ABSTRACT
In complex nervous systems, neurons must identify their correct partners to form synaptic connections. The prevailing model to ensure correct recognition posits that cell-surface proteins (CSPs) in individual neurons act as identification tags. Thus, knowing what cells express which CSPs would provide insights into neural development, synaptic connectivity, and nervous system evolution. Here, we investigated expression of Dpr and DIP genes, two CSP subfamilies belonging to the immunoglobulin superfamily, in Drosophila larval motor neurons (MNs), muscles, glia and sensory neurons (SNs) using a collection of GAL4 driver lines. We found that Dpr genes are more broadly expressed than DIP genes in MNs and SNs, and each examined neuron expresses a unique combination of Dpr and DIP genes. Interestingly, many Dpr and DIP genes are not robustly expressed, but are found instead in gradient and temporal expression patterns. In addition, the unique expression patterns of Dpr and DIP genes revealed three uncharacterized MNs. This study sets the stage for exploring the functions of Dpr and DIP genes in Drosophila MNs and SNs and provides genetic access to subsets of neurons.
KEY WORDS: Dpr, DIP, Motor neuron, Sensory neuron, Synaptic recognition
Summary: Using a range of reporter lines, expression maps of synaptic recognition molecules have been generated, which may provide insights into their function in Drosophila nervous system development.
INTRODUCTION
During nervous system development, neurons contact thousands of cells but only form synapses with a small subset. Precise neural wiring is achieved through a series of steps, including axon pathfinding, partner recognition and synaptic pruning (Sanes and Zipursky, 2020; Zarin and Labrador, 2019). Although the mechanisms underlying these processes are not completely understood, one prevailing model proposes that cell-surface proteins (CSPs) instruct chemoattraction and -repulsion, self-avoidance, and synaptic partner recognition (Honig and Shapiro, 2020; Wit and Ghosh, 2016). CSPs fall into several protein families, including the immunoglobin superfamily (IgSF), the cadherin protein family (Cdhs), the leucine-rich repeat protein family (LRRs), the receptor tyrosine kinases (RTKs) and many more (Bali et al., 2022; Jontes, 2017; Kurusu et al., 2008; Sanes and Zipursky, 2020; Zinn and Özkan, 2017). In vitro biochemical studies have shown that subsets of these CSPs interact in a homo- or heterophilic manner, and many of these interactions are implicated in synaptic connectivity in both vertebrates and invertebrates (Cheng et al., 2019; Honig and Shapiro, 2020; Özkan et al., 2013; Wit and Ghosh, 2016).
The genetically tractable Drosophila melanogaster is an excellent model in which to study CSP expression and function owing to its stereotyped neurogenesis and circuit wiring as well as the availability of an extensive range of genetic tools. Neurons in the fly brain assemble into highly complex circuits similar to those observed in vertebrates but numerically less daunting. In the fly mushroom body, neurons rely on different isoforms of Dscam1 to discriminate self from non-self (Hattori et al., 2009; Wang et al., 2004; Zhan et al., 2004). In the olfactory system, the epidermal growth factor (EGF) repeat-containing transmembrane Teneurin proteins Ten-m and Ten-a are required for the one-to-one matching between olfactory receptor neurons and projection neurons (Hong et al., 2012).
Specific challenges are also encountered in the Drosophila embryonic/larval neuromuscular system; 33 motor neurons (MNs) within each neuromere in the ventral nerve cord (VNC) send their projections to the periphery where they follow defined paths and ultimately choose specific muscle(s) to innervate among 30 potential targets (Grueber et al., 2007; Hoang and Chiba, 2001; Menon et al., 2013). Each efferent motor neuron can be recognized by its stereotypical innervation pattern and morphology. Utilizing the neuromuscular system, many CSPs have been identified as recognition cues between MNs and muscles, including Toll proteins (Inaki et al., 2010; Rose et al., 1997), Connectin (Nose et al., 1992, 1997) and Capricious (Kurusu et al., 2008; Shishido et al., 1998) from the LRR family, and Fasciclin 2 (Davis et al., 1997; Winberg et al., 1998) and Fasciclin 3 (Chiba et al., 1995; Kose et al., 1997) from the IgSF. In contrast, the afferent neurons of the sensory nervous system are localized in the periphery and send their projections to the VNC. Forty-two sensory neurons (SNs) are stereotypically distributed throughout each hemisegment of the larval body wall and establish synaptic connections with interneurons (Orgogozo and Grueber, 2005). Studies from dendritic arborization (da) neurons identified several CSPs that are required for self-avoidance, such as Dscam1 and semaphorin proteins (Meltzer et al., 2016; Miura et al., 2013; Soba et al., 2007). Thus, the unambiguous identification of cells in the motor and sensory circuits provides an ideal system in which to examine the genes and mechanisms that underlie synaptic specificity and development.
In a previous ‘interactome’ screen, we and others identified two subfamilies of the Drosophila IgSF, the Defective proboscis response proteins (Dprs; 21 members) and the Dpr-interacting proteins (DIPs; 11 members) (Carrillo et al., 2015; Özkan et al., 2013). Dprs and DIPs provide a vast repertoire of unique combinations for synaptic specificity. Interactions between Dprs and DIPs have been implicated in synaptic connectivity, cell survival and synaptic growth (Ashley et al., 2019; Bornstein et al., 2021; Carrillo et al., 2015; Courgeon and Desplan, 2019; Menon et al., 2019; Sanes and Zipursky, 2020; Venkatasubramanian et al., 2019; Xu et al., 2018, 2019). However, most studies focused on Dprs and DIPs have implicated only a small subset, likely due to low-penetrance targeting defects and molecular redundancy. For example, in the larval neuromuscular circuit, loss of DIP-α leads to complete loss of muscle 4 innervation by a specific motor neuron; however, most neuromuscular junctions (NMJs) on other muscles formed by the same neuron are unaffected, suggesting that different synaptic recognitions utilize different pairs of CSPs even within the same neuron (Ashley et al., 2019). Thus, obtaining a complete expression map of families of CSPs in individual neurons within specific circuits would facilitate subsequent functional studies.
In this study, we interrogate the expression patterns of Dpr and DIP genes. We generated a collection of GAL4 lines and utilized different UAS reporters to examine expression of Dpr and DIP genes in Drosophila larval neuromuscular and sensory circuits. We showed expression maps of Dpr and DIP genes in MNs, SNs and muscles, and found that each MN and SN expresses a unique subset of Dpr and DIP genes. Utilizing hierarchical clustering, we found that the same class of SNs expresses similar Dpr and DIP genes, suggesting roles in identifying overlapping synaptic partners. Finally, the highly distinct expression patterns of Dpr and DIP genes in MNs revealed previously unidentified MNs. The expression analyses generated by this study will benefit future functional studies of Dprs and DIPs in the motor and sensory circuits. The genetic tools and pipeline provided here will facilitate expression studies of Dprs and DIPs, and other CSPs, in other Drosophila neural circuits to promote the discovery of identification tags utilized for circuit assembly.
RESULTS
Generating a GAL4 collection of Dpr and DIP genes
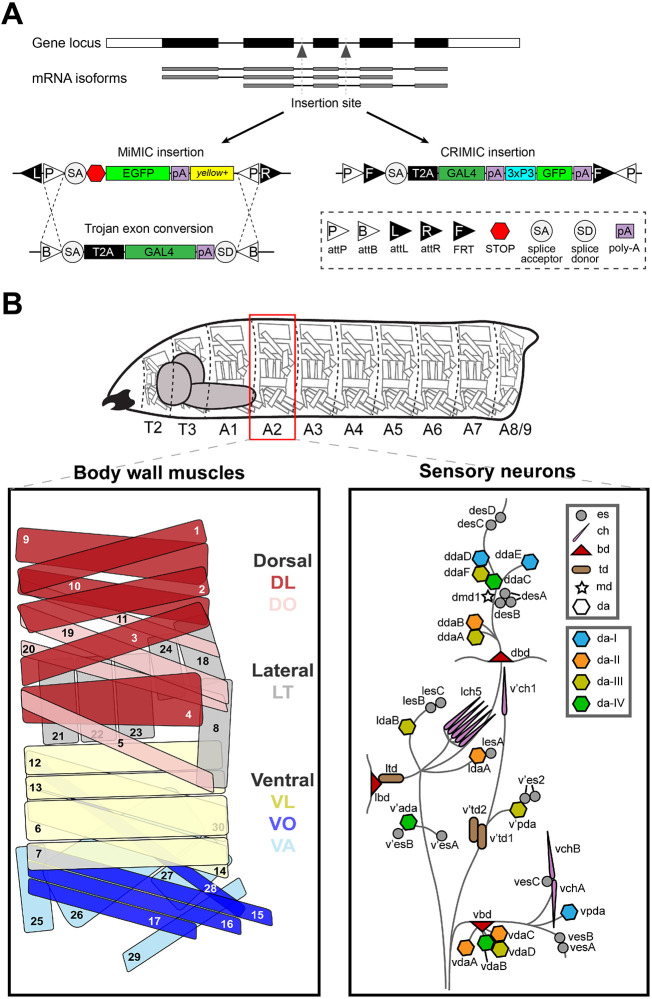
Using Drosophila Minos-mediated integration cassette (MiMIC) insertions followed by Trojan conversion, and CRISPR-mediated integration cassette (CRIMIC) insertions, we and others generated a collection of GAL4 lines of all DIP genes and dpr1-dpr19 (Diao et al., 2015; Kanca et al., 2019; Lee et al., 2018; Nagarkar-Jaiswal et al., 2015a; Venken et al., 2011) (Fig. 1A). For each dpr- and DIP-GAL4, the cassette is inserted into a common intron or the 5′UTR shared by all isoforms (Fig. 1A, Table S1). Therefore, GAL4 should report the expression of all isoforms of each gene. Insertion of the SA-T2A-GAL4-PolyA should generate truncated transcripts because of the presence of the PolyA tail (Logan et al., 1987; Zhang et al., 2015). In addition, the presence of a T2A-GAL4 leads to an arrest during translation at the T2A site followed by a re-initiation of translation at the GAL4 sequence (Diao et al., 2015; Szymczak-Workman et al., 2012). To confirm the disruption of the gene of interest, we measured transcript expression by qRT-PCR using primers downstream of the insertion site and confirmed that most GAL4 lines are loss-of-function alleles. For example, in homozygous viable GAL4 lines, DIP-α-GAL4 and DIP-ζ-GAL4 showed no detectable DIP-α and DIP-ζ mRNA, respectively (Fig. S1A), suggesting they are null alleles. Several GAL4 lines, such as DIP-β-GAL4 and dpr15-GAL4, showed a reduction in mRNA levels, whereas some lines, such as DIP-ι-GAL4 and dpr16-GAL4, showed no change in mRNA expression. Although these GAL4 lines do not show a significant loss of transcription, the T2A sequence should still disrupt translation and generate mutant proteins. For homozygous lethal lines, we examined mRNA levels in heterozygous animals and found that most GAL4 lines show expression near 50% (Fig. S1B), suggesting these GAL4 lines are severe loss-of-function alleles. The qRT-PCR results are summarized in Table S2. In summary, approximately 70% of the insertions caused a severe disruption of transcription.
Fig. 1.
Schematic of GAL4 insertion and larval body plan. (A) MiMIC or CRIMIC cassettes were inserted into a common intron or 5′UTR to capture the expression of all isoforms for each Dpr and DIP gene. MiMIC insertions were flanked by two attP sites which are later swapped by a GAL4 exon or T2A-GAL4 trojan exon. (B) Drosophila larvae are divided into three thoracic segments and nine abdominal segments, with repeated muscles, MNs and SNs. Muscles are divided into three main groups, the ventral, lateral and dorsal muscles. MNs innervating these muscles are not shown in this diagram. SNs are divided into six main classes: the es neurons, ch neurons, bd neurons, td neurons, md neuron and da neurons (Orgogozo and Grueber, 2005). In addition, da neurons are further divided into da-I, da-II, da-III and da-IV subclasses.
Because most GAL4 insertions are mutants, we used heterozygotes to map Dpr and DIP gene expression. Loss of a single copy of any Dpr and DIP gene did not affect gross viability, cell survival or synaptic connectivity in heterozygotes as revealed by presence of a postsynaptic marker, Discs Large (DLG), and a presynaptic marker, anti-horseradish peroxidase (HRP; a marker for all neuronal membranes; Jan and Jan, 1982) (see Materials and Methods). Thus, the dpr/DIP-GAL4 driver lines should faithfully report the cells that express Dpr and DIP genes (Lee et al., 2018; Nagarkar-Jaiswal et al., 2015b).
Expression of Dpr and DIP genes in MNs
The larval body wall is segmented, and each abdominal hemisegment consists of 30 muscles that are grouped into three major muscle groups: ventral, lateral and dorsal (Fig. 1B) (Bate, 1990; Hooper, 1986; Zarin et al., 2019). Innervating those muscles are 33 MNs classified as type-I (29), type-II (3) and type-III (1) based on their terminal morphology and neurotransmitter type (Choi et al., 2004; Hoang and Chiba, 2001; Landgraf et al., 1997; Zarin et al., 2019). All MN axon terminals contain strings of bead-like structures called boutons which house the active zones. Type-I MNs are excitatory glutamatergic neurons, and they are further subdivided into type-I big (Ib) and type-I small (Is) based on their bouton size and innervation patterns: Ib MNs (in the larva named MN1-Ib to MN30-Ib corresponding to the muscle number) generally have larger boutons and innervate single muscle fibers whereas Is MNs have smaller boutons and innervate muscle groups (Choi et al., 2004; Lnenicka and Keshishian, 2000). The Is MN that innervates ventral muscles is referred to as the ventral common exciter (vCE), RP5 or MNISNb/d-Is, and the Is MN that innervates dorsal muscles is called the dorsal common exciter (dCE), RP2 or MNISN-Is (Broadus et al., 1995; Doe et al., 1988; Takizawa et al., 2007). Similarly, three neuromodulatory type-II MNs innervate the ventral, lateral and dorsal muscle groups, and the single type-III MN primarily innervates m12 (Hoang and Chiba, 2001; Schmid et al., 1999). Based on these distinguishing features – terminal morphology and innervation patterns – we can unambiguously identify MNs that express each Dpr and DIP gene.
To examine the expression of Dpr and DIP genes in MNs, we first crossed each GAL4 line to a fluorescent reporter line and monitored reporter expression at third instar NMJs (Fig. 2A). GAL4 lines derived from MiMIC insertions were crossed to a GFP reporter, whereas CRIMIC GAL4 lines were crossed to an mCherry reporter as CRIMIC insertions carry a 3XP3-GFP marker that expresses in glial cells and the lateral bipolar dendrite (lbd) neuron (Fig. S2). To identify all NMJs, we labeled preparations with antibodies against DLG and HRP and confirmed that the gross muscle innervation was normal in dpr/DIP-GAL4 heterozygous lines. GFP or RFP labeling of NMJs revealed the corresponding MNs that express each Dpr and DIP gene. We followed this pipeline for each dpr/DIP-GAL4 to record expression in all MNs.
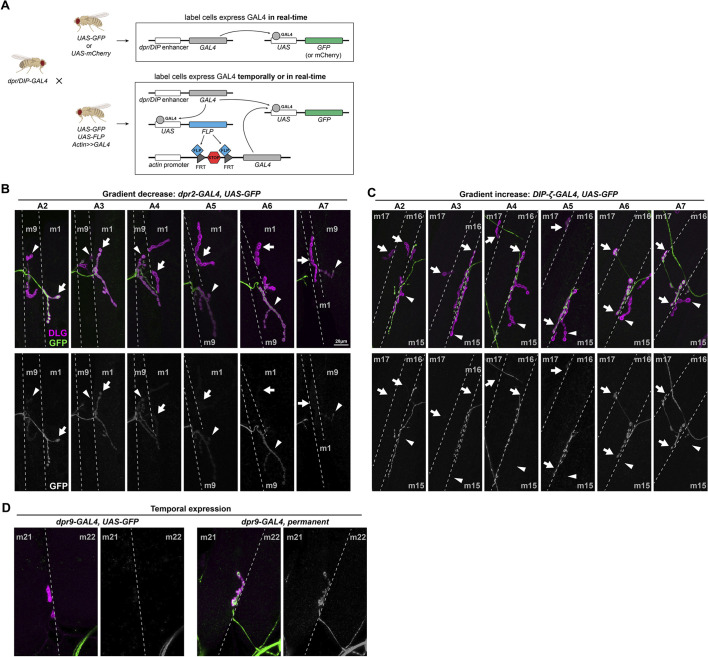
Fig. 2.
Dpr and DIP genes are expressed in various patterns in MNs. (A) Schematic showing the experimental procedure. Each dpr/DIP-GAL4 line was crossed to a real-time reporter (UAS-GFP or UAS-mCherry) and a permanent reporter [UAS-GFP, UAS-FLP, actin-(FRT.STOP)-GAL4] to reveal the dynamic expression of Dpr and DIP genes. (B) Example of a decrease in expression of dpr2-GAL4 in MN1-Ib (arrows) from anterior hemisegment A2 to posterior hemisegment A7. Note that the expression in nearby MN9-Ib (arrowheads) is also not robust as it was not expressed in A2 and A3 but was expressed in A4 to A7. (C) Example of an increase in expression of DIP-ζ-GAL4 in MN16/17-Ib (arrows) from anterior hemisegment A2 to posterior hemisegment A7. Note that the expression in nearby MN15/16-Ib (arrowheads) was always absent. (D) Example of temporal expression of dpr9-GAL4 in MN21-Ib. MN21-Ib was not labeled by dpr9-GAL4>GFP animals, but 50% of MN21-Ib were labeled in the cross to the permanent reporter. Dashed lines indicate muscle boundaries.
We first confirmed some previously observed expression patterns (Ashley et al., 2019; Carrillo et al., 2015); for example, DIP-α was selectively expressed in Is MNs but not in Ib MNs (Fig. S3A). Interestingly, we found that several Dpr and DIP genes are not always expressed at the same level in a specific MN. For example, DIP-δ-GAL4 only labeled 22% of MN12-Ib (ten out of 45 MN12-Ib examined; Fig. S3B). Similarly, dpr16-GAL4 was expressed in ten out of 25 MN30-Ib and nine out of 25 MN14-Ib examined. In addition to Ib and Is MNs, we also revealed expression in type-II and type-III MNs. For example, dpr16-GAL4 was expressed in type-II MNs but not the type-III MN (Fig. S4A,B), whereas DIP-κ-GAL4 was expressed in the type-III MN but not in type-II MNs (Fig. S4C,D). Surprisingly, we also found that some Dpr and DIP genes are expressed in a gradient along the anterior to posterior axis. For example, dpr2 showed high expression in MN1-Ib in the anterior but became undetectable from abdominal segment 4 (A4) to the posterior (Fig. 2B). DIP-ζ-GAL4, by contrast, labeled anterior MN16/17-Ib weakly, but was much stronger in the posterior (Fig. 2C). These complex expression patterns suggest intricate regulatory mechanisms of Dpr and DIP genes.
Work from our lab and others suggested that Dprs and DIPs are synaptic recognition molecules. In the fly neuromuscular circuit, MN axons explore the musculature field beginning at embryonic stage 14 and synaptic markers are observed at stage 16 (Yoshihara et al., 1997). A regular UAS reporter will only report real-time expression and will not reveal if a Dpr or DIP gene is temporally expressed earlier in development. To capture the temporal expression patterns of Dpr and DIP genes, we utilized a permanent labeling reporter to constantly label the GAL4-expressing neuron (Fig. 2A). Interestingly, we observed only a few Dpr and DIP genes that are temporally expressed in MNs. For example, MN21/22-Ib is not labeled when dpr9-GAL4 is crossed to UAS-GFP, but with the permanent labeling reporter, the same neuron showed strong expression (Fig. 2D). The temporal expression of only a small subset of Dpr and DIP genes suggests that most of these CSPs may function in different steps of nervous system development. It is noteworthy that the CRIMIC cassettes are excisable by Flippase as well due to the presence of flanking FRT sites (Fig. 1A). However, because of the activation of the permanent actin-GAL4, the excision of CRIMIC cassettes does not pose a technical issue.
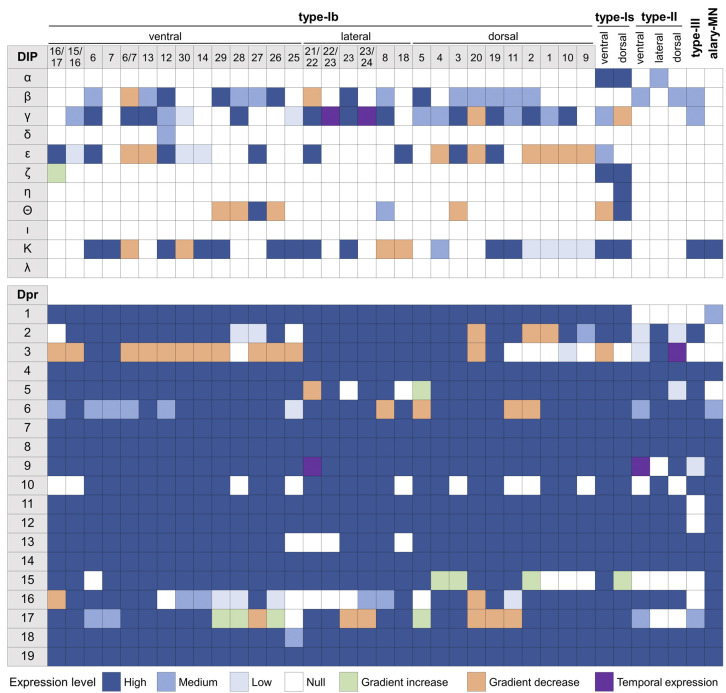
The expression of Dpr and DIP genes in all MNs is summarized in Fig. 3. Here, we included variable expression patterns (defined by how frequently a cell expresses the reporter) and gradient and temporal expression patterns. Criteria for each expression category is described in Materials and Methods and in Fig. S5. In general, Dpr genes are expressed in many MNs whereas DIP genes are expressed much more selectively. Each MN expresses at least one DIP gene, and, overall, each MN has a unique Dpr and DIP gene expression signature. Taken together, we generated an expression map of Dpr and DIP genes in all larval MNs and found that each MN expresses a unique subset.
Fig. 3.
Expression map of Dpr and DIP genes in all larval MNs. Each column represents an MN including type-Ib, type-Is, type-II, type-III and the alary MN. Expression of each gene in each MN is characterized into a specific category as indicated in the key. Note that MN16/17-Ib was named as MN15/16/17-Ib by Hoang and Chiba (2001); MN6-Ib is represented only in A2 hemisegments (see further characterization below); MN7-Ib is represented only in A2 hemisegments (see further characterization below); and MN23-Ib is a newly identified neuron (see further characterization below).
Expression of Dpr and DIP genes in muscles
In a previous study, we observed dpr10 expression in ventral and dorsal muscles and its interacting partner, DIP-α, in Is MNs (Ashley et al., 2019). To label muscle nuclei and report both temporal and real-time gene expression, we used the G-TRACE system (Evans et al., 2009), which takes advantage of FLP-FRT and GAL4/UAS (Fig. 4A). If a GAL4 is transiently expressed, then cell nuclei will be GFP positive. However, if the cell nuclei are labeled by both GFP and RFP, this may suggest the GAL4 is consistently expressed. We utilized the G-TRACE system and confirmed expression of dpr10 in all longitudinal muscles, but not in oblique or transverse muscles (Fig. 4B,C). This expression pattern suggests distinct transcriptional regulation programs between muscle groups (Bate, 1990). In addition, we found that a small subset of muscles, such as m5 and m8, showed inconsistent expression of dpr10 (Fig. 4B, arrow). Also, all muscle nuclei co-labeled with GFP and RFP in dpr10-GAL4>G-TRACE, suggesting that dpr10 expression is maintained throughout larval development.
Fig. 4.
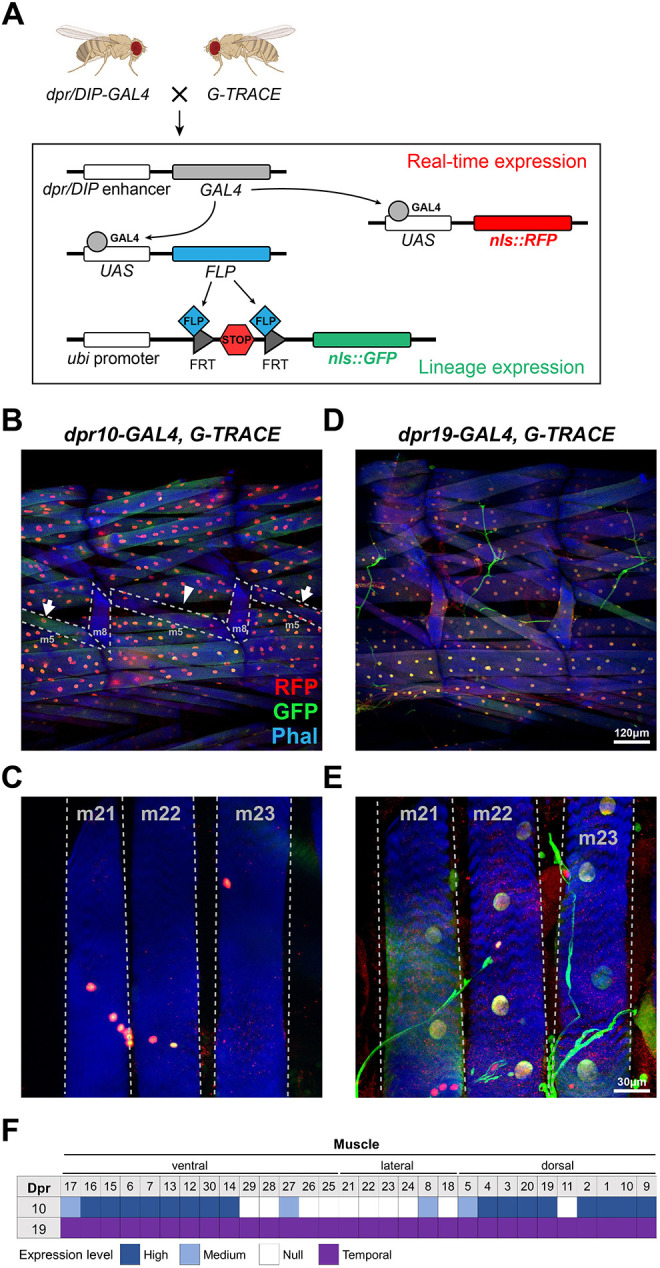
Using the G-TRACE system to probe expression of Dpr and DIP genes in muscles and glial cells. (A) Schematic showing the cross between dpr/DIP-GAL4 and the G-TRACE reporter. Red signal represents real-time GAL4 expression and green signal represents earlier GAL4 expression. (B,C) dpr10 is consistently expressed in most muscles (B) but absent in transverse muscles (C) and some deeper ventral muscles. Expression in some muscles is not consistent. For example, in some hemisegments m5 nuclei are not labeled (arrowhead), but an adjacent hemisegment shows labeling of m5 nuclei (arrows). dpr10 expression is maintained throughout development as revealed by co-labeling with GFP and RFP. (D,E) dpr19 is expressed in all muscles (D), including transverse muscles (E). Compared with dpr10, these nuclei have less RFP intensity, which may indicate that dpr19 is temporally expressed in early development and turned off later. (F) Expression map of dpr10 and dpr19 in muscles.
We examined other Dpr and DIP genes and found that dpr19 is expressed in all muscles (Fig. 4D), including the oblique and transverse muscles (Fig. 4E). Unlike dpr10, most muscle nuclei in dpr19-GAL4>G-TRACE were only GFP positive, suggesting that dpr19 is temporally expressed and turned off in late larval stages. To confirm this temporal expression, we examined dpr19-GAL4>mCherry first instar larva and observed a high level of muscle expression, which we did not observe in third instar (Fig. S6). Taken together, we showed that muscles express many fewer Dpr and DIP genes compared with motor and sensory neurons (Fig. 4F). These results suggest that a subset of Dpr and DIP genes may function in MN-muscle recognition and others in premotor neuron-MN recognition.
Expression of Dpr and DIP genes in glial cells
In the Drosophila larval peripheral nervous system, glia play important roles in nervous system development and extensively interact with MN axons (Kottmeier et al., 2020). Therefore, we examined the glial expression of Dpr and DIP genes.
To distinguish glial and neuronal expression unambiguously, we used G-TRACE to probe glial expression. We crossed each dpr/DIP-GAL4 line to the G-TRACE reporter and found that dpr1 is expressed in glia (Fig. S7). Additionally, dpr1 expression is highly dynamic as some glia temporarily express dpr1 whereas others maintain dpr1 expression. This result suggests that Dpr1 in glia and its interacting partners DIP-η, DIP-θ and DIP-ι in MNs (Fig. 3) may be involved in glia-neuron interactions to guide various processes, including neuronal development, axon pathfinding and synaptic homeostasis (Bittern et al., 2020; Yildirim et al., 2019).
Expression of Dpr and DIP genes in SNs
Next, we examined expression of Dpr and DIP genes in larval SNs using a similar approach to the MN analyses. Two morphologically distinct types of SNs can be classified in the larval body wall (Fig. 1B) (Orgogozo and Grueber, 2005; Veling et al., 2019). Type-I SNs project a single dendrite that associates with chordotonal (ch) organs or external sensory (es) organs to detect mechanical and chemical stimuli. Type-II SNs are multidendritic (md) neurons that transmit proprioceptive information. Type-II SNs can be further classified into bipolar dendrite (bd) neurons, tracheal dendrite (td) neurons and da neurons. The da neurons are then subdivided into four classes based on the complexity of their dendrite morphology (da-I, da-II, da-III and da-IV) (Fig. 1B) (Grueber et al., 2002).
To examine expression of Dpr and DIP genes in SNs, we labeled larvae with anti-HRP to locate the cell bodies of SNs. The Dpr and DIP gene expression map in all SNs is shown in Fig. 5. Similar to MNs, DIP genes are more sparsely expressed in SNs compared with Dpr genes, which are broadly expressed. However, several Dpr genes (dpr14, dpr15 and dpr17) are only expressed in a subset of SNs, unlike their broad expression pattern in MNs. We also observed that some Dpr and DIP genes are temporally expressed. For example, the dorsal da neurons (ddaA, -C, -F and -D) are labeled when dpr5-GAL4 is crossed to the permanent labeling reporter, but not in dpr5-GAL4>UAS-GFP animals (Fig. S8). Taken together, we generated expression data for Dpr and DIP genes in SNs (Fig. 5) and showed that each SN expresses a unique subset of Dpr and DIP genes, providing support for their roles as identification tags.
Fig. 5.
Expression map of Dpr and DIP genes in all larval SNs. Each column represents an SN. Expression of each gene in each SN is characterized into a specific category as indicated in the key.
SNs in the same class express similar subsets of Dpr and DIP genes
Larval SNs can be divided into types based on their morphology and function, including ch, es, bd, td and da neurons. Although SNs from the same type are distributed throughout the body wall and project their afferent axons through different trajectories, their axon terminals innervate the same region in the VNC and contact common interneuron partners (Grueber et al., 2007; Landgraf et al., 2003a; Merritt and Murphey, 1992; Murphey et al., 1989). For example, the ventral, ventral’ and lateral mechanosensory ch neurons all project to the ventral medial region of the VNC and share synapses with some interneurons (Heckscher et al., 2015; Valdes-Aleman et al., 2021). Similarly, different classes of da neurons innervate unique sections of the VNC (Grueber et al., 2007; Merritt and Whitington, 1995; Schrader and Merritt, 2000). Overall, these innervation patterns suggest that SNs from the same class may share similar identification tags to wire with common interneurons.
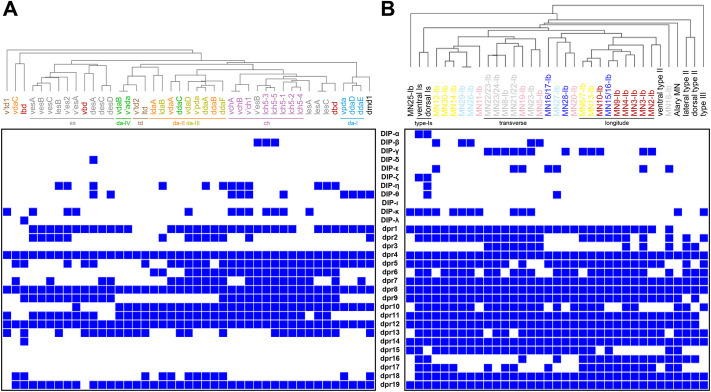
Dprs and DIPs have been implicated in synaptic partner recognition so we hypothesized that shared Dpr and DIP gene expression may be utilized by the same type/class of neurons to instruct connectivity. To test this model, we generated an unbiased hierarchical clustering of SNs based on their Dpr and DIP gene expression (Fig. 6A). Surprisingly, we found a high correlation between SN types/classes and expression of Dpr and DIP genes. For example, most es neurons are grouped together, as well as all ch neurons, indicating that these two subclasses of type-I SNs can be distinguished by their expression of Dpr and DIP genes. Similarly, subclasses of da neurons are clustered separately. We found that da-I neurons are identifiable by expression of DIP-θ and the lack of dpr2, dpr6, dpr9, dpr11 and dpr13, and da-II/da-III neurons are grouped by expression of dpr2 and dpr18, and the lack of dpr9 (Fig. 6A). These results suggest that SNs in the same type/class may utilize similar sets of Dpr and DIP genes to recognize common interneuron targets.
Fig. 6.
Hierarchical clustering of SNs and MNs reveals shared expression patterns of Dpr and DIP genes in neurons from the same class. (A) SNs from the same class are clustered together based on their expression pattern of Dpr and DIP genes. For example, most es neurons (gray), all chordotonal neurons (purple), and da neurons fall into distinct clusters. (B) Modulatory MNs (II and III) and type-Is MNs are distinct from the main type-Ib cluster. However, individual type-Ib MNs are not easily distinguished based on their expression of Dpr and DIP genes.
Expression of Dpr and DIP genes is more diversified in MNs
Next, we examined whether MNs that project to the same muscle groups also share the same expression patterns of Dpr and DIP genes. Muscles are grouped into three main spatial and functional groups – ventral, lateral, dorsal – and further divided into six subgroups based on their orientation – dorsal longitudinal (DL), dorsal oblique (DO), ventral longitudinal (VL), ventral oblique (VO), ventral acute (VA) and lateral transverse (LT) (Fig. 1B) (Bate, 1990; Hooper, 1986; Zarin et al., 2019). Each muscle is normally innervated by one Ib MN and previous studies showed that Ib MNs innervating a muscle group project their dendrites to the same region in the VNC neuropil where they receive input from common premotor neurons (Kim et al., 2009; Landgraf and Thor, 2006; Landgraf et al., 1997, 2003b; Mauss et al., 2009; Zarin et al., 2019). Thus, if Ib MNs of the same muscle group share premotor neuron partners, they may share wiring molecules. We generated an unbiased hierarchical clustering based on expression of Dpr and DIP genes for all MNs (Fig. 6B). Type-Is, type-II and type-III MNs form independent clusters and are distinct from Ib MNs. For example, DIP-α, DIP-ζ, dpr6 and dpr16 are expressed in type-Is MNs, and lateral and dorsal type-II MNs are identified by the lack of DIP-κ, dpr15 and dpr17 and the expression of dpr3 and dpr16 (Fig. 6B). However, within Ib MNs, only the MNs innervating LT and DL muscles are clustered together, whereas the other MNs appear randomly distributed. These results suggest that, based on the expression patterns of Dpr and DIP genes, MNs can be clustered by their type, but Ib MNs cannot be further clustered by the muscles they innervate. MNs must identify distinct pre- and postsynaptic partners, which may explain the inability to cluster Ib MNs based on their expression patterns of Dpr and DIP genes. Therefore, more complex identification codes may be necessary for MNs to distinguish both pre- and postsynaptic partners.
Dpr and DIP gene expression maps reveal additional MNs
Alary muscle MN
In addition to the muscles required for larval locomotion, larvae have another segmentally repeated muscle, the alary muscle, which attaches to the trachea along the larval heart tube (Bataillé et al., 2015). The alary muscle MN axon resides in the transverse nerve (TN) and projects along m8 towards the alary muscle. Here, we mapped the expression of Dpr and DIP genes in the alary muscle MN. As previously observed, the dendrite of the lbd neuron travels in parallel with the alary muscle MN axon within the TN (Gorczyca et al., 1994; Macleod et al., 2003; Thor and Thomas, 1997). Thus, if a Dpr or DIP gene is expressed in both the alary muscle MN and lbd, we would be unable to distinguish them in the nerve. Therefore, we monitored the colocalization of DLG and the fluorescent reporter on the alary muscle to assign expression unambiguously (Fig. S9). We observed that alary muscle MN NMJs share features of type-I boutons, including the size and DLG labeling surrounding the boutons. Using the same criteria described for MNs and SNs, we found one DIP gene and many Dpr genes that are expressed in the alary muscle MN, including DIP-κ, dpr4 and dprs7-19. These expression data and driver lines will facilitate future characterization of this MN.
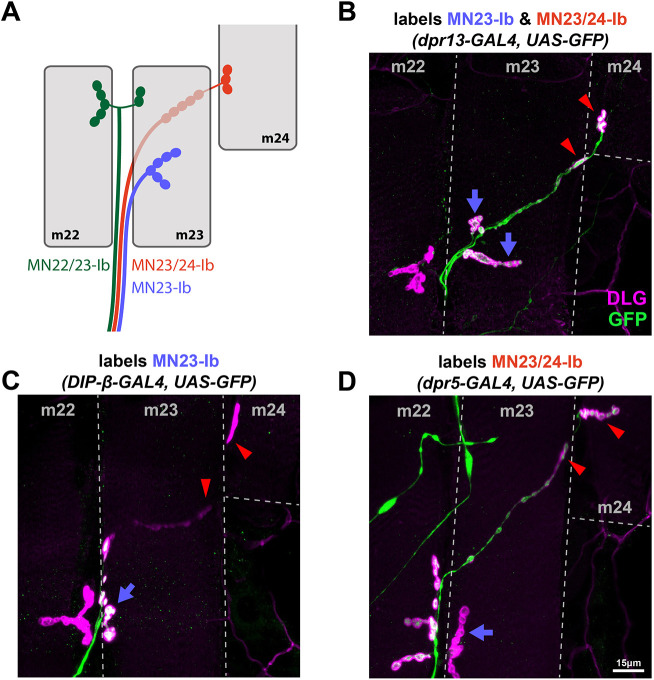
MN23-Ib
Most Ib MNs have a single muscle target. However, some Ib MNs innervate two muscles in close proximity, likely due to shared recognition cues. For example, a previous study found that Ib MNs innervating the lateral muscles can synapse with neighboring muscles and thus named these neurons MN21/22-Ib, MN22/23-Ib and MN23/24-Ib (Fig. 7A) (Hoang and Chiba, 2001). These innervation patterns were later confirmed by MARCM (mosaic analysis with a repressible cell marker) analysis (Kim et al., 2009).
Fig. 7.
Differentially expressed Dpr and DIP genes reveal an MN that solely innervates m23. (A) Schematic of transverse muscles 22, 23 and 24 (gray) with previously identified MN22/23-Ib (green), MN23/24-Ib (red) and newly identified MN23-Ib (blue). (B) Representative image showing dpr13-GAL4 expression in both MN23/24-Ib (red arrowheads) and MN23-Ib (blue arrows). Thus, all boutons on m23 and m24 are labeled by GFP. (C) Representative image showing DIP-β-GAL4 expression in MN23-Ib (blue arrow). Boutons underneath m23 and boutons from m22, m24 (red arrowheads) are not labeled by GFP, thus DIP-β-GAL4 is not expressed in MN22/23-Ib and MN23/24-Ib. (D) Representative image showing dpr5-GAL4 expression in MN22/23-Ib and MN23/24-Ib (red arrowheads), but not in MN23-Ib (blue arrow). The lack of GFP in the arbor on m23 indicated the existence of an MN that solely innervates m23.
In our analyses, we observed that m23 has several Ib NMJ branches and m24 has only one NMJ (Fig. 7B). Although a single MN can form several branches on a muscle, we found some dpr/DIP-GAL4 lines that only label one Ib branch on m23 and no other branches on lateral muscles (Fig. 7C). These data suggest the existence of an additional MN that solely innervates m23, and we named it MN23-Ib. The bouton size and DLG labeling intensity of M23-Ib boutons indicates that it is a type-Ib NMJ. DIP-β and DIP-κ are expressed in MN23-Ib and not in the nearby MN22/23-Ib and MN23/24-Ib (Fig. 7C). Note that MN23/24-Ib forms long, linear Ib NMJs on the underside of m23 before it reaches m24 (Fig. 7A,C). We also found that dpr5 was expressed in MN23/24-Ib and nearby MN22/23-Ib, but not in MN23-Ib (Fig. 7D), providing further evidence for an additional Ib MN solely innervating m23. Additional Dpr and DIP genes are expressed in both MN23-Ib and MN23/24-Ib (Fig. 7B). Thus, we describe a previously unidentified Ib MN that innervates m23.
MN6-Ib and MN7-Ib in A2
Another example of a dual-targeting Ib MN is MN6/7-Ib (also known as RP3 in the embryo) (Schmid et al., 1999; Sink and Whitington, 1991a,b). The innervation pattern of MN6/7-Ib was initially identified by dye-fill labeling and MARCM (Hoang and Chiba, 2001; Kim et al., 2009; Sink and Whitington, 1991b). Owing to the ease of accessibility of m6 and m7, MN6/7-Ib is extensively used for studies of synaptic connectivity, synaptic growth and synaptic homeostasis.
Based on these previous studies, we predicted that if a Dpr or DIP gene were expressed in MN6/7-Ib, the Ib NMJs on both m6 and m7 would be completely fluorescently labeled (Fig. 8A, right). Surprisingly, in A2, we observed several dpr/DIP-GAL4s that label Ib MNs that have large NMJs on m6 and others that label mainly the Ib NMJs on m7 (Fig. 8A, left). For example, DIP-β, DIP-γ and DIP-ε were expressed in an MN that mainly innervates m6 (Fig. 8B), whereas dpr15 was expressed in an MN that mainly innervates m7 (Fig. 8C). These expression patterns suggested that two Ib MNs innervate m6 and m7 in A2. Hereafter, we named these MNs as MN6-Ib and MN7-Ib. Prior studies hinted at the possibility of two MNs based on the larger synaptic terminal area on m6/7 in A2 compared with A3-A6 (Lnenicka and Keshishian, 2000). However, it was thought that the larger synaptic area was due to a large NMJ from a single Ib MN. In the larval neuromuscular circuit, the number of boutons reflects the size of the NMJ. We quantified the m6 and m7 Ib NMJs and observed a significantly larger arbor in A2 compared with A3 (Ib NMJ on m6: 34.2 on A2 and 18.5 on A3; Ib on m7: 23.1 on A2 and 11.7 on A3) (Fig. S10). Taken together, we conclude that m6 and m7 in A2 are innervated by two Ib MNs.
Fig. 8.
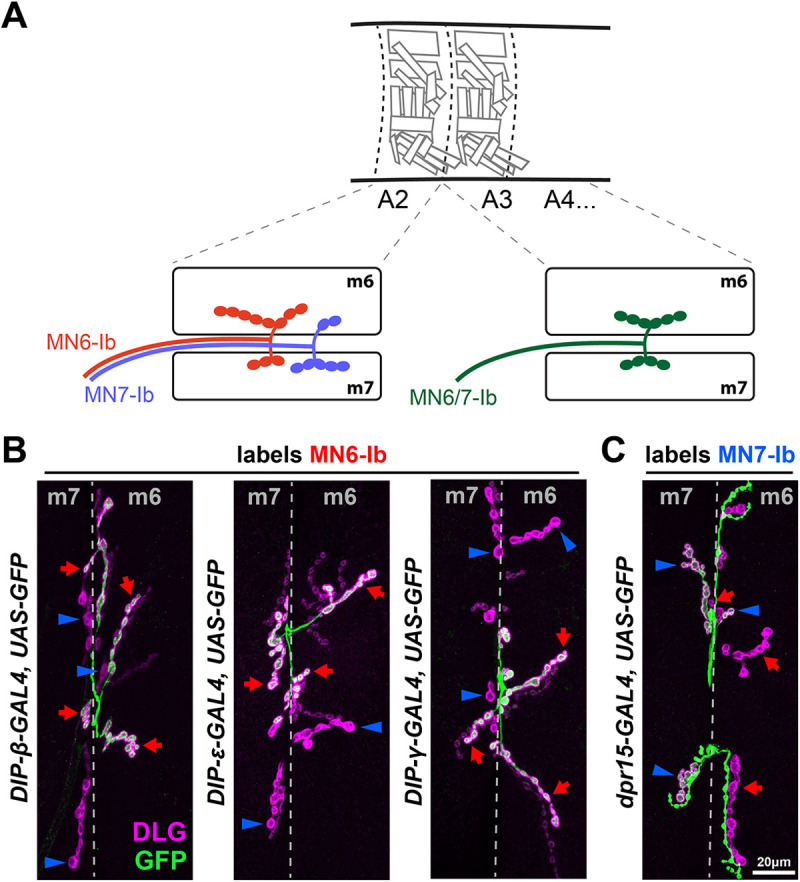
Differentially expressed Dpr and DIP genes reveal MN6-Ib and MN7-Ib in segment A2. (A) Schematic of MN6-Ib (red) and MN7-Ib (blue) in segment A2, and MN6/7-Ib in A3-A7 (green). MN6-Ib preferentially innervates m6 but also forms a small NMJ on m7, whereas MN7-Ib prefers m7 but also forms a small NMJ on m6. (B) Representative images showing that DIP-β, DIP-ε and DIP-γ are specifically expressed in MN6-Ib (red arrows), but not in MN7-Ib (blue arrowheads). Note that MN6-Ib forms boutons with both m6 and m7, as there is a small GFP-positive type-Ib NMJ on m7 (red arrows on m7). Conversely, the lack of GFP in most m7 type-Ib NMJ and the small m6 type-Ib NMJ (blue arrowheads) also indicate dual innervation of both muscles by MN7-Ib. (C) Representative image showing that dpr15 is specifically expressed in MN7-Ib (blue arrowheads) but not in MN6-Ib (red arrows). MN6-Ib and MN7-Ib also show dual innervation patterns in this genetic background.
Characterization of MN6-Ib and MN7-Ib
A recent study reported a GAL4 driver (GMR79H07-GAL4) that labels MN6-Ib in A2 (Aponte-Santiago et al., 2020). We tested this driver and confirmed MN6-Ib expression; however, it sometimes labels MN7-Ib NMJs or both MN6-Ib and MN7-Ib (Fig. S11). We examined MN6-Ib and MN7-Ib further in order to understand better their innervation patterns and dendritic projections. Interestingly, we found that MN6-Ib and MN7-Ib preferentially innervate their corresponding muscle, but sometimes these MNs also form minor NMJs on the neighboring muscle (Fig. 8A). Next, we monitored the frequency of dual innervation of each MN using GMR79H07-GAL4 and found that 68.2% of MN6-Ib and 72.7% of MN7-Ib innervate both muscles (Fig. 9A). We also determined the size of each NMJ by counting Ib boutons and found that on average MN6-Ib forms 48.6 boutons on m6 and 5.9 boutons on m7, whereas MN7-Ib forms 3.1 and 30.8 boutons on m6 and m7, respectively (Fig. 9B).
Fig. 9.
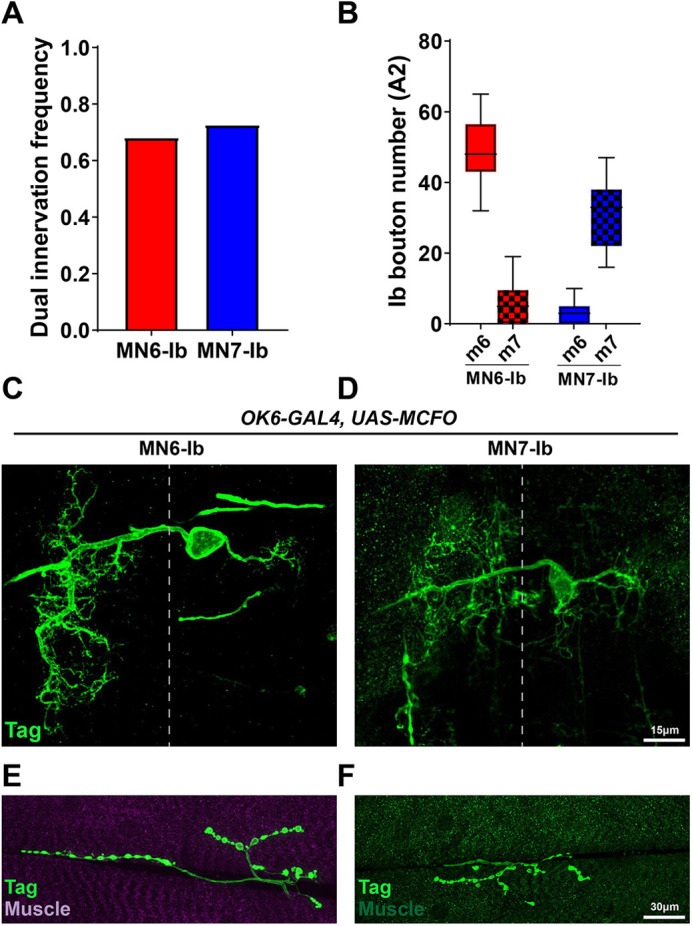
Further characterization of MN6-Ib and MN7-Ib. (A) Quantification of the dual innervation frequencies of MN6-Ib and MN7-Ib: 68.2% of MN6-Ib also innervate m7 and 72.7% of MN7-Ib also innervate m6 (n=21 hemisegments). (B) Quantification of MN6-Ib and MN7-Ib NMJ sizes on both muscles (n=21 hemisegments). (C,D) A pan-MN driver OK6-GAL4 driving MCFO revealed the dendritic morphology of MN6-Ib and MN7-Ib in the VNC. (E,F) Corresponding NMJ images from the same neuron shown in C (MN6-Ib) and D (MN7-Ib).
MN6/7-Ib (RP3) is derived from neuroblast 3-1 (NB3-1) (Schmid et al., 1999; Sink and Whitington, 1991a,b). To visualize the dendritic projections of MN6-Ib and MN7-Ib in A2, we examined their cell body position in the VNC and dendrite morphology using a pan-MN driver, OK6-GAL4, with multi-color FLP-out (MCFO) (Nern et al., 2015). We found that the cell bodies of MN6-Ib and MN7-Ib are both localized at the dorsal neuropil and project axons to the contralateral hemineuromere. They also extend a small dendritic arbor to the ipsilateral side (Fig. 9C-F). These features are shared with RP3 (MN6/7-Ib) (Kim et al., 2009). These data suggest that these two MNs likely both originated from NB3-1. Overall, we identified and confirmed the presence of two Ib MNs in A2 that preferentially innervate m6 or m7.
DISCUSSION
Dprs and DIPs play important roles in nervous system development, and they are widely expressed across many neural circuits. Several groups have utilized the GAL4/UAS system to visualize expression of Dpr and DIP genes in olfactory neurons (Barish et al., 2018), adult leg MNs and SNs (Venkatasubramanian et al., 2019), optic lobe neurons (Cosmanescu et al., 2018; Tan et al., 2015) and fru P1 neurons (Brovero et al., 2021). Although these studies revealed unique Dpr and DIP gene expression in the respective neurons, the depth of the expression maps was limited due to the less complete GAL4 collection at the time, and some studies only focused on a global expression pattern without characterization of individual cell types.
Here, we reported a collection of GAL4 enhancer trap lines for all DIP genes and 19 Dpr genes, and examined their expression in larval MNs, muscles, peripheral glia and SNs. Interestingly, we found that many Dpr and DIP genes are expressed in patterns including different expression levels, anterior-posterior gradients and temporal expression. Our expression analyses also revealed previously uncharacterized larval MNs that differentially express Dpr and DIP genes. The Dpr and DIP gene expression maps identified here, along with the GAL4 lines that are also hypomorphs or loss-of-function alleles, will facilitate examination of Dpr-DIP interactions in development of motor, sensory, and many other circuits.
Insights from Dpr and DIP gene expression maps to aid functional studies
The goal of developing expression maps for Dpr and DIP genes in MNs and SNs is to instruct the functional study of Dpr-DIP interactions. Here, we discuss testable hypotheses based on our expression maps that may serve as an entry point for future research.
Based on our expression map, all muscles express dpr19 and most also express dpr10. Dpr10 and Dpr19 interact with DIP-α/β/λ and DIP-ε/ζ, respectively, and a majority of MNs express at least one of these DIPs. Thus, Dpr-DIP interactions could instruct MN-muscle recognition and/or act combinatorially with other synaptic connectivity molecules. However, some MNs do not express any of these DIPs, suggesting that other pairs of CSPs are involved in MN-muscle recognition. Alternatively, muscles may express unknown Dpr or DIP interactors not tested in the previous biochemical screen. In addition, we found many Dpr-DIP interactors that were co-expressed in the same MNs. For example, DIP-β and its interacting partners, dpr6/8/9/11, are co-expressed in MN12-Ib, suggesting that Dpr-DIP cis interactions may contribute to NMJ development and connectivity.
Another way to approach the function of Dpr-DIP interaction is focusing on the commonly or differentially expressed Dpr and DIP genes. Hierarchical clustering analyses of SNs grouped SNs from the same class together based on the expression of Dpr and DIP genes, suggesting that similar SNs have common Dpr and DIP genes. Future studies could determine the Dpr and DIP gene expression maps in the downstream interneurons to identify synaptic partners that express cognate Dpr-DIP pairs. However, one should also note that cluster analysis based solely on binary Dpr and DIP gene expression ignores expression levels and localization of proteins, which are important determinants for circuit wiring. Combining these with other parameters, such as transcription factor expression, can refine the clustering results and reduce unlikely correlations.
Instead of commonly expressed genes, differentially expressed Dpr and DIP genes in similar projecting neurons could shed light on connectivity mechanisms. For example, MN6-Ib and MN7-Ib, identified in this study, have similar morphology and innervation patterns, but with a preference for m6 and m7, respectively. One interesting question is how these neurons distinguish their muscle targets to generate such preference. Based on the expression map, MN6-Ib and MN7-Ib co-express a large subset of Dpr and DIP genes, but DIP-β, DIP-γ, DIP-ε and dpr15 are selectively expressed. These differentially expressed genes are excellent candidates to explore the recognition mechanism of these MNs. Similar approaches could be adapted to other MNs that innervate neighboring muscles.
The Dpr-DIP interactome (Carrillo et al., 2015; Cosmanescu et al., 2018; Özkan et al., 2013) revealed binding promiscuity in the interactions and our expression maps showed that many cells co-express many Dpr and DIP genes, suggesting redundant mechanisms for synaptic recognition. Several subfamilies of CSPs are implicated in recognition, but loss-of-function mutants rarely are 100% penetrant. For example, loss of Teneurin signaling causes a 90% decrease of MN3-Ib innervation (Hong et al., 2012), and Toll null mutants revealed defects in 35% of MN6/7-Ib (Rose et al., 1997). These data suggested that other CSPs are required in the recognition between MNs and their respective muscles. Utilizing the Dpr and DIP gene expression maps, co-expressed Dpr and DIP genes could be simultaneously knocked out in specific MN or SN to examine redundancy. For example, the dorsal Is MN expresses six DIPs, and DIP-α is required for Is innervation of m4 but only partially required for Is innervation of other muscles. If redundant DIP codes are required for specific innervations, a sextuple DIP mutant should reveal complete loss of dorsal Is NMJs.
CSP expression patterns in the fly nervous system
CSPs can serve several functions in nervous system development, including molecular codes for partner recognition and self-avoidance. CSP expression patterns can suggest different functions; the expression of CSPs could be deterministic to instruct stereotyped synaptic connectivity or stochastic to avoid dendritic overlap and self-synapses. For example, Capricious is robustly expressed in MN12-Ib and some dorsal MNs (Nose, 2012; Shishido et al., 1998), and loss-of-function and gain-of-function approaches have revealed neuromuscular wiring defects, suggesting that the robust expression of Capricious instructs synaptic partner recognition. In our study, we showed that many Dpr and DIP genes are robustly expressed in SNs and MNs, indicating their potential roles in synaptic wiring.
By contrast, some CSPs are stochastically expressed in subsets of cells. For example, probabilistic splicing of Dscam1 generates random isoform expression in SNs to mediate dendritic self-avoidance by inhibitory homophilic interactions (Miura et al., 2013). Interestingly, we found that many Dpr and DIP genes are also stochastically expressed in MNs and SNs. Such irregular expression patterns may suggest additional functions of Dpr and DIP genes in circuit formation.
In this study, we also uncovered some Dpr and DIP genes that are expressed in a gradient along the anterior-to-posterior axis. Such patterns are reminiscent of the expression of several Hox genes in the VNC. For example, Ubx and Abd-A are highly expressed in anterior segments whereas Abd-B is mainly in the posterior (Estacio-Gómez and Díaz-Benjumea, 2013; Meng and Heckscher, 2020). These transcriptional factors were proposed to set up segmental cues in the nervous system, but the downstream genes and pathways are not completely understood. The similar expression patterns suggest that gradient transcriptional factors may regulate segmental development, in part, through Dpr and DIP genes.
dpr/DIP-GAL4 collection to enable neuron identification and manipulation
The maps of Drosophila MNs and SNs was established decades ago using dye backfills (Broadus et al., 1995; Hoang and Chiba, 2001; Landgraf et al., 2003a). However, fluorescent dyes have some technical limitations as they do not always flow into every terminal structure, which may have resulted in some neurons being overlooked. In this study, we used a genetic approach to probe individual neurons and revealed three uncharacterized MNs: MN23-Ib, MN6-Ib (A2) and MN7-Ib (A2).
In addition, the GAL4 lines in this study provide genetic access to manipulate subsets of neurons. In the Drosophila motor circuit, several studies have identified reporters that are expressed in subsets of motor neurons, muscles and interneurons (Aponte-Santiago et al., 2020; Li et al., 2014; Pérez-Moreno and O'Kane, 2018; Wang et al., 2021). However, the coverage of these reporters is very limited (i.e. only a small number of cells can be targeted). To generate new genetic tools for targeting subsets of MNs, the Dpr and DIP gene expression maps could be inspected for partially overlapping or non-overlapping dpr/DIP-GAL4 expression and converted to split-GAL4 or GAL80, respectively. Thus, the expression data in the present study and the MiMIC/CRIMIC lines provide a pipeline to expand the genetic toolbox and to label and manipulate neurons in a highly specific manner.
Using the Dpr/DIP gene code to annotate single-cell RNA sequencing data
Recent advances in single-cell RNA sequencing (scRNAseq) provide a powerful, high-throughput approach to identify large-scale gene expression patterns. Various Drosophila neural tissues have been analyzed by scRNAseq (Li, 2020). However, most studies report the transcriptome of large cell clusters, including MNs, ganglion cells, neuroblasts and glial cells because of the difficulty of matching single-cell reads to a specific cell type and identity, impeding detailed analyses from scRNAseq data.
One method to deconvolve these large cell clusters is sorting cells before performing scRNAseq. Researchers may also use the scRNAseq data to identify specific drivers, and then identify which neuron expresses this driver (Simon and Konstantinides, 2021). However, this approach reduces the scale because only a few cell types can be identified in this manner. Utilizing the expression of a gene family known to be differentially expressed within a specific subset of cells can provide a more complete examination. For example, the Dpr and DIP gene expression maps would generate a cell-specific atlas to annotate clusters in scRNAseq data and help to identify individual MNs from an MN cluster in a larval VNC sample (Nguyen et al., 2021; Vicidomini et al., 2021). In addition to Dpr and DIP genes, other CSP subfamilies have been reported in several scRNAseq datasets, suggesting that expression maps of other subfamilies and even combinations of subfamilies can be utilized to refine cell types in datasets (Kurmangaliyev et al., 2020; Ma et al., 2021; Xie et al., 2021).
Limitations of using GAL4 lines to profile expression patterns
In the current study, we present expression maps of Dpr and DIP genes in a variety of cells using a GAL4 collection. However, several caveats exist. First, using a GAL4/UAS approach will not provide spatial information about where Dprs and DIPs are localized subcellularly, e.g. in axons or dendrites. Future work will generate endogenously tagged versions of, or antibodies against, Dprs and DIPs. In addition, using the current GAL4/UAS pipeline, we cannot unequivocally identify specific interneurons that express Dpr and DIP genes because of their indistinguishable cell morphologies in the densely packed VNC. Transcription factor staining and generation of split GAL4s can reveal interneurons identities but at relatively low throughput. Finally, utilizing a lineage-tracing system, we uncovered temporally expressed Dpr and DIP genes. However, neither of our approaches revealed when Dpr and DIP genes were first expressed. Embryo or early-stage larval dissection will provide more temporal resolution.
MATERIALS AND METHODS
Drosophila lines
All dpr/DIP-GAL4 lines are listed in Table S1. Other driver lines were: OK6-GAL4 (Bloomington Drosophila Stock Center, 64199), GMR79H07-GAL4 (gift from Troy Littleton, MIT, MA, USA), MHC-GAL80 (gift from Timothy Mosca, Thomas Jefferson University, PA, USA). Reporter lines were: 10XUAS-mCD8::GFP (Bloomington Drosophila Stock Center, 32184), 20XUAS-mCherry (Bloomington Drosophila Stock Center, 52268), UAS-2XEGFP; actin-(FRT.STOP)-GAL4,UAS-FLP (permanent reporter, gift from Ellie Heckscher, University of Chicago, IL, USA), UAS-RedStinger, UAS-FLP, Ubi-p63E(FRT.STOP)-Stinger (G-TRACE; Bloomington Drosophila Stock Center, 28280), R57C10-FLP;;UAS-MCFO (Bloomington Drosophila Stock Center, 64089). Lines used to generate Trojan-GAL4 were: yw; Sp/CyO; loxP(Trojan-GAL4)x3 (Bloomington Drosophila Stock Center, 60311), yw; loxP(Trojan-GAL4)x3; Dr/TM3,Sb,Ser (Bloomington Drosophila Stock Center, 60310), yw,hs-Cre,vas-phiC31:int (Bloomington Drosophila Stock Center, 60299).
Antibodies
Primary antibodies used in this study were: rabbit anti-GFP (1:40,000; gift from Michael Glozter, University of Chicago, IL, USA), rabbit anti-HA (1:1000; Cell Signaling Technology, C29F4), mouse anti-DLG (1:100; Developmental Studies Hybridoma Bank, 4F3), mouse anti-Repo (1:100; Developmental Studies Hybridoma Bank, 8D12), mouse anti-Myosin (1:100; Invitrogen, A31466), chicken anti-GFP (1:500; Invitrogen, A10262), chicken anti-RFP (1:500; Novus Biologicals, NBP2-25158), chicken anti-V5 (1:500; Bethyl Laboratories, A190-118A), rat anti-Flag (1:200; Novus Biologicals, NBP1-06712).
Secondary antibodies used were: goat anti-rabbit Alexa 488 (1:500; Invitrogen, A11008), goat anti-rabbit Alexa 568 (1:500; Invitrogen, A11036), goat anti-mouse Alexa 568 (1:500; Invitrogen, A11031), goat anti-mouse Alexa 647 (1:500; Invitrogen, A32728), goat anti-chicken Alexa 488 (1:500; Invitrogen, A11039), donkey anti-chicken Cy3 (1:500; Jackson ImmunoResearch, 703-165-155), goat anti-rat Alexa 647 (1:500; Invitrogen, A21247), goat anti-HRP Alexa 647 (1:100; Jackson ImmunoResearch, 123-605-021), goat anti-Phalloidin Alexa 405 (1:100; Invitrogen, A30104).
Fly genetics
When examining available dpr/DIP-GAL4 lines to confirm the GAL4 insertion sites and the version of GAL4 used, we found that the original dpr13-GAL4 no longer contained the GAL4 sequence (Barish et al., 2018; Brovero et al., 2021). Therefore, we generated new dpr13-GAL4 and dpr8-GAL4 from respective MiMIC insertion lines using Trojan exons (Diao et al., 2015). To generate DIP-λ CRIMIC insertions, gRNA (5′-AGCATCTATCGCTTGTGAAAGGG-3′) was designed to target the coding intron. The insertion sites and GAL4 versions are indicated in Table S1.
qRT-PCR
Five larvae per genotype were collected and homogenized using pellet pestles (Fisher Scientific). All samples tested contained a mix of males and females, except for dpr8-GAL4, for which only females were used due to its location on the X chromosome and homozygous lethality. RNA was extracted using RNAqueous Total RNA Isolation Kit (Thermo Fisher, AM1912) and subsequently treated with DNaseI for 30 min at 37°C to remove genomic DNA. cDNA was generated from 1 μg of RNA using random hexamers and SuperScript IV First-Strand Synthesis System (Thermo Fisher, 18091050) and RNA was removed using RNase H at 37°C for 20 min. Primers were designed to be 18-23 bp long, amplify 100-200 bp, and have a melting temperature of ∼60°C (Table S2). All primer locations are downstream of mapped GAL4 insertion sites and were validated with control cDNA. qRT-PCR was performed with Power SYBR Green PCR Master Mix (Bio-Rad, 4368577) and run on a QuantStudio 3 real-time PCR system (Thermo Fisher). All reactions were normalized to the housekeeping gene RpL32 and control flies, yielding ΔΔCt values (Ponton et al., 2011). Relative fold change was calculated as 2^-ΔΔCt. Each reaction was run in technical and biological triplicate.
Dissection and immunocytochemistry
Larval dissections and immunostaining were performed as previously described (Ashley et al., 2019). Briefly, wandering third instar larvae were dissected along the dorsal midline in PBS on a Sylgard plate and stretched out with insect pins. To visualize alary muscles, larva was dissected from the ventral side. Dissected body walls were washed once with PBS and fixed for 30 min with 4% paraformaldehyde. Samples were then washed three times with PBT (PBS+0.05% Triton X-100). Samples were incubated with primary antibody at 4°C overnight, washed three times with PBT, and then incubated in secondary antibody at room temperature for 2 h. Samples were finally mounted in 30 μl vectashield (Vector Laboratories). Representative images were taken with a Zeiss LSM800 confocal microscope with a 40× Plan-Neofluar 1.3 NA objective and processed with ImageJ.
Examining expression of Dpr and DIP genes in MNs and SNs
We dissected six third instar larvae from each cross and immunostained for GFP/RFP, DLG and HRP. Mounted samples were examined under a Zeiss AxioImager M2 with a Lumen light engine with a 20× Plan Apo 0.8 NA objective. Each sample was examined twice with the same criteria to reduce human error. To map the expression of Dpr and DIP genes in MNs, NMJs were identified by labeling for DLG or HRP, and then examined for GFP/RFP colocalization. For expression in SNs, SN cell bodies were located by HRP labeling, and then examined for GFP/RFP colocalization. We counted all MNs and SNs from anterior to posterior hemisegments (abdominal segments A2-A7) to gain a full Dpr and DIP gene expression map across the body wall. Note that we did not observe the third type-Is MN (MNSNa-Is) described by Hoang and Chiba (2001). The pipeline and criteria of determining the expression level is described below (Fig. S4).
In dpr/DIP-GAL4>GFP/RFP animals, if the reporter gene was expressed constantly in a specific MN/SN in all hemisegments, then this GAL4 line was counted as ‘high expression level’ in this MN/SN. If the fluorescent reporter was not expressed consistently in a specific MN/SN, then: (1) if the fluorescent reporter showed a gradient increase or decrease along the anterior to posterior axis, then the expression of this GAL4 line was reported as ‘gradient increase’ or ‘gradient decrease’, respectively; or (2) if the reporter gene was not expressed in a gradient, but was randomly expressed in a specific MN/SN, then the expression was counted as ‘medium expression level’ in this MN/SN. Note that we did not record gradient expression for SNs, because the reporter expression was more variable in SNs compared with MNs.
In the cross between dpr/DIP-GAL4 and the permanent labeling reporter, we first confirmed the high, medium and gradient expression level described above. Then, if a GAL4 line showed no expression in the cross to UAS-GFP/RFP but did show expression in the cross to the permanent labeling reporter, we counted how frequently this MN/SN was labeled: (1) if the labeling frequency was lower than 30% across all hemisegments, then this GAL4 was recorded as ‘low expression level’ in this MN/SN because the expression could be too low to detect in the cross to UAS-GFP/RFP but sufficient to trigger some FLP-out; (2) if the labeling frequency was between 30% and 60%, then this GAL4 expression was recorded as ‘medium expression level’ in this MN/SN; (3) if the labeling frequency was higher than 60%, then this GAL4 expression was considered as ‘temporal expression’ as it indicates a high GAL4 expression level temporally in early developmental stages because it triggers high frequency FLP-out. Finally, if a GAL4 was not expressed in both the cross to UAS-GFP/RFP or permanent reporter, it was recorded as ‘null expression’.
dpr10-GAL4 was crossed to UAS-GFP together with MHC-GAL80 to prevent muscle GFP expression, because a high level of muscle GFP would mask NMJs and SN cell bodies. In addition, Dpr genes with muscle expression (dpr10 and dpr19) were not crossed to the permanent labeling reporter.
Examining expression of Dpr and DIP genes in glia and muscles
We examined expression of Dpr and DIP genes in glia and muscles with the G-TRACE reporter (Evans et al., 2009). We dissected six larvae from each cross and immunostained for GFP, RFP, HRP and Repo. Glial expression was confirmed by GFP/RFP colocalization with Repo. Muscle expression was confirmed by GFP/RFP-positive muscle nuclei. Although the cross to UAS-GFP/RFP and the permanent labeling line also showed muscle expression, the diffusible GFP signal impeded the clear distinction of muscle boundaries.
Hierarchical clustering using Dpr and DIP gene expression
To perform hierarchical clustering, the expression of Dpr and DIP genes were first converted to binary values of ‘0’ and ‘1’. Robust expression, including high expression and temporal expression, were considered as ‘1’, whereas medium and low expression, and gradient expression were considered as ‘0’. We reasoned that robust expression of Dpr and DIP genes may suggest more a significant role in the respective cell. Binary data were subjected to hierarchical analysis using Morpheus (Broad Institute) (Metric: Cosine Similarity; Method: Average). Figures were exported and color coded in Adobe Illustrator to indicate different types of MNs and SNs.
Bouton number and dual innervation counting
To quantify m6 and m7 NMJs in wild-type animals, we located Ib NMJs by DLG labeling and counted bouton number by HRP labeling. To measure the MN6-Ib or MN7-Ib NMJ sizes in GMR79H07-GAL4>GFP animals, we first looked for GFP colocalization with DLG to distinguish MN6-Ib and MN7-Ib. For example, if the major Ib arbor on m6 is GFP positive, then it is formed by MN6-Ib, and the GFP-negative boutons are formed by MN7-Ib. We then counted the bouton numbers of each Ib arbor by HRP labeling. Unpaired, two-tailed Student's t-test was used for comparison between two groups (followed by Welch's correction in cases of unequal variance, Prism 8). Error bars indicate s.e.m.
Supplementary Material
Acknowledgements
We thank the Drosophila Gene Disruption Project for generating MiMIC and CRIMIC insertion lines. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. The hybridomas 4F3 and 8D12 were developed by Corey Goodman, and obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. We thank Troy Littleton (MIT), Lawrence Zipursky (UCLA) and Michelle Arbeitman (FSU) for sharing fly lines. We also thank Kai Zinn, Edwin ‘Chip’ Ferguson, Richard Fehon, Ellie Heckscher, David Pincus, Martha Plutarco Jr and members from the Carrillo laboratory for valuable discussions and comments. Some figure components were created with BioRender.com.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.W., J.A., R.A.C.; Formal analysis: Y.W., M.L.-R.; Investigation: Y.W., M.L.-R., J.A., V.A., P.C.; Resources: Y.W., J.A., H.J.B., O.K.; Writing - original draft preparation: Y.W., R.A.C.; Writing - review and editing: Y.W., M.L.-R., J.A., H.J.B., O.K., R.A.C.; Supervision: R.A.C.; Funding acquisition: M.L.-R., H.J.B., R.A.C.
Funding
This work was supported by the National Institute of Neurological Disorders and Stroke (R01 NS123439 01), the National Science Foundation (IOS-2048080), and a University of Chicago Faculty Diversity Grant and Pilot Project Grant to R.A.C. and the National Institute of General Medical Sciences (T32 GM007183) and National Institute of Neurological Disorders and Stroke (F31NS120458) to M.L.-R. This work is also supported by funds from the University of Chicago Biological Science Division, Committee of Developmental Biology and Department of Molecular Genetics and Cellular Biology. Further support came from the Office of Research Infrastructure Programs, National Institutes of Health (awards R24 OD022005 and R24 OD031447 to H.J.B.). Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.200355.
References
- Aponte-Santiago, N. A., Ormerod, K. G., Akbergenova, Y. and Littleton, J. T. (2020). Synaptic plasticity induced by differential manipulation of tonic and phasic motoneurons in Drosophila. J. Neurosci. 40, 6270-6288. 10.1523/JNEUROSCI.0925-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley, J., Sorrentino, V., Lobb-Rabe, M., Nagarkar-Jaiswal, S., Tan, L., Xu, S., Xiao, Q., Zinn, K. and Carrillo, R. A. (2019). Transsynaptic interactions between IgSF proteins DIP-α and Dpr10 are required for motor neuron targeting specificity. eLife 8, e42690. 10.7554/eLife.42690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali, N., Lee, H.-K. P. and Zinn, K. (2022). Sticks and stones, a conserved cell surface ligand for the Type IIa RPTP Lar, regulates neural circuit wiring in Drosophila. Elife 11, e71469. 10.7554/eLife.71469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish, S., Nuss, S., Strunilin, I., Bao, S., Mukherjee, S., Jones, C. D. and Volkan, P. C. (2018). Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila. PLoS Genet. 14, e1007560. 10.1371/journal.pgen.1007560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillé, L., Frendo, J.-L. and Vincent, A. (2015). Hox control of Drosophila larval anatomy; the alary and thoracic alary-related muscles. Mech. Dev. 138, 170-176. 10.1016/j.mod.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Bate, M. (1990). The embryonic development of larval muscles in Drosophila. Dev. Camb. Engl. 110, 791-804. 10.1242/dev.110.3.791 [DOI] [PubMed] [Google Scholar]
- Bittern, J., Pogodalla, N., Ohm, H., Brüser, L., Kottmeier, R., Schirmeier, S. and Klämbt, C. (2020). Neuron–Glia Interaction in the Drosophila nervous system. Dev. Neurobiol. 81, 438-452. 10.1002/dneu.22737 [DOI] [PubMed] [Google Scholar]
- Bornstein, B., Meltzer, H., Adler, R., Alyagor, I., Berkun, V., Cummings, G., Reh, F., Keren–Shaul, H., David, E., Riemensperger, T.et al. (2021). Transneuronal Dpr12/DIP–δ interactions facilitate compartmentalized dopaminergic innervation of Drosophila mushroom body axons. EMBO J. 40, e105763. 10.15252/embj.2020105763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus, J., Skeath, J. B., Spana, E. P., Bossing, T., Technau, G. and Doe, C. Q. (1995). New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech. Dev. 53, 393-402. 10.1016/0925-4773(95)00454-8 [DOI] [PubMed] [Google Scholar]
- Brovero, S. G., Fortier, J. C., Hu, H., Lovejoy, P. C., Newell, N. R., Palmateer, C. M., Tzeng, R.-Y., Lee, P.-T., Zinn, K. and Arbeitman, M. N. (2021). Investigation of Drosophila fruitless neurons that express Dpr/DIP cell adhesion molecules. Elife 10, e63101. 10.7554/eLife.63101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo, R. A., Özkan, E., Menon, K. P., Nagarkar-Jaiswal, S., Lee, P.-T., Jeon, M., Birnbaum, M. E., Bellen, H. J., Garcia, K. C. and Zinn, K. (2015). Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins. Cell 163, 1770-1782. 10.1016/j.cell.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S., Park, Y., Kurleto, J. D., Jeon, M., Zinn, K., Thornton, J. W. and Özkan, E. (2019). Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence. Proc. Natl. Acad. Sci. USA 116, 201818631. 10.1073/pnas.1818631116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, A., Snow, P., Keshishian, H. and Hotta, Y. (1995). Fasciclin III as a synaptic target recognition molecule in Drosophila. Nature 374, 166-168. 10.1038/374166a0 [DOI] [PubMed] [Google Scholar]
- Choi, J. C., Park, D. and Griffith, L. C. (2004). Electrophysiological and morphological characterization of identified motor neurons in the drosophila third instar larva central nervous system. J. Neurophysiol. 91, 2353-2365. 10.1152/jn.01115.2003 [DOI] [PubMed] [Google Scholar]
- Cosmanescu, F., Katsamba, P. S., Sergeeva, A. P., Ahlsen, G., Patel, S. D., Brewer, J. J., Tan, L., Xu, S., Xiao, Q., Nagarkar-Jaiswal, S.et al. (2018). Neuron-subtype-specific expression, interaction affinities, and specificity determinants of DIP/Dpr cell recognition proteins. Neuron 100, 1385-1400.e6. 10.1016/j.neuron.2018.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgeon, M. and Desplan, C. (2019). Coordination between stochastic and deterministic specification in the Drosophila visual system. Science 366, eaay6727. 10.1126/science.aay6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, G. W., Schuster, C. M. and Goodman, C. S. (1997). Genetic analysis of the mechanisms controlling target selection: target-derived fasciclin II regulates the pattern of synapse formation. Neuron 19, 561-573. 10.1016/S0896-6273(00)80372-4 [DOI] [PubMed] [Google Scholar]
- Diao, F., Ironfield, H., Luan, H., Diao, F., Shropshire, W. C., Ewer, J., Marr, E., Potter, C. J., Landgraf, M. and White, B. H. (2015). Plug-and-play genetic access to Drosophila cell types using exchangeable exon cassettes. Cell Rep. 10, 1410-1421. 10.1016/j.celrep.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C. Q., Smouse, D. and Goodman, C. S. (1988). Control of neuronal fate by the Drosophila segmentation gene even-skipped. Nature 333, 376-378. 10.1038/333376a0 [DOI] [PubMed] [Google Scholar]
- Estacio-Gómez, A. and Díaz-Benjumea, F. J. (2013). Roles of Hox genes in the patterning of the central nervous system of Drosophila. Fly 8, 26-32. 10.4161/fly.27424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, C. J., Olson, J. M., Ngo, K. T., Kim, E., Lee, N. E., Kuoy, E., Patananan, A. N., Sitz, D., Tran, P., Do, M.-T.et al. (2009). G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods 6, 603-605. 10.1038/nmeth.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca, M. G., Phillis, R. W. and Budnik, V. (1994). The role of tinman, a mesodermal cell fate gene, in axon pathfinding during the development of the transverse nerve in Drosophila. Development 120, 2143-2152. 10.1242/dev.120.8.2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber, W. B., Jan, L. Y. and Jan, Y. N. (2002). Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129, 2867-2878. 10.1242/dev.129.12.2867 [DOI] [PubMed] [Google Scholar]
- Grueber, W. B., Ye, B., Yang, C.-H., Younger, S., Borden, K., Jan, L. Y. and Jan, Y.-N. (2007). Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development 134, 55-64. 10.1242/dev.02666 [DOI] [PubMed] [Google Scholar]
- Hattori, D., Chen, Y., Matthews, B. J., Salwinski, L., Sabatti, C., Grueber, W. B. and Zipursky, S. L. (2009). Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature 461, 644-648. 10.1038/nature08431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckscher, E. S., Zarin, A. A., Faumont, S., Clark, M. Q., Manning, L., Fushiki, A., Schneider-Mizell, C. M., Fetter, R. D., Truman, J. W., Zwart, M. F.et al. (2015). Even-Skipped+ interneurons are core components of a sensorimotor circuit that maintains left-right symmetric muscle contraction amplitude. Neuron 88, 314-329. 10.1016/j.neuron.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, B. and Chiba, A. (2001). Single-cell analysis of Drosophila larval neuromuscular synapses. Dev. Biol. 229, 55-70. 10.1006/dbio.2000.9983 [DOI] [PubMed] [Google Scholar]
- Hong, W., Mosca, T. J. and Luo, L. (2012). Teneurins instruct synaptic partner matching in an olfactory map. Nature 484, 201-207. 10.1038/nature10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig, B. and Shapiro, L. (2020). Adhesion protein structure, molecular affinities, and principles of cell-cell recognition. Cell 181, 520-535. 10.1016/j.cell.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, J. E. (1986). Homeotic gene function in the muscles of Drosophila larvae. EMBO J. 5, 2321-2329. 10.1002/j.1460-2075.1986.tb04500.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaki, M., Shinza-Kameda, M., Ismat, A., Frasch, M. and Nose, A. (2010). Drosophila Tey represses transcription of the repulsive cue Toll and generates neuromuscular target specificity. Development 137, 2139-2146. 10.1242/dev.046672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, L. Y. and Jan, Y. N. (1982). Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc. Natl. Acad. Sci. USA 79, 2700-2704. 10.1073/pnas.79.8.2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jontes, J. D. (2017). The cadherin superfamily in neural circuit assembly. Csh. Perspect Biol. 10, a029306. 10.1101/cshperspect.a029306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanca, O., Zirin, J., Garcia-Marques, J., Knight, S. M., Yang-Zhou, D., Amador, G., Chung, H., Zuo, Z., Ma, L., He, Y.et al. (2019). An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms. Elife 8, e51539. 10.7554/eLife.51539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. D., Wen, Y. and Jan, Y.-N. (2009). Patterning and organization of motor neuron dendrites in the Drosophila larva. Dev. Biol. 336, 213-221. 10.1016/j.ydbio.2009.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose, H., Rose, D., Zhu, X. and Chiba, A. (1997). Homophilic synaptic target recognition mediated by immunoglobulin-like cell adhesion molecule Fasciclin III. Development 124, 4143-4152. 10.1242/dev.124.20.4143 [DOI] [PubMed] [Google Scholar]
- Kottmeier, R., Bittern, J., Schoofs, A., Scheiwe, F., Matzat, T., Pankratz, M. and Klämbt, C. (2020). Wrapping glia regulates neuronal signaling speed and precision in the peripheral nervous system of Drosophila. Nat. Commun. 11, 4491. 10.1038/s41467-020-18291-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmangaliyev, Y. Z., Yoo, J., Valdes-Aleman, J., Sanfilippo, P. and Zipursky, S. L. (2020). Transcriptional programs of circuit assembly in the Drosophila visual system. Neuron 108, 1045-1057.e6. 10.1016/j.neuron.2020.10.006 [DOI] [PubMed] [Google Scholar]
- Kurusu, M., Cording, A., Taniguchi, M., Menon, K., Suzuki, E. and Zinn, K. (2008). A screen of cell-surface molecules identifies leucine-rich repeat proteins as key mediators of synaptic target selection. Neuron 59, 972-985. 10.1016/j.neuron.2008.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf, M. and Thor, S. (2006). Development of Drosophila motoneurons: Specification and morphology. Semin. Cell Dev. Biol. 17, 3-11. 10.1016/j.semcdb.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Landgraf, M., Bossing, T., Technau, G. M. and Bate, M. (1997). The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J. Neurosci. 17, 9642-9655. 10.1523/JNEUROSCI.17-24-09642.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf, M., Sánchez-Soriano, N., Technau, G. M., Urban, J. and Prokop, A. (2003a). Charting the Drosophila neuropile: a strategy for the standardised characterisation of genetically amenable neurites. Dev. Biol. 260, 207-225. 10.1016/S0012-1606(03)00215-X [DOI] [PubMed] [Google Scholar]
- Landgraf, M., Jeffrey, V., Fujioka, M., Jaynes, J. B. and Bate, M. (2003b). Embryonic origins of a motor system: motor dendrites form a myotopic map in Drosophila. PLoS Biol. 1, e41. 10.1371/journal.pbio.0000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P.-T., Zirin, J., Kanca, O., Lin, W.-W., Schulze, K. L., Li-Kroeger, D., Tao, R., Devereaux, C., Hu, Y., Chung, V.et al. (2018). A gene-specific T2A-GAL4 library for Drosophila. Elife 7, e35574. 10.7554/eLife.35574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2020). Single–cell RNA sequencing in Drosophila: technologies and applications. Wiley Interdiscip Rev. Dev. Biol. 10, e396. 10.1002/wdev.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.-H., Kroll, J. R., Lennox, S. M., Ogundeyi, O., Jeter, J., Depasquale, G. and Truman, J. W. (2014). A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Rep. 8, 897-908. 10.1016/j.celrep.2014.06.065 [DOI] [PubMed] [Google Scholar]
- Lnenicka, G. A. and Keshishian, H. (2000). Identified motor terminals in Drosophila larvae show distinct differences in morphology and physiology. J. Neurobiol. 43, 186-197. [DOI] [PubMed] [Google Scholar]
- Logan, J., Falck-Pedersen, E., Darnell, J. E. and Shenk, T. (1987). A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl. Acad. Sci. USA 84, 8306-8310. 10.1073/pnas.84.23.8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D., Przybylski, D., Abruzzi, K. C., Schlichting, M., Li, Q., Long, X. and Rosbash, M. (2021). A transcriptomic taxonomy of Drosophila circadian neurons around the clock. Elife 10, e63056. 10.7554/eLife.63056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod, G. T., Suster, M. L., Charlton, M. P. and Atwood, H. L. (2003). Single neuron activity in the Drosophila larval CNS detected with calcium indicators. J. Neurosci. Meth. 127, 167-178. 10.1016/S0165-0270(03)00127-4 [DOI] [PubMed] [Google Scholar]
- Mauss, A., Tripodi, M., Evers, J. F. and Landgraf, M. (2009). Midline signalling systems direct the formation of a neural map by dendritic targeting in the Drosophila motor system. PLoS Biol. 7, e1000200. 10.1371/journal.pbio.1000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer, S., Yadav, S., Lee, J., Soba, P., Younger, S. H., Jin, P., Zhang, W., Parrish, J., Jan, L. Y. and Jan, Y.-N. (2016). Epidermis-derived semaphorin promotes dendrite self-avoidance by regulating dendrite-substrate adhesion in Drosophila sensory neurons. Neuron 89, 741-755. 10.1016/j.neuron.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, J. L. and Heckscher, E. S. (2020). Development of motor circuits: from neuronal stem cells and neuronal diversity to motor circuit assembly. Curr. Top. Dev. Biol. 142, 409-442. 10.1016/bs.ctdb.2020.11.010 [DOI] [PubMed] [Google Scholar]
- Menon, K. P., Carrillo, R. A. and Zinn, K. (2013). Development and plasticity of the Drosophila larval neuromuscular junction. Wiley Interdiscip Rev. Dev. Biol. 2, 647-670. 10.1002/wdev.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, K. P., Kulkarni, V., Takemura, S., Anaya, M. and Zinn, K. (2019). Interactions between Dpr11 and DIP-γ control selection of amacrine neurons in Drosophila color vision circuits. Elife 8, e48935. 10.7554/eLife.48935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, D. J. and Murphey, R. K. (1992). Projections of leg proprioceptors within the CNS of the fly Phormia in relation to the generalized insect ganglion. J. Comp. Neurol. 322, 16-34. 10.1002/cne.903220103 [DOI] [PubMed] [Google Scholar]
- Merritt, D. and Whitington, P. (1995). Central projections of sensory neurons in the Drosophila embryo correlate with sensory modality, soma position, and proneural gene function. J. Neurosci. 15, 1755-1767. 10.1523/JNEUROSCI.15-03-01755.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, S. K., Martins, A., Zhang, K. X., Graveley, B. R. and Zipursky, S. L. (2013). Probabilistic splicing of Dscam1 establishes identity at the level of single neurons. Cell 155, 1166-1177. 10.1016/j.cell.2013.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey, R. K., Possidente, D., Pollack, G. and Merritt, D. J. (1989). Modality–specific axonal projections in the CNS of the flies Phormia and Drosophila. J. Comp. Neurol. 290, 185-200. 10.1002/cne.902900203 [DOI] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal, S., Lee, P.-T., Campbell, M. E., Chen, K., Anguiano-Zarate, S., Gutierrez, M. C., Busby, T., Lin, W.-W., He, Y., Schulze, K. L.et al. (2015a). A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife 4, e05338. 10.7554/eLife.05338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal, S., DeLuca, S. Z., Lee, P.-T., Lin, W.-W., Pan, H., Zuo, Z., Lv, J., Spradling, A. C. and Bellen, H. J. (2015b). A genetic toolkit for tagging intronic MiMIC containing genes. Elife 4, e08469. 10.7554/eLife.08469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern, A., Pfeiffer, B. D. and Rubin, G. M. (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112, E2967-E2976. 10.1073/pnas.1506763112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. H., Vicidomini, R., Choudhury, S. D., Coon, S. L., Iben, J., Brody, T. and Serpe, M. (2021). Single–cell RNA sequencing analysis of the drosophila larval ventral cord. Curr. Protoc. 1, e38. 10.1002/cpz1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose, A. (2012). Generation of neuromuscular specificity in Drosophila: novel mechanisms revealed by new technologies. Front Mol Neurosci 5, 62. 10.3389/fnmol.2012.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose, A., Mahajan, V. B. and Goodman, C. S. (1992). Connectin: a homophilic cell adhesion molecule expressed on a subset of muscles and the motoneurons that innervate them in Drosophila. Cell 70, 553-567. 10.1016/0092-8674(92)90426-D [DOI] [PubMed] [Google Scholar]
- Nose, A., Umeda, T. and Takeichi, M. (1997). Neuromuscular target recognition by a homophilic interaction of connectin cell adhesion molecules in Drosophila. Development 124, 1433-1441. 10.1242/dev.124.8.1433 [DOI] [PubMed] [Google Scholar]
- Orgogozo, V. and Grueber, W. B. (2005). FlyPNS, a database of the Drosophila embryonic and larval peripheral nervous system. BMC Dev. Biol. 5, 4. 10.1186/1471-213X-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan, E., Carrillo, R. A., Eastman, C. L., Weiszmann, R., Waghray, D., Johnson, K. G., Zinn, K., Celniker, S. E. and Garcia, K. C. (2013). An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154, 228-239. 10.1016/j.cell.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Moreno, J. J. and O'Kane, C. J. (2018). GAL4 drivers specific for type Ib and type is motor neurons in Drosophila. G3 (Bethesda) 9: 453-462. 10.1534/g3.118.200809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponton, F., Chapuis, M.-P., Pernice, M., Sword, G. A. and Simpson, S. J. (2011). Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57, 840-850. 10.1016/j.jinsphys.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Rose, D., Zhu, X., Kose, H., Hoang, B., Cho, J. and Chiba, A. (1997). Toll, a muscle cell surface molecule, locally inhibits synaptic initiation of the RP3 motoneuron growth cone in Drosophila. Development 124, 1561-1571. 10.1242/dev.124.8.1561 [DOI] [PubMed] [Google Scholar]
- Sanes, J. R. and Zipursky, S. L. (2020). Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell 181, 536-556. 10.1016/j.cell.2020.04.008 [DOI] [PubMed] [Google Scholar]
- Schmid, A., Chiba, A. and Doe, C. Q. (1999). Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development 126, 4653-4689. 10.1242/dev.126.21.4653 [DOI] [PubMed] [Google Scholar]
- Schrader, S. and Merritt, D. J. (2000). Central projections of Drosophila sensory neurons in the transition from embryo to larva. J. Comp. Neurol. 425, 34-44. [DOI] [PubMed] [Google Scholar]
- Shishido, E., Takeichi, M. and Nose, A. (1998). Drosophila synapse formation: regulation by transmembrane protein with leu-rich repeats, CAPRICIOUS. Science 280, 2118-2121. 10.1126/science.280.5372.2118 [DOI] [PubMed] [Google Scholar]
- Simon, F. and Konstantinides, N. (2021). Single-cell transcriptomics in the Drosophila visual system: Advances and perspectives on cell identity regulation, connectivity, and neuronal diversity evolution. Dev. Biol. 479, 107-122. 10.1016/j.ydbio.2021.08.001 [DOI] [PubMed] [Google Scholar]
- Sink, H. and Whitington, P. (1991a). Pathfinding in the central nervous system and periphery by identified embryonic Drosophila motor axons. Development (Cambridge, England) 112, 307-316. 10.1242/dev.112.1.307 [DOI] [PubMed] [Google Scholar]
- Sink, H. and Whitington, P. M. (1991b). Location and connectivity of abdominal motoneurons in the embryo and larva of Drosophila melanogaster. J. Neurobiol. 22, 298-311. 10.1002/neu.480220309 [DOI] [PubMed] [Google Scholar]
- Soba, P., Zhu, S., Emoto, K., Younger, S., Yang, S.-J., Yu, H.-H., Lee, T., Jan, L. Y. and Jan, Y.-N. (2007). Drosophila sensory neurons require dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 54, 403-416. 10.1016/j.neuron.2007.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak-Workman, A. L., Vignali, K. M. and Vignali, D. A. A. (2012). Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb. Protoc. 2012, 199-204. 10.1101/pdb.ip067876 [DOI] [PubMed] [Google Scholar]
- Takizawa, E., Komatsu, A. and Tsujimura, H. (2007). Identification of common excitatory motoneurons in Drosophila melanogaster larvae. Zool. Sci. 24, 504-513. 10.2108/zsj.24.504 [DOI] [PubMed] [Google Scholar]
- Tan, L., Zhang, K. X., Pecot, M. Y., Nagarkar-Jaiswal, S., Lee, P.-T., Takemura, S., McEwen, J. M., Nern, A., Xu, S., Tadros, W.et al. (2015). Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell 163, 1756-1769. 10.1016/j.cell.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor, S. and Thomas, J. B. (1997). The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron 18, 397-409. 10.1016/S0896-6273(00)81241-6 [DOI] [PubMed] [Google Scholar]
- Valdes-Aleman, J., Fetter, R. D., Sales, E. C., Heckman, E. L., Venkatasubramanian, L., Doe, C. Q., Landgraf, M., Cardona, A. and Zlatic, M. (2021). Comparative connectomics reveals how partner identity, location, and activity specify synaptic connectivity in Drosophila. Neuron 109, 105-122.e7. 10.1016/j.neuron.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veling, M. W., Li, Y., Veling, M. T., Litts, C., Michki, N., Liu, H., Ye, B. and Cai, D. (2019). Identification of neuronal lineages in the drosophila peripheral nervous system with a “Digital” multi-spectral lineage tracing system. Cell Rep. 29, 3303-3312.e3. 10.1016/j.celrep.2019.10.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubramanian, L., Guo, Z., Xu, S., Tan, L., Xiao, Q., Nagarkar-Jaiswal, S. and Mann, R. S. (2019). Stereotyped terminal axon branching of leg motor neurons mediated by IgSF proteins DIP-α and Dpr10. Elife 8, e42692. 10.7554/eLife.42692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken, K. J. T., Schulze, K. L., Haelterman, N. A., Pan, H., He, Y., Evans-Holm, M., Carlson, J. W., Levis, R. W., Spradling, A. C., Hoskins, R. A.et al. (2011). MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8, 737-743. 10.1038/nmeth.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicidomini, R., Nguyen, T. H., Choudhury, S. D., Brody, T. and Serpe, M. (2021). Assembly and exploration of a single cell atlas of the drosophila larval ventral cord. Identification of rare cell types. Curr. Protoc. 1, e37. 10.1002/cpz1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Ma, X., Yang, J. S., Zheng, X., Zugates, C. T., Lee, C.-H. J. and Lee, T. (2004). Transmembrane/juxtamembrane domain-dependent dscam distribution and function during mushroom body neuronal morphogenesis. Neuron 43, 663-672. 10.1016/j.neuron.2004.06.033 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Lobb-Rabe, M., Ashley, J., Anand, V. and Carrillo, R. A. (2021). Structural and functional synaptic plasticity induced by convergent synapse loss in the Drosophila neuromuscular circuit. J. Neurosci. 41, 1401-1417. 10.1523/JNEUROSCI.1492-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg, M. L., Mitchell, K. J. and Goodman, C. S. (1998). Genetic analysis of the mechanisms controlling target selection: complementary and combinatorial functions of netrins, semaphorins, and IgCAMs. Cell 93, 581-591. 10.1016/S0092-8674(00)81187-3 [DOI] [PubMed] [Google Scholar]
- de Wit, J., and Ghosh, A. (2016). Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 17, 4. 10.1038/nrn.2015.3 [DOI] [PubMed] [Google Scholar]
- Xie, Q., Brbic, M., Horns, F., Kolluru, S. S., Jones, R. C., Li, J., Reddy, A. R., Xie, A., Kohani, S., Li, Z.et al. (2021). Temporal evolution of single-cell transcriptomes of Drosophila olfactory projection neurons. Elife 10, e63450. 10.7554/eLife.63450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., Xiao, Q., Cosmanescu, F., Sergeeva, A. P., Yoo, J., Lin, Y., Katsamba, P. S., Ahlsen, G., Kaufman, J., Linaval, N. T.et al. (2018). Interactions between the Ig-superfamily proteins DIP-α and Dpr6/10 regulate assembly of neural circuits. Neuron 100, 1369-1384.e6. 10.1016/j.neuron.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C., Theisen, E., Maloney, R., Peng, J., Santiago, I., Yapp, C., Werkhoven, Z., Rumbaut, E., Shum, B., Tarnogorska, D.et al. (2019). Control of synaptic specificity by establishing a relative preference for synaptic partners. Neuron 103, 865-877.e7. 10.1016/j.neuron.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim, K., Petri, J., Kottmeier, R. and Klämbt, C. (2019). Drosophila glia: few cell types and many conserved functions. Glia 67, 5-26. 10.1002/glia.23459 [DOI] [PubMed] [Google Scholar]
- Yoshihara, M., Rheuben, M. B. and Kidokoro, Y. (1997). Transition from growth cone to functional motor nerve terminal in Drosophila embryos. J. Neurosci. 17, 8408-8426. 10.1523/JNEUROSCI.17-21-08408.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarin, A. A. and Labrador, J.-P. (2019). Motor axon guidance in Drosophila. Semin. Cell Dev. Biol. 85, 36-47. 10.1016/j.semcdb.2017.11.013 [DOI] [PubMed] [Google Scholar]
- Zarin, A. A., Mark, B., Cardona, A., Litwin-Kumar, A. and Doe, C. Q. (2019). A multilayer circuit architecture for the generation of distinct locomotor behaviors in Drosophila. Elife 8, e51781. 10.7554/eLife.51781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, X.-L., Clemens, J. C., Neves, G., Hattori, D., Flanagan, J. J., Hummel, T., Vasconcelos, M. L., Chess, A. and Zipursky, S. L. (2004). Analysis of dscam diversity in regulating axon guidance in drosophila mushroom bodies. Neuron 43, 673-686. 10.1016/j.neuron.2004.07.020 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Rigo, F. and Martinson, H. G. (2015). Poly(A) signal-dependent transcription termination occurs through a conformational change mechanism that does not require cleavage at the poly(A) site. Mol. Cell 59, 437-448. 10.1016/j.molcel.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Zinn, K. and Özkan, E. (2017). Neural immunoglobulin superfamily interaction networks. Curr. Opin. Neurobiol. 45, 99-105. 10.1016/j.conb.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.