Abstract
Hepatocellular carcinoma (HCC) is one of the deadliest malignancies in the world. There is a lack of effective treatment. Previous studies have shown that myocyte enhancer factor 2D (MEF2D) promotes the progression of HCC. Underlying mechanisms have not been fully elucidated. In this study, we reported experimental results obtained using double luciferase. Our results showed that AMOTL2, a negative regulator of Hippo/YAP signaling, and the MEF2 cis-acting element in the upstream region of its promoter bind to MEF2D, inhibiting its transcriptional expression. Studies confirmed that MEF2D affected the protein expression level of AMOTL2 and the YAP signaling activation. It promoted the migration and proliferation of hepatoma cells. We found that luteolin, a natural flavonoid, has anti-tumor activity in HCC cells by affecting YAP signaling transduction. In conclusion, we demonstrated that AMOTL2/YAP signaling is associated with MEF2D-related HCC progression. Luteolin is a promising anti-HCC compound for regulating this signaling.
Keywords: Myocyte enhancer factor 2D, hepatocellular carcinoma, YAP, AMOTL2, luteolin
Introduction
Hepatocellular carcinoma (HCC) is a type of malignant tumor arising from mutated hepatocytes. It is characterized by therapy resistance and poor prognosis [1,2]. One of the major problems hampering HCC treatment is that the molecular mechanism underlying HCC formation and progression is far from being completely understood [3].
As a member of the MEF2 family, the myocyte enhancer factor 2D (MEF2D) is a transcription factor that was initially identified in heart and muscle cells. It affects biological activity by regulating the transcription of its downstream target genes [4,5]. MEF2D is also associated with diseases such as liver cancer [4,6]. Although the abnormal cell cycle is related to MEF2D’s role in HCC, its underlying molecular mechanism has not been completely elucidated [6,7].
Hippo/YAP signaling plays a critical role during development [8,9] and has also been implicated in the progression of human disease [10-13], including HCC [14]. Some natural compounds have been shown to suppress HCC by affecting the YAP pathway [15]. The upstream factors triggering the overactivation of YAP signaling in HCC have not been defined.
We aimed to study if MEF2D exerts its tumor-promoting effect by enhancing the activation of YAP signaling and if interfering with this signaling can suppress HCC.
Materials and methods
Cell lines and reagents
HepG2, Huh7, PLC/PRF/5, and 293T cell lines were cultured in DMEM medium (BI, Cat#2112078) with 10% serum and 1% penicillin-streptomycin double antibody (Gibco, Cat#2289326). The concentration of CO2 was 5%, and the incubator was set to 37°C. After 2-3 days of culture, the fresh medium was replaced, or 0.25% trypsin (Gibco, Cat#2185855) solution was used for digestion and subculture. Luteolin (purity > 99%) was purchased from MCE (Cat#HY-N0162). Puromycin was purchased from Meilunbio (Cat#MB2005). Polybrene was purchased from Santa Cruz (Cat#sc-134220).
PCR and plasmid construction
The upstream and downstream primers of MEF2D were designed by Gene Bank. The coding region of MEF2D was amplified by high fidelity KOD enzyme (TOYOBO LIFE SCIENCE, Cat#KOD-401). cDNA was extracted from hepatoma cells. PCR amplified the AMOTL2 promoter region sequence and the mutant AMOTL2 promoter region sequence. The amplified DNA fragment was purified by gel extraction and then ligated into dual-luciferase reporter plasmid pGL3.
Lentivirus production and transfection
293T cells were inoculated into a 10 cm dish. The packaging plasmid and target plasmid: pMD2G, psPAX2, and pLenti-CrisprV2 were transfected with polyethyleneimine (PEI) in the proportion of 2 μg, 4 μg, and 4 μg. We changed the fresh medium after 6-8 hours and collected the virus solution once every 24 hours and 48 hours. We filtered the mixture with 0.45 μm microporous membrane (sartorius stedim, Cat#16533-K) and store at -80°C.
Liver cancer cells were inoculated into 6-well plates. The cell coverage rate was maintained at 50-60% during transfection. We mixed Lipofectamine 2000 (Invitrogen, Cat#2307486), Opti-mem (Gibco, Cat#31985070), and plasmids according to the Lipofectamine 2000 transfection kit instructions. They were mixed with the medium and placed in a 37°C and 5% CO2 incubator for culture. After 6-8 hours, the medium was replaced with a fresh one. Follow up experiments were conducted after 48 hours.
Cell proliferation experiment
A total of 5×103 cells were plated and cultured with DMEM containing 10% bovine calf serum for 24 hours. After discarding the culture medium, we added a fresh culture medium containing 10% CCK8 enhancer and placed it in an incubator at 37°C without carbon dioxide for 1-2 hours. The absorbance was detected at the wavelength of 450 nm. The data represent the means ± S.D. of three independent experiments.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted by Trizol reagent (Life Technologies, Cat#15596026) and corresponding experimental steps. The cDNA synthesis reverse transcribed the previously extracted total RNA using a reverse transcription kit (TRAN, Cat#AE311). qRT-PCR was performed using the 2× SYBR Green qPCR Master Mix (Bimake, Cat#B21202). Information about the primers is provided in Table 1.
Table 1.
qRT-PCR primer Sequence
| Primer | Forward Sequence | Reverse Sequence |
|---|---|---|
| β-actin (Human) | AAGAGAGGCATCCTGACCCT | TACATGGCTGGGGTGTTGAA |
| MEF2D (Human) | CAGCAGCCAGCACTACAGAG | ACTTGGCAGGGATGACTTTG |
| AMOTL2 (Human) | AGTGAGCGACAAACAGCAGACG | ATCTCTGCTCCCGTGTTTGGCA |
Luciferase reporter assay
A total of 1.5×105 cells were inoculated in 24 well plates. After the cells adhered to the wall, they were co-transfected with GFP-pGL3-AMOTL2-pro + Renilla, GFP-pGL3-AMOTL2-pro-mut + Renilla, MEF2D-pGL3-amotl2-pro + Renilla, and MEF2D-pGL3-AMOTL2-pro-mut + Renilla. The fresh medium was changed every 6-8 hours. After 48 hours, the cells were collected and lysed. The cell lysate was mixed with luciferase detection reagent II. The luciferase activity of firefly was detected by full wavelength enzyme labeling instrument chemiluminescence method. The same volume of Stop & Glo reagent was added to detect the luciferase activity of Renilla luciferase. The results were analyzed by statistical software. The above experiments were conductor for three independent experiments. Information about the primers is provided in Table 2.
Table 2.
primer Sequence
| Primer | Sequence (5’-3’) |
|---|---|
| AMOTL2-pro-F (XhoI) | CTAGCTAGCTTCAGAAGGGCAGACCAGAAAGAG |
| AMOTL2-pro-R (NheI) | CCGCTCGAGTCAGAAAAGGTGACAGATGAGAGGC |
| AMOTL2-pro-mut-F (XhoI) | GTCACCTGGAGGCCCCGCTCCTGTTTG |
| AMOTL2-pro-mut-R (NheI) | CAGTGGACCTCCGGGGCGAGGACAAAC |
Western blotting
Hepatoma cells were lysed with a lysis buffer added with phosphatase inhibitor and protease inhibitor. After high-speed centrifugation, the supernatant was obtained for protein quantification. The protein was isolated with 10% polyacrylamide gel electrophoresis and then transferred to the polyvinylidene difluoride (PVDF) membrane (Immobilon-P, Cat#IPVH00010). The primary antibodies were MEF2D (BD, Cat#610775), AMOTL2 (ABclonal, Cat#A16723), YAP (Cell Signaling Technology, Cat#13008S), Phospho-YAP (Cell Signaling Technology, Cat#13008S), β-actin (Abcam, Cat#ab8227). The secondary antibody used was HRP-conjugated anti-mouse and anti-rabbit IgG (ABclonal, Cat#AS014). The ECL luminescent solution (Millipore, Cat#WBKLS0500) was covered with the PVDF membrane, observed, and recorded on the gel imager.
Wound healing and transwell migration assays
The 5×105 hepatoma cells were inoculated into 6-well plates. After one night, a horizontal line was drawn vertically with the gun head. They were washed with PBS buffer and cultured in serum-free medium with different drug concentrations. Photographs were captured with an inverted microscope at 0 and 24 hours, respectively. Three different visual fields were selected for measurement. The scratch distance was measured with Image J. Cell mobility = (initial scratch distance - scratch distance after test)/initial scratch distance.
The liver cancer cells were starved overnight to eliminate the effects of the serum. The hepatoma cells were resuspended in a serum-free medium with different drug concentrations and plated transwell chambers (JET BioFIL, Cat#TCS-003-024). The migrated cells were stained with 0.1% crystal violet stain solution (Solarbio, Cat#g1063) and photographed under a microscope. Three different visual fields were selected, and the cells were counted by Image J.
Statistical analysis
Data were presented as mean ± SD from at least three independent experiments. Data between categorical variables were analyzed by the χ2 and Fisher’s exact tests. The differences between the two groups or among groups were analyzed by two-tailed student’s t test or one-way ANOVA by GraphPad Prism 7, respectively. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 was statistically significant.
Results
MEF2D negatively regulates the level of AMOTL2 in HCC cells
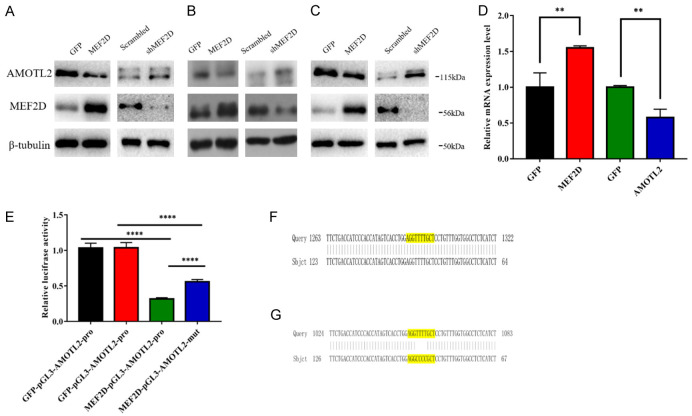
In our previous study, we found that AMOTL2, a well-known YAP suppressor, was listed as one of the MEF2D target genes. We employed western blotting to examine the level of AMOTL2 protein in HCC cell lines, which were transfected with a MEF2D overexpression vector or MEF2D shRNA. The results showed that AMOTL2 level was reduced when MEF2D was overexpressed. Its expression level was elevated under MEF2D shRNA treatment (Figure 1A-C). A qPCR assay confirmed that MEF2D overexpression also suppressed AMOTL2 mRNA (Figure 1D).
Figure 1.
MEF2D inhibited the expression of AMOTL2 and bound to the promoter of AMOTL2 in HCC cells. A. The expression levels of MEF2D and AMOTL2 were identified by western blot in HepG2 cells with overexpression or silencing of MEF2D and AMOTL2. B. The expression levels of MEF2D and AMOTL2 were identified by western blot in PLC/PRF/5 cells with overexpression or silencing of MEF2D and AMOTL2. C. The expression levels of MEF2D and AMOTL2 were identified by western blot in Huh7 cells with overexpression or silencing of MEF2D and AMOTL2. D. MEF2D and AMOTL2 mRNA were detected by qRT-PCR in PLC/PRF/5 cells overexpressing MEF2D. The data represent the means ± SDs of triplicate experiments. **, P < 0.01. E. pGL3 vectors controlled by wild type AMOTL2 promoter (pGL3-AMOTL2-pro) or MEF2-site-mutant ones (pGL3-AMOTL2-mut) was subjected to luciferase assay, under the overexpression of MEF2D or GFP. The data represent the mean ± SDs of three independent experiments. ****, P < 0.0001. F. Partial sequencing of pGL3-AMOTL2-pro vector showed that the yellow marker was MEF2 binding site. G. Partial sequencing of pGL3-AMOTL2-mut vector showed that the yellow marker was MEF2 binding site and mutant MEF2 binding sequence.
MEF2D suppresses the transcription driven by AMOTL2 promoter
Based on our results that MEF2D negatively regulates the level of AMOTL2, which is a candidate MEF2D target, we constructed a luciferase reporter driven by an AMOTL2 promoter (pGL3-AMOTL2-pro) and one driven by mutants (pGL3-AMOTL2-mut). Our results showed that MEF2D overexpression could reduce the expression of pGL3-AMOTL2-pro to a more potent extent than pGL3-AMOTL2-mut (Figure 1E). This finding suggested that the existence of MEF2 site (Figure 1F, 1G), a well-documented element responsive to MEF2D and located within AMOTL2 promoter, contributes to the inhibitory effect of MEF2D on AMOTL2 transcription.
MEF2D overexpression facilitates the activation of YAP signaling in HCC cells
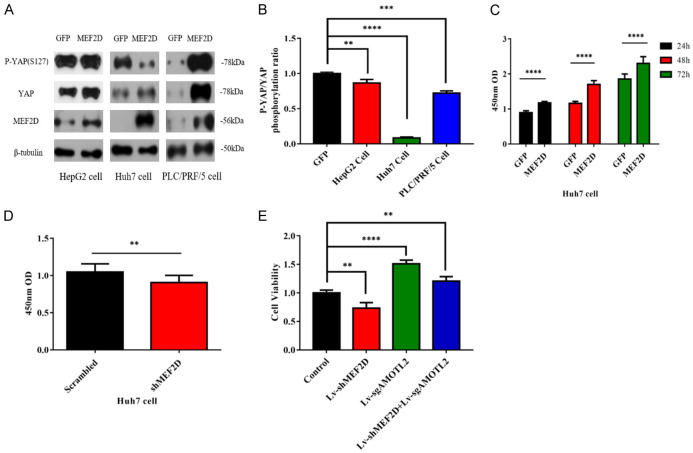
Since AMOTL2 is a negative regulator of YAP signaling, we tested whether MEF2D overexpression affects the activity of this signaling in HCC cells. Western blot data indicated that MEF2D overexpression reduced the ratio between phosphorylated and total YAP (Figure 2A) even though the values varied in the tested three HCC cell lines (Figure 2B). The above data confirmed that MEF2D promotes the activation of YAP signaling in HCC.
Figure 2.
MEF2D promotes YAP activation and the proliferation of HCC cells. A. The expression levels of MEF2D, total YAP and phosphorylated YAP in HepG2, Huh7, and PLC/PRF/5 cells were detected by western blot. B. The relative ratios of phosphorylated and total YAP protein were determined in the above HCC cells. The data represent the means ± SDs of triplicate experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. C. The absorbance of Huh7 cells transfected with MEF2D or GFP vector were determined by CCK8 assay at the indicated time points. D. The absorbance of Huh7 cells transfected with shMEF2D or scrambled vector were also determined by CCK8 assay. E. The cell viability results of the indicated groups at 24 hours were tested by CCK8 experiment in HepG2 cells. The above experimental results were obtained through three independent experiments. The data represent the means ± SDs of triplicate experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
MEF2D facilitates the proliferation of HCC cells
YAP signaling promotes the proliferation of HCC cells [16]. We were interested in the effect of MEF2D overexpression and reduction on the proliferation rate of liver cancer cells based on our finding that MEF2D contributes to YAP signaling activation. A CCK8 assay was conducted to determine the proliferation rate of HCC cells under different conditions. The data indicated that MEF2D overexpression caused HCC cells to proliferate more quickly (Figure 2C). MEF2D silencing resulted in a lower proliferation rate (Figure 2D). In the MEF2D and AMOTL2 double knockout experiments, we found that MEF2D inhibited the expression of AMOTL2 and promoted the proliferation of hepatoma cells (Figure 2E).
Luteolin suppressed the activation of YAP pathway in HCC cells
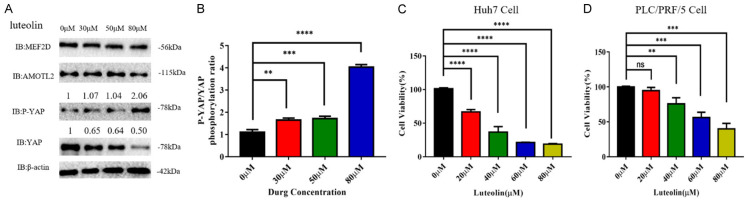
Regarding the activation of YAP signaling in HCC cells, we aimed to study if targeting this pathway exerts some anti-tumor effect. We examined luteolin, a natural flavonoid compound, and its inhibitory effect on YAP signaling. Luteolin was found to suppress the activation of YAP signaling, as evidenced by the elevated relative phosphorylation level of YAP protein (Figure 3A and 3B). These results showed that luteolin could act as a YAP-targeting compound.
Figure 3.
Luteolin inhibited the activation of Hippo/YAP signaling pathway and cell proliferation in HCC cells. A. The protein levels of MEF2D, AMOTL2, phosphorylated, and total YAPs were detected in HCC cells under the exposure with different doses of luteolin. B. The relative ratios of phosphorylated and total YAPs protein were determined in the above HCC cells. The data represent the means ± SDs of triplicate experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. C, D. Huh7 and PLC/PRF/5 cells were treated with luteolin at the specified concentration (0-80 μM) for 24 hours. The cell viability was detected by CCK8 test. This experimental result was obtained through more than three independent experiments. The data represent the means ± SDs of triplicate experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Luteolin inhibited the proliferation and migration of hepatoma cells
We designed experiments to determine if luteolin affected the proliferation and migration of liver cancer cells. After treating HCC cell lines with luteolin at the indicated doses, we found that this natural compound significantly suppresses the proliferation rates of Huh7 and PLC/PRF/5 cell lines (Figure 3C and 3D). Huh7 cells appeared to be more sensitive to luteolin than PLC/PRF/5 cells (Figure 3C and 3D).
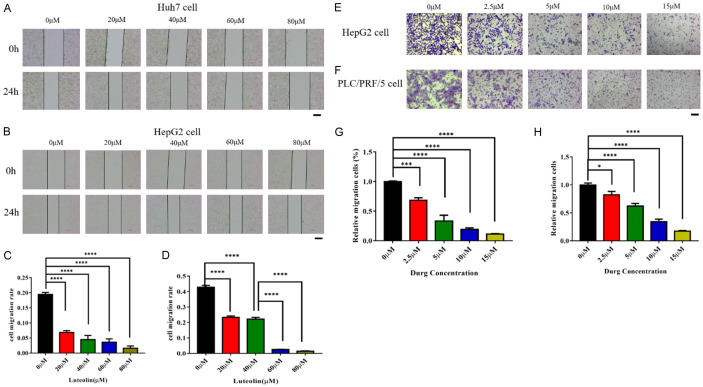
We detected if luteolin influenced the migration ability of liver cancer cells. The wound healing migration assay indicated that the luteolin treatment weakened the migration of Huh7 and HepG2 cell lines (Figure 4A-D). The transwell assay also showed that luteolin inhibited the migration of HepG2 and PLC/PRF/5 cell lines (Figure 4E-H).
Figure 4.
Inhibitory effect of luteolin on migration of hepatoma cell. A, B. The representative images of wound healing test of Huh7 and HepG2 cells at specified luteolin concentration (0-80 μM). C, D. The relative migration distance of cells in three different visual fields. This experimental result was obtained through more than three independent experiments. The data represent the means ± SDs of triplicate experiments. ****, P < 0.0001. E, F. The transwell migration experiments and specific representative images of HepG2 cells and PLC/PRF/5 cell at specific luteolin concentrations. G, H. Quantitative measurement of numbers of migrated cells in three different visual fields are shown for 24 h. This experimental result was obtained through more than three independent experiments. Scale bar = 200 μm. The data represent the means ± SDs of triplicate experiments. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Discussion
The role of MEF2D in various types of cancers, including HCC, is well-documented. There are no publication reports showing that MEF2D is associated with YAP signaling. Our study was the first to focus on the relationship between MEF2D and the YAP pathway. Our evidence showed that MEF2D inhibited the expression of AMOTL2, a well-defined YAP suppressor, by directly binding its promoter region and reducing its transcription activity [17]. It has been proven that USP9X activates the expression of YAP by inhibiting the mono-ubiquitination of AMOTL2. It has also been proven that ubiquitinated AMOTL2 can bind to the UBA domain of LATS kinase, which is necessary for the function of lights. LATS is an effective Yes-related protein inhibitor [18,19]. The activation of the YAP pathway is triggered by MEF2D overexpression in HCC cells. HCC cells gain advantages in the form of proliferation and migration capabilities.
Luteolin plays an anti-tumor and proliferative role in many cancers, such as breast cancer [20], liver cancer [21], and pancreatic cancer [22]. Luteolin has been widely studied in liver cancer. Luteolin induces cell cycle arrest and apoptosis of liver cancer cells through TGF-β1, P53, Fas/Fas ligand, and other signaling pathways [23]. Luteolin can also induce endoplasmic reticulum stress in a p53-independent manner [24]. It has been reported that luteolin enhances JNK-mediated apoptosis in liver cancer cells combined with sorafenib [25]. Although luteolin displayed some anti-tumor activity in liver cancer cells [24,26,27], improving the efficacy of drug delivery is necessary. Mannosylated mixed micelles were shown to deliver anti-tumor drugs as a vehicle in an effective fashion [28]. Our future study will focus on developing an efficient delivery system for HCC treatment.
Based on the molecular mechanism underlying HCC progression described above, we must find compounds that can target and interfere with MEF2D/YAP signaling to achieve effective treatment. In our study, luteolin was found to suppress the YAP pathway activation and the proliferation and migration of HCC cells (Figure 5). These data indicated that luteolin may be a promising YAP-targeting compound for use in HCC treatment.
Figure 5.
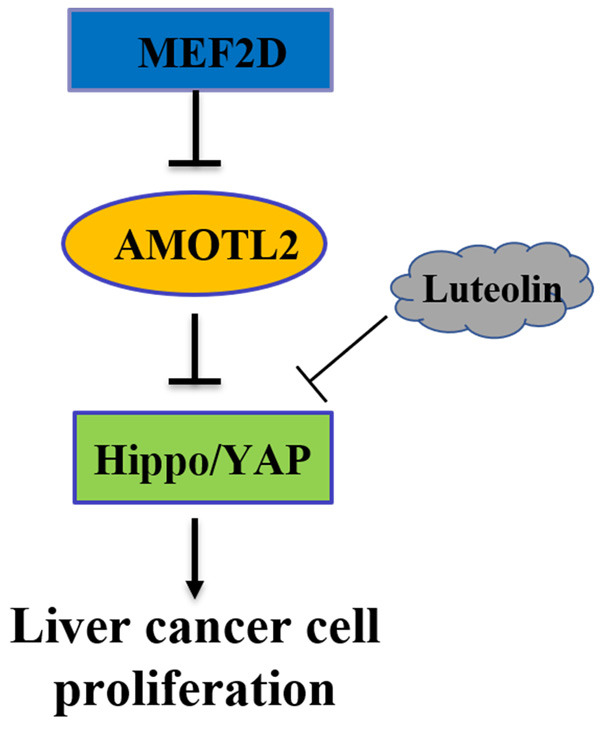
Summary of the mechanisms of MEF2D/YAP signaling.
Disclosure of conflict of interest
None.
References
- 1.Raees A, Kamran M, Ozkan H, Jafri W. Updates on the diagnosis and management of hepatocellular carcinoma. Euroasian J Hepatogastroenterol. 2021;11:32–40. doi: 10.5005/jp-journals-10018-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–159. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Elpek GO. Molecular pathways in viral hepatitis-associated liver carcinogenesis: an update. World J Clin Cases. 2021;9:4890–4917. doi: 10.12998/wjcc.v9.i19.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Gao B, Ponnusamy M, Lin Z, Liu J. MEF2 signaling and human diseases. Oncotarget. 2017;8:112152–112165. doi: 10.18632/oncotarget.22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins M, Potier D, Romanelli L, Jacobs J, Mach J, Hamaratoglu F, Aerts S, Halder G. An ectopic network of transcription factors regulated by Hippo signaling drives growth and invasion of a malignant tumor model. Curr Biol. 2016;26:2101–2113. doi: 10.1016/j.cub.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Liu J, Liu L, Duan G, Wang Q, Xu Y, Xia F, Shan J, Shen J, Yang Z, Bie P, Cui Y, Bian XW, Prieto J, Avila MA, Qian C. Overexpression of the transcription factor MEF2D in hepatocellular carcinoma sustains malignant character by suppressing G2-M transition genes. Cancer Res. 2014;74:1452–1462. doi: 10.1158/0008-5472.CAN-13-2171. [DOI] [PubMed] [Google Scholar]
- 7.Di Giorgio E, Hancock WW, Brancolini C. MEF2 and the tumorigenic process, hic sunt leones. Biochim Biophys Acta Rev Cancer. 2018;1870:261–273. doi: 10.1016/j.bbcan.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Ma S, Meng Z, Chen R, Guan KL. The Hippo pathway: biology and pathophysiology. Annu Rev Biochem. 2019;88:577–604. doi: 10.1146/annurev-biochem-013118-111829. [DOI] [PubMed] [Google Scholar]
- 9.Vanyai HK, Prin F, Guillermin O, Marzook B, Boeing S, Howson A, Saunders RE, Snoeks T, Howell M, Mohun TJ, Thompson B. Control of skeletal morphogenesis by the Hippo-YAP/TAZ pathway. Development. 2020;147:dev187187. doi: 10.1242/dev.187187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Zhou J, Li Q, Xu J, Qi J, Bian H. The inhibition of Hippo/Yap signaling pathway is required for magnesium isoglycyrrhizinate to ameliorate hepatic stellate cell inflammation and activation. Biomed Pharmacother. 2018;106:83–91. doi: 10.1016/j.biopha.2018.06.102. [DOI] [PubMed] [Google Scholar]
- 11.Pan X, Wu B, Fan X, Xu G, Ou C, Chen M. YAP accelerates vascular senescence via blocking autophagic flux and activating mTOR. J Cell Mol Med. 2021;25:170–183. doi: 10.1111/jcmm.15902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M, Yan M, Lv H, Wang B, Lv X, Zhang H, Xiang S, Du J, Liu T, Tian Y, Zhang X, Zhou F, Cheng T, Zhu Y, Jiang H, Cao Y, Ai D. Macrophage K63-linked ubiquitination of YAP promotes its nuclear localization and exacerbates atherosclerosis. Cell Rep. 2020;32:107990. doi: 10.1016/j.celrep.2020.107990. [DOI] [PubMed] [Google Scholar]
- 13.Gokey JJ, Patel SD, Kropski JA. The role of Hippo/YAP signaling in alveolar repair and pulmonary fibrosis. Front Med (Lausanne) 2021;8:752316. doi: 10.3389/fmed.2021.752316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Zhou D. Role of the transcriptional coactivators YAP/TAZ in liver cancer. Curr Opin Cell Biol. 2019;61:64–71. doi: 10.1016/j.ceb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Wang J, Cao Y, Li W, Wang Y, Xu J, Xu G. Molecular evidence for better efficacy of hypocrellin A and oleanolic acid combination in suppression of HCC growth. Eur J Pharmacol. 2019;842:281–290. doi: 10.1016/j.ejphar.2018.10.042. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y, Yang R, Ding J, Zhu F, Zhu C, Xu Q, Cai J. KAT6A is associated with sorafenib resistance and contributes to progression of hepatocellular carcinoma by targeting YAP. Biochem Biophys Res Commun. 2021;585:185–190. doi: 10.1016/j.bbrc.2021.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M, Kim M, Park SJ, Lee C, Lim DS. Role of angiomotin-like 2 mono-ubiquitination on YAP inhibition. EMBO Rep. 2016;17:64–78. doi: 10.15252/embr.201540809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paramasivam M, Sarkeshik A, Yates JR 3rd, Fernandes MJ, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu HT, Lin J, Liu YE, Chen HF, Hsu KW, Lin SH, Peng KY, Lin KJ, Hsieh CC, Chen DR. Luteolin suppresses androgen receptor-positive triple-negative breast cancer cell proliferation and metastasis by epigenetic regulation of MMP9 expression via the AKT/mTOR signaling pathway. Phytomedicine. 2021;81:153437. doi: 10.1016/j.phymed.2020.153437. [DOI] [PubMed] [Google Scholar]
- 21.Sagawa H, Naiki-Ito A, Kato H, Naiki T, Yamashita Y, Suzuki S, Sato S, Shiomi K, Kato A, Kuno T, Matsuo Y, Kimura M, Takeyama H, Takahashi S. Connexin 32 and luteolin play protective roles in non-alcoholic steatohepatitis development and its related hepatocarcinogenesis in rats. Carcinogenesis. 2015;36:1539–1549. doi: 10.1093/carcin/bgv143. [DOI] [PubMed] [Google Scholar]
- 22.Kato H, Naiki-Ito A, Suzuki S, Inaguma S, Komura M, Nakao K, Naiki T, Kachi K, Kato A, Matsuo Y, Takahashi S. DPYD, down-regulated by the potentially chemopreventive agent luteolin, interacts with STAT3 in pancreatic cancer. Carcinogenesis. 2021;42:940–950. doi: 10.1093/carcin/bgab017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yee SB, Choi HJ, Chung SW, Park DH, Sung B, Chung HY, Kim ND. Growth inhibition of luteolin on HepG2 cells is induced via p53 and Fas/Fas-ligand besides the TGF-β pathway. Int J Oncol. 2015;47:747–754. doi: 10.3892/ijo.2015.3053. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y, Kwon YH. Regulation of apoptosis and autophagy by luteolin in human hepatocellular cancer Hep3B cells. Biochem Biophys Res Commun. 2019;517:617–622. doi: 10.1016/j.bbrc.2019.07.073. [DOI] [PubMed] [Google Scholar]
- 25.Feng XQ, Rong LW, Wang RX, Zheng XL, Zhang L, Zhang L, Lin Y, Wang X, Li ZP. Luteolin and sorafenib combination kills human hepatocellular carcinoma cells through apoptosis potentiation and JNK activation. Oncol Lett. 2018;16:648–653. doi: 10.3892/ol.2018.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang PW, Lu ZY, Pan Q, Chen TT, Feng XJ, Wang SM, Pan YC, Zhu MH, Zhang SH. MicroRNA-6809-5p mediates luteolin-induced anticancer effects against hepatoma by targeting flotillin 1. Phytomedicine. 2019;57:18–29. doi: 10.1016/j.phymed.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 27.Cai S, Bai Y, Wang H, Zhao Z, Ding X, Zhang H, Zhang X, Liu Y, Jia Y, Li Y, Chen S, Zhou H, Liu H, Yang C, Sun T. Knockdown of THOC1 reduces the proliferation of hepatocellular carcinoma and increases the sensitivity to cisplatin. J Exp Clin Cancer Res. 2020;39:135. doi: 10.1186/s13046-020-01634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Zang X, Qiao M, Zhao X, Hu H, Chen D. Targeted delivery of dasatinib to deplete tumor-associated macrophages by mannosylated mixed micelles for tumor immunotherapy. ACS Biomater Sci Eng. 2020;6:5675–5684. doi: 10.1021/acsbiomaterials.0c01046. [DOI] [PubMed] [Google Scholar]