Abstract
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) provides a simple and scalable way to detect the unique chromatin landscape associated with a cell type and how it may be altered by perturbation or disease. ATAC-seq requires a relatively small number of input cells and does not require a priori knowledge of the epigenetic marks or transcription factors governing the dynamics of the system. Here, we describe an updated and optimized protocol for ATAC-seq, called Omni-ATAC, that is applicable across a broad range of cell and tissue types. The ATAC-seq workflow has five main steps: sample preparation, transposition, library preparation, sequencing, and data analysis. This protocol details the steps to generate and sequence ATAC-seq libraries, with recommendations for sample preparation and downstream bioinformatic analysis. ATAC-seq libraries for ~12 samples can be generated in 10 hours by someone familiar with basic molecular biology and downstream sequencing analysis can be implemented using benchmarked pipelines by someone with basic bioinformatics skills and access to a high-performance computing environment.
Keywords: ATAC-seq, chromatin accessibility, epigenetics, gene regulation
INTRODUCTION
Mapping alterations in cell states is a key aspect of understanding biological systems. Whether in development, differentiation, or disease, cell state is governed by changes in gene expression which are, in turn, orchestrated by changes in gene regulatory programs. In recent years, it has become increasingly clear that these gene regulatory programs are established and controlled by the activity of transcription factors (TFs) that both interpret and alter the underlying epigenetic state of chromatin. The epigenetic state of chromatin can be regulated by a variety of mechanisms, including chemical modification of both DNA and histone proteins which, in turn, alter chromatin dynamics and high-dimensional chromatin structure. We now recognize that chromatin can exist in several different states1,2 which are defined by combinations of different epigenetic modifications and are associated with particular gene regulatory patterns. At the two ends of the spectrum are (i) active gene regulatory elements such as enhancers, promoters, and insulators, which are bound by DNA binding proteins and (ii) inactive regions of silenced or poised chromatin, which are generally refractory to gene expression machinery3. Understanding the epigenetic state of chromatin in a certain biological context can shed light onto the molecular mechanisms underlying the observed gene expression patterns.
The epigenome can be assayed in a variety of ways, ranging in specificity. The most specific methods assay the location and abundance of a particular histone modification, DNA modification, or TF using an antibody-based pull-down method and refinements of the classical chromatin immunoprecipitation-sequencing (ChIP-seq) technique4. Antibodies against the target of interest are used to enrich genomic segments from bulk chromatin. These marked fragments are then prepared into a library, sequenced using high-throughput sequencing, and aligned to the genome. Genome regions where many reads overlap appear as peaks, indicating that the target of interest was present at that particular locus in a large proportion of the cells. Several refinements have been made to this classic technique5,6. Recently, the sensitivity of these antibody-based techniques to determine DNA-protein interactions has been improved with the development of chromatin immunocleavage techniques (ChIC)7, including CUT&RUN8 and CUT&TAG9, which tether a nuclease or Tn5 transposase to the targeting antibody via the use of Protein-A. These modifications increase the resolution of the resulting protein binding site, remove the need for an immunoprecipitation step, and decrease the amount of input material required. These techniques, and other derivations of similar technologies such as ChIPmentation10, CoBATCH11, ChIL-seq12,13 and scChIC-seq14, were recently adapted to work on single-cells11,15-17. Such targeted profiling methods can provide key insights into the epigenetic changes driving a particular cell state but require a priori knowledge of the expected mechanism.
In some cases, it can be more helpful to obtain a broader picture of the gene regulatory landscape, particularly when a phenomenon is observed but the specific nature of the epigenetic changes remains unknown. For this reason, alternate strategies have been developed to probe the gene regulatory landscape by mapping all TF binding sites, agnostic to the precise TF identities, for a cell type or tissue. A variety of such agnostic profiling techniques have been developed, including Deoxyribonuclease I hypersensitivity sequencing (DNase-seq)18-20, Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE-seq)21,micrococcal nuclease digestion with sequencing (MNase-seq)22,23, and Nucleosome Occupancy and Methylome Sequencing (NOMe-seq)24 and were recently reviewed in detail25. Among the first of these assays to be developed was DNase-seq20, which applied high-throughput sequencing to the classic technique of DNase digestion to measure different chromatin states26. The activity of DNase is obstructed at sites bound by TFs or nucleosomes, thus protecting these DNA fragments and allowing them to be identified via sequencing. Simultaneously, the binding of TFs creates adjacent nucleosome-free regions of DNA which are hypersensitive to enzymatic digestion by DNase. Thus, mapping of these hypersensitive sites has been used extensively to identify putative gene regulatory elements in an unbiased manner. In particular, DNase-seq has become the gold-standard technique for TF footprinting27-29. Similarly, MNase-seq is based on the use of MNase, an endo-exonuclease that cleaves regions of DNA that are not protected by nucleosomes or DNA binding proteins23. It can be thought of as an orthogonal approach to DNase-seq in that it measures nucleosome occupied regions and is most often used to map nucleosome occupancy quantitatively genome wide30,31.
The original DNase-seq and MNase-seq assays traditionally had complex, time-consuming library preparation protocols and required large numbers of cells as starting material. To address some of these limitations, while keeping the agnostic profiling of chromatin, the assay for transposase-accessible chromatin using sequencing (ATAC-seq) was developed32. ATAC-seq uses the activity of an engineered, hyperactive Tn5 transposase33 preloaded with sequencing adapters to determine the sites of accessible chromatin. The development of ATAC-seq was based on two observations: (i) a transposase had previously been used to generate “tagmentation” libraries, in which a Tn5 transposase was preloaded with sequencing adapters and used to simultaneously fragment and tag genomic DNA for high-throughput sequencing library preparation34 and (ii) the observation that in vivo Tn5 could efficiently insert into nucleosome-free regions35. ATAC-seq generates genome-wide regulatory maps that are highly similar to those derived from DNase-seq and MNase-seq32, while reducing library preparation complexity and hands-on time. ATAC-seq has been widely adopted due to its low input material requirements (< 50,000 cells) and the fast-processing time scale which facilitates data generation from large numbers of samples. Here, we describe an updated version of the ATAC-seq protocol, based on our previous Omni-ATAC protocol36, and provide helpful guidelines and benchmarks to ensure high-quality data generation from most, if not all, input cell types.
Applications of ATAC-seq
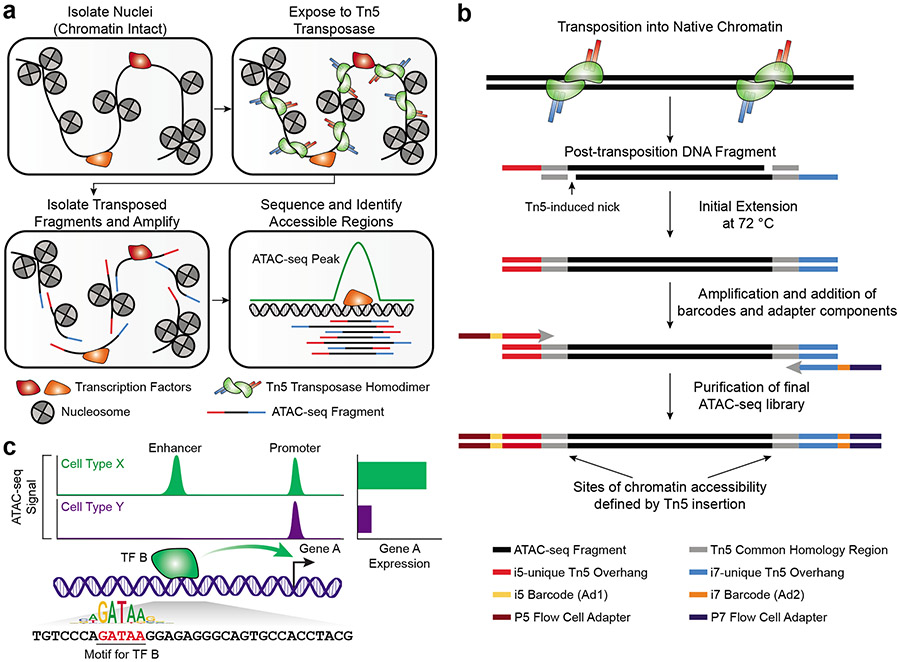
ATAC-seq provides a simple and scalable way to assay the regions of the genome that are bound by TFs, and to compare how these landscapes change between particular contexts or perturbations. This is accomplished using in vitro transposition of sequencing adapters into native chromatin (Fig. 1a). Each unique transposition event, termed an "insertion,” marks a location in the genome where a Tn5 transposase dimer is able to access DNA and perform a cut-and-paste reaction. The transposase simultaneously fragments the DNA and inserts sequence handles that are then used for amplification during library preparation. A sequenceable ATAC-seq DNA fragment is created by two separate transposase insertion events (Fig. 1b). The precise biochemical interactions that govern Tn5 transposition at these sites is not yet fully understood. It is generally accepted that the binding of a TF to DNA is associated with the creation of adjacent nucleosome-free regions and that these are associated with increased Tn5 transposition. We discuss several nuances to interpreting the resulting Tn5- chromatin accessibility in Supplementary Note 1.
Figure 1: Schematic of the ATAC-seq transposition reaction and library preparation.
a. Overview of the different steps in ATAC-seq. Nuclei are isolated from cells, maintaining the chromatin structure and any associated DNA binding proteins, including nucleosomes and TFs, intact. This chromatin is then exposed to the Tn5 transposase, which acts as a homodimer to simultaneously fragment the chromatin and insert sequences containing PCR handles that enable downstream amplification with i5/P5 and i7/P7 ATAC-seq adapters. Only fragments that receive an i5/P5 adapter at one end and an i7/P7 adapter at the other end will be properly amplified and sequenced. After sequence analysis of the library fragments, genomic regions enriched for many Tn5 transposition events are designated as peaks of chromatin accessibility, or ATAC-seq peaks. b. Detailed schematic of the fragments generated by transposition into native chromatin in Step 10 of the protocol. After Tn5 insertion of the PCR handles, the nicks left behind by the transposase are filled in during the initial 72 °C extension in the first step of the barcoding PCR. Then, the fragments are barcoded and prepared for sequencing. c. A toy example of what hypothetical ATAC-seq data might look like. Cell types X and Y both have ATAC-seq peaks at the promoter of Gene A. However, cell type X has a cell type-specific enhancer upstream of the TSS, which drives higher levels of expression of Gene A. In the ATAC-seq data, this would appear as a cell type-specific ATAC-seq peak, which would be differentially accessible between cell types X and Y. In this toy example, the differential accessibility at the enhancer is driven by TF B, which binds to the cell type-specific enhancer and increases gene expression. ATAC-seq data can also help reveal the TF binding motif associated with such differential peaks, here depicted as “GATAA”.
Extensive profiling efforts have shown that regions of Tn5-accessbile chromatin can be found at promoters, located proximal to the transcription start site (TSS), and at intergenic regions of the genome largely corresponding to enhancers, insulators, or silencers32,37,38. These patterns and locations of Tn5-accessibility, especially those at distal elements, are often cell type- or cell state-specific37. Thus, ATAC-seq presents a valuable tool to understand how cells control gene expression, by mapping the location of putative gene regulatory elements. After processing and alignment of ATAC-seq fragments, enrichment of Tn5 transposition events at specific genomic regions is used to identify peaks of Tn5-accessible chromatin in each sample. These are often termed “ATAC-seq peaks.” Chromatin accessibility signal within these peak regions can be compared between different sample types using established pipelines (see the Data Analysis section of Experimental Design), and serve as the starting point for a variety of downstream analyses. For example, peaks can be linked to putative gene targets by using orthogonal chromatin conformation capture datasets or by naively assigning each peak to the nearest gene. These predicted gene regulatory interactions can provide a hint as to the functional importance of a given peak. Often, genes with several ATAC-seq peaks in their promoter and gene body are inferred to be actively expressed in that cell type. While gene expression is more accurately measured by RNA-sequencing, ATAC-seq can explain the mechanism behind how gene expression is regulated or why it might be different between two cell types or conditions.
A common application of ATAC-seq is to identify novel enhancers or gene regulatory regions for a given cell type or cell context of interest. For example, observing an ATAC-seq peak in a region 5 kb upstream of Gene A’s TSS in cell type X but not in cell type Y suggests the hypothesis that a cell type-specific enhancer may regulate Gene A’s expression in cell type X (Fig. 1c). Such observations have been made comparing cells before and after exposure to a variety of stimuli39-41 or between different cell types or developmental time points42-46. Differential activity of regulatory elements has been found in a wide array of diseases38,47-50 and such findings motivate efforts to use ATAC-seq to characterize gene regulatory landscapes in large patient cohorts38,51,52. ATAC-seq has also been used to fine-map disease-associated genetic variants identified through genome-wide association studies (GWASs), thus enabling prediction of putative functional noncoding alterations53-57. Historically, GWASs have identified single-nucleotide polymorphisms (SNPs) that often reside in noncoding regions, making it difficult to interpret their functional impact. Intersecting regulatory regions identified through ATAC-seq in disease-relevant cell types with GWAS SNPs can help build hypotheses about which of those SNPs may affect gene expression and thus mediate disease-associated phenotypes.
ATAC-seq peaks can also be annotated for the presence of various TF motif sequences and enrichment tests can be used to predict the drivers of differential chromatin accessibility. Such motif-based analyses can be useful for comparing different cell types or disease states, understanding differentiation or developmental trajectories45,58, or comparing how in vitro derived cells compare to their in vivo equivalents59. In the toy example above, the cell type X-specific ATAC-seq peak 5 kb upstream of Gene A might be bound by TF B, which is not expressed in cell type Y (Fig. 1c). Such a cell type-specific difference in TF expression might result in many peaks throughout the genome being bound by TF B, leading to a significant enrichment of peaks harboring the motif for TF B within the subset of peaks that are differentially accessible between cell types X and Y. These types of analyses have been used to show changes in TF usage between different cell states – for example during metastasis in small cell lung cancer60, before and after exposure to inflammatory stimuli40, or during reprogramming of fibroblasts into neurons59. ATAC-seq data can also be used to infer the positions of nucleosomes61, providing insights into chromatin regulation beyond TF binding. In summary, ATAC-seq is an effective technique for uncovering the gene regulatory changes that govern why cells express certain genes and how gene expression changes are mediated.
Comparison with other chromatin profiling methods
The diversity of available techniques to map DNA regulatory elements can make it challenging to determine which technique is the most appropriate and informative for a specific application. In Table 1, we compare some technical and experimental aspects of the most commonly used techniques for mapping DNA regulatory elements: ATAC-seq32,36, DNase-seq19, MNase-seq25, ChIP-seq5, and targeted CUT&TAG62, to help new users decide which assay is best for their particular application. We recommend basing this decision on (i) what kind of information is necessary to answer the specific research question and (ii) what kind of input material is available. In general, epigenomic profiling is appropriate to answer how or why a cell type or tissue might exhibit gene regulatory changes. For questions dealing predominantly with what is changing, we suggest beginning with RNA-sequencing63.
Table 1:
Comparison to other chromatin profiling protocols
| ATAC-seq | DNase-seq | MNase-seq | CUT&TAG or related ChIC techniques |
|
|---|---|---|---|---|
| Enzyme type | Tn5 | endonuclease | endonuclease and exonuclease | Tn5 conjugated to an antibody via Protein A. |
| Sequence bias? | Yes; complex, Tn5 insertion bias, with preference for A/Ts in insertion site and C/Gs flanking133-135 | Yes; complex, partially dependent on enzyme concentration and on methylation status of CpGs85,136 | Yes; preferential cutting upstream of A/T compared to G/C137,138 | Yes; dictated by antibody used to guide Tn5 and by Tn5 bias. |
| Number of input cells/nuclei for standard bulk assays. | 500-50,000 | 1-10 million | 10,000-100,000 | 100,000-500,000 |
| Low-input/single-cell methods available? | Yes86,87; commercial solutions available. | Yes67 | Yes66 | Yes62,64,139-141 |
| Sample type | Fresh or cryopreserved cells or nuclei. Fresh or frozen tissues. | Fresh or cryopreserved cells or nuclei. Fresh or frozen tissues. Formaldehyde cross-linked or formalin-fixed paraffin-embedded samples. | Fresh or cryopreserved cells or nuclei. Fresh or frozen tissues. Formaldehyde cross-linked samples. | Fresh or cryopreserved cells or nuclei. Fresh or frozen tissues. |
| Library preparation time | ~10 hours for 12 samples (this protocol) | 1-3 days | ~ 2-days | 1-2 days |
| Technical considerations | Library quality is highly dependent on cell viability. Protocol alterations are required for use on fixed cells and data quality is often reduced for those samples. | Enzyme concentration and digestion duration may need to be optimized to sample type. Size of fragments selected affects downstream analysis.28 |
Enzyme concentration and digestion duration may need to be optimized to sample type. Apparent nucleosome occupancy is a function of MNase concentration. |
The amount of antibody used must be titrated for the cell type or sample. This will be a function of the strength of the antibody and the abundance of the target protein. The assay is as specific as the primary antibody used. Additionally, this is a targeted technique, so additional libraries must be made of each modification or protein tested. |
| Sequencing type | Paired-end | Single-end | Single-end | Single-end or paired-end |
| Sequencing depth | Low; 10 million read-pairs per sample with Omni-ATAC. | Medium/high: 20-50 million uniquely mapping reads per sample; 200 million for TF footprinting. | High; 150-200 million reads per sample (human)142 | Very low; 3 million read-pairs per sample. |
| Data produced | Tn5-accessible chromatin; | DNase-accessible chromatin; TF footprinting. | Nucleosome positioning, inaccessible chromatin. | Location of target on DNA. |
| Major advantage | Links labeling of accessible regions and NGS library preparation, making preparation of library straightforward. | Footprinting analysis. | Method of choice for nucleosome positioning and quantitative nucleosome dynamics. | Enables mapping of specific TF or histone modification in low cell numbers. Some histone modifications, like H3K27ac, can be used to look for active enhancers. |
To decide which epigenomic profiling technique is right for a specific application, the first decision to make is whether a broad or targeted approach is appropriate. If the question involves the location of a specific TF, DNA binding factor, or histone modification, TF ChIP-seq or related technologies would be most appropriate5. ChIP-seq of histone modifications such as H3K27ac, H3K4me1, or H3K4me3 has been widely used to map DNA regulatory elements across a variety of cell and tissue types. For example, H3K27ac predominantly marks active promoters and enhancers. Thus, by combining genome-wide maps of one or more of these modifications, an overall picture of the epigenome can be formed. However, in general, these assays are best interpreted in combination, and a single mark may not capture all types of DNA regulatory elements (i.e. promoters, enhancers and silencers). Alternatively, if the epigenomic profiling will predominantly serve for hypothesis generation or the desired information is better captured by measuring global patterns in regulatory element activity, techniques such as ATAC-seq, DNase-seq, or MNase-seq may be more appropriate. These techniques have the advantage of being able to capture many different types of regulatory information with a single assay.
Compared to DNase-seq and MNase-seq, ATAC-seq has two major advantages for “omics” users: (i) the library preparation is fast and easy with the ability to generate sequence-ready libraries from >12 samples in a single work day, and (ii) the assay requires little-to-no tuning or optimization of the reaction parameters for different cell types. We find that this protocol works without modification on a variety of cell lines and primary cell/tissue types. In contrast, the DNase-seq and MNase-seq assays often require optimization of the enzyme concentration and the reaction time for the specific input material to avoid over- or under-digestion of chromatin which influences the types of hypersensitive sites detected28. ATAC-seq is a non-catalytic enzymatic reaction, where each Tn5 molecule can only perform a single transposition reaction, thus reducing the risk of chromatin over digestion inherent in endonuclease assays. The Tn5 enzyme is commercially available with preloaded adapters, making assay set up simple and with little to no calibration required (see the Tn5-to-cell Ratio section of Experimental Design for more details). Since their inception, all of these methods have been adapted to the low input or single-cell scale 64-67, no longer limiting their applications to highly abundant cell types. ATAC-seq is a highly portable technique which can be applied to many different experimental scenarios and in any laboratory, regardless of previous experience with epigenomics, to answer general questions about the gene regulatory landscape. However, there are instances where ATAC-seq may not be the optimal assay of choice, notably for TF footprinting, for analyzing dynamics of nucleosome lability, or when dealing with formalin-fixed paraffin-embedded (FFPE) treated samples.
Comparison with previous ATAC-seq methods
The initial development of ATAC-seq32 made the study of gene regulation accessible to a much larger community of researchers. The assay was quickly applied to a variety of cell lines and primary cell types, enabling applications that were previously impossible. Despite this success, multiple shortcomings were noted in the original method. For example, since mitochondrial DNA is not chromatinized, the presence of lysed mitochondria in an ATAC-seq reaction can lead to a very high fraction of ATAC-seq reads mapping to mitochondrial DNA. In some applications, greater than 97% of all reads mapped to mitochondrial DNA44, making it difficult and costly to capture sufficient reads mapping to the desired nuclear genome. Additionally, low signal-to-background ratios in many cell types and contexts made application of ATAC-seq to certain experimental systems difficult or impossible. To overcome these challenges, researchers developed cell type-specific optimizations including the use of different detergents for cellular lysis such as digitonin37 or Triton X-10068, the addition of more transposase enzyme to the assay69, or the use of clathrin inhibitors such as Pitstop 270. However, these optimizations were targeted for specific applications and do not necessarily extrapolate well to a wide variety of cell types, highlighting the need for more broadly applicable protocol optimizations. We have previously developed a generalizable and optimized version of the ATAC-seq method, called Omni-ATAC36, that removed many of the cell- or context-specific problems limiting broad application of ATAC-seq.
Development of the Omni-ATAC protocol
The Omni-ATAC protocol improved upon the original ATAC-seq methodology by reducing reads mapping to mitochondrial DNA and increasing signal-to-background ratios across diverse cell lines, tissues, and frozen samples36. This was achieved through improvements to cell lysis, nuclei isolation, and transposition. The optimizations in the Omni-ATAC protocol enable lysis of diverse cell types by including Tween-20 and digitonin in addition to Nonidet P40 (NP40). NP40 serves as the primary permeabilization reagent, allowing permeabilization of both plasma and nuclear membranes. Digitonin is a steroidal saponin that binds to cholesterol and related molecules that are highly enriched in the plasma membrane compared to intracellular membranes. Because of this, digitonin efficiently permeabilizes the plasma membrane but not the nuclear or mitochondrial membranes. Tween-20, which on its own is a weak permeabilization agent, serves primarily to prevent non-specific binding of mitochondria to the nuclear membrane. In combination, these detergents provide broad-spectrum lysis of diverse cell types without over lysis. The introduction of a post-lysis nuclei wash step with Tween-20 serves to remove mitochondria from the transposition reaction, thereby increasing library complexity and reducing sequencing costs. Omni-ATAC also includes Tween-20 and digitonin in the transposition reaction at 37°C, ensuring adequate lysis of harder-to-lyse cell types. Finally, the addition of PBS to the transposition reaction simultaneously increases the signal-to-background ratio and the total number of unique fragments recovered. We believe that this effect is driven by the chaotropic nature of PBS which may make the chromatin slightly more accessible for transposition. In combination, these optimizations enable the identification of more transposase-accessible peaks, improve data quality when profiling low cell numbers (>500 cells), and establish a single protocol that can be applied to a wide variety of cell types, including many that could not be assayed using the original ATAC-seq protocol. The protocol described below represents an updated version of the Omni-ATAC protocol, incorporating a more modern polymerase, reducing the amount of primer used in amplification, changing how pre-amplification is performed, and providing more explicit guidelines for library preparation and proper amplification of ATAC-seq libraries (Supplementary Fig. 1a-c).
Experimental Design
Brief overview
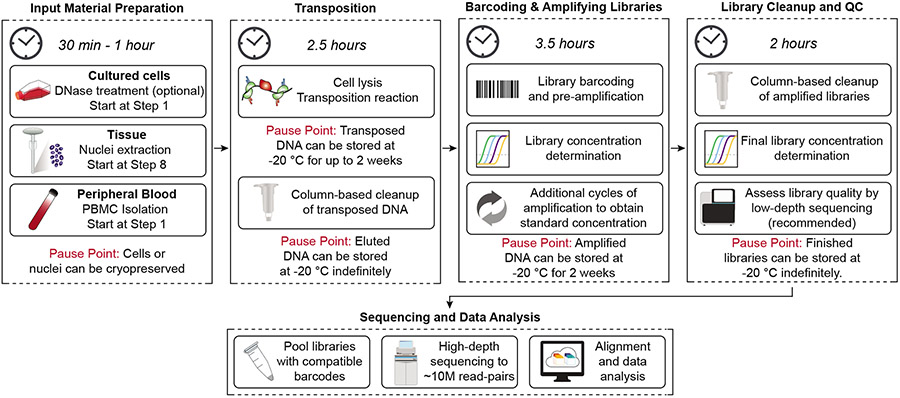
ATAC-seq can be summarized in five stages: input material preparation, transposition, library preparation, sequencing, and data analysis. This protocol focuses in detail on the transposition and library preparation stages - with guidance for how to obtain the proper input material and how finalized libraries should be sequenced and analyzed. We refer readers to several established bioinformatics pipelines and software tools for ATAC-seq analysis for this final stage 71-73. See the Data Analysis section for an overview of the analysis workflow. An overview of the experimental wet-lab protocol, with possible pause points, is presented in Fig. 2.
Figure 2: Schematic overview of ATAC-seq protocol.
The primary protocol steps are shown in sequence, along with approximate timing and pause points.
Briefly, ATAC-seq library preparation involves (i) transposition into native chromatin, (ii) barcoding and amplification of transposed DNA, and (iii) purification and quantification of libraries for sequencing. After sample collection (see Input material preparation section), cells are lysed, releasing nuclei. The nuclear membrane is permeabilized in a buffer containing NP40, Tween-20, and digitonin. Transposition, using Tn5 enzymes complexed with double-stranded oligos containing PCR-compatible handles (Supplementary Fig. 2), is performed at 37°C and the resulting DNA fragments are purified and barcoded by PCR. Finally, to ensure optimal amplification of all samples across conditions, the concentration of the pre-amplified library is quantified by qPCR and the necessary number of extra PCR cycles is determined. After this final amplification, ATAC-seq DNA is purified and ready for high-throughput sequencing. Below we detail some aspects of the experimental design to consider when setting up an ATAC-seq experiment.
Input material preparation
One of the most important considerations for a successful ATAC-seq experiment is the quality of the input material. The Omni-ATAC protocol is robust to a variety of mammalian cell and tissue types; however, some optimization of sample preparation may be necessary. Input material can be as low as 500 cells (or nuclei), although we find that optimal results are obtained with 50,000 cells and a comfortable lower limit for most researchers would be 5,000 cells. This protocol starts from fresh or cryopreserved whole cells or nuclei. Flash-frozen cell pellets or formaldehyde-crosslinked cells generally give lower-quality libraries, thus input cells should either be freshly cultured or stored frozen in a cryopreservative, such as BAM Banker (see Reagents list). Other similar, serum-free cryopreservatives would also be suitable. We describe the most common input material types and provide general recommendations for handling. Note that the recommendations below are for mammalian cells, although ATAC-seq, with some modifications, has been applied to plants74,75, whole Caenorhabditis elegans45, Drosophila embryos76, yeast61, and many other non-mammalian systems.
Cultured cells:
If using freshly cultured mammalian cells, ensure that the cells are viable – dead cells often have large amounts of unchromatinized DNA which is highly accessible to Tn5 and thus increases background noise. If the cells are >85% viable, pre-treatment with DNase can remove any free-floating DNA (see Box 1). For samples with lower viability, we recommend using a ficoll gradient to remove dead cells (see Reagents list for product and associated protocol) , or fluorescence-activated cell sorting (FACS) against a marker of dead cells such as 4′,6-diamidino-2-phenylindole (DAPI), propidium iodide (PI), or Annexin V77. Adherent cells can be lifted off the plate with Trypsin or via other methods that are suitable for the cell type of interest while retaining viability. Suspension cells can be collected directly by centrifugation. Cells can also be cryopreserved and thawed at a later date for use in ATAC-seq. We recommend cryopreservation in aliquots of ~60,000 cells (accounting for some loss) in 100 μL of cryopreservative (see Cryopreserved cells or nuclei).
Box 1: DNase treatment of cultured cells.
For optimal data quality, ensure that the cells are viable prior to performing ATAC-seq. We recommend viability above 90%, preferably around 95%. To deplete highly viable cultures (85-95% viable) of free-floating DNA that originates from dead cells, cells can be treated with DNase, which may increase data quality. Cultured cells can be treated directly on the plate, adding DNase to a final concentration of 200 Kunitz units/mL directly to the cell culture media followed by incubation at 37°C for 30 minutes in a cell culture incubator. Be sure that the culture media lacks EDTA, as DNase needs divalent cations for its activity. Remove 65,000 cells per ATAC-seq reaction (assuming a 30% loss of cells with spinning and washing) and place in a 1.5 mL LoBind tube. Fill the tube with sterile PBS. Centrifuge the cells at room temperature for 5 minutes at 350 g. Remove the supernatant and resuspend the cells in 1 mL of sterile PBS. Centrifuge again at room temperature for 5 minutes at 350 g. The resulting cell pellet can be used as input to the ATAC-seq protocol, starting at Step 9. If clear cell pellets are not obtained in PBS, try adding Tween-20 to a final concentration of 0.1% (wt/vol) or BSA to a final concentration of 0.5% (wt/vol).
Whole Blood:
Peripheral blood mononuclear cells (PBMCs) should be isolated from whole blood, either by a ficoll gradient or similar approaches. Because of their unique chromatin structure78 and high levels of endogenous proteases, neutrophils present a challenge in ATAC-seq. We recommend ensuring the complete removal of neutrophils prior to performing ATAC-seq as it has been shown to greatly improve ATAC-seq data quality79. If using cryopreserved PBMCs, we recommend pre-treating the thawed cells with DNase prior to ficoll or FACS as described in Box 1.
Tissues:
For complex tissues, including flash-frozen tissue, we recommend extracting nuclei from the tissue prior to starting the transposition reaction, as direct tissue lysis with the ATAC-seq Lysis Buffer is not efficient. We have previously established a nuclei isolation protocol which performs well for a variety of human and non-human tissues36,80 and we include a detailed version of this protocol in Supplementary Protocol 1. When collection of samples makes the use of fresh tissue impossible, tissues should be flash-frozen or stored in cryopreservative, prior to nuclei extraction. A picture of nuclei input material is given in Supplementary Fig. 3a. If using Supplementary Protocol 1 for nuclei extraction, the resulting high-quality nuclei can be pelleted and treated as the input material for transposition, beginning at Step 8 of the main Procedure. Because these nuclei were isolated in the presence of NP40, there is no need for additional lysis. Nuclei can also be cryopreserved. We recommend cryopreservation in aliquots of ~60,000 nuclei (accounting for some loss) in 100 μL of cryopreservative which can be used as described below (see Cryopreserved cells or nuclei and Supplementary Protocol 1). We note that many nuclei isolation protocols have been published and certain tissue types require specialized procedures for nuclei isolation. Table 2 provides a non-exhaustive list of references for a variety of nuclei isolation protocols that could be considered as alternatives.
Table 2:
Published nuclei extraction protocols
| Input material | Method of isolation & purification |
Reference | Notes |
|---|---|---|---|
| Cells, tissues | Douncing; iodixanol gradient centrifugation | Supplementary Protocol 1 | Validated for use with ATAC-seq for a wide range of tissues, especially those with low extracellular matrix content (ECM). Protocol is derived from several methods36,38,143. |
| Tissues with high ECM or vasculature content | Incubation in lysis buffer and gentle trituration with 5-mL serological pipette | Yang et al. 2021144 | Good for difficult-to-dissociate tissues, validated for choroid plexus scRNA-seq; can capture vascular cells often lost in other isolation methods. |
| Tissues; enteric neurons | Chopping with scissors on ice + FACS | Drokhlyansky et al. 2020145 | Validated for mouse enteric neurons in scRNA-seq; retains a larger percentage of ER associated RNAs. |
| Tagged nuclei (INTACT) | Expression of nuclear targeting fusion protein; bead purification. | Deal and Henikoff 2010 75,146. | Developed originally for plants, but has been applied to many systems. |
| Fibrous tissues (PAN-INTACT); validated for heart and kidney | Douncing; filtration; sucrose cushion centrifugation | Bhattacharyya et al. 2019147 | Show that changing the concentrations of the sucrose gradient can enrich for different cell types. |
| PBMCs, cell lines | Lysis and centrifugation | 10x Demonstrated Protocols148 | Validated for use with the scATAC 10x system. |
| Plant cells | Sucrose sedimentation; streptavidin bead separation of tagged nuclei | Bajic et al. 201874 | Has been used in several plant types, including Arabidopsis, tomato, and rice. INTACT system can be applied to other non-plant systems, supposing that they have a nuclear tag. For example, the use of the SUN1-GFP tag in mammalian cells149. |
| Yeast | Spheroplast centrifugation or homogenization or manual rupture of frozen cells | Kiseleva et al 2007150 | Several options given; similar to other published protocols61,151 |
| Tissues, especially brain tissue. | Mild fixation, douncing, sucrose centrifugation and FACS. | Nott et al. 2021152 | Validated for brain tissues; FACS sorting to obtain cell type specific nuclei (e.g. NeuN, PU.1 or OLIG2 staining). Great for selecting a target cell of interest in the brain. Similar strategy used in other papers, without fixation153,154. |
| Drosophila embryos | Freeze, lysis buffer and crush | Haines and Eisen 2018155,155 | Based on protocols similar to the one described in this publication, but with embryo specific lysis steps. |
| Caenorhabditis elegans | Based on the INTACT methods. Biotin-labeling of nuclear envelope. | Steiner et al. 2012156 | Especially for muscle tissue. Also used for Drosophila. Another method for C. elegans germ cells157 |
Cryopreserved cells or nuclei:
If cryopreserving cells, it is important to ensure that the cells are viable prior to cryopreservation. Generally speaking, it is the cell state at the time of cryopreservation that will be assessed by ATAC-seq. Upon thaw, some plasma membrane lysis will occur but the nuclear membrane and chromatin will be preserved if cells or nuclei are maintained at 4 °C. We therefore do not worry about cells “dying” during the thaw. To thaw aliquots of ~60,000 cryopreserved cells/nuclei, we recommend adding 1 mL of cold ATAC-seq Wash Buffer directly to the 100 μL of frozen cryopreservative. Incubate on ice until the cryopreservative has thawed, and proceed to Step 3 for cells or Step 8 for nuclei. We use one aliquot of ~60,000 cell/nuclei for a single ATAC-seq reaction and assume that this results in ~50,000 cells/nuclei accounting for the extra handling and centrifugation steps involved in freezing and thawing. To thaw cells/nuclei which have been cryopreserved in aliquots of more than ~60,000 cells/nuclei, we recommend thawing the cryopreservative on ice, counting cell density if needed, removing the desired number of cells/nuclei, washing them in 1 mL of cold ATAC-seq Wash Buffer, and proceeding to Step 3 for cells or Step 8 for nuclei. We find that nuclei stored in cryopreservative can be freeze-thawed multiple times with only very moderate effects on downstream data quality.
Input types that do not work for ATAC-seq:
As of the writing of this manuscript, application of ATAC-seq to formalin-fixed paraffin-embedded (FFPE) tissue is not widely established, though published methods have been developed 81. However, even when using methods that have been tailored to FFPE, ATAC-seq library quality from FFPE tissues remains much lower than other input material types. Alternative non-ATAC-seq methods for profiling chromatin accessibility in FFPE tissues have also been developed82. Nevertheless, we recommend avoiding the use of flash-frozen cell pellets, formaldehyde-crosslinked cells, or FFPE tissues, if possible, as these methods yield lower-quality ATAC-seq data and are not well-suited as input to the Omni-ATAC protocol.
Biological versus technical replicates
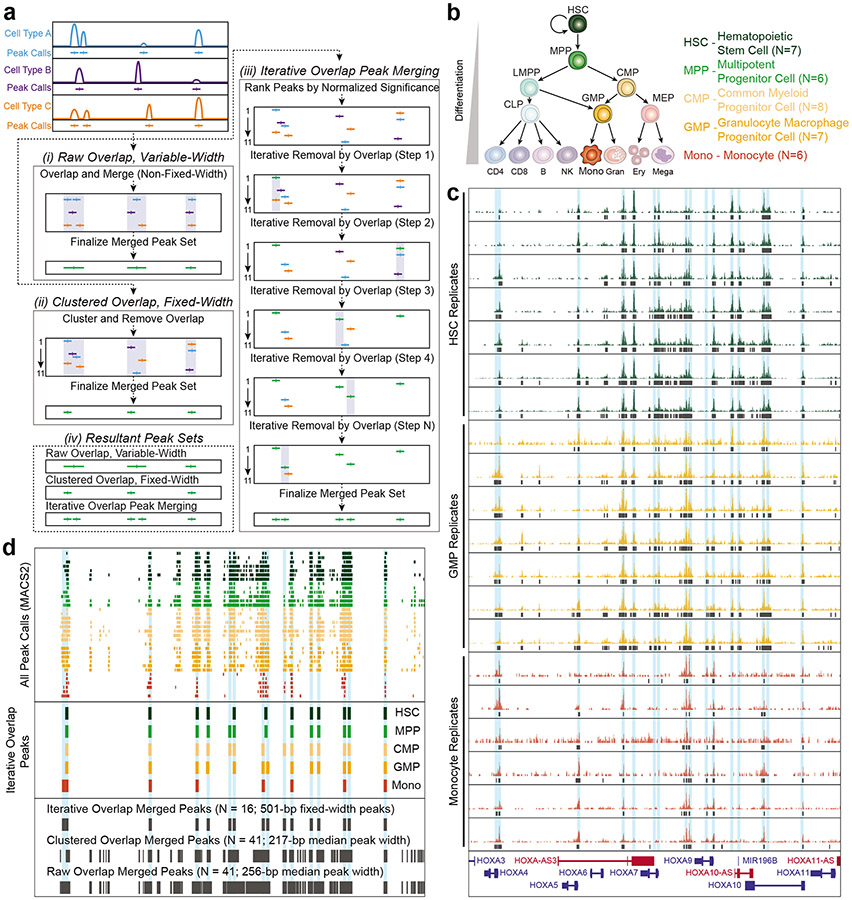
We generally advise the use of biological rather than technical replicates, when resources are constraining. When the number of available biological replicates is constraining, it may be advantageous to perform 2-3 technical replicates to ensure reproducible peak capture. Technical replicates, i.e. those derived from the same biological material, should be combined at the peak merging step of analysis (see Peak calling, merging and counting and annotation). Additionally, it is important to note that many differential accessibility tools will expect at least two biological samples per condition for accurate modeling and statistics.
Tn5-to-cell Ratio
Recent work has shown that the ratio of Tn5 to cells has an effect on data quality83. To standardize the transposition reaction, we suggest keeping the ratio of cells to Tn5 constant across all samples by standardizing the number of cells or nuclei in the ATAC-seq reactions. This helps to ensure similar TSS scores across different input amounts (Supplementary Fig. 4). The Tn5 ratio used in the main protocol assumes the use of 50,000 cells in the reaction. Refer to Supplementary Note 2 if using more or fewer than 50,000 cells.
Quality control of ATAC-seq libraries
We strongly recommend determining the quality of final ATAC-seq libraries through low-depth sequencing (50,000 – 100,000 read-pairs per sample). The success of ATAC-seq library generation is determined by four key factors: (i) the enrichment of transposase insertions in regions of known chromatin accessibility (signal-to-background ratio), (ii) the total number of unique fragments (library complexity), (iii) the ratio of sequencing reads mapping to the nuclear genome (desired) versus those mapping to the mitochondrial genome (undesired), and (iv) the fragment size distribution.
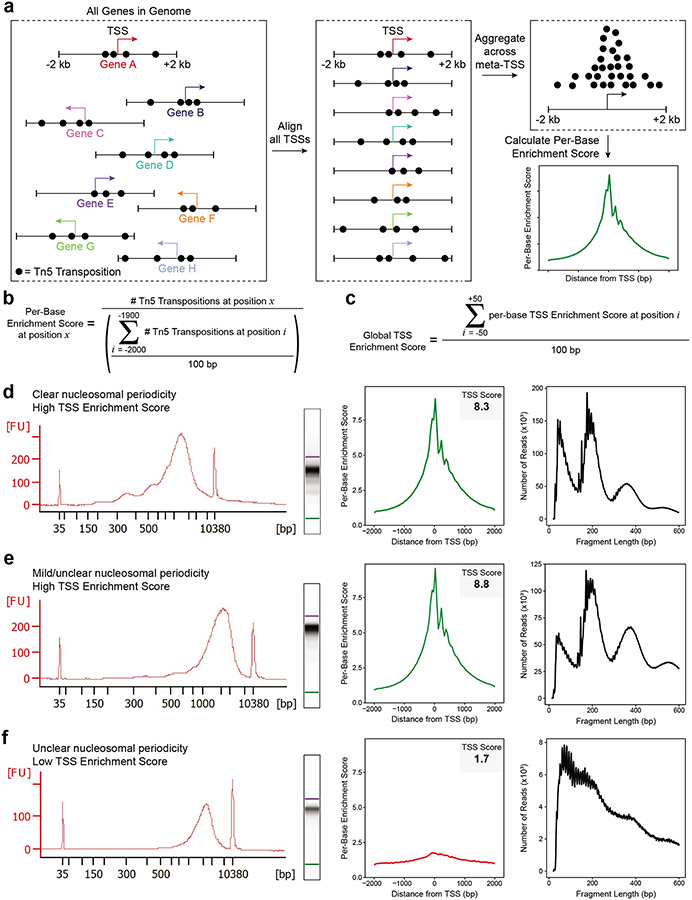
Of these, the signal-to-background ratio is the single-most important quality control metric for ATAC-seq. Other epigenomic assays such as ChIP-seq have often used the fraction of reads in peaks (FRiP) to determine signal-to-background ratios. This requires a priori knowledge of the location of peak regions or sufficient sequencing depth to call peak regions on a per-sample basis, the latter of which is flawed as it yields incomparable results across samples of differing sequencing depths. Instead, we summarize this signal-to-background ratio using a TSS Enrichment Score which quantifies the relative enrichment of signal in regions surrounding transcription start sites (TSSs) (Fig. 3a-c) because these promoter regions are known to be consistently enriched in accessible chromatin32. Accurate TSS Enrichment Scores for an ATAC-seq library can be obtained from as few as 50,000 read-pairs. TSS Enrichment Scores were introduced in the original description of ATAC-seq and have been adopted as the standard by multiple ATAC-seq pipelines72,73,84 (see Data Analysis section). It is important to note that the numeric value of the TSS Enrichment Score is dependent on the set of TSSs used in the calculation, which means that scores may not be directly comparable across different tools or species and each pipeline will provide recommendations for passable scores.
Figure 3: Assessing ATAC-seq library quality.
a. A schematic showing transposition events at a set of genes. The transcriptional start site (TSS) is depicted as an arrow with the direction of the arrow indicating whether the gene is present on the plus or minus strand. Each transposition event is shown as a black circle. Each gene has been clipped to include the region +/− 2 kb from the annotated TSS, and this region will be used in the calculation of the TSS Enrichment Score. Next, the TSS location and direction for every gene in the genome are aligned and the reads are aggregated across all of these sites into a ‘meta-TSS’. This then allows for the calculation of a per-base enrichment score via the equation in (b), which can be plotted along the +/− 2 kb of the meta-TSS, as shown in the TSS Enrichment Score graph in the bottom right. b. The equation, implemented by the PEPATAC pipeline, for calculating the per-base enrichment score. c. The equation, implemented by the PEPTATAC pipeline, for calculating the global TSS Enrichment Score for the whole library. d-f. From left to right: Bioanalyzer electropherogram trace and digitally rendered gel (left), TSS enrichment plot (middle), and fragment size distribution (right). Shown for (d) a successful ATAC-seq library with a high TSS Enrichment Score (8.3) and clear nucleosomal periodicity in the Bioanalyzer trace, (e) a successful ATAC-seq library with a high TSS Enrichment Score (8.8) but minimal observed nucleosomal periodicity in the Bioanalyzer trace, and (f) an unsuccessful ATAC-seq library with a low TSS Enrichment Score (1.7) and no clear nucleosomal periodicity in the Bioanalyzer trace. Note that all three libraries (d-f) have abundant high molecular weight fragments (>2000 bp) on the Bioanalyzer trace (left) but these fragments do not cluster on the sequencer and thus are not represented in the fragment size distributions derived from the corresponding sequencing data (right). In (e), despite not being easily observed on the Bioanalyzer trace, the characteristic fragment size distribution is observed using low-depth sequencing. In (f), the TSS Enrichment Score and fragment size distribution plots on the right indicate a poor signal-to-background ratio and low sample quality, respectively, marking this library a poor candidate for high-depth sequencing.
Low-depth sequencing can also be used to assess the ratio of sequencing reads mapping to the nuclear and mitochondrial genomes and the fragment size distribution. All of these metrics are standard outputs of the ATAC-seq pipelines recommended below. If low-depth sequencing is not readily available, the shape of a Bioanalyzer or TapeStation trace can be used to determine the fragment size distribution. While we do not recommend using this as a proxy for quality, it is often true that the library is of passable quality if it shows some nucleosomal periodicity (Fig. 3a). However, we would caution that some good quality libraries do not have the clear nucleosomal periodicity banding and this does not necessarily imply that they have been under transposed. For example, two libraries with similar TSS Enrichment Scores (8.3 and 8.8, respectively) have different Bioanalyzer traces, and the relative absence of the nucleosomal banding alone does not imply a library has a lower TSS Enrichment Score (Fig. 3d-e). Compared to the original ATAC-seq method, the Omni-ATAC protocol generates ATAC-seq libraries that are biased towards higher molecular weight fragments, likely due to the inclusion of PBS in the reaction. Occasionally, the ratio of higher molecular weight fragments to lower molecular weight fragments is sufficiently high to obscure the nucleosomal periodicity of the lower molecular weight fragments. However, the presence of these high molecular weight fragments does not imply that the library is under-transposed. These larger fragments do not cluster on Illumina sequencers and can be safely ignored (Fig. 3d-f).
Sequencing of final ATAC-seq libraries
ATAC-seq data should always be sequenced using paired-end sequencing because each end of an ATAC-seq fragment corresponds to a unique Tn5 transposition event (Fig. 1b). Therefore, using single-end sequencing effectively ignores 50% of the data. For high-depth sequencing of final libraries we recommend targeting 10 million read-pairs (10 million clusters passing filter). This is sufficient for most routine analyses including differential accessibility analysis and motif enrichment analysis. However, when genotyping of the ATAC-seq data will be performed, longer reads can be used to capture more DNA sequence. Detailed guidelines are provided in Table 3. Because both ends of the ATAC-seq fragments have 8-bp barcodes, two 8-bp indexing reads should be performed. We provide adapter and barcode sequences in Supplementary Table 2. The minimum read length will be dependent on the particular application of the ATAC-seq library; for most standard applications, 36 base pairs is sufficient and provides equivalent mapping efficiency to longer reads from a variety of input materials (Supplementary Table 1). Mapping rates and TSS scores for a variety of different read lengths are provided in Supplementary Table 1.
Table 3:
Guidelines for sequencing parameters
| Application | Insight gained | Minimum Read Length† |
Index Length* |
Paired- or single- end |
Read- pairs per sample |
|---|---|---|---|---|---|
| Gene regulatory landscape profiling | Peaks, differential peaks between samples, motif analysis of peaks | 36 bp | 8 | Paired | 10 million |
| Genotyping | Gene regulatory landscape + genotype of sample; useful for patient samples and to determine if sequence variants affect a peak. | 100 bp | 8 | Paired | 10 million |
| Footprinting Analysis | Footprinting of different TFs to determine binding sequence at base-pair resolution | 36 bp | 8 | Paired | 200 million |
| Nucleosome occupancy | Location of nucleosomes along DNA | 36 bp | 8 | Paired | 60 million |
Longer read lengths can be used but may not provide additional gains. We find that 36 bp is sufficient to map transposition sites. Given that most ATAC-seq fragments are less than 200 bp in length, sequencing reads longer than 100 bp do not provide much additional data.
Index length assuming the barcodes listed in Supplementary Table 2 are used.
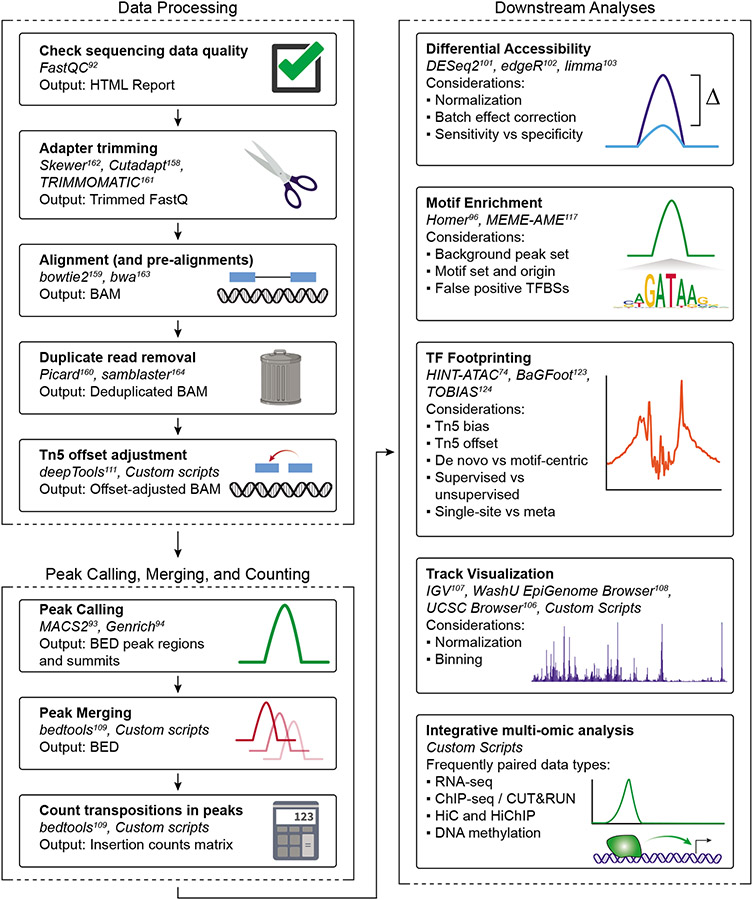
Data Analysis
After sequencing, we recommend using publicly available pipelines to perform alignment and downstream analysis (see Anticipated Results section). PEPATAC73 is a highly portable and user-friendly pipeline that provides multiple quality metrics including the TSS Enrichment Score. ENCODE72 and nf-core84 also provide similarly robust pipelines for ATAC-seq analysis. Any of these pipelines can be used for analysis of low-depth quality control data or high-depth sequencing data. Regardless of the pipeline, ATAC-seq data analysis follows these general steps: pre-alignment sequence quality control, trimming of adapter sequences, alignment to the reference genome of interest, removal of reads mapping to mitochondrial DNA and PCR duplicates, and peak calling. This is often followed by more application-specific analyses such as differential accessibility testing, motif enrichment analysis, and integration with other data types such as RNA-seq, ChIP-seq, or chromatin conformation capture.
Single-cell ATAC-seq
The Omni-ATAC protocol described here was designed for bulk ATAC-seq. However, the commercially available scATAC-seq products from 10x Genomics utilize similar lysis and transposition reaction conditions. For researchers interested in performing scATAC-seq, we recommend following the manufacturer instructions for standard cell types. For frozen tissues, we provide recommendations for how to isolate nuclei to use as input to the 10x Genomics scATAC-seq kit in our nuclei isolation protocol(Supplementary Protocol 1)80.
Necessary expertise
Basic molecular biology skills are necessary to perform ATAC-seq. Sequencing of the final ATAC-seq libraries requires the use of high-throughput sequencing platforms usually found in a genomics core facility. For data analysis, access to a computational server or high-performance computing environment may be necessary, as processing data on this scale can be computationally intensive for most standard computers. Basic familiarity with command-line tools and bioinformatics is necessary for pipeline implementation.
Limitations
As discussed above, ATAC-seq gives a broad picture of the epigenetic landscape of a sample; however, it cannot provide specific details about the exact mechanism of action (i.e. what chromatin marks or TFs are present on the region of interest). As such, ATAC-seq data often benefits from correlation with other data types for interpretation. Additionally, ATAC-seq is not very well suited for TF footprinting of an individual genomic locus, which is used to identify the precise base-pair resolution sequence bound by a TF. Such goals would require very high-depth sequencing data and may be better served by other techniques such as DNase-seq27-29. Similar to DNase-seq28,85, ATAC-seq is known to have sequence biases, based on the insertion preferences of Tn532. Additionally, the protocol detailed here obtains ATAC-seq data from bulk cell populations, and thus presupposes that there is limited intercellular heterogeneity or that this heterogeneity is not of interest. If the input material is highly heterogeneous, for example a complex tissue, then the resultant ATAC-seq profile will represent the average signal of all the cells and cell types, thus lacking signatures of rare cell types (<20% of the total cells)53. To adequately capture this heterogeneity, it is either necessary to perform single-cell/single-nucleus ATAC-seq (snATAC-seq)86,87 or to FACS-purify the desired population of cells and perform bulk ATAC-seq on that purified population. Finally, it is important to note that ATAC-seq can only help give regulatory insights for phenomena that are regulated at the transcriptional level and will inherently miss any post-transcriptional changes such as RNA-decay or modification, export, or translation.
MATERIALS
Biological Materials
Cell line(s) or input material of interest.
Reagents
Phosphate-Buffered Saline (Thermo Fisher Scientific, cat. no. 10010049)
1M Tris-HCl pH 7.5 (Thermo Fisher Scientific, cat no. 15-567-027)
5M NaCl (Corning, cat. no. 46-032-CV)
1M MgCl2 (Invitrogen, cat. no. AM9530G)
UltraPure DNase/RNase-Free Distilled Water (Thermo Fisher Scientific, cat. no. 10977015)
-
2% Digitonin wt/vol (Promega, cat. no. G9441)
CAUTION: Digitonin solutions can cause skin and eye irritation. Handle using appropriate protective gloves.
CRITICAL: Digitonin solutions can be difficult to make and the dry reagent is acutely toxic. For best results, purchase 2% digitonin and dilute as described in the reagent setup section.
-
10% Tween-20 wt/vol (Millipore-Sigma, cat. no. 11332465001)
CRITICAL: 100% Tween-20 can be difficult to dilute accurately. For best results, purchase 10% Tween-20.
-
10% Nonidet P40 Substitute wt/vol (Millipore-Sigma, cat. no. 11332473001)
CAUTION: NP40 can cause skin and eye irritation. Handle using appropriate protective gloves.
CRITICAL: 100% Nonidet P40 Substitute can be difficult to dilute accurately. For best results, purchase 10% NP40.
Sequencing Adapters (IDT, see Supplementary Table 2)
-
NEBNext Ultra II Q5 2x Master Mix (New England Biolabs, cat. no. M0544S)
CRITICAL: We have optimized the ATAC-seq library preparation using the NEBNext Ultra II Q5 2x Master Mix. Other PCR master mixes may be substituted, but optimization may be necessary.
-
Tagment DNA TDE1 Enzyme and Buffer Kit (Illumina, cat. no. 20034197 [48 reactions] or 20034198 [96 reactions])
CRTICAL: We have optimized the volume of Tn5 transposase per ATAC-seq reaction based on products purchased from Illumina. As the manufacturer does not provide an enzyme concentration, it is difficult to translate this to other Tn5 preparations. Thus, although other suppliers can be used, this will require additional optimization. It is also possible to produce and purify Tn5 enzyme in the laboratory (see Supplementary Note 3).
DNA Clean and Concentrator-5 Kit (Zymo Research, cat. no. D4014)
NEBNext Library Quant Kit (New England Biolabs, cat. no. E7630)
BAM Banker Cryopreservative (Fisher Scientific, cat. no. NC9582225)
DNase (optional; Worthington, cat. no. LS002007)
Ficoll Paque Plus (optional; Cytvia Life Sciences cat. no. 17144002)
Hanks Balanced Salt Solution, with calcium and magnesium, no phenol red (for DNase, optional; Thermo Fisher Scientific, cat no. 14025092)
Qubit 1X dsDNA HS Assay Kit (optional; Thermo Fisher Scientific, cat. no. Q33231)
Equipment
2 μL, 20 μL, 200 μL, and 1,000 μL single-channel Pipettes (Rainin, cat. nos. 17014393, 17014392, 17014391, and 17014382)
20 μL, 200 μL, and 1,000 μL filter tips (Rainin, cat. nos. 17014961, 17014963, and 17014967)
5 mL, 10 mL, 25 mL, and 50 mL Serological pipettes (Thermo Fisher Scientific, cat. nos. 07-200-573, 07-200-574, 07-200-575, and 07-200-576)
Pipet-Aid XP (Drummond, cat. no. 4-000-101)
-
1.5 mL DNA LoBind tubes (Eppendorf, cat. no. 4043-1021)
CRITICAL: LoBind tubes should be used to maximize sample recovery of nucleic acids as they reduce sample-to-surface binding.
8-strip DNase-free PCR tubes (USA Scientific, cat. no. 1402-4700)
Vortex mixer (VWR, cat. no. 97043-562)
CFX Opus 384 Real-Time PCR System (Bio-Rad, cat. no. 12011452)
C1000 Touch thermal cycler (Bio-Rad, cat. no. 1851148)
MyFuge 12 tabletop centrifuge (Benchmark Scientific, cat. no. C1012)
Microcentrifuge (Eppendorf, cat. no. 2231000768)
Refrigerated microcentrifuge (Eppendorf, cat. no. 5404000413)
15 mL and 50 mL conical tubes (Thermo Fisher Scientific, cat. nos. 05-538-53F and 05-538-55A)
1.5 mL Microcentrifuge tubes (Thermo Fisher Scientific, cat. no. 14-222-155)
384-well plates (Bio-Rad, cat. no. HSP3905)
Microseal ‘B’ PCR plate sealing film (Bio-Rad, cat. no. MSB1001)
MPS 1000 plate centrifuge (Labnet International, cat. no. C1000)
Thermomixer (Eppendorf, cat. no. 2231000680)
0.22 μm 250 mL cellulose nitrate sterilizing filter (Corning, cat. no. 430756)
0.22 μm 50 mL PVDF Steriflip-GV Sterile Centrifuge Tube Top Filter Unit (for DNase, optional; Millipore-Sigma, cat. no. SE1M179M6)
Qubit 4 fluorometer (optional; Thermo Fisher Scientific, cat. no. Q33238)
Vacuum manifold (optional; QIAGEN, cat. no. 19413)
VacConnectors sterile adapters for vacuum manifold (optional; QIAGEN, cat. no. 19407)
High-performance computing environment or similar
Software
Several options – see the Anticipated Results section and Table 4 for options.
Iterative overlap peak merging script88 – see Code Availability section.
Table 4:
Data analysis pipelines available for ATAC-seq
| Step/Process | ENCODE ATAC-seq | PEPATAC | nf-core atacseq |
|---|---|---|---|
| Version used in comparison | v1.10.0 | v0.10.0 | v1.2.1 |
| Environment | Cromwell/caper | Pypiper | Nextflow |
| Trimming, Alignment, and Deduplication | Cutadapt158 bowtie2159 Picard160 | TRIMMOMATIC161 or skewer162 bowtie2159 or BWA163 samblaster164 or Picard | TrimGalore!165 BWA Picard |
| Tn5 offset correction | Yes | Yes | No |
| Mitochondrial filter | Yes | Yes | Yes |
| Peak calling method | MACS293 | MACS2 (default), F-seq166, or Genrich94 | MACS2 |
| Peak merging method | Based on the irreproducible discovery rate (IDR) for replicates – does not merge for a whole set of samples | Fixed-width, iterative overlap | Raw peak overlap using bedtools109 merge |
| Output provided | BAM files, bigwig files (one representing fold enrichment over expected background and the other representing statistical significance), BED file of peaks for each file and for the merged peak set | QC plots including alignment scoring, TSS scores and library complexity, BED peaks and counts, bam files, bigwig files (nucleotide resolution and smoothed) | QC html report, bam files, normalized bigwig files, BED peaks, annotation of peaks (HOMER), merged peak set, differential accessibility (DESeq2), IGV output. |
| Code repository | https://github.com/ENCODE-DCC/atac-seq-pipeline | https://github.com/databio/pepatac | https://github.com/nf-core/atacseq |
REAGENT SETUP
ATAC-seq Resuspension Buffer (ATAC-RSB)
For 100 mL, combine 1 mL of 1M Tris-HCl pH 7.5, 200 μL of 5M NaCl, 300 μL of 1M MgCl2, and 98.5 mL of UltraPure Distilled Water. Filter sterilize using a 0.22 μm filter. The final composition of ATAC-RSB is 10 mM Tris-HCl pH 7.5, 10 mM NaCl, and 3 mM MgCl2 in water. Store at 4 °C for up to 6 months.
Digitonin
The recommended digitonin from Promega is supplied at a 2% wt/vol concentration in DMSO. Dilute this digitonin 1:1 with water to make a 1% wt/vol (100x) stock solution. This DMSO-water mixture will no longer freeze at −20 °C. Store at −20 °C for up to 6 months.
Tn5 Transposase Enzyme and 2x Tagment DNA (TD) Buffer
The Tn5 transposase enzyme pre-complexed with adapters (TDE1) and the 2x Tagment DNA (TD) Buffer should be purchased from Illumina (see Materials). We strongly recommend purchasing these reagents as they are the core components of the reaction. However, protocols for producing, purifying, and complexing the Tn5 transposase enzyme have been published89 and are discussed briefly in Supplementary Note 3.
NEBNext Library Quantification Kit
Following manufacturer instructions, add the primer to the enzyme mix. If using ROX for normalization, add ROX as directed by the manufacturer.
Barcoding Adapter Resuspension
If ordering the barcoding adapters (Supplementary Table 2) reconstitute the oligonucleotides to 100 μM stock solutions in water and make 5 μM working stock solutions with water. Both solutions can be stored at −20 °C for five years or more.
DNase Resuspension
If cells will be pre-treated with DNase (optional), lyophilized DNase should be resuspended in Hanks Balanced Salt Solution to 20,000 Kunitz units/ml for a 100x stock solution. Filter sterilize this solution using a 0.22 μm PVDF membrane. Aliquot in 100 μL – 1 mL volumes (according to the application) and store at −20 °C for up to 2 years. Once thawed, an aliquot can be stored at 4 °C for up to 1 month but should not be refrozen.
PROCEDURE
Transposition (Timing: 2.5h for ~12 samples)
-
Prepare samples according to the Input material preparationsection above. Here, we assume that 50,000 freshly cultured cells have been collected in a 1.5 mL LoBind tube for each sample. For the purposes of this protocol, we refer to the starting material as cells but the same protocol applies to nuclei. See Box 1 for optional DNase treatment.
CRITICAL STEP: We recommend piloting the cell/nuclei isolation ahead of time to make sure that visible pellets can be obtained after centrifugation and to practice supernatant aspiration in Step 4. If using the recommended 50,000 cells/nuclei, a small pellet should be visible.
?TROUBLESHOOTING
Prior to starting, make the ATAC-seq Lysis Buffer and ATAC-seq Wash Buffer and keep them on ice. Be sure to use freshly made ATAC-seq Lysis Buffer and ATAC-seq Wash Buffer each time.
Pellet 50,000 viable cells at 500 g for 5 min at 4 °C in a fixed-angle microcentrifuge. Orient the tubes in a consistent fashion so that the pellet will be visible on the outer surface of the tube towards the bottom.
-
Aspirate all the supernatant using two pipetting steps. First, aspirate down to 100 μL with a p1000 pipette. Then, remove the final 100 μL with a p200 pipette.
CRITICAL STEP: Make sure to avoid the visible cell pellet when pipetting. Optimal removal of supernatant and minimal disruption of the cell pellet is attained when the removal of the final 100 μL is performed in a consistent and fluid motion without starting and stopping.
Resuspend the cell pellet in 50 μL of ATAC-seq Lysis Buffer by pipetting up and down three times. ATAC-seq Lysis Buffer should be made fresh each time and mixed thoroughly prior to use.
ATAC-seq Lysis Buffer
| Reagent | Volume per sample (μL) | Final conc. |
|---|---|---|
| Cold ATAC-RSB | 48.5 | |
| 10% NP40 wt/vol | 0.5 | 0.1% wt/vol |
| 10% Tween-20 wt/vol | 0.5 | 0.1% wt/vol |
| 1% Digitonin wt/vol | 0.5 | 0.01% wt/vol |
| Total volume | 50 |
-
6
Incubate on ice for 3 minutes. If lysing multiple samples, make sure that all samples are lysed for the same total amount of time by proceeding to Step 7 after 3 minutes. An example image of lysed GM12878 cells is given in Supplementary Fig. 3b.
-
7
Add 1 mL of ATAC-seq Wash Buffer to dilute the lysis reagents. Invert the tube 5 times to mix. ATAC-seq Wash Buffer should be made fresh each time and mixed thoroughly prior to use.
ATAC-seq Wash Buffer
| Reagent | Volume per sample (μL) | Final conc. |
|---|---|---|
| Cold ATAC-RSB | 990 | |
| 10% Tween-20 wt/vol | 10 | 0.1% wt/vol |
| Total volume | 1000 |
-
8
Pellet nuclei at 500 g for 10 min at 4 °C in a fixed-angle microcentrifuge. Orient the tubes in a consistent fashion so that the pellet will end up in the same location. If using nuclei isolated from frozen tissues as described in the Input material preparation section above, start the ATAC-seq protocol here using 50,000 nuclei resuspended in 1 mL of ATAC-seq Wash Buffer.
-
9
Aspirate all supernatant using two pipetting steps as above. First, aspirate down to 100 μL with a p1000 pipette. Then, remove the final 100 μL with a p200 pipette.
CRITICAL STEP: Make sure to avoid the visible cell pellet when pipetting. Optimal removal of supernatant and minimal disruption of the cell pellet is attained when the removal of the final 100 μL is performed in a consistent and fluid motion without starting and stopping.
-
10
Resuspend the cell pellet in 50 μL of Transposition Mix by pipetting up and down 6 times. Transposition Mix should be made fresh each time and mixed thoroughly prior to use.
Transposition Mix
| Reagent | Volume per sample (μL) | Final conc. |
|---|---|---|
| 2x TD Buffer | 25 | 1x |
| PBS | 16.5 | |
| UltraPure Distilled H2O | 5 | |
| 1% Digitonin wt/vol | 0.5 | 0.01% wt/vol |
| 10% Tween-20 wt/vol | 0.5 | 0.1% wt/vol |
| TDE1 Tagment DNA Enzyme (Tn5 Transposase) | 2.5 | |
| Total volume | 50 |
-
11
Incubate reaction at 37 °C for 30 minutes in a thermomixer with 1000 RPM mixing.
-
12
Remove the tubes from the thermomixer and immediately terminate the transposition reaction by adding 250 μL (5 volumes) of DNA Binding Buffer from the DNA Clean and Concentrator-5 Kit and mix well by pipetting or inversion.
CRITICAL STEP: We strongly recommend the use of different kits/reagents for the cleanup of pre- and post-amplification products so as to not contaminate post-amplification products into pre-amplification samples.
-
13
Pulse centrifuge to collect solution in the bottom of the tube.
PAUSE POINT: This solution can be stored at −20 °C for up to 2 weeks. Allow this mixture to warm back to room temperature (22 °C) and mix thoroughly before proceeding.
-
14
Clean up the transposition reaction using the DNA Clean and Concentrator-5 Kit. If using the vacuum manifold, we recommend using sterile single-use VacConnectors to prevent cross-contamination. Transfer each sample, mixed with the DNA Binding Buffer, to a Zymo-Spin Column in a collection tube. Centrifuge at room temperature for 30 seconds at 10,000 g and discard the flow through.
-
15
Add 200 μL of DNA Wash Buffer to the column and centrifuge at room temperature for 30 seconds at 10,000 g.
-
16
Repeat this wash for a total of 2 wash steps.
-
17
Perform a final “dry spin” after the second wash step to remove any traces of residual wash buffer from the column membrane. To do this, remove any flowthrough from the collection tube and centrifuge the column and collection tube at room temperature for 1 minute at >13,000 g.
-
18
Transfer the column to a clean pre-labeled 1.5 mL LoBind tube. Pipette 21 μL of Elution Buffer directly onto the column membrane and wait for 1 minute.
-
19
Centrifuge the column at room temperature for 1 minute at 13,000 g to elute the DNA. This elution volume typically results in 20 μL of product.
PAUSE POINT: This solution can be stored at −20 °C for as long as necessary.
Barcoding of transposed fragments (Timing: 30m)
-
20
Assign each sample in the study to a unique combination of the Adapter 1 and Adapter 2 sequences found in Supplementary Table 2 and record this information. These combinations will be used to assign each read-pair to its appropriate sample designation.
CRITICAL STEP: The adapters contain the dual-indexing sample-specific barcodes; therefore, each individual sample should receive a unique combination of Ad1 and Ad2. More explicitly, it is ok if two different samples share the same Ad1 as long as they can be differentiated by their Ad2, or vice versa. Samples with identical combinations of Ad1 and Ad2 cannot be sequenced together as the index reads derived from each sample will be indistinguishable.
-
21
Transfer each cleaned-up transposed DNA sample to a 200 μL PCR tube.
-
22
Add 25 μL of NEBNext Ultra II Q5 2x Master Mix to each tube.
-
23
Add in 2.5 μL of the corresponding Adapter 1 to each sample.
-
24
Add in 2.5 μL of the corresponding Adapter 2 to each sample.
-
25
Cap tubes, vortex and spin down to collect all liquid at the bottom of the tube. When completed, each reaction should contain the following:
Barcoding PCR Reaction (per sample)
| Reagent | Volume (μL) | Final conc. |
|---|---|---|
| Transposed sample | 20 | |
| NEBNext Ultra II Q5 2x Master Mix | 25 | 1x |
| 5 μM Adapter Ad1* | 2.5 | 0.25 μM |
| 5 μM Adapter Ad2* | 2.5 | 0.25 μM |
| Total volume | 50 |
See Supplementary Table 2 for adapter sequences.
-
26Run the barcoding PCR reactions according to the following cycling conditions:
Cycle no. Denature Anneal Extend 1 72 °C, 5 min 2 98 °C, 30 s 3-5 (3 cycles) 98 °C, 10 s 65 °C, 30 s 65 °C, 45 s Hold at 4 °C
-
27
Remove tubes from the thermocycler and store on ice. Proceed to the next step immediately.
CRITICAL STEP: Additional cycles of amplification will be performed directly in this reaction tube, with the same reagents, so it is critical that samples remain cold and that the next step is performed immediately.
Library quantification and amplification (Timing: 3h)
-
28
Determine how the library concentrations will be quantified. Steps 29-32 detail an optimized method of quantification using the NEBNext Library Quant Kit, although other comparable products could be utilized following the manufacturer’s recommendations. When preparing very few ATAC-seq libraries (fewer than 4), an alternative quantification method using the Qubit fluorometer (Invitrogen) may prove faster, though less precise. See Box 2.
-
29
Make a sufficient volume of 1x NEB Dilution Buffer by diluting the 10x NEB Dilution Buffer in water (~100 μL per sample).
-
30
Dilute 1 μL of pre-amplified sample with 99 μL of 1x NEB Dilution Buffer for a 1:100 dilution and mix thoroughly.
-
31
Make 10 μL qPCR reactions using the NEBNext Library Quant Kit in a 384-well plate. Run all samples, the 4 standards, and a no template control in technical duplicate.
Box 2: An alternative method for determining ATAC-seq library concentration after initial barcoding PCR.
We recommend using qPCR to determine the concentration of ATAC-seq libraries. This provides an accurate estimate of how many additional cycles of amplification should be performed to obtain an optimally amplified library. This optimal amplification paradigm ensures that enough DNA is available for sequencing but avoids over-amplification, which can introduce bias. It is also possible to estimate the number of additional cycles required via qPCR without a standard curve, as demonstrated previously90.
However, as the additional number of cycles to amplify is ultimately an estimate, a Qubit fluorometer can be used in lieu of qPCR. We find that this these estimates are best for samples using 25,000 to 100,000 nuclei/cells. For samples with fewer than 25,000 cells/nuclei, we strongly recommend using qPCR quantification, as the Qubit is often inaccurate.
To perform Qubit library quantification, after the library barcoding PCR in Step 27, use 1 μL of undiluted product as input to the Qubit dsDNA HS Assay Kit following manufacturer instructions. Determine the concentration and estimate the number of additional cycles required using the equation below (see Supplementary Fig. 5 for data detailing how it was derived). We recommend performing two individual Qubit readings per ATAC-seq reaction for increased accuracy.
Round the number of cycles to the nearest whole integer. On average, Qubit readings underestimate library concentration. This problem can be exacerbated at very low library concentrations. See an example calculation in Box 3.
Library Quantification qPCR Mix
| Reagent | Volume per sample (μL) |
|---|---|
| NEBNext Library Quant Master Mix (with Primer) | 6 |
| Diluted pre-amplified sample OR standard OR H2O control | 2 |
| UltraPure Distilled H2O | 2 |
| Total volume | 10 |
-
32Seal the plate, mix thoroughly by vortexing, pulse spin to collect sample at the bottom of each well, and run according to the following cycling conditions:
Cycle no. Denature Anneal Extend 1 95 °C, 1 min 2-36 (35 cycles) 95 °C, 15 s 63 °C, 45 s
-
33
After the qPCR is complete, use the standard curve to determine the concentration of DNA in the pre-amplified sample. New England Biolabs provides an online tool to assist in this calculation if needed at http://nebiocalculator.neb.com/. We have not found it necessary to adjust the resultant concentration based on the average library fragment size. Typical library concentrations are between 0.7 nM – 2 nM when using ~50,000 cells as input. However, the concentration will depend on many factors including input cell/nuclei quantity, viability, and the proportion of cells/nuclei lost during processing.
?TROUBLESHOOTING
-
34Use the concentration obtained in the previous step to estimate how many additional cycles are required to obtain a final yield of 240 fmoles after column-based cleanup (20 μL at 12 nM). Round the fractional cycle number to the nearest whole integer. This PCR is highly efficient so we assume a perfect doubling in concentration with each cycle. Note that we add one extra cycle in the below equation to ensure that sufficient library concentrations are obtained for all samples. See Box 3 for an example calculation.
Box 3: Example calculation for additional library amplification cycles.
Here we detail an example calculation for how to determine the additional number of cycles either by qPCR quantification (recommended) or by Qubit reading. These two examples represent the results of qPCR and Qubit performed on the same pre-amplified ATAC-seq library. The "Additional cycles” column gives the number of cycles that would be used in Step 35.
| Measurement type | Concentration | Fractional cycles | Additional cycles |
|---|---|---|---|
| qPCR (nM) | 0.34 | 4.85 | 5 |
| Qubit (ng/ μL) | 1.5 | 5.69 | 6 |
-
35Place the tubes containing the pre-amplification reaction (now containing 49 μL) back in a thermocycler without addition of any more reagents and run the required number of additional cycles for each individual sample.
Cycle no. Denature Anneal Extend 1 98 °C, 30 s 2 or more cycles (may vary between samples) 98 °C, 10 s 65 °C, 30 s 65 °C, 45 s Final cycle 65 °C, 3 min Hold at 4 °C
-
36
Remove the PCR tubes from the thermocycler and transfer the 49 μL of amplified sample to a clean pre-labeled 1.5 mL LoBind tube.
-
37
Add 245 μL of DNA Binding Buffer (from the Zymo DNA Clean and Concentrator-5 kit) to each tube and mix well by pipetting.
CRITICAL STEP: We strongly recommend the use of different kits/reagents for the cleanup of pre- and post-amplification products so as to not cross contaminate post-amplification product into pre-amplification samples.
-
38
Transfer each sample, mixed with the DNA Binding Buffer, to a Zymo-Spin Column in a collection tube. Centrifuge at room temperature for 30 seconds at 10,000 g and discard the flow through.
-
39
Add 200 μL of DNA Wash Buffer to the column and centrifuge at room temperature for 30 seconds at 10,000 g.
-
40
Repeat this wash for a total of 2 wash steps.
-
41
Perform a final “dry spin” after the second wash step to remove any traces of residual wash buffer from the column membrane. To do this, remove any flowthrough from the collection tube and centrifuge the column and collection tube at room temperature for 1 minute at >13,000 g.
-
42
Transfer the column to a clean pre-labeled 1.5 mL LoBind tube. Pipette 21 μL of Elution Buffer directly onto the column membrane and wait for 1 minute.
-
43
Centrifuge the column at room temperature for 1 minute at 13,000 g to elute the DNA. This elution volume typically results in 20 μL of product.
PAUSE POINT: This solution can be stored at −20 °C for as long as necessary.
Final library concentration determination (Timing: 2h)
CRITICAL Here, we describe the final library quantification by qPCR, which we find to be the most reliable method for determining the concentration of ATAC-seq libraries. Alternatively, Qubit can also be used to obtain an estimated library concentration for pooling samples.
-
44
Make a sufficient volume of 1x NEB Dilution Buffer by diluting the 10x NEB Dilution Buffer in water (~220 μL per sample).
-
45
Dilute the samples from Step 43 4000-fold so that they fall within the concentration range of the standards (0.01 pM to 10 pM). To do this, first dilute 40x by adding 0.5 μL library to 19.5 μL of 1x NEB Dilution Buffer. Mix well and then dilute 2 μL of this 40x diluted mixture into 198 μL of 1x NEB Dilution Buffer for a 100x dilution, creating a combined 4000x dilution.
?TROUBLESHOOTING
-
46
Make 10 μL qPCR reactions using the NEBNext Library Quant Kit in a 384-well plate. Run all diluted samples, the 4 standards, and a no template control in technical duplicate.
Library Quantification qPCR Mix
| Reagent | Volume per sample (μL) |
|---|---|
| Primer + NEBNext Library Quant Master Mix | 6 |
| Diluted pre-amplified sample OR standard OR H2O control | 2 |
| UltraPure Distilled H2O | 2 |
| Total volume | 10 |
-
47Seal the plate, mix thoroughly by vortexing, pulse spin to collect sample at the bottom of each well, and run according to the following cycling conditions:
Cycle no. Denature Anneal Extend 1 95 °C, 1 min 2-36 (35 cycles) 95 °C, 15 s 63 °C, 45 s
-
48
After the qPCR is complete, use the standard curve to determine the concentration of DNA in each of the final ATAC-seq libraries. New England Biolabs provides an online tool to assist in this calculation if needed at http://nebiocalculator.neb.com/. We have not found it necessary to adjust the resultant concentration based on the average library fragment size. Ideal library concentrations are between 15 nM – 25 nM but can range between 2 nM – 100 nM depending on the accuracy of the estimation of additional required PCR cycles.
-
49
If desired, dilute each library to 8 nM with UltraPure Distilled Water (or as low as 2 nM if needed). This facilitates pooling at equimolar quantities for sequencing. Samples are now ready for high-throughput sequencing.
-
50
We recommend using low-depth sequencing to check sample quality prior to high-depth sequencing. This will allow for calculation of the TSS Enrichment Score, observation of the fragment size distribution, and determination of the percent of reads mapping to mitochondrial DNA. See the Quality control of ATAC-seq libraries section of the Experimental Design for more details. The PEPATAC pipeline can be used to calculate the TSS Enrichment Score. Alternatively, a Bioanalyzer trace can be used to roughly estimate library quality and concentration.
?TROUBLESHOOTING
-
51After quality assessment, libraries should be sent for high-depth sequencing. If using a sequencing core facility, ask for the following cycling parameters:
Read Read 1 i7 index i5 index Read 2 Purpose ATAC-seq fragment Sample barcode Sample barcode ATAC-seq fragment Length >=36 * 8 8 >=36 * *See the Experimental Design section for ATAC-seq applications that may require longer reads.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 5.
Table 5.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1 | The cell or nuclei pellet is hard to visualize after centrifugation. | Nuclei and certain cell types can be hard to properly pellet. Pellets of 50,000 cells, even for small cell types such as B cells, should be easily visualized. | Add sterile BSA to a final concentration of 0.5% wt/vol or Tween-20 to a final concentration of 0.1% wt/vol to help cells/nuclei pellet properly. |
| 33 | There is no amplification of the ATAC-seq libraries. | the incorrect barcode/adapter sequences were used when performing the barcoding step. | Ensure that compatible barcodes were added, as detailed in Supplementary Table 2. |
| 33 | There is no amplification of the ATAC-seq libraries. | Excess ethanol from the column may not have been properly removed using a dry spin prior to elution of transposed DNA. | Ensure that the additional dry spin after the second wash step of the DNA Clean and Concentrator-5 kit is performed. |
| 45 | The ATAC-seq libraries are outside the standard curve of the NEB Quant kit. | The libraries may be either too dilute or too concentrated. | We make dilutions in the 2000-4000-fold range. Adjust the dilutions of the final libraries as needed. |
| 50 | Low-depth sequencing shows a low TSS Enrichment Score. | Low signal-to-background ratios are often caused by unhealthy or otherwise non-ideal input material. | Consider pre-treating cells with DNase or using flow cytometry to sort viable cells. |
| There is no nucleosomal periodicity in the Bioanalyzer traces. | Not all ATAC-seq libraries show nucleosomal periodicity on Bioanalyzer. | This may not be a problem. See the section on Quality control of ATAC-seq libraries. |
TIMING
Timing information is for ~12 samples.
Steps 1-19, Transposition, 2.5 h
Steps 20-27, Barcoding of transposed fragments, 30 m
Steps 28-43, Library quantification and amplification, 3 h
Steps 44-49, Final library concentration determination, 2 h
Steps 50-51, Low and/or high-depth sequencing of library, variable
ANTICIPATED RESULTS
Brief overview of ATAC-seq analysis:
ATAC-seq analysis can be broadly divided into three main stages: (1) data processing, (2) peak calling, merging, and counting, and (3) downstream analysis. These major steps are detailed in Fig. 4. Data processing refers to all the steps required to take the raw FASTQ files from the sequencer and prepare them for alignment to the genome and adjustment for the Tn5 offset (Supplementary Fig. 6; described below). Following this, the cleaned and adjusted reads are then used for peak calling on a sample-by-sample basis. These peaks are then merged across all samples that are being compared in the experiment, forming the union peak set, and the number of insertion events in each peak of the union peak set is counted across all samples. This insertion counts matrix and union peak set can be used for a variety of downstream analyses, including differential accessibility analysis to determine peaks that are significantly more or less accessible and motif enrichment on relevant sets of peaks. These stages, along with detailed descriptions of possible analysis tools, are described in greater detail below. In general, we recommend the use of the PEPATAC pipeline73 as it performs many of the steps described below automatically - however, each use case may require tailoring of the precise tools used.
Figure 4: Overview of the steps of ATAC-seq data analysis.
A schematic overview of the necessary steps in the ATAC-seq data analysis workflow, starting with the raw FASTQ files generated by the sequencer. Broadly, ATAC-seq analysis is composed of three major steps: (i) data processing that cleans and aligns the raw reads, (ii) peak calling, merging, and insertion counting that determines the locations of Tn5 accessible chromatin and the relative signal within each accessible region, and (iii) the downstream analysis that can help assign putative functions and pathways to the called peaks. Each step, designated by a box, includes the possible bioinformatics tools that can be used to perform the analysis, as well as the expected output file type that should be utilized for the next step. Many pipelines can manage several aspects of this workflow automatically – see Table 4.
Computational resources needed to process ATAC-seq data:
In general, the quantity of data generated from ATAC-seq libraries is too large to be analyzed on a standard personal laptop or desktop in a timely manner. We recommend the use of a high-performance computing environment (HPCE), often available at many institutions. The analysis pipelines depicted here can also be accomplished on smaller dedicated servers with fewer computational resources, for example, 32 cores and 128 GB of RAM. If an HPCE is not available, such services can also be purchased on a pay-per-use bases from cloud computing providers such as Amazon Web Services (AWS).
Pipelines to process ATAC-seq data:
ATAC-seq data analysis consists of a variety of different steps, which are detailed in Fig. 4 and in the sections below. Often, each step requires a different tool and it can be complex for novice uses to navigate between different input and output files. For these reasons, a variety of different pipelines have been developed for ATAC-seq data analysis, combining several or all of the steps of analysis together into an easy-to-use package. With these pipelines, the user specifies the input data and a predetermined set of analytical steps are performed to create the desired output files, including important quality control metrics. Pipelines are highly recommended for beginners and experts alike, as they are robust, easily portable, and easy to replicate. Several options exist, including, PEPATAC73, the ENCODE ATAC-seq pipeline, and nf-core71,84. More ATAC-seq pipelines and analysis protocols are reviewed in Smith et al 202091. In general, most pipelines will take raw FASTQ files as input, calculate QC metrics, clean-up and align the reads, and provide ATAC-seq peaks as the final output. However, some pipelines do provide other features such as differential accessibility testing and motif enrichment determination. In Table 4, we compare the capabilities of three of the most commonly used pipelines. Each pipeline has its own advantages and disadvantages. For example, the ENCODE ATAC-seq Pipeline was used to generate all of the ATAC-seq data files available from the ENCODE consortium, making it a good choice for users who aim to compare directly with those datasets. PEPATAC and nf-core provide a high degree of flexibility in choosing different parameters for analysis, without having to directly interface with each tool. PEPATAC was used for the analysis of the data presented in this protocol.
Data processing after deep sequencing:
The final output of the ATAC-seq protocol is typically FASTQ-formatted sequencing data files for each sample that was transposed, with each library having a Read 1 and Read 2 file, corresponding to the paired-end reads of either side of the ATAC-seq fragment (Fig. 1b). Each of these reads results from a unique Tn5 transposition event. The overall sequencing quality of each library can be checked using FastQC92, which provides an HTML report with per-base quality scores, as well as any observed biases in the data.
Standard ATAC-seq pipelines will take the FASTQ files as input and perform a series of quality control and data cleaning steps followed by alignment to a reference genome. First, any bases corresponding to the ATAC-seq adapter sequence will be removed (Fig. 4). This is important because ATAC-seq fragments can be shorter than the sequencing read length, resulting in read-through into the adapter sequence at the end of the read. Because this exogenous sequence interferes with proper alignment, it must be removed. Reads originating from mitochondrial DNA, which result from transposition into the un-chromatinized mitochondrial genome and are considered “contaminating”, should be removed, either before full genome alignment, or after. In general, with the Omni-ATAC protocol, we expect to see less than 15% of all reads mapping to the mitochondrial genome. Higher percentages may indicate over lysis or insufficient removal of mitochondria during the wash step with ATAC-seq Wash Buffer (Step 7) and may require the libraries to be sequenced at higher depth to have sufficient unique reads mapping to the nuclear genome. The remaining reads are mapped to the reference genome and filtered for low-quality alignment.
Next, PCR duplicates are marked and removed based on identical start and end positions of the sequence fragment (Fig. 4). In general, we find that ATAC-seq libraries generated with this protocol from 50,000 cells require minimal amplification (3-7 extra cycles in Step 34). This level of amplification is associated with a library complexity that typically results in less than 10% of aligned reads marked as duplicates; however, it is important to note that the percent of reads marked as duplicates depends on many factors including the total sequencing depth and we do not use this metric as a way to determine library quality. For example, samples with high levels of dead cells will result in libraries with high library complexity and low duplicate read percentages because their DNA is largely de-chromatinized and thus is fully accessible to the transposase; however, this would not be considered a high quality ATAC-seq library. Nevertheless, a high percentage of duplicate reads (>25%), especially when libraries are made from 50,000 cells and sequenced to ~10 million read-pairs, may suggest that the transposition was inefficient and could reflect poor or insufficient input material. We find that most ATAC-seq libraries have >80% of the total reads mapping to the nuclear genome, after this series of QC steps. As described above, the TSS Enrichment Score is the single most important QC metric and should be recalculated on the deep sequencing data (Fig. 3a-c).
Finally, the ATAC-seq fragments are adjusted for the “Tn5 offset”. This Tn5 offset accounts for the molecular mechanism of binding and transposition by the Tn5 transposase dimer. Tn5 binds to DNA as a homodimer, with a 9-bp section of DNA between the two Tn5 molecules. Thus, each homodimer binding event creates two insertions, nine base-pairs apart, and the central 9 bp are duplicated and present on each of the corresponding fragments. The true center point of this Tn5 binding event is in the center of this 9-bp region. To account for this, the field has adopted the convention of adjusting the start position of each read by adding 4 bp to plus-stranded insertions and −5 bp to minus-stranded insertions32 (Supplementary Fig. 6).
Peak calling, merging and counting and annotation:
After alignment, the next analysis step is to identify “peaks” of chromatin accessibility – i.e. regions of the genome that are enriched for Tn5 insertions, suggesting that many cells in that sample had Tn5-accessible chromatin at that site. The location, and in some respects the magnitude, of those peaks, can be informative in understanding the gene regulatory landscape.
Many programs exist to identify these peak regions, though MACS2 93, and more recently Genrich94, are two of the most frequently used (Fig. 4). HMMRATAC95 was specifically designed for calling peaks in ATAC-seq data, and Genrich offers an ATAC-seq specific mode as well. As MACS2 is the most commonly used peak caller by ATAC-seq pipelines (Table 4), we focus on it here. MACS or Model-based Analysis of ChIP-Seq is a popular peak calling tool for a variety of datasets, including ChIP-seq, ChIC-seq, DNAse-seq and ATAC-seq. MACS2 calls peaks using a sliding window method, in which a user-provided window size is used to slide across the genome to find those regions with a fold enrichment (also user provided) relative to the background signal. Because MACS2 was not designed specifically for ATAC-seq, certain parameter adjustments are crucial. For example, to accurately represent the individual Tn5 insertion events in MACS2, the “shift” and “extsize” parameters should be used to ensure that the single-base position that represents the Tn5 insertion is directly in the middle of the “read” provided to MACS2. To this end, the PEPATAC pipeline described above, performs peak calling with MACS2 with the following settings as default: --shift −75 --extsize 150 --nomodel --call-summits --nolambda --keep-dup all -p 0.01. It is important to note that peak calling in MACS2 is strongly affected by sequencing depth. Thus, the more reads in a dataset, the more peaks will be called and the significance of each peak will be greater. Therefore, it is not appropriate to compare raw peak calls across samples. In the past, we have advocated for the use of a normalized peak score which can be obtained by normalizing the individual peak score (−log10(p-value)) to the sum total of all peak scores from the peaks identified in that sample38.
Peak sets should first be identified on a sample-specific, and possibly technical-replicate-specific, basis and should then be merged between all samples that will be compared, creating what we refer to as the union peak set (Fig. 5). For example, if comparing cells treated with a compound to cells treated with a vehicle control, peaks should first be called for each technical replicate, then merged between all technical replicates in the group (e.g., all vehicle controls) and finally between the two experimental groups (e.g. treatment and vehicle). While this peak merging sounds straightforward, there are many ways to call and merge peaks and the field has not yet come to a consensus on which is the most appropriate. We advocate for the use of non-overlapping fixed-width 501-bp peaks, centered on the peak summit, as a standard because most DNA regulatory elements are less than 500-bp in length38. Fixed-width peaks make downstream computation easier because peak length does not need to be normalized, as compared to methods that use variable-width peaks. To merge peaks across samples, we advocate for the use of an iterative overlap procedure that maintains a fixed peak width while avoiding bias favoring deeper sequenced libraries or differences in the number of replicates for a given sample type. In the iterative overlap, peaks are first ranked by their normalized significance (based on MACS2 scoring), then the most significant peak is retained, and any overlapping peaks are removed, iteratively, until no overlapping peaks remain. Iterative overlapping avoids two common problems of peak merging: 1) the creation of ever-larger peaks when multiple samples are compared to each other, since each sample-specific peak set will not overlap exactly, and 2) the loss of sensitivity that occurs when clustered overlap techniques are used, which take all the peaks that cluster together and keep a single winner to summarize the cluster. To facilitate the use of this iterative overlap peak merging approach with any set of MACS2 peak calls, we have made the peak merging script used in previous studies38 available (https://github.com/corceslab/ATAC_IterativeOverlapPeakMerging).
Figure 5: Schematic of peak merging strategies and the resulting merged peak sets.
a. Schematic of the three possible peak merging options that have been frequently used for ATAC-seq data: raw overlap with variable-width peaks, clustered overlap with fixed-width peaks, and iterative overlap with fixed-width peaks. This panel has been directly reproduced from Granja & Corces et al. 2021132. (i) In the raw overlap with variable-width peaks approach, any peaks that overlap with each other are merged together into a single, larger peak. This type of peak merging approach is implemented using the bedtools merge command, and will result in peaks that are of variable widths and often span multiple distinct regulatory elements. In this example, the raw overlap approach results in 41 peaks with a median peak width of 256 base pairs (bp) (+/− 408-bp standard deviation). (ii) In the clustered overlap with fixed-width peaks approach, clustered peaks are taken together and a single winner is chosen among them. This is typically implemented using the bedtools cluster command. The resulting merged peak set contains fixed-width peaks and has a tendency to under-represent regulatory elements that are located in close proximity. In this example, the clustered overlap approach results in 41 peaks with a median peak width of 217 bp (+/− 326-bp standard deviation). (iii) In the iterative overlap with fixed-width peaks approach, first introduced in Corces & Granja et al. 201838, fixed-width peaks are first ranked by their normalized significance. Once ranked, the most significant peak is retained, and any peaks directly overlapping with that peak are removed. This ranking and removal are iterated until there are no more overlapping peaks. The resulting merged peak set contains fixed-width peaks. In this example, the iterative overlap approach results in 16 peaks with a fixed peak width of 501 bp (0-bp standard deviation). (iv) Comparison of the resulting merged peak sets made using methods i-iii. b. Diagram of the hematopoietic differentiation hierarchy; to the right is number of samples used in panels (c) and (d) for each cell type. c. ATAC-seq signal tracks of the data from three distinct hematopoietic cell types from Corces & Buenrostro et al. 201637. MPP and CMP data were excluded to improve figure legibility. Each track represents a different human donor. MACS2 peak calls are shown as black boxes below each signal track. (d) Comparison of the MACS2 peak calls and the peak merging approaches for the tracks shown in (c). (top) All MACS2 peak calls from (c) colored by their respective cell type. (middle) The cell type-specific peak sets derived from the first round of the iterative overlap approach are shown after merging peaks from each of the biological replicates. (bottom) Final merged peak set for all biological replicates across all cell types using the three methods described in (a). The number of resulting peaks, and their summary statistics, are shown.
Once a union peak set has been created, the number of transposition events per peak per sample is compiled into an insertion counts matrix. This raw matrix should be normalized for the total number of reads in peaks and then log transformed, though these operations are typically performed as part of standard differential analysis workflows. Peak functionality can partially be inferred by annotating peaks to the nearest gene. A variety of tools exist to perform this analysis, including HOMER (HOMER annotatePeaks.pl)96, ChiPseeker97, and ChIPpeakAnno98. While the nearest gene provides a simple way to annotate peak function, it is important to remember that distal regulatory elements can act across large genomic distances and that the nearest gene often is not the true target for every peak38,99.
Assessing the consistency of samples and technical replicates:
After an insertion counts matrix has been obtained, the correlation between technical or biological replicates should be assessed. The precise Pearson correlation coefficient expected of biological replicates is largely dependent on the origin of the replicates. For example, one might expect CD4+ T cells isolated from 3 individual mice of the same inbred background to have a Pearson correlation value of >0.9, whereas the same cell type isolated from 3 different human donors might have a Pearson correlation value of >0.8 due to the larger natural variation between humans compared to inbred mice housed together. Regardless of the sample origin, we expect technical replicates (i.e. two ATAC-seq reactions performed side-by-side at the same time using different aliquots of the same starting material) to have a Pearson correlation >0.9 and ideally >0.95. These relationships between samples are typically visualized using a one-to-one plot or a Pearson correlogram.
Differentially Accessible Regions:
Identification of differentially accessible regions (DARs) can be performed in a variety of different ways (Fig. 4) which have been well discussed previously100. The choice of which workflow and tool to use for differential accessibility testing should be based on the goals of the project, and the amount of validation or follow-up analysis that will be performed. In general, the results from Gontarz et al 100 suggest that DESeq and DESeq2101 are the best choice for researchers who want to minimize false positives - i.e. minimize the false discovery rate (FDR). This may be appropriate for applications that will have little orthogonal validation and are designed to nominate possible high-likelihood candidate regions for further study. Conversely, for applications that aim to detect as many potential DARs as possible with tolerance to false-positives, edgeR102 or limma103 are a good choice for high-sensitivity, with limma giving the best results for regions with lower signal – such as distal enhancers. High-sensitivity techniques may be useful for studies leveraging integrative analysis with other sequencing techniques, which could lend additional confidence to DARs. An in-depth discussion of several different applications of ATAC-seq data, and the variety of tools that are designed to accomplish them, is also reviewed in Yan et al. 104.
Batch effects:
As with most ‘omics methods, batch effects can occur at multiple steps throughout the ATAC-seq library generation process and may impact downstream analyses, especially when comparing ATAC-seq datasets obtained at different time points or from different groups. In general, best practice rules should be used when generating ATAC-seq data within a specific experimental context to limit variation due to technical artifacts. However, some sources of batch effects may be challenging to control for as they can also originate from upstream sample attributes, for example the post-mortem interval of human tissues. To remove these sources of unwanted variation between different ATAC experiments, one can use the built-in batch correction tools from edgeR or DESeq2100. Both of these programs will allow for the inclusion of batch as a covariate in the experimental design. In edgeR, the batch is regressed out using a negative binomial generalized linear model (GLM) for the batch and experimental conditions and performing the likelihood test as a generalization of the paired samples t-test102. In a similar approach, DESeq2 also fits a negative binomial GLM and uses the Wald test to determine the significance of the experimental condition101. An alternate approach to these two programs is RUVseq105, which is predominantly used for RNA-seq data, but has also been applied to ATAC-seq data100. It performs factor analysis on the upper quartile normalized counts using residuals calculated by edgeR105.
Track visualization:
Regardless of the workflow used to identify DARs, we recommend visual inspection of high-importance peaks using properly normalized sequencing tracks. This ensures that the difference identified through differential analysis is in line with expectation. A common way to do this is to create normalized bigWig (.bw) files that can be uploaded to genome browser tools such as the UCSC Genome Browser106, Integrative Genomics Viewer (IGV)107, or the WashU Epigenome Browser108. BigWig files provide a streamlined way to visualize genome-wide alignments, and in the case of ATAC-seq data, genome-wide patterns in chromatin accessibility. Normalization can be performed using bedtools genomecov109,110 or deepTools bamCoverage111 by applying a scale factor based on the number of reads in TSS regions. This approach is analogous to normalizing based on reads in peaks, enabling simultaneous normalization for both sequencing depth and data quality. We recommend using reads in TSS regions because these regions are invariant across samples whereas use of reads in peaks requires re-normalization any time the peak set changes. We also recommend using a standardized bin size when creating the bigWig files. The larger the bin size, the lower the resolution and the smaller the resulting bigWig file. We recommend a bin size of 100 bp for most applications.
Motif Enrichment Analysis:
Motif enrichment analysis is based on searching for TF binding motifs within a given peak set. Motif information, from both experimental observations as well as computational predictions, has been collated in a variety of large databases, such as JASPAR112, CIS-BP113, and ENCODE114. Each motif is stored as a position-weight matrix (PWM), and a variety of tools are able to scan the DNA sequence of each peak for the presence of each motif, including HOMER96, TFBSTools115, motifmatchr116, and MEME117. Enrichment for the presence of a given motif within the peak set of interest compared to a set of background regions implies that the given TF may be important in driving the accessibility at those peak regions. For example, when comparing differentiated to undifferentiated cells, these motif enrichment tools would identify motifs that are statistically enriched in the peaks gained after differentiation, compared to those shared between the differentiated and undifferentiated cells. This could provide a hypothesis about the TF(s) responsible for driving differentiation. Various statistical tests have been used to identify motif enrichment including the hypergeometric test (HOMER) and the rank sum test (MEME). When performing these analyses, it is important to select a meaningful set of regions or peaks to serve as the background for enrichment testing. It is often not correct to use the entire genome as the background and a matched subset of peaks or the whole peak set should be used instead, depending on the particular application.
While powerful tools, it is important to remember that peak scanning-based motif enrichment methods can contain many false positives, as multiple different motifs may be present within a peak, and often TFs from the same family share similar motifs. Additionally, motif enrichment is not direct evidence for TF binding. One way to increase the biological meaning of motif enrichment results is to refine potential TF mediators based on their expression in the cell type or tissue of interest. Another way to analyze differences in motifs is to calculate the difference in observed versus expected accessibility at peak sets. This type of analysis is predicated on the idea that greater accessibility, i.e. insertion counts, is correlated with greater TF activity. Thus, a difference in fragment counts can be taken as a change in the activity of that TF. Two programs have been implemented to do this type of analysis – chromVAR118 and diffTF119. ChromVAR, which was originally developed for single-cell ATAC-seq data, calculates an accessibility deviation across multiple conditions for each motif. diffTF, which is conceptually similar to chromVAR, was designed to work with bulk ATAC-seq data and can also take matched RNA-seq data as input, for its “classification” mode to determine the mode of action for a given TF (i.e. repressor or activator)119. diffTF is run from within a Snakemake pipeline workflow and uses aligned BAM files as input.
Transcription Factor Footprinting analysis:
TF regulation of the accessible peaks can also be determined by footprinting analysis. Footprinting analysis originates from the classic DNase footprint27,28, and was later adapted to sequencing-based analysis (sometimes termed digital genomic footprinting120). Within the context of ATAC-seq, TF footprints would occur when a TF binds to DNA and prevents Tn5 insertions specifically at the DNA bases that are directly bound. This leaves a section of DNA, or a “footprint”, with a dearth of insertions compared to the adjacent nucleosome-free (and TF-free) regions (Fig. 4). Thus, the goal of footprinting algorithms is to first identify these locations of “dipped” Tn5 insertions and second, to determine which TF might have been bound there by examining the sequence of the central bases inferred to be directly bound by the TF. Identifying TF footprints can aid in the reconstruction of gene regulatory networks, and may be more specific than motif scanning in detecting the presence of a TF. However, there are many challenges to applying footprinting analysis to ATAC-seq data, and the interpretation can often be confounded by Tn5’s sequence bias28. Classically, TF footprinting was designed to predict TF binding at a single site in the genome. This requires many fragments mapping to that particular locus to provide sufficient observations to identify the protected bases. In the context of ATAC-seq performed on 50,000 cells, it is often the case that insufficient depth is obtained to perform single-site footprinting, even at high sequencing depth. Because of this, many researchers have performed “meta” footprinting where hundreds or thousands of genomic locations harboring the motif for a given TF are collated into a single footprint. It is important to note that this is not the classical use of footprinting and its results have more in common with motif enrichment than they do with single-site footprinting. As with other ATAC-seq methods, TF footprinting analysis should adjust fragments for the Tn5 offset32,121 (Supplementary Fig. 6) and account for the Tn5 insertion bias122. Several approaches for single-site footprinting, including HINT-ATAC74, BaGFoot123 and TOBIAS124, are discussed in Supplementary Note 4.
Nucleosome positioning:
ATAC-seq data has also been used to look at nucleosome positioning61, although these techniques are still in development, and require very high sequencing depth to be accurately calculated. Nucleosome positioning refers to techniques which attempt to measure differences in the organization of nucleosomes at certain locations, including their positions and occupancy125. Changes in nucleosome profiles have been associated with changes in gene regulation and expression levels at a given promoter126. Classically, nucleosome positioning has been performed with MNase-seq data, and several tools have been developed for that type of data. However, NucleoATAC61 and HMMRATAC95 were developed specifically for predicting nucleosome positions from ATAC-seq data. In general, for the sequencing depth of the average ATAC-seq library, applying nucleosome profiling techniques is not advised.
Integrating ATAC-seq with other ‘omic profiling techniques:
As mentioned in the introduction, ATAC-seq gives a general landscape of chromatin accessibility across the genome. As such, it can be interesting to compare the chromatin accessibility landscapes from ATAC-seq with other matched genomic, transcriptional, or epigenomic profiles. Intersecting these datasets may help to interpret the function of a given ATAC-seq peak. For example, ATAC-seq can be intersected with H3K27ac ChIP-seq (or other ChIC techniques) with the expectation that a subset of ATAC-seq peaks would overlap with the active enhancers and promoters marked by H3K27ac. With a large enough dataset, cross-sample correlations between ATAC-seq chromatin accessibility and nearby gene expression can be used to identify peak-to-gene links that represent putative gene regulatory interactions. Such interactions could be orthogonally supported using chromosome conformation capture techniques such as HiC127 or HiChIP128.
Similarly, ATAC-seq provides a unique window into the effects of sequence changes in the noncoding genome. By combining ATAC-seq data with whole-genome sequencing data, one can identify regions of allelic imbalance whereby the two different alleles show differential accessibility, implying differential TF binding129. This has been used to identify chromatin accessibility quantitative trait loci129 and to begin to assign function to noncoding polymorphisms identified through genome-wide association studies53,130,131. These types of analyses can help to assign relevance or function to specific putative regulatory elements identified through ATAC-seq.
Preparing for publication:
ATAC-seq data included in publications should be deposited in publicly available repositories, such as the Gene Expression Omnibus (GEO), the Sequencing Read Archive (SRA), or the European Nucleotide Archive (ENA). Instructions specific to each repository can be found on their websites. However, it is most common to include the following files: 1) all raw FASTQ files (Read 1 and Read 2) prior to QC and adapter trimming, 2) an insertion counts matrix for the union peak set of all samples analyzed in the study, including the genomic coordinates of the peaks, 3) normalized bigWig files that can be easily visualized on a genome browser, and 4) a list of differentially accessible regions (DARs) if applicable. We also encourage researchers to include a supplementary table reporting the QC metrics, as detailed above, for each of their sequenced samples, especially the TSS Enrichment Score.
Supplementary Material
Supplementary Figure 1: Validation of alterations to Omni-ATAC protocol
Supplementary Figure 2: Sequences of ATAC-seq adapters and barcodes
Supplementary Figure 3: Images of nuclei prior to ATAC-seq
Supplementary Figure 4: Scaling of Tn5 for different quantities of cells
Supplementary Figure 5: Derivation and validation of Qubit equation
Supplementary Figure 6: Schematic of Tn5 offset
Supplementary Note 1: Transposition, chromatin accessibility, and TF dynamics
Supplementary Note 2: Calculating Tn5-to-cell ratio
Supplementary Note 3: Adapters for transposition reaction
Supplementary Note 4: Details about footprinting analysis
Supplementary Protocol 1: Isolation of Nuclei from frozen tissues
Supplementary Table 1: Length of ATAC-seq read sequencing versus mapping efficiency
Supplementary Table 2: Adapters for transposition reaction
Acknowledgements
This work was supported by NIH R00-AG059918, U01-AG072573, P01-AG073082, UM1-HG012076 and a gift from the Ray and Dagmar Dolby Family Fund (to the Gladstone Institutes). FCG is an Alan Kaganov Scholar. MRC is additionally supported by the Farmer Family Foundation Parkinson’s Research Initiative and an American Society of Hematology Scholar Award.
Footnotes
Code availability
The source code for the iterative overlap is freely available at https://github.com/corceslab/ATAC_IterativeOverlapPeakMerging 88. All other ATAC-seq data analysis for the figures used in this protocol were generated using PEPATAC73 with Bulker (container version 1.0.8), available at http://pepatac.databio.org/en/latest/.
Competing interests
The authors declare that they have no competing financial interests.
Data availability
The ATAC-seq datasets generated for the protocol optimizations detailed in Supplementary Figs. 1, 4, and 5 are available on GEO under accession number GSE188797. The data used in Fig. 5 are taken from Corces et al. 201838. All analyses were performed using the hg38 human genome.
REFERENCES
- 1.Ernst J & Kellis M Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol 28, 817–825 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst J et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klemm SL, Shipony Z & Greenleaf WJ Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Johnson DS, Mortazavi A, Myers RM & Wold B Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science 316, 1497–1502 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Furey TS ChIP–seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nat. Rev. Genet 13, 840–852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakato R & Sakata T Methods for ChIP-seq analysis: A practical workflow and advanced applications. Methods 187, 44–53 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Schmid M, Durussel T & Laemmli UK ChIC and ChEC: Genomic Mapping of Chromatin Proteins. Mol. Cell 16, 147–157 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Skene PJ & Henikoff S An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaya-Okur HS et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidl C, Rendeiro AF, Sheffield NC & Bock C ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q et al. CoBATCH for High-Throughput Single-Cell Epigenomic Profiling. Mol. Cell 76, 206–216.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Handa T et al. Chromatin integration labeling for mapping DNA-binding proteins and modifications with low input. Nat. Protoc 15, 3334–3360 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Harada A et al. A chromatin integration labelling method enables epigenomic profiling with lower input. Nat. Cell Biol 21, 287–296 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Ku WL et al. Single cell chromatin immunocleavage sequencing (scChIC-Seq) to profile histone modification. Nat. Methods 16, 323–325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X-Y & Gehring M Low-input chromatin profiling in Arabidopsis endosperm using CUT&RUN. Plant Reprod. 32, 63–75 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Hainer SJ, Bošković A, McCannell KN, Rando OJ & Fazzio TG Profiling of Pluripotency Factors in Single Cells and Early Embryos. Cell 177, 1319–1329.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skene PJ, Henikoff JG & Henikoff S Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc 13, 1006–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Crawford GE et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16, 123–131 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song L & Crawford GE DNase-seq: A High-Resolution Technique for Mapping Active Gene Regulatory Elements across the Genome from Mammalian Cells. Cold Spring Harb. Protoc 2010, pdb.prot5384 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle AP et al. High-Resolution Mapping and Characterization of Open Chromatin across the Genome. Cell 132, 311–322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giresi PG, Kim J, McDaniell RM, Iyer VR & Lieb JD FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui K & Zhao K Genome-Wide Approaches to Determining Nucleosome Occupancy in Metazoans Using MNase-Seq. in Chromatin Remodeling: Methods and Protocols (ed. Morse RH) 413–419 (Humana Press, 2012). doi: 10.1007/978-1-61779-477-3_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schones DE et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly TK et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 22, 2497–2506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minnoye L et al. Chromatin accessibility profiling methods. Nat. Rev. Methods Primer 1, 1–24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weintraub H & Groudine M Chromosomal subunits in active genes have an altered conformation. Science 193, 848–856 (1976). [DOI] [PubMed] [Google Scholar]
- 27.Galas DJ & Schmitz A DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5, 3157–3170 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He HH et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods 11, 73–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung M-H, Baek S & Hager GL Genome-wide footprinting: ready for prime time? Nat. Methods 13, 222–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mieczkowski J et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun 7, 11485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chereji RV, Bryson TD & Henikoff S Quantitative MNase-seq accurately maps nucleosome occupancy levels. Genome Biol. 20, 198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goryshin IY & Reznikoff WS Tn5 in vitro transposition. J. Biol. Chem 273, 7367–7374 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Adey A et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 11, R119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gangadharan S, Mularoni L, Fain-Thornton J, Wheelan SJ & Craig NL DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc. Natl. Acad. Sci 107, 21966–21972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corces MR et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corces MR et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corces MR et al. The chromatin accessibility landscape of primary human cancers. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calderon D et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet 51, 1494–1505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naik S et al. Inflammatory Memory Sensitizes Skin Epithelial Stem Cells to Tissue Damage. Nature 550, 475–480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marco A et al. Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat. Neurosci 23, 1606–1617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D et al. Chromatin Accessibility Dynamics during iPSC Reprogramming. Cell Stem Cell 21, 819–833.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Guo J et al. Chromatin and Single-Cell RNA-Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development. Cell Stem Cell 21, 533–546.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Daugherty AC et al. Chromatin accessibility dynamics reveal novel functional enhancers in C. elegans. Genome Res. 27, 2096–2107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gury-BenAri M et al. The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell 166, 1231–1246.e13 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Liu Q et al. Chromatin accessibility landscapes of skin cells in systemic sclerosis nominate dendritic cells in disease pathogenesis. Nat. Commun 11, 5843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y et al. Chromatin accessibility landscape of articular knee cartilage reveals aberrant enhancer regulation in osteoarthritis. Sci. Rep 8, 15499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenwald WW et al. Pancreatic islet chromatin accessibility and conformation reveals distal enhancer networks of type 2 diabetes risk. Nat. Commun 10, 2078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J et al. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature 572, 335–340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidl C et al. Combined chemosensitivity and chromatin profiling prioritizes drug combinations in CLL. Nat. Chem. Biol 15, 232–240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scharer CD et al. Epigenetic programming underpins B cell dysfunction in human SLE. Nat. Immunol 20, 1071–1082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corces MR et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet 52, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banovich NE et al. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Res. 28, 122–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forrest MP et al. Open Chromatin Profiling in hiPSC-Derived Neurons Prioritizes Functional Noncoding Psychiatric Risk Variants and Highlights Neurodevelopmental Loci. Cell Stem Cell 21, 305–318.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiou J et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nott A et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q et al. Genome-Wide Temporal Profiling of Transcriptome and Open-Chromatin of Early Cardiomyocyte Differentiation Derived From hiPSCs and hESCs. Circ. Res 121, 376–391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wapinski OL et al. Rapid Chromatin Switch in the Direct Reprogramming of Fibroblasts to Neurons. Cell Rep. 20, 3236–3247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denny SK et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 166, 328–342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schep AN et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaya-Okur HS, Janssens DH, Henikoff JG, Ahmad K & Henikoff S Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc 15, 3264–3283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stark R, Grzelak M & Hadfield J RNA sequencing: the teenage years. Nat. Rev. Genet 20, 631–656 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Bartosovic M, Kabbe M & Castelo-Branco G Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol 39, 825–835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao W, Lai B, Ni B & Zhao K Genome-wide profiling of nucleosome position and chromatin accessibility in single cells using scMNase-seq. Nat. Protoc 15, 68–85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai B et al. Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 562, 281–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin W et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takaku M et al. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 17, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujiwara S, Baek S, Varticovski L, Kim S & Hager GL High Quality ATAC-Seq Data Recovered from Cryopreserved Breast Cell Lines and Tissue. Sci. Rep 9, 516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mulqueen RM et al. Improved single-cell ATAC-seq reveals chromatin dynamics of in vitro corticogenesis. bioRxiv 637256 (2019) doi: 10.1101/637256. [DOI] [Google Scholar]
- 71.nf-core/atacseq. (nf-core, 2021). doi: 10.5281/zenodo.2634132. [DOI] [Google Scholar]
- 72.ATAC-seq Data Standards and Processing Pipeline – ENCODE. https://www.encodeproject.org/atac-seq/. [Google Scholar]
- 73.Smith JP et al. PEPATAC: an optimized pipeline for ATAC-seq data analysis with serial alignments. NAR Genomics Bioinforma. 3, lqab101 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bajic M, Maher KA & Deal RB Identification of Open Chromatin Regions in Plant Genomes Using ATAC-Seq. Methods Mol. Biol. Clifton NJ 1675, 183–201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deal RB & Henikoff S The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc 6, 56–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haines JE & Eisen MB Patterns of chromatin accessibility along the anterior-posterior axis in the early Drosophila embryo. PLOS Genet. 14, e1007367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson S, Nguyen V & Coder D Assessment of Cell Viability. Curr. Protoc. Cytom 64, 9.2.1–9.2.26 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Chen X et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 13, 1013–1020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swanson E et al. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. eLife 10, e63632 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Corces R Isolation of nuclei from frozen tissue for ATAC-seq and other epigenomic assays. (2019) doi: 10.17504/protocols.io.6t8herw. [DOI] [Google Scholar]
- 81.Polavarapu VK et al. Profiling chromatin accessibility in formalin-fixed paraffin-embedded samples. Genome Res. gr.275269.121 (2021) doi: 10.1101/gr.275269.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chin HG et al. Universal NicE-seq for high-resolution accessible chromatin profiling for formaldehyde-fixed and FFPE tissues. Clin. Epigenetics 12, 143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Orchard P, Kyono Y, Hensley J, Kitzman JO & Parker SCJ Quantification, Dynamic Visualization, and Validation of Bias in ATAC-Seq Data with ataqv. Cell Syst. 10, 298–306.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ewels PA et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278 (2020). [DOI] [PubMed] [Google Scholar]
- 85.Koohy H, Down TA & Hubbard TJ Chromatin Accessibility Data Sets Show Bias Due to Sequence Specificity of the DNase I Enzyme. PLOS ONE 8, e69853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Satpathy AT et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol 37, 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cusanovich DA et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Granja JM & Corces MR ATAC_IterativeOverlapPeakMerging. (2022). DOI: 10.5281/zenodo.5903680. [DOI] [Google Scholar]
- 89.Picelli S et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith JP & Sheffield NC Analytical Approaches for ATAC-seq Data Analysis. Curr. Protoc. Hum. Genet 106, e101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bioinformatics Babraham - FastQC A Quality Control tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- 93.Zhang Y et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaspar JM Genrich: detecting sites of genomic enrichment. (2021). [Google Scholar]
- 95.Tarbell ED & Liu T HMMRATAC: a Hidden Markov ModeleR for ATAC-seq. Nucleic Acids Res. 47, e91–e91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heinz S et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu G, Wang L-G & He Q-Y ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 98.Zhu LJ et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mumbach MR et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet 49, 1602–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gontarz P et al. Comparison of differential accessibility analysis strategies for ATAC-seq data. Sci. Rep 10, 10150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yan F, Powell DR, Curtis DJ & Wong NC From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis. Genome Biol. 21, 22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Risso D, Ngai J, Speed TP & Dudoit S Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol 32, 896–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.UCSC Genome Browser Home. https://genome.ucsc.edu/index.html. [Google Scholar]
- 107.Robinson JT et al. Integrative genomics viewer. Nat. Biotechnol 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.WashU Epigenome Browser. http://epigenomegateway.wustl.edu/browser/. [Google Scholar]
- 109.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.genomecov — bedtools 2.30.0 documentation. https://bedtools.readthedocs.io/en/latest/content/tools/genomecov.html.
- 111.bamCoverage — deepTools 3.5.0 documentation. https://deeptools.readthedocs.io/en/develop/content/tools/bamCoverage.html. [Google Scholar]
- 112.Khan A et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weirauch MT et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kheradpour P & Kellis M Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42, 2976–2987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tan G & Lenhard B TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32, 1555–1556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fast Motif Matching in R • motifmatchr. https://greenleaflab.github.io/motifmatchr/index.html. [Google Scholar]
- 117.Bailey TL et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schep AN, Wu B, Buenrostro JD & Greenleaf WJ chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berest I et al. Quantification of Differential Transcription Factor Activity and Multiomics-Based Classification into Activators and Repressors: diffTF. Cell Rep. 29, 3147–3159.e12 (2019). [DOI] [PubMed] [Google Scholar]
- 120.Hesselberth JR et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martins AL, Walavalkar NM, Anderson WD, Zang C & Guertin MJ Universal correction of enzymatic sequence bias reveals molecular signatures of protein/DNA interactions. Nucleic Acids Res. 46, e9–e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baek S, Goldstein I & Hager GL Bivariate Genomic Footprinting Detects Changes in Transcription Factor Activity. Cell Rep. 19, 1710–1722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bentsen M et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun 11, 4267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Singh AK & Mueller-Planitz F Nucleosome Positioning and Spacing: From Mechanism to Function. J. Mol. Biol 433, 166847 (2021). [DOI] [PubMed] [Google Scholar]
- 126.Lai WKM & Pugh BF Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol 18, 548–562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Belton J-M et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods San Diego Calif 58, 268–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mumbach MR et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kumasaka N, Knights AJ & Gaffney DJ Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat. Genet 48, 206–213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gate RE et al. Genetic determinants of co-accessible chromatin regions in activated T cells across humans. Nat. Genet 50, 1140–1150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Örd T et al. Single-Cell Epigenomics and Functional Fine-Mapping of Atherosclerosis GWAS Loci. Circ. Res 129, 240–258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Granja JM et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet 53, 403–411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kia A et al. Improved genome sequencing using an engineered transposase. BMC Biotechnol. 17, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Green B, Bouchier C, Fairhead C, Craig NL & Cormack BP Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob. DNA 3, 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ason B & Reznikoff WS DNA Sequence Bias During Tn5 Transposition. J. Mol. Biol 335, 1213–1225 (2004). [DOI] [PubMed] [Google Scholar]
- 136.Lazarovici A et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci 110, 6376–6381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dingwall C, Lomonossoff GP & Laskey RA High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 9, 2659–2673 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hörz W & Altenburger W Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 9, 2643–2658 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu SJ et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol 39, 819–824 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Patty BJ & Hainer SJ Transcription factor chromatin profiling genome-wide using uliCUT&RUN in single cells and individual blastocysts. Nat. Protoc 16, 2633–2666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carter B et al. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat. Commun 10, 3747 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chang P, Gohain M, Yen M-R & Chen P-Y Computational Methods for Assessing Chromatin Hierarchy. Comput. Struct. Biotechnol. J 16, 43–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mo A et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yang AC et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 595, 565–571 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Drokhlyansky E et al. The Human and Mouse Enteric Nervous System at Single-Cell Resolution. Cell 182, 1606–1622.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Deal RB & Henikoff S A Simple Method for Gene Expression and Chromatin Profiling of Individual Cell Types within a Tissue. Dev. Cell 18, 1030–1040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bhattacharyya S, Sathe AA, Bhakta M, Xing C & Munshi NV PAN-INTACT enables direct isolation of lineage-specific nuclei from fibrous tissues. PLOS ONE 14, e0214677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nuclei Isolation for Single Cell ATAC Sequencing -Demonstrated Protocol -Sample Prep -Single Cell ATAC -Official 10x Genomics Support. https://support.10xgenomics.com/single-cell-atac/sample-prep/doc/demonstrated-protocol-nuclei-isolation-for-single-cell-atac-sequencing. [Google Scholar]
- 149.Lawler AJ et al. Cell Type-Specific Oxidative Stress Genomic Signatures in the Globus Pallidus of Dopamine-Depleted Mice. J. Neurosci 40, 9772–9783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kiseleva E et al. A protocol for isolation and visualization of yeast nuclei by scanning electron microscopy (SEM). Nat. Protoc 2, 1943–1953 (2007). [DOI] [PubMed] [Google Scholar]
- 151.Niepel M, Farr JC, Rout MP & Strambio-De-Castillia C Rapid isolation of functionally intact nuclei from the yeast Saccharomyces. 162388 https://www.biorxiv.org/content/10.1101/162388v1 (2017) doi: 10.1101/162388. [DOI] [Google Scholar]
- 152.Nott A, Schlachetzki JCM, Fixsen BR & Glass CK Nuclei isolation of multiple brain cell types for omics interrogation. Nat. Protoc 16, 1629–1646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fullard JF et al. An atlas of chromatin accessibility in the adult human brain. Genome Res. 28, 1243–1252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hauberg ME et al. Common schizophrenia risk variants are enriched in open chromatin regions of human glutamatergic neurons. Nat. Commun 11, 5581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Haines J ATAC-seq on nuclei from frozen, sliced, Drosophila melanogaster embryo halves. (2017) doi: 10.17504/protocols.io.kj5cuq6. [DOI] [Google Scholar]
- 156.Steiner FA, Talbert PB, Kasinathan S, Deal RB & Henikoff S Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 22, 766–777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Han M, Wei G, McManus CE, Hillier LW & Reinke V Isolated C. elegans germ nuclei exhibit distinct genomic profiles of histone modification and gene expression. BMC Genomics 20, 500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 159.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.broadinstitute/picard. (Broad Institute, 2021) https://github.com/broadinstitute/picard. [Google Scholar]
- 161.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jiang H, Lei R, Ding S-W & Zhu S Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.SAMBLASTER: fast duplicate marking and structural variant read extraction ∣ Bioinformatics ∣ Oxford Academic. https://academic.oup.com/bioinformatics/article/30/17/2503/2748175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Krueger F Trim Galore. (2021). [Google Scholar]
- 166.Boyle AP, Guinney J, Crawford GE & Furey TS F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24, 2537–2538 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Validation of alterations to Omni-ATAC protocol
Supplementary Figure 2: Sequences of ATAC-seq adapters and barcodes
Supplementary Figure 3: Images of nuclei prior to ATAC-seq
Supplementary Figure 4: Scaling of Tn5 for different quantities of cells
Supplementary Figure 5: Derivation and validation of Qubit equation
Supplementary Figure 6: Schematic of Tn5 offset
Supplementary Note 1: Transposition, chromatin accessibility, and TF dynamics
Supplementary Note 2: Calculating Tn5-to-cell ratio
Supplementary Note 3: Adapters for transposition reaction
Supplementary Note 4: Details about footprinting analysis
Supplementary Protocol 1: Isolation of Nuclei from frozen tissues
Supplementary Table 1: Length of ATAC-seq read sequencing versus mapping efficiency
Supplementary Table 2: Adapters for transposition reaction
Data Availability Statement
The ATAC-seq datasets generated for the protocol optimizations detailed in Supplementary Figs. 1, 4, and 5 are available on GEO under accession number GSE188797. The data used in Fig. 5 are taken from Corces et al. 201838. All analyses were performed using the hg38 human genome.