Abstract
Vestimentiferan tubeworms thriving in sulfidic deep-sea hydrothermal vents and cold seeps are constrained by their nutritional reliance on chemoautotrophic endosymbionts. In a recent phylogenetic study using 16S ribosomal DNA, we found that endosymbionts from vent and seep habitats form two distinct clades with little variation within each clade. In the present study, we used two different approaches to assess the genetic variation among biogeographically distinct vestimentiferan symbionts. DNA sequences were obtained for the noncoding, internal transcribed spacer (ITS) regions of the rRNA operons of symbionts associated with six different genera of vestimentiferan tubeworms. ITS sequences from endosymbionts of host genera collected from different habitats and widely distributed vent sites were surprisingly conserved. Because the ITS region was not sufficient for distinguishing endosymbionts from different habitats or locations, we used a DNA fingerprinting technique, repetitive-extragenic-palindrome PCR (REP-PCR), to reveal differences in the distribution of repetitive sequences in the genomes of the bacterial endosymbionts. Most of the endosymbionts displayed unique REP-PCR patterns. A cladogram generated from these fingerprints reflected relationships that may be influenced by a variety of factors, including host genera, geographic location, and bottom type.
Invertebrates that are endemic to the highly sulfidic, reducing environments at deep-sea hydrothermal vents and cold seeps are commonly associated with chemosynthetic endosymbiotic bacteria (7, 18, 19). These bacteria oxidize the reduced sulfur compounds that are abundant in hydrothermal fluid (8, 36, 46). The resultant energy produced by the endosymbiont is coupled to the production of carbon sources that support the growth and maintenance of the invertebrate hosts (6, 8, 37, 48). The metabolic link between the invertebrate hosts and their endosymbionts has obvious implications for the dispersal and colonization strategies employed by the host organisms.
Vent endemic host organisms may employ one of three different mechanisms for transmission of their endosymbionts to the next generation. Previous studies have shown that the Vesicomyid bivalves form species-specific associations with endosymbiotic bacteria (12, 13, 16). Host specificity is maintained in these associations by vertical transmission of the bacteria through the egg from the parent to the offspring (3, 35). The endosymbiont and host phylogenies are congruent, which is consistent with a vertical mode of symbiont transmission (3, 4, 12, 35). In contrast, results from several studies suggest that endosymbiont transmission in vestimentiferan tubeworms occurs horizontally (5, 17), possibly through ingestion of bacteria upon larval settlement (28, 42). It is also possible that horizontal transmission in vestimentiferan-bacterial symbioses could occur through infection of new recruits with bacteria previously associated with established congeners (20), although potential mechanisms for this transfer have not been determined. If horizontal transmission is indeed the mechanism for bacterial acquisition in vestimentiferans, we expect that biogeographic variation would exist among symbionts acquired from the same host species collected in geographically isolated vent and seep sites. Conversely, if vestimentiferan larvae settle in the same locations where their symbionts are acquired, we expect that vestimentiferans living at the same site would harbor identical symbionts.
In a recent study, genetic variation in vestimentiferan endosymbionts was investigated by comparing sequences of the 16S rRNA genes of bacteria collected from various species of hydrothermal vent and cold seep tubeworms (20). The results showed a marked phylogenetic distinction between vestimentiferan endosymbionts collected from hydrothermal vents and soft-bottom, cold seep environments. This study suggested that vestimentiferans acquire one of two distinct species of free-living bacteria depending on whether they settle on basaltic, hydrothermal vent sites or sedimented, cold seep sites. A third, unique bacterial species was discovered in a tubeworm that inhabited a sedimented whale fall. Significant genetic variation was not found among symbionts found within each of the habitat types. Symbionts from three different host genera collected from five different hydrothermal vent sites along the East Pacific Rise (EPR) had identical 16S rRNA gene sequences.
The observation of so little sequence variation among endosymbiont 16S rRNA genes suggests that all tubeworm endosymbionts may belong to a single species. However, the conservative nature of the 16S rRNA gene often renders it inadequate for distinguishing among conspecific strains of bacteria (50). Therefore, it is not surprising that Feldman et al. (20) did not find significant genetic variation within the habitat clades. It is possible that significant genetic variation exists between endosymbionts of geographically distributed vestimentiferan hosts but that these species- or strain-level differences have not been detected by approaches that have been employed to date.
The goal of our study was to resolve the question of genetic variation among widely distributed vestimentiferan endosymbionts by utilizing two molecular techniques that have historically proven useful for comparing closely related strains of bacteria. Our first approach entailed sequencing of the internal transcribed spacer (ITS) region, a noncoding region within the rRNA that often contains genetic variation sufficient for differentiating species of prokaryotes (2). Our second approach involved using a DNA fingerprinting technique that utilizes repetitive extragenic palindromes (REPs) as priming sites to generate diagnostic banding patterns from prokaryotic genomic DNA (25, 45). The REP-PCR technique is particularly useful for investigations of bacterial symbiosis because REP sequences have been found only in bacterial DNA; phage and eukaryotes do not contain REP elements (25). By virtue of these repetitive elements, symbiont DNA can be selectively amplified from a mixed population of symbiont and host DNA to allow analysis of bacterial population diversity. Our results provide information on the extent of genetic variation among the symbionts, as well as further evidence to support a horizontal mode of symbiont acquisition among the vestimentiferan tubeworms.
MATERIALS AND METHODS
Sample collection.
Vestimentiferans were collected with the aid of submersibles from several locations, including two sites in the Western Pacific Ocean, four sites along the EPR, two sites in the Pacific Northwest, one site along the Galapagos Rift (GR), two sites along the western coast of California and Mexico, and two locations in the Gulf of Mexico (GOM) (Table 1). Samples included in this study were chosen to represent the entire range of host vestimentiferan distribution. Habitat types included hydrothermal vent sites with a basaltic substrate, cold seep habitats with a sedimented bottom, and one sedimented site associated with a whale carcass (Table 1). All samples were carried to the surface in chilled seawater (0.5 to 2°C).
TABLE 1.
Collection sites of vestimentiferan symbionts included in this studya
| Host species | Collection site | Latitude; longitude | Substrate, community type | Depth (m) | Reference |
|---|---|---|---|---|---|
| R. pachyptila | 18°S, EPR (18S) | 18°36′S; 113°24′W | Basaltic, vent | 2,637 | 20 |
| R. pachyptila | 9°N, EPR (9N) | 9°49′N; 104°17′W | Basaltic, vent | 2,516 | 20 |
| R. pachyptila | Guaymas Basin vent (GY-V) | 26°59′N; 111°24′W | Basaltic, vent | 2,016 | 20 |
| R. pachyptila | GR | 0°48′N; 86°13′N | Basaltic, vent | 2,461 | 20 |
| R. pachyptila | 13°N, EPR (13N) | 12°49′N; 103°56′W | Basaltic, vent | 2,630 | This study |
| Oasisia alvinae | 21°N, EPR (21N) | 20°47′N; 109°08′W | Basaltic, vent | 2,577 | 20 |
| T. jerichonana | 13°N, EPR (13N) | 12°49′N; 103°56′W | Basaltic, vent | 2,630 | This study |
| T. jerichonana | 9°N, EPR (9N) | 9°49′N; 104°17′W | Basaltic, vent | 2,516 | 20 |
| Ridgeia piscesae | JDF | 45°58′N; 130°01′W | Basaltic, vent | 1,550 | This study |
| Lamellibrachia sp. | Green Canyon, GOM | 27°41′N; 91°32′W | Sedimented, seep | 700 | 5 |
| Lamellibrachia sp. | Gorda Ridge (Gorda) | 42°45′N; 126°42′W | Basaltic, vent | 2,847 | 20 |
| E. laminata | WFE | 26°02′N; 84°55′W | Sedimented, seep | 3,243 | 20 |
| E. spicata | Santa Catalina Basin (whale) | 33°12′N; 118°30′W | Sedimented, whale fall | 1,240 | 20 |
| E. spicata | Guaymas Basin vent (GY-V) | 26°59′N; 111°24′W | Basaltic, vent | 2,016 | This study |
| E. spicata | Guaymas Basin seep (GY-S) | 26°59′N; 111°24′W | Sedimented, seep | 2,016 | This study |
| L. columna | Lau Basin, Hine Hina (Lau-Fiji) | 22°32′S; 176°43′W | Sedimented, seep | 1,859 | 42 |
| Undescribed species | Nikko Seamount (Nikko) | 23°05′N; 142°20′E | Basaltic, vent? | 433 | This study |
See the work of Feldman et al. (20) for a map of the sample distribution.
DNA extraction.
The symbiont-containing trophosome tissue from each vestimentiferan host was aseptically removed, and a small portion of it was homogenized in 5 M guanidinium isothiocyanate. Prior to DNA extraction, 25 μl of a 20% Chelex 100 (47) solution was added to the homogenate and incubated for 1 h at 4°C. The samples were briefly spun to remove the Chelex 100. Total bacterial DNA was extracted from 100 μl of guanidinium isothiocyanate homogenate using the IsoQuick DNA purification kit (Orca, Bothell, Wash.) according to the manufacturer's instructions and quantified spectrophotometrically. In addition to symbiont DNA, host DNA from three different genera was extracted from vestimentum tissue (non-symbiont-containing tissue) using the same extraction protocol to serve as controls. A number of the DNA samples used in this study (e.g., 18°S Riftia pachyptila, Guaymas Vent and Seep [GY-V and GY-S, respectively] Escarpia spicata, whale E. spicata, Gorda Lamellibrachia sp., 21°N Oasisia alvinae, and GY-V R. pachyptila) were prepared separately at Rutgers University according to previously published protocols (20).
16S rRNA gene characterization.
The methods used to PCR amplify, purify, and sequence the 16S rRNA symbiont genes were as described in the work of Feldman et al. (20). DNA sequence alignments were initially constructed using PileUp and then optimized (minimizing overall alignment differences) by eye in the SeqLab environment (Genetics Computing Group). Insertions and deletions were eliminated from the alignment before phylogenetic trees were determined. Phylogenetic trees for the symbiont 16S rRNA genes were computed using fastDNAml and bootstrapped 100 times using fastDNAml boot (21, 34). For the maximum likelihood analyses, operational taxonomic unit input order was randomized and global rearrangements were performed at each bootstrap replication.
ITS region characterization.
PCRs amplified the ITS region of the symbiont ribosomal DNA (rDNA) using two ITS-specific primers, ITS16F-G1 (5′-GAAGTCGTAACAAGG-3′) (27) and ITS23R-L1 (5′-CAAGGCATCCACCGT-3′) (27). ITS16F-G1 is nested about 30 to 40 nucleotides upstream from the spacer boundary in the 3′ end of the 16S rRNA gene (positions 1491 to 1505, Escherichia coli 16S rRNA gene). ITS23R-L1 is nested in the 5′ end of the 23S rRNA gene, approximately 20 bases downstream from the spacer boundary (positions 21 to 35, E. coli 23S rRNA gene). PCR mixtures (50-μl total volume) contained final concentrations of the following: 50 ng of symbiont DNA, 1× PCR buffer, 0.2 mM (each) deoxynucleoside triphosphates, 10 pmol of each primer, 1.5 mM MgCl2, 5% (vol/vol) acetamide, and 1.25 U of Taq polymerase (Promega). PCR conditions were as follows: 35 cycles, each consisting of denaturation at 92°C for 1 min, hybridization at 55°C for 2 min, and elongation at 72°C for 2 min with 5 s added to each extension per cycle. A hot start (9) with denaturation at 95°C for 2 min was used at the beginning of the reaction to heighten reaction specificity. In addition, acetamide was added to the PCR to encourage more efficient amplification of GC-rich templates (38). All PCRs were performed on an MJ-Minicycler (MJ Research, Inc., Watertown, Mass.).
Because our attempts to directly sequence the smaller ITS PCR amplicon yielded poor results, amplified ITS regions were cloned to facilitate sequencing. Amplification products were pooled from three separate PCRs and cloned directly using the TA cloning kit with the pCR-II or the pCR-2.1-TOPO cloning vectors (Invitrogen, San Diego, Calif.) according to the manufacturer's instructions. Plasmid preparations were made using a standard alkaline-lysis preparation (39). The sizes of the inserts were verified before sequencing by restriction analysis. The plasmid DNA was further purified for sequencing using the Plasmid MiniKit (Qiagen, Inc., Valencia, Calif.) according to the manufacturer's instructions. Two clones from each amplified ITS region were cycle sequenced using the Perkin-Elmer (Foster City, Calif.) ABI BigDye dye termination cycle sequencing ready reaction kit with Ampli Taq DNA polymerase FS according to the manufacturer's instructions. Sequencing was performed on an ABI PRISM 310 genetic analyzer.
The ITS regions were bidirectionally sequenced and confirmed prior to their alignment using the Sequence Navigator and AutoAssembler programs (Applied Biosystems, Inc.). Only sequences that overlapped with 0% ambiguity were included in the analysis. Final alignments were generated in the Genetic Data Environment, version 3.2. Phylogenetic relationships were determined from these aligned sequences using PHYLIP version 3.572 (22). A Kimura two-parameter distance matrix was constructed, from which neighbor-joining trees with 100 bootstrap replications were generated. A second distance matrix, based on pairwise differences in the geographic distribution between samples, was generated from latitude-longitude information for each sample site using GeoDist version 3.01e, R package (32). A third distance matrix, based on pairwise comparisons of the sample habitat types, was created by assigning “0” values to pairs of samples that were taken from similar habitat types (both vent or both seep) and “1” values to pairs of samples that were taken from different habitat types. Mantel correlation tests were performed to compare ITS sequence distance with geographic and habitat distance (14, 33). Two- and three-way Mantel tests were performed using different combinations of the three distance matrices to assess whether ITS genetic distance significantly correlates with the geographic distances between endosymbiont collection sites or their habitat types (41). These Mantel tests were performed using the R package (32).
REP elements.
REP sequences were amplified from each of the vestimentiferan symbionts with PCR using two universal REP primers, REPIR-I (5′-IIIICGICGICATCIGGC-3′) (45) and REP2-I (5′-ICGICTTATCIGGCCTAC-3′) (44). Each 25-μl REP-PCR mixture contained 50 ng of symbiont or host DNA and final concentrations of 1× Gitschier buffer [16.6 mM (NH4)2SO4, 67 mM Tris-HCl, 6.7 mM MgCl2, 6.7 μM EDTA, 30 mM β-mercaptoethanol (29)], 160 μg of DNA-grade bovine serum albumin per ml, 10% dimethyl sulfoxide, 1.25 mM (each) four deoxynucleoside triphosphates, 10 pmol of each REP primer, and 2 U of Taq polymerase (Gibco). One cycle of 95°C for 7 min was performed to denature the genomic DNA, followed by 30 cycles of 94°C for 1 min, 44°C for 1 min, and 65°C for 8 min, and a final extension at 65°C for 15 min. PCRs were performed with an MJ-Minicycler (MJ Research, Inc.). Reaction products (7.5 μl) were analyzed via agarose gel electrophoresis.
A digital image of each gel was captured and stored using the Alpha Imager 2000 Documentation and Analysis System (AlphaInnotech Corp., San Leandro, Calif.). Each gel image was analyzed using the software package GelCompar (version 4; Applied Maths, Kortrijk, Belgium). This software was used to normalize the molecular weight of each of the bands in each gel to all of the others based on the inclusion of the same set of molecular weight markers on each gel. Once normalized, all of the fingerprints in the database were compared to each other using an unweighted-pair-group-method-with-averages clustering algorithm and Jaccard coefficient.
Nucleotide sequence accession numbers.
GenBank accession numbers for the new ITS sequences reported in this paper are as follows: AF076795, Ridgeia piscesae endosymbiont, Juan de Fuca Ridge (JDF); AF076796, Escarpia laminata endosymbiont, West Florida Escarpment (WFE); AF076797, Lamellibrachia sp. endosymbiont, GOM-10; AF076798, E. spicata endosymbiont, GY-S; AF076799, R. pachyptila endosymbiont, GY-V; AF076800, Tevnia jerichonana endosymbiont, 9N; AF076801, R. pachyptila endosymbiont, 9N; AF076802, T. jerichonana endosymbiont, 13N; AF076803, R. pachyptila endosymbiont, 13N; AF076804, Lamellibrachia sp. endosymbiont, Gorda; AF076805, R. pachyptila endosymbiont, GR; AF076806, R. pachyptila endosymbiont, 18S; AF076807, O. alvinae endosymbiont, 21N; AF076808, E. spicata endosymbiont, whale; AF076809, undescribed species endosymbiont, Nikko Seamount; AF076810, L. columna endosymbiont, Lau-Fiji; AF076811, Lamellibrachia sp. endosymbiont, GOM-12; AF076812, E. spicata endosymbiont, GY-V; AF076813, Lucina floridana endosymbiont. Accession numbers for the three 16S rRNA sequences generated for this study are as follows: AF165909, E. spicata endosymbiont, GY-V; AF165907, undescribed species endosymbiont, Nikko Seamount; and AF165908, E. spicata endosymbiont, GY-S.
RESULTS AND DISCUSSION
16S rDNA sequence variation among vestimentiferan endosymbionts.
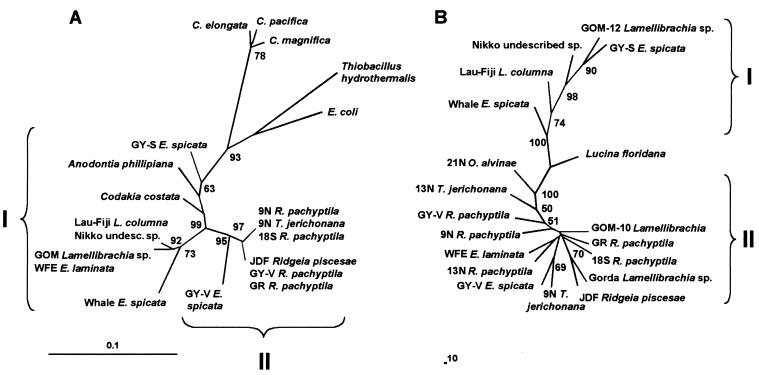
In order to maintain a comparable data set, many of the endosymbiont DNA samples used in our ITS and REP-PCR analyses are the same DNA samples used in the previous 16S rDNA study of geographically distinct endosymbionts (20). Our study also includes additional samples to expand the representation of symbiont distribution. In concordance with our expanded data set, a new 16S rDNA phylogenetic tree was generated to include three of the endosymbionts that were added: GY-V E. spicata, GY-S E. spicata, and the undescribed species from the Nikko Seamount (Fig. 1A). In this new analysis of 16S rDNA sequences, the symbionts continue to cluster into two main groups: group I includes all of the symbionts from seep- or sediment-dwelling host organisms, while group II includes all of the symbionts from vent-dwelling host fauna. These relationships are in accordance with our previous findings (20). Interestingly, the difference between vent and seep symbionts is maintained between symbionts from the same host, E. spicata, collected from vent (GY-V) and seep (GY-S) habitats at a single site in Guaymas Basin. Likewise, symbionts from two different vestimentiferan hosts are closely related when collected from the same habitat type (e.g., 9N R. pachyptila and T. jerichonana symbionts, both from vent habitats).
FIG. 1.
(A) Dendrogram based on 16S rDNA sequences of vestimentiferan symbionts. Phylogenetic relationships were computed using fastDNAml and bootstrapped 100 times using fastDNAml boot (21, 34). For the maximum likelihood analyses, operational taxonomic unit input order was randomized and global rearrangements were performed at each bootstrap replication. Two major groups of endosymbionts are indicated. Scale bar represents 10% sequence dissimilarity. Reference sequences used in this 16S study were obtained from the GenBank database and include the following accession numbers: L25712, Codakia costata symbiont; L25711, Anodontia phillipiana symbiont; M99446, Calyptogena magnifica symbiont; L25710, Calyptogena pacifica symbiont; L25718 and L25719, Calyptogena elongata symbiont; M90662, Thiobacillus hydrothermalis; V00348, E. coli; U77478, 9N and 18S R. pachyptila endosymbiont and 9N T. jerichonana endosymbiont; U77479, GOM Lamellibrachia sp. endosymbiont and WFE E. laminata endosymbiont; U77480, JDF Ridgeia piscesae endosymbiont and GY-V and GR R. pachyptila endosymbiont; U77481, Lau-Fiji L. columna endosymbiont; U77482, whale E. spicata endosymbiont. (B) Dendrogram showing the relationships of vestimentiferan symbionts based on ITS sequences. The tree was rooted with the symbiont from a marine bivalve, L. floridana, as an outgroup. The tree was constructed using the neighbor-joining method with 100 bootstrap replicates. Each number on a branch indicates the number of times (out of 100) that the node was supported by the bootstrap analysis. Only bootstrap values of ≥50% are reported. Two major clusters of endosymbionts are indicated. Scale bar represents 10% sequence dissimilarity.
ITS sequence variation among vestimentiferan symbionts.
ITS amplifications of vestimentiferan symbionts were generally 550 bp in length, although some individuals yielded a second, larger PCR product of roughly 800 bp in length (data not shown). The larger amplicon did not match any ITS sequences in the database, and we found that its intensity in the total PCR product was reduced by the use of higher annealing temperatures. For these reasons, we believe that the larger product represented an artifact of the PCR rather than rRNA operon heterogeneity. To separate these two distinct amplicons, we cloned the ITS PCR amplification products and used only the smaller amplicon from each symbiont for sequence analysis. As in many bacteria, all of the endosymbiont ITS regions contained putative sequences for the tRNA genes for alanine and isoleucine (Fig. 2). Five of the symbionts contained prominent insertion sequences (approximately base position 312 in ITS region) that totaled approximately 50 bp (Fig. 2).
FIG. 2.
Alignment of a portion of the ITS region (bp 240 to 480). Putative sequences for the tRNA genes for alanine and isoleucine are indicated by the boxes. A 50-bp insertion that is present in five of the sequences is highlighted by boldface. Identical bases are indicated by dots, while gaps in the sequence alignment corresponding to insertions or deletions are indicated with dashes. Lamell., Lamellibrachia.
ITS sequences of endosymbionts collected from several vestimentiferan host genera were compared phylogenetically using the corresponding ITS sequence of the symbiont of the lucinid bivalve L. floridana as an outgroup. A similarity matrix of ITS sequences was constructed based upon a Kimura two-parameter distance estimate. Genetic distances were calculated based on 500 bp of aligned sequences after the nucleotide insertions and deletions were removed. The sequences were surprisingly conserved overall, with percent sequence similarity ranging from 87.0 to 100% (data not shown). The symbionts from R. pachyptila hosts collected from Guaymas Basin, 9°N, and 18°S displayed 100% sequence similarity.
Symbiont relationships were depicted as a cladogram based upon ITS sequence diversity (Fig. 1B). Because of the high similarity among the ITS sequences, many of the bootstrap values at branch points in the neighbor-joining analysis were low or insignificant. Only bootstrap values of ≥50% were reported. The ITS cladogram showed a prominent break within the symbionts: group I included the symbionts of E. spicata from the whale carcass, L. columna from the Lau-Fiji basin, the undescribed species from a vent site at the Nikko Seamount, one of the two Lamellibrachia spp. collected from a seep site in the GOM (GOM-12), and E. laminata from the seep site at Guaymas Basin; group II included the rest of the symbionts (Fig. 1B). Apart from differing in sequence from the members of group II, the symbionts in group I contained prominent insertion sequences that totaled approximately 50 bp (Fig. 2). The sequences of these insertions were nearly identical in all of the symbionts in which they were found. Groups I and II in the 16S rDNA cladogram (Fig. 1A) correlate well with groups I and II in the ITS cladogram (Fig. 1B), with one exception: E. laminata symbiont, WFE, appears in group I according to its 16S rDNA sequence and in group II according to its ITS sequence (Fig. 1).
To determine if there was a significant correlation between ITS sequence diversity and either geography or habitat type, we performed two- and three-way Mantel tests to compare distance matrices generated from pairwise differences between ITS sequences, geographic distances, or habitat types. The results of these Mantel tests are shown in Table 2. Despite the remarkable similarity among the ITS sequences, the genetic distances among ITS regions correlate significantly with geographic distances. Although none of the correlations were particularly strong (r ≤ −0.334 for all tests), P values from two different Mantel tests indicated that ITS sequence similarity and geography were significantly correlated (P = 0.0349, test 3; P = 0.0480, test 4). In contrast, P values for a two-way Mantel test of ITS sequence similarity versus habitat type alone (test 1) or a three-way test versus habitat type with geography factored out (test 5) were not statistically significant (P = 0.146 and 0.102, respectively). Thus, ITS sequence similarity was significantly correlated with broad-scale endosymbiont geography (P ≤ 0.05) but not significantly correlated with habitat type. In light of the fact that the ITS sequences for 9°N, 18°S, and Guaymas R. pachyptila were identical, our results suggest that the ITS region is variable enough to show broad-scale biogeographic variation but is not variable enough to distinguish among symbionts located along the same ridge axis. These results are in contrast to the results of Feldman et al. (20) and the new analysis of 16S rDNA relationships presented in this study, which suggest that symbiont sequence distances correlate with habitat type (basaltic, vent versus sedimented, seep substrates). Although our data suggest that the geographic distribution of the hosts may be a dominant influence in determining genetic variation of the endosymbionts, it is clear that several factors affect endosymbiont diversity.
TABLE 2.
Mantel tests of correlation among geographic, habitat, and ITS sequence similarity matrices for vestimentiferan symbiontsa
| Test | Matrix A | Matrix B | Conditional matrix C | r | t | P |
|---|---|---|---|---|---|---|
| Two-way | ||||||
| 1 | ITS sim | Habitat | −0.133 | −1.402 | 0.1459 | |
| 2 | Geography | Habitat | 0.006 | 0.053 | 0.4555 | |
| 3 | ITS sim | Geography | −0.332 | −2.291 | 0.0349 | |
| Three-way | ||||||
| 4 | Geography | ITS sim | Habitat | −0.334 | −2.349 | 0.0480 |
| 5 | Habitat | ITS sim | Geography | −0.139 | −1.508 | 0.1019 |
sim, similarity.
Historically, the ITS region has been useful for discriminating among strains of several bacterial species, including a hyperthermophilic archaeon (11), Rhizobium sp. (30), Bifidobacterium sp. (31), infectious Pseudomonas sp. (43), and Trichodesmium sp. (49). In the present study, we found that the ITS region was much more conserved than expected. Although the ITS region is usually very informative at the subspecies and/or strain level (11, 30, 31, 49), it has been unreliable in some cases. In studies of the Mycobacterium tuberculosis complex (23) and with several Listeria isolates (15), the ITS region failed to differentiate between species and/or strains.
REP-PCR fingerprints of the vestimentiferan symbionts.
The REP-PCR technique is particularly well suited for investigations of bacterial symbiosis, in that it allows investigators to survey the symbiont genome in the presence of host DNA because the REP priming sites are exclusively bacterial (25). To confirm that REP sequences were not present in the genomes of the vestimentiferan hosts, we performed REP-PCR amplifications on DNA that was aseptically acquired from vestimentum (non-symbiont-containing) tissue from three different host genera (Escarpia, Riftia, and Tevnia). Most of these samples failed to amplify at all (data not shown). The exception was E. laminata, which produced a single band that did not correspond with any of the bands seen in the REP fingerprint of its endosymbiont.
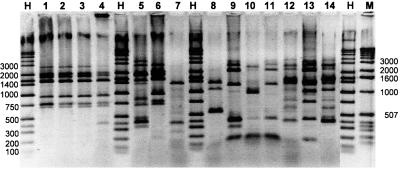
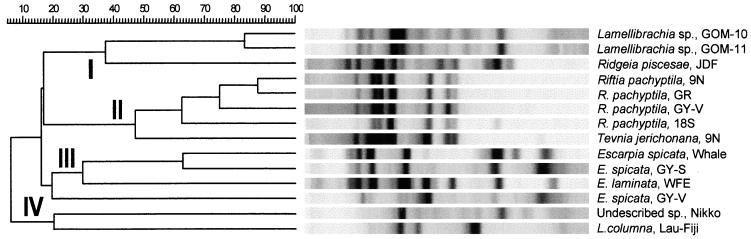
Fingerprint patterns of the vestimentiferan endosymbiont DNA displayed remarkable variation (Fig. 3). Relationships based upon the number of shared bands among the fingerprint patterns were compared using GelCompar software and depicted as a cladogram with a corresponding, computer-generated gel image (Fig. 4). All of the symbionts included in the REP analysis displayed unique fingerprints, with the exception of 9°N R. pachyptila, and GR R. pachyptila, which exhibited indistinguishable patterns (Fig. 3 and 4). The REP analysis seems to divide the symbionts into four distinct groups, or clades (Fig. 4). Group I includes both Lamellibrachia symbionts from the GOM and the symbiont from Ridgeia piscesae from the JDF. Group II includes the R. pachyptila symbionts and the symbiont from T. jerichonana. Group III includes all of the Escarpia symbionts, from both E. spicata and E. laminata. Group IV includes the endosymbionts from the undescribed species at Nikko Seamount and L. columna from the Lau-Fiji Basin, two distantly located sites in the western Pacific Ocean.
FIG. 3.
Digital image of the REP-PCR fingerprint patterns of the vestimentiferan symbionts. Shown are the REP-PCR products generated by using chromosomal DNA of endosymbionts extracted from the following vestimentiferans: R. pachyptila, GY-V (lane 1); R. pachyptila, 9N (lane 2); R. pachyptila, GR (lane 3); R. pachyptila, 18S (lane 4); Ridgeia piscesae, JDF (lane 5); T. jerichonana, 9N (lane 6); undescribed sp., Nikko Seamount (lane 7); L. columna, Lau-Fiji (lane 8); E. spicata, whale (lane 9); E. spicata, GY-V (lane 10); E. spicata, GY-S (lane 11); Lamellibrachia sp., GOM-10 (lane 12); E. laminata, WFE (lane 13); Lamellibrachia sp., GOM-11 (lane 14). Lanes H, Hi Low DNA marker; lane M, 1-kb DNA ladder. Molecular sizes (base pairs) of selected bands in the DNA markers are indicated. The photo represents a composite of a larger gel that was altered in Adobe Photoshop version 5.0 to include only the individuals presented in this study.
FIG. 4.
Cladogram depicting relationships of vestimentiferan endosymbionts based upon their REP-PCR fingerprints. The degree of genetic similarity is indicated above the cladogram in percentage units. To facilitate visual comparison, a computer-generated gel that depicts the lanes reloaded according to their relationships in the dendrogram has been included. The symbionts separated into four groups based upon the percent similarity (measured as number of shared bands) among their REP fingerprint patterns. Group I, GOM Lamellibrachia sp. symbionts and JDF Ridgeia piscesae symbiont; group II, R. pachyptila and T. jerichonana symbionts; group III, Escarpia spp. symbionts; group IV, Nikko Seamount and Lau-Fiji Basin L. columna symbionts. Slight visual differences from the original gel lane images in Fig. 3 result from the normalization and analysis process of the GelCompar software.
Upon closer examination, the REP analysis seems to reflect three different influences on endosymbiont differentiation: host genus, bottom or substrate type, and geography. Host genus seems to be the primary source of differentiation, as most of the clades break up into distinct groupings of congeners. The only clade where this breaks down is group II, which includes symbionts from Riftia and Tevnia host genera. This clade, however, supports previous studies using DNA-DNA hybridization and in situ hybridization that suggest that the endosymbionts of Riftia and Tevnia are closely related (5, 17).
The secondary influences for symbiont genetic differentiation seem to be geographic location and substrate type. Within clades of congeners, it appears that symbionts differ based upon where they are collected and/or the habitat type (basaltic or sedimented). In group II, all of the R. pachyptila symbionts collected from the hosts at northern EPR sites (i.e. GY-V, 9N, and GR) are less closely related to the R. pachyptila symbiont collected from a site 18°S along the southern EPR. Within the Escarpia symbiont clade (group III), both geographic location and substrate type seem to be at work: symbionts collected from hosts at the whale fall in the Santa Catalina Basin (whale), Guaymas Basin seep (GY-S), and WFE, all soft-bottom substrate types, clade together and group away from the symbionts collected from the Guaymas Basin vent (GY-V). Within the soft-bottom grouping of Escarpia symbionts, it appears that the symbionts from California and Guaymas (both on the western coast of North America) group away from the symbiont from Florida (on the eastern coast). The symbionts from the Nikko Seamount and Lau-Fiji Basin are clearly different from the rest of the symbionts, presumably because they are so geographically removed from the rest of the collection sites.
REP-PCR has been useful for genomic fingerprinting of various strains of bacteria, including Actinobacillus (1), Rhizobium (10, 30), Legionella (24), Streptococcus (44), Bacillus (26), and Citrobacter diversus (51) strains. In the present study, we found that the REP-PCR fingerprint analysis was sufficiently fine-scaled to reveal some interesting strain-level genetic variation among vestimentiferan symbionts. Presumably, this is because the REP analysis targets the whole genome, rather than a specific, relatively conservative region of the DNA. This physically maximizes the probability of finding strain-level variation. It should be noted that the symbiont of O. alvinae was not included in the REP analysis, because a reliable REP-PCR amplification and subsequent fingerprint pattern were not obtainable with this DNA. When the DNA was run on an agarose gel, it appeared sheared, and although it was sufficiently large enough for performance of the 550-bp ITS amplification, there was insufficient high-molecular-weight genomic DNA to support the REP analysis.
Implications for symbiont relatedness and acquisition.
Previous studies have suggested that vestimentiferans acquire their symbionts through ingestion of free-living bacteria upon larval settlement (28, 42). If this mechanism alone is employed, one would expect that symbionts from two different vestimentiferan host species living at the same location would be identical or related more closely to each other than they are to other endosymbionts collected from distant vent sites. This, in fact, was not the case, as symbionts from 9°N T. jerichonana and 9°N R. pachyptila (both collected from the same rock) did not display identical ITS sequences or REP-PCR fingerprints. Instead, the symbionts grouped according to their respective host species as revealed through comparisons of shared bands in their REP-PCR fingerprints.
One explanation for this is that larvae could acquire their symbionts prior to attachment, either from a different substrate than where they eventually colonize or from the water column. This could enable two vestimentiferans that reside in the same site to harbor different symbionts. Alternatively, symbiont acquisition may be related to ecological succession. T. jerichonana is the first colonizer of new hydrothermal vent fields, in areas of the most intense diffuse flow (40). Presumably, this is because T. jerichonana is more tolerant (than R. pachyptila) of the high temperatures and elevated concentrations of H2S that are present in newly formed vent sites. It is only after T. jerichonana has become established and the levels of reduced chemicals have attenuated that R. pachyptila begins to colonize a new hydrothermal vent site (40). The T. jerichonana and R. pachyptila hosts may simply acquire a different subset of a free-living chemoautotrophic community due to temporal and/or spatial consequences of ecological succession.
Specific host recognition mechanisms may exist that allow certain strains of endosymbiont bacteria to survive in specific vestimentiferan hosts. Although these mechanisms have not been demonstrated in vestimentiferans, they could explain how two different hosts living on the same rock could harbor different symbionts. Alternatively, the larvae may acquire bacteria that have been released from congeners residing at the site of larval settlement, potentially through host decomposition. These mechanisms might yield REP-PCR fingerprint patterns that imply close relationships between endosymbionts of the same host genus. Because only one individual from each vestimentiferan genus was sampled from most of the locations, we cannot determine the extent of ITS sequence variation among endosymbionts at each site.
The extent of vestimentiferan symbiont relatedness and the host transmission mechanisms used to maintain these symbioses have remained ongoing questions. The results presented here demonstrate the existence of significant strain-level variation between endosymbionts of different vestimentiferan hosts collected from geographically separated areas. Further work to pinpoint the stage during vestimentiferan development when the symbiosis is first established will shed light on symbiont transmission and help to explain the genetic diversity among symbiont populations.
ACKNOWLEDGMENTS
We thank the crews and pilots of the R/V Atlantis II and the deep-submergence vehicle Alvin, the Nautile, and the U.S. Navy Advanced Tethered Vehicle for their assistance in sample collection. Samples of L. columna from the Lau-Fiji Basin were generously provided by A. M. Alayse. Samples of Lamellibrachia sp. from Green Canyon, GOM, were graciously supplied by Craig Young and Adele Pile. ITS primers and sequences for 13°N Tevnia and Riftia, Gorda Lamellibrachia, and Galapagos Riftia were contributed by Jeff Stein. We thank Barbara Campbell for assistance with REP-PCR fingerprinting and Kathy Coyne for critically reviewing the manuscript. We also thank Ken Halanych for thoughtful discussion. The manuscript was greatly improved by the helpful comments of two anonymous reviewers.
This work was supported by two NSF grants to S.C.C. (OCE-9314595 and OCE-9596082). The whale carcass E. spicata sample was collected on a cruise led by Craig Smith and funded by an NSF grant to R.C.V. (OCE-9633131).
REFERENCES
- 1.Appuhamy S, Coote J G, Low J C, Parton R. PCR methods for rapid identification and characterization of Actinobacillus seminis strains. J Clin Microbiol. 1998;36:814–817. doi: 10.1128/jcm.36.3.814-817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry T, Colleran G, Glennon M, Dunican L, Gannon F. The 16S/23S ribosomal spacer as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1:51–56. doi: 10.1101/gr.1.1.51. [DOI] [PubMed] [Google Scholar]
- 3.Cary S C. Vertical transmission of a chemoautotrophic symbiont in the protobranch bivalve, Solemya reidi. Mol Mar Biol Biotechnol. 1994;3:121–130. [PubMed] [Google Scholar]
- 4.Cary S C, Giovannoni S J. Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. Proc Natl Acad Sci USA. 1993;90:5695–5699. doi: 10.1073/pnas.90.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cary S C, Warren W, Anderson E, Giovannoni S J. Identification and localization of bacterial endosymbionts in hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol Mar Biol Biotechnol. 1993;2:51–82. [PubMed] [Google Scholar]
- 6.Cavanaugh C M. Symbioses of chemoautotrophic bacteria and marine invertebrates from hydrothermal vents and reducing sediments. Bull Biol Soc Wash. 1985;6:373–388. [Google Scholar]
- 7.Cavanaugh C M, Gardiner S L, Jones M L, Jannasch H W, Waterbury J B. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science. 1981;213:340–341. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- 8.Dando P R, Southward A J, Terwilliger N B, Terwilliger R C. Sulphur-oxidizing bacteria and haemoglobin in gills of the bivalve mollusc Myrtea spirifera. Mar Ecol Prog Ser. 1985;23:85–98. [Google Scholar]
- 9.D'Aquila R T, Bechtel L J, Videler J A, Eron J J, Gorczyca P, Kaplan J C. Maximizing sensitivity and specificity of PCR by preamplification heating. Nucleic Acids Res. 1991;19:3749. doi: 10.1093/nar/19.13.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Bruijn F J. Use of repetitive (repetitive extragenic element and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiRuggiero J, Tuttle J H, Robb F T. Rapid differentiation of hyperthermophilic Archaea by restriction mapping of the intergenic spacer regions of the ribosomal RNA operons. Mol Mar Biol Biotechnol. 1995;4:123–127. [PubMed] [Google Scholar]
- 12.Distel D L, Felbeck H, Cavanaugh C M. Evidence for phylogenetic congruence among sulfur-oxidizing chemoautotrophic bacterial endosymbionts and their bivalve hosts. J Mol Evol. 1994;38:533–542. [Google Scholar]
- 13.Distel D L, Lane D J, Olsen G J, Giovannoni S J, Pace B, Pace N R, Stahl D A, Felbeck H. Sulfur-oxidizing bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J Bacteriol. 1988;170:2506–2510. doi: 10.1128/jb.170.6.2506-2510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas M E, Endler J A. Quantitative matrix comparisons in ecological and evolutionary investigations. J Theor Biol. 1982;99:777–795. [Google Scholar]
- 15.Drebot M, Neal S, Schlech W, Rozee K. Differentiation of Listeria isolates by PCR amplicon profiling and sequence analysis of 16S-23S rRNA internal transcribed spacer loci. J Appl Bacteriol. 1996;80:174–178. doi: 10.1111/j.1365-2672.1996.tb03206.x. [DOI] [PubMed] [Google Scholar]
- 16.Durand P, Gros O, Frenkiel L, Prieur D. Phylogenetic characterization of sulfur-oxidizing bacterial endosymbionts in three tropical Lucinidae by 16S rDNA sequence analysis. Mol Mar Biol Biotechnol. 1996;5:37–42. [Google Scholar]
- 17.Edwards D B, Nelson D C. DNA-DNA solution hybridization studies of the bacterial symbionts of hydrothermal vent tube worms (Riftia pachyptila and Tevnia jerichonana) Appl Environ Microbiol. 1991;57:1082–1088. doi: 10.1128/aem.57.4.1082-1088.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felbeck H. Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera) Science. 1981;213:336–338. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- 19.Felbeck H. CO2 fixation in the hydrothermal vent tube worm Riftia pachyptila Jones. Physiol Zool. 1985;58:272–281. [Google Scholar]
- 20.Feldman R A, Black M B, Cary C S, Lutz R A, Vrijenhoek R C. Molecular phylogenetics of bacterial endosymbionts and their vestimentiferan hosts. Mol Mar Biol Biotechnol. 1997;6:268–277. [PubMed] [Google Scholar]
- 21.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. PHYLIP manual. Berkeley: University Herbarium, University of California; 1990. [Google Scholar]
- 23.Frothingham R, Hills H G, Wilson K H. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J Clin Microbiol. 1994;32:1639–1643. doi: 10.1128/jcm.32.7.1639-1643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georghiou P R, Doggett A M, Kielhogner M A, Stout J E, Watson D A, Lupski J R, Hamill R J. Molecular fingerprinting of Legionella species by repetitive element PCR. J Clin Microbiol. 1994;32:2989–2994. doi: 10.1128/jcm.32.12.2989-2994.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilson E, Clement J M, Perrin D, Hofnung M. Palindromic units: a case of highly repetitive DNA sequences in bacteria. Trends Genet. 1987;3:226–230. [Google Scholar]
- 26.Herman L, Heyndrickx M, Waes G. Typing of Bacillus sporothermodurans and other Bacillus species isolated from milk by repetitive element sequence based PCR. Lett Appl Microbiol. 1998;26:183–188. doi: 10.1046/j.1472-765x.1998.00314.x. [DOI] [PubMed] [Google Scholar]
- 27.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones M L, Gardiner S L. Evidence for a transient digestive tract in Vestimentifera. Proc Biol Soc Wash. 1988;101:423–433. [Google Scholar]
- 29.Kogan S, Doherty M, Gitschier J. An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences. Application to hemophilia A. N Engl J Med. 1987;317:985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- 30.Laguerre G, Mavingui P, Allard M, Charnay M, Louvrier P, Mazurier S, Rigottier-Gois L, Amarger N. Typing of Rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol. 1996;62:2029–2036. doi: 10.1128/aem.62.6.2029-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leblond-Bourget N, Philippe H, Mangin I, Decaris B. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int J Syst Bacteriol. 1996;46:102–111. doi: 10.1099/00207713-46-1-102. [DOI] [PubMed] [Google Scholar]
- 32.Legendre P, Vaudor A. The R-package: multidimensional analysis, spatial analysis. Montreal, Canada: Département de Sciences Biologiques, Université de Montréal; 1991. [Google Scholar]
- 33.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 34.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 35.Peek A S, Feldman R A, Lutz R A, Vrijenhoek R C. Cospeciation of chemoautotrophic bacteria and deep sea clams. Proc Natl Acad Sci USA. 1998;95:9962–9966. doi: 10.1073/pnas.95.17.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell M A, Somero G N. Adaptations to sulfide by hydrothermal vent animals: sites and mechanisms of detoxification and metabolism. Biol Bull. 1986;171:274–290. [Google Scholar]
- 37.Rau G H, Hedges J I. Carbon-13 depletion in a hydrothermal vent mussel: suggestion of a chemosynthetic food source. Science. 1979;203:648–649. doi: 10.1126/science.203.4381.648. [DOI] [PubMed] [Google Scholar]
- 38.Reysenbach A L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Shank T M, Fornari D J, Von Damm K L, Lilley M D, Haymon R M, Lutz R A. Temporal and spatial patterns of biological community development at nascent deep-sea hydrothermal vents (950′N, East Pacific Rise) Deep-Sea Res II. 1998;45:465–515. [Google Scholar]
- 41.Smouse P E, Long J C, Sokal R R. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst Zool. 1986;35:627–632. [Google Scholar]
- 42.Southward E C. Development of the gut and segmentation of newly-settled stages of Ridgeia (Vestimentifera): implications for relationships between vestimentifera and pogonophora. J Mar Biol Assoc U K. 1988;68:465–487. [Google Scholar]
- 43.Tyler S D, Strathdee C A, Rozee K R, Johnson W M. Oligonucleotide primers designed to differentiate pathogenic pseudomonads on the basis of the sequencing of genes coding for 16S-23S rRNA internal transcribed spacers. Clin Diagn Lab Immunol. 1995;2:448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Versalovic J, Kapur V, Mason E O, Jr, Shah U, Koeuth T, Lupski J R, Muser J M. Penicillin resistant Streptococcus pneumoniae strains recovered in Houston, Texas: identification and molecular characterization of multiple clones. J Infect Dis. 1993;167:850–856. doi: 10.1093/infdis/167.4.850. [DOI] [PubMed] [Google Scholar]
- 45.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacteria genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vetter R. Elemental sulfur in the gills of three species of clams containing chemoautotrophic symbiotic bacteria: a possible inorganic energy storage compound. Mar Biol. 1985;88:33–42. [Google Scholar]
- 47.Walsh P S, Metzger D A, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 48.Williams P M, Smith K L, Druffel E M, Linick T W. Dietary carbon sources of mussels and tubeworms from Galapagos hydrothermal vents determined from tissue 14C activity. Nature. 1981;292:448–449. [Google Scholar]
- 49.Wilmotte A, Neefs J, De Wachter R. Evolutionary affiliation of the marine nitrogen-fixing cyanobacterium Trichodesmium sp. strain NIBB 1067, derived by 16S ribosomal RNA sequence analysis. Microbiology. 1994;140:2159–2164. doi: 10.1099/13500872-140-8-2159. [DOI] [PubMed] [Google Scholar]
- 50.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woods C R, Versalovic J, Koeuth T, Lupski J R. Analysis of relationships among isolates of Citrobacter diversus using DNA fingerprints generated by repetitive sequence-based primers in the polymerase chain reaction. J Clin Microbiol. 1992;30:2921–2929. doi: 10.1128/jcm.30.11.2921-2929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]