Summary
Knowledge on the population history of endangered species is critical for conservation, but whole-genome data on chimpanzees (Pan troglodytes) is geographically sparse. Here, we produced the first non-invasive geolocalized catalog of genomic diversity by capturing chromosome 21 from 828 non-invasive samples collected at 48 sampling sites across Africa. The four recognized subspecies show clear genetic differentiation correlating with known barriers, while previously undescribed genetic exchange suggests that these have been permeable on a local scale. We obtained a detailed reconstruction of population stratification and fine-scale patterns of isolation, migration, and connectivity, including a comprehensive picture of admixture with bonobos (Pan paniscus). Unlike humans, chimpanzees did not experience extended episodes of long-distance migrations, which might have limited cultural transmission. Finally, based on local rare variation, we implement a fine-grained geolocalization approach demonstrating improved precision in determining the origin of confiscated chimpanzees.
Keywords: chimpanzee, non-invasive samples, fecal samples, hybridization capture, population genetics, conservation genomics, geolocalization, population dynamics, chimpanzee demography
Graphical abstract
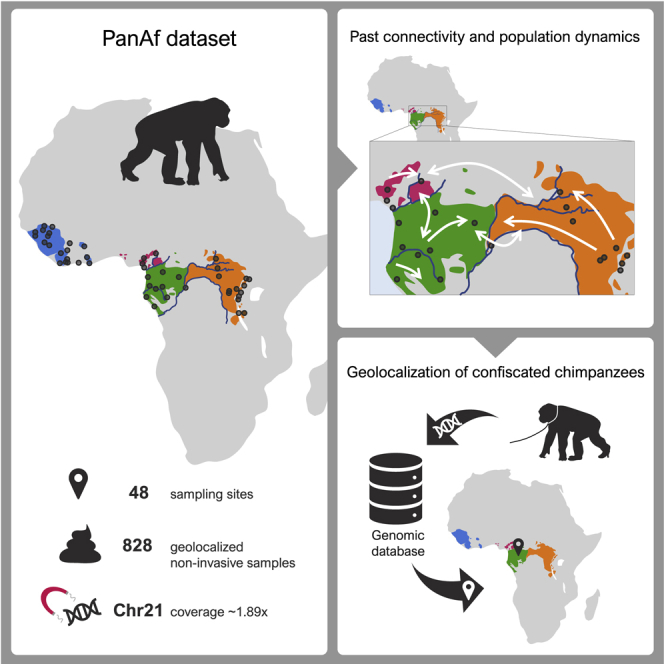
Highlights
-
•
Chromosome 21 capture of geolocalized non-invasive chimpanzee samples
-
•
Support for four differentiated subspecies with local population structure
-
•
Fine-scale description of gene flow barriers and corridors since Late Pleistocene
-
•
Power to infer geographical origin of confiscated chimpanzees
Fontsere et al. captured and sequenced chromosome 21 from 828 non-invasively collected chimpanzee samples, providing an extensive catalog of genomic diversity for wild chimpanzee populations. The authors describe patterns of isolation and connectivity between localities and implement a fine-grained geolocalization approach to infer the origin of confiscated chimpanzees.
Introduction
Genetic data on chimpanzee (Pan troglodytes) populations have been used to study the species’ diversity and population structure, as well as to characterize their demographic history and patterns of admixture at a broad subspecies level1, 2, 3, 4, 5, 6, 7 and with their sister species, bonobos (Pan paniscus).6,8 Due to a limited fossil record and absence of ancient DNA record, chimpanzee population genetics is inherently restricted to modern-day individuals.9
Four chimpanzee subspecies are currently recognized (western -P. t. verus-, Nigeria-Cameroon -P. t. ellioti-, central -P. t. troglodytes-, and eastern -P. t. schweinfurthii-, Figure 1A) but conflicting hypotheses still exist about whether genetic diversity in central and eastern chimpanzee populations reflects two distinctly separated subspecies,6 or a cline of variation under isolation-by-distance.1,10,11 This long-standing question also relates to the degree of connectivity among subspecies over time, which requires a fine-scaled reconstruction of the demographic history of chimpanzee populations after their split more than ∼100 thousand years ago (kya)6 and their inter-connectivity since the Last Glacial Maximum (LGM). Identifying genetic connections between present-day chimpanzee communities and the role of past environmental change in shaping these12,13 may be linked to behavioral variation in chimpanzee communities,14 similar to what has been explored extensively in humans as a strongly migratory species.15 Also, it will provide crucial tools for the development of conservation strategies for an endangered species that has suffered a dramatic decline in the last decades.16,17 A comprehensive genomic knowledge of a threatened species18 can guide conservation plans both in situ and ex situ.19 Furthermore, genetic information has proven useful to infer the populations of origin of confiscated individuals from illegal trade, detect poaching hotspots,20,21 and guide repatriation planning.22,23
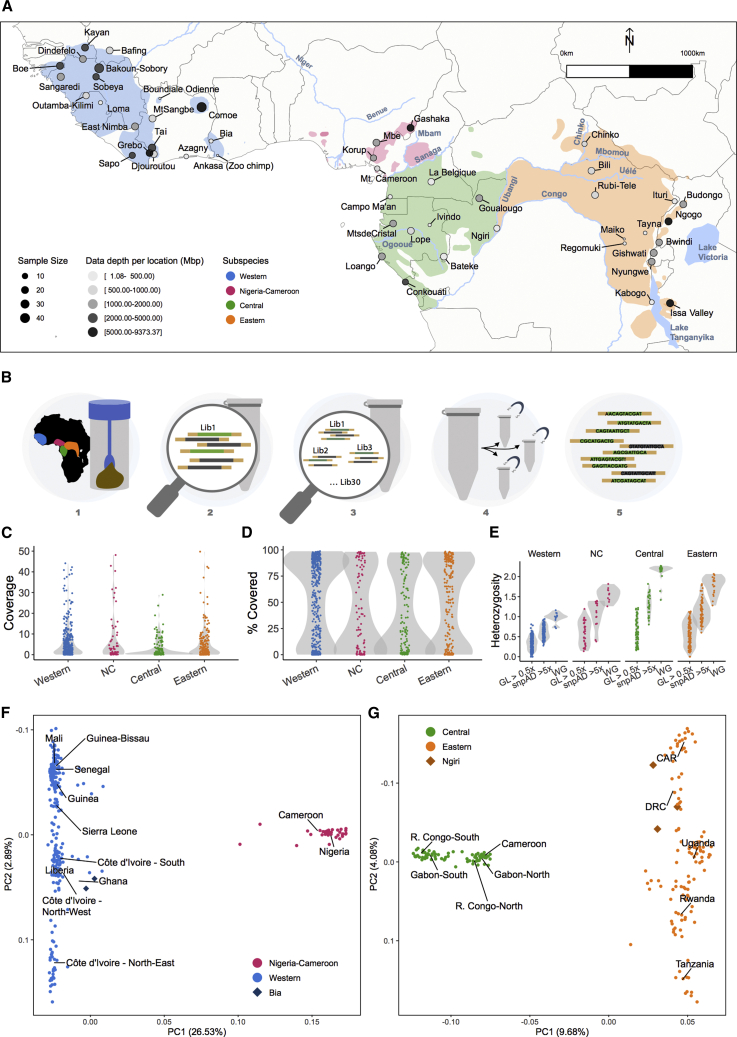
Figure 1.
Overview on sampling, capture, and chimpanzee population history
(A) Geographic distribution of chimpanzee subspecies and PanAf sampling locations. The western chimpanzee range is shown in blue, Nigeria-Cameroon in pink, central in green, and eastern in orange. The size of the dots represents the number of sequenced samples (n = 828) and color intensity represents the amount of chimpanzee genetic data generated (mega-base pairs of mapped sequence) from each sampling site.
(B) Experimental pipeline. (1) Samples were collected from 48 sampling sites, DNA extracted and screened for amplification success, uniqueness, and relatedness using microsatellites;11 (2) one library per individual24 was prepared; (3) between 10 and 30 libraries were pooled equi-endogenously;25 (4) enrichment for chromosome 21 with target capture methods, between three and five times per library;25 (5) sequencing data were generated with Illumina.
(C) Average coverage on the target region of chromosome 21 for each sample.
(D) Percentage of the target space covered by at least one read.
(E) Heterozygosity estimates per subspecies derived from ANGSD genotype likelihood on PanAf samples with more than 0.5-fold coverage (GL > 0.5×), from snpAD genotype calls on PanAf samples with more than 5-fold coverage, and from GATK genotype calls on previously published whole-genome (WG) chimpanzee samples.6
(F) PCA of western (blue) and Nigeria-Cameroon (pink) chimpanzee subspecies. Dark blue diamonds, Bia sampling site in Ghana at the eastern fringe of the extant western chimpanzee range.
(G) PCA of central (green) and eastern (orange) chimpanzee subspecies. Dark orange diamonds, Ngiri sampling site at the western fringe of the eastern chimpanzee distribution. CAR, Central African Republic; DRC, Democratic Republic of Congo; R. Congo, Republic of Congo. See also Figures S3–S13, S29–S39, and Table S4
For a detailed reconstruction of chimpanzee population structure and demographic history, it is crucial to gather data from a large number of individuals covering the current range of the species and of sufficient data depth. Since practical and ethical concerns impede the collection of blood samples from wild ape populations, non-invasive samples, such as feces,26,27 are a promising alternative, although low quality and quantities of host DNA (hDNA)27 have typically precluded population data analysis using single-nucleotide polymorphisms (SNPs). However, in the last years, several technical advances in target capture methods have allowed the use of non-invasive samples in large-scale genomic studies.25, 27, 28, 29 Here, we take advantage of these advances to generate an extensive dataset on genomic variation in georeferenced chimpanzees to infer their demographic history and develop a tool for the geolocation of chimpanzee samples.
Results
Capturing the diversity of wild chimpanzees
A total of 828 unique individuals were identified from non-invasive samples collected from 48 sampling sites across the chimpanzee range (Figure 1A) as part of the PanAfrican Program: The Cultured Chimpanzee.11 Using previously developed methods, we captured chromosome 21 from chimpanzee fecal DNA11,25, 28, 29 (Figure 1B) and generated sequencing data to a median coverage of 1.89-fold (0- to 90.14-fold) in the target space (Figure 1C), covering on average 12.9 million positions per sample (STAR Methods; Notes S1 and S2; Table S1).
Numerous samples have high levels of sequencing reads mapping to other primate species than chimpanzee (n = 100, Figures S9, S10, S14, and S15; Note S3), likely due to the inclusion of sympatric primate species in the diet, a well-known phenomenon,30,31 or sample misidentification during collection of feces.11,32 We also assessed human contamination among the remaining 728 samples using an approach very sensitive in low-coverage data,33 finding 36 of those with more than 1% of such contamination (Figure S16). This is similar to patterns of contamination observed in ancient DNA studies on humans.34 There is also large variation in coverage and hDNA content according to the sampling site, suggesting that environmental and/or dietary factors influence DNA quantity and preservation (Figures 1C, 1D, S4, and S7). Heterozygosity estimates, after careful quality assessment (Figures S21–S29; Note S3), are consistent with known patterns from high-coverage samples:3,6 highest in central chimpanzees, followed by eastern, Nigeria-Cameroon, and western chimpanzee subspecies (Figure 1E).
The history of Pan populations during the Middle Pleistocene
We deemed samples with more than 0.5-fold average coverage (on the target regions of chr21) and low levels of contamination to be of sufficient data depth and quality (n = 555) (Figures S9–S13; Note S3) for a PCA from genotype likelihoods,35,36 and found that these cluster according to the four described subspecies (Figures 1F, 1G, and S13) that were previously estimated to have diverged during the Middle Pleistocene (139–633 kya), after the split from bonobos (<2 million years ago [mya]) (Figure 2A).6 Low levels of ancient introgression from bonobos into the non-western chimpanzee subspecies (<1%) had previously been identified, most likely as the result of bonobo admixture into the ancestral population of eastern and central chimpanzees more than 200 kya6,37 (Figure 2A), possibly associated with a reduction of the Congo River discharge, the natural barrier separating both species.38 On chromosome 21, we did not observe a significant enrichment of allele sharing (F statistics)39 between bonobos and chimpanzees, likely due to limitations in the data, a small extent of admixture, and the small number of independent loci (Note S8). However, given the information from previous models based on whole genomes, we sought to determine introgressed fragments on chromosome 21 with the larger number of individuals used in this study. To this end, we inferred bonobo introgression using admixfrog,40 a method developed to reliably detect introgressed fragments even in low-coverage ancient genomes. With this hidden Markov model we inferred local ancestry for each sample (target) using different sources, which represent the admixing population (bonobo and all chimpanzee subspecies) from the reference panel6 (STAR Methods; Note S8). We found that all central chimpanzee communities sampled south of the Ogooué River (Figure 2A) (Loango, Lopé, Conkouati, and Batéké) harbor significantly more bonobo-like genomic fragments than those north of the river (two-sided Wilcoxon rank-sum test, Benjamini-Hochberg adjusted p value = 5.735 × 10−8), or any other chimpanzee population (adjusted p value < 0.01; Figure S64; Table S5; Note S8), with some of the individuals from the Lopé and Loango sampling sites showing the highest bonobo ancestry (Figure S64). These fragments are also longer than in other chimpanzee populations (Table S5; Figure S66J), which may hint at a separate, more recent admixture event, although this observation was not significant (two-sided Wilcoxon rank-sum test).
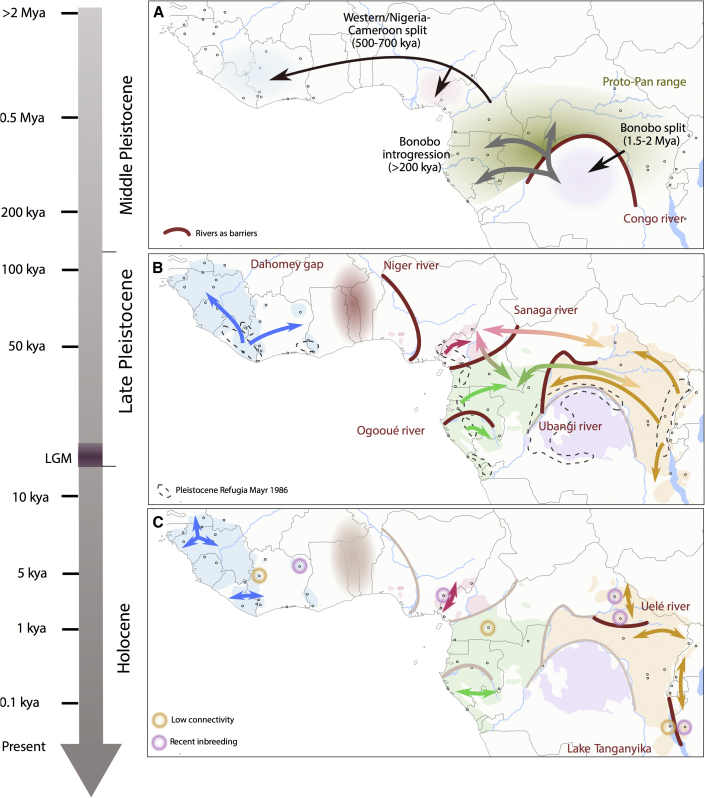
Figure 2.
Reconstruction of chimpanzee genetic history
Major rivers and lakes (red lines) and the Dahomey gap (red shading) represent geographical barriers separating populations at different timescales.
(A) Formation of and migration between Pan species (chimpanzees and bonobos) and subspecies formation during the Middle Pleistocene; separation and migration events inferred in previous studies,6,8,38 additional gene flow into southern central populations was inferred here using admixfrog.
(B) Corridors of gene flow (arrows) during the Late Pleistocene and after the Last Glacial Maximum (LGM), when chimpanzee populations expanded from refugia;12,14 within subspecies based on migration surfaces obtained with EEMS and shared rare variation, between subspecies based on short IBD-like tracts (<0.5 Mbp) and shared fragments of ancestry inferred with admixfrog.
(C) Population connectivity and isolation during the Holocene; connectivity was determined by long (>0.5 Mbp) IBD-like fragments between sampling locations within subspecies and supported by presence or absence of shared rare variation; signatures of recent inbreeding are represented by long regions of homozygosity in individuals from a given sampling location. See also Figures S40–S53, S57–S60, S64, S66, S76–S79, S82–S85, and S92.
Within chimpanzees, our dense sampling approach, including communities at the border between subspecies (eastern Ngiri in DRC, on the eastern bank of the Ubangi River) and thousands of markers (Table S1), allows us to assess the relationship between central and eastern chimpanzee subspecies. Despite Ngiri being geographically closer to Goualougo (a central chimpanzee sampling site, ∼280 km) than to any eastern chimpanzee location in our dataset (Rubi-Télé, ∼845 km, and Bili, ∼900 km) (Figure 1A), individuals from Ngiri clearly fall within the genetic diversity of eastern chimpanzees in the PCA (Figures 1G and S33), pointing to a clear long-term separation of these subspecies. These findings support an unequivocal separation of central and eastern chimpanzee subspecies over a large evolutionary time. However, subsequent recent interbreeding has been suggested by other studies.6,11
Long-term subspecies differentiation and genetic exchange during the Late Pleistocene
The sustained genetic differentiation of chimpanzee subspecies can be interpreted in the context of geographical barriers impeding gene flow, especially the major rivers in tropical Africa.41 We applied the EEMS method42 to analyze long-term migration landscapes during the Late Pleistocene and Early Holocene43 (Figures 2B and S82). We found evidence for regions of reduced effective migration that overlap with geographic barriers, such as the Sanaga River (separating Nigeria-Cameroon and central chimpanzees) and the Ubangi River (separating central and eastern chimpanzees) (Figures 2B and S82; Note S10). These patterns of stratification and shared drift were also supported by FST and f3 statistics (Figures S45, and S54; Notes S6 and S7).
Previous evidence suggested that some chimpanzee subspecies have not been fully isolated since their separation, but rather experienced migration events.3,6,11,44 To analyze the permeability of subspecies barriers to gene flow, we used two methods designed to capture signatures of gene flow at different timescales. First, we used identical-by-descent-like (IBD-like) segments detected between individuals from different subspecies using IBDseq,45 i.e., regions of the chromosome where two individuals share variation. Since the detected segments are smaller than 0.5 mega-base pairs (Mbp) between subspecies, they represent genetic exchange that happened more than approximately 5 kya, assuming an exponential decay of fragment length due to recombination (STAR Methods; Note S10; Figure S89; Table S7). Second, we inferred shorter introgressed fragments between chimpanzee subspecies with the aforementioned method admixfrog,40 using four genomes of each chimpanzee subspecies from the reference panel as sources6 to partition genomic regions into the subspecies state they resemble most (STAR Methods; Note S8). We found evidence of gene flow between the central, eastern, and Nigeria-Cameroon subspecies with both methods (Figures 2B, 3, and S66; Table S7), indicating low levels of genetic exchange at different timescales despite their long-term separation. We observed that Nigeria-Cameroon Gashaka individuals carry more fragments of central and eastern chimpanzee ancestry than other Nigeria-Cameroon chimpanzees, while the central Goualougo individuals carry more eastern and Nigeria-Cameroon chimpanzee fragments than the other central communities (Figure S66). This indicates gene flow between these local populations, which is also supported by an analysis of shared rare alleles, which are likely to have emerged more recently46 and whose sharing patterns are informative on recent admixture (Figures S76, and S77; Note S9). The observation of a northern area of past genetic exchange between the three subspecies is broadly consistent with conclusions from microsatellite data,11 and with previous studies suggesting a hybrid zone between central and Nigeria-Cameroon chimpanzees in central Cameroon.44,47
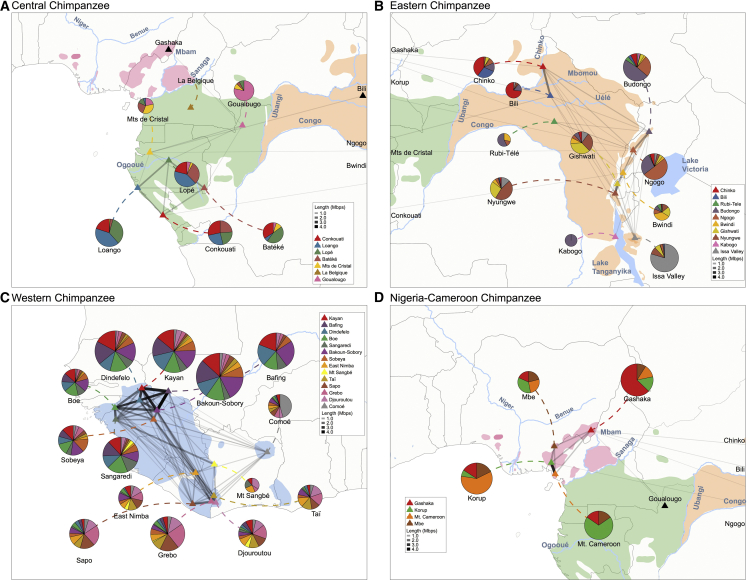
Figure 3.
Recent connectivity between chimpanzee populations
(A–D) The size of the pie charts represents the pairwise number of shared fragments, normalized by the number of pairs. Thickness of lines indicates the average length of IBD-like tracts (in Mbp). Triangles show the location of sites. Colors in pies indicate the origin of IBD-like tracts, including comparison between samples from the same site. (A) Central chimpanzees, (B) eastern chimpanzees, (C) western chimpanzees, and (D) Nigeria-Cameroon chimpanzees. Note also few and short IBD-like fragment connections between central, eastern, and Nigeria-Cameroon subspecies. See also Figures S86–S94.
Recent history between communities since the LGM
Local population stratification within subspecies, probably arising during the Late Pleistocene, has been partially explored previously for eastern and central chimpanzees using whole genomes, but with a much smaller sample size and sampling density.6 Here, for the first time, we can explore the fine-scale population structure and recent connectivity across the whole geographic range since the LGM and into the Holocene for all subspecies, partially down to the specific site level (Figure S31). To do this, we combine information from different methods that can specifically identify connectivity and isolation at different timescales, specifically EEMS42 (more than 6 kya), shared rare alleles (∼1.5–15 kya, Note S9),48 long (>0.5 Mbp) IBD-like tracts shared between communities of the same subspecies (less than 5 kya; please see more on possible caveats to this approach in the Limitations of the study and Note S10),49 as well as recent inbreeding with regions of homozygosity (RoH) (Figures S40–S42; Note S6). This yields a comprehensive and detailed picture of genetic connectivity across the chimpanzee range and within subspecies, beyond the broad genetic clines in eastern and western chimpanzees (Figures 1F, 1G, and S31–S34; Note S5).
Overall, western chimpanzees exhibit higher levels of connectivity across their range and across timescales than the other subspecies, as detected with IBD-like shared fragments, rare variation, and EEMS (Figures 2B, 2C, 3C, S79, S82, and S84; Note S10). Remarkably, for the same geographic distances, western chimpanzee sampling sites share more and longer IBD-like tracts than the other subspecies (Figures 3C and S90), especially within their northern range (Senegal, Mali, northern Guinea, and Guinea-Bissau). Also, shared rare variation resembles the results from IBD-like shared fragments (Figure S79). It is important to note that western chimpanzees have the lowest diversity and likely suffered a strong bottleneck,6 so our results could support two different scenarios: either high levels of recent connectivity between persisting populations during the past ∼780 years (according to the IBD-like tract length; range 117–2,200 years) (Table S9), or a range expansion into the fringe areas of the chimpanzee habitat within the same time frame, resulting in a very recent separation of these populations50 (Figure 2C). However, at this stage we cannot distinguish these scenarios based on genetic data only. All four sampling sites of Nigeria-Cameroon chimpanzees seem to have been connected within the past 2,500 years (mean 1,600 until 1,000 years ago), indicated by both IBD-like segments and rare allele connectivity (Figures 3D and S77; Note S9). Furthermore, a signature of recent inbreeding in Mbe (i.e., long RoH) suggests that this population was strongly isolated only very recently51 (Figure S40).
Eastern chimpanzee sampling sites largely follow a pattern of isolation-by-distance, shown as an exponential decay of IBD-like fragment length (Figure S90) along a genetic North-South cline also found in the PCA (Figure 1G). However, we observed three clusters of recent connectivity reflected in a higher number and longer IBD-like segments (Figure 3B, thicker lines between Chinko-Bili, Budongo-Ngogo, and Gishwati-Nyungwe). Also, the Uéle River and Lake Tanganyika likely acted as isolation barriers in eastern chimpanzee populations in recent times, which is supported by IBD-like segments and shared rare variation (Figures 2C, 3B, S76–S79, and S92; Table S3; Notes S9 and S10). Dispersal corridors suggested for populations in western Uganda52 and between western Uganda and the eastern DRC53 (Figure 2B) are supported by these types of analyses (Figures 3B and S78). Finally, all eastern chimpanzee populations share rare variation with those communities living in the area of previously proposed Pleistocene refugia12,14 (Budongo, Bwindi, Gishwati, Ngogo, and Nyungwe), suggesting an expansion into the southeast (Issa Valley54), central and southwest (Regomuki), and northwest (Rubi-Télé, Bili, Chinko, and Ngiri) after the LGM (Figures 2B and S78; Note S9). In central chimpanzees, we detected two strongly differentiated population clusters rather than a cline (Figures 2B,S76, S82, and S84; Notes S6, S7, and S10), separated by the Ogooué River in Gabon, which appears to have been a barrier reducing migration between these regions at least since the LGM, and maintained through the Holocene. Meanwhile, connectivity was higher within each central chimpanzee cluster, indicated by IBD-like tracts, rare allele sharing, and the EEMS surface (Figures 3A, S76, and S84; Notes S9 and S10). The southern cluster also matches with those populations that show a larger amount of bonobo-like introgressed fragments (Figure S64).
Geolocalization of chimpanzees using rare alleles
Our unique sampling breadth allowed the discovery of ∼50% more new genetic variants on chromosome 21 (Figure S29) in comparison with previously published chimpanzee whole genomes.6 In particular, rare variation likely emerged recently (during few hundreds to thousands of years46), and will be geographically structured. Hence, rare alleles are particularly useful for geolocalization because the chimpanzee groups studied here do show local stratification in the sharing of these alleles (Figures S76–S79; Note S9). Here, we developed a strategy to use rare variants (STAR Methods; Figures S67–S75; Note S9) to infer the geographic origin of samples. In brief, we used a reference panel of 434 samples of sufficient quality (Note S3) across 38 sampling locations, obtained the derived frequency of each SNP within each population, and retained SNPs that were observed at one given sampling location but at low cumulative frequency (lower than 1) across all other locations (STAR Methods; Note S9). We then tested samples by calculating their proportion of matching genotypes, across all such positions, to each reference population, and applied a spatial interpolation (kriging) across the chimpanzee range, allowing the visualization of regions of putative origin (e.g., Figure 4A; Data S1, Figure S96).
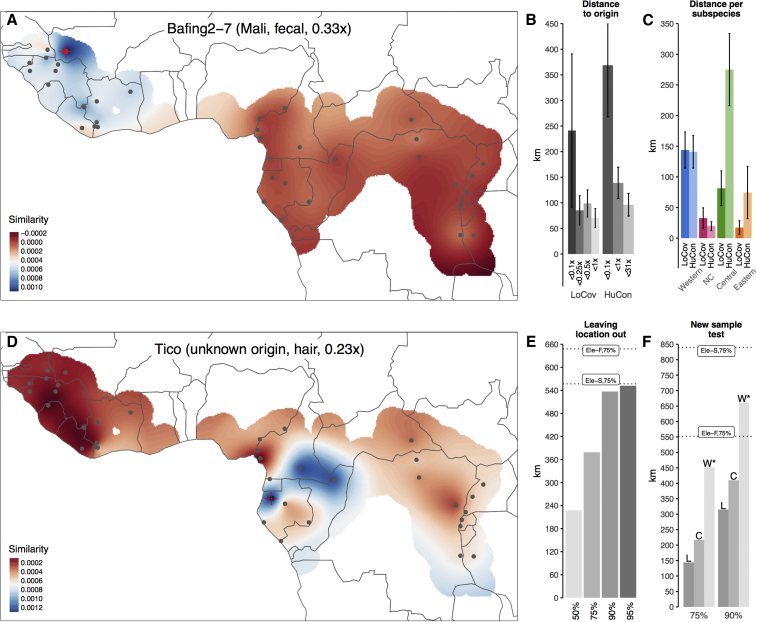
Figure 4.
Chimpanzee geolocalization based on rare variation
(A) Spatial model of shared rare alleles with 38 sampling locations. Red indicates lower amounts, while blue indicates larger amounts of shared rare alleles. Black dots, locations included in the reference panel; red dot, known place of origin (low-coverage sample Baf2-7 from Bafing, Mali); red cross, inferred origin. Red cross and dot overlap in this correctly assigned sample.
(B) Average distance (km) of best matching to true location in bins of coverage for samples with low coverage and contamination (LoCov) (n = 99) and samples with human contamination of more than 0.5% (HuCon) (n = 139). Error bars represent the SEM.
(C) Average distance of best matching to true location per subspecies, stratified by low-coverage and human-contaminated samples; note that the Nigeria-Cameroon chimpanzee range is smaller than that of other subspecies, thus resulting in smaller distances. Error bars represent the SEM.
(D) Geolocalization of the chimpanzee Tico from a rescue center in Spain, here assigned to Gabon or Equatorial Guinea.
(E) Assignment accuracy when leaving full locations out (n = 434), with 50th, 75th, 90th, and 95th percentiles for the distance of inferred to true origin; for comparison, best 75th percentiles for geolocalization of elephants21 are shown as dotted lines (Ele-S, Savanna elephant; Ele-F, forest elephant).
(F) Assignment accuracy for samples not included in the reference panel (L, low coverage; C, contaminated; W, whole-genome data6); for comparison, the 75th percentiles of the single sample test in elephants are shown as dotted lines; the asterisk marks that the origin for whole genomes may be different from the place of confiscation reported for these individuals. See also Figures S67–S75, S80, and S81.
First, we applied this strategy to 99 samples excluded from the reference panel due to low coverage (<1-fold), as well as 139 samples with human contamination (>0.5%) (Data S1, Figures S96 and S97). At a coverage of more than 0.1-fold, samples are, on average, located 81 km (0–502 km) from their true origin (Figure 4B). In the presence of human contamination (>0.5%, coverage >0.1-fold), this average increases to 139 km, on average, mostly due to central chimpanzee samples (Figure 4C). Samples from locations not included in the reference panel are assigned to nearby regions of the corresponding subspecies (Data S1, Figure S98).
We assessed the accuracy of our method using an approach from a previous study in elephants21 by inferring the origin of samples when leaving their sampling location out of the reference panel. We find that 75% of the samples are inferred to originate from within 379 km of their sampling location (Figure 4E), considerably closer than the closest 75th percentile in elephants (557 km for sample groups of savannah elephants21). Remarkably, when comparing the closest 75th percentile of testing single samples where the sampling location was included in elephants (552 km for forest elephants), in chimpanzees we find that this distance from the true location is less than half for the low-coverage (144 km) and contaminated (217 km) samples (Figure 4E). Our geographically dense reference panel with thousands of markers, likely enhanced by a lower overall mobility of chimpanzees compared with elephants, makes our methodology outperform the elephant one, even though genotype data are extremely patchy and incomplete. Also, the approximate origin of previously published chimpanzee whole genomes6 (Note S9; Data S1, Figure S102) is closer to the known place of origin or confiscation (75th percentile: 452 km; Figure 4F) than what has been found for elephants of known origin. Finally, we used this strategy to estimate the most probable origin of 20 chimpanzees from two Spanish rescue centers (Fundació Mona and Fundación Rainfer), which were sequenced at low coverage from hair and blood samples (median 0.35-fold coverage, ranging from 0.15- to 4.3-fold) (Figures S80 and S81; Note S9). Hence, with our method even shallow sequencing (without target capture) provides enough information for the approximate geolocalization of chimpanzees with unknown or low confidence origin information (e.g., Figures 4D, S80, and S81; Note S9).
Discussion
Our study shows how non-invasive samples can be used as a source of genomic DNA for population and conservation genomic purposes. Here, we have implemented target capture on chimpanzee fecal samples, although it is worth noting that the same approach could be applied to other great ape and primate species, broadening their application from a few autosomal, sex-linked, or mtDNA markers to an entire chromosome. Precisely, by target capturing a complete chromosome we have the power to discover variation previously unreported and detect contiguous segments of DNA that are inherited together. We found evidence supporting the genetic differentiation of the four recognized subspecies of chimpanzee populations,3,6 whose differentiation could be linked to historical geographical barriers, in particular the Sanaga River and Ubangi River. Such barriers of gene flow41,55 have been proposed before, particularly the Congo river separating bonobos from chimpanzees. However, rivers have not been immutable throughout history, and a reduction of river discharge during glaciation periods likely opened corridors for migration;38 for example, allowing ancient introgression from bonobos into non-western chimpanzees6 and also between chimpanzee subspecies.44 Here, we detected differential amounts of ancient introgression from bonobos to central chimpanzee populations north and south of the Ogooué River. This could be explained either by multiple phases of genetic exchange between chimpanzees and bonobos, as has been suggested previously,6 or by a dilution of bonobo ancestry due to admixture with other chimpanzee populations, as supported by a higher Nigeria-Cameroon and eastern chimpanzee ancestry in the central chimpanzee populations north of the Ogooué River. However, these scenarios are not mutually exclusive, and need to be further investigated using multiple whole genomes from these different regions.
Importantly, this dataset is useful to study the population history and connectivity of wild chimpanzee communities in more recent times. Population stratification in chimpanzee populations can be explained by isolation-by-distance to some degree,11 but known ecological or geographical barriers have also reduced gene flow between certain populations for extended periods of time, leading to substantial substructure in chimpanzees. This is the case for the Ogooué River acting as a barrier between northern and southern central chimpanzee populations, or Lake Tanganyika separating eastern chimpanzee populations in the south.53 The Uélé River, isolating eastern chimpanzees since the LGM in the north, is concordant with observed behavioral differences to its north and south.56,57 Corridors of gene flow between non-western chimpanzee subspecies have been suggested previously,3,6,11 and we restrict these events mainly to specific areas between central, Nigeria-Cameroon, and eastern chimpanzee populations in the north of their range, particularly between Goualougo and Gashaka, located at the northern fringe of the distribution of these subspecies. However, due to the lack of sampling in eastern Cameroon, we propose that a historical corridor may have reached from the northern range of central chimpanzees to Gashaka through central Cameroon, in concordance with previous results on mtDNA.44
These patterns of isolation-by-distance over tens of thousands of years, with genetic interactions occurring on a local scale, stand in apparent contrast to the demographic history of most human populations during the same time frame, which is characterized by high levels of migration.46 We speculate that chimpanzee’s comparably lower migration pattern might be related to a lower extent of information transmission, which is a fundamental difference between them and humans.58 We speculate that limited genetic and cultural exchange in chimpanzees compared with humans might be a consequence of the social structure of chimpanzees.59 The higher inter-connectivity of western chimpanzees may also help to explain their larger behavioral diversity compared with non-western chimpanzee populations. A large degree of sharing of IBD-like fragments in the northwestern range of western chimpanzees, resulting from either recent expansion or high recent connectivity, might reflect population movements from Pleistocene refugia in the south (Liberia, Côte d’Ivoire) after the LGM (Figure 2B),12,13 possibly related to the proposed cultural expansion in western chimpanzees.14 However, the Comoé sites in the east of Côte d’Ivoire are genetically closer to forest populations in the south (Figures S45 and S54), despite seemingly being behaviorally similar to the north-eastern mosaic woodland habitat populations.60 We also find genomic support for an expansion from Pleistocene refugia in eastern chimpanzee populations to the south, west, and northwest after the LGM (Figure 2B).
Using our knowledge of genetic diversity linked to geographical locations, we present a strategy for geolocalization with improved accuracy and precision, even when using low-coverage or contaminated samples (Figure 4). Geolocalization of chimpanzees has direct conservation applications: first, it can help ensure that confiscated chimpanzees from illegal pet trade61 are placed into sanctuaries in their countries of origin as mandated by the international standards.62 Second, when sequencing confiscated individuals or wildlife products (e.g., bushmeat), it can allow for the detection of poaching hotspots, so relevant authorities can enforce national and international laws enacted for protected species.21,63 Successful methods have been developed for African elephants,21,64 but past attempts in chimpanzees did not provide sufficient spatial resolution;19,20 while, unfortunately, microsatellite data do not yield a sufficient degree of genetic structure in chimpanzees.11 However, some geographic regions are not well resolved, resulting in different possible countries of origin, as is the case for other species.21 Considering that samples are assigned to nearby locations when their sampling site is not covered (Data S1; Figure S99), this is likely to be improved with yet better sampling. Our strategy is based on low-coverage shotgun sequencing, with lower costs but requiring state-of-the-art laboratory facilities and bioinformatic know-how to process and identify the origin of a confiscated individual, which is not accessible for in situ genotyping.65,66 However, new optimizations on sequencing technologies, such as Oxford Nanopore Technologies,67 might be helpful to obtain genotype information on-site, to ascertain the origins of confiscated wildlife and products.
In conclusion, using the capture of chromosome 21 on hundreds of chimpanzee fecal samples, we presented the first geographically linked catalog of genomic diversity in extant wild chimpanzee populations. This resource allows for the determination of fine-scale population structure, past and recent gene flow, and migration events, and the construction of a geo-genetic map for the geolocalization of orphaned chimpanzees and confiscated bushmeat.
Limitations of the study
The use of non-invasive samples for population genomics is still limited by their low quality and low proportions of hDNA. Under these circumstances, whole-genome sequencing, which would provide stronger support in many analyses, is prohibited by both low library complexity and economical constraints. Since sequencing was limited to a portion of the genome, we could not reach enough confidence to resolve the origin of the differential amount of bonobo introgression in central chimpanzees, and we cannot apply the standard methods to study gene flow. The nature of our dataset also impedes the reconstruction of recent connectivity using IBD-like segments since the accuracy to detect those segments is directly limited by the missingness inherent in low-coverage datasets. Therefore, the timing of the events using the length of the IBD-like fragments can encompass large confidence intervals since the low coverage and high missingness in the data could result in underestimating their length, leading to inaccurate timings.
Fecal samples may be subject to contamination from mammalian or primate DNA from species included in the diet of the chimpanzee. Even though we used a very thorough quality control, due to our limited coverage we cannot discard small remnants of contamination in our dataset.
Finally, the geolocalization approach is based on rare variation, and relies on having a dense georeferenced panel of samples; even after our extensive sampling effort there are some under-represented areas where future studies should focus on gathering samples to fill in the current gaps.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Chimpanzee hair sample | This Study | AZA-01-01 |
| Chimpanzee hair sample | This Study | AZA-01-02 |
| Chimpanzee hair sample | This Study | AZA-01-03 |
| Chimpanzee hair sample | This Study | AZA-01-06 |
| Chimpanzee hair sample | This Study | AZA-01-08 |
| Chimpanzee hair sample | This Study | AZA-01-09 |
| Chimpanzee hair sample | This Study | AZA-01-10 |
| Chimpanzee hair sample | This Study | AZA-01-11 |
| Chimpanzee hair sample | This Study | AZA-01-12 |
| Chimpanzee fecal sample | This Study | Baf1-12 |
| Chimpanzee fecal sample | This Study | Baf1-13 |
| Chimpanzee fecal sample | This Study | Baf1-16 |
| Chimpanzee fecal sample | This Study | Baf1-17 |
| Chimpanzee fecal sample | This Study | Baf1-19 |
| Chimpanzee fecal sample | This Study | Baf1-2 |
| Chimpanzee fecal sample | This Study | Baf1-5 |
| Chimpanzee fecal sample | This Study | Baf2-1 |
| Chimpanzee fecal sample | This Study | Baf2-26 |
| Chimpanzee fecal sample | This Study | Baf2-27 |
| Chimpanzee fecal sample | This Study | Baf2-4 |
| Chimpanzee fecal sample | This Study | Baf2-42 |
| Chimpanzee fecal sample | This Study | Baf2-43 |
| Chimpanzee fecal sample | This Study | Baf2-44 |
| Chimpanzee fecal sample | This Study | Baf2-46 |
| Chimpanzee fecal sample | This Study | Baf2-68 |
| Chimpanzee fecal sample | This Study | Baf2-7 |
| Chimpanzee fecal sample | This Study | Baf2-73 |
| Chimpanzee fecal sample | This Study | Baf2-74 |
| Chimpanzee fecal sample | This Study | Baf2-75 |
| Chimpanzee fecal sample | This Study | Bat1-1 |
| Chimpanzee fecal sample | This Study | Bat1-10 |
| Chimpanzee fecal sample | This Study | Bat1-11 |
| Chimpanzee fecal sample | This Study | Bat1-14 |
| Chimpanzee fecal sample | This Study | Bat1-15 |
| Chimpanzee fecal sample | This Study | Bat1-16 |
| Chimpanzee fecal sample | This Study | Bat1-18 |
| Chimpanzee fecal sample | This Study | Bat1-19 |
| Chimpanzee fecal sample | This Study | Bat1-20 |
| Chimpanzee fecal sample | This Study | Bat1-22 |
| Chimpanzee fecal sample | This Study | Bat1-24 |
| Chimpanzee fecal sample | This Study | Bat1-3 |
| Chimpanzee fecal sample | This Study | Bat1-37 |
| Chimpanzee fecal sample | This Study | Bat1-39 |
| Chimpanzee fecal sample | This Study | Bat1-4 |
| Chimpanzee fecal sample | This Study | Bat1-42 |
| Chimpanzee fecal sample | This Study | Bat1-48 |
| Chimpanzee fecal sample | This Study | Bat1-5 |
| Chimpanzee fecal sample | This Study | Bat1-50 |
| Chimpanzee fecal sample | This Study | Bat1-6 |
| Chimpanzee fecal sample | This Study | Bil1-1 |
| Chimpanzee fecal sample | This Study | Bil1-10 |
| Chimpanzee fecal sample | This Study | Bil1-15 |
| Chimpanzee fecal sample | This Study | Bil1-20 |
| Chimpanzee fecal sample | This Study | Bil1-24 |
| Chimpanzee fecal sample | This Study | Bil1-25 |
| Chimpanzee fecal sample | This Study | Bil1-28 |
| Chimpanzee fecal sample | This Study | Bil1-3 |
| Chimpanzee fecal sample | This Study | Bil1-30 |
| Chimpanzee fecal sample | This Study | Bil1-37 |
| Chimpanzee fecal sample | This Study | Bil1-38 |
| Chimpanzee fecal sample | This Study | Bil1-41 |
| Chimpanzee fecal sample | This Study | Bil1-46 |
| Chimpanzee fecal sample | This Study | Bil1-47 |
| Chimpanzee fecal sample | This Study | Bil1-50 |
| Chimpanzee fecal sample | This Study | Bil1-51 |
| Chimpanzee fecal sample | This Study | Bil1-7 |
| Chimpanzee fecal sample | This Study | Bil1-8 |
| Chimpanzee fecal sample | This Study | Bili2-5 |
| Chimpanzee fecal sample | This Study | Bili2-7 |
| Chimpanzee fecal sample | This Study | Boe1-30 |
| Chimpanzee fecal sample | This Study | Boe1-32 |
| Chimpanzee fecal sample | This Study | Boe1-33 |
| Chimpanzee fecal sample | This Study | Boe1-36 |
| Chimpanzee fecal sample | This Study | Boe1-43 |
| Chimpanzee fecal sample | This Study | Boe1-44 |
| Chimpanzee fecal sample | This Study | Boe1-57 |
| Chimpanzee fecal sample | This Study | Boe1-60 |
| Chimpanzee fecal sample | This Study | Boe1-71 |
| Chimpanzee fecal sample | This Study | Boe1-75 |
| Chimpanzee fecal sample | This Study | Boe1-77 |
| Chimpanzee fecal sample | This Study | Boe2-10 |
| Chimpanzee fecal sample | This Study | Boe2-11 |
| Chimpanzee fecal sample | This Study | Boe2-18 |
| Chimpanzee fecal sample | This Study | Boe2-22 |
| Chimpanzee fecal sample | This Study | Boe2-26 |
| Chimpanzee fecal sample | This Study | Boe2-30 |
| Chimpanzee fecal sample | This Study | Boe2-33 |
| Chimpanzee fecal sample | This Study | Boe2-39 |
| Chimpanzee fecal sample | This Study | Boe2-6 |
| Chimpanzee fecal sample | This Study | Bou1-1 |
| Chimpanzee fecal sample | This Study | Bud1-1 |
| Chimpanzee fecal sample | This Study | Bud1-10 |
| Chimpanzee fecal sample | This Study | Bud1-12 |
| Chimpanzee fecal sample | This Study | Bud1-15 |
| Chimpanzee fecal sample | This Study | Bud1-17 |
| Chimpanzee fecal sample | This Study | Bud1-2 |
| Chimpanzee fecal sample | This Study | Bud1-21 |
| Chimpanzee fecal sample | This Study | Bud1-6 |
| Chimpanzee fecal sample | This Study | Bud1-7 |
| Chimpanzee fecal sample | This Study | Bud1-9 |
| Chimpanzee fecal sample | This Study | Bud2-29 |
| Chimpanzee fecal sample | This Study | Bud2-45 |
| Chimpanzee fecal sample | This Study | Bud2-48 |
| Chimpanzee fecal sample | This Study | Bud2-52 |
| Chimpanzee fecal sample | This Study | Bud2-54 |
| Chimpanzee fecal sample | This Study | Bud2-73 |
| Chimpanzee fecal sample | This Study | Bud2-81 |
| Chimpanzee fecal sample | This Study | Bud2-86 |
| Chimpanzee fecal sample | This Study | Bud3-13 |
| Chimpanzee fecal sample | This Study | Bud3-30 |
| Chimpanzee fecal sample | This Study | Bwi-2-26 |
| Chimpanzee fecal sample | This Study | Bwi-2-39 |
| Chimpanzee fecal sample | This Study | Bwi1-10 |
| Chimpanzee fecal sample | This Study | Bwi1-16 |
| Chimpanzee fecal sample | This Study | Bwi1-29 |
| Chimpanzee fecal sample | This Study | Bwi1-3 |
| Chimpanzee fecal sample | This Study | Bwi1-36 |
| Chimpanzee fecal sample | This Study | Bwi1-4 |
| Chimpanzee fecal sample | This Study | Bwi1-55 |
| Chimpanzee fecal sample | This Study | Bwi1-58 |
| Chimpanzee fecal sample | This Study | Bwi1-6 |
| Chimpanzee fecal sample | This Study | Bwi1-61 |
| Chimpanzee fecal sample | This Study | Bwi1-64 |
| Chimpanzee fecal sample | This Study | Bwi1-66 |
| Chimpanzee fecal sample | This Study | Bwi1-69 |
| Chimpanzee fecal sample | This Study | Bwi1-7 |
| Chimpanzee fecal sample | This Study | Bwi1-71 |
| Chimpanzee fecal sample | This Study | Bwi1-74 |
| Chimpanzee fecal sample | This Study | Bwi1-78 |
| Chimpanzee fecal sample | This Study | Bwi1-90 |
| Chimpanzee fecal sample | This Study | Cam1-13 |
| Chimpanzee fecal sample | This Study | Cam1-14 |
| Chimpanzee fecal sample | This Study | Cam1-18 |
| Chimpanzee fecal sample | This Study | Cam1-2 |
| Chimpanzee fecal sample | This Study | Cam1-21 |
| Chimpanzee fecal sample | This Study | Cam1-26 |
| Chimpanzee fecal sample | This Study | Cam1-27 |
| Chimpanzee fecal sample | This Study | Cam1-29 |
| Chimpanzee fecal sample | This Study | Cam1-44 |
| Chimpanzee fecal sample | This Study | Cam1-49 |
| Chimpanzee fecal sample | This Study | Cam1-50 |
| Chimpanzee fecal sample | This Study | Cam1-7 |
| Chimpanzee fecal sample | This Study | Cam1-71 |
| Chimpanzee fecal sample | This Study | Cam1-74 |
| Chimpanzee fecal sample | This Study | Cam1-78 |
| Chimpanzee fecal sample | This Study | Cam2-77 |
| Chimpanzee fecal sample | This Study | Cam3-29 |
| Chimpanzee fecal sample | This Study | Cam3-40 |
| Chimpanzee fecal sample | This Study | Cam3-41 |
| Chimpanzee fecal sample | This Study | Cam3-45 |
| Chimpanzee fecal sample | This Study | Chinko-1 |
| Chimpanzee fecal sample | This Study | Chinko-10 |
| Chimpanzee fecal sample | This Study | Chinko-12 |
| Chimpanzee fecal sample | This Study | Chinko-13 |
| Chimpanzee fecal sample | This Study | Chinko-14 |
| Chimpanzee fecal sample | This Study | Chinko-16 |
| Chimpanzee fecal sample | This Study | Chinko-3 |
| Chimpanzee fecal sample | This Study | Chinko-5 |
| Chimpanzee fecal sample | This Study | Chinko-8 |
| Chimpanzee fecal sample | This Study | CMNP1-19 |
| Chimpanzee fecal sample | This Study | CMNP1-24 |
| Chimpanzee fecal sample | This Study | CMNP1-43 |
| Chimpanzee fecal sample | This Study | CMNP1-8 |
| Chimpanzee fecal sample | This Study | CMNP2-1_B |
| Chimpanzee fecal sample | This Study | CMNP2-5 |
| Chimpanzee fecal sample | This Study | CMNP2-6 |
| Chimpanzee fecal sample | This Study | Cnp1-1 |
| Chimpanzee fecal sample | This Study | Cnp1-14 |
| Chimpanzee fecal sample | This Study | Cnp1-2 |
| Chimpanzee fecal sample | This Study | Cnp1-36 |
| Chimpanzee fecal sample | This Study | Cnp1-37 |
| Chimpanzee fecal sample | This Study | Cnp1-47 |
| Chimpanzee fecal sample | This Study | Cnp1-63 |
| Chimpanzee fecal sample | This Study | Cnp1-70 |
| Chimpanzee fecal sample | This Study | Cnp1-75 |
| Chimpanzee fecal sample | This Study | CNPE1-1 |
| Chimpanzee fecal sample | This Study | CNPE1-12 |
| Chimpanzee fecal sample | This Study | CNPE1-2 |
| Chimpanzee fecal sample | This Study | CNPE1-22 |
| Chimpanzee fecal sample | This Study | CNPE1-26 |
| Chimpanzee fecal sample | This Study | CNPE1-3 |
| Chimpanzee fecal sample | This Study | CNPE1-31 |
| Chimpanzee fecal sample | This Study | CNPE1-36 |
| Chimpanzee fecal sample | This Study | CNPE1-6 |
| Chimpanzee fecal sample | This Study | CNPE1-7 |
| Chimpanzee fecal sample | This Study | CNPN1-20 |
| Chimpanzee fecal sample | This Study | CNPN1-35 |
| Chimpanzee fecal sample | This Study | CNPN1-63 |
| Chimpanzee fecal sample | This Study | CNPW1-13 |
| Chimpanzee fecal sample | This Study | CNPW1-16_2 |
| Chimpanzee fecal sample | This Study | CNPW1-17 |
| Chimpanzee fecal sample | This Study | CNPW1-2 |
| Chimpanzee fecal sample | This Study | CNPW1-40 |
| Chimpanzee fecal sample | This Study | CNPW1-7 |
| Chimpanzee fecal sample | This Study | CNPW2-29 |
| Chimpanzee fecal sample | This Study | CNPW2-43 |
| Chimpanzee fecal sample | This Study | Con1-12 |
| Chimpanzee fecal sample | This Study | Con2-23 |
| Chimpanzee fecal sample | This Study | Con2-25 |
| Chimpanzee fecal sample | This Study | Con2-27 |
| Chimpanzee fecal sample | This Study | Con2-38 |
| Chimpanzee fecal sample | This Study | Con2-48 |
| Chimpanzee fecal sample | This Study | Con2-49 |
| Chimpanzee fecal sample | This Study | Con2-50 |
| Chimpanzee fecal sample | This Study | Con2-53 |
| Chimpanzee fecal sample | This Study | Con2-56 |
| Chimpanzee fecal sample | This Study | Con2-57 |
| Chimpanzee fecal sample | This Study | Con2-64 |
| Chimpanzee fecal sample | This Study | Con2-66 |
| Chimpanzee fecal sample | This Study | Con2-67 |
| Chimpanzee fecal sample | This Study | Con2-71 |
| Chimpanzee fecal sample | This Study | Con2-80 |
| Chimpanzee fecal sample | This Study | Con3-10 |
| Chimpanzee fecal sample | This Study | Con3-8 |
| Chimpanzee fecal sample | This Study | Din1-10 |
| Chimpanzee fecal sample | This Study | Din1-22 |
| Chimpanzee fecal sample | This Study | Din1-26 |
| Chimpanzee fecal sample | This Study | Din1-3 |
| Chimpanzee fecal sample | This Study | Din1-4 |
| Chimpanzee fecal sample | This Study | Din1-53 |
| Chimpanzee fecal sample | This Study | Din1-6 |
| Chimpanzee fecal sample | This Study | Din1-68 |
| Chimpanzee fecal sample | This Study | Din1-7 |
| Chimpanzee fecal sample | This Study | Din2-22 |
| Chimpanzee fecal sample | This Study | Din2-29 |
| Chimpanzee fecal sample | This Study | Din2-3 |
| Chimpanzee fecal sample | This Study | Din2-38 |
| Chimpanzee fecal sample | This Study | Din2-43 |
| Chimpanzee fecal sample | This Study | Din2-79 |
| Chimpanzee fecal sample | This Study | Din2-83 |
| Chimpanzee fecal sample | This Study | Din3-5 |
| Chimpanzee fecal sample | This Study | Din3-7 |
| Chimpanzee fecal sample | This Study | Din3-8 |
| Chimpanzee fecal sample | This Study | Din3-9 |
| Chimpanzee fecal sample | This Study | Dja1-16 |
| Chimpanzee fecal sample | This Study | Dja1-17 |
| Chimpanzee fecal sample | This Study | Dja1-23 |
| Chimpanzee fecal sample | This Study | Dja1-8 |
| Chimpanzee fecal sample | This Study | Dja2-20 |
| Chimpanzee fecal sample | This Study | Dja2-21 |
| Chimpanzee fecal sample | This Study | Dja2-22 |
| Chimpanzee fecal sample | This Study | Dja2-23 |
| Chimpanzee fecal sample | This Study | Dja2-25 |
| Chimpanzee fecal sample | This Study | Dja2-27 |
| Chimpanzee fecal sample | This Study | Dja2-30 |
| Chimpanzee fecal sample | This Study | Dja2-36 |
| Chimpanzee fecal sample | This Study | Dja2-39 |
| Chimpanzee fecal sample | This Study | Dja2-42 |
| Chimpanzee fecal sample | This Study | Dja2-57 |
| Chimpanzee fecal sample | This Study | Dja3-19 |
| Chimpanzee fecal sample | This Study | Dja3-20 |
| Chimpanzee fecal sample | This Study | Dja3-21 |
| Chimpanzee fecal sample | This Study | Dja3-6 |
| Chimpanzee fecal sample | This Study | Dja3-7 |
| Chimpanzee fecal sample | This Study | Djo1-13 |
| Chimpanzee fecal sample | This Study | Djo1-14 |
| Chimpanzee fecal sample | This Study | Djo1-2 |
| Chimpanzee fecal sample | This Study | Djo1-20 |
| Chimpanzee fecal sample | This Study | Djo1-22 |
| Chimpanzee fecal sample | This Study | Djo1-37 |
| Chimpanzee fecal sample | This Study | Djo1-5 |
| Chimpanzee fecal sample | This Study | Djo1-50 |
| Chimpanzee fecal sample | This Study | Djo1-54 |
| Chimpanzee fecal sample | This Study | Djo1-6 |
| Chimpanzee fecal sample | This Study | Djo1-60 |
| Chimpanzee fecal sample | This Study | Djo1-66 |
| Chimpanzee fecal sample | This Study | Djo2-29 |
| Chimpanzee fecal sample | This Study | Djo2-4 |
| Chimpanzee fecal sample | This Study | Djo2-5 |
| Chimpanzee fecal sample | This Study | Djo2-50 |
| Chimpanzee fecal sample | This Study | Djo2-68 |
| Chimpanzee fecal sample | This Study | Djo2-8 |
| Chimpanzee fecal sample | This Study | Djo3-1 |
| Chimpanzee fecal sample | This Study | Djo3-2 |
| Chimpanzee fecal sample | This Study | El3-16 |
| Chimpanzee fecal sample | This Study | El3-17 |
| Chimpanzee fecal sample | This Study | El3-18 |
| Chimpanzee fecal sample | This Study | El3-4 |
| Chimpanzee fecal sample | This Study | El3-5 |
| Chimpanzee fecal sample | This Study | El3-6 |
| Chimpanzee fecal sample | This Study | Fjn1-10 |
| Chimpanzee fecal sample | This Study | Fjn1-20 |
| Chimpanzee fecal sample | This Study | Fjn1-21 |
| Chimpanzee fecal sample | This Study | Fjn1-22 |
| Chimpanzee fecal sample | This Study | Fjn1-42 |
| Chimpanzee fecal sample | This Study | Fjn2-13 |
| Chimpanzee fecal sample | This Study | Fjn2-50 |
| Chimpanzee fecal sample | This Study | Fjn2-52 |
| Chimpanzee fecal sample | This Study | Fjn2-62 |
| Chimpanzee fecal sample | This Study | Fjn2-7 |
| Chimpanzee fecal sample | This Study | Fjn2-9 |
| Chimpanzee fecal sample | This Study | Fjn3-24 |
| Chimpanzee fecal sample | This Study | Fjn3-43 |
| Chimpanzee fecal sample | This Study | Fjn3-53 |
| Chimpanzee fecal sample | This Study | Fjn3-54 |
| Chimpanzee fecal sample | This Study | Fjn3-56 |
| Chimpanzee fecal sample | This Study | Fjn3-68 |
| Chimpanzee fecal sample | This Study | Fjn3-84 |
| Chimpanzee fecal sample | This Study | Fouta1-10 |
| Chimpanzee fecal sample | This Study | Fouta1-17 |
| Chimpanzee fecal sample | This Study | Fouta1-6 |
| Chimpanzee fecal sample | This Study | Fouta2-8 |
| Chimpanzee fecal sample | This Study | Fouta3-1 |
| Chimpanzee fecal sample | This Study | Fouta3-15 |
| Chimpanzee fecal sample | This Study | Fouta3-25 |
| Chimpanzee fecal sample | This Study | Fouta3-29 |
| Chimpanzee fecal sample | This Study | Fouta3-30 |
| Chimpanzee fecal sample | This Study | Fouta3-32 |
| Chimpanzee fecal sample | This Study | Fouta3-34 |
| Chimpanzee fecal sample | This Study | Fouta3-35 |
| Chimpanzee fecal sample | This Study | Fouta3-37 |
| Chimpanzee fecal sample | This Study | Fouta3-38 |
| Chimpanzee fecal sample | This Study | Fouta3-40 |
| Chimpanzee fecal sample | This Study | Fouta3-51 |
| Chimpanzee fecal sample | This Study | Fouta3-55 |
| Chimpanzee fecal sample | This Study | Fouta3-80 |
| Chimpanzee fecal sample | This Study | Fouta3-82 |
| Chimpanzee fecal sample | This Study | Fouta3-87 |
| Chimpanzee fecal sample | This Study | Gas1-10 |
| Chimpanzee fecal sample | This Study | Gas1-17 |
| Chimpanzee fecal sample | This Study | Gas1-22 |
| Chimpanzee fecal sample | This Study | Gas1-23 |
| Chimpanzee fecal sample | This Study | Gas1-26 |
| Chimpanzee fecal sample | This Study | Gas1-27 |
| Chimpanzee fecal sample | This Study | Gas1-36 |
| Chimpanzee fecal sample | This Study | Gas1-5 |
| Chimpanzee fecal sample | This Study | Gas2-14 |
| Chimpanzee fecal sample | This Study | Gas2-19 |
| Chimpanzee fecal sample | This Study | Gas2-23 |
| Chimpanzee fecal sample | This Study | Gas2-28 |
| Chimpanzee fecal sample | This Study | Gas2-29 |
| Chimpanzee fecal sample | This Study | Gas2-34 |
| Chimpanzee fecal sample | This Study | Gas2-37 |
| Chimpanzee fecal sample | This Study | Gas2-4 |
| Chimpanzee fecal sample | This Study | Gas2-40 |
| Chimpanzee fecal sample | This Study | Gas2-55 |
| Chimpanzee fecal sample | This Study | Gas2-67 |
| Chimpanzee fecal sample | This Study | Gas2-7 |
| Chimpanzee fecal sample | This Study | GB-10-03 |
| Chimpanzee fecal sample | This Study | GB-11-10 |
| Chimpanzee fecal sample | This Study | GB-11-11 |
| Chimpanzee fecal sample | This Study | GB-13-13 |
| Chimpanzee fecal sample | This Study | GB-13-21 |
| Chimpanzee fecal sample | This Study | GB-14-05 |
| Chimpanzee fecal sample | This Study | GB-22-06 |
| Chimpanzee fecal sample | This Study | GB-25-02 |
| Chimpanzee fecal sample | This Study | GB-25-05 |
| Chimpanzee fecal sample | This Study | GB-28-02 |
| Chimpanzee fecal sample | This Study | GB-29-06 |
| Chimpanzee fecal sample | This Study | GB-30-11 |
| Chimpanzee fecal sample | This Study | GB-34-16 |
| Chimpanzee fecal sample | This Study | GB-34-22 |
| Chimpanzee fecal sample | This Study | GB-36-07 |
| Chimpanzee fecal sample | This Study | GB-36-16 |
| Chimpanzee fecal sample | This Study | GB-37-04 |
| Chimpanzee fecal sample | This Study | GB-37-09 |
| Chimpanzee fecal sample | This Study | Gbo1-10 |
| Chimpanzee fecal sample | This Study | Gbo1-13 |
| Chimpanzee fecal sample | This Study | Gbo1-15 |
| Chimpanzee fecal sample | This Study | Gbo1-27 |
| Chimpanzee fecal sample | This Study | Gbo1-41 |
| Chimpanzee fecal sample | This Study | Gbo1-53 |
| Chimpanzee fecal sample | This Study | Gbo1-86 |
| Chimpanzee fecal sample | This Study | Gbo2-10 |
| Chimpanzee fecal sample | This Study | Gbo2-2 |
| Chimpanzee fecal sample | This Study | Gbo2-25 |
| Chimpanzee fecal sample | This Study | Gbo2-43 |
| Chimpanzee fecal sample | This Study | Gbo2-48 |
| Chimpanzee fecal sample | This Study | Gbo2-57 |
| Chimpanzee fecal sample | This Study | Gbo2-59 |
| Chimpanzee fecal sample | This Study | Gbo2-63 |
| Chimpanzee fecal sample | This Study | Gbo2-66 |
| Chimpanzee fecal sample | This Study | Gbo2-85 |
| Chimpanzee fecal sample | This Study | Gbo3-17 |
| Chimpanzee fecal sample | This Study | Gbo3-2 |
| Chimpanzee fecal sample | This Study | Gco1-25 |
| Chimpanzee fecal sample | This Study | Gco1-32 |
| Chimpanzee fecal sample | This Study | Gco1-33_2 |
| Chimpanzee fecal sample | This Study | Gco1-37 |
| Chimpanzee fecal sample | This Study | Gco1-39 |
| Chimpanzee fecal sample | This Study | Gco1-42_2 |
| Chimpanzee fecal sample | This Study | Gco1-43 |
| Chimpanzee fecal sample | This Study | Gco1-44 |
| Chimpanzee fecal sample | This Study | Gco1-48 |
| Chimpanzee fecal sample | This Study | Gco1-5 |
| Chimpanzee fecal sample | This Study | Gco1-50 |
| Chimpanzee fecal sample | This Study | Gco1-51 |
| Chimpanzee fecal sample | This Study | Gco1-55 |
| Chimpanzee fecal sample | This Study | Gco1-56 |
| Chimpanzee fecal sample | This Study | Gco1-60 |
| Chimpanzee fecal sample | This Study | Gco1-61 |
| Chimpanzee fecal sample | This Study | Gco1-8 |
| Chimpanzee fecal sample | This Study | Gco2-5 |
| Chimpanzee fecal sample | This Study | Gco2-7 |
| Chimpanzee fecal sample | This Study | Gco2-8 |
| Chimpanzee fecal sample | This Study | Gco2-9 |
| Chimpanzee fecal sample | This Study | Gco4-2 |
| Chimpanzee fecal sample | This Study | Gep1-21 |
| Chimpanzee fecal sample | This Study | Gep1-23 |
| Chimpanzee fecal sample | This Study | Gep1-25 |
| Chimpanzee fecal sample | This Study | Gep1-26 |
| Chimpanzee fecal sample | This Study | Gep1-62 |
| Chimpanzee fecal sample | This Study | Gep1-65 |
| Chimpanzee fecal sample | This Study | Gep2-10 |
| Chimpanzee fecal sample | This Study | Gep2-20 |
| Chimpanzee fecal sample | This Study | Gep2-28 |
| Chimpanzee fecal sample | This Study | Gep2-29 |
| Chimpanzee fecal sample | This Study | Gep2-30 |
| Chimpanzee fecal sample | This Study | Gep2-37 |
| Chimpanzee fecal sample | This Study | Gep2-40 |
| Chimpanzee fecal sample | This Study | Gep2-41 |
| Chimpanzee fecal sample | This Study | Gep2-45 |
| Chimpanzee fecal sample | This Study | Gep2-48 |
| Chimpanzee fecal sample | This Study | Gep2-52 |
| Chimpanzee fecal sample | This Study | Gep2-53 |
| Chimpanzee fecal sample | This Study | Gep2-61 |
| Chimpanzee fecal sample | This Study | Gha-01-01 |
| Chimpanzee fecal sample | This Study | Gha-01-04 |
| Chimpanzee fecal sample | This Study | Gha-01-05 |
| Chimpanzee fecal sample | This Study | Gha-01-06 |
| Chimpanzee fecal sample | This Study | Gha-01-07 |
| Chimpanzee fecal sample | This Study | Gha-01-08 |
| Chimpanzee fecal sample | This Study | Gha-01-11 |
| Chimpanzee fecal sample | This Study | Gis1-1 |
| Chimpanzee fecal sample | This Study | Gis1-10 |
| Chimpanzee fecal sample | This Study | Gis1-11 |
| Chimpanzee fecal sample | This Study | Gis1-13 |
| Chimpanzee fecal sample | This Study | Gis1-17 |
| Chimpanzee fecal sample | This Study | Gis1-20 |
| Chimpanzee fecal sample | This Study | Gis1-21 |
| Chimpanzee fecal sample | This Study | Gis1-23 |
| Chimpanzee fecal sample | This Study | Gis1-24 |
| Chimpanzee fecal sample | This Study | Gis1-25 |
| Chimpanzee fecal sample | This Study | Gis1-4 |
| Chimpanzee fecal sample | This Study | Gis1-47 |
| Chimpanzee fecal sample | This Study | Gis1-5 |
| Chimpanzee fecal sample | This Study | Gis1-59 |
| Chimpanzee fecal sample | This Study | Gis1-6 |
| Chimpanzee fecal sample | This Study | Gis1-70 |
| Chimpanzee fecal sample | This Study | Gis1-8 |
| Chimpanzee fecal sample | This Study | Gis2-2 |
| Chimpanzee fecal sample | This Study | Gis2-50 |
| Chimpanzee fecal sample | This Study | Gis2-7 |
| Chimpanzee fecal sample | This Study | Gou1-14 |
| Chimpanzee fecal sample | This Study | Gou1-15 |
| Chimpanzee fecal sample | This Study | Gou1-18 |
| Chimpanzee fecal sample | This Study | Gou1-20 |
| Chimpanzee fecal sample | This Study | Gou1-21 |
| Chimpanzee fecal sample | This Study | Gou1-23 |
| Chimpanzee fecal sample | This Study | Gou1-24 |
| Chimpanzee fecal sample | This Study | Gou1-27 |
| Chimpanzee fecal sample | This Study | Gou1-38 |
| Chimpanzee fecal sample | This Study | Gou1-4 |
| Chimpanzee fecal sample | This Study | Gou1-40 |
| Chimpanzee fecal sample | This Study | Gou1-51 |
| Chimpanzee fecal sample | This Study | Gou1-58 |
| Chimpanzee fecal sample | This Study | Gou1-61 |
| Chimpanzee fecal sample | This Study | Gou1-66 |
| Chimpanzee fecal sample | This Study | Gou1-7 |
| Chimpanzee fecal sample | This Study | Gou1-70 |
| Chimpanzee fecal sample | This Study | Gou1-75 |
| Chimpanzee fecal sample | This Study | Gou1-8 |
| Chimpanzee fecal sample | This Study | Gou1-9 |
| Chimpanzee fecal sample | This Study | Itu-01-01 |
| Chimpanzee fecal sample | This Study | Itu-01-02 |
| Chimpanzee fecal sample | This Study | Itu-01-03 |
| Chimpanzee fecal sample | This Study | Itu-01-04 |
| Chimpanzee fecal sample | This Study | Itu-01-05 |
| Chimpanzee fecal sample | This Study | Itu-01-06 |
| Chimpanzee fecal sample | This Study | Itu-01-07 |
| Chimpanzee fecal sample | This Study | Itu-01-08 |
| Chimpanzee fecal sample | This Study | Itu-01-09 |
| Chimpanzee fecal sample | This Study | Itu-01-10 |
| Chimpanzee fecal sample | This Study | Itu-01-11 |
| Chimpanzee fecal sample | This Study | Itu-01-12 |
| Chimpanzee fecal sample | This Study | Ivi1-1 |
| Chimpanzee fecal sample | This Study | Ivi1-2 |
| Chimpanzee fecal sample | This Study | Kab1-1 |
| Chimpanzee fecal sample | This Study | Kab1-2 |
| Chimpanzee fecal sample | This Study | Kab1-3 |
| Chimpanzee fecal sample | This Study | Kab1-4 |
| Chimpanzee fecal sample | This Study | Kab1-5 |
| Chimpanzee fecal sample | This Study | Kab2-1 |
| Chimpanzee fecal sample | This Study | Kab2-4 |
| Chimpanzee fecal sample | This Study | Kab2-5 |
| Chimpanzee fecal sample | This Study | Kay1-12 |
| Chimpanzee fecal sample | This Study | Kay1-13 |
| Chimpanzee fecal sample | This Study | Kay1-15 |
| Chimpanzee fecal sample | This Study | Kay1-16 |
| Chimpanzee fecal sample | This Study | Kay1-17 |
| Chimpanzee fecal sample | This Study | Kay1-20 |
| Chimpanzee fecal sample | This Study | Kay1-23 |
| Chimpanzee fecal sample | This Study | Kay1-4 |
| Chimpanzee fecal sample | This Study | Kay2-20 |
| Chimpanzee fecal sample | This Study | Kay2-24 |
| Chimpanzee fecal sample | This Study | Kay2-25 |
| Chimpanzee fecal sample | This Study | Kay2-26 |
| Chimpanzee fecal sample | This Study | Kay2-29 |
| Chimpanzee fecal sample | This Study | Kay2-3 |
| Chimpanzee fecal sample | This Study | Kay2-32 |
| Chimpanzee fecal sample | This Study | Kay2-4 |
| Chimpanzee fecal sample | This Study | Kay2-41 |
| Chimpanzee fecal sample | This Study | Kay2-49 |
| Chimpanzee fecal sample | This Study | Kay2-52 |
| Chimpanzee fecal sample | This Study | Kay2-54 |
| Chimpanzee fecal sample | This Study | Kor1-12 |
| Chimpanzee fecal sample | This Study | Kor1-14 |
| Chimpanzee fecal sample | This Study | Kor1-15 |
| Chimpanzee fecal sample | This Study | Kor1-24 |
| Chimpanzee fecal sample | This Study | Kor1-25 |
| Chimpanzee fecal sample | This Study | Kor1-27 |
| Chimpanzee fecal sample | This Study | Kor1-34 |
| Chimpanzee fecal sample | This Study | Kor1-35 |
| Chimpanzee fecal sample | This Study | Kor1-65 |
| Chimpanzee fecal sample | This Study | Kor1-79 |
| Chimpanzee fecal sample | This Study | Kor1-8 |
| Chimpanzee fecal sample | This Study | Kor1-84 |
| Chimpanzee fecal sample | This Study | Kor2-1 |
| Chimpanzee fecal sample | This Study | Kor2-14 |
| Chimpanzee fecal sample | This Study | Kor2-17 |
| Chimpanzee fecal sample | This Study | Kor2-26 |
| Chimpanzee fecal sample | This Study | Kor2-35 |
| Chimpanzee fecal sample | This Study | Kor2-5 |
| Chimpanzee fecal sample | This Study | Kor2-8 |
| Chimpanzee fecal sample | This Study | LCA-3-10 |
| Chimpanzee fecal sample | This Study | LCA-3-12 |
| Chimpanzee fecal sample | This Study | Lib1-25D |
| Chimpanzee fecal sample | This Study | Lib1-6-D |
| Chimpanzee fecal sample | This Study | Lib2-10 |
| Chimpanzee fecal sample | This Study | Lib2-14 |
| Chimpanzee fecal sample | This Study | Lib2-15 |
| Chimpanzee fecal sample | This Study | Lib2-17 |
| Chimpanzee fecal sample | This Study | Lib2-2 |
| Chimpanzee fecal sample | This Study | Lib2-23 |
| Chimpanzee fecal sample | This Study | Lib2-26 |
| Chimpanzee fecal sample | This Study | Lib2-27 |
| Chimpanzee fecal sample | This Study | Lib2-28 |
| Chimpanzee fecal sample | This Study | Lib2-48 |
| Chimpanzee fecal sample | This Study | Lib2-62 |
| Chimpanzee fecal sample | This Study | Lib2-66 |
| Chimpanzee fecal sample | This Study | Lib3-34 |
| Chimpanzee fecal sample | This Study | Loma2-1 |
| Chimpanzee fecal sample | This Study | Loma2-2 |
| Chimpanzee fecal sample | This Study | Loma2-3 |
| Chimpanzee fecal sample | This Study | Loma2-4 |
| Chimpanzee fecal sample | This Study | Loma2-5 |
| Chimpanzee fecal sample | This Study | Loma2-6 |
| Chimpanzee fecal sample | This Study | Loma2-7 |
| Chimpanzee fecal sample | This Study | Lop1-13 |
| Chimpanzee fecal sample | This Study | Lop1-14 |
| Chimpanzee fecal sample | This Study | Lop1-23 |
| Chimpanzee fecal sample | This Study | Lop1-24 |
| Chimpanzee fecal sample | This Study | Lop1-25 |
| Chimpanzee fecal sample | This Study | Lop2-11 |
| Chimpanzee fecal sample | This Study | Lop2-16 |
| Chimpanzee fecal sample | This Study | Lop2-3 |
| Chimpanzee fecal sample | This Study | Lop2-34 |
| Chimpanzee fecal sample | This Study | Lop2-35 |
| Chimpanzee fecal sample | This Study | Lop2-43 |
| Chimpanzee fecal sample | This Study | Lop2-45 |
| Chimpanzee fecal sample | This Study | Lop2-76 |
| Chimpanzee fecal sample | This Study | Lop2-77 |
| Chimpanzee fecal sample | This Study | Lop2-80 |
| Chimpanzee fecal sample | This Study | Lop2-82 |
| Chimpanzee fecal sample | This Study | Lop2-88 |
| Chimpanzee fecal sample | This Study | Lop3-11 |
| Chimpanzee fecal sample | This Study | Lop3-14 |
| Chimpanzee fecal sample | This Study | Lop3-20 |
| Chimpanzee fecal sample | This Study | Mbe-02-01 |
| Chimpanzee fecal sample | This Study | Mbe-02-04 |
| Chimpanzee fecal sample | This Study | Mbe-02-05 |
| Chimpanzee fecal sample | This Study | Mbe-02-07 |
| Chimpanzee fecal sample | This Study | Mbe-02-09 |
| Chimpanzee fecal sample | This Study | Mbe-02-12 |
| Chimpanzee fecal sample | This Study | Mbe-02-13 |
| Chimpanzee fecal sample | This Study | Mbe1-10 |
| Chimpanzee fecal sample | This Study | Mbe1-12 |
| Chimpanzee fecal sample | This Study | Mbe1-13 |
| Chimpanzee fecal sample | This Study | Mbe1-15 |
| Chimpanzee fecal sample | This Study | Mbe1-16 |
| Chimpanzee fecal sample | This Study | Mbe1-17 |
| Chimpanzee fecal sample | This Study | Mbe1-18 |
| Chimpanzee fecal sample | This Study | Mbe1-19 |
| Chimpanzee fecal sample | This Study | Mbe1-2 |
| Chimpanzee fecal sample | This Study | Mbe1-20 |
| Chimpanzee fecal sample | This Study | Mbe1-21 |
| Chimpanzee fecal sample | This Study | Mbe1-22_2 |
| Chimpanzee fecal sample | This Study | Mbe1-23 |
| Chimpanzee fecal sample | This Study | Mbe1-24 |
| Chimpanzee fecal sample | This Study | Mbe1-25 |
| Chimpanzee fecal sample | This Study | Mbe1-26 |
| Chimpanzee fecal sample | This Study | Mbe1-4 |
| Chimpanzee fecal sample | This Study | Mbe1-5 |
| Chimpanzee fecal sample | This Study | Mbe1-7 |
| Chimpanzee fecal sample | This Study | Mbe1-9 |
| Chimpanzee fecal sample | This Study | Mtc1-26 |
| Chimpanzee fecal sample | This Study | Mtc1-40 |
| Chimpanzee fecal sample | This Study | Mtc1-43 |
| Chimpanzee fecal sample | This Study | Mtc1-54 |
| Chimpanzee fecal sample | This Study | Mtc1-55 |
| Chimpanzee fecal sample | This Study | Mtc1-56 |
| Chimpanzee fecal sample | This Study | Mtc1-58 |
| Chimpanzee fecal sample | This Study | Mtc1-63 |
| Chimpanzee fecal sample | This Study | Mtc1-66 |
| Chimpanzee fecal sample | This Study | Mtc1-67 |
| Chimpanzee fecal sample | This Study | Mtc1-71 |
| Chimpanzee fecal sample | This Study | Mtc1-72 |
| Chimpanzee fecal sample | This Study | MTC2-24 |
| Chimpanzee fecal sample | This Study | MTC2-31 |
| Chimpanzee fecal sample | This Study | MTC2-33 |
| Chimpanzee fecal sample | This Study | MTC2-40 |
| Chimpanzee fecal sample | This Study | MTC2-42 |
| Chimpanzee fecal sample | This Study | MTC2-5 |
| Chimpanzee fecal sample | This Study | MTC2-6 |
| Chimpanzee fecal sample | This Study | MTC2-7 |
| Chimpanzee fecal sample | This Study | N173-11 |
| Chimpanzee fecal sample | This Study | N173-14 |
| Chimpanzee fecal sample | This Study | N173-17 |
| Chimpanzee fecal sample | This Study | N181-11 |
| Chimpanzee fecal sample | This Study | N181-14 |
| Chimpanzee fecal sample | This Study | N182-2 |
| Chimpanzee fecal sample | This Study | N183-5 |
| Chimpanzee fecal sample | This Study | N183-6 |
| Chimpanzee fecal sample | This Study | N186-8 |
| Chimpanzee fecal sample | This Study | N186-9 |
| Chimpanzee fecal sample | This Study | N190-3 |
| Chimpanzee fecal sample | This Study | N259-5 |
| Chimpanzee fecal sample | This Study | N259-6 |
| Chimpanzee fecal sample | This Study | N259-8 |
| Chimpanzee fecal sample | This Study | N260-10 |
| Chimpanzee fecal sample | This Study | N260-6 |
| Chimpanzee fecal sample | This Study | N260-8 |
| Chimpanzee fecal sample | This Study | N261-3 |
| Chimpanzee fecal sample | This Study | N261-5 |
| Chimpanzee fecal sample | This Study | N262-4 |
| Chimpanzee fecal sample | This Study | Ngi1-1 |
| Chimpanzee fecal sample | This Study | Ngi1-2 |
| Chimpanzee fecal sample | This Study | Ngi1-3 |
| Chimpanzee fecal sample | This Study | Ngi1-4 |
| Chimpanzee fecal sample | This Study | Ngi1-5 |
| Chimpanzee fecal sample | This Study | Ngi1-7 |
| Chimpanzee fecal sample | This Study | Ngi1-8 |
| Chimpanzee fecal sample | This Study | Ngi2-1 |
| Chimpanzee fecal sample | This Study | Ngi2-3 |
| Chimpanzee fecal sample | This Study | Ngi2-4 |
| Chimpanzee fecal sample | This Study | Ngi2-5 |
| Chimpanzee fecal sample | This Study | Ngi2-6 |
| Chimpanzee fecal sample | This Study | Ngi2-7 |
| Chimpanzee fecal sample | This Study | Ngi2-8 |
| Chimpanzee fecal sample | This Study | Nim1-10 |
| Chimpanzee fecal sample | This Study | Nim1-2 |
| Chimpanzee fecal sample | This Study | Nim1-3 |
| Chimpanzee fecal sample | This Study | Nim1-47 |
| Chimpanzee fecal sample | This Study | Nim1-49 |
| Chimpanzee fecal sample | This Study | Nim1-5 |
| Chimpanzee fecal sample | This Study | Nim1-51 |
| Chimpanzee fecal sample | This Study | Nim1-52 |
| Chimpanzee fecal sample | This Study | Nim1-7 |
| Chimpanzee fecal sample | This Study | Nim1-77 |
| Chimpanzee fecal sample | This Study | Nim1-78 |
| Chimpanzee fecal sample | This Study | Nim1-79 |
| Chimpanzee fecal sample | This Study | Nim2-12 |
| Chimpanzee fecal sample | This Study | Nim2-17 |
| Chimpanzee fecal sample | This Study | Nim2-3 |
| Chimpanzee fecal sample | This Study | Nim2-33 |
| Chimpanzee fecal sample | This Study | Nim2-34 |
| Chimpanzee fecal sample | This Study | Nim2-35 |
| Chimpanzee fecal sample | This Study | Nim2-44 |
| Chimpanzee fecal sample | This Study | Nim2-58 |
| Chimpanzee fecal sample | This Study | NNP1-11 |
| Chimpanzee fecal sample | This Study | NNP1-15 |
| Chimpanzee fecal sample | This Study | NNP1-26 |
| Chimpanzee fecal sample | This Study | NNP1-34 |
| Chimpanzee fecal sample | This Study | NNP1-40 |
| Chimpanzee fecal sample | This Study | NNP1-44 |
| Chimpanzee fecal sample | This Study | NNP1-54 |
| Chimpanzee fecal sample | This Study | NNP1-57 |
| Chimpanzee fecal sample | This Study | NNP1-77 |
| Chimpanzee fecal sample | This Study | NNP1-86 |
| Chimpanzee fecal sample | This Study | NNP2-2 |
| Chimpanzee fecal sample | This Study | NNP2-35 |
| Chimpanzee fecal sample | This Study | NNP2-4 |
| Chimpanzee fecal sample | This Study | NNP2-54 |
| Chimpanzee fecal sample | This Study | NNP2-55 |
| Chimpanzee fecal sample | This Study | NNP2-67 |
| Chimpanzee fecal sample | This Study | NNP2-68 |
| Chimpanzee fecal sample | This Study | NNP2-74 |
| Chimpanzee fecal sample | This Study | NNP2-79 |
| Chimpanzee fecal sample | This Study | NNP3-14 |
| Chimpanzee fecal sample | This Study | Onp1-11 |
| Chimpanzee fecal sample | This Study | Onp1-12 |
| Chimpanzee fecal sample | This Study | Onp1-2 |
| Chimpanzee fecal sample | This Study | Onp1-20 |
| Chimpanzee fecal sample | This Study | Onp1-21 |
| Chimpanzee fecal sample | This Study | Onp1-24 |
| Chimpanzee fecal sample | This Study | Onp1-25 |
| Chimpanzee fecal sample | This Study | Onp1-26 |
| Chimpanzee fecal sample | This Study | Onp1-27 |
| Chimpanzee fecal sample | This Study | Onp1-28 |
| Chimpanzee fecal sample | This Study | Onp1-29 |
| Chimpanzee fecal sample | This Study | Onp1-31 |
| Chimpanzee fecal sample | This Study | Onp1-32 |
| Chimpanzee fecal sample | This Study | Onp1-34 |
| Chimpanzee fecal sample | This Study | Onp1-35 |
| Chimpanzee fecal sample | This Study | Onp1-39 |
| Chimpanzee fecal sample | This Study | Onp1-6 |
| Chimpanzee fecal sample | This Study | Onp1-7 |
| Chimpanzee fecal sample | This Study | Onp1-8 |
| Chimpanzee fecal sample | This Study | Onp1-9 |
| Chimpanzee fecal sample | This Study | Rt1-1 |
| Chimpanzee fecal sample | This Study | Rt1-2 |
| Chimpanzee fecal sample | This Study | Rt1-5 |
| Chimpanzee fecal sample | This Study | Rt1-7 |
| Chimpanzee fecal sample | This Study | Rt1-8 |
| Chimpanzee fecal sample | This Study | Rt2-1 |
| Chimpanzee fecal sample | This Study | Rt2-14 |
| Chimpanzee fecal sample | This Study | Rt2-21 |
| Chimpanzee fecal sample | This Study | Rt2-22 |
| Chimpanzee fecal sample | This Study | Rt2-24 |
| Chimpanzee fecal sample | This Study | Rt2-25 |
| Chimpanzee fecal sample | This Study | Rt2-26_2 |
| Chimpanzee fecal sample | This Study | Rt2-31 |
| Chimpanzee fecal sample | This Study | Rt2-35 |
| Chimpanzee fecal sample | This Study | Rt2-37 |
| Chimpanzee fecal sample | This Study | Rt2-38 |
| Chimpanzee fecal sample | This Study | Rt2-41 |
| Chimpanzee fecal sample | This Study | Rt2-6 |
| Chimpanzee fecal sample | This Study | Rt2-7 |
| Chimpanzee fecal sample | This Study | Rt2-8 |
| Chimpanzee fecal sample | This Study | San1-13 |
| Chimpanzee fecal sample | This Study | San1-17 |
| Chimpanzee fecal sample | This Study | San1-19 |
| Chimpanzee fecal sample | This Study | San1-2 |
| Chimpanzee fecal sample | This Study | San1-20 |
| Chimpanzee fecal sample | This Study | San1-22 |
| Chimpanzee fecal sample | This Study | San1-3 |
| Chimpanzee fecal sample | This Study | San1-32 |
| Chimpanzee fecal sample | This Study | San1-39 |
| Chimpanzee fecal sample | This Study | San1-4 |
| Chimpanzee fecal sample | This Study | San2-1 |
| Chimpanzee fecal sample | This Study | San2-10 |
| Chimpanzee fecal sample | This Study | San2-13 |
| Chimpanzee fecal sample | This Study | San2-16 |
| Chimpanzee fecal sample | This Study | San2-20 |
| Chimpanzee fecal sample | This Study | San2-26 |
| Chimpanzee fecal sample | This Study | San2-48 |
| Chimpanzee fecal sample | This Study | San2-49 |
| Chimpanzee fecal sample | This Study | San2-53 |
| Chimpanzee fecal sample | This Study | San2-59 |
| Chimpanzee fecal sample | This Study | Sob1-24 |
| Chimpanzee fecal sample | This Study | Sob1-27 |
| Chimpanzee fecal sample | This Study | Sob1-31 |
| Chimpanzee fecal sample | This Study | Sob1-32 |
| Chimpanzee fecal sample | This Study | Sob1-33 |
| Chimpanzee fecal sample | This Study | Sob1-4 |
| Chimpanzee fecal sample | This Study | Sob1-47 |
| Chimpanzee fecal sample | This Study | Sob1-5 |
| Chimpanzee fecal sample | This Study | Sob1-56 |
| Chimpanzee fecal sample | This Study | Sob1-57 |
| Chimpanzee fecal sample | This Study | Sob1-6 |
| Chimpanzee fecal sample | This Study | Sob1-7 |
| Chimpanzee fecal sample | This Study | Sob1-77 |
| Chimpanzee fecal sample | This Study | Sob1-83_2 |
| Chimpanzee fecal sample | This Study | Sob1-84 |
| Chimpanzee fecal sample | This Study | Sob2-12 |
| Chimpanzee fecal sample | This Study | Sob2-3 |
| Chimpanzee fecal sample | This Study | Sob2-37 |
| Chimpanzee fecal sample | This Study | Sob2-43 |
| Chimpanzee fecal sample | This Study | Sob2-5 |
| Chimpanzee fecal sample | This Study | Tai_R1-23 |
| Chimpanzee fecal sample | This Study | Tai_R1-26 |
| Chimpanzee fecal sample | This Study | Tai_R1-28 |
| Chimpanzee fecal sample | This Study | Tai_R1-4 |
| Chimpanzee fecal sample | This Study | Tai_R2-13 |
| Chimpanzee fecal sample | This Study | Tai_R2-15 |
| Chimpanzee fecal sample | This Study | Tai_R2-16 |
| Chimpanzee fecal sample | This Study | Tai_R2-18 |
| Chimpanzee fecal sample | This Study | Tai_R2-22 |
| Chimpanzee fecal sample | This Study | Tai_R2-30 |
| Chimpanzee fecal sample | This Study | Tai_R2-4 |
| Chimpanzee fecal sample | This Study | Tai_R2-43 |
| Chimpanzee fecal sample | This Study | Tai_R2-5 |
| Chimpanzee fecal sample | This Study | Tai_R2-52 |
| Chimpanzee fecal sample | This Study | Tai_R2-57 |
| Chimpanzee fecal sample | This Study | Tai_R2-6 |
| Chimpanzee fecal sample | This Study | Tai_R2-8 |
| Chimpanzee fecal sample | This Study | Tai_R2-80 |
| Chimpanzee fecal sample | This Study | Tai_R2-88 |
| Chimpanzee fecal sample | This Study | Tai_R2-9 |
| Chimpanzee fecal sample | This Study | Tai-E1-13 |
| Chimpanzee fecal sample | This Study | Tai-E1-42 |
| Chimpanzee fecal sample | This Study | Tai-E1-50 |
| Chimpanzee fecal sample | This Study | Tai-E1-52 |
| Chimpanzee fecal sample | This Study | Tai-E1-54 |
| Chimpanzee fecal sample | This Study | Tai-E1-55 |
| Chimpanzee fecal sample | This Study | Tai-E1-56 |
| Chimpanzee fecal sample | This Study | Tai-E1-58 |
| Chimpanzee fecal sample | This Study | Tai-E1-60 |
| Chimpanzee fecal sample | This Study | Tai-E1-7 |
| Chimpanzee fecal sample | This Study | Tai-E1-8 |
| Chimpanzee fecal sample | This Study | Tai-E2-11 |
| Chimpanzee fecal sample | This Study | Tai-E2-18 |
| Chimpanzee fecal sample | This Study | Tai-E2-29 |
| Chimpanzee fecal sample | This Study | Tai-E2-31 |
| Chimpanzee fecal sample | This Study | Tai-E2-35 |
| Chimpanzee fecal sample | This Study | Tai-E2-48 |
| Chimpanzee fecal sample | This Study | Tai-E2-51 |
| Chimpanzee fecal sample | This Study | Tai-E2-8 |
| Chimpanzee fecal sample | This Study | Uga1-1 |
| Chimpanzee fecal sample | This Study | Uga1-11 |
| Chimpanzee fecal sample | This Study | Uga1-12 |
| Chimpanzee fecal sample | This Study | Uga1-17 |
| Chimpanzee fecal sample | This Study | Uga1-22 |
| Chimpanzee fecal sample | This Study | Uga1-34 |
| Chimpanzee fecal sample | This Study | Uga1-9 |
| Chimpanzee fecal sample | This Study | Uga2-1 |
| Chimpanzee fecal sample | This Study | Uga2-22 |
| Chimpanzee fecal sample | This Study | Uga2-28 |
| Chimpanzee fecal sample | This Study | Uga2-31 |
| Chimpanzee fecal sample | This Study | Uga2-41 |
| Chimpanzee fecal sample | This Study | Uga2-44 |
| Chimpanzee fecal sample | This Study | Uga2-46 |
| Chimpanzee fecal sample | This Study | Uga2-49 |
| Chimpanzee fecal sample | This Study | Uga2-53 |
| Chimpanzee fecal sample | This Study | Uga2-73 |
| Chimpanzee fecal sample | This Study | Uga2-74 |
| Chimpanzee fecal sample | This Study | Uga2-81 |
| Chimpanzee fecal sample | This Study | Uga3-29 |
| Chimpanzee hair sample | This Study | Africa_Mona |
| Chimpanzee hair sample | This Study | Bea_Mona |
| Chimpanzee hair sample | This Study | Charly_Mona |
| Chimpanzee blood sample | This Study | Cheeta_Mona |
| Chimpanzee hair sample | This Study | Cheeta_Rainfer |
| Chimpanzee hair sample | This Study | Coco_Mona |
| Chimpanzee hair sample | This Study | Gombe_Rainfer |
| Chimpanzee blood sample | This Study | Guille_Rainfer |
| Chimpanzee blood sample | This Study | Iván_Rainfer |
| Chimpanzee blood sample | This Study | Jackie_Rainfer |
| Chimpanzee blood sample | This Study | Judi_Rainfer |
| Chimpanzee hair sample | This Study | Lulú_Rainfer |
| Chimpanzee hair sample | This Study | Marco_Mona |
| Chimpanzee blood sample | This Study | Maxi_Rainfer |
| Chimpanzee hair sample | This Study | Nico_Mona |
| Chimpanzee hair sample | This Study | Sammy_Rainfer |
| Chimpanzee hair sample | This Study | Sandy_Rainfer |
| Chimpanzee hair sample | This Study | Tico_Mona |
| Chimpanzee hair sample | This Study | Toni_Mona |
| Chimpanzee blood sample | This Study | Toti_Rainfer |
| Critical commercial assays | ||
| QIAamp Fast DNA Stool Mini Kit | Qiagen | cat#51604 |
| High Sensitivity Genomic DNA 50Kb Analysis kit | Advanced Analytical | cat#DNF-488 |
| Bioanalyzer Agilent DNA 7500 kit | Agilent | cat#5067-1506 |
| Bioanalyzer High Sensitivity DNA Analysis | Agilent | cat#5067-4626 |
| Deposited data | ||
| Raw sequencing reads | This study | ENA: PRJEB46115 |
| Chimpanzee genomes | de Manuel et al., 20166 | ENA: PRJEB15086 |
| Chimpanzee genomes | Prado-Martinez et al., 20133 | SRA: PRJNA18943 and SRP018689 |
| Homo sapiens reference genome (Hg19 or GRCh37) | Church et al., 201168 | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ |
| Pan troglodytes reference genome (panTro6) | Kronenberg et al., 201869 | https://www.ncbi.nlm.nih.gov/assembly/GCF_002880755.1/ |
| Papio anubis reference genome (Panu_3.0) | Roger et al., 201970 | https://www.ncbi.nlm.nih.gov/assembly/GCF_000264685.3/ |
| green monkey (Chlorocebus_sabeus_1.1) | Warren et al., 201571 | https://www.ncbi.nlm.nih.gov/assembly/GCF_000409795.2/ |
| Colobus angolensis palliatus reference genome (Cang.pa_1.0) | Genereux et al., 202072 | https://www.ncbi.nlm.nih.gov/assembly/GCF_000951035.1/ |
| Cercocebus atys reference genome (Caty_1.0) | Genereux et al., 202072 | https://www.ncbi.nlm.nih.gov/assembly/GCF_000955945.1/ |
| Gorilla gorilla gorilla reference genome (gorGor4) | Scally et al., 201273 | https://www.ncbi.nlm.nih.gov/assembly/GCF_000151905.2/ |
| Mandrillus leucophaeus reference genome (Mleu.le_1.0) | Genereux et al., 202072 | https://www.ncbi.nlm.nih.gov/assembly/GCF_000951045.1 |
| Erythrocebus patas reference genome (EryPat_v1_BIUU) | Genereux et al., 202072 | https://www.ncbi.nlm.nih.gov/assembly/GCA_004027335.1/ |
| Cercopithecus neglectus reference genome (CertNeg_v1_BIUU) | Genereux et al., 202072 | https://www.ncbi.nlm.nih.gov/assembly/GCA_004027615.1 |
| Mandrillus sphinx (BGI_mandrill_1.0) | Yin et al., 202074 | https://www.ncbi.nlm.nih.gov/assembly/GCA_004802615.1/ |
| Oligonucleotides | ||
| Univ_Block_P7: 5′-AGATCGGAAGAGCACACG TCTGAACTCCAGTCAC-Pho-3′ |
Rohland and Reich, 2012;75 Sigma-Aldrich | N/A |
| Univ_Block_P5: 5′-AGATCGGAAGAGCGTCGT GTAGGGAAAG-Pho-3′ |
Rohland and Reich, 2012;75 Sigma-Aldrich | N/A |
| P5_Indexing_Primer: 5′-AATGATACGGCGACCA CCGAGATCTACACNNNNNNNACACTCTTTCCC TACACGACGCTCTT-3′ |
Meyer and Kircher, 2010;76 Sigma-Aldrich |
N/A |
| P7_Indexing_Primer: 3′-TGTGCAGACTTGAGGT CAGTGNNNNNNNTAGAGCATACGGCAGAAGA CGAAC-5′ |
Meyer and Kircher, 2010;76 Sigma-Aldrich |
N/A |
| PreHyb_P5_F 5′-CTTTCCCTACACGACGCTCTTC-3′ | Meyer and Kircher, 2010;76 Sigma-Aldrich |
N/A |
| PreHyb_P7_R 3′-GTGTGCAGACTTGAGGTCAGTG-5’ | Meyer and Kircher, 2010;76 Sigma-Aldrich |
N/A |
| F_P5_7nt_XX Indexed Adapter: 5′-CTTTCCCTACAC GACGCTCTTCCGATCTNNNNNNN-3′ |
Rohland and Reich, 2012;75 Teknokroma | N/A |
| F_P7_7nt_XX Indexed Adapter: 5′-GTGACTGGAGT TCAGACGTGTGCTCTTCCGATCTNNNNNNN-3′ |
Rohland and Reich, 2012;75 Teknokroma | N/A |
| R_P5/P7_7nt_XX Indexed Common Adapter: 5′-NN NNNNNAGATCGGAA-3′ |
Rohland and Reich, 2012;75 Teknokroma | N/A |
| Ns represent indexes | N/A | N/A |
| Software and algorithms | ||
| Admixfrog | Peter, 202140 | https://github.com/BenjaminPeter/admixfrog |
| AdmixTools | Patterson et al., 201277 | https://github.com/DReichLab/AdmixTools |
| ANGSD v0.916 | Meisner and Albrechtsen, 201836 | http://www.popgen.dk/angsd/index.php/ANGSD |
| BBsplit | N/A | https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/ |
| BEDtools v2.22.1 | Quinlan and Hall, 201078 | https://bedtools.readthedocs.io/en/latest/ |
| BWA-mem v0.7.12 | Li and Durbin, 200979 | https://bio-bwa.sourceforge.net/ |
| EEMS | Petkova et al., 201642 | https://github.com/dipetkov/eems |
| FastMe v2.1.5 | Lefort et al., 201580 | https://www.atgc-montpellier.fr/fastme/ |
| GATK v3.7 | McKenna et al., 201081 | https://gatk.broadinstitute.org/hc/en-us |
| HuConTest | Kuhlwilm et al., 202133 | https://github.com/kuhlwilm/HuConTest |
| IBDseq | Browning and Browning, 201345 | https://faculty.washington.edu/browning/ibdseq.html |
| Mapdamage v2.0 | Jónsson et al., 201382 | https://ginolhac.github.io/mapDamage/ |
| NGSAdmix | Skotte et al., 201383 | https://www.popgen.dk/software/index.php/NgsAdmix |
| ngsDist v1.0.2 | Vieira et al.,201684 | https://github.com/fgvieira/ngsDist |
| NgsRelate | Korneliussen and Moltke, 201385 | https://github.com/ANGSD/NgsRelate |
| PCAngsd V0.8 | Meisner and Albrechtsen, 201836 | https://www.popgen.dk/software/index.php/PCAngsd |
| Picard v1.95 | N/A | https://broadinstitute.github.io/picard |
| PLINK v1.9 | Purcell et al., 200786 | https://www.cog-genomics.org/plink/ |
| R pacakge maptools | Bivand and Lewin-Koh, 201387 | https://cran.r-project.org/web/packages/maptools/index.html |
| R package admixr | Petr et al., 201988 | https://cran.r-project.org/web/packages/admixr/index.html |
| R package ape v5.4-1 | Paradis and Schliep, 201989 | https://cran.r-project.org/package=ape |
| R package Phangorn | Schliep, 201190 | https://cran.r-project.org/web/packages/phangorn/index.html |
| R package phytools | Revell, 201291 | https://cran.r-project.org/web/packages/phytools/index.html |
| R package rEEMSplots | Petkova et al., 201642 | https://github.com/dipetkov/eems |
| R package sf | Pebesma, 201892 | https://cran.r-project.org/web/packages/sf/index.html |
| R package sp | Pebesma and Bivand, 200593 | https://cran.r-project.org/web/packages/sp/index.html |
| R package Vegan | Oksanen et al., 202094 | https://CRAN.R-project.org/package=vegan |
| R v3.6.3 | R Core Team95 | https://www.R-project.org/ |
| rareCAGA | This study |
https://github.com/kuhlwilm/rareCAGA https://doi.org/10.5281/zenodo.6199201 |
| RAxML-NG v0.9.0 | Kozlov et al., 201996 | https://github.com/amkozlov/raxml-ng |
| realSFS v0.916 | Nielsen et al., 201297 | http://www.popgen.dk/angsd/index.php/RealSFS |
| Sabre | N/A | https://github.com/najoshi/sabre |
| Samtools v1.5 | Li et al., 200998 | https://www.htslib.org/ |
| snpAD v0.3.2 | Prüfer, 201899 | https://bioinf.eva.mpg.de/snpAD/ |
| TreeMix v1.12 | Pickrell and Pritchard, 2012100 | https://bitbucket.org/nygcresearch/treemix/wiki/Home |
| Trimmomatic v0.36 | Bolger et al., 2014101 | http://www.usadellab.org/cms/?page=trimmomatic |
| vcftools v0.1.12b | Danecek, 2011102 | http://vcftools.sourceforge.net/ |
| Other | ||
| SureSelect Custom Array (chr21) | Agilent | N/A |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact, Tomas Marques-Bonet (tomas.marques@upf.edu) or Mimi Arandjelovic (arandjel@eva.mpg.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All fecal and shed hair samples from wild chimpanzees included in this study were collected in a non-invasive manner, following standard practices and with no animal contact and no direct observation of the animals under study. Fecal samples from a zoo chimpanzee in Ghana were also obtained non-invasively. Full research approval, sample collection approval and research and sample permits of national ministries and protected area authorities were obtained in all countries of study. Sample export was also done with all necessary certificates, export and import permits. Fecal samples are exempt from the Convention on the Trade in Endangered Species of Wild Fauna and Flora (CITES), CITES permits were obtained for all hair samples. For all PanAf samples and research sites, research permits, veterinary certificates, certificates of origin, national export and import (German) permits and CITES import and export permits (when needed by export countries) were obtained by the PanAf through the Max Planck Institute for Evolutionary Anthropology, Department of Primatology, reviewed by the designated department officer and approved by national export and import officials. All documents are permanently stored with the PanAf and copies are electronically archived with the Max Planck Society. Hair and blood samples from chimpanzees in the Spanish rescue centers (Fundació Mona and Rainfer) were collected during a routine veterinary check of the animals. All laboratory work conforms to the relevant regulatory standards of the Max Planck Society, Germany and University Pompeu Fabra, Spain. No experiments were undertaken with live animals.
Method details
Sample selection and sequencing
Fecal DNA was extracted from a total of 5,397 PanAf samples and screened with a microsatellite genotyping assay,11 leaving only non-related samples, from which a minimum of 20 samples per location were selected for further sequencing whenever possible (Figure S1). Samples were randomized in batches of 24–48 samples and processed on different days for library preparation. A unique double-inline barcoded library was prepared for each sample following the BEST protocol with minor modifications.25,24 Pooling for capture was devised based on the host DNA content (fraction of chimpanzee DNA, relative to gut microbial and exogenous DNA) (Supplemental Note 1, Figures S3, S4, Tables S1–S3).28,25 Each pool was divided into several aliquots to perform multiple hybridizations (Figure S5). Afterwards, with predesigned RNA baits (SureSelect Agilent), we captured the non-repetitive regions of chromosome 21 following the protocol provided by the Agilent Sureselect Custom Array, adding two consecutive hybridization rounds for pools containing samples with <5% host DNA. Captured libraries were sequenced on the HiSeq 4000 Illumina platform with 2 × 100 paired-end reads.
Data processing and filtering
We processed the data to demultiplex libraries belonging to the same hybridization pool using Sabre (https://github.com/najoshi/sabre) and reads were trimmed with Trimmomatic (version 0.36).101 Paired-end reads were then aligned to the human genome Hg19 (GRCh37, Feb.2009 (GCA_000001405.1))68 using BWA (version 0.7.12).79 Duplicates were removed using PicardTools (version 1.95) (http://broadinstitute.github.io/picard/) and further filtering of the reads was done using samtools (version 1.5).98 To retrieve the on-target reads we used intersectBed from the BEDTOOLS package (version 2.22.1).78 Average coverage of the target space was calculated as the number of bases in the target region divided by the size of the target space (Figures S6 and S7). We obtained genotype likelihoods using ANGSD35 version 0.916 and genotype calls using snpAD99 v0.3.2, a software that takes DNA damage and biases into account for genotype calling (Figure S2). Analyses of error damage patterns and genotype discovery can be found in Supplemental Note 3 (Figures S20–S28). Principal Component Analysis (PCA) was performed using PCAngsd36 (Supplemental Note 3, Figures S9–S13 and S30–S34). Sources of primate contamination in fecal samples were determined using BBsplit (https://sourceforge.net/projects/bbmap/), mapping to 11 different primate genomes (Supplemental Note 3, Figures S14 and S15). Human contamination was estimated as the fraction of the number of observations of human-like alleles across all positions where chimpanzees and humans consistently differ, using the available script HuConTest33 which has been designed and tested for this purpose, and shown to work on fecal samples at very low coverage (Supplemental Note 3, Figures S16 and S17). Although samples had been screened prior to library preparation with a microsatellite assay,103,104 we used NgsRelate105 (Supplemental Note 3) to identify and remove identical or putative first order relative individuals (Figures S18 and S19). Due to the high variation of sample qualities and specific requirements for the application of different methods, a variety of filtering procedures was applied. In most analyses, samples with evidence of contamination from either human (>1% or >0.5%) or other primate species were removed, as well as first degree relatives and identical samples (n = 89), as well as samples that were found to be most likely mislabeled (n = 2) (Supplemental Note 5, Table S1, Figure S30). Finally, we used samples with different coverage cutoffs for different analyses (0.5-fold, 1-fold or 5-fold), as indicated for each method. The minimum coverage cutoff for the initial PCA was decided at 0.5-fold, so at least half of the chr21 would be covered on average by at least 1 read, which would provide sufficient expected overlap of variants between individuals.
Quantification and statistical analysis
Population genetics
To obtain pairwise FST estimates between sampling sites, we computed the 2-dimensional SFS (2d SFS) between each pair of geographical sites with ANGSD -doSaf 1 and realSFS.35 The genetic relationships between populations were used to build a matrix, from which we constructed a neighbor-joining tree using the ape package89 in R (version 3.5.2) (Figures S45–S48). F3 outgroup statistics were calculated between sampling sites using qp3Pop77 and taking an orangutan (Pongo pygmaeus) as the outgroup (pygmaeus_ERS1986511);106 this also ensures that low remaining amounts of human contamination would not influence the analysis on genotypes called on the human genome (Figures S54–S56). Regions of Homozygosity (RoH) were defined as heterozygous positions with a distance larger than 100 kbp, irrespective of missing information in between, for individuals with more than 5-fold average coverage in the target space (Figures S40–S42). We defined short RoHs as those between 10 and 100 kbp, and long RoHs as those longer than 100 kbp, following a previous approach.51 Long-term effective migration rates were calculated using EEMS42 with samples of more than 5-fold coverage (Supplemental Note 10, Figures S82–S85). The same dataset was used to obtain IBD-like tracts using the IBDseq software, which does not require phasing of the data.45 To increase the power to detect IBD-like fragments in such sparse dataset, we restricted the analysis on samples with >5-fold coverage, we included the genotype data on the chromosome 21 of 59 previously published whole-genome chimpanzees,6 and we kept only genotypes with a depth of at least eight reads (Supplemental Note 10, Figures S86–S88). We observed an exponential decay of IBD-like tract lengths with geographical distance within eastern and western chimpanzees (Figures S89 and S90), as expected for isolation-by-distance. The number of shared IBD-like tracts is likely the consequence of recent migration events or the shared population history between geographic sites or areas (Supplemental Note 10, Figures S91 and S93). The length of the shared segments is correlated with the time of such genetic exchange, with more recent migration resulting in longer IBD-like tracts. Therefore, a way to estimate the age of an IBD-like segment is by using its length. When the time (in generations g) to the most recent common ancestor (MRCA) is known, the total length (in cM) of a shared IBD segment follows an exponential distribution with rate 100/2g. Therefore, to time the events, we followed this rate of g = 100/(2∗cM),49,107 with cM being the length of the fragments, and g the number of generations. The length in cM was estimated from the length in Mbps by applying the western chimpanzee recombination map108 to the same subspecies and assuming an effective population size of Ne = 17,378.6 For the rest of the subspecies we used the Nigeria-Cameroon recombination map,109 with the following effective population sizes for each subspecies: central Ne = 47,314, eastern Ne = 32,492 and Nigeria-Cameroon Ne = 27,795.6 For timing the events between subspecies, we assumed a constant recombination map of 1cM/1Mbp since the recombination maps differ substantially between subspecies. We assumed a generation time of 25 years to calculate the time.110 We took the maximum IBD-like tract length per pair of individuals between sites to estimate the time frame of connectivity per site, and calculated average, maximum and minimum for each subspecies. Our reported expected time to the MRCA, derived from the length of the IBD-like fragments, can encompass large confidence intervals as other factors (technical and biological) could modify it. The log connectivity ratio of each sampling site was calculated as the sum of IBD-like tract counts (normalized by the number of pairwise sample comparisons between sites) that each site shares with the other sites, over the median global average of normalized IBD-like tract counts between all sampling sites (Figures S92, S94, Table S10). Subspecies ancestry introgressed fragments and bonobo introgression were determined with admixfrog.40 Admixfrog is a newly developed method to reliably infer ancestry fragments even from low-coverage and contaminated data. It uses a Hidden Markov Model (HMM) to infer local ancestry in a target individual from different sources which represent the admixing populations. Here, we use as potential sources of admixture 10 bonobo genomes and 16 chimpanzees (4 of each subspecies) from previous publications. The reference panel on chromosome 21 (source) was built using an equal number of individual genomes of each chimpanzee subspecies (16 genomes), 10 bonobo genomes,6 two human genomes111 (to serve as potential source of contamination and remove its effect) and 1 orangutan106 (as ancestral state). We recovered the global simulated runs of ancestry (from .res2 file) (Figures S61 and S63–S65) and the called runs of ancestry (from.rle file) (Figures S62 and S66), following the instructions of the method (link to https://github.com/BenjaminPeter/admixfrog). A Wilcoxon ranked test was performed in R, correcting for multiple testing with p.adjust (method = ”BH”) in R (Supplemental Note 8).
Rare alleles
Rare variation was used to assess connectivity between geographic regions in the recent past (1.5kya-15kya), and to estimate the most probable origin of chimpanzee fecal samples (Supplemental Note 9). For each sampling location (38 locations) with at least one individual at more than 1-fold coverage and less than 0.5% human contamination, we determined positions that were derived at the location itself and observed with a cumulative frequency of less than 1 across all other sampling locations (434 individuals, on average 11 per location; 963,656 SNPs, on average 26,671 per location) (Figures S67–S72, Table S6). Observation of a quality genotype (Supplemental Note 3) in either allele state (ancestral or derived) was required for at least 2 sampling locations, while missing data was ignored. The proportion of shared near-private sites of all observed near-private sites for each reference population was calculated, with heterozygous and homozygous derived positions equally treated as derived. Spatial modeling and kriging to the chimpanzee range were performed using the R package gstat112 to create a surface of rare allele sharing (R version 3.5.0). Accuracy was assessed by leaving one location out, calculating rare alleles for the remaining 37 locations, and applying the test to the individuals from the 38th location, analogous to the “leave-location-out” cross validation in Wasser et al., 2015.21 For comparison with previous work, we calculated the 75th percentiles of distances to the true origin (Figure S73). We applied this method to all remaining samples from this study (low coverage, substantial human contamination, PCA outliers), as well as chromosome 21 from great ape whole genomes6[3, 106, 113] and shallow sequencing data (median 0.25-fold coverage, ranging from 0.15-fold to 4.3-fold, Table S8) of blood and hair samples from 20 rescued chimpanzees from two Spanish rescue centers (Supplemental Note 9, Figures S74, S75, S80, S81, Data S1 – Figures S95–S105). Since the rare variants used in our approach are not necessarily fixed at a given location, but can be present at other locations, the pattern of shared rare variants is informative on past connectivity (Figures S76–S79, Data S1 – Figure S106). We calculated the proportion of derived variants in a given population shared with all other populations. We then used these data points to infer a landscape of sharing with other populations, and applied the kriging procedure described above, where we left out the test population from the landscape.
Acknowledgments
We would like to thank Marc de Manuel, Juan Antonio Rodriguez, Sojung Han, Christopher Barratt, Richard McElreath, and Lauren White for helpful discussion, and Alan Riedel, Katharina Madl, Veronika Staedele, and Amy Heilman for assistance in the laboratory. We thank Martijn Ter Heegde, Nadege Wangue Njomen, Joshua M. Linder, John Hart, Thurston Cleveland Hicks, Arcel Bamba, Richard Tshombe, Bruno Perodeau, Gita Chelluri, Theophile Desarmeaux, Vianet Mihindou, David Fine, Laura Kehoe, Lucy D’Auvergne, Nuria Maldonado, Luz Calia Miramontes Sequeiros, Theo Freeman, Hilde Vanleeuwe, Jean Claude Dengui, Paul Telfer, Michael Masozera, Nicolas Ntare, Alhaji Malikie Siaka, Henk Eshuis, Jill Pruetz, Geoffrey Muhanguzi, Karsten Dierks, Marcel Ketchan Eyong, Manasseh Eno-Nku, Abel Nzeheke, Yasmin Moebius, Floris Aubert, Matthieu Bonnet, Gregory Brazzola, Chloe Cipoletta, Katherine Corogenes, Charlotte Coupland, Bryan Curran, Emmanuel Dilambaka, Dervla Dowd, Kathryn J. Jeffery, Mohamed Kambi, Vincent Lapeyre, Vera Leinert, Giovanna Maretti, Rumen Martin, Amelia Meier, Protais Niyigabae, Robinson Orume, Jodie Preece, Sebastien Regnaut, Emilien Terrade, and Alexander Tickle for assistance in field site coordination and sample collection. We thank the field assistants and volunteers from the Jane Goodall Institute Spain and Senegal for their help with sample collection in Dindefelo. We thank the team of Rescue & Rehabilitation of the Fundació Mona. We thank Agilent for their collaboration in the project.
We thank the following government agencies for their support in conducting field research in their countries: Ministère de la Recherche Scientifique et de l'Innovation, Cameroon; Ministère des Forêts et de la Faune, Cameroon; Ministère des Eaux et Forêts, Cote d’Ivoire; Ministère de l’Enseignement Supérieur et de la Recherche Scientifique in Côte d’Ivoire; Institut Congolais pour la Conservation de la Nature, DR-Congo; Ministère de la Recherche Scientifique, DR-Congo; Agence Nationale des Parcs Nationaux, Gabon; Center National de la Recherche Scientifique (CENAREST), Gabon; Société Equatoriale d’Exploitation Forestière (SEEF), Gabon; Department of Wildlife and Range Management, Ghana; Forestry Commission, Ghana; Ministère de l'Agriculture de l'Elevage et des Eaux et Forets, Guinea; Instituto da Biodiversidade e das Áreas Protegidas (IBAP), Guinea-Bissau; Ministro da Agricultura e Desenvolvimento Rural, Guinea-Bissau; Forestry Development Authority, Liberia; Eaux et Forets, Mali; Ministre de l'Environnement et de l'Assainissement et du Developpement Durable du Mali; Conservation Society of Mbe Mountains (CAMM), Nigeria; National Park Service, Nigeria; Ministère de l’Economie Forestière, R-Congo; Ministère de le Recherche Scientifique et Technologique, R-Congo; Ministry of Education, Rwanda; Rwanda Development Board, Rwanda; Direction des Eaux, Forêts, et Chasses, Senegal; Reserve Naturelle Communautaire de Dindefelo, Senegal; Ministry of Agriculture, Forestry, and Food Security, Sierra Leone; National Protected Area Authority, Sierra Leone; Tanzania Commission for Science and Technology, Tanzania; Tanzania Wildlife Research Institute, Tanzania; Makerere University Biological Field Station (MUBFS), Uganda; Uganda National Council for Science and Technology (UNCST), Uganda; Uganda Wildlife Authority, Uganda; National Forestry Authority, Uganda; Agence Congolaise de la Faune et des Aires Protégées; and The Wild Chimpanzee Foundation (WCF).
Funding: “la Caixa” Foundation doctoral fellowship program LCF/BQ/DE15/10360006 (to C.F.). FPI (Formación de Personal Investigador) PRE2018-083966 from Ministerio de Ciencia, Universidades e Investigación (to M.A.-E.) and CGL2017-82654-P (MINECO/FEDER, UE) (to E.L.). “la Caixa” Foundation (ID 100010434), fellowship code LCF/BQ/PR19/11700002 (to M.K.). Vienna Science and Technology Fund (WWTF) and the City of Vienna project VRG20-001 (to M.K.). The European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 864203) (to T.M.-B.). BFU2017-86471-P (MINECO/FEDER, UE) (to T.M.-B.). “Unidad de Excelencia María de Maeztu”, funded by the AEI (CEX2018-000792-M) (to T.M.-B.). Howard Hughes International Early Career (to T.M.-B.). NIH 1R01HG010898-01A1 (to T.M.-B.). Secretaria d’Universitats i Recerca and CERCA Program del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880) (to T.M.-B.). UCL’s Wellcome Trust ISSF3 award 204841/Z/16/Z (to A.M.A. and J.M.S.). Generalitat de Catalunya (2017 SGR-1040) (to M. Llorente). Wellcome Trust Investigator Award 202802/Z/16/Z (to D.A.H.). The Pan African Program: The Cultured Chimpanzee (PanAf) is generously funded by the Max Planck Society, the Max Planck Society Innovation Fund, and the Heinz L. Krekeler Foundation.
Author contributions
T.M.-B. and M.A. conceived and supervised the study. M.A., C.B., and H.S.K. direct the Pan African Program: The Cultured Chimpanzee. P.D., T.A., P.A.-V., A.A., S.A., A.K.A., E.A.A., E.B., D.B., M.B., A.C.-A., R.C., H.C., E.D., T.D., A.D., J.D., V.E.E., O.F., A.G., A.C.G., J.H., D.H., V.H., R.A.H.A., I.I., SJ., J.J., P.K., M.K., M.V.K., A.K.K., I.K., D.K., K.L., J.L., B.L., A.L., K.L., M. Llana, M. Llorente, S.M., D.M., F.M., M.M., E.N., S. Nicholl, S. Nixon, E.N., C.O., L.J.O., L.P., A.P., L.R., M.M.R., A.R., C.S., L.S., V.S., F.A.S., N.T., L.R.T., E.T., J.v.S., V.V., E.G.W., J.W., R.M.W., Y.G.Y., K.Y., and K.Z. supervised, conducted fieldwork, and collected samples. C.F., M.A.-E., J.D.L., J.H., M.A., and E.L. performed experimental laboratory work. C.F. and M.K. performed the analysis. C.M.-S., P.G., J.M.S., L.V., A.M.A., D.A.H., H.S.H., E.L., M.A., and T.M.-B. provided analytical support. C.F. and M.K. wrote the manuscript with input from all coauthors.
Declaration of interest
The authors declare no competing interests.
Published: June 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2022.100133.
Contributor Information
Claudia Fontsere, Email: claudia.fontsere@upf.edu.
Mimi Arandjelovic, Email: arandjel@eva.mpg.de.
Tomas Marques-Bonet, Email: tomas.marques@upf.edu.
Supplemental information
(A) Adjusted p values (method “BH”) for comparison of the amount of bonobo ancestry in each group.
(B) Mean fragment length per subspecies group.
Data and code availability
All genomic data generated is available at a public repository (ENA) under the accession code ENA: PRJEB46115. Code for geolocalization is available on a public repository (https://github.com/kuhlwilm/rareCAGA). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Fischer A., Pollack J., Thalmann O., Nickel B., Pääbo S. Demographic history and genetic differentiation in apes. Curr. Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 2.Becquet C., Przeworski M. A new approach to estimate parameters of speciation models with application to apes. Genome Res. 2007;17:1505–1519. doi: 10.1101/gr.6409707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prado-Martinez J., Sudmant P.H., Kidd J.M., Li H., Kelley J.L., Lorente-Galdos B., Veeramah K.R., Woerner A.E., O’Connor T.D., Santpere G., et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lobon I., Tucci S., De Manuel M., Ghirotto S., Benazzo A., Prado-Martinez J., Lorente-Galdos B., Nam K., Dabad M., Hernandez-Rodriguez J., et al. Demographic history of the genus Pan inferred from whole mitochondrial genome reconstructions. Genome Biol. Evol. 2016;8:2020–2030. doi: 10.1093/gbe/evw124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallast P., Maisano Delser P., Batini C., Zadik D., Rocchi M., Schempp W., Tyler-Smith C., Jobling M.A. Great ape Y Chromosome and mitochondrial DNA phylogenies reflect subspecies structure and patterns of mating and dispersal. Genome Res. 2016;26:427–439. doi: 10.1101/gr.198754.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Manuel M., Kuhlwilm M., Frandsen P., Sousa V.C., Desai T., Prado-Martinez J., Hernandez-Rodriguez J., Dupanloup I., Lao O., Hallast P., et al. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science. 2016;354:477–481. doi: 10.1126/science.aag2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlwilm M., de Manuel M., Nater A., Greminger M.P., Krützen M., Marques-Bonet T. Evolution and demography of the great apes. Curr. Opin. Genet. Dev. 2016;41:124–129. doi: 10.1016/j.gde.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Kuhlwilm M., Han S., Sousa V.C., Excoffier L., Marques-Bonet T. Ancient admixture from an extinct ape lineage into bonobos. Nat. Ecol. Evol. 2019;3:957–965. doi: 10.1038/s41559-019-0881-7. [DOI] [PubMed] [Google Scholar]
- 9.McBrearty S., Jablonski N.G. First fossil chimpanzee. Nature. 2005;437:105–108. doi: 10.1038/nature04008. [DOI] [PubMed] [Google Scholar]
- 10.Fünfstück T., Arandjelovic M., Morgan D.B., Sanz C., Reed P., Olson S.H., Cameron K., Ondzie A., Peeters M., Vigilant L. The sampling scheme matters: Pan troglodytes troglodytes and P. t schweinfurthii are characterized by clinal genetic variation rather than a strong subspecies break. Am. J. Phys. Anthropol. 2015;156:181–191. doi: 10.1002/ajpa.22638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lester J.D., Vigilant L., Gratton P., McCarthy M.S., Barratt C.D., Dieguez P., Agbor A., Álvarez-Varona P., Angedakin S., Ayimisin E.A., et al. Recent genetic connectivity and clinal variation in chimpanzees. Commun. Biol. 2021;4:283. doi: 10.1038/s42003-021-01806-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayr E., O’Hara R.J. The biogeographic evidence supporting the Pleistocene forest refuge hypothesis. Evolution (N. Y). 1986;40:55–67. doi: 10.1111/j.1558-5646.1986.tb05717.x. [DOI] [PubMed] [Google Scholar]
- 13.Barratt C., Lester J., Gratton P., Onstein R., Kalan A., McCarthy M., Bocksberger G., White L., Vigilant L., Dieguez P., et al. Late Quaternary habitat suitability models for chimpanzees ( Pan troglodytes ) since the Last Interglacial (120,000 BP) bioRxiv. 2020 doi: 10.1101/2020.05.15.066662. Preprint at. [DOI] [PubMed] [Google Scholar]
- 14.Kalan A.K., Kulik L., Arandjelovic M., Boesch C., Haas F., Dieguez P., Barratt C.D., Abwe E.E., Agbor A., Angedakin S., et al. Environmental variability supports chimpanzee behavioural diversity. Nat. Commun. 2020;11:4451. doi: 10.1038/s41467-020-18176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts P., Stewart B.A. Defining the ‘generalist specialist’ niche for Pleistocene Homo sapiens. Nat. Hum. Behav. 2018;2:542–550. doi: 10.1038/s41562-018-0394-4. [DOI] [PubMed] [Google Scholar]
- 16.Humle, T., Maisels, F., Oates, J.F., Plumptre, A., and Williamson, E.A. (2016). Pan troglodytes (Errata Version Published in 2018). IUCN Red List Threat. Species, e.T15933A129038584. 10.2305/IUCN.UK.2016-2RLTS.T15933A17964454.en.
- 17.Kühl H.S., Sop T., Williamson E.A., Mundry R., Brugière D., Campbell G., Cohen H., Danquah E., Ginn L., Herbinger I., et al. The Critically Endangered western chimpanzee declines by 80% Am. J. Primatol. 2017;79:e22681. doi: 10.1002/ajp.22681. [DOI] [PubMed] [Google Scholar]
- 18.Supple M.A., Shapiro B. Conservation of biodiversity in the genomics era. Genome Biol. 2018;19:131. doi: 10.1186/s13059-018-1520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frandsen P., Fontsere C., Nielsen S.V., Hanghøj K., Castejon-Fernandez N., Lizano E., Hughes D., Hernandez-Rodriguez J., Korneliussen T.S., Carlsen F., et al. Targeted conservation genetics of the endangered chimpanzee. Heredity. 2020;125:15–27. doi: 10.1038/s41437-020-0313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghobrial L., Lankester F., Kiyang J.A., Akih A.E., de Vries S., Fotso R., Gadsby E.L., Jenkins P.D., Gonder M.K. Tracing the origins of rescued chimpanzees reveals widespread chimpanzee hunting in Cameroon. BMC Ecol. 2010;10:2. doi: 10.1186/1472-6785-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasser S.K., Brown L., Mailand C., Mondol S., Clark W., Laurie C., Weir B.S. Genetic assignment of large seizures of elephant ivory reveals Africa’s major poaching hotspots. Science. 2015;349:84–87. doi: 10.1126/science.aaa2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banes G.L., Galdikas B.M.F., Vigilant L. Reintroduction of confiscated and displaced mammals risks outbreeding and introgression in natural populations, as evidenced by orang-utans of divergent subspecies. Sci. Rep. 2016;6:22026. doi: 10.1038/srep22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oklander L.I., Caputo M., Solari A., Corach D. Genetic assignment of illegally trafficked neotropical primates and implications for reintroduction programs. Sci. Rep. 2020;10:3676. doi: 10.1038/s41598-020-60569-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carøe C., Gopalakrishnan S., Vinner L., Mak S.S.T., Sinding M.H.S., Samaniego J.A., Wales N., Sicheritz-Pontén T., Gilbert M.T.P. Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 2018;9:410–419. doi: 10.1111/2041-210x.12871. [DOI] [Google Scholar]
- 25.Fontsere C., Alvarez-Estape M., Lester J., Arandjelovic M., Kuhlwilm M., Dieguez P., Agbor A., Angedakin S., Ayuk Ayimisin E., Bessone M., et al. Maximizing the acquisition of unique reads in noninvasive capture sequencing experiments. Mol. Ecol. Resour. 2021;21:745–761. doi: 10.1111/1755-0998.13300. [DOI] [PubMed] [Google Scholar]
- 26.Vigilant L., Guschanski K. Using genetics to understand the dynamics of wild primate populations. Primates. 2009;50:105–120. doi: 10.1007/s10329-008-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perry G.H., Marioni J.C., Melsted P., Gilad Y. Genomic-scale capture and sequencing of endogenous DNA from feces. Mol. Ecol. 2010;19:5332–5344. doi: 10.1111/j.1365-294x.2010.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez-Rodriguez J., Arandjelovic M., Lester J., de Filippo C., Weihmann A., Meyer M., Angedakin S., Casals F., Navarro A., Vigilant L., et al. The impact of endogenous content, replicates and pooling on genome capture from faecal samples. Mol. Ecol. Resour. 2018;18:319–333. doi: 10.1111/1755-0998.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White L.C., Fontsere C., Lizano E., Hughes D.A., Angedakin S., Arandjelovic M., Granjon A., Hans J.B., Lester J.D., Rabanus-Wallace M.T., et al. A roadmap for high-throughput sequencing studies of wild animal populations using noninvasive samples and hybridization capture. Mol. Ecol. Resour. 2019;19:609–622. doi: 10.1111/1755-0998.12993. [DOI] [PubMed] [Google Scholar]
- 30.Boesch C., Boesch H. Hunting behavior of wild chimpanzees in the taï national park. Am. J. Phys. Anthropol. 1989;78:547–573. doi: 10.1002/ajpa.1330780410. [DOI] [PubMed] [Google Scholar]
- 31.Stanford C.B., Wallis J., Matama H., Goodall J. Patterns of predation by chimpanzees on red colobus monkeys in gombe national park, 1982–1991. Am. J. Phys. Anthropol. 1994;94:213–228. doi: 10.1002/ajpa.1330940206. [DOI] [PubMed] [Google Scholar]
- 32.Arandjelovic M., Head J., Kühl H., Boesch C., Robbins M.M., Maisels F., Vigilant L. Effective non-invasive genetic monitoring of multiple wild western gorilla groups. Biol. Conserv. 2010;143:1780–1791. doi: 10.1016/j.biocon.2010.04.030. [DOI] [Google Scholar]
- 33.Kuhlwilm M., Fontsere C., Han S., Alvarez-Estape M., Marques-Bonet T. HuConTest: testing human contamination in great ape samples. Genome Biol. Evol. 2021;13:evab117. doi: 10.1093/gbe/evab117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peyrégne S., Peter B.M. AuthentiCT: a model of ancient DNA damage to estimate the proportion of present-day DNA contamination. Genome Biol. 2020;21:246. doi: 10.1186/s13059-020-02123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korneliussen T.S., Albrechtsen A., Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinf. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meisner J., Albrechtsen A. Inferring population structure and admixture proportions in low-depth NGS data. Genetics. 2018;210:719–731. doi: 10.1534/genetics.118.301336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nye J., Laayouni H., Kuhlwilm M., Mondal M., Marques-Bonet T., Bertranpetit J. Selection in the introgressed regions of the chimpanzee genome. Genome Biol. Evol. 2018;10:1132–1138. doi: 10.1093/gbe/evy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takemoto H., Kawamoto Y., Furuichi T. How did bonobos come to range south of the Congo river? Reconsideration of the divergence of Pan paniscus from other Pan populations. Evol. Anthropol. Issues News Rev. 2015;24:170–184. doi: 10.1002/evan.21456. [DOI] [PubMed] [Google Scholar]
- 39.Peter B.M. Admixture, population structure, and f-statistics. Genetics. 2016;202:1485–1501. doi: 10.1534/genetics.115.183913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peter B. 100,000 years of gene flow between Neandertals and Denisovans in the Altai mountains. bioRxiv. 2020 doi: 10.1101/2020.03.13.990523. Preprint at. [DOI] [Google Scholar]
- 41.Mitchell M.W., Locatelli S., Sesink Clee P.R., Thomassen H.A., Gonder M.K. Environmental variation and rivers govern the structure of chimpanzee genetic diversity in a biodiversity hotspot. BMC Evol. Biol. 2015;15:1. doi: 10.1186/s12862-014-0274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petkova D., Novembre J., Stephens M. Visualizing spatial population structure with estimated effective migration surfaces. Nat. Genet. 2016;48:94–100. doi: 10.1038/ng.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Asadi H., Petkova D., Stephens M., Novembre J. Estimating recent migration and population-size surfaces. PLoS Genet. 2019;15:e1007908. doi: 10.1371/journal.pgen.1007908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell M.W., Locatelli S., Ghobrial L., Pokempner A.A., Sesink Clee P.R., Abwe E.E., Nicholas A., Nkembi L., Anthony N.M., Morgan B.J., et al. The population genetics of wild chimpanzees in Cameroon and Nigeria suggests a positive role for selection in the evolution of chimpanzee subspecies. BMC Evol. Biol. 2015;15:3. doi: 10.1186/s12862-014-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browning B.L., Browning S.R. Detecting identity by descent and estimating genotype error rates in sequence data. Am. J. Hum. Genet. 2013;93:840–851. doi: 10.1016/j.ajhg.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen R., Akey J.M., Jakobsson M., Pritchard J.K., Tishkoff S., Willerslev E. Tracing the peopling of the world through genomics. Nature. 2017;541:302–310. doi: 10.1038/nature21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gagneux P., Gonder M.K., Goldberg T.L., Morin P.A. Philosophical Transactions of the Royal Society B: Biological Sciences; 2001. Gene Flow in Wild Chimpanzee Populations: What Genetic Data Tell Us about Chimpanzee Movement over Space and Time; pp. 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiffels S., Haak W., Paajanen P., Llamas B., Popescu E., Loe L., Clarke R., Lyons A., Mortimer R., Sayer D., et al. Iron age and Anglo-saxon genomes from east england reveal British migration history. Nat. Commun. 2016;7:10408. doi: 10.1038/ncomms10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson E.A. Identity by descent: variation in meiosis, across genomes, and in populations. Genetics. 2013;194:301–326. doi: 10.1534/genetics.112.148825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wessling E.G., Kühl H.S., Mundry R., Deschner T., Pruetz J.D. The costs of living at the edge: seasonal stress in wild savanna-dwelling chimpanzees. J. Hum. Evol. 2018;121:1–11. doi: 10.1016/j.jhevol.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Kuhlwilm M., Gronau I., Hubisz M.J., De Filippo C., Prado-Martinez J., Kircher M., Fu Q., Burbano H.A., Lalueza-Fox C., De La Rasilla M., et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 2016;530:429–433. doi: 10.1038/nature16544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCarthy M.S., Lester J.D., Howe E.J., Arandjelovic M., Stanford C.B., Vigilant L. Genetic censusing identifies an unexpectedly sizeable population of an endangered large mammal in a fragmented forest landscape. BMC Ecol. 2015;15:21. doi: 10.1186/s12898-015-0052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rich A.M., Wasserman M.D., Hunt K.D., Kaestle F.A. Chimpanzee (Pan troglodytes schweinfurthii) population spans multiple protected areas in the albertine rift. Folia Primatol. 2020;91:595–609. doi: 10.1159/000508073. [DOI] [PubMed] [Google Scholar]
- 54.Moore D.L., Vigilant L. Genetic diversity at the edge: comparative assessment of Y-chromosome and autosomal diversity in eastern chimpanzees (Pan troglodytes schweinfurthii) of Ugalla, Tanzania. Conserv. Genet. 2014;15:495–507. doi: 10.1007/s10592-013-0556-x. [DOI] [Google Scholar]
- 55.Brunke J., Radespiel U., Russo I.R., Bruford M.W., Goossens B. Messing about on the river: the role of geographic barriers in shaping the genetic structure of Bornean small mammals in a fragmented landscape. Conserv. Genet. 2019;20:691–704. doi: 10.1007/s10592-019-01159-3. [DOI] [Google Scholar]
- 56.Hicks T.C., Tranquilli S., Kuehl H., Campbell G., Swinkels J., Darby L., Boesch C., Hart J., Menken S.B.J. Absence of evidence is not evidence of absence: discovery of a large, continuous population of Pan troglodytes schweinfurthii in the Central Uele region of northern DRC. Biol. Conserv. 2014;171:107–113. doi: 10.1016/j.biocon.2014.01.002. [DOI] [Google Scholar]
- 57.Hicks T.C., Kühl H.S., Boesch C., Dieguez P., Ayimisin A.E., Fernandez R.M., Zungawa D.B., Kambere M., Swinkels J., Menken S.B.J., et al. Bili-uéré: a chimpanzee behavioural realm in northern democratic republic of Congo. Folia Primatol. 2019;90:3–64. doi: 10.1159/000492998. [DOI] [PubMed] [Google Scholar]
- 58.Boesch C., Tomasello M. Chimpanzee and human cultures. Curr. Anthropol. 1998;39:591–614. doi: 10.1086/204785. [DOI] [Google Scholar]
- 59.Langergraber K.E., Siedel H., Mitani J.C., Wrangham R.W., Reynolds V., Hunt K., Vigilant L. The genetic signature of sex-biased migration in patrilocal chimpanzees and humans. PLoS One. 2007;2:e973. doi: 10.1371/journal.pone.0000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kühl H.S., Kalan A.K., Arandjelovic M., Aubert F., D’Auvergne L., Goedmakers A., Jones S., Kehoe L., Regnaut S., Tickle A., et al. Chimpanzee accumulative stone throwing. Sci. Rep. 2016;6:22219. doi: 10.1038/srep22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stiles D., Redmond I., Cress D., Nellemann C., Formo R.K. In: Stolen Apes - The Illicit Trade in Chimpanzees, Gorillas, Bonobos and Orangutans. Stiles R.K., Redmond I., Cress D., Nellemann C., Formo, editors. United Nations Environment Programme, GRID-Arendal; 2013. pp. 1–56. [Google Scholar]
- 62.PASA (2016). Pan African Sanctuary Alliance. Operation Manual.
- 63.Gouda S., Kerry R.G., Das A., Chauhan N.S. Wildlife forensics: a boon for species identification and conservation implications. Forensic Sci. Int. 2020;317:110530. doi: 10.1016/j.forsciint.2020.110530. [DOI] [PubMed] [Google Scholar]
- 64.Wasser S.K., Torkelson A., Winters M., Horeaux Y., Tucker S., Otiende M.Y., Sitam F.A.T., Buckleton J., Weir B.S. Combating transnational organized crime by linking multiple large ivory seizures to the same dealer. Sci. Adv. 2018;4:eaat0625. doi: 10.1126/sciadv.aat0625. eaat0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mcmahon B.J., Teeling E.C., Höglund J. How and why should we implement genomics into conservation? Evol. Appl. 2014;7:999–1007. doi: 10.1111/eva.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R Taylor H., Dussex N., van Heezik Y. Bridging the conservation genetics gap by identifying barriers to implementation for conservation practitioners. Glob. Ecol. Conserv. 2017;10:231–242. doi: 10.1016/j.gecco.2017.04.001. [DOI] [Google Scholar]
- 67.Krehenwinkel H., Pomerantz A., Henderson J.B., Kennedy S.R., Lim J.Y., Swamy V., Shoobridge J.D., Graham N., Patel N.H., Gillespie R.G., Prost S. Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. Gigascience. 2019;8:giz006. doi: 10.1093/gigascience/giz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Church D.M., Schneider V.A., Graves T., Auger K., Cunningham F., Bouk N., Chen H.C., Agarwala R., McLaren W.M., Ritchie G.R.S., et al. Modernizing reference genome assemblies. Plos Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kronenberg Z.N., Fiddes I.T., Gordon D., Murali S., Cantsilieris S., Meyerson O.S., Underwood J.G., Nelson B.J., Chaisson M.J.P., Dougherty M.L., et al. High-resolution comparative analysis of great ape genomes. Science. 2018;360:eaar6343. doi: 10.1126/science.aar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogers J., Raveendran M., Harris R.A., Mailund T., Leppälä K., Athanasiadis G., Schierup M.H., Cheng J., Munch K., Walker J.A., et al. Baboon Genome Analysis Consortium The comparative genomics and complex population history of Papio baboons. Sci. Adv. 2019;5:eaau6947. doi: 10.1126/sciadv.aau6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warren W.C., Jasinska A.J., García-Pérez R., Svardal H., Tomlinson C., Rocchi M., Archidiacono N., Capozzi O., Minx P., Montague M.J., et al. The genome of the vervet (Chlorocebus æthiops sabæus) Genome Res. 2015;25:1921–1933. doi: 10.1101/gr.192922.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Genereux D.P., Serres A., Armstrong J., Johnson J., Marinescu V.D., Murén E., Juan D., Bejerano G., Casewell N.R., Chemnick L.G., et al. A comparative genomics multitool for scientific discovery and conservation. Nature. 2020;587:240–245. doi: 10.1038/s41586-020-2876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scally A., Dutheil J.Y., Hillier L.W., Jordan G.E., Goodhead I., Herrero J., Hobolth A., Lappalainen T., Mailund T., Marques-Bonet T., et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin Y., Yang T., Liu H., Huang Z., Zhang Y., Song Y., Wang W., Guang X., Sahu S.K., Kristiansen K. The draft genome of mandrill (Mandrillus sphinx): an Old World monkey. Sci. Rep. 2020;10:2431. doi: 10.1038/s41598-020-59110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rohland N., Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer M., Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 77.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lefort V., Desper R., Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program: table 1. Mol. Biol. Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jónsson H., Ginolhac A.A., Schubert M., Johnson P.L.F.F., Orlando L., Jonsson H., Ginolhac A.A., Schubert M., Johnson P.L.F.F., Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Skotte L., Korneliussen T.S., Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vieira F.G., Lassalle F., Korneliussen T.S., Fumagalli M. Improving the estimation of genetic distances from Next-Generation Sequencing data. Biol. J. Linn. Soc. 2016;117:139–149. doi: 10.1111/bij.12511. [DOI] [Google Scholar]
- 85.Korneliussen T.S., Moltke I., Albrechtsen A., Nielsen R. Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinf. 2013;14:289. doi: 10.1186/1471-2105-14-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., De Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bivand, R., and Lewin-Koh, N. (2013). Maptools: Tools for Handling Spatial Objects. R Packag. version 0.8.
- 88.Petr M., Vernot B., Kelso J. Admixr-R package for reproducible analyses using ADMIXTOOLS. Bioinformatics. 2019;35:3194–3195. doi: 10.1093/bioinformatics/btz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 90.Schliep K.P. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Revell L.J. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol. Evol. 2012;3:217–223. doi: 10.1111/j.2041-210x.2011.00169.x. [DOI] [Google Scholar]
- 92.Pebesma E. Simple features for R: standardized support for spatial vector data. R. J. 2018;10:439–446. doi: 10.32614/rj-2018-009. [DOI] [Google Scholar]
- 93.Pebesma E.J., Bivand R.S. Classes and methods for spatial data in {R. R. News. 2005;5:9–13. [Google Scholar]
- 94.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’Hara R.B., Simpson G.L., Solymos P., et al. 2020. Vegan: Community Ecology Package. [Google Scholar]
- 95.R Core Team A Language and Environment for Statistical Computing. R. Found. Stat. Comput. 2020;2 https://www.R-project.org [Google Scholar]
- 96.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nielsen R., Korneliussen T., Albrechtsen A., Li Y., Wang J. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS One. 2012;7:e37558. doi: 10.1371/journal.pone.0037558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prüfer K. SNPAD: an ancient DNA genotype caller. Bioinformatics. 2018;34:4165–4171. doi: 10.1093/bioinformatics/bty507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pickrell J.K., Pritchard J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arandjelovic M., Guschanski K., Schubert G., Harris T.R., Thalmann O., Siedel H., Vigilant L. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol. Ecol. Resour. 2009;9:28–36. doi: 10.1111/j.1755-0998.2008.02387.x. [DOI] [PubMed] [Google Scholar]
- 104.Arandjelovic M., Head J., Rabanal L.I., Schubert G., Mettke E., Boesch C., Robbins M.M., Vigilant L. Non-invasive genetic monitoring of wild central chimpanzees. PLoS One. 2011;6:e14761. doi: 10.1371/journal.pone.0014761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Korneliussen T.S., Moltke I. NgsRelate: a software tool for estimating pairwise relatedness from next-generation sequencing data. Bioinformatics. 2015;31 doi: 10.1093/bioinformatics/btv509. btv509–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nater A., Mattle-Greminger M.P., Nurcahyo A., Nowak M.G., de Manuel M., Desai T., Groves C., Pybus M., Sonay T.B., Roos C., et al. Morphometric, behavioral, and genomic evidence for a new orangutan species. Curr. Biol. 2017;27:3487–3498.e10. doi: 10.1016/j.cub.2017.09.047.e10. [DOI] [PubMed] [Google Scholar]
- 107.Nait Saada J., Kalantzis G., Shyr D., Cooper F., Robinson M., Gusev A., Palamara P.F. Identity-by-descent detection across 487,409 British samples reveals fine scale population structure and ultra-rare variant associations. Nat. Commun. 2020;11:6130. doi: 10.1038/s41467-020-19588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Auton A., Fledel-Alon A., Pfeifer S., Venn O., Ségurel L., Street T., Leffler E.M., Bowden R., Aneas I., Broxholme J., et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stevison L.S., Woerner A.E., Kidd J.M., Kelley J.L., Veeramah K.R., McManus K.F., Bustamante C.D., Hammer M.F., Wall J.D., Lorente-Galdos B., et al. The time scale of recombination rate evolution in great apes. Mol. Biol. Evol. 2016;33:928–945. doi: 10.1093/molbev/msv331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Langergraber K.E., Prüfer K., Rowney C., Boesch C., Crockford C., Fawcett K., Inoue E., Inoue-Muruyama M., Mitani J.C., Muller M.N., et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl. Acad. Sci. U S A. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A., et al. The simons genome diversity project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pebesma E., Gräler B. Spatio-temporal geostatistics using gstat. R. J. 2014;8:204–218. [Google Scholar]
- 113.Xue Y., Prado-Martinez J., Sudmant P.H., Narasimhan V., Ayub Q., Szpak M., Frandsen P., Chen Y., Yngvadottir B., Cooper D.N., et al. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science. 2015;348:242–245. doi: 10.1126/science.aaa3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Adjusted p values (method “BH”) for comparison of the amount of bonobo ancestry in each group.
(B) Mean fragment length per subspecies group.
Data Availability Statement
All genomic data generated is available at a public repository (ENA) under the accession code ENA: PRJEB46115. Code for geolocalization is available on a public repository (https://github.com/kuhlwilm/rareCAGA). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.