Abstract
This study investigated the prevalence of human papillomavirus (HPV) infection in oral squamous cell carcinoma (OSCC) cases, as well as the association between HPV presence and p16INK4a expression, in Thai patients with OSCC. Eighty-one formalin-fixed paraffin-embedded specimens of OSCC were obtained. DNA extraction was performed; this was followed by nested polymerase chain reaction analysis to determine HPV DNA status, using consensus primers for the L1 region of HPV. HPV subtypes were determined by DNA sequencing. HPV-positive specimens and HPV-negative specimens from age- and sex-matched patients were subjected to immunohistochemical analysis to determine p16INK4a expression status. Of the 81 OSCC specimens, eight (9.9%) exhibited HPV DNA; DNA sequencing confirmed that the viral subtype was HPV-18 in all eight specimens. These eight HPV-positive specimens, as well as eight HPV-negative specimens from age- and sex-matched patients, were subjected to immunohistochemical analysis to determine p16INK4a expression status. Three of eight (37.8%) HPV-positive specimens and three of eight (37.8%) HPV-negative specimens showed positive p16INK4a expression findings. However, we did not find a significant association between HPV status and p16INK4a expression status in our OSCC samples. In conclusion, the prevalence of high-risk HPV was low in this group of OSCC patients; no association between HPV status and p16INK4a expression status was identified.
Keywords: HPV, Oral cancer, OSCC, PCR, p16INK4a, Thai
Introduction
Oral cancer is the sixth most common cancer worldwide, and more than 90% of oral cancer cases comprise oral squamous cell carcinoma (OSCC) [1]. OSCC is a malignant neoplasm that arises from the mucosal epithelium [2]. Tobacco smoking, alcohol consumption, and betel nut chewing habits are major risk factors for OSCC. Nevertheless, OSCC occurs in patients that have not been exposed to these risk factors. Other causes (e.g., genetic predisposition, diet, and oncogenic virus infection) are putative contributing factors for OSCC [3].
Oncogenic viruses, such as human papillomavirus (HPV) and Epstein–Barr virus, cause approximately 20% of human cancers [4]. HPVs (e.g., HPV-16 and HPV-18) have been linked to the development of OSCC in patients without traditional risk factors [4, 5]. After oncogenic viral infection, the virus integrates its genetic material into the host genome, leading to the expression of viral proteins, as well as promotion of the growth and proliferative abilities of virus-infected cells. During HPV infection, E6 and E7 viral oncoproteins inactivate the tumor suppressor proteins p53 and pRb, respectively; thus, they have important roles in cellular immortalization, transformation, and carcinogenesis [5].
In the Asia-Pacific region, the rate of HPV-positivity in OSCC is reportedly 37.55%, and the highest prevalence (48.61%) is observed in Southeast Asia [5]. In Thailand, the association between HPV infection and oral cancer varies among different parts of the country. Previous studies in Thailand revealed that the prevalence of HPV-positivity in OSCC ranged from 0 to 52.6%, depending on the geographic region [6–12]. A low prevalence of HPV-positivity was detected in the central part of the country, particularly in Bangkok; in the northern and northeastern parts of the country, higher prevalences of HPV-positivity were found.
The p16INK4a tumor suppressor protein is a member of the cyclin-dependent kinase (CDK) inhibitor family of proteins, which block CDK4 and CDK6 phosphorylation of pRb; these effects on CDK4 and CDK6 result in cell cycle inhibition. In normal cells, p16INK4a expression is epigenetically silenced by polycomb repressive complexes; thus, the steady state level of p16INK4a is low [13]. A previous study showed that high-risk HPV-infected cancer cells overexpress p16INK4a because of E7-induced oncogenesis [13]. The high-risk HPV E7 proteins, which trigger p16INK4a expression, have evolved to overcome the cell cycle inhibition response by targeting pRB for degradation. The overexpression of p16INK4a has been proposed as a marker for HPV-related cancer, particularly in cervical and oropharyngeal cancers [13]. Furthermore, p16INK4a overexpression was detected in HPV-related OSCC, while overexpression of p16INK4a was less frequently observed in HPV-unrelated OSCC [14]. However, there have been some contradictory results concerning the use of p16INK4a as a marker for HPV-related OSCC [15, 16]. Therefore, it remains unclear whether p16INK4a can be used as a marker for HPV-positive OSCC.
To our knowledge, few reports have examined the prevalence of HPV-related OSCC and the relationship with p16INK4a expression status in Thailand. The aim of this study was to investigate the prevalence of HPV infection in OSCC cases, as well as p16INK4a expression status in HPV-positive and -negative OSCC tissues, in a group of Thai patients.
Materials and Methods
Tissue Samples
This retrospective study was approved by the Faculty of Dentistry/Faculty of Pharmacy, Mahidol University, Institutional Review Board (COA.No.MU-DT/PY-IRB 2019/037.1806 and MU-DT/PY-IRB 2019/041.0307) and the Center for Ethics in Human Research, Khon Kaen University (IRB00001189). The ethical guidelines of the Declaration of Helsinki were followed in this study.
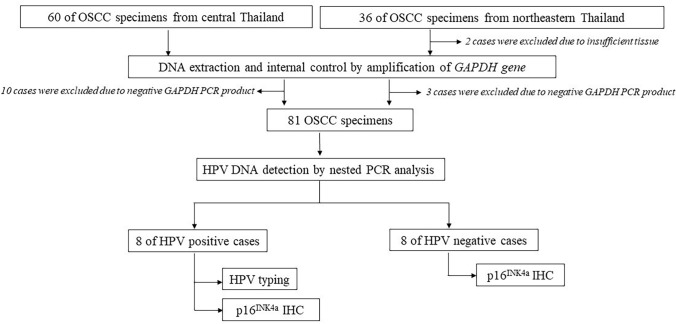
The study flow and sample selection procedure is illustrated in Fig. 1. Formalin-fixed paraffin-embedded (FFPE) OSCC specimens were retrieved from the archives of the Department of Oral and Maxillofacial Pathology, Faculty of Dentistry, Mahidol University, Bangkok, Thailand (60 specimens); the Department of Oral Biomedical Sciences, Faculty of Dentistry, Khon Kaen University, Khon Kaen, northeastern Thailand (24 specimens); and during oral cancer screening for residents in Buriram, Chaiyaphum, Nakhorn-Ratchasima, and Surin, northeastern Thailand (12 specimens). All samples were collected between 2013 and 2019. Clinical information for all cases, including patient age, sex, marital status, tumor location, risk factors (e.g., smoking status, alcohol consumption, and betel nut chewing status), and tumor TNM stage, was obtained from either pathological request forms or patients’ clinical records.
Fig. 1.
Schematic of study flow
Histopathological diagnosis of each case was confirmed, and histological grade was determined by one of the qualified oral pathologists (PL, PK, or AS). The 2017 World Health Organization histological criteria for OSCC were used in this study [2]. In well differentiated tumors, the malignant cells resemble normal epithelial cells. Intercellular bridges are remarkable whereas nuclear hyperchromatism and pleomorphism are unobvious. Keratin pearls and dyskeratotic cells are frequently observed. In moderately differentiated tumors, the malignant cells are less differentiated and demonstrate less keratin production than do well differentiated tumors. In addition, nuclear pleomorphism, mitotic activity, and abnormal mitoses are present in the malignant cells. In poorly differentiated tumors, the malignant epithelial cells exhibit minimal or no squamous differentiation and almost no keratin production. Mitotic figures, abnormal mitoses and nuclear pleomorphism are conspicuous [2].
DNA Extraction, Nested PCR, and HPV Typing
DNA was extracted from FFPE samples using deparaffinization solution and the QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany), in accordance with the manufacturer’s instructions. The concentration and purity of DNA were determined using a NanoDrop™ (Thermo Scientific, MA, USA). Polymerase chain reaction (PCR) analysis of the GAPDH house-keeping gene was used for DNA quality control, as described in a previous study [17].
PCR primer sequences, conditions, thermocycling, and PCR product length are listed in Table 1. For HPV DNA detection, nested PCR was performed using the HotStarTaq DNA Polymerase Kit (Qiagen). Two sets of primers, MY09/MY11 (outer set) and HPV1003/HPV1004 (inner set), were used as described in a previous study [18]. The MY09/MY11 primers detect the L1 regions of HPV-6, -11, -16, -18, -31, -33, -35, -39, -45, -51, -52, -58, and -68. The HPV1003/1004 primers detect the L1 regions of HPV-1, -6, -8, -11, -13, -16, -18, -30, -31, -32, and -33 [18]. DNA extracted from HeLa cells was used as a positive control; deionized sterile water was used as a negative control. After PCR amplification, 5 µl of each PCR product was subjected to 2% agarose gel electrophoresis, along with a 100-bp DNA ladder (Thermo Scientific). The gel was stained with RedSafe (iNtRON Biotechnology, Jungwon-Gu, South Korea) and photographed under ultraviolet light (GelDoc, Bio-Rad, CA, USA).
Table 1.
Primer sequences and PCR conditions for nested PCR
| Primer | Sequence | Product size (base pairs) | Target region | Thermocycler protocol |
|---|---|---|---|---|
|
MY09 MY11 |
CGTCCAAGGGGAAACTGATC | 450 | L1 (outer) |
95 °C (15 min) 35 cycles of [95 °C (30 s) 54 °C (30 s) 72 °C (30 s)] 72 °C (3 min) |
| GCCCAAGGACATAACAATGG | ||||
|
HPV 1003 HPV 1004 |
TTTGTTACTGTGGTAGATA | 150 | L1 (inner) |
95 °C (10 min) 35 cycles of [95 °C (30 s) 54 °C (30 s) 72 °C (30 s)] 72 °C (3 min) |
| GAAAAATAAACTGTAAATC | ||||
|
GAPDH-F GAPDH-R |
TGAGGCTCCCACCTTTCTCATC | 150 | GAPDH |
95 °C (1 min) 45 cycles of [95 °C (45 s) 56 °C (45 s) 72 °C (45 s)] 72 °C (7 min) |
| TGAGGCCCTGCAGCGTACTC |
For HPV DNA typing, 20 µl of the final PCR product at a concentration of 100 ng/µl was subjected to DNA sequencing (Macrogen, Seoul, South Korea). The resulting sequences were analyzed using the Basic Local Alignment Search Tool to determine HPV subtypes.
Immunohistochemistry (IHC)
Eight HPV-positive OSCC samples and eight HPV-negative samples from age- and sex-matched patients were subjected to p16INK4a immunohistochemical analysis. Four-micrometer-thick tissue sections were mounted on 3-aminopropyltriethoxysilane-coated slides and incubated at 56 °C overnight. Tissue sections were deparaffinized in xylene and rehydrated using a graded alcohol series. Antigen retrieval was performed by microwaving the tissue sections in Target Retrieval Solution (S1699, Dako, Carpinteria, CA, USA). After sections had been cooled, they were treated with 3% hydrogen peroxide to block endogenous peroxidase activity. The samples were rinsed with phosphate-buffered saline, then incubated with mouse monoclonal anti-p16INK4a antibody (dilution 1:60, #Z2117, Zeta Corporation, Arcadia, CA, USA) overnight at 4 °C. The sections were washed and incubated with the labeled polymer (EnVision Detection Systems Peroxidase/DAB, Rabbit/Mouse, HRP Rabbit/Mouse [DAB+], Dako, Glostrup, Denmark). Color was developed using diaminobenzidine (Dako Liquid DAB + Substrate Chromogen System, Dako). Sections were counterstained with Mayer’s hematoxylin. A tissue section of cervical squamous cell carcinoma served as a positive control; sections that had not been treated with primary antibody served as negative controls.
Evaluation of p16INK4a Expression
p16INK4a expression findings were considered positive when samples showed nuclear and cytoplasmic staining of moderate to strong intensity in ≥ 70% of the tumor cells, in accordance with established criteria [15, 19].
Statistical Analysis
Statistical analysis was performed using PASW® statistics version 18 (IBM, NY, USA). Comparison of clinicopathological data between HPV-positive and HPV-negative OSCC cases was performed using Fisher’s exact test. p16INK4a expression findings in HPV-positive specimens and HPV-negative specimens from age- and sex-matched patients were compared using McNemar’s test. p-values < 0.05 were considered statistically significant.
Results
Patient Characteristics, Clinicopathological Data, and HPV DNA Status
For this study, we obtained 60 and 36 FFPE OSCC specimens from the central and northeastern regions of Thailand, respectively (Fig. 1). After the exclusion of 10 and five OSCC samples from the central and northeastern parts of Thailand, respectively, because of poor DNA integrity and insufficient tissue availability, 81 OSCC samples were included in our analysis of HPV DNA. The characteristics of the OSCC patients who provided the 81 samples in this study are listed in Table 2. Most included patients were women (60.5%), married (75.3%), and aged > 40 years (96.3%).
Table 2.
Characteristics of patients with OSCC according to HPV status
| HPV-positive samples (n = 8) | HPV-negative samples (n = 73) | Total (n = 81) | p-value | |
|---|---|---|---|---|
| Sex | ||||
| Male | 1 (12.5%) | 31 (42.5%) | 32 (39.5%) | 0.099 |
| Female | 7 (87.5%) | 42 (57.5%) | 49 (60.5%) | |
| Marital status | ||||
| Single | 2 (25%) | 9 (12.3%) | 11 (13.6%) | 0.353 |
| Married | 6 (75%) | 55 (75.4%) | 61 (75.3%) | |
| N/A | 0 | 9 (12.3%) | 9 (11.1%) | |
| Age | ||||
| ≤ 40 years | 1 (12.5%) | 2 (2.7%) | 3 (3.7%) | 0.271 |
| > 40 years | 7 (87.5%) | 71 (97.3%) | 78 (96.3 %) | |
| Region | ||||
| Central Thailand | 5 (62.5%) | 45 (61.6%) | 50 (61.7%) | 0.639 |
| Northeastern Thailand | 3 (37.5%) | 28 (38.4%) | 31 (38.3%) | |
| Tumor location | ||||
| Gingiva | 3 (37.5%) | 23 (31.5%) | 26 (32.1%) | 0.489 |
| Mobile tongue | 2 (25%) | 22 (30.2%) | 24 (29.6%) | |
| Buccal mucosa | 1 (12.5%) | 10 (13.7%) | 11 (13.6%) | |
| Hard palate | 0 | 6 (8.2%) | 6 (7.4%) | |
| Retromolar trigone | 0 | 5 (6.8%) | 5 (6.2%) | |
| Lip | 2 (25%) | 3 (4.1%) | 5 (6.2%) | |
| Floor of mouth | 0 | 4 (5.5%) | 4 (4.9%) | |
| Risk factors | ||||
| Yes | 3 (37.5%) | 27 (37%) | 30 (37%) | 0.633 |
| No | 3 (37.5%) | 29 (39.8%) | 32 (39.5%) | |
| N/A | 2 (25%) | 17 (23.2%) | 19 (23.5%) | |
| Tobacco | ||||
| Smoker (current/former) | 1 (12.5%) | 18 (24.7%) | 19 (23.5%) | 0.397 |
| Non-smoker (never) | 5 (62.5%) | 38 (52.1%) | 43 (53%) | |
| N/A | 2 (25%) | 17 (23.2%) | 19 (23.5%) | |
| Alcohol | ||||
| Drinker (current/former) | 2 (25%) | 13 (17.9%) | 15 (18.5%) | 0.451 |
| Non-drinker (never) | 4 (50%) | 43 (58.9%) | 47 (58%) | |
| N/A | 2 (25%) | 17 (23.2%) | 19 (23.5%) | |
| Betel nut | ||||
| Chewer (current/former) | 1 (12.5%) | 8 (11%) | 9 (11.1%) | 0.627 |
| Non-chewer (never) | 5 (62.5%) | 48 (65.8%) | 53 (65.4%) | |
| N/A | 2 (25%) | 17 (23.3%) | 19 (23.5%) | |
| Histological grade | ||||
| Well differentiated | 3 (37.5%) | 50 (68.4%) | 53 (65.4%) | 0.102 |
| Moderately differentiated | 3 (37.5%) | 19 (26%) | 22 (27.2%) | |
| Poorly differentiated | 1 (12.5%) | 1 (1.4%) | 2 (2.5%) | |
| Papillary squamous cell carcinoma | 1 (12.5%) | 2 (2.8%) | 3 (3.7%) | |
| Acantholytic squamous cell carcinoma | 0 | 1 (1.4%) | 1 (1.2%) | |
| TNM stage | ||||
| I | 5 (62.5%) | 23 (31.5%) | 28 (34.6%) | 0.265 |
| II | 0 | 12 (16.4%) | 12 (14.8%) | |
| III | 2 (25%) | 15 (20.5%) | 17 (21%) | |
| IVA | 0 | 16 (22%) | 16 (19.8%) | |
| IVB | 0 | 1 (1.4%) | 1 (1.2%) | |
| IVC | 0 | 0 | 0 | |
| N/A | 1 (12.5%) | 6 (8.2%) | 7 (8.6%) |
HPV DNA was detected in eight of 81 (9.9%) OSCC samples, which included five OSCC samples from central Thailand (5/50, 10%) and three OSCC samples from northeastern Thailand (3/31, 9.7%). Representative PCR analysis results are shown in Fig. 2. The viral subtype was HPV-18 in all HPV-positive OSCC samples. Importantly, we found no significant associations between clinicopathological data and HPV status.
Fig. 2.
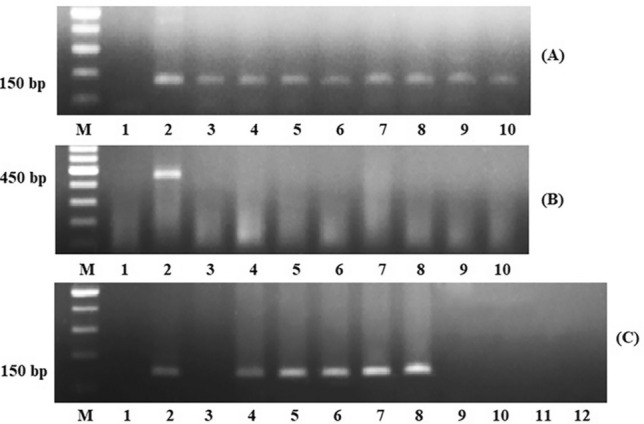
Representative images of PCR products. PCR analysis of GAPDH gene (A). Lane M: 100-bp DNA ladder marker, lane 1: negative control, lane 2: positive control, lanes 3–10: OSCC samples with GAPDH amplification. First step of nested PCR using MY09-MY011 primers (B). Lane M: 100-bp DNA ladder marker, lane 1: negative control, lane 2: positive control, lanes 3–10: representative OSCC samples. Second step of nested PCR using HPV1003-HPV1004 primers (C). Lane M: 100-bp DNA ladder marker, lane 1: negative control, lane 2: positive control, lane 3: PCR product of the first step negative control, lane 4: PCR product of the first step positive control, lanes 5–8: OSCC samples with positive results, lanes 9–12: OSCC samples with negative results
Immunohistochemical Analysis of p16INK4a Expression
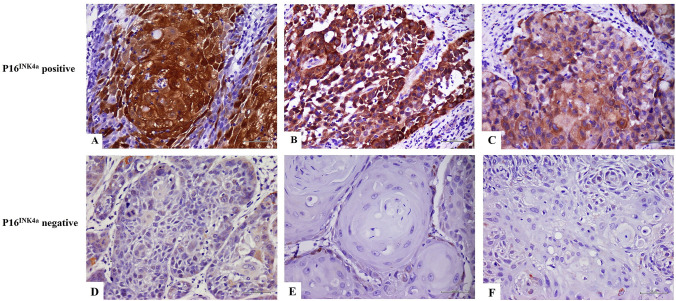
The eight HPV-positive specimens and eight HPV-negative specimens from age- and sex-matched patients were subjected to immunohistochemical analysis to determine p16INK4a expression status. Representative immunostaining images of p16INK4a expression findings are shown in Fig. 3. In accordance with established criteria, p16INK4a expression findings were considered positive when samples showed nuclear and cytoplasmic staining of moderate to strong intensity in ≥ 70% of the tumor cells [15]. Among the eight HPV-positive OSCC samples, three (37.5%) specimens exhibited moderate to strong p16INK4a expression and were considered to have positive p16INK4a expression findings. In the eight HPV-negative OSCC samples, three samples also showed moderate to strong p16INK4a expression and were thus considered to have positive p16INK4a expression findings. Notably, there was no significant association between p16INK4a expression and HPV status (p = 1.000).
Fig. 3.
Representative images of p16INK4a immunohistochemical findings. Strong and diffuse p16INK4a nuclear and cytoplasmic immunostaining is present in ≥ 70% of tumor cells in p16INK4a-positive cases; original magnification 400× (A and B). Moderate and diffuse p16INK4a nuclear and cytoplasmic immunostaining is present in ≥ 70% of tumor cells in p16INK4a-positive cases; original magnification ×400 (C). Focal mild p16INK4a nuclear and cytoplasmic immunostaining is present in p16INK4a-negative cases; original magnification ×400 (D, E, and F)
Histological Grades of Specimens Subjected to p16INK4a Immunohistochemical Analysis
Among the three HPV-positive specimens with positive p16INK4a expression findings, one specimen exhibited well-differentiated OSCC and two specimens exhibited moderately differentiated OSCC. Among the p16INK4a-positive specimens that lacked HPV DNA, two specimens exhibited well-differentiated OSCC and one specimen exhibited moderately differentiated OSCC. Of the 10 p16INK4a-negative specimens, four specimens exhibited well-differentiated OSCC and six specimens exhibited moderately differentiated OSCC.
Discussion
HPV has been linked to increased risks of head and neck cancers, particularly oropharyngeal squamous cell carcinoma (OPSCC) and OSCC. Previous reports indicated a prevalence of HPV-related OSCC in Thai populations that ranged from 0 to 56.2% [6–12]. In the present study, 9.9% of OSCC samples exhibited HPV DNA, which is consistent with the findings of previous studies in Thailand. The variations in HPV prevalence among previous studies may have resulted from many factors, such as racial and geographical differences, differences in sample collection techniques, sample preparation, and HPV detection methods.
Most samples (61.7%) in this study came from the central region of Thailand, particularly the Bangkok metropolitan area; HPV DNA was detected in 10% of these samples from the central region. In our previous study, also conducted in Bangkok, we observed a prevalence of 3.12% [6]. These different HPV prevalences in the same region might be related to the DNA detection methods: we used conventional PCR analysis in the previous study and nested PCR in the present study. Nested PCR has been shown to provide good efficacy with more sensitivity for the detection of HPV in oral and oropharyngeal tissues, compared with conventional PCR analysis [20].
Regarding the HPV prevalences in different regions of Thailand, we found that 10% of OSCC samples from central Thailand and 9.7% of samples from northeastern Thailand exhibited positive HPV DNA findings. Previous studies revealed a lower prevalence (0–5.13%) of HPV DNA in central Thailand, compared with the prevalence in northeastern Thailand (52.6%) [6–9]. These different prevalences of HPV between the central region and other parts of the country might be related to ethno-geographical differences, diagnostic technique sensitivity, specimen type, sample sources (e.g., fresh frozen or FFPE biopsy), and HPV detection procedure (e.g., in situ hybridization, Southern blot hybridization, or high-specificity PCR assay). Two studies conducted in Bangkok, the central part of Thailand, used conventional PCR analysis of fresh frozen tissues; they reported positive HPV findings in 0% (zero of 117 specimens) and 3.12% (one of 32 specimens) of cases [6, 8]. Another study in Bangkok used FFPE and real-time PCR to detect HPV-16 and -18; the authors reported that 5.13% of patients with oral cavity squamous cell carcinoma exhibited positive HPV-16 and -18 findings [9]. In contrast, a study conducted in the northeastern part of Thailand using conventional PCR analysis of FFPE specimens showed that 56.2% of the samples exhibited positive HPV DNA findings [7]. In that study, the authors also performed in situ hybridization; they found that 43.8% of FFPE specimens from the northeastern area exhibited positive HPV E6/E7 mRNA findings [7]. Although these data suggested a difference in HPV prevalence between the central and northeastern regions of Thailand, the present study showed no significant difference in HPV prevalence between these two areas.
Among the known HPV subtypes, HPV-16 and HPV-18 are most frequently involved in HPV-associated OSCC worldwide [5]. In the Asia Pacific region, HPV-16 was found in 90% of HPV-associated head and neck squamous cell carcinoma and 50–68% of oral cavity cancers [5]. HPV-18 was the second most prevalent oncogenic HPV detected in OSCC. In our study, HPV-18 was detected in all HPV-positive OSCC samples. This result is important because most studies in Thailand reported that HPV-16 was the predominant subtype found in OSCC. However, our findings are consistent with the results of a recent multicenter study conducted in several parts of Thailand; in that study, HPV-18 was detected in 57.1% of oral cancer specimens, HPV-16 was detected in 14.3% of specimens, and co-infection of HPV-16 and -18 was detected in 5.8% of specimens [12]. The highest prevalence of HPV-16 and -18 (20%) was found in oral cancer specimens retrieved from the northern part of Thailand. The authors of the multicenter study concluded that the prevalences of HPV-16 and -18 in OSCC exhibited geographical diversity among regions.
p16INK4a overexpression has been used as a surrogate marker for HPV-associated malignancies, particularly in cervical and oropharyngeal cancers. In this study, 37.5% of HPV-positive OSCC specimens exhibited p16INK4a expression findings. However, an equal proportion (37.5%) of HPV-negative OSCC specimens showed positive p16INK4a expression findings. This observation is unsurprising because previous studies reported a weak correlation between p16INK4a expression status and the presence of HPV DNA in OSCC [12, 21]. A study by Zafereo and colleagues indicated a weak correlation between p16INK4a immunohistochemical findings and the presence of HPV-16 and -18, as determined by PCR assays with primers specific for the E6 and E7 regions [21]. Thus, Zafereo and colleagues concluded that p16INK4a status is a poor surrogate marker for high-risk HPV infection of tumors in the oral cavity, which is consistent with our results.
Regarding histologic criteria, the p16INK4a-positive OSCC specimens in our study exhibited well or moderately differentiated morphologies. A previous study reported that p16INK4a-positive OPSCC tends to constitute non-keratinizing squamous cell carcinoma; the non-keratinizing morphology was an excellent predictor of high-risk HPV infection in OPSCC [22]. However, tumor histology and p16INK4a immunostaining findings had limited predictive value for high-risk HPV in OSCC; thus, histologic criteria or p16INK4a immunohistochemical findings were not recommended for the prediction of high-risk HPV infection in OSCC.
p16INK4a protein accumulation is usually detected in senescent cells, and a lack of p16INK4a expression can be more observed in cancer cells [23]. Of the 16 examined OSCC specimens in this study, a lack of p16INK4a expression was observed in 10 specimens. The lack of p16INK4a expression might be explained by the effects of various risk factors (e.g., smoking or heavy alcohol consumption), which can lead to p16INK4a gene deletion or promoter methylation and loss of p16INK4a expression [24]. The oral cavity can be exposed to various carcinogenic factors, which may explain the weak association between p16INK4a expression and HPV status.
We acknowledge that the detection of HPV DNA by PCR alone cannot distinguish between HPV-driven malignancies and bystander HPV infection [24]. Transcriptionally-active high-risk HPV is an important contributing factor in HPV-driven head and neck squamous cell carcinoma; therefore, RNA in situ hybridization for high-risk HPV E6/E7 mRNA is the recommended method to detect HPV-driven malignant transformation [13]. However, this method is expensive and has limited accessibility, compared with PCR and IHC. p16INK4a can be overexpressed in high-risk HPV-infected tumor cells because of E7 viral oncoprotein activity, which causes pRb degradation that leads to the de-repression of p16INK4a expression [13]. In this study, we used a high cut-off point for p16INK4a positivity (≥ 70% of tumor cells) and selected only strong and moderate staining intensities for p16INK4a-positive cases to ensure that these tumors were HPV-associated malignancies, rather than tumors with bystander HPV infection. Singhi and colleagues established a robust algorithm for the identification of genuine HPV infection in OPSCC. According to this algorithm, tumors are considered HPV-positive if they exhibit positive p16INK4a IHC and HPV in situ hybridization findings or positive p16INK4a IHC and HPV PCR analysis findings [25]. Based on the combined presence of high-risk HPV DNA and overexpression of p16INK4a, we speculate that the three cases of p16INK4a-positive and HPV-positive OSCC involved translationally active viruses that drove oral malignancy. Therefore, the combination of p16INK4a immunostaining findings and the presence of high-risk HPV (determined by PCR analysis or in situ hybridization) should be encouraged to distinguish between HPV-driven malignancy and bystander HPV infection.
In conclusion, the prevalence of HPV infection in our cohort was 9.9% in OSCC cases; HPV-18 was the only detected HPV subtype. We did not find a significant association between HPV status and p16INK4a expression in our OSCC samples. Based on this low prevalence of HPV infection, we presume that HPV did not have a major role in OSCC onset in this group of Thai patients.
Acknowledgements
The authors thank Mr. Eakapong Tamboon, Ms. Jintana Pankam, Dr. Khin Mya Tun, and Mr. Dusit Bumalee for technical assistance. We also thank Gabrielle White Wolf, Ph.D. and Ryan Chastain-Gross, Ph.D., from Edanz (https://www.edanz.com/ac) for editing a draft of this manuscript.
Abbreviations
- HPV
Human papillomavirus
- OSCC
Oral squamous cell carcinoma
- CDK
Cyclin-dependent kinase
- FFPE
Formalin-fixed paraffin-embedded
- IHC
Immunohistochemistry
- PCR
Polymerase chain reaction
- OPSCC
Oropharyngeal squamous cell carcinoma
Author Contributions
DR: performed all experiments, performed data analysis, and wrote the manuscript. NP: reviewed the manuscript for important intellectual content. PL: prepared specimens, performed some experiments, and reviewed the manuscript for important intellectual content. NK: supplied some materials and reviewed the manuscript for important intellectual content. PK: prepared some OSCC specimens and provided clinical data. AS: prepared some OSCC specimens and provided clinical data. VS: supplied research funding. BK: supplied research funding, prepared some OSCC specimens, and provided clinical data. SPK: conceptualized the overall research aims, designed the experiments, prepared the manuscript for publication, and supplied research funding.
Funding
This work was supported by Mahidol University research grant (051/2562) to Siribang-on Piboonniyom Khovidhunkit, Boworn Klongnoi, and Vanvisa Sresumatchai under the Development of Disease Management Model for Oral Cancer with an Integration Network of Screening, Surveillance, and Treatment in Northeast Health District project.
Data Availability
The data presented in this study are available on request from the corresponding author. The data are not publicly available because of ethical restrictions.
Code Availability
Not applicable.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Consent to Participate
No informed consent was required for this study because only pathology slides were used.
Consent for Publication
Not applicable.
Ethical Approval
The study was performed in accordance with the principles of the Declaration of Helsinki. This study was approved by the Faculty of Dentistry/Faculty of Pharmacy, Mahidol University, Institutional Review Board (COA.No.MU-DT/PY-IRB 2019/037.1806 and MU-DT/PY-IRB 2019/041.0307) and the Center for Ethics in Human Research, Khon Kaen University (IRB00001189).
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–16. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 2.El-Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ. World Health Organization classification of head and neck tumours. 4. Lyon: IARC; 2017. [Google Scholar]
- 3.Metgud R, Astekar M, Verma M, Sharma A. Role of viruses in oral squamous cell carcinoma. Oncol Rev. 2012;6:e21. doi: 10.4081/oncol.2012.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo GG, Ou JH. Oncogenic viruses and cancer. Virol Sin. 2015;30:83–4. doi: 10.1007/s12250-015-3599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaikh MH, McMillan NA, Johnson NW. HPV-associated head and neck cancers in the Asia Pacific: a critical literature review & meta-analysis. Cancer Epidemiol. 2015;39:923–38. doi: 10.1016/j.canep.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Khovidhunkit SO, Buajeeb W, Sanguansin S, Poomsawat S, Weerapradist W. Detection of human papillomavirus in oral squamous cell carcinoma, leukoplakia and lichen planus in Thai patients. Asian Pac J Cancer Prev. 2008;9:771–5. [PubMed] [Google Scholar]
- 7.Phusingha P, Ekalaksananan T, Vatanasapt P, Loyha K, Promthet S, Kongyingyoes B, et al. Human papillomavirus (HPV) infection in a case-control study of oral squamous cell carcinoma and its increasing trend in northeastern Thailand. J Med Virol. 2017;89:1096–101. doi: 10.1002/jmv.24744. [DOI] [PubMed] [Google Scholar]
- 8.Chaisrisawadisuk S, Sa-Nguanraksa D, Thumrongtaradol T, O-charoenrat P. Prevalence of human papilloma virus in head and neck cancer in Thai population. J Med Assoc Thai. 2020;103:37–41. [Google Scholar]
- 9.Potaporn M, Apipan P, Suphanpayak S. Prevalence of human papillomavirus 16&18 in oral cavity and oropharyngeal squamous cell carcinoma in Rajavithi hospital. J Med Assoc Thai. 2018;101:1–8. [Google Scholar]
- 10.Sritippho T, Pongsiriwet S, Lertprasertsuke N, Buddhachat K, Sastraruji T, Iamaroon A. p16 - a possible surrogate marker for high-risk human papillomaviruses in oral cancer? Asian Pac J Cancer Prev. 2016;17:4049–57. [PubMed] [Google Scholar]
- 11.Chotipanich A, Siriarechakul S, Mungkung OO. Role of high-risk human papillomavirus in the etiology of oral and oropharyngeal cancers in Thailand: a case-control study. SAGE Open Med. 2018;6:2050312118765604. doi: 10.1177/2050312118765604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komolmalai N, Pongsiriwet S, Lertprasertsuke N, Lekwanavijit S, Kintarak S, Phattarataratip E, et al. Human papillomavirus 16 and 18 infection in oral cancer in Thailand: a multicenter study. Asian Pac J Cancer Prev. 2020;21:3349–55. doi: 10.31557/APJCP.2020.21.11.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JS., Jr Human papillomavirus testing in head and neck squamous cell carcinoma in 2020: where are we now and where are we going? Head Neck Pathol. 2020;14:321–9. doi: 10.1007/s12105-019-01117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pannone G, Rodolico V, Santoro A, Lo Muzio L, Franco R, Botti G, et al. Evaluation of a combined triple method to detect causative HPV in oral and oropharyngeal squamous cell carcinomas: p16 immunohistochemistry, consensus PCR HPV-DNA, and in situ hybridization. Infect Agent Cancer. 2012;7:4. doi: 10.1186/1750-9378-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez BY, Lynch CF, Chan OTM, Goodman MT, Unger ER, Steinau M, et al. Human papillomavirus DNA detection, p16(INK4a), and oral cavity cancer in a U.S. population. Oral Oncol. 2019;91:92–6. doi: 10.1016/j.oraloncology.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minami K, Kogashiwa Y, Ebihara Y, Nakahira M, Sugasawa M, Fujino T, et al. Human papillomavirus and p16 protein expression as prognostic biomarkers in mobile tongue cancer. Acta Otolaryngol. 2017;137:1121–6. doi: 10.1080/00016489.2017.1339327. [DOI] [PubMed] [Google Scholar]
- 17.Bumalee D, Lapthanasupkul P, Tamboon E, Aittiwarapoj A, Klongnoi B, Kitkumthorn N. Low frequency of human papillomavirus and Epstein-Barr virus DNA in ameloblastoma of Thai patients. World J Dent. 2020;11:446–50. doi: 10.5005/jp-journals-10015-1771. [DOI] [Google Scholar]
- 18.Boonmongkolraksa P, Doungudomdacha S, Surarit R, Khovidhunkit SO. Survey of human papilloma prevalence in the oral cavity of Thai dental patients. Southeast Asian J Trop Med Public Health. 2020;51:640–9. [Google Scholar]
- 19.Lewis JS, Jr, Beadle B, Bishop JA, Chernock RD, Colasacco C, Lacchetti C, et al. Human papillomavirus testing in head and neck carcinomas: guideline from the College of American Pathologists. Arch Pathol Lab Med. 2018;142:559–97. doi: 10.5858/arpa.2017-0286-CP. [DOI] [PubMed] [Google Scholar]
- 20.Jalouli M, Jalouli J, Ibrahim SO, Hirsch JM, Sand L. Comparison between single PCR and nested PCR in detection of human papilloma viruses in paraffin-embedded OSCC and fresh oral mucosa. Vivo. 2015;29:65–70. [PubMed] [Google Scholar]
- 21.Zafereo ME, Xu L, Dahlstrom KR, Viamonte CA, El-Naggar AK, Wei Q, et al. Squamous cell carcinoma of the oral cavity often overexpresses p16 but is rarely driven by human papillomavirus. Oral Oncol. 2016;56:47–53. doi: 10.1016/j.oraloncology.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nopmaneepaisarn T, Tangjaturonrasme N, Rawangban W, Vinayanuwattikun C, Keelawat S, Bychkov A. Low prevalence of p16-positive HPV-related head-neck cancers in Thailand: tertiary referral center experience. BMC Cancer. 2019;19:1050. doi: 10.1186/s12885-019-6266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klingenberg B, Hafkamp HC, Haesevoets A, Manni JJ, Slootweg PJ, Weissenborn SJ, et al. p16 INK4A overexpression is frequently detected in tumour-free tonsil tissue without association with HPV. Histopathology. 2010;56:957–67. doi: 10.1111/j.1365-2559.2010.03576.x. [DOI] [PubMed] [Google Scholar]
- 24.Evans MF, Matthews A, Kandil D, Adamson CS, Trotman WE, Cooper K. Discrimination of ‘driver’ and ‘passenger’ HPV in tonsillar carcinomas by the polymerase chain reaction, chromogenic in situ hybridization, and p16(INK4a) immunohistochemistry. Head Neck Pathol. 2011;5:344–8. doi: 10.1007/s12105-011-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116:2166–73. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available because of ethical restrictions.
Not applicable.