Abstract
Background
Late bedtime is a common form of unhealthy sleep pattern in adulthood, which influences circadian rhythm, and negatively affects health. However, little is known about the effect of regular late bedtime on skin characteristics, particularly on skin microbiome.
Objective
To investigate the changes and effects of the regular late bedtime on skin physiological parameters and facial bacterial microbiome of 219 cases of Chinese women aged 18–38 years living in Shanghai.
Methods
Based on the Self-Evaluation Questionnaire, bedtime was categorized as 11:00 PM; thus, the volunteers were divided into early bedtime group (S0) and late bedtime group (S1). The physiological parameters of facial skin were measured by non-invasive instrumental methods, and the skin microbiome was analyzed by 16S rRNA high-throughput sequencing.
Results
The skin physiological parameters of the late bedtime group exhibited significant decrease in skin hydration content, skin firmness (F4) and elasticity (R2), while TEWL, sebum and wrinkle significantly increased. The result indicated that late bedtime significantly impaired the integrity of skin barrier, damaged skin structure, and disrupted water–oil balance. Furthermore, the analysis of α-diversity, Sobs, Ace and Chao index were found to significantly decrease (P < 0.05) in the late bedtime group, suggesting that late bedtime reduced both the abundance and the diversity of facial bacterial microbiota. Moreover, the abundance of Pseudomonas increased significantly, while Streptococcus, Stenotrophomonas, Acinetobacter, Haemophilus, Actinomyces and Neisseria decreased significantly. In addition, Spearman correlation analysis revealed strong correlations between the microbiota and the physiological parameters. Notably, the abundance of Pseudomonas significantly positively correlated with skin firmness and elasticity, but significantly negatively correlated with skin hemoglobin content, melanin content and skin hydration.
Conclusion
Bedtime is an important factor in maintaining skin health. Regular late bedtime not only damages the skin barrier and skin structure but also reduces the diversity and composition of facial bacterial microbiome.
Keywords: sleep, late bedtime, skin biophysical property, skin barrier, skin microbiota
Introduction
In modern society, sleep pattern is highly influenced by a large number of social and individual factors, such as employment, gender, age, culture, race and ethnicity, and has great impact on health.1 Late bedtime has developed into a new kind of sleep habit, and short sleep duration and low sleep quality have been found to be strongly linked with both physical and mental problems,2–4 such as anxiety symptoms, dental caries, obesity, heart disease, high blood pressure and diabetes.5–7
It has been reported that low sleep efficiency and chronically poor sleep quality are significantly associated with TEWL, a direct indicator of skin barrier function.8,9 Meanwhile, contradictory results have been presented about whether sleep deprivation changes facial appearances measured both objectively and subjectively.10–13 Despite the fact that bedtime procrastination, which was suggested to induce insufficient sleep,14 exists in a large proportion of the general population15 and causes physical illness,16 little is known regarding how regular late bedtime affects skin status.
Skin surface is covered with numerous microorganisms, and together with the physical barrier, they protect us from the invasion of pathogens.17,18 When the delicate balance within this symbiotic system is disturbed, unfavorable alteration of skin conditions or even skin diseases could happen, although it is still rather unclear which happens first, the disturbance of such balance or the change of the skin status.19–21
Hence, in the current study, we aimed to investigate the underlying correlation between self-reported bedtime and variability in skin physiological properties and skin microbiota. The results will further our knowledge of microbiota–host interactions in the skin.
Materials and Methods
Subjects and Inclusion Criteria
This study was based on the investigation of the correlation between skin health status and skin and gut microbiota of qualified volunteers living in Shanghai (aged 18–60, n = 494). This research was abided by the ethical guidelines of the Declaration of Helsinki. Ethics approval was provided by the Institutional Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital South Campus (Shanghai Fengxian District Central Hospital, Approval No.: 2021-KY-15). A written informed consent was obtained from each subject prior to sample collection. Volunteers had no skin diseases and no antibiotics uptake within the last 3 months prior to the study. All volunteers needed to carefully finish the Self-Evaluation Questionnaire, which included questions about their skin condition, life habits and physiology. All skin parameters and microbial samples were collected in July 2021.
Considering that skin physiological parameters and skin microbiota are significantly influenced by gender, age and skin site,22,23 data on the biophysical parameters and microbiota of the cheek from a total of 219 healthy women aged 18–38 years were included in this study. The sleep patterns of the volunteers were obtained through the Self-Evaluation Questionnaire using questions including “At what time do you usually fall asleep on weekdays or workdays (hour, AM or PM)?”, “At what time do you usually fall asleep on weekends or your non-work days (hour, AM or PM)?”, “How much time do you usually sleep on weekdays and weekends (hour)?” and “Do you have sleep problems?”. According to several sleep-related studies,7,24 it is generally believed that falling asleep after 11:00 PM belongs to late sleep time, so in our study bedtime was classified as 11:00 PM, then the volunteers were divided into early bedtime group (S0) and late bedtime group (S1). The basic information of the subjects in each group is summarized in Table 1.
Table 1.
Basic Information of the Subjects in Each Group
| Characteristics | Early Bedtime Group (S0) | Late Bedtime Group (S1) | P |
|---|---|---|---|
| Total number of subjects, n | 98(44.75%) | 121(55.25%) | |
| Distribution of age (yr) Min, Max | 18–38 | 18–38 | |
| Age (yr), mean±SD | 26.92±1.96 | 25.76±2.33 | 0.132 |
| BMI (KG/m2), mean±SD | 21.25±3.30 | 20.36±2.99 | 0.118 |
| Skin type no. (%) | |||
| Dry | 7(7.14) | 6(4.96) | |
| Neutral | 23(23.46) | 9(7.44) | |
| Oily | 14(14.29) | 18(14.8) | |
| Mixed | 54(55.10) | 88(72.73) | |
| Sleep duration per day (h) | 7.50±0.83 | 7.16±.75 | 0.002** |
Note: **P < 0.01.
Skin Physiological Parameters Collection
All subjects were required not to use skin care products and cosmetics after washing their faces the night before the measurement and the day of the test. The measurements of the skin parameters were conducted in controlled conditions (Temperature: 20–22°C; Relative humidity: 40–60%), and the subjects were required to sit in such conditions for 30 minutes prior to the tests.
The skin parameters were measured by different instruments (Courage + Khazaka electronic GmbH, Germany) as follows: skin hydration in the stratum corneum (Corneometer CM 825); TEWL (Tewameter TM 300); skin sebum (Sebumeter SM 815); skin pH (Skin-pH-Meter pH 905); skin glossiness (Glossymeter GL200); skin melanin and hemoglobin (Mexameter MX 18); skin wrinkle, smoothness and roughness (Visioscan VC 98), skin elasticity (R2) and firmness (F4) (Cutometer dual MPA580).
Skin Sample Collection
All subjects were not allowed to wash their faces on the day of skin microbial sample collection, and an 8-hour interval between face washing and sample collection was required. Samples were collected from an area of 3*3 cm2 on the cheeks by the swabs moistened with sterile collection solution (containing 0.9% NaCl and 0.1% Tween-20).25 The sampling regions were swabbed approximately 30 times for at least 20 seconds. After collection, the samples were immediately stored at −80°C for subsequent DNA extraction.
DNA Extraction and Illumina MiSeq Sequencing
Microbial genomic DNA was extracted from skin swabs using the FastDNA Spin Kit (MP Biomedicals, USA) according to the kit instructions. The V3–V4 hypervariable region in the 16S rRNA gene was amplified with upstream primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and downstream primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR products were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, USA) according to manufacturer’s instructions. Libraries were constructed using the NEXTFLEX Rapid DNA-Seq Kit (Bioo Scientific, USA), and sequencing was performed using the MiSeq Reagent Kit (Illumina, USA) with the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
Processing of Sequencing Data
The raw 16S rRNA gene sequencing reads were demultiplexed and quality-filtered by fastp (https://github.com/OpenGene/fastp, version 0.20.0) and merged by FLASH (http://www.cbcb.umd.edu/software/flash, version 1.2.7). Operational taxonomic units (OTUs) with 97% similarity cutoff were clustered using UPARSE (http://drive5.com/uparse/, version 7.1), and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier (http://rdp.cme.msu.edu/, version 2.2) against the 16S rRNA database (eg Silva v138) using confidence threshold of 0.7.26
The raw data from high-throughput sequencing were collated and filtered, and the validated sequences were obtained for subsequent analysis. Approximately 5,694,129 valid sequences were obtained. All samples were flattened out according to the minimum sequence number, and then in turn a total of 13,539 OTUs were obtained, belonging to 61 phyla, 1963 genera and 4389 species.
Statistical Analysis
All data are represented as mean ± standard deviation (X±SD) unless otherwise indicated. The statistical significance level was 0.05 unless otherwise noted. One-way ANOVA (if the data present normal distribution) and non-parametric Wilcoxon rank-sum test (if the data did not show normal distribution) were performed to compare biophysical parameters between the two groups of subjects using SPSS 26.0 software (IBM, USA). Meanwhile, a non-parametric Wilcoxon rank-sum test was used to compare microbial diversity and composition between the two groups. To compare the β-diversity between two groups, Jaccard, Bray-Curtis, Unweighted distance metrics and Weighted distance metrics were used. The Spearman rank correlation test was used to determine the Spearman correlation between the biophysical parameters and the abundance of bacteria. These data were analyzed on the online platform of MajorbioI-Sanger Cloud Platform (www.i-sanger.com).
Results
Characteristics of the Subjects
As we have known in Table 1, the proportion of late sleepers was up to 55.25% in our female volunteers. Obviously, late bedtime is common in modern life. In our survey, volunteers had no sleep problems or disorders. The average age and BMI between group S0 and S1 did not differ significantly. The distribution of skin types (dry, neutral, oily and mixed) obtained from Self-Evaluation Questionnaire in each group was analyzed. Among participants, mixed skin accounted for the most, and there was no obvious difference in the proportion of dry skin and oily skin between the two groups. The proportion of mixed skin was higher in the S1 group (72.73%) than that in the S0 group (55.10%), while the S0 group (23.46%) had more neutral skin type compared to the S1 group (7.44%). Average sleep durations in the two groups were both more than 7 hours, which meant volunteers had relatively sufficient sleep time. Moreover, S0 group had significantly longer sleep time than that in S1 group (P=0.002), so our study was different from previous studies, which utilized short-term or long-term sleep deprivation method.10,12,13
Skin Physiological Parameters are Significantly Affected by Bedtime
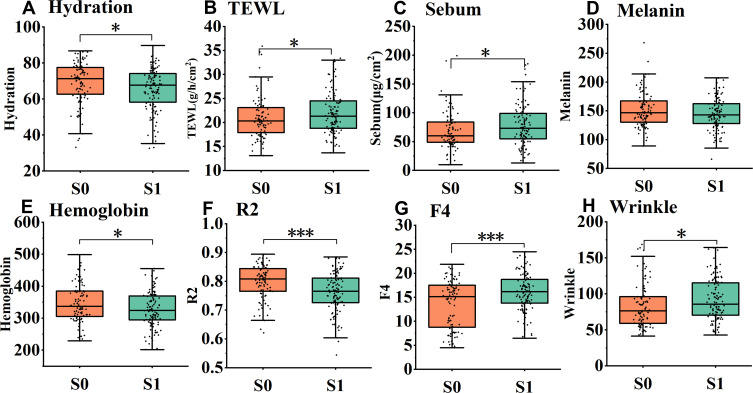
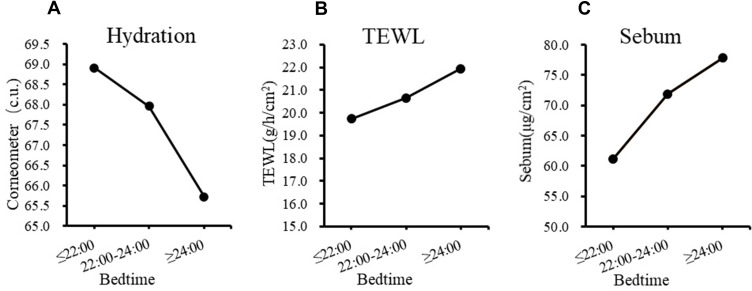
The physiological parameters of the volunteers in groups S0 and S1 were measured and analyzed (Figure 1). Skin hydration in the stratum corneum decreased significantly (Figure 1A, P = 0.023) in the late bedtime group, while TEWL (P = 0.043) and sebum content (P = 0.045) increased significantly (Figure 1B and C). These demonstrate that late bedtime could lead to skin barrier damage. Indeed, further analysis by dividing the subjects into 3 groups based on their bedtime (at and before 22:00, n = 24; between 22:00 and 24:00, n = 96; and at and after 24:00, n = 99) showed that, along with the delay of bedtime, skin hydration dropped progressively, as TEWL and sebum content displayed a gradual ascending trend (Figure 2). Meanwhile, there was no difference in skin pH (5.14 ± 0.43 vs 5.11 ± 0.40, P = 0.501) between the two groups, which suggested that regular late bedtime would not lead to acid–base imbalance on the skin surface.
Figure 1.
Skin physiological parameters in early bedtime group (S0) and late bedtime group (S1). Significant differences were shown in the figure. (A) Hydration, (B) TEWL, (C) Sebum, (D) Melanin, (E) Hemoglobin, (F) R2, (G) F4 and (H) Wrinkle. *P < 0.05, ***P < 0.001.
Figure 2.
The influence of bedtime on skin barrier related parameters. (A) Hydration, (B) TEWL and (C) Sebum.
In the late bedtime group, elasticity (R2) and firmness (F4) values declined significantly (Figure 1F and G, P < 0.001), while skin wrinkle (Figure 1H, P = 0.019) and skin roughness (3.37 ± 1.38 vs 4.30 ± 4.99, P = 0.013) elevated significantly. Nevertheless, skin scale (0.61 ± 0.15 vs 0.58 ± 0.17, P = 0.392), glossiness (9.31 ± 2.49 vs 9.35 ± 2.71, P = 0.957) and smoothness (206.36 ± 69.59 vs 216.4 ± 62.78, P = 0.101) had no significant change. These results implied that late bedtime would cause skin aging by damaging the skin tissue structure. Moreover, the content of hemoglobin decreased significantly in volunteers that regularly go to bed late (Figure 1E, P = 0.049), but there was no dramatic change in the melanin content (Figure 1D, P = 0.120). Hence, it seems that late bedtime would cause insufficient or poor blood flow but probably would not substantially affect skin color.
Late Bedtime Leads to Alterations in Skin Microbiome
Late Bedtime Reduces the Number of OTUs
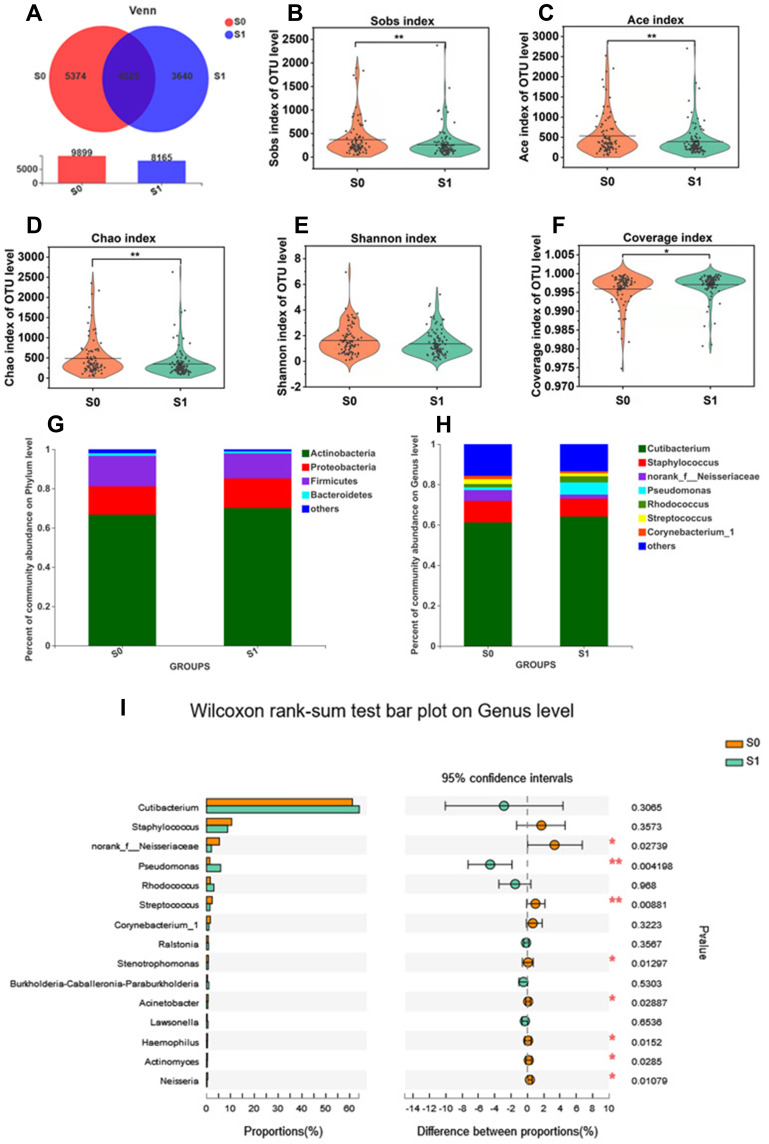
Venn diagram showed the numbers of common and unique OTUs in group S0 and S1 (Figure 3A). There were in total 9899 OTUs in S0 and 8165 OTUs in S1. Also, there were 4525 common OTUs between the two groups, while 5374 OTUs and 3640 OTUs were found only in S0 and S1, respectively. The results indicated that late bedtime would decrease the total number of bacterial species on the cheek skin.
Figure 3.
Bacterial diversity and composition analysis.(A) Venn diagram on OTU level; (B) the Alpha diversity evaluated index:Sobs index; (C) Ace index; (D) Chao index; (E) Shannon index; (F) Coverage index; (G) Species abundance on phylum level of S0, S1 (H) Species abundance on genus level of S0, S1; (I) Differences of species abundance on genus level of S0, S1; S0: early bedtime group; S1: late bedtime group; difference test method: Wilcoxon rank-sum test, P< 0.05 means significant difference, *P < 0.05, **P < 0.01.
Late Bedtime Decreases the α-Diversity
The α-diversity between S0 and S1 groups was analyzed by Wilcoxon rank-sum test, and the results are shown in Figure 3B–F. At the OTU level, the Sobs index (P = 0.003; Figure 3B), Ace index (P = 0.029; Figure 3C) and Chao index (P = 0.003; Figure 3D) of the late bedtime group were significantly lower than those of the early bedtime group, indicating a notable reduction in bacterial community richness in people with late bedtime. Also, albeit lack of statistical significance, the Shannon index (P = 0.053; Figure 3E) and the Simpson index (P = 0.143) showed a decreasing and increasing tendency in S1, respectively, indicating that the bacterial community diversity in the late bedtime group was lower. The Coverage index was close to 1 in both groups (Figure 3F), indicating that the sequencing results could represent the real situation of microorganisms in the sample. In other words, a late bedtime would reduce the community richness and diversity of the skin microbiota.
No Difference of β-Diversity Between the Two Sleep Groups
In order to compare the β-diversity between people who went to bed early and late, Jaccard, Bray-Curtis, Unweighted distance metrics and Weighted distance metrics were calculated (data not shown). Nonmetric multidimensional scaling (NMDS) was plotted based on the metrics, and analysis of similarity (ANOSIM) was used to evaluate the difference between the two groups. Using different calculation methods, the difference between the two groups was not significant, indicating that the overall species composition and relative abundance of bacteria of early sleepers and late sleepers was similar.
Differences in Taxonomic Profiles of Skin Bacteria Between the Two Sleep Groups
The bacterial composition of the two sleep groups was analyzed at different taxonomic levels. The major phyla were similar between the two groups, and were mainly composed of Actinobacteria, Proteobacteria, Firmicutes and Bacteroidetes, accounting for more than 97% of the bacterial community (Figure 3G). Although the relative abundance of these major phyla did not appear to be different, the less abundant phyla, such as Cyanobacteria (P = 0.007), Fusobacteria (P = 0.000), Patescibacteria (P = 0.004) and Deinococcus-Thermus (P = 0.010), significantly declined in the late bedtime group.
Cutibacterium, Staphylococcus, Pseudomonas, Rhodococcus, norank_f_Neisseriaceae, Streptococcus and Corynebacterium were the dominant bacterial genera in the two groups (Figure 3H). The comparison analysis between group S0 and S1 showed that, among the 15 most abundant genera, the bacterial abundance of 8 genera significantly varied (Figure 3I). More specifically, the relative abundance of Pseudomonas significantly elevated in the late bedtime group (P = 0.004), but the abundance of norank_f_Neisseriaceae (P = 0.027), Streptococcus (P = 0.009), Stenotrophomonas (P = 0.013), Acinetobacter (P = 0.029), Haemophilus (P = 0.015), Actinomyces (P = 0.028) and Neisseria (P = 0.011) all decreased significantly.
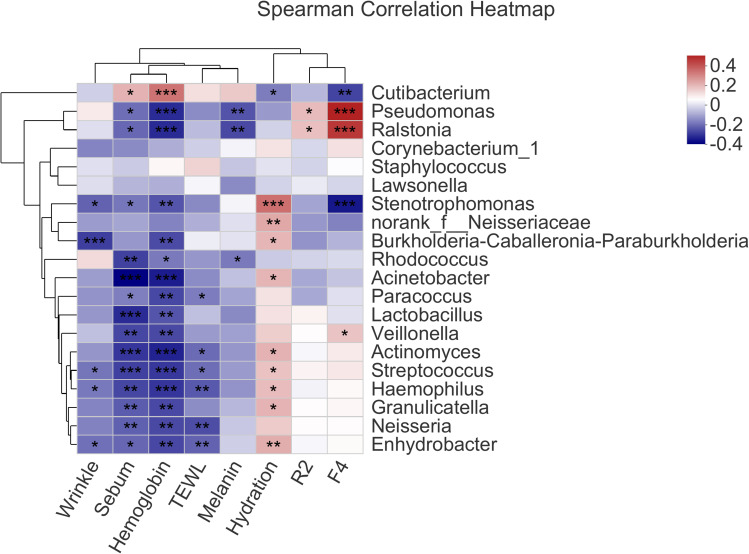
Correlation Between Skin Bacteria and Physiological Parameters
The Spearman correlation between skin physiological parameters (sebum, hydration, TEWL, F4, R2, hemoglobin, melanin and wrinkle) and the skin bacteria was analyzed and summarized into a Spearman correlation heatmap (Figure 4). The relative abundance of Cutibacterium had significantly positive correlation with sebum content (r = 0.175, P = 0.013), skin hemoglobin content (r = 0.282, P = 0.000), and significantly negative correlation with skin hydration (r = −0.143, P = 0.043) and F4 (r = −0.226, P = 0.001) (Figure 4). In addition, with statistical significance, the relative abundance of Pseudomonas positively correlated with F4 (r = 0.417, P = 0.000) and R2 (r = 0.161, P = 0.023), and negatively correlated with skin hemoglobin content (r = −0.262, P = 0.000), melanin content (r = −0.203, P = 0.003) and sebum level (r = −0.164, P = 0.020) (Figure 4).
Figure 4.
Spearman correlations between facial bacterial abundance and skin physiological parameters; only the top 20 genera in abundance were shown. *P < 0.05, **P < 0.01, ***P < 0.001.
Moreover, both Streptococcus and Haemophilus had significantly positive correlation with skin hydration (r = 0.149, P = 0.034, and r = 0.158, P = 0.025, respectively), but significantly negative correlation with TEWL (r = −0.163, P = 0.034, and r = −0.196, P = 0.005, respectively), sebum content (r = −0.231, P = 0.001, and r = −0.222, P = 0.002, respectively) and hemoglobin content (r = −0.243, P = 0.000, and r = −0.246, P = 0.000, respectively) (Figure 4). Furthermore, Neisseria was negatively correlated with physiological parameters such as sebum (r = −0.185, P = 0.009), hemoglobin (r = −0.218, P = 0.002) and TEWL (r = −0.212, P = 0.003) (Figure 4).
It is worthy noticing that Norank_f_Neisseriaceae (r = 0.193, P = 0.006), Streptococcus (r = 0.149, P = 0.034), Stenotrophomonas (r = 0.289, P = 0.000), Haemophilus (r = 0.158, P = 0.025), Actinomyces (r = 0.176, P = 0.012) and Acinetobacter (r = 0.171, P = 0.015) were all significantly positively correlated with skin hydration (Figure 4). Indeed, this was consistent with the fact that the skin hydration decreased significantly in the late bedtime group (Figure 1A), while the abundance of these genera also significantly dropped (Figure 3I).
Discussion
In our investigation, the proportion of late sleepers was up to 55.25% in the female volunteers. Obviously, late bedtime has developed into a new sleep habit. To date, current evidence has been focusing on the effects of insufficient sleep or sleep deprivation on skin characteristics,11,13,27,28 yet little is known about whether bedtime has any impact on the skin status, particularly on the skin microbiome. Thus, to the best of our knowledge, the current study is the first attempt to investigate the effects of regular late bedtime on facial skin and skin bacterial microbiota in healthy women.
By measuring and comparing skin biophysical parameters, we first found that regular late bedtime would damage skin barrier function and alter skin structure. In the late bedtime group, the physiological indicators of skin barrier property, including skin hydration content, TEWL and sebum content, were significantly altered, which was also observed in many other researches on the relationship between sleep conditions, such as poor sleep, short-term or long-term sleep deprivation, and skin physiological characteristics.11–13,27,28 We argued that the underlying mechanism that caused this impairment of the skin barrier was likely to be associated with circadian rhythms, which have been demonstrated previously to regulate and affect skin function.29,30 It was reported that epidermal and hair follicle keratinocytes, dermal fibroblasts and melanocytes present circadian oscillators, which affect cell migration and proliferation, wound healing and susceptibility of the tissues to damage.31 Circadian rhythms were also shown to influence facial skin surface lipid excretion content, TEWL and pH.32,33 In addition, recent studies have revealed that circadian rhythms would affect lipid metabolism of the skin, produce different lipid products, and then in turn affect TEWL and pH of the skin,34 thus affecting the barrier function of the skin and inducing skin problems such as acne.35–37
Circadian rhythms also influenced elastin and elastic fiber-associated proteins.30,38 Indeed, recent studies have also linked the circadian clock to psoriasis, sensitive skin and atopic dermatitis.39–42 Moreover, it was reported that poor sleep quality deregulated the immune system and impaired the integrity of collagen fibers,43,44 which might another reason for the reduction of skin elasticity and firmness. Therefore, we speculated that the significant reduction of skin firmness and elasticity in the late bedtime group was probably also linked with altered circadian rhythms caused by regular late bedtime.
The diversity and abundance of skin microbiota vary with intrinsic factors, including ethnicity, gender, age and body site,21,23,45–47 as well as external factors, such as climate, air quality, living environment and personal hygiene habits.23,48–52 While previous studies have already demonstrated that sleep habit affects circadian rhythms, intestine microbiota, brain and skin health,29,30,53,54 our results revealed that bedtime also affected the diversity and composition of skin microorganisms.
Previous studies have revealed that when the skin is in a bad condition or even in the disease state, the diversity of skin microbiota would change.21,55,56 Coincidentally, we observed that the OTU numbers of the late bedtime group decreased, compared to the early bedtime group. Further analysis of α-diversity proved significant decrease in the community richness and diversity of facial bacteria in the late bedtime group. However, there was no significant difference between the two groups by ß-diversity analysis, indicating that the species composition of early sleepers and late sleepers was similar. Similar results were observed in other similar studies. For example, the β-diversity of sensitive skin and different skin types did not exhibit significant differences from healthy skin.26,57,58 Hence, we inferred that it could possibly take a remarkable change in the skin physiological status to shift the overall species composition and relative abundance of bacteria, like psoriasis or other diseases.59,60
Our results indicated that late bedtime induced changes in the abundance of bacteria. There were some significant differences in the phyla with low abundance, although the main bacteria remained the same. At the genus level, the most abundant genera of facial skin microbiota were Cutibacterium and Staphylococcus in both groups, which was consistent with previous reports.21,48,50 In addition, although it is worth mentioning that no statistically significant difference was found in the abundance of Cutibacterium and Staphylococcus between early and late sleepers, it was still interesting to see that the abundance of Cutibacterium increased when the abundance of Staphylococcus decreased in the late bedtime group, and this led to the elevation of the ratio between Cutibacterium and Staphylococcus. It is believed that alteration of the ratio between these two main genera of the facial microbiota would affect the skin homeostasis.59,61,62 Indeed, Cutibacterium acne and Staphylococcus epidermidis, which are the main species on the facial skin, are also known as sentinel bacteria.61 The relative abundance of Cutibacterium acne affects skin health and plays a crucial role in maintaining skin homeostasis.63 Also, the abnormal variation in the abundance of Staphylococcus species (S. epidermidis and S. aureus) has been suggested to cause atopic dermatitis (AD), sensitive skin, acne vulgaris and dandruff.20,58,63–65 Therefore, we thought that regularly going to bed late would cause shifts in the major skin bacteria, which is potentially detrimental to skin homeostasis and health.
We discovered that, in people who went to bed late regularly, the relative abundance of Pseudomonas increased significantly, while with significant correlation, hemoglobin and melanin contents decreased in the late bedtime group. Pseudomonas, belonging to the commensal bacteria on the skin, has been found to possess relatively high abundance in the elderly, normal young adults and people living in high-altitude environment.47,66,67 Under normal circumstances, Pseudomonas would not cause skin infection but instead could induce keratinocyte cells to produce antibacterial proteins, but when the skin barrier was not integrity, it could aggravate the damage to the skin barrier.68 Therefore, it was our speculation that elevated abundance of Pseudomonas at least partially contributed to the damage to skin barrier in late sleepers, but how Pseudomonas and its metabolites affect skin physiology requires further research.
Meanwhile, our results revealed that the abundance of other commensal bacteria decreased significantly, including norank_f_Neisseriaceae, Streptococcus, Stenotrophomonas, Acinetobacter, Haemophilus and Neisseria. As all of these bacteria exhibited significant correlation with physiological parameters, we are inclined to believe that, when the skin microenvironment changes, manifested as variations in skin hydration, sebum and pH value, the abundance of these bacteria alters too; however, it is still unclear what is the direction of causality between these two events, if there was one. Interestingly, Gihyeon Kim et al69 reported that Streptococcus had a relatively high abundance in young women and found that Streptococcus-secreted spermidine contributed to the recovery of skin structure and barrier function. Together with our findings, this implied that the deterioration of the skin barrier integrity was possibly correlated with Streptococcus at least.
The survival of skin bacteria is inseparable from the external environment, which is determined by the skin physiological state and is directly reflected by the physiological parameters. Indeed, the abundance of skin bacteria is closely related to skin physiological parameters.47 We found that, in the late bedtime group, sebum content increased significantly, while skin hydration level decreased significantly. Coincidentally, the relative abundance of Cutibacterium, which was most likely Cutibacterium acne, a lipophilic species,61 also increased. On the contrary, the relative abundance of Neisseria, Streptococcus, Stenotrophomonas, Haemophilus and Acinetobacter, which preferred humid environment, decreased. Our results suggested that other bacteria with low relative abundance were affected by changes in high abundance bacteria, such as Cutibacterium, Staphylococcus, and strongly linked to the variation of physiological parameters like sebum and hydration level. Investigation into these relatively less abundant bacteria is also warranted, as low or rare abundance bacteria have been proven to help in maintaining an overall microbial community structure.70,71
As we have known, skin flora is easily disturbed by a variety of environmental factors.23 Using pillowcase with antibacterial textiles or filled with functional plant ingredients in pillow, which would have a certain impact on our facial skin flora, but everyone’s skin flora is relatively stable over time.72 Human skin harbors a diverse milieu of commensals, including bacteria, fungi, viruses and mites, which live together as an intricate ecological community. In this study, we mainly focused on the impact of late bedtime on the cutaneous bacterial abundance and diversity, but did not extend our research to fungi and skin mites. It has proven that fungi, viruses and mites also have important implications for skin health.56,73 For example, Malassezia species, and more recently Candida species, have been implicated in acne pathology, causing well-documented cases of folliculitis.74–76 In addition, the Demodex mites, such as Demodex folliculorum and Demodex brevis, are significantly increased in the skin of rosacea patients,77 causing sensitive skin,78 and the endosymbionts of Demodex Folliculorum like Bacillus Oleronius induced proinflammatory cytokine.77 Our study revealed that late bedtime could lead to significantly elevated skin sebum, so we could speculate that the abundance of lipophilic fungi and mites would also increase, which might affect the microbial homeostasis and further the function of the skin. More studies are needed to reveal the effects of late bedtime on fungi and mites, and to get an insight into the association between late bedtime and the skin microecology.
In conclusion, the results in this study demonstrated that regular late bedtime could damage the skin barrier and skin structure, and also reduce the diversity and composition of facial bacterial microbiome. In particular, the effect of the relative abundance of different species, such as Pseudomonas and Streptococcus, on the skin barrier and structure is worthy of further investigation. Findings from this study suggest that bedtime is an important factor in maintaining skin health. We think that the data on skin characteristics and microbiome obtained from this study are useful for the development of personal care products and has furthered our knowledge on the relationship between microbiota and our skin.
Acknowledgments
The authors sincerely acknowledge all the study participants who provided skin physiological parameters and specimens for the study.
Funding Statement
This work was funded by Shandong Freda Biotech Co., Ltd. and Shanghai Collaborative Innovation Center of Fragrance, Flavor and Cosmetics (1021ZK202002008).
Data Sharing Statement
The sequence dataset has been deposited on the NCBI Sequence Reads Archive (SRA) Database (Accession Number: SRP330206).
Disclosure
The authors declare no competing interests.
References
- 1.Grandner A. Sleep, health, and society. Sleep Med Clin. 2017;12(1):1–22. doi: 10.1016/j.jsmc.2016.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St-Onge MP, Grandner MA, Brown D, et al. Sleep duration and quality: impact on lifestyle behaviors and cardiometabolic health: a scientific statement from the American Heart Association. Circulation. 2016;134(18):e367–e386. doi: 10.1161/CIR.0000000000000444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smiley A, King D, Bidulescu A. The association between sleep duration and metabolic syndrome: the NHANES 2013/2014. Nutrients. 2019;11(11):2582. doi: 10.3390/nu11112582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi L, Chen SJ, Ma MY, et al. Sleep disturbances increase the risk of dementia: a systematic review and meta-analysis. Sleep Med Rev. 2018;40:4–16. doi: 10.1016/j.smrv.2017.06.010 [DOI] [PubMed] [Google Scholar]
- 5.Alqaderi H, Redline S, Tavares M, Goodson JM. Effect of late bedtime on salivary glucose and abdominal obesity in children. Sleep Biol Rhythms. 2017;15(3):227–233. doi: 10.1007/s41105-017-0105-y [DOI] [Google Scholar]
- 6.Alqaderi H, Tavares M, Al-Mulla F, Al-Ozairi E, Goodson JM. Late bedtime and dental caries incidence in Kuwaiti children: a longitudinal multilevel analysis. Community Dent Oral Epidemiol. 2020;48(3):181–187. doi: 10.1111/cdoe.12523 [DOI] [PubMed] [Google Scholar]
- 7.Yan B, Fan Y, Zhao B, et al. Association between late bedtime and diabetes mellitus: a large community-based study. J Clin Sleep Med. 2019;15(11):1621–1627. doi: 10.5664/jcsm.8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oyetakin-White P, Suggs KB, Matsui MS A, Yarosh D, Cooper KDB. Does poor sleep quality affect skin ageing? Clin Exp Dermatol. 2015;40(1):17–22. doi: 10.1111/ced.12455 [DOI] [PubMed] [Google Scholar]
- 9.Yoshizaki T, Kimira Y, Mano H, et al. Association between skin condition and sleep efficiency in Japanese young adults. J Nutr Sci Vitaminol. 2017;63(1):15–20. doi: 10.3177/jnsv.63.15 [DOI] [PubMed] [Google Scholar]
- 10.Sundelin T, Lekander M, Sorjonen K, Axelsson J. Negative effects of restricted sleep on facial appearance and social appeal. Royal Soc Open Sci. 2017;4(5):160918. doi: 10.1098/rsos.160918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holding BC, Sundelin T, Cairns P, Perrett DI, Axelsson J. The effect of sleep deprivation on objective and subjective measures of facial appearance. J Sleep Res. 2019;28(6):e12860. doi: 10.1111/jsr.12860 [DOI] [PubMed] [Google Scholar]
- 12.Min AK, Kim EJ, Kang BY, Lee HK. The effects of sleep deprivation on the biophysical properties of facial skin. J Cosmet Dermatol Sci Appl. 2017;7(1):34–47. doi: 10.4236/jcdsa.2017.71004 [DOI] [Google Scholar]
- 13.Sue I, Myeongryeol J L, Jiyeon H, et al. A study of skin characteristics with long-term sleep restriction in Korean women in their 40s. Skin Res Technol. 2020;26(2):193–199. doi: 10.1111/srt.12797 [DOI] [PubMed] [Google Scholar]
- 14.Kroese FM, Ridder D, Evers C, Adriaanse MA. Bedtime procrastination: introducing a new area of procrastination. Front Psychol. 2014;5:611. doi: 10.3389/fpsyg.2014.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroese FM, Evers C, Adriaanse MA, Ridder DD. Bedtime procrastination: a self-regulation perspective on sleep insufficiency in the general population. J Health Psychol. 2016;21(5):853–862. doi: 10.1177/1359105314540014 [DOI] [PubMed] [Google Scholar]
- 16.Yang T, Peng S, Barnett R, Wu D, Feng X, Oliffe JL. Association between bedtime and self-reported illness among college students: a representative nationwide study of China. Public Health. 2018;159:67–69. doi: 10.1016/j.puhe.2018.01.035 [DOI] [PubMed] [Google Scholar]
- 17.Prescott SL, Larcombe DL, Logan AC, et al. The skin microbiome: impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ J. 2017;10(1):1–16. doi: 10.1186/s40413-017-0160-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacy A, Belkaid Y. Microbial guardians of skin health. Science. 2019;363(6424):227–228. doi: 10.1126/science.aat4326 [DOI] [PubMed] [Google Scholar]
- 19.SPaller AS, Kong HH, Seed P, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(1):26–35. doi: 10.1016/j.jaci.2018.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grice EA. The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin Cutan Med Surg. 2014;33(2):98–103. doi: 10.12788/j.sder.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16(3):143–155. doi: 10.1038/nrmicro.2017.157 [DOI] [PubMed] [Google Scholar]
- 22.Park S. Biochemical, structural and physical changes in aging human skin, and their relationship. Biogerontology. 2022;23(1):1–14. doi: 10.1007/s10522-021-09945-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimitriu PA, Iker B, Malik K, Leung H, Hillebrand GG. New insights into the intrinsic and extrinsic factors that shape the human skin microbiome. mBio. 2019;10(4):e00839–19. doi: 10.1128/mBio.00839-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao C, Wang X, Mao Y, et al. Variation of biophysical parameters of the skin with age, gender, and lifestyles. J Cosmet Dermatol. 2021;20(1):249–255. doi: 10.1111/jocd.13453 [DOI] [PubMed] [Google Scholar]
- 25.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci. 2008;105(46):17994–17999. doi: 10.1073/pnas.0807920105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y, Liang H, Zhou M, Liya S, He C. Skin bacterial structure of young females in China: the relationship between skin bacterial structure and facial skin types. Exp Dermatol. 2021;30(10):1366–1374. doi: 10.1111/exd.14105 [DOI] [PubMed] [Google Scholar]
- 27.Sundelin T, Lekander M, Kecklund G, Van Someren EJ, Olsson A, Axelsson J. Cues of fatigue: effects of sleep deprivation on facial appearance. Sleep. 2013;36(9):1355–1360. doi: 10.5665/sleep.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang SI, Jung Y, Lee M, et al. Evaluation of changes in skin characteristics due to the poor quality of sleep caused by smartphone usage. J Cosmet Dermatol. 2022;21(4):1656–1665. doi: 10.1111/jocd.14265 [DOI] [PubMed] [Google Scholar]
- 29.Matsui MS, Pelle E, Dong K, Pernodet N. Biological rhythms in the skin. Int J Mol Sci. 2016;17(6):801. doi: 10.3390/ijms17060801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyons AB, Moy L, Moy R, Tung R. Circadian rhythm and the skin: a review of the literature. J Clin Aesthet Dermatol. 2019;12(9):42–45. [PMC free article] [PubMed] [Google Scholar]
- 31.Sherratt MJ, Hopkinson L, Naven M, et al. Circadian rhythms in skin and other elastic tissues. Matrix Biol. 2019;84:97–110. doi: 10.1016/j.matbio.2019.08.004 [DOI] [PubMed] [Google Scholar]
- 32.Yosipovitch G, Xiong GL, Haus E, Sackett-Lundeen L, Ashkenazi I, Maibach HI. Time-dependent variations of the skin barrier function in humans: transepidermal water loss, stratum corneum hydration, skin surface pH, and skin temperature. J Invest Dermatol. 1998;110(1):20–23. doi: 10.1046/j.1523-1747.1998.00069.x [DOI] [PubMed] [Google Scholar]
- 33.Le Fur I, Reinberg A, Lopez S, Morizot F, Mechkouri M, Tschachler E. Analysis of circadian and ultradian rhythms of skin surface properties of face and forearm of healthy women. J Invest Dermatol. 2001;117(3):718–724. doi: 10.1046/j.0022-202x.2001.01433.x [DOI] [PubMed] [Google Scholar]
- 34.Jia Y, Zhou M, Huang H, Gan Y, Yang M, Ding R. Characterization of circadian human facial surface lipid composition. Exp Dermatol. 2019;28(7):858–862. doi: 10.1111/exd.13933 [DOI] [PubMed] [Google Scholar]
- 35.Zhou M, Yang M, Zheng Y, et al. Skin surface lipidomics revealed the correlation between lipidomic profile and grade in adolescent acne. J Cosmet Dermatol. 2020;19(12):3349–3356. doi: 10.1111/jocd.13374 [DOI] [PubMed] [Google Scholar]
- 36.Zhou M, Wang H, Yang M, et al. Lipidomic analysis of facial skin surface lipids reveals an altered lipid profile in infant acne. Br J Dermatol. 2020;182(3):817–818. doi: 10.1111/bjd.18474 [DOI] [PubMed] [Google Scholar]
- 37.Yang M, Zhou M, Li Y, Huang H, Jia Y. Lipidomic analysis of facial skin surface lipid reveals the causes of pregnancy-related skin barrier weakness. Sci Rep. 2021;11(1):1–10. doi: 10.1038/s41598-020-79139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plikus MV, Van Spyk EN, Pham K, et al. The circadian clock in skin: implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J Biol Rhythms. 2015;30(3):163–182. doi: 10.1177/0748730414563537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Németh V, Horváth S, Kinyó Á, Gyulai R, Lengyel Z. Expression patterns of clock gene mRNAs and clock proteins in human psoriatic skin samples. Int J Mol Sci. 2021;23(1):121. doi: 10.3390/ijms23010121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farage MA. The prevalence of sensitive skin. Front Med. 2019;6:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duan J, Greenberg EN, Karri SS, Andersen B. The circadian clock and diseases of the skin. FEBS Lett. 2021;595(19):2413–2436. doi: 10.1002/1873-3468.14192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaughn AR, Clark AK, Sivamani RK, Shi VY. Circadian rhythm in atopic dermatitis-pathophysiology and implications for chronotherapy. Pediatr Dermatol. 2018;35(1):152–157. doi: 10.1111/pde.13364 [DOI] [PubMed] [Google Scholar]
- 43.Romera-Vilchez M, Montero-Vilchez T, Herrero-Fernandez M, et al. Impact of exposome factors on epidermal barrier function in patients with obstructive sleep apnea syndrome. Int J Environ Res Public Health. 2022;19(2):659. doi: 10.3390/ijerph19020659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kahan V, Andersen M, Tomimori J, Tufik S. Stress, immunity and skin collagen integrity: evidence from animal models and clinical conditions. Brain Behav Immun. 2009;23(8):1089–1095. doi: 10.1016/j.bbi.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 45.Jugé R, Rouaud‐Tinguely P, Breugnot J, et al. Shift in skin microbiota of Western European women across aging. J Appl Microbiol. 2018;125(3):907–916. doi: 10.1111/jam.13929 [DOI] [PubMed] [Google Scholar]
- 46.Kim H-J, Kim JJ, Myeong NR, et al. Segregation of age-related skin microbiome characteristics by functionality. Sci Rep. 2019;9(1):1–11. doi: 10.1038/s41598-018-37186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee S, Mitra R, Maitra A, et al. Sebum and hydration levels in specific regions of human face significantly predict the nature and diversity of facial skin microbiome. Sci Rep. 2016;6(1):1–11. doi: 10.1038/srep36062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung M, Tong X, Bastien P, et al. Changes of the human skin microbiota upon chronic exposure to polycyclic aromatic hydrocarbon pollutants. Microbiome. 2020;8(100):1–17. doi: 10.1186/s40168-020-00874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung M, Tong X, Wilkins D, Cheung H, Lee P. Individual and household attributes influence the dynamics of the personal skin microbiota and its association network. Microbiome. 2018;6(26):1–15. doi: 10.1186/s40168-018-0412-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, Bai X, Peng T, Yi X, Hu D. New insights into the skin microbial communities and skin aging. Front Microbiol. 2020;11:565549. doi: 10.3389/fmicb.2020.565549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moskovicz V, Gross A, Mizrahi B. Extrinsic factors shaping the skin microbiome. Microorganisms. 2020;8(7):1023. doi: 10.3390/microorganisms8071023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y, Wang Z, Zhang Y, Ruan L, Li A, Liu X. Microbiome in healthy women between two districts with different air quality index. Front Microbiol. 2020;2020:2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parekh PJ, Oldfield EC, Johnson IVDA. The effects of sleep on the commensal microbiota: eyes wide open? J Clin Gastroenterol. 2018;52(3):204–209. doi: 10.1097/MCG.0000000000000965 [DOI] [PubMed] [Google Scholar]
- 54.Shochat T, Magzal F, Tamir S, Agmon M, Haimov I. Sleep timing is associated with gut microbiota composition in older adults with insomnia. Sleep Med. 2019;64:S350. doi: 10.1016/j.sleep.2019.11.976 [DOI] [Google Scholar]
- 55.Wang R, Farhat M, Na J, Li R, Wu Y. Bacterial and fungal microbiome characterization in patients with rosacea and healthy controls. Br J Dermatol. 2020;183(6):1112–1114. doi: 10.1111/bjd.19315 [DOI] [PubMed] [Google Scholar]
- 56.Schommer NN, Gallo RL. Structure and function of the human skin microbiome. Trends Microbiol. 2013;21(12):660–668. doi: 10.1016/j.tim.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bai Y, Wang Y, Zheng H, Tan F, Yuan C. Correlation between facial skin microbiota and skin barriers in a Chinese female population with sensitive skin. Infect Drug Resist. 2021;14:219. doi: 10.2147/IDR.S287844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng Y, Liang H, Li Z, Tang M, Song L. Skin microbiome in sensitive skin: the decrease of Staphylococcus epidermidis seems to be related to female lactic acid sting test sensitive skin. J Dermatol Sci. 2019;97(3):225–228. doi: 10.1016/j.jdermsci.2019.12.004 [DOI] [PubMed] [Google Scholar]
- 59.Fahlén A, Engstrand L, Baker BS, Powles A, Fry L. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch Dermatol Res. 2012;304(1):15–22. [DOI] [PubMed] [Google Scholar]
- 60.Bjerre R, Bandier J, Skov L, Engstrand L, Johansen J. The role of the skin microbiome in atopic dermatitis: a systematic review. Br J Dermatol. 2017;177(5):1272–1278. doi: 10.1111/bjd.15390 [DOI] [PubMed] [Google Scholar]
- 61.Fournière M, Latire T, Souak D, Feuilloley MG, Bedoux G. Staphylococcus epidermidis and Cutibacterium acnes: two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms. 2020;8(11):1752. doi: 10.3390/microorganisms8111752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu Z, Wang Z, Yuan C, et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci Rep. 2016;6(1):1–9. doi: 10.1038/s41598-016-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rozas M, Ruijter A, Fabrega MJ, Zorgani A, Brillet F. From dysbiosis to healthy skin: major contributions of Cutibacterium acnes to skin homeostasis. Microorganisms. 2021;9(3):628. doi: 10.3390/microorganisms9030628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakatsuji T, Chen TH, Narala S, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9(378):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Claudel J-P, Auffret N, Leccia M-T, Poli F, Corvec S, Dréno B. Staphylococcus epidermidis: a potential new player in the physiopathology of acne? Dermatology. 2019;235(4):287–294. doi: 10.1159/000499858 [DOI] [PubMed] [Google Scholar]
- 66.Wilantho A, Deekaew P, Srisuttiyakorn C, Tongsima S, Somboonna N. Diversity of bacterial communities on the facial skin of different age-group Thai males. PeerJ. 2017;5(11):e4084. doi: 10.7717/peerj.4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng B, Zhao J, Guo W, et al. High-altitude living shapes the skin microbiome in humans and pigs. Front Microbiol. 1929;2017(8):1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansmann B, Schröder J-M, Gerstel U. Skin-derived C-terminal filaggrin-2 fragments are Pseudomonas aeruginosa-directed antimicrobials targeting bacterial replication. PLoS Pathog. 2015;11(9):e1005159. doi: 10.1371/journal.ppat.1005159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim G, Kim M, Kim M, et al. Spermidine-induced recovery of human dermal structure and barrier function by skin microbiome. Commun Biol. 2021;4(1):1–11. doi: 10.1038/s42003-020-01566-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shade A, Gilbert JA. Temporal patterns of rarity provide a more complete view of microbial diversity. Trends Microbiol. 2015;23(6):335–340. doi: 10.1016/j.tim.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 71.Kong HH, Oh J. State of residency: microbial strain diversity in the skin. J Invest Dermatol. 2022;142(5):1260–1264. doi: 10.1016/j.jid.2021.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oh J, Byrd AL, Park M, Kong HH, Segre JA, Program NCS. Temporal stability of the human skin microbiome. Cell. 2016;165(4):854–866. doi: 10.1016/j.cell.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pessemier BD, Grine L, Debaere M, Maes A, Paetzold B, Callewaert C. Gut–skin axis: current knowledge of the interrelationship between microbial dysbiosis and skin conditions. Microorganisms. 2021;9(2):353. doi: 10.3390/microorganisms9020353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akaza N, Akamatsu H, Numata S, et al. Microorganisms inhabiting follicular contents of facial acne are not only Propionibacterium but also Malassezia spp. J Dermatol. 2016;43(8):906–911. doi: 10.1111/1346-8138.13245 [DOI] [PubMed] [Google Scholar]
- 75.Song HS, Kim SK, Kim YC. Comparison between Malassezia folliculitis and non-Malassezia folliculitis. Ann Dermatol. 2014;26(5):598–602. doi: 10.5021/ad.2014.26.5.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prohic A, Jovovic Sadikovic T, Krupalija‐Fazlic M, Kuskunovic‐Vlahovljak S. Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol. 2016;55(5):494–504. doi: 10.1111/ijd.13116 [DOI] [PubMed] [Google Scholar]
- 77.Jarmuda S, O’Reilly N, Żaba R, Jakubowicz O, Szkaradkiewicz A, Kavanagh K. Potential role of Demodex mites and bacteria in the induction of rosacea. J Med Microbiol. 2012;61(11):1504–1510. doi: 10.1099/jmm.0.048090-0 [DOI] [PubMed] [Google Scholar]
- 78.Yasak Guner R, Tosun M, AkyolS M, Hayta B. Demodex infestation as a cause of sensitive skin in a dermatology outpatient clinic. J Cosmet Dermatol. 2022;21(4):1610–1615. doi: 10.1111/jocd.14246 [DOI] [PubMed] [Google Scholar]