Abstract
Neurodevelopmental disorders (NDDs), including autism spectrum disorder (ASD) and intellectual disability (ID), are pervasive, often lifelong disorders, lacking evidence‐based interventions for core symptoms. With no established biological markers, diagnoses are defined by behavioral criteria. Thus, preclinical in vivo animal models of NDDs must be optimally utilized. For this reason, experts in the field of behavioral neuroscience convened a workshop with the goals of reviewing current behavioral studies, reports, and assessments in rodent models. Goals included: (a) identifying the maximal utility and limitations of behavior in animal models with construct validity; (b) providing recommendations for phenotyping animal models; and (c) guidelines on how in vivo models should be used and reported reliably and rigorously while acknowledging their limitations. We concluded by recommending minimal criteria for reporting in manuscripts going forward. The workshop elucidated a consensus of potential solutions to several problems, including revisiting claims made about animal model links to ASD (and related conditions). Specific conclusions included: mice (or other rodent or preclinical models) are models of the neurodevelopmental insult, not specifically any disorder (e.g., ASD); a model that perfectly recapitulates a disorder such as ASD is untenable; and greater attention needs be given to validation of behavioral testing methods, data analysis, and critical interpretation.
Keywords: autism, behavior, developmental, genetic, genetic disorder, intellectual disability, models, mouse models, neurodevelopmental disorder, social, syndrome
1. INTRODUCTION
1.1. Why are animal models for neurodevelopmental disorders so important?
Neurodevelopmental disorders (NDDs), including autism spectrum disorder (ASD), and intellectual disability (ID), are pervasive, typically lifelong disorders, for which effective, evidence‐based interventions for core symptoms are not universally available. ASD reportedly affects a significant number of individuals and significantly overlaps with ID, a disorder with a prevalence rate of approximately 1:44 or 2%. 1 , 2 Diagnostic criteria for ASD, outlined by the Diagnostic and Statistical Manual of Mental Disorders, 5th edition,(DSM 5) 3 are purely behavioral, with symptoms including impairments in social communication and interaction along with repetitive behaviors, restricted interests, and behavioral inflexibility. The current consensus is that causes, including genetic and environmental etiologies, and clinical behavioral presentations of NDDs are highly heterogeneous. ASD and ID often co‐occur with each other and with other conditions including seizure disorders, motor problems, and numerous other psychiatric diagnoses.
Genetic work in ASD (also relevant for ID) has identified a broad collection of potential risk genes, nearing ~900 in total, (see https://gene.sfari.org/database/gene-scoring/) with varying level of confidence about their specificity to ASD. Findings from large‐scale whole exome and whole genome studies continue to add to the growing list of high confidence risk genes for ASD. Current estimates include 102 genes, with 26 reaching the highest confidence threshold. 4 , 5 Genes described include de novo and inherited variants, as well as autosomal recessive and X‐linked variants. 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 Although forging definitive links between genetic variants and behavioral impairments is challenging, numerous behavioral assays relevant to the diagnostic domains of ASD and ID have provided researchers with tools to gain insight into a specific genetic variant's impact on behavioral features. To date, the most frequently employed animal models are made in mice with a mutation in one of the many ASD risk genes. 5 , 14 , 15 , 16 , 17 , 18 , 19 Although, given rats' exceptionally higher signal of social play, compared with the more commonly used mouse, some are beginning to highlight the value and potential cross‐species convergence and divergence between mouse and rat rodent models. 20 , 21 , 22 , 23
Many efforts to date have addressed challenges relating to developing and testing model systems of NDDs. 24 These efforts have identified problems in experimental design and study interpretation and have offered suggested solutions. 25 However, some of these recurring issues have not been addressed sufficiently. These issues include: reliability, construct validity, convergent validity, criterion validity, discriminant validity, face validity, predictive validity, translatability and rigor. 24 Of particular importance and highlighted by our workshop discussion are the concepts of face validity and construct validity. Face validity, or “the degree of phenotypic similarity to disease‐specific symptoms”, as defined by the APA, is an important aspect of validity, due to the complex behaviors that are delayed or deficient in NDDs, but limitations are evident, 24 including that ASD is a uniquely human disorder. Construct validity, defined by the APA, as the “degree to which a model system is capable of measuring a concept or trait”, is the more preferred method of validation. Animal models with the strongest construct validity are currently the high confidence risk genetic models. While ASD etiology is multi‐factorial, making construct validity “slippery” at times, 26 these high confidence rare genetic variants may contribute up to 27% of ASD risk. 27 Now that soon we model nearly a third of ASD with strong construct validity, the field is in critical need of a re‐examination of the way behavioral assays are used, interpreted and the findings reported. To allow a discussion of these issues, the Autism Science Foundation convened a workshop that included two virtual meetings in October and December 2020. Goals of the workshop were to: (a) identify and discuss needs for maximal use/utility of behavior in animal models for ASD and NDDs, (b) provide recommendations to prevent over‐reaching claims regarding animal models, and (c) suggest guidelines on how behavior in models should be used reliably and rigorously while acknowledging their limitations.
Based on discussions held at these meetings, we outline the scope of current problems with reporting of behavioral assays used in ASD/NDD research and offer potential solutions.
1.1.1. Scope of the problem
We started by focusing on construct validity as it pertains to ASD, which ideally mimicks the molecular and/or structural basis of the disorder. 24 After initial studies of a handful of genetic conditions relevant to ASD, an outpouring of novel genetic models were of substantial focus, including PTEN, neuroligins, neurexins, and shanks. 21 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 While there have now been many publications of so‐called “mouse models of ASD" (see for example, reporting of 74 different genetic models), 45 only a portion of these are based on genes highly associated with ASD (others are based on environmental exposure or those with low or no association to evidence‐based risk). Unfortunately, despite this plethora of potential models, there are still only two FDA approved therapeutics with indications for irritability and aggression in ASD and no approved compounds for the core features, impaired social communication and repetitive, restricted behaviors. 46 Our goal was to focus on the employment of strategies with rigorous approaches, collaboration, and harmonization, which will ultimately speed translation into therapeutics that can be used in humans. The FDA requires some in vivo animal model efficacy for new drug applications, ideally data illustrating functional improvements. However, many behavioral assays are rudimentary, do not engage similar neural circuitry as in humans, and lack translational face validity. 47 , 48 Given behaviors in a model system with apparent face validity to symptoms of ASD (specifically social communication deficits together with the presence of restrictive and repetitive behaviors) are never going to look the same as those displayed in a human, it is imperative that researchers avoid exaggerating the model system's relevance by recognizing and stating that not every behavioral feature in an animal model should be expected to completely phenocopy the complex and heterogeneous features of NDDs in humans. Thus, to enhance the likelihood of developing efficacious therapeutics, rigorous, reliable, objective, and quantifiable preclinical behavioral outcome measures must be available. This led us to ask the following question:
1.2. What is the current state of behavioral assays in genetic mouse models of NDDs?
To understand the diverse practices of scientists investigating model systems, we reviewed the literature on factors critical to experimental design that influenced interpretation of 69 models from a total of 148 preclinical models specified to ASD (which are relevant to NDDs more generally but were reported as specific to ASD). These 69 models included one of the 26 genes (highest statistical significance of the total 102 identified by Satterstrom et al., 5 ). The goal was not to be comprehensive of the existing reported models but, rather, to focus on construct valid genetic models by utilizing high‐confidence risk genes to gather key, reproducible, valid metrics on commonly used behavioral assays. We focused on genetic rather than environmental factors because of their higher potential for inter‐laboratory reproducibility and their known construct validity. We limited the search to the past 5 years, resulting in 69 publications. Our analysis is detailed in Figure 1. These studies included only rare genetic variants, not copy number variants (CNVs), which are also critical to ASD etiology, see Hyman 2021, 49 polygenic cases, or gene‐by‐environment or ‐immune interaction models of ASD. Of note, SFARI has published an outstanding, highly detailed summary of a larger array of mouse models, which goes beyond this report to include the specific behavioral assays utilized here: https://gene.sfari.org/database/animal-models/genetic-animal-models/.
FIGURE 1.
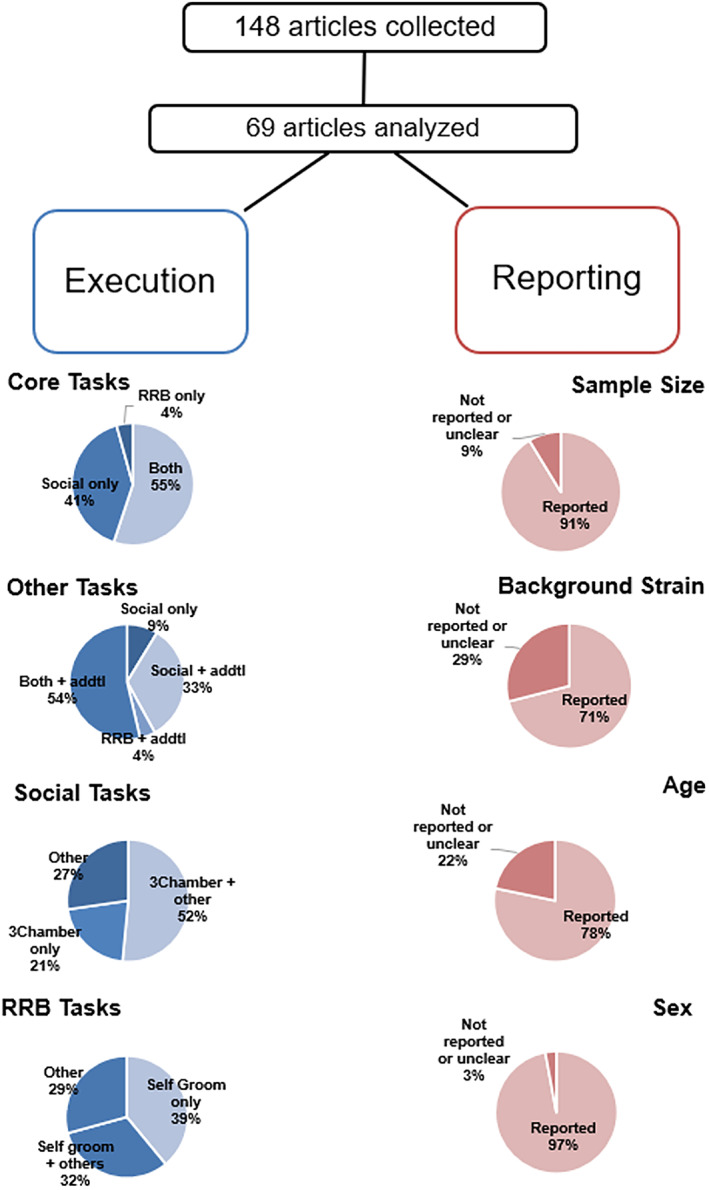
Literature review of behavioral phenotyping of the so‐called "autism spectrum disorder (ASD) mouse models" reveals inconsistent patterns in behavior execution and reporting. A literature review of the so‐called "mouse models of ASD" was conducted to observe trends in behavioral phenotyping execution and reporting. We analyzed how often core diagnostic criteria of ASD were assessed in mouse models, how often these criteria were paired with other behavioral domains, and how often common assays used in social and repetitive and restricted behaviors were used as singular outcomes for each domain. We also highlighted how often four key features of experimental design were not clearly reported to highlight gaps in reporting that may inhibit or prohibit successful interpretation and reproduction of data published
Results from our analysis revealed: clear omissions in some parameters from behavioral assays, failure to consistently report background strains and/or breeding schema, scarce level of detail in testing protocols (e.g., order of tasks in a testing battery, age at time of tests, sample sizes, and statistical approaches). We also investigated the use of behavioral tests reported to measure core ASD symptoms, how the assays were performed, how they were reported, and any other data that was collected. Findings observed included: a lack of adequate statistical power, differing reporting practices, varying procedures for testing (even within the same named assay), variability in assays to assess different types of social or repetitive behaviors, utilization of behavioral assays for broad interpretations of complex behavior, and other sources of variability which undoubtedly influence and result in failures to reproduce. These findings indicate a need for detailed and clarified reporting practices, as outlined in Table 1.
TABLE 1.
Variables for consideration, key to behavioral outcomes
| Guidance | Addresses | Recommend | References |
|---|---|---|---|
| Sample size blinding randomization | Power false reporting | N = 15–20 per genotype/treatment/sex | 50, 51, 52 |
| Age and sex | Age‐dependent effects | One sex cannot be tested; genders cannot be combined with powered Ns and statistics showing no difference; Animals of wildly different ages cannot be compared | 53, 54, 55 |
| Sex‐specific effects | |||
| Breeding schema | Developmental environment | WT × WT and KO × KO is unacceptable; a KO's maternal behavior is likely different than WT; thus, het by het breeding recommended | 51, 52, 55, 56, 57, 58, 59 |
| Housing conditions | Developmental environment | Housed within genotype/treatment or mixed genotype/treatment; house isolated or grouped | 51, 52, 58, 60 |
| Background strain | Background genetics | Consistent reporting/congenic when possible | 55, 61, 62 |
| Order of testing | Test‐re test influencing on behavior | Non stressful to stressful | 63, 64, 65 |
| Littermates or litter effects | Developmental environment consistency | Reporting and accounting for appropriate control subjects accounting for maternal care; litter size | 66, 67, 68, 69 |
| Task validation | Tests must be validated to measure what you are assessing. | Examples include a benzodiazepine will change behavior in an anxiety test and/or a sedative lower motor activity | 57 |
| Reproducibility | Chance findings/type 1 error | For rigorous, reliable behavioral outcome measures | 59, 70 |
1.3. What we learned and agreed upon
1.3.1. Statement #1: Complete face validity should not be expected to be fully apparent
While workshop participants agreed that many aspects of validity are critical to animal models, including construct, face, and predictive validity, 61 , 71 , 72 , 73 , 74 some aspects of validity need to be prioritized over others. For example, face validity is the way in which a model system can “look” like an NDD; in other words, are the behavioral features similar? However, we discussed that relying solely on face validity for ASD models will undoubtedly lead to misinterpretation of mechanisms. In addition, given the limitations of the rodent visual system and the lack of species emphasis on the visual system for sensory information, deficient eye contact cannot be studied directly in rodents. Further, given the limitations in complexity and the lack of a generalized use of expressive vocalization in mice, several social communication impairments, including pragmatic language deficits, are completely unable to be studied. Two major conclusions were that the workshop participants agreed that construct validity was more important than face validity, at this point in time 49 , 75 and that exhibiting the triad or DSM 5‐termed dyad of ASD core behavioral impairments were not key for an useful ASD model. Another issue we discovered, as a group of common reviewers of manuscripts in submission for moderate‐top tier journals with models based solely face validity, was the lack of clarity missing in the descriptions of methods and results. Even the most common, standardized, and well‐described 39 , 76 , 77 , 78 , 79 measure reported, the 3 chamber social approach task, varied across studies in the exact procedures used for this task. In turn, these variations are strong influencers to findings, because they may result in the measurement of different constructs within social interaction (i.e., olfactory communication versus recognition).
Therefore, we recommend that: (a) behavioral neuroscientists continually and iteratively work with clinical scientists, and (b) clinician scientists should advise behavioral neuroscientists on areas such as appropriate motivators for different types of social behaviors, the developmental trajectory of behavioral aberrations, and the unique features of specific genetic syndromes. There is a balance between high‐throughput screening and the in‐depth comprehensive analysis involved in behavioral phenotyping. With extensive input upfront on nuanced observations and the use of an open access database framework, we can increase aspects of face validity and provide accurate inputs for future research. Methods for doing this will increasingly include automated and machine learning observation algorithms. 80 , 81 , 82
1.3.2. Statement #2: Construct validity creates informative model systems from broad NDDs to specific ASDs, and requires rigorous testing via multiple behavioral assays
As stated, there was a consensus to increase transparency and interpretability of ongoing studies, priority should be placed on construct validity. Given that behavioral assays conducted with mouse models may measure more than one construct or risk factor, some of which may or may not be specific to NDDs, the field should stop labeling these as models of a specific disorder without a strong, accepted etiologic cause, such as “models of ASD”, per se. Instead, those experiments that analyze domains relevant to multiple neurological and neurodevelopmental behaviors should refer to NDDs broadly. One example of this might be elevations in prenatal neuroinflammation in addition to a de novo genetic mutation. Another example is when authors claim an “animal model of ASD” without construct or face validity for NDDs (i.e., based solely on disrupted behaviors that are broadly neuropsychiatric (e.g., anxiety‐like)). These models should be placed or named as models of the specific behavior (e.g., deletion of this gene results in pronounced anxiety‐like behavior) rather than ASD or NDD. 83 For NDDs, it would be more appropriate to focus on various behavioral domains taken together as a group rather than singled out individually. The approach of examining only one core feature, or one single behavioral experiment, will not shed light on pathogenesis of any particular NDD or ASD, a solely behaviorally‐defined disorder, but these models may yield useful scientific information about the roles of certain chemicals and pathways in the development and maintenance of social behavior in general. Construct validity, therefore, remains a key component to providing evidence for a strong candidate model relevant to NDDs and ASD that can be utilized for therapeutic development.
1.3.3. Statement #3: The complexity of behavior in animal models should not be underestimated
Outcomes of specific behavioral tasks do not define “sociability” or “learning and memory” but, rather, simply measure a group of subject's performance in each task, often resulting in more than one potential explanation. It is often the case that alterations in motor or olfactory behavior can easily explain the results being labeled as social or cognitive deficits. Because each task is just that, one task, we urge all translational researchers to stay away from "a single task" approach that appears to have face validity and attempts to make the novel model system relevant to a specific NDD, co‐morbid to ASD.
1.3.4. Moving forward
How does the field rethink the utility of model systems, 74 recognizing their limitations but appreciating their unique contribution to preclinical research leading directly to viable therapeutics to help families? Below are challenges and solutions discussed from the workshop.
Challenge #1: Given that mice are not people, how do we translate?
Redefine the utility of rodent models, accept and report their limitations. Validated tools for social communication, translationally relevant learning and memory, and other ASD symptom domains of sophisticated functional outcomes in animal models remain underdeveloped. Of utmost importance, behavioral assays investigating NDDs including ASD should not be limited to simply lack of social interaction/behavior and repetitive behavior. Sensory function, cognitive ability, anxiety, and other features of NDDs that may serve as alternate explanations for behavioral profiles that “look” like specific disorders (e.g., ASD, ID, ADHD, schizophrenia) need to be included. Very few studies in our review included behavioral tests, such as motor assays, which might rule out confounding findings, to understand the specificity of a model to a specific NDD versus findings of alternative deficits.
There is a desperate need for robust phenotypes even if not specific to a particular NDD, since individuals with specific NDDs often show deficits in development or behaviors that are not part of the ASD core symptoms but are nevertheless quality of life‐impairing and in need of interventions. The 3‐chamber social approach task ignited the field to study social motivation, social interaction, and olfactory recognition in rodents. However, as the field grows, this task alone is not sufficient for an animal model to be considered ASD‐specific. Other behavioral assays share this same challenge.
To date, we do not understand the full dynamic range of social behaviors, in humans and in other species. Use of computational methods to analyze data across multiple time points, capturing regression and/or progression and decline, is one solution to enhancing analysis of social behavior. Shifts to research using a more naturalistic, ethologically relevant environment is another. Scientists should be encouraged to improve upon existing behavioral assays, integrate multiple facets of social behavior along with features of NDDs that are not always tested (i.e., sensory sensitivities or insensitivities, cognitive delays, and motor problems including gait analysis), and consider implementing more sophisticated communication assays in rat over mouse models. 22
Challenge #2: Laboratories use different background strains for various reasons
Outcomes, causal to the gene mutation, will present different behavioral results because of genetic background. 84 , 85 Agree on a reporting standard that includes background strain and breeding strategies. For the past two decades, mice gained prevalence in preclinical model basic research over rats due to their ability to be manipulated efficiently using sophisticated technology, from turning genes on and off to limiting gene expression to regions and cell types. At least 90% of the mouse genome map is orthologous to the human genome, indicating a high degree of genetic conservation between the species. 86 , 87 This conservation of genes has led to mice being the most commonly used animal model for studying human disease. 88 Newer genetic technologies allow mice to be manipulated to “model” virtually any human condition with a known genetic variant. Mice also have phenotypic advantages that are not present in invertebrates or in vitro (i.e., behavioral outcome measures with clinical relevance). In addition to genetics, advantages include size, ability to reproduce robustly and quickly, easy handling, transportability, and the length of lifespan of a mouse. All of these logistical positives allow for the opportunity to investigate the results of genetic insults relevant from neurodevelopment to neurodegeneration. 89 Due to the benefits of using mice in disease modeling, various inbred strains have been created, intentionally or unintentionally via genetic drift, yielding genetically un‐identical strains from vendors and adding variance. Inadvertantly, mouse background strain has added variability and a loss of rigor. In addition to behavioral phenotyping uniformly, genetic background and age of testing of the mouse models have varied among most behavioral reports of numerous genetic models. 90 In Figure 1, 29% of the articles reviewed either did not report or unclearly reported the congenicity of the background strains in their mice, making reproducibility impossible. It should be noted that if a genetic mutation causes a deficit in one strain but not the other and behavioral deficits do not generalize, there are other intervening variables that need to be studied, which may moderate the penetrance of that gene on the behavior.
Challenge #3: There are differing methodologies, data transformations, and metrics reported across laboratories using the same task
Standardization is key for transparency and reproducibility. The field of behavioral neuroscience made a strong case for the standardization of behavioral assessments and multi‐task batteries that comprehensively phenotype genetic lines, with substantial evidence that this strategy will improve reproducibility. Variability in experimental and laboratory environmental conditions are unavoidable. A rigorous, reliable result should be detectable despite the nuances of that setting. 39 , 56 , 57 , 61 , 63 , 78 , 91 , 92 , 93 , 94 , 95 One possible solution is automated software systems as a replacement to manual coding. Automated systems combine video tracking and machine learning to automatically detect and score innate social behaviors, such as aggression, mating, and social investigation, between mice in a home‐cage environment or arenas. These technologies have the potential to have transformative impacts on high‐throughput screening of behavioral outcomes. However, the use of automated systems does not take the place of the requirement to provide detailed methodologies and report standardizable components.
Challenge #4: Lack of inter‐laboratory reproducibility
Behavioral phenotyping can be well‐reproduced both intra‐ and inter‐laboratory by executing at least two corroborating behavioral assays in each domain studied (such as those relevant to ASD) and using gold‐standard methods in at least two independent cohorts. 35 , 39 , 40 , 57 , 70 , 96 , 97 , 98 , 99 Assays should be blinded, unbiased, and highly powered with appropriate age‐ and sex‐matched littermate controls. Statistical power for most behavioral assays requires at least N = 15–20 per genotype per sex for two independent cohorts, from multiple litters, to adequately assess behavioral abnormalities with sufficient statistical power. Other relevant biological variables such as sex, age, genetic background, and circadian rhythm/time of day should be carefully controlled, considered, and described in detail in the methods text. The importance of procedural and environmental differences often complicate direct comparisons of phenotypic data. However, these points are not insurmountable. We and others have reported replicability intra‐ and inter‐ laboratories, time‐zones, countries, and seasons in numerous genetic mouse models. 35 , 39 , 57 , 90 , 96 , 99 , 100 , 101 , 102 When findings do not replicate, it may not be the behavior itself that is not reproducible. It may be the absence of the validation of the behavioral task in the reporting environment, the lack of technical proficiency, inadequate statistical power, and/or a difference in genetic background that prohibits reproducibility.
Challenge #5: Complex statistical analyses and/or chosen statistical tests are often unclear, undescribed, or incorrectly applied
Bring in behavioral neuroscientists and statisticians at all stages of experimental design and analysis. Encourage reporting of negative data. Often, due to time and financial restrictions, genetically modified mice are in short supply. While the sample size may be adequate for each behavioral assay, the same cohort of animals and their wildtype or heterozygous littermates might be put through weeks and weeks of behavioral assays. If the particular testing order is reiterated every time, this may not be a huge adverse effect on the data. In fact, it allows for cross behavioral domain correlation and behavior and molecular correlation. However, this design results in practice or test–retest effects, 63 , 64 , 65 , 103 this design brings up the issue of how to handle statistical analysis and proper use of multiple comparisons, as a broad variety of domains: cognitive ability, anxiety, olfactory function, motor skills, social assessment, and so forth, may be parts of the behavioral battery. For those mice for which a different cohort can be used for each domain, the statistical analyses may be rigorous and clear. But for those that require repeated testing across domains, how should the study be designed and analyzed? Our discussion concluded that use of different metrics to capture the same behavior (e.g., number of bouts, time spent engaged), should be held to higher statistical rigor than across behavioral assays capturing different behaviors entirely. If there is no influence from one task to the other, multiple comparisons are unnecessary. For example, it should be expected that multiple indices of gait analysis (stride length, stance width, stride frequency) need a more rigorous post‐hoc statistical analysis than measures of gait analysis versus indices of social interaction. Behavioral batteries can be utilized without multiple comparison correction, unless aging or time is the independent variable of interest, or if the test order shows a test–retest effect. 65 Statistical corrections should be employed when many indices are capturing the same exact behavior. A few remaining questions on a uniform methodology for the appropriate multiple comparisons statistics is currently in debate. 104 , 105
If the tasks are completely different and not within similar domains, they may be analyzed separately but it is critical to include negative data in reporting to both accurately represent the model system and alleviate the statistical and interpretative hesitations in the results. 106 , 107 In fact, reporting of negative data may also improve replication, despite the fact that many groups are discouraged from reporting this data and assume that findings are spurious when they are actually stable. Unfortunately, high‐impact journals are less likely to publish negative data, 108 which may introduce bias. However, scientific journals and stakeholders must continue to push and require the publication of both positive and negative data. It is important to state if a model does not replicate behavior, initially or upon replication. 109 A genetic model is still a model of a gene mutation, regardless of the exact behavior observed from the mouse model. Such models can still teach us about gene function in the brain. The multi‐laboratory, multi‐model, variable behavioral results following the synaptic cell adhesion protein Neuroligin‐4 (Nlgn4) and the chromatin remodeling protein Chd8 (Chd8) are excellent examples of this point. 96 , 98 , 110 , 111 , 112
Challenge #6: ASD and NDDs change trajectory over time, but most papers report a single testing time point
In an ideal world, testing would be performed at different time periods to examine developmental changes (progressive decline, regression, stabilization) within and between cohorts. NDDs are, by definition, observed and present during development and progress and regress and exist across the lifespan. In NDDs, pathways of developmental delay may be apparent as early as 1–2 years of age, typically in the form of plateauing or declining abilities and loss of milestones after they are initially achieved within the very early developmental period (akin to the first 3 year of life in humans). 113 Given the variable onset patterns related to various rare genetic conditions associated with NDDs, the early developmental period may be viewed as a critical time for both ontology and plasticity for trajectory change.
Very few animal studies embark on a developmental perspective, perhaps because rodents are altricial (i.e., many pups, underdeveloped, requiring minimal resources) compared with humans, who are both altricial and born precocial (fully formed, large resource investment from dam). Too often, behavioral assays in model systems are performed at adulthood, missing the critical windows of development seen in NDDs and assume that behavioral development is linear, over variable. 114 There are many different metrics that could be used to translate the biological age of a mouse to a human. Despite the similarities, mice have a diminutive lifespan compared with humans. In this study, we found that one human year is equivalent to ~9 mice days. 115 Weaning occurs between 21 and 28 days, while humans take approximately 180 days. Hormonal changes and secondary sexual characteristics associated with adolescence develop around 42 days in mice and 11.5 years in humans, making 1 mouse day equivalent to about 100 human days up until adolescence. This relationship is maintained until adulthood, upon which mouse aging slows relative to human aging. In female mice, reproductive function is lost at 12–15 months, while in women, menopause occurs at an average age of 50, making one mouse day equivalent to around 41 human days. 116 This “age matching” to the human lifespan approach also assumes that a given phenotype is stable, which is faulty, since we know that human symptoms are not stable throughout childhood and into adulthood. There is large variation in age of testing across laboratories, including a lack of consistency, since, more or less, any time after 6 weeks is considered adulthood. Finally, and importantly, regression or progression of phenotypes on a developmental timeline is overall lacking in NDD animal model behavioral research. A developmental perspective is paramount; it is as crucial to determine the onset and trajectories of phenotypes as it is to identify them in adulthood.
Solution 6a: The earlier the better: Studies have demonstrated significant heterogeneity in NDD clinical phenotypes in the first 10 years of life. 15 , 117 , 118 , 119 , 120 We need to consider how appropriate rodent behavioral tasks are and to what human ages they correspond. For example, tracking of ultrasonic vocalization (USV) production across a neonatal time course will likely be more informative about communication deficits than a singular chosen day of USV collection. 121 , 122 , 123 , 124 Similarly, developmental delay of motor skills in rodents can be tracked to capture onset and genesis of phenotypes. These early developmental phenotypes also rely on stable and innate behaviors, such as USV calling when a pup is isolated from a dam. USVs are not the only solution yet they expand the horizon of neurodevelopmental phenotypes.
Solution 6b: Lifespan approaches: Both common and rare variant genetic work in NDDs point to temporal–spatial heterogeneity, meaning that gene expression is altering brain development and activity at various points both prenatally and postnatally. 125 , 126 , 127 There are numerous other clues to the importance of a developmental approach in humans and animal models. These include findings about both structural and functional brain changes and electrical wave activity patterns that appear to differ based on developmental period. 128 , 129 , 130 , 131 , 132 , 133 , 134 , 135 , 136 , 137 Behavioral research in animal models of NDDs should include assays at times that correspond to neuroanatomical/neurophysiological differences found early as well as later in life. In an ideal world, an established database of translational phenotypes such as sleep, EEG, MRI neuroanatomy, the development of motor skills, and others would be available to researchers so that typical rodent development and the natural aging processes would be known for these phenotypes; currently they are not. Relating earlier‐developing behaviors may provide important insight into the appearance, or delay, of later‐developing behaviors. Given the emphasis both on early behavioral intervention as well as the potential for early pharmacological or even mechanism‐modifying therapeutics (e.g., gene therapy), it is critical that a greater understanding of how to test models over the lifespan, but particularly during a period akin to early childhood, be prioritized.
Challenge #7: Can mice represent the full repertoire of complex behaviors?
Use more than one model system, and try to validate across these systems or observe a phenotype within two species. Perhaps the most popular recommendation from the workshop was the importance of cross‐species comparisons. Using a focused approach across species and leveraging the strengths of each species could give insight into how social information is processed by various organisms. For example, rodents are olfactory creatures while non‐human primates are visual. Rats use auditory ultrasonic communication as pups, juveniles and adults while mice use it mainly during neonatal period (for survival) and during mating (as instinctual). Rats use auditory ultrasonic calls as a more sophisticated behavior. For example, their repertoire includes social contact calls, playful calls, tickling calls, anticipation calls, fear‐related alarm calls and warning of predator calls. Adult rats emit alarm calls to warn their conspecifics. This is not the case in mice. 22 , 138 , 139 , 140
Non‐human primates (NHP) represent a species with an even wider repertoire of social behaviors, acoustic vocalizations, but the cost, gestation periods, push back from activists, are making NHP research and its feasibility teneous, at best. On the other hand of the evolutionary scale, zebrafish and drosophila have a limited repertoire of behavior compared with rodents, and the origin of these behaviors (innate, instinctual, or intentional) are unknown. As most of the molecular basis for learning came from early drosophila research, 141 , 142 , 143 , 144 these species could be used as complements to other model systems. Utilizing species like zebrafish and drosophila can identify specific circuitry involved in behavior. Using lower order species could be tailored to the species‐specific unique strengths (i.e., high‐throughput, genetics, visible neuroanatomical development) and not limited to examining behavioral outcomes for similarities to NDDs. Although, studying the relationship between neurodevelopment and behavior in these model systems is also inherently important and may be able to address the gaps in developmental rodent research discussed earlier.
Challenge #8. NDDs are complex and must be characterized via multiple domains
ASD behavioral phenotypes are not a monolith of two behavioral domains, and two tasks do not solely determine the validity and utility of a novel animal model. When evaluating a novel, construct‐valid model, a broad capture approach may be less comprehensive substantially to limited tailored behavioral phenotyping. In addition to ensuring that both of the DSM‐5 categories (social‐communication and restrictive and repetitive behavior) are included, assessing multiple assays across multiple domains of behavior, such as social, motor, sensory and cognition, is a stronger strategy for finding ASD/NDD‐relevant phenotypes, eliminating and/or discovering potential confounding behaviors. Finding robust, reliable, reproducible phenotypes in “non‐core” domains, such as anxiety or sensorimotor, and investigating it in‐depth should not be punished by reviewers of manuscripts but, rather, encouraged. Again, reporting negative phenotypes in behavioral tasks of any kind should be encouraged. A scientific environment welcoming the reports focused on strong, rigorous, and accurate behavioral phenotyping, outside the common perception of ASD, should be encouraged, as it will improve predictive validity. A list of required controls, minimum sample sizes for power, sex controls, breeding schemas, implementing guidelines for Animal Research: Reporting of In Vivo Experiments (ARRIVE), and other points for consideration are given in Table 1.
1.4. Here we provide a summary and a request for meticulous reporting of experimental design, detailed methods and materials, and results with apropos statistics
Cognitive and motor abilities are understudied and undervalued. Social interaction and communication deficits are of high interest and relevance (particularly to ASD) but are highly complex and require careful controls and interpretations. ID is often a predominant feature of specific genetic ASD conditions 145 , 146 associated with NDDs, and yet, experimental assays targeting cognition are often passed up in favor of those testing behaviors seen as relevant to core ASD features. Ignoring cognitive and motor abilities result in a lack of generalizability of findings. Cognitive function, learning, and memory assays should be more accepted as part of ASD‐related phenotypes because they are general neurodevelopmental phenotypes. Further, these phenotypes should be studied in parallel, while motor abilities should be assessed for unconfounded interpretation. Behavioral neuroscientists have many gold‐standard, validated assays to assess many kinds of cognition, learning, and memory in rodent models, and we need to expand the accepted umbrella of NDD‐related behavioral phenotypes to include them. Relatedly, motor abilities contribute to virtually any complex behavioral assay conducted in mice, and they are inextricably linked to murine social abilities.
Specific NDDs, such as ASD, are multi‐faceted, and core features are rarely the sole features presented in the clinic in humans. Comorbidities of disorders such as ASD span a spectrum of other behavioral domains including anxiety, motor disability, ID, epilepsy, sleep, attentional and behavioral disruptions. Thus, animal models of NDDs may reflect those features as unique phenotypic indices.
We need to keep in mind the role of olfaction in driving social interactions in rodents, while vision is the driver in humans. Control assays that test not only for intact olfactory ability, but also olfactory discrimination and sensitivity, should be employed with rigor and become standardized. 50
Missing in many social tasks are controls for motivations. It is important to combine and study all motivations, external, such as food reward after food restriction, and observe these against internal motivators, such as social stimulus, to introduce a more dynamic observation of social behavior and social motivation.
2. SUMMARY AND RECOMMENDATIONS
NDDs, such as ASD and ID, continue to be diagnosed behaviorally and it is critical that alteration of appropriate behavioral outcomes in valid models continue to screen therapeutics, which will improve the lives of individuals and their families. However, scientists must be cautious in the behaviors they choose and responsible in the analysis of the data and the protocols they design and implement. There must be some level of validation and/or a positive/negative control group. Scientists must continue to be open‐minded to reconsidering methodology and interpretation. We summarize by again recommending removing the phrase “animal model of ASD”, since they are also modeling other neurodevelopmental and neuropsychiatric conditions associated with genetic variances, and to a high degree include ID. Few model systems to date have embraced the idea of expanding investigation beyond the two core domains of ASD. In addition to treatment development, proper use of behavioral standardization, control groups, and validation cohorts will help better define, describe, and establish underlying processes and neurobiological mechanisms that are common across measures. Eventually, these systems can be leveraged to pinpoint specific neural networks which regulate behaviors related to NDDs.
We ask that attention be paid to minimum reporting requirements. These include: type and description of behavioral test used, sample sizes used (in the body of the main text and not as an appendix), a description of the statistical test used, utilization of more than one measure of core behaviors (for ASD, both social communication and restrictive and repetitive behavior), demonstration in the same or a different study of the neural mechanisms affected by the gene or genetic/environmental combination under study, agreement on standard background strains to be used, and, above all, constant collaboration with behavioral neuroscientists, who are trained not only to design but also to help interpret the results and suggest further experiments before premature publication. This may also include additional control groups; examples are in Table 1 but can be found by following ARRIVE guidelines. 51 , 52 , 58 , 147
Studying social behavior in a mouse is relevant, and studying related neural circuitry in a mouse is necessary but not sufficient. 148 Using behavioral assays in in vivo models are essential but need substantially more additional rigor and reproducibility. A behavioral phenotype, much less a rescue of that phenotype, should not preclude an otherwise well‐executed developmental, physiological, cellular, and molecular discovery from impactful publication. Behaviors exhibited by construct‐valid mice do not have to be all‐or‐nothing. The time for multi‐disciplinary tried and true TEAM science is NOW and must be embraced if gains and progress are to be made fruitful for NDD and ASD stakeholders.
ACKNOWLEDGMENTS
This work was supported by generous funding from the Autism Science Foundation, NINDS, (R01NS097808; Stela P. Petkova, Jill L. Silverman), NIMH Intramural Research Program (1ZICMH002961), and the MIND Institute's Intellectual and Developmental Disabilities Resource Center (NIH P50 HD103526; Jill L. Silverman; Melissa D. Bauman), University of Iowa Hawkeye Intellectual and Developmental Disorders Research Center P50 HD 103556 (TA), Intellectual and Developmental Disabilities Research Center NIH/NICHD P50 HD105354 at CHOP/Penn (ESB) and Markus Wöhr (DFG WO 1732/4‐2; Markus Wöhr). We gratefully acknowledge Dr. Craig M. Powell, Chair, Department of Neurobiology, University of Alabama at Birmingham, Birmingham, AL and Civitan International Research Center at UAB, Birmingham, AL, and Dr. Alice Luo Clayton of the Simon's Foundation who participated in the 2 days zoom workshop.
Silverman JL, Thurm A, Ethridge SB, et al. Reconsidering animal models used to study autism spectrum disorder: Current state and optimizing future. Genes, Brain and Behavior. 2022;21(5):e12803. doi: 10.1111/gbb.12803
Funding information Autism Science Foundation; DFG, Grant/Award Number: WO 1732/4‐2; National Institute of Child Health and Human Development; National Institute of Mental Health Intramural Research Program, Grant/Award Number: 1ZICMH002961; National Institute of Neurological Disorders and Stroke, Grant/Award Number: R01NS097808; MIND Institute's Intellectual and Developmental Disabilities Resource Center, Grant/Award Number: P50 HD103526; University of Iowa Hawkeye Intellectual and Developmental Disorders Research Center, Grant/Award Number: P50 HD 103556; CHOP/Penn Intellectual and Developmental Disabilities Research Center, Grant/Award Number: NIH/NICHD P50 HD105354
DATA AVAILABILITY STATEMENT
Data sharing not applicable ‐ no new data generated.
REFERENCES
- 1. Christensen DL, Baio J, Van Naarden BK, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill Summ. 2016;65(3):1‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Christensen DL, Bilder DA, Zahorodny W, et al. Prevalence and characteristics of autism spectrum disorder among 4‐year‐old children in the autism and developmental disabilities monitoring network. J Dev Behav Pediatr. 2016;37(1):1‐8. [DOI] [PubMed] [Google Scholar]
- 3. Association AP . Diagnostic and Statistical Manual of Mental Disorders. 5th ed.; American Psychiatric Association; 2013. [Google Scholar]
- 4. Wilfert AB, Turner TN, Murali SC, et al. Recent ultra‐rare inherited variants implicate new autism candidate risk genes. Nat Genet. 2021;53(8):1125‐1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Satterstrom FK, Kosmicki JA, Wang J, et al. Large‐scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180(3):568‐584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70(5):898‐907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Celestino‐Soper PB, Violante S, Crawford EL, et al. A common X‐linked inborn error of carnitine biosynthesis may be a risk factor for nondysmorphic autism. Proc Natl Acad Sci. 2012;109(21):7974‐7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Des Portes V. X‐linked mental deficiency. Handb Clin Neurol. 2013;111:297‐306. [DOI] [PubMed] [Google Scholar]
- 9. Guarnieri FC, Pozzi D, Raimondi A, et al. A novel SYN1 missense mutation in non‐syndromic X‐linked intellectual disability affects synaptic vesicle life cycle, clustering and mobility. Hum Mol Genet. 2017;26(23):4699‐4714. [DOI] [PubMed] [Google Scholar]
- 10. Kaya N, Colak D, Albakheet A, et al. A novel X‐linked disorder with developmental delay and autistic features. Ann Neurol. 2012;71(4):498‐508. [DOI] [PubMed] [Google Scholar]
- 11. Laumonnier F, Bonnet‐Brilhault F, Gomot M, et al. X‐linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74(3):552‐557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poo‐Arguelles P, Arias A, Vilaseca MA, et al. X‐linked creatine transporter deficiency in two patients with severe mental retardation and autism. J Inherit Metab Dis. 2006;29(1):220‐223. [DOI] [PubMed] [Google Scholar]
- 13. Skuse D. X‐linked genes and the neural basis of social cognition. Novartis Found Symp. 2003;251:84‐98. [PubMed] [Google Scholar]
- 14. Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome Biol. 2012;13(7):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chaste P, Roeder K, Devlin B. The Yin and Yang of autism genetics: how rare de novo and common variations affect liability. Annu Rev Genomics Hum Genet. 2017;18:167‐187. [DOI] [PubMed] [Google Scholar]
- 16. Vorstman JAS, Parr JR, Moreno‐De‐Luca D, Anney RJL, Nurnberger JI Jr, Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet. 2017;18(6):362‐376. [DOI] [PubMed] [Google Scholar]
- 17. Yin J, Schaaf CP. Autism genetics ‐ an overview. Prenat Diagn. 2017;37(1):14‐30. [DOI] [PubMed] [Google Scholar]
- 18. Grove J, Ripke S, Als TD, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51(3):431‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Satterstrom FK, Walters RK, Singh T, et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein‐truncating variants. Nat Neurosci. 2019;22(12):1961‐1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Till SM, Hickson RDL, Kind PC. Cross‐species considerations in models of neurodevelopmental disorders. Trends Neurosci. 2022;45(3):171‐172. doi: 10.1007/BFb0072152 [DOI] [PubMed] [Google Scholar]
- 21. Berg EL, Copping NA, Rivera JK, et al. Developmental social communication deficits in the Shank3 rat model of phelan‐mcdermid syndrome and autism spectrum disorder. Autism Res. 2018;11(4):587‐601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berg EL, Jami SA, Petkova SP, et al. Excessive laughter‐like vocalizations, microcephaly, and translational outcomes in the Ube3a deletion rat model of Angelman syndrome. J Neurosci. 2021;41(42):8801‐8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berg EL, Pride MC, Petkova SP, et al. Translational outcomes in a full gene deletion of ubiquitin protein ligase E3A rat model of Angelman syndrome. Transl Psych. 2020;10(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Medicine. Io Improving the utility and translation of animal models for nervous system disorders: Workshop Summary 2013. [PubMed]
- 25. Crusio WE, Goldowitz D, Holmes A, Wolfer D. Standards for the publication of mouse mutant studies. Genes Brain Behav. 2009;8(1):1‐4. [DOI] [PubMed] [Google Scholar]
- 26. Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13(10):1161‐1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahjani B, De Rubeis S, Gustavsson Mahjani C, et al. Prevalence and phenotypic impact of rare potentially damaging variants in autism spectrum disorder. Mol Autism. 2021;12(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blundell J, Tabuchi K, Bolliger MF, et al. Increased anxiety‐like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 2009;8(1):114‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tabuchi K, Blundell J, Etherton MR, et al. A neuroligin‐3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318(5847):71‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou J, Blundell J, Ogawa S, et al. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural‐specific Pten knock‐out mice. J Neurosci. 2009;29(6):1773‐1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kwon CH, Luikart BW, Powell CM, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50(3):377‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ogawa S, Kwon CH, Zhou J, Koovakkattu D, Parada LF, Sinton CM. A seizure‐prone phenotype is associated with altered free‐running rhythm in Pten mutant mice. Brain Res. 2007;1168:112‐123. [DOI] [PubMed] [Google Scholar]
- 33. Zhou J, Parada LF. PTEN signaling in autism spectrum disorders. Curr Opin Neurobiol. 2012;22(5):873‐879. [DOI] [PubMed] [Google Scholar]
- 34. Bozdagi O, Sakurai T, Papapetrou D, et al. Haploinsufficiency of the autism‐associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang M, Bozdagi O, Scattoni ML, et al. Reduced excitatory neurotransmission and mild autism‐relevant phenotypes in adolescent Shank3 null mutant mice. J Neurosci. 2012;32(19):6525‐6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Etherton M, Foldy C, Sharma M, et al. Autism‐linked neuroligin‐3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc Natl Acad Sci USA. 2011;108(33):13764‐13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin‐1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106(42):17998‐18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Copping NA, Berg EL, Foley GM, et al. Touchscreen learning deficits and normal social approach behavior in the Shank3B model of Phelan‐McDermid syndrome and autism. Neuroscience. 2017;345:155‐165. doi: 10.1016/j.neuroscience.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dhamne SC, Silverman JL, Super CE, et al. Replicable in vivo physiological and behavioral phenotypes of the Shank3B null mutant mouse model of autism. Mol Autism. 2017;8:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kouser M, Speed HE, Dewey CM, et al. Loss of predominant Shank3 isoforms results in hippocampus‐dependent impairments in behavior and synaptic transmission. J Neurosci. 2013;33(47):18448‐18468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Speed HE, Kouser M, Xuan Z, et al. Autism‐associated insertion mutation (InsG) of Shank3 exon 21 causes impaired synaptic transmission and behavioral deficits. J Neurosci. 2015;35(26):9648‐9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Drapeau E, Riad M, Kajiwara Y, Buxbaum JD. Behavioral phenotyping of an improved mouse model of Phelan‐McDermid syndrome with a complete deletion of the Shank3 gene. eNeuro. 2018;5(3):ENEURO.0046‐18.2018. doi: 10.1523/eneuro.0046-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peca J, Feliciano C, Ting JT, et al. Shank3 mutant mice display autistic‐like behaviours and striatal dysfunction. Nature. 2011;472(7344):437‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Speed HE, Kouser M, Xuan Z, Liu S, Duong A, Powell CM. Apparent genetic rescue of adult Shank3 exon 21 insertion mutation mice tempered by appropriate control experiments. eNeuro. 2019;6(5):ENEURO.0317‐19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Provenzano G, Chelini G, Bozzi Y. Genetic control of social behavior: lessons from mutant mice. Behav Brain Res. 2017;325:237‐250. [DOI] [PubMed] [Google Scholar]
- 46. McCracken JT, Anagnostou E, Arango C, et al. Drug development for autism spectrum disorder (ASD): Progress, challenges, and future directions. Eur Neuropsychopharmacol. 2021;48:3‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gauthier J, Spiegelman D, Piton A, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150(3):421‐424. [DOI] [PubMed] [Google Scholar]
- 48. Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81(6):1289‐1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hyman SE. Use of mouse models to investigate the contributions of CNVs associated with schizophrenia and autism to disease mechanisms. Curr Opin Genet Dev. 2021;68:99‐105. [DOI] [PubMed] [Google Scholar]
- 50. Macleod MR, Lawson McLean A, Kyriakopoulou A, et al. Risk of bias in reports of in vivo research: a focus for improvement. PLoS Biol. 2015;13(10):e1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Percie du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. J Physiol. 2020;598(18):3793‐3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sprague W. Arrive Guidelines 2.0. Vet Clin Pathol. 2020;49(3):378‐379. [DOI] [PubMed] [Google Scholar]
- 53. McCarthy MM. Sex differences in the developing brain as a source of inherent risk. Dialogs Clin Neurosci. 2016;18(4):361‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McCarthy MM, Arnold AP, Ball GF, Blaustein JD, De Vries GJ. Sex differences in the brain: the not so inconvenient truth. J Neurosci Off J Soc Neurosci. 2012;32(7):2241‐2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sukoff Rizzo SJ, McTighe S, McKinzie DL. Genetic background and sex: impact on generalizability of research findings in pharmacology studies. Handb Exp Pharmacol. 2020;257:147‐162. [DOI] [PubMed] [Google Scholar]
- 56. Silverman JL, Nithianantharajah J, Der‐Avakian A, Young JW, Sukoff Rizzo SJ. Lost in translation: at the crossroads of face validity and translational utility of behavioral assays in animal models for the development of therapeutics. Neurosci Biobehav Rev. 2020;116:452‐453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sukoff Rizzo SJ, Silverman JL. Methodological considerations for optimizing and validating behavioral assays. Curr Protoc Mouse Biol. 2016;6(4):364‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Percie du Sert N, Ahluwalia A, Alam S, et al. Reporting animal research: explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020;18(7):e3000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silverman JL, Yang M, Lord C, Crawley JN. Behavioral phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11(7):490‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang M, Lewis F, Foley G, Crawley JN. In tribute to bob Blanchard: divergent behavioral phenotypes of 16p11.2 deletion mice reared in same‐genotype versus mixed‐genotype cages. Physiol Behav. 2015;146:16‐27. [DOI] [PubMed] [Google Scholar]
- 61. Moy SS, Nadler JJ, Young NB, et al. Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains. Behav Brain Res. 2007;176(1):4‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Copping NA, Adhikari A, Petkova SP, Silverman JL. Genetic backgrounds have unique seizure response profiles and behavioral outcomes following convulsant administration. Epilepsy Behav. 2019;101(Pt A):106547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Crawley JN, Paylor R. A proposed test battery and constellations of specific behavioral paradigms to investigate the behavioral phenotypes of transgenic and knockout mice. Horm Behav. 1997;31(3):197‐211. [DOI] [PubMed] [Google Scholar]
- 64. McIlwain KL, Merriweather MY, Yuva‐Paylor LA, Paylor R. The use of behavioral test batteries: effects of training history. Physiol Behav. 2001;73(5):705‐717. [DOI] [PubMed] [Google Scholar]
- 65. Paylor R, Spencer CM, Yuva‐Paylor LA, Pieke‐Dahl S. The use of behavioral test batteries, II: effect of test interval. Physiol Behav. 2006;87(1):95‐102. [DOI] [PubMed] [Google Scholar]
- 66. Payne‐Sturges DC, Cory‐Slechta DA, Puett RC, Thomas SB, Hammond R, Hovmand PS. Defining and intervening on cumulative environmental neurodevelopmental risks: introducing a complex systems approach. Environ Health Perspect. 2021;129(3):35001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hollander JA, Cory‐Slechta DA, Jacka FN, et al. Beyond the looking glass: recent advances in understanding the impact of environmental exposures on neuropsychiatric disease. Neuropsychopharmacology. 2020;45(7):1086‐1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sobolewski M, Singh G, Schneider JS, Cory‐Slechta DA. Different behavioral experiences produce distinctive parallel changes in, and correlate with, frontal cortex and hippocampal global post‐translational histone levels. Front Integr Neurosci. 2018;12:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lazic SE, Essioux L. Improving basic and translational science by accounting for litter‐tolitter variation in animal models. BMC Neurosci. 2013;14:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Silverman JL, Ellegood J. Behavioral and neuroanatomical approaches in models of neurodevelopmental disorders: opportunities for translation. Curr Opin Neurol. 2018;31(2):126‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. McGraw CM, Ward CS, Samaco RC. Genetic rodent models of brain disorders: perspectives on experimental approaches and therapeutic strategies. Am J Med Genet C Semin Med Genet. 2017;175(3):368‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Crawley JN. Mouse behavioral assays relevant to the symptoms of autism. Brain Pathol. 2007;17(4):448‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Crawley JN. Medicine. Testing hypotheses about autism. Science. 2007;318(5847):56‐57. [DOI] [PubMed] [Google Scholar]
- 74. Pankevich DE, Altevogt BM, Dunlop J, Gage FH, Hyman SE. Improving and accelerating drug development for nervous system disorders. Neuron. 2014;84(3):546‐553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chadman KK. Animal models for autism in 2017 and the consequential implications to drug discovery. Expert Opin Drug Discov. 2017;12(12):1187‐1194. [DOI] [PubMed] [Google Scholar]
- 76. Ellegood J, Crawley JN. Behavioral and neuroanatomical phenotypes in mouse models of autism. Neurotherapeutics. 2015;12(3):521‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Silverman JL, Pride MC, Hayes JE, et al. GABAB receptor agonist R‐baclofen reverses social deficits and reduces repetitive behavior in two mouse models of autism. Neuropsychopharmacology. 2015;40(9):2228‐2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11(7):490‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang M, Silverman JL, Crawley JN. Automated three‐chambered social approach task for mice. Curr Protoc Neurosci. 2011;56(1):1‐16. doi: 10.1002/0471142301.ns0826s56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schaefer IC, Siebel AM, Piato AL, Bonan CD, Vianna MR, Lara DR. The side‐by‐side exploratory test: a simple automated protocol for the evaluation of adult zebrafish behavior simultaneously with social interaction. Behav Pharmacol. 2015;26(7):691‐696. [DOI] [PubMed] [Google Scholar]
- 81. Shams S, Amlani S, Scicluna M, Gerlai R. Argus: an open‐source and flexible software application for automated quantification of behavior during social interaction in adult zebrafish. Behav Res Methods. 2019;51(2):727‐746. [DOI] [PubMed] [Google Scholar]
- 82. Thanos PK, Restif C, O'Rourke JR, Lam CY, Metaxas D. Mouse social interaction test (MoST): a quantitative computer automated analysis of behavior. J Neural Transm. 2017;124(1):3‐11. [DOI] [PubMed] [Google Scholar]
- 83. Sartori SB, Landgraf R, Singewald N. The clinical implications of mouse models of enhanced anxiety. Future Neurol. 2011;6(4):531‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bryant CD, Zhang NN, Sokoloff G, et al. Behavioral differences among C57BL/6 substrains: implications for transgenic and knockout studies. J Neurogenet. 2008;22(4):315‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sittig LJ, Carbonetto P, Engel KA, Krauss KS, Barrios‐Camacho CM, Palmer AA. Genetic background limits generalizability of genotype‐phenotype relationships. Neuron. 2016;91(6):1253‐1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Emes RD, Goodstadt L, Winter EE, Ponting CP. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum Mol Genet. 2003;12(7):701‐709. [DOI] [PubMed] [Google Scholar]
- 87. Waterston RH, Lindblad‐Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520‐562. [DOI] [PubMed] [Google Scholar]
- 88. Ericsson AC, Crim MJ, Franklin CL. A brief history of animal modeling. Mol Med. 2013;110(3):201‐205. [PMC free article] [PubMed] [Google Scholar]
- 89. Bryda EC. The mighty mouse: the impact of rodents on advances in biomedical research. Mol Med. 2013;110(3):207‐211. [PMC free article] [PubMed] [Google Scholar]
- 90. Sukoff Rizzo SJ, Anderson LC, Green TL, McGarr T, Wells G, Winter SS. Assessing healthspan and lifespan measures in aging mice: optimization of testing protocols, replicability, and rater reliability. Curr Protoc Mouse Biol. 2018;8(2):e45. [DOI] [PubMed] [Google Scholar]
- 91. Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57(6):809‐818. [DOI] [PubMed] [Google Scholar]
- 92. Gulinello M, Mitchell HA, Chang Q, et al. Rigor and reproducibility in rodent behavioral research. Neurobiol Learn Mem. 2019;165:106780. doi: 10.1016/j.nlm.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Moy SS, Nadler JJ. Advances in behavioral genetics: mouse models of autism. Mol Psychiatry. 2008;13(1):4‐26. [DOI] [PubMed] [Google Scholar]
- 94. Bohlen M, Hayes ER, Bohlen B, Bailoo JD, Crabbe JC, Wahlsten D. Experimenter effects on behavioral test scores of eight inbred mouse strains under the influence of ethanol. Behav Brain Res. 2014;272:46‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: interactions with laboratory environment. Science. 1999;284(5420):1670‐1672. [DOI] [PubMed] [Google Scholar]
- 96. Ey E, Yang M, Katz AM, et al. Absence of deficits in social behaviors and ultrasonic vocalizations in later generations of mice lacking neuroligin4. Genes Brain Behav. 2012;11(8):928‐941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Copping NA, Christian SGB, Ritter DJ, et al. Neuronal overexpression of Ube3a isoform 2 causes behavioral impairments and neuroanatomical pathology relevant to 15q11.2‐q13.3 duplication syndrome. Hum Mol Genet. 2017;26(20):3995‐4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gompers AL, Su‐Feher L, Ellegood J, et al. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat Neurosci. 2017;20(8):1062‐1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Silverman JL, Smith DG, Rizzo SJ, et al. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci Transl Med. 2012;4(131):131ra151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Berg, Pride , Petkova Pride, et al.. Developmental outcomes in a full gene deletion of E6 Ubiquitin Ligase genetic rat model of Angelman Syndrome. Paper presented at: Society for Neuroscience 2018; San Diego, CA.
- 101. Born HA, Martinez LA, Levine AT, et al. Early developmental EEG and seizure phenotypes in a full gene deletion of ubiquitin protein ligase E3A rat model of Angelman syndrome. eNeuro. 2021;8(2):ENEURO.0345‐20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Genestine M, Lin L, Durens M, et al. Engrailed‐2 (En2) deletion produces multiple neurodevelopmental defects in monoamine systems, forebrain structures and neurogenesis and behavior. Hum Mol Genet. 2015;24(20):5805‐5827. doi: 10.1093/hmg/ddv301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Johnson PD, Besselsen DG. Practical aspects of experimental design in animal research. ILAR J. 2002;43(4):202‐206. [DOI] [PubMed] [Google Scholar]
- 104. Jimenez JA, Zylka MJ. Controlling litter effects to enhance rigor and reproducibility with rodent models of neurodevelopmental disorders. J Neurodev Disord. 2021;13(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stukalin Y, Einat H. Analyzing test batteries in animal models of psychopathology with multivariate analysis of variance (MANOVA): one possible approach to increase external validity. Pharmacol Biochem Behav. 2019;178:51‐55. [DOI] [PubMed] [Google Scholar]
- 106. Fanelli D. Do pressures to publish increase scientists' bias? An empirical support from US states data. PLoS One. 2010;5(4):e10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Fanelli D. "Positive" results increase down the hierarchy of the sciences. PLoS One. 2010;5(4):e10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Littner Y, Mimouni FB, Dollberg S, Mandel D. Negative results and impact factor: a lesson from neonatology. Arch Pediatr Adolesc Med. 2005;159(11):1036‐1037. [DOI] [PubMed] [Google Scholar]
- 109. Berg EL, Petkova SP, Born HA, Adhikari A, Anderson AE, Silverman JL. Insulin‐like growth factor‐2 does not improve behavioral deficits in mouse and rat models of Angelman syndrome. Mol Autism. 2021;12(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Katayama Y, Nishiyama M, Shoji H, et al. CHD8 haploinsufficiency results in autistic‐like phenotypes in mice. Nature. 2016;537(7622):675‐679. [DOI] [PubMed] [Google Scholar]
- 111. Platt RJ, Zhou Y, Slaymaker IM, et al. Chd8 mutation leads to autistic‐like behaviors and impaired striatal circuits. Cell Rep. 2017;19(2):335‐350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jamain S, Radyushkin K, Hammerschmidt K, et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci USA. 2008;105(5):1710‐1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Rice D, Barone S Jr. Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ Health Perspect. 2000;108(3):511‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lord C, Charman T, Havdahl A, et al. The lancet commission on the future of care and clinical research in autism. Lancet. 2022;399(10321):271‐334. doi: 10.1016/s0140-6736(21)01541-5 [DOI] [PubMed] [Google Scholar]
- 115. Dutta S, Sengupta P. Men and mice: relating their ages. Life Sci. 2016;152:244‐248. [DOI] [PubMed] [Google Scholar]
- 116. Jackson SJ, Andrews N, Ball D, et al. Does age matter? The impact of rodent age on study outcomes. Lab Anim. 2017;51(2):160‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ozonoff S, Young GS, Brian J, et al. Diagnosis of autism spectrum disorder after age 5 in children evaluated longitudinally since infancy. J Am Acad Child Adolesc Psychiatry. 2018;57(11):849‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lord C. Recognising the heterogeneity of autism. Lancet Psychiatry. 2019;6(7):551‐552. [DOI] [PubMed] [Google Scholar]
- 119. Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000;28(2):355‐363. [DOI] [PubMed] [Google Scholar]
- 120. Chaste P, Klei L, Sanders SJ, et al. A genome‐wide association study of autism using the Simons simplex collection: does reducing phenotypic heterogeneity in autism increase genetic homogeneity? Biol Psychiatry. 2015;77(9):775‐784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Schwarting RK, Jegan N, Wohr M. Situational factors, conditions and individual variables which can determine ultrasonic vocalizations in male adult Wistar rats. Behav Brain Res. 2007;182(2):208‐222. [DOI] [PubMed] [Google Scholar]
- 122. Sungur AO, Schwarting RK, Wohr M. Early communication deficits in the Shank1 knockout mouse model for autism spectrum disorder: developmental aspects and effects of social context. Autism Res. 2016;9(6):696‐709. [DOI] [PubMed] [Google Scholar]
- 123. Wohr M, Schwarting RK. Affective communication in rodents: ultrasonic vocalizations as a tool for research on emotion and motivation. Cell Tissue Res. 2013;354(1):81‐97. [DOI] [PubMed] [Google Scholar]
- 124. Wohr M, Seffer D, Schwarting RK. Studying socio‐affective communication in rats through playback of ultrasonic vocalizations. Curr Protoc Neurosci. 2016;75:31‐38. [DOI] [PubMed] [Google Scholar]
- 125. Arnett AB, Trinh S, Bernier RA. The state of research on the genetics of autism spectrum disorder: methodological, clinical and conceptual progress. Curr Opin Psychol. 2018;27:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Goin‐Kochel RP, Trinh S, Barber S, Bernier R. Gene disrupting mutations associated with regression in autism Spectrum disorder. J Autism Dev Disord. 2017;47(11):3600‐3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Trinh S, Arnett A, Kurtz‐Nelson E, Beighley J, Picoto M, Bernier R. Transcriptional subtyping explains phenotypic variability in genetic subtypes of autism spectrum disorder. Dev Psychopathol. 2020;32(4):1353‐1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Shen MD, Nordahl CW, Young GS, et al. Early brain enlargement and elevated extra‐axial fluid in infants who develop autism spectrum disorder. Brain. 2013;136(9):2825‐2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hazlett HC, Gu H, Munsell BC, et al. Early brain development in infants at high risk for autism spectrum disorder. Nature. 2017;542(7641):348‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Shen MD, Piven J. Brain and behavior development in autism from birth through infancy. Dialogues Clin Neurosci. 2017;19(4):325‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wolff JJ, Swanson MR, Elison JT, et al. Neural circuitry at age 6 months associated with later repetitive behavior and sensory responsiveness in autism. Mol Autism. 2017;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31(3):137‐145. [DOI] [PubMed] [Google Scholar]
- 133. Nordahl CW, Dierker D, Mostafavi I, et al. Cortical folding abnormalities in autism revealed by surface‐based morphometry. J Neurosci. 2007;27(43):11725‐11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ohta H, Nordahl CW, Iosif AM, Lee A, Rogers S, Amaral DG. Increased surface area, but not cortical thickness, in a subset of Young boys with autism spectrum disorder. Autism Res. 2015;9(2):232‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Smith E, Thurm A, Greenstein D, et al. Cortical thickness change in autism during early childhood. Hum Brain Mapp. 2016;37(7):2616‐2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Adhikari A, Copping NA, Beegle J, et al. Functional rescue in an Angelman syndrome model following treatment with lentivector transduced hematopoietic stem cells. Hum Mol Genet. 2021;30(12):1067‐1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Silva‐Santos S, van Woerden GM, Bruinsma CF, et al. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J Clin Invest. 2015;125(5):2069‐2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wohr M, van Gaalen MM, Schwarting RK. Affective communication in rodents: serotonin and its modulating role in ultrasonic vocalizations. Behav Pharmacol. 2015;26(6):506‐521. [DOI] [PubMed] [Google Scholar]
- 139. Abbott A. Laboratory animals: the renaissance rat. Nature. 2004;428(6982):464‐466. [DOI] [PubMed] [Google Scholar]
- 140. Homberg JR, Wohr M, Alenina N. Comeback of the rat in biomedical research. ACS Chem Nerosci. 2017;8(5):900‐903. [DOI] [PubMed] [Google Scholar]
- 141. Tully T. Discovery of genes involved with learning and memory: an experimental synthesis of Hirschian and Benzerian perspectives. Proc Natl Acad Sci USA. 1996;93(24):13460‐13467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Tully T, Bolwig G, Christensen J, et al. A return to genetic dissection of memory in drosophila. Cold Spring Harb Symp Quant Biol. 1996;61:207‐218. [PubMed] [Google Scholar]
- 143. Tully T, Bolwig G, Christensen J, et al. Genetic dissection of memory in drosophila. J Physiol Paris. 1996;90(5–6):383. [DOI] [PubMed] [Google Scholar]
- 144. Quinn WG, Harris WA, Benzer S. Conditioned behavior in Drosophila melanogaster . Proc Natl Acad Sci USA. 1974;71(3):708‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Thurm A, Kelleher B, Wheeler A. Outcome measures for core symptoms of intellectual disability: state of the field. Am J Intellect Dev Disabil. 2020;125(6):418‐433. [DOI] [PubMed] [Google Scholar]
- 146. Myers SM, Challman TD, Bernier R, et al. Insufficient evidence for "autism‐specific" genes. Am J Hum Genet. 2020;106(5):587‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Katsnelson A. Arriving again: 10 years on, ARRIVE 2.0 offers a simplified set of reporting guidelines. Lab Anim. 2020;49(10):267. [DOI] [PubMed] [Google Scholar]
- 148. Lewis AS, Calipari ES, Siciliano CA. Toward standardized guidelines for investigating neural circuit control of behavior in animal research. eNeuro. 2021;8(2):ENEURO.0498‐20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable ‐ no new data generated.


