Abstract
Background
Microbial cell-free DNA (mcfDNA) sequencing of plasma can identify the presence of a pathogen in a host. In this study, we evaluated the duration of pathogen detection by mcfDNA sequencing vs conventional blood culture in patients with bacteremia.
Methods
Blood samples from patients with culture-confirmed bloodstream infection were collected within 24 hours of the index positive blood culture and 48 to 72 hours thereafter. mcfDNA was extracted from plasma, and next-generation sequencing was applied. Reads were aligned against a curated pathogen database. Statistical significance was defined with Bonferroni adjustment for multiple comparisons (P < .0033).
Results
A total of 175 patients with Staphylococcus aureus bacteremia (n = 66), gram-negative bacteremia (n = 74), or noninfected controls (n = 35) were enrolled. The overall sensitivity of mcfDNA sequencing compared with index blood culture was 89.3% (125 of 140), and the specificity was 74.3%. Among patients with bacteremia, pathogen-specific mcfDNA remained detectable for significantly longer than conventional blood cultures (median 15 days vs 2 days; P < .0001). Each additional day of mcfDNA detection significantly increased the odds of metastatic infection (odds ratio, 2.89; 95% confidence interval, 1.53–5.46; P = .0011).
Conclusions
Pathogen mcfDNA identified the bacterial etiology of bloodstream infection for a significantly longer interval than conventional cultures, and its duration of detection was associated with increased risk for metastatic infection. mcfDNA could play a role in the diagnosis of partially treated endovascular infections.
Keywords: microbial cell, free diagnostics, bacteremia
Among patients with bacteremia, microbial cell-free DNA was detectable for significantly longer than conventional blood cultures and duration of detection was associated with metastatic infections.
Accurate microbial diagnosis of a bloodstream infection is critical [1, 2]. While blood culture remains the gold standard for diagnosis of bloodstream infection, its sensitivity may be as low as 38% in individuals with severe sepsis [3, 4]. Conventional blood culture systems require viable organisms in the bloodstream; however, antibiotic therapy rapidly diminishes the likelihood that blood cultures will identify a pathogen. A diagnostic platform that can identify bloodstream pathogens even when conventional culture systems fail to do so could improve patient management, especially in syndromes such as culture-negative endocarditis.
Recent studies indicate that microbial cell-free DNA (mcfDNA) may be a valuable diagnostic platform for the identification of pathogens in severely ill patients with bloodstream infections [5, 6]. In this study, we evaluated the presence and duration of mcfDNA in patients treated for culture-confirmed bacterial bloodstream infections.
METHODS
Study Population
This was a prospective, single-center, observational, cohort study of hospitalized patients with Staphylococcus aureus bacteremia (SAB) or gram-negative bacteremia (GNB) from July 2016 to April 2018. Patients were eligible if they were aged ≥18 years, were hospitalized at Duke University Hospital, had a positive blood culture for either S. aureus or a gram-negative pathogen, and had a blood sample collected in a K2-EDTA tube to be used for mcfDNA testing within 24 to 48 hours of the positive blood culture. Additional mcfDNA testing was performed for each patient every 2–3 days for up to a total of 5 samples (including the index sample). An uninfected control group consisting of patients hospitalized during the study period with no clinical suspicion of infection, no current or past history of a blood or nonblood culture, and no positive cultures during their current admission was also enrolled.
Patients were ineligible for the study if they failed to understand instructions or comply with study-related procedures or if the treating physician believed the patient to have any condition that would prevent the patient from completing the study. The Duke Institutional Review Board (IRB) approved the study. Patients (or legally authorized representatives) provided written informed consent. If a patient died prior to notification of blood culture results, the patient was enrolled using an IRB-approved notification of decedent research.
Clinical Data Abstraction
Clinical data were collected from each participant’s electronic medical record using a standardized case report form. Information including demographics, comorbidities, hospitalization, and clinical outcomes was obtained for all in-patient participants. Route of infection was defined as hospital acquired, community acquired, or healthcare associated community acquired, as previously defined [7]. Time to blood culture positivity was determined by calculating the interval between the time blood culture was collected and the time blood culture demonstrated any growth. Duration between antibiotic initiation and diagnostic testing was determined by calculating the days between antibiotic initiation and blood culture collection and between antibiotic initiation and mcfDNA test collection. Sepsis was defined as having a positive blood culture and having met at least 2 of the following systemic inflammatory response syndrome (SIRS) criteria: temperature >38°C or <36°C, heart rate >90 beats per minute, respiratory rate >20 or partial pressure of carbon dioxide <32, white blood cell count >12 000 cells/mm3 or <4000 cells/mm3, or having >10% immature neutrophil forms (band neutrophils) [8]. Septic shock was defined as sepsis with hypotension (systolic blood pressure ≤90 mm Hg) and perfusion abnormalities, as previously described [8]. Acute Physiology and Chronic Health Evaluation II scores, including Acute Physiology Scores, were calculated on the day of the index positive blood culture [9]. Acute renal failure was defined as serum creatinine >1.5 times the baseline creatinine or increasing by >0.3 mg/dL within 48 hours [10]. Persistent bacteremia was defined as the presence of repeat positive blood cultures following appropriate antimicrobial therapy after ≥5 days for SAB patients [11] and ≥3 days for patients with GNB [12]. Patients were considered to have metastatic infection if they developed any of the following: infective endocarditis, septic emboli, septic thrombophlebitis, vertebral osteomyelitis, septic arthritis, a metastatic abscess, or other deep tissue abscess, as previously defined [11]. Infective endocarditis was defined based on modified Duke criteria [13].
Laboratory Studies
Bacterial isolates were speciated by the Duke Clinical Microbiology Laboratory using standard techniques. Minimum inhibitory concentration (MIC) values were determined using the MicroScan Walkaway system (microbroth dilution method). The MIC breakpoint values for each antibiotic were defined according to the most recent Clinical and Laboratory Standards Institute guidelines [14].
mcfDNA Sequencing
All methods and materials for mcfDNA sequencing have been previously described [15]. Briefly, the Karius test is a laboratory-developed test performed in a Karius’s Clinical Laboratory Improvement amendments (CLIA)-certified and CAP-accredited laboratory that detects mcfDNA in plasma. After mcfDNA is extracted and next-generation sequencing is performed, human reads are removed and the remaining sequences are aligned to a curated database of >1400 organisms. mcfDNA from organisms present above a statistical threshold are reported and quantified in molecules per microliter. See Supplementary Methods for full details.
Statistical Analyses
The distributions of continuous measures are presented as medians and quartiles, and categorical variables are evaluated using counts and percentages. Statistical comparisons between groups were made with a 1-sided McNemar test for binary diagnostic test result concordance (blood culture vs mcfDNA), the Kruskal-Wallis test for continuous variables, and the Fisher exact test for categorical variables. Statistical significance was set at P < .05. Each mcfDNA test and blood culture time point that detected the index blood culture pathogen was used for longitudinal analysis. Day“0 is defined as the day the index blood culture was collected; duration of positivity was calculated using the day of sample collection. Survival curves for duration of positivity were generated using GraphPad Prism (San Diego, CA). Duration of positivity curves for each test method (blood culture vs mcfDNA test) was compared using a log-rank (Mantel-Cox) test. Multiple testing correction was performed for all log-rank tests with the Bonferroni method with desired P = .05 with m = 15; the final P value for statistical significance after multiple comparisons adjustment was .0033. All comparisons for which the Bonferroni method was applied are indicated with an asterisk throughout the text. For all remaining comparisons, statistical significance was defined as P < .05.
Logistic regression prediction models were fit using the rms R package (version 6.1–0) in R (version 4.0.3). Model summary statistics were formatted using Stargazer (version 5.2.2), and logistic regression plots were created using ggplot2 (version 3.3.3). Additional methods can be found in the Supplementary Methods and Supplementary Table 4.
Exploratory Analyses of mcfDNA Results Obtained From the Uninfected Control Group
We performed additional exploratory analyses to consider the significance of mcfDNA false-positive results in the uninfected control group (eg, bacteria identified by mcfDNA in the blood of patients without active clinical infection who had no blood cultures performed). In these exploratory analyses, we interpreted the clinical significance of mcfDNA results in the uninfected control group as “false positive” if the Karius test identified any pathogen included on the list of the most common bacterial causes of bloodstream infection from the SENTRY Antimicrobial Surveillance Program [16] and as “true negative” otherwise.
RESULTS
Clinical Characteristics
A total of 140 patients with positive blood cultures (SAB n = 66; GNB n = 74) were included in the study (Table 1). Additional microbiological characteristics are listed in Table 2. Patients with SAB and GNB differed by route of infection (P = .002), median time to positive blood culture results (SAB 21.2 hours vs GNB 18.9 hours; P = .031), rates of metastatic infection (SAB 43.9% vs GNB 12.2%; P < .001*), and rates of persistent bacteremia (SAB 27.3% vs GNB 4.1%; P < .001*). Frequency of all-cause mortality between the 2 groups was similar (SAB 19.7% vs GNB 24.3%; P = .510). The average time from initiating antibiotics was significantly shorter for conventional blood cultures than for initial mcfDNA test collection (blood culture 0.16 days ± 2.07 days vs mcfDNA 1.60 days ± 2.72 days; P < .001*).
Table 1.
Clinical Characteristics of the Study Population
| Characteristic | Overall Bacteremia Cohort, N = 140 |
Staphylococcus aureus Bacteremia, N = 66 |
Gram-Negative Bacteremia, N = 74 |
P Value | Controls, N = 35 |
|---|---|---|---|---|---|
| Demographics | |||||
| Age, median (Q1, Q3), y | 62.0 (50.8, 71.0) | 61.5 (52.3, 69.8) | 63.0 (48.5, 71.0) | .759 | 61 (52.5, 68.0) |
| Sex (female) | 56 (40) | 22 (33.3) | 34 (45.9) | .167 | 16 (45.7) |
| Race | .035 | ||||
| Black | 45 (32.1) | 26 (29.4) | 19 (25.7) | 8 (22.9) | |
| White | 86 (61.4) | 39 (59.1) | 47 (63.5) | 25 (71.4) | |
| Other | 9 (6.4) | 1 (1.5) | 8 (10.8) | 2 (5.7) | |
| Comorbidities | |||||
| Dialysis dependent | 19 (13.6) | 11 (16.7) | 8 (10.8) | .334 | |
| Diabetes mellitus | 64 (45.7) | 31 (47.0) | 33 (44.6) | .865 | |
| Neoplasm | 32 (22.9) | 12 (18.2) | 20 (27.0) | .233 | |
| Living with human immunodeficiency virus | 2 (1.4) | 2 (3.0) | 0 (0) | .221 | |
| Transplant recipient | 21 (17.6) | 6 (9.1) | 15 (20.3) | .096 | |
| Injection drug use | 8 (6.3) | 5 (7.6) | 3 (4.1) | .475 | |
| Corticosteroid use (30 day) | 38 (27.1) | 17 (25.8) | 21 (28.4) | .849 | |
| Acute Physiology and Chronic Health Evaluation II, median (Q1, Q3) | 17 (13.0, 18.7) | 17.5 (13.0, 22.0) | 17.0 (13.0, 25.8) | .643 | |
| Acute Physiology Score, median (Q1, Q3) | 11 (7.0, 16.0) | 10 (7.0, 14.8) | 11.5 (7.0, 17.0) | .454 | |
| Route of infection | .002 | ||||
| Hospital acquired | 27 (19.3) | 6 (9.1) | 21 (28.4) | ||
| Healthcare associated, community acquired | 65 (46.4) | 40 (60.6) | 25 (33.8) | ||
| Nonhealthcare associated, community acquired | 48 (34.3) | 20 (30.3) | 28 (37.8) | ||
| Symptoms | |||||
| Fever at presentation | 95 (67.9) | 46 (69.7) | 49 (66.2) | .719 | |
| Fever >72 h | 17 (12.1) | 10 (15.2) | 7 (9.5) | .316 | |
| White blood cell count ×109/L, median (Q1, Q3) | 11.7 (8.2, 16.2) | 13.0 (9.3, 18.8) | 11.0 (6.4, 14.8) | .016 | |
| Time to blood culture positivity,a median (Q1, Q3), hours | 20.2 (16.7, 24.3) | 21.2 (18.0, 24.4) | 18.9 (15.4, 23.4) | .031 | |
| Complications | |||||
| Hospital length of stay median (Q1, Q3), d | 11.0 (7.0, 20.5) | 12.0 (9.0, 24.8) | 8.0 (5.0, 14.0) | <.001 | |
| Any metastatic infection | 38 (27.1) | 29 (43.9) | 9 (12.2) | <.001 | |
| Metastatic abscess | 13 (9.3) | 9 (13.6) | 4 (5.4) | .689 | |
| Septic emboli | 11 (7.9) | 9 (13.6) | 2 (2.7) | 1.000 | |
| Metastatic vertebral osteomyelitis | 6 (4.3) | 5 (7.6) | 1 (1.4) | 1.000 | |
| Metastatic nonvertebral osteomyelitis | 3 (2.1) | 2 (3.0) | 1 (1.4) | 1.000 | |
| Metastatic arthritis | 2 (1.4) | 2 (3.0) | 0 (0) | 1.000 | |
| Metastatic psoas abscess | 1 (0.7) | 1 (1.5) | 0 (0) | 1.000 | |
| Other metastatic infection | 4 (2.9) | 4 (6.1) | 0 (0) | .555 | |
| Infective endocarditis | 11 (7.9) | 10 (15.1) | 1 (1.4) | .237 | |
| Sepsis | 114 (81.4) | 53 (80.3) | 61 (82.4) | .829 | |
| Septic shock | 16 (11.4) | 8 (12.1) | 8 (10.8) | 1.000 | |
| Acute respiratory distress | 16 (11.4) | 9 (13.6) | 7 (9.5) | .596 | |
| Acute renal failure | 49 (35.0) | 21 (31.8) | 28 (37.8) | .483 | |
| Persistent bacteremia | 21 (15.0) | 18 (27.3) | 3 (4.1) | <.001 | |
| Outcomes | |||||
| Recurrent infection | 6 (4.3) | 3 (4.5) | 3 (4.1) | 1.000 | |
| Attributable mortality | 12 (8.6) | 6 (9.1) | 6 (8.1) | 1.000 | |
| All-cause mortality | 31 (22.1) | 13 (19.7) | 18 (24.3) | .547 |
Abbreviations: Q1, first quartile; Q3, third quartile.
aCalculated by time of blood culture flagged positive for growth minus time of blood culture collection.
Table 2.
Microbiologic Characteristics of the Study Cohort
| Microbiologic Characteristic | Overall Bacteremia Cohort, N = 140 (%) |
|---|---|
|
Staphylococcus aureus bacteremia |
66 |
| Methicillin-resistant S. aureus | 30 (45.5) |
| Methicillin-susceptible S. aureus | 36 (54.5) |
| Gram-negative bacteremia |
74 |
| Escherichia coli | 29 (39.2) |
| Klebsiella pneumoniae | 11 (14.9) |
| Pseudomonas aeruginosa | 11 (14.9) |
| Enterobacter cloacae/asburiae | 6 (8.1) |
| Serratia marcescens | 3 (4.1) |
| Enterobacter aerogenes | 2 (2.7) |
| Haemophilus influenzae | 2 (2.7) |
| Proteus mirabilis | 2 (2.7) |
| Other speciesa | 8 (10.8) |
aSpecies with 1 organism only: Acinetobacter baumannii, Burkholderia spp., Citrobacter amalonaticus, Enterobacter gergoviae, Morganella morganii, Providencia stuartii, Salmonella spp., polymicrobic with E. coli and K. pneumoniae.
Assay Performance
Sensitivity
The overall sensitivity of mcfDNA sequencing compared with the index blood culture for all patients with bacteremia was 89.3% (125 of 140 true positives and 9 of 35 false positives, McNemar P < .31), including 86.4% (57 of 66 true positives and 1 of 35 false positives; P < .027) for SAB and 91.9% (68 of 74 true positives and 5 of 35 false positives; P < 1.0) for GNB (Table 3, Table 4). False-negative index mcfDNA tests are delineated in Supplementary Table 1. Positive agreement between the index mcfDNA test and the index blood culture of all patients with bacteremia was 84.7% (116 of 137 true positives and 9 of 35 false positives; P < .044), including 78.1% (50 of 64 true positives and 1 of 35 false positives; P < .002) for SAB and 90.4% (66 of 73 true positives and 5 of 35 false positives; P < .77) for GNB.
Table 3.
Performance of Microbial Cell-Free DNA Test Versus Index Blood Culture
| True Positive | False Negative | False Positive | True Negative | Total | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|---|
| Index mcfDNA | 116 | 21 | 9 | 26 | 172a | 84.7% | 74.3%/88.6%b |
| Any mcfDNA | 125 | 15 | 13 | 22 | 175 | 89.3% | Not applicable |
Sensitivity and specificity of either the index mcfDNA test or any mcfDNA obtained for each case vs the index blood culture. Listed by type of bacteremia with cumulative n = 172 in the index mcfDNA vs index blood culture analysis, n = 175 in the all mcfDNA vs index blood culture analysis.
aExcludes 3 index samples that were QNS. Specificity based on 35 negative controls admitted but no suspicion of an infection.
bSpecificity calculation based on the most common causes of bloodstream infection by the SENTRY Antimicrobial Surveillance Program.
Abbreviations: mcfDNA, microbial cell free DNA; QNS, quantity not sufficient.
Table 4.
Performance of Microbial Cell-Free DNA Test Versus Index Blood Culture by Pathogen Type
| Any mcfDNA Versus Index Blood Culture, N = 140 | Index mcfDNA Versus Index Blood Culture, N = 137 | |||||
|---|---|---|---|---|---|---|
| Study Group | mcfDNA Positive | mcfDNA Negative | Agreement, % | mcfDNA Positive | mcfDNA Negative | Agreement, % |
| Gram-negative bacteremia | 68 | 6 | 91.9 | 66 | 7 | 90.4 |
| Staphylococcus aureus bacteremia | 57 | 9 | 86.4 | 50 | 14 | 78.1 |
| Methicillin-resistant S. aureus | 29 | 2 | 93.5 | 28 | 4 | 87.5 |
| Methicillin-susceptible S. aureus | 28 | 7 | 80 | 22 | 10 | 68.8 |
| Cumulative | 125 | 15 | 89.3 | 116 | 21 | 84.7 |
| Controla | n/a | n/a | n/a | 9 | 26 | n/a |
Percent agreement of any of the mcfDNA tests (n = 140) and index mcfDNA (n = 137) obtained for each case vs the index blood culture for all patients with positive blood cultures, listed by type of bacteremia.
Abbreviations: mcfDNA, microbial cell-free DNA; n/a, not applicable.
a The control group consisted of patients who did not have a suspicion for infection during their admission and did not have blood cultures performed; thus, no agreement could be calculated.
Specificity
Overall specificity of the mcfDNA assay in this study was 74.3%. This overall specificity is due to the fact that mcfDNA identified an organism in 9 of 35 uninfected controls. In post hoc exploratory analyses, we considered the significance of these 9 false-positive results. Five of 9 false-positive pathogen detections were not common causes of bloodstream infection characterized by the SENTRY Antimicrobial Surveillance Program [16]. Four of these 5 false-positive uncommon causes of bloodstream infection included organisms that were not included in the study cohort (eg, neither S. aureus nor gram-negative bacteria). Thus, when we included the remaining 4 false-positive common causes of bloodstream infection in a post hoc calculation, specificity for mcfDNA was 88.6% (Supplementary Table 2).
mcfDNA Duration of Positivity
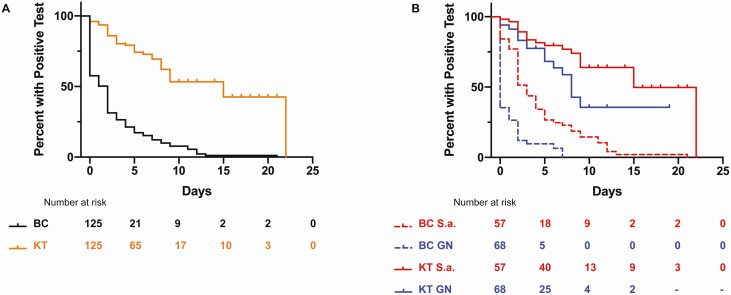
The median duration of pathogen identification by mcfDNA was significantly longer than identification by conventional blood cultures in the overall cohort with bacteremia (15 days vs 2 days; P < .0001* by log-rank Mantel-Cox test; Figure 1A). This finding persisted for the subgroups of SAB (15 days vs 3 days; P < .0001* log-rank Mantel-Cox test) and GNB (8 days vs 0 days; P < .0001* log-rank Mantel-Cox test; Figure 1B).
Figure 1.
Duration of microbial cell-free DNA (mcfDNA) positivity compared with blood culture for Staphylococcus aureus and GN bacteremia. A, Kaplan-Meier curve showing duration of positivity for KT vs BC for all bloodstream infections. The median duration of positivity was significantly different in mcfDNA at 15 days vs BC at 2 days (P < .0001 by log-rank test). B, Kaplan-Meier curve showing duration of positivity for KT and BC for S.a., the median duration of positivity was significantly different in mcfDNA at 15 days vs BC at 3 days (P < .0001 by log-rank test). For GN infections, the median duration of positivity was significantly different in mcfDNA at 8 days vs BC at 0 day (P < .0001 by log-rank test). Abbreviations: BC, blood culture; GN, gram-negative; KT, Karius test; S.a., Staphylococcus aureus.
Duration of Detectable mcfDNA in Metastatic Infection
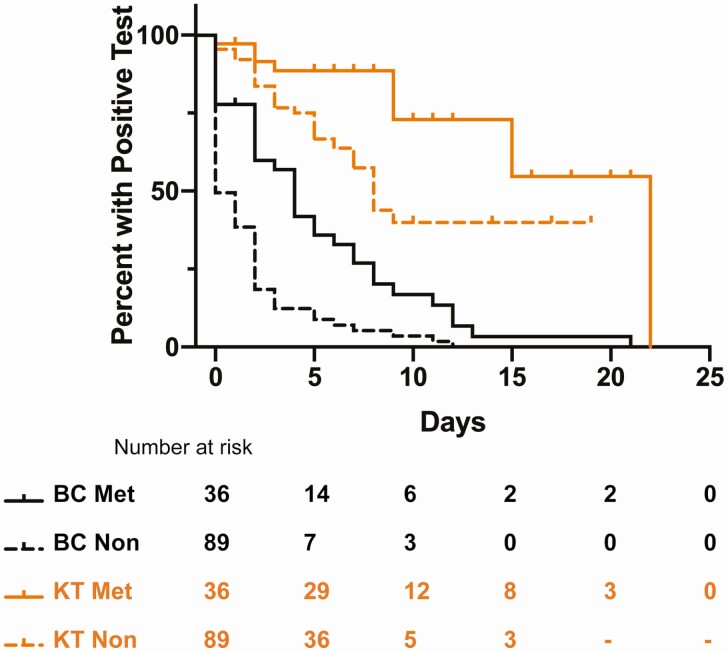
Median duration of detectable mcfDNA was longer in bacteremic patients with metastatic infection than in bacteremic patients without metastatic infection (22 days vs 8 days; P = .0054; Figure 2, Supplementary Figure 1A). Next, we examined pre- and post-procedure mcfDNA levels in patients with metastatic infection who underwent procedures to achieve source control (eg, surgical debridement, device removal; Supplementary Figure 2). Those with definitive source control tended to exhibit a more rapid decrease in their mcfDNA, whereas those with persistent foci of infection had a slower decay of mcfDNA. These differences in decay did not reach statistical significance following multiple comparisons adjustment. Additional information regarding the nature of these procedures and any remaining foci of infection is provided in Supplementary Table 3.
Figure 2.
Duration of microbial cell-free DNA positivity in patients with metastatic infection. Kaplan-Meier curves showing duration of positivity for KT and BC in patients with metastatic vs nonmetastatic infection. BC metastatic median duration of positivity was 4 days. BC nonmetastatic median duration of positivity was 0 days. KT metastatic duration of positivity was 22 days. KT nonmetastatic duration of positivity was 8 days. P = .0054 by log-rank (Mantel-Cox) test. Abbreviations: BC, blood culture; KT, Karius test; Met, metastatic; Non, nonmetastatic.
Longitudinal mcfDNA Detection and Presence of Metastatic Infection
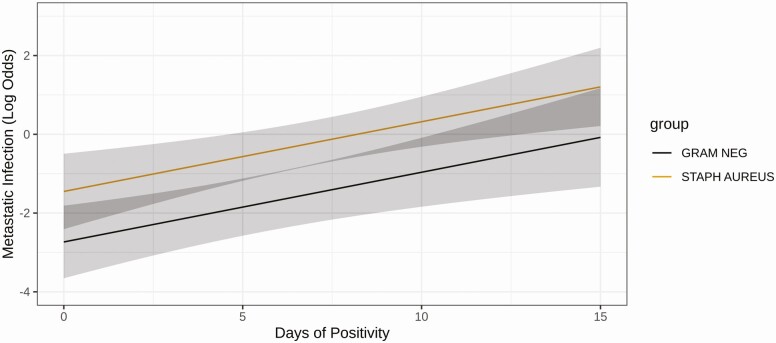
Logistic regression was used to assess associations between the duration of detection of mcfDNA and the class of infection (SAB or GNB) with the presence of metastatic infection in the source patient from whom the samples were drawn. Model coefficients used for the regression analysis are provided in Supplementary Table 4. The additive 2-predictor model possessed a pseudo r-squared value of 0.315 (resampling corrected index = 0.29), a c-index of 0.791, and a Brier score of 0.16 (resampling corrected index = 0.16). Each additional day of mcfDNA detection significantly increased the odds of metastatic infection (odds ratio [OR], 2.89; 95% confidence interval [CI], 1.53–5.46; P = .0011*). Detection of S. aureus mcfDNA was associated with a higher risk of metastatic infection compared with detection of gram-negative bacteria by mcfDNA (OR, 3.6; 95% CI, 1.41–9.26; P = .0077; Figure 3).
Figure 3.
Prediction of Staphylococcus aureus metastatic infection using duration of microbial cell-free DNA (mcfDNA) detection/positivity. Logistic regression predictions of metastatic infection conditioned on days of detectable plasma mcfDNA and type of infection (S. aureus bacteremia or gram-negative bacteremia). Briefly, 125 patients were used to construct an additive 2-parameter model (pseudo r-squared 0.315, Brier score = 0.16, Harrell’s concordance index = 0.791). The predicted log of odds from the fitted model (y-axis) using days of detectable mcfDNA (x-axis; odds ratio [OR], 2.89; 95% confidence interval [CI], 1.53–5.46 for each additional day) and class of infection (OR, 3.61; 95% CI, 1.40–9.26 for S. aureus infection) are shown. Abbreviations: Gram Neg, gram-negative; Staph aureus, Staphylococcus aureus.
DISCUSSION
For more than a century, the diagnosis of bacteremia has hinged on culturing live organisms from blood. mcfDNA represents a potential paradigm shift in the diagnosis of bacterial bloodstream infection. In the current study, we found that mcfDNA testing could identify the etiology and suggest the presence of metastatic manifestations in patients with bloodstream infection. By evaluating both conventional blood cultures and mcfDNA longitudinally in patients with confirmed bloodstream infection, we made 2 key observations.
First, we found that mcfDNA from the causative pathogen was detected almost 2 weeks after conventional blood cultures became negative. This finding is consistent with previous studies [5, 6] and suggests that mcfDNA can enhance our ability to accurately diagnose and treat patients with culture-negative endovascular infections caused by prior antibiotic therapy (Supplementary Figure 1B). For example, a specific bacterial diagnosis with mcfDNA could help to avoid the broad-spectrum, toxic empiric treatment recommended [17] for culture-negative endocarditis.
Second, we found that duration of mcfDNA detection in patients with bloodstream infection was associated with the presence of metastatic infection. Our results indicated that each additional day of detectable mcfDNA increased the odds of metastatic infection by almost 3. This finding suggests that the duration of mcfDNA detection may eventually provide clinicians with an ability to individualize antibiotic therapy for bloodstream infections. For example, patients treated for bloodstream infections with a known pathogen whose mcfDNA assay converts to nondetectable during their antibiotic course could be considered for abbreviated durations of treatment or for step-down to oral antibiotics. Conversely, patients with persistent levels of detectable mcfDNA could be identified as high risk for metastatic infection and would benefit from additional diagnostic imaging and evaluation (eg, transesophageal echocardiography, positron emission tomography scans) [18]. Such metastatic foci may often be occult [19, 20], yet generally require longer durations of therapy and/or surgical source control [11, 18, 21].
While not statistically significant, we also observed a general pattern of decreasing mcfDNA levels in patients with metastatic infections who underwent debridement procedures (Supplementary Figure 2, Supplementary Table 3). One example is patient KA1099, who achieved definitive source control with amputation. There were 12 patients who exhibited persistent mcfDNA levels after attempted source control, 11 of whom had clinical or radiographic evidence of additional or remaining infectious foci of infection. Additional analyses demonstrated a predictable decline in mcfDNA abundance for both SAB and GNB in response to antibiotic treatment (Supplementary Methods, Supplementary Figure 3). Further prospective studies are needed to evaluate the utility of mcfDNA in estimating the burden of infection in patients with complicated bloodstream infections.
Finally, our results are consistent with prior published reports of the sensitivity of mcfDNA compared with conventional blood culture [15, 22, 23]. Additionally, the majority (17 of 21) of the false-negative samples had evidence of the causative pathogen in the raw data (Supplementary Table 1). Unlike the evaluation of mcfDNA in our unambiguous clinical categories of SAB and GNB, the clinical consequences of organisms identified by mcfDNA in our uninfected control patients, whose selection was based on the clinical absence of active infection, is unknown. False-positive mcfDNA results in a cohort of hospitalized, uninfected patients may represent source contamination, commensalism, mucosal barrier injury (MBI), or event occult bacteremia related to minor procedures such as toothbrushing [24]. Comparison of the organisms found by sequencing in the uninfected control group against common causes of bloodstream infection can provide additional context into potential contaminants, commensals, or those resulting from MBI. In order to contextualize the false positives in the uninfected control group, we explored specificity using various parameters (including comparison against common causes of bloodstream infection defined by the SENTRY Antimicrobial Surveillance Program), and the range was 74.3% to 88.6% [16]. This approach was taken by Goggin and colleagues in their study on mcfDNA in bloodstream infections in immunocompromised hosts [25]. Similar methods of adjudication were undertaken by the Centers for Disease Control and Prevention/National Healthcare Safety Network to prevent misclassification of bloodstream infections caused by MBI in immunocompromised hosts [26].
Our study has limitations. First, our uninfected control group consisted of hospitalized individuals without clinical suspicion of infection. Given its ability to identify pathogens in culture-negative infections, it is possible that some controls who had detectable bacteria by mcfDNA had occult bacteremia or mucosal barrier injury [15]. In addition, the use of remnant K2-EDTA plasma from routine clinical care may have introduced potential contamination. Second, the mean time from antibiotic initiation to mcfDNA collection (1.6 days ± 2.72) was significantly longer than the mean time from antibiotic initiation to blood culture collection (0.16 ± 2.07). This collection delay of the investigational mcfDNA relative to conventional blood cultures collected as a matter of routine clinical care may have impacted its sensitivity compared with blood culture. Finally, the maximum of 5 samples that were collected per patient may not have provided sufficient duration to fully demarcate the persistence of mcfDNA in each patient.
CONCLUSIONS
Despite these limitations, we demonstrate that a novel mcfDNA sequencing assay can accurately identify the etiology of causative pathogens in patients with bloodstream infection. Future studies are needed to evaluate the potential role of mcfDNA in managing patients with bacteremia, including diagnosing the etiology of culture-negative endocarditis, guiding decision-making on duration and route of antibiotic therapy, and identifying patients at high risk for underlying metastatic infections who require additional diagnostic procedures. The results of this study support the hypothesis that mcfDNA may ultimately offer an innovative method to personalize the management of bacterial bloodstream infection.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by funding from Karius Inc. Additionally, this work was supported by a grant to V. G. F. from the National Institutes of Health (NIH; K24-AI093969).
Potential conflicts of interests. V. G. F. reports personal fees from Novartis, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, Medicines Co, Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Basilea, Affinergy, Janssen, xBiotech, Contrafect, Regeneron, Basilea, Destiny, Amphliphi Biosciences, and Integrated Biotherapeutics; C3J, grants from NIH, MedImmune, Cerexa/Forest/Actavis/Allergan, Pfizer, Advanced Liquid Logics, Theravance, Novartis, Cubist/Merck, Medical Biosurfaces, Locus, Affinergy, Contrafect, Karius, Genentech, Regeneron, Basilea, and Janssen; educational fees from Green Cross, Cubist, Cerexa, Durata, Theravance, and Debiopharm; royalties from UpToDate; stock options from Valanbio; and a patent sepsis diagnostics pending. A. A. A. reports being an employee/the medical director at Karius Inc. D. H. H., N. D., and C. R. D. are employees at Karius Inc. E. R. S. is an employee at Karius Inc and reports personal fees from Sema4. L. B. is an employee at Karius Inc and reports personal fees from Karius Inc during the time of the study and outside of the submitted work. T. A. B. is an employee of Karius Inc, reports personal fees outside the submitted work, has a patent US20170016048A1 pending, a patent US20170275691A1 issued, and a patent US 20190256891A1 pending. H. S. reports being a former employee of Karius Inc. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Emily M Eichenberger, Division of Infectious Diseases, Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
Christiaan R de Vries, Karius Inc, Redwood City, California, USA.
Felicia Ruffin, Division of Infectious Diseases, Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
Batu Sharma-Kuinkel, Division of Infectious Diseases, Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
Lawrence Park, Division of Infectious Diseases, Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
David Hong, Karius Inc, Redwood City, California, USA.
Erick R Scott, Karius Inc, Redwood City, California, USA.
Lily Blair, Karius Inc, Redwood City, California, USA.
Nicholas Degner, Karius Inc, Redwood City, California, USA.
Desiree H Hollemon, Karius Inc, Redwood City, California, USA.
Timothy A Blauwkamp, Karius Inc, Redwood City, California, USA.
Carine Ho, Karius Inc, Redwood City, California, USA.
Hon Seng, Karius Inc, Redwood City, California, USA.
Pratik Shah, Department of Medicine, Nova Southeastern University, Fort Lauderdale, Florida, USA.
Lisa Wanda, Department of Medicine, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA; Department of Medicine, University of North Carolina Gillings School of Global Public Health, Chapel Hill, North Carolina, USA.
Vance G Fowler, Jr, Division of Infectious Diseases, Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
Asim A Ahmed, Karius Inc, Redwood City, California, USA.
References
- 1. Scheer CS, Fuchs C, Gründling M, et al. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: a prospective clinical cohort study. Clin Microbiol Infect 2019; 25:326–31. [DOI] [PubMed] [Google Scholar]
- 2. Garnacho-Montero J, Gutiérrez-Pizarraya A, Escoresca-Ortega A, et al. De-escalation of empirical therapy is associated with lower mortality in patients with severe sepsis and septic shock. Intensive Care Med 2014; 40:32–40. [DOI] [PubMed] [Google Scholar]
- 3. Coburn B, Morris AM, Tomlinson G, Detsky AS. Does this adult patient with suspected bacteremia require blood cultures? JAMA 2012; 308:502–11. [DOI] [PubMed] [Google Scholar]
- 4. Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med 2013; 41: 580–637. [DOI] [PubMed] [Google Scholar]
- 5. Grumaz S, Grumaz C, Vainshtein Y, et al. Enhanced performance of next-generation sequencing diagnostics compared with standard of care microbiological diagnostics in patients suffering from septic shock. Crit Care Med 2019; 47:e394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grumaz S, Stevens P, Grumaz C, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med 2016; 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friedman ND, Kaye KS, Stout JE, et al. Health care–associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 2002; 137:791–7. [DOI] [PubMed] [Google Scholar]
- 8. American College of Chest Physicians. Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med 1992; 20:864–74. [PubMed] [Google Scholar]
- 9. Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med 1985; 13:818–29. [PubMed] [Google Scholar]
- 10. Kellum JA, Lameire N; KDIGO AKI Guideline Work Group. . Diagnosis, evaluation, and management of acute kidney injury: a KDIGO summary (part 1). Crit Care 2013; 17:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Souli M, Ruffin F, Choi SH, et al. Changing characteristics of Staphylococcus aureus bacteremia: results from a 21-year, prospective, longitudinal study. Clin Infect Dis 2019; 69:1868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris JA, Cobbs CG. Persistent gram-negative bacteremia. Observations in twenty patients. Am J Surg 1973; 125:705–17. [DOI] [PubMed] [Google Scholar]
- 13. Li JS, Sexton DJ, Mick N, et al. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis 2000; 30:633–8. [DOI] [PubMed] [Google Scholar]
- 14. Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed. CLSI Document M100. Wayne, PA: Clinical Laboratory Standards Institute, 2017. [Google Scholar]
- 15. Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 2019; 4:663–74. [DOI] [PubMed] [Google Scholar]
- 16. Diekema DJ, Hsueh PR, Mendes RE, et al. The microbiology of bloodstream infection: 20-year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 2019; 63:e00355–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baddour LM, Wilson WR, Bayer AS, et al. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: a scientific statement for healthcare professionals from the American Heart Association. Circulation 2015; 132:1435–86. [DOI] [PubMed] [Google Scholar]
- 18. Kuehl R, Morata L, Boeing C, et al. Defining persistent Staphylococcus aureus bacteraemia: secondary analysis of a prospective cohort study. Lancet Infect Dis 2020; 20:1409–17. [DOI] [PubMed] [Google Scholar]
- 19. Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 2015; 28:603–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holland TL, Raad I, Boucher HW, et al. ; Staphylococcal Bacteremia Investigators. . Effect of algorithm-based therapy vs usual care on clinical success and serious adverse events in patients with staphylococcal bacteremia: a randomized clinical trial. JAMA 2018; 320:1249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gopal AK, Fowler VG Jr, Shah M, et al. Prospective analysis of Staphylococcus aureus bacteremia in nonneutropenic adults with malignancy. J Clin Oncol 2000; 18:1110–5. [DOI] [PubMed] [Google Scholar]
- 22. Farnaes L, Wilke J, Ryan Loker K, et al. Community-acquired pneumonia in children: cell-free plasma sequencing for diagnosis and management. Diagn Microbiol Infect Dis 2019; 94:188–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rossoff J, Chaudhury S, Soneji M, et al. Noninvasive diagnosis of infection using plasma next-generation sequencing: a single-center experience. Open Forum Infect Dis 2019; 6:ofz327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lockhart PB, Brennan MT, Sasser HC, Fox PC, Paster BJ, Bahrani-Mougeot FK. Bacteremia associated with toothbrushing and dental extraction. Circulation 2008; 117:3118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goggin KP, Gonzalez-Pena V, Inaba Y, et al. Evaluation of plasma microbial cell-free DNA sequencing to predict bloodstream infection in pediatric patients with relapsed or refractory cancer. JAMA Oncol 2020; 6:552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steinberg JP, Coffin SE. Improving the central line-associated bloodstream infection surveillance definition: a work in progress. Infect Control Hosp Epidemiol 2013; 34:777–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.