Abstract
Objective
To summarize applications of natural language processing (NLP) in model informed drug development (MIDD) and identify potential areas of improvement.
Materials and Methods
Publications found on PubMed and Google Scholar, websites and GitHub repositories for NLP libraries and models. Publications describing applications of NLP in MIDD were reviewed. The applications were stratified into 3 stages: drug discovery, clinical trials, and pharmacovigilance. Key NLP functionalities used for these applications were assessed. Programming libraries and open-source resources for the implementation of NLP functionalities in MIDD were identified.
Results
NLP has been utilized to aid various processes in drug development lifecycle such as gene-disease mapping, biomarker discovery, patient-trial matching, adverse drug events detection, etc. These applications commonly use NLP functionalities of named entity recognition, word embeddings, entity resolution, assertion status detection, relation extraction, and topic modeling. The current state-of-the-art for implementing these functionalities in MIDD applications are transformer models that utilize transfer learning for enhanced performance. Various libraries in python, R, and Java like huggingface, sparkNLP, and KoRpus as well as open-source platforms such as DisGeNet, DeepEnroll, and Transmol have enabled convenient implementation of NLP models to MIDD applications.
Discussion
Challenges such as reproducibility, explainability, fairness, limited data, limited language-support, and security need to be overcome to ensure wider adoption of NLP in MIDD landscape. There are opportunities to improve the performance of existing models and expand the use of NLP in newer areas of MIDD.
Conclusions
This review provides an overview of the potential and pitfalls of current NLP approaches in MIDD.
Keywords: NLP, machine learning, deep learning, drug development
INTRODUCTION
Natural language processing (NLP) is an artificial intelligence (AI) technique to process and analyze human-generated spoken or written data. It utilizes syntactic and semantic analysis to analyze text data. NLP has evolved over the last decade and advanced to a level where it has become an integral part of our life—it is being used for email filters, voice assistants, language translation, digital phone calls, and text analytics.1
The rise of big data in the healthcare industry is setting the stage for AI tools such as NLP to assist with improving the delivery of care.2 One of the big problems of healthcare fields is that about 80% of medical data remains unstructured (eg, text, image, signal, etc.) and untapped after it is created.3 NLP has shown high potential in healthcare and model informed drug development (MIDD)4 to overcome the challenges that exist with natural language data utilization and generation.3 NLP has enabled the shift from time-consuming manual and siloed curation of natural language data to automated, large scale and standard processes for analyzing text and speech data.
MIDD involves leveraging quantitative models to inform decision-making in drug development.5 In the field of MIDD, NLP can be leveraged to extract information out of structured (eg, electronic health records [EHRs]) and unstructured (eg, research documents) data to optimize and/or accelerate various processes in the drug development lifecycle, eg, determining drug–target interaction6 and drug–drug interaction,7 biomarker discovery,8 drug repurposing,9,10 patient-trial matching,11 model-based meta-analysis,12 disease progression modeling,13 and others.14 NLP platforms perform the role of assessing potential associations between chemical/drug entities, their target proteins, and novel disease-related pathways by extensive analysis of scientific literature. NLP can also accelerate repurposing of approved drugs for new diseases which enables pharmacologists to address new market at a fraction of cost and time. NLP contribution in future drug safety is an important aspect of leveraging text mining automation to unveil valuable information invisible among aggregation of unstructured data. NLP usage for matching participants to clinical trials is a crucial application in this area.6 NLP and AI provide a suitable solution to handle this problem to save time.
Papers in the past have identified the potential of AI in drug discovery and development fields.4,14,15 Some literature has focused on the drug discovery processes.16–18 This review focuses on current NLP applications in the field of drug discovery and development and provides a comprehensive overview of NLP in MIDD. We highlight the technical aspects of various tools utilized to develop the existing language models. We also provide information on various easily accessible resources which can be deployed to develop an NLP model for MIDD applications. Lastly, this article gives insights into potential opportunities that currently exist to expand and carry NLP in MIDD forward.
METHODS
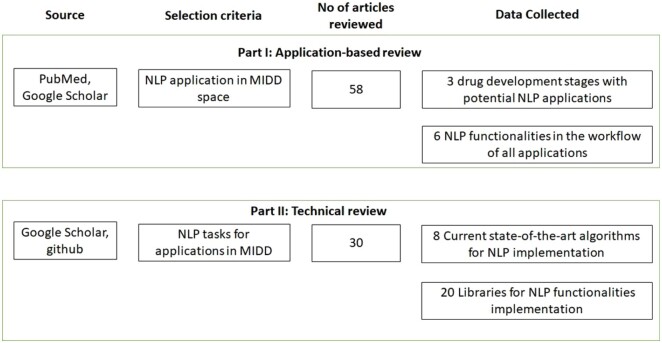
The review process was divided into 2 parts: review of the applications of NLP algorithms in different stages of drug development lifecycle and review of technical aspects of various NLP algorithms (Figure 1).
Figure 1.
Entire review process workflow. The review process was divided into 2 parts: (1) review of applications NLP in MIDD space and (2) technical review of state-of-the-art methods for implementation of various NLP functionalities most used in MIDD space.
Firstly, all the papers identified on PubMed and Google Scholar with use of NLP techniques in different stages of drug discovery and development were reviewed based on the inclusion and exclusion criteria. The drug development process was stratified into 3 stages: (1) Discovery, (2) Clinical Trials, and (3) Pharmacovigilance. Papers highlighting the use of NLP were classified into 1 of the 3 stages. For each application, key NLP functionalities in the workflow were identified.
In the next step, a technical review of all the identified NLP functionalities was carried out. For each functionality, the implementation pipeline was analyzed. Furthermore, the current state-of-the-art for the functionalities were identified. Biomedical application specific AI-based models and libraries for implementation of those functionalities were reviewed from sources which include publications and GitHub or websites for the specific models (Figure 2). This information on various models and libraries was used to populate the 2 inventories presented in this article (Tables 2 and 3). Various NLP-task-based features of the libraries such as ability to perform text preprocessing, named entity recognition (NER), relation extraction, sentiment analysis etc. were included in the inventory (Table 2). Additional features for the libraries in the inventory include the availability of pre-trained neural models for direct implementation using transfer learning and support for multiple languages. The current state-of-the-art model inventory (Table 3) incorporates information about transformer-based models that were recently developed and can be used for carrying out various NLP tasks in the MIDD space. These models have been pretrained on biomedical literature and are known to produce state-of-the-art results on various tasks.
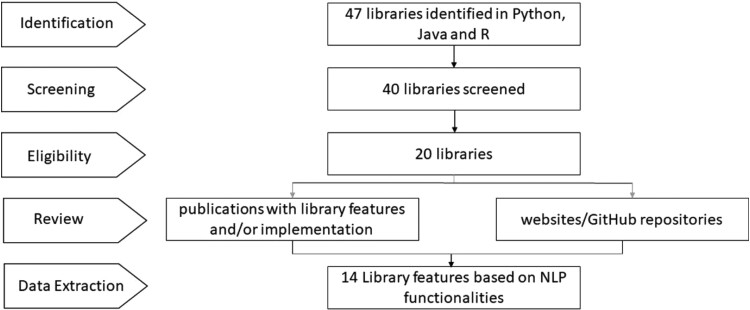
Figure 2.
Process flow for NLP libraries inventory. The figure describes the review process followed for developing the “NLP libraries inventory for drug discovery and development.” A total of 47 libraries were identified from Google scholar resources. Out of these, 7 libraries for speech processing were excluded from further screening. Out of the remaining 40 libraries, 20 were found to be used in different biomedical or biochemical applications. The websites, github repositories, and publications on the libraries were reviewed and the libraries were analyzed for the presence or absence of 14 features. These features were selected based on the most used NLP functionalities in the drug discovery and development space.
Table 2.
NLP libraries for MIDD
| Library name | Features |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Programming language | Pretrained neural network models | Word embeddings | Multi-language support | Tokenization | Part-of-speech tagging | Stemming/lemmatization | Named entity recognition | Entity resolution | Sentiment analysis | Relation extraction | Assertion status detection | Topic modeling | |
| Spacy43 | Python | x | x | x | x | x | x | x | x | x | |||
| Gensim44 | Python | x | x | x | x | x | x | x | |||||
| NLTK45 | Python | x | x | x | x | x | x | x | x | ||||
| CoreNLP46 | Java | x | x | x | x | x | x | x | x | x | |||
| Scispacy47 | Python | x | x | x | x | x | x | x | x | x | |||
| SparkNLP48 | Python, Java, Scala, R | x | x | x | x | x | x | x | x | x | x | x | x |
| SparkNLP for healthcare49 | Python, Java, Scala, R | x | x | x | x | x | x | x | x | x | x | ||
| Torchtext50 | Python | x | x | x | x | ||||||||
| KoRpus51 | R | x | x | x | |||||||||
| Tensorflow52 | Python | x | x | x | x | x | x | x | x | x | x | ||
| Scikit learn53 | Python | x | x | x | |||||||||
| Textblob54 | Python | x | x | x | x | ||||||||
| Pattern55 | Python, R | x | x | x | x | x | |||||||
| Hugging face56 | Python | x | x | x | x | x | |||||||
| Allen NLP57 | Python | x | x | x | x | x | x | x | x | x | |||
| Fasttext21 | Python | x | x | x | x | x | x | ||||||
| Stanza58 | Python | x | x | x | x | x | x | x | |||||
| Flair59 | Python | x | x | x | x | x | x | ||||||
| Fastai60 | Python | x | x | x | x | ||||||||
| Spacyr61 | R | x | x | x | x | x | x | ||||||
Table 3.
NLP models for MIDD
| Model | Full form | Pretrained on | Architecture | Built on | Performance | Year |
|---|---|---|---|---|---|---|
| BioBERT62,63 | Bio-Bidirectional Encoder Representations from Transformers | PubMed and PMC | Transformer | BERT | Outperforms state-of-the-art (SOTA) for named entity recognition, relation extraction, question answering | September 19 |
| SciBERT64,65 | Science—Bidirectional Encoder Representations from Transformers | Semantic Scholar | Transformer | BERT | Outperforms SOTA for named entity recognition, relation extraction, patient enrollment task | November 19 |
| ClinicalBERT66,67 | Clinical Bidirectional Encoder Representations from Transformers | MIMIC III | Transformer | BERT | Outperforms deep language model for clinical prediction | November 20 |
| BioClinicalBERT68,69 | Bio-Clinical Bidirectional Encoder Representations from Transformers | MIMIC III | Transformer | BioBERT | Outperforms BERT and BioBERT on named entity recognition and natural language inference | June 19 |
| BioMed-RoBERTa70,71 | BioMedical Robustly optimized Bidirectional Encoder Representations from Transformers | Semantic Scholar | Transformer | RoBERTa | Outperforms RoBERTa on text classification, relation extraction and named entity recognition | May 20 |
| Bio Discharge Summary BERT69,72 | Bio Discharge Summary Bidirectional Encoder Representations from Transformers | MIMIC III discharge summaries | Transformer | BioBERT | Outperforms BERT and BioBERT on named entity recognition and natural language inference | June 19 |
| BioALBERT73 | Bio-A Lite Bidirectional Encoder Representations from Transformers | PubMed, PMC, MIMIC III | Transformer | ALBERT | Outperforms SOTA for named entity recognition, relation extraction, question answering, sentence similarity, document classification | July 21 |
| ChemBERTa74,75 | Chem-Bidirectional Encoder Representations from Transformers | PubChem | Transformer | RoBERTa | Outperforms baseline on one task of molecular property prediction | October 20 |
Inclusion criteria
The included articles must be published between 2010 and February 2022.
Articles which discussed the most recent development (until February 2022) or current state-of-the-art algorithm that outperforms baseline for various NLP functionalities.
Articles which highlighted applications of NLP algorithms in various stages of drug development including drug discovery, clinical trial, and pharmacovigilance.
Recently launched transformer-based current state-of-the-art models for biomedical applications were included for the model inventory.
Articles which highlighted NLP algorithm implementation in drug discovery and development areas using any open-source pre-trained models.
Libraries used for Biomedical NLP applications in Python, Java, R, and Scala were included for the library inventory.
Exclusion criteria
Any NLP implementation libraries in languages other than python, java, R, Scala, and C++ were excluded from the review.
NLP transformer models that were trained on datasets other than biomedical datasets such as PubMed, ChemProt, NCBI-diseases etc.
NLP systems involving speech analysis or generation.
RESULTS
NLP aims to transform text information into structured data with the purpose of enhancing the usability of the data and quality of decisions made based on that data. Looking into the specific field of drug discovery and development, a plethora of NLP approaches have been utilized in the previous few years to make use of the huge amount of unstructured data that has been generated and is available in the domain. NLP offers several functionalities that enable analysis of unstructured text data for drug discovery and development applications. Some of the most used NLP functionalities for drug development are listed and explained in the next section (Table 1) and Supplementary Material.
Table 1.
Relevant NLP key concepts
| NLP concept | Definition | Methodology | Biomedical or biochemical applications | MIDD-specific open-source resources |
|---|---|---|---|---|
| Word embedding | A class of techniques where individual words are represented as real-valued vectors, often tens or hundreds of dimensions in a predefined vector space. | It uses language models and feature extraction methods to map words to vectors capturing their context and meaning. Generic pre-trained models such as GloVe,19 word2vec,20 and fastText21 have become prevalent. | Biomedical NLP encompasses use of word embeddings as feature input to downstream ML or DL models. Different textual resources like EHR, clinical notes, biomedical publications, Wikipedia, news etc. are utilized to train these word embeddings. | BioWordVec and BioSentVec22 |
| Named Entity Recognition (NER) | A sequence-labeling task that encompasses locating and categorizing important nouns and proper nouns in text which carry key information in a sentence. | It utilizes either 1 or a combination of the 2 underlying methods: (1) Rule-based method which uses a set of handcrafted grammatical and syntactic rules, and dictionaries to extract the named entities. (2) Machine learning (ML) or deep learning (DL) based method that utilizes a feature-based representation of the observed data.23 | It is used in the clinical domain to extract names of drugs, protein, disease, and genes from radiology reports, discharge summaries, problem lists, nursing documentation, medical education documents, and scientific literature. | MedLEE,24 MetaMap,25 KnowledgeMap,26 cTAKES,27 HiTEX,28 MedTagger,29 and ChemSpot30 |
| Assertion status detection | Status detection in medical assertions as “present,” “absent,” “conditional,” or “associated with someone else,” | Given an entity in a medical text, it classifies its asserted class from the context as being present, absent, or possible in the patient.31 In recent years, assertion detection models have been developed using Convolutional neural networks (CNNs), Long-short term memory network (LSTMs) and attention techniques.32 | In bio-clinical NLP, it is primarily used for assertion status detection for disease modeling. The meaning of clinical entities is heavily affected by assertion modifiers such as negation, uncertain, hypothetical, experiencer, and so on. | MITRE system33 |
| Entity resolution | It is the practice of linking data records that represent the same entity in the absence of a join key. | The process is comprised of the following steps: (1) Blocking—categorizing entities into blocks based on their descriptions. (2) Block processing—removing redundancies within blocks. (3) Matching—matching within a block based on entity descriptions. (4) Clustering—grouping of identified matches together. | In biomedical applications, it is used in record linkage by taking domain-specific knowledge into consideration to avoid domain-general assumptions that do not hold in this domain (eg, overlap in names of chemical compounds).34 | DeepER35 and Bell et al.’s rule-based sieve architecture34 |
| Relation extraction | It is the task of extracting structured information and semantic relations from natural language text between 2 or more entities of a certain type like person, organization, or location. | It uses co-occurrence, pattern matching, machine learning, deep learning, knowledge-driven methods,36 or transfer learning. | In the drug discovery and development domain, it is relevant in extraction of drug–disease, gene–disease, drug–target, and drug–drug relationships. | BioReI37 and DocRBERT38 |
| Topic modeling | It is an unsupervised approach used for finding and classifying various topics embedded within a document or a piece of text. | It is based on the idea that a document is a mixture of topics which are a probability distribution over words. Term frequency-inverse document frequency, non-negative matrix factorization, Latent Dirichlet Allocation, Latent Semantic Analysis,39 attention,39 and generative adversarial networks40 are some of the methods used for implementing it. | In the biomedical domain, topic modeling has been applied to use-cases beyond documents and words, eg, to classify genomic sequences, to classify drugs according to safety and therapeutic use and to find links between genes and diseases.41 | Gensim, Stanford topic modling toolbox and MALLET42 |
One or more of these functionalities can be utilized to build a text processing pipeline to accomplish various drug discovery and development application objectives such as mining EHR data to detect adverse drug reactions or extracting information for potential drug targets from scientific literature. A typical NLP pipeline in drug development can also include text pre-processing methodologies such as tokenization, stemming, lemmatization, and part-of-speech tagging followed by a combination of various NLP functionalities.
NLP model inventory
The NLP landscape is promising, as several techniques have been developed to optimize the performance of various NLP functionalities. With the emergence of transfer learning and transformers in NLP, the efficiency of the process has increased while reducing the dependence on large amount of training data. Several libraries in most-commonly used languages such as python, R, and Java have made the implementation easier with the availability of pre-trained state-of-the-art neural network models. With the increasing use of various NLP algorithms in the healthcare sector, transformer models have been developed specific to healthcare applications such as BioBERT, SciBERT, etc. by training the basic BERT model on biomedical data corpus like PubMed, MIMIC-III, etc. Numerous libraries have made such models accessible for implementation.
We summarized our findings on the NLP in MIDD libraries and models in Tables 2 and 3, respectively. Table 2 provides features of all the state-of-the-art libraries in python, R, and Java for biomedical applications. The features in the inventory are crucial NLP functionalities in different phases of drug discovery and development which were extracted from our literature search and highlighted in the section above. Table 3 provides an overview of state-of-the-art NLP models useful in MIDD. These inventories aim to help researchers in choosing the resources for implementing pre-trained neural models or NLP techniques for their respective NLP application.
NLP in the lifecycle of drug development
There are several facets of the lifecycle of drug development (DD) in which Real World Data (RWD) and NLP algorithms have been implemented with the aim of improving outcomes. To guide the structure of the model inventory, we reviewed literature and use-cases in the following areas:
Drug discovery
Clinical trials
Pharmacovigilance
In each of the cases, different forms of NLP were applied to textual data to derive novel insights in all stages of drug development that would previously have been difficult or even impossible to capture.
Drug discovery
In the drug discovery process, understanding gene–disease associations, pathways, and systems is critical. Much of the data that can aid in extracting this information is in unstructured text.76 Furthermore, most new targets are derived from novel biological discoveries first appearing in scientific literature from academic sources.77 NLP-based text mining has provided a solution which has been widely utilized for applications in gene–disease mapping, target identification, biomarker discovery, and drug repurposing efforts.78 NLP has also been utilized to analyze text-based representations of molecular structures for discovery and design of novel drugs.16 Instead of relying on disparate manually curated sources, an NLP system can mine and extract relevant and valuable knowledge from all these sources at once. In the sections below, the uses of NLP in various drug discovery areas are highlighted.
Gene–disease mapping
Analyzing gene–disease association is a crucial step for target identification and biomarker discovery in the drug discovery pipeline. Experimental methods for identifying gene–disease associations, such as genome-wide association studies and linkage analysis can be expensive and time-consuming.79 Hence, researchers have turned to various insilico methods in the past few years which utilize text-mining, crowdsourcing, network and semantic-similarity-based algorithms.79,80 Mining of biomedical literature is key to extracting actionable information present in free-text data. NLP comes into play in the process by enabling automated text-mining with techniques such as NER81 and relation extraction.82 A few examples of such systems include DisGeNET,83 BeFREE,81 a co-occurrence interaction network presented in Al-Aamri et al80 and a BioBERT-based model introduced in Deng et al.84
Drug–target interaction prediction
Predicting drug–target interaction aims to identify binding of new drug candidate compounds to protein targets. A few approaches have been recently developed to address this problem using NLP techniques. These techniques use word embeddings to represent chemical structures of the drug molecule and the binding protein from an un-labeled biomedical literature.85 Raw data such as simplified molecular-input line-entry system (SMILES) strings for molecules and protein sequences are vectorized in this process of feature representation.86 One common approach for feature representation is using CNN-based models.87 However, it fails to take into account the relationship between different atoms in the molecules.88 Hence, self-attention88 and transformer-based embedding models85 are used to overcome this challenge. Further steps in the process involve machine learning or deep learning models to predict the affinity between the drug molecule and target protein.89,90 Features such as the biological, topological and physio-chemical properties of the drugs/target are considered for making these predictions.91
Biomarker discovery
With the rise of the technologies to extract valuable information available in biomedical big data—electronic medical records (EMRs) and biomedical literature, there is an increased hope of discovering novel biomarkers that can be used to diagnose, predict, and monitor the important aspects of a disease. Biomarkers also serve as surrogate endpoints in early-phase trials.8,92,93 Biomarker and disease names are identified in free-text data using NER and the frequency of their co-occurrences. The relationship between disease and biomarkers can be understood using word embedding and similarity approaches.93 Singh et al8 presents a big data mining approach from EHR data using NER and assertion status detection techniques along with machine learning to facilitate biomarker discovery. Holmes et al25,94 introduced a new method to extract high quality, contextual biomarker information from pathology reports using MetaMap.
Drug repurposing
Drug repurposing is discovering new therapeutic opportunities for existing drugs. It can ensure a faster drug development and approval process, safer treatment and reduced healthcare cost. Computational approaches like virtual screening, molecular docking, deep learning and NLP play a vital role in many of the drug repurposing studies.10,95–102 The drug–disease treatment pairs that are extracted using NLP from literature, EHR, clinical notes, and real-world sources can be used for drug repurposing in 2 ways: the extracted pairs being used themselves or a drug or disease’s similarity with candidate drug or disease respectively is used to hypothesize a new therapeutic indication for a given drug.103 Subramanian et al95 used SciBERT for drug-cancer association classification. In another study, the researchers carried out a drug-wide association study for COVID-19 drug-repurposing using MedXN104 NLP platform for drug information extraction.105 Relation extraction and entity linking are other key NLP techniques that help capture complex relationships in unstructured text.96
Drug design
Drug design in the initial stages of drug discovery is rendered as an optimization problem to search for the optimal combination of building blocks to find the most stable structure in the given conditions. De novo design of molecules has recently benefited from deep generative models, various NLP techniques and transfer learning.106 The task of generating more SMILES strings having an input string is viewed as a language modeling task. To this end, Transmol was developed as a vanilla transformer language model for SMILES sequence generation.107,108 In another study, ULMFit model is used to leverage transfer learning to generate new molecular sequences.107 A recent study introduced Seq2Mol, a method conditioned on the protein target sequence to generate de novo SMILES strings of molecules that are relevant to the target using a deep bi-directional language model ELMo.109
Clinical trials
Approximately 5.6% of clinical trials in the clinicaltrials.gov database have been terminated prematurely (2021). A failed trial sinks not only the investment into the trial itself but also the preclinical development costs, rendering the loss per failed clinical trial at 800 million to 1.4 billion USD.110 Failure to optimize clinical trial design, inefficient enrollment processes, and poor retention rates are one of the main reasons for premature trial closure.111 NLP has been utilized to overcome these issues and help improve the clinical trial process.
Patient-trial matching
Identification of suitable patients can be resource-intensive, often relying on manual review of clinical notes to identify potentially eligible patients, where the information may be split over different systems. Researchers have utilized various NLP techniques for automating clinical trial eligibility pre-screening for patients, increasing the efficiency of the patient selection and recruitment process. NER, assertion status detection, relation extraction, and entity linking features have been primarily used to extract relevant fields from clinical trial eligibility criteria.112–116 These were mapped against relevant fields extracted from unstructured patient EHRs using the same techniques for efficient patient-cohort matching.114,117–120 Recently, advanced models such as Criteria2Query121 which uses an Information Extraction pipeline integrated with a Natural Language Interface, DeepEnroll122 which uses hierarchical embeddings and COMPOSE123 which uses word embeddings on clinical trials eligibility criteria along with a pseudo-Siamese network have provided significant improvement in the patient-trial matching process.
Pharmacokinetic/Pharmacodynamic (PK/PD) studies
PK/PD studies are crucial to determine the dosing and schedule during a clinical trial.124 Post-marketing PK/PD analyses are used to evaluate drug response in patients in real-world setting. These studies require longitudinal dose, outcomes, and potential covariates information. Mining EHRs for this data can be a potential solution. NLP has been leveraged by researchers to automate the process of real-world data extraction from EHRs.125 Existing NLP data mining tools such as MedEx,126 MedXN,104 and medExtractR127 were utilized.
Document preparation for regulatory submissions
NLP is being used to accelerate document preparation with tools that can perform parallel search, document creation, data integrity review and rapidly assembling briefing documents for regulatory submissions.128
Pharmacovigilance
According to the Center for Disease Control and Prevention (CDC), adverse drug events (ADEs) cause approximately 1.3 million emergency department visits each year. It is extremely vital to assess the safety of a drug to avoid any potential adverse events resulting from it. Additionally, active post-marketing surveillance is crucial to account for all side effects that can result from the drug in a larger population over the duration of its usage. EHR and NLP have enabled a more accurate detection of such adverse events compared to the conventional manual methods.
Adverse drug event detection
ADEs are unexpected medical occurrences resulting from drug related intervention. The current method for ADE detection involves manual retrospective record review of medical data stored within EMRs in structured and free-text form. Over the past few years, researchers have utilized various NLP techniques to automate this process by including free-text data from EHRs. The workflow includes identifying and extracting the relationship between a drug and ADE from unstructured EHR data, incident reporting systems, or social media. NER identifies medications and their attributes (dosage, route, duration, and frequency), indications, ADEs, and severity.129,130 Word Sense Disambiguation is used to further filter the identified entities and confirm their contextual sense. The relation extraction task identifies relations between the named entities: medication-indication and medication-ADE.129 Word embeddings are utilized to vectorize the input for training an ML130 or DL131 model to identify and classify ADEs. Numerous publicly available NLP systems have been extended to perform ADE detection tasks, including MedLEE,132 MetaMap,25 cTAKES,27 MedEx,126 and GATE.133 Wu et al134 introduced an NLP system with multi-head self-attention to detect adverse drug reactions from tweets using pre-trained word embeddings, text pre-processing, part of speech embeddings, and sentiment embeddings.
Drug–drug interaction prediction
In cases where 2 or more drugs are co-administered, drug-drug interaction detection becomes a critical part of post-marketing surveillance. Interactions between 2 drugs may lead to side-effects, increased or decreased impact or an adverse reaction. Since there are numerous combinations of drugs available, it is difficult and time-consuming to manually collect all the drug–drug interaction events of patients from reports and scientific literature. To overcome this, several efforts have been made to automate the process by using different text-mining approaches incorporating NLP techniques such as NER, relation extraction, and word embeddings.135,136
EHR data de-identification
To facilitate the use of EHR data without compromising patient privacy multiple NLP methods are being explored.137 These methods include Rule-based extraction, feature-based ML, and Neural methods. The goal is to be both effective in detecting protected health information (PHI) and efficient in processing the data.
DISCUSSION
With the rapid advances in the field of NLP in the past few years, it has found applications in several industries to automate time-consuming manual processing of human-generated natural language. Drug discovery and development is one such field which can leverage the promising future of NLP to its advantage. Our literature review results presented in this article highlight some of the most promising avenues of NLP applications in the journey of a drug from molecule to market.
We found that several researchers have utilized NLP techniques such as NER, relation extraction, word embeddings, assertion status detection, topic modeling, natural language generation, and entity resolution for drug discovery and development applications. Table 2 lists all the current state-of-the-art library resources in python, Java, R, and Scala that can be used to develop models for one or more of the mentioned tasks. The table also includes bio- and clinical-specific libraries that can be utilized to achieve better performance in drug discovery and development applications. The state-of-the-art performance is attributed to the availability of pre-trained neural network models within these libraries that have been trained on biomedical literature. The neural language model-based approaches have been proven to achieve better performance.
With further improvements in the deep learning space, NLP models have moved from Recurrent neural network (RNN) and LSTM to attention-based models and transformers. The added feature of transfer learning with the transformers has led to even higher accuracies. Table 3 captures the trends in the evolution of the state-of-the-art transformer models pretrained on biochemical and biomedical literature. Many of the libraries listed in table 2 utilize these models for enhanced performance. Both these tables provide insights into the technical aspects of various NLP algorithms and tools that are available to easily access those algorithms for drug development implementation that are highlighted in Figure 3.
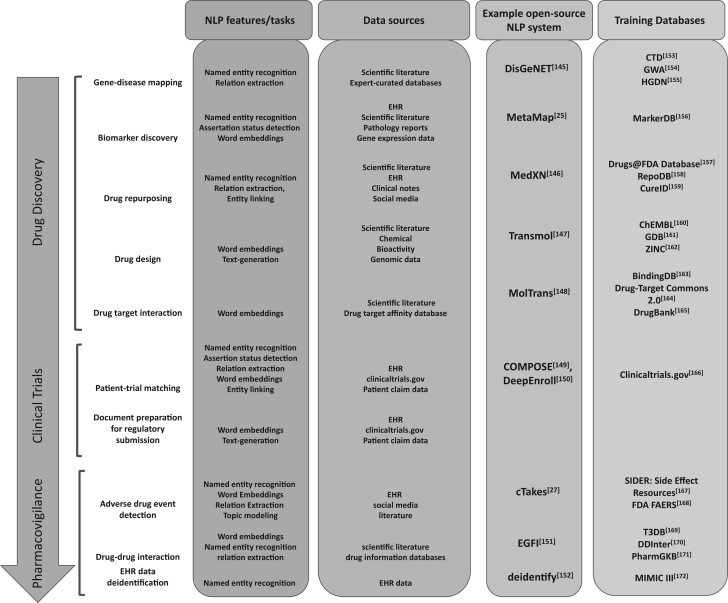
Figure 3.
NLP in stages of drug development. The figure shows NLP functionalities used for applications in 3 stages of drug development process: (1) drug discovery, (2) clinical trials, and (3) pharmacovigilance. The data sources utilized for NLP implementation in these applications are also listed. We also provide some examples of open-source systems for these applications along with links to training datasets.
Figure 3 provides a comprehensive overview of the applicability of various NLP tasks for drug discovery and development use-cases. The figure summarizes NLP use cases in MIDD with examples for drug discovery, clinical trials and pharmacovigilance. The figure ties together the findings of the 2 parts of the literature review—applications and technical aspects of various NLP techniques. It depicts the techniques used for each application of NLP in the drug development domain.
As we saw in our results, there has been a shift from rule-based approaches to increasingly complex neural language models leading to achieving state-of-the-art results. However, this shift has come with a performance-explainability trade-off. The traditional NLP techniques using rule-based or statistical methods are inherently explainable but the prevalence of deep learning models and word embedding techniques have given rise to the need of incorporating explainability as a feature in the models. Some work has been done in recent years to expand the field of explainable and interpretable NLP models.138–141 Biological and chemical interpretability and explainability of NLP models remains a challenge to be addressed in the field of drug discovery and development. Further exploration and application of explainability and interpretability for NLP neural models in drug discovery and development is crucial at the moment to improve NLP acceptability by researchers as well as regulators. Further issues like bias in NLP models stemming from bias in data and algorithm design, security issues surrounding PHI, reproducibility of results are some of the limitations that are hindering wider adoption of these advance techniques for drug development applications.
In order to ensure better adoption of NLP to MIDD, we identified the following opportunities in the field as a result of our research:
The drug discovery and development fields present several opportunities to researchers to apply NLP to further improve the performance of the already existing models.
In order to enable wider adoption of NLP in MIDD, additional work is required in the field to make the models more explainable, interpretable, fair, reproducible, and to overcome issues of security (discussed further in Supplementary File).
Within drug discovery and development, applications in improving clinical trials and pharmacovigilance can be critical for cost savings. It is evident from our review that opportunities exist to explore the fields of document preparation for regulatory submissions, PK/PD modeling and EHR deidentification as not much work has been done on these applications.
Current NLP approaches are limited to a few languages like English or Dutch.142,143 The research can be expanded to include other languages to make the best use of the plethora of available data in regional languages. This can be useful in expanding the reach of NLP systems and improving the performance of the current state-of-the-art algorithms.
Another avenue of interest can be the use of few-shot learning in NLP144 to overcome the challenge of limited data, eg, in the case of drug discovery for rare diseases.
CONCLUSION
Our review focuses on how NLP’s use is evolving in the drug development space. It highlights several functionalities of NLP that aid in automation of MIDD processes in favor of increased efficiency. The article also mentions some resources that can be useful in developing an NLP pipeline using current state-of-the-art methods for MIDD applications. Lastly, it provides insights into how it can be taken forward by addressing some of the unmet needs in the field.
FUNDING
Critical Path Institute is supported by the Food and Drug Administration (FDA) of the U.S. Department of Health and Human Services (HHS) and is 54.2% funded by the FDA/HHS, totaling $13 239 950, and 45.8% funded by non-government source(s), totaling $11 196 634. The contents are those of the author(s) and do not necessarily represent the official views of, nor an endorsement by, FDA/HHS or the U.S. Government.
For more information, please visit FDA.gov.
AUTHOR CONTRIBUTIONS
RB was involved in the conception and design of the work and acquisition, analysis and interpretation of the data. JTP, SS, and MB were involved in the conception and design of the work.
SUPPLEMENTARY MATERIAL
Supplementary material is available at JAMIA Open online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge Grace Erhart, Senior Project Coordinator at the Critical Path Institute, for assistance during manuscript preparation. We would also like to thank Nick Henscheid, Quantitative Medicine Scientist and Mike Pauley, Quantitative Medicine Developer at the Critical Path Institute for reviewing the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
DATA AVAILABILITY STATEMENT
No original data was generated in support of this study.
Contributor Information
Roopal Bhatnagar, Data Science, Data Collaboration Center, Critical Path Institute, Tucson, Arizona, USA.
Sakshi Sardar, Quantitative Medicine, Critical Path Institute, Tucson, Arizona, USA.
Maedeh Beheshti, Quantitative Medicine, Critical Path Institute, Tucson, Arizona, USA.
Jagdeep T Podichetty, Quantitative Medicine, Critical Path Institute, Tucson, Arizona, USA.
REFERENCE
- 1. Khurana D, Koli A, Khatter K, et al. Natural language processing: state of the art, current trends and challenges. arXiv:170805148 [cs]. Published Online First: 17 August 2017. http://arxiv.org/abs/1708.05148. Accessed January 11, 2022.
- 2. Olaronke I, Olaleke J.. A systematic review of natural language processing in healthcare. Int J Inf Technol Comput Sci 2015; 7 (8): 44–50. [Google Scholar]
- 3. Kong H-J. Managing unstructured big data in healthcare system. Healthc Inform Res 2019; 25 (1): 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Z, Liu X, Hogan W, et al. Applications of artificial intelligence in drug development using real-world data. Drug Discov Today 2021; 26 (5): 1256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y, Zhu H, Madabushi R, et al. Model-informed drug development: current US regulatory practice and future considerations. Clin Pharmacol Ther 2019; 105 (4): 899–911. [DOI] [PubMed] [Google Scholar]
- 6. Thafar MA, Olayan RS, Albaradei S, et al. DTi2Vec: drug–target interaction prediction using network embedding and ensemble learning. J Cheminform 2021; 13 (1): 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayes AL, Smith S. Predicting Drug-Drug Interactions from Text using NLP and Privileged Information. ACM 2016; doi: 10.475/123_4.
- 8. Singh S. Big dreams with big data! Use of clinical informatics to inform biomarker discovery. Clin Transl Gastroenterol 2019; 10 (3): e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Subramanian S, Baldini I, Ravichandran S, et al. A natural language processing system for extracting evidence of drug repurposing from scientific publications. AAAI 2020; 34 (08): 13369–81. [Google Scholar]
- 10. Issa NT, Stathias V, Schürer S, et al. Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol 2021; 68: 132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang K, Demner-Fushman D.. Automated classification of eligibility criteria in clinical trials to facilitate patient-trial matching for specific patient populations. J Am Med Inform Assoc 2017; 24 (4): 781–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan P, Peskov K, Song X.. Applications of model-based meta-analysis in drug development. Pharm Res Published Online First: 16 February 2022;doi:10.1007/s11095-022-03201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barrett JS, Nicholas T, Azer K, et al. Role of disease progression models in drug development. Pharm Res Published Online First: 11 April 2022;doi:10.1007/s11095-022-03257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Z, Roberts RA, Lal-Nag M, et al. AI-based language models powering drug discovery and development. Drug Discov Today 2021; 26 (11): 2593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vamathevan J, Clark D, Czodrowski P, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 2019; 18 (6): 463–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Öztürk H, Özgür A, Schwaller P, et al. Exploring chemical space using natural language processing methodologies for drug discovery. Drug Discov Today 2020; 25 (4): 689–705. [DOI] [PubMed] [Google Scholar]
- 17. Chen H, Engkvist O, Wang Y, et al. The rise of deep learning in drug discovery. Drug Discov Today 2018; 23 (6): 1241–50. [DOI] [PubMed] [Google Scholar]
- 18. Gupta R, Srivastava D, Sahu M, et al. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers 2021; 25 (3): 1315–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GloVe: Global Vectors for Word Representation. https://nlp.stanford.edu/projects/glove/. Accessed February 8, 2022.
- 20. Mikolov T, Chen K, Corrado G, et al. Efficient estimation of word representations in vector space. arXiv:13013781 [cs]. Published Online First: 6 September 2013. http://arxiv.org/abs/1301.3781. Accessed February 8, 2022.
- 21. fastText. https://fasttext.cc/index.html. Accessed February 8, 2022.
- 22.BioWordVec & BioSentVec: pre-trained embeddings for biomedical words and sentences. NLM/NCBI BioNLP Research Group (PI: Zhiyong Lu). 2022. https://github.com/ncbi-nlp/BioSentVec. Accessed February 8, 2022.
- 23. Eltyeb S, Salim N.. Chemical named entities recognition: a review on approaches and applications. J Cheminform 2014; 6: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A Natural Language Processing Resource. https://people.dbmi.columbia.edu/~friedma/Projects/nlp.html. Accessed February 8, 2022.
- 25.MetaMap. https://lhncbc.nlm.nih.gov/ii/tools/MetaMap.html. Accessed February 8, 2022.
- 26. Denny JC, Irani PR, Wehbe FH, et al. The KnowledgeMap Project: development of a concept-based medical school curriculum database. AMIA Annu Symp Proc 2003; 2003: 195–9. [PMC free article] [PubMed] [Google Scholar]
- 27.Apache cTAKES™—clinical Text Analysis Knowledge Extraction System. https://ctakes.apache.org/. Accessed February 8, 2022.
- 28. HITEx Manual. https://www.i2b2.org/software/projects/hitex/hitex_manual.html. Accessed February 8, 2022.
- 29.GitHub—medtagger/MedTagger: a collaborative framework for annotating medical datasets using crowdsourcing. https://github.com/medtagger/MedTagger. Accessed February 8, 2022.
- 30. Rocktäschel T, Weidlich M, Leser U.. ChemSpot: a hybrid system for chemical named entity recognition. Bioinformatics 2012; 28 (12): 1633–40. [DOI] [PubMed] [Google Scholar]
- 31. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trajanovska I. Assertion Detection in Electronic Health Records. https://prof.bht-berlin.de/fileadmin/prof/aloeser/master_thesis_ivana_trajanovska_2020.pdf. Accessed February 8, 2022.
- 33. Clark C, Aberdeen J, Coarr M, et al. Determining Assertion Status for Medical Problems in Clinical Records. 2011.
- 34. Bell D, Hahn-Powell G, Valenzuela-Escarcega MA, et al. Sieve-based Coreference Resolution in the Biomedical Domain. In: Proceedings of the Tenth International Conference on Language Resources and Evaluation (LREC'16). Portorož, Slovenia: European Language Resources Association (ELRA); 2016: 177–83. [Google Scholar]
- 35. Ebraheem M, Thirumuruganathan S, Joty S, et al. Distributed representations of tuples for entity resolution. Proc VLDB Endow 2018; 11 (11): 1454–67. [Google Scholar]
- 36. Wei C-H, Peng Y, Leaman R, et al. Assessing the state of the art in biomedical relation extraction: overview of the BioCreative V chemical-disease relation (CDR) task. Database 2016; 2016: baw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xing R, Luo J, Song T.. BioRel: towards large-scale biomedical relation extraction. BMC Bioinformatics 2020; 21 (Suppl 16): 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peng Y, Yan S, Lu Z. Transfer learning in biomedical natural language processing: an evaluation of BERT and ELMo on ten benchmarking datasets. arXiv:190605474 [cs] Published Online First: 18 June 2019. http://arxiv.org/abs/1906.05474. Accessed February 15, 2022.
- 39. Vayansky I, Kumar SAP.. A review of topic modeling methods. Inf Syst 2020; 94: 101582. [Google Scholar]
- 40. Wang R, Zhou D, He Y.. ATM: Adversarial-neural Topic Model. Inf Process Manage 2019; 56 (6): 102098. [Google Scholar]
- 41. ElShal S, Mathad M, Simm J, et al. Topic modeling of biomedical text. In: IEEE International Conference on Bioinformatics and Biomedicine (BIBM); Shenzhen, China; 2016: 712–6. doi: 10.1109/BIBM.2016.7822606. [DOI]
- 42. Liu L, Tang L, Dong W, et al. An overview of topic modeling and its current applications in bioinformatics. Springerplus 2016; 5 (1): 1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.spaCy Industrial-strength Natural Language Processing in Python. https://spacy.io/. Accessed February 15, 2022.
- 44. Řehůřek R, Sojka P.. Software Framework for Topic Modelling with Large Corpora. Valetta, MT: University of Malta; 2010. http://is.muni.cz/publication/884893/en. Accessed February 15, 2022. [Google Scholar]
- 45. NLTK :: Natural Language Toolkit. https://www.nltk.org/. Accessed February 15, 2022.
- 46. CoreNLP. CoreNLP. https://stanfordnlp.github.io/CoreNLP/. Accessed February 15, 2022.
- 47. scispacy. scispacy. https://allenai.github.io/scispacy/. Accessed February 15, 2022.
- 48. John Snow Labs—Spark NLP. Spark NLP. https://nlp.johnsnowlabs.com/. Accessed February 15, 2022.
- 49. Spark NLP for Healthcare | Award Winning Medical NLP | John Snow Labs. Spark NLP for Healthcare. https://www.johnsnowlabs.com/spark-nlp-health/. accessed February 15, 2022.
- 50. RoBERTa: An optimized method for pretraining self-supervised NLP systems. https://ai.facebook.com/blog/roberta-an-optimized-method-for-pretraining-self-supervised-nlp-systems/. Accessed February 15, 2022.
- 51. koRpus. https://cran.r-project.org/web/packages/koRpus/vignettes/koRpus_vignette.html. Accessed February 16, 2022.
- 52.ALBERT: A Lite BERT for Self-Supervised Learning of Language Representations. Google AI Blog. http://ai.googleblog.com/2019/12/albert-lite-bert-for-self-supervised.html. Accessed February 16, 2022.
- 53.scikit-learn: machine learning in Python—scikit-learn 1.0.2 documentation. https://scikit-learn.org/stable/. Accessed February 16, 2022.
- 54.TextBlob: Simplified Text Processing—TextBlob 0.16.0 documentation. https://textblob.readthedocs.io/en/dev/. Accessed February 16, 2022.
- 55. Pattern. Computational Linguistics Research Group 2022. https://github.com/clips/pattern. Accessed February 16, 2022.
- 56.Hugging Face—The AI community building the future. https://huggingface.co/. Accessed February 16, 2022.
- 57.AllenNLP—Allen Institute for AI. https://allenai.org/allennlp. Accessed February 16, 2022.
- 58. Stanza. Stanza. https://stanfordnlp.github.io/stanza/. Accessed February 16, 2022.
- 59. flairNLP/flair. flair 2022. https://github.com/flairNLP/flair. Accessed February 16, 2022.
- 60. fastai. fastai. https://docs.fast.ai/. Accessed February 16, 2022.
- 61. A Guide to Using spacyr. https://cran.r-project.org/web/packages/spacyr/vignettes/using_spacyr.html. Accessed February 16, 2022.
- 62. BioBERT. DMIS Laboratory—Korea University 2022. https://github.com/dmis-lab/biobert. Accessed February 16, 2022.
- 63. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020; 36 (4): 1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. SciBERT. AI2 2022. https://github.com/allenai/scibert. Accessed February 16, 2022.
- 65. Beltagy I, Lo K, Cohan A. SciBERT: a pretrained language model for scientific text. arXiv:190310676 [cs] Published Online First: 10 September 2019. http://arxiv.org/abs/1903.10676. Accessed February 16, 2022.
- 66. Huang K. ClinicalBERT 2022. https://github.com/kexinhuang12345/clinicalBERT. Accessed February 16, 2022.
- 67. Huang K, Altosaar J, Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. arXiv:190405342 [cs] Published Online First: 28 November 2020. http://arxiv.org/abs/1904.05342. Accessed February 16, 2022.
- 68.Bio_ClinicalBERT at main. https://huggingface.co/emilyalsentzer/Bio_ClinicalBERT/tree/main. Accessed February 16, 2022.
- 69. Alsentzer E, Murphy J, Boag W, et al. Publicly Available Clinical BERT Embeddings. In: Proceedings of the 2nd Clinical Natural Language Processing Workshop. Minneapolis, Minnesota, USA: Association for Computational Linguistics; 2019. 72–8. doi: 10.18653/v1/W19-1909. [DOI]
- 70.allenai/biomed_roberta_base at main. https://huggingface.co/allenai/biomed_roberta_base/tree/main. Accessed February 16, 2022.
- 71. Gururangan S, Marasović A, Swayamdipta S, et al. Don’t stop pretraining: adapt language models to domains and tasks. arXiv:200410964 [cs] Published Online First: 5 May 2020. http://arxiv.org/abs/2004.10964. Accessed February 16, 2022.
- 72.Bio_Discharge_Summary_BERT at main. https://huggingface.co/emilyalsentzer/Bio_Discharge_Summary_BERT/tree/main. Accessed February 20, 2022.
- 73.M@@N. BioALBERT. 2022. https://github.com/usmaann/BioALBERT. Accessed February 20, 2022.
- 74. Chithrananda S. ChemBERTa. 2022. https://github.com/seyonechithrananda/bert-loves-chemistry. Accessed February 20, 2022.
- 75. Chithrananda S, Grand G, Ramsundar B. ChemBERTa: large-scale self-supervised pretraining for molecular property prediction. arXiv:201009885 [physics, q-bio] Published Online First: 23 October 2020.http://arxiv.org/abs/2010.09885. Accessed February 20, 2022.
- 76. Leyens L, Reumann M, Malats N, et al. Use of big data for drug development and for public and personal health and care. Genet Epidemiol 2017; 41 (1): 51–60. [DOI] [PubMed] [Google Scholar]
- 77. Mohs RC, Greig NH.. Drug discovery and development: role of basic biological research. Alzheimers Dement (N Y) 2017; 3 (4): 651–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zheng S, Dharssi S, Wu M, et al. Text mining for drug discovery. In: Larson RS, Oprea TI, eds. Bioinformatics and Drug Discovery. New York, NY: Springer; 2019:231–52. doi: 10.1007/978-1-4939-9089-4_13. [DOI] [PubMed] [Google Scholar]
- 79. Opap K, Mulder N.. Recent advances in predicting gene–disease associations. F1000Res 2017; 6: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Al-Aamri A, Taha K, Al-Hammadi Y, et al. Analyzing a co-occurrence gene-interaction network to identify disease-gene association. BMC Bioinformatics 2019; 20 (1): 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bravo À, Piñero J, Queralt-Rosinach N, et al. Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research. BMC Bioinformatics 2015; 16: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ben Abdessalem Karaa W, Alkhammash EH, Bchir A.. Drug disease relation extraction from biomedical literature using NLP and machine learning. Mobile Inf Syst 2021; 2021: 1–10. [Google Scholar]
- 83. Pinero J, Queralt-Rosinach N, Bravo A, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015; 2015: bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Deng C, Zou J, Deng J, et al. Extraction of gene-disease association from literature using BioBERT. In: The 2nd International Conference on Computing and Data Science. New York, NY, USA: Association for Computing Machinery; 2021:1–4. doi: 10.1145/3448734.3450772. [DOI]
- 85. Huang K, Xiao C, Glass LM, et al. MolTrans: molecular interaction transformer for drug–target interaction prediction. Bioinformatics 2021; 37 (6): 830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang Y-F, Wang X, Kaushik AC, et al. SPVec: a Word2vec-inspired feature representation method for drug-target interaction prediction. Front Chem 2019; 7: 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Öztürk H, Özgür A, Ozkirimli E.. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics 2018; 34 (17): i821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shin B, Park S, Kang K, et al. Self-attention based molecule representation for predicting drug-target interaction. Published Online First: 15 August 2019. https://arxiv.org/abs/1908.06760v1. Accessed October 10, 2021.
- 89.Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model—ScienceDirect. https://www.sciencedirect.com/science/article/pii/S2001037020300490. Accessed October 10, 2021. [DOI] [PMC free article] [PubMed]
- 90. Abbasi K, Razzaghi P, Poso A, et al. Deep learning in drug target interaction prediction: current and future perspectives. Curr Med Chem 2021; 28 (11): 2100–13. [DOI] [PubMed] [Google Scholar]
- 91. Bagherian M, Sabeti E, Wang K, et al. Machine learning approaches and databases for prediction of drug–target interaction: a survey paper. Brief Bioinform 2021; 22 (1): 247–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang F, Wu X, Chen J. Computational Biomarker Discovery. In: Chen M, Hofestädt R, eds. Approaches in Integrative Bioinformatics. Berlin, Heidelberg: Springer; 2014:355–86. doi: 10.1007/978-3-642-41281-3_13. [DOI]
- 93. Song H-J, Yoon B-H, Youn Y-S, et al. A method of inferring the relationship between biomedical entities through correlation analysis on text. Biomed Eng Online 2018; 17 (Suppl 2): 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Holmes B, Chitale D, Loving J, et al. Customizable natural language processing biomarker extraction tool. JCO Clin Cancer Inf 2021; (5): 833–41. [DOI] [PubMed] [Google Scholar]
- 95. Subramanian S, Baldini I, Ravichandran S, et al. Drug repurposing for cancer: an NLP approach to identify low-cost therapies. arXiv:191107819 [cs, stat] Published Online First: 5 December 2019. http://arxiv.org/abs/1911.07819. Accessed January 13, 2022.
- 96. Baldini I, Bernagozzi M, Aggarwal S, et al. Exploring the efficacy of generic drugs in treating cancer. Proc AAAI Conf Artif Intell2021;35:15988–90. [Google Scholar]
- 97. Sosa DN, Derry A, Guo M, et al. A literature-based knowledge graph embedding method for identifying drug repurposing opportunities in rare diseases. Pac Symp Biocomput 2020; 25: 463–74. [PMC free article] [PubMed] [Google Scholar]
- 98. Bloom B. Recent successes and future predictions on drug repurposing for rare diseases. Expert Opin Orphan Drugs 2016; 4 (1): 1–4. [Google Scholar]
- 99. Roessler HI, Knoers NVAM, van Haelst MM, et al. Drug repurposing for rare diseases. Trends Pharmacol Sci 2021; 42 (4): 255–67. [DOI] [PubMed] [Google Scholar]
- 100. Singh TU, Parida S, Lingaraju MC, et al. Drug repurposing approach to fight COVID-19. Pharmacol Rep 2020; 72 (6): 1479–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Senanayake SL. Drug repurposing strategies for COVID-19. Future Drug Discovery 2020; 2 (2). doi: 10.4155/fdd-2020-0010. [DOI] [Google Scholar]
- 102. Dotolo S, Marabotti A, Facchiano A, et al. A review on drug repurposing applicable to COVID-19. Brief Bioinform2020; 22: bbaa288. doi: 10.1093/bib/bbaa288. [DOI] [PMC free article] [PubMed]
- 103. Xu R, Wang Q.. Large-scale extraction of accurate drug-disease treatment pairs from biomedical literature for drug repurposing. BMC Bioinformatics 2013; 14: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sohn S, Clark C, Halgrim SR, et al. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc 2014; 21 (5): 858–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bejan CA, Cahill KN, Staso PJ, et al. DrugWAS: drug-wide association studies for COVID-19 drug repurposing. Clin Pharmacol Ther 2021; 110 (6): 1537–46. doi: 10.1002/cpt.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu X, IJzerman A, Westen G. Computational approaches for de novo drug design: past, present, and future. In: Cartwright H, ed. Artificial Neural Networks. Methods in Molecular Biology (Clifton, N.J.). vol. 2190. New York, NY: Humana; 2020: 139–65. doi: 10.1007/978-1-0716-0826-5_6. [DOI] [PubMed]
- 107. Zhumagambetov R, Molnár FA, Peshkov V, et al. Transmol: repurposing a language model for molecular generation. RSC Adv 2021; 11 (42): 25921–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Santana MVS, Silva FP Jr.. De novo design and bioactivity prediction of SARS-CoV-2 main protease inhibitors using recurrent neural network-based transfer learning. BMC Chem 2021; 15 (1): 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ghanbarpour A, Lill MA. Seq2Mol: Automatic design of de novo molecules conditioned by the target protein sequences through deep neural networks. arXiv:201015900 [q-bio] Published Online First: 29 October 2020. http://arxiv.org/abs/2010.15900. Accessed February 20, 2022.
- 110. Harrer S, Shah P, Antony B, et al. Artificial intelligence for clinical trial design. Trends Pharmacol Sci 2019; 40 (8): 577–91. [DOI] [PubMed] [Google Scholar]
- 111. Fogel DB. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: a review. Contemp Clin Trials Commun 2018; 11: 156–64. doi: 10.1016/j.conctc.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Liu H, Chi Y, Butler A, et al. A knowledge base of clinical trial eligibility criteria. J Biomed Inform 2021; 117: 103771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kang T, Zhang S, Tang Y, et al. EliIE: an open-source information extraction system for clinical trial eligibility criteria. J Am Med Inform Assoc 2017; 24 (6): 1062–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bompelli A, Silverman G, Finzel R, et al. Comparing NLP systems to extract entities of eligibility criteria in dietary supplements clinical trials using NLP-ADAPT. In: Artificial Intelligence in Medicine—18th International Conference on Artificial Intelligence in Medicine, AIME 2020, Proceedings. Springer Science and Business Media Deutschland GmbH; 2020:67–77. doi: 10.1007/978-3-030-59137-3_7. [DOI]
- 115. Hao T, Chen X, Huang G. Discovering commonly shared semantic concepts of eligibility criteria for learning clinical trial design. In: Li FWB, Klamma R, Laanpere M, et al., eds. Advances in Web-Based Learning—ICWL 2015. Cham: Springer International Publishing; 2015:3–13. doi: 10.1007/978-3-319-25515-6_1. [DOI]
- 116. Tseo Y, Salkola MI, Mohamed A, et al. Information extraction of clinical trial eligibility criteria. arXiv:200607296 [cs] Published Online First: 28 July 2020. http://arxiv.org/abs/2006.07296. Accessed August 28, 2021.
- 117. Tissot HC, Shah AD, Brealey D, et al. Natural language processing for mimicking clinical trial recruitment in critical care: a semi-automated simulation based on the LeoPARDS trial. IEEE J Biomed Health Inform 2020; 24 (10): 2950–9. [DOI] [PubMed] [Google Scholar]
- 118. Ni Y, Kennebeck S, Dexheimer JW, et al. Automated clinical trial eligibility prescreening: increasing the efficiency of patient identification for clinical trials in the emergency department. J Am Med Inform Assoc 2015; 22 (1): 166–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chen L, Gu Y, Ji X, et al. Clinical trial cohort selection based on multi-level rule-based natural language processing system. J Am Med Inform Assoc 2019; 26 (11): 1218–26. doi: 10.1093/jamia/ocz109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Raghavan P, Chen JL, Fosler-Lussier E, et al. How essential are unstructured clinical narratives and information fusion to clinical trial recruitment? AMIA Jt Summits Transl Sci Proc2014;2014:218–23. [PMC free article] [PubMed] [Google Scholar]
- 121. Yuan C, Ryan PB, Ta C, et al. Criteria2Query: a natural language interface to clinical databases for cohort definition. J Am Med Inform Assoc 2019; 26 (4): 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhang X, Xiao C, Glass LM, et al. DeepEnroll: patient-trial matching with deep embedding and entailment prediction. arXiv:200108179 [cs] Published Online First: 22 January 2020. http://arxiv.org/abs/2001.08179. Accessed Auguast 28, 2021.
- 123. Gao J, Xiao C, Glass LM, et al. COMPOSE: cross-modal pseudo-siamese network for patient trial matching. arXiv:200608765 [cs] Published Online First: 15 June 2020. http://arxiv.org/abs/2006.08765. Accessed August 28, 2021.
- 124. Roberts JA, Taccone FS, Lipman J.. Understanding PK/PD. Intensive Care Med 2016; 42 (11): 1797–800. [DOI] [PubMed] [Google Scholar]
- 125. Choi L, Beck C, McNeer E, et al. Development of a system for postmarketing population pharmacokinetic and pharmacodynamic studies using real-world data from electronic health records. Clin Pharmacol Ther 2020; 107 (4): 934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010; 17 (1): 19–24. doi: 10.1197/jamia.M3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Weeks HL, Beck C, McNeer E, et al. medExtractR: a medication extraction algorithm for electronic health records using the R programming language. 2019:19007286. doi: 10.1101/19007286. [DOI]
- 128. Viswanath S, Fennell JW, Balar K, et al. An industrial approach to using artificial intelligence and natural language processing for accelerated document preparation in drug development. J Pharm Innov 2021; 16 (2): 302–16. [Google Scholar]
- 129. Jagannatha A, Liu F, Liu W, et al. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 1.0). Drug Saf 2019; 42 (1): 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ujiie S, Yada S, Wakamiya S, et al. Identification of adverse drug event–related japanese articles: natural language processing analysis. JMIR Med Inform 2020; 8 (11): e22661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Cocos A, Fiks AG, Masino AJ.. Deep learning for pharmacovigilance: recurrent neural network architectures for labeling adverse drug reactions in Twitter posts. J Am Med Inform Assoc 2017; 24 (4): 813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004; 11 (5): 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.GATE.ac.uk—index.html. https://gate.ac.uk/. Accessed February 8, 2022.
- 134. Wu C, Wu F, Liu J, et al. Detecting tweets mentioning drug name and adverse drug reaction with hierarchical tweet representation and multi-head self-Attention. In: Proceedings of the 2018 EMNLP Workshop SMM4H: The 3rd Social Media Mining for Health Applications Workshop & Shared Task. Brussels, Belgium: Association for Computational Linguistics; 2018:34–7. doi: 10.18653/v1/W18-5909 [DOI]
- 135. Wu H-Y, Chiang C-W, Li L.. Text mining for drug–drug interaction. Methods Mol Biol 2014; 1159: 47–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lim S, Lee K, Kang J.. Drug drug interaction extraction from the literature using a recursive neural network. PLoS One 2018; 13 (1): e0190926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Trienes J. De-identification of electronic health records using NLP. Nedap. 2020. https://medium.com/nedap/de-identification-of-ehr-using-nlp-a270d40fc442. Accessed September 7, 2021.
- 138. Naylor M, French C, Terker S, et al. Quantifying explainability in NLP and analyzing algorithms for performance-explainability tradeoff. Published Online First: 12 July 2021. https://arxiv.org/abs/2107.05693v1. Accessed January 13, 2022.
- 139. Gao K, Fokoue A, Luo H, et al. Interpretable drug target prediction using deep neural representation. In: International Joint Conference on Artificial Intelligence; 2018: 3371–7. doi: 10.24963/ijcai.2018/468. [DOI]
- 140. Goh G, Hodas N, Siegel C, et al. SMILES2Vec: an interpretable general-purpose deep neural network for predicting chemical properties. ArXiv abs/1712.020342017.
- 141. Feldhus N, Schwarzenberg R, Möller S. Thermostat: a large collection of NLP model explanations and analysis tools. Published Online First: 31 August 2021. https://arxiv.org/abs/2108.13961v1. Accessed January 13, 2022.
- 142. Olthof AW, Shouche P, Fennema EM, et al. Machine learning based natural language processing of radiology reports in orthopaedic trauma. Comput Methods Programs Biomed 2021; 208: 106304. [DOI] [PubMed] [Google Scholar]
- 143. Névéol A, Dalianis H, Velupillai S, et al. Clinical natural language processing in languages other than English: opportunities and challenges. J Biomed Semantics 2018; 9 (1): 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Hofer M, Kormilitzin A, Goldberg P, et al. Few-shot learning for named entity recognition in medical text. Published Online First: 13 November 2018. https://arxiv.org/abs/1811.05468v1. Accessed January 13, 2022.
- 145. Piñero J, Saüch J, Sanz F, et al. The DisGeNET cytoscape app: exploring and visualizing disease genomics data. Comput Struct Biotechnol J 2021; 19: 2960–7. doi: 10.1016/j.csbj.2021.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. OHNLP. MedXN: Medication Extraction and Normalization for Clinical Text. Open Health Natural Language Processing; 2022. https://github.com/OHNLP/MedXN. Accessed February 15, 2022. [Google Scholar]
- 147. saulhazelius. TRANSMOL. 2021. https://github.com/saulhazelius/transmol. Accessed February 20, 2022.
- 148. Huang K. MolTrans: Molecular Interaction Transformer for Drug Target Interaction Prediction. 2022. https://github.com/kexinhuang12345/MolTrans. Accessed February 20, 2022. [DOI] [PMC free article] [PubMed]
- 149. v1xerunt. COMPOSE: Cross-Modal Pseudo-Siamese Network for Patient Trial Matching. 2021. https://github.com/v1xerunt/COMPOSE. Accessed February 20, 2022.
- 150. deepenroll. deepenroll/DeepEnroll. 2021. https://github.com/deepenroll/DeepEnroll. Accessed February 20, 2022.
- 151. Layne_Huang. EGFI. 2022. https://github.com/Layne-Huang/EGFI. Accessed February 28, 2022.
- 152. deidentify. Nedap N.V. 2022. https://github.com/nedap/deidentify. Accessed February 28, 2022.
- 153.The Comparative Toxicogenomics Database | CTD. http://ctdbase.org/. Accessed February 28, 2022.
- 154. GWAS Catalog. https://www.ebi.ac.uk/gwas/. Accessed February 28, 2022.
- 155. Literature-derived Human Gene-Disease Network. https://www.dbs.ifi.lmu.de/~bundschu/LHGDN.html. Accessed February 28, 2022.
- 156. MarkerDB. https://markerdb.ca/. Accessed February 28, 2022.
- 157.Drugs@FDA: FDA-Approved Drugs. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm. Accessed February 28, 2022.
- 158. repoDB. http://apps.chiragjpgroup.org/repoDB/. Accessed February 28, 2022.
- 159. CURE ID. https://cure.ncats.io/introduction. Accessed February 28, 2022.
- 160. ChEMBL Database. https://www.ebi.ac.uk/chembl/. Accessed February 28, 2022.
- 161. GDB Databases. https://gdb.unibe.ch/downloads/. Accessed February 28, 2022.
- 162. Sterling T, Irwin JJ.. ZINC 15—ligand discovery for everyone. J Chem Inf Model 2015; 55 (11): 2324–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Liu T, Lin Y, Wen X, et al. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res 2007; 35 (Database issue): D198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Tanoli Z, Alam Z, Vähä-Koskela M, et al. Drug Target Commons 2.0: a community platform for systematic analysis of drug–target interaction profiles. Database (Oxford) 2018; 2018: bay083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018; 46 (D1): D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. ClinicalTrials.gov. https://www.clinicaltrials.gov/. Accessed February 28, 2022.
- 167. SIDER Side Effect Resource. http://sideeffects.embl.de/. Accessed February 28, 2022.
- 168.Research C for DE and FDA Adverse Event Reporting System (FAERS) Public Dashboard. FDA Published Online First: 22 October 2021. https://www.fda.gov/drugs/questions-and-answers-fdas-adverse-event-reporting-system-faers/fda-adverse-event-reporting-system-faers-public-dashboard. Accessed February 28, 2022.
- 169. T3DB. http://www.t3db.ca/. Accessed February 28, 2022.
- 170. DDinter. http://ddinter.scbdd.com/. Accessed February 28, 2022.
- 171. PharmGKB. PharmGKB. https://www.pharmgkb.org/. Accessed February 28, 2022.
- 172. Johnson A, Pollard T, Mark R. MIMIC-III Clinical Database. 2015. doi: 10.13026/C2XW26. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No original data was generated in support of this study.