Abstract
Lysosomes play major roles in growth regulation and catabolism, and are recognized as critical mediators of cellular remodeling. An emerging theme is how the lysosome is itself subjected to extensive remodeling in order to perform specific tasks that meet the changing demands of the cell. Accordingly, lysosomes can sustain physical damage and undergo dramatic changes in composition following pathogen infection, accumulation of protein aggregates or cellular transformation, necessitating dedicated pathways for their repair, remodeling and restoration. In this review, we focus on emerging molecular mechanisms for piecemeal remodeling of lysosomal components, and wholesale repair and discuss their implications in physiological and pathogenic challenges such as cancer, neurodegeneration and pathogen infection.
Keywords: Lysosome, neurodegeneration, cancer, infection, repair, membrane damage
Key properties of the lysosome
In order to grow, proliferate, differentiate and overcome stress, cells continuously change their internal composition of macromolecules and metabolites through both bulk and targeted degradative processes. A major player in cellular remodeling is the lysosome, a catabolic organelle that is the end-point of several degradative pathways including endocytosis, phagocytosis and autophagy [1, 2]. The lysosome is distinguished by unique features that enable its catabolic functions. These include a low internal pH (4.5–5.0) established by an ATP-driven proton pump, and a set of approximately 60 hydrolases that degrade polypeptides, nucleic acids, complex lipids and sugars, and which work optimally at acidic pH [1, 3]. A set of transporters and permeases spanning the lysosomal limiting membrane enable the export of digestion products to the cytoplasm, whereas ion channels help maintain biophysical properties such as the internal pH and membrane potential of this organelle [4].
The outer (cytoplasm-facing) surface of the lysosome is also a highly active compartment, particularly due to the presence of protein complexes that direct the intracellular trafficking of lysosomes and their fusion with incoming endocytic and autophagic vesicles. Moreover, important signaling complexes associate with the outer lysosomal surface, particularly the master growth regulator, mechanistic Target of Rapamycin Complex 1 (mTORC1) kinase, which, upon integrating nutrients, growth factor and energy signals through its association with the lysosomal membrane, triggers signals that regulate the balance between anabolism and catabolism [5, 6].
As a key mediator of cellular remodeling, lysosomes represent a highly dynamic compartment that continuously adjusts its number, size and composition in response to both internal and external stressors. Lysosomal remodeling relies on dedicated signaling pathways, which detect lysosomal stress and dysfunction and trigger both transcriptional and posttranslational programs that adjust lysosomal function in a compensatory manner [1, 2, 6]. Stressors that trigger lysosomal adaptation are diverse and plentiful, and include accumulation of undigested substrates within the lumen, changes in lipid composition of the limiting and internal membranes, loss of pH and/or membrane potential and outright rupturing of the lysosomal limiting membrane by mechanical or chemical insults [7].
Prompt compensation and repair of lysosomal injuries is important for numerous cell types and especially essential for neuronal cells, which invariably die when lysosomal function is compromised [6, 8]. Conversely, enhanced lysosomal biogenesis and plasticity are emerging as key adaptive mechanisms of certain cancer types that thrive in challenging, nutrient- and oxygen-poor microenvironments [9]. In this review we describe how a deeper understanding of the molecular underpinnings of lysosomal remodeling and repair impacts normal cell physiology as well as pathogenic processes from neurodegeneration to cancer.
Maintaining lysosomal homeostasis
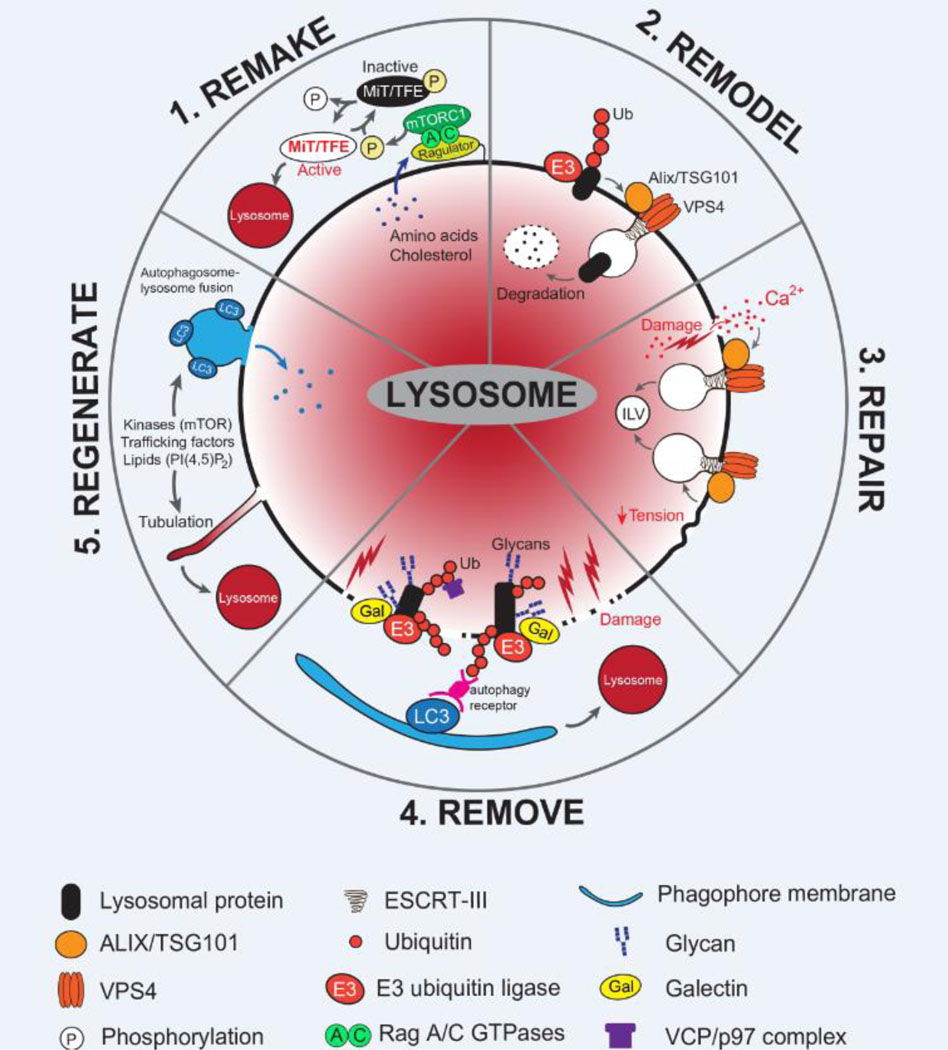
Maintaining the number and integrity of lysosomes within the cell is critical for enabling stress adaptation and general homeostasis. A fine balance between biogenesis, repair and clearance helps to ensure continued functionality of the lysosome system in response to a diverse array of stimuli and stressors (Figure 1).
Figure 1: Molecular mechanisms of lysosomal remodeling and repair.
Lysosomes undergo dynamic alterations in their number, composition, distribution and function in response to numerous stimuli. Mechanisms mediating lysosome homeostasis are critical for maintaining degradative capacity and cellular homeostasis. These include 1. The ability to ‘REMAKE’ new lysosomes via the activity of the master lysosomal transcription factors, MiT/TFE, the activity of which are regulated via posttranslational modification (e.g. Phosphorylation by mTORC1 mediates cytoplasmic retention and inactivation); 2. The lysosomal membrane can be ‘REMODELed’ via ubiquitylation of select membrane associated proteins, which are subsequently internalized and degraded in an ESCRT-dependent manner; 3. In response to acute damage, the lysosome membrane can undergo ‘REPAIR’ via the action of ALIX/TSG101 mediated recruitment of ESCRT-III proteins. Membrane remodeling in this instance is thought to generate intra-lumenal vesicles (ILV) that effectively remove the damaged portion of the membrane; 4. In the event that lysosomal membrane damage is prolonged or irreversible, dysfunctional lysosomes can be ‘REMOVEd’ via lysophagy. Here, exposed glycans present on lysosomal lumenal proteins recruit Galectins, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases, which modify them with ubiquitin. Autophagy receptors recognize ubiquitylated proteins and recruit the autophagy machinery to facilitate engulfment; 5. Following autophagosome-lysosome fusion the resulting ‘autolysosome’ can undergo tubulation to ‘REGENERATE’ new ‘proto-lysosomes’ in a process known as autophagic lysosome reformation (ALR). Through ALR, key components are recycled from hybrid autolysosomes to the new lysosomal vesicles, thereby ensuring cellular lysosome number is maintained.
Lysosomal biogenesis
Transcriptional, post-translational and trafficking processes cooperate to maintain a functional lysosome pool within cells. Lysosomes are derived from progressive maturation and acidification of vesicular intermediates along the endocytic pathway. Key proteins and enzymes are delivered to the lysosome either via direct transport from the golgi to the lysosome, or via routing from the plasma membrane and early endosomes [1, 2, 6].
At the transcriptional level, lysosome biogenesis is primarily regulated by the microphthalmia/transcription factor E (MiT/TFE) family of transcription factors (TFs) - MITF, TFEB, TFE3, and TFEC [9, 10]. These basic-helix-loop helix transcription factors belong to the Myc Superfamily and recognize a palindromic 10-base pair motif (GTCACGTGAC) closely related to canonical E-Box elements [11]. This motif, termed the coordinated lysosomal expression and regulation (CLEAR) element is present in the promoter region of numerous lysosomal genes including those that encode for hydrolases, lysosomal membrane permeases, and associated proteins and is necessary for MiT/TFE mediated activation of gene expression [11–15]. Given the high level of homology of the DNA binding domain of MiT/TFE factors, it is predicated that all family members are capable of recognizing and binding to the CLEAR element. Accordingly, both TFEB and TFE3 have been shown to directly bind to the CLEAR element [9, 11, 16]. The MiT/TFE proteins also regulate several autophagy genes and therefore are considered master regulators of catabolism through coordinated capture, via autophagy, and subsequent degradation in the lysosome, of cellular macromolecules (Figure 1).
In healthy cells the pro-catabolic activity of MiT/TFE factors is triggered by upstream stimuli that broadly lead to an acute decrease in available nutrients such as starvation, fasting and exercise [17–20]. Post-translational modification of MiT/TFE factors are key for their nuclear translocation, binding to CLEAR elements and transcriptional activity to ensure coordinated induction of autophagy and lysosome gene programs. For instance, phosphorylation by mTORC1 serves to retain MiT/TFE proteins in the cytoplasm and therefore inactive when nutrients are abundant [6, 21, 22]. Under starvation conditions, the simultaneous inactivation of mTORC1 and dephosphorylation of MiT/TFE proteins by the phosphatase Calcineurin permits their nuclear entry [23]. Additional growth-regulating kinases such as MEK/ERK and glycogen synthase kinase 3 (GSK3) were shown to influence MiT/TFE nuclear trans-localization [24–26]. Alterations in acetylation [27, 28], SUMOylation [29, 30], oxidation [31], and ubiquitylation [32] are similarly proposed to regulate nuclear localization, DNA binding and transcriptional activity of the MiT/TFE TFs. Additional cellular stress conditions (endoplasmic reticulum (ER) stress, mitochondrial and DNA damage, oxidative stress), or tissue and organism level stress (inflammation, exercise, aging, neurodegeneration, cancer) are also associated with alterations in MiT/TFE factor activity [33–36]. Collectively these studies highlight a key role for lysosome biogenesis and function in regulation of metabolic adaptation, both at the cellular and the organismal level in response to diverse stimuli.
Additional transcription-independent pathways for lysosome biogenesis include autophagic lysosome reformation (ALR), which occurs during the final stages of autophagy to replenish the lysosome pool [37]. Completion of autophagy requires fusion of autophagosomes with lysosomes leading to the generation of transient hybrid autolysosomes. Subsequent restoration of the lysosomal pool involves the generation of tubules extending from autolysosomes which pinch off and give rise to “proto-lysosomes” that ultimately mature into functional lysosomes [37]. This process was shown to coincide with reactivation of mTORC1 following completion of autophagy. Thus, conditions which compromise mTOR signaling, autophagy [38] or lysosomal function [39, 40] would also likely compromise ALR. Analogous pathways that involve tubulation of endosomes (endocytic lysosome reformation) and phagosomes (phagocytic lysosome reformation) also participate in lysosome reformation in specific cell types [41].
Repair
Lysosomal damage is broadly defined as disruption of membrane integrity. Several chemical, physical or pathogenic agents can lead to lysosomal damage - accumulation of undigestible intra-lumenal content that mechanically ruptures the membrane, pathogen induced modification of lysosome membrane permeability and integrity or changes in membrane lipid composition associated with aging or lysosome storage disease (discussed below) (Figure 1) [7]. Experimentally, endo-lysosomal membrane damage can be induced by using lysosomotropic agents such as L-leucyl-L-leucine methyl ester (LLOME) or glycyl-Lphenylalanine 2-naphthylamide (GPN), which are converted by cathepsin C into membranolytic peptides within the lysosome lumen [42, 43]. Treatment with these agents is sufficient to cause leakage of small lysosomal accumulated dyes and cause proton leakage leading to lysosomal pH neutralization. Importantly, these effects are reversible upon washout of the damaging agent indicating that repair mechanisms are in place to “seal holes” caused by puncturing agents such as LLOME and GPN.
Two recent studies uncovered a new role for the endosomal sorting complex required for transport (ESCRT) machinery in direct lysosome membrane repair [44, 45]. ESCRT proteins include complexes ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III and the AAA+ ATPase, VPS4 and are critical mediators of membrane remodeling during viral budding, intra-luminal vesicle (ILV) biogenesis, cargo sorting and cytokinesis [46]. Hence a role for these proteins in lysosome membrane repair would be consistent with their broader role in membrane remodeling. Accordingly, ESCRT-I, ESCRT-III and ALIX proteins were shown to rapidly localized to the lysosome membrane following treatment with LLOME or GPN [44, 45]. Importantly, ESCRT proteins were recruited within 1min of LLOME treatment suggesting that this pathway is an early acting response to disruption of membrane integrity that may prevent further damage from occurring. Consistent with this idea, depletion of ALIX, TSG101 or ESCRT-III components blocks lysosome recovery from LLOME treatment upon washout [45]. ESCRT proteins were similarly recruited to endo-lysosomes following damage with silica crystals or following accumulation of endocytosed protein aggregates. These important studies identified a previously unrecognized role for ESCRT proteins as an early acting pathway for maintaining lysosome membrane integrity in response to acute damage.
Mechanistically, lysosomal calcium release following damage was shown to be required for ESCRT protein recruitment, possibly via an intermediate calcium-binding protein [45, 47]. ESCRT recruitment to endo-lysosome membranes was also recently shown to occur following hypertonic shock and loss of membrane tension [48]. Under these conditions, endo-lysosomal localization of the ESCRT-III protein CHMP4B triggered increased ILV formation and restoration of membrane curvature. Collectively, these studies suggest that the intrinsic ability of ESCRT proteins to form spiral filaments that lead to inward constriction of membrane edges is responsible for resealing punctures or restoring curvature. Alternatively, it is also possible that progressive assembly of ESCRT-III filaments may protect a membrane wound from further expansion and provide the necessary time for spontaneous membrane resealing or recruitment of additional unknown machinery that directly repair membrane [49].
Removal
When the duration and extent of lysosomal membrane damage exceed the repair ability of ESCRT-III, a form of selective autophagy known as lysophagy is initiated to clear permanently damaged lysosomes [50] (Figure 1). First, prolonged damage leads to exposure of carbohydrates found on lumenal lysosomal proteins which triggers recruitment of the carbohydrate binding proteins known as Galectins [51, 52]. Unlike rapid Ca2+ efflux or changes in membrane tension that recruit ESCRT proteins to facilitate repair, effective exposure of lumenal glycoproteins is thought to require extensive and long-lasting lysosomal membrane rupture. Accordingly, lysosomal recruitment of Galectins occurs later than ESCRTs, under conditions when ESCRT-mediated repair is either not possible or fails, and instead triggers a cascade of reactions that initiate clearance via lysophagy. Upon binding, Galectins, cooperate with a series of additional proteins, including E3 ubiquitin (Ub) ligases and autophagy cargo receptors to initiate clearance. For instance, Galectin-3 recruits the atypical tripartite motif family protein TRIM16 to damaged lysosomes, to facilitate ubiquitylation of lysosomal proteins [53]. Similarly, the E2 ligase, UBE2QL1 [54] and the E3 ligase complex SKP1/CUL1/F-Box (SCF) [55] have been shown to localize to damaged lysosomes leading to progressive ubiquitylation of lysosomal membrane proteins, (though it remains unclear whether specific lysosomal proteins are preferentially ubiquitylated). The activities of E2 and E3 ligases are in turn required for recruitment of autophagy receptors (eg. SQSTM1/p62, TAX1BP1) [54, 56, 57] and regulators (ULK1, Beclin1 and ATG16L1) [56], which enable recognition of ubiquitylated substrates. Depending on the damaging agent, different autophagy receptors may be recruited. For instance, the autophagy receptor Optineurin is recruited to Ub-tagged lysosomes following damage by α-synuclein fibrils [58], whereas NDP52 is recruited in response to Salmonella-induced endomembrane damage in a Ub-independent manner [59, 60]. Damaged lysosomes can also trigger direct recruitment of the AAA+ATPase, p97/VCP which functions together with cofactors, UBXD1, PLAA and the deubiquitylating enzyme YOD1 [57] to facilitate clearance. This complex was shown to preferentially remove Lysine-48 (K48)-linked Ub in favor of Lysine-63 (K63)-linked conjugates to promote autophagosome mediated recognition and clearance of damaged lysosomes.
It is worth noting that lysophagy-mediated clearance of damaged lysosomes can only occur if an active undamaged pool of lysosomes is still present following onset of the insult, or if biogenesis is triggered under chronic conditions. For instance, inactivation of mTORC1 and activation of AMPK signaling pathways are known to occur during late stages of lysosome damage and lysophagy, and the resulting induction of de novo lysosomal biogenesis via the MiT/TFE factors may play a key role in repair [61, 62].
Mechanisms of lysosomal membrane protein degradation and turnover
In addition to wholesale lysosome repair, specific proteins located on the lysosomal limiting membrane are subject to turnover in response to variations in intracellular nutrient levels as well as damage (Figure 1). In the vacuole of the budding yeast, Saccharomyces Cerevisiae the permease Ypq1, homologous to the Arg/Lys transporter PQLC2 in mammalian lysosomes, was shown to be internalized and degraded inside the vacuole in response to withdrawal of Lys from growth media. Starvation-dependent degradation of Ypq1 required its ubiquitylation at the vacuolar membrane by the Rsp5 E3 ligase, followed by ESCRT-dependent sorting into intralumenal vesicles [63]. Intriguingly, Rsp5-dependent ubiquitylation involves recognition of the arrested (non-transporting) conformation of Ypq1 by the vacuole-localized Ssh4 adaptor, thus directly coupling transport activity to protein stability [64].
The Ypq1 paradigm was shown to extend, albeit with mechanistic differences, to other vacuolar membrane permeases, including transporters implicated in homeostatic maintenance of cytosolic levels of zinc, iron and various amino acids [65–67]. For instance, low Zn2+ triggered the degradation of the Cot1 permease, which sequesters this ion from the cytosol into the vacuolar lumen. In contrast high Zn2+ triggered degradation of Zrt3, a cytosol-directed lysosomal Zn2+ exporter, presumably to avoid excess Zn2+ in the cytoplasm. Both Cot1 and Zrt3 degradation required their ubiquitylation by the Defective for SREBP Cleavage (DSC) E3 ubiquitin ligase complex, and the activity of the Cdc48/p97 AAA+ATPase [65].
Selective degradation of lysosomal/vacuolar membrane proteins is coupled to the overall metabolic state of the cell via TORC1 signaling. Rapamycin-mediated TORC1 inactivation enhances the degradation of several permeases. This effect is due, at least in part, to TORC1-dependent repression of the expression or activity of E3 ligases involved in vacuolar membrane ubiquitylation when nutrient levels are high, including an assembly factor for the vacuolar DSC complex [67]. Similar principles of selective ubiquitin-dependent degradation could apply to mammalian lysosomal membrane proteins, although more studies are needed to elucidate the client proteins and physiological conditions under which this process occurs [68].
Lysosomal remodeling and repair in neurodegeneration
Optimal lysosomal function is essential for the homeostasis and survival of neuronal cells and, accordingly, defective lysosomal remodeling and damage repair are emerging as key factors in multiple neurodegenerative conditions [6, 69] (Figure 2). The most evident alterations in lysosomal composition and function occur in a class of diseases known as lysosomal storage disorders (LSDs) [70]. In LSDs, mutations in over 60 different genes encoding for hydrolases involved in substrate degradation, or permeases that export degradation products to the cytoplasm, lead to buildup of the respective substrates within the lysosomal lumen. These substrates include all the main classes of macromolecules degraded and exported by the lysosome, including glycosphingolipids, sterols, amino acids, nucleotides and sugars.
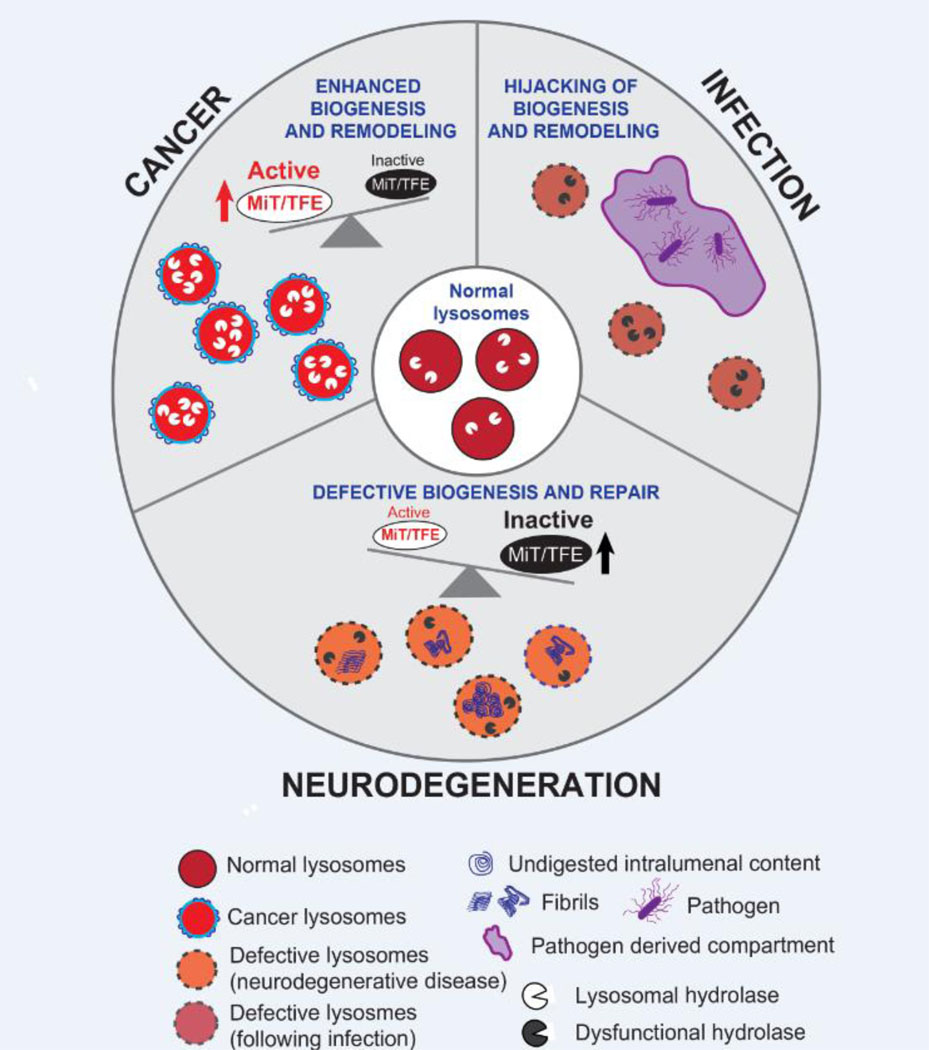
Figure 2: Lysosomal remodeling and repair in disease.
Alternations in lysosome function associated with enhanced or defective biogenesis, repair or remodeling are linked to disease including cancer, neurodegeneration and infection. In cancer (top left), elevated MiT/TFE activity ensures increased levels of lysosome biogenesis and activity (eg. higher levels of hydrolase expression). Additional lysosome membrane proteins and potential alterations in lipid composition may favor enhanced activity and increased resistance to damage in the context of cancer. On the other hand, neurodegenerative disorders are characterized by defects in lysosome biogenesis and function, including defective membrane repair (bottom). Accumulation of undigested intralumenal context is a hallmark of these disorders. Lysosome dysfunction has broader effects on cellular physiology including defects in trafficking, signaling and downstream quality control processes such as mitophagy and misfolded protein clearance. Ultimately, these pathogenic features compound to cause the death of critical cellular populations in the central nervous system. Intracellular pathogens subvert the endo-lysosomal system in order to gain control of and proliferate within the host cell (top right). Pathogens can secrete specialized factors, which reprogram or subvert host defense mechanisms – such as autophagy and lysosome mediated degradation - while simultaneously promoting host mediated remodeling of the compartment in which they reside.
Substrate accumulation within the lysosome has major morphological and functional impacts: in most LSDs, lysosomes are grossly enlarged, internally disorganized and exhibit trafficking defects including inability to fuse with incoming endosomes and autophagosomes. Along with internal organization and trafficking, lysosome-associated processes such as mTORC1 signaling are often disrupted [71–76]. Moreover, downstream quality control processes such as mitophagy and misfolded protein clearance are compromised. Ultimately, these pathogenic features compound to cause the death of critical cellular populations in the central nervous system and elsewhere, leading to the neurological impairment, stunted growth and early lethality that characterize most LSDs.
How substrate accumulation within LSD lysosomes ultimately triggers this cascade of pathogenic events is poorly understood. Recent studies employing lysosomal immuno-isolation and mass spectrometry-based proteomics have begun to shed light on this key question. One example is Niemann-Pick type C (NPC), a devastating LSD characterized by massive cholesterol storage within lysosomes due to the loss of the NPC1 cholesterol transporters [70]. Lysosome immuno-capture and proteomic analysis showed that, relative to their wild-type counterparts, NPC1-defective lysosomes exhibit decreased levels of multiple hydrolases, resulting in defective processing of protein and lipid substrates [75, 77]. Newly synthesized hydrolases may fail to reach NPC1-defective lysosomes due to trafficking defects observed in NPC cells. In addition, proteomics of NPC lysosomes revealed that these organelles are highly susceptible to damage, as shown by increased recruitment of both ESCRT-III components and Gal3 upon treatment with lysosome membrane destabilizing agents [75, 78]. Increased lysosomal membrane permeability could provide a route for multiple luminal hydrolases to ‘leak’ into the cytosol, directly contributing to the defective storage of secondary metabolites detected in NPC lysosomes [75, 77].
Newly synthesized hydrolases are continuously supplied to lysosomes to maintain the degradative capacity of this compartment. The CLN6 and CLN8 proteins were recently shown to bind to each other in a complex that promotes the export of different classes of hydrolases from the ER-Golgi for subsequent Mannose 6-Phosphate Receptor (M6PR)-dependent delivery to lysosomes [79, 80]. Mutations in CLN6 and CLN8 trigger, respectively, Neuronal Ceroid Lipofuscinosis (NLC) 6 and 8 (also known as Batten disease), fatal neurodegenerative disorders characterized by accumulation of auto-fluorescent pigments (‘lipofuscins’) within lysosomes [70]. CLN6-CLN8 may be especially critical under conditions that trigger TFEB activation, when TFEB-induced hydrolases must be delivered to the lysosome for stress adaptation and survival.
Efficient lysosomal remodeling and repair may play key roles in both onset and progression of protein misfolding diseases. In Alzheimer’s Disease (AD) and Frontotemporal Degeneration (FTD), small aggregates of the microtubule-associated protein Tau are thought to propagate from cell to cell and to ‘seed’ the formation of larger Tau aggregates in the cytoplasm, ultimately killing neuronal cells. A genome-wide screen for factors that regulate Tau seed propagation identified endo-lysosomal escape as a key step in this process [81]. In particular, several ESCRT-III subunits such as CHMP6, CHMP2A and CHMP2B were required to limit Tau exit into the cytoplasm following its endocytic uptake [81]. These results are consistent with genetic evidence linking defective ESCRT-III-dependent membrane repair to neurodegeneration. For instance, inherited mutations in CHMP2B cause a familial form of FTD [82, 83].
The connection between endo-lysosomal repair and neurodegeneration likely extends beyond tauopathies. Recently, the leucine-rich repeat kinase 2 (LRRK2) kinase, mutations in which cause a subset of familial Parkinson’s Disease, was shown to be activated upon endolysosomal membrane damage in macrophages, and to favor the assembly of ESCRT-III on damaged organelles through phosphorylation of its substrate, the small GTPase RAB8a [84]. Studies in astrocytes indicate that LRRK2 may also contribute to membrane repair by promoting tubulation and sorting from damaged endo-lysosomes [85].
A further connection between endolysosomal repair and cellular homeostasis is supported by the reported role for VCP/p97 in lysophagy [57]. Mutations in VPC/p97 give rise to multisystem proteinopathy, a protein aggregation disease involving the brain, bone and skeletal muscle [86], suggesting potential links between proteotoxic aggregate clearance and lysosomal repair in cellular homeostasis.
Lysosomal remodeling and repair in cancer
Alterations in lysosome number, distribution and composition have long been associated with malignant transformation of cells (Figure 2) [87]. Changes in the expression of several classes of lysosomal hydrolases have been detected in a variety of tumor types, correlate with prognosis and are targets for drug development [88]. However, their precise role in promoting versus suppressing tumor growth is likely context and tissue dependent [89]. For instance, expression levels of specific cathepsin family cysteine proteases have been associated with both tumor progression [90] and with tumor suppression [91, 92]. Several cathepsins are secreted, and can also function extracellularly to break down basement membrane and promote invasion and metastasis [90, 93]. Enhanced cathepsin secretion by cancer cells could be due to their overexpression, which overwhelms M6PR-mediated capture, leading to extracellular release [94]. Alternatively, increased lysosomal exocytosis could promote bulk release of multiple hydrolases to the extracellular space [95].
In addition to cathepsin proteases, aberrant expression of the lysosomal salidase Neu1, which is responsible for removal of sialic acids on glycans, was also shown to be pro-tumorigenic in non-small cell lung cancer [96] and hepatocellular carcinoma [97] but anti-tumorigenic in human colon cancer [98] and bladder cancer cell lines [99]. It is likely that context dependent and perhaps tissue specific expression and activity of lysosomal hydrolases dictate whether they function in a pro- or anti-tumorigenic manner. Additional factors such as the amount and type of cargo delivered to the lysosome, expression of permeases that transport digestion products out of the lysosome, or expression of catabolic co-factors such as the saposins may in turn influence the abundance and activity of the corresponding hydrolases [70].
Changes in lysosome biogenesis rates and their distribution are also noted across different cancers [9, 100]. For example, studies in pancreatic ductal adenocarcinoma (PDA) [101], Non-small cell lung cancer [102] and Melanoma [25, 103] show elevated lysosome biogenesis is a hallmark of disease and correlates with pro-tumorigenic phenotypes. In PDA, lysosome biogenesis is elevated due to constitutive nuclear localization of MiT/TFE TFs [101]. These cells also display heighted autophagy [101] and macropinocytosis [104] which converge on the lysosome for processing and recycling of intra- and extra-cellular cargo, respectively. Accordingly, increased lysosome biogenesis serves to facilitate efficient clearance of incoming cargo delivered to the lysosome via these trafficking routes and to fuel the metabolic needs of the cell.
Studies noting a more peripheral distribution of lysosomes upon malignant transformation suggested that lysosomal repositioning may serve to enhance invasion and metastatic potential [100]. Peripheral transport of lysosomes was recently shown to be regulated by the BORC protein complex consisting of myrlysin, lyspersin, diaskedin, snapin, BLOS1, BLOS2 and KXD1 [105].The BORC complex recruits the Arf GTPase, Arl8b to the lysosome membrane to drive microtubule-associated kinesin dependent anterograde transport of lysosomes towards the cell periphery. Inhibition of BORC causes dissociation of Arl8b from lysosomes, leading to perinuclear clustering of lysosomes and inhibited cell spreading and motility without compromising lysosomal degradative function. Increased TFEB activity was also shown to promote lysosomal repositioning towards the cell periphery in order to mediate increased rates of exocytosis in response to changes in intracellular Ca2+ [106]. Thus, the combination of dynamic changes in lysosome number, composition (eg. hydrolase abundance and activity) and distribution in the cell likely has a critical role in establishing how the lysosome contributes to tumorigenesis.
The enhanced demand placed on cancer lysosomes also suggests that specialized mechanisms that prolong lysosome health may exist. Consistent with this idea, a recent study showed that PDA lysosomes are more resistant to lysosomal membrane damage relative to lysosomes in normal epithelial cells [107]. Treatment of PDA cells with LLOME, GPN or silica led to delayed loss of lysosomal dyes and delayed recruitment of ESCRT and Gal3 relative to non-cancer cells. Proteomics analysis of isolated lysosomes uncovered selective enrichment of the Ferlin family of membrane repair proteins on the membrane of PDA lysosomes. The Ferlin proteins – Myoferlin (MYOF) and Dysferlin (DYSF) in particular – have previously been shown to aid in plasma membrane repair in muscle cells [108–110]. Accordingly, PDA cells may co-opt the Ferlin repair mechanism to retain lysosomal membrane integrity and sustain enhanced functionality. Consistent with this hypothesis loss of MYOF led to constitutive lysosomal recruitment of ESCRT proteins and profound lysosome dysfunction ultimately leading to decreased PDA cell viability [107]. The discovery of MYOF as a putative lysosomal repair factor suggests that unique protective mechanisms may be in place to prevent damage or dysfunction in the context of cancer. Additional changes in the lipid composition of the lysosomal membrane have also been implicated in lysosome response to damage in the context of cancer [87]. For instance, increased abundance of lipid kinases which regulate phosphatidylinositol abundance on the lysosome membrane can increase the efficiency of autophagosome to lysosome fusion [111, 112]. Future studies incorporating lysosomal proteomics and metabolomics in the context of progressive malignant transformation coupled to analysis of lysosomal positional information may uncover additional cancer specific lysosomal features which could be exploited to therapeutically target the lysosome in cancer.
Lysosomal remodeling and repair during pathogen infection
Intracellular pathogens subvert the endo-lysosomal system in order to gain control of and proliferate within the host cell (Figure 2). This invasion process can serve multiple goals, including: 1- to gain entry into the cytoplasm from the extracellular space 2- to avoid delivery of the pathogen to the lysosomal lumen, where it may be destroyed and 3- to hijack and redirect metabolic and signaling processes to aid pathogen growth. These actions rely on the secretion of specialized factors, which reprogram or subvert host defense mechanisms while simultaneously promoting host mediated remodeling of the compartment in which they reside. For instance, the ESX-1 type VII secretion system (T7SS) utilized by M.Tuberculosis is capable of blunting host ESCRT mediated endomembrane repair [113, 114]. By disabling ESCRT function, the bacterium can continue to perforate the phagosome to gain access to nutrients and host cytosolic factors while also preventing phagolysosome maturation or fusion with lysosomes. Accordingly, host cells lacking ESCRT proteins [113–115] or Gal3 [53] are more susceptible to pathogen infection.
M.Tuberculosis and Salmonella can also manipulate lysosomal signaling. Upon cell entry, an acute defense response involving upregulation of autophagy limits the intracellular spread of these pathogens [53, 115–117]. In the case of Salmonella, this response may be based, at least in part, on endomembrane damage caused by pathogen entry, such that the subsequent leakage of amino acids inhibits mTORC1 and favors induction of autophagy [118]. In order to escape autophagic degradation and delivery to lysosomes, Salmonella secretes several effectors that reactivate mTORC1 signaling by redirecting the lysosomal mTORC1 scaffolding complex, composed of Rag GTPases and Ragulator complex, to the surface of the pathogen-containing vacuole [118].
Beta-Coronaviruses, which include severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), enter the cytoplasm via fusion with either the plasma membrane or, when endocytic uptake occurs, with the endo-lysosomal limiting membrane [119–121]. Subsequently, viral proteins translated within the host cytosol induce the rearrangement of cellular membranes to form double-membrane vesicles (DMVs), where replication of the viral genomes occurs. The resulting genomic RNAs are assembled together with structural proteins into mature virions at the ER-Golgi intermediate complex (ERGIC). An unexpected finding is that exit of newly assembled SARS-CoV2 virions from infected cells involves their loading into lysosomes, followed by fusion of virion-loaded lysosomes with the plasma membrane [122]. Virion loading disrupts the lysosome, which become de-acidified, lose proteolytic enzymes and, in infected macrophages, lose the ability to cross-present antigens [122]. Thus, SARSCoV-2 transforms lysosomes into mere vessels to enable its rapid egress and spreading.
In response to infection, immune cells can also initiate profound remodeling of their endo-lysosomal system to increase their ability to capture and destroy the invading pathogens. For instance, activation of mouse RAW 267.4 macrophages with Lipopolysaccharide (LPS) was shown to increase resistance to endo-lysosomal damage mediated by silica, suggesting that mediators of macrophage activation may confer protection against pathogen induced endomembrane damage [123]. Accordingly, initial stimulation of macrophages with either Toll-Like Receptor ligands, Interferon-γ, Tumor Necrosis Factor-α or intact bacteria is sufficient to limit endomembrane damage induced by silica or subsequent bacterial infection [124]. These findings suggest that an activated cell state may change the composition of endo-lysosomal membrane compartments to make them more resistant to damage. Consistent with this idea, proteomics analysis of isolated latex-bead laden late endosomes/lysosomes from immature versus mature dendritic cells (DC) revealed changes in lysosome composition, including increased abundance of membrane transporters and ESCRT-III proteins in mature, activated DCs [125]. Thus, lysosomal damage and remodeling are at the center of a tug-of-war between pathogens and immune cells, justifying their targeting in therapeutic settings.
Concluding remarks
Despite increasingly clear evidence of key roles for lysosomal remodeling and repair in multiple pathophysiological processes, basic mechanistic questions about its regulation and implementation still remain (see ‘Outstanding Questions box’).
Highlights:
The lysosome is a catabolic organelle that is the end-point of degradative pathways including endocytosis, phagocytosis and autophagy.
Lysosomal damage can result from accumulation of undigested substrates within the lumen, changes in lipid composition of the membrane, loss of pH and/or membrane potential and outright rupturing of the lysosomal limiting membrane by mechanical or chemical insults.
Lysosomes are subject to continuous remodeling and repair via the action of dedicated signaling pathways, which detect lysosomal stress and dysfunction and trigger both transcriptional and post-translational programs that adjust lysosomal function in a compensatory manner.
Prompt repair of lysosomal injury is important for numerous cell types and especially essential for neuronal cells, while enhanced biogenesis and plasticity are emerging as key adaptive mechanisms in cancer cells.
Outstanding Questions:
What are the factors that specifically recruit ESCRT-III to damaged endo-lysosomes? While Ca2+ and the ALIX subunit are both required for ESCRT-III recruitment, their relationship remains to be clarified, and the possible role of additional protein and/or phospholipid intermediates is unknown.
How does ESCRT-III assembly for endo-lysosomal repair relate to or differ from the Multivesicular body biogenesis pathway, or to other ESCRT-III-dependent processes such nuclear envelope reformation and repair?
What is the relationship between the fast-acting, ESCRT-III-dependent mechanism and the slower and late-acting lysophagy pathway? Is there a clear damage threshold past which lysophagy is activated? How does lysophagy restore functional lysosomes?
What is the role of accessory repair pathways that can complement or substitute for ESCRT-III and lysophagy, such as lysosomal exocytosis, membrane contact sites, and repurposing of membrane stabilization factors such as the Ferlins? Further investigation into these pathways will shed light on alternative, context and tissue dependent mechanisms for maintaining lysosomal homeostasis.
The progressive decline in lysosomal integrity associated with neurodegeneration and aging suggests that strategies aimed at its augmentation via chemical or genetic means could be beneficial to maintain neuronal health. A possible avenue could involve modulating the activity and subcellular targeting of neuroprotective proteins implicated in lysosomal repair, such as LRRK2 and VCP/p97 [57, 84]. One can also envision the use of molecular glues that selectively direct the assembly of ESCRT-III or other stabilizing/repair factors to the endolysosomal membrane [107, 126].
Along with enhanced repair, boosting lysosome biogenesis is a viable strategy to maintain neuronal homeostasis in the face of proteotoxic challenges. Aided by our rapidly increasing knowledge of the complex regulatory mechanisms of the MiT/TFE factors [127–129], strategies to boost their nuclear localization and activity may hopefully reach the clinic in the coming years.
On the other hand, enhanced lysosome activity in cancer cells may represent a potential Achilles heel that, if targeted effectively, would lead to selective cancer cell death. Current strategies to target the lysosome include treatment with lysosomotropic agents such as the anti-malarial hydroxychloroquine, which was shown to have significant anti-tumor effects across several in vitro and in vivo cancer models [130]. New-generation inhibitors targeting unique cancer-associated lysosomal proteins may enable cancer cell-selective lysosomal destabilization, leading to increased efficacy and reduced normal tissue toxicity.
ACKNOWLEDGEMENTS
R.Z acknowledges NIH R01GM127763, a University of Notre Dame/APMRF grant and an Edward J. Mallinckrodt Jr Foundation Scholarship.
R.M.P acknowledges NIH R01CA260249, the Ed Marra Passion to Win Fund and the Shorenstein Fund.
We apologize to colleagues whose work we were unable to cite due to space limitations.
R.Z. is a co-founder, shareholder, and consultant for Frontier Medicines Corp.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ballabio A. and Bonifacino JS (2020) Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol 21 (2), 101–118. [DOI] [PubMed] [Google Scholar]
- 2.Perera RM and Zoncu R. (2016) The Lysosome as a Regulatory Hub. Annu Rev Cell Dev Biol 32, 223–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasanthakumar T. and Rubinstein JL (2020) Structure and Roles of V-type ATPases. Trends Biochem Sci 45 (4), 295–307. [DOI] [PubMed] [Google Scholar]
- 4.Xu H. and Ren D. (2015) Lysosomal physiology. Annu Rev Physiol 77, 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu GY and Sabatini DM (2020) Author Correction: mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21 (4), 246. [DOI] [PubMed] [Google Scholar]
- 6.Shin HR and Zoncu R. (2020) The Lysosome at the Intersection of Cellular Growth and Destruction. Dev Cell 54 (2), 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhen Y. et al. (2021) Sealing holes in cellular membranes. EMBO J 40 (7), e106922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma J. et al. (2018) Lysosomes and Brain Health. Annu Rev Neurosci 41, 255–276. [DOI] [PubMed] [Google Scholar]
- 9.Perera RM et al. (2019) MiT/TFE Family of Transcription Factors, Lysosomes, and Cancer. Annu Rev Cancer Biol 3, 203–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemesath TJ et al. (1994) microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 8 (22), 2770–80. [DOI] [PubMed] [Google Scholar]
- 11.Palmieri M. et al. (2011) Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20 (19), 3852–66. [DOI] [PubMed] [Google Scholar]
- 12.Martina JA et al. (2014) Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cell Mol Life Sci 71 (13), 2483–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Napolitano G. and Ballabio A. (2016) TFEB at a glance. J Cell Sci 129 (13), 2475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sardiello M. et al. (2009) A gene network regulating lysosomal biogenesis and function. Science 325 (5939), 473–7. [DOI] [PubMed] [Google Scholar]
- 15.Settembre C. et al. (2013) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14 (5), 283–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martina JA et al. (2014) The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7 (309), ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansueto G. et al. (2017) Transcription Factor EB Controls Metabolic Flexibility during Exercise. Cell Metab 25 (1), 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Rourke EJ and Ruvkun G. (2013) MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 15 (6), 668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Settembre C. and Ballabio A. (2014) Lysosome: regulator of lipid degradation pathways. Trends Cell Biol 24 (12), 743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Settembre C. et al. (2013) TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15 (6), 647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goding CR and Arnheiter H. (2019) MITF-the first 25 years. Genes Dev 33 (15–16), 983–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puertollano R. et al. (2018) The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J 37 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina DL et al. (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17 (3), 288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchand B. et al. (2015) Glycogen synthase kinase-3 (GSK3) inhibition induces prosurvival autophagic signals in human pancreatic cancer cells. J Biol Chem 290 (9), 5592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ploper D. et al. (2015) MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci U S A 112 (5), E420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Settembre C. et al. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332 (6036), 1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y. et al. (2020) Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep 21 (1), e48335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J. et al. (2018) Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy 14 (6), 1043–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller AJ et al. (2005) Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem 280 (1), 146–55. [DOI] [PubMed] [Google Scholar]
- 30.Murakami H. and Arnheiter H. (2005) Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res 18 (4), 265–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H. et al. (2020) Oxidation of multiple MiT/TFE transcription factors links oxidative stress to transcriptional control of autophagy and lysosome biogenesis. Autophagy 16 (9), 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vu HN et al. (2021) User guide to MiT-TFE isoforms and post-translational modifications. Pigment Cell Melanoma Res 34 (1), 13–27. [DOI] [PubMed] [Google Scholar]
- 33.Lapierre LR et al. (2013) The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 4, 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martina JA et al. (2016) TFEB and TFE3 are novel components of the integrated stress response. EMBO J 35 (5), 479–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pastore N. et al. (2019) Nutrient-sensitive transcription factors TFEB and TFE3 couple autophagy and metabolism to the peripheral clock. EMBO J 38 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santaguida S. et al. (2015) Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev 29 (19), 2010–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu L. et al. (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465 (7300), 942–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munson MJ et al. (2015) mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J 34 (17), 2272–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magalhaes J. et al. (2016) Autophagic lysosome reformation dysfunction in glucocerebrosidase deficient cells: relevance to Parkinson disease. Hum Mol Genet 25 (16), 3432–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong Y. et al. (2011) Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci U S A 108 (19), 7826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C. and Wang X. (2021) Lysosome biogenesis: Regulation and functions. J Cell Biol 220 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bright NA et al. (2016) Endolysosomes Are the Principal Intracellular Sites of Acid Hydrolase Activity. Curr Biol 26 (17), 2233–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiele DL and Lipsky PE (1990) Mechanism of L-leucyl-L-leucine methyl ester-mediated killing of cytotoxic lymphocytes: dependence on a lysosomal thiol protease, dipeptidyl peptidase I, that is enriched in these cells. Proc Natl Acad Sci U S A 87 (1), 83–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radulovic M. et al. (2018) ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J 37 (21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skowyra ML et al. (2018) Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360 (6384). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vietri M. et al. (2020) The many functions of ESCRTs. Nat Rev Mol Cell Biol 21 (1), 25–42. [DOI] [PubMed] [Google Scholar]
- 47.Scheffer LL et al. (2014) Mechanism of Ca(2)(+)-triggered ESCRT assembly and regulation of cell membrane repair. Nat Commun 5, 5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercier V. et al. (2020) Endosomal membrane tension regulates ESCRT-III-dependent intralumenal vesicle formation. Nat Cell Biol 22 (8), 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bohannon KP and Hanson PI (2020) ESCRT puts its thumb on the nanoscale: Fixing tiny holes in endolysosomes. Curr Opin Cell Biol 65, 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maejima I. et al. (2013) Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 32 (17), 2336–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aits S. et al. (2015) Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11 (8), 1408–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johannes L. et al. (2018) Galectins at a glance. J Cell Sci 131 (9). [DOI] [PubMed] [Google Scholar]
- 53.Chauhan S. et al. (2016) TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Codirect Autophagy in Endomembrane Damage Homeostasis. Dev Cell 39 (1), 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koerver L. et al. (2019) The ubiquitin-conjugating enzyme UBE2QL1 coordinates lysophagy in response to endolysosomal damage. EMBO Rep 20 (10), e48014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida Y. et al. (2017) Ubiquitination of exposed glycoproteins by SCF(FBXO27) directs damaged lysosomes for autophagy. Proc Natl Acad Sci U S A 114 (32), 8574–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujita N. et al. (2013) Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol 203 (1), 115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papadopoulos C. et al. (2017) VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J 36 (2), 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bussi C. et al. (2018) Alpha-synuclein fibrils recruit TBK1 and OPTN to lysosomal damage sites and induce autophagy in microglial cells. J Cell Sci 131 (23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravenhill BJ et al. (2019) The Cargo Receptor NDP52 Initiates Selective Autophagy by Recruiting the ULK Complex to Cytosol-Invading Bacteria. Mol Cell 74 (2), 320–329 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thurston TL et al. (2016) Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2dependent antibacterial autophagy. EMBO J 35 (16), 1779–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jia J. et al. (2018) Galectins Control mTOR in Response to Endomembrane Damage. Mol Cell 70 (1), 120–135 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jia J. et al. (2020) AMPK is activated during lysosomal damage via a galectin-ubiquitin signal transduction system. Autophagy 16 (8), 1550–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li M. et al. (2015) Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol Cell 57 (3), 467–78. [DOI] [PubMed] [Google Scholar]
- 64.Arines FM et al. (2021) A selective transmembrane recognition mechanism by a membrane-anchored ubiquitin ligase adaptor. J Cell Biol 220 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M. et al. (2015) Membrane-anchored ubiquitin ligase complex is required for the turnover of lysosomal membrane proteins. J Cell Biol 211 (3), 639–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang X. et al. (2021) ESCRT, not intralumenal fragments, sorts ubiquitinated vacuole membrane proteins for degradation. J Cell Biol 220 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang X. et al. (2020) TORC1 regulates vacuole membrane composition through ubiquitin- and ESCRT-dependent microautophagy. J Cell Biol 219 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang W. et al. (2021) A conserved ubiquitin- and ESCRT-dependent pathway internalizes human lysosomal membrane proteins for degradation. PLoS Biol 19 (7), e3001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bajaj L. et al. (2019) Lysosome biogenesis in health and disease. J Neurochem 148 (5), 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Platt FM et al. (2018) Lysosomal storage diseases. Nat Rev Dis Primers 4 (1), 27. [DOI] [PubMed] [Google Scholar]
- 71.Andrzejewska Z. et al. (2016) Cystinosin is a Component of the Vacuolar H+-ATPase-Ragulator-Rag Complex Controlling Mammalian Target of Rapamycin Complex 1 Signaling. J Am Soc Nephrol 27 (6), 1678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartolomeo R. et al. (2017) mTORC1 hyperactivation arrests bone growth in lysosomal storage disorders by suppressing autophagy. J Clin Invest 127 (10), 3717–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berger Z. et al. (2006) Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet 15 (3), 433–42. [DOI] [PubMed] [Google Scholar]
- 74.Brown RA et al. (2019) mTOR hyperactivity mediates lysosomal dysfunction in Gaucher’s disease iPSC-neuronal cells. Dis Model Mech 12 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davis OB et al. (2021) NPC1-mTORC1 Signaling Couples Cholesterol Sensing to Organelle Homeostasis and Is a Targetable Pathway in Niemann-Pick Type C. Dev Cell 56 (3), 260–276 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ivanova EA et al. (2016) Altered mTOR signalling in nephropathic cystinosis. J Inherit Metab Dis 39 (3), 457–464. [DOI] [PubMed] [Google Scholar]
- 77.Tharkeshwar AK et al. (2017) A novel approach to analyze lysosomal dysfunctions through subcellular proteomics and lipidomics: the case of NPC1 deficiency. Sci Rep 7, 41408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu EA et al. (2020) Fbxo2 mediates clearance of damaged lysosomes and modifies neurodegeneration in the Niemann-Pick C brain. JCI Insight 5 (20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bajaj L. et al. (2020) A CLN6-CLN8 complex recruits lysosomal enzymes at the ER for Golgi transfer. J Clin Invest 130 (8), 4118–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.di Ronza A. et al. (2018) CLN8 is an endoplasmic reticulum cargo receptor that regulates lysosome biogenesis. Nat Cell Biol 20 (12), 1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen JJ et al. (2019) Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J Biol Chem 294 (50), 18952–18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clayton EL et al. (2015) Frontotemporal dementia caused by CHMP2B mutation is characterised by neuronal lysosomal storage pathology. Acta Neuropathol 130 (4), 511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Urwin H. et al. (2010) Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet 19 (11), 2228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Herbst S. et al. (2020) LRRK2 activation controls the repair of damaged endomembranes in macrophages. EMBO J 39 (18), e104494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bonet-Ponce L. et al. (2020) LRRK2 mediates tubulation and vesicle sorting from lysosomes. Sci Adv 6 (46). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van den Boom J. and Meyer H. (2018) VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol Cell 69 (2), 182–194. [DOI] [PubMed] [Google Scholar]
- 87.Kallunki T. et al. (2013) Cancer-associated lysosomal changes: friends or foes? Oncogene 32 (16), 1995–2004. [DOI] [PubMed] [Google Scholar]
- 88.Dheer D. et al. (2019) Cathepsin-sensitive nanoscale drug delivery systems for cancer therapy and other diseases. Adv Drug Deliv Rev 151–152, 130–151. [DOI] [PubMed] [Google Scholar]
- 89.Olson OC and Joyce JA (2015) Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer 15 (12), 712–29. [DOI] [PubMed] [Google Scholar]
- 90.Mohamed MM and Sloane BF (2006) Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer 6 (10), 764–75. [DOI] [PubMed] [Google Scholar]
- 91.Boudreau F. et al. (2007) Loss of cathepsin L activity promotes claudin-1 overexpression and intestinal neoplasia. FASEB J 21 (14), 3853–65. [DOI] [PubMed] [Google Scholar]
- 92.Dennemarker J. et al. (2010) Deficiency for the cysteine protease cathepsin L promotes tumor progression in mouse epidermis. Oncogene 29 (11), 1611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gocheva V. and Joyce JA (2007) Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle 6 (1), 60–4. [DOI] [PubMed] [Google Scholar]
- 94.Braulke T. and Bonifacino JS (2009) Sorting of lysosomal proteins. Biochim Biophys Acta 1793 (4), 605–14. [DOI] [PubMed] [Google Scholar]
- 95.Machado E. et al. (2015) Regulated lysosomal exocytosis mediates cancer progression. Sci Adv 1 (11), e1500603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lv T. et al. (2020) p53-R273H promotes cancer cell migration via upregulation of neuraminidase-1. J Cancer 11 (23), 6874–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hou G. et al. (2016) Neuraminidase 1 (NEU1) promotes proliferation and migration as a diagnostic and prognostic biomarker of hepatocellular carcinoma. Oncotarget 7 (40), 64957–64966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Uemura T. et al. (2009) Contribution of sialidase NEU1 to suppression of metastasis of human colon cancer cells through desialylation of integrin beta4. Oncogene 28 (9), 1218–29. [DOI] [PubMed] [Google Scholar]
- 99.Zhou X. et al. (2020) Sialidase NEU1 suppresses progression of human bladder cancer cells by inhibiting fibronectin-integrin alpha5beta1 interaction and Akt signaling pathway. Cell Commun Signal 18 (1), 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hamalisto S. and Jaattela M. (2016) Lysosomes in cancer-living on the edge (of the cell). Curr Opin Cell Biol 39, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Perera RM et al. (2015) Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524 (7565), 361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eichner LJ et al. (2019) Genetic Analysis Reveals AMPK Is Required to Support Tumor Growth in Murine Kras-Dependent Lung Cancer Models. Cell Metab 29 (2), 285–302 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Du J. et al. (2019) YY1 cooperates with TFEB to regulate autophagy and lysosomal biogenesis in melanoma. Mol Carcinog 58 (11), 2149–2160. [DOI] [PubMed] [Google Scholar]
- 104.Commisso C. et al. (2013) Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497 (7451), 633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pu J. et al. (2015) BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell 33 (2), 176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Medina DL et al. (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21 (3), 421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gupta S. et al. (2021) Lysosomal retargeting of Myoferlin mitigates membrane stress to enable pancreatic cancer growth. Nat Cell Biol 23 (3), 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bansal D. and Campbell KP (2004) Dysferlin and the plasma membrane repair in muscular dystrophy. Trends Cell Biol 14 (4), 206–13. [DOI] [PubMed] [Google Scholar]
- 109.Bansal D. et al. (2003) Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 423 (6936), 168–72. [DOI] [PubMed] [Google Scholar]
- 110.Davis DB et al. (2000) Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum Mol Genet 9 (2), 217–26. [DOI] [PubMed] [Google Scholar]
- 111.Arora GK et al. (2021) Expanding role of PI5P4Ks in cancer: A promising druggable target. FEBS Lett. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lundquist MR et al. (2018) Phosphatidylinositol-5-Phosphate 4-Kinases Regulate Cellular Lipid Metabolism By Facilitating Autophagy. Mol Cell 70 (3), 531–544 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mittal E. et al. (2018) Mycobacterium tuberculosis Type VII Secretion System Effectors Differentially Impact the ESCRT Endomembrane Damage Response. mBio 9 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Philips JA et al. (2008) ESCRT factors restrict mycobacterial growth. Proc Natl Acad Sci U S A 105 (8), 3070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cheng LW et al. (2005) Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc Natl Acad Sci U S A 102 (38), 13646–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Otten EG et al. (2021) Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature 594 (7861), 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Watson RO et al. (2012) Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150 (4), 803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tattoli I. et al. (2012) Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11 (6), 563–75. [DOI] [PubMed] [Google Scholar]
- 119.Baggen J. et al. (2021) Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet 53 (4), 435–444. [DOI] [PubMed] [Google Scholar]
- 120.Fung TS and Liu DX (2019) Human Coronavirus: Host-Pathogen Interaction. Annu Rev Microbiol 73, 529–557. [DOI] [PubMed] [Google Scholar]
- 121.Walls AC et al. (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181 (2), 281–292 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ghosh S. et al. (2020) beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 183 (6), 1520–1535 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Davis MJ and Swanson JA (2010) Technical advance: Caspase-1 activation and IL-1beta release correlate with the degree of lysosome damage, as illustrated by a novel imaging method to quantify phagolysosome damage. J Leukoc Biol 88 (4), 813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Davis MJ et al. (2012) Inducible renitence limits Listeria monocytogenes escape from vacuoles in macrophages. J Immunol 189 (9), 4488–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Duclos S. et al. (2011) The endosomal proteome of macrophage and dendritic cells. Proteomics 11 (5), 854–64. [DOI] [PubMed] [Google Scholar]
- 126.Maniaci C. and Ciulli A. (2019) Bifunctional chemical probes inducing protein-protein interactions. Curr Opin Chem Biol 52, 145–156. [DOI] [PubMed] [Google Scholar]
- 127.Lawrence RE et al. (2019) Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science 366 (6468), 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Napolitano G. et al. (2020) A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dube syndrome. Nature 585 (7826), 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Napolitano G. et al. (2018) mTOR-dependent phosphorylation controls TFEB nuclear export. Nat Commun 9 (1), 3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Piao S. and Amaravadi RK (2016) Targeting the lysosome in cancer. Ann N Y Acad Sci 1371 (1), 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]