Abstract
Background
Risk of tumors of the breast, ovary, and meninges has been associated with hormonal factors and with one another. Genome-wide association studies (GWAS) identified a meningioma risk locus on 10p12 near previous GWAS hits for breast and ovarian cancers, raising the possibility of genetic pleiotropy.
Methods
We performed imputation-based fine-mapping in three case-control datasets of meningioma (927 cases, 790 controls), female breast cancer (28 108 cases, 22 209 controls), and ovarian cancer (25 509 cases, 40 941 controls). Analyses were stratified by sex (meningioma), estrogen receptor (ER) status (breast), and histotype (ovarian), then combined using subset-based meta-analysis in ASSET. Lead variants were assessed for association with additional traits in UK Biobank to identify potential effect-mediators.
Results
Two-sided subset-based meta-analysis identified rs7084454, an expression quantitative trait locus (eQTL) near the MLLT10 promoter, as lead variant (5.7 × 10–14). The minor allele was associated with increased risk of meningioma in females (odds ratio (OR) = 1.42, 95% Confidence Interval (95%CI):1.20–1.69), but not males (OR = 1.19, 95%CI: 0.91–1.57). It was positively associated with ovarian (OR = 1.09, 95%CI:1.06–1.12) and ER+ breast (OR = 1.05, 95%CI: 1.02–1.08) cancers, and negatively associated with ER– breast cancer (OR = 0.91, 95%CI: 0.86–0.96). It was also associated with several adiposity traits (P < 5.0 × 10–8), but adjusting for body mass index did not attenuate its association with meningioma. MLLT10 and ESR1 expression were positively correlated in normal meninges (P = .058) and meningioma tumors (P = .0065).
Conclusions
We identify a MLLT10 eQTL positively associated with risk of female meningioma, ER+ breast cancer, ovarian cancer, and obesity, and implicate a potential estrogenic mechanism underlying this pleiotropy.
Keywords: breast cancer, meningioma, MLLT10, ovarian cancer, pleiotropy
Key Points.
Risk of tumors of the breast, ovary, and meninges have been associated with hormonal factors and with one another.
We identify a MLLT10 eQTL positively associated with risk of female meningioma, ER+ breast cancer, ovarian cancer, and obesity, and implicate a potential estrogenic mechanism underlying this pleiotropy.
Importance of the Study.
Risk of tumors of the breast, ovary, and meninges have been associated with hormonal factors and with one another but the exact nature of this relationship remains unclear. We performed imputation-based fine-mapping in three case-control datasets of meningioma (927 cases, 790 controls), female breast cancer (28 108 cases, 22 209 controls), and ovarian cancer (25 509 cases, 40 941 controls). Analyses were stratified by sex (meningioma), estrogen receptor (ER) status (breast), and histotype (ovarian), then combined using subset-based meta-analysis in ASSET. Lead variants were assessed for association with additional traits in UK Biobank to identify potential effect-mediators. We identify a MLLT10 eQTL positively associated with risk of female meningioma, ER+ breast cancer, ovarian cancer, and obesity, and implicate a potential estrogenic mechanism underlying this pleiotropy.
Meningioma is the most common primary brain tumor, accounting for approximately 36% of all intracranial tumors and occurring in approximately 1% of individuals.1,2 Females have a two-fold to three-fold increased risk of meningioma relative to men, with the female-to-male risk-ratio peaking prior to menopause and decreasing thereafter.3 The higher incidence in women, as well as associations with additional hormonal factors,3–14 have led to the speculation that hormonal factors are associated with meningioma risk. Similar to the associations with meningioma, tumors of the breast and ovary have been widely associated with endogenous and exogenous hormonal factors.15,16 Breast cancers are classified according to estrogen receptor (ER) and progesterone receptor (PR) status, with treatment and outcome varying across receptor subtype.17
Family history of meningioma in a first-degree relative is associated with a 2-fold increased risk, implicating genetic susceptibility in meningioma etiology18; similar familial risks have been reported for breast19 and ovarian cancer.20 Although the results are not entirely consistent, elevated associations between meningioma risk and a personal history of breast9,18 or ovarian cancer,18 as well as a family history of breast or ovarian cancer have been reported18 and suggest possible shared genetic or environmental risk factors across tumor type. Recent genome-wide association studies (GWAS) have identified common variation in a region of chromosome 10 near MLLT10 as a meningioma risk locus.21–23 Interestingly, this region has also been implicated as a risk locus for both breast24,25 and ovarian26 cancers by previous GWAS. However, the lead SNPs from each study differ and it remains unclear whether specific MLLT10 variants may display genetic pleiotropy—conferring risk of diverse tumor types possibly via a common mediating factor—including those with a hormonal component.
To elucidate the role of MLLT10 variation in contributing to risk of meningioma, breast cancer, and ovarian cancer and to examine potential mediating factors, we performed imputation-based fine-mapping across a 1.1 Mb region of chromosome 10p12 in three case-control datasets from 1) the US-based Meningioma Consortium Case-Control Study,22 2) the DRIVE Oncoarray Breast Cancer case-control study and 3) the Ovarian Cancer Association Consortium (OCAC). Datasets were analyzed individually with results combined using traditional meta-analysis as well as association analysis based on subsets (ASSET) approaches. Lead variants from the integrated analyses were queried for association with more than 700 traits using a phenome-wide association study (PheWAS)27 approach to identify potential intermediate phenotypes that could mediate the association between MLLT10 variants and risk of diverse neoplasms.
Methods
Meningioma Case-Control Subjects
The US meningioma case-control dataset (Meningioma Consortium) comprised 927 cases (661 female) and 790 controls (572 female) diagnosed between the ages of 20 and 79.22 Case patients eligible for the study included all persons diagnosed between 2006 and 2013 with a histologically confirmed intracranial meningioma among residents of the states of California, Connecticut, Massachusetts, North Carolina, and Texas. Controls were obtained through random-digit dialing and were frequency matched with case patients by 5-year age interval, sex, and state of residence. Participants were genotyped using the Affymetrix Axiom EUR array with quality control as described previously.22
Breast Cancer Case-Control Subjects
The Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) project was initiated in 2010 as part of the National Cancer Institute’s (NCI) Genetic Associations and Mechanisms in Oncology (GAME-ON) initiative to translate promising research leads derived from cancer GWAS. The DRIVE data included here are 28108 female breast cancer cases and 22209 female controls. Estrogen but not progesterone receptor data are available for these subjects. Participants in DRIVE underwent genotyping on the Illumina Oncoarray.28 Data were downloaded from dbGaP Study Accession phs001265.v1.p1.
Ovarian Cancer Case-Control Subjects
OCAC genotype data from the combined iCOGS and Oncoarray GWAS meta-analyses28 were used for these analyses. The OCAC data comprise 63 case-control sets that have been previously described in detail.29 SNP QC was carried out according to OncoArray Guidelines.28 Following quality-control filters and limiting to subjects of European-ancestry, 40941 controls and 25509 cases (22406 invasive and 3103 borderline) were included.
Targeted Genotype Imputation
We performed imputation separately for each genotyping project data set. We imputed genotypes into the reference panel from the 1000 Genomes Project v3 (October 2014).30 We imputed a 1.1Mb region of chromosome 10p12 from 21.515 Mb to 22.615 Mb using single-step imputation, without prephasing, using the IMPUTE2 software (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html). Imputation employed 90 MCMC iterations (15 used as burn-in), a buffer region of 500kb on either side of the region boundaries, and 100 haplotypes used as templates when phasing observed genotypes. SNPs were excluded from association analyses if their imputation accuracy (INFO) score was <0.60 or their minor allele frequency among cases was <0.01.
Association Analyses
The association between genotype and disease was evaluated using the imputed genotype dosage in logistic regression models, separately for meningioma, breast cancer, and ovarian cancer case-control datasets. Regression was performed with SNPTESTv2 (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html), under an allelic
additive model, using probabilistic genotype dosages (ie, method expected). All analyses were adjusted for population substructure by including the eigenvectors of project-specific principal components as covariates (range 2–10 PCs). Meningioma case-control analyses were adjusted for subject sex, and in mediation analyses were also adjusted for BMI. Breast cancer case-control analyses were stratified by ER status (dbgaP variable accession phv00292705.v1.p1) and additional case-only analyses were performed comparing ER+ breast cancer patients to ER– breast cancer patients. For OCAC data, case-control comparisons were also adjusted for project/study and stratified by histotype.
Traditional and Subset-Based Meta-Analyses
Meta-analysis was performed using ASSET,31 a suite of statistical tools specifically designed for pooling association signals across multiple studies when true effects may exist in only a subset of these studies, potentially with allelic effects in opposite directions across traits. Final test-statistics are obtained by maximizing the subset-specific test-statistics over all possible subsets and after adjustment for multiple testing. The method returns a P-value for significance for the overall evidence of association at a SNP using traditional fixed-effects meta-analyses approaches, and also outputs the “best subset” containing only the studies that contribute to an overall association signal.
In addition to standard fixed-effects meta-analysis, a one-sided subset search was performed with ASSET, maximizing the standard fixed-effect meta-analysis test-statistics over all possible subsets to detect the best possible association signals. The P-value returned for the maximum test-statistics penalizes for multiple testing due to subset search and can be interpreted as evidence of an overall association for the SNP across the k subsets, identifying only those subsets that have associations in the same direction (ie, same risk allele). A two-sided subset search was also performed with ASSET, which applies the one-side subset search separately for positively and negatively associated traits at a given SNP and then combines the association signals from two directions into a single combined Χ 2-type statistic. The method is sensitive in detecting alleles that may be associated with different traits in different directions. By performing ASSET-based meta-analysis, candidate “causal” SNPs can be prioritized even if alleles have opposite directions of effect across tumor type.
Meta-analyses, including subset-based meta-analyses, were performed with three different groupings of subsets. Grouping 1 included three subsets: all breast cancer cases versus breast cancer controls, ovarian cancer cases versus ovarian cancer controls, and meningioma cases versus meningioma controls. Grouping 2 included four subsets: ER+ breast cancer cases versus breast cancer controls, ER– breast cancer cases versus breast cancer controls, ovarian cancer cases versus ovarian cancer controls, and meningioma cases versus meningioma controls. ASSET accounts for the use of shared controls across these two breast cancer subsets when determining statistical significance. Grouping 3 included three subsets: ER+ breast cancer cases versus ER– breast cancer cases, ovarian cancer cases versus ovarian cancer controls, and meningioma cases versus meningioma controls. Further sensitivity analyses were performed using additional subsets, including: female meningioma cases versus female meningioma controls, male meningioma cases versus male meningioma controls, and five ovarian cancer histotypes (low grade serous, high grade serous, clear cell, mucinous, and endometrioid) compared to the same pooled set of ovarian cancer controls, accounting for the use of shared controls in ASSET.
In-Silico Analysis of SNP Regulatory Effects
We investigated the functional implications of lead 10p12 variants using HaploReg,32 RegulomeDB (http://regulomedb.org/), the UCSC Genome Browser (https://genome.ucsc.edu/) and the Epigenome Browser (https://epigenomegateway.wustl.edu/). We assessed whether SNPs were expression quantitative trait loci (eQTL) using the Genotype-Tissue Expression (GTEx) Project (https://www.gtexportal.org/home/).
UK Biobank GeneATLAS and PheWAS Analyses
The atlas of genetic associations from the UK Biobank (GeneATLAS; http://geneatlas.roslin.ed.au.uk/) was constructed by genotyping roughly 450 000 European-ancestry individuals for 805 426 genetic variants, performing genome-wide SNP imputation and quality-control, then linking the genetic data to electronic health record data (https://www.ukbiobank.ac.uk/). GeneATLAS contains data for 778 traits (118 quantitative, 660 binary) and their association with 9 113 133 genetic variants (genotyped and imputed after quality-control) across 452 264 individuals.33 We queried the GeneATLAS database for lead MLLT10 SNPs associated with pleiotropic risk of meningioma, breast cancer, and ovarian cancer to identify any associations of these SNPs with the 778 phenotypes included in the database in order to detect pleiotropic associations with additional traits, or to identify potential intermediate phenotypes that may mediate the association between the SNP and risk of these neoplasms.34
Gene Expression in Normal Meninges and Meningioma Tumors
RNA expression was measured in 16 adult meninges and 96 grade I meningiomas using the Illumina HumanHT-12 V4.0 expression beadchip, as previously described (https://www.ncbi.nlm.nih.gov/geo/).35 Transcripts of interest included MLLT10 (ILMN_1743538), C10orf114 (ILMN_2159300), ESR1 (ILMN_1678535), and ESR2 (ILMN_1740045), and correlations were calculated with adjustment for batch.
Results
Analyses included 927 meningioma patients (71% female) and 790 meningioma controls (72% female), 28 108 female breast cancer patients (69% ER+, 11% ER–, 19% unknown), and 22 209 female breast cancer controls, as well as 25 509 ovarian cancer patients and 40 941 ovarian cancer controls. Ovarian histotypes are 1954 (7.7%) serous borderline, 1149 (4.5%) mucinous borderline, 1012 (4%) low grade serous, 13 037 (51.1%) high grade serous, 2810 (11%) endometrioid, 1366 (5.3%) clear cell, 1417 (5.5%) mucinous, and 2764 (10.8%) other epithelial ovarian cancer. A total of 1805 SNPs in a 1.1 Mb region of 10p12 passed genotyping and imputation quality-control metrics in all three datasets.
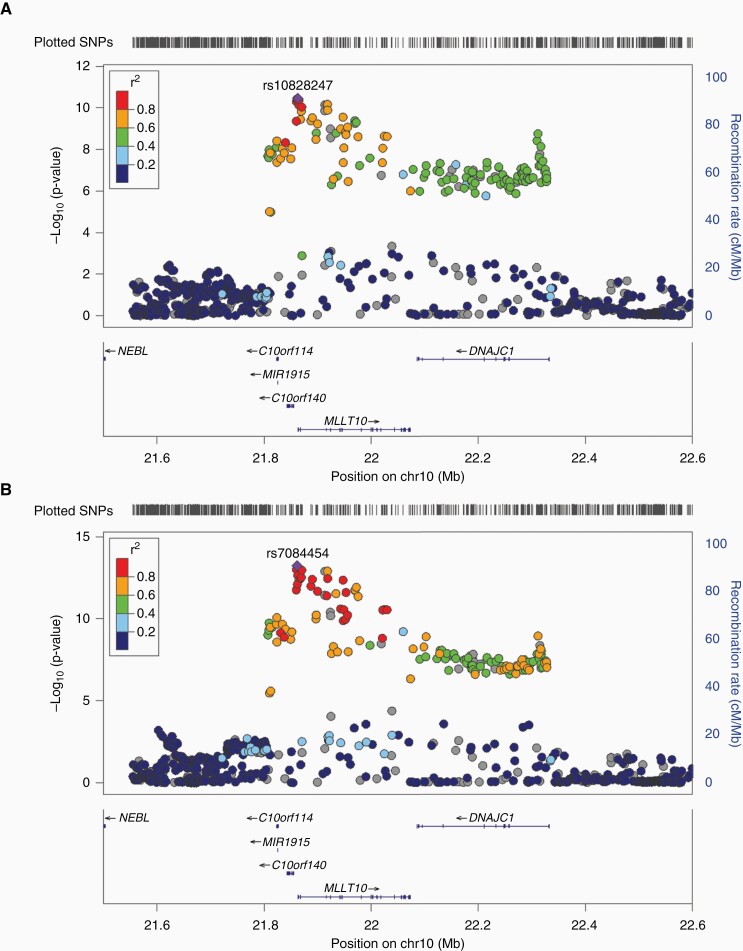
Meta-analysis of Grouping 1 identified rs10828247, a noncoding SNP located 245bp upstream of the MLLT10 transcription start site, as the most significantly associated variant in all models (Table 1). In the two-sided subset-based meta-analysis of Grouping 1, the G allele of rs10828247 was associated with increased neoplasm risk in all three subsets (OR = 1.07; 95% CI = 1.05–1.09; P = 3.4 × 10–11) (Table 1; Figure 1A). Within subsets, rs10828247 was associated with risk of meningioma (OR = 1.28; 95% CI = 1.11–1.49; P = 7.1 × 10–4), breast cancer (OR = 1.04; 95% CI = 1.01–1.07; P = 2.5 × 10–3), and ovarian cancer (OR = 1.09; 95% CI = 1.06–1.12; P = 1.1 × 10–9).
Table 1.
Lead Variants From ASSET-Based Meta-Analysis of Meningioma, Breast Cancer, and Ovarian Cancer Risk Across the Subsets Included in Analyses
| Subsets modeleda | Analysis | Top signal | OR (95% CI) | P-value | Subsets selected by ASSET |
|---|---|---|---|---|---|
| Grouping 1 | Meta-analysis | rs10828247 | 1.07 (1.07-1.07) | 1.2 × 10–11 | NAb |
| Grouping 1 | 1-sided subset | rs10828247 | 1.07 (1.05-1.09) | 2.2 × 10–11 | meningioma, breast (all), ovarian |
| Grouping 1 | 2-sided subset (combined) | rs10828247 | 1.07 (1.05-1.09) | 3.4 × 10–11 | meningioma, breast (all), ovarian |
| Positive | 1.07 (1.05-1.09) | 3.4 × 10–11 | meningioma, breast (all), ovarian | ||
| Negative | - | - | - | ||
| Grouping 2 | Meta-analysis | rs10828247 | 1.05 (1.05-1.05) | 1.2 × 10–8 | NAb |
| Grouping 2 | 1-sided subset | rs1416901 | 1.07 (1.05-1.09) | 1.7 × 10–12 | meningioma, breast (ER+), ovarian |
| Grouping 2 | 2-sided subset (combined) | rs7084454 | NAc | 5.7 × 10–14 | meningioma, breast (ER+), breast (ER–), ovarian |
| Positive | 1.07 (1.05-1.10) | 1.7 × 10–12 | meningioma, breast (ER+), ovarian | ||
| Negative | 0.91 (0.86-0.96) | 8.9 × 10–4 | breast (ER–) | ||
| Grouping 3 | Meta-analysis | rs7084454 | 1.11 (1.11-1.11) | 5.4 × 10–17 | NAb |
| Grouping 3 | 1-sided subset | rs7084454 | 1.11 (1.08-1.38) | 1.1 × 10–15 | meningioma, breast (ER+ vs. ER–), ovarian |
| Grouping 3 | 2-sided subset (combined) | rs7084454 | 1.11 (1.08-1.14) | 2.2 × 10–15 | meningioma, breast (ER+ vs. ER–), ovarian |
| Positive | 1.11 (1.08-1.14) | 2.2 × 10–15 | meningioma, breast (ER+ vs. ER–), ovarian | ||
| Negative | - | - | - |
aGrouping 1 includes association data from: meningioma cases versus controls, breast cancer cases (all) versus controls, and ovarian cancer cases versus controls. Grouping 2 includes association data from: meningioma cases versus controls, breast cancer cases (ER+) versus controls, breast cancer cases (ER–) versus controls, and ovarian cancer cases versus controls. Grouping 3 includes association data from: meningioma cases versus controls, ER+ breast cancer cases versus ER– breast cancer cases, and ovarian cancer cases versus controls.
bStandard fixed-effect meta-analysis from ASSET includes all subsets, without conducting a subset search, and has optimal power when an effect is observed across all subsets with the same direction of effect.
cSummary odds ratio not calculated because effects in different directions across subsets.
Fig. 1.
Chromosome 10p12 loci associated with risk of meningioma and estrogen-mediated cancers using two-sided ASSET meta-analysis. A) Two-sided ASSET meta-analysis of meningioma patients (N = 927) and controls (N = 790), female breast cancer patients (N = 28 108) and controls (N = 22 209), and ovarian cancer patients (N = 25 509) and controls (N = 40 941). All subsets were included in meta-analysis of lead SNP rs10828247 (Pmeta = 3.4 × 10–11). B) Two-sided ASSET meta-analysis of meningioma patients (N = 927) and controls (N = 790), female ER+ breast cancer patients (N = 19 436) and controls (N = 22 209), female ER– breast cancer patients (N = 3217) and controls (N = 22 209), and ovarian cancer patients (N = 25 509) and controls (N = 40 941). All subsets were included in the meta-analysis for lead SNP rs7084454 (Pmeta = 5.7 × 10–14), but the risk allele was flipped for ER– breast cancer. Manhattan plots generated using LocusZoom.
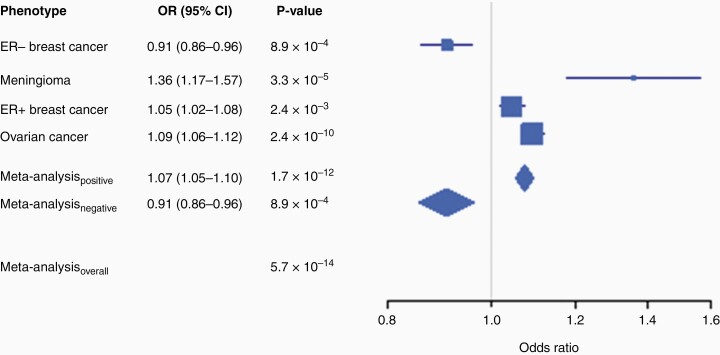
Meta-analysis of Grouping 2 again identified rs10828247 as the most significantly associated variant in fixed-effects meta-analysis, but significant heterogeneity in effect was observed (PHet < 0.001). One-sided ASSET meta-analysis identified rs1416901 as the lead variant (OR = 1.07; 95% CI = 1.05–1.09; P = 1.7 × 10–12) but omitted ER– breast cancer associations when selecting subsets (Table 1). The reason for this omission became clear when two-sided ASSET meta-analysis was performed, as a new lead SNP was identified at rs7084454 (P = 5.7 × 10–14) (Figure 1B). The rs7084454 “A” allele was associated with increased risk of meningioma, ER+ breast cancer, and ovarian cancer (OR = 1.07; 95% CI = 1.05–1.10; P = 1.7 × 10–12) and decreased risk of ER– breast cancer (Table 1; Figure 2). Within each subset, the rs7084454 “A” allele was associated with increased risk of meningioma (OR = 1.36; 95% CI = 1.17–1.57; P = 3.3 × 10–5), ER+ breast cancer (OR = 1.05; 95% CI = 1.02–1.08; P = 2.4 × 10–3), and ovarian cancer (OR = 1.09; 95% CI = 1.06–1.12; P = 2.4 × 10–10), and decreased risk of ER– breast cancer (OR = 0.91; 95% CI = 0.86–0.96; P = 8.9 × 10–4) (Table 2).
Fig. 2.
Association of lead SNP rs7084454 with risk of ER– breast cancer (3217 cases, 22 209 controls), meningioma (N = 927 cases, 790 controls), ER+ breast cancer (19 436 cases, 22 209 controls), and ovarian cancer (25 509 cases, 40 941 controls) using two-sided ASSET meta-analysis. Odds ratios are for each additional copy of the minor (A) allele, under an allelic additive model. No odds ratio is presented for the overall meta-analysis because of differing directions of effect across subsets.
Table 2.
Individual Subset Associations With Lead Pleiotropic SNP rs7084454
| Cancer | EAFa | OR (95% CI) | P-value |
|---|---|---|---|
| Meningioma | 0.34 | 1.36 (1.17–1.57) | 3.3 × 10–5 |
| Females | 1.42 (1.20–1.69) | 3.9 × 10–5 | |
| Males | 1.19 (0.91–1.57) | 0.20 | |
| Breast (all) | 0.34 | 1.03 (1.01–1.06) | 0.015 |
| ER+ | 1.05 (1.02–1.08) | 2.4 × 10–3 | |
| ER– | 0.91 (0.86–0.96) | 8.9 × 10–4 | |
| Ovarian | 0.33 | 1.09 (1.06–1.12) | 2.4 × 10–10 |
| Serous high grade | 1.10 (1.07–1.14) | 2.8 × 10–9 | |
| Serous low grade | 1.10 (1.00–1.22) | 0.049 | |
| Mucinous | 0.99 (0.91–1.08) | 0.86 | |
| Clear cell | 1.12 (1.03–1.21) | 9.0 × 10–3 | |
| Endometrioid | 1.06 (1.00–1.13) | 0.048 |
aEAF: Effect allele frequency in controls, where “A” is the effect allele and “G” is the alternate allele.
When meningioma case-control analyses were stratified by sex, rs7084454 had a larger magnitude of effect in women (OR = 1.42; 95% CI = 1.20–1.69; P = 3.9 × 10–5) than in men (OR = 1.19; 95% CI = 0.91–1.57; P = 0.20), but significant effect modification by sex was not detected (PHET = 0.28, I2 = 13.1). Additionally, rerunning two-sided ASSET meta-analysis with sex-stratified meningioma associations again identified rs7084454 as the lead variant and selected both the male and female meningioma subsets for inclusion. Stratifying by ovarian cancer histotype did not reveal significant heterogeneity of association across strata and identified significant positive associations for serous high grade, serous low grade, clear cell, and endometrioid ovarian cancers (Table 2). No association was observed for mucinous ovarian cancer (OR = 0.99; 95% CI = 0.91–1.08; P = 0.86), but two-sided ASSET meta-analysis again identified rs7084454 as the lead variant when modeling histotype-specific associations.
Meta-analysis of Grouping 3 complemented the two-sided ASSET analysis of Grouping 2, identifying rs7084454 as the most significantly associated variant in all models (Table 1). In case-only analysis of breast cancer patients, the “A” allele of rs7084454 was associated with 1.15-fold greater odds of having an ER+ tumor (95% CI = 1.09–1.22; P = 1.3 × 10–6).
To detect pleiotropic associations with additional traits, or to identify potential intermediate phenotypes that may mediate the association between MLLT10 SNPs and risk of various tumor types, we undertook a PheWAS approach using data from the U.K. Biobank. Data on rs10828247 could not be retrieved, but a total of 37 traits were associated with rs7084454 at genome-wide statistical significance (ie, <5.0 × 10–8), including: body fat percentage (P = 7.4 × 10–25), waist circumference (P = 9.2 × 10–25), body mass index (BMI) (P = 7.5 × 10–22), and weight (P = 1.0 × 10–17) (Table 3). The rs7084454 risk allele for meningioma, ovarian cancer, and ER+ breast cancer from our analyses (A) was associated with increases in all of these measures of adiposity. The SNP was also associated with “malignant neoplasms of the breast” (P = 1.6 × 10–4, OR = 1.06) and with “malignant neoplasm of the ovary” (P = 0.029, OR = 1.12) in UK Biobank data. In addition to anthropometric and cancer traits, the PheWAS also identified significant associations with several behavioral traits, including: greater time spent watching television (P = 2.2 × 10–13) and decreased intake of salad/raw vegetables (P = 5.7 × 10–10).
Table 3.
Association of Lead Pleiotropic SNP rs7084454 With Additional Traits in PheWAS Analysis of UK Biobank Data (N = 452 264)
| Trait | Betaa | P-valueb |
|---|---|---|
| Leg fat percentagec | 0.11 | 1.7 × 10–30 |
| Leg fat massc | 0.029 | 2.6 × 10–25 |
| Body fat percentage | 0.12 | 7.4 × 10–25 |
| Waist circumference | 0.23 | 9.2 × 10–25 |
| Whole body fat mass | 0.17 | 2.5 × 10–23 |
| Trunk fat mass | 0.090 | 2.5 × 10–22 |
| Body mass index (BMI) | 0.082 | 7.5 × 10–22 |
| Trunk fat percentage | 0.13 | 8.9 × 10–22 |
| Oily fish intake | –0.018 | 3.5 × 10–21 |
| Hip circumference | 0.16 | 2.2 × 10–20 |
| Fresh fruit intake | –0.028 | 5.9 × 10–20 |
| Arm fat percentagec | 0.12 | 3.3 × 10–19 |
| Arm fat massc | 0.0098 | 2.6 × 10–18 |
| Weight | 0.21 | 1.0 × 10–17 |
| Time spent watching television (TV) | 0.024 | 2.2 × 10–13 |
| Waist circumference/ Hip circumference | 0.00092 | 5.8 × 10–13 |
| Essential hypertension | 0.0052 | 2.3 × 10–11 |
| Hypertensive diseases | 0.0052 | 2.5 × 10–11 |
| Number of operations, self-reported | 0.021 | 3.5 × 10–11 |
| Monocyte count | 0.0019 | 1.2 × 10–10 |
| Dried fruit intake | –0.019 | 3.2 × 10–10 |
| Usual walking pace | –0.0078 | 4.6 × 10–10 |
| Salad/ raw vegetable intake | –0.025 | 5.7 × 10–10 |
| Arm fat-free massc | 0.0049 | 3.0 × 10–9 |
| Arm predicted massc | 0.0045 | 4.9 × 10–9 |
| Number of treatments/medications taken | 0.031 | 6.7 × 10–9 |
| Basal metabolic rate | 8.1 | 1.8 × 10–8 |
| Leg fat-free mass (left) | 0.012 | 2.3 × 10–8 |
| Impedance of armc | –0.37 | 3.9 × 10–8 |
| Leg predicted mass (left) | 0.011 | 4.0 × 10–8 |
aBeta values correspond to the direction of effect associated with each additional copy of the rs7084454 “A” allele, which is the allele associated with increased risk of meningioma, ovarian cancer and ER+ breast cancer, and decreased risk of ER– breast cancer, in our analyses.
bSeven hundred and seventy-eight total traits were queried. Those displayed in Table 3 had P-values < 5.0 × 10–8, a canonical cutoff for genome-wide statistical significance.
cA number of traits in UK Biobank are stratified by laterality (eg, “leg fat percentage, left” and “leg fat percentage, right”). When each of a pair of lateral traits reached genome-wide significance, only one member of the pair is listed in Table 3.
Because obesity is a known risk factor for meningioma,36,37 we assessed whether adiposity may serve as a mediating factor connecting variation at rs7084454 to meningioma risk by testing the association between rs7084454 and meningioma case-control status after adjusting for subject BMI. The odds ratio increased modestly from 1.36 without adjustment for BMI to 1.38 with adjustment for BMI, indicating that rs7084454 may contribute independently to both adiposity traits and meningioma risk through one or more additional mechanisms.
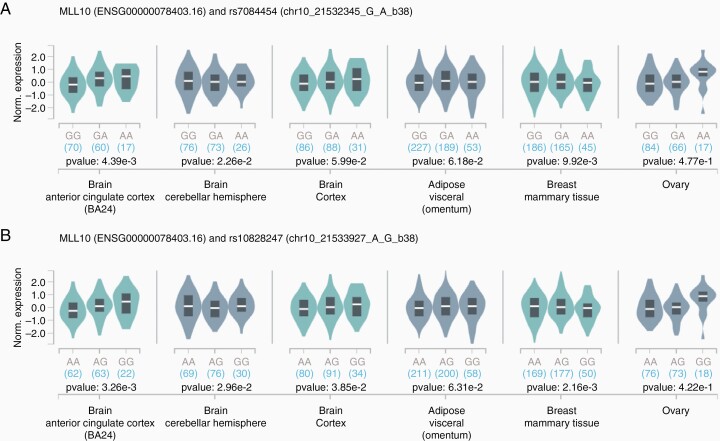
The two lead pleiotropic SNPs from our analyses, rs7084454 and rs10828247, are in strong LD in European-ancestry subjects from the 1000 Genomes project (R2 = 0.90).38 They are less tightly linked in African-ancestry populations from 1000 Genomes (R2 = 0.69), but have higher risk allele frequencies (RAF) in these populations (41% versus 34% and 47% versus 35% for rs7084454 and rs10828247, respectively). Additionally, these variants are uncommon in Asian populations (RAF < 0.05). Both variants map to DNAse hypersensitivity sites, indicating regions of open chromatin, across a diverse set of tissues including brain and breast tissues.39 Further, rs7084454 maps to both promoter and enhancer histone marks across multiple tissues.39 We queried GTEx data and observed that both SNPs showed evidence of acting as expression quantitative trait loci (eQTLs) for MLLT10 in brain, adipose, and breast tissues, with the strongest association observed in anterior cingulate cortex tissue (Figure 3).40 The risk allele for meningioma, ER+ breast cancer, and ovarian cancer was generally associated with modest increases in MLLT10 expression, although allelic effects in breast mammary tissue went in the opposite direction.
Fig. 3.
Expression of MLLT10 in selected tissues, across strata of genotype at rs7084454 (A) and rs10828247 (B). The expression quantitative trait analyses were conducted using data from the Genotype-Tissue Expression (GTEx) Project (https://www.gtexportal.org/home/).
MLLT10 encodes the AF10 transcription factor, which interacts with histone methyltransferase DOT1L and is essential for enabling H3K79 dimethylation of histone H3,41,42 a mark of active promoters.43 Although we were most interested in looking at the effect of risk alleles on expression levels in normal tissue, where they may predispose to malignancy, tissue from normal meninges are not included in current eQTL databases and MLLT10 expression itself may have important downstream effects on transcriptional regulation of other genes. We, therefore, assessed associations between MLLT10 transcript expression and expression of ESR1 and ESR2 in normal meninges and meningioma tumor samples. MLLT10 expression was positively correlated with ESR1 expression in sixteen samples of normal meninges (R = 0.48; P = 0.058), but showed no correlation with expression of ESR2 (R = 0.044, P = 0.87). In 96 meningioma samples, MLLT10 expression was positively correlated with expression of ESR1 (R = 0.28; P = 0.0065) but again did not correlate with expression of ESR2 (R = 0.050, P = 0.85). We observed no differences in the correlation of MLLT10 and ESR1 when stratifying by NF2 status, the most recurrently mutated gene in meningioma tumors (R = 0.27 in 27 NF2-altered tumors, R = 0.28 in 69 NF2-WT tumors).
Lead SNP rs7084454 was also observed to function as a cis-eQTL for C10orf114 (also referred to as MIR1915HG), which is an uncharacterized gene located upstream of MLLT10. Expression of C10orf114 was weakly positively correlated with expression of MLLT10 in normal meninges (r = 0.25; P = 0.35) and in meningioma (r = 0.080; P = 0.44). It was also positively correlated with expression of ESR1 in normal meninges (r = 0.36; P = 0.17), and this association reached statistical significance in the larger sample of meningioma tumor samples (r = 0.31; P = 0.0024), indicating a potential role for multiple 10p12 genes in trans-regulation of ESR1 expression.
Discussion
Our ASSET-based meta-analyses identified the “A” allele of rs7084454, near the promoter of MLLT10, as a lead pleiotropic variant that confers risk of meningioma, ovarian cancer, and ER+ breast cancer, but was significantly protective against ER– breast cancer. in silico functional analyses suggested that rs7084454 is a MLLT10 eQTL, with the risk allele associated with higher RNA expression in multiple brain tissues. This may be mediated by promoter or enhancer activity, as rs7084454 is located in promoter and enhancer histone marks across multiple tissues. The rs7084454 “A” allele was also associated with a number of additional traits in our PheWAS analysis of UK Biobank data, most notably with increases in several adiposity-related traits and with poorer physical and dietary health behaviors. Although higher BMI has been associated with increased risk of numerous neoplasms—including meningioma, breast, and ovarian cancer44—our meningioma case-control analyses indicated that rs7084454 likely does not confer meningioma risk via its effects on adiposity, suggesting that this variant instead impacts one or more additional traits that may independently increase meningioma risk and adiposity (eg, estrogenic signaling).
MLLT10 (MLLT10 histone lysine methyltransferase DOT1L cofactor) encodes the AF10 protein, which is known to interact with DOT1L (DOT1-like histone H3K79 methyltransferase). This interaction is essential for H3K79 dimethylation of histone H3,41,42 a mark of active promoters.43 Given its role as both a putative transcription factor and in methylating histone H3 to activate gene expression, there are myriad downstream genes that may be upregulated by increases in MLLT10 expression. Indeed, several MLLT10-activating chromosomal translocations have been associated with broad transcriptional changes in acute myeloid and acute lymphoblastic leukemias.45
Given the association of our lead variant with increased risk of ER+ breast cancer, decreased risk of ER– breast cancer, and increased risk of meningioma independent of BMI, we hypothesized that expression of genes on 10p12 might impact expression of estrogen receptor genes ESR1 (ERα) and ESR2 (ERβ). We observed a positive correlation of both MLLT10 and C10orf114 gene expression with ESR1 levels in normal meninges and in meningioma tumors, although the products of these genes are transcriptional activators and directionality of the associations could not be resolved. Nevertheless, it is intriguing to consider that a germline polymorphism associated with increased MLLT10 and C10orf114 expression might also increase ERα activity in a manner that could simultaneously impact risk of several hormonally-mediated cancers and adiposity-related traits.
The analyses here include only persons of European ancestry and thus may not apply to persons from other racial/ethnic groups. The meningioma risk allele frequency at rs7084454 and rs10828247 were notably higher in African-ancestry subjects than in European-ancestry subjects in 1000 Genomes data, and were lowest in Asian populations. In the U.S., meningioma risk is two-fold higher in African-Americans than non-Hispanic whites, and is lowest in Asians.2 In addition to racial variation in overall risk, the prevalence of many hormonal factors associated with risk of meningioma, breast, and ovarian cancers vary by race and the female-to-male ratio (meningioma and breast) also varies by race.3 While dozens of SNPs have been associated with breast and ovarian cancer risk in recent GWAS, only two common meningioma risk loci have been identified to-date.21–26,29 Thus, population-level differences in meningioma incidence may reflect underlying ancestry-related differences in MLLT10 risk allele frequency across groups. Interestingly, the 10p12 region that includes MLLT10 was recently found to contain a risk locus for pituitary adenoma,46 a nonmalignant brain tumor frequently associated with hormone oversecretion and which also varies in incidence by sex and race/ethnicity.2
We used both traditional and subset-based meta-analyses for these data. One caveat to such an approach is that relative to standard fixed or random effects analyses, subset-based meta-analysis has reduced power when the majority of underlying associations are in one direction. While a traditional meta-analysis approach identified rs10828247 as the lead SNP across subsets, applying two-sided subset-based meta-analysis to the same subsets enabled us to identify an association at rs7084454 that was five orders of magnitude more statistically significant and which revealed meaningful biological differences in breast cancer etiology across strata of ER–status.
We confirm evidence of a MLLT10 eQTL associated with increased risk of meningioma, ER+ breast cancer, ovarian cancer, and adiposity, and with decreased risk of ER– breast cancer, suggesting a possible estrogenic mechanism underlying this pleiotropy. Future analyses would benefit from the inclusion of additional numbers of male meningioma and breast cancer patients, receptor status of meningioma patients, consideration of additional hormone receptors including those related to progesterone, inclusion of patients of additional race/ethnicities, and exploration of potential gene-environment interactions with hormone-related exposures.
Contributor Information
Kyle M Walsh, Department of Neurosurgery and Duke Cancer Institute, Duke University School of Medicine. Durham, North Carolina, USA; Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, California, USA.
Chenan Zhang, Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, California, USA.
Lisa Calvocoressi, School of Public Health, Yale University, New Haven, Connecticut, USA.
Helen M Hansen, Department of Neurological Surgery, University of California, San Francisco, San Francisco, California, USA.
Andrew Berchuck, Department of Obstetrics and Gynecology and Duke Cancer Institute, Duke University School of Medicine. Durham, North Carolina, USA.
Joellen M Schildkraut, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA.
Melissa L Bondy, Department of Epidemiology and Population Health, Stanford University, Palo Alto, California, USA.
Margaret Wrensch, Department of Neurological Surgery, University of California, San Francisco, San Francisco, California, USA.
Joseph L Wiemels, Center for Genetic Epidemiology, University of Southern California, Los Angeles, California, USA.
Elizabeth B Claus, School of Public Health, Yale University, New Haven, Connecticut, USA; Department of Neurosurgery, Brigham and Women’s Hospital, Boston, Massachusetts, USA.
Funding
This work was supported by National Institutes of Health R01 Grants CA109468, CA109461, CA109745, CA109473, CA109745, CA052689, and CA151933, National Institutes of Health T32 Grant CA151022, as well as by the Brain Science Foundation, the Meningioma Mommas and The Sontag Foundation.
References
- 1. Vernooij MW, Ikram MA, Tanghe HL, et al. Incidental findings on brain MRI in the general population. N Engl J Med. 2007;357:1821–1828. [DOI] [PubMed] [Google Scholar]
- 2. Ostrom QT, Gittleman H, Truitt G, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol 2018;20:iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wiemels J, Wrensch M, Claus EB. Epidemiology and etiology of meningioma. J Neurooncol. 2010;99:307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Claus EB, Black PM, Bondy ML, et al. Exogenous hormone use and meningioma risk: what do we tell our patients? Cancer 2007;110:471–476. [DOI] [PubMed] [Google Scholar]
- 5. Custer B, LongstrethWT, Jr., Phillips LE, Koepsell TD, Van Belle G. Hormonal exposures and the risk of intracranial meningioma in women: a population-based case-control study. BMC Cancer 2006;6:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cea-Soriano L, Blenk T, Wallander MA, Rodriguez LA. Hormonal therapies and meningioma: is there a link? Cancer Epidemiol 2012;36:198–205. [DOI] [PubMed] [Google Scholar]
- 7. Claus EB, Calvocoressi L, Bondy ML, et al. Exogenous hormone use, reproductive factors, and risk of intracranial meningioma in females. J Neurosurg. 2013;118:649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cowppli-Bony A, Bouvier G, Rue M, et al. Brain tumors and hormonal factors: review of the epidemiological literature. Cancer Causes Control 2011;22:697–714. [DOI] [PubMed] [Google Scholar]
- 9. Custer BS, Koepsell TD, Mueller BA. The association between breast carcinoma and meningioma in women. Cancer 2002;94:1626–1635. [DOI] [PubMed] [Google Scholar]
- 10. Jhawar BS, Fuchs CS, Colditz GA, Stampfer MJ. Sex steroid hormone exposures and risk for meningioma. J Neurosurg. 2003;99:848–853. [DOI] [PubMed] [Google Scholar]
- 11. Johnson DR, Olson JE, Vierkant RA, et al. Risk factors for meningioma in postmenopausal women: results from the Iowa Women’s Health Study. Neuro Oncol 2011;13:1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michaud DS, Gallo V, Schlehofer B, et al. Reproductive factors and exogenous hormone use in relation to risk of glioma and meningioma in a large European cohort study. Cancer Epidemiol Biomarkers Prev. 2010;19:2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schildkraut JM, Calvocoressi L, Wang F, et al. Endogenous and exogenous hormone exposure and the risk of meningioma in men. J Neurosurg. 2014;120:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wigertz A, Lonn S, Mathiesen T, et al. Risk of brain tumors associated with exposure to exogenous female sex hormones. Am J Epidemiol. 2006;164:629–636. [DOI] [PubMed] [Google Scholar]
- 15. Horn J, Vatten LJ. Reproductive and hormonal risk factors of breast cancer: a historical perspective. Int J Womens Health 2017;9:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moorman PG, Calingaert B, Palmieri RT, et al. Hormonal risk factors for ovarian cancer in premenopausal and postmenopausal women. Am J Epidemiol. 2008;167:1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Claus EB, Calvocoressi L, Bondy ML, et al. Family and personal medical history and risk of meningioma. J Neurosurg. 2011;115:1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Claus EB, Risch N, Thompson WD. Autosomal dominant inheritance of early-onset breast cancer. Implications for risk prediction. Cancer 1994;73:643–651. [DOI] [PubMed] [Google Scholar]
- 20. Schildkraut JM, Thompson WD. Familial ovarian cancer: a population-based case-control study. Am J Epidemiol. 1988;128:456–466. [DOI] [PubMed] [Google Scholar]
- 21. Dobbins SE, Broderick P, Melin B, et al. Common variation at 10p12.31 near MLLT10 influences meningioma risk. Nat Genet. 2011;43:825–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Claus EB, Cornish AJ, Broderick P, et al. Genome-wide association analysis identifies a meningioma risk locus at 11p15.5. Neuro Oncol 2018;20:1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Egan KM, Baskin R, Nabors LB, et al. Brain tumor risk according to germ-line variation in the MLLT10 locus. Eur J Hum Genet. 2015;23:132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghoussaini M, Pharoah PDP, Easton DF. Inherited genetic susceptibility to breast cancer: the beginning of the end or the end of the beginning? Am J Pathol. 2013;183:1038–1051. [DOI] [PubMed] [Google Scholar]
- 25. Michailidou K, Hall P, Gonzalez-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61, 361e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pharoah PD, Tsai YY, Ramus SJ, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–70, 370e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diogo D, Tian C, Franklin CS, et al. Phenome-wide association studies across large population cohorts support drug target validation. Nat Commun. 2018;9:4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amos CI, Dennis J, Wang Z, et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol Biomarkers Prevent. 2017;26:126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Phelan CM, Kuchenbaecker KB, Tyrer JP, et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017;49:680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhattacharjee S, Rajaraman P, Jacobs KB, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90:821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Semmes EC, Vijayakrishnan J, Zhang C, et al. Leveraging genome and phenome-wide association studies to investigate genetic risk of acute lymphoblastic leukemia. Cancer Epidemiol Biomarkers Prev. 2020;29:1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clark VE, Harmanci AS, Bai H, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48:1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walsh KM. Epidemiology of meningiomas. Handb Clin Neurol 2020;169:3–15. [DOI] [PubMed] [Google Scholar]
- 37. Takahashi H, Cornish AJ, Sud A, et al. Mendelian randomization provides support for obesity as a risk factor for meningioma. Sci Rep. 2019;9:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alexander TA, Machiela MJ. LDpop: an interactive online tool to calculate and visualize geographic LD patterns. BMC Bioinf. 2020;21:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Farooq Z, Banday S, Pandita TK, Altaf M. The many faces of histone H3K79 methylation. Mutat Res Rev Mutat Res. 2016;768:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vlaming H, van Leeuwen F. The upstreams and downstreams of H3K79 methylation by DOT1L. Chromosoma. 2016;125:593–605. [DOI] [PubMed] [Google Scholar]
- 43. Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007;129:823–837. [DOI] [PubMed] [Google Scholar]
- 44. Kyrgiou M, Kalliala I, Markozannes G, et al. Adiposity and cancer at major anatomical sites: umbrella review of the literature. BMJ 2017;356:j477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peterson JF, Sukov WR, Pitel BA, et al. Acute leukemias harboring KMT2A/MLLT10 fusion: a 10-year experience from a single genomics laboratory. Genes Chromosomes Cancer 2019;58:567–577. [DOI] [PubMed] [Google Scholar]
- 46. Ye Z, Li Z, Wang Y, et al. Common variants at 10p12.31, 10q21.1 and 13q12.13 are associated with sporadic pituitary adenoma. Nat Genet. 2015;47:793–797. [DOI] [PubMed] [Google Scholar]