Abstract
Rosacea is a chronic skin disease characterized by photosensitivity, abnormal dermal vascular behavior, inflammation and enhanced expression of the antimicrobial peptide LL-37. We observed that dermal endothelial cells in rosacea had increased expression of VCAM1 and hypothesized that LL-37 could be responsible for this response. Digestion of dsRNA from keratinocytes exposed to ultraviolet B radiation (UVB) blocked the capacity of these cells to induce adhesion molecules on dermal microvascular endothelial cells. However, a synthetic non-coding snoU1RNA was only capable of increasing adhesion molecules on endothelial cells in the presence of LL-37, suggesting that the capacity of UVB exposure to promote both dsRNA and LL-37 was responsible for the endothelial response to keratinocytes. Sequencing of RNA from endothelial cells uncovered activation of gene ontology pathways relevant to the human disease such as type I and II interferon signaling, cell-cell adhesion, leukocyte chemotaxis and angiogenesis. Functional relevance was demonstrated as dsRNA and LL-37 promoted adhesion and transmigration of monocytes across endothelial cell monolayers. Gene knock down of TLR3, RIGI or IRF1 decreased monocyte adhesion endothelial cells, confirming the role of dsRNA recognition pathways. These observations show how expression of LL-37 can lead to enhanced sensitivity to UVB radiation in rosacea.
Keywords: Adhesion Molecules, Inflammation, endothelial dysfunction, nucleic acids, Double-stranded RNA, cathelicidin
INTRODUCTION
Rosacea is a common centro-facial inflammatory disorder of the skin affecting up to 5 % of the world population (Gether et al. 2018). Persistent facial erythema due to chronic recurrent inflammation significantly affects the quality of life of rosacea patients (Zeichner et al. 2018). Rosacea skin is characterized by hyperproliferation of blood vessels known as telangiectasia, with increased recruitment of pro-inflammatory immunocytes to the sites of telangiectasia. Exposure to ultraviolet radiation B (UVB) is one of the most frequently reported triggers of flares in rosacea (Buddenkotte and Steinhoff 2018). The mechanism responsible for increased sensitivity to ultraviolet radiation in rosacea is unknown.
It is well known that the endothelial cells lining the microvascular system actively participate in regulating blood flow, protein uptake and local inflammation (Michiels 2003). LL-37, a cathelicidin antimicrobial peptide, is overexpressed in facial skin of rosacea patients, and promotes vascular proliferation in vivo (Koczulla et al. 2003; Yamasaki et al. 2007) . LL-37 has been implicated in vascular dysfunction by enhancing the expression of adhesion molecule Intracellular Adhesion Molecule-1 (ICAM1)and Monocyte chemoattractant protein −1 on endothelial cells in patients with atherosclerosis and may contribute to cardiovascular disease progression (Edfeldt et al. 2006). In this study, we sought to understand the effect of epidermal photodamage on dermal microvascular endothelial cells and determine if the innate immune dysfunction leading to excess LL-37 in rosacea skin might contribute to the photosensitivity observed in these patients. Our observations uncover a direct link between UVR damage to keratinocytes and the subsequent increase in adhesion molecule expression observed on microvascular endothelial cells.
RESULTS:
Adhesion molecule expression is increased in rosacea and can be modeled by UV damage to keratinocytes
Immunohistochemical analysis of facial skin biopsies from patients with rosacea revealed that adhesion molecule VCAM1, ICAM1 and E-selectin along with antimicrobial peptide LL-37 was highly expressed (Figure 1a). Since the inflammation observed in patients with rosacea is exacerbated by UV exposure (D’Orazio et al. 2013; Murphy 2004), we hypothesized that UVB exposure of keratinocytes might promote the expression of adhesion molecules in the adjacent endothelial cells. Normal human keratinocytes (NHEKs) were exposed to UVB (25 mJ/cm2) and then cell extracts were transferred onto human dermal microvascular endothelial cells (HDMECs). Extracts of non-irradiated NHEKs were used as a control. NHEKs exposed to UVB showed an increase in necrotic cells, immediately following exposure (Figure S1 a,b). HDMECs exposed to the irradiated keratinocyte extract showed increased expression of adhesion molecules VCAM1, ICAM1 and SELE, and TNF, CCL5, CCL10, CX3CL1 and IFNB1 (Figure 1b and Figure S1c).
Figure 1: LL-37 enables UVB and dsRNA to increase adhesion molecule expression on HDMECs.
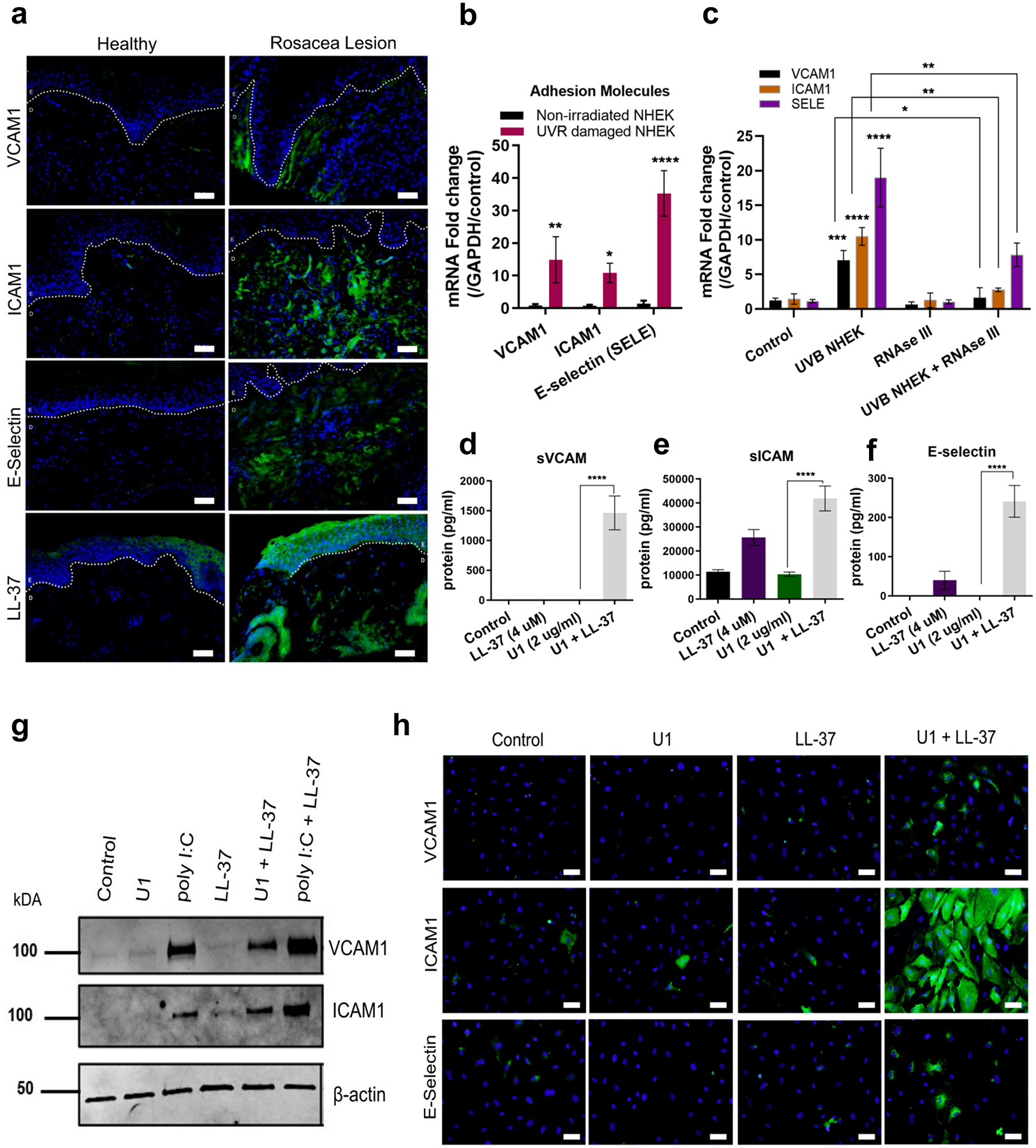
(a) Facial skin from healthy and rosacea subjects were stained VCAM1, ICAM1, E-Selectin and LL-37 antibody (green) and nuclear stain DAPI (blue). E=Epidermis and D=Dermis, Scale= 20 μM (b) UVB exposed and non-irradiated NHEKs extract were added onto HDMECs (12 h). Gene expression of adhesion molecules VCAM1, ICAM1 and SELE was assessed with q-RT PCR. (n=3) (c) UVB NHEKs were treated with RNAse III, then extracts co-cultured with HDMECs and mRNA expression of VCAM1, ICAM1 and SELE was assessed. (n=3) (d, e, f) Secreted adhesion molecules was analyzed by ELISA. (n=3). (g, h) Immunoblot (g) and immunofluorescence (h) analysis for adhesion molecules. Scale = 20 μM. (*, p< 0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001)
UVB promotes release of small non-coding dsRNA from keratinocytes (Bernard et al. 2012a). We hypothesized therefore that the release of dsRNA was necessary for the response of the HDMECs to UVB NHEK extracts. Digestion of dsRNA with RNAse III inhibited expression of adhesion molecules ICAM1, VCAM1 and SELE and pro-inflammatory proteins TNF, CXCL10, CCL5 and IFNB1 (Figure 1c and Figure S1d, e, f, g).
We hypothesized that LL-37 produced by irradiated NHEK also contributed to the response of endothelial cells to dsRNA. When added individually, LL-37, or a synthetic snoU1 dsRNA (U1) caused minimal change in adhesion molecule expression on HDMECs. However, co-treatment of HDMECs with LL-37 and U1 enhanced expression of VCAM, ICAM1 and E-Selectin (Figure 1d, e, f). An increase in expression of adhesion molecules was dependent on time of exposure (Figure S2a, b, c), and dose of LL-37 (Figure S2 d). VCAM1 and ICAM1 protein were also increased as seen by immunoblot from cultured HDMECs (Figure 1 g). PolyI:C, a viral double-stranded RNA mimetic (Alexopoulou et al. 2001; Matsumoto and Seya 2008; Uematsu and Akira 2007), enhanced VCAM1 and ICAM1 protein expression by itself and was further enhanced by addition of LL-37 (Figure 1g). Immunohistochemical staining of cultured HDMECs further confirmed enhanced expression of VCAM1, ICAM1 and E-Selectin by co-treatment with U1 and LL-37 (Figure 1h).
Distinct pathways activated in endothelial cells exposed to LL-37 and U1
A global transcriptomic profile was obtained by direct RNA sequencing of HDMECs exposed to vehicle, U1, LL-37 or LL-37+U1. Principal Component Analysis (PCA) plot showed that the combination of LL-37 and U1 had a uniquely different profile (Figure 2a). Of all genes increased by 1.5-fold or more, 40.6 % genes were uniquely up-regulated by co-treatment with U1 and LL-37 (Figure 2b). Gene ontology (GO) analysis showed enrichment of several pathways including: type 1 interferon signaling; cytokine signaling; antigen processing and presentation via MHC class I; leukocyte chemotaxis; and adhesion and angiogenesis (Figure 2c). Reconstruction of protein-protein interaction networks from genes up-regulated upon co-treatment with U1 and LL-37 showed upregulation of type I interferon signaling pathway regulated genes STAT1 and ISG15, MHC Class I regulated gene PSMB10, and MYD88 (Figure 2d). Furthermore, hierarchical clustering and heat map visualization showed co-treatment with U1 and LL-37 led to increased expression of: pattern recognition receptors TLR3 and DDX58 (encoding RIGI); chemokines CX3CL1, CXCL11, CXCL10, CXCL15 and CCL5; MYD88; genes involved in antigen processing and presentation HLA-B and HLA-C; and type 1 interferon regulated genes STAT1, IRF1, IRF9, STAT3 among others (Figure 2e). A overlap in up-regulated innate immune pathways was seen when HDMECs were co-treated with poly I:C (0.1 μg/ml) and LL-37 (2.5 μM), compared to cells co-treated with U1 (2 μg/ml) and LL-37 (2.5 μM) (Figure S2f). Overall, the RNA sequencing data showed LL-37 enabling U1 to activate several unique innate immunity signaling pathways in HDMECs.
Figure 2: Transcriptional profiling shows LL-37 enables U1 to trigger activation of distinct innate immune pathways.
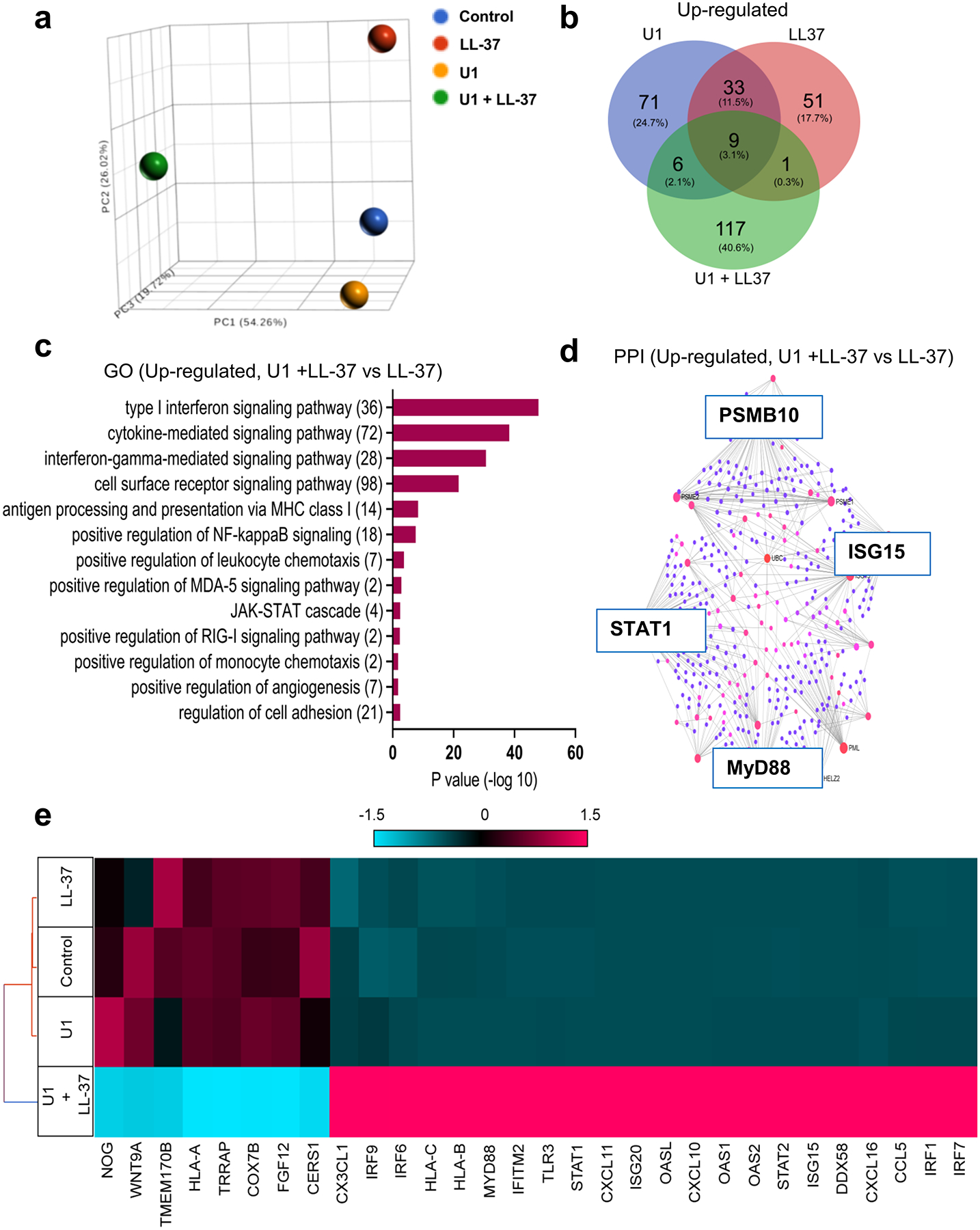
(a) A principal component analysis (PCA) plot for total transcriptional profile (b) Venn Diagram for up-regulated gene set from HDMECs treated with U1 and/or LL-37 (c) Gene ontology (GO) pathway analysis of genes up-regulated in cells co-treated with U1 and LL-37 (d) Protein-protein interaction (PPI) networks derived from network analysis for genes up-regulated in U1 andLL-37 co-treated gene set as compared to LL-37 treatment are shown. The red and blue color nodes represent their betweenness centrality values, red being the highest and blue lowest. (e) Hierarchical clustering and Heatmap visualization of selected genes from GO enriched pathways (1.5-fold change)
Mechanisms that promote adhesion molecule expression by RNA and LL-37
To understand how dsRNA and LL-37 activate HDMECs, we explored known RNA recognition systems including TLR3 and the cytoplasmic dsRNA helicase RIGI and its co-receptor MAVS. The type 1 interferon pathway was also of interest since it regulates VCAM1 expression in human umbilical vein endothelial cells (Ivashkiv and Donlin 2014; Neish et al. 1995a). Expression of TLR3, RIGI, MAVS and IRF1 was significantly up-regulated after co-treatment with U1 and LL-37 in HDMECs (Figure 3b). STAT1 phosphorylation was also increased, indicating activation of type 1 interferon pathway (Figure 3a). Transient knockdown of TLR3, RIGI, MAVS and IRF1 by small interfering RNA (siRNA) was performed (Figure 3c) and inhibition of each of these genes significantly suppressed endothelial expression of VCAM-1 and ICAM-1 after exposure to LL-37 and U1 (Figure 3d, e). Thus, these results indicate that LL-37 mediated sensing of U1 by several dsRNA sensing mechanisms.
Figure 3: TLR3, RIGI and IRF1 regulate U1 and LL-37 induced adhesion molecule expression.
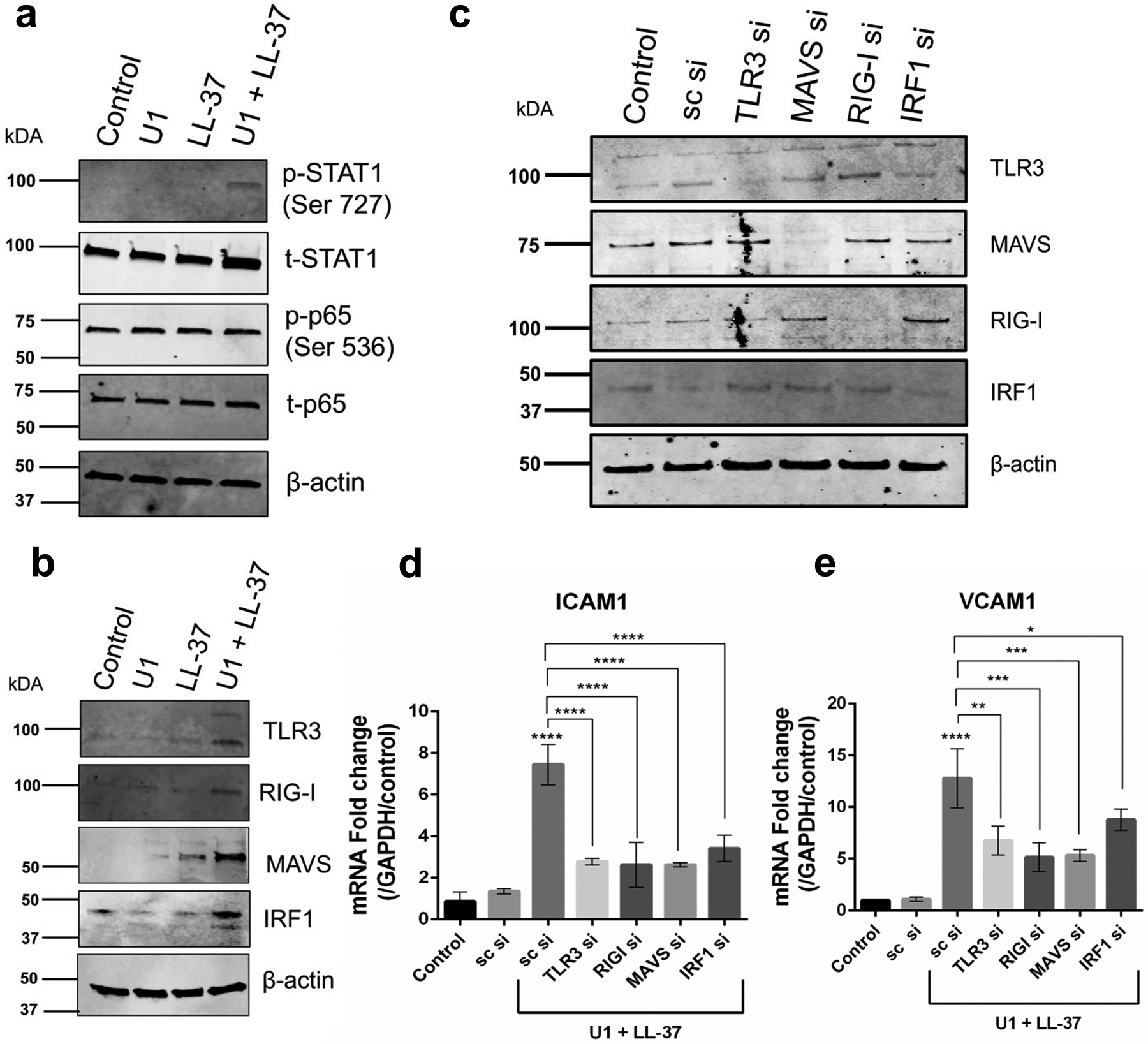
(a, b) HDMECs were treated with U1 (2 μg/ml) and/or LL-37 (4 μM) Protein expression of p-STAT1, STAT1, p-p65, p65 (a) and TLR3, RIGI, MAVS and IRF1 (b) and was assessed with immunoblot. β-actin was used as a loading control. (c) Gene knockdown of TLR3, RIGI, RIGI co-receptor MAVS and IRF1 was confirmed with immunoblot. Scrambled siRNA (sc si) was used as a negative siRNA control. β-actin used as a loading control. (d, e) HDMECs with gene knockdown for TLR3, RIGI, MAVS and IRF2 were co-treated with U1 (2 μg/ml) and LL-37 (4 μM) for 24 h. Gene expression of adhesion molecules ICAM1 and VCAM1 was assessed (n=3) (*, p< 0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001)
LL-37 and dsRNA promote leukocyte adhesion and transmigration
VCAM1 and ICAM1 participate in adhesion and transmigration of leukocytes across endothelial cells (Carlos and Harlan 1994). Enhanced adhesion of THP-1 monocytes to HDMECs was observed in HDMECs exposed to extracts of UVB-damaged keratinocytes (Figure 4a). RNAse III significantly inhibited adherence to HDMECs, thus supporting the contribution of dsRNA to monocyte adhesion (Figure 4a). Similarly, LL-37 enhanced adhesion of leukocytes by facilitating recognition of U1 (Figure 4b,c). THP-1 adhesion to HDMEC also increased in after co-treatment with U1 and LL-37 (Figure 4b,c). HDMECs co-treated with poly I:C and LL-37 showed similar results (Figure S2g). The capacity of THP-1 monocytes, PBMCs and CD14+ monocytes to adhere to endothelial cells was dependent on LL-37 concentration (Figure 4d,e,f). Co-treatment with LL-37 and U1 also enhanced transmigration of leukocytes across the endothelial layer (Figure 4g).
Figure 4: LL-37 and dsRNA enhance adhesion and migration of leukocytes across HDMECs.
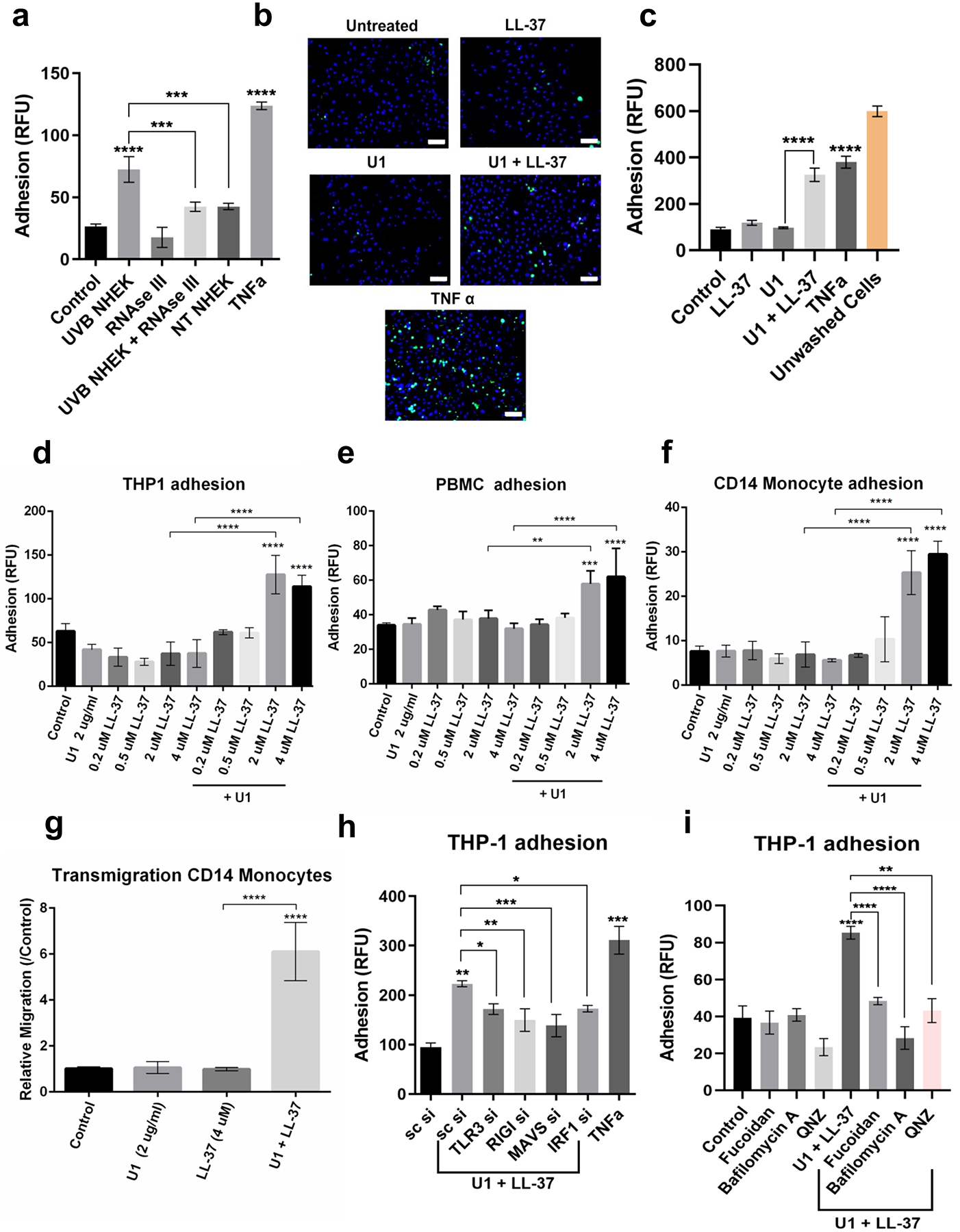
(a) Non-treated and RNAse III treated UVB NHEK extracts added onto HDMECs for 12 hours and adhesion assay was performed with Calcein labeled THP-1 cells. (n=3). (b) Adhesion assay with THP-1 cells (green) was performed for HDMECs treated with U1 and/or LL-37 for 24 h. TNFα is positive control. DAPI (blue). Scale=50 μM. (c) Quantification of (b). (d, e, f) HDMECs treated with U1 (2 μg/ml) and/or LL-37 (0.2 μM-4 μM) for 24 h. Adhesion of labeled THP-1 monocytes (d), PBMCs (e) and CD14+ primary monocytes (f) to HDMECs was assessed. (n=3) (g) Trans-endothelial migration of Calcein labeled CD14+ primary monocytes (n=3) (h) siRNAs against TLR3, RIGI, MAVS and IRF1 in HDMECs, followed by treatment with U1 and LL-37. Adhesion assay with labeled THP-1 monocytes (n=3). (i) THP1 adhesion assay for HDMECs pre-treated with fucoidan (50 μg/ml), Bafilomycin A1 (100 nM) and QNZ (100 nM) for 60 min, followed by U1 and LL-37 treatment for 24 h. (*, p< 0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001)
TLR3, RIGI, MAVS and IRF1 knockdown inhibited U1 and LL-37 mediated adhesion of THP-1 monocytes to endothelial cells (Figure 4h). Inhibition of endosomal acidification with Bafilomycin A1 also suppressed THP-1 adhesion to HDMECs (Figure 4i), a finding consistent with the role of TLR3 in activation (Singh et al. 2014). Inhibition of NFkB with chemical inhibitor QNZ, also inhibited adhesion of THP-1 monocytes to HDMECs co-treated with U1 and LL-37 (Figure 4i). Thus, multiple signaling regulatory mechanisms consistent with dsRNA recognition were involved in controlling leukocyte adhesion to dermal endothelial cells.
Scavenger receptors form a tripartite complex with U1 and LL-37 to facilitate internalization and activation of cells (Takahashi et al. 2018). Consistent with this mechanism, treatment with fucoidan, a pan-scavenger receptor inhibitor, inhibited adhesion of THP-1 monocytes to HDMECs (Figure 4i). The scavenger receptor CD36 is abundantly expressed on endothelial cells and regulates their innate immune responses (Silverstein and Febbraio 2009). Proximity Ligation Assay that detects in situ protein-protein interactions showed colocalization of CD36 and U1 when HDMECs were co-treated with U1 and LL-37, thus confirming CD36 is among the receptors interacting with LL37 and U1 (Figure S3).
U1 and LL-37 enhance adhesion molecule expression in mice
To demonstrate a role for LL-37 in U1-mediated expression of VCAM1 in vivo, 8-week-old BALB/c female mice were injected intradermally with U1 alone, LL-37 alone, or co-injected with both U1 and LL-37. Enhanced ear thickness, epidermal hyperplasia and immune cell infiltration into the dermis was seen after co-treatment with U1 and LL-37 (Figure 5a,b and Figure S4a,b,d). An increase in monocytes/macrophages and neutrophils was observed in mice co-treated with U1 and LL-37 (Figure S4d). Similarly, the viral dsRNA mimic poly I:C caused inflammation (Figure S5c, d, e). Western blot analysis of tissue extracts, and immunohistochemistry, showed injection of U1 and LL-37 could increase VCAM1, ICAM1 and E-Selectin protein expression (Figure 5d). U1 and LL-37 injection co-injection enhanced VCAM1 and ICAM1 expression on CD31 positive endothelial cells (Figure 5c and Figure S4d). Since, U1 treatment alone did not affect adhesion molecule expression in vitro, we hypothesized that mouse cathelicidin related antimicrobial peptide (Cramp) may aid in recognition of U1. To investigate this, we injected U1 into Camp −/− mice, but this did not abrogate the adhesion molecule expression induced by U1 alone (Figure S5 a, b). We also tested a transgenic mouse carrying the human CAMP gene crossed onto the Camp−/− mouse background (CAMP +/+, Camp−/−). Minimal additional expression was observed for VCAM1 and ear inflammation in the CAMP+/+ Camp −/− mice as compared to the Camp −/− mice (Figure S5 b). Overall, our in vivo results confirmed that the combination of U1 and LL-37 enhanced VCAM1 expression, but in contrast to results with isolated cells, U1 or LL-37 alone in vivo could also induce a response.
Figure 5: U1 and LL-37 treatment enhances adhesion molecule VCAM1 expression in vivo.
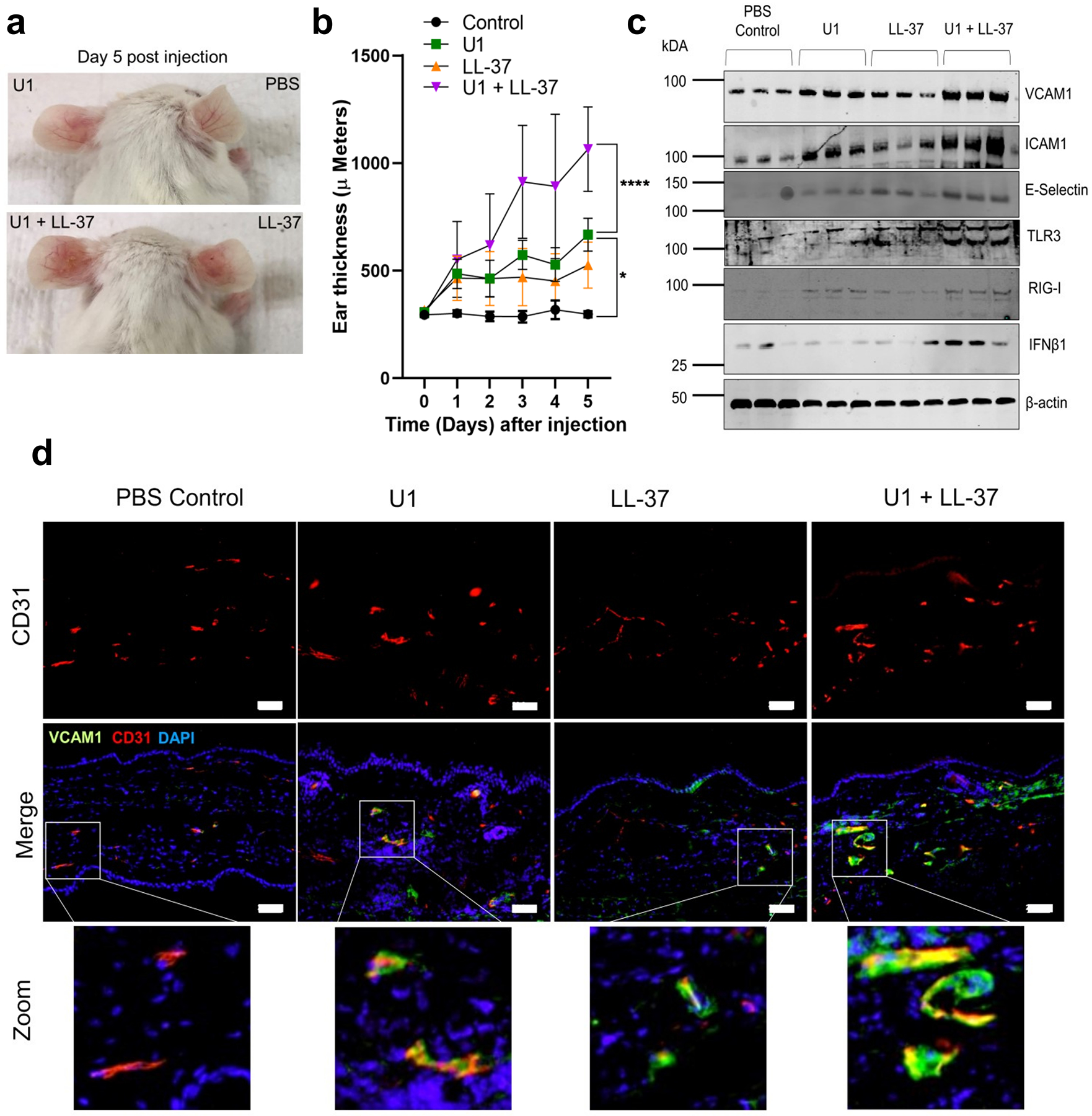
(a, b, c) BALB/c (n=5) were injected intradermally into ears with U1 (25 ug) and/or LL-37 (32 μM). (a) Representative mouse images injected with U1 and LL-37 showing erythema on the ear. (b) Temporal changes in mouse ear thickness (Day 0–5). (c) Western blot of tissue extracts stained for VCAM1, ICAM1, E-selectin, TLR3, RIGI and IFNB1. β-actin used as a loading control. (d) Tissue sections stained with antibodies against VCAM1 (green) and CD31 (Red). DAPI (blue) is nuclear stain. Scale= 20 μM.
DISCUSSION:
Rosacea is a facial skin disorder characterized by endothelial cell proliferation, vascular dilatation and epidermal hyperplasia (Gallo et al. 2018). It is also associated with significant co-morbidities like cardiovascular and neurological disease (Haber and El Gemayel 2018). An increase in the abundance of LL-37, a cationic antimicrobial peptide, is considered an important contributor to the pathophysiology of this disease (Yamasaki et al. 2007). In the current study we sought insight into why exposure to UV radiation promotes vascular inflammation in rosacea (Murphy 2004). We now report a potential explanation of this phenomenon. First, subjects with rosacea have increased expression of the adhesion molecules VCAM1, ICAM1 and E-Selectin. Secondly, we show that the cathelicidin antimicrobial peptide LL-37 enables this expression in response to dsRNA released after UV damage of epidermal keratinocytes. Since UV exposure of keratinocytes can also lead to increased CAMP expression (Kim et al. 2005), and LL-37 alone can promote inflammation, UV exposure likely has several mechanisms of promoting disease.
UVB light does not penetrate deeply into the dermis (D’Orazio et al. 2013) and therefore we hypothesized that dermal endothelial cells might indirectly respond to UV damaged epidermal keratinocytes. Supporting this, we observed that extracts from UV damaged keratinocytes, promoted activation of dermal endothelial cells. This response was decreased when dsRNA was digested from keratinocytes after UV exposure. This suggested that dsRNA from the keratinocyte was responsible for the endothelial cell response. Although several forms of dsRNA could promote this response in endothelial cells, the small non-coding RNA U1 served as a model of endogenous human mRNAs that contain double stranded hairpin loops (Bernard et al. 2012a). The combination of U1 and LL-37 greatly enhanced gene and protein expression of multiple adhesion molecules and chemokines on human cultured endothelial cells and on dermal vessels in mice. Thus, it is reasonable to hypothesize that patients with rosacea who express high amounts of LL-37 are poised to respond to photo-damaged keratinocytes.
Recent work has demonstrated an important role for LL-37 as a molecule that promotes recognition of self-nucleic acids released upon infection and autoinflammatory responses. This phenomenon was first observed with recognition of extracellular self-DNA and self-RNA as recognized by plasmacytoid dendritic cells via the toll like receptors (TLR9 and TLR7/8, respectively), triggering a pro-inflammatory response and type 1 interferon production in psoriasis patients (Chamilos et al. 2012; Ganguly et al. 2009). Type 1 interferon signalling was also activated in endothelial cells co-treated with U1 and LL-37 (Figure 2 and Supplemental Figure S3). It will be of interest to determine if the local effects of LL-37 on vascular function may be relevant to the systemic co-morbidities associated with rosacea (Haber and El Gemayel 2018).
LL-37 aids in recognition of U1 via multiple scavenger receptors that mediate endocytic uptake and downstream activation of endosomal pattern recognition receptor TLR3 and cytoplasmic RNA helicase RIGI in human keratinocytes (Takahashi et al. 2018; Zhang et al. 2016). IRF1 and nuclear transcription factor NF-kB co-operatively regulate the expression of adhesion molecule VCAM1 and an IRF1 binding site has been mapped on the VCAM1 promoter (Lechleitner et al. 1998; Neish et al. 1995b). Co-treatment of dermal endothelial cells with U1 and LL-37 increased protein expression of TLR3, RIGI, MAVS and downstream transcription factor IRF1. siRNA mediated knockdown of these pattern recognition receptors inhibited adhesion molecule expression and leukocyte adhesion to endothelial cells in vitro. Also, inhibition of scavenger receptors and NF-kB with chemical inhibitors also interfered with THP-1 monocyte adhesion to endothelial cells co-treated with U1 and LL-37. This finding suggests that a similar mechanism exists in endothelial cells as in keratinocytes, and that LL-37 facilitates association with scavenger receptors on the endothelial cell surface. This “innate immune vetting” phenomenon for LL-37 may be a critical regulator of the capacity of endothelial cells to respond to tissue damage.
Endothelial cell activation leads to leukocyte adhesion and transmigration, a critical step in the inflammatory response (Pober and Sessa 2007). Adhesion molecule expression has been shown to be regulated by pro-inflammatory chemokines and growth factors including IL-1, TNFα, VEGFA, CX3CL1 (Chen et al. 2001; Kim et al. 2001; Yang et al. 2007). Since co-treatment of human dermal endothelial cells with U1 and LL-37 enhanced gene expression of cytokines, chemokines and growth factors including TNF, CX3CL1 and VEGFA, all of which may contribute to regulation of adhesion molecule expression via distinct activation of downstream pathways, it is possible that autocrine chemokine response contributes to the increase in adhesion molecule expression. Furthermore, we show that expression of VCAM1, ICAM1 and E-Selectin in mice was enhanced by direct administration of either U1 or LL-37. Interestingly, there was activity by U1 alone even in the Camp−/− mice. This finding suggests other molecules may also be acting in vivo in a manner similar to cathelicidin and compensate for its loss. Observations that IL26, beta-defensins and lysozyme can promote DNA recognition support this conclusion (Lande et al. 2011; Meller et al. 2015; Poli et al. 2017). Thus, although the mechanisms responsible for the regulation of inflammation in rosacea are not completely elucidated, these observations of adhesion molecule expression in rosacea represent an important new step in understanding the pathogenesis of this disease. Clinical relevance may be seen when considering that topical application of Brimonidine, an α2-adrenergic receptor agonist, has also been shown to alleviate vascular inflammation by inhibiting leukocyte adhesion to the endothelial vessels in a in vivo mouse model (Bertino et al. 2018). Clinical significance may be demonstrated in the future by blocking endothelial adhesion molecule interactions with their cognate ligands on leukocytes as a strategy to improve the pro-inflammatory microenvironment in patients with multiple photosensitive disorders.
METHODS AND MATERIALS:
Cell culture and chemicals
HDMECs (Promocell, USA), NHEKs (Life Technologies, USA), fresh peripheral blood mononuclear cells (PBMCs) and frozen CD14 + monocytes (iXcell Biotechnologies, USA) were grown according to manufacturer’s instructions. Poly I:C (Invivogen, San Diego, CA), LL-37 (Genemed, USA), RNAse III (Life Technologies, USA), fucoidan (Sigma Aldrich, USA), Bafilomycin A (Sigma Aldrich, USA) and QNZ (Santa cruz Biotechnologies, USA) were reconstituted according to manufactures instructions. U1 dsRNA and its biotinylated form was synthesized as described previously (Takahashi et al. 2018).
Human skin and mice
Informed consent was obtained from five healthy subjects and rosacea patients with approval (UCSD Human Research Institutional Review Board Approval Number:140144). After local anesthesia, skin biopsies were fixed in 4 %. Paraformaldehyde. All mouse procedures were approved by the UCSD Institutional Animal Care and Use Program (Protocol Number: S09074). Eight-week old female BALB/c wild type mice, C57BL/6 mouse cathelicidin knockout (Camp −/−) (Nizet et al. 2001) and human cathelicidin transgenic C57BL/6 mice (CAMP +/+) bred against C57BL/6 Camp −/− background (CAMP +/+ Camp−/−) were used for the animal studies. (Jiang et al. 2018). BALB/c and C57BL/6 mice ears were injected intradermally with U1, poly I:C and/or LL-37. The injections were performed once every alternate day for a total three times. Twenty-four hours after the last injection, mice were euthanized with CO2 asphyxiation and ears excised for whole tissue immunoblot, immunofluorescence staining and immunohistochemistry.
UVB irradiation of NHEKs and RNAse III treatment
NHEKs were exposed to 25 mJ/cm2 narrowband Ultraviolet B (UVB) irradiation (Spectronics handheld UVB lamp UVB irradiated or non -irradiated NHEKs were immediately scrapped, briefly sonicated and extracts were added on to HDMEC cells. For dsRNA digestion, UVB irradiated NHEKs were treated with RNAse III cells as previously described (Zhang et al. 2016).
Enzyme-linked immunosorbent assay (ELISA)
Cell culture supernatant was frozen at −80°C until use for analysis. Multiplex ELISA was performed with human premixed multi-analyte kit (RandD Systems, USA) on a MAGPIX instrument (Luminex, USA). Data was analyzed with the xPONENT 4.2 software (Luminex, USA).
RNA-sequencing
RNA was extracted using a Purelink RNA mini kit (Life technolgies, USA). Isolated RNA was submitted to the UCSD IGM Genomics Center for RNA-sequencing performed on a high-output run V4 platform (Illumina, USA) with a single read 100 cycle runs. Data alignment was done on Partek flow software (Partek, USA) with Tophat2 (version 2.0.8) Gene ontology (GO) enrichment analysis was performed on differentially regulated genes (≥1.5-fold) using DAVID 6.8 (Huang et al. 2007). The protein-protein interaction network was generated in the InnateDB database (Breuer et al. 2013).
Quantitative real-time PCR
RNA extracted from cells (Pure Link RNA isolation kit, Life Technologies, USA) was quantified on the nanodrop 2000/200c spectrophotometer (ThermoFischer, USA). 500 ng of purified RNA was used to synthesize cDNA with iScript cDNA Synthesis Kit (Bio-Rad, USA). Primes used are listed in supplementary excel file 1. Pre-developed Taqman (Thermofisher, USA) and SYBR (Integrated DNA technologies, USA) gene expression Assays were used to evaluate mRNA transcript levels. STAT1 primers were used as described previously (Zhang et al. 2016).Taqman and SYBR cycling conditions were used as previously described (Kulkarni et al. 2017; Zhang et al. 2016). All experiments were performed on a CFX96 system (Biorad, USA). Fold change relative to GAPDH was calculated using the Livak ΔΔCt method (Livak and Schmittgen 2001).
Immunoblot
Cells were lysed in in complete RIPA buffer with added 1X protease and phosphatase inhibitor cocktail (Life Technologies, USA). The lysate was centrifuged at 4°C, 13000 rpm for 20 min and total cytoplasmic supernatant fraction was stored at −80 °C until future use. For mouse ear whole tissue immunoblot tissue was homogenized in 1X complete RIPA buffer. Total protein was quantified for each treatment with biorad protein assay (Biorad, USA). Ten μg of total protein was loaded onto a 4–20% Mini-PROTEAN TGX gel (Bio-Rad), transferred to a polyvinylidene difluoride membrane, and probed with primary antibodies. All primary antibodies are listed in supplementary excel file 1. β-actin was used as loading control. IRDye conjugated anti-rabbit and anti-mouse secondary antibodies (IRDye800CW; Licor,USA) were used. The images were acquired on an Odyssey CLx Imaging System (Licor, USA).
Immunohistochemistry
Immunohistochemistry was performed as previously described (Kulkarni et al. 2017; Takahashi et al. 2018). Refer to supplementary material for detailed method.
Leukocyte adhesion and transmigration assay
THP-1 monocytes, PBMCs and CD14+ monocytes (150,000 cells/ml) were fluorescently labeled with 5 μM Calcein AM dye according to manufacturer’s instructions (Life technologies, USA) HDMECs were cultured in 48 well culture plates (50,000 cells/well) to 80% confluence. Labeled leukocytes allowed to adhere onto the HDMECs for 60 min at 37 °C after treatment. Following incubation, cells were gently washed with 1X PBS and permeabilized with 1% SDS. Fluorescence was quantified on Spectramax fluorimeter (Molecular Devices, USA) and is represented as Relative Fluorescence Units (RFU). For the transmigration assay, HDMECs were cultured on 3.0 μM trans-well permeable support (Fluoroblock, Corning, USA). Calcein AM labeled CD14 + monocytes were allowed to migrate from the upper chamber of the trans-well to the lower chamber across the HDMEC cell layer for 8 h. The monocytes that had migrated to the lower chamber were read on a Spectramax fluorimeter (Molecular Devices, USA) as RFU.
Proximity ligation assay (PLA)
PLA was performed as described previously (Takahashi et al. 2018). Refer to supplementary material for detailed method.
siRNA transfection
siRNAs against TLR3 and MAVS were purchased from Thermofisher (Smartpool OnTarget Plus). RIGI and IRF1 siRNAs were purchased from Ambion (Silencer Select). Scrambled siRNA control (sc siRNA) was purchased from Life Technologies (On Targetplus). siRNA transfection was performed on HDMECs as previously described for NHEKs (Takahashi et al. 2018).
Statistical analysis
Normally distributed results are represented as means and standard deviation of the means. For comparison of multiple groups either one-way ANOVA (Tukey post hoc) or two-way ANOVA (Bonferroni post hoc) was used, as indicated in figure legends. All the statistical analyses were performed on Prism 8 (Graph Pad, San Diego, USA).
Supplementary Material
Supplemental Figure S1: dsRNA released from UVB treated NHEKs enhance pro-inflammatory gene expression in HDMECs. (a) Apoptosis and necrosis were accessed in NHEKs was accessed post UVB expoure (25 mJ/cm2) by flow cytometry (n=3). (b) Percent necrotic cells from (a). (c) UVB (25 mJ/cm2) and non-irradiated NHEKs extracts were added to cultures of HDMECs for 12 hours. The mRNA expression of IFNB1, and chemokines TNF, CCL5, CXCL10 and CX3CL1 as assessed with q-RT PCR (n=3). (d, e, f, g) UVB NHEKs were treated with RNAse III, then extracts co-cultured with HDMECs and mRNA expression of TNF, CXCL10, CCL5 and IFNB1 was assessed (n=3). (*, p< 0.05; **, p< 0.01; ****, p<0.0001)
Supplemental Figure S2: LL-37 enhances innate immune pathway activation in dermal endothelial cells treated with dsRNA and promotes monocyte adhesion. (a, b, c) Time response of U1 (2 μg/ml) and LL-37 (4 μM) treated HDMECs (1 h- 36h) Gene expression for ICAM1, VCAM1 and SELE was assessed (n=3). (d) HDMECs were co-treated with U1 (2 μg/ml) and increasing concentrations of LL-37 (0.2 μM-4 μM) at increasing time points (1 h- 36h). Gene expression of IFNB1, STAT1, CCL5, CXCL10, CX3CL1, ICAM1, VCAM1, TNF and HLAB was assessed with q-RT PCR. Data are represented as a heat map. (n=3) (e) HDMECs were treated with U1 (2 μg/ml) and LL-37 (4 μM) at increasing time (5 min-360 min). Protein expression of p-STAT1, STAT1, p-p65, p65, p- IRF7 and IRF7 was assessed with immunoblot. β-actin was used as a loading control. (f) HDMECs were treated with LL-37 (2.5 μM) and poly I:C (0.1 μ μg/ml) or LL-37 (2.5 μM) and U1 (2 μg/ml) for 24 h. Gene ontology analysis from the overlapping gene set showed pathways related to type 1 interferon signaling, angiogenesis, cytokine and NF-kb signaling were enriched. The GO gene set is represented with REVIGO. Color coding represents enriched gene sets with regards to p value. (g) HDMEC cells were treated with poly I:C (0.1 μg/ml) and/or LL-37 (4 μM) for 24 h. Calcein AM labeled THP-1 monocytes (green) were allowed to adhere onto treated HDMECs for 60 min. THP-1 cells adhering to HDMECs were quantified and represented as RFU. (n=3, One-way ANOVA). (**, p<0.01; ****, p<0.0001)
Supplemental Figure S3: LL-37 enables U1 association with endothelial scavenger receptor CD36 (a) Proximity ligation assay (PLA) for U1 dsRNA and scavenger receptor CD36. HDMECs were treated with LL-37 (4 μM) for 15 min, followed by treatment with biotinylated U1 (2 μg/ml) for 60 min at 4 °C. Physical proximity of U1 and CD36 was determined with fluorescence-based PLA that produces a red fluorescent signal. DAPI (blue) was used as a nuclear counterstain. Scale bar= 50μm. (b) Signal counts of (a) from five visual fields were used for PLA signal quantification (n=3, One-way ANOVA). (***, p<0.001)
Supplemental Figure S4: U1 and LL-37 enhance inflammatory response in vivo (a) Tissue sections (5 μM) from mice ears injected with U1 (25 μg) and/or LL-37 (32 μM) were stained with hematoxylin and eosin (H and E), Scale= 20 μM (b) Epidermal hyperplasia (acanthosis) was measured at day 5 post injection with U1 (25 μg) and/or LL-37 (32 μM). (c, d) Tissue sections stained with antibodies against VCAM1 (green) and CD31 (Red) (c) and against immunocytes monocytes/macrophages (moma-2), neutrophils (neutrophil marker) and pan-T cells (CD3) (green) (d). DAPI (blue) is nuclear stain. Scale= 20 μM.
Supplemental Figure S5: In vivo dsRNA and LL-37 treatment enhances VCAM1 expression.
(a, b) C57BL/6 Camp−/− and C57BL/6 CAMP +/+ Camp−/− transgenic mice were injected with U1 dsRNA (25 ug) (n=3). (a) H and E staining, Scale= 50 μM (b) Western blot against VCAM1 and LL37. β-actin is loading control. hCAP-18, the pro-form of LL-37 was mainly observed. (c, d, e). BALB/c (n=5) were injected intradermally in the ears with poly I:C (10 μg) and/or LL-37 (320 μM). Ear tissue was processed for immunoblot and immunohistochemistry analyses. (c) Ear tissue sections (5 μM) were stained with hematoxylin and eosin (H and E), Scale= 50 μM (d) Extracts of mouse ear tissue assessed by Western blot for blood vessel marker CD31, adhesion molecule VCAM1, pattern recognition receptors TLR3 and RIGI and IFNB1. β-actin was used as a loading control. (e) Ears stained with antibodies against VCAM1 (green) and CD31 (Red). Nuclei were stained with DAPI (blue) Scale= 50 μM.
ACKNOWLEDGEMENTS
This study was supported by NIH R01AR052728, R01AR064781 R01AI116576 and R01AI052453 (RLG). We would like to thank Dr. Michael R Williams for help with synthesis of U1 dsRNA. Dr. Tatsuya Dokoshi, Dr. Alan O’ Niell, Dr. Christopher A Adase, Dr. Ling Juan Zhang for technical help and Gallo lab members for helpful discussions on the project. Carlos Aguilera maintained and genotyped the mouse strains.
Footnotes
CONFLICT OF INTERESTS
Richard L Gallo is a consultant and has equity interest in MatriSys and Sente, Inc.
Data availability
Datasets related to this article can be found at “https://www.ncbi.nlm.nih.gov/geo/” with accession numbers GSE129152 and GSE129160
REFERENCES:
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. Nature Publishing Group; 2001;413(6857):732–8 Available from: http://www.nature.com/articles/35099560 [DOI] [PubMed] [Google Scholar]
- Bernard JJ, Cowing-Zitron C, Nakatsuji T, Muehleisen B, Muto J, Borkowski AW, et al. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat. Med NIH Public Access; 2012a;18(8):1286–90 Available from: http://www.ncbi.nlm.nih.gov/pubmed/22772463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard JJ, Cowing-Zitron C, Nakatsuji T, Muehleisen B, Muto J, Borkowski AW, et al. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat. Med 2012b;18(8):1286–90 Available from: http://www.ncbi.nlm.nih.gov/pubmed/22772463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertino B, Blanchet-Réthoré S, Thibaut de Ménonville S, Reynier P, Méhul B, Bogouch A, et al. Brimonidine displays anti-inflammatory properties in the skin through the modulation of the vascular barrier function. Exp. Dermatol John Wiley & Sons, Ltd (10.1111); 2018;27(12):1378–87 Available from: http://doi.wiley.com/10.1111/exd.13793 [DOI] [PubMed] [Google Scholar]
- Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res Oxford University Press; 2013;41(Database issue):D1228–33 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23180781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddenkotte J, Steinhoff M. Recent advances in understanding and managing rosacea. F1000Research. 2018;7:1885 Available from: http://www.ncbi.nlm.nih.gov/pubmed/30631431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos T, Harlan J. Leukocyte-endothelial adhesion molecules. Blood. 1994;84(7) Available from: http://www.bloodjournal.org/content/84/7/2068.long?sso-checked=true [PubMed] [Google Scholar]
- Chamilos G, Gregorio J, Meller S, Lande R, Kontoyiannis DP, Modlin RL, et al. Cytosolic sensing of extracellular self-DNA transported into monocytes by the antimicrobial peptide LL37. Blood. American Society of Hematology; 2012;120(18):3699–707 Available from: http://www.ncbi.nlm.nih.gov/pubmed/22927244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Liu C, Sun S, Mei Y, Tong E. Cytokine-induced cell surface expression of adhesion molecules in vascular endothelial cells in vitro. J. Tongji Med. Univ 2001;21(1):68–71 Available from: http://www.ncbi.nlm.nih.gov/pubmed/11523254 [DOI] [PubMed] [Google Scholar]
- D’Orazio J, Jarrett S, Amaro-Ortiz A, Scott T. UV radiation and the skin. Int. J. Mol. Sci Multidisciplinary Digital Publishing Institute (MDPI); 2013;14(6):12222–48 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23749111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edfeldt K, Agerberth B, Rottenberg ME, Gudmundsson GH, Wang X-B, Mandal K, et al. Involvement of the Antimicrobial Peptide LL-37 in Human Atherosclerosis. Arterioscler. Thromb. Vasc. Biol 2006;26(7):1551–7 Available from: http://www.ncbi.nlm.nih.gov/pubmed/16645154 [DOI] [PubMed] [Google Scholar]
- Gallo RL, Granstein RD, Kang S, Mannis M, Steinhoff M, Tan J, et al. Standard classification and pathophysiology of rosacea: The 2017 update by the National Rosacea Society Expert Committee. J. Am. Acad. Dermatol Mosby; 2018;78(1):148–55 Available from: https://www.sciencedirect.com/science/article/pii/S0190962217322971?via%3Dihub [DOI] [PubMed] [Google Scholar]
- Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, et al. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J. Exp. Med Rockefeller University Press; 2009;206(9):1983–94 Available from: http://www.ncbi.nlm.nih.gov/pubmed/19703986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether L, Overgaard LK, Egeberg A, Thyssen JP. Incidence and prevalence of rosacea: a systematic review and meta-analysis. Br. J. Dermatol John Wiley & Sons, Ltd (10.1111); 2018;179(2):282–9 Available from: http://doi.wiley.com/10.1111/bjd.16481 [DOI] [PubMed] [Google Scholar]
- Haber R, El Gemayel M. Comorbidities in rosacea: A systematic review and update. J. Am. Acad. Dermatol Mosby; 2018;78(4):786–792.e8 Available from: https://www.sciencedirect.com/science/article/pii/S0190962217323423?via%3Dihub [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. BioMed Central; 2007;8(9):R183 Available from: http://www.ncbi.nlm.nih.gov/pubmed/17784955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat. Rev. Immunol Nature Publishing Group; 2014;14(1):36–49 Available from: http://www.nature.com/articles/nri3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhang Y, Indra AK, Ganguli-Indra G, Le MN, Wang H, et al. 1α,25-dihydroxyvitamin D 3 -eluting nanofibrous dressings induce endogenous antimicrobial peptide expression. Nanomedicine. Future Medicine Ltd; London, UK; 2018;13(12):1417–32 Available from: https://www.futuremedicine.com/doi/10.2217/nnm-2018-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Kim BJ, Jeong MS, Seo SJ, Kim MN, Hong CK, et al. Expression and Modulation of LL-37 in Normal Human Keratinocytes, HaCaT cells, and Inflammatory Skin Diseases. J. Korean Med. Sci 2005;20(4):649 Available from: http://www.ncbi.nlm.nih.gov/pubmed/16100459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Moon SO, Kim SH, Kim HJ, Koh YS, Koh GY. Vascular endothelial growth factor expression of intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), and E-selectin through nuclear factor-kappa B activation in endothelial cells. J. Biol. Chem American Society for Biochemistry and Molecular Biology; 2001;276(10):7614–20 Available from: http://www.ncbi.nlm.nih.gov/pubmed/11108718 [DOI] [PubMed] [Google Scholar]
- Koczulla R, von Degenfeld G, Kupatt C, Krötz F, Zahler S, Gloe T, et al. An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. J. Clin. Invest 2003;111(11):1665–72 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=156109&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni NN, Adase CA, Zhang L-J, Borkowski AW, Li F, Sanford JA, et al. IL-1 Receptor-Knockout Mice Develop Epidermal Cysts and Show an Altered Innate Immune Response after Exposure to UVB Radiation. J. Invest. Dermatol NIH Public Access; 2017;137(11):2417–26 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28754339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci. Transl. Med American Association for the Advancement of Science; 2011;3(73):73ra19 Available from: http://www.ncbi.nlm.nih.gov/pubmed/21389263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechleitner S, Gille J, Johnson DR, Petzelbauer P. Interferon Enhances Tumor Necrosis Factor–induced Vascular Cell Adhesion Molecule 1 (CD106) Expression in Human Endothelial Cells by an Interferon-related Factor 1–dependent Pathway. J. Exp. Med Rockefeller University Press; 1998;187(12):2023–30 Available from: http://www.ncbi.nlm.nih.gov/pubmed/9625761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–8 Available from: http://www.ncbi.nlm.nih.gov/pubmed/11846609 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Seya T. TLR3: Interferon induction by double-stranded RNA including poly(I:C). Adv. Drug Deliv. Rev Elsevier; 2008;60(7):805–12 Available from: https://www.sciencedirect.com/science/article/pii/S0169409X07003833?via%3Dihub [DOI] [PubMed] [Google Scholar]
- Meller S, Di Domizio J, Voo KS, Friedrich HC, Chamilos G, Ganguly D, et al. TH17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat. Immunol Nature Publishing Group; 2015;16(9):970–9 Available from: http://www.nature.com/articles/ni.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels C Endothelial cell functions. J. Cell. Physiol 2003;196(3):430–43 Available from: http://www.ncbi.nlm.nih.gov/pubmed/12891700 [DOI] [PubMed] [Google Scholar]
- Murphy G Ultraviolet light and rosacea. Cutis. 2004;74(3 Suppl):13–6, 32–4 Available from: http://www.ncbi.nlm.nih.gov/pubmed/15499753 [PubMed] [Google Scholar]
- Neish AS, Read MA, Thanos D, Pine R, Maniatis T, Collins T. Endothelial interferon regulatory factor 1 cooperates with NF-kappa B as a transcriptional activator of vascular cell adhesion molecule 1. Mol. Cell. Biol 1995a;15(5):2558–69 Available from: http://www.ncbi.nlm.nih.gov/pubmed/7537851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish AS, Read MA, Thanos D, Pine R, Maniatis T, Collins T. Endothelial interferon regulatory factor 1 cooperates with NF-kappa B as a transcriptional activator of vascular cell adhesion molecule 1. Mol. Cell. Biol American Society for Microbiology (ASM); 1995b;15(5):2558–69 Available from: http://www.ncbi.nlm.nih.gov/pubmed/7537851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, et al. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. Nature Publishing Group; 2001;414(6862):454–7 Available from: http://www.nature.com/articles/35106587 [DOI] [PubMed] [Google Scholar]
- Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol Nature Publishing Group; 2007;7(10):803–15 Available from: http://www.nature.com/articles/nri2171 [DOI] [PubMed] [Google Scholar]
- Poli C, Augusto JF, Dauvé J, Adam C, Preisser L, Larochette V, et al. IL-26 Confers Proinflammatory Properties to Extracellular DNA. J. Immunol American Association of Immunologists; 2017;198(9):3650–61 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28356384 [DOI] [PubMed] [Google Scholar]
- Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal American Association for the Advancement of Science; 2009;2(72):re3 Available from: http://www.ncbi.nlm.nih.gov/pubmed/19471024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Vaughan R, Kao CC. LL-37 peptide enhancement of signal transduction by Toll-like receptor 3 is regulated by pH: identification of a peptide antagonist of LL-37. J. Biol. Chem American Society for Biochemistry and Molecular Biology; 2014;289(40):27614–24 Available from: http://www.ncbi.nlm.nih.gov/pubmed/25092290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Kulkarni NN, Lee EY, Zhang L, Wong GCL, Gallo RL. Cathelicidin promotes inflammation by enabling binding of self-RNA to cell surface scavenger receptors. Sci. Rep Nature Publishing Group; 2018;8(1):4032 Available from: http://www.nature.com/articles/s41598-018-22409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S, Akira S. Toll-like receptors and Type I interferons. J. Biol. Chem American Society for Biochemistry and Molecular Biology; 2007;282(21):15319–23 Available from: http://www.ncbi.nlm.nih.gov/pubmed/17395581 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Di Nardo A, Bardan A, Murakami M, Ohtake T, Coda A, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat. Med 2007;13(8):975–80 Available from: http://www.ncbi.nlm.nih.gov/pubmed/17676051 [DOI] [PubMed] [Google Scholar]
- Yang XP, Mattagajasingh S, Su S, Chen G, Cai Z, Fox-Talbot K, et al. Fractalkine Upregulates Intercellular Adhesion Molecule-1 in Endothelial Cells Through CX3CR1 and the Jak–Stat5 Pathway. Circ. Res 2007;101(10):1001–8 Available from: http://www.ncbi.nlm.nih.gov/pubmed/17885215 [DOI] [PubMed] [Google Scholar]
- Zeichner JA, Eichenfield LF, Feldman SR, Kasteler JS, Ferrusi IL. Quality of Life in Individuals with Erythematotelangiectatic and Papulopustular Rosacea: Findings From a Web-based Survey. J. Clin. Aesthet. Dermatol 2018;11(2):47–52 Available from: http://www.ncbi.nlm.nih.gov/pubmed/29552276 [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Sen GL, Ward NL, Johnston A, Chun K, Chen Y, et al. Antimicrobial Peptide LL37 and MAVS Signaling Drive Interferon-β Production by Epidermal Keratinocytes during Skin Injury. Immunity. Elsevier; 2016;45(1):119–30 Available from: https://linkinghub.elsevier.com/retrieve/pii/S1074761316302436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: dsRNA released from UVB treated NHEKs enhance pro-inflammatory gene expression in HDMECs. (a) Apoptosis and necrosis were accessed in NHEKs was accessed post UVB expoure (25 mJ/cm2) by flow cytometry (n=3). (b) Percent necrotic cells from (a). (c) UVB (25 mJ/cm2) and non-irradiated NHEKs extracts were added to cultures of HDMECs for 12 hours. The mRNA expression of IFNB1, and chemokines TNF, CCL5, CXCL10 and CX3CL1 as assessed with q-RT PCR (n=3). (d, e, f, g) UVB NHEKs were treated with RNAse III, then extracts co-cultured with HDMECs and mRNA expression of TNF, CXCL10, CCL5 and IFNB1 was assessed (n=3). (*, p< 0.05; **, p< 0.01; ****, p<0.0001)
Supplemental Figure S2: LL-37 enhances innate immune pathway activation in dermal endothelial cells treated with dsRNA and promotes monocyte adhesion. (a, b, c) Time response of U1 (2 μg/ml) and LL-37 (4 μM) treated HDMECs (1 h- 36h) Gene expression for ICAM1, VCAM1 and SELE was assessed (n=3). (d) HDMECs were co-treated with U1 (2 μg/ml) and increasing concentrations of LL-37 (0.2 μM-4 μM) at increasing time points (1 h- 36h). Gene expression of IFNB1, STAT1, CCL5, CXCL10, CX3CL1, ICAM1, VCAM1, TNF and HLAB was assessed with q-RT PCR. Data are represented as a heat map. (n=3) (e) HDMECs were treated with U1 (2 μg/ml) and LL-37 (4 μM) at increasing time (5 min-360 min). Protein expression of p-STAT1, STAT1, p-p65, p65, p- IRF7 and IRF7 was assessed with immunoblot. β-actin was used as a loading control. (f) HDMECs were treated with LL-37 (2.5 μM) and poly I:C (0.1 μ μg/ml) or LL-37 (2.5 μM) and U1 (2 μg/ml) for 24 h. Gene ontology analysis from the overlapping gene set showed pathways related to type 1 interferon signaling, angiogenesis, cytokine and NF-kb signaling were enriched. The GO gene set is represented with REVIGO. Color coding represents enriched gene sets with regards to p value. (g) HDMEC cells were treated with poly I:C (0.1 μg/ml) and/or LL-37 (4 μM) for 24 h. Calcein AM labeled THP-1 monocytes (green) were allowed to adhere onto treated HDMECs for 60 min. THP-1 cells adhering to HDMECs were quantified and represented as RFU. (n=3, One-way ANOVA). (**, p<0.01; ****, p<0.0001)
Supplemental Figure S3: LL-37 enables U1 association with endothelial scavenger receptor CD36 (a) Proximity ligation assay (PLA) for U1 dsRNA and scavenger receptor CD36. HDMECs were treated with LL-37 (4 μM) for 15 min, followed by treatment with biotinylated U1 (2 μg/ml) for 60 min at 4 °C. Physical proximity of U1 and CD36 was determined with fluorescence-based PLA that produces a red fluorescent signal. DAPI (blue) was used as a nuclear counterstain. Scale bar= 50μm. (b) Signal counts of (a) from five visual fields were used for PLA signal quantification (n=3, One-way ANOVA). (***, p<0.001)
Supplemental Figure S4: U1 and LL-37 enhance inflammatory response in vivo (a) Tissue sections (5 μM) from mice ears injected with U1 (25 μg) and/or LL-37 (32 μM) were stained with hematoxylin and eosin (H and E), Scale= 20 μM (b) Epidermal hyperplasia (acanthosis) was measured at day 5 post injection with U1 (25 μg) and/or LL-37 (32 μM). (c, d) Tissue sections stained with antibodies against VCAM1 (green) and CD31 (Red) (c) and against immunocytes monocytes/macrophages (moma-2), neutrophils (neutrophil marker) and pan-T cells (CD3) (green) (d). DAPI (blue) is nuclear stain. Scale= 20 μM.
Supplemental Figure S5: In vivo dsRNA and LL-37 treatment enhances VCAM1 expression.
(a, b) C57BL/6 Camp−/− and C57BL/6 CAMP +/+ Camp−/− transgenic mice were injected with U1 dsRNA (25 ug) (n=3). (a) H and E staining, Scale= 50 μM (b) Western blot against VCAM1 and LL37. β-actin is loading control. hCAP-18, the pro-form of LL-37 was mainly observed. (c, d, e). BALB/c (n=5) were injected intradermally in the ears with poly I:C (10 μg) and/or LL-37 (320 μM). Ear tissue was processed for immunoblot and immunohistochemistry analyses. (c) Ear tissue sections (5 μM) were stained with hematoxylin and eosin (H and E), Scale= 50 μM (d) Extracts of mouse ear tissue assessed by Western blot for blood vessel marker CD31, adhesion molecule VCAM1, pattern recognition receptors TLR3 and RIGI and IFNB1. β-actin was used as a loading control. (e) Ears stained with antibodies against VCAM1 (green) and CD31 (Red). Nuclei were stained with DAPI (blue) Scale= 50 μM.
Data Availability Statement
Datasets related to this article can be found at “https://www.ncbi.nlm.nih.gov/geo/” with accession numbers GSE129152 and GSE129160


