Graphical abstract
Keywords: SARS-CoV-2, Predictive prognosis, Machine learning, Mutation identification, Temporal benchmarking
Abbreviations: SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; VoC, Variant of Concern; ML, Machine Learning; HLA, Human Leukocyte Antigens; MHC, Major Histocompatibility Complex; SHAP, SHapley Additive exPlanations; GISAID, Global Initiative on Sharing All Influenza Data; ROC, Receiver Operator Characteristic; ROC AUC, Area Under the ROC Curve; t-SNE, t-distributed Stochastic Neighbor Embedding; PCA, Principal Component Analysis; UMAP, Uniform Manifold Approximation and Projection; XGBoost, eXtreme Gradient Boosting; VREs, Variants of Reference Epitopes
Abstract
Motivation
Continuous emergence of new variants through appearance/accumulation/disappearance of mutations is a hallmark of many viral diseases. SARS-CoV-2 variants have particularly exerted tremendous pressure on global healthcare system owing to their life threatening and debilitating implications. The sheer plurality of variants and huge scale of genomic data have added to the challenges of tracing the mutations/variants and their relationship to infection severity (if any).
Results
We explored the suitability of virus-genotype guided machine-learning in infection prognosis and identification of features/mutations-of-interest. Total 199,519 outcome-traced genomes, representing 45,625 nucleotide-mutations, were employed. Among these, post data-cleaning, Low and High severity genomes were classified using an integrated model (employing virus genotype, epitopic-influence and patient-age) with consistently high ROC-AUC (Asia:0.97 ± 0.01, Europe:0.94 ± 0.01, N.America:0.92 ± 0.02, Africa:0.94 ± 0.07, S.America:0.93 ± 03). Although virus-genotype alone could enable high predictivity (0.97 ± 0.01, 0.89 ± 0.02, 0.86 ± 0.04, 0.95 ± 0.06, 0.9 ± 0.04), the performance was not found to be consistent and the models for a few geographies displayed significant improvement in predictivity when the influence of age and/or epitope was incorporated with virus-genotype (Wilcoxon p_BH < 0.05). Neither age or epitopic-influence or clade information could out-perform the integrated features. A sparse model (6 features), developed using patient-age and epitopic-influence of the mutations, performed reasonably well (>0.87 ± 0.03, 0.91 ± 0.01, 0.87 ± 0.03, 0.84 ± 0.08, 0.89 ± 0.05). High-performance models were employed for inferring the important mutations-of-interest using Shapley Additive exPlanations (SHAP). The changes in HLA interactions of the mutated epitopes of reference SARS-CoV-2 were then subsequently probed. Notably, we also describe the significance of a ‘temporal-modeling approach’ to benchmark the models linked with continuously evolving pathogens. We conclude that while machine learning can play a vital role in identifying relevant mutations and factors driving the severity, caution should be exercised in using the genotypic signatures for predictive prognosis.
Introduction
Continuous evolution of SARS-CoV-2 and emergence of virulent variants have burdened the global healthcare system at unprecedented levels. With more than 485 million cumulative reported cases and over 6 million causalities (worldwide), Covid-19 continues to challenge the adequacy of global healthcare infrastructure (https://covid19.who.int/, accessed 1 April 2022). This has been further complicated by the lack of knowledge pertaining to the factors driving the severity of the SARS-CoV-2 infection. Previous attempts have indicated variable success in predicting the infection prognosis using machine learning and deep learning (interpretable as well as black-box) methods based on the symptom profile, co-morbidities, blood biomarkers, chromosomal-scale length variation, epitope profiling of infected individuals.1, 2, 3, 4, 5, 6 Such efforts are important as they lay the ground for a much-needed thought towards predictive prognosis which may aid in mitigating the potential burden on healthcare system. Mutations in the SARS-CoV-2 genome have a link to the Covid-19 virulence. While the severity of an infection is rightly attributed to host immunity, it is well founded that certain variants of concern (VoCs) are more infectious owing to their mutational peculiarity.7 Identification of the key mutations, their functional relevance or physiological consequence (infection severity) and emergence of concerning variants of SARS-CoV-2 has therefore become one of the major goals of global genome sequencing efforts.8 The latter has been exceptional in the entire history of infectious diseases as close to 10 million genome sequences have already been deposited to public repositories like Global initiative on sharing all influenza data (GISAID) (https://www.gisaid.org/, accessed 1 April 2022). The traceability of health status of sequencing sample donor is also appreciable, which is reflected in the large cohort of more than 200,000 such samples (and corresponding sequence data) deposited globally with GISAID alone (https://www.epicov.org/epi3/, accessed 1 April 2022). Given the large scale of such ‘labelled’ datasets, an ample ground for obtaining clinical intelligence by employing biology-informed data science methods is eminent.9, 10 Supervised machine learning approaches can potentially learn important mutation signatures from these labelled sequences of SARS-CoV-2 genomes and guide prediction of infection severity based on observed mutation signatures.11 Concerted efforts are therefore required to utilize not only the existing methods rooted in biology (e.g., employing the symptom profile, family history, genetic predisposition, sequence analysis, phylogenetics, structural biology, etc.), but also apply unconventional data driven approaches that have conventionally and consistently been proven to yield actionable intelligence in a domain agnostic fashion.12, 13, 14 As rightly quoted in a news piece published in Nature last year, “scientists can spot mutations faster than they can make sense of them”.15 This situation has only aggravated further with identification of hundreds of thousands of unique mutations in over 9 million SARS-CoV-2 genome sequences shared by researchers from across the globe through GISAID as on 1 April, 2022.16
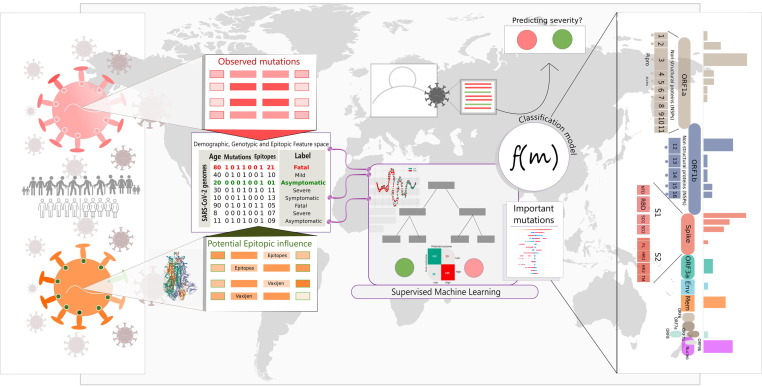
Like humans, machines or computers can learn from experience. For machines, this experience is derived from the data, which could be labelled or unlabeled. While labelled data refers to the data which is well annotated (e.g., blood biochemistry of diseased and healthy individuals), unlabeled data refers to a data without any ancillary information (e.g., blood biochemistry of unknown samples). These two available forms of data drive the two important types of machine learning approaches, namely, unsupervised and supervised machine learning methods.17 Unsupervised algorithms, like Principal Component Analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE), aim to decipher unobserved patterns in the unlabeled data and potentially group the input data points based on patterns of similarity. On the other hand, supervised algorithms, like decision trees and logistic regression, are built on an assumption that there exists a relationship between the input data and their labels, and are therefore aimed at inferring the said relationship. The latter class of machine learning algorithms are therefore cornerstone of predictive analytics and through this article we intend to highlight the possibilities and bottlenecks of predictive prognosis of Covid-19 infection by exploiting the large scale ‘labelled genome sequence’ data. Importantly, we highlight the applicability of a now emerging facet of machine learning – ‘explainable machine learning’18, 19, 20 in identifying the mutations of interest, which can significantly aid the global efforts in understanding the molecular evolution of SARS-CoV-2. Figure 1 provides a graphical summary of the underlying idea of (machine) learning the labelled genome sequence (and mutation) data of SARS-CoV-2, developing severity predictor(s) and using explainable machine learning to identify mutations of interest.
Figure 1.
Exploring utility of machine learning in capturing the mutation signatures of SARS-CoV-2 for predicting severity outcomes. Mutation profile in a given SARS-CoV-2 genome is an important feature that may drive the course of infection. This can be engineered into derived features like epitope load created by the features. A numeric encoded (presence-absence) matrix of observed mutations along with patient age/gender/geography information for each genome can serve as an input data for machine learning (ML) methods. Supervised machine learning may therefore potentially enable prediction of infection severity by analyzing the patterns of important mutations in the large number of sequenced genomes and in the process, particularly through explainable machine learning techniques, enable identification of key features including mutations that drive the prediction.
However, caution must be exercised in reporting the accuracies and clinical applicability of predictive models, especially where model features (mutations or symptoms) are not expected to exhibit a temporally stable profile.11, 13, 14 Current approaches, in addition to over speculating the goals of predictive model development, under-utilize the large label space for infection outcomes.11 While the former leads to over-ambitious speculation on clinical applicability of machine learnt models (trained using reported mutations or symptoms in the past) in predicting infection severity; the latter (under-utilized label space) under-estimates the span of significant mutations of concern. It is therefore prudent to acknowledge the limitations of a predictive prognosis exercise while trusting its ability to guide the prediction goals by identifying the mutations of interests from reported severity labelled genomes of SARS-CoV-2.
The main goals of our present work are (i) exploring the utility of machine learning towards viral-genotype based predictive model development for infection prognosis, using the patient health status labelled genome sequences of SARS-CoV-2 and (ii) contributing fundamental knowledge towards the utility of machine learning in deciphering the latent molecular signatures and other factors driving the severity. Notably, these goals, rooted in machine learning, are different from probing the hypothesis tested association of viral clades or individual mutations to different severity outcomes. The latter (class comparison through hypothesis testing) is fundamentally different from class prediction (through machine learning) but has previously yielded interesting insights into the relationship of infection severity with the genotype of evolving virus (captured in the phylogenetic clades).21 Corroborating the previous observations across geography/country specific viral clades and their potential severity association (e.g. GR clade by GISAID clade definition being predominantly observed in low severity cases and GK clade in low severity cases of Europe and North America), through Pearson’s Chi-squared test of independence for observed distributions (Supplementary Table 1,2), we try to dig deeper by probing for the ability to predict the infection severity using supervised machine learning and, in the process infer mutations and features of interest (including predictive contribution of clades).
We adopted a graduated approach of building multiple machine learning classifiers to gauge the predictive power of SARS-CoV-2 mutation signatures towards prognosis of a Covid-19 infection. It involved an initial (data cleaning) exercise of manually curating the patient health status into incremental severity levels namely, (i) Asymptomatic (ii) Mild (iii) Moderate (iv) Severe and (v) Fatal. These were grouped into two primary classes of Low and High severity genomes as well. The entire mutational landscape of the genomes in the cleaned data was analyzed for quantifying its potential epitopic influence (e.g. epitopic load and VaxiJen score,22 as detailed in the Methods section). Patient age, viral genotype and epitopic influence informed integrated feature space based binary classifiers (models) for Low and High severity health statuses, were able to correctly recall genome sequences causing high severity symptoms with greater than 0.92 ROC AUC in all cross folds across all major geographical regions of the world. Same was observed for an age informed genotypic model. Notably, age or epitopic influence alone couldn’t yield models as robust as the combined feature space of age/epitopic influence and genotype together. Furthermore, across all geographies (except Africa with sample insufficiency), the Asymptomatic vs Fatal binary classifier using integrated feature space was able to consistently classify the target classes with > 0.94 ROC AUC across all cross folds. The observations of model performances were gender agnostic. An interesting observation however pertained to the ability of models, developed using only age and five quantified metrics for potential ‘epitopic and potential antigenicity’ influence of mutations (Methods section 2.1), to predict severity with good accuracy/ROC AUC (>0.8 ROC AUC) as described later. This, we opine, opens possibilities for tracing severity outcome by transforming the entire mutational space of the genomes (at any time point in evolution) into potential metrics of antigenicity/epitopic consequence (derived from total epitopic load and VaxiJen scores in our research).
The important mutations were inferred from the high accuracy genotype incorporated models using SHapley Additive exPlanations (SHAP), a concept adopted from coalitional game-theory but frequently being employed for interpretable machine learning.20 In order to arrive at mutations of interest among these machine-learnt mutations, analysis of (statistically significant) influence on HLA interactions was probed.3, 23 In addition, these were surveyed against the literature evidences pertaining to mutations observed in variants of concern (VoCs). Many of the identified mutations of interest from the machine learning exercise were observed to have significant impact on epitopic load and consequent interactions with population specific HLA alleles. An additional temporal modeling exercise benchmarked the suitability of non-temporal validation strategies which are currently being adopted to report mutation based predictive prognosis (ML based) methods. We argue that while non-temporal machine learning methods are well adapt for identifying the mutation signatures, their applicability for predictive prognosis, when using genotype (which can potentially change with the evolving virus) should be cautiously reported (and adopted).
Methods
Mutation profiles
A total of 199519SARS-CoV-2 sequences labeled with patient status information were obtained from Global Initiative on Sharing Avian Influenza Data (GISAID). Details of downloaded genomes are provided in acknowledgement section (as per GISAID data sharing policy). The complete genome sequence of coronavirus-2 isolate (Wuhan-Hu-1) corresponding to NCBI Genbank accession NC_045512 (GISAID ID EPI_ISL_402125) was employed as the reference (REF.fa) for the purpose of mutation profiling. Fasta files corresponding to each of the downloaded individual genomes (INPUT.fa) were mapped on the reference genome using minimap224 with the following flags:
minimap2 --cs -cx asm5 INPUT.fa REF.fa > OUT.paf
The generated PAF (pairwise alignment format) files were subsequently used for variant calling through the paftools.js module in minimap2 package using the following command in a Linux environment:
Sort -k6,6 -k8,8n OUT.paf | paftools.js call -l 200 -L 200 -q 30 -f REF.fa > input.vcf
Amino acid changes corresponding to the identified nucleotide variations were predicted using BCFtools/csq program.25 In total, 45,625 unique nucleotide mutations were identified in 199,519 high quality genome sequences downloaded from GISAID (fulfilling the high coverage, complete sequence and low coverage exclusion criteria of GISAID).
The mutation vector for all the genomes was numeric encoded to create a 199519 × 45,625 matrix of nucleotide mutation data. This matrix was processed using a prevalence filter to trim the mutations that occur in less than 10 genomes across the entire data. This resulted in a 198935 × 14885 matrix. Subsequently, in order to capture the contextual neighborhood (e.g., codon context) and functional consequence (e.g., synonymous, missense, UTR, stop codon, etc.) we transformed the entire dimension of mutations into N-grams (monograms, bi-grams and tri-grams) along with the annotation of functional consequence (e.g., TTTTTGGGTG21980T_inframe_deletion_Spike, G28280C_A28281T_T28282A_missense_Nucleocapsid, G28881A_G28882A_missense_Nucleocapsid, etc.). This resulted in a mutation consequence and context informed space of corresponding genomes, represented in the 198935 × 15313 matrix of mutation data, post which genomes were filtered based on the label quality (described below).
Data cleaning and choice of target outcomes for prediction
Based on the goal of predicting Covid-19 severity, we sought to initially identify unambiguous patient health status labels among the ∼ 200,000 genome sequences. This was intended to ensure definitive assessment of severity levels without noise which would be important for developing reliable models (avoiding the ‘garbage in, garbage out’). Consequently, we ignored genomes pertaining to ambiguous labels (like Hospitalized, Inpatient, Outpatient, Released, etc.), as they do not provide conclusive indication of health status. Our stringent criteria of data selection ensured retention of 30,556 genomes after removing ambiguous or noisy labels for the study. Consequently, the mutational data was filtered down to a matrix of size 30556 × 12477. Since it was crucial to perform training on a consistent set of genomes for statistical comparability of performance of different models, we further omitted the genomes without age information, thereby resulting in a 21301 × 12117 matrix. Age imputation for missing data points was avoided to ensure confidence in model training.
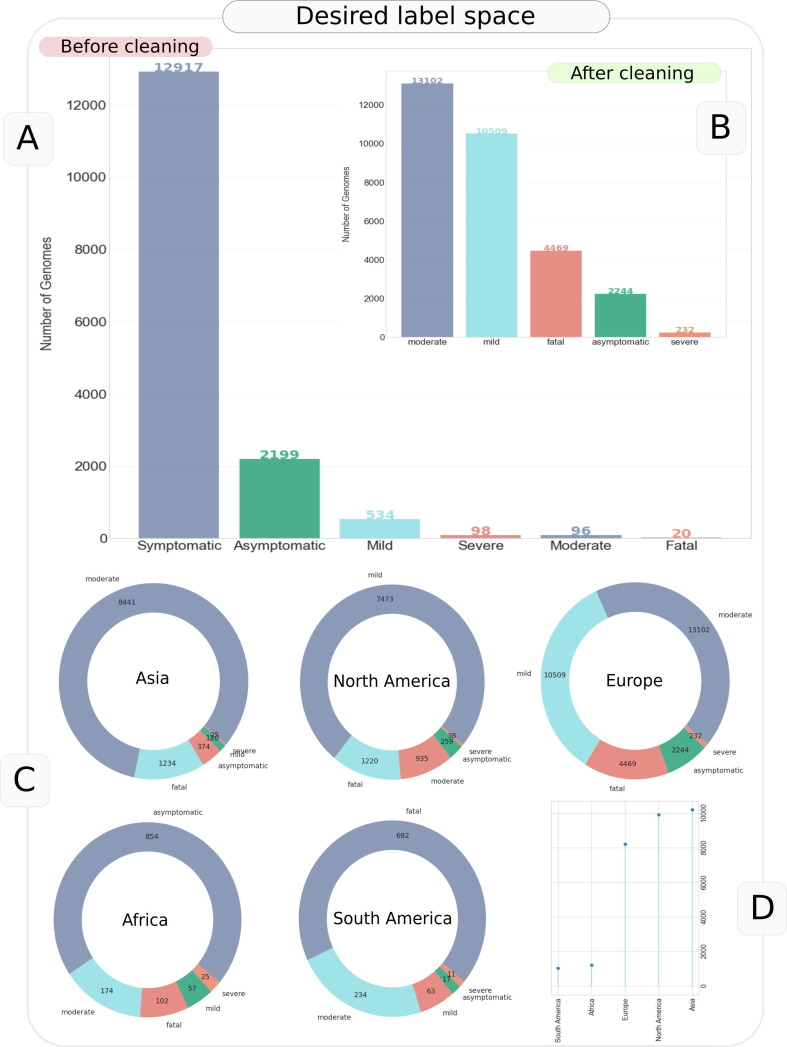
Given our goal to assess prediction of incremental severity levels, SARS-CoV-2 genome sequences pertaining to 5 unambiguous target outcomes namely, Asymptomatic, Mild, Moderate, Severe and Fatal, were stratified for the study. These chosen genomes were then segregated into Low and High severity categories based on the reported health status (e.g. status pertaining to mild or asymptomatic or symptomatic, moderate symptom associated genomes was tagged as Low severity while those leading to fatality or very severe symptoms were all labelled as High severity genomes). It may be noted that ‘Symptomatic’ label was the only apparently ambiguous class of sample that was employed (in later phases of study, post data analysis). This was done after observing a high classification accuracy between Symptomatic class and the unambiguous labels (especially Asymptomatic, Mild and Fatal outcomes), hinting towards a potential employability of this class as Moderate outcome (a label which has not been used often in the patient status data). We however report results by omitting the genomes labelled with ‘Symptomatic’ status as well. This led to the total target space of incremental severity prediction to five labels or disease outcomes: (I) Asymptomatic (II) Mild (III) Moderate (IV) Severe and (V) Fatal (Figure 2 ). Caution however must be exercised in conclusive interpretation of ‘Symptomatic’ label as moderate, and it is recommended that unambiguous labeling be preferred over ambiguous labels.
Figure 2.
Summary of the label cleaning exercise and the size of desired label space for SARS-CoV-2 genomes. The figure represents statistics before trimming the genomes that lacked patient-age information.
Choice of machine learning strategy
Unsupervised machine learning exercise
In addition to the primary goal of exploring supervised machine learning based mutation inference and predictive prognosis, a preliminary unsupervised learning of segmentation between genome groups (based on associated disease outcome) was attempted using the non-linear t-Distributed Stochastic Neighbor Embedding (t-SNE), Principal Component Analysis (PCA) and Uniform Manifold Approximation and Projection (UMAP).26, 27 The three methods were chosen to explore the consistency in observed patterns (if any). This was done for all datatypes (as described later) from all geographical regions for which quality filtered and unambiguously labelled genomes were available (i.e., Europe, North America, Asia, Africa and South America) to minimize any geography driven confounding effect. Separate analysis was also performed for the available gender information of the patients to account for the effect of gender as a confounder. The purpose of this exercise was to explore and visualize the large number of genomes and to obtain an initial intuition for the role of mutation signatures in segregating genomes in the space of reduced dimensions. All three methods were adopted from the implementations available in the yellow brick library of python (Supplementary File 1).
Supervised machine learning exercise
Given the non-linear nature of the label encoded mutation data, we chose decision tree learning approach for the machine learnt model development and used the well-founded highly efficient gradient boosted tree system of XGboost algorithm.28 The choice of XGboost algorithm, apart from its efficiency, flexibility and portability, is also rooted in the optimized and fast integration of Shapley20 value assessment for feature importance extraction from the gradient boosted models of XgBoost (https://github.com/slundberg/shap/). The latter, as introduced later, is critical for inferring important mutations in order to guide subsequent identification of mutations of interest, which is a key goal of this study. It may however be noted that a preliminary exercise of testing the performance of different machine learning algorithms, including random forest, support vector classifier, logistic regression, chain classifier and AdaBoost was also performed. However the performance was comparable XgBoost which is highly time efficient for SHAP coupled analysis. The gain of performance through hyper-parameter tuning was also monitored and was observed to be comparable to default XgBoost parameters. The results of the trials of different algorithms and tuning approaches have not been included in this report for brevity. Notably, as performed for unsupervised machine learning, all models of supervised machine learning approach were developed independently for various geographical regions and gender groups to avoid confounder bias arising due to these underlying differentiators of the patients.
Having chosen XgBoost for machine learning, it was prudent to adopt the classification strategy. Given the binary (Low and High severity) and multi-class (five incremental outcomes) nature of patient-status labels, we adopted two approaches for developing unified model(s) to probe predictive power for disease outcomes:
-
a)
Binary classification of non severe and severe outcomes
The initial broad categorization of data into Low and High severity outcomes was rationally processed through a binary classification routine using XgBoost. The single model developed using this data therefore aimed at predicting a non-specific severity level by scanning the input feature space associated with the patients (virus genotype, age, epitopic influence, etc.).
-
b)
Multi-classification using the One-vs-One and One-vs-Rest approach
We employed the two commonly adopted strategies for arriving at multi-class predictors of five chosen target outcomes.29 The first strategy used One-vs-One (OVO) approach, wherein discriminant functions were developed for all possible binary combinations of classes (n(n − 1)/2 or 5(5–1)/2 = 10 models in present case for each datatype (i.e., age/genotype/integrated etc.), as well for each confounder (i.e., geography/gender). On the other hand, in the second strategy of utilizing One-vs-Rest (OVR) approach, discriminant functions were developed for each individual class by treating rest of the data as opposing single-class of samples (n or 5 models in the present study for each datatype/confounder). Both these approaches aimed at developing a single model for predicting one among all the target incremental classes by ensembling the underlying binary predictors.
The datatypes employed for each exercise of model development (across geographies/genders) were of three types (i) patient age (ii) virus genotype (mutation profile) and (iii) five metrics of potential antigenicity or epitopic influence of mutations (details in the next section – 2.3.3). Each of these data types were employed for supervised machine learning in three settings (i) Independent (ii) Coupled (iii) Integrated. The independent setting employed only one datatype for developing the predictive models. Coupled setting employed a pair of datatypes (e.g., patient age and virus-genotype) and integrated setting employed all datatypes for model development. Additionally, GISAID clade information of the genomes was also employed for probing utility in severity prediction through machine learnt model development. Clade information was not integrated into genotype data in order to avoid bias arising out of already embedded mutational information in clade definitions. It was however coupled with age to probe predictive-performance changes. The statistical significance of the differences observed in model performances was computed for each model pair.
Engineering a reduced functional feature space from entire mutational dimension
Given the plurality of mutations fed to the genotype driven models, it may be expected that the resultant model, even if efficient, may or may not be simple and general enough for capturing signatures of severity. This is particularly true for mutations which may appear as a result of the evolution and which were never captured during training. It is therefore important to use or engineer features which are more functional and less structural or compositional in nature. To this end, we attempted to reduce the entire mutational dimension of the labelled genomes into their epitopic influence and tried to compute potential metrics of functional consequence of the mutations in each genome. The underlying assumptions, concept and methodology of the approach is as follows:
Viral epitopes that are crucial for immune response against SARS-CoV-2 can be altered by mutations causing changes in their amino acid signature. These changes may lead to an alteration in their antigenicity which can ultimately affect the immune response against the virus. Such alterations were examined by mapping mutation profile of SARS-CoV-2 genomes to a set of epitopes predicted using worldwide pool of HLA alleles. 487 predicted T-cell epitopes (from reference SARS-CoV2 genome) were obtained from the study by Bose et al.23 Mis-sense mutations observed in the ∼ 30000 genomes obtained from GISAID were mapped with these epitopes to gauge changes, if any, in the said epitopic regions due to the observed mutations in all these genomes. The reference epitopes as well as the peptides (potential variants of reference epitopes), obtained as a result of the mutations, were subjected to prediction of their potential antigenicity (indexed by the VaxiJen score) using VaxiJen server (https://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html). The peptides with length less than eight amino acids were not able to generate a VaxiJen score and hence were not considered. Difference between the VaxiJen score for each (mutated) peptide as well as the corresponding reference epitope was computed. Total epitopes affected by the mutations present in each genome (Epitopes_n), number of epitopes with a potential increase in antigenicity (i.e. increased VaxiJen score with respect to the reference, Epitopes_pos), number of epitopes with a potential decrease in antigenicity (i.e., reduced VaxiJen score as compared to the reference, Epitopes_neg) and cumulative excess Vaxijen score of Epitopes_pos (Pos_Vax) as well as cumulative reduced VaxiJen score of Epitopes_neg (Neg_vax), were calculated for each genome. This derived feature space (Epitopes_n, Epitopes_pos, Epitopes_neg, Pos_Vax and Neg_Vax) was subsequently employed to train models in various settings as described previously. Supplementary Table 3 represents a sample of the total employable feature space for machine learning exercise.
Training and evaluation
Throughout the study, it was ensured that sample size distribution was equated to the size of minority class population during the model development process in order to avoid unfair learning due to bias arising from skewed sample sizes (unbalanced classes). The data with equal proportion of all classes was split into training and testing sets using stratified splitting into 80:20 proportion. In other words, while models were built using 80% of the data, testing of models were performed based on the remaining 20% held out testing set. A stratified 10-fold cross validation was also performed for each model (using the 80% training data) to evaluate the model performance and to ensure that models are not overfitted. Accuracy (average accuracy for cross validation), Precision, Recall, ROC AUC, F1-score and the confusion matrix were assessed to evaluate the models in terms of quantifiable metrics. Classification reports were generated for each of the model consisting of important features (mutations) contributing to model accuracy, confusion matrix, precision-recall-f1 report for each outcome and AUC ROC plot. Each individual model’s mean ROC value was subject to one sample Wilcoxon test to test the null hypothesis (μ = 0.5 ROC) that there is no discriminative power in the models whose performance is reported. BH correction was employed for adjusting the observed p-values given the multiple-testing.
Comparison of models
As there were three primary feature types (age, virus-genotype and epitopic influence), it was prudent to note the statistical significance of model performance enhancement (or depreciation) in presence or absence of one or more of the feature types with respect to each other. Towards this goal, provisions for Wilcoxon signed rank test were enabled (through retention of state of the function calls for data splits) while performing stratified k-fold cross-validation of every model in the study. Wilcoxon signed rank addresses the violation of independence between individual observations of model performances during cross-validation. Since a paired t-test on cross-validation performance of two models is known to have high type 1 error but avoids type 2 error, it was prudent to use it (as well) for testing the hypothesis that there is no difference between models being compared.30 For both the testing strategies, Benjamini-Hochberg (BH) procedure was employed for correcting the p-values. The latter, notably, corrects for type 1 error.
Identifying mutations that guide the prediction
Inferring mutations of interest first requires identification of important mutations (features) that contribute towards the outcome of the model. For this purpose, manually developed (outside the one-vs-one framework of sci-kit learn library of python) individual binary models/classifiers (using genotype and age informed genotype feature space) for all possible pairs of disease outcomes were employed. We employed a three-step strategy to identify important mutations for each of the models. The first strategy included creation of a union of model reported important mutations from each iteration of 10-fold cross-validation, in which multiple models were developed across 10 iterations. A union of mutations with non-zero model linked importance helped in identifying a significantly smaller but important set of features that control the predictive capability of the final model. Once a sparse set of mutations were identified, in the second strategy, Shapley20 values for each of the features were computed. The concept of Shapley values is originally from coalitional game theory for optimal distribution of game-payout to the team players.20 However, this concept has grown popular in the domain of machine learning for assigning outcome contributions to the constituting features (players) of the model towards a given prediction (model payout).18, 19, 20, 31, 32 Shapley values > 0 were therefore used for identifying the features (mutations) contributing to the positive outcome (High in case of Low vs High severity prediction) and values < 0 were used to get important features contributing to the negative outcome (Low in case of Low vs High). Subsequently, all SHAP values of the model linked mutations were plotted using a Bee-swarm plot for visual inspection of the contribution of each mutation to the disease outcome. Among these, top 20 mutations from each model were retained for performing a union with mutations from all trained models. Given that recognition of epitopes by the Human Leukocyte Antigen (HLA) system plays a vital role in T-cell immune response against pathogens, potential epitope variants (peptides) resulting from the union of mutations were selected for assessing the consequent change in immune recognition ability of HLA alleles to recognize SARS-CoV2. For this, all potential variants of SARS-CoV2 epitopes corresponding to MHC class I (CD8) and MHC class II (CD4) were subjected to binding affinity prediction with the set of 342 most frequent HLAs present in worldwide populations.23 Epitope prediction tools NetMHCpan 4.1a (https://www.cbs.dtu.dk/services/NetMHCpan/index_4.1a.php) and NetMHCIIpan 4.0 (https://www.cbs.dtu.dk/services/NetMHCIIpan/) were used for MHC I and MHC II variants respectively to obtain the prediction score of these peptides with the set of target HLA alleles. Default parameters of the tools were used for this process. Peptides with high prediction score (>0.95) were retained and identified as variants of reference epitopes (VREs).
To get estimates of allele frequencies of each geography, geography-wise cumulative allele frequencies of the chosen 342 alleles were computed by taking population-wise frequencies of the alleles from the study by Bose T. et al [Supplementary Table 5 of Bose T. et al].23 The data depicted in the cited study was obtained by combining frequency data from the Allele Frequency Net Database and the 1000 Genomes Project.33 Weighted allele frequencies for the four geographies of interest (Asia, Europe, Africa, North America) were calculated by summing up the product of frequencies of each allele in a population (belonging to the respective geographical region of interest) with the total number of samples present in that population. Resulting value was then divided by the sum of samples present in each population of a geographical region to obtain the final cumulative frequency for each geography. Combined frequency of reference and variant alleles was then computed by combining the cumulative frequencies of the alleles of each geography that were able to recognize the listed epitopes or VREs. At every step of mutation set filtration (from creating union till finding high affinity VRE causing mutations) the cumulated sum of the occurrence of each mutation, in each geography (as well across the globe), in low and high severity causing genomes, was counted. This distribution of frequencies was subjected to Pearson’s chi-square test for significance of observed association of distributions. We also subsequently performed a genome wide association study using treeWAS34 on training datasets with ordered mutational profile for genomes of individual geographies employed for Low/High severity prediction. This was intended for performing an unbiased search for mining significant associations between the Low/High severity outcome associated with each genome and its genotype. Notably though, the goal of this exercise was to report common signatures picked by ML exercise and GWAS exercise. Neither of these are intended to support or oppose the fundamentally different methodologies rooted in the said exercises. It is therefore prudent to appreciate that statistical significance doesn’t necessarily imply predictivity and vice versa. Features which are observed to be statistically significant may or may not have high predictive value and features perfected by the ML methods (e.g., rigorous gradient boosting routines of XGBoost) for making a reliable prediction may not necessarily pass the standard measurements and thresholds of p-values of hypothesis testing. Additionally, feature tables and GFFs for each individual protein in the reference SARS-CoV-2 genome (https://www.ncbi.nlm.nih.gov/nuccore/NC_045512) were downloaded and curated to create a map of the mutation loci against the genomic region specific to the protein(s) for the mutations of interest. This ensured that the fundamental information pertaining to the genomic/ structural context of the identified mutations is available. This was followed by the literature mining of the SARS-CoV-2 case reports as well as mutations observed in variants of concern (VoCs).
Temporal validation
As discussed previously, a reliable model for predictive prognosis should ideally be benchmarked on unobserved ‘chronologically recent’ data which was not included in the training of the model. This would validate the suitability of proposed models for clinical implementation wherein the viral genome is expected to continuously evolve and accumulate new mutations. Given that it is well founded that SARS-CoV-2 has been evolving with time, an unconditional applicability of models learnt on past data (mutation profiles) must not be assumed.
We therefore devised a chronological data sampling technique with incrementally increasing time windows to test models trained on historical data against a held out unobserved data from a future time-period. For this purpose, entire data (specific to the target outcomes) was first sorted according to the date of collection of samples (only those samples were selected for which complete date of collection was available – including the day, month and year of collection) and multiple held-out test-datasets were created using the chronologically recent subset of data windows. The incremental time window approach was used to create the future test data for observing the effect of time-gap on model performance (where time gap refers to the time-duration between the sample collection day of latest data record used in the training data and the oldest data record of test data). Increase in time gap was approximated by reducing the number of old samples (close to the date of collection of the most recent training sample) in the test data without changing the size of training data. Each test window was however kept the same size as previous window. It was important not to change the size and content of the training data to ensure that variations in the performance of model are only time driven (and not training data or size driven). Given the high performance observed for Asymptomatic and Fatal outcomes in non-chronological data sampling approach, binary model specific to Asymptomatic-Fatal combination was employed for temporal validation across Asia, Europe and North America (Africa and South America were not considered due to sample insufficiency). Accuracy and ROC AUC values for each time-gap based model development exercise were compared for assessing the importance of time as a confounding factor in developing accurate models of predicting the prognosis of SARS-CoV-2 infection using mutation signatures. The data splits were created to ensure that three held out test-datasets, each of ∼ 50% the size of training data each were created. Each test data had a greater average time gap with respect to the most recent sample in the training dataset.
Databases, tools and implementation
Supplementary Table 4 provides the details of various key resources including databases, tools and packages employed in this study. Additionally, in an attempt to conform with TRIPOD guidelines, the workflow employed has been provided in the Supplementary File 1. This can seamlessly be plugged to any machine learning framework to enable development of customized models, updating existing models and testing thereof using a simple tabulated form of mutation lists (provided by platforms like GISAID). All machine learning related analyses were performed on an AMD Ryzen 5 laptop with 4 cores and 8 logical processors, 2.1 GHz and 8 GB RAM. The genome wide association study using TreeWAS was performed on a 20 core Xeon 51 series 2.4 GHz machine with 64 GB RAM to accommodate the memory and computational requirements. The latter was observed to fail due to resource constraints of the standard laptop used for ML exercise.
Results
Unsupervised learning provides cues to key factors discriminating the disease outcome
It was interesting to observe that while virus-genotype displayed partial signs of discriminative power (through t-SNE, PCA and UMAP based spatial distribution of genomes – Supplementary Figure 1 and 2(a-j)) towards low/high severity outcome, the discrimination ability was evidently distinct when age was integrated to the genotypic data, in a geography and gender agnostic manner (Supplementary Figure 1 and 2(a-j)). Age of patient and epitopic consequence information, when coupled to virus genotype (creating an integrated data), were consistently able to spatially segregate the genomes according to their severity association (Supplementary Figure 2(a-j)). The engineered feature space of epitopic influence alone displayed some signs of discrimination, but the same were not comparable to segregation achieved by genotype, age informed genotype, or integrated data (Supplementary Figure 2(a-j)). Interestingly, age informed epitopic influence alone (total six features) provided encouraging evidence of spatial segregation providing cues to the suitability of a transformed mutational space (especially pertaining to epitopic load or influence) when coupled to patient age for predicting severity of an infection (Supplementary Figure 2(a-j)). Clade information was observed to exhibit some power of discrimination (albeit not as evident as genotypic information), which improved when coupled with the age information of the patients (Supplementary Figure 2(a-j)). Expectedly, as evident in Supplementary Figure 3, multiclass labeled samples were segregated in a manner that genomes pertaining to fatal class were clearly distinct as compared to other severity levels (i.e., asymptomatic, mild and moderate). The genomes pertaining to severe class (sample insufficiency in each geography) were omitted in multi-class case, as our approach aimed at stratified and balanced sampling of genomes from each class of severity label.
Supervised machine learning for unified multi-class classifier shows limited success, binary models show encouraging signals of discriminative power
Mutations and their consequence indeed drive the Low-High severity outcomes
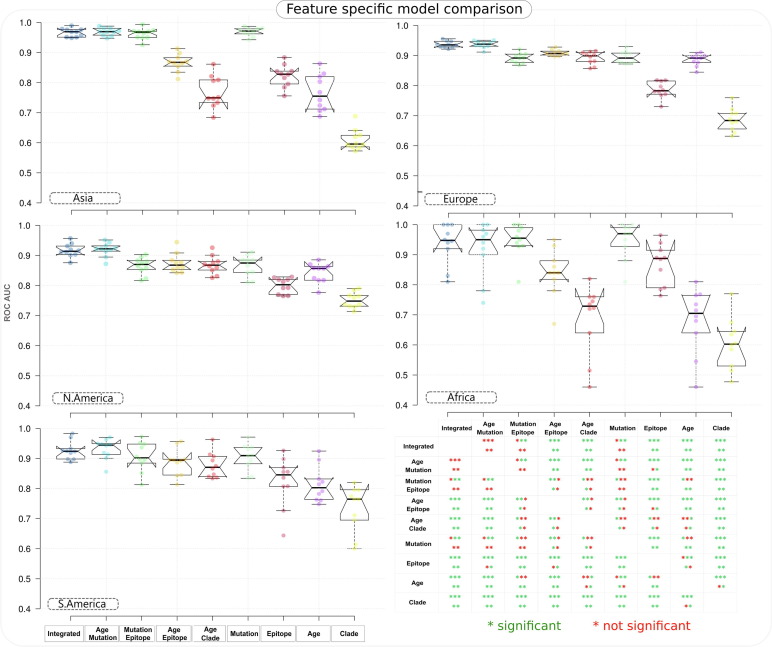
Binary models developed using SARS-CoV-2 genome sequences associated with Low and High severity of infection revealed high but differential classification accuracy across 10 fold cross-validations (0.90 ± 0.02, 0.81 ± 0.03, 0.77 ± 0.04, 0.9 ± 0.09, 0.84 ± 0.05)) and an encouraging ROC AUC 0.97 ± 0.01, 0.89 ± 0.02, 0.86 ± 0.04, 0.95 ± 0.06, 0.90 ± 0.04 using the observed virus genotype alone (mutation data) in the different geographical regions of Asia, Europe, North America, Africa and South America, respectively. Enhancement of feature space by including patient age and/or epitopic influence of the mutations marginally improved the predictive performance and consistency in high predictivity across all geographies (Figure 3 , Supplementary Table 5–7). The gain in performance was not statistically significant though (Wilcoxon paired p > 0.05, BH corrected, Supplementary Table 7). Notably, neither of the age or epitopic consequence alone could outperform the performance of an integrated or in-silo model (Wilcoxon paired p < 0.05, BH corrected) based on virus-genotype (untransformed mutational space) as summarized in Figure 3 and Supplementary Table 7. Importantly, the integrated model consistently outperformed (or performed as good as) other models across all geographies. These trends were consistent not only across geographies, but also for gender (Boxplots in Supplementary File 2,3, Supplementary Table 8,9). Clade information alone displayed better classification ability than a random classifier, however the ROC AUC and accuracy were quite lower than genotype-based models (Figure 3). Inclusion of age information with the clade improved the performance and was found to be significantly lower than the integrated model and were also seen to perform as good as genotype-alone (mutations) model for Europe and North America. A detailed comparison of the key model pairs for Low vs High severity prediction is provided in Figure 3, while Supplementary Table 7–9 provides the comparison summaries of all possible pairs of models. It may be noted that the performance of individual models for Low vs High severity prediction (Supplementary Table 5,6) was consistently observed to be statistically significantly (p < 0.05, BH corrected, Wilcoxon one sample statistic) better than a random classifier (μ = 0.5 ROC for null hypothesis) except in many cases for the graduated outcome models developed using only age (especially in Africa) and in some cases only epitopic influence as the composite feature of the models (Supplementary Table 5,6).
Figure 3.
Box plots and significance map for comparison of Low vs High severity model performance (ROC AUC value) observed in cross validation results of all feature-specific models and combinations. Bottom right panel represents the results of statistical significance test on ROC AUC values performed for each pair of models, where a green star represents rejection of null hypothesis at p < 0.05 (significant performance difference), while a red star indicates the contrary. Wilcoxon signed rank test was performed for each comparison and p-values were corrected through BH correction. Order of the stars (each indicating statistical significance of comparison in a given geography) maps with the geographical regions namely Asia, Europe, North America, Africa and South America.
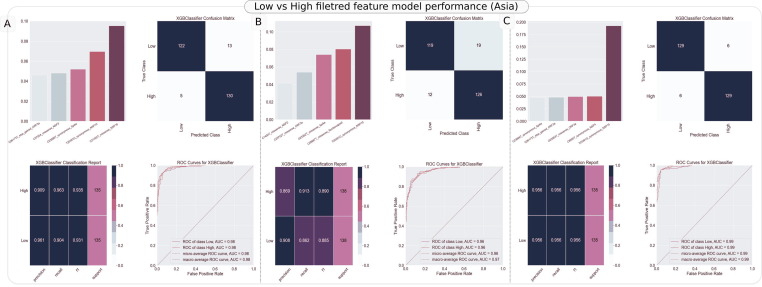
Notably, the held-out test performance (ROC AUC) of genotype-based models (e.g. integrated model) was also consistently greater than 0.93 across geographies with sample sufficiency (Asia: 0.97, Europe: 0.94 and North America: 0.93) (Supplementary Table 10). These results were observed without any mutation filtration or elimination, i.e. using the entire corpus of nucleotide mutations in each geography specific samples pertaining to low (asymptomatic, mild, moderate) and high (severe, fatal) class of severity. Results were however equally encouraging (as presented for held-out test results, as an example, in Figure 4 for Asia – Panel (A) Integrated model (B) Age informed genotype (c) Genotype alone, wherein feature space was significantly reduced from the original corpus (4172, 4167, 4166) to a reduced set (187, 182, 181), after eliminating features with consistently null importance across 10-fold cross validations.
Figure 4.
Classification reports generated for the binary model trained using Low vs High severity status linked SARS-CoV-2 genomic sequences from Asia. In each of the models, filtered set of features were employed. The report consists of important features (mutations) contributing to model accuracy (top-left), confusion matrix (top-right), precision-recall-f1 report (bottom left) for each outcome and AUC ROC plot (bottom right) in each panel. Panel A refers to the report for model developed using integrated feature space. Panel B refers to the model developed using age informed genotype model and Panel C represents classification report for model developed using virus genotype alone.
As apparent, while the accuracies and ROC AUC are indicative of good discriminative power of mutations, the high recall of > 91% for high severity cases using virus genotype linked models points towards suitability of developing mutation signature-based severity estimators for SARS-CoV-2 infection. Binary models were developed for each possible pair of graduated outcomes as well using all possible features and their combinations (Supplementary File 2,3 and Supplementary Table 11,12). Among these, while asymptomatic vs fatal model consistently yielded very high performance using a combination of ‘age, genotype and/or derived features’ (e.g. accuracy: 0.89 ± 0.04 and AUC ROC 0.96 ± 0.02 in Asia using Integrated model), models targeting ‘severe’ as one of the outcomes, especially severe vs fatal, were frequently observed to be weak learners due to consistent sample insufficiency as observed in Figure 2 (e.g. severe vs fatal, accuracy: 0.40 ± 0.13 and AUC ROC: 0.39 ± 0.19 in North America using Integrated model). Among the features, as observed for Low vs High severity model development, weakest models (no better than a random classifier) were developed if only age, only clade or only epitopic influence were considered as composite features of the models (Supplementary File 2,3 and Supplementary Table 11,12). Coupling of features like age with epitopic influence (1 + 5 features), age with mutations and all integrated feature space performed statistically significantly better than any model developed using former two features in silos (Supplementary File 2,3 and Supplementary Table 11,12). Sample of data structure for all feature types (mutations, age, epitopic influence, clade etc) is provided in Supplementary File 4. These were obtained by processing the labelled genome sequences (Supplementary File 5) downloaded from GISAID (as described in methods section).
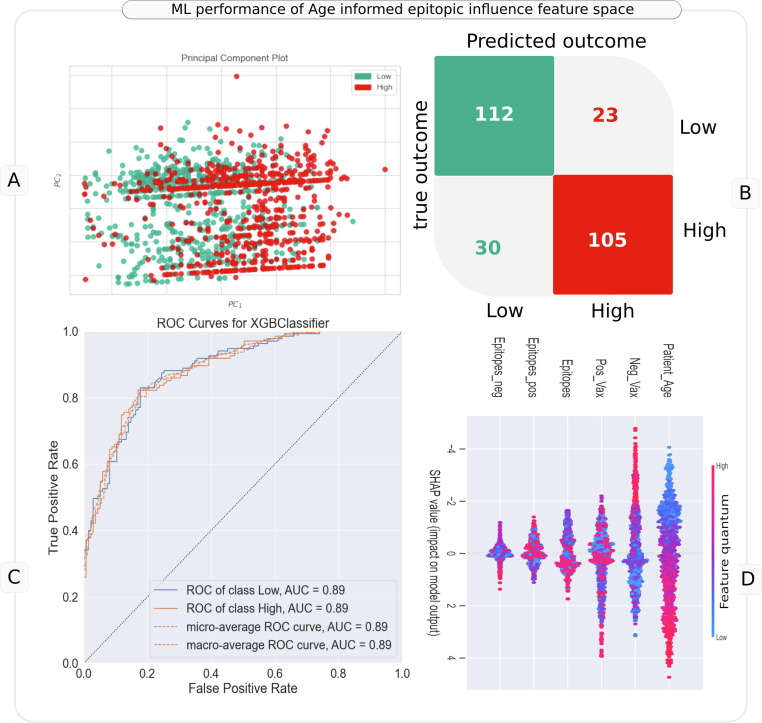
It was interesting to observe that the sparse AgeEpitope model constituted by 6 features, i.e patient age and five derived features (indicating potential antigenicity influence of each mutation) from the entire mutational landscape performed consistently well (∼0.8 AUC for Low vs High severity across geographies, Figure 3). It was therefore prudent to probe the model developed using these features. As a representative example, we discuss the results for Asia (Low vs High AUC 0.87 ± 0.03). Initial signs of discriminative power in the age informed epitopic influence feature space (total 6 features) was observed during unsupervised machine learning (t-SNE, PCA and UMAP), wherein genomes corresponding to Low and High severity were observed to be segregated across all geographies in a gender agnostic manner, exhibiting good scope for a fair decision boundary (Supplementary Figure 2(a-j), Figure 5 (A)). The classification model developed using these features corroborated the same wherein the model was observed to have an AUC of 0.87 ± 0.03 (p < 0.05, BH corrected Wilcoxon) and a decent performance (AUC: 0.89, Figure 5(B,C)) on held out test data (270 samples, 135 corresponding to each of Low and High severity class). Given the good performance of the model, interpretation of the same using SHAP values was prudent. It was observed that a higher patient age, total epitopic load (Epitopes) and very high value of cumulative positive VaxiJen score (Pos_Vax) contributed by the mutations were driving the direction of the decision towards high severity (Figure 5(D)). On the other hand, a very high value of cumulative negative VaxiJen score (Neg_Vax) for genomes was driving the decision towards low severity outcome. However, there was high interdependence of all the features in driving the overall decision of the model (as indicated by the dispersion in the SHAP values of each feature). As described in methods section, VaxiJen score serves as an index of potential antigenicity of a target peptide. We had computed the positive scores (Pos_Vax) and negative scores (Neg_Vax) by cumulating the difference between VaxiJen scores of reference and mutated epitope linked to each mutation. For example, mutated epitopes with a reduced potential antigenicity were counted as Epitopes_neg, and sum of their reduced VaxiJen scores as Neg_Vax. indexing high potential antigenicity and negative scores. Interestingly, this derived feature space (including patient age) were observed to have significantly different mean value between low and high severity linked genomes across all major geographies (Welch’s t test BH pval < 0.05, Supplementary Table 14). Africa was an exception where difference between mean Epitopes_neg, Neg_Vax and Pos_Vax was not observed to be statistically significant. Notably, given this analysis considered only the mutations that mutated reference epitopes in a non-synonymous manner, it doesn’t capture the impact of every mutation (including those in UTRs). Nevertheless, corroborating previous reports at various geographic levels, it points towards the rational utility of viewing mutations from the point of view of their immunological consequence (e.g., number of reference epitopes mutated, their immunogenicity potential, etc.). A possibility for further improving this approach of transformation of mutational space into potential epitotpic-influence dimensions, for inferring severity outcomes may be useful, in absence of comprehensive medical, genetic, and other meta-information about patients.
Figure 5.
Utility of age informed epitopic influence feature space in machine learning the severity outcomes in Asia. Panel A represents PCA plot generated using these features with overlay of Low and High severity labels. Panel B represents the confusion matrix of predictions made by the model (for a held-out test data) developed using the six features of age informed epitopic influence space. Panel C represents the ROC curve for AUC estimation. Panel D represent the bee-swarm (global SHAP value) plot for model interpretation.
Importantly, in a similar exercise of model interpretation, key mutations driving the performance of each of the models, can also be developed using genotype-based data. This can aid identification of severity specific important mutations which may be further be probed for their relevance, and are described later.
Also, it is prudent to point that given these observations pertain to random sampling of entire dataset, without consideration to the continuous evolution of the virus over time, conclusions cannot (and should not) be made without temporal benchmarking of machine learning exercise for proposing any potential models for clinical predictive prognosis (described later in Temporal validation section).
Multi-class classification, repurposed-regression and validations thereof
A single (Integrated) model developed using One-vs-Rest (OvR) approach for all five incremental severity based target classes (Asymptomatic, Mild, Moderate, Severe and Fatal) didn’t yield a high accuracy (e.g. 0.39 ± 0.16, using 10-fold cross validation for Asia). Accuracy however cannot be considered as a perfect metric for a multi-class (5 classes in this case) predictor. Specifically, this classifier was trained (100 samples) and tested (25) using 25 samples from each class (total 125 samples, as the size of each class was reduced to the minority (Severe: 25) class size in Asia) to enable an unbiased/stratified learning. Nevertheless, it was encouraging to observe that the majority prediction for each of the target outcomes was not significantly skewed (confusion matrix in Supplementary Figure 4). An ROC AUC macro-average of 0.69 ± 0.01 through 10-fold cross validation indicated a fair degree of discrimination of individual classes from rest of the data (One Vs Rest) even after having used a very small sample size, which was also indicated in the held-out testing where the model was able to separate fatal class from the rest with an ROC AUC of 0.93 (Supplementary Figure 4). ROC AUC plots in Supplementary Figure 4 were plotted using the held-out test data (20% of 125).
These results pertain to unfiltered features with no feature selection routines. Filtered features gave comparable results as well. Given the expectation that Severe class (25 samples) could be driving the sub-optimal training of otherwise large data size for each of the other target classes (greater than 100 samples each in Asia, Supplementary Figure 5), we therefore sought to ask if it was possible to improve the learning of discriminating function by omitting this minority class. The OvR classifier was therefore re-trained using only Asymptomatic, Mild, Moderate and Fatal classes (Supplementary Figure 5). Omitting Severe class, yielded significantly improved accuracy (0.60 ± 0.05) and macro-average of ROC AUC (0.82 ± 0.03). The held-out testing also yielded high ROC AUC for individual classes against rest of the samples (e.g. Fatal vs Rest ROC AUC: 0.97, Supplementary Figure 5) Model developed using One-vs-One (OvO) approach yielded comparable results (Supplementary Figure 6) on held-out data as well as 10-fold cross validation accuracy. While these results indicated a need for caution while developing an ambitious ‘single multiclass model’ to predict multiple incremental outcomes of Covid-19 severity, it was encouraging to observe latent signals of mutation peculiarity in the average ROC AUC (>0.8) from the contributing models of the unified OvR model (Supplementary Figure 5), i.e., Rest Vs Asymptomatic, Rest Vs Mild, Rest vs Moderate and particularly Rest vs Fatal.
Identification of the important mutations is crucial to trace the evolution of the virus without missing the hitherto unobserved variants through traditional exercise of variant tracing rooted in epidemiology and phylogenetics. Can biology informed machine learning potentially aid this task? We explore this further.
Identification of mutations and features of interest
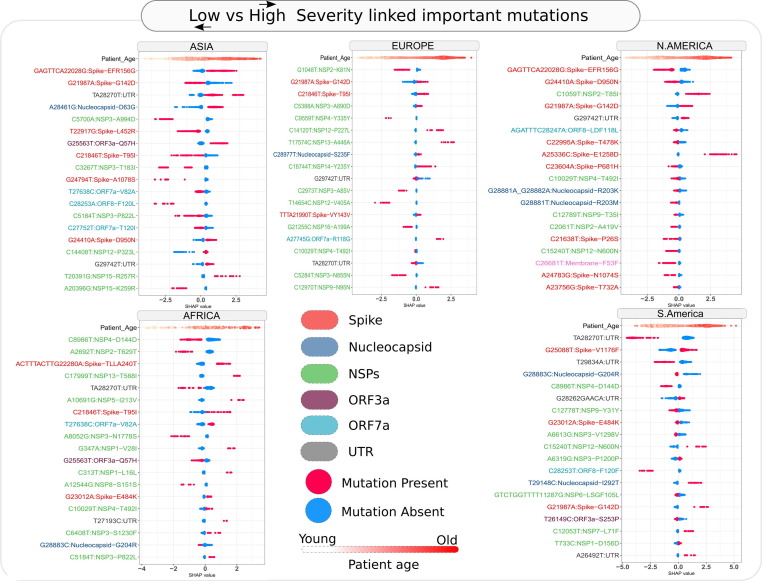
The SHAP value-based contribution inclination of each of the composite feature among top 20 for Low vs High severity models across geographies is summarized in the Bee-swarm plots of Figure 6 . The density of genomes (dots in the plot) with SHAP > 0 presents contribution of the large/small (e.g., for age) quantum of features or presence/absence (e.g., for mutations) of features, indicated by red/blue dots respectively to the higher severity outcome and SHAP < 0 indicates contribution towards lower severe outcome. Across all the geographies, a higher age was consistently observed to contribute to the prediction of high severity outcome by the model. A universally consistent pattern was however not apparent for effect of the mutations (except for few common signatures) on severity outcome in different geographies. Each geography in fact had a rather peculiar signature of top mutations contributing to the severity predictions. Consensus mutations if any were observed to have opposite effects in different geographies. This underscores the importance of probing the individual and population specific factors like variations in human leukocyte antigen (HLA) alleles that are known to affect the severity (and even susceptibility) of infections. For example, how (and if) the epitopes resulted by the mutations in the SARS-CoV-2 interact with the individual alleles may decide the outcome of the infection. For example, while a deletion in N-terminal domain (NTD) of Spike protein (GAGTTCA22028G, i.e., EFR156G) was observed to be consistently contributing to the high severity predictions in Asia, its contribution was completely but consistently reversed in North America. A missense substitution in Spike NTD, G21987A (i.e., G142D) was another such example of a mutation observed to contribute to severe outcome prediction in both Europe and North America, but the effect was reversed in Asia. Factors like co-morbidities, prior or ongoing pharmaceutical/ non-pharmaceutical interventions, environmental factors and more can also affect the way an infection would manifest in an individual, thereby mandating that the signatures and predicted severities may have scope for further improvement before they can be utilized clinically. The intelligence generated by the predictive models and the interpreted mutations, however, seek to help reduce the combinatorial complexity of a large size of mutational landscape that the scientific community is trying to decipher. In order to infer mutations of interest from these model specific corpora, we carried out an exercise of inferring the impact of the observed mutations on generation of epitopes that might interact with HLA alleles with strong binding affinity (refer Methods section 2.6).
Figure 6.
Mutations of interest selected using model importance coupled SHAP value assessment for Low vs High severity prediction across important geographies of the world. Bee-swarm plot indicates the contribution of presence/absence of a mutation towards High or Low severity outcome of XgBoost based classification model. Values greater than zero indicate contribution towards a severe outcome, while SHAP values less than zero indicate contribution to less severe outcome of the model. Additionally, the labels of the mutations have been colored according to the protein in which the said mutations appear.
We also employed treeWAS to probe whether a rigorous genome wide association analysis can reveal statistically significant signs of mutational association with severity outcomes. Signs of mutational association were indicated only in Asia and North America, while Europe, South America and Africa were not observed to have statistically significant signs of mutational association to severity outcomes (i.e., Low and High severity). Synonymous mutation C313T and codon linked mutations G28881A, G28882A and G28883C were observed to have severity implications by treeWAS (Supplementary File 6). In North America, association with low severity were attributed to A25336C, GAGTTCA22028G, G24410A_missense_Spike and AGATTTC28247A (Supplementary File 7). Mutational contribution to phylogenetic clustering of genomes according to their severity affiliation were apparent in Asia and North America.
As a similar exercise of SHAP value assessment (e.g., as exemplified in Figure 6) on all virus-genotype based models, a total of 254 mutations were obtained from the union of top 20 SHAP value based important mutations (Supplementary Table 13). Among these, a total of 76 synonymous, 13 UTR, 155 missense, 6 in-frame deletions and two stop gained mutations were present. 155 missense and 6 in-frame deletion mutations positions were considered for mapping on the epitopic regions present on the reference genome. Out of these, 44 mutations were mapped to 73 reference epitopes (REs) resulting in 74 (49 CD8 + 25 CD4) potential variants of the REs. Further, binding affinity prediction of the potential variants (with the set of 342 most frequent HLAs present in worldwide populations23 resulted in 28 mutations corresponding to 44 VREs that were able to be recognized by the chosen pool of HLA alleles (>0.95 prediction score). 16 mutations were found to cause a possible escape of the 23 potential VREs from recognition by the entire set of HLA alleles (i.e., no significant binding affinity observed) (Supplementary Table 13). We additionally mapped these 28 mutations (resulting in high binding affinity VREs) to the Low vs High age informed genotype-based models to identify 11 unique nucleotide mutations associated with either of the outcomes, potentially due to the underlying allelic interactions (in addition to host age and other unknown factors) as summarized in Supplementary Table 13.
Studies have reported that a coordinated SARS-CoV2 specific CD8 + T, CD4 + T cells and adaptive immunity response to be linked with protective immunity against the disease.35 On the other hand, a dysregulation among these can influence severe inflammatory immune response, thereby leading to organ damage. The HLA profiling of SARS-CoV2 epitopes indicated mixed outcomes in terms of association between disease severity and T cell based immune recognition ability of the individuals of each geographical region. For example, Mutation NSP3_A85Vwhich was identified as a key mutation in European region, was found to be affecting two SARS-CoV2 epitopes, both of which showed potential increase in immune recognition after mutation. Earlier Epitope 235 was identified by one HLA allele (HLA-B*18:01) which increased to 4 alleles (HLA-B*18:01, HLA-B*18:02, HLA-B*18:03, HLA-B*18:05) predicted to be identifying its variant (VRE). Epitope 329 could not be identified by the European population earlier, as HLA-B*18:02 was not found to be present in European population. However, after this mutation, it was predicted to be recognized by around 7% of the population. Disease outcome as predicted by the ML model indicated that the absence of this mutation is associated with high severity, which leads to the speculation that because of less immune recognition of the wild type/reference epitope, a compromised immune response may have been generated against the infection. A study done by Wilson et al. 36 has also indicated the possibility of inverse correlation between total population epitope load and death rates which supports this speculation.
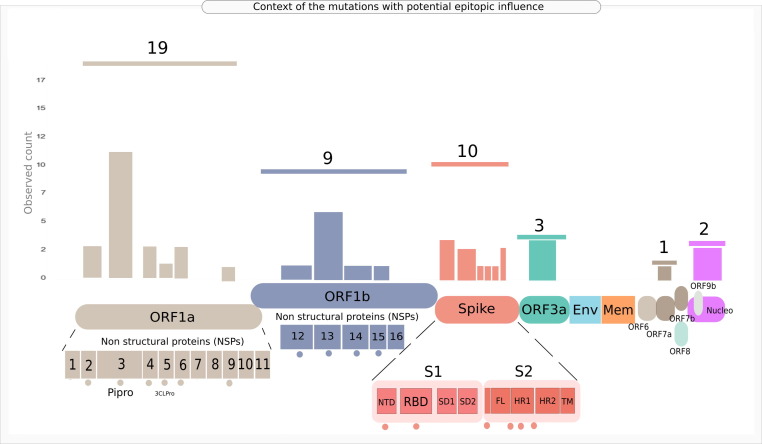
On the contrary, the mutation NSP3_A994D, affecting six epitopic regions on the SARS-CoV2 proteome, resulted in potential loss of significant (with > 0.95 prediction score) epitope-allele interaction in three VREs after the mutation. This indicated the possibility of VREs being not recognized by as many alleles as compared to the reference epitopes in the Asian population with the percentage of population potentially recognizing Epitope 113 dropping from around 21% to 0. This raises a speculation whether it potentially indicates low T-cell based immune recognition of the population after mutation. Although theoretically, the presence of this mutation could be associated with a high disease severity, the prediction model indicated the opposite. It is worth further investigation to evaluate if there is a direct connection between number of epitopes/ VREs identified by the individuals of a geographical region and the observed COVID-19 severity in that region. A few mutations, as we discussed earlier (like NSP4_T492I) were noted to show different disease outcomes with respect to different geographical regions (absence indicating low severity in Africa and same indicating high severity in North America). This observation might be because of the varied HLA signature present in the population of these two regions. While HLA-A*24:03 potentially recognizing Epitope 195 is present in 0.07% of the African population, 0.4% of North American population were found to have this allele. These speculations, notably, do not account for the immigration driven ethnicity mixtures, which should be considered for more comprehensive research (once such metadata are available). It must also be noted that in our approach of inferring mutations of interest, only one arm of immune recognition, namely, T-cell epitope recognition, has been assessed using in-silico techniques. In case of any infection, several other players of the human immune system come into play, B-cell antibodies being one of them. A conclusive statement can only be provided after analyzing the effect of these mutations on the overall human immune system validated experimentally by immune assays. Figure 7 provides the observed genomic context of the 44 genic mutations whose epitopic influence was profiled for significant HLA allelic interactions (Supplementary Table 13), indicating a high prevalence of mutations of concern in Spike, ORF1a and ORF1b regions.
Figure 7.
Group chart embedded illustration of the genomic context of the mutations of concern identified through SHAP based explainable machine learning followed by potential epitopic influence profiling. Horizontal lines above the bars indicate aggregate observed count for the mutations identified in the given protein or genic region. Individual bars pertain to the observed mutation count for the domains/regions/sites specific to the modular protein/genic region. Dots under the genic regions indicate that the bars pertain to the said sites or regions or domains in the given protein.
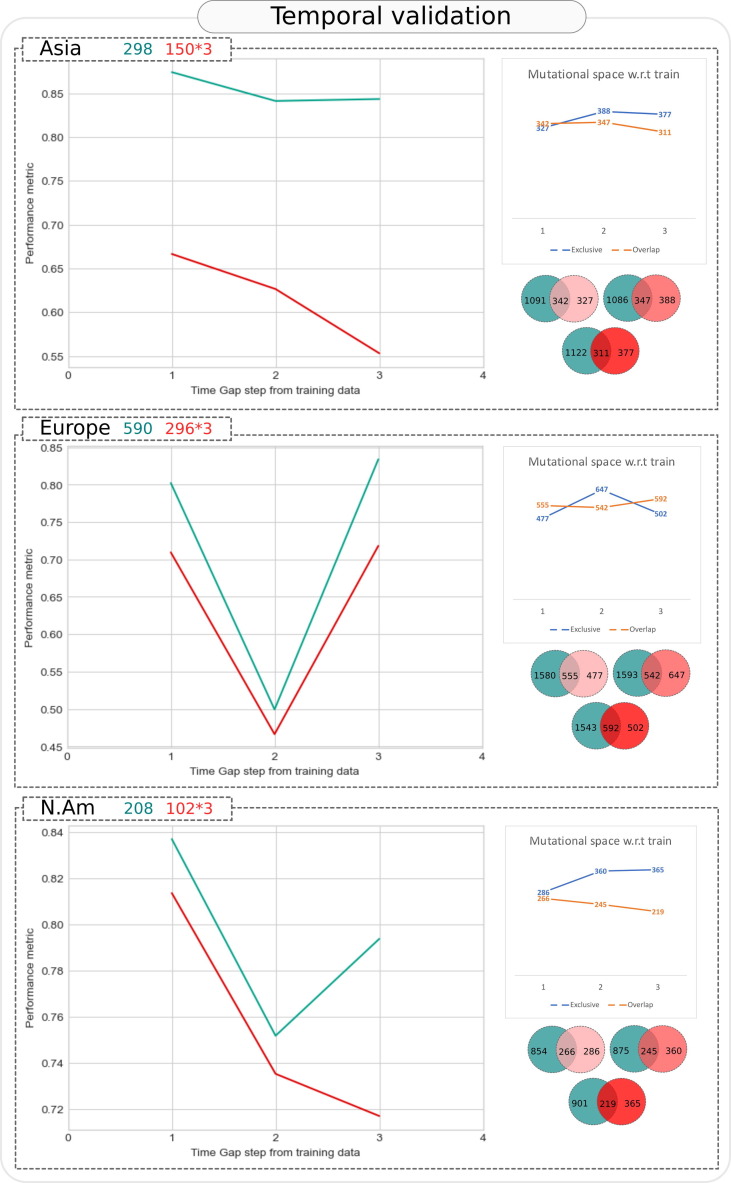
Temporal validation
The incremental time window approach of creating three chronologically recent held-out test datasets (for which complete dates of sample collection were available) revealed the rational limitation of mutation based predictive prognosis models. With increase in the time gap between the constant training data and the chronological test datasets, model performance was observed to drop for the pathogen (SARS-CoV-2) which evolved to accumulate new mutations leading to lesser overlap with the learnt mutation corpus and greater size of exclusive mutation corpus (Figure 8 ). These observations were consistent across all available geographies and gender information. For example, as shown in Figure 8, in Asia, a model trained using mutational profile of ∼ 300 SARS-CoV-2 genomes/samples and tested on chronologically distinct 3 separate windows of 150 samples each, had highest accuracy and ROC AUC in Window 1 (closest to training data records). The performance dropped in Window 2 (chronologically distant from training data). This drop may be attributed to appearance of more exclusive or new mutations (388 in window 2 as compared to 327 in window 1) and similar number of overlapping mutations (347 in window 2 as compared to 342 in window 1). Model performance worsened further in window 3 where overlapping mutations were very less (311) even though the new or exclusive mutations were slightly lesser than window 2 (377). This dependency of model performance on mutational space is further revealed in results of temporal validation for Europe where the model performance was found to recover in Window 3 due to greater overlap and smaller exclusive mutation set as compared to previous window. This leads to the below mentioned three important inferences –.
-
•
As long as the virus is mutating, it may be over-speculative to propose models rooted in mutation signature for prognosis in a clinical setting.
-
•
A judicious use of predictive models can however take place where reliability of the prediction is indexed by the fraction of mutations that are already accounted for in the model (as indicated in the Venn diagrams and grouped line plots for mutational space in Figure 8).
-
•
The role of machine learning in identifying the important mutations among the large existing corpus of SARS-CoV-2 mutations should not be ignored as this can significantly aid the ongoing activities of tracing variants of concern. Importantly, it is apparent that machine learning can yield high predictivity models when mutational space is recent. It can do so even in the absence of available information of patient symptoms (which is possible in early diagnosis or when disease hasn’t already progressed to advanced stage). The predictive prognosis exercises can in such cases turn fruitful provided models are continuously updated with available genomic records. The proposed temporal benchmarking would prove useful for such use cases.
Figure 8.
Temporal validation of Asymptomatic-Fatal predictive model. The trend of ROC AUC and accuracy at various time gap windows (as shown on X axis), wherein window 1 contained test samples which were chronologically closest to the most recent SARS-CoV-2 genome in training data, while window 3 contained most distant samples (number of samples employed in training and each test window is depicted in the header of each geography specific panel). Y axis represents the value of two performance metrices namely, ROC AUC (turquoise line) and accuracy (red line) of the model. The total number of unique (exclusive) mutations and common (overlapping) mutations in each test window, with respect to training data are plotted in grouped line charts above the Venn diagrams. Number of exclusive and overlapping mutations between training data and each tested window are also presented through the Venn diagrams.
Caveats and Conclusions
Machine learnt models are rarely perfect. The imperfection is attributed to fractional representation of information in the chosen datasets (i.e., complete data for any case/event/population is rarely available). Consequently, there is always a scope for improving the learnt models by incorporating new data to the machine learning framework. This limitation is particularly pronounced for viral genomes which are continuously evolving. New data will always be useful in updating the mutation feature profile of the models which will help in improving the accuracy of the prospective predictions. Development of well streamlined machine learning frameworks can potentially simplify the process of accommodating new data, updating the predictive models and for obtaining quick insights into the newfound mutations of concern. Through this study, we attempted to provide evidence towards suitability of using SARS-CoV-2 mutation data (as well as using a highly sparse but reliable transformation of entire mutational space into epitopic influence) to develop machine learning methods of severity classification. By profiling the epitopic load and HLA interactions enabled by mutations identified through interpretable machine learning, we also explored a potential approach towards identification of mutations of interest. Our demonstration of a temporal validation strategy further seeks to attract the attention of the community towards methods to avoid over-speculation for predictive prognosis approaches, especially when it pertains to continuously evolving pathogens (like SARS-CoV-2 in this case). We propose that the while caution should be exercised for clinical implementation of models learnt on past molecular signatures of a pathogen, reporting of metrics indicating reliability of the model can improve acceptance of the predictive prognosis exercises. This reliability can be scored based on the observed overlap or exclusivity of feature space in the tested samples, as compared to the feature space employed in the trained model(s). The issue of unsuitability of an otherwise useful technique may therefore be avoided. We emphasize that the need for concerted efforts in the direction of building dynamic machine learning workflows should also not be ignored, as that can aid in updating the previously learnt models as and when new mutation data is available, thereby making the entire approach more acceptable. This can greatly support the ongoing efforts of deducing the mutational landscape/relevance of SARS-CoV-2 and potentially, help in predictive prognosis.
CRediT authorship contribution statement
Sunil Nagpal: Conceptualization, Methodology, Investigation, Data curation, Software, Formal analysis, Validation, Writing – original draft, Writing – review & editing, Visualization. Nishal Kumar Pinna: Data curation, Formal analysis, Visualization, Writing - review and editing. Namrata Pant: Data curation, Investigation, Writing - review and editing. Rohan Singh: Formal analysis, Validation. Divyanshu Srivastava: Formal analysis, Validation. Sharmila S. Mande: Supervision, Formal analysis, Writing – review & editing.
Acknowledgments
Acknowledgements
We gratefully acknowledge all the Authors from the Originating laboratories responsible for obtaining the specimens and the Submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this research is based. Genome sequences and meta-data should be downloaded from https://www.gisaid.org. A sample file for integrated data profiles generated for this research has been provided in Supplementary File 4. The original annotated genome data can be downloaded from the GISAID initiative. We gratefully acknowledge the original contributors of the virus genome sequences used in this study in Supplementary File 5. The image of the protein structure presented in the graphical abstract (and Figure 1) is obtained from the Protein Data Bank37 (6VXX: Structure of the SARS-CoV-2 spike glycoprotein, closed state). SN would like to thank Dr. Bhupesh Taneja (PhD supervisor) for encouraging the work on building ML frameworks for important feature recognition in biological datasets.
Funding
Authors are salaried research employees of TCS Research, Tata Consultancy Services Ltd, Pune, India. SN is an industry sponsored PhD fellow at CSIR-IGIB. TCS Research or CSIR-IGIB had no role in writing of the manuscript or the decision to submit it for publication.
Conflict of Interest
Authors are salaried scientists at TCS Research. No conflicting interest declared.
Edited by James Zou
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2022.167684.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data Availability
Data sources have been mentioned in the manuscript and data contributors are acknowledged in Supplementary File 3 as well as acknowledgements section
References
- 1.Mottaqi M.S., Mohammadipanah F., Sajedi H. Contribution of machine learning approaches in response to SARS-CoV-2 infection. Informat. Med. Unlocked. 2021;23 doi: 10.1016/J.IMU.2021.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kivrak M., Guldogan E., Colak C. Prediction of death status on the course of treatment in SARS-COV-2 patients with deep learning and machine learning methods. Comput. Meth. Prog. Biomed. 2021;201 doi: 10.1016/J.CMPB.2021.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science (1979) 2020;370 doi: 10.1126/SCIENCE.ABD4250/SUPPL_FILE/ABD4250_TABLE_S8.GZ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., et al. An interpretable mortality prediction model for COVID-19 patients. Nature Mach. Intell. 2020;2(5):283–288. doi: 10.1038/s42256-020-0180-7. [DOI] [Google Scholar]
- 5.Toh C., Brody J.P. Evaluation of a genetic risk score for severity of COVID-19 using human chromosomal-scale length variation. Hum. Genom. 2020;14:36. doi: 10.1186/S40246-020-00288-Y/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoabi Y., Deri-Rozov S., Shomron N. Machine learning-based prediction of COVID-19 diagnosis based on symptoms. Npj Digital Med. 2021;4 doi: 10.1038/s41746-020-00372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanyaolu A., Okorie C., Marinkovic A., Haider N., Abbasi A.F., Jaferi U., Prakash S., Balendra V. The emerging SARS-CoV-2 variants of concern. Therap. Adv. Infect. Dis. 2021;8 doi: 10.1177/20499361211024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rochman N.D., Wolf Y.I., Faure G., Mutz P., Zhang F., Koonin E.V. Ongoing global and regional adaptive evolution of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2104241118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zahn L.M. Natural language predicts viral escape. Science (1979) 2021;371 doi: 10.1126/SCIENCE.371.6526.248-Q. [DOI] [Google Scholar]
- 10.Nagpal S., Srivastava D., Mande S.S. What if we perceive SARS-CoV-2 genomes as documents? Topic modelling using Latent Dirichlet Allocation to identify mutation signatures and classify SARS-CoV-2 genomes (preprint) BioRxiv. 2020 [Google Scholar]
- 11.Nagy Á., Ligeti B., Szebeni J., Pongor S., Györffy B. COVIDOUTCOME - Estimating COVID severity based on mutation signatures in the SARS-CoV-2 genome. Database. 2021;2021 doi: 10.1093/database/baab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho D.V., Pereira E.M., Cardoso J.S. Machine learning interpretability: A survey on methods and metrics. Electron. (Switzerland) 2019;8 doi: 10.3390/electronics808. [DOI] [Google Scholar]
- 13.Collins G.S., Reitsma J.B., Altman D.G., Moons K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. Eur. Urol. 2015;67 doi: 10.1016/j.eururo.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Yadaw A.S., Chak Li Y., Bose S., Iyengar R., Bunyavanich S., Pandey G. Clinical features of COVID-19 mortality: development and validation of a clinical prediction model. Lancet Digital Health. 2020;2 doi: 10.1016/S2589-7500(20)30217-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callaway E. The coronavirus is mutating - does it matter? Nature. 2020;585 doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- 16.Shu Y., McCauley J. GISAID, Global initiative on sharing all influenza data - from vision to reality. Euro Surveillance: Bull. Eur. Sur. Les Maladies Transmissibles = Eur. Commun. Disease Bull. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu J., Wu Q., Ding G., Xu Y., Feng S. A survey of machine learning for big data processing. Eurasip. J. Adv. Signal Process. 2016;2016 doi: 10.1186/s13634-016-0355-x. [DOI] [Google Scholar]
- 18.Messalas, A., Kanellopoulos, Y., & Makris, C. (2019). Model-Agnostic Interpretability with Shapley Values. In 10th International Conference on Information, Intelligence, Systems and Applications, IISA 2019. https://doi.org/10.1109/IISA.2019.8900669.
- 19.Roscher R., Bohn B., Duarte M.F., Garcke J. Explainable Machine Learning for Scientific Insights and Discoveries. IEEE Access. 2020;8 doi: 10.1109/ACCESS.2020.2976199. [DOI] [Google Scholar]
- 20.Lundberg S.M., Lee S.I. A unified approach to interpreting model predictions. Adv. Neural Informat. Process. Syst. 2017 [Google Scholar]
- 21.Nakamichi K., Shen J.Z., Lee C.S., Lee A., Roberts E.A., Simonson P.D., Roychoudhury P., Andriesen J., et al. Hospitalization and mortality associated with SARS-CoV-2 viral clades in COVID-19. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-82850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doytchinova I.A., Flower D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformat. 2007;8:1–7. doi: 10.1186/1471-2105-8-4/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bose T., Pant N., Pinna N.K., Bhar S., Dutta A., Mande S.S. Does immune recognition of SARS-CoV2 epitopes vary between different ethnic groups? Virus Res. 2021;305 doi: 10.1016/J.VIRUSRES.2021.198579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34 doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danecek P., McCarthy S.A. BCFtools/csq: Haplotype-aware variant consequences. Bioinformatics. 2017;33 doi: 10.1093/bioinformatics/btx100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Maaten L., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9 [Google Scholar]
- 27.McInnes L., Healy J., Saul N., Großberger L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018;3 doi: 10.21105/joss.00861. [DOI] [Google Scholar]
- 28.Chen, T. & Guestrin, C. (2016). XGBoost. https://doi.org/10.1145/2939672.2939785.
- 29.Student S., Fujarewicz K. Stable feature selection and classification algorithms for multiclass microarray data. Biol. Direct. 2012;7 doi: 10.1186/1745-6150-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietterich T.G. Approximate Statistical Tests for Comparing Supervised Classification Learning Algorithms. Neural Comput. 1998;10:1895–1923. doi: 10.1162/089976698300017197. [DOI] [PubMed] [Google Scholar]
- 31.Elshawi R., Al-Mallah M.H., Sakr S. On the interpretability of machine learning-based model for predicting hypertension. BMC Med. Informat. Decis. Mak. 2019;19 doi: 10.1186/s12911-019-0874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Pérez R., Bajorath J. Interpretation of machine learning models using shapley values: application to compound potency and multi-target activity predictions. J. Comput.-Aided Mol. Des. 2020;34 doi: 10.1007/s10822-020-00314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke L., Zheng-Bradley X., Smith R., Kulesha E., Xiao C., Toneva I., Vaughan B., Preuss D., et al. The 1000 Genomes Project: data management and community access. Nat. Meth. 2012;9(5):459–462. doi: 10.1038/nmeth.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins C., Didelot X. A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLOS Comput. Biol. 2018;14 doi: 10.1371/JOURNAL.PCBI.1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., Kato Y., Crotty E.G., Kim C., Rawlings S.A., Mateus J., Tse L.P.V., Frazier A., Baric R., Peters B., Greenbaum J., Ollmann Saphire E., Smith D.M., Sette A., Crotty S. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996. doi: 10.1016/J.CELL.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson E.A., Hirneise G., Singharoy A., Anderson K.S. Total predicted MHC-I epitope load is inversely associated with population mortality from SARS-CoV-2. Cell Rep Med. 2021;2 doi: 10.1016/J.XCRM.2021.100221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sources have been mentioned in the manuscript and data contributors are acknowledged in Supplementary File 3 as well as acknowledgements section