Summary
The cytokine interleukin-3 (IL-3) acts on early hematopoietic precursor cells. In humans, Treg cells secrete IL-3 and repress inflammatory cells except for basophils. The present study aims to elucidate the contribution of IL-3 in the development and the course of allergic asthma. We therefore analyzed the secretion of IL-3 in PBMCs and total blood cells in two cohorts of pre-school children with and without asthma. In a murine model of allergic asthma, we analyzed the phenotype of IL-3−/− mice compared to wild-type mice. PBMCs from asthmatic children showed increased IL-3 secretion, which directly correlated with improved lung function. IL-3−/− asthmatic mice showed increased asthmatic traits. Moreover, IL-3-deficient mice had a defect in T regulatory cells in the lung. In conclusion, IL-3 downregulation was found associated with more severe allergic asthma in pre-school children. Consistently, targeting IL-3 resulted in an induced pathophysiological response in a murine model of allergic asthma.
Subject areas: Biological sciences, Immunology, immune response
Graphical abstract
Highlights
-
•
The cytokine interleukin-3 (IL-3) acts on early hematopoietic precursor cells
-
•
PHA-stimulated PBMCs from asthmatic children showed increased IL-3 secretion
-
•
IL-3 from PBMCs from asthmatic children correlated with improved lung function
-
•
Targeting IL-3 resulted in an induced pathophysiological response in asthma model
Biological sciences; Immunology; Immune response
Introduction
Asthma bronchiale is a chronic inflammatory disease of the airways that affects millions of people worldwide. Due to the airway hyperresponsiveness (AHR), the contact with otherwise harmless antigens, like pollen, house dust mite, or ovalbumin (OVA), triggers an inflammation of the bronchial mucosa, associated with increased mucus production, airway remodeling, and bronchospasms. The underlying immunological mechanisms are heterogeneous, complex, and vary depending on the form and endotype of the asthma (Lambrecht and Hammad, 2015; Wenzel, 2012; Taher et al., 2010).
During the early phase reaction, allergen re-exposure triggers the cross-linking of IgE antibodies, which are bound to their specific Fcε receptor on mast cells and basophils. This leads to the release of pre-formed inflammatory mediators such as histamine, leukotrienes, tryptases, and cytokines, like interleukin (IL)-3, IL-4, and IL-5 (Gauvreau et al., 2015; van de Veen and Akdis, 2019). The type I hypersensitivity reaction affected by these mediators implicates the recruitment of T helper 2 (TH2) cells as well as other inflammatory cells, mainly eosinophils, into the site of allergen and causes the characteristic lung inflammation (Holgate et al., 2015; Akdis, 2013; Barnes, 2011).
The cytokine IL-3 is predominantly released by activated T cells, as well as from basophils, eosinophils, and mast cells (Arai et al., 1990; Celestin et al., 2001; Asquith et al., 2008; Akdis et al., 2016). IL-3 in general acts as a hematopoietic growth factor in the early phases of the hematopoiesis (Auclair et al., 2014; Akdis et al., 2016). In particular, IL-3 stimulates the growth, the differentiation, and the release of mediators from mast cells and basophils, such as histamine, IL-4, and IL-6, as well as the growth of eosinophils, macrophages, and dendritic cells (Schrader, 1997; Lantz et al., 2008; Broughton et al., 2012; Voehringer, 2012; Auclair et al., 2014). Thereby, IL-3 signals through a heterodimeric receptor complex, consisting of an IL-3-specific α-chain (IL3Rα, CD123) and the common β-chain (IL3Rβ, CD131), which IL-3 shares together with IL-5 and granulocyte-macrophage colony-stimulating factor (GM-CSF) (Schrader, 1997; Asquith et al., 2008; Broughton et al., 2012; Esnault and Kelly, 2016). Both receptor chains are expressed on different blood cells, like basophils, mast cells, eosinophils, and monocytes (Renner et al., 2018).
However, less is known about the contribution of IL-3 in the development and the course of allergic asthma. Different studies showed that the increased expression of IL-3 in the context of bronchial infections in early childhood (Bertrand et al., 2015), as well as certain polymorphisms in the IL3 gene (Park et al., 2004), are associated with an increased risk of developing asthma later in life. Additionally, increased IL-3 expression in the lung of asthmatic patients has been observed (Robinson et al., 1992). Along with the results from further studies on different inflammatory diseases (Lourenço et al., 2008; Renner et al., 2015; Auclair et al., 2014; Lantz et al., 1998), these findings suggest a role of IL-3 in the immunological response of asthma. To investigate this hypothesis, we analyzed the secretion of IL-3 and its receptor expression in pre-school children with and without asthma and studied IL-3−/− mice in an experimental model of allergic asthma.
Results
IL-3 is associated with amelioration of asthma in pre-school children
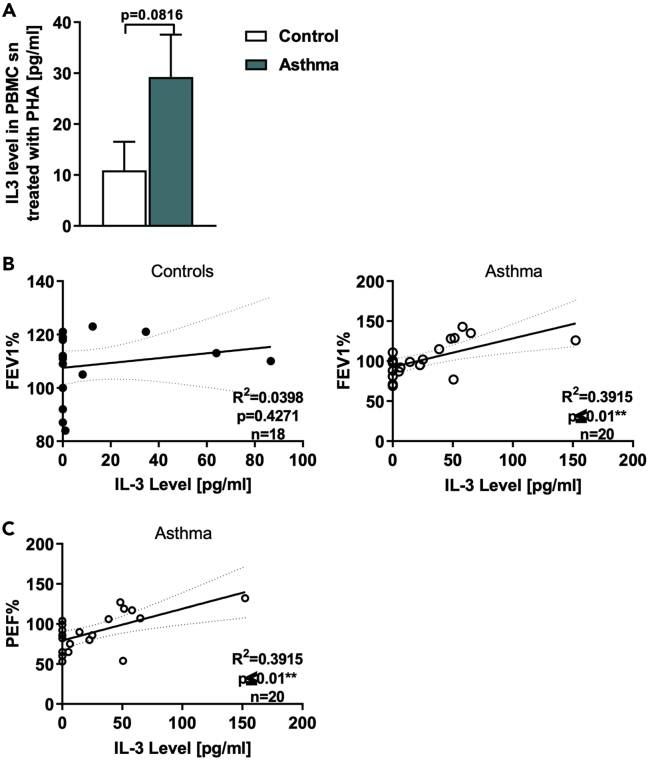
In this study, we analyzed two cohorts of pre-school children with and without asthma. IL-3 is known to be released by activated T cells (Mosmann et al., 1986), thus PBMCs were challenged with the lectin phytohemagglutinin (PHA). Here, we found an increased secretion of IL-3 in the PBMCs of asthmatic children compared to healthy children (Figure 1A). Furthermore, we found that the IL-3 secretion in asthmatic children correlated directly with the corresponding FEV1%, predicted and the PEF% (peak expiratory flow, predicted) (Figures 1B and 1C), but not in healthy control children (Figures 1C and S1A). FEV1%, predicted is the best validated value for asthma also in children. Thereby, children with a higher FEV1%, predicted released more IL-3 than children with a lower FEV1%, predicted, indicating that IL-3 is accompanied with the amelioration of asthma in children.
Figure 1.
IL-3 induction is associated with ameliorated asthma in children
(A) ELISA analysis of IL-3 levels in the supernatant from peripheral blood mononuclear cells (PBMCs) isolated from healthy control and asthmatic children and cultured with PHA for 24h (n = 19/21).
(B) Correlation between the IL-3 quantified by ELISA analysis in the supernatant from PHA-stimulated PBMCs isolated from healthy control (n = 18: IL-3 n = 20; FEV1% predicted n = 19) and asthmatic children and the predicted FEV1%, predicted (FEV1% predicted n = 23; IL-3: n = 20; correlation FEV1% predicted and IL-3: n = 20).
(C) the PEF% predicted (n = 20; IL-3 n = 20 and PEF% n = 23) at the baseline visit. Data are presented as means ± SEMs. Two-tailed Student’s t test was used to calculate statistical significance. ∗p ≤ 0.05; ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. For the Correlations, the Pearson coefficient was used after data normalization.
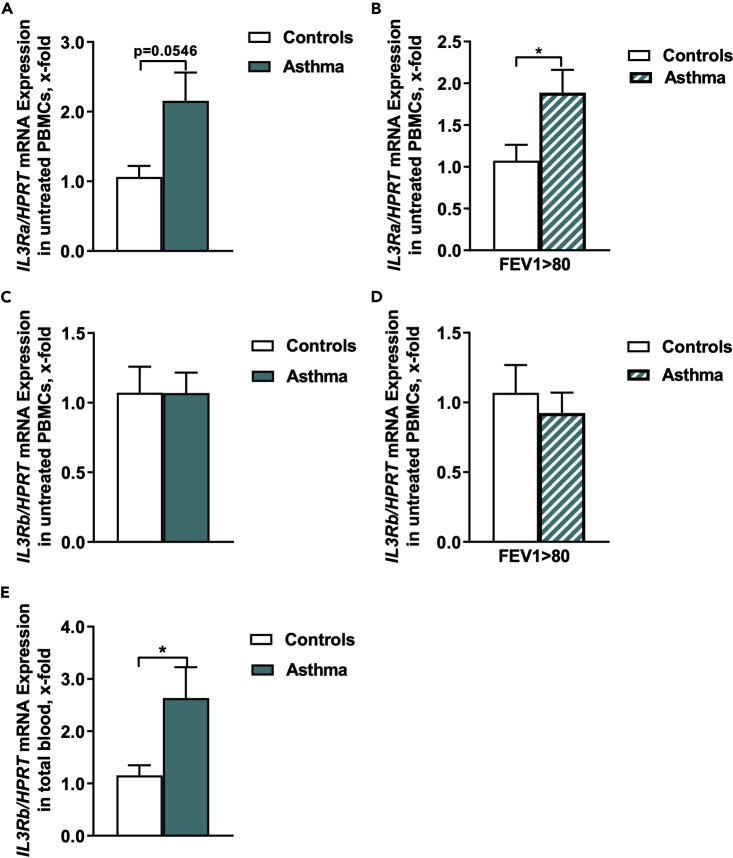
IL3Ra expression in PBMCs is associated with the resolution of asthma in children
We next analyzed the mRNA expression of both IL-3 receptor (IL3R) chains in untreated PBMCs and found an increased IL3Ra/HPRT mRNA expression in asthmatic children compared to healthy children (Figure 2A). Regarding the IL3Ra/HPRT mRNA expression and the FEV1%, we found a statistically significant increased gene expression in asthmatic children with an FEV1% higher than 80 (Figure 2B). We next looked at the IL3Ra/GAPDH mRNA expression and the FEV1%, predicted in total blood cells and found it upregulated by trend in asthmatic children (Figure S1B).
Figure 2.
IL-3Ra induction is associated with ameliorated asthma in children
(A) IL3Ra/HPRT mRNA expression in PBMCs of healthy control and asthmatic children at the baseline visit (n = 7/11).
(B) IL3Ra/HPRT mRNA expression in PBMCs of healthy control and asthmatic children at the baseline visit with a FEV1%, predicted above 80 (n = 6/9).
(C) IL3Rb/HPRT mRNA expression in PBMCs of healthy control and asthmatic children at the baseline visit (n = 7/11).
(D) IL3Rb/HPRT mRNA expression in PBMCs of healthy control and asthmatic children at the baseline visit with a FEV1%, predicted above 80 (n = 6/9).
(E) IL3Rb/HPRT mRNA expression in total blood cells of healthy control and asthmatic children at the baseline visit (n = 12/16). Data are presented as means ± SEMs. Two-tailed Student’s t test was used to calculate statistical significance. ∗p ≤ 0.05; ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
IL3Rb expression in total blood is associated with asthma in children
We next looked at IL3R common (IL-5, GM-CSF) beta chain expression in PBMCs of our cohort but no difference between controls and asthmatics was observed (Figures 2C and 2D). We therefore reasoned that IL3R beta chain might have been regulated in other blood cells and thus further analyzed the IL3R gene expression in total blood samples and this time we found an upregulation of the IL3Rb/HPRT (Figure 2E) but similar IL3Rb/GAPDH mRNA expression in asthmatic children compared to healthy children (Figure S1C). Taken together, we reasoned that the IL3Rα is expressed and regulated in PBMCs and the IL3Rβ chain is probably regulated in blood cells other than PBMCs in children with asthma. Further studies in bigger cohorts of patients should be performed to confirm these findings.
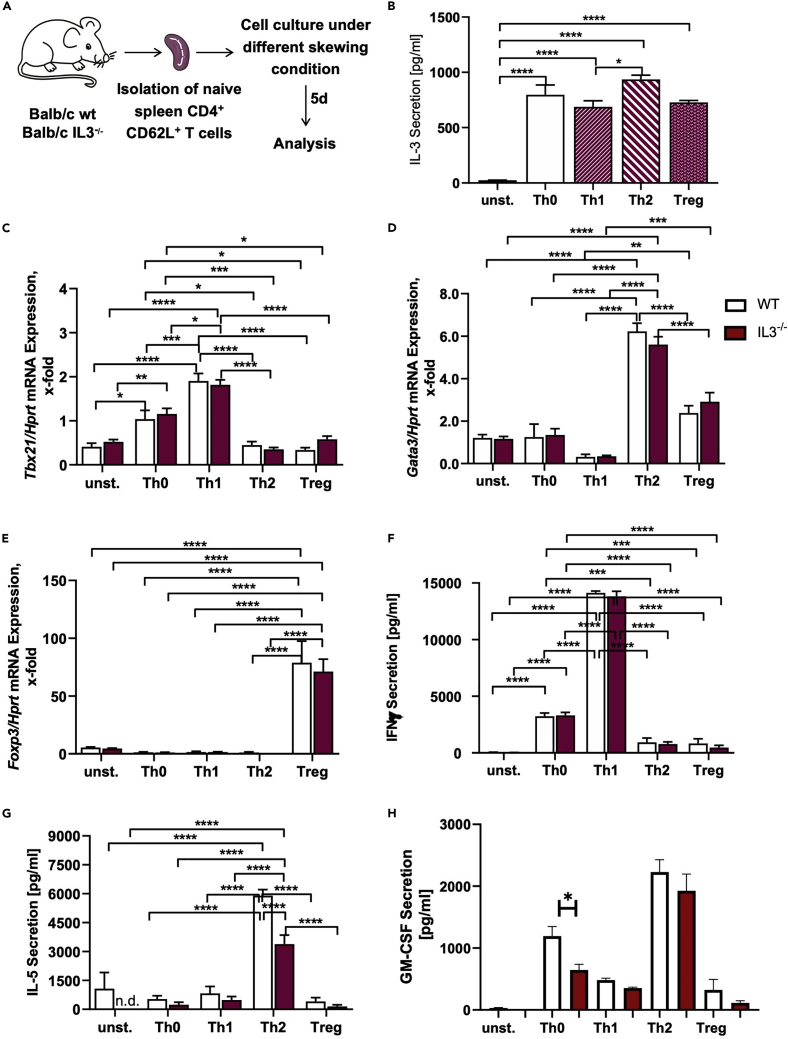
Decreased IL-5 secretion by IL-3-deficient TH2 cells
As mentioned before, IL-3 is mainly produced by activated T cells. Thus, we analyzed if a deficiency in IL-3 has any influence on the development of different T cell subpopulations (Figures 3A and S2). However, we could not observe any differences in the development of TH1, TH2, as well as Treg cells from the spleen between WT and IL-3−/− mice (Figures S3A and S3B). We then analyzed the IL-3 secretion under these different T cell subtypes skewing conditions and found that TH2 cells exhibited the highest IL-3 secretion compared to TH1 and Treg cells (Figure 3B). To analyze the phenotype of these differentiated cells in more detail, we performed different gene expression and protein analysis and found that the deficiency of IL-3 has no influence on the mRNA expression of Tbx21 (T-box transcription factor 21), GATA3 (GATA-binding protein 3), and Foxp3 (Forkhead-box protein 3) (Figures 3C–3E, respectively). Furthermore, we could not observe any differences in the expression of interferon (IFN) γ, which is predominately secreted by TH1 cells (Figure 3F). Instead of this, we found a markedly inhibition of the IL-5 secretion in IL-3-deficient TH2 cells (Figure 3G). Also, GM-CSF was found downregulated in the absence of IL-3 in TH0 cells isolated from IL-3−/− naive mice (Figure 3H). Furthermore, we could not observe any differences in the expression of the IL3Rα chain on these cells (Figure S3C).
Figure 3.
In vitro differentiation of TH1, TH2, and Treg cells in the presence and absence of IL-3
(A) Experimental design for the in vitro differentiation of T cell subpopulations.
(B) ELISA analysis of the IL-3 level in the cell culture supernatant of in vitro differentiated T cell subtypes and unstimulated spleen cells of WT mice (n = 2 (unstimulated), n = 4 (TH0, TH1, TH2, and Treg).
(C–E) Tbx21/Hprt, Gata3/Hprt, and Foxp3/Hprt mRNA expression in in vitro differentiated T cell subtypes as well as naive spleen cells of WT and IL-3−/− mice (n = 3/4 (naive, TH0), n = 4/3 (TH1), n = 4/4 (TH2, Treg).
(F–H) ELISA analysis of the IFNγ, IL-5, and GM-CSF level in the cell culture supernatant of in vitro differentiated T cell subtypes and unstimulated spleen cells of WT and IL-3−/− mice (n = 2/2 (unstimulated), n = 4/4 (TH0), n = 4/3 (TH1), n = 4/4 (TH2, Treg). Data are presented as means ± SEMs. Two-tailed Student’s t test, two-way ANOVA or ordinary One-way ANOVA was used to calculate statistical significance. ∗p ≤ 0.05; ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
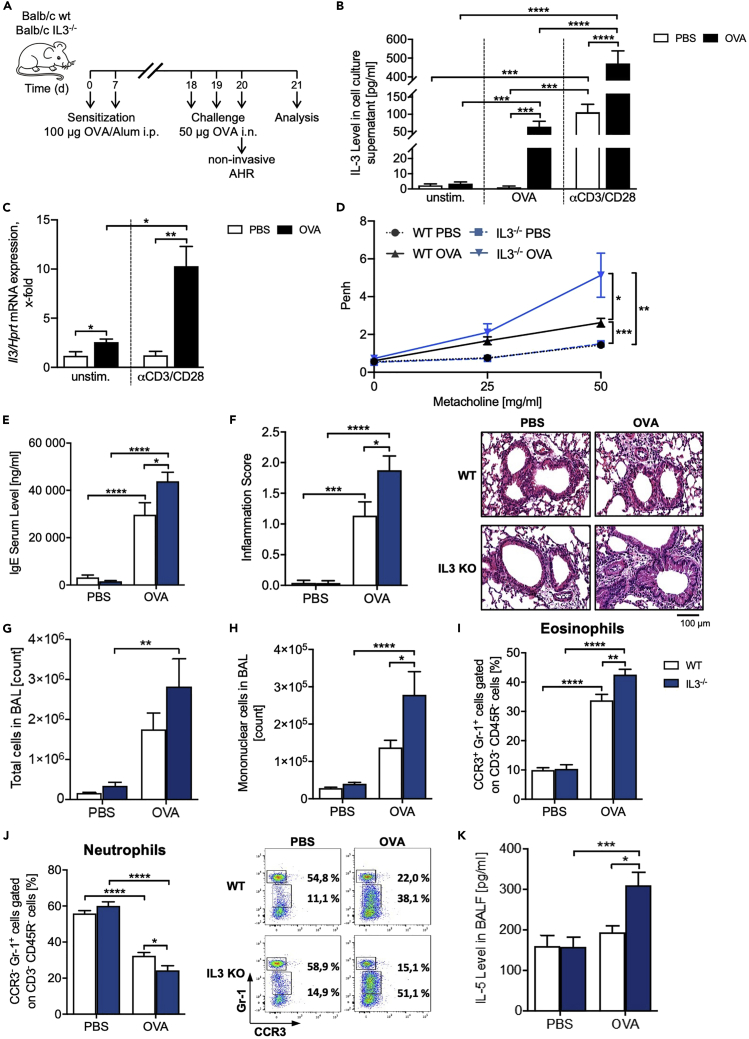
IL-3-deficient mice have increased asthmatic traits
As we demonstrated that IL-3 as well as its two receptor chains is differentially regulated in allergic asthma, we started to investigate the role of IL-3 in a murine model of allergic asthma (Figure 4A). First, we found an upregulation of the IL-3 secretion, as well as Il3/Hprt mRNA expression in OVA and especially αCD3/αCD28 antibodies-treated total lung cells of asthmatic WT mice compared to untreated littermates (Figures 4B and 4C), indicating that the source of IL-3 in the lung of asthmatic mice is activated T cells. In the presence of immunologic sensitization and challenge with OVA, IL-3−/− mice showed significantly increased response to methacholine compared to WT mice, as measured by using non-invasive whole body plethysmography (Figure 4D). We further observed an increased IgE serum level (Figure 4E). The increased AHR was found accompanied with an increased lung inflammation characterized by increased peribronchial and perivascular induction of inflammatory cells (Figure 4F). Moreover, we observed increased numbers of total cells (Figure 4G) as well as mononuclear cells (Figure 4H) in the BALF of IL-3 −/− mice as compared to the WT littermates. In addition, we observed increased amounts of lung eosinophils and decreased amounts of neutrophils in the absence of IL-3 in total lung cells from these mice (Figures 4I and 4J), whereas the analysis of the BALF cells did not reveal differences in eosinophils and neutrophils between WT and IL-3−/− mice (Figures S4A and S4B). Moreover, there were no differences in the lung mucus production after Periodic acid-Schiff (PAS) staining between asthmatic OVA-treated WT and IL-3−/− mice (Figure S4C). Finally, we detected an increase of lymphocytes (Figure S4D) as well as IL-5 secretion in the BALF (Figure 4K) of asthmatics OVA-treated IL-3−/− mice compared to the WT asthmatic littermates.
Figure 4.
IL-3-deficient mice have increased asthmatic trait
(A) Experimental design for the induction of allergic asthma.
(B) ELISA analysis of the IL-3 level in the supernatant obtained from total lung cells (n = 13/12 (unst., OVA), n = 14/13 (αCD3/CD28)).
(C) Il3/Hprt mRNA expression in total lung cells of BALB/c WT mice with and without asthma (n = 4/4 (unst.), n = 4/5 (αCD3/CD28)).
(D) Airway resistance to increasing doses of methacholine measured by non-invasive plethysmography. Airway resistance was analyzed as Penh (n = 16/16/17/18).
(E) IgE serum level of mice with and without asthma measured by ELISA (n = 13/13/13/12).
(F) Pathological score of the lung inflammation of mice with and without asthma (n = 12/13/11/12). A representative light microscopic picture of the H&E staining of lung sections is shown for each group.
(G and H) Total cell count in the BAL of WT and IL3−/− mice with and without asthma (n = 11/11/11/13), and (h) cell count of mononuclear cells in the BALF of mice with and without asthma (n = 14/14/12/13).
(I, and J) Flow cytometry analysis of CCR3+ Gr-1+ eosinophils (i) and CCR3- Gr-1+ neutrophils (j) in total lung cells (n = 9/9/9/10). A representative dot plot is shown for each group.
(K) ELISA analysis of the IL-5 level in the BALF (n = 7/10/9/11). Data are presented as means ± SEMs. Two-tailed Student’s t test or two-way ANOVA was used to calculate statistical significance. ∗p ≤ 0.05; ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
The lack of IL-3 is accompanied by decreased Foxp-3 T regulatory cells in the lung
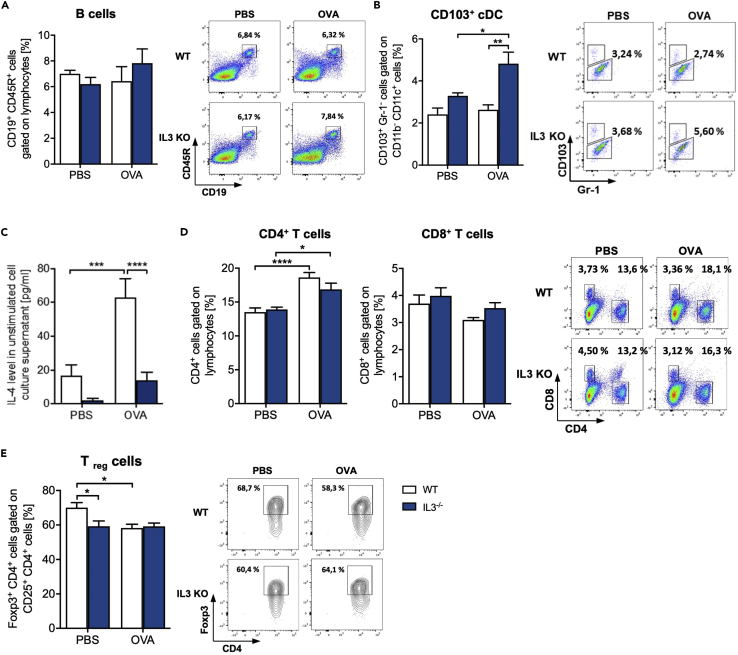
Because targeted deletion of IL-3 resulted in TH2 cytokine hyperproduction, we wanted to understand if the number of T and B cells was changed in the absence of IL-3 in the lung. The B cell number was not found differently regulated in WT and IL-3−/− mice (Figure 5A). Because antigen-presenting cells play an important role in the immune-pathogenesis of allergic asthma inducing cytokines that activate T cells, we next analyzed this heterogenic cell population and found increased amounts of CD103+ conventional dendritic cells (cDC) and decreased amounts of CD103- DCs in OVA-treated IL-3−/− mice when compared to their WT littermates (Figures 5B, S4E and S4F). Although it is known that IL-3 is an important growth factor for mast cells and basophils, the deficiency of IL-3 has no influence on the in vivo development of these cells under physiological conditions as well as after OVA treatment, whereby the amount of both cell populations is increased in asthmatic mice (Figures S5A and S5B, respectively). In addition, we found a defect of the basophil-produced cytokine IL-4 (Figure 5C). Finally, CD4+ and CD8+ T cells were not differently regulated between the two genotypes (Figure 5D). However, the CD4+ T cells were induced in asthma in both genotypes (Figure 5D). Moreover, we detected an inhibition of Treg cell number in naive IL-3-deficient mice (Figure 5E). Although the number of Treg was downregulated in the wild-type asthmatic mice as compared to naive wild-type mice, the Treg level in IL-3-deficient asthmatic mice was not changed (Figure 5E). In conclusion, IL-3 inhibited the development of T regulatory cells in the lung of naive mice.
Figure 5.
Analysis of T and B cells in total lung cells of OVA-treated IL-3−/− mice
(A andB) Flow cytometry analysis of CD19+ CD45R+ B cells (n = 5) (a) and CD103+ cDCs (n = 4/4/5/5) (b) cells in total lung cells isolated from BALB/c WT and IL3−/− mice with and without asthma.
(C) ELISA analysis of the IL-4 level in cell culture supernatants obtained from total lung cells of mice with and without asthma (n = 14/14/15/15).
(D and E) CD4+ and CD8+ T cells (d) and Foxp3+ CD25+ CD4+ Treg (e) (n = 10 (CD4+ cells), n = 5 (CD8+, Treg)). A representative dot plot is shown for each group. Data are presented as means ± SEMs. two-way ANOVA was used to calculate statistical significance. ∗p ≤ 0.05; ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
Increased IL-5, IL-13, and GM CSF in the lung of IL-3-deficient asthmatic mice
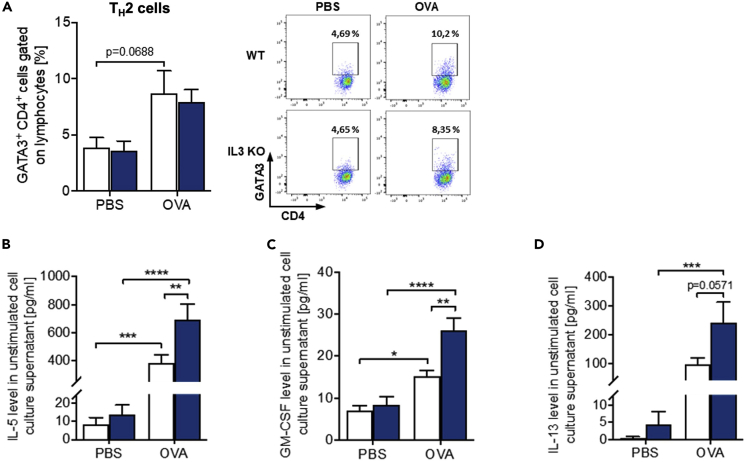
We next asked about TH2 induction in the lung in the absence of IL-3. Asthmatic mice showed an upregulation of GATA3+ TH2 cell number (Figure 6A). Consistent with the IL-5 upregulation in the BALF of IL-3-deficient mice, and with a compensatory role of IL-5 and GM-CSF on IL-3, we found an increased secretion of IL-5, GM-CSF, and IL-13 in total lung cells isolated from IL-3−/− mice compared to WT mice in asthma (Figures 6B–6D). Furthermore, OVA-treated IL-3−/− mice showed increased IL-10 levels (Figure S5C). Altogether, these data suggest a novel role of IL-3 in the inhibition of allergic asthma exacerbations.
Figure 6.
Increased TH2 cytokines in the lung of IL-3−/− asthmatic mice
(A) Flow cytometry analysis of GATA3+ CD4+ TH2 cells and in cell culture supernatants obtained from total lung cells of mice with and without asthma (n = 5). A representative dotplot is shown for each group.
(B–D) ELISA analysis of the IL-5, GM-CSF, and IL-13 level in cell culture supernatants obtained from total lung cells of mice with and without asthma (n = 14/14/15/15 (IL-5 and IL-13); n = 13/14/14/14 (GM-CSF)). Data are presented as means ± SEMs. two-way ANOVA was used to calculate statistical significance. ∗p ≤ 0.05; ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
Discussion
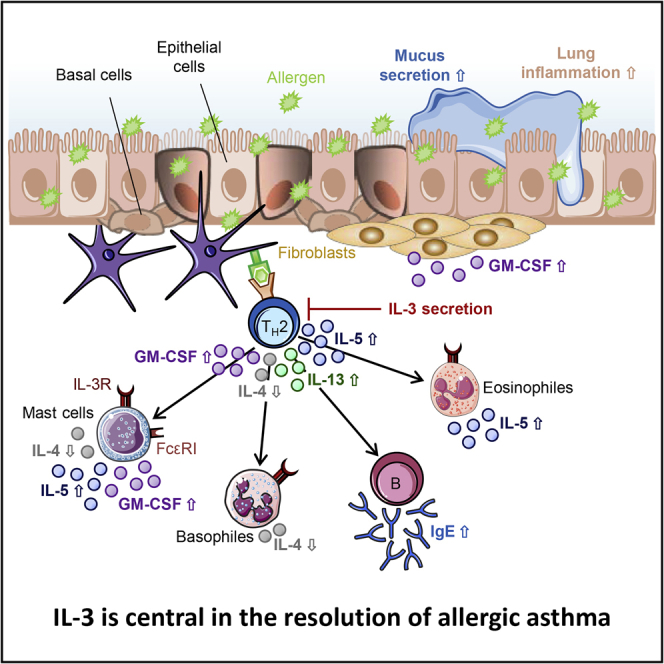
The cytokine IL-3 stimulates especially the growth and differentiation of mast cells and basophils, as well as the release of mediators of these cells, such as histamine, IL-4, and IL-6. Thus, it is assumed that IL-3 plays also an important role in allergic diseases (Auclair et al., 2014; Lantz et al., 1998; Voehringer, 2012) as well as in allergic asthma (Bertrand et al., 2015; Park et al., 2004). Hence, in the present study, we determined the role of IL-3 in allergic asthma in more detail.
In our human study, we describe an increased secretion of IL-3 in PHA-stimulated PBMCs of asthmatic children, whereby the amount of IL-3 correlates directly with better lung function in asthmatic children and thus with the resolution of the disease. Although we are only at the beginning to understand why patients with asthma in general have higher levels of IL-3 than normal individuals, we recently published a manuscript showing that among the pre-school asthmatic children analyzed, only those treated with steroids and with controlled and partially controlled asthma had higher IL-3 (Krammer et al., 2022). These data are extended in the current manuscript considering the lung function. Taken together, the increased IL-3 in asthma could be associated with asthmatic children treated with steroids and thus with better lung function. Further analysis in bigger cohorts will be performed in the future to further prove these findings. This observation is in line with different previous human studies (Park et al., 2004; Bertrand et al., 2015; Robinson et al., 1992; Lai et al., 1996). We could further show that the expression of the IL3Ra is associated with the resolution of asthma in these children. A similar trend could be observed by previous studies, in which it could be shown that an increased IL-3 concentration leads to an increased IL3Ra expression (Gregory et al., 2003; Esnault et al., 2015). In summary, these data indicate that IL-3 could be important in allergic asthma.
In support of this hypothesis, in a murine model of allergic asthma, we found that IL-3−/− mice have increased asthmatic traits, such as AHR, lung inflammation, local eosinophil infiltration and IgE, as well as IL-5, GM-CSF, and IL-13 secretion. Considering that IL-5 and GM-CSF use the same beta chain of the receptor as IL-3, these data indicated a compensatory mechanism mediated by IL-5 in the absence of IL-3 in asthma but also an inhibitory function of IL-3 on IL-5 and GM-CSF which was found to be associated with the asthmatic condition. IL-5 and GM-CSF are responsible for the recruitment, maturation, and survival of eosinophils. These data are consistent with the upregulation of the IL-3Rβ chain in the blood and not in the PBMCs of the asthmatic children analyzed.
While IL-5 contributes crucially to eosinophilia and AHR in allergic asthma (Mauser et al., 1995; Foster et al., 1996; Shi et al., 1998), earlier studies showed an increased GM-CSF concentration in patients with acute severe asthma (Brown et al., 1991; Davies et al., 1997). Various human studies have already shown that the expression of the specific α-subunit of the common beta cytokines IL-3, IL-5, and GM-CSF are regulated differently by themselves with an induction of the IL3Rα expression by IL-3 (Wang et al., 1998; Liu et al., 2002; Gregory et al., 2003; Kumar et al., 2020; Esnault et al., 2015). Since this has not yet been investigated in a murine model, we assume that the expression of both receptor chains in mice is also regulated by IL-3 itself (Brown et al., 1991; Davies et al., 1997). Contrary to this, we found that the secretion of IL-4 is strongly inhibited in the absence of IL-3, which is in line with a study from Rignault-Bricard et al. (2018). The secretion of IL-4 takes place mainly by basophils, whereby it is mainly induced via the signaling pathway of IL-3 (Sullivan et al., 2011; Broughton et al., 2012). These results along with the observation of increased serum IgE in the absence of IL-3 might indicate a compensatory effect of IL-13 on IgE production in the presence of low IL-4. Another potential alternate sources of IL-3 are mast cells which are also dependent from IL-3 and therefore are possibly missing in the IL-3-deficient mice and thus this could be a potential confounding issue in the data interpretation (Itakura et al., 2001).
Further experiments in these directions are needed. To look further the mechanism of induction of AHR and inflammation in the absence of IL-3, we took a look at T regulatory cells. These immunosuppressive cells were found downregulated in the absence of IL-3, indicating an immunoregulatory role of IL-3 in the lung. This T regulatory cell defect might be responsible of the induced inflammation and airway hyperresponsibility in the IL-3-deficient asthmatic mice. However, this hypothesis needs further experiments in which mice are reconstituted with T regulatory cells. In summary, our data from the pediatric cohort support a role of IL-3 in the amelioration of allergic asthma as we found increased IL-3 production in activated peripheral lymphocytes associated with better lung function in asthmatic children. In addition, IL-3-deficient mice have increased airway hyperresponsiveness and inflammation as compared to wild-type littermates. Based on these observations, we hypothesized that increased IL-3 might be associated with amelioration of asthma. At the beginning, there is a homeostatic state; the lung is dominated by a tolerogenic environment with T regulatory cells. After allergen sensitization and challenge, one of the first cytokine released by the TH0, TR1 activated T cells is IL-3. We hypothesized that in this milieu the TH2 cells are not yet developed and thus IL-3 probably is released by TH1-TH0 cells, as it was previously reported by Mosmann T et al. (Mosmann et al., 1986). Thus our data unreveal a not yet appreciated role of IL-3 in the resolution of asthma that needs further investigation.
Limitations of the study
One caveat of the human study is to evaluate lung function in small children (4–6 year old). The lung function parameters can be measured, but the doctor needs also information from the parents, auscultation of the lungs, and other clinical parameters as the level of therapy or the degree of asthma control. Most of children in this age group with “asthma” show parameters which indicate obstruction of the upper and the smaller airways.
The second caveat for the human studies is the limitation of the samples size obtained from small children. The use of a single housekeeping gene for human qPCR studies must be carefully interpreted as a potential weakness.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| V450 Hamster anti-Mouse CD3e | BD Bioscience, Heidelberg, Germany | Cat# 560801, RRID: AB_2034005 |
| BV421 Rat Anti-Mouse CD4 | BD Bioscience, Heidelberg, Germany | Cat# 740007, RRID: AB_2739779 |
| BV510 Rat Anti-Mouse CD8a | BD Bioscience, Heidelberg, Germany | Cat# 563068, RRID: AB_2687548 |
| V450 Rat anti-CD11b | BD Bioscience, Heidelberg, Germany | Cat# 560455, RRID: AB_1645266 |
| APC/Cyanine7 anti-mouse CD11c Antibody | BioLegend, San Diego, CA, USA | Cat#117323, RRID: AB_830646 |
| CD19 Monoclonal Antibody, anti-human/mouse, APC | eBioscience™, Thermo Fisher Scientific, Waltham, USA | Cat#17-0193-82, RRID: AB_1659676 |
| APC-Cy™7 Rat Anti-Mouse CD25 | BD Bioscience, Heidelberg, Germany | Cat# 557658, RRID: AB_396773 |
| PerCP-Cy™5.5 Rat Anti-Mouse CD25 | BD Bioscience, Heidelberg, Germany | Cat# 561112, RRID: AB_394031 |
| CD45R (B220) Antibody, anti-mouse, FITC, REAfinity™ | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany | Cat# 130-110-845, RRID: AB_2658274 |
| FITC Rat Anti-Mouse CD62L | BD Bioscience, Heidelberg, Germany | Cat# 561917, RRID: AB_10893197 |
| CD103 Antibody, anti-mouse, FITC | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany | Cat# 130-118-681, RRID: AB_2751541 |
| CD117 (c-Kit) Monoclonal Antibody, anti-human/mouse, APC | eBioscience™, Thermo Fisher Scientific, Waltham, USA | Cat# 17-1171-82, RRID: AB_469430 |
| CD123 Antibody, anti-mouse, PE, REAfinity™ | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany | Cat# 130-102-583, RRID: AB_2654775 |
| CD123 Antibody, anti-mouse, PE-Vio®770, REAfinity™ | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany | Cat# 130-102-323, RRID: AB_2654775 |
| Alexa Fluor® 647 Rat Anti-Mouse CD193 | BD Bioscience, Heidelberg, Germany | Cat# 557974, RRID: AB_396967 |
| FceR1 alpha Monoclonal Antibody, anti-human, FITC | eBioscience™, Thermo Fisher Scientific, Waltham, USA | Cat# 11-5899-42, RRID: AB_10732835 |
| FoxP3 Antibody, anti-human/mouse, APC | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany | Cat# 130-113-470, RRID: AB_2733420 |
| Alexa Fluor® 488 Mouse anti-GATA3 | BD Bioscience, Heidelberg, Germany | Cat# 560077, RRID: AB_1645303 |
| PE Rat Anti-Mouse Ly-6G and Ly-6C | BD Bioscience, Heidelberg, Germany | Cat# 553128, RRID: AB_394644 |
| PE-CF594 Mouse Anti-T-bet | BD Bioscience, Heidelberg, Germany | Cat# 562467, RRID: AB_2737621 |
| Purified NA/LE Hamster Anti-Mouse CD3e | BD Bioscience, Heidelberg, Germany | Cat# 553057, RRID: AB_394590 |
| Purified anti-mouse CD28 Antibody | BioLegend, San Diego, CA, USA | Cat# 102101, RRID: AB_312866 |
| Purified anti-mouse IL-4 Antibody | BioLegend, San Diego, CA, USA | Cat# 504101, RRID: AB_315315 |
| IFN-γ Antibody, anti-mouse, pure-functional grade | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany | Cat# 130-095-729, RRID: AB_2784369 |
| Chemicals, peptides, and recombinant proteins | ||
| Human TGF-β1 | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany | Cat# 130-095-067 |
| Mouse rIL-2 | ImmunoTools, Friesoythe, Germany | Cat# 12340026 |
| Mouse rIL-4 | ImmunoTools, Friesoythe, Germany | Cat# 12340045 |
| Mouse rIL-12 | ImmunoTools, Friesoythe, Germany | N/A |
| Ovalbumin (Albumin chicken egg, 5x crystalline) | Calbiochem, Merck KGaA, Darmstadt, Germany | N/A |
| Phytohemagglutinin-M (PHA-M) | Sigma-Aldrich Chemie GmbH, München, Germany | Cat# 11082132001 |
| QIAzol Lysis Reagent | Qiagen, Hilden, Germany | Cat# 79306 |
| SsoFast EvaGreen® Supermix | Bio-Rad Laboratories GmbH, Feldkirchen, Germany | Cat# 1725200 |
| PeqGold RNAPure™ | PeqLab Biotechnologie, VWR International GmbH, Darmstadt | Cat# 30-1010 |
| Critical commercial assays | ||
| Human IL-3 DuoSet ELISA | R&D Systems GmbH, Wiesbaden, Germany | Cat# DY203 |
| Mouse IL-13 DuoSet ELISA | R&D Systems GmbH, Wiesbaden, Germany | Cat# DY413 |
| Mouse GM-CSF DuoSet ELISA | R&D Systems GmbH, Wiesbaden, Germany | Cat# DY415 |
| Mouse IL-3 OptEIA™ ELISA | BD Bioscience, Heidelberg, Germany | Cat# 555228; RRID: AB_2869046 |
| Mouse IL-4 OptEIA™ ELISA | BD Bioscience, Heidelberg, Germany | Cat# 555232; RRID: AB_2869047 |
| Mouse IL-5 OptEIA™ ELISA | BD Bioscience, Heidelberg, Germany | Cat# 555236; RRID: AB_2869048 |
| Mouse-IL-10 OptEIA™ ELISA | BD Bioscience, Heidelberg, Germany | Cat# 555252; RRID: AB_2869052 |
| Mouse IFNγ OptEIA™ ELISA | BD Bioscience, Heidelberg, Germany | Cat# 555138; RRID: AB_2869028 |
| Mouse IgE OptEIA™ ELISA | BD Bioscience, Heidelberg, Germany | Cat# 555248; RRID: AB_2869051 |
| CD4+ CD62L+ T Cell Isolation Kit, mouse | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany | Cat# 130-106-643 |
| MagMAX™ for Stabilized Blood Tubes RNA Isolation Kit | Invitrogen, Thermo Fisher Scientific, Waltham, USA | Cat# 4451893 |
| Foxp3 / Transcription Factor Fixation / Permeabilization Concentrate and Diluent | eBioscience™, Thermo Fisher Scientific, Waltham, USA | Cat# 00-5521-00 |
| Permeabilization Buffer (10X) | eBioscience™, Thermo Fisher Scientific, Waltham, USA | Cat# 00-8333-56 |
| RevertAid RT Reverse Transcription Kit | Invitrogen, Thermo Fisher Scientific, Waltham, USA | Cat # K1691 |
| Experimental models: Organisms/Strains | ||
| BALB/cAnNRj (WT) mice | Janvier Labs, Saint-Berthevin, France | N/A |
| Balb/c-Il3tm1Glli (IL-3 KO) mice | RIKEN BRC Laboratories, Japan | RBRC02298 |
| Oligonucleotides | ||
| hCD131 (5‘-CCA CGG CCA ATA CAT CGT CT-3‘, 5‘-GGG CCA TCT GGA TGT TCA CT-3‘) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| hHPRT (5‘-TGA CAC TGG CAA AAC AAT G CA-3‘, 5‘-GGT CCT TTT CAC CAG CAA GCT-3‘) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| hGAPDH (5‘-AAA TCA AGT GGG GCG ATG CT, 5‘ CAA ATG AGC CCC AGC CTT CT) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| mFoxp3 (5‘-AGA GCC CTC ACA ACC AGC TA-3‘, 5‘-CCA GAT GTT GTG GGT GAG TG-3‘) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| mGata3 (5‘-GTC ATC CCT GAG CCA CAT CT-3‘, 5‘-TAG AAG GGG TCG GAG GAA CT-3‘) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| mHprt (5‘-GCC CCA AAA TGG TTA AGG TT-3‘, 5‘-TTG CGC TCA TCT TAG GCT TT-3‘) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| mIl3 (5‘-GAA GCT CCC AGA ACC TGA A CT-3‘, 5‘-TCT CCT TGG CTT TCC ACG AAT-3‘) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| mTbx21 (5‘-CCT GGA CCC AAC TGT CAA CT-3‘, 5‘-AAC TGT GTT CCC GAG GTG TC-3‘) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| hCD123 (5'-ACT CCA CCC AAC ATGACT GC-3′, 5'-GGTTCTGTCTCTGACCTGTTCT-3′) |
Eurofins Genomics, Ebersberg, Germany | Custom |
| Software and algorithms | ||
| GraphPad Prism 7 | GraphPad Software Inc., San Diego, USA | https://www.graphpad.com/ |
| CaseViewer 2.0 | 3D Histech Ltd., Budapest, Hungary | https://www.3dhistech.com/solutions/caseviewer/ |
| FlowJo 7.6.1, V10 | Treestar Inc., SanJose, USA | https://www.flowjo.com/ |
| FACS Diva | BD Biosciences, Heidelberg, Germany | https://www.bdbiosciences.com/en-eu/products/software/instrument-software/bd-facsdiva-software |
| FinePoint, V2.1.0.9 | Data Sciences International (DSI), St. Paul, USA | https://www.datasci.com/products/software/finepointe-software |
| Revelation Quicklink | Dynex Technologies GmbH, Denkendorf | N/A |
Resource availability
Lead contact
Further information and requests for resources, methods and reagents should be directed to and will be fulfilled by the lead contact, Prof. Dr. Dr. Susetta Finotto (susetta.finotto@uk-erlangen.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
All mice used in this study are on a Balb/c genetic background. The Balb/c wildtype (WT) mice were obtained from Janvier Labs (Saint-Berthevin, France). The IL-3 deficient mice (Balb/c-Il3tm1Glli, IL-3−/−, IL-3 KO) were a generous gift from PD Dr. Georg Weber (Department of Surgery, University Hospital Erlangen) and originally purchased from RIKEN BRC Laboratories, Japan. The experiments were performed with both, female and male mice. All mice were maintained under specific pathogen free conditions and had free access to food and water. All experiments were performed in accordance with the German and European laws for animal protection and were approved by the government of Unterfranken, Bavaria (Az. 54-2532.1-2/10).
Human study
In the European study PreDicta (Post-infectious immune reprogramming and its association with persistence and chronicity of respiratory allergic diseases) we examined two cohorts of children with (n = 24) and without asthma (n = 21) at the age of 4–6 years. In general, boys are consistently reported to have more prevalent wheeze and asthma than girls, although this pattern changes in the adolescence. In this study the cohorts of children recruited in Erlangen was composed of 38.1% females and 61.9% males for the control group and 37,5% females and 62,5% male for the asthma group. The study was performed in collaboration with the children hospital in Erlangen and approved by the ethics committee of the Friedrich-Alexander University Erlangen-Nürnberg, Germany (Re-No 4435). The study is registered in the German Clinical Trials Register (www.germanctr.de: DRKS00004914). One aim of PreDicta is to identify altered host-pathogen interactions, molecules and pathways that mediate the establishment and persistence of chronic inflammation in allergic diseases, thus to develop new preventive, diagnostic and therapeutic strategies. Therefore, we and the other study centres established and followed up a cohort of preschool children with and without asthma for two years. At the time of recruitment, whole blood was drawn from healthy and asthmatic children for the isolation of PBMCs.
One inclusion criteria for cases and controls was the need of a written informed consent from the child's parents or guardians. The gestational age had to be 36 weeks or above. Another criterion for inclusion for cases was a diagnosis of asthma within the last two years, confirmed by a doctor of the Children's Hospital in Erlangen. The kind of asthma should be mild to moderate persistent severity according to the GINA guidelines (2005). There must be three episodes in the preceding 12 months and one of them in the last six months. In addition, the child had to be able to perform at least one Peak Expiratory flow (PEF) manoeuvre. The controls should not have a history of asthma or wheezing and atopic illness. Furthermore, the legal custodian must have the verbal, writing and mental ability to understand the intent and character of the study. Severe or brittle asthma was an exclusion criterion for the cases of this study. Furthermore, the child should not receive an immunotherapy. Children with more than six courses of oral steroids during the previous 12 months were also excluded. Children with other chronic respiratory diseases (cystic fibrosis, bronchopulmonary dysplasia, immunodeficiencies) except allergic rhinitis were not be included in the study as well as children with other chronic diseases or chronic medication use except atopic eczema. Atopy was proven by at least one positive skin prick test, while asthma was defined in accordance to a physician’s diagnosis of mucus production, bronchial hyperesponsiveness and dyspnoea. The healthy control subjects did not have a history of atopy or asthma. Every single participant of the study was assigned to a specific number. Only the clinical investigators and study nurses of the Children's Hospital had access to the full name.
The recruitment of the two cohorts of preschool children, the timescale for the clinical visits, the inclusion and exclusion criteria as well as the data collection were described recently (Bergauer et al., 2017; Bielor et al., 2017; Graser et al., 2016; Hentschke et al., 2017; Haag et al., 2018; Übel et al., 2014) and are reported in other form in Tables S1 and S2 along with the relevant clinical aspects and characteristics.
At the Baseline visit the FEV1 (Forced expiratory volume in 1 s) of all children was measured by using spirometry. Further, blood was collected for the isolation and culture of peripheral blood mononuclear cells (PBMCs) and subsequent analysis, as well as for gene expression analysis of total blood cells.
Method details
Isolation of human peripheral blood mononuclear cells (PBMCs) and cell culture
Heparinized blood was collected from children with and without asthma at the Baseline Visit of the PreDicta study and subsequently PBMCs were isolated with Ficoll using density centrifugation. Afterwards one part of the PBMCs were used for extraction of total mRNA by using QIAzol Lysis Reagent (Qiagen, Hilden, Germany), according to the manufacturer’s protocol, and subsequently gene expression analysis as described below. In the Key resources table the primer sequences are reported. The remaining cells were cultured in a concentration of 1 x 106 cells/ml for 48 h in RPMI 1640 medium supplemented with 25 mmol/L HEPES, 100 IU/mL penicillin, 100 μg/mL streptomycin, 1% L-glutamine (200 mmol/L) (all anprotec, Bruckberg, Germany), 50 μmol/L β-mercaptoethanol, 1% MEM Vitamin (all Sigma-Aldrich, Steinheim, Germany), 1% non-essential amino acids, 1% sodium pyruvate (all Gibco®, Thermo Fisher Scientific, Waltham, USA), and 10% FBS (Biochrom GmbH, Berlin, Germany) at 37°C and 5% CO2 and stimulated with 10 μg/mL Phytoheamagluttinin (PHA) (Sigma-Aldrich, Steinheim, Germany).
Human IL-3 was detected in the cell-culture supernatants by using IL-3 DuoSet ELISA kit from R&D (Wiesbaden, Germany) according to the manufacturer’s protocol.
Spirometry
Lung function measurements were done with a whole body plethysmograph from Jaeger Comp. Würzburg. The results were calculated and listed with a computer system. Spirometry was carried out according to the ATS/ERS criteria. Measurement that did not met the quality criteria were excluded. At the Baseline visit (B0) the FEV1 (Forced expiratory volume in 1 s), FVC (Forced vital capacity) and PEF (Peak expiratory flow) of all children were measured by using spirometry. For the measurement of the FEV1 the participant should inhale maximal, immediately followed by maximal and fast exhalation. Thereby the volume exhaled in 1 s is defined as the FEV1, whereas the total exhaled volume is indicated as the FVC. The FEV1%, predicted is the best validated value for asthma also in children and defined as FEV1% of the patient divided by the average FEV1% in the population for any person of similar age, gender, and body composition. The PEF is defined as the largest expiratory flow, which is achieved with a maximum forced effort after maximum inspiration. The specified PEF% and FVC% can be calculated from the measured PEF and FVC values, respectively, and the corresponding age and gender-dependent target values (Criée et al., 2015). FVC is not an important value for asthma as it prescribes pulmonary restriction and not obstruction. The PEF it is not as robust as the FEV1%. The spirometry data of all children are listed in Table S3.
Human RNA isolation and quantitative real-time PCR
For gene expression analysis total blood was collected from the children at the baseline visit in Tempus® Blood RNA Tubes (Life Technologies™, GmbH, Darmstadt, Germany) and total mRNA was extracted with the MagMax for Stabilized Blood Tubes RNA Isolation Kit, according to the manufacturer’s protocol. For reverse transcription of RNA (1 μg) from total blood cells and PBMCs we used the RevertAid RT kit (ThermoFisher Scientific, Waltham, USA) in accordance to the manufacturer’s protocol. The resulting template cDNA was amplified by quantitative real-time PCR (qPCR) using SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Feldkirchen, Germany). The qPCR itself was performed with a cycle of 2 min 98°C, 50 cycles at 5 s 95°C, 10 s 60°C, followed by 5 s 65°C and 5 s 95° in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories). The primer and sequences used for the analysis are listed in the Key resources table. The mRNA of the genes of interest was normalized using the housekeeping gene HPRT (Hypoxanthine Guanine Phosphoribosyl Transferase).
Mouse naïve CD4+ CD62L+ T cell isolation and in vitro differentiation into T cell subsets
Spleens were removed from naïve Balb/c WT and IL-3−/− mice at the age of 10 weeks. Single cell suspension was prepared by pushing the spleen through a 40 μm cell strainer, lysing with 10 mL of an Ammonium-Chloride-Potassium (ACK)-Lysis buffer (0.15 M NH4Cl, 0.1 mM KHCO3, 0.1 mM Na2-EDTA dissolved in ddH2O) and washing with PBS. Spleen CD4+ CD62L+ T cells were positively sorted by magnetic bead isolation using the mouse CD4+ CD62L+ T cell isolation kit (MACS, Miltenyi Biotec, Bergisch-Gladbach, Germany) in accordance to the manufacturer’s protocol. Cells were platted at a concentration of 1 x 106 cells/ml and cultured in RPMI 1640 medium containing 100 IU/mL penicillin, 100 μg/mL streptomycin (all anprotec, Bruckberg, Germany), and 10% FCS (Biochrom GmbH, Berlin, Germany) at 37 °C and 5% CO2. For the unstimulated condition naiïve CD4+ CD62L+ T cells were cultured without any further stimulant, whereas for all other conditions the naïve T cells were activated by plate bound anti-CD3 (0.5 μg/mL; BD Biosciences, Heidelberg, Germany) and soluble anti-CD28 (1 μg/mL; Biolegend, San Diego, USA). Furthermore different cytokines (rmIL-2, rmIL-4, and rmIL-12 from Immunotools, Friesoythe, Germany; rhTGFβ from Miltenyi Biotec, Bergisch-Gladbach, Germany) and antibodies (anti-IL-4 from Biolegend, San Diego, USA; anti-IFNγ from Miltenyi Biotec, Bergisch-Gladbach, Germany) were added to the cell culture medium to generate TH0 cells (20 ng/mL rmIL-2), TH1 cells (20 ng/mL rmIL-2, 20 ng/mL rmIL-12, 10 μg/mL anti-IL-4), TH2 cells (20 ng/mL rmIL-2, 20 ng/mL rmIL-4, 10 μg/mL anti-IFNγ) and Treg cells (100 ng/mL rmIL-2, 30 ng/mL rhTGFβ). After three days the cells were restimulated with the same conditions. On day 5 cells and cell culture supernatants were harvested for analysis (Figure S2).
Experimental murine asthma model after OVA sensitization and challenge
For the induction of OVA-dependent allergic asthma WT and IL-3−/− mice at the age of 6-8 weeks were sensitized twice at day 0 and 7 with intraperitoneal (i.p.) injections of 100 μg ovalbumin (OVA; Calbiochem, San Diego, USA) complexed with 10% aluminum potassium sulfate (Sigma Aldrich, Steinheim, Germany). Thereafter, the mice were challenged intranasally (i.n.) with 50 μg OVA, dissolved in PBS, on days 18, 19 and 20. On day 20 airway hyperresponsiveness was measured performing a non-invasive whole-body plethysmography as described below. On day 21 the animals were sacrificed to obtain bronchoalveolar lavage and to isolate and analyse total lung cells.
Airway hyperresponsiveness
The airway hyperresponsiveness was measured on day 20 performing a non-invasive whole-body plethysmography at least 2 h after the last challenge with OVA by using a Buxco Electronics apparatus (Buxco Research Systems, Wilmington, NC). Thereby mice were challenged with increasing doses of nebulized methacholine (MCh) in an exposure chamber, and Penh responses were measured as previously described (Menachery et al., 2015). Thereby the mice were undergoing an acclimatization period of 6 min, before raising doses of methacholine (0 mg/mL, 25 mg/mL and 50 mg/mL methacholine in PBS) were nebulized for 90 s. The response time to the methacholine challenge was 6 min, followed by 1 min of recovery to go back to baseline after each dose. The data are expressed as mean values of enhanced pause ± SEMs.
Collection and analysis of the BAL
Bronchoalveolar lavage (BAL) was obtained on day 21 by intratracheally injection and aspiration of 0.8 mL Sterofundin (B. Braun Melsungen AG, Melsungen, Germany) twice. After its collection the BALF was centrifuged for 5 min at 1500 rpm. The BALF supernatants were frozen (−20°C) and subsequently analysed by using ELISA. The remaining cell pellet was resuspended in PBS, counted with a hemocytometer and subsequently used for flow cytometric analysis as described below.
Serum collection
Whole blood was collected from the heart on day 21. After a 30-min incubation at room temperature (RT), the blood was centrifuged (2500 rpm, 30 min, RT) and the serum was collected for further analysis.
Lung histology
Lungs were removed and the middle lobar of the right lung were fixed in 4% formaldehyde, dehydrated and embedded in paraffin. Serial paraffin sections (3 μm) were stained with hematoxylin and eosin (H&E) for semi quantification of inflammation. The scoring of the perivascular and peribronchial inflammation was gathered blindly by a pathologist using a semi-quantitative scoring system with a range pending between 1 (mild) and 4 (severe) as defined by Doganci et al. (Doganci et al. (2008). For the quantification of mucus production, lung sections were prepared as described above and stained with periodic acid-Schiff (PAS). For the analysis, the number of PAS positive cells per micrometer of bronchus diameter was determined (Hausding et al., 2011).
Isolation and culture of murine lung cells
Lungs were removed on day 21 of the asthma protocol, and total cells were isolated according to the protocol of Sauer et al. (Sauer et al. (2006). Briefly, lung tissue was cut into small pieces by a scalpel and digested with 300 U/ml Collagenase Typ Ia and 0.015% DNase (10 mg/mL) in PBS at 37°C for 1h. Digested lung was then pushed through a 40 μm cell strainer and subsequently lysed with an Ammonium-Chloride-Potassium (ACK)-Lysis buffer (0.15 M NH4Cl, 0.1 mM KHCO3, 0.1 mM Na2-EDTA dissolved in ddH2O). Finally, the lung cells were washed with PBS and counted using a hemocytometer. Cells were platted at a concentration of 1 x 106 cells/ml and cultured in RPMI 1640 medium containing 100 IU/mL penicillin, 100 μg/mL streptomycin (all anprotec, Bruckberg, Germany), and 10% FCS (Biochrom GmbH, Berlin, Germany) at 37°C and 5% CO2. For the unstimulated condition total lung cells were cultured without any further stimulant, whereas for the OVA condition cells were restimulated with 500 μg/mL OVA. For T cell stimulation total lung cells were activated by plate bound anti-CD3 (0.5 μg/mL; BD Biosciences, Heidelberg, Germany) and soluble anti-CD28 (1 μg/mL; Biolegend, San Diego, USA). After 20 h cells and cell culture supernatants were harvested and analysed by quantitative RT-PCR and ELISA, respectively.
Enzyme-linked immunosorbent assay (ELISA)
Serum levels of IgE were measured using the OptEIA™ sandwich ELISA kit from BD Bioscience (Heidelberg, Germany). To detect mouse IL-3, IL-4, IL-5, IL10 and IFNγ in BALF and cell culture supernatants OptEIA™ sandwich ELISA kit from BD Bioscience (Heidelberg, Germany) were used. Mouse IL-13 and GM-CSF were detected in cell culture supernatants and BALF by using DuoSet sandwich ELISA kits from R&D System (Wiesbaden, Germany). Each ELISA was performed according to the manufacturer’s instructions.
Flow cytometry analysis of murine cells
Single cell suspensions from murine lung, spleen or BALF cells were incubated with the respective mix of surface antibodies dissolved in 50 μL FACS buffer (1x PBS (anprotec, Bruckberg, Germany), 1% FCS (Biochrom GmbH, Berlin), 0.1% NaN3 (Carl Roth GmbH & Co. KG, Karlsruhe, Germany)) for 30 min in the dark at 4°C. After washing the cells once in 200 μL FACS buffer, cells were fixed in 200 μL 2% PFA (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) for flow cytometric analysis. For subsequently intracellular staining, cells were fixed and permeabilized with 150 μL Fixation/Permeabilization buffer (eBioscience, Thermo Fisher Scientific, Waltham, USA) in accordance to the manufacture’s protocol for 35 min in the dark at 4°C followed by intracellular staining with different transcriptions markers dissolved in 60 μL 1x Permeabilization buffer (eBioscience, Thermo Fisher Scientific, Waltham, USA) in the dark at 4 °C for 30 min. Afterwards cells were washed with 200 μL 1x Permeabilization buffer and resuspended in 200 μL FACS buffer for analysis.
All antibodies used for flow cytometry analysis are listed in the Key resources table. Flow cytometry was performed on FACS Canto II (BD Biosciences, Heidelberg, Germany) and flow cytometry data were analysed by FlowJo v10 (Treestar Inc., SanJose, USA).
Gating strategies used in FACS analysis
Eosinophils and neutrophils
Singlets gated on all cells (FSC-H ↳ FSC-A) → Non-Lymphocytes gated on Singlets (SSC-A ↳ FSC-A) → CD3− CD45R− cells gated on Non-Lymphocytes (CD45R FITC ↳ CD3 V450) → Eosinophils: Gr-1+ CCR3+ cells gated on CD3− CD45R− cells, Neutrophils: Gr-1+ CCR3- cells gated on CD3− CD45R− cells (Gr-1 PE ↳ CCR3 Alexa Fluor 647)
B cells
Lymphocytes gated on all cells (SSC-A ↳ FSC-A) → CD19+ CD45R+ cells gated on Lymphocytes (CD45R FITC ↳ CD19 APC)
CD4 and CD8+ T cells
Lymphocytes gated on all cells (SSC-A ↳ FSC-A) → CD4+ T cells: CD4+ CD8− cells gated on Lymphocytes, CD8+ T cells: CD4− CD8+ cells gated on Lymphocytes (CD8 BV510 ↳ CD4 BV421).
Tregs
Lymphocytes gated on all cells (SSC-A ↳ FSC-A) → CD4+ cells gated on Lymphocytes (SSC-A ↳ CD4 BV421) → CD4+ CD25+ cells gated on CD4+ cells (CD25 APC-Cy7 ↳ CD4 BV421) → CD4+ Foxp3+ cells gated on CD4+ CD25+ cells (Foxp3 APC ↳ CD4 BV421)
DCs
Singlets gated on all cells (FSC-H ↳ FSC-A) → Non-Lymphocytes gated on Singlets (SSC-A ↳ FSC-A) → CD11b− CD11c+ cells gated on Non-Lymphocytes (CD11c APC-Cy7 ↳ CD11b V450) → CD103+ Gr-1- cells gated on CD11b− CD11c+ cells (CD103 FITC ↳ Gr-1 PE)
TH2
Lymphocytes gated on all cells (SSC-A ↳ FSC-A) → CD4+ cells gated on Lymphocytes (SSC-A ↳ CD4 BV421) → GATA3+ CD4+ cells gated on CD4+ cells (GATA3 AlexaFluor 488 ↳ CD4 BV421)
Murine RNA isolation and quantitative real-time PCR
To extract RNA from murine lung, spleen and bone marrow cells we used PeqGold RNA Pure according to the manufacturer’s protocol (PeqLab, Erlangen, Germany). For reverse transcription of mRNA (1 μg) we used the RevertAid RT kit (ThermoFisher Scientific, Waltham, USA) as described in the manufactures protocol. The resulting template cDNA was then amplified by quantitative real-time PCR (qPCR) as described above. The primer sequences used for the real time PCR are listed in the Key resources table. The mRNA of the genes of interest was normalized using the housekeeping gene Hprt.
Quantification and statistical analysis
All statistical analysis were performed using GraphPad Prism v7 for Windows (GraphPad, La Jolla, CA). Statistical significances were calculated using two-tailed Student t test for the analysis of two-group comparisons and one- or two-way ANOVA for multiple comparisons to generate P-value data (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001). For post-hoc analysis we used the Tukey method test (one-way ANOVA) and the Sidak method test (two-way ANOVA), respectively. Unless otherwise indicated, data are presented as mean ± SEM. For correlation analysis, data were normalized before correlation was performed. In this case Pearson correlation was used.
Additional resources
The human study PreDicta is registered in the German Clinical Trials Register (www.germanctr.de: DRKS00004914).
Acknowledgments
The authors thank the whole team at the Department of Molecular Pneumology, especially S. Mittler, S. Trump, A. Geiger, and E. Nendel for their excellent technical support. Furthermore, we thank all children and their parents/guardians who took part in our Europe-wide study PreDicta. The authors are grateful to A. Graser, S. Koch at the Department of Molecular Pneumology, and to E. Muschiol and I. Jawa at the Pediatric Pneumology-Allergology, Department of Pediatrics and Adolescent Medicine, Universitätsklinikum Erlangen for their technical support in the PreDicta study. Moreover, the authors thank Drs. M. Wölfel, M.O. Melichar, C. Reinhardt, A. Neubert, and Prof. Dr. W. Rascher from the Department of Pediatrics and Adolescent Medicine, Universitätsklinikum Erlangen for their assistance and support in the PreDicta UKER WP-1 study. Funding Sources: This work was supported by the European Grant PreDicta (Collaborative Project grant 260895) awarded to S.F. in Erlangen and in the other European Centers to N.G.P. in Athens. Furthermore, the work was supported by the Department of Molecular Pneumology and the SFB grant CRC1181/TP08N (Checkpoints for Resolution of Inflammation) awarded to S.F.
Author contributions
J.K. performed all the murine experiments described in this manuscript. The human study Predicta was performed by previous member of the group at the Molecular Pneumology in Collaboration with the Children Hospital in Erlangen and J.K. did the IL-3 human ELISA and analyzed the human data with S.F. T.Z. and A.K. are the pediatricians that saw most of the children in Predicta WP1-UKER and made the medical diagnosis. N.G.P. designed the WP1 project Predicta with the help of P.X. and N.G.P. was the coordinator of Predicta. R.J.R. did the histology and histological analysis. S.Z. provided expertize with the IL-3-deficient mice and edited the manuscript. S.F. supervised the entire work, contributed to manuscript and figure generation, and performed the revision of this manuscript.
Declaration of interests
The authors declare no conflict of interest on the matter described in this manuscript.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104440.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact if required upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Akdis M. In: Global Atlas of Asthma. AKDIS C.A., editor. European Academy of Allergy and Clinical Immunology; 2013. The pathogenesis of asthma; pp. 28–30. [Google Scholar]
- Akdis M., Aab A., Altunbulakli C., Azkur K., Costa R.A., Crameri R., Duan S., Eiwegger T., Eljaszewicz A., Ferstl R., et al. Interleukins (from IL-1 to IL-38), interferons, transforming growth factor β, and TNF-α: receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2016;138:984–1010. doi: 10.1016/j.jaci.2016.06.033. [DOI] [PubMed] [Google Scholar]
- Arai K.I., Lee F., Miyajima A., Miyatake S., Arai N., Yokota T. Cytokines: coordinators of immune and inflammatory responses. Annu. Rev. Biochem. 1990;59:783–836. doi: 10.1146/annurev.bi.59.070190.004031. [DOI] [PubMed] [Google Scholar]
- Asquith K.L., Ramshaw H.S., Hansbro P.M., Beagley K.W., Lopez A.F., Foster P.S. The IL-3/IL-5/GM-CSF common receptor plays a pivotal role in the regulation of Th2 immunity and allergic airway inflammation. J. Immunol. 2008;180:1199–1206. doi: 10.4049/jimmunol.180.2.1199. [DOI] [PubMed] [Google Scholar]
- Auclair S.R., Roth K.E., Saunders B.L., Ogborn K.M., Sheikh A.A., Naples J., Young A.M.P., Boisen D.K., Tavangar A.T., Welch J.E., Lantz C.S. Interleukin-3-deficient mice have increased resistance to blood-stage malaria. Infect. Immun. 2014;82:1308–1314. doi: 10.1128/IAI.01140-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P.J. Pathophysiology of allergic inflammation. Immunol. Rev. 2011;242:31–50. doi: 10.1111/j.1600-065X.2011.01020.x. [DOI] [PubMed] [Google Scholar]
- Bergauer A., Sopel N., Kroß B., Vuorinen T., Xepapadaki P., Weiss S.T., Blau A., Sharma H., Kraus C., Springel R., et al. Rhinovirus species/genotypes and interferon-lambda: subtypes, receptor and polymorphisms - missing pieces of the puzzle of childhood asthma? Eur. Respir. J. 2017;49:1700265. doi: 10.1183/13993003.00265-2017. [DOI] [PubMed] [Google Scholar]
- Bertrand P., Lay M.K., Piedimonte G., Brockmann P.E., Palavecino C.E., Hernández J., León M.A., Kalergis A.M., Bueno S.M. Elevated IL-3 and IL-12p40 levels in the lower airway of infants with RSV-induced bronchiolitis correlate with recurrent wheezing. Cytokine. 2015;76:417–423. doi: 10.1016/j.cyto.2015.07.017. [DOI] [PubMed] [Google Scholar]
- Bielor C., Sopel N., Maier A., Blau A., Sharma H., Vuorinen T., Kroß B., Mittler S., Graser A., Mousset S., et al. Role of TGF-beta in anti-rhinovirus immune responses in asthmatic patients. J. Allergy Clin. Immunol. 2017;140:283–286.e10. doi: 10.1016/j.jaci.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton S.E., Dhagat U., Hercus T.R., Nero T.L., Grimbaldeston M.A., Bonder C.S., Lopez A.F., Parker M.W. The GM-CSF/IL-3/IL-5 cytokine receptor family: from ligand recognition to initiation of signaling. Immunol. Rev. 2012;250:277–302. doi: 10.1111/j.1600-065X.2012.01164.x. [DOI] [PubMed] [Google Scholar]
- Brown P.H., Crompton G.K., Greening A.P. Proinflammatory cytokines in acute asthma. Lancet. 1991;338:590–593. doi: 10.1016/0140-6736(91)90605-o. [DOI] [PubMed] [Google Scholar]
- Celestin J., Rotschke O., Falk K., Ramesh N., Jabara H., Strominger J., Geha R.S. IL-3 induces B7.2 (CD86) expression and costimulatory activity in human eosinophils. J. Immunol. 2001;167:6097–6104. doi: 10.4049/jimmunol.167.11.6097. [DOI] [PubMed] [Google Scholar]
- Criée C.P., Baur X., Berdel D., Bösch D., Gappa M., Haidl P., Husemann K., Jörres R.A., Kabitz H.J., Kardos P., et al. [Standardization of spirometry: 2015 update. Published by German Atemwegsliga, German respiratory society and German society of occupational and environmental medicine] Pneumologie. 2015;69:147–164. doi: 10.1055/s-0034-1391345. [DOI] [PubMed] [Google Scholar]
- Davies R.J., Wang J., Abdelaziz M.M., Calderon M.A., Khair O., Devalia J.L., Rusznak C. New insights into the understanding of asthma. Chest. 1997;111 doi: 10.1378/chest.111.2_supplement.2s. 2S-10S-10s. [DOI] [PubMed] [Google Scholar]
- Doganci A., Karwot R., Maxeiner J.H., Scholtes P., Schmitt E., Neurath M.F., Lehr H.A., Ho I.C., Finotto S. IL-2 receptor beta-chain signaling controls immunosuppressive CD4+ T cells in the draining lymph nodes and lung during allergic airway inflammation in vivo. J. Immunol. 2008;181:1917–1926. doi: 10.4049/jimmunol.181.3.1917. [DOI] [PubMed] [Google Scholar]
- Esnault S., Kelly E.A. Essential mechanisms of differential activation of eosinophils by IL-3 compared to GM-CSF and IL-5. Crit. Rev. Immunol. 2016;36:429–444. doi: 10.1615/CritRevImmunol.2017020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault S., Kelly E.A.B., Shen Z.J., Johansson M.W., Malter J.S., Jarjour N.N. IL-3 maintains activation of the p90S6K/RPS6 pathway and increases translation in human eosinophils. J. Immunol. 2015;195:2529–2539. doi: 10.4049/jimmunol.1500871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P.S., Hogan S.P., Ramsay A.J., Matthaei K.I., Young I.G. Interleukin 5 deficiency abolishes eosinophilia, airways hyperreactivity, and lung damage in a mouse asthma model. J. Exp. Med. 1996;183:195–201. doi: 10.1084/jem.183.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauvreau G.M., El-Gammal A.I., O'Byrne P.M. Allergen-induced airway responses. Eur. Respir. J. 2015;46:819–831. doi: 10.1183/13993003.00536-2015. [DOI] [PubMed] [Google Scholar]
- Graser A., Ekici A.B., Sopel N., Melichar V.O., Zimmermann T., Papadopoulos N.G., Taka S., Ferrazzi F., Vuorinen T., Finotto S. Rhinovirus inhibits IL-17A and the downstream immune responses in allergic asthma. Mucosal Immunol. 2016;9:1183–1192. doi: 10.1038/mi.2015.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory B., Kirchem A., Phipps S., Gevaert P., Pridgeon C., Rankin S.M., Robinson D.S. Differential regulation of human eosinophil IL-3, IL-5, and GM-CSF receptor alpha-chain expression by cytokines: IL-3, IL-5, and GM-CSF down-regulate IL-5 receptor alpha expression with loss of IL-5 responsiveness, but up-regulate IL-3 receptor alpha expression. J. Immunol. 2003;170:5359–5366. doi: 10.4049/jimmunol.170.11.5359. [DOI] [PubMed] [Google Scholar]
- Haag P., Sharma H., Rauh M., Zimmermann T., Vuorinen T., Papadopoulos N.G., Weiss S.T., Finotto S. Soluble ST2 regulation by rhinovirus and 25(OH)-vitamin D3 in the blood of asthmatic children. Clin. Exp. Immunol. 2018;193:207–220. doi: 10.1111/cei.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausding M., Tepe M., Ubel C., Lehr H.A., Röhrig B., Höhn Y., Pautz A., Eigenbrod T., Anke T., Kleinert H., et al. Induction of tolerogenic lung CD4+ T cells by local treatment with a pSTAT-3 and pSTAT-5 inhibitor ameliorated experimental allergic asthma. Int. Immunol. 2011;23:1–15. doi: 10.1093/intimm/dxq451. [DOI] [PubMed] [Google Scholar]
- Hentschke I., Graser A., Melichar V.O., Kiefer A., Zimmermann T., Kroß B., Haag P., Xepapadaki P., Papadopoulos N.G., Bogdan C., Finotto S. IL-33/ST2 immune responses to respiratory bacteria in pediatric asthma. Sci. Rep. 2017;7:43426. doi: 10.1038/srep43426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holgate S.T., Wenzel S., Postma D.S., Weiss S.T., Renz H., Sly P.D. Asthma. Nat. Rev. Dis. Primers. 2015;1:15025. doi: 10.1038/nrdp.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura A., Miura Y., Hikasa Y., Kiso Y., Matsuda H. Interleukin-3 and stem cell factor modulate cell cycle regulatory factors in mast cells: negative regulation of p27Kip1 in proliferation of mast cells induced by interleukin-3 but not stem cell factor. Exp. Hematol. 2001;29:803–811. doi: 10.1016/s0301-472x(01)00659-2. [DOI] [PubMed] [Google Scholar]
- Krammer S., Yang Z., Zimmermann T., Xepapadaki P., Geppert C.I., Papadopoulos N.G., Finotto S. An immunoregulatory role of interleukin-3 in allergic asthma. Front. Immunol. 2022;13:821658. doi: 10.3389/fimmu.2022.821658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Rani L., Mhaske S.T., Pote S.T., Behera S., Mishra G.C., Wani M.R. IL-3 receptor expression on activated human Th cells is regulated by IL-4, and IL-3 synergizes with IL-4 to enhance Th2 cell differentiation. J. Immunol. 2020;204:819–831. doi: 10.4049/jimmunol.1801629. [DOI] [PubMed] [Google Scholar]
- Lai C.K., Ho S.S., Chan C.H., Leung R., Lai K.N. Gene expression of interleukin-3 and granulocyte macrophage colony-stimulating factor in circulating CD4+ T cells in acute severe asthma. Clin. Exp. Allergy. 1996;26:138–146. doi: 10.1111/j.1365-2222.1996.tb00072.x. [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., Hammad H. The immunology of asthma. Nat. Immunol. 2015;16:45–56. doi: 10.1038/ni.3049. [DOI] [PubMed] [Google Scholar]
- Lantz C.S., Boesiger J., Song C.H., Mach N., Kobayashi T., Mulligan R.C., Nawa Y., Dranoff G., Galli S.J. Role for interleukin-3 in mast-cell and basophil development and in immunity to parasites. Nature. 1998;392:90–93. doi: 10.1038/32190. [DOI] [PubMed] [Google Scholar]
- Lantz C.S., Min B., Tsai M., Chatterjea D., Dranoff G., Galli S.J. IL-3 is required for increases in blood basophils in nematode infection in mice and can enhance IgE-dependent IL-4 production by basophils in vitro. Lab. Invest. 2008;88:1134–1142. doi: 10.1038/labinvest.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.Y., Sedgwick J.B., Bates M.E., Vrtis R.F., Gern J.E., Kita H., Jarjour N.N., Busse W.W., Kelly E.A.B. Decreased expression of membrane IL-5 receptor alpha on human eosinophils: I. Loss of membrane IL-5 receptor alpha on airway eosinophils and increased soluble IL-5 receptor alpha in the airway after allergen challenge. J. Immunol. 2002;169:6452–6458. doi: 10.4049/jimmunol.169.11.6452. [DOI] [PubMed] [Google Scholar]
- Lourenço F.D., Azor M.H., Santos J.C., Prearo E., Maruta C.W., Rivitti E.A., Duarte A.J., Sato M.N. Activated status of basophils in chronic urticaria leads to interleukin-3 hyper-responsiveness and enhancement of histamine release induced by anti-IgE stimulus. Br. J. Dermatol. 2008;158:979–986. doi: 10.1111/j.1365-2133.2008.08499.x. [DOI] [PubMed] [Google Scholar]
- Mauser P.J., Pitman A.M., Fernandez X., Foran S.K., Adams G.K., 3rd, Kreutner W., Egan R.W., Chapman R.W. Effects of an antibody to interleukin-5 in a monkey model of asthma. Am. J. Respir. Crit. Care Med. 1995;152:467–472. doi: 10.1164/ajrccm.152.2.7633694. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Baric R.S., Ferris M.T. New metrics for evaluating viral respiratory pathogenesis. PLoS One. 2015;10:e0131451. doi: 10.1371/journal.pone.0131451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T.R., Cherwinski H., Bond M.W., Giedlin M.A., Coffman R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- Park B.L., Kim L.H., Choi Y.H., Lee J.H., Rhim T., Lee Y.M., Uh S.T., Park H.S., Choi B.W., Hong S.J., et al. Interleukin 3 (IL3) polymorphisms associated with decreased risk of asthma and atopy. J. Hum. Genet. 2004;49:517–527. doi: 10.1007/s10038-004-0184-x. [DOI] [PubMed] [Google Scholar]
- Renner K., Hermann F.J., Schmidbauer K., Talke Y., Rodriguez Gomez M., Schiechl G., Schlossmann J., Brühl H., Anders H.J., Mack M. IL-3 contributes to development of lupus nephritis in MRL/lpr mice. Kidney Int. 2015;88:1088–1098. doi: 10.1038/ki.2015.196. [DOI] [PubMed] [Google Scholar]
- Renner K., Metz S., Metzger A.M., Neumayer S., Schmidbauer K., Talke Y., Buchtler S., Halbritter D., Mack M. Expression of IL-3 receptors and impact of IL-3 on human T and B cells. Cell. Immunol. 2018;334:49–60. doi: 10.1016/j.cellimm.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Rignault-Bricard R., Machavoine F., Mecheri S., Hermine O., Schneider E., Dy M., Leite-de-Moraes M. IL-3-producing basophils are required to exacerbate airway hyperresponsiveness in a murine inflammatory model. Allergy. 2018;73:2342–2351. doi: 10.1111/all.13480. [DOI] [PubMed] [Google Scholar]
- Robinson D.S., Hamid Q., Ying S., Tsicopoulos A., Barkans J., Bentley A.M., Corrigan C., Durham S.R., Kay A.B. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N. Engl. J. Med. 1992;326:298–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- Sauer K.A., Scholtes P., Karwot R., Finotto S. Isolation of CD4+ T cells from murine lungs: a method to analyze ongoing immune responses in the lung. Nat. Protoc. 2006;1:2870–2875. doi: 10.1038/nprot.2006.435. [DOI] [PubMed] [Google Scholar]
- Schrader J.W. Interleukin-3. Growth Factors and Cytokines in Health and Disease. 1997;2:49–84. [Google Scholar]
- Shi H.Z., Xiao C.Q., Zhong D., Qin S.M., Liu Y., Liang G.R., Xu H., Chen Y.Q., Long X.M., Xie Z.F. Effect of inhaled interleukin-5 on airway hyperreactivity and eosinophilia in asthmatics. Am. J. Respir. Crit. Care Med. 1998;157:204–209. doi: 10.1164/ajrccm.157.1.9703027. [DOI] [PubMed] [Google Scholar]
- Sullivan B.M., Liang H.E., Bando J.K., Wu D., Cheng L.E., Mckerrow J.K., Allen C.D.C., Locksley R.M. Genetic analysis of basophil function in vivo. Nat. Immunol. 2011;12:527–535. doi: 10.1038/ni.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher Y.A., Henricks P.A.J., van Oosterhout A.J.M. Allergen-specific subcutaneous immunotherapy in allergic asthma: immunologic mechanisms and improvement. Libyan J. Med. 2010;5 doi: 10.3402/ljm.v5i0.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Übel C., Sopel N., Graser A., Hildner K., Reinhardt C., Zimmermann T., Rieker R.J., Maier A., Neurath M.F., Murphy K.M., Finotto S. The activating protein 1 transcription factor basic leucine zipper transcription factor, ATF-like (BATF), regulates lymphocyte- and mast cell-driven immune responses in the setting of allergic asthma. J. Allergy Clin. Immunol. 2014;133:198–206.e1-9. doi: 10.1016/j.jaci.2013.09.049. [DOI] [PubMed] [Google Scholar]
- van de Veen W., Akdis M. The use of biologics for immune modulation in allergic disease. J. Clin. Invest. 2019;129:1452–1462. doi: 10.1172/JCI124607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D. Basophil modulation by cytokine instruction. Eur. J. Immunol. 2012;42:2544–2550. doi: 10.1002/eji.201142318. [DOI] [PubMed] [Google Scholar]
- Wang P., Wu P., Cheewatrakoolpong B., Myers J.G., Egan R.W., Billah M.M. Selective inhibition of IL-5 receptor alpha-chain gene transcription by IL-5, IL-3, and granulocyte-macrophage colony-stimulating factor in human blood eosinophils. J. Immunol. 1998;160:4427–4432. [PubMed] [Google Scholar]
- Wenzel S.E. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact if required upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.