the replication complex by one of two daughter copies during λ plasmid replication in Escherichia coli. J Mol Biol. 1992;226:681–688. doi: 10.1016/0022-2836(92)90625-t. [DOI] [PubMed] [Google Scholar]
- 27.Węgrzyn G, Neubauer P, Krueger S, Hecker M, Taylor K. Stringent control of replication of plasmids derived from coliphage λ. Mol Gen Genet. 1991;225:94–98. doi: 10.1007/BF00282646. [DOI] [PubMed] [Google Scholar]