Abstract
Benzene, toluene, xylenes, phenol, naphthalene, and biphenyl are among a group of compounds that have at least one reported pathway for biodegradation involving catechol 2,3-dioxygenase enzymes. Thus, detection of the corresponding catechol 2,3-dioxygenase genes can serve as a basis for identifying and quantifying bacteria that have these catabolic abilities. Primers that can successfully amplify a 238-bp catechol 2,3-dioxygenase gene fragment from eight different bacteria are described. The identities of the amplicons were confirmed by hybridization with a 238-bp catechol 2,3-dioxygenase probe. The detection limit was 102 to 103 gene copies, which was lowered to 100 to 101 gene copies by hybridization. Using the dioxygenase-specific primers, an increase in catechol 2,3-dioxygenase genes was detected in petroleum-amended soils. The dioxygenase genes were enumerated by competitive quantitative PCR with a 163-bp competitor that was amplified using the same primers. Target and competitor sequences had identical amplification kinetics. Potential PCR inhibitors that could coextract with DNA, nonamplifying DNA, soil factors (humics), and soil pollutants (toluene) did not impact enumeration. Therefore, this technique can be used to accurately and reproducibly quantify catechol 2,3-dioxygenase genes in complex environments such as petroleum-contaminated soil. Direct, non-cultivation-based molecular techniques for detecting and enumerating microbial pollutant-biodegrading genes in environmental samples are powerful tools for monitoring bioremediation and developing field evidence in support of natural attenuation.
Bioremediation is a low-cost treatment alternative for the cleanup of petroleum-contaminated soils and groundwater. Monitored natural attenuation (MNA) is one form of bioremediation where natural processes are used to treat petroleum contamination. In order to establish whether MNA is feasible, several lines of evidence must be evaluated to demonstrate the types of in situ attenuation mechanisms active onsite (37). Precise and accurate enumeration of aromatic-hydrocarbon-degrading microorganisms would provide such evidence.
Despite the well-known biases of cultivation-based techniques, standard culture methods are used for site evaluation to determine whether indigenous bacteria are capable of degrading the contaminants. Molecular genetic techniques allow researchers to examine microbial communities without cultivation using universal 16S rRNA gene primers (5). PCR has been particularly useful for detecting genes involved in the degradation of xenobiotic compounds (13, 18, 23, 24). There are potential biases associated with molecular techniques (32, 38). However, conditions and experiments can be designed to minimize such biases.
In order to enumerate gene copy number, competitive quantitative PCR techniques have been developed. Competitive quantitative PCR techniques were initially used in medicinal research to measure viral loads in humans (15, 31). More recently these techniques have been used to measure numbers of plant pathogens (20), fungal populations (2), 4-chlorobiphenyl degraders (10), and uncultivated bacterial strains in soils (27).
Competitive quantitative PCR would be a significant improvement over cultivation-based techniques for monitoring bioremediation. Greater catabolic gene copy numbers within a contaminated area (relative to those in uncontaminated soils) could be used as evidence of natural attenuation or of the effectiveness of exogenously supplied growth amendments in engineered bioremediation. Bacteria that aerobically degrade aromatic hydrocarbons use dioxygenase enzymes to activate and cleave the aromatic ring (3, 7); therefore, the corresponding genes are excellent targets on which to base a competitive quantitative PCR assay. Most aerobic aromatic-hydrocarbon biodegradation pathways converge through catechol-like intermediates that are typically cleaved by ortho- or meta-cleavage dioxygenases (7). Meta-cleavage dioxygenases, or catechol 2,3-dioxygenases (C23DO), are believed to be more capable than ortho-cleavage dioxygenases of degrading alkyl-substituted aromatics such as xylenes (9). C23DO genes also have a well-characterized phylogeny (12) that allows for the systematic design of dioxygenase-specific primers. The I.2.A dioxygenase subfamily is involved in the degradation of benzene, toluene, xylenes (BTX), and naphthalene. Due to their toxicity and mobility in the environment, BTX cleanup endpoints are more stringent than those of other petroleum constituents. The ability to specifically and accurately detect BTX-biodegrading bacteria in the environment is desirable to establish the feasibility of MNA.
In this study we describe a single set of C23DO-specific primers and use them to detect and enumerate the genes that make up the I.2.A subfamily of C23DO genes (12). This subfamily contains a broad spectrum of C23DO genes that are involved in the biodegradation of aromatic compounds of environmental concern. Nonamplifying DNA, toluene, and soil factors such as humics, which are potential inhibitors of PCR, are shown to have no effect on dioxygenase gene enumeration.
(A portion of this work was presented at the 99th General Meeting of the American Society for Microbiology [M. Mesarch, C. Nakatsu, and L. Nies, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. Q-429, p. 615, 1999].)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The organisms and their carbon sources used in this study are listed in Table 1. Cells were grown overnight at room temperature in sterile defined minimal medium (29). Biphenyl or naphthalene was added as solid crystals to the liquid medium or on the inverted lids of petri plates. Phenol or meta-toluic acid (for toluene-degraders) was added to a final concentration of 0.250 g liter−1. The medium was solidified with 15 g of agar liter−1.
TABLE 1.
Organisms used to design C23DO-specific primers and their carbon sources
| Organism | Growth substrate | % Identity to HS1 probe | Reference |
|---|---|---|---|
| Pseudomonas putida HS1 | Toluene | 100 | 26 |
| Pseudomonas putida mt-2 | Toluene | 95 | 42 |
| Pseudomonas putida H | Phenol | 88 | 21 |
| Pseudomonas putida P35X | Phenol | 87 | 19 |
| Pseudomonas sp. strain CF600 | Phenol | 86 | 14 |
| Pseudomonas sp. strain IC | Biphenyl | 89 | 6 |
| Pseudomonas aeruginosa JI104 | Biphenyl or benzene | 89 | 44 |
| Pseudomonas sp. strain PpG7 | Naphthalene | 84 | 11 |
DNA extraction.
Genomic DNAs were extracted from pure cultures of all eight isolates using a total genomic DNA isolation method (28) or a FastDNA Kit (Bio 101). Soil DNA extractions were performed using the FastPrep System and the FastDNA Spin Kit for Soil (Bio 101). DNA was quantified by fluorometry using a model TKO100 DNA fluorometer (Hoefer Scientific Instruments, San Francisco, Calif.) calibrated with calf thymus DNA.
PCR primer design.
PCR primers (Table 2) were constructed based upon conserved amino acid sequences (12) and nucleic acid alignments of these regions (DNAman version 2.7). The primers 23CAT-F and 23CAT-R were selected based upon conserved nucleotide regions for six of the eight isolates listed in Table 1. Pseudomonas sp. strain CF600 had one mismatch with 23CAT-R, and Pseudomonas sp. strain PpG7 had two mismatches with 23CAT-F and three with 23CAT-R. Primers DEG-F and DEG-R are identical to 23CAT-F and 23CAT-R except for five positions where degenerate bases were used to account for primer-target mismatches with Pseudomonas sp. strain PpG7 (Table 2). We searched GenBank and found that the primer sequences matched only other C23DO sequences, from Pseudomonas stutzeri AN10 (AF039534) and OM1 (AB001722), which fit into the I.2.A subfamily of dioxygenase genes. Primer QUANT-F was designed for use as the competitor to amplify a 163-bp sequence from Pseudomonas putida HS1 or mt-2 when it was used with primer 23CAT-R or DEG-R. Primers were synthesized at the Laboratory for Macromolecular Structure, Purdue University (23CAT-F and 23CAT-R), and Integrated DNA Technologies, Inc., Coralville, Iowa (DEG-F, DEG-R, and QUANT-F).
TABLE 2.
Primers developed to enumerate dioxygenase gene copy numbera
| Primer | Sequence |
|---|---|
| 23CAT-F | 5′-CGACCTGATCTCCATGACCGA-3′ |
| 23CAT-R | 5′-TCAGGTCAGCACGGTCA-3′ |
| DEG-F | 5′-CGACCTGATC(AT)(CG)CATGACCGA-3′ |
| DEG-R | 5′-T(CT)AGGTCA(GT)(AC)ACGGTCA-3′ |
| QUANT-F | 5′-CGACCTGATCTCCATGACCGATAACCGCAACGAAGTGTTCTG-3′ |
Letters in parentheses indicate positions of degenerate bases.
PCR conditions.
Optimization of PCR conditions using primers DEG-F and DEG-R were tested for P. putida HS1 and Pseudomonas sp. strain PpG7 because they were either identical to the nondegenerate primers (HS1) or had the most mismatches (PpG7). Annealing temperatures of 52 to 63°C were tested using a Robocycler Gradient 96 thermal cycler (Stratagene, La Jolla, Calif.). The PCR temperature program began with an initial 5-min denaturation step at 95°C; 30 cycles of 94°C for 1 min, 52 to 63°C for 1 min, and 72°C for 2 min; and a final 10-min extension step at 72°C. All reaction mixtures were held at 4°C until analyzed. Magnesium chloride (Promega, Madison, Wis.) concentrations (1.5, 3.0, 4.0, and 5.0 mM), primer concentrations (0.125, 0.250, and 0.375 μM), and template DNA concentrations (10, 1, and 0.1 ng) were tested individually. All reaction mixtures also included 1× PCR buffer (Promega), 0.2 mM each deoxynucleoside triphosphate (Amersham Pharmacia, Piscataway, N.J.), and 1 U of Taq DNA polymerase. Following optimization, all reactions were performed in a PTC-100 programmable thermal cycler (MJ Research, Inc.). All experiments included controls without any added DNA. Ten microliters of each PCR mixture was run on a 1.5% agarose gel (Bio-Rad, Richmond, Calif.) in 1× Tris-acetate-EDTA (TAE) buffer stained with ethidium bromide (0.0001%) and visualized under UV light. PCR was performed on DNAs extracted from all eight isolates using both primer sets: DEG-F and DEG-R and CAT-F and CAT-R (Table 1). To determine the detection limit of this protocol, a 10-fold dilution series of P. putida HS1 and Pseudomonas sp. strain PpG7 (from 1 ng to 1 fg of DNA per reaction) was created and amplified using DEG-F and DEG-R. For competitive quantitative PCR experiments both target and competitor DNAs were added to reaction tubes.
Hybridization experiments.
Confirmation of the eight amplicons as dioxygenase fragments was made through hybridization with a digoxigenin-labeled probe. The probe was a 238-bp P. putida HS1 dioxygenase gene fragment (hereafter described as an HS1 probe) labeled by PCR incorporation of digoxigenin-labeled dUTP (Roche Molecular Biochemicals, Indianapolis, Ind.). The probe was also tested on a dilution series of HS1 or PpG7 DNA amplified with the dioxygenase-specific primers to determine if further amplification occurred that was not visible on an agarose gel.
Southern transfer, hybridization, and detection were carried out according to the instructions of the manufacturer (Roche Molecular Biochemicals). Hybridization was performed under low-, medium-, and high-stringency conditions by varying the temperatures of posthybridization washes (65, 80, and 85°C, respectively). Wash solutions contained 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (34).
Detection of C23DO genes in petroleum-amended soils.
To determine whether C23DO genes can be detected in environmental samples, DNAs were extracted from unamended and petroleum-amended soils (<2% organic carbon). Uncontaminated soil was amended with 30,000 mg of diesel fuel kg−1, 20,000 mg of motor oil kg−1, and 200 mg (each) of naphthalene, phenanthrene, anthracene, pyrene, and chrysene kg−1 and aged for 12 months. PCR was performed on the soil DNA extracts (500 mg of soil extracted, 10 ng of DNA used per reaction) using primers DEG-F and DEG-R.
Construction of competitor.
The competitor was constructed based upon the approach of Jin et al. (22). A 163-bp sequence was amplified from P. putida HS1 or mt-2 using QUANT-F and DEG-R. The 163-bp amplicon was purified using a MERmaid prep kit (Bio 101). The purified amplicon was ligated with pGEM-T (Promega) and transformed into competent Escherichia coli DH5α (34). The presence of the correctly sized insert was confirmed by PCR (using DEG-F and DEG-R), plasmid isolation, and restriction enzyme (SphI and NdeI) digestion (34). By multiplying the competitor plasmid concentration (in nanograms per microliter) by the fraction of plasmid constituted by the competitor fragment (163 bp of a total of 3,166 bp) and performing the appropriate unit conversions, we were able to calculate the competitor gene copy number used in each competitive quantitative PCR experiment. All quantitative competitive PCR results were subsequently expressed in terms of C23DO gene copy number.
Evaluation of competitive quantitative PCR protocol.
Competitive PCR was first performed with pure cultures using a 10-fold dilution series of competitor plasmid ranging from 1 ng to 1 fg of DNA per reaction as standards. PCR products were separated on 1.5% agarose gels. Target and competitor band intensities were analyzed and compared using Scion Image software (Scion Corp., Frederick, Md.). To kinetically validate the quantitative PCR protocol, equal amounts of target and competitor were amplified in the same reaction mixture in triplicate for 20, 25, 30, and 35 cycles. Log ratios of target to competitor band intensities were plotted as a function of cycle number, and the data were analyzed using linear regression.
To evaluate how well this protocol might work on field samples, the effects of three potential PCR inhibitors were examined. In order to simulate the effects of nonamplifying DNA (such as DNA extracted from a microbial community) on competitive PCR, E. coli DH5α DNA was spiked into each reaction tube and PCR was performed. E. coli DNA/HS1 DNA ratios of 1:0, 1:1, 10:1, 100:1, 1000:1, and 10,000:1 were tested.
To determine the effects of soil factors on competitive quantitative PCR, soil (Chalmers silty loam, 4% organic carbon, sieved with a 2-mm screen) was sterilized by autoclaving. PpG7 grown in liquid culture was concentrated by centrifugation for 2 min at 14,000 × g, resuspended in 100 μl of sterile Tris-EDTA (to minimize changes to soil moisture content), added to 500 mg of sterile soil, and allowed to sit for 30 min. Uninoculated sterile soil was used as a control. Following incubation, total DNA was extracted and DNA extraction efficiency was determined by comparing concentrations of soil DNA extracts to the concentration of DNA extracted from an equivalent amount of cells not inoculated in soil. Quantitative PCR was then performed on soil DNA extracts and pure-culture DNA extracts. All extractions were performed in triplicate. A serial dilution of cells was also plated onto minimal medium plates, with naphthalene crystals added as a sole source of carbon. Plate counts were also performed on soils that had already been extracted to determine the lysis efficiency of the DNA extraction protocol. Cell lysis efficiency was determined by comparing plate counts of uninoculated pure cultures to plate counts of the extracted soil.
Environmental contaminants are also potential PCR inhibitors. Therefore, the effects of high levels of one potential inhibitor, toluene, in soil were examined. Toluene was spiked into sterile soils to a concentration of 10,000 mg kg−1. These soils were spiked with PpG7 cells as described above, allowed to incubate 30 min, and processed identically to the unpolluted soil samples.
RESULTS AND DISCUSSION
PCR conditions.
Primers 23CAT-F and 23CAT-R were capable of amplifying the expected 238-bp product from seven of the eight isolates, including Pseudomonas sp. strain CF600, which had a one base pair mismatch with 23CAT-R. The only isolate not detected was Pseudomonas sp. strain PpG7, which contained the most mismatches with 23CAT-F and 23CAT-R. DEG-F and DEG-R were able to amplify the expected 238-bp fragment from pure cultures of all eight isolates, including PpG7 (Fig. 1A). Amplification using a gradient of annealing temperatures indicated optimal annealing temperatures of 55 and 57°C for PpG7 and HS1, respectively. An annealing temperature of 56°C was used in all subsequent experiments. Based on PCR product intensity, optimal amplification was achieved using concentrations of 1 ng of DNA, 3.0 mM MgCl2, and 0.250 μM each primer using either HS1 DNA or PpG7 DNA. Until now, PCR conditions have not been optimized for an entire subfamily of C23DO genes. Optimization using C23DO genes from HS1 and PpG7, which have the greatest sequence difference from one another in their respective primer sequences, indicated that identical PCR conditions can be used to monitor dioxygenase gene copy number in all I.2.A C23DO subfamily members without sacrificing accuracy.
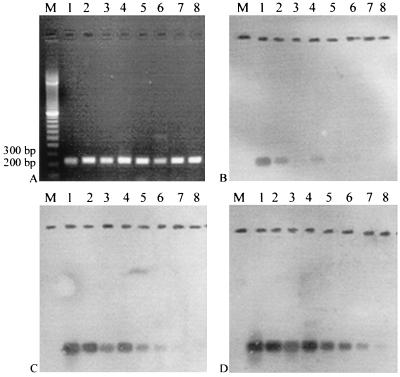
FIG. 1.
Detection of 238-bp dioxygenase amplicons by PCR and hybridization using primers DEG-F and DEG-R. Lane M, 100- to 3,000-bp marker (Bio-Rad); lanes 1 to 8, P. putida HS1, P. putida mt-2, P. aeruginosa JI104, Pseudomonas sp. strain IC, P. putida P35X, Pseudomonas sp. strain CF600, P. putida H, and Pseudomonas sp. strain PpG7, respectively. Agarose gel (1.5% agarose) of the amplicons with approximately equal amounts of DNA in each lane; (B, C, and D) membranes hybridized under high (B)-, medium (C)-, and low (D)-stringency conditions using an exposure time of 2 to 3 h.
The broad target spectrum of our primers allows identification and enumeration of dioxygenase genes involved in the degradation of a variety of aromatic hydrocarbons, which allows site investigators to detect catabolic genes from organisms with both single-ring (HS1, mt-2, H, P35X, and CF600) and double-ring (IC, JI104, and PpG7) substrate specificities. Often primers are designed for site-specific needs such as identification of genes involved in the degradation of polychlorinated biphenyls (13), 2,4-dichlorophenoxyacetic acid (4), or naphthalene (17). While such primers proved useful in investigating the distribution of specific catabolic genes, their exclusive nature limits their usefulness in characterizing the bacteria in petroleum-contaminated environments that contain several different environmentally regulated aromatic compounds.
Other dioxygenase-specific primers have previously been described for C23DO genes. However, none of the previously described C23DO primers were developed to detect members of the entire I.2.A C23DO subfamily. A set of C23DO primers has been described for quantifying organisms harboring this gene in toluene-contaminated soils, but they can detect only isolates mt-2, JI104, and IC (16). A different set of primers used to monitor phenol-degrading P. putida strains in an industrial wastewater process would be able to detect only isolates mt-2 and JI104 (39). C23DO primers identical to mt-2 genes alone have been used as a molecular marker for the identification of a fish pathogen in lakewater using PCR (30). Primers based upon an alignment of six I.2.A C23DO genes were successful in detecting the three organisms in the I.2.A subfamily on which they were tested (41). However, these primers required more degenerate positions than primers DEG-F and DEG-R yet still contained a mismatch with two of the I.2.A subfamily members.
Hybridization experiments.
Hybridization of the digoxigenin-labeled probe to 238-bp amplicons from all eight isolates under high-stringency conditions detected P. putida HS1 (100% identical to probe), P. putida mt-2 (95% identical), P. aeruginosa JI104 (89% identical), and Pseudomonas sp. strain IC (89% identical) (Fig. 1B, lanes 1 to 4), although the signals were very weak from JI104 and IC. These amplicons were more easily seen under medium-stringency conditions (80°C). In addition to JI104 and IC, P. putida P35X (87% identical), Pseudomonas sp. strain CF600 (86% identical), and P. putida H (90% identical) were detected (Fig. 1C, lanes 5 to 7) under medium-stringency conditions. Amplicons from all eight isolates were detected under low-stringency conditions (65°C) (Fig. 1D, lanes 1 to 8), confirming the identities of all amplicons as C23DO fragments.
Detection limit.
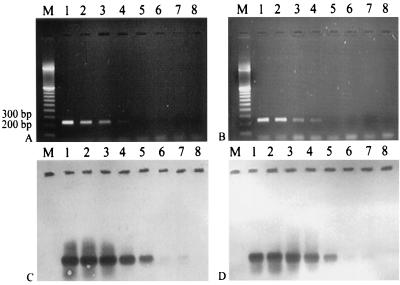
We were capable of detecting from 102 to 103 dioxygenase gene copies from either HS1 or PpG7 DNA (Fig. 2A and B). Hybridization with a C23DO probe allowed us to detect PCR products resulting from the use of as few as 100 to 101 dioxygenase gene copies per reaction (Fig. 2C and D), which compares well with literature values for similar systems (1, 16). A single detection limit for multiple organisms ensures that PCR results will be accurate regardless of the target sequence amplified. This is not true for all primer sets; primers for the gyrB gene had different detection limits for two different phenol-degrading organisms (40). The low detection limit of our primers will also allow for more sensitive detection of C23DO-harboring organisms in uncontaminated or extreme environments, where their numbers are apt to be low, and can provide ecologists with information regarding their distribution in these systems.
FIG. 2.
PCR detection limit. Lane M, 100- to 3,000-bp marker; lanes 1 to 8, amplification products resulting from the use of 1 ng (containing 106 copies of C23DO), 0.1 ng (105 copies), 0.01 ng (104 copies), 0.001 ng (103 copies), 0.0001 ng (102 copies), 0.00001 ng (101 copies), 0.000001 ng (100 copies), and no DNA, respectively. (A and B) Agarose gels (1.5% agarose) of PCR products of P. putida HS1 and Pseudomonas sp. strain PpG7, respectively, amplified with primers DEG-F and DEG-R; (C and D) membranes corresponding to the gels in panels A and B, respectively, hybridized with a C23DO probe under low-stringency conditions using an exposure time of 18 to 20 h.
Detection of C23DO genes in petroleum-amended soils.
Using the DEG-F and DEG-R primers, amplification of DNA extracted from the petroleum-amended soils resulted in the anticipated 238-bp product (data not shown). No PCR products were seen from DNA samples extracted from the unamended soils as determined by visualization on agarose gels (detection limit of 102 to 103 copies). This demonstrates that our C23DO primers will detect the expected enrichment of aromatic-hydrocarbon degraders upon exposure to petroleum. This detection is direct evidence that C23DO genes can be amplified from native soil organisms and that they represent good marker genes for monitoring bioremediation in soils.
Competitive quantitative PCR.
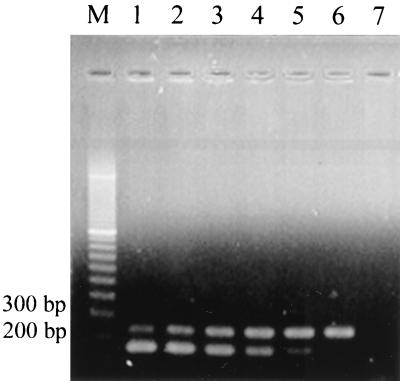
Construction of competitor DNA was confirmed by restriction enzyme digestion of the recombinant plasmid and PCR amplification of the expected 163-bp competitor fragment. Quantitative PCR was first run with equal concentrations of competitor and target DNA from HS1 or PpG7. The logarithms of the ratios of target to competitor band intensities were approximately zero and did not vary with cycle number (n = 12 samples, r2 = 0.01), indicating that target and competitor sequences amplified equally regardless of cycle number or DNA source used (PpG7 or HS1). The identical amplification kinetics exhibited by the target and competitor sequences allows quantification of dioxygenase gene copy number to be based solely upon the ratio of target and competitor sequences (27).
A number of factors can render results of competitive quantitative PCR inaccurate. These factors can originate from the sensitivity of the PCR protocol itself, where inhibition due to high concentrations of nonamplifying DNA relative to target DNA can occur (1). E. coli DNA was used to dilute HS1 DNA in order to simulate background DNA that would be coextracted from environmental samples. No PCR products resulted from E. coli DNA amplified alone using primers DEG-F and DEG-R. Enumeration of dioxygenase copy number of HS1 DNA was not affected by dilution with E. coli DNA using E. coli DNA/HS1 DNA ratios as high as 10,000:1 (approximately 10 ng of total DNA per reaction mixture).
Other potential sources of PCR inhibition include impurities such as humic acids that can coextract with DNA (36) and interfere with reaction components (43) or bind DNA (8) and organic chemicals such as toluene, although there has been no examination as to their potential for inhibition. PpG7 cells spiked into sterile soils were accurately enumerated using the quantitative PCR protocol (Fig. 3). Soil DNA extracts were clear and appeared free of humic material. No DNA was measured in extracts from autoclaved soils by fluorometry, and amplification was not evident using primers DEG-F and DEG-R. In seeded soils the DNA extraction procedure recovered 99% of the DNA and was successful in lysing 94% of seeded cells in clean soils and 96% of seeded cells in toluene-amended soils. Using seeded soils, high extraction efficiencies such as ours obtained by the same DNA extraction procedure have been previously reported (5). There was no significant difference between the lysis efficiencies of toluene-amended and unamended soils or between the DNA extraction efficiencies (P = 0.05). The dioxygenase gene copy number in seeded soils did not differ significantly (P = 0.05) from the dioxygenase gene copy number in toluene-spiked, seeded soils and pure-culture extracts (Table 3). This result indicates that the DNA extraction technique prevented potentially inhibitory compounds such as toluene and soil factors such as humic acids from coextracting with DNA. Additional structurally similar compounds should behave in the same manner as toluene. The number of cells as determined by competitive quantitative PCR were greater by a factor of 10 than plate counts of an equivalent preparation of cells (Table 3).
FIG. 3.
Competitive quantitative PCR performed on Pseudomonas sp. strain PpG7 cells using primers DEG-F and DEG-R. Lane M, 100- to 3,000-bp marker; lanes 1 to 6, 1 ng of target DNA (each lane) plus 1.1 × 108, 1.1 × 107, 1.1 × 106, 1.1 × 105, 1.1 × 104, and 1.1 × 103 copies of competitor per reaction mixture, respectively. Lane 7 is a control (no DNA) lane.
TABLE 3.
Comparison of C23DO gene copy numbers and plate counts for Pseudomonas sp. strain PpG7
| Parameter | Avg | SD |
|---|---|---|
| Clean soil extracta | 9.56 × 107 | 7.08 × 107 |
| Toluene-spiked soil extracta | 3.54 × 108 | 1.97 × 108 |
| Pure-culture extracta | 3.26 × 108 | 2.26 × 108 |
| Pure-culture plate countb | 1.41 × 107 | 1.62 × 106 |
Competitive quantitative PCR results are expressed as C23DO gene copy number g of soil−1. Approximately 1.4 × 107 Pseudomonas sp. strain PpG7 cells were seeded per g of soil.
Plate counts are expressed as CFU g of soil−1.
The C23DO-specific primers we have described are capable of detecting bacteria that can degrade both one- and two-ringed aromatic hydrocarbons, including BTX. Thus, they can detect a broader subset of environmentally important aromatic-hydrocarbon-degrading bacteria than previously described primers while retaining their specificity for dioxygenase genes. When coupled with a quantitative competitive PCR approach these primers can be used to accurately quantify aromatic-hydrocarbon-degrading bacteria in soils, which makes it an ideal method for determining the feasibility of MNA. Due to the current available phylogenetic information about C23DO, all of the organisms tested are of the Pseudomonas genera. Some research has suggested that the importance of Pseudomonas in soil is overestimated (5). However, evidence using culture-based methods (25, 33) and culture-independent methods (35) indicates that Pseudomonas organisms are present in contaminated environments and are important in the in situ biodegradation of environmental contaminants. It is true that the presence of a single gene does not ensure that the entire catabolic pathway will be present or that these genes will be expressed. However, primers for other genes in these catabolic pathways are being developed in our laboratory and can be used to determine whether multiple pathway genes are present. We are also experimenting with mRNA extraction techniques to try to detect gene expression in environmental samples. Given the known nucleotide sequence diversity of meta-cleavage dioxygenase genes (12), it is also possible that uncultured organisms other than Pseudomonas contain dioxygenases with identical or sufficiently similar sequences that could be amplified with DEG-F and DEG-R. Future research will allow development of primers outside of the I.2.A subfamily of C23DO genes.
ACKNOWLEDGMENTS
We thank Daniel Kunz, Michael Roberts, Heidrun Hermann, Victoria Shingler, and Atsushi Kitayama for generously providing the organisms used in this research and Bob Kim for the amended and unamended soils.
This work was funded by the Joint Transportation Research Program of the School of Civil Engineering, Purdue University, and the Indiana Department of Transportation, and the Showalter Trust.
REFERENCES
- 1.Alvarez A J, Buttner M P, Stetzenbach L D. PCR for bioaerosol monitoring: sensitivity and environmental interference. Appl Environ Microbiol. 1995;61:3639–3644. doi: 10.1128/aem.61.10.3639-3644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek J, Kenerley C M. Detection and enumeration of a genetically modified fungus in soil environments by quantitative competitive polymerase chain reaction. FEMS Microbiol Ecol. 1998;25:419–428. [Google Scholar]
- 3.Bayly R C, Barbour M G. The degradation of aromatic compounds by the meta and gentisate pathways. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 253–294. [Google Scholar]
- 4.Berthelet M, Greer C. Detection of catabolic genes in the soil using the polymerase chain reaction. In: Moo-Young M, editor. Environmental biotechnology: principles and applications. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 635–644. [Google Scholar]
- 5.Borneman J, Skroch P W, O'Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrington B, Lowe A, Shaw L E, Williams P A. The lower pathway operon for benzoate catabolism in biphenyl-utilizing Pseudomonas sp. strain IC and the nucleotide sequence of the bphE gene for catechol 2,3-dioxygenase. Microbiology. 1994;140:499–508. doi: 10.1099/00221287-140-3-499. [DOI] [PubMed] [Google Scholar]
- 7.Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons. Adv Appl Microbiol. 1984;30:31–71. doi: 10.1016/s0065-2164(08)70052-2. [DOI] [PubMed] [Google Scholar]
- 8.Crecchio C, Stotzky G. Binding of DNA on humic acids: effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol Biochem. 1998;30:1061–1067. [Google Scholar]
- 9.Dagley S. Biochemistry of aromatic hydrocarbon degradation in pseudomonads. In: Sokatch J R, Ornston L N, editors. The bacteria. New York, N.Y: Academic Press, Inc.; 1986. pp. 527–555. [Google Scholar]
- 10.Ducrocq V, Pandard P, Hallier-Soulier S, Thybaud E, Truffaut N. The use of quantitative PCR, plant and earthworm bioassays, plating and chemical analysis to monitor 4-chlorobiphenyl biodegradation in soil microcosms. Appl Soil Ecol. 1999;12:15–27. [Google Scholar]
- 11.Dunn N W, Gunsalus I C. Transmissable plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973;114:974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eltis L D, Bolin J T. Evolutionary relationships among extradiol dioxygenases. J Bacteriol. 1996;178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erb R W, Wagner-Döbler I. Detection of polychlorinated biphenyl degradation genes in polluted sediments by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1993;59:4065–4073. doi: 10.1128/aem.59.12.4065-4073.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey J, Bagdasarian M, Feiss D, Franklin F C, Deshusses J. Stable cosmid vectors that enable the introduction of cloned fragments into a wide range of Gram-negative bacteria. Gene. 1983;24:299–308. doi: 10.1016/0378-1119(83)90090-2. [DOI] [PubMed] [Google Scholar]
- 15.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallier-Soulier S, Ducrocq V, Mazure N, Truffaut N. Detection and quantification of degradative genes in soils contaminated by toluene. FEMS Microbiol Ecol. 1996;20:121–133. [Google Scholar]
- 17.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrick J B, Madsen E L, Batt C A, Ghiorse W C. Polymerase chain reaction amplification of naphthalene-catabolic and 16S rRNA gene sequences from indigenous sediment bacteria. Appl Environ Microbiol. 1993;59:687–694. doi: 10.1128/aem.59.3.687-694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopper D J, Chapman P J, Dagley S. The enzymatic degradation of alkyl-substituted gentisates, maleates, and malates. Biochem J. 1971;122:29–40. doi: 10.1042/bj1220029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Lai F-M, Reddy S N, Ishimaru C A. Quantitative detection of Clavibacter michiganensis subsp. sepedonicus by competitive polymerase chain reaction. Phytopathology. 1995;85:1468–1473. [Google Scholar]
- 21.Janke D, Pohl R, Fritsche W. Regulation of phenol degradation in Pseudomonas putida. Z Allg Mikrobiol. 1981;21:295–303. doi: 10.1002/jobm.3630210405. [DOI] [PubMed] [Google Scholar]
- 22.Jin C, Mata M, Fink D J. Rapid construction of deleted DNA fragments for use as internal standards in competitive PCR. PCR Methods Appl. 1994;3:252–255. doi: 10.1101/gr.3.4.252. [DOI] [PubMed] [Google Scholar]
- 23.Joshi B, Walia S. PCR amplification of catechol 2,3-dioxygenase gene sequences from naturally occurring hydrocarbon degrading bacteria isolated from petroleum hydrocarbon contaminated groundwater. FEMS Microbiol Ecol. 1996;19:5–15. [Google Scholar]
- 24.Joshi B, Walia S K. Detection of metapyrocatechase homologous genes in petroleum hydrocarbon contaminated groundwater by polymerase chain reaction. J Microbiol Methods. 1996;27:121–128. [Google Scholar]
- 25.Kämpfer P, Steiof M, Dott M. Microbiological characterization of a fuel-oil contaminated site including numerical identification of heterotrophic water and soil bacteria. Microb Ecol. 1991;21:227–251. doi: 10.1007/BF02539156. [DOI] [PubMed] [Google Scholar]
- 26.Kunz D A, Chapman P J. Isolation and characterization of spontaneously occurring TOL plasmid mutants of Pseudomonas putida HS1. J Bacteriol. 1981;146:952–964. doi: 10.1128/jb.146.3.952-964.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:209–218. [Google Scholar]
- 29.Mesarch M B, Nies L. Modification of heterotrophic plate counts for assessing the bioremediation potential of petroleum-contaminated soils. Environ Technol. 1997;18:639–646. [Google Scholar]
- 30.Morgan J A W, Rhodes G, Pickup R W. Survival of nonculturable Aeromonas salmonicida in lake water. Appl Environ Microbiol. 1993;59:874–880. doi: 10.1128/aem.59.3.874-880.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piatak M, Jr, Luk K, Williams B, Lifson J D. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques. 1993;14:70–80. [PubMed] [Google Scholar]
- 32.Power M, van der Meer J R, Tchelet R, Egli T, Eggen R. Molecular-based methods can contribute to assessments of toxicological risks and bioremediation strategies. J Microbiol Methods. 1998;32:107–119. [Google Scholar]
- 33.Ridgway H F, Safarik J, Phipps D, Carl P, Clark D. Identification and catabolic activity of well-derived gasoline-degrading bacteria from a contaminated aquifer. Appl Environ Microbiol. 1990;56:3565–3575. doi: 10.1128/aem.56.11.3565-3575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Shen Y, Stehmeier L G, Voordouw G. Identification of hydrocarbon-degrading bacteria in soil by reverse sample genome probing. Appl Environ Microbiol. 1998;64:637–645. doi: 10.1128/aem.64.2.637-645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai Y, Olson B H. Rapid method for separation of bacterial DNA from humic substances for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.U.S. Environmental Protection Agency. Use of monitored natural attenuation at Superfund, RCRA corrective action, and underground storage tank sites. Directive 9200.4-17P. U.S. Washington, D.C.: Environmental Protection Agency; 1999. [Google Scholar]
- 38.van Elsas J D, Duarte G F, Rosado A S, Smalla K. Microbiological and molecular biological methods for monitoring microbial inoculants and their effects in the soil environment. J Microbiol Methods. 1998;32:133–154. [Google Scholar]
- 39.Wand H, Laht T, Peters M, Becker P M, Stottmeister U, Heinaru A. Monitoring of biodegradative Pseudomonas putida strains in aquatic environments using molecular techniques. Microb Ecol. 1997;33:124–133. doi: 10.1007/s002489900014. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe K, Yamamoto S, Hino S, Harayama S. Population dynamics of phenol-degrading bacteria in activated sludge determined by gyrB-targeted quantitative PCR. Appl Environ Microbiol. 1998;64:1203–1209. doi: 10.1128/aem.64.4.1203-1209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wikström P, Wiklund A, Andersson A, Forsman M. DNA recovery and PCR quantitation of catechol 2,3-dioxygenase genes from different soil types. J Biotechnol. 1996;52:107–120. doi: 10.1016/s0168-1656(96)01635-5. [DOI] [PubMed] [Google Scholar]
- 42.Williams P A, Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (Arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974;120:416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yarmoff J, Kawakami Y, Yago T, Maruo H, Nishimura H. cis-benzeneglycol production using a mutant Pseudomonas strain. J Ferment Bioeng. 1988;66:305–312. [Google Scholar]