Abstract
Twelve wild collections and one commercial strain were used to characterize breeding systems and to develop molecular identities in the Arvenses section of the genus Agaricus, which includes the “horse mushroom” A. arvensis. Two morphotypes were identified based on macro- and micromorphological features. However, not all collections could be delimited by conventional taxonomic characters. Sequencing of the small subunit intergenic spacer (ITS) region (368 to 370 bp) of the rRNA genes clearly resolved the 13 collections into two clusters consistent with the identified morphotypes. Single-spore progenies and mating type testers were established and used to test intra- and interstock compatibility. The two compatibility groups identified were consistent with ITS clusters. Compatibility group I stocks readily interbred within the constraints of a unifactorial heterothallic system with a multiallelic mating type factor. Compatibility group II had a more restricted breeding pattern, and interactions were difficult to predict on the basis of mating type. Morphological data, ITS sequences, and the ability to interbreed suggest that these collections are part of a complex of interrelated species. Single-spore, homokaryotic isolates from both compatibility groups were able to fruit in compost culture, and two of the collections may represent natural homokaryotic fruiting. We conclude that species from the section Arvenses have versatile unifactorial heterothallic life cycles that permit both interbreeding and homokaryotic fruiting.
The genus Agaricus has a worldwide distribution, with up to 90 species recorded in Europe (2, 10) and more than 40 species recorded in the United Kingdom (13). Estimates for the worldwide totals of Agaricus species vary, but are likely to exceed 200. These species, which include the cultivated white button mushroom Agaricus bisporus (Lange) Imbach, exhibit a variety of breeding systems. Examples of both homothallic and heterothallic life cycles have been found (7, 20). Mushrooms from the Arvenses section of Flavescentes (10) have medium to large white sporophores with a yellowing surface, double pendent annular ring, and an aniseed or almond-like odor (10, 28). The section Arvenses contains 19 defined species within six subgroups: Aestivalis, Arvensis, Augustus, Macrosporus, Spissicaulis, and Sylvicola (10). These morphologically similar species include A. sylvicola (Vitt.) Sacc., the wood mushroom; A. arvensis Schff. ex. Fr., the horse mushroom; A. nivescens (Möll.) Möll.; A. macrocarpus (Möll.) Pilat; and A. essettei Bon. (syn. A. abruptibulbus Peck sensu auct. europ) (10). Several species from the Arvenses section have commercial potential (23, 25–27). Morphological features used to distinguish taxa within the Arvenses section include sporophore color, the presence of a “cogwheel” structure on the lower surface of the double ring, and a swollen base to the stipe (e.g., A. abrubtibulbus), the size of spores and/or cheilocystidia, and habitat of the specimen (10, 15, 28).
Identification within the Arvenses section is problematic. Single-spore isolates from a collection identified as A. arvensis produced mating reactions among single-spore progeny, but were later shown to interact with several other Agaricus collections, including A. bitorquis (1, 43). The tentative nomenclature of such collections in previously published work, which does not include detailed taxonomic data, should therefore be regarded with caution.
Breeding systems within the section Arvenses are poorly defined, and there has been considerable disparity in reports of mating interactions and self-fertility of single-spore progeny. In some cases, authors have described heterothallic behavior with matings between single-spore isolates (17, 18, 43, 50) and fruiting of heterokaryons in compost culture (19, 50). With other collections, successful pairings could not be established (1, 17, 43), and occasional fruiting of single-spore isolates was described (17, 50). It was hypothesized that the partial fruiting response of single-spore progeny could represent homokaryotic fruiting of a heterothallic species (17).
Nuclear numbers have been used to differentiate homokaryons and heterokaryons. For example, heterokaryons of Agaricus bitorquis (Quel.) Sacc. are predominately binucleate, while homokaryons are multinucleate (30, 42). However, there is some disparity over the utility of this character in collections from the section Arvenses (30, 50).
In this article, we describe molecular, morphological, and interbreeding characteristics of 12 wild collections from the section Arvenses and a commercial “horse mushroom” strain. We demonstrate that the measurement of morphological features is insufficient to fully characterize collections. Our results confirm the predominance of heterothallic breeding systems in the section Arvenses and test the hypothesis that self-fertility of single-spore progeny is the result of homokaryotic fruiting.
MATERIALS AND METHODS
Mycological media.
Agaricus cultures were maintained on either complete yeast extract medium (CYM) (44) or a compost extract medium (CE/CYM) which was prepared by a modification of the procedure described by Xu et al. (56). Fresh pasteurized mushroom compost was heated for 3 to 4 h at 120°C in thin layers to assist rapid drying. Dried compost was ground to a fine powder with a Cyclotec 1093 sample mill (Foss UK Ltd., Didcot, United Kingdom). To prepare compost extract, 138 g of dried powdered compost was placed in 1 liter of boiling water and simmered for 1 h. After adjustment for water loss, the extract was filtered through Miracloth (Calbiochem-Novabiochem Co., Nottingham, United Kingdom) and then centrifuged at 4,000 × g for 20 min at 5°C. The supernatant was stored at −20°C. To prepare CE/CYM, 200 ml of compost extract was added to 20 ml of 5× CYM stock and made up to 1 liter with ultrapure water (ca. 18 MΩ). Media were autoclaved for 20 min at 121°C and, where appropriate, solidified by the addition of 1.2% (wt/vol) technical agar no. 3 (Oxoid Ltd., Basingstoke, United Kingdom).
Agaricus collections: macro- and micromorphology.
Specimens from the Arvenses section were collected from a range of habitats and localities in the United Kingdom, continental Europe, and North America (Table 1). Mycelial cultures were established through tissue culture of wild fruit bodies and from a grain spawn culture for the commercial strain R20 (Sylvan, Ltd., Peterborough, United Kingdom). Cultures were incubated at 25°C for mycelial growth and basidiospore germination. Single-spore isolates were obtained by either dilution on CYM (39) or micromanipulation of basidiospores from gill surfaces (17). Spore germination was stimulated in the presence of A. bisporus mycelia (16). Cultures were maintained at 4°C (short-term storage) or immersed in liquid nitrogen (11).
TABLE 1.
Culture sources of wild and cultivated specimens from the Agaricus section Arvenses
| Collection | Collector | Location/origin | Date isolated | Habitat notes |
|---|---|---|---|---|
| AA0373 | M. Loftus | Meder St. Park, Santa Cruz, Calif. | December 1994 | Parkland grass |
| AA0390 | L. Gai | De La Veaga Park, Santa Cruz, Calif. | March 1996 | Parkland grass |
| AR1 | C. Ducatillon | Antibes, France | May 1995 | Beneath Sequoia sempervirens |
| R20 | Commercial “horse mushroom” strain | |||
| W3B | T. Elliott | Littlehampton, West Sussex, England | June 1974 | Grass, near Cupressus tree |
| W6I (ATCC 38044) | T. Elliott | Rustington, West Sussex, England | October 1975 | Edge of ploughed field, beneath Castanea |
| 93.7 | H. M. Grogan | Littlehampton, West Sussex, England | September 1993 | Leaf litter, beneath Pinus sylvestris |
| 93.9 | H. M. Grogan | Littlehampton, West Sussex, England | October 1993 | Leaf litter, beneath Cupressus |
| 93.10 | R. H. Gaze | Bedham, West Sussex, England | October 1993 | Composting grass clippings, upon turf |
| 94.1 | Z. Kwasek | Kielce, Poland | August 1994 | Grass, beneath Quercus robur |
| 94.22 | J. F. Smith | Welford-on-Avon, Warwick, England | October 1994 | Grass, beneath Cupressus |
| 94.31 | R. Noble | Ferring, West Sussex, England | November 1994 | Grass, beneath Sarothamnus scoparius |
| 94.33 | J. T. Fletcher | Olantigh, Kent, England | November 1994 | Beneath Castanea sativa |
Micromorphological observations to determine the numbers of spores per basidium and the dimensions of spores (n = 40) and cheilocystidia were performed as previously described (39). The statistical significance of differences in spore dimensions was determined by t tests.
Mating interactions.
Pairs of mating type testers were established through intrastock crosses and used for interstock compatibility analysis. Pairings between single-spore isolates were performed as previously described (17, 42), but with CE/CYM agar. Junction zone interactions were initially classified into one of three morphological types (56): (i) positive, with vigorous, highly dense, fluffy mycelia; (ii) cryptic, with distinct but less dense mycelia; and (iii) negative, with no visible interaction. Junction zone transfers were made from all positive, all cryptic, and some negative interactions. Visual comparisons of mycelial morphology and vigor were made between putative heterokaryons and their component single-spore isolates. Final classifications were based on the ability to isolate mycelia, which were morphologically distinct from the paired components. Pairings with reproducible visible interactions from which heterokaryons could not be isolated were classified as cryptic matings.
Fluorescence cytology.
Nuclei of homokaryons and heterokaryons were stained with 5 μg of bisbenzimide (Hoechst 33258) per ml as described by Kangatharalingam and Ferguson (31). Mycelia were grown onto the surface of glass coverslips on CE/CYM agar. For observation, coverslips were placed on slides, flooded with 100 μl of bisbenzimide, covered with a second slip, and held at ambient temperature in the dark for 15 min. Preparations were viewed with a Leitz (Wetzlar, Germany) Dialux 20 research microscope at a ×400 magnification, Ploemopak 2.4 vertical UV illuminator, and Leitz excitation filter block A (no. 513596). The numbers of nuclei from duplicate preparations were counted in 40 hyphal compartments. Differences in nuclear numbers were assessed with t tests.
Fruiting tests.
Grain spawn inocula and compost substrates were prepared as previously described (21, 39). Compost was inoculated with the appropriate culture grain spawn (2% [wt/wt]). Substrate was maintained in the dark at 25°C with 95% relative humidity and 0.4 to 0.5% CO2 in a controlled environment chamber until completely colonized. The compost was then covered with a peat-sugar beet lime (4:1 [vol/vol]) casing layer to a depth of ca. 30 mm. When mycelium became visible at the casing surface, environmental conditions were changed to favor initiation and fruit body development: air temperature, relative humidity, and CO2 concentration were reduced to levels of 16.5 to 17.5°C, 90 to 92%, and 0.06 to 0.7%, respectively. Cropping chambers were illuminated with white fluorescent tubular lamps (1.5 W/m2, 12-h day). The casing was maintained at 66 to 69% (wt/vol) moisture by regular, light watering at least every 4 days. Containers were cropped for up to 40 days. A minimum of two replicates was used for each fruiting test. At least five single-spore isolates were tested from each parental stock.
RAPD analysis.
Genomic DNA for randomly amplified polymorphic DNA (RAPD) analyses was isolated as previously described (12), purified with QIAprep8 mini-prep kits coupled with a QIAvac 6S manifold (Qiagen Ltd., Crawley, United Kingdom), and eluted in 1 mM Tris-HCl (pH 8).
Amplifications were performed in 40-μl volumes with an OmniGene thermal cycler (Hybaid, Ltd., Teddington, United Kingdom) with tube control and 1 of 10 oligonucleotides from Operon kit A (Operon Technologies, Inc., Alameda, Calif.): OPA01 (CAGGCCCTTC), OPA03 (AGTCAGCCAC), OPA05 (AGGGGTCTTG), OPA07 (GAAACGGGTG), OPA10 (GTGATCGCAG), OPA12 (TCGGCGATAG), OPA15 (TTCCGAACCC), OPA17 (GACCGCTTGT), OPA18 (AGGTGACCGT), or OPA20 (GTTGCGATCC). The reaction components were 5 μl of genomic DNA (ca. 75 ng), 25 ng of primer, 100 μM each deoxynucleoside triphosphate (dNTP; Finnzymes Oy, Espoo, Finland), and 1 U of DyNAzyme II DNA polymerase (Finnzymes Oy) with the supplied reaction buffer containing 1.5 mM MgCl2. The cycling conditions were a preliminary denaturation of 94°C for 1 min, followed by 35 cycles of 92°C for 1 min, 35°C for 1 min, and 72°C for 2 min. Negative water controls were performed in all cycling experiments. Products were separated on ethidium bromide-agarose gels (1.5% [wt/vol], 3 V/cm, 4 h) with molecular markers III and VI (125 to 21,226 bp; Roche Diagnostics, Ltd., Lewes, United Kingdom) and sized by the method of Schaffer and Sederoff (46).
ITS amplification and sequencing.
DNA for intergenic spacer (ITS) amplification was prepared by a modification of the method described by Screenivasaprasad (47). Actively growing mycelia were scratched from the surface of five 10-mm-diameter agar discs, placed into a microcentrifuge tube, frozen in liquid nitrogen, and ground with a plastic inoculation needle. Macerates were mixed with 100 μl of lysis buffer (200 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1 mM EDTA, and 1% [wt/vol] sodium dodecyl sulfate), frozen in liquid nitrogen, and subjected to three alternate, 1-min, freeze-boil treatments, with a final boiling for 10 min. Samples were microcentrifuged (11,000 × g, 15 min) at room temperature. Supernatants were purified with QIAquick PCR spin columns (Qiagen) according to the manufacturer's instructions. Genomic DNA was eluted in 50 μl of 1 mM Tris-HCl (pH 8) and stored at −20°C.
Ribosomal DNA (rDNA) repeat units (small and large subunits) were amplified as a single product by using extended versions of the ITS primers described by White et al. (55): ITS1extB (AACAAGGTTTCCGTAGGTGAACCTGC) and ITS4extA (TTCTTTTCCTCCGCTTATTGATATGC). Amplifications were performed in 50-μl volumes with the OmniGene and simulated tube control. The reaction components were 10 μl of genomic DNA (ca. 50 to 100 ng), 850 pmol of each primer, 40 μM each dNTP, and 1 U of DyNAzyme II polymerase with the 1.5 mM MgCl2 reaction buffer. The cycling conditions were a preliminary denaturation of 95°C for 2 min, followed by 25 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min and a final extension of 72°C for 5 min. Negative water controls were performed in all ITS amplifications. In addition to templates from the section Arvenses, rRNA genes were amplified from Coprinus cinereus (R348 [ATCC 96627]) and A. bisporus (W25) genomic DNAs prepared as described above. Amplified ITS products were purified with the QIAquick spin columns and eluted in 50 μl of 1 mM Tris-HCl. Band purity and DNA concentration were estimated by ethidium bromide staining and electrophoresis through 1.5% (wt/vol) agarose (45).
Double-stranded sequence was generated for the small subunit ITS region with the nested primers ITS1 and ITS2, described by White et al. (55). Cycle-sequencing reactions (20 μl) were performed with ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kits with AmpliTaq DNA polymerase FS (Applied Biosystems, Perkin-Elmer Corp., North Warrington, United Kingdom). Protocols were followed according to the manufacturer's instructions with 30 to 90 ng of template DNA, 6 μl of Terminator Ready Reaction mix, 3.2 pmol of primer, and 25 thermal cycles of 96°C for 30 s, 50°C for 15 s, and 60°C for 4 min, with a final hold at 20°C. Cycle sequencing products were purified by precipitation with ethanol and sodium acetate according to ABI recommendations. Sequencing gels were run by the Sequencing Services at the University of Durham, Durham, United Kingdom, or on an Applied Biosystems ABI 377 sequencer.
ITS data analysis.
Double-stranded DNA sequences were assembled by using the SeqMan II sequence analysis package (Lasergene software; DNAstar, Inc., Madison, Wis.). The MegAlign package (DNAstar) was used to prepare multiple sequence alignment files (MSF) via the Clustal V algorithm (29) and to calculate percent pairwise similarities. MSF alignments were analyzed by using the Distances (Jukes-Cantor algorithm) and GrowTree packages (UPGMA method) within the GCG Wisconsin suite (Genetics Computer Group, Inc., Madison, Wis.). Alternative distance algorithms and neighbor-joining methodologies were tested but found not to affect tree topologies. Distance trees were prepared by using TreeView (40). Bootstrap values (n = 1,000) were determined with the Clustal X package (52).
Nucleotide sequence accession number.
ITS sequence data from this program of work have been deposited within the EMBL database under accession no. AJ250588 to AJ250602.
RESULTS
Sporophore morphology and fruiting tests.
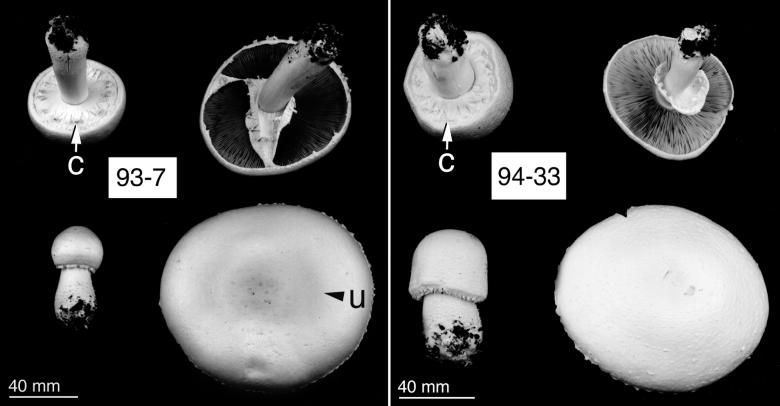
We observed two morphotypes. Type I, which included AA0373, AA0390, W3B, R20, 93.7, 93.9, and 93.10, had pale yellow pilei and distinct umbonate centers (Fig. 1). Morphotype II included AR1 and 94.33 and had uniformly white, nonumbonate pilei (Fig. 1). Four collections, W6I, 94.1, 94.22, and 94.31, had intermediate macromorphologies and could not be clearly categorized as either morphotype. The pileus diameters of all collections were generally 80 to 130 mm, with stipe lengths of 60 to 120 mm and stipe widths of 13 to 22 mm, swelling to 23 to 30 mm near the base. All of the collections had an aniseed odor and displayed distinct cogwheel formations of the lower annulus, which became pendent upon opening (Fig. 1). Lamellae were pale brown, turning to chocolate brown with age.
FIG. 1.
Macromorphology of Agaricus collections from section Arvenses as grown in compost culture. Morphotype I fruit bodies had pale yellow and, as indicated (u), distinctly umbonate pilei (e.g., 93.7). Morphotype II fruit bodies had uniform, white, nonumbonate pilei (e.g., 94.33). Lower annulus cogwheel formation is indicated (c). Note gross differences in immature sporophore morphology. The photographs of fungi were edited with PhotoShop (v. 4.0; Adobe Systems, Edinburgh, United Kingdom).
All wild collections and cultivated sporophores had four-spored basidia. Spore lengths were significantly (P < 0.01) greater in AA0390, R20, 93.7, 93.9, and 93.10 (all morphotype I) than for AR1, 94.33 (morphotype II), or 94.1 and 94.22 (intermediate morphotypes). Spore lengths in the 94.31 (parental), W6I, and the morphotype I AA0373 collection were intermediate (Table 2). Cheilocystidia of all collections were within the general range of 14 to 19 by 8 to 13 μm, but varied considerably within isolates. There were no significant differences between the micromorphologies of wild and cultivated fruited specimens or between parental isolates and single-spore progeny, except for single-spore isolate 94.31-5, which had significantly (P < 0.001) larger spores than the parental isolate, 94.31.
TABLE 2.
Spore dimensions in the Agaricus section Arvenses
| Wild collection or cultivated sporophore | Single-spore isolate | Mean length in μm (± SE) | Mean width in μm (± SE) |
|---|---|---|---|
| 93.7 (wild) | 7.8 (±0.10) | 5.5 (±0.09) | |
| 93.7 | 7.9 (±0.08) | 5.2 (±0.04) | |
| 93.9 (wild) | 8.2 (±0.07) | 5.3 (±0.06) | |
| 93.9 | 8.0 (±0.11) | 5.1 (±0.04) | |
| 93.9-1 | 7.8 (±0.04) | 5.2 (±0.08) | |
| 93.9-19 | 8.1 (±0.05) | 5.1 (±0.03) | |
| 93.10 (wild) | 8.0 (±0.10) | 5.4 (±0.08) | |
| 93.10 | 7.8 (±0.05) | 5.0 (±0.04) | |
| R20 | 7.5 (±0.06) | 5.1 (±0.05) | |
| AA0390 | 7.6 (±0.06) | 5.1 (±0.05) | |
| 94.33 | 6.2 (±0.03) | 5.1 (±0.03) | |
| AR1 | 6.4 (±0.06) | 4.9 (±0.06) | |
| 94.1 | 6.6 (±0.07) | 4.7 (±0.06) | |
| 94.22 | 6.6 (±0.05) | 5.0 (±0.05) | |
| 94.22-19 | 6.8 (±0.09) | 4.4 (±0.08) | |
| 94.31 | 7.2 (±0.08) | 4.6 (±0.07) | |
| 94.31-5 | 8.1 (±0.10) | 5.1 (±0.06) | |
| W6I | 7.2 (±0.05) | 4.8 (±0.04) | |
| W6I-148 | 7.3 (±0.04) | 4.9 (±0.03) | |
| AA0373 | 7.3 (±0.05) | 5.0 (±0.04) | |
| A. arvensisa | 6.5–8 | 4–5 | |
| A. essetteia | 6–8 | 4–5 | |
| A. macrocarpusa | 7–8 | 4.5–5 | |
| A. nivescensa | 5–7 | 4–5 | |
| A. sylvicolaa | 5–6 | 3–4 |
Spore data from Capelli (10) wild collections.
All of the parental collections fruited in compost culture. Single-spore progeny from AA0390, W3B, R20, 93.7, 93.9, and 93.10 were all self-fertile. Within collections AA0373, AR1, W6 I, 94.1, 94.22, 94.31, and 94.33, between 33 and 80% of single-spore progeny fruited in compost culture. Single-spore isolates yielded numbers of fruit bodies similar to those produced by the original parental collections and ranged from 1 to 10 sporophores per kg of compost.
Self-fertility of single-spore progeny implied homokaryotic or homothallic fruiting. We examined 20 single-spore progeny from a fruiting single-spore isolate of 93.7 (93.7a) with RAPDs, which have previously been used to characterize genetic variation in segregating single-spore progeny of heterothallic Agaricus species (8, 9). From 10 primers, we scored 53 RAPD products, ranging from 560 to 3,700 bp, but no polymorphisms were identified, and all 20 single-spore isolates were the same. When paired in all possible pairwise combinations, the 93.7a isolates were incompatible. The single-spore isolates of 93.7a mated with specific W3B testers in a pattern consistent with intrastock matings of parental isolate 93.7a and indicated that all of the progeny had the same mating type. In addition, six out of seven single-spore isolates from fruiting homokaryon R20-2 were self-fertile in compost culture. Single-spore isolates regularly produced fruiting initials when grown on CE/CYM agar, but these failed to differentiate into mature fruit bodies.
The self-fertility of single-spore isolates, lack of RAPD variation in sampled progeny, and the observed mating interactions from such isolates all support the hypothesis that fruiting was homokaryotic rather than homothallic.
Inter- and intrastock mating of single-spore isolates.
Mating type interactions within and between the 13 different stocks are summarized in Table 3. Mating type testers were readily established for all stocks except 94.1 and 94.22, in which none of the single-spore progeny were intracompatible. Comprehensive mating type designations were confounded (Table 3) by various pairings within and between collections that were predicted to give positive interactions but failed to do so. Specifically, in 93.7, one mating factor could not be differentiated between A3 and A4, while 94.22 and 94.33 each had a single factor that was not resolved. Summarized interactions (Table 3) were scored as positive if any positive pairings were observed. Full records of the mating data are available in reference 8. The 13 collections fell into two interstock compatibility groups (Table 3). In compatibility group I, all collections exhibited unrestricted interbreeding potential and were intercompatible with appropriate tester strains (Fig. 2). At least eight compatibility group I mating types were detected, and some of these were common to collections from both Europe and the United States (Table 3). In compatibility group II, interbreeding was restricted, and even though at least five mating types were detected, interstock crosses gave less predictable results. Some crosses were clearly positive (e.g., W6I paired with AR1 or 93.9 [Table 3]), while in other cases, either stable heterokaryons could not be isolated despite junction zone (cryptic) interactions, or the pairings were incompatible. Collections 94.1 and 94.33 were largely incompatible in interstock crosses, and only the latter formed stable heterokaryons in pairings with AR1.
TABLE 3.
Classification of inter- and intrastock matings between Agaricus section Arvenses single-spore isolates and assignment of mating types
| Compatibility classificationa
|
Collection (no. of single-spore isolates) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I (mating type)b
|
Group II (mating type)b
|
||||||||||||
| AA0390 (A3, A4) | AA0373 (A5, A6) | 94.31 (A2, A8) | W3B (A1, A2) | R20 (A5, A6) | 93.7 (A3/4, A7) | 93.10 (A1, A6) | AR1 (A2, A3) | 93.9 (A4) | W6I (A1, A2) | 94.22 (A5, A?) | 94.33 (A2, A?) | 94.1 (A4) | |
| * | + | + | + | + | + | + | + | + | ± | − | ± | ± | AA0390 (5) |
| * | + | + | + | + | + | ± | ± | ± | − | − | ± | AA0373 (8) | |
| * | + | + | + | + | − | + | − | − | − | − | 94.31 (10) | ||
| * | + | + | + | − | − | − | − | − | − | W3B (20) | |||
| * | + | + | + | ± | ± | ± | − | − | R20 (5) | ||||
| * | + | + | ± | ± | ± | − | ± | 93.7 (5) | |||||
| * | ± | ± | ± | ± | − | ± | 93.10 (8) | ||||||
| * | ± | + | ± | + | ± | AR1 (8) | |||||||
| = | + | ± | − | − | 93.9 (6) | ||||||||
| * | + | − | ± | W6I (20) | |||||||||
| * | ± | ± | 94.22 (8) | ||||||||||
| * | − | 94.33 (10) | |||||||||||
| = | 94.1 (8) | ||||||||||||
Intrastock classifications were compatible (*) or incompatible (=). Interstock classifications were positive (+) with isolation of stable heterokaryons, cryptic (±) where, although junction zone interactions were observed stable heterokaryons could not be established, or incompatible (−).
Mating type assignments are not equivalent in different compatibility groups.
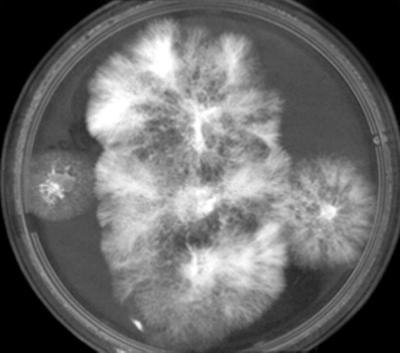
FIG. 2.
Comparison of heterokaryon (center) and component homokaryons AA0390-12 and R20-4 (left and right). The video image captured with Grabber software (v. 2.00; Phoretix International, Newcastle upon Tyne, United Kingdom) was edited with Adobe PhotoShop.
Stable heterokaryons within and between compatibility groups were confirmed by using RAPD markers, and examples can be found in reference 8. Some incompatibility reactions exhibited distinct barrage reactions at the junction zone (characterized by a browning and thinning of approaching mycelia). This reaction was most notable in pairings of W3B with AR1, W6I, and 93.9 and in some mating interactions between W6I and 93.10 or 94.1. One to 2 weeks after hyphal intermingling, the brown coloration became more intense.
Cytology.
Nuclear numbers were analyzed for five putative heterokaryons and their component homokaryons. Four of the heterokaryons (93.9 × AA0373, 93.9 × AA0390, 93.10 × W6I, and 94.22 × W6I) had significantly higher mean numbers of nuclei (8 to 12 per cell) than the component homokaryons (6 to 8 per cell). A fifth heterokaryon (93.10 × A158) did not have elevated nuclear numbers.
ITS sequences.
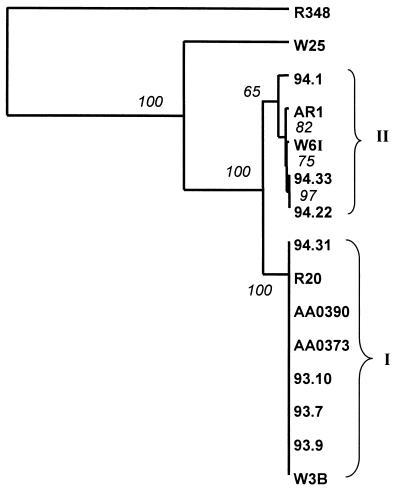
We compared small subunit ITS sequences of the 13 section Arvenses collections (Fig. 3). The percent identity between C. cinereus and A. bisporus sequences was 50.3%. With the species from section Arvenses and C. cinereus, identities ranged from 46% (e.g., 94.1) to 52% (e.g., W3B). Percent identities of A. bisporus with the Arvenses collections were higher, at between 81% (e.g., AA0390) and 82% (AR1).
FIG. 3.
Distance tree comparison of small subunit rDNA ITS regions from Agaricus section Arvenses collections. The tree was rooted by using a Coprinus cinereus (R348) outgroup. W25 is A. bisporus. Bootstrap values (in italics) are assigned to branch points. Cluster I values ranged between 57 and 69%.
Two principal clusters were resolved (Fig. 3). The largest cluster (I) comprised AA0373, AA0390, W3B, R20, 94.31, 93.9, 93.7, and 93.10. Sequences within cluster I were all 368 bp in length and were identical. Cluster II sequences (AR1, W6I, 94.1, 94.33, and 94.22) were all 370 bp in length and were 98% or more similar. Between the two clusters, up to 4.3% sequence variation was observed. Clusters I and II were in accord with compatibility groups, except for 93.9. Single-base-pair substitutions were observed along the length of the ITS sequences (40 to 315 bp). Changes were often, but not always, conserved within a cluster: e.g., three of the four cluster II strains exhibited GA insertions at 299 to 300 bp. A few changes were confined to individual strains: e.g., transitions G to A in W6I (308 bp) and T to C in 94.1 (100 bp). A single transversion, A to T, was observed within 94.1 (301 bp).
Data summary.
A comparison of morphotypes, compatibility groups, and ITS clusters is summarized in Table 4.
TABLE 4.
Summary of morphological features, compatibility, and ITS characterization of collections from the Agaricus section Arvenses
| Macromorphology | Micromorphology | Compatibility grouping | ITS sequence |
|---|---|---|---|
| Type I; pale yellow pilei, umbonate center; AA0373, AA0390, R20, W3B, 93.7, 93.9, 93.10 | Type I; greater spore length; AA0373, R20, 93.7, 93.9, 93.10 | Group I; high interbreeding; AA0373, AA0390, R20, W3B, 93.7, 93.10, 94.31 | Cluster I; identical 368-bp sequences; AA0373, AA0390, R20, W3B, 93.7, 93.9, 93.10, 94.31 |
| Type II; uniformly white, nonumbonate pilei; AR1, 94.33 | Type II; lesser spore length; AR1, 94.1, 94.22, 94.33 | Group II; restricted interbreeding; AR1, W6I, (93.9),a 94.1, 94.22, 94.33 | Cluster II; 370 bp; ≥98% similarity; AR1, W6I, 94.1, 94.22, 94.33 |
| Intermediate (W6I, 94.1, 94.22, 94.31)a | Intermediate (AA0373, W6I, 94.31)a |
Collections in parentheses were anomalous with regard to ITS clusters.
DISCUSSION
An analysis of macro- and micromorphological features in our 13 collections revealed two morphotypes. However, some stocks (W6I and 94.31) could not be unambiguously categorized by morphological criteria alone. Micro- and macromorphological measurements, which have been traditionally used in Agaricus taxonomy, are subject to variation and can be affected by environmental conditions (32, 49).
Sequences of small subunit ITS rDNAs also defined two clusters that were consistent with our morphotypes and placed W6I and 94.31 into groups I and II, respectively. These observations indicate the utility of molecular data to support, underpin, or even resolve taxonomic difficulties within Agaricus.
ITS regions tend to be highly polymorphic and have been used extensively in fungal taxonomic and phylogenetic studies (5, 55). Within the genus Agaricus, Bunyard et al. (6) used restriction analysis of 26S and 5S rRNA genes and the intergenic spacer regions to develop a phylogeny. We have previously found that restriction analysis could not detect polymorphisms in Agaricus ITS sequences (A. Richards and M. P. Challen, unpublished data). The restriction enzymes used could not discriminate between single-base changes characterized in this study, small changes which have previously been shown to be of significance in ITS differentiation of fungal taxa (38).
The ITS data reported here form part of an ongoing study of molecular phylogenies across the genus Agaricus. The sequence variation within different collections from the Arvenses section (ca. 4%) is higher than that seen between some discreet species of Agaricus, e.g., A. bisporus and A. subfloccosus (2%). Despite this divergence, diverse collections from the section Arvenses can still interbreed.
Single-spore progeny from the 13 collections were regularly self-fertile and could mate both within and between different stocks. Two compatibility groups were identified and, with the exception of 93.9, were consistent with the two morphological and ITS types. In compatibility group I, mating type testers interbred within the constraints of unifactorial mating type alleles. In compatibility group II, interbreeding was more restricted, and positive interactions could not be predicted simply on the basis of mating type alleles. Compatibility group II isolates often exhibited a barrage in incompatible pairings. Similar incompatibility interactions have been described for other Agaricus species (1, 17, 37).
In two wild collections (93.9 and 94.1), only a single mating type was identified. The behavior of these two collections was consistent with our observations on homokaryotic fruiting in a single-spore isolate, 93.7a. We believe that these two collections are naturally occurring self-fertile homokaryons. Homokaryotic fruiting was first described in Schizophyllum commune (53) and appears to be relatively widespread in the fungi (22, 51). The genetic basis of this trait in this basidiomycete is complex (33, 34), and considerable variability has been observed in natural populations (35). Our single-spore isolates regularly fruited in compost and also formed initials on agar, a feature consistent with homokaryotic fruiting (54). We previously described homokaryotic fruiting from both laboratory and wild collections of A. bitorquis (36), and it may be that the phenomenon is more widespread in the genus Agaricus than is generally appreciated. Homokaryotic fruiting followed by the loss of mating ability could occur in the evolution of homothallism. Such a process might, in part, account for the single disparity (isolate 93.9) between compatibility groups and ITS clusters.
Cytological observations of nuclear numbers were largely consistent with previous investigations of related species (30, 50). Elevated numbers of nuclei were observed in some, but not all, heterokaryons. Although there may be some regulation of nuclear numbers in the section Arvenses, it is far less clear than that described for A. bitorquis (30, 42).
The variability in morphology, ITS sequences, and mating behavior suggests that the Agaricus section Arvenses harbors morphologically and genetically diverse species. If sharing the same gene pool is the criterion for species status (14), then collections within compatibility group I are conspecific. The situation in compatibility group II is more complex. These collections have variable ITS sequences, show partial interbreeding, and had limited compatibility with group I stocks. Collections within group II could be regarded as individual species. Interpretation of the relationships between the collections is further complicated by the fact that, although mating reactions within and between collections are consistent and reproducible, they were restricted to various numbers of single-spore isolates. Perkins (41) discussed these issues and stressed the need for precision when describing interactions between sibling species and mating populations. An unresolved question remains about the interactions between the Arvenses collections that we describe here. Is the ability to form conspicuous junction zone heterokaryons indicative of conspecificity, or would sporophore production linked to meiotic segregation be a more realistic measure? For some of the collections, this distinction could only be determined by further protracted study.
The sexuality of the collections from section Arvenses as described here does not readily accord with the generalized homothallic and heterothallic life cycles of Blakeslee (4). Such disparities are not unique. Biggs (3) reported that the unifactorial Peniophora ludovinciana exhibited “abnormal” mating behavior; single-spore isolates were self-fertile in culture and were compatible with strains of other mating type specificities. The term “amphithallism,” introduced by Lange (24), does not adequately describe the mating behavior of Agaricus section Arvenses. In amphithallic species, both homothallic and heterothallic spore progeny are recovered from a single fruit body (48). Our study demonstrates that species from the Agaricus section Arvenses combine a trend for homokaryotic fruiting within a predominately unifactorial, heterothallic life cycle. This combination is a significant departure from previously defined life cycles for the Agaricus species. It allows recombination to generate variation, and yet enables rapid homokaryotic spore dispersal in appropriate environmental conditions. Such versatility provides unequivocal advantages.
ACKNOWLEDGMENTS
Some of the work described here formed part of the Ph.D. project for L. Calvo-Bado, who thanks C. F. Thurston, Kings College, London, for supervision. We thank Ruth Finch for help with sequencing at Horticultural Research International (HRI). We also thank Rongchun Li for cytological observations and Helen Grogan and Pat Edwards, who performed some of the macro- and micromorphological comparisons.
This work was funded by grants to HRI from MAFF, BBSRC, and the Horticultural Development Council. L.C.-B. thanks the Consejo Nacional de Ciencia y Tecnología, Mexico, for funding.
REFERENCES
- 1.Anderson J B, Petsche D M, Herr F B, Horgen P A. Breeding relationships among several species of Agaricus. Can J Bot. 1984;62:1884–1889. [Google Scholar]
- 2.Bas C. A short introduction to the ecology, taxonomy and nomenclature of the genus Agaricus. In: Van Griensven LJLD, editor. Genetics and breeding of Agaricus. Wageningen, The Netherlands: Pudoc; 1991. pp. 21–24. [Google Scholar]
- 3.Biggs R. Cultural studies in the Thelephoraceae and related fungi. Mycologia. 1938;30:64–78. [Google Scholar]
- 4.Blakeslee A F. Sexual reproduction in the Mucorinae. Proc Am Acad Sci. 1904;40:205–319. [Google Scholar]
- 5.Bridge P D, Arora D K. Interpretation of PCR methods for species definition. In: Bridge P D, Arora D K, Reddy C A, Elander R P, editors. Applications of PCR in mycology. London, United Kingdom: CAB International; 1998. pp. 63–84. [Google Scholar]
- 6.Bunyard B A, Nicholson M S, Royse D J. Phylogeny of the genus Agaricus inferred from restriction analysis of enzymatically amplified ribosomal DNA. Fungal Genet Biol. 1996;20:243–253. doi: 10.1006/fgbi.1996.0039. [DOI] [PubMed] [Google Scholar]
- 7.Callac P, Hocquart S, Imbernon M, Desmerger C, Olivier J-M. Bsn-t alleles from French field strains of Agaricus bisporus. Appl Environ Microbiol. 1998;64:2105–2110. doi: 10.1128/aem.64.6.2105-2110.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo-Bado L A. Sexuality in wild Agaricus. Classical and molecular analysis. Ph.D. thesis. London, United Kingdom: Kings College; 1999. [Google Scholar]
- 9.Calvo-Bado L, Challen M P, Elliott T J. Classical and molecular characterisation of the genus Agaricus. In: Van Griensven L J L D, Visser J, editors. Proceedings of the Fourth Meeting on the Genetics and Cellular Biology of the Basidiomycetes, Nijmegan, The Netherlands. Horst, The Netherlands: Mushroom Experimental Station; 1998. p. 163. [Google Scholar]
- 10.Capelli A. Agaricus. L.: Fr. (Psalliota Fr.). Saronno, Italy: Liberia editrice Bella Giovanna; 1984. [Google Scholar]
- 11.Challen M P, Elliott T J. Polypropylene straw ampoules for the storage of microorganisms in liquid nitrogen. J Microbiol Methods. 1986;5:11–23. [Google Scholar]
- 12.Challen M P, Martinez-Carrera D, Moore A J. Facile extraction and purification of filamentous fungal DNA. BioTechniques. 1995;18:975–978. [PubMed] [Google Scholar]
- 13.Dennis R W G, Orton P D, Hora F B. New check list of British agaric and boletes. Trans Br Mycol Soc Suppl. 1960;43:1–225. [Google Scholar]
- 14.Dobzhansky T. Mendelian populations and their evolution. Am Nat. 1950;84:401–418. [Google Scholar]
- 15.Edwards P J. Effects of the fairy ring fungus Agaricus arvensis on nutrient availability in grassland. New Phytol. 1988;110:377–381. [Google Scholar]
- 16.Elliott T J. Sex and the single spore. Mushroom Sci. 1972;8:11–18. [Google Scholar]
- 17.Elliott T J. Comparative sexuality in Agaricus species. J Gen Microbiol. 1978;107:113–122. [Google Scholar]
- 18.Elliott T J. Sexuality in the genus Agaricus. Mushroom Sci. 1979;10:41–50. [Google Scholar]
- 19.Elliott T J. Mushroom investigations: other Agaricus spp. Rep Glasshouse Crops Res Inst. 1981;1980:137. [Google Scholar]
- 20.Elliott T J. The genetics and breeding of species of Agaricus. In: Flegg P B, Spencer D M, Wood D A, editors. The biology and technology of the cultivated mushroom. Chichester, United Kingdom: John Wiley & Sons; 1985. pp. 111–129. [Google Scholar]
- 21.Elliott T J. Spawn-making and spawns. In: Flegg P B, Spencer D M, Wood D A, editors. The biology and technology of the cultivated mushroom. Chichester, United Kingdom: John Wiley & Sons; 1985. pp. 131–139. [Google Scholar]
- 22.Elliott T J. Developmental genetics—from spore to sporophore. In: Moore D, Casselton L A, Wood D A, Frankland J C, editors. Developmental biology of higher fungi. British Mycological Society Symposium 10. Cambridge, United Kingdom: Cambridge University Press; 1985. pp. 451–465. [Google Scholar]
- 23.Fermore T R. Agaricus macrosporus: an edible fungus with commercial potential. Sci Horticult. 1982;16:273–282. [Google Scholar]
- 24.Fincham J R S, Day P R. Genetic control of sexual development. In: Fincham J R S, Day P R, editors. Fungal genetics. 3rd ed. Oxford, England: Blackwell Scientific Publications; 1971. pp. 283–313. [Google Scholar]
- 25.Fritsche G. Tests on breeding with Agaricus arvensis. Mushroom Sci. 1979;10:91–101. [Google Scholar]
- 26.Fritsche G. Ontwikkelingswerk met de Akkerchampignon (Agaricus arvensis Schaeffer ex Secr) Champignon. 1989;33:7–13. [Google Scholar]
- 27.Gramss G. Ein champignon für den kommerziellen anbau auf mischsubstraten. Champignon. 1976;181:13–16. [Google Scholar]
- 28.Heinemann P. Essai d'une de determination des genres Agaricus et Macropsalliota. Sydowia. 1977;30:6–37. [Google Scholar]
- 29.Higgins D G, Sharp P M. CLUSTAL V: a package for performing multiple sequence alignment on a micro computer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 30.Hou H H, Elliott T J. Comparative cytology in the genus Agaricus. Mushroom Sci. 1979;10:51–62. [Google Scholar]
- 31.Kangatharalingam N, Ferguson W. A simple and rapid technique for fluorescence staining of fungal nuclei. Curr Microbiol. 1984;10:99–104. [Google Scholar]
- 32.Kerrigan R W. What's in a name? The chaetaceous case of the chaste champignon. Dev Crop Sci. 1987;10:141–154. [Google Scholar]
- 33.Leslie J F, Leonard T J. Monokaryotic fruiting in Schizophyllum commune: genetic control of the response to mechanical injury. Mol Gen Genet. 1979;175:5–12. [Google Scholar]
- 34.Leslie J F, Leonard T J. Three independent genetic systems that control initiation of a fungal fruiting body. Mol Gen Genet. 1979;175:257–260. [Google Scholar]
- 35.Leslie J F, Leonard T J. Monokaryotic fruiting in Schizophyllum commune: survey of a population from Wisconsin. Am Mid Nat. 1980;103:367–374. [Google Scholar]
- 36.Martinez-Carrera D, Challen M P, Smith J F, Elliott T J, Thurston C F. Homokaryotic fruiting in Agaricus bitorquis: a new approach. Mushroom Sci. 1995;14:37–44. [Google Scholar]
- 37.Martinez-Carrera D, Smith J F, Challen M P, Elliott T J, Thurston C F. Evolutionary trends in Agaricus bitorquis complex and their relevance for breeding. Mushroom Sci. 1995;14:29–36. [Google Scholar]
- 38.Muthumeenakshi S. Molecular taxonomy of the genus Trichoderma. Ph.D. thesis. Belfast, N. Ireland: The Queens University of Belfast; 1996. [Google Scholar]
- 39.Noble R, Grogan H M, Elliott T J. Variation in morphology, growth and fructification of isolates in the Agaricus subfloccosus complex. Mycol Res. 1995;99:1453–1461. [Google Scholar]
- 40.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 41.Perkins D D. How should the infertility of interspecies crosses be designated? Mycologia. 1994;86:758–761. [Google Scholar]
- 42.Raper C A. Sexuality and life-cycle of the edible, wild Agaricus bitorquis. J Gen Microbiol. 1976;95:54–66. [Google Scholar]
- 43.Raper C A, Kaye G. Sexual and other relationships in the genus Agaricus. J Gen Microbiol. 1978;105:135–151. [Google Scholar]
- 44.Raper C A, Raper J R, Miller R E. Genetic analysis of the life-cycle of Agaricus bisporus. Mycologia. 1972;64:1088–1117. [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Schaffer H E, Sederoff R R. Improved estimation of DNA fragment length from agarose gels. Anal Biochem. 1981;115:113–122. doi: 10.1016/0003-2697(81)90533-9. [DOI] [PubMed] [Google Scholar]
- 47.Screenivasaprasad, S. Isolation of fungal nucleic acids. In R. Rapley, and J. M. Walker (ed.), The nucleic acids handbook, in press. Humana Press, Totowa, N.Y.
- 48.Sequiera L. Nuclear phenomena in the basidia and basidiospores of Omphalia flavida. Mycologia. 1954;46:470–483. [Google Scholar]
- 49.Smith J F, Love M E. Investigations into the cultural requirements of a brown capped Agaricus strain (W4 II) isolated from Cupressus leaf litter. J Hortic Sci. 1995;70:963–974. [Google Scholar]
- 50.Sonnenberg A S M, Fritsche G. Cytological observations in Agaricus arvensis. Mushroom Sci. 1989;12:101–107. [Google Scholar]
- 51.Stahl U, Esser K. Genetics of fruit body production in higher basidiomycetes. I. Monokaryotic fruiting and its correlation with dikaryotic fruiting in Polyporus ciliatus. Mol Gen Genet. 1976;148:183–197. [Google Scholar]
- 52.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wakefield E M. Uber die bedingungen der fruchtkörperbildung, sowie das auftreten fertiler und steriler stamme bei hymenomyceten. Natwiss Zeitschr Forst Landwirt. 1909;7:521–551. [Google Scholar]
- 54.Wessels J G H. Fruiting in the higher fungi. Adv Microb Physiol. 1993;34:147–202. doi: 10.1016/s0065-2911(08)60029-6. [DOI] [PubMed] [Google Scholar]
- 55.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 56.Xu J, Horgen P A, Anderson J B. Media and temperature effects on mating interactions of Agaricus bisporus. Cult Mushroom Res Newsl. 1993;1:25–32. [Google Scholar]