Abstract
Clinical evidence suggests that some patients diagnosed with coronavirus disease 2019 (COVID-19) experience a variety of complications associated with significant morbidity, especially in severe cases during the initial spread of the pandemic. To support early interventions, we propose a machine learning system that predicts the risk of developing multiple complications. We processed data collected from 3,352 patient encounters admitted to 18 facilities between April 1 and April 30, 2020, in Abu Dhabi (AD), United Arab Emirates. Using data collected during the first 24 h of admission, we trained machine learning models to predict the risk of developing any of three complications after 24 h of admission. The complications include Secondary Bacterial Infection (SBI), Acute Kidney Injury (AKI), and Acute Respiratory Distress Syndrome (ARDS). The hospitals were grouped based on geographical proximity to assess the proposed system's learning generalizability, AD Middle region and AD Western & Eastern regions, A and B, respectively. The overall system includes a data filtering criterion, hyperparameter tuning, and model selection. In test set A, consisting of 587 patient encounters (mean age: 45.5), the system achieved a good area under the receiver operating curve (AUROC) for the prediction of SBI (0.902 AUROC), AKI (0.906 AUROC), and ARDS (0.854 AUROC). Similarly, in test set B, consisting of 225 patient encounters (mean age: 42.7), the system performed well for the prediction of SBI (0.859 AUROC), AKI (0.891 AUROC), and ARDS (0.827 AUROC). The performance results and feature importance analysis highlight the system's generalizability and interpretability. The findings illustrate how machine learning models can achieve a strong performance even when using a limited set of routine input variables. Since our proposed system is data-driven, we believe it can be easily repurposed for different outcomes considering the changes in COVID-19 variants over time.
1. Introduction
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has led to a global health emergency since the emergence of the coronavirus disease 2019 (COVID-19) [1]. Despite containment efforts, more than 491 million confirmed cases have been reported globally, including 892,170 cases in the United Arab Emirates (UAE) as of April 4, 2022 [1]. Due to unexpected burdens on healthcare systems, identifying high risk groups using prognostic models has become vital to support patient triage and resource allocation.
Most of the published prognostic models for patients with COVID-19 focus on predicting mortality or the need for intubation [2]. While the prediction of such adverse events is important for patient triage, clinical evidence suggests that patients with COVID-19 may also experience a variety of complications in organ systems that could lead to severe morbidity and mortality [3,4], especially amongst severe cases during the early waves of the pandemic. In this study, we identified three such complications associated with poor patient outcomes based on clinical evidence, prior to the emergence of the less severe variants [5]: Acute Respiratory Distress Syndrome (ARDS) [6], Acute Kidney Injury (AKI) [7], and Secondary Bacterial Infection (SBI) [8].
ARDS-related pneumonia has been reported as a major complication among patients with COVID-19 that have poor prognosis [9] and was a major cause of ventilator shortages worldwide [6,10,11]. In a Chinese study published in 2020, 31.0% of patients developed ARDS within a median of 12 days from the onset of COVID-19, and ARDS was the second most frequently observed complication after sepsis [10]. Additionally, only a few patients manifest clear clinical symptoms in the early stages of developing ARDS [6,12], so it is difficult to suspect ARDS unless it occurs. Hence, we identified early prediction of the risk of developing ARDS, prior to its onset, of high importance, since ARDS was considered as one of the main risk factors of death among hospitalized patients with COVID-19 [13].
Although COVID-19 primarily emerged as a respiratory disease, some patients with COVID-19 experience both respiratory and extra-respiratory complications including renal complications such as AKI [7,14]. Patients with AKI require special care and resources such as renal replacement therapy and dialysis [15]. It was estimated that AKI developed in 36.6% of patients admitted with COVID-19 in metropolitan New York in 2020, of which 35% had died [15]. Therefore, risk prediction of AKI can help in initiating preventive interventions in order to avoid quite poor patient prognosis.
Moreover, several studies reported alarming percentages of hospitalized patients with COVID-19 who develop SBI [10]. SBI is known for poor outcomes in several respiratory viral infections. Hence, it led to increased burdens on hospitals in the 1918 influenza pandemic, 2009 H1N1 influenza pandemic, and in seasonal flu [[16], [17], [18]]. Patients with COVID-19 who developed SBI have shown worse outcomes, including admission to the Intensive Care Unit (ICU) and mortality, compared to those who did not develop SBI [19]. Therefore, early prediction of SBI can potentially improve patient prognosis, such as by taking aseptic procedures especially when hospitals get crowded [8].
In recent years, machine learning gained popularity for the development of algorithms for clinical decision support tools [[20], [21], [22]]. In the context of COVID-19, most machine learning studies have focused either on diagnosis or prognosis based on adverse events, mostly mortality and intubation [2,23,24]. We summarize a few examples in Table 1 . Since ARDS is considered a major manifestation of the COVID-19 disease, some studies focused on developing machine learning models to predict ARDS as an outcome [6,12], such as by using a large set of hematological and biochemical markers [12]. One limitation of such approaches is that they rely on laboratory-test results that may not be routinely measured. In another study, the authors used both statistical machine learning models and deep neural networks for the prediction of ARDS, by combining a large feature set of chest Computed Tomography (CT) findings, demographics, epidemiology, clinical symptoms, and laboratory-test results [6]. Similarly, for AKI prediction amongst patients with COVID-19, a multivariate logistic regression was developed using findings of CT imaging, laboratory-test results, vital-sign measurements, and patient demographics [25]. While recent work on SBI mainly focused on its clinical manifestations and occurrence [16,26,27], one study investigated sepsis risk prediction among patients with COVID-19 using hematological parameters and other biomarkers [28]. To summarize, existing work tends to predict a single complication at a time, which is less informative than predicting multiple complications known to be common among patients with COVID-19, use costly input features that may not be readily available, or rely on training deep neural networks that require high computational resources and large training datasets.
Table 1.
Examples of machine learning studies that aim to predict various outcomes for in-patients with confirmed COVID-19 diagnosis. We refer the readers to extensive published literature reviews [2,23,24].
| Reference | Outcome | Input Data | Models | Study Location |
|---|---|---|---|---|
| [29] |
Deterioration (intubation or ICU admission or mortality) |
Chest X-ray images and clinical data (patient demographics, seven vital-sign variables, and 24 laboratory-test results) |
Convolutional neural network for chest X-ray images and gradient boosting model for clinical data |
United States |
| [30] |
Mortality |
Five laboratory-test results |
Support vector machine |
United States |
| [31] |
Severe progression (high oxygen flow rate, mechanical ventiliation or mortality) |
Chest CT scans, patient demographics, five vital-sign variables, symptoms, comorbidities, 14 laboratory-test results, and chest CT radiology report findings |
Deep neural network and logistic regression |
France |
| [32] |
Prognostication (intubation or hospital admission, or mortality) |
Chest X-ray images, two vital-sign variables, and nine laboratory-test results |
Convolutional neural network |
United States |
| [28] |
Sepsis |
Eight laboratory-test results |
Gradient boosting model |
China |
| [25] |
AKI |
Findings of abdominal CT scans, demographics, vital signs, comorbidities, and three laboratory-test results |
Logistic regression |
United States |
| [33] | ARDS | Demgraphics, interventions, comorbidities, 17 laboratory-test results, and eight vital signs | Gradient boosting model | United States |
Therefore, there is a pressing need for a low-cost predictive system that uses routine clinical data to predict complications and support patient management. In this work, we address this need by developing and evaluating a machine learning system that predicts the risk of ARDS, SBI, and AKI among patients with COVID-19 admitted to the Abu Dhabi Health Services (SEHA) facilities, UAE, from April 1st, 2020 to April 30th, 2020, during the first wave of the pandemic. While we focus on three complications only, namely because their occurrence could be identified retrospectively using clinical criteria, the system and proposed training framework can be scaled to incorporate predictions of other complications, and can be fine-tuned using datasets of other patient cohorts. An overview of the pipeline is shown in Fig. 1 . Next, we describe our methodology in Section 2 and the performance and explainability results in Section 3. We then discuss the limitations and strengths of the study in Section 4, and conclude by highlighting the potential of our system in clinical settings in Section 5. To allow for reproducibility and external validation, we made our code and one of the evaluation test sets publicly available at: https://github.com/nyuad-cai/COVID19Complications.
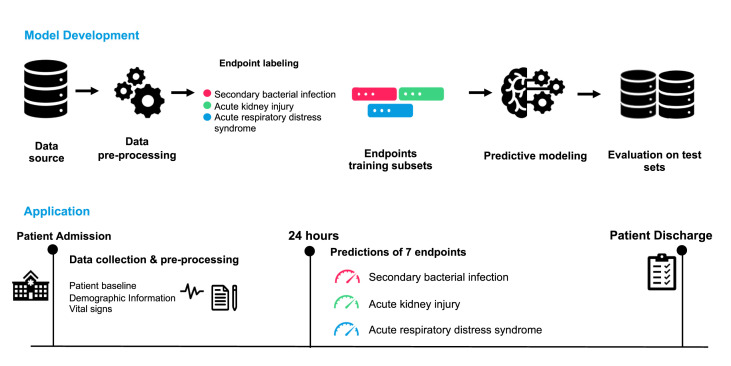
Fig. 1.
Overview of our proposed model development approach and expected application in practice. As shown in the first row, we develop our complication-specific models by first preprocessing the data, identifying the occurrences of the complications based on the criteria shown in Table 2, training and selecting the best-performing models on the validation set, and then evaluating the performance on the test set, retrospectively. As for the application (second row), we expect our system to predict the risk of developing any of the three complications for any patient after 24 h of admission.
2. Methods
This study is reported following the TRIPOD guidance [34].
2.1. Data source
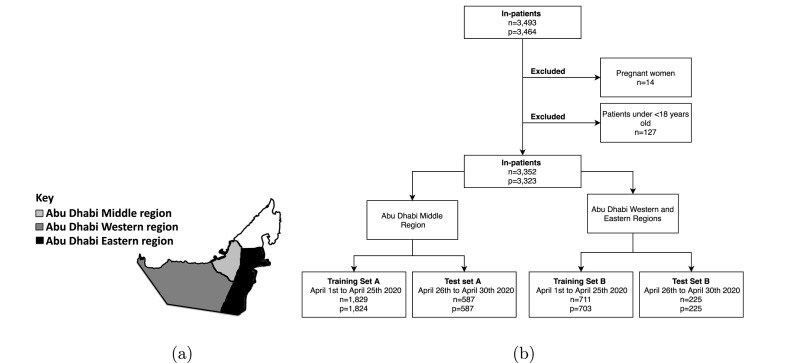
This study is a retrospective multicentre study that includes anonymized data recorded within 3,493 COVID-19 hospital encounters at 18 Abu Dhabi Health Services (SEHA) healthcare facilities in Abu Dhabi, United Arab Emirates. The study received approval by the Institutional Review Board (IRB) from the Department of Health (Ref: DOH/CVDC/2020/1125) and New York University Abu Dhabi (Ref: HRPP-2020-70). Informed consent was not required for this study as it was determined as exempt. All methods were performed in accordance with the relevant guidelines and regulations. There were nine facilities in the Middle region, which includes the capital city, and nine facilities in the Eastern and Western regions. Those regions are highlighted in Fig. 2 (a). Fig. 2(b) shows the flowchart of how the exclusion criteria was applied to obtain the final data splits. We excluded 127 non-adult encounters and 14 pregnant encounters and split the dataset into training and test sets. The training sets were used for model training and selection, while the test sets were used for evaluation. Training set A consisted of 1,829 encounters recorded in the Middle region between April 1, 2020 and April 25, 2020. To evaluate for temporal generalizability, test set A included 587 encounters recorded in the Middle region between April 26, 2020 and April 30, 2020. Training set B included 711 encounters admitted to the Eastern and Western regions between April 1, 2020 and April 25, 2020 and test set B included 225 encounters admitted to the same hospitals between April 26, 2020, and April 30, 2020.
Fig. 2.
(a) The UAE map showcasing the location of the healthcare facilities included in this study. (b) Flowchart for the overall dataset showing how the inclusion and exclusion criteria were applied to obtain the final training and test sets, where n represents the number of patient encounters, and p represents the number of unique patients.
2.2. Outcomes
Based on clinical evidence and in collaboration with clinical experts, we focused on predicting three clinically diagnosed events, SBI, AKI [35] and ARDS [36] that are associated with poor patient prognosis. For each patient encounter in the training and test sets, we identified the first occurrence (i.e., date and time), if any, of each complication based on the criteria shown in Table 2 . SBI is defined based on positive cultures within 24 h of sample collection, AKI is defined based on the KDIGO classification criteria [35], and ARDS is defined based on the Berlin definition [36], which required the processing of free-text chest radiology reports. Further details on the processing of those reports is described in Supplementary Section A.
Table 2.
Criteria used to define the occurrence of complications.
| Complication | Definition | Reference |
|---|---|---|
| SBI |
Positive blood, urine, throat or sputum cultures within 24 h of sample collection |
a |
| AKI | Based on the Kidney Disease Improving Global Guidelines (KDIGO) classification, increase in Serum Creatinine by ≥ 0.3 mg/dl within 48 h | [35] |
| OR | ||
| Increase in Serum Creatinine by ≥ to 1.5 times | ||
| OR | ||
| Urine volume 0.5 ml/kg/h for 6 hb | ||
| ARDS | Based on the Berlin definition, presence of bilateral opacity in radiology reports | [36] |
| AND | ||
| Oxygenation: PaO2/FiO2 ≤ 300 mm Hg | ||
| AND | ||
| Timing: ≤ one week | ||
| AND | ||
| Origin: pulmonary |
Based on SEHA's clinical standards.
Urine output was not measured in our dataset because it is collected in the intensive care unit.
2.3. Input features
We considered data recorded within the first 24 h of admission as input features for the predictive models. This data included continuous and categorical features related to the patient baseline information, demographics, and vital signs. Within the patient's baseline and demographic information, age and Body Mass Index (BMI) were treated as continuous features, whereas pre-existing medical conditions (i.e., hypertension, diabetes, chronic kidney disease, and cancer), symptoms recorded at admission (i.e., cough, fever, shortness of breath, sore throat, and rash) and patient sex were treated as binary features. As for the vital signs, we included seven continuous features, including systolic blood pressure, diastolic blood pressure, respiratory rate, peripheral pulse rate, oxygen saturation, auxiliary temperature, and the Glasgow Coma Score. We selected those features as they are commonly used in early warning score systems [37]. All vital signs measurements were processed into minimum, maximum, and mean statistics. We summarized patient demographics, prevalence of the complications, and the distributions of the input features across the training and test sets.
2.4. Predictive modeling
The proposed system predicts the risk of developing each of the three complications during the patient's stay after 24 h of admission. This is represented by a vector y consisting of three predictions, where each prediction is computed by a complication-specific model, such that
where y complication ∈ [0, 1].
The overall workflow of the model development is depicted in Fig. 1. For each complication-specific model, we excluded from its training and test sets patients who developed that complication prior to the time of prediction. For AKI, we also excluded patients with chronic kidney disease. Then for each complication, our system trains four model ensembles based on four types of base learners: logistic regression (LR), k-nearest neighbors (KNN), support vector machine (SVM) and a light gradient boosting model (LGBM). Missing data was imputed using median imputation for all models except for LGBM, which can natively learn from missing data, and the data was further scaled using min-max scaling for LR and standard scaling for SVM and KNN.
For each type of base learner, the system performs a stratified k-folds cross-validation using the complication's respective training set with k = 3. We performed random hyperparameter search for each base learner [38] with 30 iterations, resulting in three trained models for each hyperparameter set selected per iteration. The choice of random search was motivated by its relative simplicity, and high efficiency and performance compared to other hyperparameter tuning methods [38]. The hyperparameter search ranges are summarized in Supplementary Section B. The ranges were defined based on initial experiments with manually chosen hyperparameters.
We then selected the top two hyperparameter sets whose models achieved the highest average area under the receiving operator characteristic curve (AUROC) on the validation sets, resulting in six trained models. We created an ensemble of those six models, and each model within the ensemble was further calibrated using isotonic regression on its respective validation set to ensure non-harmful decision making [39], except for the LR models. Isotonic regression takes a trained model's raw predictions as inputs, and computes well-calibrated output probabilities. This is done by grouping the raw predictions into bins associated with estimates of empirical probabilities [40]. The final prediction of each complication consisted of an average of the calibrated predictions of all models within an ensemble. All analysis was performed using Python (version 3.7.3). The LR, KNN, and SVM models were implemented using the Python scikit-learn package and the LGBM models were implemented using the LightGBM package [41].
2.5. Model interpretability
We performed post-hoc feature importance analysis using the SHapley Additive exPlanations (SHAP) [42,43]. SHAP values are indicative of the relative importance of the input variables and their impact on the predictions. The analysis was conducted using the open-source SHAP package [43], where we obtained the mean absolute SHAP values of the features for the six models per ensemble. For each feature, the six SHAP values were averaged and then ranked to reveal the overall importance of the features with respect to the ensembled prediction. We present the four top ranked features per complication ensemble for each test set using bar plots.
2.6. Performance assessment
We evaluated each complication ensemble using the AUROC and the area under the precision-recall curve (AUPRC) on the test set. The AUROC is a measure of the model's ability to discriminate between positive (complications) and negative cases (no complication) [44], while the AUPRC is a measure of model robustness when dealing with imbalanced datasets, i.e. unequal distribution of positive and negative cases [45]. The closer the AUROC and AUPRC are to 1, the better the performance of the model. Confidence intervals for all of the evaluation metrics were computed using bootstrapping with 1,000 iterations [46]. We also assessed the calibration of the ensemble, after post-hoc calibration of its trained models, using reliability plots and reported calibration intercepts and slopes [39].
3. Results
A total of 3,352 encounters were included in the study and the statistics of the characteristics of the final data splits are presented in Table 3 . Across all the data splits, the mean age ranges between 39.3 and 45.5 years and the proportion of males ranges between 84.8% and 88.9%. The mortality rate was also less than 4% across all data splits, ranging between 1.3% and 3.7%. ARDS was the most prevalent complication developed in the first 24 h of admission across all datasets. The incidence of the complications developed after 24 h were higher in the test sets than in their respective training sets. The distributions of the vital signs and demographics in terms of the mean and interquartile ranges, are shown in Table 4 .
Table 3.
Summary of the baseline characteristics of the patient cohort in the training sets and test sets and the prevalence of the predicted complications. Note that n represents the total number of patients while % is the proportion of patients within the respective dataset.
| Training set A | Test set A | Training set B | Test set B | |
|---|---|---|---|---|
|
Patient Cohort | ||||
| Encounters, n | 1829 | 587 | 711 | 225 |
| Age, mean (IQR) | 41.7 (17.0) | 45.5 (18.0) | 39.3 (17.0) | 42.7 (20.0) |
| Male, n (%) | 1582 (86.5) | 522 (88.9) | 622 (87.5) | 191 (84.8) |
| Arab, n (%) | 295 (16.1) | 89 (15.2) | 120 (16.9) | 43 (19.1) |
| Non-Arab, n (%) | 1534 (83.9) | 498 (84.8) | 591 (83.1) | 182 (80.9) |
| Mortality, n (%) |
36 (2.0) |
22 (3.7) |
9 (1.3) |
3 (1.3) |
|
Complications | ||||
| SBI, n (%) | 92 (5.0) | 45 (7.7) | 23 (3.2) | 17 (7.6) |
| Developed within 24 h from admission, n (%) | 1 (0.1) | 3 (0.5) | 1 (0.1) | 1 (0.4) |
| Developed after 24 h from admission, n (%) |
91 (5.0) |
42 (7.2) |
22 (3.1) |
16 (7.1) |
| AKI, n (%) | 126 (6.9) | 52 (8.9) | 32 (4.5) | 16 (7.1) |
| Developed within 24 h from admission, n (%) | 28 (1.5) | 9 (1.5) | 14 (2.0) | 3 (1.3) |
| Developed after 24 h from admission, n (%) |
98 (5.4) |
43 (7.3) |
18 (2.5) |
13 (5.8) |
| ARDS, n (%) | 117 (6.4) | 57 (9.7) | 45 (6.3) | 24 (10.7) |
| Developed within 24 h from admission, n (%) | 61 (3.3) | 26 (4.4) | 23 (3.2) | 13 (5.8) |
| Developed after 24 h from admission, n (%) | 56 (3.1) | 31 (5.3) | 22 (3.1) | 11 (4.9) |
Table 4.
Characteristics of the variables that were used as input features to our models. The mean and interquartile ranges are shown for the demographic features, and vital-sign measurements. For the comorbidities and symptoms admission, n denotes the number of patients and % denotes the percentage of patients per the respective dataset.
| Variable, unit | Training set A | Test set A | Training set B | Test set B |
|---|---|---|---|---|
|
Demographics, mean (IQR) | ||||
| Age | 41.7 (17.0) | 45.5 (18.0) | 39.3 (17.0) | 42.7 (20.0) |
| BMI | 26.9 (5.2) | 26.7 (5.7) | 26.5 (5.7) | 27.9 (6.2) |
| Male, n (%) |
1582 (86.5) |
522 (88.9) |
622 (87.5) |
191 (84.8) |
|
Comorbidities, n (%) | ||||
| Hypertension | 550 (30.1) | 213 (36.3) | 168 (23.6) | 71 (31.6) |
| Diabetes | 427 (23.3) | 221 (37.6) | 121 (17.0) | 73 (32.4) |
| Chronic kidney disease | 68 (3.7) | 30 (5.1) | 20 (2.8) | 7 (3.1) |
| Cancer |
30 (1.6) |
7 (1.2) |
12 (1.7) |
8 (3.6) |
|
Symptoms at admission, n (%) | ||||
| Cough | 851 (46.5) | 338 (57.6) | 259 (36.4) | 99 (44.0) |
| Fever | 28 (1.5) | 20 (3.4) | 3 (0.4) | 3 (1.3) |
| Shortness of breath | 190 (10.4) | 99 (16.9) | 71 (10.0) | 34 (15.1) |
| Sore throat | 238 (13.0) | 89 (15.2) | 118 (16.6) | 28 (12.4) |
| Rash |
29 (1.6) |
10 (1.7) |
15 (2.1) |
5 (2.2) |
|
Vital-sign measurements, mean (IQR) | ||||
| Systolic blood pressure, mmHg | 126.3 (15.0) | 126.8 (16.0) | 128.8 (15.5) | 128.2 (15.7) |
| Diastolic blood pressure, mmHg | 77.5 (9.8) | 76.9 (9.9) | 77.9 (10.3) | 77.5 (10.7) |
| Respiratory rate, breaths per minute | 18.9 (1.0) | 20.2 (2.5) | 18.1 (0.7) | 18.7 (0.8) |
| Peripheral pulse rate, beats per minute | 82.6 (11.5) | 85.4 (11.6) | 81.7 (13.4) | 82.5 (12.5) |
| Oxygen saturation, % | 98.4 (1.6) | 97.5 (2.1) | 98.5 (1.0) | 98.2 (1.4) |
| Temperature auxiliary, °C | 36.9 (0.4) | 37.0 (0.7) | 36.9 (0.4) | 37.1 (0.6) |
| Glasgow Coma Score | 14.8 (0.0) | 15.0 (0.0) | 14.8 (0.0) | 14.8 (0.0) |
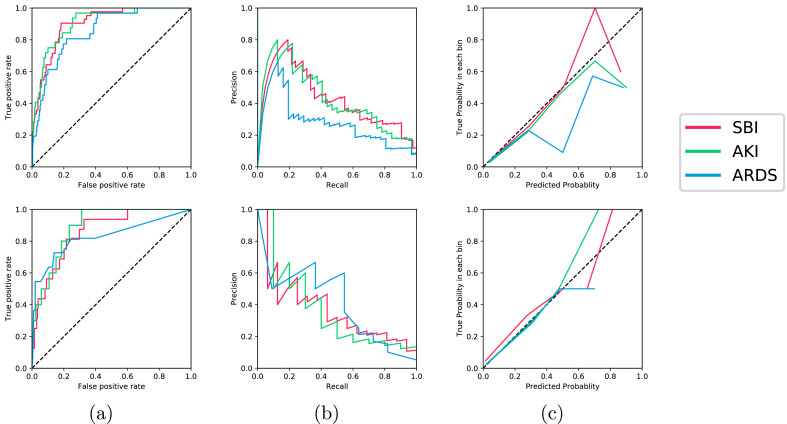
The performance results of the models selected by our system across the two test sets in terms of the AUROC and AUPRC are shown in Table 5 . The Receiver Operating Characteristic curve (ROC), Precesion Recall Curve (PRC), and reliability plots are also visualized in Fig. 3 (a) and (b), and 3(c), respectively. Across both test sets, our data-driven approach achieved good performance (0.82 AUROC) for all of the complications. In test set A, AKI was the best discriminated endpoint at 24 h from admission, with 0.906 AUROC. This is followed by SBI (0.902 AUROC), and SBI (0.854 AUROC). In test set B, AKI was the best discriminated endpoint with 0.891 AUROC, followed by SBI (0.859 AUROC), and ARDS (0.827 AUROC).
Table 5.
Performance evaluation of the best performing models on test sets A & B, which were selected based on the average AUROC performance on the validation sets, as shown in Supplementary Section C. Model type indicates the type of the base learners within the final selected ensemble. All the metrics were computed using bootstrapping with 1,000 iterations [46].
| Complication | Result | Test Set A | Test Set B |
|---|---|---|---|
| SBI | Model Type | LR | LR |
| AUROC | 0.902 (0.862, 0.939) | 0.859 (0.762, 0.932) | |
| AUPRC | 0.436 (0.297, 0.609) | 0.387 (0.188, 0.623) | |
| Calibration Slope | 0.933 (0.321, 1.370) | 1.031 (−0.066, 1.550) | |
| Calibration Intercept |
0.031 (−0.111, 0.213) |
0.010 (−0.164, 0.273) |
|
| AKI | Model Type | LR | LR |
| AUROC | 0.906 (0.856, 0.948) | 0.891 (0.804, 0.961) | |
| AUPRC | 0.436 (0.278, 0.631) | 0.387 (0.115, 0.679) | |
| Calibration Slope | 0.655 (0.043, 1.292) | 1.370 (−0.050, 2.232) | |
| Calibration Intercept |
0.059 (−0.136, 0.251) |
−0.072 (−0.183, 0.154) |
|
| ARDS | Model Type | LR | LGBM |
| AUROC | 0.854 (0.789, 0.909) | 0.827 (0.646, 0.969) | |
| AUPRC | 0.288 (0.172, 0.477) | 0.399 (0.150, 0.760) | |
| Calibration Slope | 0.598 (0.028, 1.149) | 0.742 (−0.029, 1.560) | |
| Calibration Intercept | 0.000 (−0.159, 0.164) | 0.050 (−0.166, 0.243) |
Fig. 3.
The (a) ROC curves, (b) PRC curves, and (c) calibration curves are shown for all model ensembles evaluated on test set A (top) and test set B (bottom). The color legend for all figures is shown on the right. The numerical values for the AUROC, AUPRC, calibration slopes and intercepts can be found in Table 5. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The prevalence of the predicted complications ranged between 3.2%-6.9% and 7.1%–10.7% in the training and test sets, respectively. This high class imbalance is reflected in the AUPRC results, since the AUPRC depends on the prevalence of the outcome and tends to have a low value when there is class imbalance [47].
We also observe that LR was selected as the best performing model on the validation sets for most complications, highlighting its predictive power despite its simplicity compared to the other machine learning models. LGBM was selected for ARDS in test set B, as shown in Supplementary Section C.
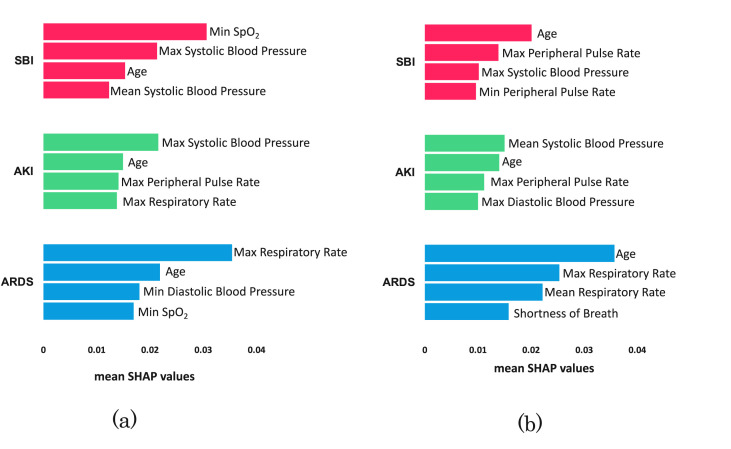
The top four important features for each complication are shown in Fig. 4 across the two test sets. Age was among the top predictive features for all the complications in both test sets. Similarly, systolic blood pressure was one of the top features for predicting SBI and AKI across both sets. Other features such as peripheral pulse rate and respiratory rate were among the top predictive features across both sets, for AKI and ARDS respectively.
Fig. 4.
The four most important features are shown for each complication in (a) test set A and (b) test set B. Feature importance was computed using the average SHAP values of the six models per ensemble.
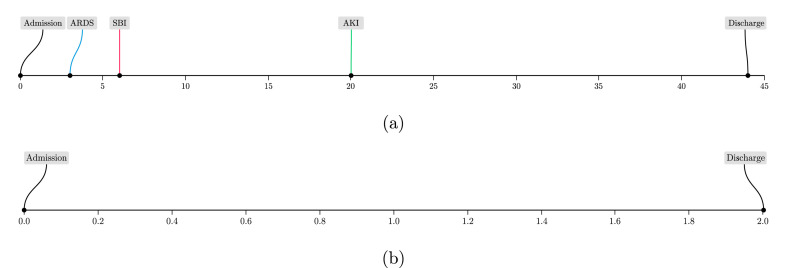
The calibration results show that our ensemble models were adequately calibrated across all complications as the calibration slopes were approximately equal to 1, as shown in Table 5 and Fig. 3(c). This is also reflected in the sample patient timelines visualized in Fig. 5 , where the predicted risks for the patient who experienced the complications were relatively higher than those predicted for the patient who did not experience any complications. In Fig. 5(a), the patient shown developed all three complications during their hospital stay of 44 days. This highlights the importance of predicting all complications simultaneously, especially for patients who may develop more than one complication. In Fig. 5(b), the patient did not develop any complications during their hospital stay of two days. To compare both patients, the system's predictions for patient (a) were relatively higher than those for patient (b). For example, the AKI predictions were 0.73 and 0.002, respectively, despite the fact that patient (a) developed AKI at around 20 days from admission. This demonstrates the value of our system in predicting the risk of developing complications early during the patient's stay.
Fig. 5.
Timeline showing the development of complications with respect to number of days from admission (x-axis) for two sample patients. (a) For [ySBI, yAKI, yARDS], our system predictions (multiplied by a 100 to obtain percentages) were [64%, 73%, 51%]. (b) This patient did not develop and complications and our model predictions were [0.2%, 0.2%, 2%].
4. Discussion
In this study, we developed an automated prognostic system to support patient assessment and triage early on during the patient's stay. We demonstrate that the system can predict the risk of multiple complications simultaneously and achieves a good performance across all complications across two geographically independent datasets. The feature importance analysis revealed that age, systolic blood pressure and respiratory rate are highly predictive of several complications across the two datasets. Since COVID-19 was predominantly a pulmonary illness especially in its early variants [48], it was not surprising that respiratory rate ranked among the highest predictive features. We also identified age and systolic blood pressure as markers for severity among patients with COVID-19, which is aligned with clinical literature [49,50]. Specifically, systolic blood pressure has been determined as an important covariate of morbidity and mortality in patients with COVID-19 [51]. This analysis demonstrates that our system's learning is clinically meaningful and relevant.
In addition, we assessed our models' calibration through reporting the calibration slopes and intercepts and visualized the calibration curves. Sufficiently large datasets are usually needed to produce stable calibration curves at model validation stage [39]. Despite the size of our dataset, we found that reporting the calibration slopes and intercepts would provide a concise summary of potential problems with our system's calibration, to avoid harmful decision-making [39].
One of the main strengths of this study is that we used multicentre data collected at 18 facilities across several regions in Abu Dhabi, UAE. COVID-19 treatment is free for all patients in the UAE, hence there were no obvious gaps in terms of access to healthcare services in our dataset. Across the training and test sets in regions A & B, 15.2%–19.1% of encounters were for Arab patients. This reflects the diversity of our dataset, since Abu Dhabi is residence for more than 200 nationalities, of which only 19.0% of the population is Emirati. This diversity makes our findings relevant to a global audience. While most previous studies have focused on European or Chinese patient cohorts [[52], [53], [54]], our study is one of few studies with large sample sizes (3,352 COVID-19 patient encounters) that focus on the patient cohort in the UAE. Compared to other international patient cohorts, our cohort is relatively younger (39.3–45.5 years across training and test sets), with a lower overall mortality rate (1.3%–3.7% across the training and test sets), suggesting that our system needs to be further validated on populations with different demographic distributions [10,55,56]. Our data-driven approach and open-access code can be easily adapted for such purposes.
Another strength is that our system predicts three complications simultaneously that are indicative of patient severity, in order to avoid poor patient outcomes. From a clinical perspective, several studies reported worse prognosis among patients with COVID-19 who had multi-organ failure, and co-infections [6,8,10,57]. Most of the existing COVID-19 prognostic studies focus on predicting mortality as an adverse event outcome [2]. The low mortality rates in our dataset strongly discouraged the development of a mortality risk prediction score, as such small sample sizes may lead to biased models [2]. An important aspect of this study is that the labeling criteria of the complications rely on renowned clinical standards and hospital-acquired data to identify the exact time of the occurrence of such complications. In collaboration with the clinical experts, this approach was considered more reliable than using International Classification of Disease (ICD) codes [58,59]. Despite the development of new ontologies [60], ICD codes are generally used for billing purposes and their derivation may vary across facilities, especially during a pandemic [61]. We also introduce new benchmark results that can be contested with other competing models on test set B. Future work should also investigate the use of multi-label deep learning classifiers for larger datasets, while accounting for the exclusion criteria during training.
Moreover, our system uses routinely collected data and does not incur high data collection costs. Other prognostic machine learning studies have also adopted this strategy to predict adverse outcomes [62,63]. By using routinely collected data rather than hematologic, cardiac, or biochemical laboratory tests that are associated with high processing times, our system is suitable for low-cost deployment. Existing studies achieved comparable performance with our system. For example, an AKI prediction model achieved 0.78 AUROC using findings of abdominal CT scans, vital-sign measurements, comorbidities, and laboratory-test results [25]. Although the results are not directly comparable due to differences in study design, our system achieved 0.91 and 0.89 AUROC in test sets A & B, respectively, without needing any imaging or laboratory-test results. In another study, an ARDS prediction model achieved 0.89 AUROC using patient demographics, interventions, comorbidities, 17 laboratory-test results and eight vital signs [33]. In comparison, our system achieved 0.85 AUROC in test set A and 0.83 AUROC in test set B. This implies that we should consider including additional variables to improve the performance of the model, such as laboratory-test results. One other study highlighted the predictive ability of eight laboratory-test results for the prediction of sepsis, where it achieved 0.93 AUROC [28]. We avoided the use of laboratory-test results to ensure that there is no overlap between the set of input features and the variables used to define the output complications (i.e. label leakage), however this is an area of future work.
Our study also has several limitations. One limitation of the labeling procedure is that it could miss patients for whom the data used in identifying a particular complication was not collected. However, this issue is more closely related to data collection practices at institutions as clinical data is often not completely missing at random. Another limitation is that since we relied on a minimal feature set, our system does not account for possible effects of treatment on the predicted outcomes and feature interactions, which is an area of future study. Moreover, the models are not perfectly calibrated due to small dataset size, which could also be attributed to the fact that the final predictions are based on model ensembles, rather than an individually calibrated model. Future work should investigate how to further improve the calibration of ensemble models. Furthermore, we utilized a dataset collected during the first wave of the pandemic, which did not include any information indicating the type of variant. Hence, the results presented here may not be directly applicable to patients with new COVID-19 variants. However, the system can be easily reused, fine-tuned, and validated using new datasets.
5. Conclusion
Our data-driven approach and results highlight the promise of machine learning in risk prediction in general and COVID-19 complications in particular. The proposed approach performs well when applied to two independent multicentre training and test sets in the UAE. The system can be easily implemented in practice due to several factors. First, the input features that our system uses are routinely collected by hospitals that accommodate patients with COVID-19 as recommended by the World Health Organization. Second, training the machine learning models within our system does not require high computational resources. Finally, through feature importance analysis, our system can offer interpretability, and is also fully automated as it does not require any manual interventions. To conclude, we propose a clinically applicable system that predicts complications among patients with COVID-19. Our system can serve as a guide to anticipate the course of patients with COVID-19 and to help initiate more targeted and complication-specific decision-making on treatment and triage.
Contributors
GOG, BA, and KWY managed and analyzed the data. FS and II extracted, anonymized, and provided the dataset for analysis. GOG, KWY, and NH developed and maintained the experimental codebase. FAK, SAJ, MAS, RA, and MAH provided clinical expertise. WZ, FS, MAH and FES designed the study. MAH and FES supervised the work. GOG, BA, KWY, and FES wrote the manuscript. All authors interpreted the results and revised and approved the final manuscript.
Funding
The work of FES, GOG, BA, KWY, RA and NH is funded by NYU Abu Dhabi, and the work of FS, II, FAK, SAJ, MAS, and MAH is funded by Abu Dhabi Health Services. This work was also supported by the NYUAD Center for Interacting Urban Networks (CITIES), funded by Tamkeen under the NYUAD Research Institute Award CG001.
Patient and public involvement
No patient involvement.
Data availability statement
To allow for reproducibility and benchmarking on our dataset, we are sharing test set B (n = 225) at https://github.com/nyuad-cai/COVID19Complications. We are unable to share the full dataset used in this study due to restrictions by the data provider. The trained models and the source code of the pipeline are also included in the repository.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Waqqas Zia and Benoit Marchand from the Dalma team at NYU Abu Dhabi for supporting access to computational resources and Philip P. Rodenbough from the NYU Abu Dhabi Writing Center for revising the manuscript. This study was supported through the data resources and staff expertise provided by Abu Dhabi Health Services.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ibmed.2022.100065.
Appendices. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Dong Ensheng, Du Hongru, Gardner Lauren. An interactive web-based dashboard to track covid-19 in real time. The Lancet Infectious Diseases. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wynants Laure, Van Calster Ben, Collins Gary S., Riley Richard D., Heinze Georg, Schuit Ewoud, Bonten Marc MJ., Dahly Darren L., Damen Johanna AA., Debray Thomas PA., et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall John C., Murthy Srinivas, Diaz Janet, Adhikari Neil, Angus Derek C., Arabi Yaseen M., Baillie Kenneth, Bauer Michael, Scott Berry, Blackwood Bronagh, et al. A minimal common outcome measure set for covid-19 clinical research. The Lancet Infectious Diseases. 2020;20(8):e192–e197. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cau Riccardo, Faa Gavino, Nardi Valentina, Balestrieri Antonella, Puig Josep, Suri Jasjit S., SanFilippo Roberto, Saba Luca. Long-covid diagnosis: from diagnostic to advanced ai-driven models. European Journal of Radiology. 2022;148 doi: 10.1016/j.ejrad.2022.110164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauring Adam S., Tenforde Mark W., Chappell James D., Gaglani Manjusha, Ginde Adit A., McNeal Tresa, Ghamande Shekhar, Douin David J., Talbot H Keipp, Casey Jonathan D., et al. Clinical severity of, and effectiveness of mrna vaccines against, covid-19 from omicron, delta, and alpha sars-cov-2 variants in the United States: prospective observational study. bmj. 2022;376 doi: 10.1136/bmj-2021-069761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Wan, Sun Nan-Nan, Gao Hai-Nv, Chen Zhi-Yuan, Yang Ya, Ju Bin, Tang Ling-Ling. Risk factors analysis of covid-19 patients with ards and prediction based on machine learning. Scientific Reports. 2021;11(1):1–12. doi: 10.1038/s41598-021-82492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yildirim Cigdem, Selcuk Ozger Hasan, Yasar Emre, Tombul Nazrin, Gulbahar Ozlem, Yildiz Mehmet, Bozdayi Gulendam, Derici Ulver, Dizbay Murat. Early predictors of acute kidney injury in covid-19 patients. Nephrology. 2021;26:513–521. doi: 10.1111/nep.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ripa Marco, Galli Laura, Poli Andrea, Oltolini Chiara, Spagnuolo Vincenzo, Mastrangelo Andrea, Muccini Camilla, Monti Giacomo, De Luca Giacomo, Landoni Giovanni, et al. Secondary infections in patients hospitalized with covid-19: incidence and predictive factors. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2021;27(3):451–457. doi: 10.1016/j.cmi.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Dawei, Hu Bo, Hu Chang, Zhu Fangfang, Liu Xing, Zhang Jing, Wang Binbin, Xiang Hui, Cheng Zhenshun, Xiong Yong, Zhao Yan, Li Yirong, Wang Xinghuan, Peng Zhiyong. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in wuhan, China. JAMA. 03 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Fei, Ting Yu, Du Ronghui, Fan Guohui, Liu Ying, Liu Zhibo, Xiang Jie, Wang Yeming, Song Bin, Gu Xiaoying, Guan Lulu, Yuan Wei, Li Hui, Wu Xudong, Xu Jiuyang, Tu Shengjin, Zhang Yi, Chen Hua, Cao Bin. Clinical course and risk factors for mortality of adult inpatients with covid-19 in wuhan, China: a retrospective cohort study. The Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar Pankaj, Kumar Malay. Management of potential ventilator shortage in India in view of on-going covid-19 pandemic. Indian Journal of Anaesthesia. 2020;64(Suppl 2):S151. doi: 10.4103/ija.IJA_342_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Xiangao, Coffee Megan, Bari Anasse, Wang Junzhang, Jiang Xinyue, Huang Jianping, Shi Jichan, Dai Jianyi, Cai Jing, Zhang Tianxiao, et al. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Computers, Materials & Continua. 2020;63(1):537–551. [Google Scholar]
- 13.Yang Xiaobo, Yuan Yu, Xu Jiqian, Shu Huaqing, Liu Hong, Wu Yongran, Zhang Lu, Yu Zhui, Fang Minghao, Ting Yu, et al. Clinical course and outcomes of critically ill patients with sars-cov-2 pneumonia in wuhan, China: a single-centered, retrospective, observational study. The Lancet Respiratory Medicine. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai Chih-Cheng, Wen-Chien Ko, Ping-Ing Lee, Shio-Shin Jean, Po-Ren Hsueh. Extra-respiratory manifestations of covid-19. International Journal of Antimicrobial Agents. 2020;56(2) doi: 10.1016/j.ijantimicag.2020.106024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch Jamie S., Ng Jia H., Ross Daniel W., Sharma Purva, Shah Hitesh H., Barnett Richard L., Hazzan Azzour D., Fishbane Steven, Jhaveri Kenar D., Abate Mersema, et al. Acute kidney injury in patients hospitalized with covid-19. Kidney International. 2020;98(1):209–218. doi: 10.1016/j.kint.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaillancourt Mylene, Jorth Peter. vaillancourt2020unrecognized MBio. 2020;11(4) doi: 10.1128/mBio.01806-20. e01806–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palacios Gustavo, Hornig Mady, Cisterna Daniel, Savji Nazir, Bussetti Ana Valeria, Kapoor Vishal, Hui Jeffrey, Tokarz Rafal, Briese Thomas, Baumeister Elsa, et al. Streptococcus pneumoniae coinfection is correlated with the severity of h1n1 pandemic influenza. PLoS One. 2009;4(12):e8540. doi: 10.1371/journal.pone.0008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chertow Daniel S., Memoli Matthew J. Bacterial coinfection in influenza: a grand rounds review. JAMA. 2013;309(3):275–282. doi: 10.1001/jama.2012.194139. [DOI] [PubMed] [Google Scholar]
- 19.Feldman Charles, Anderson Ronald. The role of co-infections and secondary infections in patients with covid-19. Pneumonia. 2021;13(1):1–15. doi: 10.1186/s41479-021-00083-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kourou Konstantina, Exarchos Themis P., Exarchos Konstantinos P., Karamouzis Michalis V., Fotiadis Dimitrios I. Machine learning applications in cancer prognosis and prediction. Computational and Structural Biotechnology Journal. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koyner Jay L., Carey Kyle A., Edelson Dana P., Churpek Matthew M. The development of a machine learning inpatient acute kidney injury prediction model. Critical Care Medicine. 2018;46(7):1070–1077. doi: 10.1097/CCM.0000000000003123. [DOI] [PubMed] [Google Scholar]
- 22.Islam Md Mohaimenul, Nasrin Tahmina, Andreas Walther Bruno, Wu Chieh-Chen, Yang Hsuan-Chia, Li Yu-Chuan. Prediction of sepsis patients using machine learning approach: a meta-analysis. Computer methods and programs in biomedicine. 2019;170:1–9. doi: 10.1016/j.cmpb.2018.12.027. [DOI] [PubMed] [Google Scholar]
- 23.Alballa Norah, Al-Turaiki Isra. Machine learning approaches in covid-19 diagnosis, mortality, and severity risk prediction: a review. Informatics in Medicine Unlocked. 2021;24 doi: 10.1016/j.imu.2021.100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montazeri Mahdieh, ZahediNasab Roxana, Ali Farahani, Mohseni Hadis, Ghasemian Fahimeh, et al. Machine learning models for image-based diagnosis and prognosis of covid-19: systematic review. JMIR medical informatics. 2021;9(4) doi: 10.2196/25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hectors Stefanie J., Riyahi Sadjad, Dev Hreedi, Krishnan Karthik, Margolis Daniel JA., Prince Martin R. Multivariate analysis of ct imaging, laboratory, and demographical features for prediction of acute kidney injury in covid-19 patients: a bi-centric analysis. Abdominal Radiology. 2021;46(4):1651–1658. doi: 10.1007/s00261-020-02823-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langford Bradley J., So Miranda, Raybardhan Sumit, Leung Valerie, Duncan Westwood, MacFadden Derek R., R Soucy Jean-Paul, Daneman Nick. Bacterial co-infection and secondary infection in patients with covid-19: a living rapid review and meta-analysis. Clinical Microbiology and Infection. 2020 doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafran Noa, Shafran Inbal, Ben-Zvi Haim, Sofer Summer, Sheena Liron, Krause Ilan, Shlomai Amir, Goldberg Elad, Sklan Ella H. Secondary bacterial infection in covid-19 patients is a stronger predictor for death compared to influenza patients. Scientific Reports. 2021;11(1):1–8. doi: 10.1038/s41598-021-92220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Guoxing, Luo Ying, Lu Feng, Li Wei, Liu Xiongcheng, Nan Yucen, Ren Yufei, Liao Xiaofei, Wu Song, Jin Hai, et al. Prediction of sepsis in covid-19 using laboratory indicators. Frontiers in Cellular and Infection Microbiology. 2020;10 doi: 10.3389/fcimb.2020.586054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamout Farah E., Shen Yiqiu, Wu Nan, Kaku Aakash, Park Jungkyu, Makino Taro, Jastrzebski Stanislaw, Jan Witowski, Wang Duo, Zhang Ben, et al. An artificial intelligence system for predicting the deterioration of covid-19 patients in the emergency department. NPJ digital medicine. 2021;4(1):1–11. doi: 10.1038/s41746-021-00453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Booth Adam L., Abels Elizabeth, McCaffrey Peter. Development of a prognostic model for mortality in covid-19 infection using machine learning. Modern Pathology. 2021;34(3):522–531. doi: 10.1038/s41379-020-00700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lassau Nathalie, Ammari Samy, Chouzenoux Emilie, Gortais Hugo, Paul Herent, Devilder Matthieu, Soliman Samer, Meyrignac Olivier, Talabard Marie-Pauline, Lamarque Jean-Philippe, et al. Integrating deep learning ct-scan model, biological and clinical variables to predict severity of covid-19 patients. Nature communications. 2021;12(1):1–11. doi: 10.1038/s41467-020-20657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon Young Joon, Toussie Danielle, Finkelstein Mark, Cedillo Mario A., Maron Samuel Z., Manna Sayan, Voutsinas Nicholas, Corey Eber, Adam Jacobi, Adam Bernheim, et al. Combining initial radiographs and clinical variables improves deep learning prognostication in patients with covid-19 from the emergency department. Radiology: Artificial Intelligence. 2020;3(2) doi: 10.1148/ryai.2020200098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singhal Lakshya, Garg Yash, Yang Philip, Tabaie Azade, Wong A Ian, Mohammed Akram, Chinthala Lokesh, Kadaria Dipen, Sodhi Amik, Holder Andre L., et al. eards: a multi-center validation of an interpretable machine learning algorithm of early onset acute respiratory distress syndrome (ards) among critically ill adults with covid-19. PloS one. 2021;16(9) doi: 10.1371/journal.pone.0257056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins Gary S., Reitsma Johannes B., Altman Douglas G., Moons Karel GM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (tripod) the tripod statement. British Journal of Surgery. 2015;102:148–158. doi: 10.1002/bjs.9736. [DOI] [PubMed] [Google Scholar]
- 35.Khwaja Arif. Kdigo clinical practice guidelines for acute kidney injury. Nephron Clinical Practice. 2012;120(4):c179–c184. doi: 10.1159/000339789. [DOI] [PubMed] [Google Scholar]
- 36.The ARDS Definition Task Force Acute respiratory distress syndrome: the Berlin definition. JAMA. 06 2012;307(23):2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 37.Prytherch David R., Smith Gary B., Schmidt Paul E., Featherstone Peter I. Views—towards a national early warning score for detecting adult inpatient deterioration. Resuscitation. 2010;81(8):932–937. doi: 10.1016/j.resuscitation.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 38.James Bergstra, Bengio Yoshua. Random search for hyper-parameter optimization. The Journal of Machine Learning Research. 2012;13(1):281–305. [Google Scholar]
- 39.Ben Van Calster, Nieboer Daan, Vergouwe Yvonne, De Cock Bavo, Pencina Michael J., Steyerberg Ewout W. A calibration hierarchy for risk models was defined: from utopia to empirical data. Journal of Clinical Epidemiology. 2016;74:167–176. doi: 10.1016/j.jclinepi.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Otto Nyberg, Klami Arto. Pacific-asia conference on knowledge discovery and data mining. Springer; 2021. Reliably calibrated isotonic regression; pp. 578–589. [Google Scholar]
- 41.Guolin Ke, Qi Meng, Finley Thomas, Wang Taifeng, Chen Wei, Ma Weidong, Ye Qiwei, Liu Tie-Yan. In: Guyon I., Luxburg U.V., Bengio S., Wallach H., Fergus R., Vishwanathan S., Garnett R., editors. vol. 30. Curran Associates, Inc.; 2017. Lightgbm: a highly efficient gradient boosting decision tree; pp. 3146–3154. (Advances in neural information processing systems). [Google Scholar]
- 42.Lundberg Scott M., Gabriel Erion, Chen Hugh, DeGrave Alex, Prutkin Jordan M., Nair Bala, Katz Ronit, Himmelfarb Jonathan, Bansal Nisha, Su-In Lee. From local explanations to global understanding with explainable ai for trees. Nature machine intelligence. 2020;2(1):2522–5839. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott Lundberg, Su-In Lee. 2017. A unified approach to interpreting model predictions. arXiv:1705.07874 [preprint] [Google Scholar]
- 44.Janssens A.Cecile JW., Martens Forike K. Reflection on modern methods: revisiting the area under the roc curve. International journal of epidemiology. 2020;49(4):1397–1403. doi: 10.1093/ije/dyz274. [DOI] [PubMed] [Google Scholar]
- 45.Saito Takaya, Rehmsmeier Marc. The precision-recall plot is more informative than the roc plot when evaluating binary classifiers on imbalanced datasets. PloS one. 2015;10(3) doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.J DiCiccio Thomas, Bradley Efron. Bootstrap confidence intervals. Statistical Science. 1996:189–212. [Google Scholar]
- 47.Ozenne Brice, Subtil Fabien, Maucort-Boulch Delphine. The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. Journal of Clinical Epidemiology. 2015;68(8):855–859. doi: 10.1016/j.jclinepi.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Alexandridi Magdalini, Mazej Julija, Palermo Enrico, Hiscott John. Cytokine & Growth Factor Reviews; 2022. The coronavirus pandemic–2022: viruses, variants & vaccines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pijls Bart G., Jolani Shahab, Atherley Anique, Derckx Raissa T., Dijkstra Janna IR., Franssen Gregor HL., Hendriks Stevie, Richters Anke, Venemans-Jellema Annemarie, Zalpuri Saurabh, et al. Demographic risk factors for covid-19 infection, severity, icu admission and death: a meta-analysis of 59 studies. BMJ open. 2021;11(1) doi: 10.1136/bmjopen-2020-044640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ran Jinjun, Song Ying, Zhuang Zian, Han Lefei, Zhao Shi, Cao Peihua, Geng Yan, Xu Lin, Qin Jing, He Daihai, et al. Blood pressure control and adverse outcomes of covid-19 infection in patients with concomitant hypertension in wuhan, China. Hypertension Research. 2020:1–10. doi: 10.1038/s41440-020-00541-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caillon Antoine, Zhao Kaiqiong, Oros Klein Kathleen, Greenwood Celia MT., Lu Zhibing, Paradis Pierre, Schiffrin Ernesto L. High systolic blood pressure at hospital admission is an important risk factor in models predicting outcome of covid-19 patients. American Journal of Hypertension. 2021;34(3):282–290. doi: 10.1093/ajh/hpaa225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Xuedi, Ng Michael, Xu Shuang, Xu Zhouming, Qiu Hui, Liu Yuwei, Lyu Jiayou, You Jiwen, Zhao Peng, Wang Shihao, et al. Development and validation of prognosis model of mortality risk in patients with covid-19. Epidemiology and Infection. 2020;148:e168. doi: 10.1017/S0950268820001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian Zhaozhi, Alaa Ahmed M., van der Schaar Mihaela. Cpas: the UK's national machine learning-based hospital capacity planning system for covid-19. Machine Learning. 2021;110(1):15–35. doi: 10.1007/s10994-020-05921-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Simin, Lin Yulan, Zhu Tong, Fan Mengjie, Xu Shicheng, Qiu Weihao, Chen Can, Li Linfeng, Wang Yao, Yan Jun, et al. Development and external evaluation of predictions models for mortality of covid-19 patients using machine learning method. Neural Computing and Applications. 2021;(1–10) doi: 10.1007/s00521-020-05592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goyal Parag, Choi Justin J., Pinheiro Laura C., Schenck Edward J., Chen Ruijun, Jabri Assem, Satlin Michael J., Campion Thomas R., Nahid Musarrat, Ringel Joanna B., Hoffman Katherine L., Alshak Mark N., Li Han A., Wehmeyer Graham T., Rajan Mangala, Reshetnyak Evgeniya, Hupert Nathaniel, Evelyn M., Horn Fernando J. Martinez, Gulick Roy M., Safford Monika M. Clinical characteristics of covid-19 in New York city. New England Journal of Medicine. 2020;382(24):2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soo Hong Kyung, Ho Lee Kwan, Hong Chung Jin, Kyeong-Cheol Shin, Young Choi Eun, Jung Jin Hyun, Geol Jang Jong, Lee Wonhwa, Hong Ahn June. Clinical features and outcomes of 98 patients hospitalized with sars-cov-2 infection in daegu, South Korea: a brief descriptive study. Yonsei Medical Journal. 2020;61(5):431. doi: 10.3349/ymj.2020.61.5.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirsch Jamie S., Ng Jia H., Ross Daniel W., Sharma Purva, Shah Hitesh H., Barnett Richard L., Hazzan Azzour D., Fishbane Steven, Jhaveri Kenar D., Abate Mersema, et al. Acute kidney injury in patients hospitalized with covid-19. Kidney International. 2020;98(1):209–218. doi: 10.1016/j.kint.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higgins Thomas L., Deshpande Abhishek, Zilberberg Marya D., Lindenauer Peter K., Imrey Peter B., Pei-Chun Yu, Haessler Sarah D., Richter Sandra S., Rothberg Michael B. Assessment of the accuracy of using icd-9 diagnosis codes to identify pneumonia etiology in patients hospitalized with pneumonia. JAMA network open. 2020;3(7) doi: 10.1001/jamanetworkopen.2020.7750. e207750–e207750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burnham Jason P., Kwon Jennie H., Babcock Hilary M., Olsen Margaret A., Kollef Marin H. Icd-9-cm coding for multidrug resistant infection correlates poorly with microbiologically confirmed multidrug resistant infection. infection control & hospital epidemiology. 2017;38(11):1381–1383. doi: 10.1017/ice.2017.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan Ling, Song Justin, He Virginia, Roman Jennifer, Whah Grace, Peng Suyuan, Zhang Luxia, He Yongqun. Development of the international classification of diseases ontology (icdo) and its application for covid-19 diagnostic data analysis. BMC bioinformatics. 2021;22(6):1–19. doi: 10.1186/s12859-021-04402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crabb Brendan T., Lyons Ann, Bale Margaret, Martin Valerie, Berger Ben, Mann Sara, West William B., Brown Alyssa, Peacock Jordan B., Leung Daniel T., et al. Comparison of international classification of diseases and related health problems, tenth revision codes with electronic medical records among patients with symptoms of coronavirus disease 2019. JAMA network open. 2020;3(8) doi: 10.1001/jamanetworkopen.2020.17703. e2017703–e2017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barton Christopher, Chettipally Uli, Zhou Yifan, Jiang Zirui, Lynn-Palevsky Anna, Le Sidney, Calvert Jacob, Das Ritankar. Evaluation of a machine learning algorithm for up to 48-hour advance prediction of sepsis using six vital signs. Computers in Biology and Medicine. 2019;109:79–84. doi: 10.1016/j.compbiomed.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Nehemiah T., Holcomb John B., Wade Charles E., Darrah Mark I., Salinas Jose. Utility of vital signs, heart rate variability and complexity, and machine learning for identifying the need for lifesaving interventions in trauma patients. Shock. 2014;42(2):108–114. doi: 10.1097/SHK.0000000000000186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
To allow for reproducibility and benchmarking on our dataset, we are sharing test set B (n = 225) at https://github.com/nyuad-cai/COVID19Complications. We are unable to share the full dataset used in this study due to restrictions by the data provider. The trained models and the source code of the pipeline are also included in the repository.