Abstract
Genomic walking PCR was used to obtained a 4,567-bp nucleotide sequence from Caldibacillus cellulovorans. Analysis of this sequence revealed that there were three open reading frames, designated ORF1, ORF2, and ORF3. Incomplete ORF1 encoded a putative C-terminal cellulose-binding domain (CBD) homologous to members of CBD family IIIb, while putative ORF3 encoded a protein of unknown function. The putative ManA protein encoded by complete manA ORF2 was an enzyme with a novel multidomain structure and was composed of four domains in the following order: a putative N-terminal domain (D1) of unknown function, an internal CBD (D2), a β-mannanase catalytic domain (D3), and a C-terminal CBD (D4). All four domains were linked via proline-threonine-rich peptides. Both of the CBDs exhibited sequence similarity to family IIIb CBDs, while the mannanase catalytic domain exhibited homology to the family 5 glycosyl hydrolases. The purified recombinant enzyme ManAd3 expressed from the cloned catalytic domain (D3) exhibited optimum activity at 85°C and pH 6.0 and was extremely thermostable at 70°C. This enzyme exhibited high specificity with the substituted galactomannan locust bean gum, while more substituted galacto- and glucomannans were poorly hydrolyzed. Preliminary studies to determine the effect of the recombinant ManAd3 and a recombinant thermostable β-xylanase on oxygen-delignified Pinus radiata kraft pulp revealed that there was an increase in the brightness of the bleached pulp.
The use of hemicellulases in the manufacture of kraft pulp has been shown to promote pulp bleaching, and in most application studies the workers have focused on using xylanases (24, 47, 48). Using mannanases was not studied until recently, when it was shown that mannanases, acting in combination with xylanases, enhance enzyme-aided bleaching of pulps in modified kraft processes (10, 40).
1,4-β-Mannans, which are some of the major constituents of hemicellulose, are hydrolyzed to mannose by endo-acting β-mannanases and exo-acting β-mannosidases. However, due to the complexity of the polysaccharide and the presence of side chain sugars, additional enzymes, including α-galactosidase, β-glucosidase, and acetylmannan esterase, are required to completely hydrolyze mannans (34). Extremely thermophilic bacteria and hyperthermophilic archaea are known to produce several types of thermostable glycosyl hydrolases (38). Few thermostable β-mannanases from thermophilic bacteria have been characterized and sequenced (14, 17–19, 33), while no archaeal β-mannanase has been described so far. The mannanases that have been sequenced belong to either glycosyl hydrolase family 5 or glycosyl hydrolase family 26 (22).
Huang et al. (X. P. Huang, J. A. Hudson, F. A. Rainey, P. D. Nichols, and H. W. Morgan, submitted for publication) described a new thermophilic, spore-forming, aerobic bacterium related to the bacilli that was able to grow on crystalline cellulose, and they named this organism Caldibacillus cellulovorans. Their isolate was related to members of the genus Alicyclobacillus, as determined by small-subunit ribosomal DNA sequence analysis, but it lacked alicyclic fatty acids in its cell membrane. It could also utilize xylan and wood pulp as substrates, which indicated that it was also hemicellulolytic. Using genomic walking PCR (GWPCR) techniques, we identified a multidomain β-mannanase gene, manA, in C. cellulovorans that codes for a thermostable enzyme, ManA, belonging to glycosyl hydrolase family 5. The ManA protein has an unusual structure; it consists of an N-terminal domain of unknown function and a catalytic domain flanked by two cellulose-binding domains (CBDs) homologous to the domains found in a variety of cellulases (45, 46). In this paper we describe cloning, sequencing, and expression of recombinant ManA, as well as biochemical properties of the purified enzyme and its action on kraft pulp.
MATERIALS AND METHODS
Bacterial strains and genomic DNA.
Escherichia coli JM101 [Δ(lac-proAB) thi-1 supE44 F′ (traD36 proAB+ lacZAM15)] was used as the bacterial host in all DNA cloning and expression studies. C. cellulovorans cells were kindly provided by Hugh W. Morgan, University of Waikato, Hamilton, New Zealand. Genomic DNA was prepared as described previously (33). The media and other reagents used have been described by Croft et al. (11).
GWPCR.
Linker assembly, linker library construction, and GWPCR were performed as described by Morris et al. (31, 33). The forward and reverse genomic walking primers used in this study are shown in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Primer | Sequence |
|---|---|
| Genomic walking primers | |
| GW3F | 5′ CGACGCAGGCCGAACAGTACTTCTG 3′ |
| GW3R | 5′ CGTCGGATCATACGAGTAGTCGTTC 3′ |
| GW4F | 5′ GCAACTTGGTCGTACAGTACCGCG 3′ |
| GW4R | 5′ CAACCGTTGCTCAGCACGATCCGC 3′ |
| GW5F | 5′ CCGGAGACCGCCGACATTCGAAGCACCGCG 3′ |
| uorf1F | 5′ GACGATAAACGGCGAATCCGGCGCGAAAACATCG 3′ |
| manA-specific primersa | |
| dom1F | 5′ TAGAGGATTTCCATGGTTCGACGGCTTATCGC 3′ |
| newcbdt1R | 5′ GTCGGCGGAATTCGGTTACGGCTCGACGCC 3′ |
| mannF | 5′ CAACGCCGACCATGGGCGGTGGACCGAACCTGAG 3′ |
| mannR | 5′ GGTGTCGGAATTCACGAGGTCCCGATCGCGTTCG 3′ |
Engineered restrictions sites are underlined.
DNA sequencing.
DNA sequencing was performed with an Applied Biosystems model 377 DNA sequencer by using Big Dye terminator chemistry. A computer analysis of sequence data was carried out by using the Genetics Computer Group software package (12).
Construction of a recombinant pJLA602 plasmid containing manA.
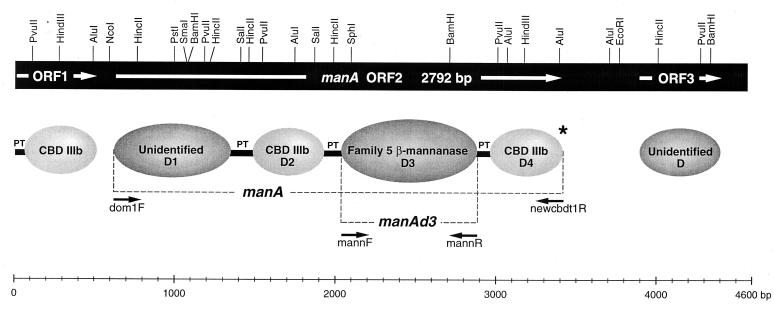
The catalytic or binding domains of ManA were numbered from the N terminus, and the linkers were ignored in this nomenclature (Fig. 1). The manA-specific primers dom1F and newcbdt1R (Table 1) were synthesized in order to PCR amplify DNA coding for the full-length manA gene from C. cellulovorans and were designed to include NcoI and EcoRI restriction enzyme sites, respectively, which allowed directional in-frame ligation of the manA PCR fragment into expression plasmid pJLA602 (36); this resulted in recombinant plasmid pSUN22. Mannanase-positive transformants were identified on plates containing galactomannan by using Congo red stain (43). Both strands of the recombinant plasmid encoding ManA were sequenced in order to confirm that there were no PCR-derived base changes in the DNA except those introduced in the engineered restrictions sites of the PCR primers (Asn→Val change in the second amino acid at the N terminus of the ManA peptide).
FIG. 1.
Diagrammatic representation and partial restriction map of ORF1, ORF3, and manA and the domains encoded by the genes. The positions of PCR primers dom1F and newcbdt1R used to amplify the manA fragment from the genomic DNA of C. cellulovorans for construction of pSUN22 are shown. The positions of PCR primers mannF and mannR used to amplify the manAd3 fragment from pSUN22 for construction of expression vector pSUN23 are also shown. The asterisk indicates the stop codon. D, domain; PT, proline-threonine linker peptide.
Construction of manAd3 recombinant pJLA602 plasmid.
The manAd3 PCR product coding for the mannanase domain of manA was amplified from recombinant plasmid pSUN22 by using the manA-specific primers mannF and mannR (Table 1). These primers were designed to include the NcoI and EcoRI restriction sites, respectively, which allowed directional in-frame ligation of the manAd3 PCR fragment into pJLA602; this resulted in recombinant plasmid pSUN23. Both strands of the recombinant plasmid encoding the ManAd3 mannanase were sequenced in order to confirm that there were no PCR-derived base changes in the DNA except those introduced in the engineered restriction sites of the PCR primers (Pro→Met and Ser→Gly changes at the beginning of the D3 peptide and introduction of a stop codon at the end of the D3 peptide).
Production and purification of ManAd3.
ManAd3 was produced as described previously (31). After host proteins were denatured at 70°C for 30 min, ManAd3 was purified further by anion-exchange chromatography by using a procedure similar to that used for XynB of Dictyoglomus thermophilum Rt46B.1 (31); a HiTrapQ column (Amersham Pharmacia Biotech) and an NaCl step gradient were used. The ManAd3 protein eluted at 75 mM NaCl.
Enzyme activity and protein determination.
β-Mannanase activity was studied by using the dinitrosalicylic acid method of Bernfeld (6). The standard assay reaction mixture contained 0.5% (wt/vol) locust bean gum (LBG) supplemented with 120 mM universal buffer (8) (pH 6.0) and enzyme, and the final volume was 0.1 ml. The reaction mixture was incubated at 85°C for 15 min. All assay reactions were linear (time versus enzyme concentration). One mannanase unit was defined as the amount of enzyme required to liberate 1 μmol of mannose per min at the assay temperature. Protein concentrations were determined by using a BCA protein quantification kit (Pierce, Rockford, Ill.).
Effects of temperature, pH, and thermostability on mannanase activity.
The effect of temperature on the reaction rate was determined by incubating purified enzyme with the substrate at temperatures ranging from 40 to 95°C under standard assay conditions. The enzyme activity of purified ManAd3 was also assayed at pH values ranging from 3.0 to 7.5 (120 mM universal buffer) and from 6.5 to 8.5 {120 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane buffer} at the optimal temperature for activity. All pH values were adjusted at 85°C.
The purified enzyme was incubated at 70 and 80°C in the absence of substrate in the thermostability experiments. Samples were removed at different times, and the residual mannanase activity was measured under standard conditions by using an assay time of 5 min.
Electrophoresis and molecular weight estimation.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed by using the method of Laemmli (26) and 10% polyacrylamide gels. Proteins were stained with Coomassie brilliant blue R-250 (Sigma Chemical Co., St. Louis, Mo.). Broad-range molecular weight standards obtained from Bio-Rad (Richmond, Calif.) were used to determine the molecular mass of the purified protein.
Substrate specificity and mode of action of ManAd3.
The substrate specificity of ManAd3 was determined by incubating the purified enzyme with different polymeric substrates under standard assay conditions. The hydrolysis products arising after the action of ManAd3 on LBG (at 70°C) were analyzed as previously described (18). In addition, mannooligosaccharide and galactosyl-mannooligosaccharide (Megazyme International, Bray, Ireland) hydrolysis was investigated by using high-performance thin-layer chromatography (HPTLC). Each hydrolysis reaction mixture contained 4 μl of 10 mM oligosaccharide substrate, 4 μl of sterile distilled water, 1 μl of 120 mM universal buffer (pH 6.0), and 1 μl of purified mannanase. The reaction mixtures were incubated at 70°C for 3 h, and then 3 μl of each reaction mixture was applied to a Silica Gel 60 HPTLC plate (Merck, Darmstadt, Germany) and separated with an n-propanol-ethanol-water (70:10:20, vol/vol/vol) solvent system. Sugars were detected after the plate was dried and dipped in a 5% sulfuric acid solution in ethanol and then incubated at 110°C for 10 min.
Enzyme treatment of oxygen-delignified kraft pulp from Pinus radiata.
Oxygen-delignified kraft pulp prepared from Pinus radiata wood was enzymatically treated with recombinant β-mannanase ManAd3 from C. cellulovorans and with recombinant xylanase XynB3 from D. thermophilum Rt46B.1 as previously described (31) and then subjected to elemental-chlorine-free (ECF) bleaching.
Nucleotide sequence accession number.
The DNA sequence described here has been deposited in the GenBank database under accession no. AF163837.
RESULTS
GWPCR and analysis of the 4,567-bp nucleotide sequence.
As part of our effort to identify a C. cellulovorans xylanase gene by using GWPCR, we obtained a product that exhibited sequence homology to β-mannanases belonging to glycosyl hydrolases family 5. This product was generated by using genomic walking primers (GW3F and GW3R [Table 1]) which bind in the sequence coding for family IIIb CBDs. Preliminary observations revealed that highly homologous CBD type IIIb domains were present in other glycosyl hydrolase structures found in C. cellulovorans (unpublished results). The nucleotide sequence data obtained for regions upstream and downstream from the GWPCR fragments were combined to generate a 4,567-bp sequence.
Analysis of the nucleotide sequence revealed that three open reading frames (ORFs), ORF1, manA, and ORF3, were present. Part of ORF1 consisted of a sequence that coded for a putative C-terminal type IIIb CBD (61 to 513 bp). Downstream of the ORF1 terminator codon, TGA, there was a 105-bp intergenic region (514 to 619 bp), and this region contained a 15-bp palindromic repeat sequence which may have acted as a transcription terminator signal.
The second ORF, manA (620 to 3,412 bp), consisted of 2,792 nucleotides that encoded a multidomain β-mannanase, ManA, whose putative size was 930 amino acids. The initiation codon, ATG, was preceded at a spacing of 5 bp by a potential ribosome-binding sequence (GAGGA). Downstream of the termination codon, TAA, there was a 481-bp intergenic region (3,413 to 3,893 bp) that contained a 19-bp palindromic repeat sequence, which also may have acted as a transcriptional terminator signal. Further downstream at position 3,894 was the putative ATG initiation codon of ORF3 (3,894 to 4,400 bp).
Amino acid sequences of the ORFs and the multidomain structure of ManA.
Figure 1 is a diagrammatic representation of the elements encoded by the three ORFs of the 4,567-bp sequence. The translated amino acid sequence of ORF3 exhibited no sequence similarity to any of the entries in the GenBank and SwissProt databases. The incomplete nucleotide sequence of ORF1 encodes a proline-threonine (PT) linker peptide followed by a C-terminal CBD homologous to family IIIb CBDs, as classified by Tormo et al. (46). The family IIIb CBD ORF1 exhibits only moderate sequence similarity to the partial ORF encoding a CBD directly upstream of Bacillus lautus CelA (level of sequence identity, 59%) (21), the internal CBD of Caldicellulosiruptor sp. strain Rt69B.1 multidomain XynC (level of sequence identity, 56%) (32), and the C-terminal CBD of Clostridium stercorarium CelY (level of sequence identity, 53%) (9).
Analysis of the sequence of ManA revealed a complex and unique multidomain structure. The N-terminal sequence contained a putative signal peptide sequence with a predicted cleavage site between position 33 (Ala) and position 34 (His). Removal of the signal peptide yielded a mature protein that had a predicted Mr of 98.1 × 103 and contained a domain with an unidentified function at its N terminus, ManAd1 (620 to 1,348 bp) (Fig. 1), followed by a 45-amino-acid PT linker peptide. ManAd1 was compared to translated DNA sequences in the GenBank database and was found to be most closely related to the N-terminal domain of the cellulose binding protein (p40) of Streptomyces halstedii (level of identity, 45%) (16). The S. halstedii p40 N-terminal domain was associated via a PT linker with a C-terminal type IIa CBD and was shown to be able to bind to Avicel. The p40 protein exhibited only very weak catalytic activity when it was incubated for 12 h with methylumbelliferyl-β-d-glucoside. Preliminary hydrolysis experiments performed with different substrates did not reveal any ManAd1 catalytic activity (unpublished results).
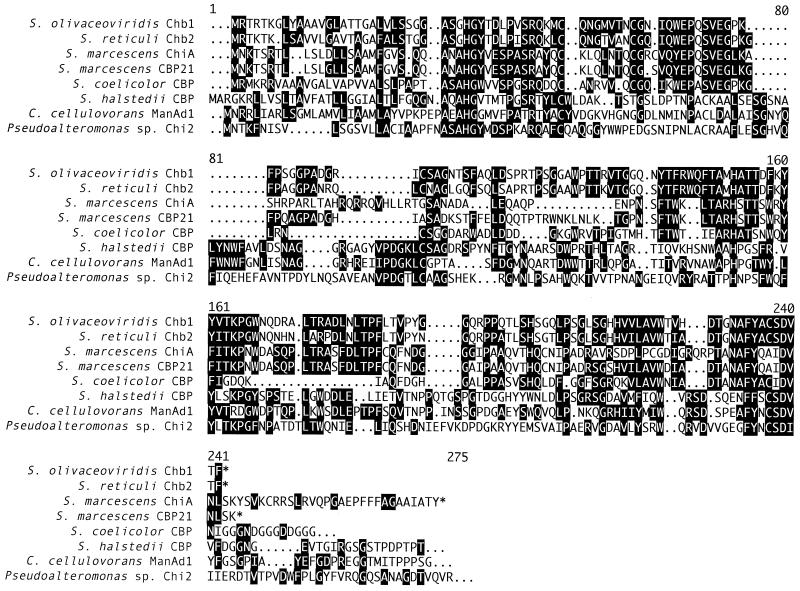
ManAd1 also exhibited some sequence similarity with a number of proteins, some of which have been shown experimentally to bind to chitin (25, 35, 37, 41, 44), and in one instance a chitinase (15). An alignment of the ManAd1 sequence with the sequences of related catalytic domains is shown in Fig. 2.
FIG. 2.
Alignment of sequences homologous to the C. cellulovorans ManAd1 sequence. Conserved residues are highlighted. The sequences used were sequences of Streptomyces olivaceoviridis Chb1 (GenBank accession no. X78535) (37), Streptomyces reticuli Chb2 (GenBank accession no. Y14315) (25), Serratia marcescens ChiA (GenBank accession no. L38384) (15), Serratia marcescens CBP21 (GenBank accession no. AB015998) (41), Streptomyces coelicolor CBP (GenBank accession no. AL031155) (35), Streptomyces halstedii CBP (GenBank accession no. U51222) (16), Caldibacillus cellulovorans ManAd1 (GenBank accession no. AF163837) (this study), and Pseudoalteromonas sp. Chi2 (GenBank accession no. AF007895) (44).
ManAd1 was followed by a second domain, ManAd2 (1,484 to 1,927 bp) (Fig. 1), which was identified as a family IIIb CBD. This CBD exhibited the highest levels of sequence similarity with the internal CBD of Caldicellulosiruptor sp. strain Rt69B.1 XynC (level of sequence identity, 45%) (32) and the C-terminal CBD of Bacillus subtilis endoglucanase EglS (level of sequence identity, 44%) (29). The internal CBD was linked through a 35-amino-acid PT linker to an internal domain, ManAd3 (2,033 to 2,884 bp) (Fig. 1), which was identified as a β-1,4-mannanase belonging to glycosyl hydrolase family 5. The three conserved active site residues (Asn-121, Glu-122, and Glu-223) and the strictly conserved residues Arg-43, His-80, His-188, Tyr-190, and Trp-248 that are characteristic of all family 5 enzymes (2, 4, 20, 28, 49) were also found in ManAd3. The mannanase domain of ManA exhibited 71% sequence identity with the mannanase catalytic domains of ManA and CelC of Caldicellulosiruptor saccharolyticus (19, 33) and 56% sequence identity with the β-mannanase ManA of Streptomyces lividans (1).
The ManAd3 catalytic domain was linked to a C-terminal family IIIb CBD, ManAd4 (2,960 to 3,412 bp) (Fig. 1), through a 25-amino-acid PT linker. This CBD was almost identical (level of sequence identity, 97%) to the C-terminal CBD of ORF1 but exhibited only 54% sequence identity to the internal ManAd2 CBD. Furthermore, ManAd4 exhibited 59, 57, and 53% sequence identity to the CBDs of ORF, XynC, and CelY of Bacillus lautus (21), Caldicellulosiruptor sp. strain Rt69B.1 (32), and Clostridium stercorarium (9), respectively.
Purification and biochemical properties of the recombinant ManAd3 enzyme.
Recombinant ManAd3 mannanase was purified from lysed E. coli cells harboring pSUN23 as described above to electrophoretic homogeneity, and the final specific activity was 1,949 mannanase units/mg. The apparent Mr of the purified enzyme was estimated to be 30.7 × 103 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown), which is consistent with the Mr of 31.3 × 103 deduced from the translated amino acid sequence of ManAd3.
Recombinant ManAd3 enzyme was active at temperatures between 40 and 95°C, and the optimal temperature for activity was 85°C (data not shown), which is 20°C above the optimum growth temperature for C. cellulovorans (Huang et al., submitted). More than 30% of the initial activity was still detected at 95°C. Thermostability experiments revealed that in the absence of the substrate (LBG), the enzyme had a half-life at 80°C of 15 min, while at 70°C the enzyme retained 95% of its initial activity after 24 h of incubation (data not shown). When assays were performed at 85°C, the purified Caldibacillus mannanase was active at pH values between 5.0 and 8.5 (data not shown). The optimal pH for activity was 6.0, and 60 and 37% of the initial mannanase activity remained at pH 8.0 and 8.5, respectively.
Substrate specificity and mode of action.
No detectable reducing sugars were released from oat spelts, birchwood, beechwood, larchwood, and purified 4-O-methyl-d-glucurono-d-xylans, β-glucan, hydroxyethyl or carboxymethyl cellulose, lichenan, laminarin, soluble starch, pullulan, and dextran. The largest amount (100%) of reducing sugars was liberated when the galactomannan LBG (mannose/galactose ratio, 4:1) was used as the substrate. Pure ivory nut mannan (99% mannose) was poorly hydrolyzed (0.7% compared to LBG), indicating that the enzyme was not able to hydrolyze unsubstituted mannan. In the presence of konjac gum glucomannan (mannose/glucose ratio, 1.5:1), only 15% of the maximum activity was observed, while replacement of LBG by the galactomannan guar gum (mannose/galactose ratio, 2:1) resulted in a reduction in the mannanase activity of more than 90%. Furthermore, no detectable reducing sugars were released during incubation with Saccharomyces cerevisiae mannan (with α-1,6, α-1,2, and α-1,3 mannosidic linkages).
The products released by the action of ManAd3 mannanase on LBG galactomannan were analyzed by anion-exchange chromatography (data not shown), and the results revealed that mannose, mannobiose, and mannotriose were the major end products of hydrolysis, which confirmed the endo-acting characteristic of the enzyme. In addition, larger oligosaccharides and trace amounts of galactose were also detected, as were unidentified products that probably represented galactosyl mannooligosaccharides. The ability of ManAd3 to hydrolyze mannooligosaccharides with different degrees of polymerization was investigated by using HPTLC (data not shown). After 3 h of incubation at 70°C, the purified recombinant β-mannanase completely hydrolyzed mannohexaose, mannopentaose, and mannotetraose, yielding mannotriose, mannobiose, and mannose. Mannotriose was only partially hydrolyzed, which produced mannobiose and traces of mannose, while mannobiose was not hydrolyzed. This result indicates that the enzyme contains an active site capable of accommodating at least six mannose residues. Furthermore, no transferase activity was observed after 3 h of incubation with mannose. Purified ManAd3 was also incubated with the galactosyl mannooligosaccharides 61-α-d-galactosyl-mannotriose and 63,64-di-α-d-galactosyl-mannopentaose. Only minimal hydrolysis of 61-α-d-galactosyl-mannotriose was observed after 3 h of incubation, and trace amounts of mannose and a second product, probably α-d-galactosyl-mannobiose, were released. 63,64-di-α-d-galactosyl-mannopentaose was not hydrolyzed by ManAd3.
Enzyme treatment of oxygen-delignified kraft pulp.
Oxygen-delignified kraft pulp was treated with two thermostable recombinant enzymes, XynB3 of D. thermophilum Rt46B.1 (31) and ManAd3 of C. cellulovorans. The results of this experiment are summarized in Table 2. The individual xylanase and mannanase treatments resulted in lower kappa numbers and greater brightness than the kappa number and brightness of the control pulp. The xylanase treatment resulted in a lower kappa number (3.4 U) than the mannanase treatment (4.0 U). However, the lowest kappa number was obtained when the pulp was treated with a mixture of the two enzymes (3.1 U). The trend in the brightness levels of the pulps after the EO stage were similar. There was a 6.8% ISO unit increase in brightness when the enzyme mixture was used (from 53.9 to 60.7% ISO compared with the control pulp); however, only 0.7% ISO unit of this increase was due to the presence of the mannanase in the enzyme mixture.
TABLE 2.
Effects of treatment with recombinant XynB3 and ManAd3 on D(EO)DD bleaching of P. radiata kraft pulp
| Prepn | Enzyme stage yield (%) | D(EO) stage
|
D1 stage brightness (% ISO) | D2 stage brightness (% ISO) | Total bleached pulp yield (%) | ||
|---|---|---|---|---|---|---|---|
| Kappa no. | Brightness (% ISO) | Yield (%) | |||||
| Control | 99.2 | 4.6 | 53.9 | 96.5 | 71.3 | 81.8 | 95.3 |
| XynB3 | 98.8 | 3.4 | 60.0 | 95.8 | 77.4 | 85.3 | 94.7 |
| ManAd3 | 99.9 | 4.0 | 55.6 | 95.9 | 72.9 | 82.8 | 95.5 |
| XynB3 + ManAd3 | 98.3 | 3.1 | 60.7 | 95.3 | 79.0 | 86.1 | 93.4 |
DISCUSSION
The modular N-terminal domain–PT linker–CBD–PT linker–catalytic domain–PT linker–CBD structure of C. cellulovorans ManA is unique. No other enzymes with type IIIb CBDs that contain both internal and C-terminal CBDs in combination with multiple auxiliary domains have been described. Previously, Tomme et al. (45) and Tormo et al. (46) described a comprehensive classification scheme for CBDs. Family IIIb CBDs have been found predominantly in association with glycosyl hydrolase family 5 and 9 endoglucanases of Bacillus, Clostridium, and Caldicellulosiruptor species, either as internal domains or as C-terminal domains of multidomain peptides. It has been observed that family IIIc CBDs are always internal and are directly associated with bacterial family 9 glycosyl hydrolases and that they appear to be required to facilitate hydrolysis of crystalline cellulose by the family 9 catalytic domain (G. K. Farrington, M. D. Gibbs, P. Anderson, J. Eldredge, P. L. Bergquist, and D. P. Williams, submitted for publication). In all Bacillus enzymes, type IIIb CBDs occur as C-terminal domains that are separated from an N-terminal catalytic domain by a PT linker. In Caldicellulosiruptor strains, type IIIb CBDs occur as internal duplicated or triplicated domains that are separated from both N- and C-terminal catalytic domains by PT linkers.
Members of glycosyl hydrolase family 26 are β-mannanases, while glycosyl hydrolase family 5 contains both endoglucanases and bacterial, fungal, and plant β-mannanases. Amino acid sequence alignments of members of family 5 rarely reveal levels of sequence identity greater than 20%, and as a result, members of family 5 have been classified in six subfamilies, subfamilies A1 to A6 (3, 27). Recently, a new alignment of the available bacterial and eukaryotic mannanase amino acid sequences resulted in description of two new subfamilies, subfamilies A7 (eukaryotic mannanases) and A8 (bacterial mannanases) (23). Members of subfamily A8 exhibit levels of sequence identity greater than 43%, while the levels of sequence identity for members of subfamily A7 and members of subfamily A8 are less than 20%. Subfamily A8 includes ManA of Thermomonospora fusca (23), ManA of S. lividans (1), ManA of Vibrio sp. (42), and the mannanase domain of CelC of C. saccharolyticus (33). Accordingly, based on amino acid sequence comparisons, the mannanase domain ManAd3 of C. cellulovorans is classified as a member of glycosyl hydrolase family 5, subfamily A8.
ManAd3 has a higher optimal temperature for activity (85°C) than most recombinant mannanases have. Thermostable β-mannanases belonging to families 5 and 26 have been described; enzymes of C. saccharolyticus (family 5) (7) and D. thermophilum Rt46B.1 (family 26) (18) have an optimal temperature for activity of 80°C, and there is no decrease in activity after incubation for 24 h at 70°C and after incubation for 16 h at 80°C, respectively. Recently, Duffaud et al. (13) described a β-mannanase of Thermotoga neapolitana 5068 which is optimally active at 92°C and has a half-life of 34 h at 85°C, but no sequence data for this enzyme is available yet.
ManAd3 of C. cellulovorans exhibited low specificity with pure ivory nut mannan and konjac gum glucomannan but high specificity with LBG galactomannan. The galactomannan specificity, however, decreased in the presence of the more substituted guar gum galactomannan, indicating that the hydrolysis yield depended on the degree of galactose substitution, as well as the nature of the substitution sugar in the mannan backbone. It has been reported that the degree of hydrolysis of galactomannan decreases with increasing substitution by galactose, which suggests that mannanase cleavage of the mannan backbone is obstructed by the sugar residue (30). Similarly, the recombinant family 5 ManA of C. saccharolyticus was found to exhibit high specificity with glucomannans and mannan, while the specificity decreased with the increase in galactose substitution in galactomannans. This result suggested that α-1,6-galactose branches may sterically hinder the mannanase, preventing enzyme-substrate binding (7).
It has been shown that using mannanases that act in association with xylanases enhances enzyme-aided bleaching of pulps obtained from modified kraft processes (10, 40). However, at this stage we do not have enough data to support the hypothesis that the activities of the two enzymes are synergistic. Additional experiments will be required to determine whether the beneficial effect of adding mannanase to xylanase could be achieved simply by increasing the dose of xylanase. The same trend was observed with the brightness levels of the pulps after the D1 and D2 stages. The total yield of bleached pulp when the mannanase was used alone was about the same as the total yield of the control. However, when the enzyme mixture was used, the mannanase was responsible for a decrease in the yield of about 1.3%. The effects of both family 5 and family 26 mannanases on pulp bleaching have been examined (5, 39). Suurnäkki et al. (39) compared the effects of mannanases from Trichoderma reesei (family 26), Aspergillus niger (probably family 5), and C. saccharolyticus (family 5) on bleaching of P. radiata kraft pulp. These authors found that only the T. reesei mannanase effectively increased the bleachability of the pulp. The effectiveness of the T. reesei mannanase was attributed to the ability of this enzyme to solubilize the pulp glucomannan. In a similar study two family 26 mannanases, one from C. saccharolyticus Rt8.B4 and one from D. thermophilum Rt46B.1, were found to be ineffective in bleaching P. radiata kraft pulp (5).
It appears that mannanases may provide only marginal improvements as bleach-boosting agents but may have other uses in the pulp and paper industry (e.g., in fiber modification).
ACKNOWLEDGMENTS
This work was supported by grants from Macquarie University Research Grants Funds and by an Australian Research Council Small Grant.
REFERENCES
- 1.Arcand N, Kluepfel D, Paradis F W, Morosoli R, Shareck F. β-Mannanase of Streptomyces lividans 66: cloning and DNA sequence of the manA gene and characterization of the enzyme. Biochem J. 1993;290:857–863. doi: 10.1042/bj2900857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baird S D, Hefford M A, Johnson D A, Sung W L, Yaguchi M, Seligy V L. The Glu residue in the conserved Asn-Glu-Pro sequence of two highly divergent endo-β-1,4-glucanases is essential for enzymatic activity. Biochem Biophys Res Commun. 1990;169:1035–1039. doi: 10.1016/0006-291x(90)91998-8. [DOI] [PubMed] [Google Scholar]
- 3.Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- 4.Belaich A, Fierobe H-P, Baty D, Busetta B, Bagnara-Tardif C, Gaudin C, Belaich J-P. The catalytic domain of endoglucanase A from Clostridium cellulolyticum: effects of arginine 79 and histidine 122 mutations on catalysis. J Bacteriol. 1992;174:4677–4682. doi: 10.1128/jb.174.14.4677-4682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergquist P L, Gibbs M D, Saul D J, Reeves R A, Morris D D, Te'o V S J. Isolation and expression of genes for hemicellulases from extremely thermophilic culturable and unculturable bacteria. Am Chem Soc Symp Ser. 1998;687:155–167. [Google Scholar]
- 6.Bernfeld P. Amylases α and β. Methods Enzymol. 1955;1:149–158. [Google Scholar]
- 7.Bicho P A, Clarke T A, Mackie K, Morgan H W, Daniel R M. The characterisation of a thermostable endo-β-1,4-mannanase cloned from “Caldocellum saccharolyticum.”. Appl Microbiol Biotechnol. 1991;36:337–343. [Google Scholar]
- 8.Britton H T S, Robinson R A. Universal buffer solutions and the dissociation constant of veronal. J Chem Soc. 1931;1931:1456–1462. [Google Scholar]
- 9.Bronnenmeier K, Kundt K, Riedel K, Schwarz W H, Staudenbauer W L. Structure of the Clostridium stercorarium gene celY encoding the exo-1,4-β-glucanase Avicelase II. Microbiology. 1997;143:891–898. doi: 10.1099/00221287-143-3-891. [DOI] [PubMed] [Google Scholar]
- 10.Buchert J, Salminen J, Siika-Aho M, Ranua M, Viikari L. The role of Trichoderma reesei xylanase and mannanase in the treatment of softwood kraft pulp prior to bleaching. Holzforschung. 1993;47:473–478. [Google Scholar]
- 11.Croft J E, Love D R, Bergquist P L. Expression of leucine genes from an extremely thermophilic bacterium in Escherichia coli. Mol Gen Genet. 1987;210:490–497. doi: 10.1007/BF00327202. [DOI] [PubMed] [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffaud G D, McCutchen C M, Leduc P, Parker K N, Kelly R M. Purification and characterization of extremely thermostable β-mannanase, β-mannosidase, and α-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. App Environ Microbiol. 1997;63:169–177. doi: 10.1128/aem.63.1.169-177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ethier N, Talbot G, Sygusch J. Gene cloning, DNA sequencing, and expression of thermostable β-mannanase from Bacillus stearothemophilus. Appl Environ Microbiol. 1998;64:4428–4432. doi: 10.1128/aem.64.11.4428-4432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gal S W, Choi J Y, Kim C Y, Cheong Y H, Choi Y J, Bahk J D, Lee S Y, Cho M J. Isolation and characterization of the 54-kDa and 22-kDa chitinase genes of Serratia marcescens KCTC2172. FEMS Microbiol Lett. 1997;151:197–204. doi: 10.1111/j.1574-6968.1997.tb12570.x. [DOI] [PubMed] [Google Scholar]
- 16.Garda A L, Fernández-Abalos J M, Sánchez P, Ruiz-Arribas A, Santamaría R I. Two genes encoding an endoglucanase and a cellulose-binding protein are clustered and co-regulated by a TTA codon in Streptomyces halstedii JM8. Biochem J. 1997;324:403–411. doi: 10.1042/bj3240403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs M D, Elinder A U, Reeves R A, Bergquist P L. Sequencing, cloning and expression of the β-1,4 mannanase gene, manA, from the extremely thermophilic anaerobic bacterium, Caldicellulosiruptor Rt8B.4. FEMS Microbiol Lett. 1996;141:37–43. doi: 10.1111/j.1574-6968.1996.tb08360.x. [DOI] [PubMed] [Google Scholar]
- 18.Gibbs M D, Reeves R A, Sunna A, Bergquist P L. Sequencing and expression of a β-mannanase gene from the extreme thermophile Dictyoglomus thermophilum Rt46B.1, and characteristics of the recombinant enzyme. Curr Microbiol. 1999;39:351–357. doi: 10.1007/s002849900471. [DOI] [PubMed] [Google Scholar]
- 19.Gibbs M D, Saul D J, Lüthi E, Bergquist P L. The β-mannanase from “Caldocellum saccharolyticum” is part of a multidomain enzyme. Appl Environ Microbiol. 1992;58:3864–3867. doi: 10.1128/aem.58.12.3864-3867.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiseppi A, Cami B, Aymeric J-L, Ball G, Creuzet N. Homology between endoglucanase Z of Erwinia chrysanthemi and endoglucanases of Bacillus subtilis and alkalophilic Bacillus. Mol Microbiol. 1988;2:159–164. doi: 10.1111/j.1365-2958.1988.tb00017.x. [DOI] [PubMed] [Google Scholar]
- 21.Hansen C K, Diderichsen B, Jørgensen P L. celA from Bacillus lautus PL236 encodes a novel cellulose-binding endo-β-1,4-glucanase. J Bacteriol. 1992;174:3522–3531. doi: 10.1128/jb.174.11.3522-3531.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilge M, Gloor S M, Rypniewski W, Sauer O, Heightman T D, Zimmermann W, Winterhalter K, Piontek K. High-resolution native and complex structures of thermostable β-mannanase from Thermomonospora fusca—substrate specificity in glycosyl hydrolase family 5. Structure. 1998;6:1433–1444. doi: 10.1016/s0969-2126(98)00142-7. [DOI] [PubMed] [Google Scholar]
- 24.Kantelinen A, Hortling B, Sundquist J, Linko M, Viikari L. Proposed mechanism of the enzymatic bleaching of kraft pulp with xylanases. Holzforschung. 1993;47:318–324. [Google Scholar]
- 25.Kolbe S, Fischer S, Becirevic A, Hinz P, Schrempf H. The Streptomyces reticuli α-chitin-binding protein CHB2 and its gene. Microbiology. 1998;144:1291–1297. doi: 10.1099/00221287-144-5-1291. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lo Leggio L, Parry N J, van Beeumen J, Claeyssens M, Bhat M K, Pickersgill R W. Crystallization and preliminary X-ray analysis of the major endoglucanase from Thermoascus aurantiacus. Acta Crystallogr Sect D. 1997;53:599–604. doi: 10.1107/S0907444997005404. [DOI] [PubMed] [Google Scholar]
- 28.Macarron R, van Beeumen J, Henrissat B, de la Mata I, Claeyssens M. Identification of an essential glutamate residue in the active site of endoglucanase III from Trichoderma reesei. FEBS Lett. 1993;316:137–140. doi: 10.1016/0014-5793(93)81202-b. [DOI] [PubMed] [Google Scholar]
- 29.MacKay R M, Lo A, Willick G, Zuker M, Baird S, Dove M, Moranelli F, Seligy V. Structure of a Bacillus subtilis endo-β-1,4-glucanase gene. Nucleic Acids Res. 1986;14:9159–9170. doi: 10.1093/nar/14.22.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCleary B V. Enzymic interactions in the hydrolysis of galactomannan in germinating guar: the role of exo-β-mannanase. Phytochemistry. 1983;22:649–658. [Google Scholar]
- 31.Morris D D, Gibbs M D, Chin C W J, Koh M-H, Wong K K Y, Allison R W, Nelson P J, Bergquist P L. Cloning of the xynB gene from Dictyoglomus thermophilum strain Rt46B.1 and action of the gene product on kraft pulp. Appl Environ Microbiol. 1998;64:1759–1765. doi: 10.1128/aem.64.5.1759-1765.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris D D, Gibbs M D, Ford M, Thomas J, Bergquist P L. Family 10 and 11 xylanase genes from Caldicellulosiruptor sp. Rt69B.1. Extremophiles. 1999;3:103–111. doi: 10.1007/s007920050105. [DOI] [PubMed] [Google Scholar]
- 33.Morris D D, Reeves R A, Gibbs M D, Saul D J, Bergquist P L. Correction of the β-mannanase domain of the celC pseudogene from Caldicellulosiruptor saccharolyticus and activity of the gene product on kraft pulp. Appl Environ Microbiol. 1995;61:2262–2269. doi: 10.1128/aem.61.6.2262-2269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puls J. Chemistry and biochemistry of hemicelluloses—relationship between hemicellulose structure and enzymes required for hydrolysis. Macromol Symp. 1997;120:183–196. [Google Scholar]
- 35.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 36.Schauder B, Blöcker H, Frank R, McCarthy J E G. Inducible expression vectors incorporating the Escherichia coli atpE transcriptional initiation region. Gene. 1987;52:279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- 37.Schnellmann J, Zeltins A, Blaak H, Schrempf H. The novel lectin-like protein CHB1 is encoded by a chitin-inducible Streptomyces olivaceoviridis gene and binds specifically to crystalline α-chitin of fungi and other organisms. Mol Microbiol. 1994;13:807–819. doi: 10.1111/j.1365-2958.1994.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 38.Sunna A, Moracci M, Rossi M, Antranikian G. Glycosyl hydrolases from hyperthermophiles. Extremophiles. 1997;1:2–13. doi: 10.1007/s007920050009. [DOI] [PubMed] [Google Scholar]
- 39.Suurnäkki A, Clark T A, Allison R W, Buchert J, Viikari L. Proceedings of the 6th International Conference on Biotechnology in the Pulp and Paper industry. Vienna, Austria: Facultas-Universitätsverlag; 1996. Mannanase-aided bleaching of softwood kraft pulps; pp. 67–74. [Google Scholar]
- 40.Suurnäkki A, Heijnesson A, Buchert J, Westermark U, Viikari L. Effect of pulp surfaces on enzyme-aided bleaching of kraft pulps. J Pulp Pap Sci. 1996;22:J91–J96. [Google Scholar]
- 41.Suzuki K, Suzuki M, Taiyoji M, Nikaidou N, Watanabe T. Chitin binding protein (CBP21) in the culture supernatant of Serratia marcescens 2170. Biosci Biotechnol Biochem. 1998;62:128–135. doi: 10.1271/bbb.62.128. [DOI] [PubMed] [Google Scholar]
- 42.Tamaru Y, Araki T, Morishita T, Kimura T, Sakka K, Ohmiya K. Cloning, DNA sequencing, and expression of the beta-1,4-mannanase gene from a marine Vibrio sp. strain MA-138. J Ferment Bioeng. 1997;83:201–205. [Google Scholar]
- 43.Teather R M, Wood P J. Use of Congo red polysaccharide interaction in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Techkarnjanaruk S, Goodman A E. Multiple genes involved in chitin degradation from the marine bacterium Pseudoalteromonas sp. strain S91. Microbiology. 1999;145:925–934. doi: 10.1099/13500872-145-4-925. [DOI] [PubMed] [Google Scholar]
- 45.Tomme P, Warren R A J, Miller R C J, Kilburn D G, Gilkes N R. Cellulose-binding domains: classification and properties. In: Saddler J N, Penner M H, editors. Enzymatic degradation of insoluble carbohydrates. Washington, D.C.: American Chemical Society; 1995. pp. 142–163. [Google Scholar]
- 46.Tormo J, Lamed R, Chirino A J, Morag E, Bayer E A, Shoham Y, Steitz T A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 47.Viikari L, Kantelinen A, Sundquist J, Linko M. Xylanases in bleaching: from an idea to the industry. FEMS Microbiol Rev. 1994;13:335–350. [Google Scholar]
- 48.Viikari L, Sundquist J, Kettunen J. Xylanase enzymes promote pulp bleaching. Paper Timber. 1991;5:384–389. [Google Scholar]
- 49.Wang Q, Tull D, Meinke A, Gilkes N R, Warren R A J, Aebersold R, Withers S G. Glu280 is the nucleophile in the active site of Clostridium thermocellum CelC, a family A endo-β-1,4-glucanase. J Biol Chem. 1993;268:14096–14102. [PubMed] [Google Scholar]