Abstract
Background
Complete signal transducer and activator of transcription 1 (STAT1) deficiency causes a rare primary immunodeficiency that is characterized by defective IFN-dependent gene expression leading to life-threatening viral and mycobacterial infections early in life.
Objective
To characterize a novel STAT1 loss-of-function variant leading to pathological infection susceptibility and hyperinflammation.
Methods
Clinical, immunologic, and genetic characterization of a patient with severe infections and hemophagocytic lymphohistiocytosis–like hyperinflammation was investigated.
Results
We reported a child of consanguineous parents who presented with multiple severe viral infections that ultimately triggered hemophagocytic lymphohistiocytosis and liver failure. Despite intensified therapy with antivirals and cytomegalovirus-specific donor cells, the child died after hematopoietic stem cell transplantation because of cytomegalovirus reactivation with acute respiratory distress syndrome. Exome sequencing revealed a homozygous STAT1 variant (p.Val339ProfsTer18), leading to loss of STAT1 protein expression. Upon type I and type II IFN stimulation, immune and nonimmune cells showed defective upregulation of IFN-stimulated genes and increased susceptibility to viral infection in vitro. Increased viral infection rates were paralleled by hyperinflammatory ex vivo cytokine responses with increased production of TNF, IL-6, and IL-18.
Conclusions
Complete STAT1 deficiency is a devastating disorder characterized by severe viral infections and ensuing hyperinflammatory responses. Early diagnosis can be made by exome sequencing and variant validation by functional testing of STAT1-dependent programmed cell death 1 ligand 1 surface expression on monocytes. Furthermore, high awareness for hyperinflammatory complications and potential targeted treatment strategies such as IL-18 binding protein could be considered. Hematopoietic stem cell transplantation is the only definitive treatment strategy but remains challenging.
Key words: Primary immune deficiency, Hyperinflammation, STAT1, Hematopoietic stem cell transplantation, Hemophagocytic lymphohistiocytosis, LOF, PD-L1, CXCL10, IL-18
Abbreviations used: ALF, Acute liver failure; AR, Autosomal recessive; CMV, Cytomegalovirus; CXCL10, C-X-C motif chemokine 10; GOF, Gain of function; HD, Healthy donor; HLH, Hemophagocytic lymphohistiocytosis; HSCT, Hematopoietic stem cell transplantation; IFNAR, IFN-α/β receptor; IFNGR, IFN-γ receptor; ISG, IFN-stimulated gene; LOF, Loss of function; PAMP, Pathogen-associated molecular pattern; PD-L1, Programmed cell death 1 ligand 1; PIC, Polyinosinic:polycytidylic acid; PRR, Pattern recognition receptor; STAT, Signal transducer and activator of transcription; VSV, Vesicular stomatitis virus; VZV, Varicella zoster virus
What is already known about this topic? Complete signal transducer and activator of transcription 1 (STAT1) loss of function (LOF) leads to severe viral infections.
What does this article add to our knowledge? Homozygous autosomal-recessive c.1011_1012delAG mutation causes STAT1 LOF. We propose a pathophysiological model for the association of viral infections and hemophagocytic lymphohistiocytosis–like hyperinflammation in complete autosomal-recessive STAT1 LOF.
How does this study impact current management guidelines? Complete STAT1 LOF can be diagnosed by defective programmed cell death 1 ligand 1 upregulation on monocytes or C-X-C motif chemokine 10 secretion via ELISA after IFN stimulation in vitro. Early functional diagnosis of STAT1 LOF and targeted therapy may improve patient outcomes.
Introduction
Signal transducer and activator of transcription (STAT) 1 is a key transcription factor downstream of IFN-α/β receptor (IFNAR) and IFN-γ receptor (IFNGR) activation.1 , 2 Binding of type I IFN (IFN-I, such as IFN-α or IFN-β), or type II IFN (IFN-II, namely, IFN-γ), to their corresponding receptors leads to receptor dimerization, Janus kinase activation, and phosphorylation of STAT1. In the case of IFNGR activation, STAT1 homodimerizes, whereas IFNAR activation induces STAT1-STAT2-IFN regulatory factor 9 complex formation. Nuclear translocation of these transcription factor complexes results in the induction of IFN-stimulated genes (ISGs) leading to an effective inflammatory and antimicrobial response.3 , 4
Several gain-of-function (GOF) as well as loss-of-function (LOF) variants of STAT1 have been described. The most common clinical manifestation of STAT1 GOF variants is chronic mucocutaneous candidiasis.5 , 6 STAT1 LOF variants increase susceptibility to viral and mycobacterial infections because of impaired cellular responses to IFN-I and IFN-II. STAT1 LOF variants can be classified as partial or complete autosomal-recessive (AR) and autosomal-dominant.7 The immunodeficiency phenotypes caused by both partial and complete AR variants are similar; however, patients with partial AR variants experience a milder clinical course as compared with patients with complete AR variants.8, 9, 10
Complete AR STAT1 deficiency was first described in 2003, and a total of 7 cases have been reported.7 , 11, 12, 13, 14 Impairment of STAT1-dependent immune responses in these patients led to life-threatening infections in the first months of life. Mortality was high and mostly attributable to viral infections. Recently, Burns et al14 described 1 patient with complete AR STAT1 deficiency who presented with 2 episodes of hyperinflammation during viral infections. However, the pathophysiology of this clinical presentation was not further investigated. Allogeneic hematopoietic stem cell transplantation (HSCT) was reported to correct the otherwise lethal phenotype in 2 patients.13 , 14
Hemophagocytic lymphohistiocytosis (HLH) is a life-threatening condition, characterized by a pathological activation of the immune system, with massive release of proinflammatory cytokines such as TNF, IL-1β, IL-6, IL-18, and IFN-γ. HLH classically can be categorized into primary and secondary forms.15 Historically, primary HLH was principally associated with genetic defects of natural killer–and T-cell cytotoxicity typically manifesting in early childhood, whereas secondary HLH was considered to reflect a response to clear triggers such as viral infection in patients without such predisposition. In recent years, however, HLH has increasingly been recognized in the context of inborn errors of immunity other than cytolytic defects, often triggered by infection. Thus, exome sequencing has revealed numerous genes related to primary immunodeficiencies or primary immune dysregulation to be linked to HLH.16
Here, we report a patient with complete STAT1 deficiency who presented with severe viral infections and developed HLH-like hyperinflammation and lethal acute respiratory distress syndrome after cytomegalovirus (CMV) reactivation after HSCT. We confirm the pivotal role of STAT1 for IFN-I and IFN-II signaling in primary immune and nonimmune cells. We establish decreased MHC-I and programmed cell death 1 ligand 1 (PD-L1) surface expression on monocytes as well as decreased C-X-C motif chemokine 10 (CXCL10) secretion after IFN-α2a and IFN-γ stimulation as diagnostic tools for STAT1 deficiency. Furthermore, we propose a pathophysiological model for the association of viral infections and HLH-like hyperinflammation in complete AR STAT1 LOF.
Methods
Subjects
The patient's legal guardians gave written informed consent according to current ethical guidelines and the Declaration of Helsinki. The local ethics committee at the Ludwig-Maximilians-Universität, Munich, approved the study (project no. 381-11).
Exome sequencing
Exome sequencing was performed at the Dr von Hauner Children's Hospital Next Generation Sequencing facility.17 Genomic DNA from whole blood was used for preparation of whole-exome libraries using the SureSelect XT Human All Exon V6+UTR kit (Agilent Technologies, Ratingen, Germany) and subsequently sequenced with a NextSeq 500 platform (Illumina, San Diego, Calif) to an average coverage depth of 90×. Bioinformatic analysis used Burrows-Wheeler Aligner 0.7.15 (http://bio-bwa.sourceforge.net/), Genome Analysis ToolKit 3.6 (Broad Institute, Cambridge, MA), and Variant Effect Predictor 89 (European Bioinformatics Institute, Hinxton, United Kingdom). Variant alleles were filtered for frequency against public (eg, Genome Aggregation Database, Exome Aggregation Consortium, and Greater Middle East) and in-house databases. Potentially causative variants were confirmed by Sanger sequencing.
Isolation, cultivation, and stimulation of PBMCs and primary fibroblasts
PBMCs were isolated from heparinized blood samples by Biocoll (Biochrom, Berlin, Germany) density gradient centrifugation and cultivated in RPMI-1640 medium (Sigma-Aldrich, Taufkirchen, Germany), supplemented with 10% FCS, 2 mM l-glutamine, 100 U/L penicillin, and 0.1 mg/mL streptomycin (all Gibco, Thermo Fisher Scientific, Karlsruhe, Germany). Primary fibroblasts were cultivated in Dulbecco modified Eagle medium high-glucose media (Sigma-Aldrich), supplemented with 10% FCS, 2 mM l-glutamine, 100 U/L penicillin, and 0.1 mg/mL streptomycin. Cells were stimulated using 10,000 U/mL recombinant human IFN-α2a (Miltenyi Biotech, Bergisch Gladbach, Germany), 10,000 U/mL recombinant human IFN-γ (Peprotech Germany, Hamburg, Germany), 200 ng/mL LPSs from Escherichia coli O55:B5 (Sigma-Aldrich), 200 ng/mL phorbol 12-myristate 13-acetate (Sigma-Aldrich), and/or 1 μg/mL ionomycin (Sigma-Aldrich), if not indicated otherwise.
RNA isolation and quantitative RT-PCR
Total cellular RNA was isolated using PeqGold Total RNA Kit (VWR International GmbH, Darmstadt, Germany). cDNA was synthesized with the SuperScript II reverse transcription kit (InvivoGen, Toulouse, France) using an equimolar mixture of random hexamer primers and oligo(dT) primers.
Quantitative RT-PCR was performed on CFX Connect Real-Time PCR Detection System (Bio-Rad, Munich, Germany) using the Biozym Probe qPCR kit (Biozym, Hessisch Oldendorf, Germany). Samples were referenced to hypoxanthine phosphoribosyltransferase 1 and glyceraldehyde 3-phosphate dehydrogenase.
Primers and hydrolysis probes were selected via the Roche Universal Probe Library assay design center.
Flow cytometry
Fc receptors were blocked using Human TruStain FcX (Biolegend, London) according to manufacturer's recommendations. Subsequently, samples were stained with Fixable Viability Dye eFluor 780 (1:5.000; eBioscience, Frankfurt, Germany), Pacific Blue, antihuman CD20 antibody (Clone 2H7), PerCP antihuman CD3 antibody (Clone OKT3), FITC antihuman CD14 antibody (Clone HCD14), Brilliant Violet 785 antihuman CD69 antibody (Clone FN50), Alexa Fluor 647 antihuman HLA-A,B,C antibody (Clone W6/32), and phycoerythrin (PE)/Cy7 antihuman CD274 (B7-H1, PD-L1) antibody (Clone 29E.2A3) (all 1:200; Biolegend).
To identify and further analyze monocytes and/or different lymphocyte populations, gates were set on the basis of morphology (forward/sideward scatter properties) and lineage marker expression (T cells: CD3+ CD20−; B cells: CD3− CD20+; monocytes: CD14+).
To analyze cytokine expression by flow cytometry, stimulation in the last 24 hours was performed in the presence of 0.5 mg/mL brefeldin A (Biolegend). Cells were stained for surface markers as described earlier, fixed and permeabilized using BD Cytofix/Cytoperm fixation and permeabilization solution (BD Biosciences, Heidelberg, Germany), followed by staining with PE or BV421 antihuman TNF antibody (Clone MAb11; 1:100; Biolegend), Alexa Fluor 647 antihuman IL-1β antibody (Clone JK1B-1; 1:50; Biolegend), PE/Dazzle 594 antihuman IL-6 antibody (Clone MQ2-13A5; 1:100; Biolegend), human IL-18/IL-1F4 propeptide PE-conjugated antibody (Clone 74801; 1:100; R&D Systems, Bio-Techne, Minneapolis, Minn), or PE/Cy7 antihuman IFN-γ antibody (Clone 4S.B3; 1:100; Biolegend).
Samples were analyzed on an LSR Fortessa flow cytometer (BD Biosciences).
Biochemical assays
PBMCs (1 × 106 PBMCs per condition) were stimulated with 10,000 U/mL recombinant human IFN-α2a (Miltenyi Biotech) for 30 minutes or left unstimulated. Cells were lysed by resuspending pelleted cells in lysis buffer containing 50 mM Tris-HCl (pH 7.8), 137 mM NaCl, 0.5 mM EDTA, 1 mM sodium orthovanadate, 10% (w/v) glycerol, 1% (w/v) NP40 supplemented with a protease inhibitor cocktail (P8340; Sigma-Aldrich), followed by 10 minutes of incubation on ice. Insoluble material was pelleted at 16,000g for 10 minutes at 4°C. Protein (35 μg) of each sample was separated by 10% denaturing SDS-PAGE followed by Western blotting on a polyvinylidene difluoride membrane using the TransBlot Turbo (Bio-Rad). STAT1 was detected using polyclonal Stat1 antibody 9172 (Cell Signaling Technologies, Leiden, The Netherlands), and β-actin was detected using β-actin antibody (C4) sc-47778 (Santa Cruz Biotechnology, Dallas, Texas).
ELISA
Cytokine levels were measured by ELISA according to manufacturer's recommendations. The following kits were used: CXCL10 (IP-10): Human IP-10 ELISA Set; TNF: Human TNF ELISA Set; IL-6: Human IL-6 ELISA Set; IL-1β: Human IL-1β ELISA Set (all BD Biosciences); and IFN-γ: DuoSet Human IFN-γ (R&D Systems).
Infection with vesicular stomatitis virus-M51R
PBMCs were infected with vesicular stomatitis virus-M51R-green fluorescent protein (VSV-M51R-GFP).18 A day (24 hours) after infection, cell-free supernatant was harvested, UV-irradiated, and subjected to ELISA as described earlier.
Statistical analysis
For comparison of 2 independent variables, the Student t test was performed after transformation of data sets using the natural logarithm to obtain normal distribution. For multiple comparisons 1-way or 2-way ANOVA followed by the Tukey multiple comparison test were performed. A P value of less than .05 was considered statistically significant. If not indicated otherwise, data are represented as mean and SD. Single data points correspond to individual results of independently conducted experiments.
Results
Complete AR STAT1 LOF segregates with the immunodeficiency and HLH-like hyperinflammation phenotype
We report on a male infant who was born to healthy consanguineous parents of West African descent. He first came to our attention at age 4 months, when returning from a visit in Guinea, where he had been treated for meningoencephalitis because of fever and generalized seizures without an identified infectious agent. He presented at our emergency department with persisting neurological impairment and recurrent seizures. He showed sinus venous thrombosis and occlusive hydrocephalus with increased intracranial pressure, which required anticoagulation and external ventricular drainage. He recovered from the putative infection; however, severe motor and sensory neurological deficits persisted. In the course of the next 5 months, the patient suffered repeated infections, mostly viral in origin (Table I ).
Table I.
Disease course before diagnosis of complete AR STAT1 LOF
| Age (mo) | Disease | Organism |
|---|---|---|
| 4 | Ventriculitis (VP shunt) | Klebsiella pneumonia |
| 5 | Peritonitis, orchitis (VP shunt) | Not tested |
| 7 | Bronchitis | Enterovirus/rhinovirus |
| 8 | Bronchitis | Respiratory syncytial virus |
| 9 | Keratoconjunctivitis | Rothia dentocariosa, Viridans streptococci |
| 11 | Viral and bacterial bronchitis | Not tested |
| 13 | VZV-like exanthema, respiratory failure, HLH | VZV vaccine strain parainfluenza virus |
VP, Ventriculoperitoneal.
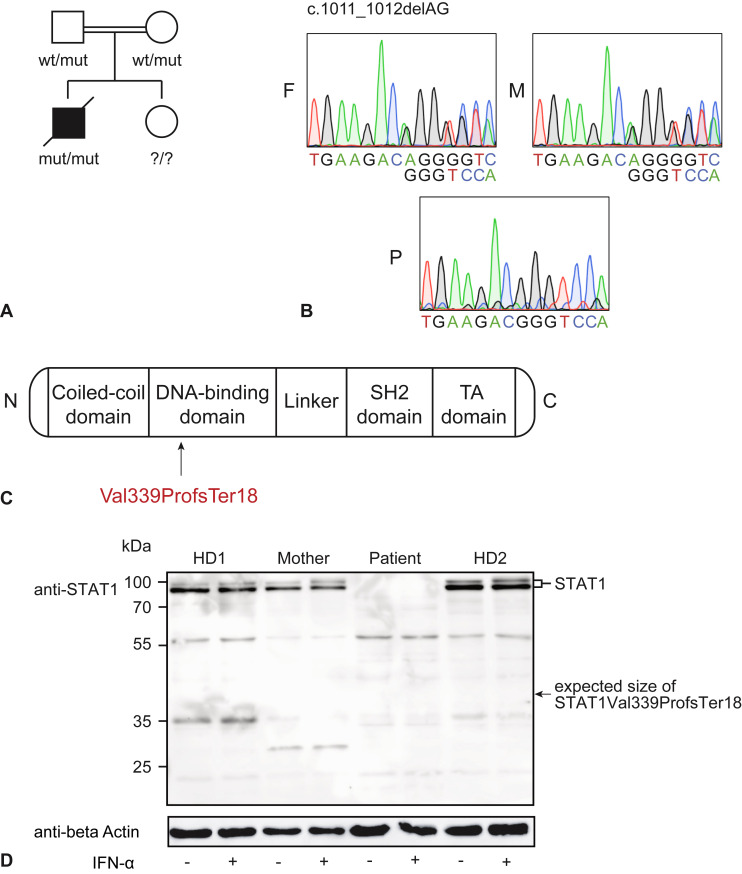
At age 13 months, he presented with fever, respiratory failure, and generalized varicella zoster virus (VZV)-like rash 1 month after VZV vaccination. Both VZV vaccine strain and parainfluenza virus were detected by PCR of vesicular fluid and tracheal secretions, respectively. During this illness he manifested hyperinflammation, fulfilling 6 of 8 HLH criteria (Table II ) together with acute liver failure (ALF) (aspartate transaminase, 2863 U/L [normal, ≤55]; alanine transaminase, 918 U/L [normal, ≤59], international normalized ratio, 1.6 [normal, 0.8-1.2]). Extensive immunologic phenotyping including natural killer–cell degranulation and perforin expression did not reveal abnormalities. Suspecting an inborn error of immunity nonetheless, we performed exome sequencing and found a homozygous novel STAT1 frameshift variant (c.1011_1012delAG, p.Val339ProfsTer18) (Figure 1 , A-C). The parents were both heterozygous carriers as determined by Sanger sequencing (Figure 1, B). A sibling's genotype was not determined because she did not show any signs or symptoms at age 5 years.
Table II.
| Criterion | Found in patient? |
|---|---|
| Fever | Yes |
| Splenomegaly | No |
| Bicytopenia | Yes: hemoglobin, 5.9 g/dL (normal, 10.7-13.4 g/dL), platelets, 8 G/L (normal, 195-464 G/L) |
| Hypertriglyceridemia or hypofibrinogenemia | Yes: fibrinogen <70 mg/dL (normal, 160-400 mg/dL) |
| Hemophagocytosis | Yes: HLH cells in bone marrow |
| Elevated ferritin | Yes: 22,935 ng/mL (normal, 12-510 ng/mL) |
| Low/absent NK-cell activity | No |
| Elevated soluble CD25 (sIL-2RA) | Yes: 15,718 kU/L (normal, 156-623 kU/L) |
NK, Natural killer.
Figure 1.
AR STAT1 Val339ProfsTer18 variant leads to complete STAT1 deficiency. (A) Pedigree of the patient's family. Double line indicates consanguinity, filled shape indicates affected, and the diagonal line crossing the solid black square indicates deceased individual. Genotype of the healthy sibling is not known. (B) Electropherograms of father (F), mother (M), and patient (P) STAT1 wt/mut and mut/mut alleles. (C) Schematic representation of the STAT1 protein domains and the localization of the p.Val339ProfsTer18 variant within the DNA-binding domain. (D) Immunoblot analysis of HD, patient (P), and mother (M) PBMCs untreated or treated with IFN-α2a for 30 minutes. Arrow indicates expected height of STAT1 Val339ProfsTer18 variant. Representative result of 2 independent experiments.
Because the variant implied a frameshift with an early stop codon in the DNA-binding domain (Figure 1, C), we tested whether it affected STAT1 protein expression. We performed STAT1 immunoblotting from PBMCs of the patient, healthy donors (HDs), and the patient's mother with and without previous stimulation with IFN-α2a using a polyclonal STAT1 antibody to detect putative truncated forms of the protein. Endogenous STAT1 expression was detectable in HD and mother PBMCs to a comparable level in both untreated and IFN-α2a–treated conditions. In contrast, patient PBMCs showed complete absence of STAT1 protein, full-length or truncated, in both experimental conditions (Figure 1, D). These results indicate that the homozygous STAT1 p.Val339ProfsTer18 variant leads to complete loss of STAT1 protein expression.
Defective ISG induction and activating surface marker expression in STAT1 LOF monocytes, T cells, and B cells
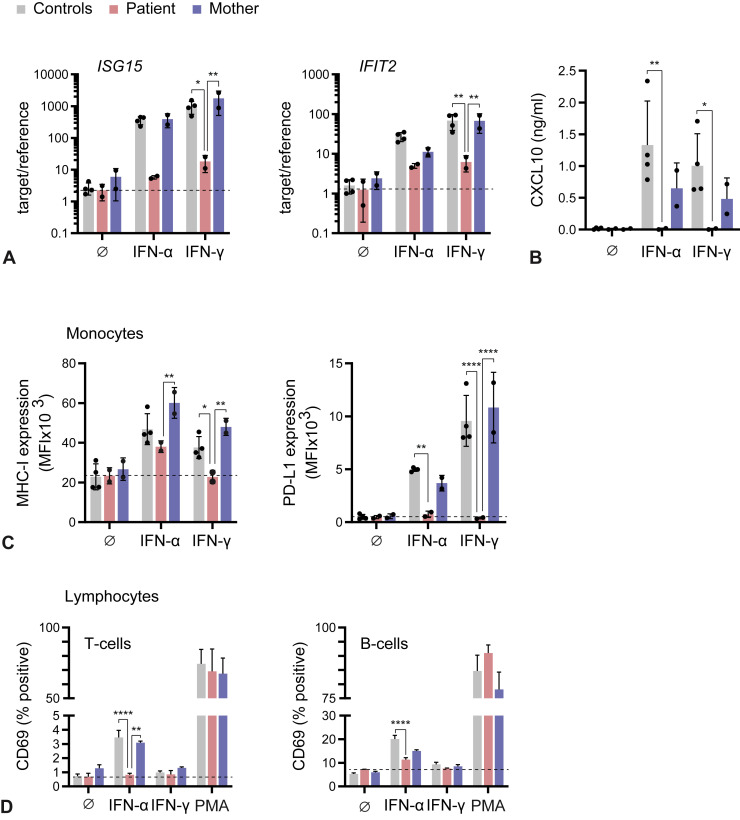
To understand the functional consequence of STAT1 p.Val339ProfsTer18 loss of protein expression in IFN-I and IFN-II signaling, we stimulated patient's, mother's, and HDs' PBMCs with recombinant human IFN-α2a and IFN-γ and analyzed the induction of ISGs by quantitative RT-PCR, ELISA, and flow cytometry. Transcriptional induction of ISG15 and interferon induced protein with tetratricopeptide repeats 2 by IFN-γ was blunted in the patient's PBMCs compared with HD and mother PBMCs (Figure 2 , A). Analysis of CXCL10 showed potent induction in HD and mother PBMCs in response to both IFN-α2a and IFN-γ treatment, whereas production by patient PBMCs remained at background level (Figure 2, B). Induction of MHC-I and PD-L1 surface expression on monocytes in response to IFN-γ treatment was also absent on patient monocytes in contrast to HD and mother monocytes (Figure 2, C). Interestingly, MHC-I induction on monocytes by IFN-α2a appeared partially independent of STAT1. However, IFN-α2a–-treated patient T and B cells displayed markedly reduced induction of CD69, an early activation marker and well-known ISG in lymphocytes (Figure 2, D).
Figure 2.
Defective ISG induction and activating surface marker expression in STAT1-deficient monocytes, T cells, and B cells. (A) Induction of ISG15 and IFIT2 measured by quantitative RT-PCR 6 hours after stimulation with IFN-α2a or IFN-γ. (B) ELISA for CXCL10 in cell-free supernatant 24 hours after stimulation. (C, D) Flow cytometric analysis of PBMCs 24 hours after stimulation as indicated. IFIT2, Interferon induced protein with tetratricopeptide repeats 2. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
Overall, these results demonstrate that STAT1 p.Val339ProfsTer18 compromises cellular responses to IFN-I and IFN-II and that IFN-α2a–and IFN-γ–induced MHC-I and PD-L1 surface expression on monocytes or secretion of CXCL10 might be used to screen for STAT1 LOF.
Defective control of VSV in STAT1 LOF monocytes and fibroblasts is associated with increased proinflammatory cytokine production
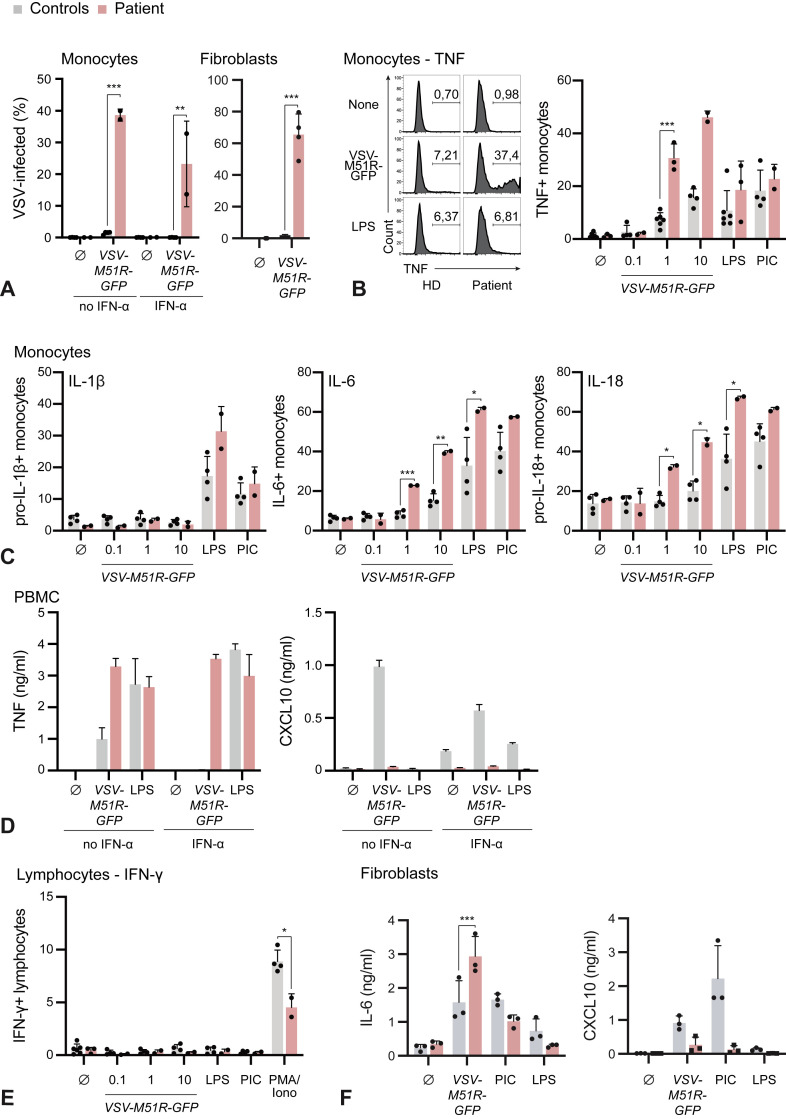
To study the effect of complete AR STAT1 LOF on viral susceptibility, we exposed patient and control cells to a GFP-tagged vesicular stomatitis virus variant carrying a mutation in the M-protein leading to high IFN-I induction (VSV-M51R-GFP).18 Nearly 40% of patient monocytes were infected after 30 hours compared with less than 5% of controls as indicated by GFP positivity (Figure 3 , A). IFN-α2a priming prevented infection of control monocytes completely but did not influence viral infection of patient monocytes (Figure 3, A). The infection rate of patient fibroblasts was nearly 60% compared with less than 2% in control fibroblasts (Figure 3, A).
Figure 3.
Defective control of VSV in STAT1-deficient monocytes and fibroblasts is associated with increased proinflammatory cytokine production. (A) Flow cytometric analysis of GFP expression 30 hours after infection with VSV-M51R-GFP (MOI: 1) with or without prestimulation with 1000 U/mL IFN-α2a overnight. (B, C) Intracellular cytokine staining of patient or HD monocytes 30 hours after infection with VSV-M51R-GFP, stimulation with LPS or PIC. (D) ELISA for TNF and CXCL10 of cell-free supernatant of patient or HD PBMCs 30 hours after stimulation as indicated in Figure 3 (A). Mean and SD of technical replicates of 1 result of 2 independent experiments. (E) Intracellular cytokine staining of patient or HD lymphocytes 30 hours after infection with VSV-M51R-GFP, stimulation with LPS or PIC. (F) ELISA for IL-6 or IP-10 of cell-free supernatant of patient or HD fibroblasts stimulated with VSV-M51R-GFP (MOI: 1), PIC (1 μg/mL), or LPS (200 ng/mL) for 24 hours. PMA, Phorbol 12-myristate 13-acetate. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Next, we turned to the pathophysiological connection between HLH-like hyperinflammation and complete AR STAT1 LOF. HLH is driven by the overproduction of proinflammatory cytokines such as TNF, IL-1β, IL-6, IL-18, and IFN-γ.20 , 21 Hence, we studied the expression of these cytokines upon viral or pathogen-associated molecular pattern (PAMP) stimulation. Intracellular cytokine staining for TNF in monocytes showed no difference in the response to LPS or the double-stranded RNA analog polyinosinic:polycytidylic acid (PIC), whereas the TNF response to VSV-M51R-GFP was highly augmented (Figure 3, B). Likewise, there was excessive production of IL-6 and pro–IL-18 by patient monocytes compared with control monocytes in response to VSV-M51R-GFP infection (Figure 3, C). Of note, pro–IL-1β was not induced by VSV infection. Interestingly, IL-6 and pro–IL-18 induction in patient monocytes was also stronger in response to LPS, when compared with control monocytes. ELISA of cell-free supernatant for TNF could confirm these findings, whereas induction of the antiviral effector chemokine CXCL10 was absent in the cells of the patient (Figure 3, D). Because IFN-γ is generally considered an important cytokine in HLH, with certain STAT1-independent effects, we assessed IFN-γ production. However, no induction could be observed under the experimental conditions chosen (Figure 3, E).
To extend these findings to nonimmune cells, we compared IL-6 and CXCL10 expression upon VSV-M51R-GFP infection as well as LPS or PIC stimulation in patient and HD fibroblasts (Figure 3, F). In line with our previous findings, CXCL10 induction by VSV-M51R-GFP, LPS or PIC was lower in the patient's fibroblasts, whereas IL-6 production after VSV-M51R-GFP infection was augmented as compared with control cells. Of note, the IL-6 response toward PAMPs such as PIC or LPS was not significantly altered in fibroblasts.
These results show that complete AR STAT1 LOF increases both susceptibility to viral infection and the ensuing production of proinflammatory cytokines such as IL-6, TNF, and IL-18. Furthermore, the proinflammatory response to PAMP stimulation per se seems to be pathologically altered in absence of STAT1, leading to overproduction of certain cytokines.
In conclusion, we propose that complete AR STAT1 LOF leads to reduced induction of antiviral defense genes through IFN-I and IFN-II upon viral infection. In turn, viral replication is increased drastically, leading to the accumulation of viral PAMPs, which may be sensed by pattern recognition receptors (PRRs) leading to excess production of proinflammatory cytokines that result in HLH-like hyperinflammation.
Therapeutic approach and clinical outcome of the complete AR STAT1 LOF patient
To protect him against viral and opportunistic infections, the patient received prophylactic immunoglobulin substitution, cotrimoxazole, and azithromycin and was isolated at home without further relevant infections for up to 4 months. Because HSCT has been reported to have achieved long-term survival in 1 case of complete AR STAT1 deficiency,22 we performed allogeneic HSCT using stem cells from his HLA-matched father. The conditioning regimen consisted of fludarabine (40 mg/m2/d on days −7 to −4), busulfan (4.8 mg/kg/d on days −7 to −3; targeted area under the curve, 81,245 ng∗h/mL), antithymocyte globulin (Grafalon; 15 mg/kg/d on days −4 to −2), and rituximab (375 mg/m2/d on day −1). Neutrophil engraftment was observed on day +14. Chimerism on day +18 was 90% to 95% donor origin. On day +25, the patient developed CMV reactivation as assessed by PCR in peripheral blood. Subsequently, treatment with ganciclovir intravenously was started but viral copy number continued to increase. The patient developed dyspnea and signs of interstitial pneumonia on chest x-ray, requiring respiratory support from day +37 onward. Intensification of antiviral therapy with anti-CMV immunoglobulin, foscarnet, and adoptive transfer of paternal CMV-specific T cells (day +47) did not lead to viral clearance. Suspecting a substantial lung inflammatory component, we additionally treated with prednisolone followed by the Janus kinase inhibitor ruxolitinib. Despite this therapy, the patient progressed to lethal respiratory failure on day +64 after HSCT, likely because of CMV pneumonitis.
Discussion
In this study, we describe a patient with a novel STAT1 variant leading to complete AR STAT1 LOF. We confirm STAT1 LOF to be a severe genetic disorder leading to life-limiting viral infections, as was observed in all 7 patients reported to date.11, 12, 13, 14 Furthermore, we substantiate the clinical finding of HLH-like hyperinflammation associated with viral infection previously reported in another child with STAT1 LOF.14 Of note, our patient developed ALF in the context of hyperinflammation, as described in a single patient by Chapgier et al.12 Therefore, STAT1 LOF can be associated not only with infection susceptibility toward viral and mycobacterial infections but also with hyperinflammation causing an HLH-like phenotype and ALF. Elevated IL-18 has been found in X-linked inhibitor of apoptosis deficiency–associated HLH, and IL-18 binding protein deficiency has been described as an inborn error of immunity leading to fulminant viral hepatitis.23 , 24 It is therefore tempting to speculate that the increased production of IL-18 observed in our study is one of the driving proinflammatory cytokines of HLH and ALF in complete AR STAT1 LOF. Given the future availability of recombinant IL-18 binding protein in phase III clinical trials to treat HLH, this observation is of particular clinical interest.
Both IFNAR and IFNGR signaling depend heavily on STAT11 and accordingly we found a broad defect in ISG upregulation in response to IFN-I and IFN-II stimulation. Although surface expression of PD-L1 was defective after stimulation with IFN-α and IFN-γ, MHC-I expression seemed to be upregulated despite STAT1 deficiency in response to IFN-α. This implies STAT1-independent IFN signaling, potentially mediated by STAT2-IFN regulatory factor 9 complexes as previously observed for other ISGs.25 , 26 Further studies are needed to determine the exact STAT1-dependent regulation in human immune and nonimmune cells.
As expected, viral infection of STAT1-deficient monocytes and fibroblasts resulted in higher infection rates and failure to induce ISGs such as CXCL10. However, virally infected patient cells produced increased amounts of the proinflammatory cytokines TNF, IL-6, and IL-18, typically induced upon sensing of viral nucleic acids by PRRs such as cGAS/STING, the RIG-I–like receptors, and Toll-like and NOD-like receptors.27 , 28 Thus, in the context of STAT1 deficiency, higher viral replication likely leads to increased amounts of ligands stimulating PRRs. However, at least for IL-1β, IL-6, and IL-18, the cytokine response toward the PAMPs LPS and PIC was also increased in patient PBMCs, implying the loss of ill-defined STAT1-dependent negative feedback. This may constitute an additional explanation for the observed hyperinflammatory HLH- and ALF-like phenotype in STAT1 LOF. However, such responses might be cell type–dependent and further investigation is needed to generally characterize cytokine responses in STAT1 deficiency. Thus, we could confirm a central, nonredundant role for STAT1 in antiviral defense in primary human immune and nonimmune cells. Furthermore, we could link STAT1 deficiency to the pathophysiological overproduction of proinflammatory cytokines as seen in HLH and ALF upon viral infection.
Two STAT1 LOF patients were transplanted successfully and 1 had reported long-term survival and full immune reconstitution.13 , 14 , 22 The STAT1 LOF patient described here reached donor chimerism of 90% to 95% on day +18, but succumbed to CMV-associated acute respiratory distress syndrome on day +64 after HSCT despite therapy with antiviral drugs and CMV-specific donor T cells. A similar course was described in another patient with lethal EBV infection and multiorgan failure upon transplantation.12 On a balance of risks, however, HSCT should be the treatment of choice and might be accompanied by targeted anti-inflammatory interventions. However, the persistence of STAT1 deficiency in the nonimmune cell compartment likely confers an ongoing elevated risk of viral infection for transplanted patients. This would be in line with observations in mice that absence of IFN-I and IFN-III signaling in the stromal compartment leads to higher susceptibility and mortality upon influenza infection.29
Conclusions
Despite our progress in understanding the mechanisms of STAT1 LOF, its consequences remain devastating. Generally, it is diagnosed only with a delay, after severe viral and mycobacterial infections have occurred. However, as in severe combined immunodeficiency, the infectious burden acquired critically has an impact on the outcome of HSCT.30 Therefore, even higher awareness toward STAT1 deficiency and related disorders is needed to facilitate early genetic testing and biochemical evaluation of STAT1 variants of unknown significance. Here, we propose determination of the IFN-α2a–and IFN-γ–induced PD-L1 surface expression on monocytes as well as CXCL10 secretion as a simple and robust test to analyze the functionality of the STAT1 signaling pathway.
Acknowledgments
We value the personal contribution of the patient and his family as well as the excellent clinical care by the nursing and medical teams. We thank Professor Sophie Hambleton (Great North Children's Hospital, Newcastle University) for critically reading the manuscript. We thank Christine Hörth and Natalie Röder for excellent technical support. We acknowledge the expert services of the sequencing facility at the Dr von Hauner Children's Hospital and the associated computational biology unit. We also acknowledge the iFlow Core Facility of the University Hospital Munich for assistance with the generation of flow cytometry data. Parts of this work have been performed for the doctoral thesis of D.F.R. Boehmer at the Ludwig-Maximilians-Universität München.
Footnotes
F.H. received funding from the Care-for-Rare Foundation (160073), the German Center for Infection Research (TTU 07.909), the Else Kröner-Fresenius-Stiftung (2017_A110), and the German Federal Ministry of Education and Research (01GM1910C). L.M.K. was supported by Else Kröner-Fresenius-Stiftung (2017_A50).
Conflicts of interest: The authors declare that they have no relevant conflicts of interest.
References
- 1.O’Shea J.J., Holland S.M., Staudt L.M. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy D.E., Darnell J.E. STATs: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L., Okada S., Kong X.-F., Kreins A.Y., Cypowyj S., Abhyankar A., et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van de Veerdonk F.L., Plantinga T.S., Hoischen A., Smeekens S.P., Joosten L.A.B., Gilissen C., et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 7.Olbrich P., Freeman A.F. STAT1 and STAT3 mutations: important lessons for clinical immunologists. Expert Rev Clin Immunol. 2018;14:1029–1041. doi: 10.1080/1744666X.2018.1531704. [DOI] [PubMed] [Google Scholar]
- 8.Kristensen I.A., Veirum J.E., Møller B.K., Christiansen M. Novel STAT1 alleles in a patient with impaired resistance to mycobacteria. J Clin Immunol. 2011;31:265–271. doi: 10.1007/s10875-010-9480-8. [DOI] [PubMed] [Google Scholar]
- 9.Chapgier A., Kong X.F., Boisson-Dupuis S., Jouanguy E., Averbuch D., Feinberg J., et al. A partial form of recessive STAT1 deficiency in humans. J Clin Invest. 2009;119:1502–1514. doi: 10.1172/JCI37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong X.F., Ciancanelli M., Al-Hajjar S., Alsina L., Zumwalt T., Bustamante J., et al. A novel form of human STAT1 deficiency impairing early but not late responses to interferons. Blood. 2010;116:5896–5906. doi: 10.1182/blood-2010-04-280586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Zaid Al-Mohsen I., Al-Jumaah S., et al. Impaired response to interferon-α/β and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 12.Chapgier A., Wynn R.F., Jouanguy E., Filipe-Santos O., Zhang S., Feinberg J., et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176:5078–5083. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 13.Vairo D., Tassone L., Tabellini G., Tamassia N., Gasperini S., Bazzoni F., et al. Severe impairment of IFN-γ and IFN-α responses in cells of a patient with a novel STAT1 splicing mutation. Blood. 2011;118:1806–1817. doi: 10.1182/blood-2011-01-330571. [DOI] [PubMed] [Google Scholar]
- 14.Burns C., Cheung A., Stark Z., Choo S., Downie L., White S., et al. A novel presentation of homozygous loss-of-function STAT-1 mutation in an infant with hyperinflammation—a case report and review of the literature. J Allergy Clin Immunol Pract. 2016;4:777–779. doi: 10.1016/j.jaip.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Henter J.I., Horne A., Aricó M., Egeler R.M., Filipovich A.H., Imashuku S., et al. HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer. 2007;48:124–131. doi: 10.1002/pbc.21039. [DOI] [PubMed] [Google Scholar]
- 16.Chinn I.K., Eckstein O.S., Peckham-Gregory E.C., Goldberg B.R., Forbes L.R., Nicholas S.K., et al. Genetic and mechanistic diversity in pediatric hemophagocytic lymphohistiocytosis. Blood. 2018;132:89–100. doi: 10.1182/blood-2017-11-814244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schober T., Magg T., Laschinger M., Rohlfs M., Linhares N.D., Puchalka J., et al. A human immunodeficiency syndrome caused by mutations in CARMIL2. Nat Commun. 2017;8:1–13. doi: 10.1038/ncomms14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stojdl D.F., Lichty B.D., tenOever B.R., Paterson J.M., Power A.T., Knowles S., et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 19.Bergsten E., Horne A.C., Aricó M., Astigarraga I., Egeler R.M., Filipovich A.H., et al. Confirmed efficacy of etoposide and dexamethasone in HLH treatment: long-term results of the cooperative HLH-2004 study. Blood. 2017;130:2728–2738. doi: 10.1182/blood-2017-06-788349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arico M., Danesino C., Pende D., Moretta L. Pathogenesis of haemophagocytic lymphohistiocytosis. Br J Haematol. 2001;114:761–769. doi: 10.1046/j.1365-2141.2001.02936.x. [DOI] [PubMed] [Google Scholar]
- 21.Filipovich A., McClain K., Grom A. Histiocytic disorders: recent insights into pathophysiology and practical guidelines. Biol Blood Marrow Transplant. 2010;16:S82–S89. doi: 10.1016/j.bbmt.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Naviglio S., Soncini E., Vairo D., Lanfranchi A., Badolato R., Porta F. Long-term survival after hematopoietic stem cell transplantation for complete STAT1 deficiency. J Clin Immunol. 2017;37:701–706. doi: 10.1007/s10875-017-0430-6. [DOI] [PubMed] [Google Scholar]
- 23.Belkaya S., Michailidis E., Korol C.B., Kabbani M., Cobat A., Bastard P., et al. Inherited IL-18BP deficiency in human fulminant viral hepatitis. J Exp Med. 2019;216:1777–1790. doi: 10.1084/jem.20190669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wada T., Kanegane H., Ohta K., Katoh F., Imamura T., Nakazawa Y., et al. Sustained elevation of serum interleukin-18 and its association with hemophagocytic lymphohistiocytosis in XIAP deficiency. Cytokine. 2014;65:74–78. doi: 10.1016/j.cyto.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Fink K., Grandvaux N. STAT2 and IRF9: beyond ISGF3. JAKSTAT. 2013;2:e27521. doi: 10.4161/jkst.27521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duerr C.U., Velazquez L.E., Decker T., Majoros A., Platanitis E., Kernbauer-Hölzl E., et al. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front Immunol. 2017;8:29. doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 28.Broz P., Dixit V.M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 29.Crotta S., Davidson S., Mahlakoiv T., Desmet C.J., Buckwalter M.R., Albert M.L., et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pai S.Y., Logan B.R., Griffith L.M., Buckley R.H., Parrott R.E., Dvorak C.C., et al. Transplantation outcomes for severe combined immunodeficiency, 2000-2009. N Engl J Med. 2014;371:434–446. doi: 10.1056/NEJMoa1401177. [DOI] [PMC free article] [PubMed] [Google Scholar]