Abstract
Purpose of Review.
Anatomical segmentation has played a major role within clinical cardiology. Novel techniques through artificial intelligence-based computer vision have revolutionized this process through both automation and novel applications. This review discusses the history and clinical context of cardiac segmentation to provide a framework for a survey of recent manuscripts in artificial intelligence and cardiac segmentation. We aim to clarify for the reader the clinical question of “Why do we segment?” in order to understand the question of “Where is current research and where should be?”.
Recent Findings.
There has been increasing research in cardiac segmentation in recent years. Segmentation models are most frequently based on a U-Net structure. Multiple innovations have been added in terms of pre-processing or connection to analysis pipelines. Cardiac MRI is the most frequently segmented modality, which is due in part to the presence of publically-available, moderately sized, computer vision competition datasets. Further progress in data availability, model explanation, and clinical integration are being pursued.
Summary.
The task of cardiac anatomical segmentation has experienced massive strides forward within the past five years due to convolutional neural networks. These advances provide a basis for streamlining image analysis, and a foundation for further analysis both by computer and human systems. While technical advances are clear, clinical benefit remains nascent. Novel approaches may improve measurement precision by decreasing inter-reader variability and appear to also have the potential for larger-reaching effects in the future within integrated analysis pipelines.
Keywords: Cardiac segmentation, computer vision, artificial intelligence, convolutional neural networks, cardiac MRI, cardiac imaging
INTRODUCTION
Anatomical segmentation, or the process of identifying local structures to determine size or shape, of medical imaging has a major role throughout clinical cardiology. All modalities of cardiac imaging incorporate this segmental information to determine critical measurements such as cardiac function, wall thickness, and other metrics of risk assessment. Artificial Intelligence (AI) based automated cardiac anatomical segmentation has received extensive interest in recent times due to the potential to automate, streamline, or improve assessment of medical imaging. At the same time, many of these projects risk falling into the concept of the “AI Chasm,” defined by Topol and Keane as scientifically sound or accurate algorithms but with limited actual clinical utility.[1, 2] In this review, we provide a context of anatomical segmentation, a survey of the past 5 years of publications regarding cardiac segmentation and AI, and a perspective on future directions of AI cardiac segmentation within clinical cardiology. One of our primary goals is to clarify for the reader the clinical question of “Why do we segment?” in order to understand the question of “Where is current research and where should be?”.
THE CLINICAL USE OF CARDIOVASCULAR SEGMENTATION:
Traditional Approaches to Cardiac Segmentation
Two-dimensional echocardiography (echo) emerged in the late 1970’s with significant advancement in our ability to assess and quantify cardiac structures. Subsequent guidelines recognized the importance of the standardization of measurements and generated the 16-segment model, as well as definitions of standard distances and measurements.[3] These segmentation models do not represent precise anatomical contouring, however, provide a framework to standardize interpretation which persists to this day. This purpose can be divided into multiple steps: 1) the use of standardized segmentation models allows for quantification of qualitative observations, 2) this creates the ability to directly compare between patients/timepoints, anatomical structures and regions such as coronary distributions versus myocardial regions across imaging, and 3) these comparisons between patients define clinical diagnoses, predict outcomes, and monitor therapies.[4]
In the early 1990’s, guidelines for standardized views and definitions within multi-modality tomographic imaging followed[5] with the explicit recognition of the need to standardize interpretation between modalities such as single photon emission computed tomographic (SPECT), positron emission tomographic (PET), cardiac magnetic resonance (CMR), and cine computed tomographic (CT) imaging. In later years, the current 17 segment American Heart Association (AHA) model was standardized within recommendations and also included echo.[6] Coronary segmentation underwent a similar growth from the 1975 original AHA segmentation[7] to cardiac CT guidelines with only minor modifications,[8] thus creating a standard nomenclature to specify locations of disease.
The ability to pursue precise anatomical contouring instead of regional segmentations has emerged with improved spatial and temporal resolution but faces unique barriers. Guidelines related to segmentation explicitly recognize limitations within anatomical segmentation methods. Increased uncertainty and lack of standardization for certain tasks are highlighted within ventricular segmentation by high-detail intra-ventricular structures such as trabeculations, papillary muscles, and the moderator band. Echo and CMR guidelines agree on the inclusion of the trabeculations and moderator band within the right ventricular (RV) cavity but disagree on the inclusions/exclusion for papillary muscles and trabeculae for left ventricular (LV) mass quantifications.[9, 10] These differences emphasize the balance between anatomical precision provided by specific modalities, as well as consistency with historically established ranges and cutoffs.
Computer Vision and Semantic Segmentation
The goal of Computer Vision was succinctly described in Simon Prince’s classic book as “to extract useful information from images.”[11] This has conceptually existed since the 1960’s but has experienced massive recent advances with the development of convolutional neural networks (CNN), exponential increases in computational processing power as described by Moore’s law,[12] and increasingly available large image datasets.[13] As a brief introduction to terminology relevant within artificial-intelligence based computer vision, neural networks are mathematical algorithms which enable machine learning. These structures are loosely based on the human brain, and in fact, the basis was initially described by McCullough and Pitts in 1943, not in the context of programming, but as a means to model neuronal activity.[14] These networks are collections of artificial “neurons”, which receives a set of inputs, passes these inputs through “hidden layers” which perform mathematical processes on the input, to result in a discrete output or set of outputs. By providing data of matched inputs and outputs, the hidden layers are “trained” by incremental adjustment of the hidden layers in order to “learn” the association between the input and output. By combining multiple neurons together, high accuracy of these networks for even complex tasks can obtained as long as the computing power to train and execute the networks are sufficient. The relations, functions, and quantity of the different elements of the neural network determine the network architecture, which in turn plays a major role in its success for a particular task.[15]
Within computer vision, there are multiple potential tasks, which include image classification, localization, detection, and modification.[16] Cardiac imaging characterization, particularly for assessing form and function, most frequently fall under the concept of “semantic segmentation”. This task classifies objects in an image to concepts, mapping each pixel in the original image to a set of constrained values, and is particularly well suited for CNNs. A convolution is a function which is performed in a stepwise fashion across an image which can used to detect imaging features. These features can be simple, such as a horizontal or vertical edge, or a gradient; by combinations of simple features, more complex features can be created to identify and classify everyday objects, such as cars, animals, or faces.[17] Within medical imaging-related semantic segmentation, the network receives the image pixels as inputs and pixel-wise labels as outputs to train the model, and the CNN analyzes the anatomical features to assign labels (e.g. myocardium, aorta, chambers, valve, everything else).[18]
The overarching term for architectures that utilize series of convolutions in order to produce the output is a CNN; however, structural changes may improve computational efficiency and accuracy. Specific well-known architectures for semantic segmentation exist.[19] Fully convolutional networks (FCN) utilize an encoder-decoder structure by downsampling in the encoder, which is an efficient way to create feature maps for semantic segmentation, with upsampling in the decoder to recover resolution to the level of the original image. This provided a significant efficiency benefit, as pixels of the same label typically localize together within a segmentation, but had difficulty creating smooth segmentation edges due to the degradation of resolution in during the downsampling.[20] Skip connections between the different resolution levels of the encoder and decoder arms was added in the U-Net structure, which has been extremely popular and broadly used. This allowed for retention of higher resolution information, which helped resolve the issue with edge resolution.[21] Other modifications have included the addition of non-segmentation modules such as residual layers which improve the ability to train networks with more layers[22] and different types of convolutions which improve spatial context (dilated/atrous convolutions).[23, 24] Architectures are provided to give clinical readers recognition of common methods; however detailed discussion of the architectures are outside of the scope of this article and can be found in these excellent reviews.[25–28]
Defining the Challenges in Human Evaluation of Medical Imaging
In order to define the optimal direction of artificial intelligence-based cardiac segmentation, we need to define the problems with human vision. Human vision’s limitations may be best conceptualized as inherent limitations and tradeoffs in realms of physical perception and time/attention. Attention-wise, with an infinite amount of time, human viewers could scrutinize all portions of an image systematically; however, clearly the needs of clinical practice make this impossible. In human evaluation of medical imaging, attention fatigue is combatted by efficiency-based techniques such as search patterns and standard frameworks. Even with these techniques, weaknesses persist. For example, inattentional blindness where priming to search for a specific finding makes us less likely to see unrelated findings but clinically relevant findings, such as incidental malignancy.[29] Computational approaches do not have the same limitations with regard to fatigue, boredom, or distraction.
From a perception standpoint, edge detection and motion tracking are typically considered relatively simple tasks for human observers,[30–34] whereas detection of contrast differences are weaker, requiring high contrast-to-noise (CNR) ratios of 3–5 per the Rose Criterion.[35] The primary challenge to anatomical contouring is less the ability to identify the appropriate location of the contours and more the time and effort required to draw them. Within this context, semi-automatic and automatic methods have been successful. Conventional approaches have included thresholding by intensity and clustering methods with minimal pre-existing inputs; methods with moderate user-inputs such as region-growing and classification; or methods with more extensive pre-existing rules/knowledge, such as deformable model and atlas-guided methods. More recently, neural networks and probabilistic models such as Markov random fields have been introduced to augment conventional approaches.[36]
A SURVEY OF CARDIOVASCULAR SEGMENTATION AND ARTIFICIAL INTELLIGENCE
Search Parameters and Results
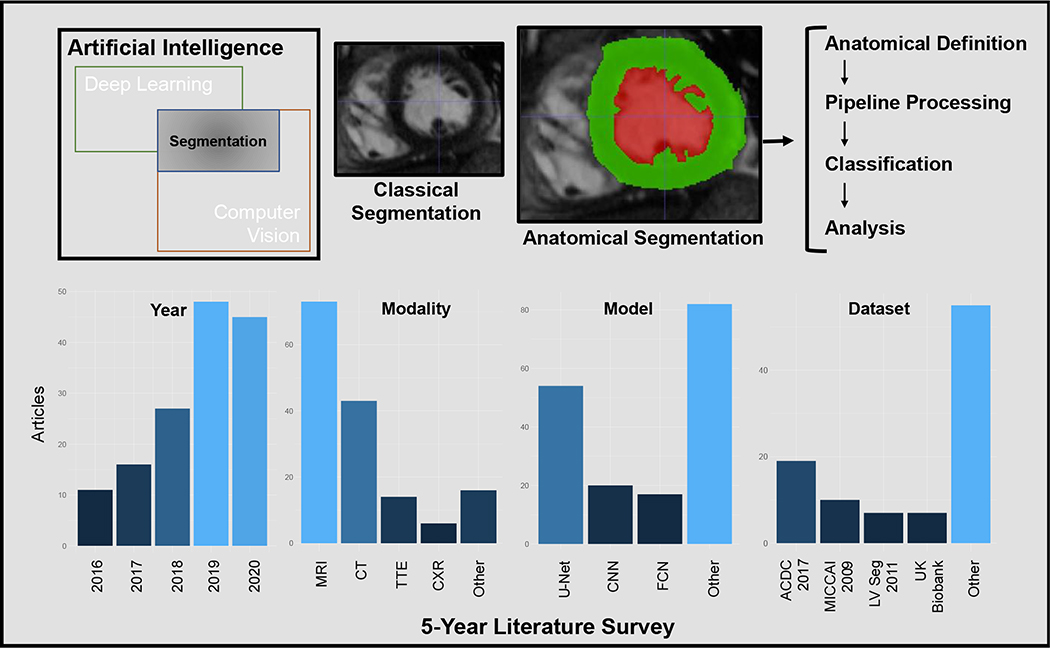
We performed a survey of papers published within the past 5 years by searching in PubMed for “cardiac segmentation” and “artificial intelligence”, limited to non-review articles, with 300 manuscripts identified (Figure 1). Manuscripts were individually reviewed and limited to cardiac anatomical segmentation (i.e. not inclusive of heart sound, electrocardiogram, or non-cardiac anatomical segmentation), which resulted in 149 papers (Supplemental Table 1). These papers were reviewed for their imaging modalities, basis architecture, number of patients used for training and testing, source of patient data, anatomy assessed, purpose of segmentation, and whether or not the code was explicitly released as noted in the manuscript. Information was not available for all papers due to either not being reported in the manuscript or limited access to the manuscript, and therefore we primarily report metrics as a percentage of the total manuscripts which were assessable within each aspect.
Figure 1: Summary figure.
Semantic segmentation exists within computer vision, and more recently has used deep learning techniques to augment traditional methods. While classical segmentation defined clinically relevant regions, anatomical segmentation follows anatomical boundaries, and enables more precise definitions of structure and function, which may be integrated into analysis pipelines. The 5-year literature survey shows recent growth in AI and cardiac segmentation, with MRI as the most common modality. External dataset use was most frequent with the ACDC 2017 set, and U-Net was most commonly used for segmentation. MRI: Magnetic resonance imaging, CT: Computed Tomography, TTE: Transthoracic echocardiography, CXR: Chest X-ray, CNN: Convolutional neural network, FCN: Fully convolutional network, ACDC 2017: Medical Image Computing and Computer Assisted Interventions (MICCAI) 2017 Automated Cardiac Diagnosis Challenge, MICCAI 2009: MICCAI/STACOM Sunnybrook Cardiac MR 2009 Left Ventricle Segmentation Challenge, LV Seg 2011: MICCAI/STACOM Left Ventricle Segmentation Dataset and Challenge from 2011
The number of AI-based cardiac segmentation manuscripts has been increasing significantly over time, with the lowest in 2016 (n=11) and increases to 48 in 2019 and 45 in 2020. Slightly over half of the manuscripts were published in clinically-oriented journals (54%), with a significant remaining portion published in technically-oriented journals. A large portion of the technically oriented journals were in IEEE journals (26%), in particular IEEE Transactions on Medical Imaging. Within the articles, the plurality focused on cardiac MRI segmentation (49%), both individually and combined with other modalities, followed by CT-based segmentation (27%) and echocardiography (13%). While most papers introduced novelties within their neural network architectures, the most frequent underlying architecture was U-Net (37%) followed by FCN (12%), though a significant portion (14%) did not report a basis architecture or stated that the architecture was an original CNN. Approximately one quarter of the papers explicitly released their code, with a direct reference in the manuscript.
Notable innovations were made in specific manuscripts. One recent paper incorporated a Monte-Carlo dropout in conjunction with a U-Net to provide uncertainty estimates for the segmentation.[37] This approach may help obviate unpredictable failure, and provide clinicians some degree of understanding of the expected quality of images. Other approaches combined images from separate views,[38] separate modalities,[39, 40] and different contrasts[41] to improve accuracy of segmentation or advantageous transfer of information from one modality to another. Serial processing such as image quality upscaling prior to segmentation,[42] or pipelines such as image processing pipelines,[43] quality control pipelines,[44] radiomics pipelines,[45] or even integration with robotics frameworks for procedures[46] were noted. Also, while diagnoses directly suggested by left ventricular mass or anatomy were proposed, we found segmentation that leads to indirect diagnosis of diseases such as pulmonary hypertension as a particularly intriguing approach as well.[47]
One particularly notable paper which balances technical innovation with a strong clinical application is presented by Augusto et al.[48] In this paper, the research team sought to automate the measurement of maximal wall thickness (MWT) measurements. A U-Net based model was used to segment the inner and outer myocardial contour, and subsequently solve a Laplace equation to determine the true MWT. The advance from anatomical segmentation to creational of a MWT line is conceptually simple but represents an important advance. Training a model to precisely mimic segmentations drawn by humans limits the maximal performance of the model to equal to or below the human-generated training data, and thus the segmentation model becomes a matter of convenience, rather than progress. The use of a segmentation model to define the potential borders of line enables the machine performance to become “superhuman” by “outworking” human observers. That is, by contouring every slice in the short axis stack, and solving the Laplace equation for every slice in order to identify the MWT, the model is systematic to a degree that would be infeasible for humans to perform during clinical reads. The authors highlight that with more precise wall thickness measurements, the sample size needed for clinical trials could be significantly smaller (2.3 times smaller to detect a 2 mm difference), which is a concrete benefit of using this measure.
Training and Testing Data Sources
The source of training and testing data varied, with slightly more than half (56%) reporting use of local institution-specific data, and a similar portion (52%) utilizing external datasets (a small proportion used both). The publicly available datasets varied in modality and implementation. Given the high proportion of MRI manuscripts, the most frequently used dataset (29% of the manuscripts using external data) was the Medical Image Computing and Computer Assisted Interventions (MICCAI) 2017 Automated Cardiac Diagnosis Challenge (ACDC), dataset which consisted of MRI images from the University Hospital of Dijon.[49] Notably, MICCAI/STACOM challenges including the Sunnybrook Cardiac MR 2009 Left Ventricle Segmentation Challenge[50] (17% of external data), and the Left Ventricle Segmentation Dataset and Challenge from 2011[51] (11% of external data) were also well-represented, with the UK-Biobank cardiac MRI dataset growing in popularity in more recent years[52] (11% of external data). Models most frequently focused on structure and function of the left ventricular myocardium and blood pool (50%), while right ventricle, and atrial segmentation was less popular. Outside of myocardial segmentation for cardiology and cardiac-imaging specific purposes, there was a notable group of applications based around anatomical planning for radiation oncology radiotherapy, though this was lower in frequency (9%). Applications outside of myocardial segmentation were even less common, however, epicardial adipose tissue,[53] coronary arterial lumen,[54] and invasive coronary imaging was also noted.[55]
It is notable that the most frequently used datasets are relatively small (100 training cases for the ACDC and MICCAI/STACOM 2011 datasets, 45 for the Sunnybrook MICCAI 2009 dataset), emphasizing the availability of data as a key bottleneck. In some ways, the popular use of smaller datasets has brought forth extra creativity in data augmentation, which should have durable benefits as limited training set sizes within medical imaging will continue into the near future. At the same time, we are beginning to see larger datasets in cardiovascular imaging, which can further advance the field. The UK Biobank and UK Digital Heart datasets contain >48,000 and ≈1200 patients’ CMRs respectively,[56, 52] and the Cardiac Acquisition for Multi-structure Ultrasound Segmentation (CAMUS) dataset[57] or the EchoNet-Dynamic dataset which contains 500 and 10,030 patients’ echocardiograms respectively. These datasets highlight the vanguard of training with larger training sets.[58]
FUTURE DIRECTIONS IN CARDIAC SEGMENTATION
Segmentation as the Primary Problem or a Pipeline Task
The popularity of the ACDC dataset highlights the major role that competition events and leaderboards have had in pushing forward the field of computer vision both within and outside of medical imaging. Within the primary publication, they summarize the accuracy of the top segmentation models from the ACDC competition.[49] These models had similar accuracy (by Dice score and Hausdorff distance) to inter/intra expert reader variability, and therefore a strong argument could be made that the problem is solved to within human accuracy. Areas of high-error rates were seen in the base and apex, which inherently have more uncertainty due to partial volume effects and variability in anatomical structure.
We view these “competition” datasets as providing a major benefit to medical computer vision. The competition approach with pre-labeled datasets help break down barriers between clinical imaging experts and non-medical computer vision experts, and provide a cross-pollination where non-medical computer vision experts benefit from access to expert labels in a curated dataset which would be otherwise inaccessible due to privacy protections, and clinical imaging experts benefit from additional computer vision expertise to determine top architectures in an appropriate context for medical imaging. This functions as a decentralized way to enable multidisciplinary collaboration, which is essential for accurate and clinically relevant models. Acknowledging the success of segmentation accuracy relative to human expert contours raises questions about how these models can be innovated further to enhance clinical benefit, affect prognostic and diagnostic assessment, and improve delivery of care. Further collaboration will be needed in order to optimally direct the field of medical computer vision.
Next Steps in Data Availability
In order to further progress towards clinical relevance and connection to clinical outcomes, issues regarding access to larger quantities of diverse data are growing in relevance. These needs are balanced against ethical implications regarding patient protection, codification of bias, data security, and ethical application of models. The ethical concerns are now being formally addressed.[59] We believe that these concerns will continue to grow with the field of machine learning in cardiac imaging. With respect to data availability, while the small ACDC dataset reported excellent results, larger datasets, have already begun to provide dividends within deep learning. In particular, the UK Biobank with expected enrollment of 100,000 patients with at least 10,000 follow up scans can be expected to provide unique benefits related to the data quantity and availability.[52] In general, large publicly-available datasets can face barriers related to protection of patient privacy, however, a new approach called “Federated Learning,” whereby the transfer of partially trained models instead of data is performed. These models can be trained on local datasets, and then transferred back to a secondary site for further training or a central site for aggregation, to avoid public release of patient data.[60, 61] This is particularly important for obtaining diversity within training datasets, both in the demographic (less-frequent racial/ethnic groups) as well as diagnostic (less frequent disease groups) as use of biased datasets will result in biased models.[62]
Next Steps in Model Explanation
In order to effectively translate technical advances into clinical application, the ability to understand a model’s decision-making process is important. Understanding our tools allows us to predict their strengths, weaknesses, and applications. With the use of complex architectures, increases in accuracy have often come at the cost of model explicability. That is, we know the training input, the model type, and can rate the output, but the actual process of “decision-making” is obscured to our understanding. Apart from overfitting to training sets, this can result in unexpected failures. There have been advances in attempting to explain computer vision models, most notably Grad-CAM, which can create an attention-based heatmap for multiple CNN types, in order to show how classification tasks are being decided.[63] While this is not directly applicable to semantic segmentation models, this movement highlights the growing understanding that both performance and interpretability is critical for broader uptake of computer vision model tasks. Within (and outside of) medical imaging, there are known examples of “specification gaming” where models literally address the stated objective but in ways that are not intended or useful by the designers.[64] Examples include identifying increased risk of skin lesion malignancy by the presence of a ruler in the picture,[65] or predicting pneumonia by identifying the location of a chest X-ray’s acquisition instead of the lungs themselves.[66] These outcomes highlight the need for model interpretability as a backstop to unpredictable behavior. These concerns are currently being addressed in the regulatory environment, with active international work through the International Medical Device Regulators Forum (IMDRF), and the US Food and Drug Administration, which continues to iterate on definitions and regulations for Software as a Medical Device.[67–69]
Future Directions: Replacement or Assistant
Clinicians recognize that we need help to deliver optimal medical care – The demands on our time relative to the accessible patient-related and evidence-related data have never been so high. At the same time, the fear of obsolescence has been historically deep-rooted in the relationship of the physician to the rapid pace of medical advancement, let alone new technology.[70–72] As these segmentation models are incorporated into more complex decision-making machines, the question arises of whether or not the resultant product will appear in a form more consistent with a clinical-decision support system to a physician, or as an autonomous decision-making system structure meant to replace physician judgement.
We believe that even in the face of technological advancement, the most optimal combination will remain the partnership of human and machine to bridge the AI chasm. This is not discounting the ability of advances in computational power, which will allow more efficient solutions. At their heart, the problems of clinical medicine are difficult to define in a way that is solvable in a formal quantitative manner while still remaining patient-focused. The imperfect nature of patient interviews, symptoms, and subjective experience create an environment which resists the development of formal rules. The abstract concept of patient preferences creates a unique solution for every case; that is, even death, or major adverse cardiac events, are not necessarily the endpoints that are most important to an individual patient. This creates a “wicked world”[73] type of scenario where the difficulty is not in solving the problem, but defining the entire question as well as appropriate benchmarks of success. The integration of clinician and computer should be the optimal direction, and AI-based segmentation provides a foundation for this interaction.
CONCLUSIONS
Visualization of cardiac structures using medical imaging has significantly improved over the last thirty years, increasing the relevance of segmentation, but also the amount of manual time spent on high-detail anatomy. The use of computer vision technologies to automate segmentation has grown out of this need, and while somewhat effective with semi-automatic statistical methods, has progressed significantly with the advent of higher computing power, data availability, and CNNs. The task of cardiac anatomical segmentation has experienced massive strides forward within the past five years, and provides a basis for streamlining image analysis, as well as a foundation for further analysis both by computer and human systems. Within the recent literature, cardiac MRI is the most common data source and U-Net is the most commonly used deep learning semantic segmentation architecture. The availability and visibility of datasets remains relevant, highlighted by the significant impact of MICCAI/STACOM challenges; however, larger volume datasets are becoming more common and will likely have a major effect. While technical advances are clear, clinical benefit remains nascent. Novel approaches may improve measurement precision by decreasing inter-reader variability and appear to also have the potential for larger-reaching effects in the future within integrated analysis pipelines. The future relationship of the physician to AI systems is uncertain, however, additional direction is needed for clinically relevant development of AI models, within which cardiac segmentation will play a major role.
Supplementary Material
Sources of Funding:
This work was supported in part by the National Institutes of Health NIH K99 HL157421-01, R01-HL134168, R01-HL131532, R01-HL143227, and the Doris Duke Charitable Foundation Grant 2020059.
Footnotes
Compliance with Ethical Standards
Conflict of Interest:
Alan C Kwan, Gerran Salto, Susan Cheng, and David Ouyang report no relevant conflicts of interest.
REFERENCES
- 1.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nature medicine. 2019;25(1):44–56. [DOI] [PubMed] [Google Scholar]
- 2.Keane PA, Topol EJ. With an eye to AI and autonomous diagnosis. npj Digital Medicine. 2018;1(1):40. doi: 10.1038/s41746-018-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiller NB, Shah PM, Crawford M, DeMaria A, Devereux R, Feigenbaum H et al. Recommendations for quantitation of the left ventricle by two-dimensional echocardiography. American Society of Echocardiography Committee on Standards, Subcommittee on Quantitation of Two-Dimensional Echocardiograms. J Am Soc Echocardiogr. 1989;2(5):358–67. doi: 10.1016/s0894-7317(89)80014-8. [DOI] [PubMed] [Google Scholar]
- 4.Lang RM, Bierig M, Devereux RB, Flachskampf FA, Foster E, Pellikka PA et al. Recommendations for chamber quantification: a report from the American Society of Echocardiography’s Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. Journal of the American Society of Echocardiography. 2005;18(12):1440–63. [DOI] [PubMed] [Google Scholar]
- 5.Bonow R, Gibbons R, Berman D, Johnson L, Rumberger J, Schwaiger M et al. Standardization of cardiac tomographic imaging. Circulation. 1992;86(1):338–9.1617787 [Google Scholar]
- 6.Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK et al. Standardized Myocardial Segmentation and Nomenclature for Tomographic Imaging of the Heart. Circulation. 2002;105(4):539–42. doi:doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 7.Austen WG, Edwards JE, Frye R, Gensini G, Gott VL, Griffith LS et al. A reporting system on patients evaluated for coronary artery disease. Report of the Ad Hoc Committee for Grading of Coronary Artery Disease, Council on Cardiovascular Surgery, American Heart Association. Circulation. 1975;51(4):5–40. [DOI] [PubMed] [Google Scholar]
- 8.Leipsic J, Abbara S, Achenbach S, Cury R, Earls JP, Mancini GJ et al. SCCT guidelines for the interpretation and reporting of coronary CT angiography: a report of the Society of Cardiovascular Computed Tomography Guidelines Committee. Journal of cardiovascular computed tomography. 2014;8(5):342–58. [DOI] [PubMed] [Google Scholar]
- 9.Lang RM, Badano LP, Mor-Avi V, Afilalo J, Armstrong A, Ernande L et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. European Heart Journal-Cardiovascular Imaging. 2015;16(3):233–71. [DOI] [PubMed] [Google Scholar]
- 10.Schulz-Menger J, Bluemke DA, Bremerich J, Flamm SD, Fogel MA, Friedrich MG et al. Standardized image interpretation and post-processing in cardiovascular magnetic resonance-2020 update. Journal of Cardiovascular Magnetic Resonance. 2020;22(1):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prince SJ. Computer vision: models, learning, and inference. Cambridge University Press; 2012. [Google Scholar]
- 12.Moore GE. Cramming more components onto integrated circuits. McGraw-Hill; New York, NY, USA:; 1965. [Google Scholar]
- 13.DOMO. Data Never Sleeps 8.0. 2020.
- 14.McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. The bulletin of mathematical biophysics. 1943;5(4):115–33. [PubMed] [Google Scholar]
- 15.Amato F, López A, Peña-Méndez EM, Vaňhara P, Hampl A, Havel J. Artificial neural networks in medical diagnosis. Elsevier; 2013. [Google Scholar]
- 16.Brownlee J Deep learning for computer vision: image classification, object detection, and face recognition in python. Machine Learning Mastery; 2019. [Google Scholar]
- 17.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S et al. Imagenet large scale visual recognition challenge. International journal of computer vision. 2015;115(3):211–52. [Google Scholar]
- 18.Wu J Introduction to convolutional neural networks. National Key Lab for Novel Software Technology Nanjing University China. 2017;5:23. [Google Scholar]
- 19.Jordan J An overview of semantic image segmentation. 2018.
- 20.Long J, Shelhamer E, Darrell T, editors. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015. [DOI] [PubMed] [Google Scholar]
- 21.Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention; 2015: Springer. [Google Scholar]
- 22.Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C. The importance of skip connections in biomedical image segmentation. Deep learning and data labeling for medical applications. Springer; 2016. p. 179–87. [Google Scholar]
- 23.Yu F, Koltun V. Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:151107122. 2015. [Google Scholar]
- 24.Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence. 2017;40(4):834–48. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Qin C, Qiu H, Tarroni G, Duan J, Bai W et al. Deep learning for cardiac image segmentation: A review. Frontiers in Cardiovascular Medicine. 2020;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai L, Gao J, Zhao D. A review of the application of deep learning in medical image classification and segmentation. Annals of translational medicine. 2020;8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asgari Taghanaki S, Abhishek K, Cohen JP, Cohen-Adad J, Hamarneh G. Deep semantic segmentation of natural and medical images: a review. Artificial Intelligence Review. 2021;54(1):137–78. doi: 10.1007/s10462-020-09854-1. [DOI] [Google Scholar]
- 28.Liu S, Wang Y, Yang X, Lei B, Liu L, Li SX et al. Deep learning in medical ultrasound analysis: a review. Engineering. 2019;5(2):261–75. [Google Scholar]
- 29.Williams L, Carrigan A, Auffermann W, Mills M, Rich A, Elmore J et al. The invisible breast cancer: Experience does not protect against inattentional blindness to clinically relevant findings in radiology. Psychonomic bulletin & review. 2020:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapley R, Tolhurst D. Edge detectors in human vision. The Journal of physiology. 1973;229(1):165–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burr DC, Morrone MC, Spinelli D. Evidence for edge and bar detectors in human vision. Vision research. 1989;29(4):419–31. [DOI] [PubMed] [Google Scholar]
- 32.Heath M, Sarkar S, Sanocki T, Bowyer K. Comparison of edge detectors: a methodology and initial study. Computer vision and image understanding. 1998;69(1):38–54. [Google Scholar]
- 33.Anderson SJ, Burr DC. Spatial and temporal selectivity of the human motion detection system. Vision research. 1985. [DOI] [PubMed] [Google Scholar]
- 34.Legge GE, Campbell F. Displacement detection in human vision. Vision research. 1981;21(2):205–13. [DOI] [PubMed] [Google Scholar]
- 35.Rose A Vision: human and electronic. Springer Science & Business Media; 2013. [Google Scholar]
- 36.Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annual review of biomedical engineering. 2000;2(1):315–37. [DOI] [PubMed] [Google Scholar]
- 37.Kim Y-C, Kim KR, Choe YH. Automatic myocardial segmentation in dynamic contrast enhanced perfusion MRI using Monte Carlo dropout in an encoder-decoder convolutional neural network. Computer methods and programs in biomedicine. 2020;185:105150. [DOI] [PubMed] [Google Scholar]
- 38.Tan LK, McLaughlin RA, Lim E, Abdul Aziz YF, Liew YM. Fully automated segmentation of the left ventricle in cine cardiac MRI using neural network regression. Journal of Magnetic Resonance Imaging. 2018;48(1):140–52. [DOI] [PubMed] [Google Scholar]
- 39.Blendowski M, Bouteldja N, Heinrich MP. Multimodal 3D medical image registration guided by shape encoder–decoder networks. International journal of computer assisted radiology and surgery. 2020;15(2):269–76. [DOI] [PubMed] [Google Scholar]
- 40.Chartsias A, Joyce T, Papanastasiou G, Semple S, Williams M, Newby DE et al. Disentangled representation learning in cardiac image analysis. Medical image analysis. 2019;58:101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang N, Yang G, Gao Z, Xu C, Zhang Y, Shi R et al. Deep learning for diagnosis of chronic myocardial infarction on nonenhanced cardiac cine MRI. Radiology. 2019;291(3):606–17. [DOI] [PubMed] [Google Scholar]
- 42.Jafari MH, Girgis H, Van Woudenberg N, Moulson N, Luong C, Fung A et al. Cardiac point-of-care to cart-based ultrasound translation using constrained CycleGAN. International journal of computer assisted radiology and surgery. 2020;15(5):877–86. [DOI] [PubMed] [Google Scholar]
- 43.Ruijsink B, Puyol-Antón E, Oksuz I, Sinclair M, Bai W, Schnabel JA et al. Fully automated, quality-controlled cardiac analysis from CMR: validation and large-scale application to characterize cardiac function. Cardiovascular Imaging. 2020;13(3):684–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarroni G, Oktay O, Bai W, Schuh A, Suzuki H, Passerat-Palmbach J et al. Learning-based quality control for cardiac MR images. IEEE transactions on medical imaging. 2018;38(5):1127–38. [DOI] [PubMed] [Google Scholar]
- 45.Kay FU, Abbara S, Joshi PH, Garg S, Khera A, Peshock RM. Identification of high-risk left ventricular hypertrophy on calcium scoring cardiac computed tomography scans: validation in the DHS. Circulation: Cardiovascular Imaging. 2020;13(2):e009678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia JDV, Navkar NV, Gui D, Morales CM, Christoforou EG, Ozcan A et al. A platform integrating acquisition, reconstruction, visualization, and manipulator control modules for MRI-guided interventions. Journal of digital imaging. 2019;32(3):420–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dawes TJ, de Marvao A, Shi W, Fletcher T, Watson GM, Wharton J et al. Machine learning of three-dimensional right ventricular motion enables outcome prediction in pulmonary hypertension: a cardiac MR imaging study. Radiology. 2017;283(2):381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Augusto JB, Davies RH, Bhuva AN, Knott KD, Seraphim A, Alfarih M et al. Diagnosis and risk stratification in hypertrophic cardiomyopathy using machine learning wall thickness measurement: a comparison with human test-retest performance. The Lancet Digital Health. 2020;3(1):e20–e8. [DOI] [PubMed] [Google Scholar]
- 49.Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng P-A et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE transactions on medical imaging. 2018;37(11):2514–25. [DOI] [PubMed] [Google Scholar]
- 50.Radau P, Lu Y, Connelly K, Paul G, Dick A, Wright G. Evaluation framework for algorithms segmenting short axis cardiac MRI. The MIDAS Journal-Cardiac MR Left Ventricle Segmentation Challenge. 2009;49. [Google Scholar]
- 51.Suinesiaputra A, Cowan BR, Al-Agamy AO, Elattar MA, Ayache N, Fahmy AS et al. A collaborative resource to build consensus for automated left ventricular segmentation of cardiac MR images. Medical image analysis. 2014;18(1):50–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raisi-Estabragh Z, Harvey NC, Neubauer S, Petersen SE. Cardiovascular magnetic resonance imaging in the UK Biobank: a major international health research resource. European Heart Journal-Cardiovascular Imaging. 2021;22(3):251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Commandeur F, Goeller M, Betancur J, Cadet S, Doris M, Chen X et al. Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE transactions on medical imaging. 2018;37(8):1835–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong Y, Commandeur F, Cadet S, Goeller M, Doris M, Chen X et al. , editors. Deep learning-based stenosis quantification from coronary CT angiography. Medical Imaging 2019: Image Processing; 2019: International Society for Optics and Photonics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyagawa M, Costa MGF, Gutierrez MA, Costa JPGF, Costa Filho CF, editors. Lumen segmentation in optical coherence tomography images using convolutional neural network. 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2018: IEEE. [DOI] [PubMed] [Google Scholar]
- 56.Oktay O, Ferrante E, Kamnitsas K, Heinrich M, Bai W, Caballero J et al. Anatomically constrained neural networks (ACNNs): application to cardiac image enhancement and segmentation. IEEE transactions on medical imaging. 2017;37(2):384–95. [DOI] [PubMed] [Google Scholar]
- 57.Leclerc S, Smistad E, Pedrosa J, Østvik A, Cervenansky F, Espinosa F et al. Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. IEEE transactions on medical imaging. 2019;38(9):2198–210. [DOI] [PubMed] [Google Scholar]
- 58.Ouyang D, He B, Ghorbani A, Yuan N, Ebinger J, Langlotz CP et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature. 2020;580(7802):252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geis JR, Brady AP, Wu CC, Spencer J, Ranschaert E, Jaremko JL et al. Ethics of artificial intelligence in radiology: summary of the joint European and North American multisociety statement. Canadian Association of Radiologists Journal. 2019;70(4):329–34. [DOI] [PubMed] [Google Scholar]
- 60.Rieke N, Hancox J, Li W, Milletari F, Roth HR, Albarqouni S et al. The future of digital health with federated learning. NPJ digital medicine. 2020;3(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheller MJ, Edwards B, Reina GA, Martin J, Pati S, Kotrotsou A et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Scientific reports. 2020;10(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential biases in machine learning algorithms using electronic health record data. JAMA internal medicine. 2018;178(11):1544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D, editors. Grad-cam: Visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE international conference on computer vision; 2017. [Google Scholar]
- 64.Krakovna V Specification gaming examples in AI. 2018.
- 65.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM et al. Dermatologist-level classification of skin cancer with deep neural networks. nature. 2017;542(7639):115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS medicine. 2018;15(11):e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Group ISW. Software as a Medical Device (SaMD): key definitions. Published online December. 2013;9:9. [Google Scholar]
- 68.US Food and Drug Administration. Evaluation of automatic class III designation (de novo) summaries. 2016.
- 69.Food US and Administration Drug. Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD) Action Plan 2021.
- 70.Miller DD, Brown EW. Artificial intelligence in medical practice: the question to the answer? The American journal of medicine. 2018;131(2):129–33. [DOI] [PubMed] [Google Scholar]
- 71.Antley MA, Antley R. Obsolescence: The physician or the diagnostician role. Academic Medicine. 1972;47(9):737–8. [PubMed] [Google Scholar]
- 72.Dubin SS. Obsolescence or lifelong education: A choice for the professional. American Psychologist. 1972;27(5):486. [Google Scholar]
- 73.Rittel HW, Webber MM. Dilemmas in a general theory of planning. Policy sciences. 1973;4(2):155–69. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.