Abstract
To explore the physiological role of tetraheme cytochrome c3 in the sulfate-reducing bacterium Desulfovibrio desulfuricans G20, the gene encoding the preapoprotein was cloned, sequenced, and mutated by plasmid insertion. The physical analysis of the DNA from the strain carrying the integrated plasmid showed that the insertion was successful. The growth rate of the mutant on lactate with sulfate was comparable to that of the wild type; however, mutant cultures did not achieve the same cell densities. Pyruvate, the oxidation product of lactate, served as a poor electron source for the mutant. Unexpectedly, the mutant was able to grow on hydrogen-sulfate medium. These data support a role for tetraheme cytochrome c3 in the electron transport pathway from pyruvate to sulfate or sulfite in D. desulfuricans G20.
The anaerobic sulfate-reducing bacteria have several low-potential c-type cytochromes that are located in the periplasm and presumably function in electron transfer events. Several members of the genus Desulfovibrio have been shown to have three cytochromes in this class, monoheme cytochrome c553, tetraheme cytochrome c3, and a high-molecular-weight cytochrome c (Hmc) with 16 hemes (20, 21). Of these cytochromes, the most abundant, and that for which the most structural information has been gathered, is tetraheme cytochrome c3 (8, 21).
The physiological role of tetraheme cytochrome c3 remains enigmatic. This cytochrome has been reported to interact effectively with an almost unrealistic list of electron donors and acceptors. In vivo, c3 has been suggested to be the redox partner of the periplasmic hydrogenases (5, 22), a role supported by the finding that this cytochrome is tightly associated with hydrogenases during purification of the latter (17). A model in which tetraheme cytochrome c3 shuttles electrons from hydrogenases to the various polyheme cytochromes (Hmc, nine-heme cytochrome, or octaheme cytochrome c3) has been proposed (2, 15, 18). In addition to having periplasmically located redox partners, tetraheme cytochrome c has been shown to form functional complexes with a number of cytosolic electron carriers, including ferredoxin (9), rubredoxin (28), and flavodoxin (16). In vitro studies have shown that tetraheme cytochrome c3 and an [NiFe] hydrogenase will mediate reduction of metals, including Fe(III), Cr(VI), and U(VI), with hydrogen as the electron donor (13).
The reactive nature of tetraheme cytochrome c3 makes biochemical characterization of redox partners and functional analysis problematic; therefore, a genetic approach was initiated. As a first step, it was necessary to determine whether this cytochrome is essential for cell growth. Construction of a strain with an interrupted gene encoding tetraheme cytochrome c3 (cycA) allowed confirmation that this cytochrome is not required for growth with lactate as the primary source of carbon and reductant and with sulfate as the terminal electron acceptor. However, the mutant grew poorly, if at all, on pyruvate-sulfate medium. We infer that the first two electrons from lactate bypass tetraheme cytochrome c3 while those from pyruvate pass through that cytochrome for sulfate reduction.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The sulfate-reducing bacterium Desulfovibrio desulfuricans G20 is a spontaneously nalidixic acid-resistant derivative of the wild-type strain G100A (34) that is also cured of the endogenous cryptic plasmid pBG1 (33). The Escherichia coli strains used as hosts for cloning and for donors in conjugations were DH5α (11) and HB101 (7). Vectors and plasmids used are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid(s) | Relevant features | Source or referencea |
|---|---|---|
| pBluescript II SK and KS | MCSb Mob− cloning vectors; Apr | Stratagene |
| pBC SK | MCS Mob− cloning vector; Cmr | Stratagene |
| pGEM-T | PCR cloning vector; Apr | Promega |
| SuperCos I | 7.6-kbp cosmid vector; Apr Kmr | Stratagene |
| pUC4KIXX | Source of kan cassette | Pharmacia |
| RSF1010 | Source of mob genes | 23 |
| pRK2013 | ColE1 replicon; Mob+ Tra+ helper plasmid; Kmr | 10 |
| pRK2073 | pRK2013::Tn7; Mob+ Tra+ helper plasmid; Spr Kms | 12 |
| pBSCYA | 1.5-kbp ApaI containing cycA in pBC SK | This work |
| pCRA | PCR amplicon from cycA in pBluescript KS | This work |
| pCOS20 | SuperCos I containing D. desulfuricans DNA with cycA | This work |
| pBGC2 | pBluescript SK with 259-bp SacII-PstI internal cycA fragment; Kmr Mob+ | This work |
Stratagene, La Jolla, Calif.; Promega, Madison, Wis.; Pharmacia, Piscataway, N.J.
MCS, multicloning site.
Growth of cells.
Routine cultures of D. desulfuricans were grown anaerobically in lactate (30 mM)-sulfate (50 mM) medium (LS medium) (24). The headspace of cultures contained the prepurified nitrogen atmosphere of the anaerobic chamber, which had small amounts of hydrogen (<4 nmol/cm3). When the lactate in the medium was replaced by pyruvate, the concentration used was 20 mM. When thiosulfate replaced sulfate, a 50 mM concentration was used. E. coli cultures were routinely grown with vigorous aeration in LC medium (1% [wt/vol] tryptone, 0.5% [wt/vol] Bacto Yeast Extract [Difco, Detroit, Mich.], and 0.5% [wt/vol] NaCl). Media were solidified by the addition of 1.5% (wt/vol) agar. Concentrations of antibiotics used for D. desulfuricans were 200 μg of nalidixic acid/ml and 175 μg of kanamycin/ml; for E. coli, kanamycin was used at 50 μg/ml, ampicillin was used at 100 μg/ml, and chloramphenicol was used at 30 μg/ml.
Growth of D. desulfuricans on hydrogen was accomplished in a defined hydrogen-sulfate medium containing 20 mM Na2SO4, 50 mM sodium acetate, 20 mM NH4Cl, 1 mM CaCl2, 3 mM MgCl2, 1.5 mM KH2PO4, and 1.5 ml of a trace element solution (25) modified to increase the FeCl2 concentration to 42 mM. After sterilization, Na2S was added to a 2.5 mM final concentration to reduce the medium. Finally, 20 ml of a sterile solution of 1 M sodium phosphate and 0.1 M NaHCO3, pH 7.0, was added per liter. Cultures to be tested for hydrogen-dependent growth were taken directly from frozen stocks and inoculated (10% [vol/vol] inoculum) into hydrogen-sulfate medium with a headspace slightly pressurized with H2 for 24 h of growth, which allowed the consumption of residual substrates. This culture was subsequently used to inoculate (10% [vol/vol] inoculum) 5 ml of the hydrogen-sulfate medium in a 70-ml serum bottle. The headspace atmosphere was made 100% H2, and the bottles were incubated horizontally at 37°C. Growth was monitored as the increase in protein. To test for suppression or reversion of the cycA mutation during hydrogen growth, 1 ml of the culture was used to inoculate LS medium. The resulting cells were used for periplasmic protein extraction and cytochrome c3 detection by Western analysis.
Analytical procedures.
Hydrogen was determined with a thermal conductivity detector on a Aerograph gas chromatograph (model 90-P; Varian, Santa Clarita, Calif.) fitted with a column packed with Molecular Sieve 5A (Supelco, Bellefonte, Pa.). Hydrogen appeared within 1 min of injection. Other gases that were present, including CO2, H2S, and N2, appeared much later and, therefore, did not interfere with hydrogen determinations. Utilization of lactate and the appearance of organic acid products were monitored by high-performance liquid chromatography. Culture filtrates were analyzed on an Aminex HPX-87H high-performance liquid chromatograph equipped with a 300- by 7.8-mm column (Bio-Rad), a Waters Associates (Milford, Mass.) chromatographic pump, and a Waters model 484 tunable absorbance detector. With 5 mM H2SO4 as the mobile phase, the organic acids lactate, pyruvate, and acetate were separated and quantified. For dry-weight determinations, 100-ml samples were harvested and washed two times with 10 mM KH2PO4 (pH 7.0). Cell pellets were resuspended in 1-ml volumes of deionized sterile water and dried until a constant weight was obtained. The growth yield of cells with lactate as substrate was calculated as the weight of cells (in grams) produced per mole of lactate consumed.
Isolation of cycA.
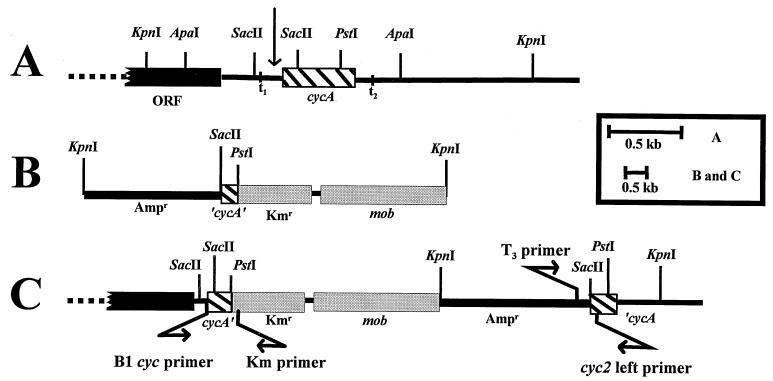
Voordouw and coworkers (32) had reported 21 residues of the N-terminal amino acid sequence of the mature cytochrome c3 purified from D. desulfuricans. From that protein sequence, degenerate primers (NT fwd and NT rev [Table 2]) were devised to generate a PCR probe for the identification of the c3 gene, cycA. An amplicon of 63 bp was produced that was cloned into SmaI-digested pBluescript II KS (Table 1). Either the excised amplicon or the entire plasmid was randomly labeled according to the protocol supplied with the Prime-a-Gene labeling system (Promega, Madison, Wis.) and used to identify homologous sequences in restriction endonuclease-digested chromosomal DNA. Sequencing of a cloned chromosomal 1.7-kbp PstI fragment that hybridized with the 63-bp amplicon showed that cycA was interrupted by the PstI site near its 3′ end. The cloned amplicon was also used to identify a cycA-containing cosmid, pCOS20, from a library of D. desulfuricans G20 DNA (S. Delgado and J. D. Wall, unpublished data). A 1.5-kbp ApaI fragment was subcloned from the cosmid into pBC SK, producing pBSCYA. Sequencing of the fragment revealed the arrangement of open reading frames (ORFs) (Fig. 1A). For the identification of cycA, the derived amino acid sequences were compared to protein sequences in GenBank. Twenty of the amino acid residues reported for the purified mature cytochrome c3 (32) were found to be part of the deduced polypeptide, confirming the identification of cycA.
TABLE 2.
PCR primers used in this studya
| Designation | Primer sequenceb | Location |
|---|---|---|
| NT fwd | 5′-GCNGARGCNCCSGCNGA-3′ | Within cycA, 66 bp from the putative initiating codon; orientation same as that of transcript |
| NT rev | 5′-CTTGTTRAAKATNAC-3′ | Within cycA, 129 bp from the putative initiating codon; orientation opposite that of transcript |
| B1 cyc | 5′-TCACGCGCAGACCCTTTG-3′ | 108 bp upstream from the putative initiating codon of cycA; orientation same as that of transcript |
| Km rev | 5′-GCTTGCGGCAGCGTGAA-3′ | Within 5′ end of kan; orientation opposite that of transcript |
| cyc2 left | 5′-GGAGTGGCTGGAGTGGTT-3′ | Within cycA, 141 bp downstream from the start codon; orientation opposite that of transcript |
| 1932 rev | 5′-GCGGAGCGGAAAACAGCAA-3′ | Beginning 114 bp beyond the termination codon of cycA; orientation opposite that of transcript |
| ORF fwd | 5′-CGCGGCACATCTTATCATA-3′ | Within upstream ORF, 23 bp from putative initiating codon; orientation same as that of transcript |
| ORF rev | 5′-GCGTTTTGCAGGCGCTTTT-3′ | Within upstream ORF, 271 bp upstream of the putative termination codon of ORF; orientation opposite that of transcript |
Oligonucleotides were synthesized by the University of Missouri—Columbia Molecular Biology Program DNA Core facility.
N, any deoxynucleotide; R, purine; K, G or T; S, C or G.
FIG. 1.
Diagrammatic representation of the D. desulfuricans G20 chromosome region encoding cycA. (A) Arrangement of genes in wild-type genomic DNA. The vertical arrow indicates the position of a putative promoter. The black bar indicates the position of a putative ORF upstream of cycA that is transcribed left to right, as is cycA in this illustration. t1 and t2 indicate the locations of possible transcriptional terminators. (B) Plasmid pBSC2 (Table 1) used for gene disruption. The SacII-PstI internal cycA fragment is designated ‘cycA’. (C) Plasmid-interrupted chromosomal cycA. The positions of PCR and sequencing primers, B1 cyc primer and Km primer for the leftward junction and T3 primer and cyc2 left primer for the rightward junction, are indicated by arrows. The left copy of cycA lacks 63 bp of the 3′ end of the coding sequence (which totals 393 bp), while the right copy lacks 71 bp of the 5′ end. Note the different scale for panel A versus that for panels B and C.
Construction of the integration plasmid.
Plasmid pBGC2 was prepared from pBluescript II SK, which does not replicate in the sulfate-reducing bacteria. A 2.7-kbp EcoRV fragment containing the mob genes from RSF1010 was inserted into pBluescript II SK at the EcoRV site. The resulting plasmid was doubly digested with SacII and PstI, and an internal 259-bp fragment from cycA generated with the same enzymes was inserted. Finally, this composite was opened at its unique EcoRI site and the kan gene from pUC4KIXX was introduced, with subsequent selection for kanamycin resistance. The resulting mutagenic plasmid was designated pBGC2 (Fig. 1B).
Conjugal transfer and selection for plasmid integration.
Conjugation was performed as described previously (1) for triparental matings with pRK2073 as the helper plasmid. Nalidixic acid was used to select against E. coli donors, and kanamycin was used to select for the D. desulfuricans recipients that had integrated the plasmid. After 5 to 7 days, the rare Kmr colonies that appeared were checked for residual E. coli contamination and streaked for isolation of single colonies. Potential mutants were grown in LS medium plus kanamycin, and samples were frozen at −80°C as stocks for characterization.
Procedures for nucleic acids.
Enzymatic DNA manipulations were carried out according to the directions of the enzyme suppliers. Plasmid DNA preparations were made with QIAprep Spin Miniprep kits (Qiagen, Inc., Santa Clara, Calif.). A Promega Wizard genomic DNA purification kit was used to obtain chromosomal DNA. In addition, when needed, CsCl purification of plasmid and chromosomal DNAs was performed as described elsewhere (4). Primer synthesis and standard dideoxy sequencing were carried out by the Molecular Biology Program DNA Core of the University of Missouri—Columbia. Southern analyses employed Zeta-Probe blotting membranes from Bio-Rad (Hercules, Calif.) and were performed in accordance with the manufacturer's recommended procedure.
To explore the integrity of the left junction of the integrated plasmid (Fig. 1C), a PCR amplicon was generated with the primer pair B1 cyc-Km rev (Table 2). The 528-bp product was sequenced directly or cloned into the pGEM-T vector and sequenced. When a PCR product was not obtained with these primers, the DNA from the putative mutant was digested with SacII (Fig. 1C) and recircularized to generate a Kmr Apr plasmid, and the cycA gene was sequenced from the T3 primer (Promega). To examine the right junction of the integrated plasmid (Fig. 1C), the T3 primer located in the plasmid and a primer in the cycA gene, cyc2 left (Table 2), were used to produce a 108-bp PCR amplicon that was cloned and sequenced. When necessary, recircularization of a KpnI digest of the mutant chromosomal DNA was performed to produce a replicon that contained the right junction, facilitating sequencing (Fig. 1C). PCRs were carried out in 50 μl of a solution containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.1% (wt/vol) Triton X-100, 2.5 mM MgCl2, 0.1 mM (each) deoxynucleoside triphosphate, 50 ng of DNA, 2.5 U of Taq DNA polymerase, and 0.4 μM (each) primer. The thermal cycling profiles consisted of three cycles of 94°C at 1 min, 1 min of ramp time, 2.5 min at 37°C, 2.5 min of ramp time, and 3 min at 72°C, followed by 35 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C.
For the preparation of RNA, 10-ml samples of culture were poured onto 3-ml portions of frozen TE buffer (10 mM Tris–1 mM EDTA, pH 8.0) to reduce the turnover of mRNA during harvesting. Then initial isolation steps were performed as described for the hot phenol method (27). The resulting RNA was then purified with an RNeasy mini kit (Qiagen). Northern analysis was performed on Magna Charge nylon transfer membranes (Micron Separations, Inc., Westborough, Mass.) by the procedures described by the company. Transfer of RNA from gels to membranes was always by capillary diffusion.
Reverse transcription-PCR (RT-PCR) was performed with a Titan One Tube RT-PCR kit (Boehringer Mannheim Corp., Indianapolis, Ind.). The RNA preparation was treated with RQ1 RNase-free DNase (Promega) as described by the manufacturer. The absence of contaminating DNA was confirmed by PCR.
Protein analyses.
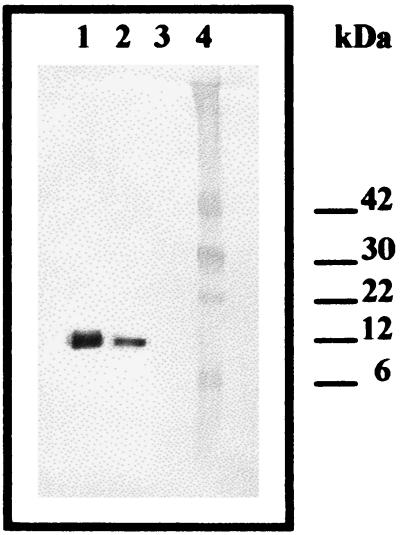
Cytochrome c3 was purified as previously described (32). Polyclonal antibodies were raised against purified cytochrome c3 in a female New Zealand White rabbit. Pure protein (50 μg) was emulsified with 0.5 ml of Freund's complete adjuvant (Sigma) and injected. The rabbits were boosted once with 50 μg of protein plus incomplete adjuvant. Western analysis showed that the antiserum interacted almost exclusively with one polypeptide in the periplasmic extracts (Fig. 2). Total protein concentrations were determined by Bradford analyses (4) with bovine serum albumin as a standard.
FIG. 2.
Immunodetection of cytochrome c3 in periplasmic extracts of D. desulfuricans. Polyclonal antibodies raised against purified G20 cytochrome c3 were used to probe periplasmic proteins separated on a denaturing 10 to 22% (wt/vol) polyacrylamide gel. Lane 1, 1 μg of purified cytochrome c3; lane 2, 15 μg of periplasmic proteins from LS-grown G20; lane 3, 15 μg of periplasmic proteins from LS-grown mutant I2; lane 4, protein molecular size standards, with the relative molecular masses (in kilodaltons) indicated on the right. This figure was digitally manipulated in Adobe Photoshop 4.0 to omit two lanes containing irrelevant samples.
The procedure of van der Westen et al. (30) was used to prepare periplasmic proteins. Briefly, 50 ml of an early-stationary-phase culture of sulfate-reducing bacterial cells was harvested and suspended in 1.0 ml of 50 mM Tris-HCl buffer (pH 9)–50 mM EDTA. The mixture was gently stirred at 0°C for 45 min; the cells were removed by centrifugation at 7,000 × g for 10 min; and the supernatant, following a second centrifugation, was used for the identification of cytochrome c3 protein by Western analysis. After polyacrylamide gel electrophoresis of the supernatant on denaturing 10 to 22% (wt/vol) gradient gels, the periplasmic proteins were transferred onto nitrocellulose membranes (BioTrace NT; Gelman Sciences, Pall Corporation, Ann Arbor, Mich.) by the method of Towbin et al. (29), with the inclusion of 0.0025% (wt/vol) sodium dodecyl sulfate in the transfer buffer. Phosphate-buffered saline (PBS)–Tween (10 mM NaH2PO4, 150 mM NaCl, 0.3% [vol/vol] Tween 20) with 2% (wt/vol) nonfat dry milk (PBS-milk) was used to block the membranes overnight prior to immunodetection. The membrane was then incubated with the primary antibody at a dilution of 1:1,000 in PBS-milk for 1.5 h and washed twice (5 min each wash) in each of three buffers: PBS-Tween, 1 M NaCl in PBS-Tween, and PBS-milk. Secondary antibodies (goat anti-mouse immunoglobulin G and goat anti-rabbit immunoglobulin G; Sigma, St. Louis, Mo.) were alkaline phosphatase conjugates that were used at a 1:1,000 dilution in PBS-milk for a 1- to 2-h incubation. Two 5-min washes with each of five solutions (PBS-Tween, 1 M NaCl in PBS-Tween, PBS-Tween, PBS, and 100 mM Tris [pH 9.5]) were performed sequentially. Color development was performed by the method of Blake et al. (6) with 100 μl each of nitroblue tetrazolium (33 mg/ml in 70% [vol/vol] dimethyl sulfoxide; Sigma) and 5-bromo-4-chloro-3-indolylphosphate (17 mg/ml in 100% [vol/vol] dimethyl sulfoxide; Sigma) per 10 ml of 100 mM Tris buffer (pH 9.5). The stop solution consisted of 10 ml of PBS-Tween containing 100 μl of 0.5 M EDTA.
Cytochrome c3 protein in periplasmic extracts was also visualized without electrophoretic separation of polypeptides, by spotting known quantities of extracted periplasmic protein directly onto nitrocellulose membranes. Processing and immunodetection were as described for Western analyses.
Reagents.
Enzymes for DNA manipulation were purchased from Boehringer Mannheim, Promega, New England Biolabs (Beverly, Mass.), and Gibco BRL (Gaithersburg, Md.) and were used according to the manufacturers' instructions. [α-32P]dCTP (3,000 Ci/mmol, 10 mCi/ml) was from NEN Life Sciences Products (Boston, Mass.). Coomassie brilliant blue G-250 for protein staining was from Eastman Kodak Co. (Rochester, N.Y.). All other chemicals and biochemicals were purchased from Fisher Scientific (Pittsburgh, Pa.) or Sigma-Aldrich (St. Louis, Mo.).
Nucleotide sequence accession number.
The sequence of the cycA gene has been deposited in the GenBank nucleotide sequence database under accession no. AF205067.
RESULTS
Cloning of cycA.
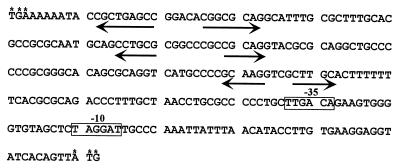
The gene encoding cytochrome c3 was cloned from D. desulfuricans G20 on a 1.5-kb ApaI fragment (Fig. 1). When an internal fragment of the cloned gene was used as a probe for chromosomal DNA, a single hybridizing band was observed, suggesting that one cytochrome c3 gene was present (data not shown). Sequencing revealed an ORF of 390 bp from which was deduced a polypeptide of 130 amino acid residues that exhibited the characteristic features of a c3 cytochrome (8) (Fig. 3). Upstream, and apparently transcribed in the same orientation, was part of an ORF whose product has similarity to a methyltransferase. In the 257-bp intergenic region (Fig. 4) were several inverted repeats that could serve as transcriptional signals. Twenty-four base pairs after the putative cytochrome stop codon was a region that when transcribed could form an exact 10-bp stem (9 of the 10 bp are GCs) with a 4-base loop that might signal transcriptional termination. No additional ORFs were detected within 160 bp of the 3′ end of the cycA coding sequence. Between the upstream ORF encoding the putative methyltransferase and cycA were found exact consensus sequences derived for Desulfovibrio promoters (21) (TTGACA and TAGGAT [the −35 and −10 sequences, respectively]). These sequences were located 44 bp upstream of the ATG start codon of cycA and were separated by 17 bp. However, RT-PCR analysis of the mRNA (data not shown) revealed that some transcripts encoding CycA extended as far upstream as 108 bp from the initiation codon of cycA. The number and location of promoters for this gene have yet to be determined.
FIG. 3.
Alignment of D. desulfuricans G20 cytochrome c3 amino acid sequences with those of other members of the genus. Boxed sequences are the four heme binding sites. The arrow indicates the apparent cleavage site for the signal peptide for the D. desulfuricans G20 proapoprotein. Asterisks mark the conserved histidines that are the sixth axial ligands to the hemes. DdG20, D. desulfuricans G20 (sequence deduced from that of the cloned gene); DvHil, D. vulgaris Hildenborough (sequence deduced from that of the cloned gene [Protein Identification Resource Database, Johns Hopkins University, PIR no. A24799]); DvMiy, D. vulgaris Miyazaki (PIR no. S33874); Ds, D. salexigens (PIR no. A00128); Dg, D. gigas (PIR no. A00126). Levels of identity between the cytochrome sequence of D. desulfuricans G20 and those of D. vulgaris Hildenborough and Miyazaki, D. salexigens, and D. gigas were 66, 69, 35, and 54%, respectively.
FIG. 4.
Sequence of the intergenic region upstream of cycA. Asterisks indicate the termination codon of the putative upstream ORF and the start codon of the cycA gene. Prominent inverted repeats are indicated by arrows. A possible promoter for cycA, the −35 and −10 regions (separated by 17 bp), is boxed.
Cytochrome features recognized in the derived polypeptide include a 23-amino-acid leader sequence for targeting the polypeptide to the periplasmic space and four heme binding motifs, two each of −CX2CH− and −CX4CH− sequences. The N-terminal 20 amino acids of the mature cytochrome c3 reported for this strain (32) correlate exactly with residues 24 through 43 of the deduced polypeptide, confirming the identity of the cycA gene. The 21st residue, reported from the protein sequencing to be a lysine (32), was found instead from the gene sequence to be a histidine, confirming the more recent protein sequencing of Aubert et al. (3). When compared with tetraheme cytochromes from several other Desulfovibrio strains (Fig. 3), conservation of the heme binding sites and the histidines providing the sixth axial ligands for the hemes was evident. The sequence of cytochrome c3 from D. desulfuricans G20 was found to be 66 and 67% identical to the homologs from Desulfovibrio vulgaris Hildenborough and Miyazaki, respectively.
Disruption of cycA by plasmid integration.
To gain insight into the role of cytochrome c3 in the metabolism of D. desulfuricans, a mutation of the cycA gene that eliminated the production of a functional protein was constructed. A plasmid unable to replicate in D. desulfuricans, pBGC2, containing a 259-bp fragment internal to cycA, whose integration into the chromosomal cycA would disrupt the gene and could be detected by the acquisition of kanamycin resistance encoded on the plasmid, was constructed (Fig. 1B). The cycA fragment lacked the coding sequences for the signal sequence at the N terminus and the attachment site for the fourth heme at the C terminus. Thus, neither of the two copies of cycA resulting from the plasmid integration encodes a proapopolypeptide that could be processed into a functional cytochrome.
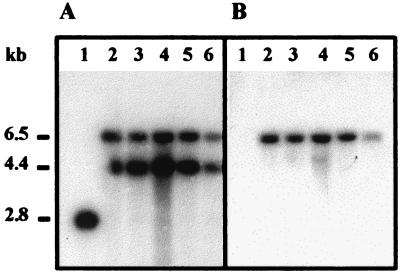
Five Kmr colonies were obtained following conjugal transfer of the plasmid into strain G20, and Southern analyses were performed to confirm the predicted chromosomal structure. KpnI digests of the DNA from the presumed mutants and the wild-type parental strain were probed sequentially with the cycA internal fragment and the Kmr gene, kan (Fig. 5). The wild-type DNA contained a single band of 2.8 kb hybridizing with the cycA probe, while each of the possible mutants had two bands, 4.4 and 6.5 kb, as predicted for an interrupted gene. As expected, the Kmr probe did not hybridize with the wild-type DNA but did hybridize with a 6.5-kb band of the mutant DNA. Free plasmid was apparently not present in the putative mutants, as evidenced by the absence of a hybridizing fragment the size of the KpnI-digested plasmid, 7.5 kb, in the Southern analysis and by the inability to recover plasmid from the exconjugants. These results confirmed integration of the plasmid into the KpnI DNA fragment containing cycA, generating D. desulfuricans mutants, I1 through I5, that were structurally identical.
FIG. 5.
Confirmation, by Southern analysis, of integration of the pBSC2 plasmid into the D. desulfuricans G20 chromosome. Lane 1, G20 total chromosomal DNA digested with KpnI; lanes 2 to 6, total chromosomal DNAs of five kanamycin-resistant strains, derived after the introduction of pBSC2, digested with KpnI. Sizes of hybridizing DNA fragments are indicated on the left (in kilobase pairs). Identical gels were probed with the pCRA amplicon insert coding for the N-terminal region of the mature cytochrome c3 (A) or with the kan cassette from pUC4KIXX (B). KpnI-digested pBSC2 migrated as a 7.5-kbp fragment (not shown). The figure is representative of six independent Southern analyses.
Northern analysis of mRNA showed that cycA transcripts of about 570 bases were abundant in the wild type but, in I3, were below the detection limit of about 10% of the wild-type level. The presence of a barely detectable transcript that was about 100 bases longer was observed in mRNA from I3 (Fig. 6). The small amount of longer transcript could be due to readthrough from the 3′-truncated (upstream) copy of the gene that no longer had the strong stem-loop structure hypothesized to terminate transcription of cycA. These data suggested that cytochrome c3 is encoded in a single-gene operon, a conclusion that is consistent with the sequence analysis. As a further test to determine whether cycA is transcribed as part of an operon, an RT-PCR to detect transcripts extending from the upstream putative methyltransferase-encoding ORF into cycA (primers [Table 2], ORF fwd and cyc2 left) was performed. In addition, a second RT-PCR to detected transcripts extending beyond the strong stem-loop structure at the 3′ end of cycA (primers, B1 cyc and 1932 rev) was carried out. Neither of these analyses gave detectable products, in contrast to RT-PCR experiments that allowed detection of the cycA (primers, B1 cyc and 1932 rev) or ORF (primers, ORF fwd and ORF rev) transcripts alone as controls (data not shown).
FIG. 6.
Transcription of cycA in D. desulfuricans G20 and constructed strain I3. Total RNA from mid-exponential-phase cultures of G20 (lane 1) and I3 (lane 2) was probed with the pCRA amplicon insert. Transcript sizes (shown to the left) were estimated relative to rRNA migration. The figure is representative of four independent analyses. This figure was digitally manipulated in Adobe Photoshop 4.0 to omit one lane containing an irrelevant sample.
Mutant instability.
We were surprised to find an abundant 13-kDa heme-staining protein in periplasmic extracts from a subculture of the cells used for the Northern analysis (data not shown). A Southern analysis of the DNA from the same cells extracted for periplasmic proteins confirmed that the plasmid was still integrated into the gene. To evaluate whether a second tetraheme cytochrome was now being produced (19), cytochromes were purified from the periplasm of this strain. Two fractions, representing a monomeric cytochrome of 13 kDa (isoelectric point, 5.8) and, presumably, a polymeric version thereof, contained 80% of the periplasmic cytochrome (A. Dolla, unpublished data). N-terminal sequencing of 21 and 12 residues from the monomeric and polymeric versions, respectively, established that the proteins were the same as that encoded by the cycA gene and previously reported as cytochrome c3 from this bacterium (32). The remainder of the cytochrome was found in fractions that corresponded to a putative high-molecular-weight cytochrome and a monoheme cytochrome c553. Clearly functional cytochrome c3 was still being produced in the constructed strain, in spite of the maintenance of the integrated plasmid.
The insertion junctions of the integrated plasmid in I3 (Fig. 1C) were sequenced to confirm the construction of an interrupted gene. The 3′ deletion in the upstream gene copy was confirmed; however, a complete copy of cycA was restored downstream. A spontaneous insertion of 235 bp containing the 5′ end of cycA, with a concomitant deletion of 209 bp of pBluescript II SK encoding the lac promoter and primer sites for sequencing, had occurred. The similarity in the sizes of the deleted and inserted material in this suppressed strain, designated B1, accounted for the inability to observe the reversion by Southern analysis. In fact, subsequent work revealed another suppressor, B5, in which a 106-bp insert restored the C-terminal coding region of the leftward copy of cycA (Fig. 1C) and was accompanied by a deletion of 101 bp to the left of the gene conferring kanamycin resistance.
To determine conditions in which the mutant might be more stable, the presence of cytochrome c3 in periplasmic extracts was monitored with polyclonal antibodies made against purified protein. These antibodies showed no cross-reactivity with other periplasmic c-type cytochromes (Fig. 2). With a detection limit of less than 10 ng of c3, and with wild-type G20 producing about 300 ng of c3/μg of periplasmic protein in early-stationary-phase cells, a 30-fold reduction in this cytochrome could be easily confirmed. After confirmation by PCR that the junctions between the integrated plasmid and the chromosomal DNA of cycA mutant I2 were as predicted, I2 was subcultured daily in LS medium plus kanamycin. Detectable c3 was not found in the periplasm (data not shown), suggesting that the majority of the population was mutant. In the absence of kanamycin, c3 protein was detectable in the third subculture and the cells were now kanamycin sensitive, as expected. Loss of the integrated plasmid in these revertants was confirmed by Southern analysis (data not shown). When the mutant culture was allowed to enter stationary phase in LS medium plus kanamycin and remain nongrowing for 4 to 5 days, the cytochrome was detected in the recovered Kmr cells, indicating that suppression had occurred. Because the cycA sequences were not deleted from the mutant chromosomes, recombination events that would be able to restore a wild-type cycA gene could be envisioned. Clearly in early work with the mutant, stationary-phase cultures maintained in kanamycin-containing medium had been subcultured for experimentation and suppressors had accumulated. Subcultures to confirm the transcription studies and all subsequent growth experiments with the mutant were started with carefully maintained exponential-phase cultures and were strictly monitored for suppression by Western analyses.
Characterization of cycA mutant I2.
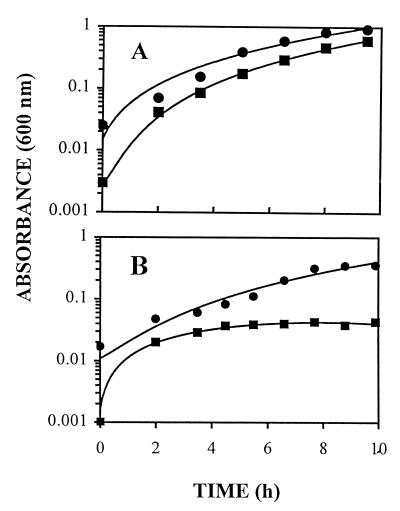
I2 maintained under growth conditions in LS medium with kanamycin appeared to be sufficiently stable for a preliminary physiological analysis. The growth rate of I2 on LS medium was not significantly different from that of the parental strain (Table 3); however, the mutant never reached the same cell density (Fig. 7A). These results were not altered when kanamycin was omitted from the I2 medium. Protein analysis of early-stationary-phase cultures established that the mutant accumulated only 70% ± 15% (four determinations) of the protein accumulated by the wild type. In preliminary analyses, the yield of exponential-phase cultures of I2 grown on lactate corresponded to about 80% of that of the wild type (5.4 g · mol−1).
TABLE 3.
Growth of D. desulfuricans wild-type strain G20 and cycA mutant I2 on selected substrates
| Growth medium | Level
of growtha
|
|
|---|---|---|
| Wild-type G20 | Mutant I2 | |
| Lactate-sulfate | 1.6 ± 0.3 | 1.6 ± 0.3 |
| Pyruvate-sulfate | 1.5 ± 0.2 | >20 |
| Hydrogen-sulfate | 100% | 89% ± 22% |
Doubling times (in hours) were determined by monitoring the absorbance at 600 nm of cultures grown at 37°C on lactate-sulfate (14 determinations) or pyruvate-sulfate (3 determinations in duplicate). Growth on hydrogen-sulfate was evaluated by measuring increases in protein of the cultures incubated with or without hydrogen in the headspace. A value of 100% for the wild type averaged 7.3 μg/ml with four independent determinations performed in duplicate.
FIG. 7.
Growth curves of D. desulfuricans wild-type strain G20 (●) and mutant strain I2 (■) on LS medium (representative of 14 trials) (A) and on pyruvate-sulfate (representative of three trials) (B). The growth temperature was 37°C.
When I2 was tested with pyruvate as the primary source of carbon and reductant and sulfate as the terminal electron acceptor, growth was barely detectable (Fig. 7B; Table 3). Curiously, preliminary experiments with pyruvate and thiosulfate as growth substrates showed that I2 grew, but two- to threefold slower than the wild type. The headspaces of cultures grown to early stationary phase in different media were tested for evolved hydrogen. Small amounts of hydrogen (0.02 μmol/mg of cell protein) were produced in lactate-sulfate cultures of the wild type, G20, but hydrogen was never detected in pyruvate cultures of G20, regardless of the electron acceptor. In contrast, 20- to 100-fold-higher hydrogen concentrations were found in cultures of I2 growing on LS medium. Even though I2 cultured with pyruvate as the electron donor grew poorly, if at all, 4 to 30 μmol of hydrogen/mg of cell protein had accumulated after 10 h of incubation. Periplasmic extracts were prepared from mutant cultures grown on each of the media to determine that suppressors or revertants did not dominate. We infer from the slower growth of the mutant on pyruvate that electron transfer from this substrate involves cytochrome c3.
To further characterize the utilization of lactate by I2 and the parental strains, the organic acid content of LS medium was monitored with growth. While both strains completely consumed the lactate, neither pyruvate nor formate accumulation could be detected in the two cultures. There was essentially a stoichiometric production of acetate as lactate was consumed (data not shown).
Growth of the mutant with hydrogen as the electron donor and sulfate as the terminal acceptor was examined. Growth, as measured by optical density or by whole-cell protein, was not abundant for the wild type; however, similar growth was detected for I2 (Table 3).
DISCUSSION
The previous determination of the N-terminal amino acid sequence of the purified tetraheme cytochrome c3 from D. desulfuricans G200 (32) allowed the isolation of the gene cycA from chromosomal DNA. The complete primary sequence of the proapoprotein was deduced from the gene sequence, revealing the presence of a signal sequence upstream of the N terminus of the mature protein that was consistent with the periplasmic location of the cytochrome. As reported for D. vulgaris Hildenborough (31), a monocistronic transcript for cycA that was consistent with the position of a consensus promoter and putative transcriptional terminator was found.
The most commonly held supposition for the function of Desulfovibrio cytochrome c3 is that it is an electron transfer partner for periplasmic hydrogenases. It was reasoned that if hydrogenase null mutants could be created, the cytochrome that couples with the hydrogenase could also be eliminated by mutation. Experiments creating knockout mutations in the [NiFe], NADP-reducing, and/or [Fe] hydrogenase gene of Desulfovibrio fructosovorans have shown that the loss of these hydrogenases, singly or in combination, is not lethal and does not impair growth on hydrogen with sulfate (14, 26; L. Casalot, M. Rousset, P. de Phillip, E. C. Hatchikian, Z. Dermoun, and J. P. Bélaich, Abstr. 5th Int. Conf. Mol. Biol. Hydrogenases, p. 113, 1997). Mutations in an [Fe] hydrogenase and in a [NiFe] hydrogenase of D. desulfuricans G20 have recently been constructed and also do not result in dramatic growth phenotypes (J. A. Ringbauer and J. D. Wall, unpublished data). A mutation in the gene encoding cytochrome c3 was constructed, but unlike mutations in the hydrogenase operons, the mutation impaired growth on organic acids but did not prevent growth on hydrogen with sulfate.
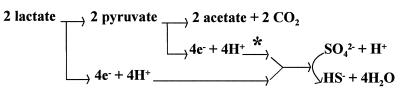
The growth phenotypes of the I2 mutant lacking cytochrome c3, wild-type growth rates on lactate but poor or no growth on pyruvate, give clues to the metabolic roles of c3. These substrates provide both carbon and reductant for the cultures (Fig. 8). Because the oxidation of lactate gives rise to pyruvate, clearly pyruvate is a ready carbon source for I2. Therefore, the lack of growth on pyruvate-sulfate must reflect a block in electron transport from pyruvate to sulfate. The accumulation of hydrogen in the headspace of the I2 cultures suggests one of three alternatives: (i) cytochrome c3 is a component of the electron transfer pathway from pyruvate to sulfate, and impaired c3 results in release of excess reductant as hydrogen (Fig. 8); (ii) electrons from pyruvate participate in intracellular hydrogen cycling (17), and cytochrome c3 functions with a periplasmic hydrogenase to allow reoxidation of the cytoplasmically produced hydrogen; or (iii) cytochrome c3 functions in both the preceding processes. We consider alternative explanation (i) to be the most likely, since growth rates on hydrogen-sulfate are not sufficient to account for the rapid growth of the wild type via recycling hydrogen from pyruvate-sulfate. Explanation (ii) is made less likely by the observation of growth of the mutant on hydrogen-sulfate. Growth of mutant I2 on pyruvate-thiosulfate, while slower than that of the wild type, did occur. Thus, cytochrome c3 may be involved in that electron pathway, but it is not essential. Although molar growth yields might be informative in distinguishing these possibilities, the quantification would be confounded by any reutilization of hydrogen, by the occurrence of suppressors, and if compensation by other c-type cytochromes occurs.
FIG. 8.
Diagrammatic representation of organic acid oxidation and electron flow to sulfate as the terminal electron acceptor for D. desulfuricans G20. The asterisk indicates the region of the metabolic pathway likely to be affected by the absence of cytochrome c3 in mutant I2, causing poor growth on pyruvate with a buildup of reductant that is released as hydrogen. Arrows do not imply single-electron-transfer components.
The inability of I2 to grow on pyruvate contributes to the lower cell yield observed when I2 is grown on lactate. Because of this altered metabolism, it is logical that suppressed derivatives would accumulate in stationary-phase LS cultures of I2, as we have observed. We conclude that cytochrome c3 is not essential for growth on lactate but is necessary for efficient use of reductant from pyruvate. Therefore, the electron transfer pathways to sulfate from these two substrates have one or more unshared components. A cycA deletion is being constructed to further characterize this pathway.
ACKNOWLEDGMENTS
We thank Nancy David, Peter Tipton, and Robert Kunz for technical advice and assistance.
This work was supported in part by the Basic Energy Research Program and the Natural and Accelerated Bioremediation Research Program of the U.S. Department of Energy through grants DE-FG02-87ER13713 and DE FG02-97ER62495, respectively; the Missouri Agricultural Experiment Station; and the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Argyle J L, Rapp-Giles B J, Wall J D. Plasmid transfer by conjugation in Desulfovibrio desulfuricans. FEMS Microbiol Lett. 1992;94:255–262. doi: 10.1016/0378-1097(92)90640-a. [DOI] [PubMed] [Google Scholar]
- 2.Aubert, C., M. Brugna, A. Dolla, M. Bruschi, and M. T. Giudici-Orticoni. A sequential electron transfer from hydrogenases to cytochromes in sulfate-reducing bacteria. Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 3.Aubert C, Leroy G, Bianco P, Forest E, Bruschi M, Dolla A. Characterization of the cytochromes C from Desulfovibrio desulfuricans. Biochem Biophys Res Commun. 1998;242:213–218. doi: 10.1006/bbrc.1997.7852. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates; 1987. [Google Scholar]
- 5.Bianco P, Haladjian J, Bruschi M, Guerlesquin F. Reactivity of [Fe] and [Ni-Fe-Se] hydrogenases with their oxido-reduction partner, the tetraheme cytochrome c3. Biochem Biophys Res Commun. 1992;189:633–639. doi: 10.1016/0006-291x(92)92247-u. [DOI] [PubMed] [Google Scholar]
- 6.Blake M S, Johnston K H, Russell-Jones G J, Gotschlich E C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 7.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 8.Coutinho I B, Xavier A V. Tetraheme cytochromes. Methods Enzymol. 1994;243:119–140. doi: 10.1016/0076-6879(94)43011-x. [DOI] [PubMed] [Google Scholar]
- 9.Dolla A, Leroy G, Guerlesquin F, Bruschi M. Identification of the site of interaction between cytochrome c3and ferredoxin using peptide mapping of the cross-linked complex. Biochim Biophys Acta. 1991;1058:171–177. doi: 10.1016/s0005-2728(05)80234-8. [DOI] [PubMed] [Google Scholar]
- 10.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Kim C H, Helinski D R, Ditta G. Overlapping transcription of the nifA regulatory gene in Rhizobium meliloti. Gene. 1986;50:141–148. doi: 10.1016/0378-1119(86)90319-7. [DOI] [PubMed] [Google Scholar]
- 13.Lovley D R. Metal-reducing microorganisms. Annu Rev Microbiol. 1993;47:263–290. doi: 10.1146/annurev.mi.47.100193.001403. [DOI] [PubMed] [Google Scholar]
- 14.Malki S, de Luca G, Fardeau M L, Rousset M, Bélaich J P, Dermoun Z. Physiological characteristics and growth behavior of single and double hydrogenase mutants of Desulfovibrio fructosovorans. Arch Microbiol. 1997;167:38–45. doi: 10.1007/s002030050414. [DOI] [PubMed] [Google Scholar]
- 15.Matias P M, Saraiva L M, Soares C M, Coelho A V, Legall J, Carrondo M A. Nine-haem cytochrome c from Desulfovibrio desulfuricans ATCC 27774: primary sequence determination, crystallographic refinement at 1.8 and modeling studies of its interaction with the tetrahaem cytochrome c3. J Biol Inorg Chem. 1999;4:478–494. doi: 10.1007/s007750050334. [DOI] [PubMed] [Google Scholar]
- 16.Palma P N, Moura I, LeGall J, van Beeumen J, Wampler J E, Moura J J G. Evidence for a ternary complex formed between flavodoxin and cytochrome c3. 1H-NMR and molecular modeling studies. Biochemistry. 1994;33:6394–6407. doi: 10.1021/bi00187a003. [DOI] [PubMed] [Google Scholar]
- 17.Peck H D., Jr . Bioenergetic strategies of the sulfate-reducing bacteria. In: Odom J M, Singleton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag; 1993. pp. 41–76. [Google Scholar]
- 18.Pereira I A C, Romao C V, Xavier A V, Legall J, Teixiera M. Electron transfer between hydrogenases and mono- and multiheme cytochromes in Desulfovibriossp. J Biol Inorg Chem. 1998;3:494–498. [Google Scholar]
- 19.Pieulle L, Haladjian J, Bonicel J, Hatchikian E C. Biochemical studies of the c-type cytochromes of the sulfate reducer Desulfovibrio africanus. Characterization of two tetraheme cytochromes c3with different specificity. Biochim Biophys Acta. 1996;1273:51–61. doi: 10.1016/0005-2728(95)00129-8. [DOI] [PubMed] [Google Scholar]
- 20.Pollock W B R, Loutfi M, Bruschi M, Rapp-Giles B J, Wall J D, Voordouw G. Cloning, sequencing, and expression of the gene encoding the high-molecular-weight cytochrome c from Desulfovibrio vulgarisHildenborough. J Bacteriol. 1991;173:220–228. doi: 10.1128/jb.173.1.220-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollock W B R, Voordouw G. Molecular biology of c-type cytochromes from Desulfovibrio vulgarisHildenborough. Biochimie. 1994;76:554–560. doi: 10.1016/0300-9084(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 22.Postgate J R. The sulphate-reducing bacteria. 2nd ed. Cambridge, United Kingdom: Cambridge University Press; 1984. [Google Scholar]
- 23.Priefer U B, Simon R, Pühler A. Extension of the host range of Escherichia colivectors by incorporation of RSF1010 replication and mobilization functions. J Bacteriol. 1985;163:324–330. doi: 10.1128/jb.163.1.324-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rapp B J, Wall J D. Genetic transfer in Desulfovibrio desulfuricans. Proc Natl Acad Sci USA. 1987;84:9128–9130. doi: 10.1073/pnas.84.24.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rousset M, Casalot L, Rapp-Giles B J, Dermoun Z, de Philip P, Bélaich J P, Wall J D. New shuttle vectors for the introduction of cloned DNA in Desulfovibrio. Plasmid. 1998;39:114–122. doi: 10.1006/plas.1997.1321. [DOI] [PubMed] [Google Scholar]
- 26.Rousset M, Dermoun Z, Chippaux M, Bélaich J P. Marker exchange mutagenesis of the hydN genes in Desulfovibrio fructosovorans. Mol Microbiol. 1991;5:1735–1740. doi: 10.1111/j.1365-2958.1991.tb01922.x. [DOI] [PubMed] [Google Scholar]
- 27.Scherrer K. Isolation and sucrose gradient analysis of RNA. In: Habel K, Salsman E, editors. Fundamental techniques in virology. New York, N.Y: Academic Press; 1969. pp. 413–432. [Google Scholar]
- 28.Stewart D E, Wampler J E. Molecular dynamics simulations of the cytochrome c3-rubredoxin complex from Desulfovibrio vulgaris. Proteins Struct Funct Genet. 1991;11:142–152. doi: 10.1002/prot.340110207. [DOI] [PubMed] [Google Scholar]
- 29.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Westen H, Mayhew S G, Veeger C. Separation of hydrogenase from intact cells of Desulfovibrio vulgaris. FEBS Lett. 1978;86:122–126. doi: 10.1016/0014-5793(78)80112-4. [DOI] [PubMed] [Google Scholar]
- 31.Voordouw G, Brenner S. Cloning and sequencing of the gene encoding cytochrome c3 from Desulfovibrio vulgarisHildenborough. Eur J Biochem. 1986;159:347–352. doi: 10.1111/j.1432-1033.1986.tb09874.x. [DOI] [PubMed] [Google Scholar]
- 32.Voordouw G, Pollock W B R, Bruschi M, Guerlesquin F, Rapp-Giles B J, Wall J D. Functional expression of Desulfovibrio vulgaris Hildenborough cytochrome c3 in Desulfovibrio desulfuricans G200 after conjugational gene transfer from Escherichia coli. J Bacteriol. 1990;172:6122–6126. doi: 10.1128/jb.172.10.6122-6126.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall J D, Rapp-Giles B J, Rousset M. Characterization of a small plasmid from Desulfovibrio desulfuricansand its use for shuttle vector construction. J Bacteriol. 1993;175:4121–4128. doi: 10.1128/jb.175.13.4121-4128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weimer P J, Van Kavelaar M J, Michel C B, Ng T K. Effect of phosphate on the corrosion of carbon steel and on the composition of corrosion products in two-stage continuous cultures of Desulfovibrio desulfuricans. Appl Environ Microbiol. 1988;54:386–396. doi: 10.1128/aem.54.2.386-396.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]