Abstract
Background
More and more evidence indicates sodium-glucose co-transporter 2 inhibitors (SGLT2is) may display clinical benefits for heart failure with preserved ejection fraction (HFpEF). However, the mechanisms of the action remain unclear.
Methods
A systematic pharmacology-based strategy was applied for predicting the potential molecular mechanisms of SGLT2is in HFpEF. The potential targets of SGLT2is and HFpEF were contained from diverse databases. After networks were constructed, Metascape was applied to functional enrichment. Moreover, the key findings were validated through molecular docking.
Results
We obtained 487 SGLT2is related targets and 1505 HFpEF related targets. The networks showed the complex relationship of HFpEF-target-HFpEF. The results of functional enrichment analysis suggested that several biological processes, including muscle system process, inflammatory response, vasculature development, heart development, regulation of MAPK cascade, positive regulation of ion transport, negative regulation of cell population proliferation, cellular response to nitrogen compound, apoptotic signaling pathway, multicellular organismal homeostasis, response to oxidative stress, regulation of cell adhesion, positive regulation of cell death, response to growth factor, and cellular response to lipid, and signaling pathways, such as cardiomyopathy, cAMP signaling pathway, cytokine-cytokine receptor interaction, apoptosis, MAPK signaling pathway, HIF-1 signaling pathway, calcium signaling pathway, and NF-kappa B signaling pathway. Finally, we validated the interactions and combinations of SGLT2is and core targets.
Conclusion
SGLT2is play the potential role of anti-HFpEF through the direct or indirect synergy of multiple targets and pathways. Our study promotes the explanation of the molecular mechanisms of SGLT2is in HFpEF.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12872-022-02693-8.
Keywords: Sodium-glucose co-transporter 2 inhibitors, Heart failure with preserved ejection fraction, Network pharmacology, Virtual screening, Molecular docking
Introduction
Heart failure (HF) is a complex clinical syndrome that results from any structural or functional impairment of ventricular filling or ejection of blood [1]. Ejection fraction (EF) is considered important in the classification of patients with HF [2] because of differing patient demographics, comorbid conditions, prognosis, and response to therapies [3] and because most clinical trials selected patients based on EF [4]. In the latest American and European guidelines, HF with preserved EF (HFpEF, EF ≥ 50%) was proposed [5, 6]. In patients with clinical HF, studies estimate that the prevalence of HFpEF is approximately 50% (range from 40 to 71%) [7]. Despite aggressive treatment, the residual risk of HFpEF remains high [8]. We have reason to believe that treatment strategies need to be continuously optimized and improved [9].
In recent years, more and more evidence shows that hypoglycemic drugs also have cardiovascular benefits [10, 11]. Sodium-glucose cotransporter-2 inhibitors (SGLT2is), including empagliflozin, dapagliflozin, canagliflozin, and ertugliflozin, show a good prospect in the treatment of HF [12–16]. EMPEROR-Preserved demonstrated the clinical benefit of empagliflozin in patients with HFpEF with or without diabetes [12]. Dapagliflozin significantly improved patient-reported symptoms, physical limitations, and exercise function and was well tolerated in chronic HFpEF [13]. Canagliflozin reduced the risk of cardiovascular death or hospitalized HF [14] and improved Kansas City Cardiomyopathy Questionnaire Total Symptom Score [15] in HFpEF patients. Among patients with diabetes and atherosclerotic cardiovascular disease, another SGLT2i, ertugliflozin was non-inferior to placebo with respect to major adverse cardiovascular events [16]. Based on these high-quality clinical trial results, we can confirm that SGLT2is have a significant therapeutic effect on HFpEF. However, the internal biological mechanisms of SGLT2is for HFpEF are unknown.
Network pharmacology is an interdisciplinary subject, its formation and development mainly benefit from artificial intelligence and big data analysis [17, 18]. The primary advantage of network pharmacology is to emphasize the integrity, systemic and biological network of the research object [19]. With the increasing application of network pharmacology, more and more drugs have been explained at the level of the molecular mechanisms and promoted in clinical practice [19, 20]. In this study, we conducted network pharmacology, which aims to construct a multilevel network through various database searches, high-throughput data analysis, and computer simulations to analyze the relationship of medicines, targets, and diseases [21], to systematically explore the targets of SGLT2is and HFpEF and further excavate the biological pathways and mechanisms of SGLT2is for HFpEF, which may lay a foundation for clinical application and in-depth mechanisms exploration of SGLT2is for HFpEF.
Material and methods
Targets screening and networks construction
The chemical structures of SGLT2is, namely empagliflozin, dapagliflozin, canagliflozin, and ertugliflozin, were obtained from PubChem [22] and imported to SwissTargetPrediction [23] for potential targets prediction. Moreover, we used DrugBank [24] and Comparative Toxicogenomics Database [25] to supplement the target information. The HFpEF-associated targets were obtained from DisGeNET [26], GeneCards [27], MalaCards [28], Therapeutic Target Database [29], Comparative Toxicogenomics Database [25], National Center for Biotechnology Information, DrugBank [24], and Online Mendelian Inheritance in Man (OMIM). All targets were transformed in the UniProt database [30]. We set up the drug-target and drug-target-disease networks successively using Cytoscape [31]. Additionally, we conducted CytoNCA plugin to calculate and evaluate analysis for several centralities of the unweighted network [32].
Functional analysis
We used Metascape [33] for gene annotation, pathway and process enrichment analysis, and protein–protein interaction enrichment analysis. For each given gene list, pathway and process enrichment analysis was carried out with the following ontology sources: Gene Ontology (GO) molecular functions, GO cellular components, GO biological processes, Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway, Reactome Gene Sets, and Pattern Gene Database (PaGenBase). Finally, KEGG Mapper was used to mapper specific signaling pathways [34]. Protein–protein interaction (PPI) enrichment analysis was carried out and the Molecular Complex Detection (MCODE) algorithm was applied to identify densely connected network components [35]. Finally, PaGenBase was conducted to analyze the cell and tissue specificity [36].
Computational validation
We conducted the receptor-ligand molecular docking to assess these interactions [37]. We chose these targets shared by all four SGLT2is and then obtained their structures from Protein Data Bank [38]. AutoDock Vina [39], PyMOL Molecular Graphics System [40], and Discovery Studio were utilized for molecular docking.
Statistical analysis
For pathway and process enrichment, the minimum overlap was set to 3, P-value was set to 0.01, and the minimum enrichment was set to 1.5. For PPI enrichment, physical core database was chosen and the minimum and the maximum network sizes were set to 3 and 500, respectively.
Results
Screening targets of SGLT2is and HFpEF
We firstly obtained the chemical structures of empagliflozin, dapagliflozin, canagliflozin, and ertugliflozin from PubChem (Table 1, Additional file 1: Figure S1). Through various databases, we got 112, 147, 114, and 114 targets in empagliflozin, dapagliflozin, canagliflozin, and ertugliflozin, respectively. We obtained 1505 HFpEF related targets.
Table 1.
Details of four SGLT2is
| SGLT2is | PubChem CID | Molecular Formula | Molecular Weight | Modify Data |
|---|---|---|---|---|
| Empagliflozin | 11,949,646 | C23H27ClO7 | 450.9 | 2021-07-17 |
| Canagliflozin | 24,812,758 | C24H25FO5S | 444.5 | 2021-07-17 |
| Dapagliflozin | 9,887,712 | C21H25ClO6 | 408.9 | 2021-07-17 |
| Ertugliflozin | 44,814,423 | C22H25ClO7 | 436.9 | 2021-07-17 |
Network construction
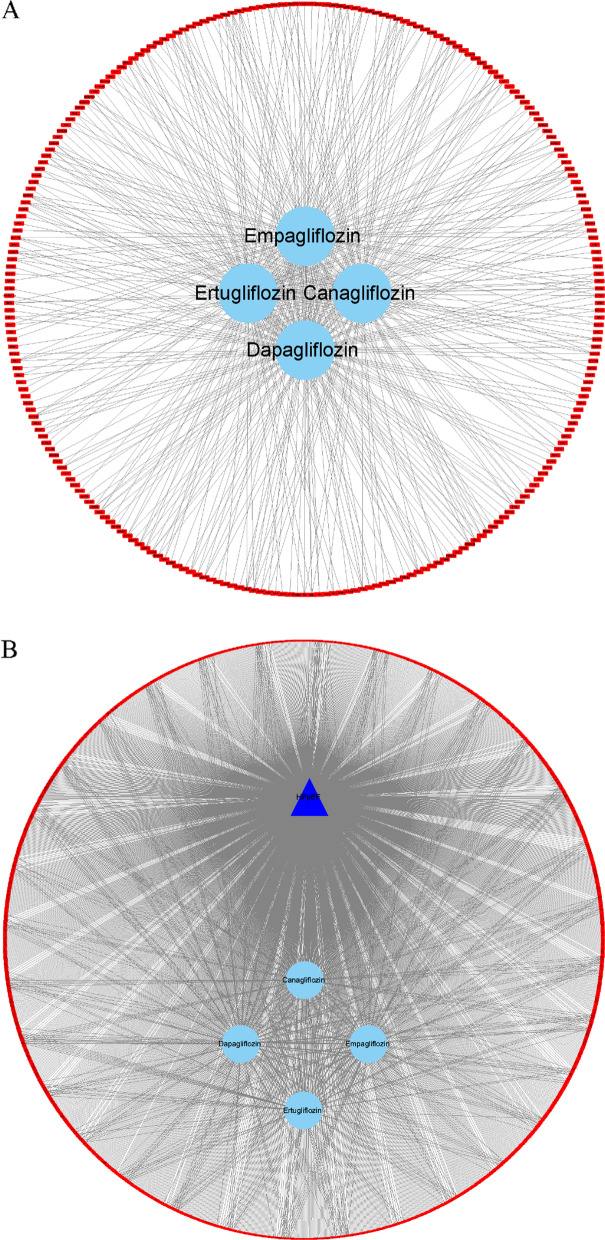
Then we constructed the drug-target network, which was composed of 258 nodes (4 SGLT2is, and 254 targets) and 479 edges (Fig. 1A). Then the drug-target-disease network, which is composed of 1647 nodes (1 HFpEF, 4 SGLT2is, and 1642 targets) and 1984 edges, was built (Fig. 1B). After evaluating by CytoNCA, we identified these targets shared by HFpEF and all SGLT2is (degree = 5), and the details are shown in Table 2.
Fig. 1.
Network construction. A The drug-target network. B The drug-target-disease network. The red rectangle nodes represent targets, sky blue ellipse nodes represent drugs, namely four SGLT2is (empagliflozin, dapagliflozin, canagliflozin, and ertugliflozin), and the navy blue triangle nodes represent disease, namely HFpEF. The edges mean that nodes can interact with each other
Table 2.
Multiple centrality measures in the drug-target-disease network (top 21)
| Nodes | Subgragh | Degree | Eigenvector | Information | Closeness | Betweenness |
|---|---|---|---|---|---|---|
| HFpEF | 4.02E + 16 | 1505 | 0.703902 | 2.108198 | 0.855509 | 2,654,751 |
| Dapagliflozin | 1.53E + 14 | 145 | 0.043505 | 2.081154 | 0.354436 | 142,937.1 |
| Canagliflozin | 8.19E + 13 | 116 | 0.031785 | 2.073808 | 0.350064 | 106,420.2 |
| Empagliflozin | 6.63E + 13 | 111 | 0.028588 | 2.072172 | 0.349321 | 115,219.2 |
| Ertugliflozin | 6.50E + 13 | 107 | 0.028322 | 2.070733 | 0.348729 | 85,940.67 |
| SLC5A2 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| SLC5A1 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| ADK | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| PDGFRB | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| ADORA2A | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| GBA | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| PDE5A | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| F3 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| CTSL | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| EGFR | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| GAPDH | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| MMP3 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| MMP1 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| P2RY12 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| JAK2 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| MME | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| MAPK1 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| ECE1 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| CDK2 | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| MGAM | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
| SI | 3.71E + 13 | 5 | 0.021382 | 1.562157 | 0.500761 | 7363.079 |
Functional enrichment analysis
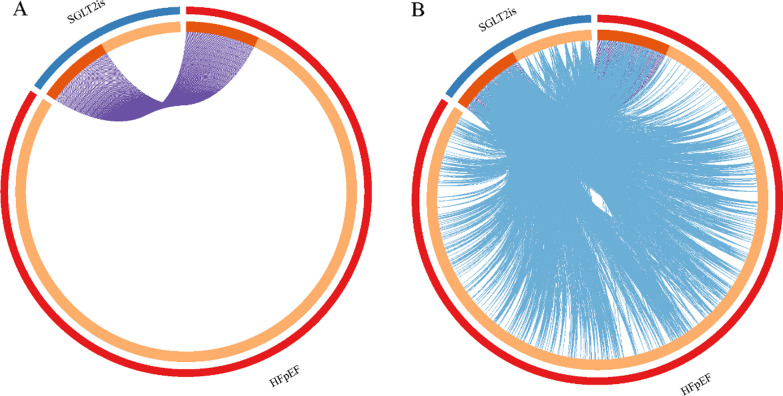
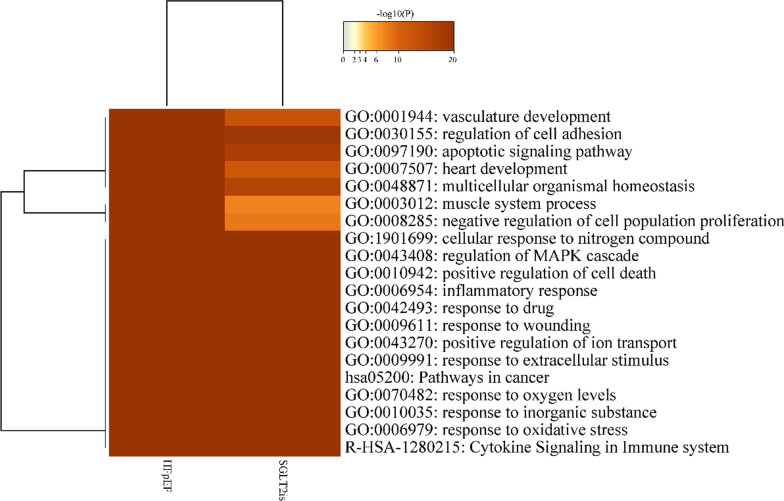
The overlaps between these targets associated with SGLT2is and HFpEF are shown in a Circos plot (Fig. 2A). Another useful representation was to overlap targets based on their functions or shared pathways. The overlaps between targets can be significantly improved by considering overlaps between genes sharing the same enriched ontology terms (Fig. 2B). From the top 20 heatmap of enriched terms across all targets (Fig. 3), we revealed that various ontology terms related to the cardiac and cardiovascular systems, such as vasculature development, response to oxidative stress, inflammatory response, positive regulation of cell death, regulation of MAPK cascade, cellular response to nitrogen compound, negative regulation of cell population proliferation, muscle system process, heart development, apoptotic signaling pathway, and regulation of cell adhesion. Up to 100 enriched clusters (Figure S2A), we viewed apoptosis, cAMP signaling pathway, regulation of lipid metabolic process, heart morphogenesis, and myofibril.
Fig. 2.
Overlap targets between HFpEF and SGLT2is. A Only at the gene level, where purple curves link identical genes. B Including the shared term level, where blue curves link genes that belong to the same enriched ontology term. The inner-circle represents gene lists, where hits are arranged along the arc. Genes that hit multiple lists are colored in dark orange, and genes unique to a list are shown in light orange
Fig. 3.
Heatmap of enriched terms across targets, colored by P-values (top 20). P-values were calculated based on the accumulative hypergeometric distribution
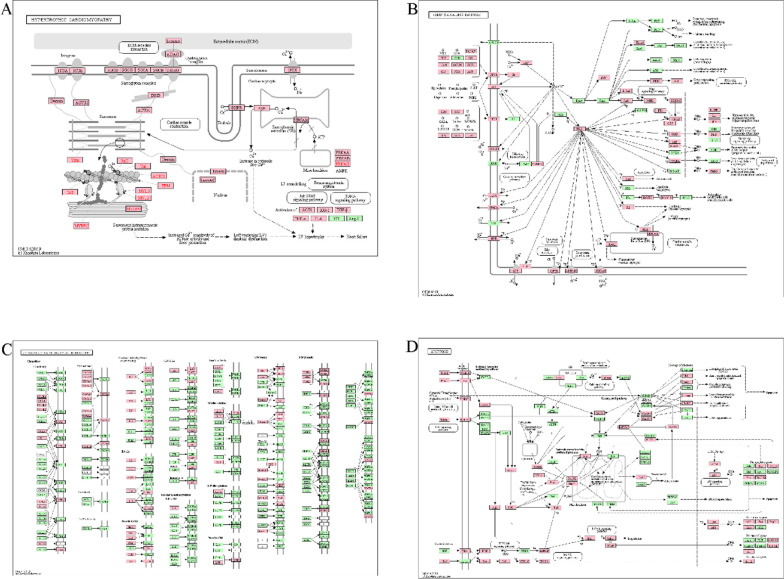
According to the GO analysis, the top 20 results were all attributed to biological processes (Table 3, Fig. 4A&B). Detailly, the top 10 results of GO molecular functions, GO cellular components, GO biological processes, and KEGG are shown in Figure S2B. Similarly, KEGG results enriched for many pathways known to be associated with cardiovascular disease (Table 4, Fig. 4C&D), such as cardiomyopathy (Fig. 5A), cAMP signaling pathway (Fig. 5B), cytokine-cytokine receptor interaction (Fig. 5C), apoptosis (Fig. 5D), MAPK signaling pathway (Figure S3A), HIF-1 signaling pathway (Figure S3B), calcium signaling pathway (Figure S3C), and NF-kappa B signaling pathway (Figure S3D). In addition, we represented the nodes in GO and KEGG networks as pie charts to particularly visualize whether the terms were shared by SGLT2is and HFpEF or unique SGLT2is or HFpEF, as well understand how these terms are associated with each other within the biological context of the meta study (Figure S4).
Table 3.
GO clusters with their representative enriched terms (top 20)
| GO | Category | Description | Count | % | Log10(P) | Log10(Q) |
|---|---|---|---|---|---|---|
| GO:0,003,012 | GO Biological Processes | Muscle system process | 192 | 12.13 | −100.00 | −96.35 |
| GO:0,006,954 | GO Biological Processes | Inflammatory response | 240 | 15.16 | −100.00 | −96.35 |
| GO:0,010,035 | GO Biological Processes | Response to inorganic substance | 188 | 11.88 | −97.16 | −93.59 |
| GO:0,009,611 | GO Biological Processes | Response to wounding | 202 | 12.76 | −93.36 | −89.85 |
| GO:0,001,944 | GO Biological Processes | Vasculature development | 217 | 13.71 | −90.37 | −86.98 |
| GO:0,007,507 | GO Biological Processes | Heart development | 183 | 11.56 | −90.06 | −86.71 |
| GO:0,043,408 | GO Biological Processes | Regulation of mapk cascade | 206 | 13.01 | −89.89 | −86.58 |
| GO:0,043,270 | GO Biological Processes | Positive regulation of ion transport | 196 | 12.38 | −87.61 | −84.34 |
| GO:0,008,285 | GO Biological Processes | Negative regulation of cell population proliferation | 198 | 13.66 | −85.08 | −81.90 |
| GO:0,009,991 | GO Biological Processes | Response to extracellular stimulus | 164 | 10.36 | −83.79 | −80.72 |
| GO:0,042,493 | GO Biological Processes | Response to drug | 141 | 8.91 | −80.81 | −77.84 |
| GO:1,901,699 | GO Biological Processes | Cellular response to nitrogen compound | 193 | 12.19 | −79.67 | −76.75 |
| GO:0,070,482 | GO Biological Processes | Response to oxygen levels | 144 | 9.10 | −79.13 | −76.26 |
| GO:0,097,190 | GO Biological Processes | Apoptotic signaling pathway | 173 | 10.93 | −77.38 | −74.53 |
| GO:0,048,871 | GO Biological Processes | Multicellular organismal homeostasis | 162 | 10.23 | −76.02 | −73.21 |
| GO:0,006,979 | GO Biological Processes | Response to oxidative stress | 149 | 9.41 | −73.85 | −71.12 |
| GO:0,030,155 | GO Biological Processes | Regulation of cell adhesion | 189 | 11.94 | −72.88 | −70.18 |
| GO:0,010,942 | GO Biological Processes | Positive regulation of cell death | 178 | 11.24 | −72.87 | −70.18 |
| GO:0,070,848 | GO Biological Processes | Response to growth factor | 189 | 11.94 | −72.67 | −69.98 |
| GO:0,071,396 | GO Biological Processes | Cellular response to lipid | 156 | 10.76 | −70.88 | −68.11 |
"Count" is the number of genes in the gene list with membership in the given ontology term. "%" is the percentage of all of the genes that are found in the given ontology term (only input genes with at least one ontology term annotation are included in the calculation). "Log10(P)" is the P-value in log base 10. "Log10(Q)" is the multi-test adjusted P-value in log base 10
Fig. 4.
Functional enrichment analysis. A GO analysis colored by cluster, where nodes that share the same cluster are typically close to each other. B GO analysis colored by P-value, where terms containing more genes tend to have a more significant P-value. C KEGG pathway enrichment analysis colored by cluster, where nodes that share the same cluster are typically close to each other. D KEGG pathway enrichment analysis colored by P-value, where terms containing more genes tend to have a more significant P-value. Only the top 20 results by P-value were shown. P-values were calculated based on the accumulative hypergeometric distribution
Table 4.
KEGG clusters with their representative enriched terms (top 20)
| GO | Category | Description | Count | % | Log10(P) | Log10(Q) |
|---|---|---|---|---|---|---|
| hsa05200 | KEGG Pathway | Pathways in cancer | 150 | 9.48 | −83.58 | −80.68 |
| ko05206 | KEGG Pathway | MicroRNAs in cancer | 109 | 6.89 | −58.39 | −56.10 |
| ko04933 | KEGG Pathway | AGE-RAGE signaling pathway in diabetic complications | 65 | 4.11 | −56.03 | −53.91 |
| hsa05161 | KEGG Pathway | Hepatitis B | 78 | 4.93 | −49.13 | −47.31 |
| hsa05410 | KEGG Pathway | Hypertrophic cardiomyopathy (HCM) | 54 | 3.72 | −48.32 | −46.54 |
| hsa04024 | KEGG Pathway | cAMP signaling pathway | 84 | 5.31 | −45.78 | −44.09 |
| hsa05166 | KEGG Pathway | HTLV-I infection | 85 | 5.37 | −41.85 | −40.27 |
| hsa05145 | KEGG Pathway | Toxoplasmosis | 58 | 3.66 | −40.54 | −39.00 |
| hsa04060 | KEGG Pathway | Cytokine-cytokine receptor interaction | 81 | 5.59 | −39.00 | −37.38 |
| hsa04210 | KEGG Pathway | Apoptosis | 60 | 3.79 | −37.47 | −36.10 |
| hsa04010 | KEGG Pathway | MAPK signaling pathway | 79 | 4.99 | −36.46 | −35.15 |
| hsa05222 | KEGG Pathway | Small cell lung cancer | 44 | 2.78 | −32.03 | −30.82 |
| ko04932 | KEGG Pathway | Non-alcoholic fatty liver disease (NAFLD) | 57 | 3.60 | −31.99 | −30.79 |
| ko05323 | KEGG Pathway | Rheumatoid arthritis | 44 | 3.03 | −31.95 | −30.61 |
| hsa04066 | KEGG Pathway | HIF-1 signaling pathway | 51 | 3.22 | −31.56 | −30.40 |
| ko05144 | KEGG Pathway | Malaria | 33 | 2.28 | −30.49 | −29.27 |
| ko05146 | KEGG Pathway | Amoebiasis | 44 | 3.03 | −30.42 | −29.21 |
| hsa04640 | KEGG Pathway | Hematopoietic cell lineage | 44 | 3.03 | −30.18 | −28.99 |
| hsa04020 | KEGG Pathway | Calcium signaling pathway | 60 | 3.79 | −29.48 | −28.38 |
| ko04064 | KEGG Pathway | NF-kappa B signaling pathway | 43 | 2.72 | −27.91 | −26.90 |
"Count" is the number of genes in the gene list with membership in the given ontology term. "%" is the percentage of all of the genes that are found in the given ontology term (only input genes with at least one ontology term annotation are included in the calculation). "Log10(P)" is the P-value in log base 10. "Log10(Q)" is the multi-test adjusted P-value in log base 10
Fig. 5.
KEGG pathway maps. A Hypertrophic cardiomyopathy. B cAMP signaling pathway. C Cytokine-cytokine receptor interaction. D Apoptosis. The pink pentagrams indicate the targets
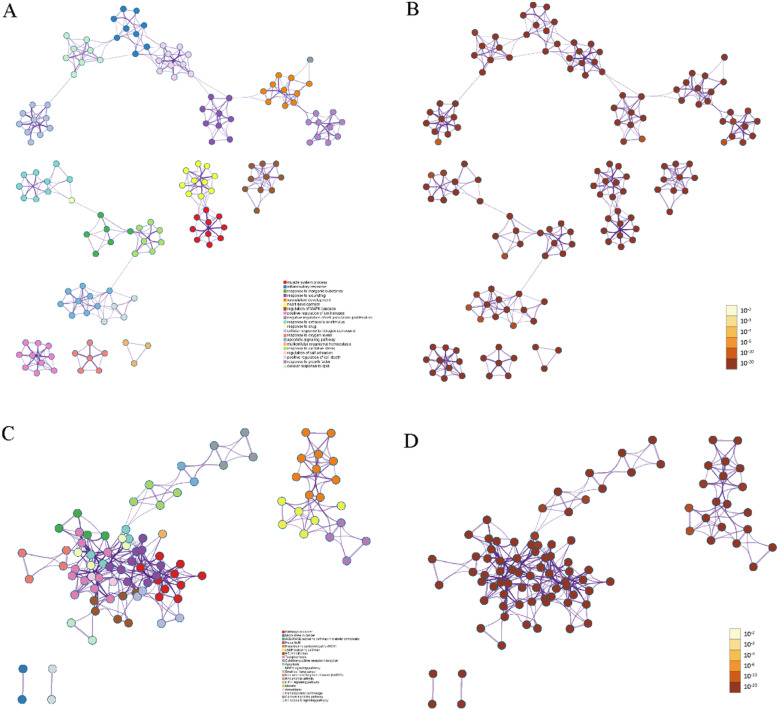
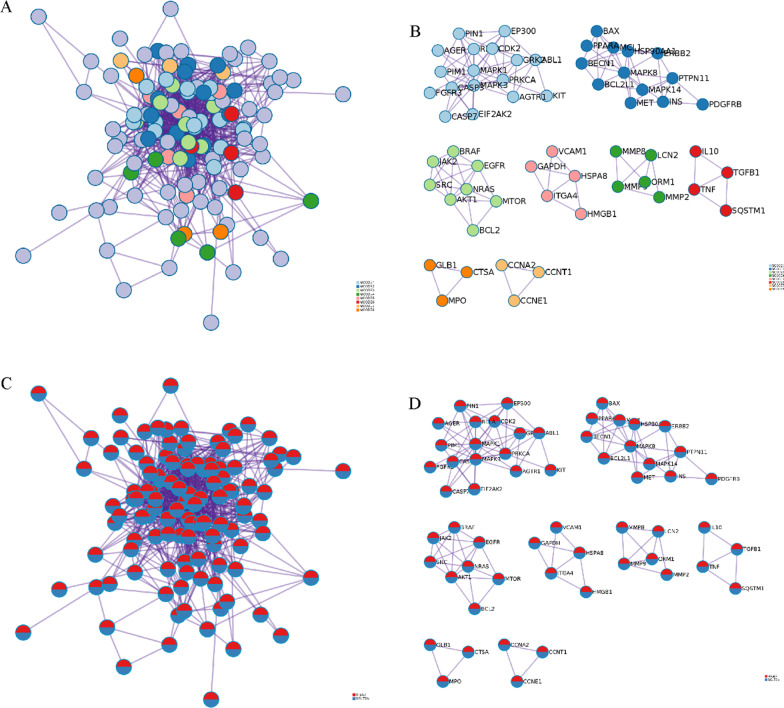
Through the PPI network, we demonstrated that targets in HFpEF were enriched to blood circulation, muscle system process, and circulatory system process (Table S1), whereas targets in SGLT2is were enriched to phosphotransferase activity, kinase activity, and protein kinase activity (Figure S5A, Table S1). A total of 11 MCODE components were enriched (Figure S5B, Table S1). Through all targets, PPI network, response to reactive oxygen species, cellular response to oxidative stress, and cellular response to chemical stress were top 3 pathway and process enrichment results (Fig. 6A&C, Table S1). A total of 8 MCODE components were retained (Fig. 6B&D, Table S1). In a word, these enrichment findings support the potential pharmacological mechanisms of SGLT2is for HFpEF.
Fig. 6.
PPI network and MCODE components. A PPI network colored by cluster, where terms containing different colors tend to have different MCODE components. B The 8 most significant MCODE components form the PPI network colored by cluster. C PPI network colored by counts, where terms containing different colors tend to have different MCODE components. D The 8 most significant MCODE components form the PPI network colored by counts
The enrichment analysis in PaGenBase demonstrated that these targets were specific in smooth muscle, heart, and blood tissue and adipocyte, human umbilical vein endothelial cells, cardiac myocytes, THY+, CD33+ myeloid cells (Figure S6, Table S2).
Computational validation
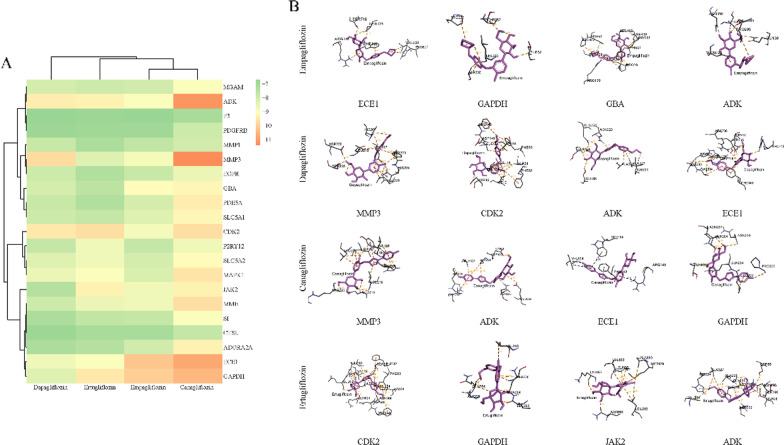
In the present study, the binding energies of four SGLT2is and 21 targets were all below -5.0 kcal/mol (Fig. 7A), indicating that these ligands and receptors could bind stability and spontaneously [41]. The local and whole docking mode between four SGLT2is and 4 key targets with the lowest binding energies are shown in Fig. 7B and Figure S7, respectively.
Fig. 7.
Molecular docking. A Heatmap of the molecular docking of 4 SGLT2is with 21 core targets. The color represents the binding energy. The oranger the color, the lower is the binding energy, and the higher is the affinity between the receptor and ligand. B Local docking mode between 4 SGLT2is and 4 key targets with the lowest binding energies
Discussion
With the release of PRESERVED-HF [13], CANVAS [14], CHIEF-HF [15], and VERTIS CV [16], SGLT2is seem a potential promising therapy for patients with HFpEF. In addition to the recently published EMPEROR-Preserved trial [12], the efficacy of SGLT2is in HFpEF is investigated in 2 additional still ongoing phase 3 trials: DELIVER [42] and CANONICAL [43]. The exact underlying mechanisms of action of SGLT2is in HFpEF have not yet been elucidated. Here, we identified the systemic mechanisms of SGLT2is in the treatment of HFpEF, and our main findings can be summarized as follows: (I) we firstly obtain 487 SGLT2is related targets and 1505 HFpEF related targets. Among SGLT2is related targets, we found there were 21 overlapping targets, which means that these 21 targets can act on four different drugs at the same time; (II) functional enrichment analysis revealed that the targets from SGLT2is were involved in various HFpEF-associated biological processes, such as muscle system process, inflammatory response, response to inorganic substance, response to wounding, vasculature development, heart development, regulation of MAPK cascade, positive regulation of ion transport, negative regulation of cell population proliferation, response to extracellular stimulus, response to drug, cellular response to nitrogen compound, response to oxygen levels, apoptotic signaling pathway, multicellular organismal homeostasis, response to oxidative stress, regulation of cell adhesion, positive regulation of cell death, response to growth factor, and cellular response to lipid; (III) KEGG results were related to hypertrophic cardiomyopathy, cAMP signaling pathway, cytokine-cytokine receptor interaction, apoptosis, MAPK signaling pathway, HIF-1 signaling pathway, calcium signaling pathway, and NF-kappa B signaling pathway, which are associated with HF.
In the present study, we included four SGLT2is, namely empagliflozin, dapagliflozin, canagliflozin, and ertugliflozin. We predicted the targets of SGLT2is in SwissTargetPrediction [23] and supplemented the targets from other databases. Similarly, we used as many databases as possible to collect targets of HFpEF to avoid missing important targets. Moreover, all targets were transformed in the UniProt database [30], which avoids the confusion caused by the aliases of the targets. Here, we identified 21 potential targets, which seem to provide references for SGLT2is in the treatment of HFpEF. SGLT1 and SGLT2, encoded by SLC5A1 and SLC5A2 respectively, are important mediators of epithelial glucose transport, and together with SI are SGLT2is targets [44]. Cardiomyocyte PDGFRB is a regulator of the compensatory cardiac response to pressure overload-induced stress [45]. MMP 3 was associated with focal fibrosis and diffuse fibrosis in HFpEF [46]. Patients and dogs with HF have increased expression of MMP1 [47, 48], suggesting progressive left ventricular remodeling. Selective PDE5A inhibition rescues left ventricular dysfunction, inflammatory immune response, and cardiac remodeling in HF [49, 50]. Studies have proved that P2RY12, MME, ADORA2A, MAPK1, EGFR, and ECE1 were potential targets for the treatment of HFpEF [51–56]. ADK inhibition augments microvascular dilator function and conducted vasodilation and prevents left ventricle diastolic dysfunction in HFpEF [57, 58], CTSL is critical for cardiac morphology and function [59]. JAK2/STAT3 pathway [60, 61], EGFR/Akt/ERK1/2 axis [62], and p27/CDK2/mTOR axis [63] linked to HF.
Different multidirectional mechanisms of SGLT2is could improve HF status [64]. However, there are few known mechanisms of SGLT2is in HFpEF. A previous study indicated that SGLT2is may upregulate the renin–angiotensin–aldosterone system [65]. Empagliflozin could improve diastolic stiffness, hence diastolic function [66], attenuate cardiac fibrosis, and improve ventricular hemodynamics [67]. Empagliflozin beneficially reduced myofilament passive stiffness by enhancing phosphorylation levels of myofilament regulatory proteins in myocardial fibers from patients and rats with HFpEF [66]. We also found that cell proliferation, apoptosis, as well as organismal homeostasis were important in the biological processes of HFpEF from the present study, which are consistent with available evidence [68–70]. Moreover, we highlighted vasculature development, heart development, and ion transport in SGLT2is treatment of HFpEF, which are also consistent with the current cognition [52, 53]. Empagliflozin reduced the activity of the cardiac Na+/H+ exchanger to possibly improve cardiac function [71, 72]. Later, it was found that dapagliflozin and canagliflozin inhibited Na+/H+ exchanger activity and reduced cytosolic Na+ [73]. Additionally, empagliflozin reduced Ca2+/calmodulin-dependent kinase II (CaMKII) activity and CaMKII-dependent sarcoplasmic reticulum Ca2+ leak [74, 75].
A large number of studies have shown that HFpEF is a syndrome of over-activation of inflammatory, oxidative stress, and autophagy [76], which is also consistent with our results. Dapagliflozin decreased hypertension and reversed left ventricle concentric remodeling in HFpEF pigs partly by restraining sympathetic tone in the aorta, leading to inhibition of the inflammatory response and NO-cGMP-PKG pathway activation [77]. Empagliflozin reduced inflammatory and oxidative stress in HFpEF and thereby improved the NO-sGC-cGMP-cascade and PKGIα activity via reduced PKGIα oxidation and polymerization [78]. Besides, canagliflozin might exert anti-inflammatory effects by inhibiting intracellular glucose metabolism and promoting autophagy [79].
Through KEGG pathways enrichment analysis, we emphasized the cAMP signaling pathway, MAPK signaling pathway, HIF-1 signaling pathway, calcium signaling pathway, and apoptosis during the occurrence and development of HFpEF. These findings are in line with previous studies. MiR-665 inhibition can stabilize the cardiac function of HF rats via the cAMP signaling pathway via upregulation of the GLP1R [58]. By switching from Gαs to Gαi2 activation, NDPK-C, a novel critical regulator of cAMP signaling and cardiac contractility, may contribute to lower cAMP levels and the related contractile dysfunction in HF [80]. MAPK has been studied in-depth about cardiac development and function [59, 60]. Elucidation of the molecular mechanisms of hypoxia signaling will greatly help us to understand the pathophysiology of cardiovascular disorders [81]. Defective cardiolipin remodeling, upon loss of the cardiolipin acyl transferase tafazzin, decreases HIF-1α signaling in hypoxia [82]. Enhanced activation of the Dyrk1A-ASF-CaMKIIδ signaling pathway may underlie the mechanisms of HF [83]. Numerous drugs can improve HF through the calcium signaling pathway. In addition, PPI network and cell and tissue specificity analysis confirmed the previous results. Importantly, we conducted molecular docking to further verify the interactions and combinations of SGLT2is and core targets. In a word, these enrichment findings support the potential pharmacological mechanisms of SGLT2is for HFpEF.
However, there were some limitations that we should pay attention to. Firstly, when we fished the targets from HFpEF, we found that some databases are not updated to HFpEF, only HF or chronic HF, which makes us inevitably lose some important targets. Moreover, we only generally analyzed the mechanisms of SGLT2is for HFpEF, but we still need to further study the single SGT2i, because the action mechanisms of different SGLT2is may not be completely consistent. In addition, the pathophysiology of HFpEF manifestations is highly heterogeneous [84, 85], more current and future endeavors are underway to evaluate the optimal methods to classify patients into phenotypically homogeneous subpopulations to facilitate better individualization of treatment [86]. Finally, this study is based on computer and biological information mining, these reliable results we obtained here still need to be verified by molecular biology experiments in the later stage.
Conclusions
In conclusion, we identified the synergistic pharmacological mechanisms of SGLT2is in HFpEF by the integrative virtual screening and network pharmacology method. Moreover, our main findings were validated with molecular docking. Our findings of the potential mechanisms of the direct or indirect synergy of multiple targets and pathways provide an optional therapy for HFpEF. However, more experimental and clinical validation is essential to reveal the effect of SGLT2is against HFpEF.
Supplementary Information
Additional file 1. Tables S1, S2 and Figure S1–S7.
Acknowledgements
We thank all individuals involved SGLT2is and HFpEF. We are also grateful to all research scientists who participated in the aforementioned databases.
Abbreviations
- CaMKII
Ca2+/calmodulin-dependent kinase II
- EF
Ejection fraction
- GO
Gene Ontology
- HF
Heart failure
- HFpEF
Heart failure with preserved ejection fraction
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein–protein interaction
- SGLT2i
Sodium-glucose cotransporter-2 inhibitor
Author contributions
BL and NG designed the research. BL and YL carried out the experiments. BL and YL performed the data analysis. BL, YL, and NG interpreted the results. BL drafted the manuscript. NG revised the manuscript. All authors have read and approved the final manuscript.
Funding
This work was partly funded by Nanjing Municipal Health Science and Technology Development Special Fund [ZKX21060].
Availability of data and materials
Chemical structures of SGLT2is were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) with PubChem CID 11,949,646 for empagliflozin, 24,812,758 for canagliflozin, 9,887,712 for dapagliflozin, and 44,814,423 for ertugliflozin.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liang B, Gu N. Liraglutide in the treatment of heart failure: insight from FIGHT and LIVE. Cardiovasc Diabetol. 2020;19(1):106. doi: 10.1186/s12933-020-01088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang B, Li R, Bai J-Y, Gu N. Bioimpedance vector analysis for heart failure: should we put it on the agenda? Front Cardiovasc Med. 2021;8:744243. doi: 10.3389/fcvm.2021.744243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fonarow GC, Stough WG, Abraham WT, Albert NM, Gheorghiade M, Greenberg BH, et al. Characteristics, treatments, and outcomes of patients with preserved systolic function hospitalized for heart failure: a report from the OPTIMIZE-HF Registry. J Am Coll Cardiol. 2007;50(8):768–777. doi: 10.1016/j.jacc.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 4.Lin Y, Fu S, Yao Y, Li Y, Zhao Y, Luo L. Heart failure with preserved ejection fraction based on aging and comorbidities. J Transl Med. 2021;19(1):291. doi: 10.1186/s12967-021-02935-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonagh TA, Metra M, Adamo M, Gardner RS, Baumbach A, Böhm M, et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. 2021;42(36):3599–3726. doi: 10.1093/eurheartj/ehab368. [DOI] [PubMed] [Google Scholar]
- 6.Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol. 2022;79(17):e263–e421. doi: 10.1016/j.jacc.2021.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Owan TE, Hodge DO, Herges RM, Jacobsen SJ, Roger VL, Redfield MM. Trends in prevalence and outcome of heart failure with preserved ejection fraction. N Engl J Med. 2006;355(3):251–259. doi: 10.1056/NEJMoa052256. [DOI] [PubMed] [Google Scholar]
- 8.Sharma K, Kass DA. Heart failure with preserved ejection fraction: mechanisms, clinical features, and therapies. Circ Res. 2014;115(1):79–96. doi: 10.1161/CIRCRESAHA.115.302922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam CSP, Voors AA, de Boer RA, Solomon SD, van Veldhuisen DJ. Heart failure with preserved ejection fraction: from mechanisms to therapies. Eur Heart J. 2018;39(30):2780–2792. doi: 10.1093/eurheartj/ehy301. [DOI] [PubMed] [Google Scholar]
- 10.Aguilar-Gallardo JS, Correa A, Contreras JP. Cardio-renal benefits of SGLT2 inhibitors in heart failure with reduced ejection fraction: mechanisms and clinical evidence. Eur Heart J Cardiovasc Pharmacother. 2021;8(3):311–321. doi: 10.1093/ehjcvp/pvab056. [DOI] [PubMed] [Google Scholar]
- 11.Liang B, Zhao Y-X, Zhang X-X, Liao H-L, Gu N. Reappraisal on pharmacological and mechanical treatments of heart failure. Cardiovasc Diabetol. 2020;19(1):55. doi: 10.1186/s12933-020-01024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anker SD, Butler J, Filippatos G, Ferreira JP, Bocchi E, Böhm M, et al. Empagliflozin in heart failure with a preserved ejection fraction. N Engl J Med. 2021;385(16):1451–1461. doi: 10.1056/NEJMoa2107038. [DOI] [PubMed] [Google Scholar]
- 13.Nassif ME, Windsor SL, Borlaug BA, Kitzman DW, Shah SJ, Tang F, et al. The SGLT2 inhibitor dapagliflozin in heart failure with preserved ejection fraction: a multicenter randomized trial. Nat Med. 2021;27(11):1954–1960. doi: 10.1038/s41591-021-01536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figtree GA, Rådholm K, Barrett TD, Perkovic V, Mahaffey KW, de Zeeuw D, et al. Effects of canagliflozin on heart failure outcomes associated with preserved and reduced ejection fraction in type 2 diabetes mellitus. Circulation. 2019;139(22):2591–2593. doi: 10.1161/CIRCULATIONAHA.119.040057. [DOI] [PubMed] [Google Scholar]
- 15.Spertus JA, Birmingham MC, Nassif M, Damaraju CV, Abbate A, Butler J, et al. The SGLT2 inhibitor canagliflozin in heart failure: the CHIEF-HF remote, patient-centered randomized trial. Nat Med. 2022;28(4):809–813. doi: 10.1038/s41591-022-01703-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon CP, Pratley R, Dagogo-Jack S, Mancuso J, Huyck S, Masiukiewicz U, et al. Cardiovascular outcomes with Ertugliflozin in type 2 diabetes. N Engl J Med. 2020;383(15):1425–1435. doi: 10.1056/NEJMoa2004967. [DOI] [PubMed] [Google Scholar]
- 17.Liang Y, Liang B, Wu X-R, Chen W, Zhao L-Z. Network pharmacology-based systematic analysis of molecular mechanisms of dingji fumai decoction for ventricular arrhythmia. Evid Based Complement Alternat Med. 2021;2021:5535480. doi: 10.1155/2021/5535480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Y, Liang B, Chen W, Wu X-R, Liu-Huo W-S, Zhao L-Z. Potential mechanism of Dingji Fumai decoction against atrial fibrillation based on network pharmacology, molecular docking, and experimental verification integration strategy. Front Cardiovasc Med. 2021;8:712398. doi: 10.3389/fcvm.2021.712398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang B, Liang Y, Li R, Zhang H, Gu N. Integrating systematic pharmacology-based strategy and experimental validation to explore the synergistic pharmacological mechanisms of Guanxin V in treating ventricular remodeling. Bioorg Chem. 2021;115:105187. doi: 10.1016/j.bioorg.2021.105187. [DOI] [PubMed] [Google Scholar]
- 20.Liang B, Zhang X-X, Gu N. Virtual screening and network pharmacology-based synergistic mechanism identification of multiple components contained in Guanxin V against coronary artery disease. BMC Complement Med Ther. 2020;20(1):345. doi: 10.1186/s12906-020-03133-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ning K, Zhao X, Poetsch A, Chen W-H, Yang J. Computational molecular networks and network pharmacology. Biomed Res Int. 2017;2017:7573904. doi: 10.1155/2017/7573904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49(D1):D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, Wiegers TC, et al. Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res. 2021;49(D1):D1138–D1143. doi: 10.1093/nar/gkaa891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinform. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 28.Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J, et al. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017;45(D1):877–887. doi: 10.1093/nar/gkw1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang S, Li F, Zhou Y, Zhang Y, Wang Z, et al. Therapeutic target database, enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031–D1041. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The UC. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, Li M, Wang J-X, Pan Y, Wu F-X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y-Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29(1):28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan J-B, Hu S-C, Shi D, Cai M-C, Li Y-B, Zou Q, et al. PaGenBase: a pattern gene database for the global and dynamic understanding of gene function. PLoS ONE. 2013;8(12):e80747. doi: 10.1371/journal.pone.0080747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang B, Li R, Liang Y, Gu N. Guanxin V acts as an antioxidant in ventricular remodeling. Front Cardiovasc Med. 2022;8:778005. doi: 10.3389/fcvm.2021.778005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodsell DS, Zardecki C, Di Costanzo L, Duarte JM, Hudson BP, Persikova I, et al. RCSB Protein Data Bank: enabling biomedical research and drug discovery. Protein Sci. 2020;29(1):52–65. doi: 10.1002/pro.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J Chem Inf Model. 2021. [DOI] [PMC free article] [PubMed]
- 40.Rigsby RE, Parker AB. Using the P y MOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ. 2016;44(5):433–437. doi: 10.1002/bmb.20966. [DOI] [PubMed] [Google Scholar]
- 41.Tong H, Yu M, Fei C, Ji D, Dong J, Su L, et al. Bioactive constituents and the molecular mechanism of Curcumae Rhizoma in the treatment of primary dysmenorrhea based on network pharmacology and molecular docking. Phytomed Int J Phytother Phytopharmacol. 2021;86:153558. doi: 10.1016/j.phymed.2021.153558. [DOI] [PubMed] [Google Scholar]
- 42.Solomon SD, Vaduganathan M, Claggett BL, de Boer RA, DeMets D, Hernandez AF, et al. Baseline characteristics of patients with HF with mildly reduced and preserved ejection fraction: DELIVER trial. JACC Heart Fail. 2022;10(3):184–197. doi: 10.1016/j.jchf.2021.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Kasama S, Masuyama T, Uemura S, Sato Y, Hiramitsu S, Masuda I, et al. Rationale and design of the CANONICAL study - randomized, open-label study to evaluate the efficacy and safety of canagliflozin for heart failure with preserved ejection fraction with type 2 diabetes mellitus. Circ Rep. 2019;1(8):347–351. doi: 10.1253/circrep.CR-19-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieg T, Vallon V. Development of SGLT1 and SGLT2 inhibitors. Diabetologia. 2018;61(10):2079–2086. doi: 10.1007/s00125-018-4654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chintalgattu V, Ai D, Langley RR, Zhang J, Bankson JA, Shih TL, et al. Cardiomyocyte PDGFR-beta signaling is an essential component of the mouse cardiac response to load-induced stress. J Clin Invest. 2010;120(2):472–484. doi: 10.1172/JCI39434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanagala P, Arnold JR, Singh A, Chan DCS, Cheng ASH, Khan JN, et al. Characterizing heart failure with preserved and reduced ejection fraction: an imaging and plasma biomarker approach. PLoS ONE. 2020;15(4):e0232280. doi: 10.1371/journal.pone.0232280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barton PJR, Birks EJ, Felkin LE, Cullen ME, Koban MU, Yacoub MH. Increased expression of extracellular matrix regulators TIMP1 and MMP1 in deteriorating heart failure. J Heart Lung Transplant. 2003;22(7):738–744. doi: 10.1016/S1053-2498(02)00557-0. [DOI] [PubMed] [Google Scholar]
- 48.Fonfara S, Tew SR, Cripps P, Dukes-McEwan J, Clegg PD. Increased blood mRNA expression of inflammatory and anti-fibrotic markers in dogs with congestive heart failure. Res Vet Sci. 2012;93(2):879–885. doi: 10.1016/j.rvsc.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 49.Liao M, Xie Q, Zhao Y, Yang C, Lin C, Wang G, et al. Main active components of Si-Miao-Yong-An decoction (SMYAD) attenuate autophagy and apoptosis via the PDE5A-AKT and TLR4-NOX4 pathways in isoproterenol (ISO)-induced heart failure models. Pharmacol Res. 2022;176:106077. doi: 10.1016/j.phrs.2022.106077. [DOI] [PubMed] [Google Scholar]
- 50.Westermann D, Becher PM, Lindner D, Savvatis K, Xia Y, Fröhlich M, et al. Selective PDE5A inhibition with sildenafil rescues left ventricular dysfunction, inflammatory immune response and cardiac remodeling in angiotensin II-induced heart failure in vivo. Basic Res Cardiol. 2012;107(6):308. doi: 10.1007/s00395-012-0308-y. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Hui J, Chen X. Preprocedural ticagrelor treatment was associated with improved early reperfusion and reduced short-term heart failure in East-Asian ST-segment elevation myocardial infarction patients undergoing primary percutaneous coronary intervention. Int J Gen Med. 2021;14:1927–1938. doi: 10.2147/IJGM.S307404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz JC, Gros C, Lecomte JM, Bralet J. Enkephalinase (EC 3.4.24.11) inhibitors: protection of endogenous ANF against inactivation and potential therapeutic applications. Life Sci. 1990;47(15):1279–97. doi: 10.1016/0024-3205(90)90192-T. [DOI] [PubMed] [Google Scholar]
- 53.Haas J, Frese KS, Park YJ, Keller A, Vogel B, Lindroth AM, et al. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol Med. 2013;5(3):413–429. doi: 10.1002/emmm.201201553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhai Y-J, Liu P, He H-R, Zheng X-W, Wang Y, Yang Q-T, et al. The association of ADORA2A and ADORA2B polymorphisms with the risk and severity of chronic heart failure: a case-control study of a northern Chinese population. Int J Mol Sci. 2015;16(2):2732–2746. doi: 10.3390/ijms16022732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y, Wang M, Xu J, Wei J, Yang H. Signature network-based survey of the effects of a traditional Chinese medicine on heart failure. J Ethnopharmacol. 2022;283:114750. doi: 10.1016/j.jep.2021.114750. [DOI] [PubMed] [Google Scholar]
- 56.Yim J, Cho H, Rabkin SW. Gene expression and gene associations during the development of heart failure with preserved ejection fraction in the Dahl salt sensitive model of hypertension. Clin Exp Hypertens. 2018;40(2):155–166. doi: 10.1080/10641963.2017.1346113. [DOI] [PubMed] [Google Scholar]
- 57.Davila A, Tian Y, Czikora I, Weissman AS, Weinand N, Dong G, et al. Adenosine kinase inhibition enhances microvascular dilator function and improves left ventricle diastolic dysfunction. Microcirculation. 2020;27(6):e12624. doi: 10.1111/micc.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davila A, Tian Y, Czikora I, Li J, Su H, Huo Y, et al. Adenosine kinase inhibition augments conducted vasodilation and prevents left ventricle diastolic dysfunction in heart failure with preserved ejection fraction. Circ Heart Fail. 2019;12(8):e005762. doi: 10.1161/CIRCHEARTFAILURE.118.005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stypmann J, Gläser K, Roth W, Tobin DJ, Petermann I, Matthias R, et al. Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin L. Proc Natl Acad Sci U S A. 2002;99(9):6234–6239. doi: 10.1073/pnas.092637699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Y, Zhang W, Lee L, Hong M, Lee M, Chou G, et al. Down-regulated microRNA-195-5p and up-regulated CXCR4 attenuates the heart function injury of heart failure mice via inactivating JAK/STAT pathway. Int Immunopharmacol. 2020;82:106225. doi: 10.1016/j.intimp.2020.106225. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Zhang L, Fan X, Yang W, Yu B, Kou J, et al. Captopril attenuates TAC-induced heart failure via inhibiting Wnt3a/β-catenin and Jak2/Stat3 pathways. Biomed Pharmacother. 2019;113:108780. doi: 10.1016/j.biopha.2019.108780. [DOI] [PubMed] [Google Scholar]
- 62.Liu L, Jin X, Hu C-F, Zhang Y-P, Ze Zhou, Li R, et al. Amphiregulin enhances cardiac fibrosis and aggravates cardiac dysfunction in mice with experimental myocardial infarction partly through activating EGFR-dependent pathway. Basic Res Cardiol. 2018;113(2):12. doi: 10.1007/s00395-018-0669-y. [DOI] [PubMed] [Google Scholar]
- 63.Su M, Wang J, Wang C, Wang X, Dong W, Qiu W, et al. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ. 2015;22(6):986–999. doi: 10.1038/cdd.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pabel S, Hamdani N, Luedde M, Sossalla S. SGLT2 inhibitors and their mode of action in heart failure-has the mystery been unravelled? Curr Heart Fail Rep. 2021;18(5):315–328. doi: 10.1007/s11897-021-00529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schork A, Saynisch J, Vosseler A, Jaghutriz BA, Heyne N, Peter A, et al. Effect of SGLT2 inhibitors on body composition, fluid status and renin-angiotensin-aldosterone system in type 2 diabetes: a prospective study using bioimpedance spectroscopy. Cardiovasc Diabetol. 2019;18(1):46. doi: 10.1186/s12933-019-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pabel S, Wagner S, Bollenberg H, Bengel P, Kovács Á, Schach C, et al. Empagliflozin directly improves diastolic function in human heart failure. Eur J Heart Fail. 2018;20(12):1690–1700. doi: 10.1002/ejhf.1328. [DOI] [PubMed] [Google Scholar]
- 67.Lee H-C, Shiou Y-L, Jhuo S-J, Chang C-Y, Liu P-L, Jhuang W-J, et al. The sodium-glucose co-transporter 2 inhibitor empagliflozin attenuates cardiac fibrosis and improves ventricular hemodynamics in hypertensive heart failure rats. Cardiovasc Diabetol. 2019;18(1):45. doi: 10.1186/s12933-019-0849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reddy YNV, Obokata M, Wiley B, Koepp KE, Jorgenson CC, Egbe A, et al. The haemodynamic basis of lung congestion during exercise in heart failure with preserved ejection fraction. Eur Heart J. 2019;40(45):3721–3730. doi: 10.1093/eurheartj/ehz713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woolley RJ, Ceelen D, Ouwerkerk W, Tromp J, Figarska SM, Anker SD, et al. Machine learning based on biomarker profiles identifies distinct subgroups of heart failure with preserved ejection fraction. Eur J Heart Fail. 2021;23(6):983–991. doi: 10.1002/ejhf.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frisk M, Le C, Shen X, Røe ÅT, Hou Y, Manfra O, et al. Etiology-dependent impairment of diastolic cardiomyocyte calcium homeostasis in heart failure with preserved ejection fraction. J Am Coll Cardiol. 2021;77(4):405–419. doi: 10.1016/j.jacc.2020.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baartscheer A, Schumacher CA, Wüst RCI, Fiolet JWT, Stienen GJM, Coronel R, et al. Empagliflozin decreases myocardial cytoplasmic Na(+) through inhibition of the cardiac Na(+)/H(+) exchanger in rats and rabbits. Diabetologia. 2017;60(3):568–573. doi: 10.1007/s00125-016-4134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trum M, Riechel J, Lebek S, Pabel S, Sossalla ST, Hirt S, et al. Empagliflozin inhibits Na+/H+ exchanger activity in human atrial cardiomyocytes. ESC Heart Fail. 2020;7(6):4429–4437. doi: 10.1002/ehf2.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uthman L, Baartscheer A, Bleijlevens B, Schumacher CA, Fiolet JWT, Koeman A, et al. Class effects of SGLT2 inhibitors in mouse cardiomyocytes and hearts: inhibition of Na(+)/H(+) exchanger, lowering of cytosolic Na(+) and vasodilation. Diabetologia. 2018;61(3):722–726. doi: 10.1007/s00125-017-4509-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mustroph J, Wagemann O, Lücht CM, Trum M, Hammer KP, Sag CM, et al. Empagliflozin reduces Ca/calmodulin-dependent kinase II activity in isolated ventricular cardiomyocytes. ESC Heart Fail. 2018;5(4):642–648. doi: 10.1002/ehf2.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cappetta D, De Angelis A, Bellocchio G, Telesca M, Cianflone E, Torella D, et al. Sodium-glucose cotransporter 2 inhibitors and heart failure: a bedside-to-bench journey. Front Cardiovasc Med. 2021;8:810791. doi: 10.3389/fcvm.2021.810791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adamczak DM, Oduah M-T, Kiebalo T, Nartowicz S, Bęben M, Pochylski M, et al. Heart failure with preserved ejection fraction-a concise review. Curr Cardiol Rep. 2020;22(9):82. doi: 10.1007/s11886-020-01349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang N, Feng B, Ma X, Sun K, Xu G, Zhou Y. Dapagliflozin improves left ventricular remodeling and aorta sympathetic tone in a pig model of heart failure with preserved ejection fraction. Cardiovasc Diabetol. 2019;18(1):107. doi: 10.1186/s12933-019-0914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kolijn D, Pabel S, Tian Y, Lódi M, Herwig M, Carrizzo A, et al. Empagliflozin improves endothelial and cardiomyocyte function in human heart failure with preserved ejection fraction via reduced pro-inflammatory-oxidative pathways and protein kinase Gα oxidation. Cardiovasc Res. 2021;117(2):495–507. doi: 10.1093/cvr/cvaa123. [DOI] [PubMed] [Google Scholar]
- 79.Xu C, Wang W, Zhong J, Lei F, Xu N, Zhang Y, et al. Canagliflozin exerts anti-inflammatory effects by inhibiting intracellular glucose metabolism and promoting autophagy in immune cells. Biochem Pharmacol. 2018;152:45–59. doi: 10.1016/j.bcp.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 80.Abu-Taha IH, Heijman J, Hippe H-J, Wolf NM, El-Armouche A, Nikolaev VO, et al. Nucleoside diphosphate kinase-C suppresses cAMP formation in human heart failure. Circulation. 2017;135(9):881–897. doi: 10.1161/CIRCULATIONAHA.116.022852. [DOI] [PubMed] [Google Scholar]
- 81.Abe H, Semba H, Takeda N. The roles of hypoxia signaling in the pathogenesis of cardiovascular diseases. J Atheroscler Thromb. 2017;24(9):884–894. doi: 10.5551/jat.RV17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chowdhury A, Aich A, Jain G, Wozny K, Lüchtenborg C, Hartmann M, et al. Defective mitochondrial cardiolipin remodeling dampens HIF-1α expression in hypoxia. Cell Rep. 2018;25(3):561–70.e6. doi: 10.1016/j.celrep.2018.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He J, Yao J, Sheng H, Zhu J. Involvement of the dual-specificity tyrosine phosphorylation-regulated kinase 1A-alternative splicing factor-calcium/calmodulin-dependent protein kinase IIδ signaling pathway in myocardial infarction-induced heart failure of rats. J Card Fail. 2015;21(9):751–760. doi: 10.1016/j.cardfail.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 84.Sotomi Y, Hikoso S, Komukai S, Sato T, Oeun B, Kitamura T, et al. Phenotyping of acute decompensated heart failure with preserved ejection fraction. Heart. 2022;heartjnl-2021-320270. [DOI] [PubMed]
- 85.Pabel S, Hamdani N, Singh J, Sossalla S. Potential mechanisms of SGLT2 inhibitors for the treatment of heart failure with preserved ejection fraction. Front Physiol. 2021;12:752370. doi: 10.3389/fphys.2021.752370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borlaug BA. Evaluation and management of heart failure with preserved ejection fraction. Nat Rev Cardiol. 2020;17(9):559–573. doi: 10.1038/s41569-020-0363-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Tables S1, S2 and Figure S1–S7.
Data Availability Statement
Chemical structures of SGLT2is were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) with PubChem CID 11,949,646 for empagliflozin, 24,812,758 for canagliflozin, 9,887,712 for dapagliflozin, and 44,814,423 for ertugliflozin.